Graphical abstract
A series of quinolone-triazole conjugates synthesized as potential antiviral agents for SARS-CoV2. Some of the conjugates are more potent than the standards.
Keywords: Quinoline, Triazole, Click chemistry, SARS-CoV-2, Molecular docking
Abstract
At present therapeutic options for severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) are very limited. We designed and synthesized three sets of small molecules using quinoline scaffolds. A series of quinoline conjugates (10a-l, 11a-c, and 12a-e) by incorporating 1,2,3-triazole were synthesized via a modified microwave-assisted click chemistry technique. Among the synthesized conjugates, 4-((1-(2-chlorophenyl)-1H-1,2,3-triazol-4-yl)methoxy)-6-fluoro-2-(trifluoromethyl)quinoline (10g) and 6-fluoro-4-(2-(1-(4-methoxyphenyl)-1H-1,2,3-triazol-4-yl)ethoxy)-2-(trifluoromethyl)quinoline (12c) show high potency against SARS-CoV-2. The selectivity index (SI) of compounds 10g and 12c also indicates the significant efficacy compared to the reference drugs.
1. Introduction:
In December 2019, an unknown viral infection was identified in a local fish and wild animal market of Wuhan city (China) [1]. Since then, the virus has rapidly spread across mainland China followed by the rest of the world [2], [3]. On Feb 11, 2020, the WHO identified the virus as the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2). The viral infection disease is called as 2019-new coronavirus disease (COVID-19), and in March 2020 declared the coronavirus outbreak a global pandemic [4]. As of April 2021, COVID-19 has affected more than 152 million people with 3.18 million deaths worldwide.
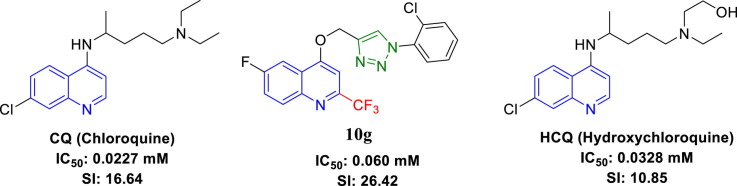
As of now, there is no potential antiviral drug available for the treatment of COVID-19. However, a few vaccines have received approval from FDA to control the pandemic. The drug repurposing approach was considered the most accessible pathway for exploring drugs to control the global pandemic. Several drugs that were considered for repurposing to control the COVID-19 pandemic include chloroquine (CQ) and hydroxychloroquine (HCQ) (Fig. 1 ). These exhibited promising responses accompanied with serious side effects (ventricular arrhythmias, retinopathy, QT prolongation, or cardiac-related toxicity) [5], [6], [7], [8], [9], [10], [11]. Recently, a few quinolone-based small molecules reported their antiviral properties against SARS-CoV-2 [12], [13], [14], [15], [16], [17], [18], [19], [20].
Fig. 1.
Examples of repurposing drugs for COVID-19.
New drug development is expensive, time-consuming, and challenging processes. In the current scenario, molecular hybridization of bioactive moieties is a powerful and attractive rational drug design strategy for new drug development because of several advantages such as a) to achieve selectivity; b) gain desired activity; c) multiple pharmacological targets; d) lower possible cytotoxicity. Our group has actively engaged in developing drug conjugates and hybrid conjugates by this approach [21], [22], [23], [24], [25], [26].
In the present study, we have designed and synthesized a set of novel quinoline-triazole conjugates with potential antiviral properties against SARS-CoV-2 using the ‘click’ chemistry approach employing various azides and the alkyne component of the propargyl linked quinolines. We considered the quinoline scaffold from the repurposed drugs (chloroquine and hydroxychloroquine) and triazole because of their importance in the drug development process and well-known for diversified biological properties [27], [28], [29], [30], [31]. Besides, the presence of electronegative fluorine atoms may alter the physicochemical properties of the molecule significantly. This may improve the stability, reducing the cytotoxicity, in addition to enhancing the overall therapeutic efficacy in our designed molecules [32].
2. Results and discussion
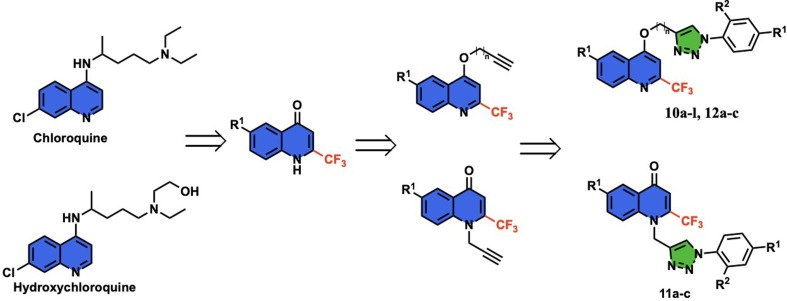
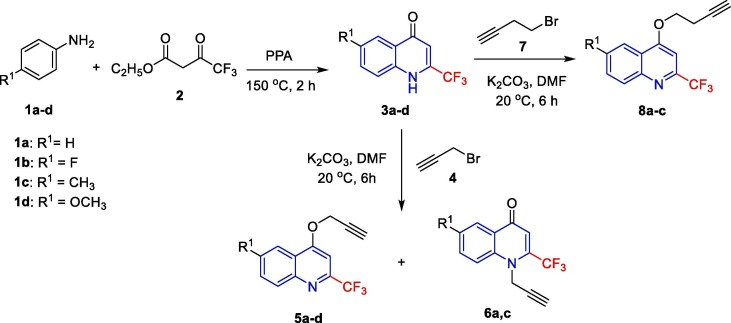
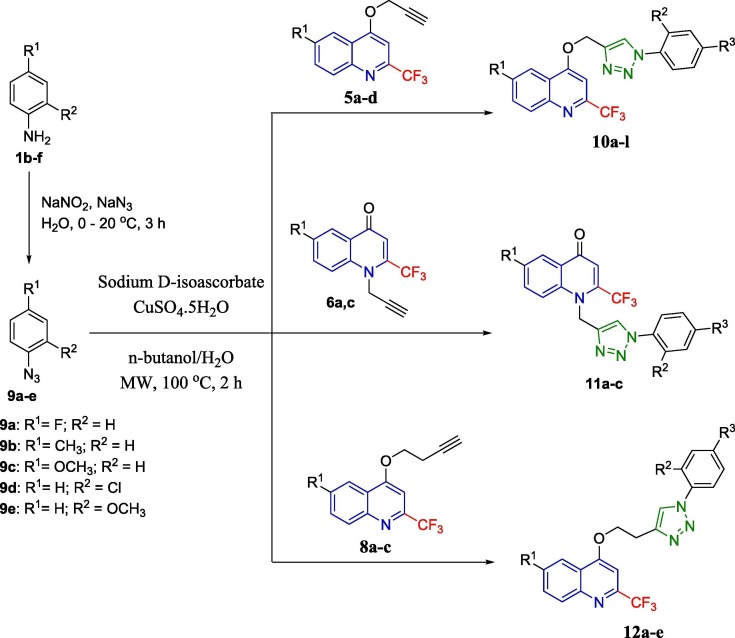
The synthetic protocol was developed for the synthesis of the targeted quinoline-triazole conjugates (Fig. 2 ) by adopting the well-established Cu-mediated click chemistry of alkynes 5, 6, and 8 with varied azides 10. The precursor alkynes 5, 6, and 8 were synthesized by treating a solution of 6-(un)substituted-2-(trifluoromethyl)quinolin-4(1H)-ones 3 in DMF with propargyl bromide (4) or 4-bromobut-1-yne (7) in the presence of potassium carbonate (K2CO3) at room temperature [33]. (Scheme 1 ). The azides of aromatic amines 1b–f were synthesized using the previously reported method [25].
