Abstract
Notochord is an embryonic midline structure that serves as mechanical support for axis elongation and the signaling center for the surrounding tissues. Precursors of notochord are initially induced in the dorsal most mesoderm region in gastrulating embryo and separate from the surrounding mesoderm/endoderm tissue to form an elongated rod-like structure, suggesting that cell adhesion molecules may play an important role in this step. In Xenopus embryo, axial protocadherin (AXPC), an orthologue of mammalian Protocadherin-1 (PCDH1), is indispensable for the assembly and separation from the surrounding tissue of the notochord cells. However, the role of PCDH1 in mammalian notochord remains unknown. We herein report that PCDH1 is expressed in the notochord of mouse embryo and that PCDH1-deficient mice form notochord normally. First, we examined the temporal expression pattern of pcdh1 and found that pcdh1 mRNA was expressed from embryonic day (E) 7.5, prior to the stage when notochord cells detach from the surrounding endoderm tissue. Second, we found that PCDH1 protein is expressed in the notochord of mouse embryos in addition to the previously reported expression in endothelial cells. To further investigate the role of PCDH1 in embryonic development, we generated PCDH1-deficient mice using the CRISPR-Cas9 system. In PCDH1-deficient embryos, notochord formation and separation from the surrounding tissue were normal. Structure and marker gene expression of notochord were also unaffected by loss of PCDH1. Major vascular patterns in PCDH1-deficient embryo were essentially normal. These results suggest that PCDH1 is dispensable for notochord formation, including the tissue separation process, in mammalian embryos. We successfully identified the evolutionary conserved expression of PCDH1 in notochord, but its function may differ among species.
Keywords: PCDH1, Notochord, Cell adhesion molecule
1. Introduction
Notochord is an embryonic mesoderm-derived, transient, midline structure common to all members of the phylum Chordata [1]. Two important functions of the notochord during embryonic development are serving as mechanical support for axis elongation and secreting signaling cues to the surrounding tissues [1,2]. The vacuolization of the notochord is required for the body axis elongation for zebrafish embryo [3], and notochord-derived signals are essential for patterning of tissues, such as the neural tube [4,5], somite [6,7], vessels [8], and endoderm [9].
In vertebrates, the notochord arises from the axial mesodermal cells, which constitute organizer or node [10]. Notochord precursors are marked by the expression of several transcription factors, such as the forkhead domain genes Foxa1 and 2 [11], the T-box domain gene Brachyury/T/no tail [12,13], and the homeobox gene floating head (flh)/Noto [14,15]. Those cells rearrange their positions during gastrulation by mutual intercalation towards the midline, migrate anteriorly, and then form an elongated stack of cells [16]. Notochord cells eventually regress in later stages of development to become the nuclei pulposi of the intervertebral discs [17]. During the cell rearrangement process, notochord precursor cells are separated from surrounding tissue, such as paraxial mesoderm or endoderm [10], suggesting that cell adhesion molecules may play an important role in this step.
Indeed, in Xenopus laevis embryo, the cell adhesion molecule Axial protocadherin (AXPC), an orthologue of mammalian Protocadherin-1 (PCDH1), is expressed in notochord precursor cells and plays an important role in the assembly of notochord cells [18,19]. In axpc knockdown Xenopus embryos, the notochord is not properly formed, and the elongation of the embryonic body axis is inhibited [19,20].
In the present study, we performed expression as well as functional analyses under the hypothesis that PCDH1 has a conserved role in mammalian notochord formation.
2. Materials and Methods
2.1. Reverse transcription polymerase chain reaction (RT-PCR)
Total RNA was isolated from embryonic day (E) 7.5, 8.5, 9.5, and 11.5 whole embryos of ICR strain using ISOGEN-LS (NIPONGENE, Tokyo, Japan), according to the manufacturer's protocol. Total RNA was reverse-transcribed with oligod(T) primers and SuperScript II Reverse Transcriptase (Thermo Fisher Scientific, Waltham, MA, USA). Quantitative PCR (qPCR) were performed using Power SYBR® Green Master Mix (Thermo Fisher Scientific). Primer sequences are shown in Supplemental Table 1.
2.2. Whole-mount in situ hybridization (WISH)
Probes to detect pcdh1 (NM_029357.3), sonic hedgehog (shh) (NM_009170.3), and platelet/endothelial cell adhesion molecule-1 (pecam-1) (NM_001032378) mRNAs were designed. Three probes with non-overlapping sequences and about 1 kb length were designed for pcdh1 mRNA (Fig. S2). Two probes for pecam-1 and one probe for shh were also designed. Multiple probes were used as mixtures. Each probe sequence was cloned into TOPO TA vector (Thermo Fisher Scientific). Digoxygenin-labeled RNA probes were transcribed with T3 or T7 RNA polymerase. WISH was performed on E9.5, 10.5, and 11.5 ICR embryos, E9.5 and 10.5 pcdh1+/+ and pcdh1Δ11/Δ11 embryos, as previously described (Shimizu et al., 2009). Primer sequences are shown in Supplemental Table 1.
2.3. Hematoxylin-eosin (HE) staining
E8.5 or E10.5 ICR embryos were fixed with 4% paraformaldehyde (PFA) for 2 h, cryoprotected by infiltration of 30% sucrose in phosphate-buffered saline (PBS) overnight and infiltration of 30% sucrose in PBS and OCT (1:1) for 1 h, and embedded in OCT embedding compound. Transverse sections (20 μm) were obtained using a cryostat. E9.5 pcdh1+/+ and pcdh1Δ11/Δ11 embryos were dissected, fixed with 4% PFA, dehydrated, cleared, and embedded into paraffin wax. Transverse sections (14 μm) were obtained using a microtome, de-waxed, hydrated, and stained with hematoxylin (Wako, Osaka, Japan) and eosin (Wako). Stained sections were dehydrated and mounted with Mount-Quick (OHMICH, Gunma, Japan).
