Abstract
Translocation is essential to the anthrax toxin mechanism. Protective antigen (PA), the binding component of this AB toxin, forms an oligomeric pore that translocates lethal factor (LF) or edema factor, the active components of the toxin, into the cell. Structural details of the translocation process have remained elusive despite their biological importance. To overcome the technical challenges of studying translocation intermediates, we developed a method to immobilize, transition, and stabilize anthrax toxin to mimic important physiological steps in the intoxication process. Here, we report a cryoEM snapshot of PApore translocating the N-terminal domain of LF (LFN). The resulting 3.3 Å structure of the complex shows density of partially unfolded LFN near the canonical PApore binding site. Interestingly, we also observe density consistent with an α helix emerging from the 100 Å β barrel channel suggesting LF secondary structural elements begin to refold in the pore channel. We conclude the anthrax toxin β barrel aids in efficient folding of its enzymatic payload prior to channel exit. Our hypothesized refolding mechanism has broader implications for pore length of other protein translocating toxins.
Subject terms: Biochemistry, Protein folding, Structural biology
Introduction
The anthrax toxin is not only a deadly Bacillus anthracis virulence factor, but also serves as a model system of protein translocation and as a peptide therapeutic delivery platform1,2. Its biological importance and biotechnology utility have spurred significant biochemical and biophysical advances in understanding the anthrax intoxication mechanism. In order to gain entry into the cell, this archetypical AB toxin must cross the endosomal membrane. Membrane penetration is accomplished by the B component of anthrax toxin, termed protective antigen (PA). PA forms a translocon pore through which lethal factor (LF) or edema factor (EF), the A component, translocates. Here, we developed an approach to elucidate the structural and mechanistic details of the anthrax toxin during translocation in an effort to understand how LF unfolds in the endosome, translocates through PA, and refolds in the cytosol.
An overview of the anthrax toxin mechanism has been reviewed by the Collier lab1 and is briefly summarized here. The first step in intoxication is the 85 kDa monomeric PA binding to host cell receptors. Then the pro-domain of PA is cleaved leaving the 63 kDa PA to oligomerize into heptameric or octameric prepore (PAprepore)3,4. Up to three LF and/or EF components can bind to the PAprepore heptamer4–6. The AB toxin complex is endocytosed through clathrin mediated endocytosis7. As the endosome acidifies, PAprepore undergoes a conformational change to a pore (PApore)8. This pore inserts into the endosomal membrane to form a channel. The low pH of the endosome and the pH gradient between the endosome and the cytosol facilitate LF or EF to unfold and rapidly translocate into the cytosol in a hypothesized Brownian ratchet mechanism9. Natively refolded LF and EF in the cytosol are then able to perform their virulent enzymatic functions10.
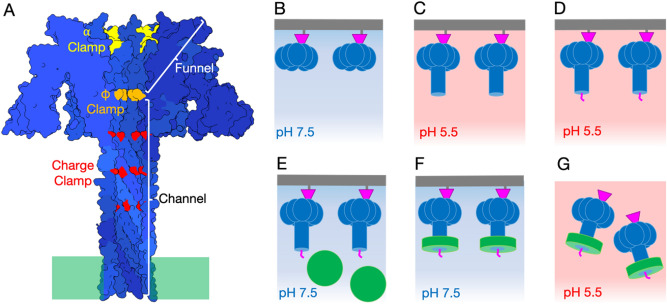
The overall structure of the PApore translocon can be divided into two regions: the funnel and the channel (Fig. 1A). The first region, the funnel, facilitates binding and unfolding of LF. LF binds to the rim of the PApore funnel and is guided down the narrowing structure. The second region of PApore is the channel, a β barrel that extends from the funnel and spans the endosomal membrane. Three nonspecific PApore clamp sites (α, Φ, and charge clamp) aid in the translocation of LF (Fig. 1A). The α clamp is located at the PA funnel rim, is formed by adjacent PA protomers, and binds helical portions of LF to position them towards the pore lumen. Heptameric PApore has seven potential α clamp binding sites. A crystal structure of the N-terminal domain of LF (LFN) bound to the PAprepore revealed the α clamp binding site11. Blocking of this α clamp binding site results in reduced translocation12. The second clamp site is the Φ clamp, a ring of seven phenylalanine residues that maintain the pH gradient between the endosome and the cytosol13. The cryoEM structure of apo PApore revealed the Φ clamp forms a narrow 6 Å diameter ring14. Secondary structural elements, such as α helices, are too wide to fit through this narrow seal. Therefore, it is hypothesized that peptide substrates must completely unfold and refold in order to translocate through the PApore and enter the cytosol of the cell14. The Φ clamp also assists in the unfolding of LF as an unfolding chaperone2. The third clamp site, the charge clamp, is located within the β barrel of PApore15,16. The charge clamp deprotonates acidic side chains of LF and ensures unidirectional movement of the polypeptide15. Interestingly, the diameter of the PApore β barrel is large enough to accomidate an α helix, which would allow for initial refolding to occur inside the pore prior to LF entering the cytosol. However, it remains unclear what structural state LF is in when interacting with the charge clamp and within the β barrel channel.
Figure 1.
Anthrax toxin immobilization, translocation, and nanodisc stabilization (TITaNS) method (A) PApore side view slice with funnel shape from α clamp (yellow) to Φ clamp (orange) and charge clamp (red) inside pore β barrel channel indicated. (B) Immobilization of LFN (magenta) PAprepore (blue) complexes on thiol sepharose beads (grey surface). (C) PAprepore transitioned to PApore. (D) Predicted translocation complex of LFN -PApore at low pH. (E) Addition of pre-nanodisc micelle (green) to complex. (F) Nanodisc formation. (G) LFN-PApore-Nanodisc translocation complexes at pH 5.5 on cryoEM grid.