Fig. 2.
Designing of quinoline-triazole conjugates.
Scheme 1.
Synthesis of alkynes 5, 6, and 8.
2.1. Synthesis of alkynes 5, 6, and 8
A solution of compounds 3a–d in DMF on reaction with propargyl bromide (4) or 4-bromobut-1-yne (7) in the presence of potassium carbonate (K2CO3) at 20 °C for 6 h gave predominately O-substituted quinolines. However, in the case of 3a and 3c on treatment with propargyl bromide 4, we got both N-substituted quinolones and O-substituted quinolines in a good ratio, which were isolated by column chromatography (5a,b, and 6a,b). We also observed the formation of N-substituted quinolones as a minor product when we treated 4-bromobut-1-yne (7) with 3a–d. The formation of O-substituted quinolones is move favored over N-substituted quinolines because of the steric and electronic properties of the CF3 group. The detailed investigation was reported by Raić‐Malić and coworkers [34].
2.2. Synthesis of triazole-quinoline conjugates
Alkynes 5, 6, and 8 were treated with aromatic azides 9 adopting a modified click chemical technique in presence of CuSO4·5H2O and sodium d-isoascorbate in n-butanol–water mixture under microwave irradiation for 2 h at 100 °C to afford the desired corresponding conjugates 10a–l, 11a–c and 12a–e in good yields (Scheme 2 ). The reactions do work at room temperature but never go to completion even after 3 days. Our optimized reaction condition able to complete the reaction in 2 h. All the synthesized compounds were fully characterized by spectroscopic techniques.
Scheme 2.
Triazole-quinoline conjugates 10a-l, 11a-c, 12a-e.
2.3. Antiviral properties
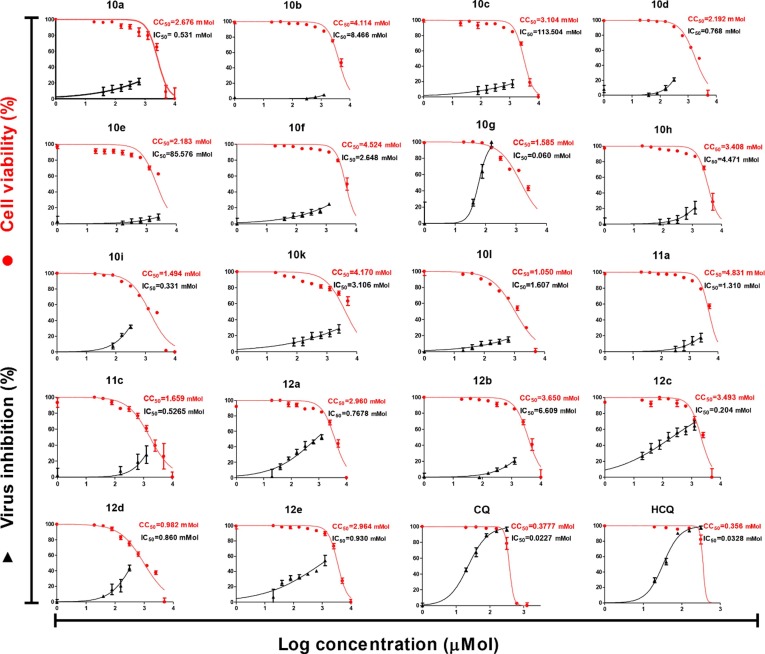
The antiviral properties of the synthesized conjugates (10, 11, and 12) against SARS-CoV-2 were determined by the standard technique [35], [36], [37]. Table 1 exhibits the antiviral properties of the tested compounds and standard references (CQ, HCQ) used expressed as IC50 (concentration necessary for 50% reduction of virus-induced cytopathic effect (CPE) compared to the virus control experiment). We have also determined the CC50 (concentration necessary for 50% growth normal cell line “VERO-E6” inhibition compared to the control experiment) for all the synthesized conjugates (Fig. 3 ). We calculated the selectivity (therapeutical) index (SI), which is the ratio of CC50 relative to IC50 of the tested conjugate. From the observed biological properties (Table 1), it has been noticed that conjugate 10g is superior among all the tested agents revealing potent antiviral inhibition with high selectivity index relative to the standard references used (IC50 = 0.060, 0.0227, 0.0328 mM; CC50 = 1.585, 0.3777, 0.356 mM, SI = 26.42, 16.64, 10.85; corresponding to 10g, CQ and HCQ, respectively). Conjugate 12c also reveals enhanced selectivity index relative to the standard references used with milder antiviral potency (IC50 = 0.204, CC50 = 3.493 mM, SI = 17.12). Additionally, compounds 10i, 11a, 11c, 12a, and 12e also show promising biological properties with a considerable selectivity index (SI = 4.51–3.15).
Table 1.
