Abstract
Artemisinin resistance (AR) emerged in South East Asia 13 years ago and the identification of the resistance conferring molecular marker, Plasmodium falciparum Kelch 13 (Pfk13), 7 years ago has provided an invaluable tool for monitoring AR in malaria endemic countries. Molecular Pfk13 surveillance revealed the resistance foci in the Greater Mekong Subregion, an independent emergence in Guyana, South America, and a low frequency of mutations in Africa. The recent identification of the R561H Pfk13 AR associated mutation in Tanzania, Uganda and in Rwanda, where it has been associated with delayed parasite clearance, should be a concern for the continent. In this review, we provide a summary of Pfk13 resistance associated propeller domain mutation frequencies across Africa from 2012 to 2020, to examine how many other countries have identified these mutations. Only four African countries reported a recent identification of the M476I, P553L, R561H, P574L, C580Y and A675V Pfk13 mutations at low frequencies and with no reports of clinical treatment failure, except for Rwanda. These mutations present a threat to malaria control across the continent, since the greatest burden of malaria remains in Africa. A rise in the frequency of these mutations and their spread would reverse the gains made in the reduction of malaria over the last 20 years, given the lack of new antimalarial treatments in the event artemisinin-based combination therapies fail. The review highlights the frequency of Pfk13 propeller domain mutations across Africa, providing an up-to-date perspective of Pfk13 mutations, and appeals for an urgent and concerted effort to monitoring antimalarial resistance markers in Africa and the efficacy of antimalarials by re-establishing sentinel surveillance systems.
Keywords: Pfk13, Artemisinin resistance, ACT, Molecular marker
Graphical abstract
Highlights
-
•
Indigenous Pfk13 resistance associated mutations have been identified in Africa
-
•
Currently, only 4 African countries have reported 6 (M476I, P553L, R561H, P574L, C580Y and A675V) of the 10 validated artemisinin resistance associated mutations
-
•
The 6 mutations though identified at low frequencies are an early warning signal for Africa, since a rise in frequency and their spread to other countries could be catastrophic for malaria control
-
•
Africa needs a concerted and systematic approach to monitoring Pfk13 mutations (beyond the few research studies published across Africa) and antimalarial efficacy through a re-establishment of sentinel surveillance platforms
1. Introduction
Efficacious antimalarial drugs are a critical component of malaria control. They rapidly clear the parasite biomass following early detection of malaria infections, thus reducing the burden of disease. Chloroquine (CQ), was a low cost treatment that was the mainstay of uncomplicated Plasmodium falciparum malaria treatment for decades. CQ resistance and clinical failure was initially observed in South East (SE) Asia in the late 1950s along the Thai-Cambodia border, that later spread into Africa in the late 1970s and an independent emergence of resistance was described in Colombia, South America also in the late 1950s (Payne 1987; Wellems and Plowe 2001). Thereafter, Sulphadoxine-Pyrimethamine (SP) became the preferred, affordable alternative to CQ. However resistance to SP quickly emerged, mirroring the spread of CQ resistance, emerging in Thailand in the mid 1960s and then spreading into Africa (Roper et al., 2004) in the 1990s, with the independent emergence of SP resistance in Africa (Pearce et al., 2009) and South America (Cortese et al., 2002; Mita et al., 2007). The widespread resistance to SP by the mid 1990s rendered the drug ineffective, prompting a policy change in the treatment of uncomplicated malaria to artemisinin-based combination therapies (ACTs). Though SP was ineffective as a radical cure, it was in use until recently in combination with artesunate (WHO Report on antimalarial efficacy, 2020) and is still in use as a chemoprophylatic agent in Intermittent Preventive Treatment in pregnancy (IPTp) (ter Kuile et al., 2007), IPT in infants (Naidoo and Roper 2011) and seasonal malaria chemoprevention (SMC) in the Sahel region (WHO, SMC, 2012). The malaria infection prevalence and disease burden significantly increased in Africa between 1985 and 2004, and although it is not possible to determine the exact contributions, it seems plausible that drug resistance was a factor (Snow et al., 2001; Snow et al., 2017). In the last 2 decades, there has been a significant decline in the malaria disease burden in Africa (WHO Malaria Report, 2020), which may be partially attributed to the successful roll out of ACTs from 2001 onwards.
2. Molecular markers of resistance
The ~40 year delay between observing CQ clinical failure to defining the molecular marker of resistance meant that tracking resistance was initially based on clinical phenotype, limiting monitoring capacities of antimalarial resistance to the laborious therapeutic efficacy studies (TES) and a >25% treatment failure by day 14 (WHO Report on antimalarial efficacy, 2020). Nevertheless, these clinical data provided valuable data that prompted the change in antimalarial treatment policy in Africa. The subsequent identification of the Plasmodium falciparum chloroquine resistance transporter (Pfcrt) gene as the marker for CQ resistance in 2000 (Fidock et al., 2000; Djimde et al., 2001), made tracking the extent of CQ resistance possible. More recently in East (Mwai et al., 2009; Wamae et al., 2019) and Southern (Frosch et al., 2014) Africa it highlighted a reversion to CQ sensitive parasite populations. In Malawi specifically, CQ clinical efficacy has also been demonstrated (Laufer et al., 2006). In contrast, SP treatment failure was observed in combination with the molecular characterization of resistance conferring mutations (i.e. Plasmodium falciparum dihydrofolate reductase (Pfdhfr) triple and Plasmodium falciparum dihydropteroate synthase (Pfdhps) double mutants (Dieckmann and Jung 1986; Cowman et al., 1988), and when both treatment failure and molecular markers of resistance were found to be widespread this prompted the change in treatment policy to ACTs. Molecular markers are therefore a valuable tool for strengthening resistance monitoring, confirming resistance, providing an early warning signal and for assessing resistance trends (WHO Report on antimalarial efficacy, 2020).