One of the many challenges in studying the anthrax toxin is that it is a dynamic membrane protein that functions under acidic conditions. Thus many questions remain, such as what path LF travels down the endosomal pore lumen from the α clamp to the Φ clamp, whether the Φ clamp adopts multiple states during translocation, and whether LF can partially refold inside the β barrel pore. To address these questions, we developed a novel toxin immobilization, translocation, and nanodisc stabilization (TITaNS) method in combination with cryoEM to structurally characterize PApore translocating the N-terminal domain of LF (LFN). This approach provides unique mechanistic insight into how LFN interacts with the three clamp sites of PApore. We observed density consistent with LFN unfolding prior to the α clamp, translocating through the dynamic Φ clamp, and beginning to refold in the channel of the PApore.
Results
Assembly of anthrax translocation complexes
In vivo, the anthrax toxin undergoes a prepore to pore conformational change under acidic conditions. Previous structural studies have generally used urea to avoid aggregation during the transition from PAprepore to PApore17–22. These approaches have limitations in that they do not account for the low pH electrostatic microenvironment in the pore lumen predicted to be important for LF-PA interactions23 and they assume similar outcomes for chaotrope and acid induced unfolding. In order to overcome these limitations we have developed a novel assembly method for toxin immobilization, translocation, and nanodisc stabilization (called TITaNS, Fig. 1B–G). TITaNS was designed to mimic important low pH physiological states during the anthrax intoxication mechanism. This approach allows for endosomal pH pore formation and imaging of individual complexes in a lipid bilayer in the biologically relevant low pH environment17,24. TITaNS can be used in combination with techniques other than cryoEM, including mass spectrometry, nuclear magnetic resonance, surface plasmon resonance, and biolayer interferometry. TITaNS also has the potential to be adapted to screen prospective pharmaceuticals that arrest or prevent endosomal membrane insertion24.
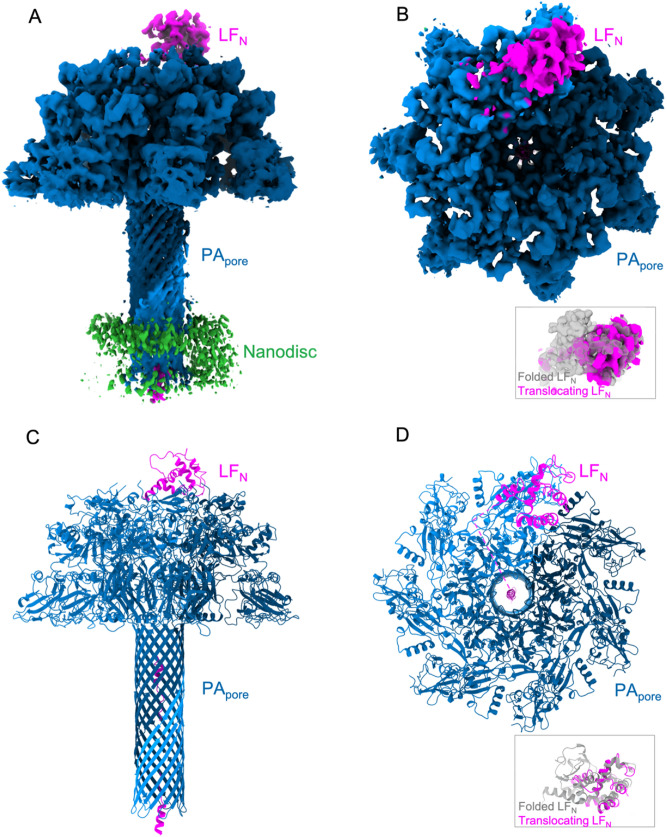
Reversible immobilization was key to the TITaNS methodology, because it allowed the stabilized complexes to be released into solution. We began with recombinantly purified, soluble forms of LFN and PAprepore that were mixed together in solution. The binary complex of LFN bound to PAprepore was then immobilized onto thiol sepharose beads by covalently coupling E126C LFN to the bead surface (Fig. 1B). The E126C immobilization site is opposite the PA binding site. The rationale for choosing this residue is that it allows for immobilization of LFN-PAprepore complexes without hindering PAprepore binding (Fig. 1B) and facilitates prepore to pore extension away from the bead surface when the complex is exposed to low pH (Fig. 1C). We predict this low pH environment initiates translocation of LFN through PApore in vitro (Fig. 1D). We base this prediction on computational and experimental evidence of early translocation events induced by low pH. Specifically, molecular simulations of anthrax toxin early translocation events predict the events are strongly influenced by the protonation state of LF and are highly favorable at low pH23. Also at low pH, the partial translocation of LF has been observed in planar lipid bilayers9. After pore formation, the next step in TITaNS was the stabilization of LFN-PApore translocation complexes using nanodisc technology25–27. Pre-nanodisc micelles were added to the bead slurry and associated with the transmembrane portion of PApore (Fig. 1E). To promote lipid bilayer formation, we dialyzed away excess detergent (Fig. 1F). The soluble LFN-PApore-nanodisc complexes were then eluted off the thiol sepharose beads using the reducing agent dithiothreitol. Eluted complexes were transferred to the cryoEM grid and the pH was dropped to pH 5.5 to capture the complex at low pH prior to blotting and plunge freezing (Fig. 1G). To prevent aggregation and migration of the nanodiscs to the air–water interface, we plunge froze the grids within 30 s of sample application. Using our TITaNS methodology, we obtained a 3.3 Å reconstruction of LFN translocating through PApore (Fig. 2A–D).