Antiviral properties of the quinine-triazole conjugates 10, 11, and 12 against SARS-CoV-2.
| Entry | Compound | R1 | R2 | R3 | IC50 (mM)a | CC50 (mM)b | SIc |
|---|---|---|---|---|---|---|---|
| 1 | 10a | H | H | F | 0.531 | 2.676 | 5.04 |
| 2 | 10b | H | H | CH3 | 8.466 | 4.114 | 0.49 |
| 3 | 10c | H | H | OCH3 | 113.504 | 3.104 | 0.03 |
| 4 | 10d | H | Cl | H | 0.768 | 2.192 | 2.85 |
| 5 | 10e | H | OCH3 | H | 85.576 | 2.183 | 0.03 |
| 6 | 10f | F | H | CH3 | 2.648 | 4.524 | 1.71 |
| 7 | 10g | F | Cl | H | 0.060 | 1.585 | 26.42 |
| 8 | 10h | F | OCH3 | H | 4.471 | 3.408 | 0.76 |
| 9 | 10i | CH3 | Cl | H | 0.331 | 1.494 | 4.51 |
| 10 | 10j | CH3 | OCH3 | H | nd | nd | nd |
| 11 | 10k | OCH3 | H | F | 3.106 | 4.170 | 1.34 |
| 12 | 10l | OCH3 | H | CH3 | 1.607 | 1.050 | 0.65 |
| 13 | 11a | H | H | OCH3 | 1.310 | 4.831 | 3.69 |
| 14 | 11b | H | OCH3 | H | nd | nd | nd |
| 15 | 11c | CH3 | H | OCH3 | 0.5265 | 1.659 | 3.15 |
| 16 | 12a | H | H | OCH3 | 0.7678 | 2.960 | 3.86 |
| 17 | 12b | F | H | F | 6.609 | 3.650 | 0.55 |
| 18 | 12c | F | H | OCH3 | 0.204 | 3.493 | 17.12 |
| 19 | 12d | F | OCH3 | H | 0.860 | 0.982 | 1.14 |
| 20 | 12e | CH3 | H | OCH3 | 0.930 | 2.964 | 3.19 |
| 21 | CQ | 0.0227 | 0.3777 | 16.64 | |||
| 22 | HCQ | 0.0328 | 0.356 | 10.85 |
nd: not done.
IC50: Concentration necessary for 50% reduction of virus-induced cytopathic effect (CPE) compared to the virus control experiment.
CC50: Half maximal cytotoxic concentration for the normal cell line (VERO-E6) inhibition compared to the control experiment.
SI (Selectivity index/therapeutical index): .
Fig. 3.
Dose-response curves for the tested compounds against SARS-CoV-2.
Some qualitative structure–activity relationship could be assigned based on observed antiviral properties. The fluoroquinolinyl conjugates show enhanced antiviral properties than the unsubstituted conjugates as exhibited in pairs 10f/10b (IC50 = 2.648, 8.466 mM), 10g/10d (IC50 = 0.060, 0.768 mM), 10h/10e (IC50 = 4.471, 85.576 mM) and 12c/12a (IC50 = 0.204, 0.7678 mM). Probably the fluorine atoms play important role in enhancing the antiviral properties. Moreover, the o-chlorophenyl triazolyl conjugates reveal higher antiviral properties than the o-methoxyphenyl analogues as shown in pairs 10d/10e (IC50 = 0.768, 85.576 mM) and 10g/10h (IC50 = 0.060, 4.471 mM). Additionally, molecular modeling can estimate the parameters/functional groups necessary for optimizing antiviral properties.
2.4. Molecular docking study
Molecular docking is one of the most important computational techniques widely accessible in medicinal chemical studies for many purposes of which, determining/predicting the mode of action of an agent and explaining the observed bio-properties. The tested compounds against SARS-CoV-2 were subjected for molecular docking study utilizing PDB ID: 6LU7 “SARS-CoV-2 main protease (Mpro) with an inhibitor N3” [38], [39]. Discovery Studio 2.5 software was used (CDOCKER, force field: CHARMm, ligand partial charge method: MMFF).
From the docking observations (Supplementary Figs. S1 and S2), it has been noticed that chloroquine and hydroxychloroquine show hydrogen bonding due to the diethylamino nitrogen interaction with GLU166 (which is one of the amino acids revealing interaction with the co-crystallized ligand “N3 inhibitor” in the PDB: 6LU7), and docking scores −46.2, −42.3 kcal mol−1 for chloroquine and hydroxychloroquine, respectively. These observations are comparable to their antiviral properties observed (IC50 = 0.0227, 0.0328 mM, respectively). Compounds 10g and 12c, which are the most effective antiviral agents synthesized (IC50 = 0.060, 0.204 mM, respectively) reveal behavior due to hydrogen bonding observation. Where the triazolyl nitrogen has hydrogen bonding interaction with the same amino acid (GLU166). Additionally, the promising agents synthesized 10d, 10i, 12a and 12e (IC50 = 0.768, 0.331, 0.7678, 0.930 mM, respectively) also exhibit similar docking observations. However, other analogs show good docking scores due to false alignment in the protein active site “interaction with amino acid(s) other than those overlapped with the co-crystallized ligand”. These findings support the experimentally observed weak antiviral properties (e.g. compounds 10c and 10e of IC50 = 113.5, 85.6 mM and docking scores = -37.9, −38.5 kcal/mol, respectively).
2.5. Conclusion
In conclusion, we have synthesized three sets of triazole incorporated quinolone conjugates in good yields by an optimized facile reaction condition. Some conjugates showed promising antiviral activity against SARS-CoV-2. Compound 10g and 12c found the most effective agents for the SARS-CoV-2. The selectivity index (SI) of compounds also indicates significant efficacy compared to the reference drugs. We believe the initial investigation and the important scaffold could be further used as resources for the development of potential drug candidates for SARS-CoV-2.
3. Experimental section
3.1. Chemistry
Melting points were determined on a capillary point apparatus equipped with a digital thermometer. NMR spectra were recorded in CDCl3, DMSO‑d 6, on Bruker NMR spectrometer operating at 500 MHz for 1H (with TMS as an internal standard) and 125 MHz for 13C. High-resolution mass spectra were recorded with TOF analyzer spectrometer by using electron spray mode. All microwave-assisted reactions were carried out with a single-mode cavity Discover Microwave Synthesizer (CEM Corporation, NC). The reaction mixtures were transferred into a 10 mL glass pressure microwave tube equipped with a magnetic stirrer bar. The tube was closed with a silicon septum and the reaction mixture was subjected to microwave irradiation (Discover mode; run time: 60 s; Power Max-cooling mode).