3. Artemisinin resistance
The rapid identification of the artemisinin resistance (AR) molecular marker, Plasmodium falciparum kelch 13 (Pfk13) (Ariey et al., 2014), only 6 years after the initial observation of ACT clinical failure once again in SE Asia along the Thai-Myanmar and Thai-Cambodia borders (Noedl et al., 2008; Dondorp et al., 2009), changed the course of surveillance in SE Asia. . The Pfk13 molecular marker made it possible to define the foci and origins of the emergence of resistance and its spread in the Greater Mekong subregion (Miotto et al., 2015). Initially, C580Y and three other loci (Y493H, R539T, I543T) were identified as the ART resistance conferring mutations associated with delayed parasite clearance (Ariey et al., 2014).
The Pfk13 gene encodes a protein containing a conserved N-terminal domain followed by a BTB/POZ domain and a 6-blade propeller domain at the C-terminal end (Adams et al., 2000; Straimer et al., 2015). The validated mutations in the propeller domain associated with ART resistance, includes F446I, N458Y, M476I, Y493H, R539T, I543T, P553L, R561H, P574L, C580Y and A675V (WWARN Genotype-Phenotype Study Group, 2019). These mutations arose independently along the Thai-Myanmar and Thai-Cambodia border regions and have since spread widely across SE Asia (Tun et al., 2015; Wang et al., 2015). Outside SE Asia, Papua New Guinea (Miotto et al., 2020) and Guyana (Mathieu et al., 2020) have reported the independent emergence of Pfk13 mutations, notably the C580Y mutation.
In South America, where ACT clinical failure has not yet been confirmed, monitoring the Pfk13 gene has allowed for the early identification of mutations that could potentially result in resistance to artemisinin. For instance, in Guyana an indegenious C580Y mutation, has been observed at a low frequency (Mathieu et al., 2020). In Africa, several studies have identified a number of low frequency Pfk13 propeller domain mutations that are the focus of this brief review that leads to a plea to mount a concerted and deliberate approach to monitor Pfk13 mutations across the continent.
4. Molecular epidemiology
Molecular epidemiology is an effective tool for the monitoring and tracking of parasite susceptibility to antimalarials, which in combination with drug efficacy trials enables prompt changes in treatment policy before clinical treatment failure negatively impacts control efforts. Given the immense success in the tracking the emergence and spread of CQ and SP resistant parasite across the globe, we have a robust framework to understand and monitor the spread of AR parasites from SE Asia. Furthermore, with a relatively small number of antimalarials in use (WHO Report on antimalarial efficacy, 2020) and the importance of ACTs to malaria control efforts in Africa, Pfk13 mutants should be closely and systematically monitored in Africa. Numerous studies have been published examining Pfk13 mutations across the continent (Kamau et al., 2015; Kayiba et al., 2020). A recent study conducted by Uwimana et al. (2020), between 2012 and 2015 in Rwanda, highlighted the local emergence of an ART resistance validated mutation, R561H. They also demonstrated through CRISPR-Cas genome editing that the 561H mutation conferred in vitro resistance to ART. In a reversal of fortune, molecular surveillance (the detection of the resistance mutation) preceded the clinical evidence of an association of the R561H resistance mutation with delayed parasite clearance (Uwimana et al., 2021). Thus, demonstrating that the utility of molecular surveillance is rapid and can be conducted at a much larger spatial scale than therapeutic efficacy studies, allowing clinical studies to be targeted in high risk locations. Unfortunately, the de novo emergence of ART resistance markers in Africa is a concern, since the spread of resistant parasites across Africa would be catastrophic.
This brief review includes a collation of data from Africa, following two recent publications that reported the R561H mutation in Rwanda (Uwimana et al., 2020) and Tanzania (Moser et al., 2020). The R561H mutation is one of the 10 validated SE Asian AR associated mutations (WWARN Genotype-Phenotype Study Group, 2019) (Table 1), raising the concern that it may spread and lead to clinical impacts. Fortunately, it currently appears at a low frequency, and is limited to three separate studies from Rwanda, Tanzania and Uganda (Table 1). We therefore examined other African studies with parasite samples collected from 2012 to date, to determine whether these studies in the continent have also identified the R561H mutation or other propeller domain mutations that would give an indication of its spread.
Table 1.
Frequencies of k13 validated mutations across Africa from 2012 to date.
| Year | Study country | Reference | Assay | N | PfK13 mutations frequency % [n] |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| F446I | M476Ia | Y493Ha | R539Ta | I543Ta | P553L | R561Ha | P574L | C580Ya | A675V | |||||
| 2019 | Angola | Dimbu et al., 2021 | Sanger | 103 | ||||||||||
| 2014 | Benin | Ogouyèmi-Hounto et al., 2016 | Sanger | 78 | ||||||||||
| 2012–2016 | Botswana | Tawe et al., 2018 | Sanger | 127 | ||||||||||
| 2014–2017 | Cameroon | Moukoko et al. 2019 | Sanger | 175 | ||||||||||
| 2017–2019 | Central African Republic | Nzoumbou-Boko et al., 2020 | Sanger | 187 | ||||||||||
| 2015–2016 | Congo | Mayengue et al., 2018 | Sanger | 127 | ||||||||||
| 2017 | Democratic Republic of Congo | Yobi et al., 2020 | Sanger | 717 | ||||||||||
| 2013–2014 | Equatorial Guinea | Li et al., 2016 | Sanger | 98 | ||||||||||
| 2014–2017 | Eritrea | Pacheco et al., 2019 | Amp-Seq | 117 | ||||||||||
| 2014 | Ethiopia | Lo et al., 2017 | Sanger | 226 | ||||||||||
| 2017–2018 | Gabon | Adegbite et al., 2019 | Sanger | 100 | ||||||||||