Figure 2.
Overview of the anthrax toxin translocation complex. CryoEM density map (A) side and (B) top view of LFN translocating through PApore. Inset: folded LFN model density (PDB 6PSN) compared to translocating LFN density. Molecular model (C) side and (D) top view of LFN translocating through PApore. Inset: folded LFN model (PDB 6PSN) compared to translocating LFN model. Discontinuous density indicated in model with dashed magenta lines.
Comparison of LFN before and during translocation
Prior to translocation, LFN is bound to the cap of PA at the interface of two PA protomers with helix α1 bound to the α clamp11,18. Our reconstruction shows a loss of LFN density in the canonical binding site above the PApore (Fig. 2A,B). Lethal factor translocates through PApore from the N-terminus to the C-terminus. Notably, our reconstruction does not show density for residues 50–135 in the canonical folded LFN position (Fig. 2B,D)11,17. However, density for residues 136–250 are apparent. Our model of unfolding, translocating LFN is consistent with a molten globular intermediate state at low pH28.
Molecular interactions of translocation complex
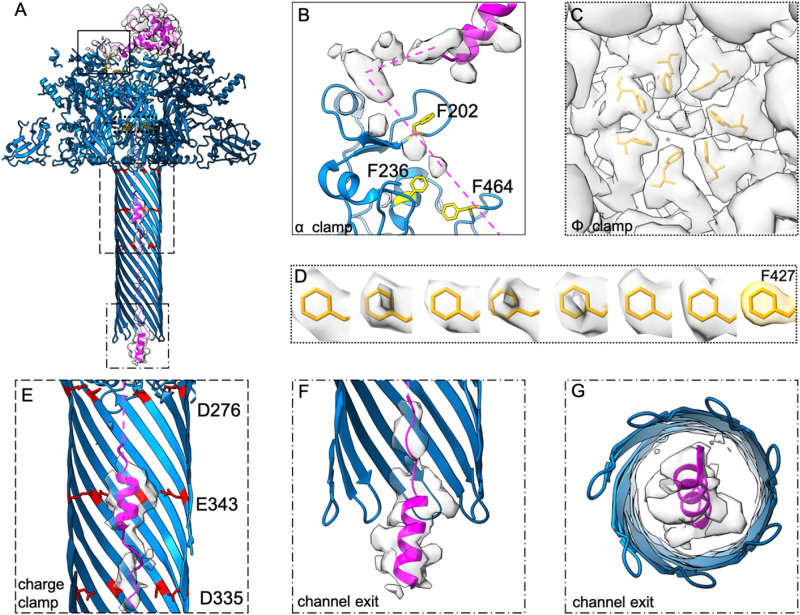
In addition to a loss of LFN density in the canonical binding site above the PApore, our reconstruction shows added density inside the pore lumen indicating a translocating complex (Fig. 3). This translocating LFN density is discontinous. Dashed lines are used to indicate regions where there is a not enough density to empirically build model (Fig. 3A). Density near the top of the PApore funnel was in proximity to several hydrophobic residues of PApore in the α clamp (Fig. 3B). F202, F236, and F464 have previously been shown to be important for early translocation. Specifically, mutagenesis of F202 and F236 decreases translocation16 while F464 has shown plasticity when binding translocation substrates18. These hydrophobic residues are predicted to aid in unfolding LF and stabilizing unfolded intermediates as they transition into the pore11. Our results are consistent with F202, F236, and F464 facilitating translocation of LFN into the pore and toward the Φ clamp in an unfolded state. We also observed added asymmetric density in and around the Φ clamp (Fig. 3C) that was not visible when we compared it to the previously published apo PApore cryoEM structure14. Specifically, there is density in the center of the Φ clamp (Fig. 3C). We attribute this density to unfolded LFN interacting with the benzyl rings of the PApore Φ clamp as LFN is translocating through the pore. In addition, the density for each of the PApore F427 residues was smeared in plane with the benzyl ring suggesting rotameric states moving up and down (Fig. 3D).
Figure 3.
Unfolding and refolding of LFN at key clamp sites during translocation through PApore. (A) Model of PApore (blue) translocating LFN (magenta) with α clamp, Φ clamp, and charge clamp residues shown in yellow, orange, and red respectively. CryoEM density of LFN shown in transparent grey. (B) Hydrophobic residues predicted to facilitate unfolding in early translocation from α clamp to Φ clamp. (C) Φ clamp ring with LFN density. (D) F427 residues for each subunit with associated cryoEM density compared to modelled density. (E) LFN model and density in β barrel charge clamp. (F) PApore channel exit into the cytosol with LFN α1 helical model and cryoEM density. (G) 90° rotation of F. Discontinuous density indicated in model with dashed magenta lines.
Translocating LFN density was also observed in the β barrel of the PApore. Focused refinement of the β barrel interior revealed density consistent with α helices along with portions of unfolded peptide (Fig. 3E–G). Notably, density located at the PApore charge clamp is consistent with an α helix and suggests the deprotonated state of LF favors helix formation within the pore. Canonical charge clamp residues D276, E343, and D335 are shown in Fig. 3E with the predicted LFN density translocating through the center of the channel. Further down the pore we observe density for helix α1 emerging from the channel exit (Fig. 3F,G). Our results provide evidence for initial refolding of LF secondary structure both inside and upon exit from the PApore. These results are consistent with ribosome exit tunnel studies that, using optical tweezers and molecular dynamics simulations, showed excluded volume effects and electrostatic interactions contribute to substrate folding29. In the future, TITaNS could be combined with site specific cross-linking between the PApore clamp sites and LFN to capture other unfolded, translocating intermediates.