2.4. Immunofluorescence staining
Immunofluorescence staining was performed as previously described [21]. In brief, cryosections were made as described as above, washed with PBS, and blocked by PBS with 10% fetal bovine serum for 1 h. Primary antibodies were diluted in PBS with 1% fetal bovine serum and incubated at 4 °C overnight. Primary antibodies were anti-PCDH1 (#H00005097-M01, 1:250; abnova, Taipei City, Taiwan), anti-CD31 (#ab28364, 1:250; abcam, Cambridge, UK), and anti-Laminin (#L9393, 1:500; Sigma-Aldrich, St. Louis, MO, USA). Slides were coverslipped with Fluoroshield Mounting Medium (ImmunoBioScience Corp., Mukilteo, WA, USA).
2.5. Animals
Pcdh1 knockout mice were generated using the double nicking approach of the CRISPR-Cas system, as previously described [22,23]. A guide RNA pair was designed with the web based design tool of genomeengeneering.org and no possible off-target site was predicted (please note that the website is no longer available at the moment of the submission of this manuscript). D10ACas9 mRNA and guide RNAs targeting pcdh1 exon 2 were microinjected into mouse zygotes obtained by mating superovulated BDF1 females and BDF1 males. The pcdh1Δ11 allele was selected from the obtained G0 mice as a founder of a PCDH1-deficient mouse line. The pcdh1Δ11/Δ11 mice used in this study were backcrossed at least three generations to C57BL/6. Mice or embryos were genotyped by a PCR analysis using genotyping primers (shown in Supplemental Table 1) and restriction enzyme treatment for genomic DNA extracted from the tail tips of pups or yolk sac of embryos. Three-to eight-week-old mice were measured for their body weight, and eight-week-old mice were measured for their body length. The mice were housed under controlled environmental conditions with free access to water and food. Light was provided between 07:00 and 19:00.
The protocols for animal experiments were approved by the Animal Care and Use Committee of the National Research Institute for Child Health and Development (Permission Number: A2004-003-C09) and Meiji University (Permission Numbers: MUIACUC-20-113).
2.6. Protein extraction from adult mice
Small piece of liver tissues was isolated from adult male pcdh1+/+ or pcdh1Δ11/Δ11 mice. The flash-frozen liver tissues were crushed into a powder using homogenizer Shakeman-3 (bio medical science, Tokyo, Japan). Lysis buffer (50 mM HEPES [pH 7.8], 200 mM NaCl, 5 mM EDTA, 1% NP40, 5% Glycerol), 1 μl of 1 M DTT, and 20 μl 50x Protease Inhibitor Cocktail (Roche, Basel, Switzerland) were added to the tissue powder. The solution was homogenized, and then the supernatant was measured for the protein concentration using a Bio-rad DC protein assay kit (#500–0111, Bio-Rad, Hercules, CA, USA) for equal loading.
2.7. Western blotting
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed using a 4%–12% precast acrylamide gel (Thermo Fisher Scientific). Extracts with equal protein amounts were loaded into each well. Proteins were transferred to an Immobilon-P (Merck Millipore, Burlington, MA, USA) using a Trans-Blot Sd Semi-Dry Transfer Cell (Bio-rad). Membranes were blocked with Blocking One (Nacalai Tesque, Kyoto, USA). The following primary antibodies were used: anti-PCDH1 (#H00005097-M01, 1:500; Abnova) and anti-GAPDH (#MAB375, 1:1000; Merk Millipore). Horseradish peroxidase conjugated anti-mouse IgG (1:2000; Sigma-Aldrich) was used as a secondary antibody. Signals were detected using Amersham ECL Western blotting Detection Reagents (GE Healthcare, Chicago, IL, USA) or SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific).
3. Results
3.1. Pcdh1 mRNA expression in mouse embryos
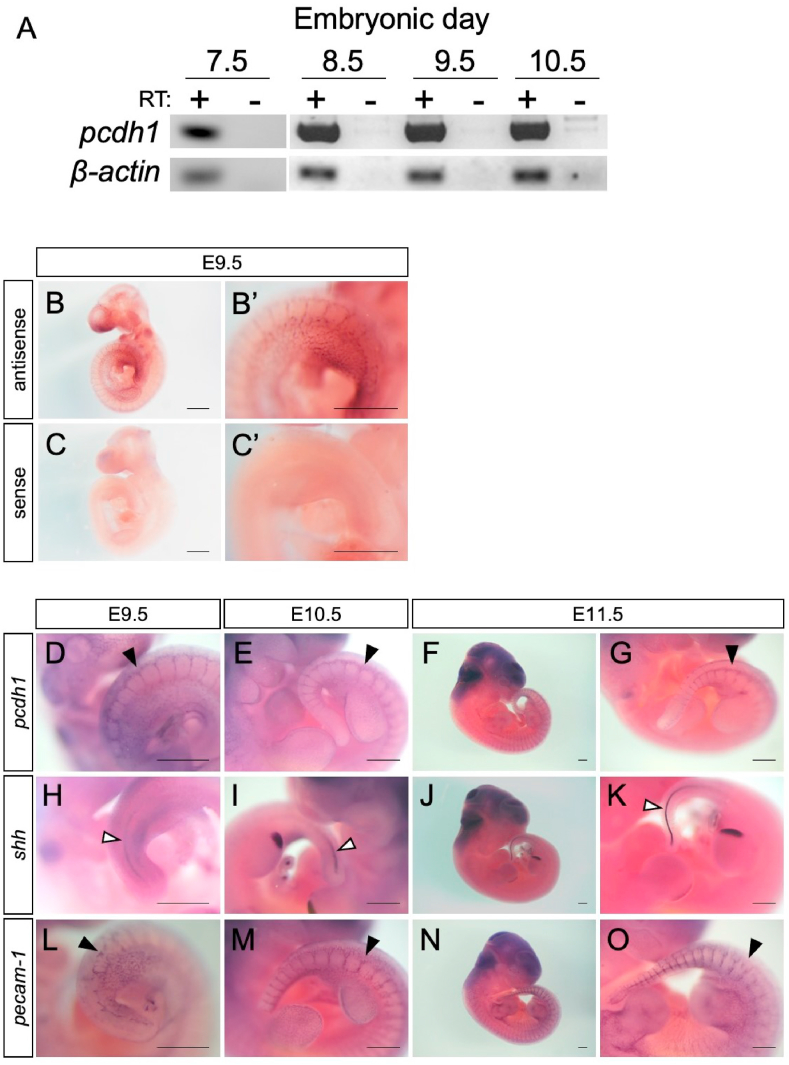
First, to explore the temporal expression of pcdh1, we performed RT-PCR on E7.5, E8.5, 9.5, and 10.5 embryos. Pcdh1 has several isoforms (see Fig. S1A), and we designed a primer pair in exons 2 and 3 shared by all of the isoforms (Fig. S1B). As a result, we detected pcdh1 mRNA in all the stages examined (Fig. 1A). Previously, pcdh1 mRNA was reported to be expressed from E9.5 [24], but in our set-up, the mRNA was detected as early as E7.5, prior to the stage when notochord cells detach from other tissue at the surface of the embryo [10]. We also detected the expression of pcdh1 mRNA in these stages with an independent primer set (Fig. S1B), indicating that the mRNA was expressed at stage of notochord formation (Fig. S2).