| 2012–2014 | Gambia | Amambua-Ngwa et al., 2017 | Taq-man/Sanger | 184 | ||||||||||
| 2014–2017 | Ghana | Mensah et al. 2020 | MIPs | 619 | 0.2 [1] | |||||||||
| 2007–2016 | Ghana | Matrevi et al. (2019) | Sanger | 854 | 0.1 [1] | 0.1 [1] | 0.1 [1] | |||||||
| 2017–2018 | Ghana | Aninagyei et al. (2020) | WGS | 84 | 3.6 [3] | |||||||||
| 2016 | Guinea | Beavogui et al., 2020 | ND | 411 | ||||||||||
| 2016 | Guinea-Bissau | Nag et al., 2019 | Amp-seq | 97 | ||||||||||
| 2014–2019 | Kenya | Omedo et al.,. Unpublished data | Amp-Seq | 284 | ||||||||||
| 2014–2017 | Liberia | Pacheco et al., 2019 | Amp-Seq | 21 | ||||||||||
| 2015–2016 | Mali | Kone et al., 2020 | WGS | 216 | ||||||||||
| 2015–2017 | Mozambique | Gupta et al., 2020 | Sanger | 206 | ||||||||||
| 2013 | Niger | Laminou et al. 2017 | Sanger | 366 | ||||||||||
| 2014–2017 | Nigeria | Pacheco et al., 2019 | Amp-Seq | 29 | 3.4 [1] | |||||||||
| 2013–2015 | Rwanda | Uwimana et al. (2020) | Sanger | 466 | 4.1 [19] | 0.2 [1] | ||||||||
| 2014–2015 | Rwanda | Tacoli et al. 2016 | Sanger | 147 | 0.7 [1] | 0.7 [1] | 0.7 [1] | |||||||
| 2015–2019 | Senegal | Delandre et al. 2020 | Sanger | 327 | ||||||||||
| 2016–2017 | Somalia | Warsame et al. 2019 | Sanger | 138 | ||||||||||
| 2018 | South Africa | Raman et al. 2019 | Sanger | 532 | ||||||||||
| 2015–2017 | Sudan | Hussien et al., 2020 | Amp-Seq | 176 | ||||||||||
| 2019 | Tanzania | Bwire et al. (2020) | Amp-Seq | 422 | 0.2 [1] | |||||||||
| 2017 | Tanzania | Moser et al. (2020) | MIPs | 764 | 0.3 [2] | |||||||||
| 2012–2013 | Togo | Dorkenoo et al. 2016 | Sanger | 500 | ||||||||||
| 2012–2016 | Uganda | Conrad et al. 2019 | Sanger | 716 | 0.1 [1] | 0.1 [1] | ||||||||
| 2016–2017 | Uganda | Asua et al. 2020 | ND | 412 | 1.7 [7] | |||||||||
| 2014–2016 | Uganda | Ikeda et al., 2020 | ND | 194 | 0.5 [1] | |||||||||
| 2018–2019 | Uganda | Asua et al. (2020) | MIPs | 796 | 0.1 [1] | 5.5 [44] | ||||||||
| 2017 | Zambia | Sitali et al. 2020 | Sanger | 70 | ||||||||||
Represents validated SE Asian artemisinin resistance mutations. N is the number of samples that were successfully genotyped per study. n is the number of samples harbouring the respective mutation. ND = could not be determined. Assay represents the genotyping assay used in the respective study i.e. Sanger - Sanger sequencing, MIP - molecular inversion probes, WGS - whole-genome sequencing, Amp-Seq - amplicon sequencing. DR Congo stands for the Democratic Republic of Congo.
5. Study selection criteria
We searched the PubMed database for peer reviewed articles from malaria endemic African countries published between January 1, 2016 and November 11, 2020, that had genotyped the Pfk13 gene, focusing on propeller domain mutations. The restriction on publication date was to enable the review of recent publications that would encompass parasite samples overlapping with the Rwandan (Uwimana et al., 2020) and Tanzanian (Moser et al., 2020) studies and recent samples, i.e. from 2012 to date. The following search terms were used: “((malaria OR falciparum) AND (molecular marker OR kelch13 OR Pfkelch13 OR K13 OR PfK13)) AND ((Africa OR Afrique OR country name [focusing on malaria endemic sub-Saharan African countries]) AND (("2016/01/01" [Date - Publication]: “2020/11/11" [Date - Publication]))”. Three individuals independently screened the articles and excluded studies based on genetically modified parasites, reviews, conference proceedings, letters of correspondence and modelling studies. Additionally, six individuals extracted the following details from the articles: the year samples were collected, country of origin, age of study participants, first line ACT recommended in the country of origin, genotyping assay, study population (asymptomatic or symptomatic), sample size and polymorphisms across the propeller domain of the Pfk13 gene.
6. Summary of the Pfk13 mutations from the extracted literature
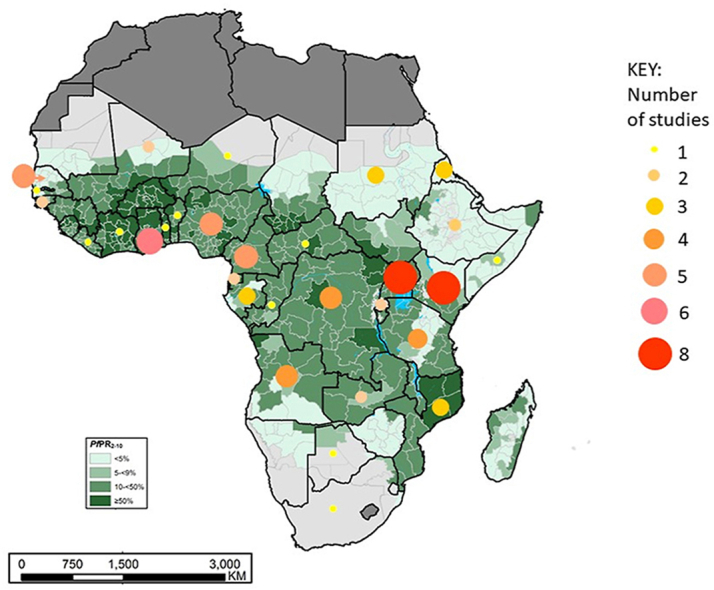
Recently, a systematic review identified Pfk13 mutations across Africa that have been associated with ART resistance, though at low frequencies (Kayiba et al., 2020). Here we present data of Pfk13 propeller domain mutations from >70 studies based on the PRISMA guidelines (Fig. 1) (Page et al., 2021) undertaken in Africa from samples obtained from 2012 to 2020, across 30 countries (Table S1, Full table accessible at Harvard Dataverse: https://doi.org/10.7910/DVN/EUPXCF). The Pfk13 genotypes detected across many African countries were primarily wild-type with only Ghana reporting the C580Y mutation (Matrevi et al., 2019; Aninagyei et al., 2020) associated with the majority of ART resistance in SE Asia. There were no Pfk13 mutations reported in Benin (2014), The Gambia (2012–2014) and Liberia (2014–2017) (Table S1). Only 3 other countries, apart from Rwanda, have reported the R561H mutation. In Tanzania, two parasites sampled from 764 were identified in 2017 (0.3%) (Moser et al., 2020) and a single parasite from 422 parasites sampled in 2019 (0.2%) (Bwire et al., 2020), similarly in Uganda one parasite was identified from 796 samples in 2018/2019 (0.1%) (Asua et al., 2020), no clinical failure was observed (consistent with the findings in Rwanda). In addition to the R561H mutation, the A675V mutation was also identified in four separate studies conducted in Uganda, showing an increase in its frequency to 5% in sample sets obtained in 2018 and 2019. The A675V mutation has also been described in a Rwandan and Nigerian study (Table 1). Only one high frequency propeller domain mutation was identified (i.e. A578S), which has been detected in 14 countries at frequencies of up to 11%. Another high frequency (>5%) Pfk13 mutation outside the propeller domain, K189T, has been identified in 8 countries. These high frequency mutations have not been associated with ART resistance or delayed parasite clearance.