Next, we wanted to determine whether there were any differences between PApore protomers that interacted directly with the translocating LFN compared to protomers that did not interact with translocating LFN. Importantly, symmetry operations were not used during single particle analysis. To compare the PApore protomers, we aligned the seven protomer chains that compose the PApore (Fig. S1A). Comparison of the chains showed little difference in the backbone or side chain rotamers for the majority of the residues at the LFN-PApore interfaces as well as the protomer-protomer interfaces. This rigidity is likely necessary for the PApore to maintain a stable β barrel and perform its function under endosomal conditions. During this analysis, we also examined the conformation of the receptor binding domain of PApore. The receptor binding domain is connected to the main body of PApore by a single loop and is responsible for anchoring the toxin to the host cell membrane prior to complex endocytosis and pore formation30. Interestingly, the receptor binding domain did show different conformations for each protomer and indicates a degree of conformational flexibility (Fig. S1A–C). This variability between receptor binding domains likely arose from the acidic conditions and lack of a receptor. Overlaying our PApore chains with the crystal structure of PAprepore bound to its receptor, capillary morphogenesis protein 2 (CMG2), revealed several conformations not conducive to receptor binding (Fig. S1C). Specifically, PA E194 was not in position to form the metal ion-dependent adhesion site (MIDAS) motif in the receptor binding pocket31. This indicates that without a receptor bound, the receptor binding domain loops adopt multiple states.
Discussion
The anthrax toxin PApore unfolds, translocates, and refolds LF, it’s enzymatic substrate. Three clamp sites aid in peptide translocation: the α clamp, the Φ clamp, and the charge clamp. We report here, cryoEM density consistent with nascent polypeptide chain translocating the length of PApore. In our model, LFN can be seen unfolding prior to the α clamp, passing through the dynamic Φ clamp, and refolding in the β barrel channel. This model is consistent with the hypothesis that LF needs to completely unfold in order to translocate. However, if the entire 90 kDa enzyme were to unfold at once, deleterious folded intermediates or aggregates would likely block the PApore translocon, especially when multiple LF are bound to PA. Therefore, in order to efficiently translocate and refold, LF unfolds from the N to C terminus32. While the low pH of the endosome destabilizes the enzyme, it does not completely unfold into its primary sequence28,33. Our results are consistent with molten globular translocation intermediates of LF being destabilized in the acidic environment of the endosome28 with the α clamp then able to apply additional unfolding force on the protein and funnel LF towards the Φ clamp2. Translocation requires step-wise unfolding and stabilization of the unfolded intermediates to prevent aggregation. When LF binds to PApore, helix α1 of LF moves away from the main body of LF and binds to the α clamp of PApore11. From here, LF has multiple paths it could take through the PApore funnel, gated by the Φ clamp. We were unable to resolve distinct unfolded LFN density passed the α clamp to the Φ clamp. However, our results do suggest a favorable path from the α to the Φ clamp would involve a series of hydrophobic residues that are amenable to unfolded translocation intermediates and likely serve as checkpoints to verify the unfolded state of LF prior to the Φ clamp (Fig. 3A–B).
The Φ clamp plays a crucial roll in translocation by acting as a hydrophobic seal between the endosome and cytosol9. Steric clashing between the narrow Φ clamp and bulkier LF side chains suggests the need for movement of the clamp34. Indeed, the dynamic nature of the Φ clamp has recently been measured using 19F NMR34. The smeared density in our structure is indicative of a dynamic clamp. Our density does not imply dilation of the clamp35, so much as a up and down motion along the pore axis. This motion could be conserted or individual F427 residues moving to accommodate various translocating side chains. The compressive and tensile forces generated by the unfolding LF in the PApore funnel above and the refolding LF in the β barrel channel below may also contribute to this movement. However, too much flexibility or dilation would cause the seal at the Φ clamp to be lost. Therefore, this dynamic motion must still maintain the pH gradient between the endosome and cytosol, while accomidating any side chain, ensuring efficient translocation. Multiple Φ clamp states have been hypothesized at pH 5.535. Our analysis did not reveal multiple distinct states, though it does not exclude the possibility. A larger dataset or different conditions may be needed to address this question.
Helix formation inside the PApore β barrel has been hypothesized but, to our knowledge, never observed14,28. We report here, evidence of concomitant α helix formation and translocation inside the β barrel of the PApore (Fig. 3E–G). We hypothesized that, along with changing the charge state of the peptide substrate, the charge clamp allows for a local folding environment within the PApore. Helical portions of LF have previously been shown to dock into the α clamp, with the periodicity of these helices aiding in efficient unfolding of LF36. We predict this periodicity is also important for hypothesized refolding of LF, beginning at the charge clamp. Our hypothesis is consistent with other anthrax toxin substrates, such as LFN fused to the catalytic chain of diphtheria toxin (LFN-DTA), which did not evolve to fold in the PApore channel. Interestingly, these non-native substrates require chaperones for enzymatic activity37 indicating the DTA portion of these proteins do not form helices in the PA channel at optimal intervals. Our model is also reminesant of the ribosome, where helix folding in the exit tunnel aids in co-translational folding of native proteins38. We predict helix folding in the PApore β barrel aids in co-translocational folding by temporally altering LF emersion from the tunnel allowing regions to fold into tertiary structures.