Fig. 1.
Thepcdh1mRNA expression in mouse embryos. (A) An RT-PCR analysis showing the expression of pcdh1 mRNA in mouse embryo. β-actin served as a loading control. RT, reverse transcription. (B, C) Whole-mount in situ hybridization using sense and antisense probes of pcdh1 with E9.5 embryos. (D–O) Whole-mount in situ hybridization using antisense probes of pcdh1, shh, and pecam-1 with E9.5, 10.5 and 11.5 embryos. Black arrowhead: intersomitic vessel. White arrowhead: notochord. Scale bar: 0.5 mm. The experiments were performed at least twice, and representative results are shown.
Next, to examine the spatial expression of pcdh1 mRNA in mouse embryos, we adopted the WISH approach. We designed three sense/antisense probes that covered most of the pcdh1 mRNA sequence (isoform NM_029357.3, Fig. S1B) and used them as a mixed probe (see the Materials and Methods for details). First, to confirm the specificity of our pcdh1 probes, WISH was performed with sense and antisense probes in E9.5 embryos. Tissue-specific signals were detected with the mix of the three antisense probes (Fig. 1B) but not with the mix of the three sense probes (Fig. 1C). We concluded that our mixed antisense probes detected pcdh1 mRNA specifically and designated the mix of the three antisense probes as “pcdh1 antisense” and that of the three sense probes as “pcdh1 sense”.
With the specificity of the probes confirmed, we examined the spatial expression pattern of pcdh1 mRNA using embryos at various stages (E9.5, 10.5, and 11.5). As we were focusing on the expression in notochord, shh, a notochord marker gene, was included as a control. Since the pcdh1 expression in blood vessels had been previously reported, pecam-1, an endothelial cell marker gene, was also included. As shown in Fig. 1D–G, pcdh1 expression was seen as lines on both sides of the dorsal midline (neural tube) as well as in the intersomitic region in all stages examined (Fig. 1D–G, black arrowheads). At E9.5 and 10.5, a mesh-like pattern of signals was detected at the surface of limb buds or branchial arches (Fig. 1D and E). These expression patterns coincided with those of pecam-1 (Fig. 1L–O), indicating that the pcdh1 mRNA was expressed in blood vessels at these stages. Although the expression of shh in notochord and the floor plate was clearly detected in E9.5–11.5 embryos (Fig. 1H–K, white arrowheads), pcdh1 mRNA was not detected in notochord in these WISH experiments (Fig. 1D–G).
3.2. PCDH1 protein expression in mouse embryos
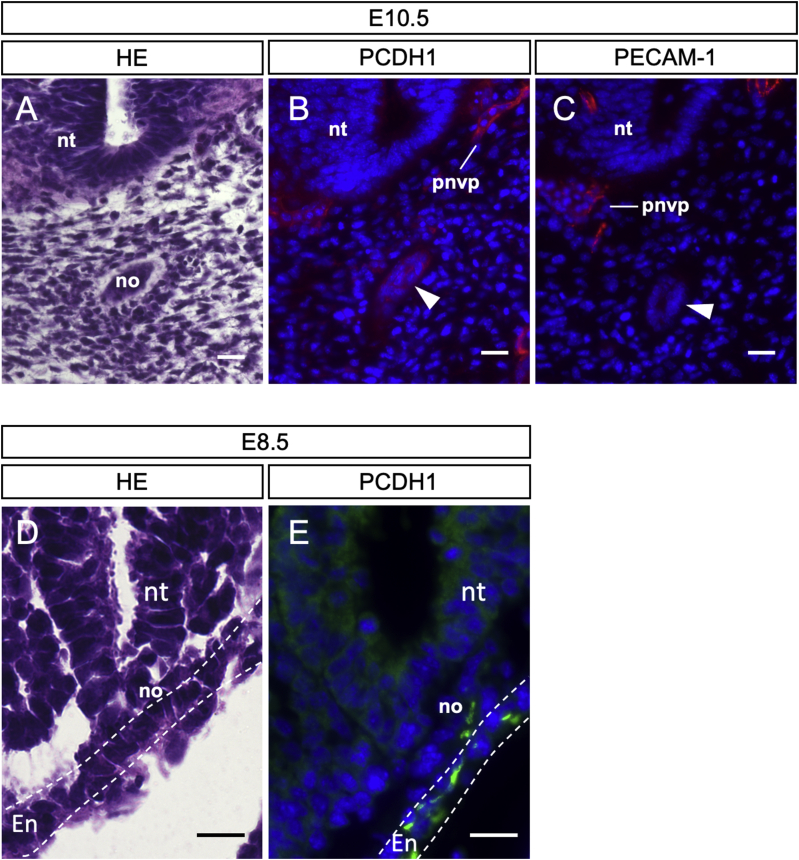
We also examined the expression of PCDH1 protein with immunofluorescence using E10.5 embryo sections. PECAM-1 was used as an endothelial cell marker. As a result, PCDH1 expression was detected in blood vessels, such as the peri-neural vascular plexus (Fig. 2B), which coincided with the PECAM-1 expression (Fig. 2C), confirming its expression in endothelial cells. In addition to the expression in the blood vessels, we detected a PCDH1 signal at the midline of the embryo, where PECAM-1 was not detected (Fig. 2B and C arrowheads). This signal corresponds with the position of the notochord structure when compared with the HE staining (Fig. 2A). These results indicate that PCDH1 is expressed in both blood vessels and notochord in mouse embryos. As the RT-PCR analysis showed the pcdh1 mRNA expression at E8.5, when the notochord formation occurs, next we examined if the PCDH1 protein is expressed in the notochord tissue. As the result, we have detected PCDH1 in a part of forming notochord cells as well as surrounding endoderm layer (Fig. 2D and E). The reason why we detected PCDH1 protein but not pcdh1 mRNA is probably due to the size and location of the mammalian notochord. The notochord in mouse embryos is a very thin tissue embedded in the middle of the embryo body, so the detection of its gene expression is more difficult with whole-mount analysis than with sections.