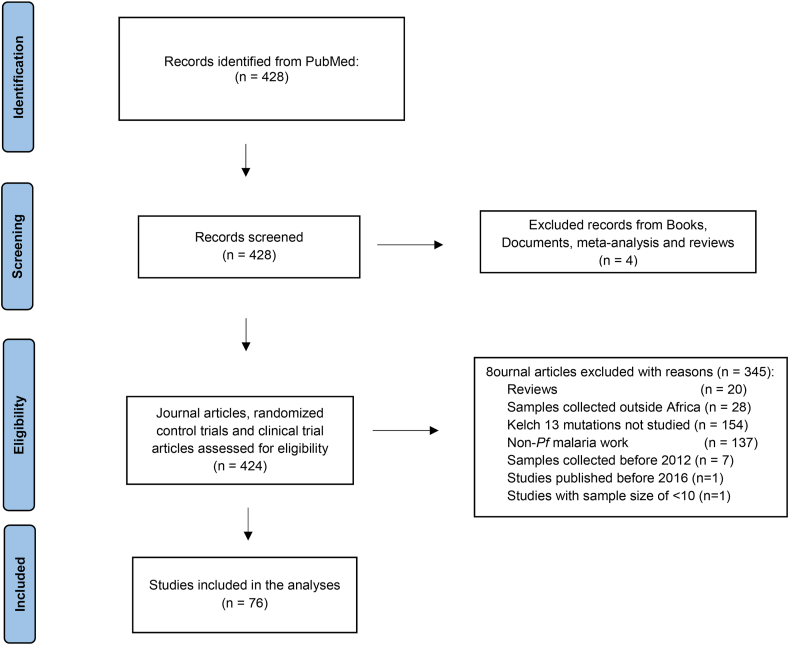
Fig. 1.
Flow diagram of study selection criteria. The diagram indicates the numbers of publications identified in PubMed, manually screened and excluded to settle on a list of 86 publications reviewed for this study. An initial screen to exclude reviews, documents, books and meta-analyses was conducted. A further selection process was done based on the following non-eligibility criteria to exclude studies that were: reviews, from outside Africa, not examining Kelch 13, non-Plasmodium falciparum (Pf), parasite samples collected before 2012, published before 2016 and a low sample size <10 to obtain 76 publications. The 86 publications that met the criteria, focusing on kelch 13 mutations in Africa in either a standard research or clinical trial article.
The collated data provides an Africa-wide perspective on the current status of the prevalence of Pfk13 mutations. The mutations known to be associated with delayed parasite clearance (i.e. M476I, P553L, R561H, P574L, C580Y and A675V) were observed at low frequencies (<5%) in four countries, Ghana, Rwanda, Uganda and Tanzania, indicating that we cannot be complacent regarding the potential spread of such mutations in Africa and that data on early signs of potential resistance to ACTs are essential. Notably, the data presented in this review is from 30 (67%) countries (Table S1) across 45 malaria endemic continental Africa and the islands of Madagascar, Sao Tome & Principe and Comoros (WHO Malaria Report, 2020), highlighting the paucity of molecular surveillance data and countries to target for future molecular surveys.
7. Conclusions
In comparison to the late identification of Pfcrt, Pfk13 was identified swiftly after artemisinin clinical failure was observed, due to the rapid advances in whole genome sequencing technologies that has revolutionized our ability to detect important mutations.
Research teams in many African countries are using a wide range of techniques from the gold standard Sanger sequencing method through to amplicon deep sequencing, molecular inversion probes (MIPs) and whole genome sequencing to provide important data on the current state of Pfk13 mutation prevalence. Molecular surveillance capacity is present in Africa to allow for the roll out of molecular surveillance tools, and African institutions have capacity to undertake the laboratory work and bioinformatics analyses across the continent. However, these studies are largely opportunistic and linked to specific research projects, rather than programmatic and linked to public health surveillance initiatives.
Based on historical evidence of antimalarial resistance to CQ and SP emerging in SE Asia and spreading to Africa, it was expected that ART resistance was likely to follow the same trajectory and hence the numerous studies to genotype Pfk13. However, the data on the de novo emergence of resistance mutations emphasize that such studies (Table S1) are important to provide a quick overview of mutation frequencies to inform policy decision making. With no new antimalarials immediately available, if ACTs fail, this threatens to reverse the significant gains made in the global reduction in malaria over the last 20 years (World Malaria report, 2020). Consequently, this recent detection of ART resistance mutations at codons R561H (recently associated with delayed parasite clearance), P574L, C580Y and A675V, serves as an early warning signal of potential resistance.
Currently, only 4 countries across malaria-endemic Africa have shown evidence of low frequency AR associated mutations, with an early indication of indigenous Pfk13 mutations in the East Africa region, suggesting that the threat of independent acquisition of resistance should be taken as seriously as the threat of imported resistance. The data generated come from independent research projects, and hence a mechanism to report regularly on findings and interact with national and international policy makers has not been defined. There is therefore an urgent need for increased, standardized and prospective antimalarial resistance molecular surveillance across Africa.