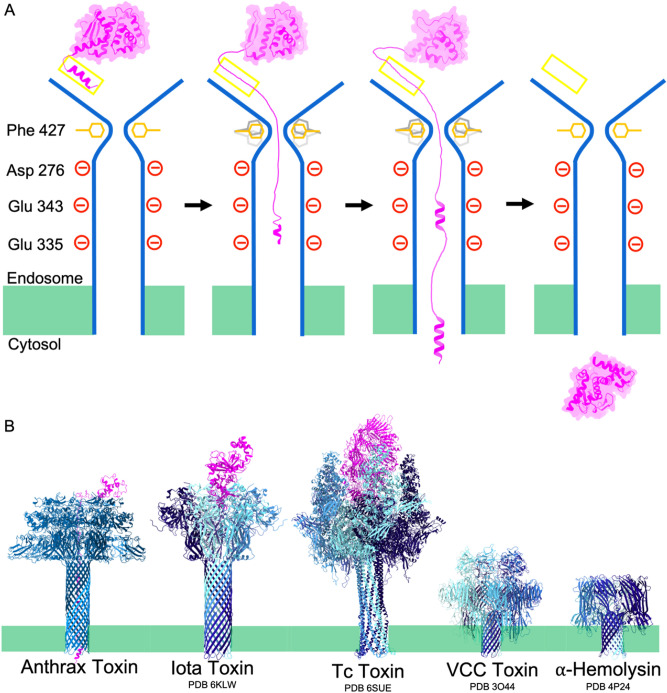
A proposed unfolding-refolding translocation model is shown in Fig. 4A starting with LFN bound to the funnel rim of PApore. LFN is unfolded and funnelled towards the Φ clamp, aided by hydrophobic residues along the funnel slope. LFN acidic residues are protonated in the acidic environment of the funnel. Completely unfolded LFN then passes the Φ clamp. This ring of F427 residues remains restrictive enough to maintain a seal while accommodating translocation. As the channel widens in the charge clamp, acidic residues are deprotonated. Folding of α helical portions places mechanical force on the translocating peptide, contributing to efficient translocation, and overcoming local energy minimum that could otherwise stall the complex. The newly formed, secondary structure favors unidirectional translocation by discouraging retrograde transfer through the narrow Φ clamp resulting in natively folded LF in the host cell cytosol.
Figure 4.
Proposed AB toxin extended pore refolding mechanism. (A) Unfolding-refolding translocation model of anthrax toxin. LFN (magenta) translocates through PApore (blue) passing the α clamp (yellow), the Φ clamp (orange and grey), and the charge clamp (red). Helical portions of LFN begin to refold in the channel ensuring proper tertiary refolding of LFN. (B) Comparison of toxin pore length between toxins that translocate proteins vs toxins that disrupt ion gradients. Membrane bilayer represented in green.
Our results have implications for other toxins. PA F427 is equivalent to F454 of Clostridium perfringens iota toxin39, F428 of Clostridium botulinum C2II binary toxin40, and W318 of Vibrio cholerae cytolysin41. We hypothesize a dynamic hydrophobic seal model is a common mechanism, applicable to these other toxins. Initial refolding in the pore channel is also likely not unique to the anthrax toxin. Indeed, other translocons, such as the iota toxin and toxin complex (Tc) toxin (Fig. 4B), have pores that extend well passed the membrane bilayer42,43. We predict these pore forming toxins have evolved extended pores to faciliate substrate refolding inside the translocon for effective intoxication. Not all pore forming toxins translocate proteins in vivo. Some, like Vibrio cholerae cytolysin (VCC) and Staphylococcus aureus α-hemolysin, form pores to distrupt ion concentrations41,44. The pore length of these toxins is noticeably shorter (Fig. 4B).
Materials and methods
Protein expression and purification
Proteins were purified as previously described17. Briefly, His6-SUMO-LFN E126C was expressed in BL21 cells, purified using anion exchange, and cleaved by small ubiquitin-related modifier protease45. Recombinant wild-type PA83 was expressed in the periplasm of Escherichia coli BL21 (DE3) and purified by ammonium precipitation and anion exchange chromatography8. After trypsin activation45, PA63 heptameric prepores were formed using anion exchange and size exclusion chromatography. Membrane scaffold protein 1D1 (MSP1D1) was expressed from the pMSP1D1 plasmid (AddGene) with an N-terminal His-tag and was purified by affinity chromatography27.
LFN-PA-nanodisc complex formation for CryoEM with TITaNS
We have previously dealt with heterogenous sample preparations of one, two, and three LFN bound to PApore. We found incubating LFN with PAprepore at sub-stoichiometric ratios prior to LFN immobilization and PA pore formation resulted in more homogenous samples17. For this work, E126C LFN and PAprepore were incubated in solution at a ratio of 1:2, respectively. Complexes were then immobilized by coupling E126C LFN to activated thiol sepharose 4B beads (GE Healthcare Bio-Sciences, Pittsburgh, PA, USA) in Assembly Buffer (50 mM Tris, 50 mM NaCl, 10 mM CaCl2 pH 7.5) at 4 °C for 12 h. Beads were washed three times with Assembly Buffer to remove any unbound PAprepore. The immobilized LFN-PAprepore complexes were then incubated in low pH buffer (10 mM acetate, 50 mM Tris, 50 mM NaCl, 10 mM CaCl2 pH 5.5) to transition the PAprepore to PApore and initiate translocation of LFN. Our previous cryoEM work used urea instead of low pH to transition PAprepore to PApore17. The beads were then washed in Assembly Buffer at neutral pH three times. Next, pre-nanodisc micelles (2.5 μM MSP1D1, 97.5 μM 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) (Avanti, Alabaster, AL, USA), 65 (POPG) in 25 mM Na-cholate (Sigma-Aldrich, St. Louis, MO, USA), 50 mM Tris, and 50 mM NaCl) were added and bound to the aggregation-prone hydrophobic transmembrane β-barrel of PApore. The micelles were collapsed into nanodiscs by removing Na-cholate using dialysis with Bio-Beads (BIO RAD, Hercules, CA, USA). Stabilized complexes were released from the thiol sepharose beads by reducing the E126C LFN-bead disulfide bond using 50 mM dithiothreitol (DTT) (Goldbio, St. Louis, MO, USA) in Assembly Buffer. Assembled complexes were initially confirmed using negative-stain TEM. Complexes were stored at -80°C prior to cryoEM grid preparation. Complex formation has also previously been confirmed using mass spectrometry and biolayer interferometry46.