3.1.1. General procedure for the synthesis of O-alkynyl-oxyquinolines 5a-d and N-alkynyl-oxyquinolines 6a,c [26], [33]
To a suspension of corresponding quinolone 3a–d (1 equiv.) in N, N-dimethylformamide (DMF; 10 mL), K2CO3 (1.5 equiv.) was added. After stirring for 30 min, propargyl bromide 4 (1 equiv.) was added and the reaction mixture was stirred for 6 h. The solvent was removed under reduced pressure and the crude residue was purified by column chromatography. These compounds were characterized by comparing their spectroscopic data to the reported ones [26], [33].
3.1.2. General procedure for the synthesis of synthesis O-alkynyl-quinolines 8a-c [33]
To a suspension of corresponding quinolone 3a–d (1 equiv.) in N, N-dimethylformamide (DMF; 10 mL), K2CO3 (1.5 equiv.) was added. After stirring for 30 min, 4-bromobut-1-yne 7 (1 equiv.) was added and the reaction mixture was subjected to microwave irradiations for 6 h. The crude mixture was diluted with water than solid formed filtered off and crystallized from ethanol. These compounds were characterized by comparing their spectroscopic data to the reported ones [28].
3.1.3. General procedure for the synthesis of quinoline-triazole conjugates
To a solution of quinoline (1 equiv.) in n-butanol/H2O (2: 1) (3 mL), CuSO4 (0.05 equiv.), and sodium d-isoascorbate monohydrate (0.15 equiv.) was added at room temperature. To this mixture, aryl azide (1.5 mmol) was added and the reaction mixture was subjected to microwave irradiations at 250 W for 2 h. The crude mixture was diluted with water then extracted with EtOAc and the combined organic layer was dried over sodium sulfate, concentrated in a vacuum, and purified through column chromatography to give the pure quinoline-triazole derivatives in good yields.
3.1.3.1. 4-((1-(4-Fluorophenyl)-1H-1,2,3-triazol-4-yl)methoxy)-2-(trifluoromethyl)quinoline (10a)
White microcrystals, m.p. 173–175 °C, yield 46% (0.28 g). IR: ν max/cm−1 3052, 2923, 1574, 1515, 1378, 1233, 831, 763; 1H NMR (CDCl3) δ: 8.22 (d, J = 8.0 Hz, 1H), 8.14 (d, J = 8.5 Hz, 1H), 8.10 (s, 1H), 7.81–7.75 (m, 1H), 7.73 (dd, J = 9.0, 4.6 Hz, 2H), 7.59 (t, J = 7.6 Hz, 1H), 7.28 (s, 1H), 7.24 (t, J = 4.0 Hz, 1H), 7.21 (s, 1H), 5.56 (s, 2H). 13C NMR (CDCl3) δ: 163.9, 162.5, 161.9, 149.4, 149.1, 148.4, 143.3, 133.3, 131.3, 129.9, 127.9, 123.0, 122.9, 122.1, 121.8, 117.2, 117.0, 97.4, 62.7. HRMS m/z for C19H12F4N4O [M+H]+ Calcd. 389.1020. Found: 389.1013.
3.1.3.2. 4-((1-(p-Tolyl)-1H-1,2,3-triazol-4-yl)methoxy)-2-(trifluoromethyl)quinoline (10b)
White microcrystals, m.p. 172–174 °C, yield 54% (0.32 g). IR: ν max/cm−1 3050, 2930, 1575, 1514, 1372, 1252, 817, 770; 1H NMR (CDCl3) δ: 8.23 (d, J = 8.3 Hz, 1H), 8.15–8.10 (m, 2H), 7.77 (t, J = 7.7 Hz, 1H), 7.64–7.54 (m, 3H), 7.32 (d, J = 8.1 Hz, 2H), 7.28 (s, 1H), 5.55 (s, 2H), 2.41 (s, 3H).13C NMR (CDCl3) δ: 162.2, 149.3, 149.1, 148.4, 139.6, 134.7, 133.0, 131.3, 130.6, 129.8, 127.9, 125.6, 122.8, 122.1, 121.8, 120.8, 120.6, 97.4, 62.8, 21.3. HRMS m/z for C20H15F3N4O [M+H]+ Calcd. 385.1271. Found: 385.1278.
3.1.3.3. 4-((1-(4-Methoxyphenyl)-1H-1,2,3-triazol-4-yl)methoxy)-2-(trifluoromethyl)quinoline (10c)
White microcrystals, m.p. 180–182 °C, yield 54% (0.32 g). IR: ν max/cm−1 3050, 2980, 1591, 1515, 1370, 1252, 840, 771; 1H NMR (CDCl3) δ: 8.19 (d, J = 8.3 Hz, 1H), 8.13–8.05 (m, 2H), 7.74 (t, J = 7.5 Hz, 1H), 7.61 (d, J = 8.5 Hz, 2H), 7.55 (t, J = 7.5 Hz, 1H), 7.25 (d, J = 6.2 Hz, 1H), 6.99 (d, J = 8.5 Hz, 2H), 5.51 (s, 2H), 3.83 (s, 3H). 13C NMR (CDCl3) δ: 162.6, 160.3, 149.2, 148.9, 148.3, 142.8, 131.3, 130.3, 129.7, 127.8, 122.7, 122.5, 122.1, 121.9, 121.8, 120.6, 115.0, 97.3, 62.8, 55.8. HRMS m/z for C20H15F3N4O2 [M+H]+ Calcd. 401.1220. Found: 401.1224.
3.1.3.4. 4-((1-(2-Chlorophenyl)-1H-1,2,3-triazol-4-yl)methoxy)-2-(trifluoromethyl)quinoline (10d)
White microcrystals, m.p. 200–202 °C, yield 44% (0.26 g). IR: ν max/cm−1 3079, 2979, 1574, 1530, 1369, 1274, 848, 788, 756; 1H NMR (CDCl3) δ: 8.40 (d, J = 8.2 Hz, 1H), 8.30 (t, J = 8.5 Hz, 1H), 8.22 (s, 1H), 8.19 (d, J = 8.4 Hz, 1H), 7.74 (t, J = 7.6 Hz, 1H), 7.65–7-62 (m, J = 10.7, 3.9 Hz, 2H), 7.57–7.44 (m, 2H), 7.33 (s, 1H), 5.65 (s, 2H).13C NMR (CDCl3) δ: 162.6, 148.5, 142.2, 132.1, 131.3, 131.1, 130.7, 129.9, 129.8, 128.3, 128.0, 127.9, 125.6, 122.2, 121.9, 121.1, 108.1, 97.5, 62.8. HRMS m/z for: C19H12ClF3N4O [M+H]+ Calcd. 405.0724. Found: 405.0724.