How to intergrate molecular surveillance and clinical effectiveness to support decision making by National Malaria Control Programmes needs defining and needs investment in order to stay ahead of the ART resistance curve. Taking lessons from historically successful networks, twenty years ago, the East African Network for Monitoring Antimalarial Treatment (EANMAT) served as a flagship collaboration between sub-regional research scientists and national ministry of health case-management implementing and policy staff (EANMAT, 2001; EANMAT, 2003). The network successfully lobbied governments and donors to switch to ACTs that were expected to last for a long time, in 2004, however EANMAT has been inactive since 2004. Re-establishing sentinel surveillance sites to formulate evidence-based antimalarial drug policies through sub-regional technical networks for monitoring antimalarial resistance such as EANMAT, West African Network for Monitoring Antimalarial Treatment (WANMAT) and Horn of Africa Network for Monitoring Antimalarial Treatment (HANMAT) (EANMAT, 2000; EANMAT, 2001; Talisuna et al., 2006; HANMAT Report 2013), would provide an effective framework to combine longitudinal, sentinel site surveillance and clinical therapeutic efficacy studies (TES). In addition, including the new recommendation of a change in drug policy if day 28 or day 42 efficacy falls below 90% in TES (WHO Report on antimalarial efficacy, 2020) to transform policy and provide a holistic view of antimalarial resistance in Africa.
Author contributions
L.N., K.M.K. and K.W. conducted the literature review and drafted the manuscript. I.O. V.O. and M.A. conducted the literature review and reviewed the manuscript. B.A., A.G., D.S.I., A.A-N., N.D-Q., S.K.T., C.K., S.T., A.A.D., and A.D. reviewed the manuscript. J.R. and R.W.S. contributed to the literature review and reviewed the manuscript. P.B. and L.I.O–O. conceived, drafted and reviewed the manuscript. All authors read and approved the final manuscript.
Declaration of competing interest
The authors declare no competing interests.
Acknowledgements
The DELTAS Africa Initiative (Grant No. DEL-15-003), an independent funding scheme of the African Academy of Sciences (AAS) Alliance for Accelerating Excellence in Science in Africa (AESA) and supported by the New Partnership for Africa's Development Planning and Coordinating Agency (NEPAD Agency) with funding from the Wellcome Trust and the UK government, DELGEME Grant 107740/Z/15/Z supports K.M.K and IDeAL Grant No. 107769/Z/10/Z supported K.W and M.A. The views expressed in this publication are those of the author(s) and not necessarily those of AAS, NEPAD Agency, Wellcome Trust or the UK government. RWS is supported as a Wellcome Trust Principal Fellow (# 212176). LN, KMK, KW, VO, MA, IO, RWS, PB and LIOO are grateful to the support of the Wellcome Trust to the Kenya Major Overseas Programme (# 203077). L.I.O–O and V.O. are supported by a Wellcome Trust Intermediate Fellowship (Grant No. 107568/Z/15/Z awarded to L.I.O–O). We also thank the Director of KEMRI for permission to publish this article.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijpddr.2021.06.001.
Appendix A. Supplementary data
The following is/are the supplementary data to this article:
References
- Adams J., Kelso R., Cooley L. The kelch repeat superfamily of proteins: propellers of cell function. Trends Cell Biol. 2000;10:17–24. doi: 10.1016/s0962-8924(99)01673-6. [DOI] [PubMed] [Google Scholar]
- Adegbite B.R., Edoa J.R., Honkpehedji Y.J., Zinsou F.J., Dejon-Agobe J.C., Mbong-Ngwese M., Lotola-Mougueni F., Koehne E., Lalremruata A., Kreidenweiss A. Monitoring of efficacy, tolerability and safety of artemether-lumefantrine and artesunate-amodiaquine for the treatment of uncomplicated Plasmodium falciparum malaria in Lambaréné, Gabon: an open-label clinical trial. Malar. J. 2019;18:1–9. doi: 10.1186/s12936-019-3015-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amambua-Ngwa A., Okebe J., Mbye H., Ceesay S., El-Fatouri F., FatouNyang J.H., Ramatoulie J., Affara M., Ahmad A. Sustained ex vivo susceptibility of Plasmodium falciparum to artemisinin derivatives but increasing tolerance to artemisinin combination therapy partner quinolines in The Gambia. Antimicrob. Agents Chemother. 2017;61(12) doi: 10.1128/AAC.00759-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aninagyei E., Duedu K.O., Rufai T., Tetteh C.D., Chandi M.G., Ampomah P., Acheampong D.O. Characterization of putative drug resistant biomarkers in Plasmodium falciparum isolated from Ghanaian blood donors. BMC Infect. Dis. 2020;20:533. doi: 10.1186/s12879-020-05266-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariey F., Witkowski B., Amaratunga C., Beghain J., Langlois A.C., Khim N., Kim S., Duru V., Bouchier C., Ma L. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;505:50–55. doi: 10.1038/nature12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asua V., Conrad M.D., Aydemir O., Duvalsaint M., Legac J., Duarte E., Tumwebaze P., Chin D.M., Cooper R.A., Yeka A. Changing prevalence of potential mediators of aminoquinoline, antifolate, and artemisinin resistance across Uganda. J. Infect. Dis. 2020;223(6):985–994. doi: 10.1093/infdis/jiaa687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beavogui A.H., Camara A., Delamou A., Diallo M.S., Doumbouya A., Kourouma K., Bouedouno P., Guilavogui T., Souza S.D.S., Kelley J. Efficacy and safety of artesunate-amodiaquine and artemether-lumefantrine and prevalence of molecular markers associated with resistance, Guinea: an open-label two-arm randomised controlled trial. Malar. J. 2020;19:1–9. doi: 10.1186/s12936-020-03290-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bwire G.M., Ngasala B., Mikomangwa W.P., Kilonzi M., Kamuhabwa A.A.R. Detection of mutations associated with artemisinin resistance at k13-propeller gene and a near complete return of chloroquine susceptible falciparum malaria in Southeast of Tanzania. Sci. Rep. 2020;10:3500. doi: 10.1038/s41598-020-60549-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortese J.F., Caraballo A., Contreras C.E., Plowe C.V. Origin and dissemination of Plasmodium falciparum drug-resistance mutations in South America. J. Infect. Dis. 2002;186:999–1006. doi: 10.1086/342946. [DOI] [PubMed] [Google Scholar]