Fig. 2.
The PCDH1 protein expression in mouse embryos. (A) HE staining of transverse sections of E10.5 embryo. (B) Immunofluorescence staining for PCDH1 with transverse sections of E10.5 embryo. (C) Immunofluorescence staining for PECAM-1 with transverse sections of E10.5 embryo. DAPI staining (blue) shows nuclear positions. Note that the sections of A-C are consecutively taken from the same sample. White arrowhead: PCDH1-positive and PECAM-1-negative structure. Diagrams illustrating the positions of notochord and vasculature are shown in Figure S3. (D) HE staining of transverse sections of E8.5 embryos. (E) Immunofluorescence staining for PCDH1 with transverse sections of E8.5 embryos. DAPI staining (blue) shows nuclear positions. Note that the sections of D and E are consecutively taken from the same sample. no: notochord. nt: neural tube. pnvp: peri-neural vessel plexus. En: endodermal layer. Scale bar: 20 μm. Three embryos were used for each experiment. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.3. Generation of pcdh1 knockout mice using the CRISPR-Cas9 system
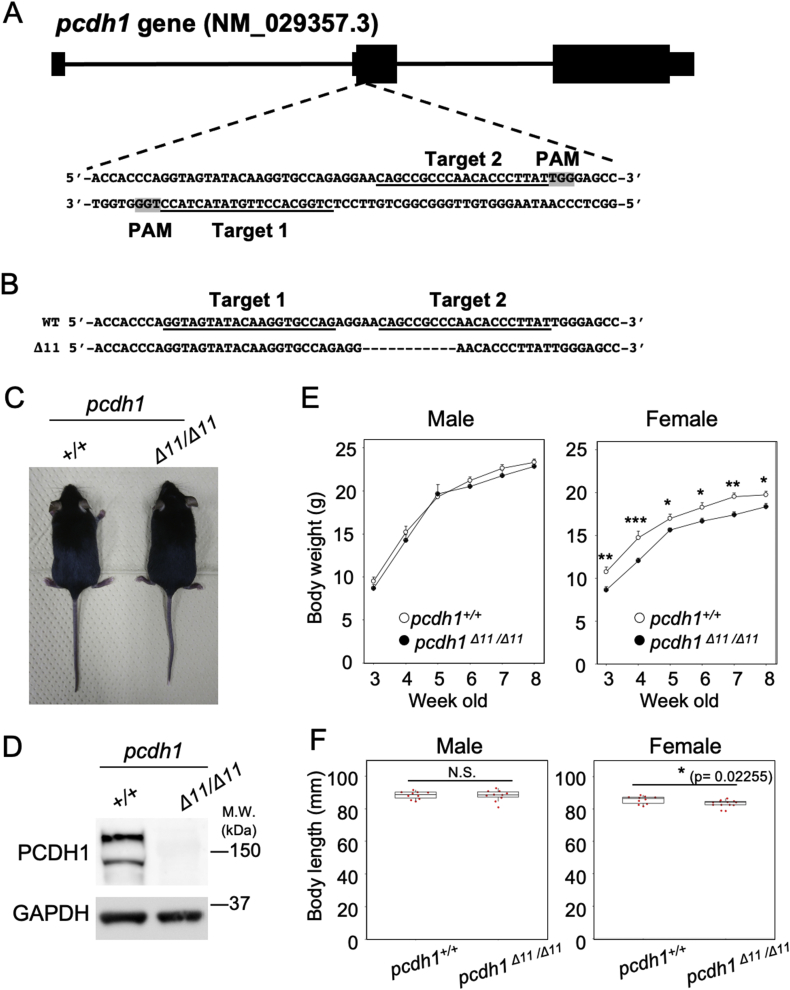
To reveal the function of PCDH1 in notochord, we generated pcdh1 gene knockout mice using the CRISPR-Cas9 system. We selected the double-nicking approach, which uses D10ACas9 and two gRNAs [23], to minimize the off-target effect. We designed a gRNA pair (target 1 and target 2) (Fig. 3A) in exon2 of the pcdh1 gene, which is common to all of the reported isoforms (Fig. S1A). We performed zygote microinjection and obtained a pup with an allele with an 11-base deletion 138 bases downstream from the start codon of the pcdh1 gene, which we hereafter call the pcdh1Δ11 allele (Fig. 3B). We successfully established a mouse line from this founder mouse.
Fig. 3.