Grid preparation for CryoEM
Complexes stored at -80°C were thawed on ice. A glow discharged Quantifoil R1.2/1.3 300 M Cu holey carbon grid was placed inside the FEI Vitrobot Mark IV humidity chamber at 100% humidity. Then, 2μl of thawed sample was applied to the grid followed by 0.5μL of 1 M acetate pH 5.5. The grids were then blotted and plunge frozen in liquid ethane. Frozen grids were stored in liquid nitrogen prior to use.
CryoEM data collection and image processing
CryoEM grids were loaded into a FEI Titan Krios electron microscope operated at 300 kV for automated image acquisition with serialEM47. cryoEM micrographs were recorded as movies on a Gatan K2 Summit direct electron detection camera using the electron counting mode in super resolution mode at × 130 K nominal magnification, a pixel size of 0.535 Å per pixel, and defocus ranging between − 1 and − 3 µm. Total dose was 50.76 e-/ Å2. Total exposure time was 9 s and fractionated into 45 frames with 200 ms exposure time for each frame. In total, 6,515 micrographs were taken in a continuous session. Frames in each movie were aligned and averaged for correction of beam-induced drift using MotionCor2 and cryoSPARC patch motion correction to generate a micrograph48,49. Micrographs generated by averaging all frames of each movie were used for defocus determination and particle picking. Micrographs obtained by averaging frames 2–36 (corresponding to ~ 40 e-/Å2) were used for two- and three-dimensional image classifications. The best 4,488 micrographs were selected for the following in-depth data processing.
Single particle analysis and density modification
Single particle analysis was performed using cryoSPARC v2.1548 (Fig. S2). A random subset of micrographs was selected for blob particle picking. These particles were subjected to 2D classification in order to obtain a set of five particle templates. Using these templates, 2,076,581 particles were selected from 4,488 micrographs. After multiple rounds of 2D classification, the remaining 671,090 ‘good’ particles were used to create an ab initio model. Heterogenous classification with four classes was then performed and the class with full length β barrel and distinct nanodisc was selected. To select particles with LFN, 3D variability analysis was performed with three orthogonal principle modes (i.e. eigenvectors of 3D covariance) and a mask of LFN bound in each of the seven possible binding sites filtered to 30 Å and gaussian blurred. 122,651 particles from three resulting clusters with potential LFN density were selected for further processing. A homogenous refinement on the per particle motion and CTF corrected particles was performed resulting in a 3.3 Å cryoEM density map. Resolution was determined using gold standard Fourier shell correlation with a cut off of 0.143. Next, local refinements and density modifications were performed (Fig. S3). To further characterize bound LFN, local refinement of the cap of the PApore was performed using a mask of LFN (PDB 3KWV) low pass filtered to 30 Å. For the β barrel interior, a cylindrical mask was used. Phenix density modification was performed on the homogenous refinement and local refinement half maps to further improve the density for PApore and LFN, respectively50. Phenix combine focused maps was then used to create a composite map (Fig. S3)51. The map showed density surrounding the transmembrane region of the beta barrel that we interpret as nanodisc density. As has been reported previously14, there was also disordered density surrounding the outside of the middle of the β barrel. This additional density was masked out of the final map using a 30 Å mask of LFN-PApore inserted into a nanodisc.
Model building and refinement
An initial model using PDB 6PSN was docked into the cryoEM map using Chimera map to model52. The LFN coarse model was adjusted manually using Coot53 to fit the density starting at the C-terminus. Model α helical assignments were based on helicity in original model, cryoEM density diameter, and consistency with previously published helical density. The PApore coarse model was refined using PHENIX real space refine51. Individual atomic model side chains were manually adjusted to fit the density map using Coot53. This process was repeated iteratively until an optimal model was obtained. Ramachandran plots and MolProbity54 were used to assess model quality. Supplementary Table 1 is a summary of cryoEM data collection and processing as well as model building and validation.
Supplementary Information
Acknowledgements
This work is dedicated to the memory of our friend, mentor, and colleague Dr. Mark T. Fisher. Dr. Fisher was an integral in the vision, design, and implementation of this project. This work was supported by National Institutes of Health (R35-GM128562 and R03-AI142361 to B.D.F.), and by University of Kansas Madison and Lila Self Graduate Fellowship to A.J.M. The authors acknowledge the use of instruments at the Electron Imaging Center for NanoMachines supported by NIH (1S10RR23057 and 1S10OD018111), NSF (DBI-1338135) and CNSI at UCLA.
Author contributions
A.J.M., M.T.F., and B.D.F. designed the study. A.J.M. performed the experiments and data analysis. A.J.M. and B.D.F. wrote the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Mark T. Fisher is deceased.
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mark T. Fisher, Email: bfreudenthal@kumc.edu
Bret D. Freudenthal, Email: bfreudenthal@kumc.edu
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-91596-3.