- Cowman A.F., Morry M.J., Biggs B.A., Cross G.A., Foote S.J. Amino acid changes linked to pyrimethamine resistance in the dihydrofolate reductase-thymidylate synthase gene of Plasmodium falciparum. Proc. Natl. Acad. Sci. U. S. A. 1988;85:9109–9113. doi: 10.1073/pnas.85.23.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieckmann A., Jung A. Mechanisms of sulfadoxine resistance in Plasmodium falciparum. Mol. Biochem. Parasitol. 1986;19:143–147. doi: 10.1016/0166-6851(86)90119-2. [DOI] [PubMed] [Google Scholar]
- Dimbu P.R., Horth R., Cândido A.L.M., Ferreira C.M., Caquece F., Garcia L.E.A, André K., Pembele G., Jandondo D., Bondo B.J. Continued low efficacy of artemether-lumefantrine in Angola, 2019. Antimicrob. Agents Chemother. 2021;65(2) doi: 10.1128/AAC.01949-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djimde A., Doumbo O.K., Cortese J.F., Kayentao K., Doumbo S., Diourte Y., Coulibaly D., Dicko A., Su X.Z., Nomura T. A molecular marker for chloroquine-resistant falciparum malaria. N. Engl. J. Med. 2001;344:257–263. doi: 10.1056/NEJM200101253440403. [DOI] [PubMed] [Google Scholar]
- Dondorp A.M., Nosten F., Yi P., Das D., Phyo A.P., Tarning J., Lwin K.M., Ariey F., Hanpithakpong W., Lee S.J. Artemisinin resistance in Plasmodium falciparum malaria. N. Engl. J. Med. 2009;361:455–467. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- East African Network for Monitoring Antimalarial Treatment (EANMAT) Monitoring antimalarial drug resistance within National Malaria Control Programmes: the EANMAT experience. Trop. Med. Int. Health. 2001;6(11) doi: 10.1046/j.1365-3156.2001.00799.x. 891–8. [DOI] [PubMed] [Google Scholar]
- East African Network for Monitoring Antimalarial Treatment (EANMAT) The efficacy of antimalarial monotherapies, sulphadoxine-pyrimethamine and amodiaquine in East Africa: implications for sub-regional policy. Trop. Med. Int. Health. 2003;8(10) doi: 10.1046/j.1360-2276.2003.01114.x. 860–7. [DOI] [PubMed] [Google Scholar]
- Fidock D.A., Nomura T., Talley A.K., Cooper R.A., Dzekunov S.M., Ferdig M.T., Ursos L.M., Sidhu A.B., Naude B., Deitsch K.W. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol Cell. 2000;6:861–871. doi: 10.1016/s1097-2765(05)00077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frosch A.E., Laufer M.K., Mathanga D.P., Takala-Harrison S., Skarbinski J., Claassen C.W., Dzinjalamala F.K., Plowe C.V. Return of widespread chloroquine-sensitive Plasmodium falciparum to Malawi. J. Infect. Dis. 2014;210:1110–1114. doi: 10.1093/infdis/jiu216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta H., Galatas B., Chidimatembue A., Huijben S., Cisteró P., Matambisso G., Nhamussua L., Simone W., Bassat Q., Menard D. Effect of mass dihydroartemisinin – piperaquine administration in southern Mozambique on the carriage of molecular markers of antimalarial resistance. PLoS ONE. 2020;15(10) doi: 10.1371/journal.pone.0240174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HANMAT Report . 21–22 February 2013. Report on the intercountry meeting of national malaria programme managers from HANMAT and PIAM-net countries, Sharm El Sheikh, Egypt.https://www.who.int/publications/i/item/WHO-EM-MAL-372-E [Google Scholar]
- Kamau E., Campino S., Amenga-Etego L., Drury E., Ishengoma D., Johnson K., Mumba D., Kekre M., Yavo W., Mead D. K13-propeller polymorphisms in Plasmodium falciparum parasites from sub-Saharan Africa. J. Infect. Dis. 2015;211:1352–1355. doi: 10.1093/infdis/jiu608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayiba N.K., Yobi D.M., Tshibangu-Kabamba E., Tuan V.P., Yamaoka Y., Devleesschauwer B., Mvumbi D.M., Okitolonda Wemakoy E., De Mol P., Mvumbi G.L. Spatial and molecular mapping of Pfkelch13 gene polymorphism in Africa in the era of emerging Plasmodium falciparum resistance to artemisinin: a systematic review. Lancet Infect. Dis. 2020;21(4):E82–E92. doi: 10.1016/S1473-3099(20)30493-X. [DOI] [PubMed] [Google Scholar]
- Kone A., Sissoko S., Fofana B., Sangare C.O., Dembele D., Haidara A.S., Diallo N., Coulibaly A., Traore A., Toure S. Different Plasmodium falciparum clearance times in two Malian villages following artesunate monotherapy. Int. J. Infect. Dis. 2020;95:399–405. doi: 10.1016/j.ijid.2020.03.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufer M.K., Thesing P.C., Eddington N.D., Masonga R., Dzinjalamala F.K., Takala S.L., Taylor T.E., Plowe C.V. Return of chloroquine antimalarial efficacy in Malawi. N. Engl. J. Med. 2006;355:1959–1966. doi: 10.1056/NEJMoa062032. [DOI] [PubMed] [Google Scholar]