Generation of pcdh1 knockout mice using the CRISPR-Cas9 system. (A) A schematic drawing showing the position and sequences of gRNAs targeting exon2 of pcdh1 gene. Exons (black boxes) and introns (black lines) indicate the structure of one of seven transcription variants of pcdh1 (NM_029357.3). Targeting sequences and PAM sequences of gRNAs are indicated with underline and a gray background, respectively. (B) Genomic sequences around the targeting site of the wild-type (WT) and Δ11 alleles. Targeting sequences of gRNAs are indicated with underline. Dashes: deleted bases. (C) Gross appearance of pcdh1+/+ and pcdh1Δ11/Δ11 female mice at 8 weeks old. (D) Western blotting to detect PCDH1 protein in liver tissue of pcdh1+/+ and pcdh1Δ11/Δ11 adult mice. GAPDH serves as a loading control. (E) Body weight of pcdh1+/+ and pcdh1Δ11/Δ11 mice at 3–8 weeks old. At least three mice were measured for each genotype. Data represent the mean ± SE. A t-test was used for the statistical analyses (*p < 0.05, **p < 0.01, and ***p < 0.001). (F) Body length of pcdh1+/+ and pcdh1Δ11/Δ11 adult male and female mice at 8 weeks old. At least three mice were measured for each genotype. Box plots show the median and 25-75th percentiles (middle line and box, respectively). Whiskers extend to the maximum and minimum data points. A t-test was used for the statistical analyses (*p < 0.05). N.S., not significant
The mice with homozygote pcdh1Δ11 allele (pcdh1Δ11/Δ11) are viable, born at a Mendelian ratio, and fertile (Fig. 3C). We performed Western blotting to confirm whether or not PCDH1 protein was depleted in pcdh1Δ11/Δ11 mice. Total protein was extracted from the liver tissues of pcdh1+/+ and pcdh1Δ11/Δ11 adult mice. The Western blotting analysis using an anti-PCDH1 antibody detected 2 bands around 150 kDa in the pcdh1+/+ (Fig. 3D) that were absent in pcdh1Δ11/Δ11 mice (Fig. 3D). Since the Δ11 mutation is in exon2 which is common in all of the reported isoforms, all PCDH1 proteins are expected to be depleted in pcdh1Δ11/Δ11 embryos and mice. We also confirmed that the PCDH1 protein was not detected by immunofluorescent in the notochord and blood vessels of pcdh1Δ11/Δ11 embryo (Fig. 4G and H). These results indicated that we had successfully generated a pcdh1 knockout mouse line.
Fig. 4.
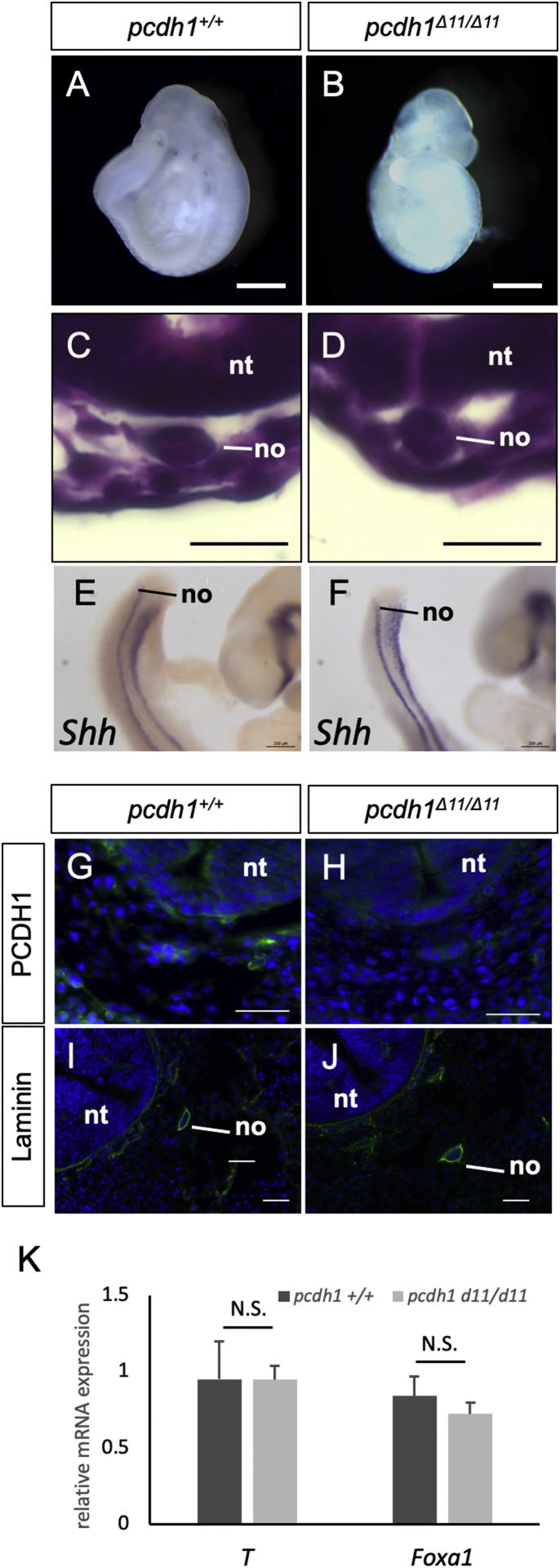
Notochord of pcdh1 knockout mice. (A, B) Gross appearance of E9.5 pcdh1+/+ and pcdh1Δ11/Δ11 embryos. Scale bars: 0.5 mm. (C, D) HE staining of the cross-sections of E9.5 pcdh1+/+ and pcdh1Δ11/Δ11 embryos. Scale bars: 20 μm. no: notochord. nt: neural tube. (E, F) In situ hybridization of E9.5 pcdh1+/+ and pcdh1Δ11/Δ11 embryos with shh probe. (G–J) Immunofluorescence of cross-sections of E10.5 pcdh1+/+ and pcdh1Δ11/Δ11 embryos. (K) qPCR analysis of E11.5 pcdh1+/+ and pcdh1Δ11/Δ11 embryos. A t-test was used for the statistical analyses (*p < 0.05). Four embryos were used for each genotype in each experiment. N.S., not significant
We measured the body weight of pcdh1+/+ and pcdh1Δ11/Δ11 mice at three to eight weeks old. As a result, the body weight of pcdh1Δ11/Δ11 female mice was significantly lower at all ages examined than wild type control (Fig. 3E), while no significant difference was observed in male mice (Fig. 3E). The body length of pcdh1Δ11/Δ11 of female mice, but not males, was also significantly lower at eight weeks old than wild type control (Fig. 3F).