3.1.3.5. 4-((1-(2-Methoxyphenyl)-1H-1,2,3-triazol-4-yl)methoxy)-2-(trifluoromethyl)quinoline (10e)
White microcrystals, m.p. 179–181 °C, yield 61% (0.37 g). IR: ν max/cm−1 3100, 2931, 1576, 1512, 1368, 1249, 831, 756; 1H NMR (CDCl3) δ: 8.23 (d, J = 8.5 Hz, 1H), 8.15 (d, J = 8.5 Hz, 1H), 8.07 (s, 1H), 7.77 (t, J = 8.1 Hz, 1H), 7.63 (d, J = 9.0 Hz, 2H), 7.58 (t, J = 7.6 Hz, 1H), 7.29 (s, 1H), 7.01 (d, J = 9.0 Hz, 2H), 5.55 (s, 2H), 3.85 (s, 3H).13C NMR (CDCl3) δ: 162.9, 151.2, 131.4, 131.3, 130.6, 129.7, 128.3, 128.0, 127.9, 126.2, 125.9, 125.7, 122.3, 122.2, 121.9, 121.6, 112.5, 97.5, 62.9, 56.2. HRMS m/z for: C20H15F3N4O2 [M+H] + Calcd. 401.1220. Found: 401.1217.
3.1.3.6. 6-Fluoro-4-((1-(p-tolyl)-1H-1,2,3-triazol-4-yl)methoxy)-2-(trifluoromethyl)quinoline (10f)
White microcrystals, m.p. 170–172 °C, yield 75% (0.46 g). IR: ν max/cm−1 3100, 2925, 1577, 1517, 1478, 1277, 815, 717; 1H NMR (CDCl3) δ: 8.18–8.09 (m, 2H), 7.81 (dd, J = 9.2, 2.7 Hz, 1H), 7.62 (d, J = 8.4 Hz, 2H), 7.52 (td, J = 9.2, 2.8 Hz, 1H), 7.38–7.26 (m, 3H), 5.55 (s, 2H), 2.42 (s, 3H).13C NMR (CDCl3) δ: 162.5, 160.5, 145.4, 142.7, 139.7, 134.7, 132.6, 132.5, 130.6, 122.9, 122.7, 121.8, 121.6, 121.4, 120.8, 106.3, 106.1, 97.9, 62.9, 21.3. HRMS m/z for C20H14F4N4O [M+H]+ Calcd. 403.1177. Found: 403.1176.
3.1.3.7. 4-((1-(2-Chlorophenyl)-1H-1,2,3-triazol-4-yl)methoxy)-6-fluoro-2-(trifluoromethyl)quinoline (10g)
White microcrystals, m.p. 174–176 °C, yield 79% (0.48 g). IR: ν max/cm−1 3050, 2930, 1600, 1574, 1518, 1478, 1233, 852, 761; 1H NMR (CDCl3) δ: 8.18–8.11 (m, 2H), 7.82 (dd, J = 9.1, 2.7 Hz, 1H), 7.66 (dd, J = 6.4, 3.0 Hz, 1H), 7.59 (dd, J = 7.3, 2.0 Hz, 1H), 7.55–7.44 (m, 3H), 7.31 (s, 1H), 5.59 (s, 2H).13C NMR (CDCl3) δ: 162.1, 160.6, 145.5, 142.0, 134.8, 132.6, 132.5, 131.3, 131.1, 128.8, 128.3, 128.0, 126.0, 121.7, 121.5, 106.3, 106.1, 97.9, 62.9. HRMS m/z for C19H11ClF4N4O [M+H]+ Calcd. 423.0630. Found: 423.0631.
3.1.3.8. 6-Fluoro-4-((1-(2-methoxyphenyl)-1H-1,2,3-triazol-4-yl)methoxy)-2-(trifluoromethyl)quinoline (10h)
White microcrystals, m.p. 154–156 °C, yield 67% (0.41 g). IR: ν max/cm−1 3100, 2980, 1599, 1506, 1478, 1249, 837; 1H NMR (CDCl3) δ: 8.31 (s, 1H), 8.12 (dd, J = 9.2, 5.2 Hz, 1H), 7.81–7.79 (m, 2H), 7.52–7.46 (m, 2H), 7.32 (s, 1H), 7.09 (dd, J = 13.0, 8.0 Hz, 2H), 5.55 (s, 2H), 3.87 (s, 3H).13C NMR (CDCl3) δ: 162.2, 160.4, 151.1, 145.3, 141.3, 132.4, 132.3, 130.6, 128.2, 127.9, 126.1, 125.9, 125.5, 121.5, 121.3, 112.5, 106.1, 97.9, 62.9, 56.2. HRMS m/z for C20H14F4N4O2 [M+H]+ Calcd. 419.1126. Found: 419.1123.
3.1.3.9. 4-((1-(2-Chlorophenyl)-1H-1,2,3-triazol-4-yl)methoxy)-6-methyl-2-(trifluoromethyl)quinoline (10i)
White microcrystals, m.p. 179–181 °C, yield 78% (0.47 g). IR: ν max/cm−1 3000, 2980, 1575, 1495, 1372, 1251, 847, 767; 1H NMR (CDCl3) δ: 8.15–8.09 (m, 2H), 7.81 (dd, J = 9.2, 2.7 Hz, 1H), 7.62 (d, J = 8.4 Hz, 2H), 7.52 (td, J = 9.2, 2.8 Hz, 1H), 7.33–7.31(m, 3H),5.55 (s, 2H), 2.42 (s, 3H). 13C NMR (CDCl3) δ: 162.0, 154.4, 147.0, 138.2, 134.9, 134.4, 133.6, 131.3, 131.1, 130.4, 129.6, 128.8, 128.3, 128.0, 125.6, 120.1, 119.7, 97.4, 62.7, 22.1. HRMS m/z for C20H14ClF3N4O [M+H]+ Calcd. 419.0881. Found: 419.0883.