References
- 1.Young JA, Collier RJ. Anthrax toxin: Receptor binding, internalization, pore formation, and translocation. Annu. Rev. Biochem. 2007;76:243–265. doi: 10.1146/annurev.biochem.75.103004.142728. [DOI] [PubMed] [Google Scholar]
- 2.Thoren KL, Krantz BA. The unfolding story of anthrax toxin translocation. Mol. Microbiol. 2011;80:588–595. doi: 10.1111/j.1365-2958.2011.07614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Santelli E, Bankston LA, Leppia SH, Liddington RC. Crystal structure of a complex between anthrax toxin and its host cell receptor. Nature. 2004;430:905. doi: 10.1038/nature02763. [DOI] [PubMed] [Google Scholar]
- 4.Kintzer AF, et al. The protective antigen component of anthrax toxin forms functional octameric complexes. J. Mol. Biol. 2009;392:614–629. doi: 10.1016/j.jmb.2009.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mogridge J, Cunningham K, Collier RJ. Stoichiometry of anthrax toxin complexes. Biochemistry. 2002;41:1079–1082. doi: 10.1021/bi015860m. [DOI] [PubMed] [Google Scholar]
- 6.C. Antoni et al., Cryo-EM structure of the fully-loaded asymmetric anthrax lethal toxin in its heptameric pre-pore state. bioRxiv (2020). [DOI] [PMC free article] [PubMed]
- 7.Abrami L, Liu S, Cosson P, Leppla SH, van der Goot FG. Anthrax toxin triggers endocytosis of its receptor via a lipid raft–mediated clathrin-dependent process. J. Cell Biol. 2003;160:321–328. doi: 10.1083/jcb.200211018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller CJ, Elliott JL, Collier RJ. Anthrax protective antigen: prepore-to-pore conversion. Biochemistry. 1999;38:10432–10441. doi: 10.1021/bi990792d. [DOI] [PubMed] [Google Scholar]
- 9.Krantz BA, Finkelstein A, Collier RJ. Protein translocation through the anthrax toxin transmembrane pore is driven by a proton gradient. J. Mol. Biol. 2006;355:968–979. doi: 10.1016/j.jmb.2005.11.030. [DOI] [PubMed] [Google Scholar]
- 10.Duesbery NS, et al. Proteolytic inactivation of MAP-kinase-kinase by anthrax lethal factor. Science. 1998;280:734–737. doi: 10.1126/science.280.5364.734. [DOI] [PubMed] [Google Scholar]
- 11.Feld GK, et al. Structural basis for the unfolding of anthrax lethal factor by protective antigen oligomers. Nat. Struct. Mol. Biol. 2010;17:1383–1390. doi: 10.1038/nsmb.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown MJ, Thoren KL, Krantz BA. Role of the α clamp in the protein translocation mechanism of anthrax toxin. J. Mol. Biol. 2015;427:3340–3349. doi: 10.1016/j.jmb.2015.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krantz BA, et al. A phenylalanine clamp catalyzes protein translocation through the anthrax toxin pore. Science. 2005;309:777–781. doi: 10.1126/science.1113380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang J, Pentelute BL, Collier RJ, Zhou ZH. Atomic structure of anthrax PA pore elucidates toxin translocation. Nature. 2015;521:545. doi: 10.1038/nature14247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wynia-Smith SL, Brown MJ, Chirichella G, Kemalyan G, Krantz BA. Electrostatic ratchet in the protective antigen channel promotes anthrax toxin translocation. J. Biol. Chem. 2012;287:43753–43764. doi: 10.1074/jbc.M112.419598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feld GK, Brown MJ, Krantz BA. Ratcheting up protein translocation with anthrax toxin. Protein Sci. 2012;21:606–624. doi: 10.1002/pro.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Machen A, et al. Asymmetric cryo-EM structure of anthrax toxin protective antigen pore with lethal factor N-terminal domain. Toxins. 2017;9:298. doi: 10.3390/toxins9100298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hardenbrook NJ, et al. Atomic structures of anthrax toxin protective antigen channels bound to partially unfolded lethal and edema factors. Nat. Commun. 2020;11:1–10. doi: 10.1038/s41467-020-14658-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akkaladevi N, et al. Assembly of anthrax toxin pore: Lethal-factor complexes into lipid nanodiscs. Protein Sci. 2013;22:492–501. doi: 10.1002/pro.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akkaladevi N, et al. Following natures lead: on the construction of membrane-inserted toxins in lipid bilayer nanodiscs. J. Membr. Biol. 2015;248:595–607. doi: 10.1007/s00232-014-9768-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gogol E, et al. Three dimensional structure of the anthrax toxin translocon–lethal factor complex by cryo-electron microscopy. Protein Sci. 2013;22:586–594. doi: 10.1002/pro.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katayama H, et al. Three-dimensional structure of the anthrax toxin pore inserted into lipid nanodiscs and lipid vesicles. Proc. Natl. Acad. Sci. 2010;107:3453–3457. doi: 10.1073/pnas.1000100107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma P, Cardenas AE, Chaudhari MI, Elber R, Rempe SB. The impact of protonation on early translocation of anthrax lethal factor: Kinetics from molecular dynamics simulations and milestoning theory. J. Am. Chem. Soc. 2017;139:14837–14840. doi: 10.1021/jacs.7b07419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.M. T. Fisher, S. Naik (2019) Systems and methods for identifying protein stabilizers. (Google Patents).