3.4. Phenotypic analyses of PCDH1-deficient mice
Having generated PCDH1-deficient mice, we examined its notochord formation. The gross appearance of pcdh1Δ11/Δ11 embryo was indistinguishable from wild-type embryos at E9.5, indicating that the axis elongation process had not been affected by the loss of PCDH1 (Fig. 4A and B). A cross section of embryos at the level of the heart showed that the notochord had formed normally in pcdh1Δ11/Δ11 embryos (Fig. 4C and D). WISH analysis with shh probe revealed that the notochord structure along embryonic anterior-posterior axis was indistinguishable between pcdh1Δ11/Δ11 and wild-type embryos (Fig. 4E and F). Immunofluorescence of Laminin showed that there was no difference in basement membrane formation of notochord of pcdh1Δ11/Δ11 and wild-type embryos (Fig. 4I and J). These results indicate that PCDH1 is dispensable for the induction of notochord and its segregation from surrounding tissue. The expression level of key transcription factors for notochord such as T or Foxa1 was similar in pcdh1Δ11/Δ11 and wild-type embryos (Fig. 4K).
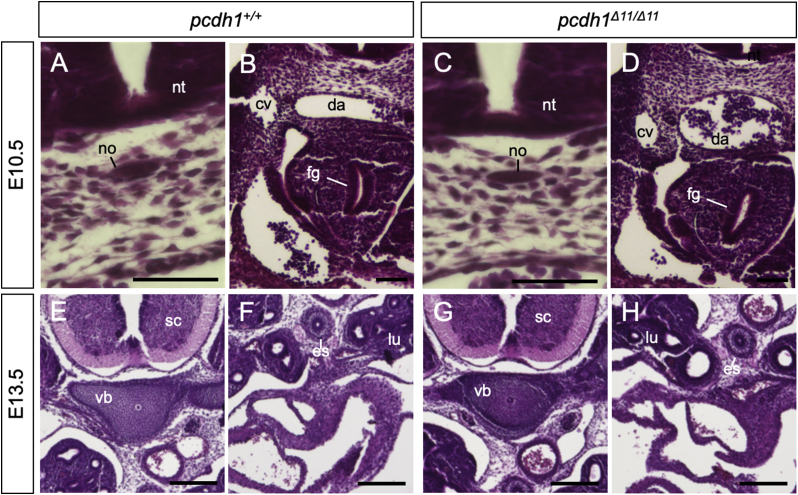
Next, to examine the notochord function as a signaling center, we observed the surrounding tissue, such as the neural tube, dorsal aorta, and foregut of E10.5 pcdh1Δ11/Δ11 embryos and the spinal code or vertebrae of E13.5 pcdh1Δ11/Δ11 embryos, and found that those tissues had formed similarly to their formation in pcdh1+/+ embryos (Fig. 5A–H). These results suggested that PCDH1 was dispensable for notochord to function as a signal source for the surrounding tissues.
Fig. 5.
Tissues surrounding notochord of pcdh1 knockout mice. (A–H) HE staining of the cross-sections of pcdh1+/+ and pcdh1Δ11/Δ11 embryos. (A–D) E10.5 embryos. Scale bars: 50 μm. (E–H) E13.5 embryos. Scale bars: 200 μm. cv: cardinal vein. da: dorsal aorta. es: esophagus. fg; foregut. lu: lung. no: notochord. nt: neural tube. sc: spinal code. vb: vertebra. Three embryos were used for each genotype in each experiment.
To investigate the effect of pcdh1 knockout on blood vessels, we compared the vessel patterns and morphology between pcdh1+/+ and pcdh1Δ11/Δ11 embryos. We performed WISH using pecam-1 probes at E10.5 and found that the major vascular patterns, including intersomitic vessels, did not differ markedly between pcdh1+/+ and pcdh1Δ11/Δ11embryos (Figs. S4A and B). In addition, we performed immunofluorescence staining for PECAM-1 in E13.5 pcdh1+/+ and pcdh1Δ11/Δ11 embryos. The expression of PECAM-1 in endothelial cells of pcdh1Δ11/Δ11 embryo was similar to that of pcdh1+/+ embryos (Figs. S4C and D). These results indicate that PCDH1 is dispensable for blood vessel formation and its embryonic patterning.
4. Discussion
In this study, we revealed that pcdh1 mRNA was expressed from E7.5, prior to the stage when notochord cells detach from other tissue, and PCDH1 protein was expressed in the notochord of mouse embryo from the stage in when notochord cells detach from other tissue. The pcdh1 knockout mice were viable and fertile with a reduced body length and weight in females. The reason for the female-specific reduction of body size is unclear at the moment, and a further study will be required to clarify the underlying mechanism. In pcdh1Δ11/Δ11 embryos, structure and gene expression of notochord and major vascular patterns were normal, indicating that PCDH1 is dispensable for notochord and blood vessel formation.
4.1. Expression pattern of mammalian pcdh1
The mouse pcdh1 gene is located on chromosome 18. Seven isoforms, including Refseq (NM_029357.3), are registered as mouse pcdh1 mRNA in the NCBI Gene data base (Fig. S1A). In a previous study, Redies et al. detected a relatively low expression of short and long isoforms of pcdh1 from E9.5 using isoform-specific primers [24]. In this study, we detected similar levels of pcdh1 mRNA in E7.5 to E10.5. The difference between our results and the previous report might be due to the primer pairs we used, which detect all seven isoforms (Fig. S1B). By immunofluorescence, we found that PCDH1 protein in notochord, endoderm layer and blood vessel (Fig. 2). While the expression of PCDH1 (AXPC) in notochord is conserved in amphibians and mammals, its expression domain is broader in mouse embryos ([19] and this study). The pcdh1 gene has isoforms with different transcription start sites (Fig. S1A). These isoforms may have different transcription regulatory region, which may account for the differences in the pcdh1 expression between amphibians and mammals. Broad expression domain of pcdh1, including notochord, endoderm, and endothelial cells, is confirmed also in the published data of single cell transcriptome analysis of E8.25 embryo [25].