- Li J., Chen J., Xie D., Eyi U.M., Matesa R.A., Obono M.O., Ehapo C.S., Yang L., Yang H., Lin M. Limited artemisinin resistance-associated polymorphisms in Plasmodium falciparum K13-propeller and PfATPase6 gene isolated from Bioko Island, Equatorial Guinea. Int. J. Parasitol. Drugs Drug Resist. 2016;6(1):54–59. doi: 10.1016/j.ijpddr.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo E., Hemming-Schroeder E., Yewhalaw D., Nguyen J., Kebede E., Zemene E., Getachew S., Tushune K., Zhong D., Zhou G. Transmission dynamics of co-endemic Plasmodium vivax and P. falciparum in Ethiopia and prevalence of antimalarial resistant genotypes. PLoS Negl. Trop. Dis. 2017;11:1–25. doi: 10.1371/journal.pntd.0005806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu L.C., Cox H., Early A.M., Mok S., Lazrek Y., Paquet J.C., Ade M.P., Lucchi N.W., Grant Q., Udhayakumar V. Local emergence in Amazonia of Plasmodium falciparum k13 C580Y mutants associated with in vitro artemisinin resistance. Elife. 2020;9 doi: 10.7554/eLife.51015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrevi S.A., Opoku-Agyeman P., Quashie N.B., Bruku S., Abuaku B., Koram K.A., Fox A., Letizia A., Duah-Quashie N.O. Plasmodium falciparum kelch propeller polymorphisms in clinical isolates from Ghana from 2007 to 2016. Antimicrob. Agents Chemother. 2019;63 doi: 10.1128/AAC.00802-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayengue P.I., Niama R.F., Kouhounina B.D., Malonga-Massanga A., Louzolo I., Bongolo C.L., Macosso L., Ottia R.I., Ngoma G.K., Dossou-Yovo L.R. No polymorphisms in K13-propeller gene associated with artemisinin resistance in Plasmodium falciparum isolated from Brazzaville, Republic of Congo. BMC Infect. Dis. 2018;18(1):538. doi: 10.1186/s12879-018-3453-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miotto O., Amato R., Ashley E.A., MacInnis B., Almagro-Garcia J., Amaratunga C., Lim P., Mead D., Oyola S.O., Dhorda M. Genetic architecture of artemisinin-resistant Plasmodium falciparum. Nat. Genet. 2015;47:226–234. doi: 10.1038/ng.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miotto O., Sekihara M., Tachibana S.I., Yamauchi M., Pearson R.D., Amato R., Goncalves S., Mehra S., Noviyanti R., Marfurt J. Emergence of artemisinin-resistant Plasmodium falciparum with kelch13 C580Y mutations on the island of New Guinea. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1009133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mita T., Tanabe K., Takahashi N., Tsukahara T., Eto H., Dysoley L., Ohmae H., Kita K., Krudsood S., Looareesuwan S. Independent evolution of pyrimethamine resistance in Plasmodium falciparum isolates in Melanesia. Antimicrob. Agents Chemother. 2007;51:1071–1077. doi: 10.1128/AAC.01186-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser K.A., Madebe R.A., Aydemir O., Chiduo M.G., Mandara C.I., Rumisha S.F., Chaky F., Denton M., Marsh P.W., Verity R. Describing the current status of Plasmodium falciparum population structure and drug resistance within mainland Tanzania using molecular inversion probes. Mol. Ecol. 2020;30(1):100–113. doi: 10.1111/mec.15706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mwai L., Ochong E., Abdirahman A., Kiara S.M., Ward S., Kokwaro G., Sasi P., Marsh K., Borrmann S., Mackinnon M. Chloroquine resistance before and after its withdrawal in Kenya. Malar. J. 2009;8:106. doi: 10.1186/1475-2875-8-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag S., Ursing J., Rodrigues A., Crespo M., Krogsgaard C., Lund O., Aarestrup F., Aligrangis M., Kofoed P. Proof of concept: used malaria rapid diagnostic tests applied for parallel sequencing for surveillance of molecular markers of anti ‑ malarial resistance in Bissau, Guinea‑Bissau during 2014 – 2017. Malar. J. 2019;18(1):252. doi: 10.1186/s12936-019-2894-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naidoo I., Roper C. Drug resistance maps to guide intermittent preventive treatment of malaria in African infants. Parasitology. 2011;138:1469–1479. doi: 10.1017/S0031182011000746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noedl H., Se Y., Schaecher K., Smith B.L., Socheat D., Fukuda M.M., Artemisinin Resistance in Cambodia 1 Study C Evidence of artemisinin-resistant malaria in western Cambodia. N. Engl. J. Med. 2008;359:2619–2620. doi: 10.1056/NEJMc0805011. [DOI] [PubMed] [Google Scholar]
- Nzoumbou-Boko R., Panté-Wockama CB.G., Ngoagoni C., Petiot N., Legrand E., Vickos U., Gody J.C., Manirakiza A., Ndoua C., Lombart J.P. Molecular assessment of kelch13 non-synonymous mutations in Plasmodium falciparum isolates from Central African Republic (2017-2019) Malar. J. 2020;19:1–7. doi: 10.1186/s12936-020-03264-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogouyèmi-Hounto A., Damien G., Deme A.B., Ndam N.T., Assohou C., Tchonlin D., Mama A., Hounkpe V.O., Moutouama J.D., Remoué F. Lack of artemisinin resistance in Plasmodium falciparum in northwest Benin after 10 years of use of artemisinin-based combination therapy. Parasite. 2016;23:28. doi: 10.1051/parasite/2016028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco M.A., Kadakia E.R., Chaudhary Z., Perkins D.J., Kelley J., Ravishankar S., Cranfield M., Talundzic E., Udhayakumar V., Escalante A.A. Evolution and genetic diversity of the k13 gene associated with artemisinin delayed parasite clearance in Plasmodium falciparum. Antimicrob. Agents Chemother. 2019;63(8) doi: 10.1128/AAC.02550-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page M.J., McKenzie J.E., Bossuyt P.M., Boutron I., Hoffmann T.C., Mulrow C.D. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ. 2021;372:n71. doi: 10.1136/bmj.n71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne D. Spread of chloroquine resistance in Plasmodium falciparum. Parasitol. Today. 1987;3:241–246. doi: 10.1016/0169-4758(87)90147-5. [DOI] [PubMed] [Google Scholar]