- 25.Bayburt TH, Sligar SG. Membrane protein assembly into Nanodiscs. FEBS Lett. 2010;584:1721–1727. doi: 10.1016/j.febslet.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Denisov I, Grinkova Y, Lazarides A, Sligar S. Directed self-assembly of monodisperse phospholipid bilayer Nanodiscs with controlled size. J. Am. Chem. Soc. 2004;126:3477–3487. doi: 10.1021/ja0393574. [DOI] [PubMed] [Google Scholar]
- 27.Ritchie T, et al. Chapter eleven-reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol. 2009;464:211–231. doi: 10.1016/S0076-6879(09)64011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krantz BA, Trivedi AD, Cunningham K, Christensen KA, Collier RJ. Acid-induced unfolding of the amino-terminal domains of the lethal and edema factors of anthrax toxin. J. Mol. Biol. 2004;344:739–756. doi: 10.1016/j.jmb.2004.09.067. [DOI] [PubMed] [Google Scholar]
- 29.F. Wruck et al., The ribosome modulates folding inside the ribosomal exit tunnel. BioRxiv (2020). [DOI] [PMC free article] [PubMed]
- 30.Sun J, Jacquez P. Roles of anthrax toxin receptor 2 in anthrax toxin membrane insertion and pore formation. Toxins. 2016;8:34. doi: 10.3390/toxins8020034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lacy DB, Wigelsworth DJ, Melnyk RA, Harrison SC, Collier RJ. Structure of heptameric protective antigen bound to an anthrax toxin receptor: a role for receptor in pH-dependent pore formation. Proc. Natl. Acad. Sci. USA. 2004;101:13147–13151. doi: 10.1073/pnas.0405405101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang S, Finkelstein A, Collier RJ. Evidence that translocation of anthrax toxin's lethal factor is initiated by entry of its N terminus into the protective antigen channel. Proc. Natl. Acad. Sci. 2004;101:16756–16761. doi: 10.1073/pnas.0405754101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gupta P, Singh S, Tiwari A, Bhat R, Bhatnagar R. Effect of pH on stability of anthrax lethal factor: correlation between denaturation and activity. Biochem. Biophys. Res. Commun. 2001;284:568–573. doi: 10.1006/bbrc.2001.5018. [DOI] [PubMed] [Google Scholar]
- 34.Gonti S, Westler WM, Miyagi M, Bann JG. Site-specific labeling and 19f nmr provide direct evidence for dynamic behavior of the anthrax toxin pore ϕ-clamp structure. Biochemistry. 2021;60:643–647. doi: 10.1021/acs.biochem.0c00833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Das D, Krantz BA. Peptide-and proton-driven allosteric clamps catalyze anthrax toxin translocation across membranes. Proc. Natl. Acad. Sci. 2016;113:9611–9616. doi: 10.1073/pnas.1600624113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Das D, Krantz BA. Secondary Structure Preferences of the Anthrax Toxin Protective Antigen Translocase. J. Mol. Biol. 2017;429:753–762. doi: 10.1016/j.jmb.2017.01.015. [DOI] [PubMed] [Google Scholar]
- 37.Dmochewitz L, et al. Role of CypA and Hsp90 in membrane translocation mediated by anthrax protective antigen. Cell. Microbiol. 2011;13:359–373. doi: 10.1111/j.1462-5822.2010.01539.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilson DN, Beckmann R. The ribosomal tunnel as a functional environment for nascent polypeptide folding and translational stalling. Curr. Opin. Struct. Biol. 2011;21:274–282. doi: 10.1016/j.sbi.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 39.Knapp O, et al. Residues involved in the pore-forming activity of the C lostridium perfringens iota toxin. Cell. Microbiol. 2015;17:288–302. doi: 10.1111/cmi.12366. [DOI] [PubMed] [Google Scholar]
- 40.Neumeyer T, et al. Clostridium botulinum C2 Toxin identification of the binding site for chloroquine and related compounds and influence of the binding site on properties of the c2ii channel. J. Biol. Chem. 2008;283:3904–3914. doi: 10.1074/jbc.M709807200. [DOI] [PubMed] [Google Scholar]
- 41.De S, Olson R. Crystal structure of the Vibrio cholerae cytolysin heptamer reveals common features among disparate pore-forming toxins. Proc. Natl. Acad. Sci. 2011;108:7385–7390. doi: 10.1073/pnas.1017442108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Piper SJ, et al. Cryo-EM structures of the pore-forming A subunit from the Yersinia entomophaga ABC toxin. Nat. Commun. 2019;10:1952. doi: 10.1038/s41467-019-09890-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamada T, et al. Cryo-EM structures reveal translocational unfolding in the clostridial binary iota toxin complex. Nat. Struct. Mol. Biol. 2020;27:288–296. doi: 10.1038/s41594-020-0388-6. [DOI] [PubMed] [Google Scholar]
- 44.Sugawara T, et al. Structural basis for pore-forming mechanism of staphylococcal α-hemolysin. Toxicon. 2015;108:226–231. doi: 10.1016/j.toxicon.2015.09.033. [DOI] [PubMed] [Google Scholar]
- 45.Wigelsworth DJ, et al. Binding stoichiometry and kinetics of the interaction of a human anthrax toxin receptor, CMG2, with protective antigen. J. Biol. Chem. 2004;279:23349–23356. doi: 10.1074/jbc.M401292200. [DOI] [PubMed] [Google Scholar]
- 46.A. J. Machen et al., Analyzing dynamic protein complexes assembled on and released from biolayer interferometry biosensor using mass spectrometry and electron microscopy. JoVE, e57902 (2018). [DOI] [PMC free article] [PubMed]
- 47.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 48.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 49.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Terwilliger TC, Ludtke SJ, Read RJ, Adams PD, Afonine PV. Improvement of cryo-EM maps by density modification. Nat. Methods. 2020;17:923–927. doi: 10.1038/s41592-020-0914-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pettersen EF, et al. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 53.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 54.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.