4.2. The role of the pcdh1 gene in mammalian development
As the deletion in pcdh1Δ11/Δ11 mice is located in exon 2, which is shared among all seven isoforms of pcdh1, the proteins of all isoforms are expected to be depleted in pcdh1Δ11/Δ11 mice. In fact, we detected two protein bands in our Western blotting analysis, both of which were depleted in pcdh1Δ11/Δ11 mice (Fig. 3D). In Syrian hamsters, pcdh1 knockout results in animals that are viable, fertile, and with no major organ abnormalities [26]. The knockout mouse phenotype in this study is consistent with this result.
The notochord of PCDH1-deficient mice was formed in the normal position. These phenotypes show clear differences compared to the phenotype of AXPC (PCDH1 orthologue)- knockdown Xenopus embryos, where the notochord is not properly formed and axis elongation is inhibited [19]. Two reasons may be responsible for this discrepancy: the structural differences in amphibian and mammalian notochord, or a functional redundant gene that might exist in the mammalian genome. In amniotes, the notochord consists of a thin, acellular sheath covering inner vacuolated cells [27,28]. In contrast, in lower vertebrates, the notochord is a multilayered structure in which large vacuolated cells are surrounded by an epithelial sheath, which is further encapsulated in extracellular matrix layers [3,29]. The diameter of the notochord in lower vertebrates is much larger than that of mammalians, suggesting a larger contribution to mechanical support in axis formation. Alternatively, PCDH1 may work redundantly with other adhesion molecules expressed in notochord, such as P-cadherin [30]. PCDH11, which forms a heterodimer with PCDH1, may also play some role with PCDH1 [31]. CDH1 and CDH2 are also expressed in notochord and endoderm layer, respectively, during the notochord formation [32]. It would be interesting to analyze mice with multiple knockout of PCDH1 and these candidate genes in a future study. Since mammalian notochord has a limited cell number, and no immortalized cell line is available, the wholistic view of the adhesion molecules expressed in the notochord remains unclear. However, newly developed technologies, such as a transcriptomics analysis [33] or induction of notochord-like cells from pluripotent stem cells [34], may provide a comprehensive understanding of notochord adhesion molecules.
In conclusion, we identified the conserved expression of PCDH1 in mammalian notochord. Phenotypes of our pcdh1Δ11/Δ11 mice suggested that the role of PCDH1 might differ among vertebrate species. It will be interesting to examine the role of PCDH1 orthologues in other vertebrate clades as well as in Cephalochordata or Urochordata in a future study.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the Yamagishi Student Project Support Program, the TTCK RAship from Yamagata prefectural government and the City of Tsuruoka, and Taikichiro Mori Memorial Research Fund (Graduate Student Researcher Development Grant) for K.F.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2021.101047.
Author contributions
H. K. and M. I. planned the study, and K. F., M. T. and N. H. performed the experiments. K. F., H. K., and M. I. planned and wrote the manuscript.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Corallo D., Trapani V., Bonaldo P. The notochord: structure and functions. Cell. Mol. Life Sci. 2015;72:2989–3008. doi: 10.1007/s00018-015-1897-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams D.S., Keller R., Koehl M.A. The mechanics of notochord elongation, straightening and stiffening in the embryo of Xenopus laevis. Development. 1990;110:115–130. doi: 10.1242/dev.110.1.115. [DOI] [PubMed] [Google Scholar]
- 3.Ellis K., Bagwell J., Bagnat M. Notochord vacuoles are lysosome-related organelles that function in Axis and spine morphogenesis. J. Cell Biol. 2013;200:667–679. doi: 10.1083/jcb.201212095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liem K.F., Jessell T.M., Briscoe J., Acampora D., Gulisano M., Simone A., Amthor H., Connolly D., Patel K., Brand-Saberi B. Regulation of the neural patterning activity of sonic hedgehog by secreted BMP inhibitors expressed by notochord and somites. Development. 2000;127:4855–4866. doi: 10.1242/dev.127.22.4855. [DOI] [PubMed] [Google Scholar]
- 5.Placzek M. The role of the notochord and floor plate in inductive interactions. Curr. Opin. Genet. Dev. 1995;5:499–506. doi: 10.1016/0959-437x(95)90055-l. [DOI] [PubMed] [Google Scholar]
- 6.Fan C.-M., Tessier-Lavigne M. Patterning of mammalian somites by surface ectoderm and notochord: evidence for sclerotome induction by a hedgehog homolog. Cell. 1994;79:1175–1186. doi: 10.1016/0092-8674(94)90009-4. [DOI] [PubMed] [Google Scholar]
- 7.Pourquié O., Coltey M., Teillet M.A., Ordahl C., Le Douarin N.M. Control of dorsoventral patterning of somitic derivatives by notochord and floor plate. Proc. Natl. Acad. Sci. U.S.A. 1993;90:5242–5246. doi: 10.1073/pnas.90.11.5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fouquet B., Weinstein B.M., Serluca F.C., Fishman M.C. Vessel patterning in the embryo of the zebrafish: guidance by notochord. Dev. Biol. 1997;183:37–48. doi: 10.1006/dbio.1996.8495. [DOI] [PubMed] [Google Scholar]
- 9.Cleaver O., Krieg P.A. Notochord patterning of the endoderm. Dev. Biol. 2001;234:1–12. doi: 10.1006/dbio.2001.0214. [DOI] [PubMed] [Google Scholar]
- 10.Balmer S., Nowotschin S., Hadjantonakis A.K. Notochord morphogenesis in mice: current understanding and open questions. Dev. Dynam. 2016;245:547–557. doi: 10.1002/dvdy.24392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sasaki H., Hogan B.L.M. Differential expression of multiple fork head related genes during gastrulation and axial pattern formation in the mouse embryo. Development. 1993;118:47–59. doi: 10.1242/dev.118.1.47. [DOI] [PubMed] [Google Scholar]