- Pearce R.J., Pota H., Evehe M.S., Ba el H., Mombo-Ngoma G., Malisa A.L., Ord R., Inojosa W., Matondo A., Diallo D.A. Multiple origins and regional dispersal of resistant dhps in African Plasmodium falciparum malaria. PLoS Med. 2009;6 doi: 10.1371/journal.pmed.1000055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roper C., Pearce R., Nair S., Sharp B., Nosten F., Anderson T. Intercontinental spread of pyrimethamine-resistant malaria. Science. 2004;305:1124. doi: 10.1126/science.1098876. [DOI] [PubMed] [Google Scholar]
- Snow R.W., Sartorius B., Kyalo D., Maina J., Amratia P., Mundia C.W., Bejon P., Noor A.M. The prevalence of Plasmodium falciparum in sub-Saharan Africa since 1900. Nature. 2017;550:515–518. doi: 10.1038/nature24059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow R.W., Trape J.F., Marsh K. The past, present and future of childhood malaria mortality in Africa. Trends Parasitol. 2001;17:593–597. doi: 10.1016/s1471-4922(01)02031-1. [DOI] [PubMed] [Google Scholar]
- Straimer J., Gnadig N.F., Witkowski B., Amaratunga C., Duru V., Ramadani A.P., Dacheux M., Khim N., Zhang L., Lam S. Drug resistance. K13-propeller mutations confer artemisinin resistance in Plasmodium falciparum clinical isolates. Science. 2015;347:428–431. doi: 10.1126/science.1260867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talisuna A.O., Staedke S.G., D’Alessandro U. Pharmacovigilance of antimalarial treatment in Africa: is it possible? Malar J. 2006;5:50. doi: 10.1186/1475-2875-5-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawe L., Menegon M., Ramatlho P., Muthoga C.W., Mutukwa N., Vurayai M., Bothudile W., Motshoge T., L’Episcopia M., Mosweunyane T. Molecular surveillance of [lasmodium falciparum drug resistance markers in clinical samples from Botswana. Am. J. Trop. Med. Hyg. 2018;99(6):1499–1503. doi: 10.4269/ajtmh.18-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Kuile F.O., van Eijk A.M., Filler S.J. Effect of sulfadoxine-pyrimethamine resistance on the efficacy of intermittent preventive therapy for malaria control during pregnancy: a systematic review. J. Am. Med. Assoc. 2007;297:2603–2616. doi: 10.1001/jama.297.23.2603. [DOI] [PubMed] [Google Scholar]
- Tun K.M., Imwong M., Lwin K.M., Win A.A., Hlaing T.M., Hlaing T., Lin K., Kyaw M.P., Plewes K., Faiz M.A. Spread of artemisinin-resistant Plasmodium falciparum in Myanmar: a cross-sectional survey of the K13 molecular marker. Lancet Infect. Dis. 2015;15:415–421. doi: 10.1016/S1473-3099(15)70032-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uwimana A., Legrand E., Stokes B.H., Ndikumana J.M., Warsame M., Umulisa N., Ngamije D., Munyaneza T., Mazarati J.B., Munguti K. Emergence and clonal expansion of in vitro artemisinin-resistant Plasmodium falciparum kelch13 R561H mutant parasites in Rwanda. Nat. Med. 2020;26:1602–1608. doi: 10.1038/s41591-020-1005-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uwimana A., Umulisa N., Venkatesan M., Svigel S.S., Zhou Z., Munyaneza T., Habimana R.M., Rucogoza A., Moriarty L.F., Sandford R., Piercefield E., Goldman I., Ezema B., Talundzic E., Pacheco M.A., Escalante A.A., Ngamije D., Mangala J.N., Kabera M., Munguti K., Murindahabi M., Brieger W., Musanabaganwa C., Mutesa L., Udhayakumar V., Mbituyumuremyi A., Halsey E.S., Lucchi N.W. Association of Plasmodium falciparum kelch13 R561H genotypes with delayed parasite clearance in Rwanda: an open-label, single-arm, multicentre, therapeutic efficacy study. Lancet Infect. Dis. 2021 doi: 10.1016/S1473-3099(21)00142-0. S1473-3099(21)00142-0, online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wamae K., Okanda D., Ndwiga L., Osoti V., Kimenyi K.M., Abdi A.I., Bejon P., Sutherland C., Ochola-Oyier L.I. No evidence of P. falciparum K13 artemisinin conferring mutations over a 24-year analysis in Coastal Kenya, but a near complete reversion to chloroquine wild type parasites. Antimicrob. Agents Chemother. 2019;63(12) doi: 10.1128/AAC.01067-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Shrestha S., Li X., Miao J., Yuan L., Cabrera M., Grube C., Yang Z., Cui L. Prevalence of K13-propeller polymorphisms in Plasmodium falciparum from China-Myanmar border in 2007-2012. Malar. J. 2015;14:168. doi: 10.1186/s12936-015-0672-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellems T.E., Plowe C.V. Chloroquine-resistant malaria. J. Infect. Dis. 2001;184:770–776. doi: 10.1086/322858. [DOI] [PubMed] [Google Scholar]
- World Malaria report . WHO; 2020. https://www.who.int/publications/i/item/9789240015791 [Google Scholar]
- Who Report on antimalarial efficacy . Global Malaria Programme, WHO; 2020. Report on Antimalarial Drug Efficacy, Resistance and Response: 10 Years of Surveillance (2010-2019)https://www.who.int/publications/i/item/9789240012813 [Google Scholar]
- Who S.M.C. World Health Organization; Geneva: 2012. Policy Recommendation: Seasonal Malaria Chemoprevention (SMC) for Plasmodium Falciparum Malaria Control in Highly Seasonal Transmission Areas of the Sahel Sub-region in Africa. [Google Scholar]
- Wwarn K13 Genotype-Phenotype Study Group Association of mutations in the Plasmodium falciparum Kelch13 gene (Pf3D7_1343700) with parasite clearance rates after artemisinin-based treatments-a WWARN individual patient data meta-analysis. BMC Med. 2019;17(1):1. doi: 10.1186/s12916-018-1207-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yobi D.M., Kayiba N.K., Mvumbi D.M., Boreux R., Bontems S., Kabututu P.Z., Mol P.D., Speybroeck N., Mvumbi G.L., Hayette M. The lack of K13-propeller mutations associated with artemisinin resistance in Plasmodium falciparum in Democratic Republic of Congo (DRC) PLoS ONE. 2020;15(8) doi: 10.1371/journal.pone.0237791. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.