3.1.3.10. 4-((1-(2-Methoxyphenyl)-1H-1,2,3-triazol-4-yl)methoxy)-6-methyl-2-(trifluoromethyl)quinoline (10j)
White microcrystals, m.p. 185–187 °C, yield 65% (0.40 g). IR: ν max/cm−1 3070, 2968, 1574, 1507, 1374, 1251, 830, 762; 1H NMR (CDCl3) δ: 8.30 (s, 1H), 8.15 (d, J = 8.6 Hz, 1H), 8.02 (s, 1H), 7.83 (dd, J = 7.9, 1.5 Hz, 1H), 7.62 (dd, J = 8.7, 1.3 Hz, 1H), 7.44 (td, J = 8.4, 1.6 Hz, 1H), 7.30 (s, 1H), 7.15–7.07 (m, 2H), 5.58 (s, 2H), 3.89 (s, 3H), 2.53 (s, 3H). 13C NMR (CDCl3) δ: 162.4, 151.2, 146.7, 141.6, 138.3, 133.7, 131.3, 131.1, 130.6, 129.3, 126.2, 125.9, 125.6, 121.9, 121.6, 121.1, 112.5, 97.5, 62.9, 56.2, 22.1. HRMS m/z for C21H17F3N4O2 [M+Na]+ Calcd. 415.1376. Found: 415.1383.
3.1.3.11. 4-((1-(4-Fluorophenyl)-1H-1,2,3-triazol-4-yl)methoxy)-6-methoxy-2-(trifluoromethyl)quinoline (10k)
White microcrystals, m.p. 190–192 °C, yield 75% (0.46 g). IR: ν max/cm−1 3100, 2923, 1590, 1513, 1438, 1373, 1229, 840, 724; 1H NMR (CDCl3) δ: 8.10 (s, 1H), 8.05–7.96 (m, 1H), 7.71 (dd, J = 8.9, 4.5 Hz, 2H), 7.42–7.34 (m, 2H), 7.22–7.20 (m, 3H), 5.52 (s, 2H), 3.89 (s, 3H). 13C NMR (CDCl3) δ: 163.8, 161.8, 161.2, 159.1, 146.7, 146.4, 144.3, 143.2, 133.2, 131.4, 123.8, 122.8, 121.9, 120.8, 117.2, 117.0, 99.9, 97.6, 62.5, 55.9. HRMS m/z for C20H14F4N4O2 [M+H]+ Calcd. 419.1126. Found: 419.1123.
3.1.3.12. 6-Methoxy-4-((1-(p-tolyl)-1H-1,2,3-triazol-4-yl)methoxy)-2-(trifluoromethyl)quinoline (10l)
White microcrystals, m.p. 181–183 °C, yield 65% (0.40 g). IR: ν max/cm−1 3138, 2927, 1578, 1505, 1482, 1378, 1232, 841, 722; 1H NMR (CDCl3) δ: 8.08 (s, 1H), 8.03 (d, J = 9.0 Hz, 1H), 7.61 (d, J = 8.3 Hz, 2H), 7.45–7.37 (m, 2H), 7.32 (d, J = 8.2 Hz, 2H), 7.26 (s, 1H), 5.55 (s, 2H), 3.91 (s, 3H), 2.42 (s, 3H). 13C NMR (CDCl3) δ: 161.3, 159.1, 146.8, 144.4, 142.9, 139.6, 134.7, 131.5, 130.6, 130.5, 123.9, 123.8, 122.9, 121.7, 120.9, 120.8, 100.0, 97.7, 62.6, 56.0, 21.4. HRMS m/z for C21H17F3N4O2 [M+H]+ Calcd. 415.1376. Found: 415.1377.
3.1.3.13. 1-((1-(4-Methoxyphenyl)-1H-1,2,3-triazol-4-yl)methyl)-2-(trifluoromethyl)quinolin-4(1H)-one (11a)
Yellow microcrystals, m.p. 195–197 °C, yield 68% (0.41 g). IR: ν max/cm−1 3000, 2865, 1664, 1598, 1519, 1269, 1150, 830, 762, 740; 1H NMR (CDCl3) δ: 8.13 (d, J = 8.6 Hz, 1H), 8.02 (s, 1H), 7.84 (d, J = 8.2 Hz, 1H), 7.69 (t, J = 7.9 Hz, 1H), 7.56 (d, J = 9.0 Hz, 2H), 7.32 (t, J = 7.7 Hz, 1H), 7.12 (s, 1H), 6.96 (d, J = 9.0 Hz, 2H), 5.66 (s, 2H), 3.83 (s, 3H). 13C NMR (CDCl3) δ: 160.8, 160.2, 140.0, 138.1, 137.9, 132.4, 130.2, 126.0, 126.0, 123.5, 122.4, 121.5, 120.9, 120.8, 116.3, 115.6, 114.9, 114.9, 55.8, 38.9. HRMS m/z for C20H15F3N4O2 [M+H]+ Calcd. 401.1220. Found: 401.1196.
3.1.3.14. 1-((1-(2-Methoxyphenyl)-1H-1,2,3-triazol-4-yl)methyl)-2-(trifluoromethyl)quinolin-4(1H)-one (11b)
White microcrystals, m.p. 184–186 °C, yield 79% (0.48 g). IR: ν max/cm−1 3138, 2980, 1662, 1598, 1483, 1235, 841, 770, 740; 1H NMR (CDCl3) δ: 8.23–8.16 (m, 2H), 7.84 (d, J = 8.2 Hz, 1H), 7.69 (dd, J = 17.1, 8.6 Hz, 2H), 7.38 (dd, J = 11.1, 4.7 Hz, 1H), 7.32 (t, J = 7.7 Hz, 1H), 7.11 (s, 1H), 7.07–6.98 (m, 2H), 5.68 (s, 2H), 3.84 (s, 3H). 13C NMR (CDCl3) δ: 170.8, 160.6, 151.1, 142.4, 139.9, 137.8, 137.5, 132.1, 130.3, 126.0, 125.7, 125.4, 123.3, 121.4, 120.8, 116.4, 115.4, 112.3, 56.0, 38.7. HRMS m/z for C20H15F3N4O2 [M+H]+ Calcd. 401.1220. Found: 401.1214.