- 12.Chesley P. Development of the short‐tailed mutant in the house mouse. J. Exp. Zool. 1935;70:429–459. [Google Scholar]
- 13.Herrmann B.G., Labeit S., Poustka A., King T.R., Lehrach H. Cloning of the T gene required in mesoderm formation in the mouse. Nature. 1990;343:617–622. doi: 10.1038/343617a0. [DOI] [PubMed] [Google Scholar]
- 14.Abdelkhalek H. Ben, Beckers A., Schuster-Gossler K., Pavlova M.N., Burkhardt H., Lickert H., Rossant J., Reinhardt R., Schalkwyk L.C., Müller I. The mouse homeobox gene Not is required for caudal notochord development and affected by the truncate mutation. Genes Dev. 2004;18:1725–1736. doi: 10.1101/gad.303504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.von Dassow G., Schmidt J.E., Kimelman D. Induction of the Xenopus organizer: expression and regulation of Xnot, a novel FGF and activin-regulated homeo box gene. Genes Dev. 1993;7:355–366. doi: 10.1101/gad.7.3.355. [DOI] [PubMed] [Google Scholar]
- 16.Stemple D.L. Structure and function of the notochord: an essential organ for chordate development. Development. 2005;132:2503–2512. doi: 10.1242/dev.01812. [DOI] [PubMed] [Google Scholar]
- 17.Nibu Y., José-Edwards D.S., Gregorio A. Di. From notochord formation to hereditary chordoma: the many roles of Brachyury. BioMed Res. Int. 2013;2013 doi: 10.1155/2013/826435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuroda H., Sakumoto H., Kinoshita K., Asashima M. Changes in the adhesive properties of dissociated and reaggregated Xenopus laevis embryo cells. Dev. Growth Differ. 1999;41:283–291. doi: 10.1046/j.1440-169x.1999.413428.x. [DOI] [PubMed] [Google Scholar]
- 19.Kuroda H., Inui M., Sugimoto K., Hayata T., Asashima M. Axial protocadherin is a mediator of prenotochord cell sorting in Xenopus. Dev. Biol. 2002;244:267–277. doi: 10.1006/dbio.2002.0589. [DOI] [PubMed] [Google Scholar]
- 20.Yoder M.D., Gumbiner B.M. Axial protocadherin (AXPC) regulates cell fate during notochordal morphogenesis. Dev. Dynam. 2011;240:2495–2504. doi: 10.1002/dvdy.22754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inui M., Tamano M., Kato T., Takada S. CRISPR/Cas9-mediated simultaneous knockout of Dmrt1 and Dmrt3 does not recapitulate the 46,XY gonadal dysgenesis observed in 9p24.3 deletion patients. Biochem. Biophys. Reports. 2017;9:238–244. doi: 10.1016/j.bbrep.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inui M., Miyado M., Igarashi M., Tamano M., Kubo A., Yamashita S., Asahara H., Fukami M., Takada S. Rapid generation of mouse models with defined point mutations by the CRISPR/Cas9 system. Sci. Rep. 2014;4:5396. doi: 10.1038/srep05396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ran F.A., Hsu P.D., Lin C.-Y., Gootenberg J.S., Konermann S., Trevino A.E., Scott D.A., Inoue A., Matoba S., Zhang Y. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Redies C., Heyder J., Kohoutek T., Staes K., Van Roy F. Expression of protocadherin-1 (Pcdh1) during mouse development. Dev. Dynam. 2008;237:2496–2505. doi: 10.1002/dvdy.21650. [DOI] [PubMed] [Google Scholar]
- 25.Ibarra-Soria X., Jawaid W., Pijuan-Sala B., Ladopoulos V., Scialdone A., Jörg D.J., Tyser R.C.V., Calero-Nieto F.J., Mulas C., Nichols J. Defining murine organogenesis at single-cell resolution reveals a role for the leukotriene pathway in regulating blood progenitor formation. Nat. Cell Biol. 2018;20:127–134. doi: 10.1038/s41556-017-0013-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jangra R.K., Herbert A.S., Li R., Jae L.T., Kleinfelter L.M., Slough M.M., Barker S.L., Guardado-Calvo P., Román-Sosa G., Dieterle M.E. Protocadherin-1 is essential for cell entry by New World hantaviruses. Nature. 2018;563:559–563. doi: 10.1038/s41586-018-0702-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi K.-S., Cohn M.J., Harfe B.D. Identification of nucleus pulposus precursor cells and notochordal remnants in the mouse: implications for disk degeneration and chordoma formation. Dev. Dynam. 2008;237:3953–3958. doi: 10.1002/dvdy.21805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ward L., Pang A.S.W., Evans S.E., Stern C.D. The role of the notochord in amniote vertebral column segmentation. Dev. Biol. 2018;439:3–18. doi: 10.1016/j.ydbio.2018.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ellis K., Hoffman B.D., Bagnat M. The vacuole within: how cellular organization dictates notochord function. BioArchitecture. 2013;3:64–68. doi: 10.4161/bioa.25503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nose A., Takeichi M. A novel cadherin cell adhesion molecule: its expression patterns associated with implantation and organogenesis of mouse embryos. J. Cell Biol. 1986;103:2649–2658. doi: 10.1083/jcb.103.6.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harrison O.J., Brasch J., Katsamba P.S., Ahlsen G., Noble A.J., Dan H., Sampogna R.V., Potter C.S., Carragher B., Honig B. Family-wide structural and biophysical analysis of binding interactions among non-clustered δ-protocadherins. Cell Rep. 2020;30:2655–2671. doi: 10.1016/j.celrep.2020.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fausett S.R., Brunet L.J., Klingensmith J. BMP antagonism by Noggin is required in presumptive notochord cells for mammalian foregut morphogenesis. Dev. Biol. 2014;391:111–124. doi: 10.1016/j.ydbio.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 33.Peck S.H., McKee K.K., Tobias J.W., Malhotra N.R., Harfe B.D., Smith L.J. Whole transcriptome analysis of notochord-derived cells during embryonic formation of the nucleus pulposus. Sci. Rep. 2017;7:10692–10695. doi: 10.1038/s41598-017-10692-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y., Zhang Z., Chen P., Ma C.Y., Li C., Au T.Y.K., Tam V., Peng Y., Wu R., Cheung K.M.C. Directed differentiation of notochord-like and nucleus pulposus-like cells using human pluripotent stem cells. Cell Rep. 2020;30:2791–2806. doi: 10.1016/j.celrep.2020.01.100. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.