3.1.3.15. 1-((1-(2-Methoxyphenyl)-1H-1,2,3-triazol-4-yl)methyl)-6-methyl-2-(trifluoromethyl)quinolin-4(1H)-one (11c)
Yellow microcrystals, m.p. 216–218 °C, yield 60% (0.37 g). IR: ν max/cm−1 3100, 2981, 1664, 1600, 1599, 1512, 1441, 1325, 1254, 830, 748; 1H NMR (CDCl3) δ: 8.19 (s, 1H), 8.08–8.03 (m, 1H), 7.66 (d, J = 7.7 Hz, 1H), 7.60 (s, 1H), 7.51 (d, J = 8.6 Hz, 1H), 7.37 (dd, J = 12.5, 4.9 Hz, 1H), 7.08 (s, 1H), 7.06–6.97 (m, 2H), 5.66 (s, 2H), 3.83 (s, 3H), 2.42 (s, 3H). 13C NMR (CDCl3) δ: 160.6, 151.3, 138.2, 133.5, 133.1, 133.1, 131.1, 130.9, 130.4, 128.0, 127.9, 126.3, 125.6, 121.3, 120.9, 116.3, 115.6, 112.4, 56.2, 38.8, 21.1. HRMS m/z for C21H17F3N4O2 [M+H]+ Calcd. 415.1330. Found: 415.1331.
3.1.3.16. 4-(2-(1-(4-Methoxyphenyl)-1H-1,2,3-triazol-4-yl)ethoxy)-2-(trifluoromethyl)-1,4-dihydroquinoline (12a)
Yellow microcrystals, m.p. 158–160 °C, yield 76% (0.46 g). IR: ν max/cm−1 3100, 2980, 1575, 1516, 1364, 1250, 838, 767, 725; 1H NMR (CDCl3) δ: 8.20 (d, J = 8.2 Hz, 1H), 8.10 (d, J = 8.2 Hz, 1H), 7.82 (s, 1H), 7.74 (t, J = 7.1 Hz, 1H), 7.60–7.52 (m, 3H), 7.05 (s, 1H), 6.96 (d, J = 8.3 Hz, 2H), 4.60 (t, J = 5.5 Hz, 2H), 3.82 (s, 3H), 3.46 (t, J = 5.5 Hz, 2H). 13C NMR (CDCl3) δ: 162.9, 160.1, 149.7, 149.4, 149.2, 148.4, 144.4, 131.2, 130.7, 130.0, 127.8, 122.4, 121.9, 121.9, 120.6, 120.4, 115.0, 97.1, 68.1, 55.8, 26.0. HRMS m/z for C21H17F3N4O2 [M+H]+ Calcd. 415.1376. Found: 415.1362.
3.1.3.17. 6-Fluoro-4-(2-(1-(4-fluorophenyl)-1H-1,2,3-triazol-4-yl)ethoxy)-2-(trifluoromethyl)-1,4-dihydroquinoline (12b)
Yellow microcrystals, m.p. 163–165 °C, yield 66% (0.40 g). IR: ν max/cm−1 3100, 2980, 1598, 1514, 1477, 1411, 1226, 841, 801, 742; 1H NMR (CDCl3) δ: 8.11 (dd, J = 9.1, 5.2 Hz, 1H), 7.87 (s, 1H), 7.76 (dd, J = 9.1, 2.3 Hz, 1H), 7.68–7.65 (m, 2H), 7.51 (t, J = 8.6 Hz, 1H), 7.2–7.17 (m, 2H),7.07 (s, 1H), 4.63 (t, J = 6.4 Hz, 2H), 3.47 (t, J = 6.4 Hz, 2H). 13C NMR (CDCl3) δ: 163.6, 162.4, 161.7, 160.4, 145.4, 144.6, 133.5, 132.6, 132.6, 122.8, 121.5, 121.3, 120.4, 117.1, 116.9, 106.0, 105.8, 97.5, 68.1, 26.0. HRMS m/z for C20H13F5N4O [M+H]+ Calcd. 421.1082. Found: 421.1066.
3.1.3.18. 6-Fluoro-4-(2-(1-(4-methoxyphenyl)-1H-1,2,3-triazol-4-yl)ethoxy)-2-(trifluoromethyl)-1,4-dihydroquinoline (12c)
Yellow microcrystals, m.p. 158–160 °C, yield 78% (0.47 g). IR: ν max/cm−1 3100, 2980, 1596, 1517, 1479, 1373, 1273, 837, 743; 1H NMR (CDCl3) δ: 8.18–8.00 (m, 1H), 7.82 (s, 1H), 7.77 (d, J = 7.9 Hz, 1H), 7.64–7.43 (m, 3H), 7.07 (s, 1H), 6.97 (d, J = 8.0 Hz, 2H), 4.61 (t, J = 5.5 Hz, 2H), 3.83 (s, 3H), 3.46 (t, J = 5.5 Hz, 2H). 13C NMR (CDCl3) δ: 162.5, 162.4, 160.4, 160.0, 145.3, 144.1, 132.6, 132.5, 130.6, 122.7, 122.4, 121.4, 121.2, 120.5, 115.0, 106.0, 105.8, 97.5, 68.2, 55.8, 26.0. HRMS m/z for C21H16F4N4O2 [M+H]+ Calcd. 433.1282. Found: 433.1263.
3.1.3.19. 6-Fluoro-4-(2-(1-(2-methoxyphenyl)-1H-1,2,3-triazol-4-yl)ethoxy)-2-(trifluoromethyl)-1,4-dihydroquinoline (12d)
Yellow microcrystals, m.p 155–157 °C, yield 62% (0.38 g). IR: ν max/cm−1 3100, 2970, 1600, 1510, 1375, 1235, 840, 766; 1H NMR (CDCl3) δ: 8.12 (dd, J = 9.2, 5.2 Hz, 1H), 8.06 (s, 1H), 7.80–7.75 (m, 2H), 7.51 (td, J = 9.2, 2.7 Hz, 1H), 7.39 (t, J = 7.9 Hz, 1H), 7.12–6.97 (m, 3H), 4.63 (t, J = 6.4 Hz, 2H), 3.83 (s, 3H), 3.48 (t, J = 6.4 Hz, 2H). 13C NMR (CDCl3) δ: 162.6, 160.4, 151.2, 148.6, 145.3, 143.0, 132.6, 132.5, 131.0, 130.3, 125.6, 124.3, 121.5, 121.4, 121.2, 112.4, 105.9, 97.5, 68.3, 56.1, 26.0. HRMS m/z for C21H16F4N4O2 [M+H]+ Calcd. 433.1282. Found: 433.1262.
3.1.3.20. 4-(2-(1-(4-Methoxyphenyl)-1H-1,2,3-triazol-4-yl)ethoxy)-6-methyl-2-(trifluoromethyl)-1,4-dihydroquinoline (12e)
Yellow microcrystals, m.p. 152–154 °C, yield 84% (0.51 g). IR: ν max/cm−1 3100, 2980, 1577, 1516, 1416, 1366, 1249, 829, 737, 715; 1H NMR (CDCl3) δ: 8.01 (d, J = 8.6 Hz, 1H), 7.96 (s, 1H), 7.81 (s, 1H), 7.69–7.50 (m, 3H), 7.04 (s, 1H), 6.98 (d, J = 9.0 Hz, 2H), 4.60 (t, J = 6.6 Hz, 2H), 3.84 (s, 3H), 3.48 (t, J = 6.6 Hz, 2H), 2.54 (s, 3H). 13C NMR (CDCl3) δ: 162.4, 160.1, 148.5, 148.2, 146.9, 144.4, 138.0, 133.4, 130.7, 129.7, 122.9, 122.4, 121.8, 121.8, 120.7, 120.4, 115.0, 97.1, 67.9, 55.8, 26.0, 22.1. HRMS m/z for C22H19F3N4O2 [M+H]+ Calcd. 429.1533. Found: 429.1526.
3.2. Biological studies
Details of biological studies are mentioned in the supplementary materials.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank the Egyptian Cultural and Educational Bureau Scholarship. The authors also thank the Center for Undergraduate Research and Scholarship (CURS), Translational Research Program (TRP) at Augusta University and the Egyptian Academy of Scientific Research and Technology (ASRT) within the program under contract number 7303 for financial support.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioorg.2021.105117.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Lu H., Stratton C.W., Tang Y.-W. Outbreak of pneumonia of unknown etiology in Wuhan, China: the mystery and the miracle. J. Med. Virol. 2020;92(4):401–402. doi: 10.1002/jmv.25678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K.S.M., Lau E.H.Y., Wong J.Y., Xing X., Xiang N., Wu Y., Li C., Chen Q.i., Li D., Liu T., Zhao J., Liu M., Tu W., Chen C., Jin L., Yang R., Wang Q.i., Zhou S., Wang R., Liu H., Luo Y., Liu Y., Shao G.e., Li H., Tao Z., Yang Y., Deng Z., Liu B., Ma Z., Zhang Y., Shi G., Lam T.T.Y., Wu J.T., Gao G.F., Cowling B.J., Yang B.o., Leung G.M., Feng Z. Early transmission dynamics in Wuhan, China, of novel coronavirus-Infected pneumonia. N. Engl. J. Med. 2020;382(13):1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Xia J., Yu T., Zhang X., Zhang L.i. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World Health Organization, Clinical management of severe acute respiratory infection when novel coronavirus (nCoV) infection is suspected: interim guidance, 2020. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance-publications.
- 5.Singh T.U., Parida S., Lingaraju M.C., Kesavan M., Kumar D., Singh R.K. Drug repurposing approach to fight COVID-19. Pharmacol Rep. 2020;72(6):1479–1508. doi: 10.1007/s43440-020-00155-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.S. Dotolo, A. Marabotti, A. Facchiano, R. Tagliaferri, A review on drug repurposing applicable to COVID-19, briefings in bioinformatics, 2020, bbaa288. 10.1093/bib/bbaa288. [DOI] [PMC free article] [PubMed]
- 7.Xiu S., Dick A., Ju H., Mirzaie S., Abdi F., Cocklin S., Zhan P., Liu X. Inhibitors of SARS-CoV-2 entry: Current and future opportunities. J. Med. Chem. 2020;63(21):12256–12274. doi: 10.1021/acs.jmedchem.0c00502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yao X., Ye F., Zhang M., Cui C., Huang B., Niu P., Liu X.u., Zhao L.i., Dong E., Song C., Zhan S., Lu R., Li H., Tan W., Liu D. In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Clin. Infect. Dis. ciaa237. 2020;71(15):732–739. doi: 10.1093/cid/ciaa237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen C.-Y., Wang F.-L., Lin C.-C. Chronic hydroxychloroquine use associated with qt prolongation and refractory ventricular arrhythmia. Clin. Toxicol. 2006;44(2):173–175. doi: 10.1080/15563650500514558. [DOI] [PubMed] [Google Scholar]
- 10.Stas P., Faes D., Noyens P. Conduction disorder and QT prolongation secondary to long-term treatment with chloroquine. Int. J. Cardiol. 2008;127:e80–e82. doi: 10.1016/j.ijcard.2007.04.055. [DOI] [PubMed] [Google Scholar]
- 11.Yaylali S.A., Sadigov F., Erbil H., Ekinci A., Akcakaya A.A. Chloroquine and hydroxychloroquine retinopathy-related risk factors in a turkish cohort. Int. Ophthalmol. 2013;33(6):627–634. doi: 10.1007/s10792-013-9748-0. [DOI] [PubMed] [Google Scholar]
- 12.Musharrafieh R., Kitamura N., Hu Y., Wang J. Development of broad-spectrum enterovirus antivirals based on quinoline scaffold. Bioorg. Chem. 2020;101:103981. doi: 10.1016/j.bioorg.2020.103981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ali A., Sepay N., Afzal M., Sepay N., Alarifi A., Shahid M., Ahmad M. Molecular designing, crystal structure determination and in silico screening of copper(II) complexes bearing 8-hydroxyquinoline derivatives as anti-COVID-19. Bioorg. Chem. 2021;110:104772. doi: 10.1016/j.bioorg.2021.104772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alshammari M.B., Ramadan M., Aly A.A., El-Sheref E.M., Bakht M.A., Ibrahim M.A.A., Shawky A.M. Synthesis of potentially new schiffbases of N -substituted-2-quinolonylacetohydrazides as anti-COVID-19 agents. J. Mol. Struct. 2021;1230:129649. doi: 10.1016/j.molstruc.2020.129649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aly A.A., Hassan A.A., Mohamed A.H., Osman E.M., Bräse S., Nieger M., Ibrahim M.A.A., Mostafa S.M. Synthesis of 3,3′-methylenebis(4-hydroxyquinolin-2(1H)-ones) of prospective anti-COVID-19 drugs. Mol. Div. 2021;25(1):461–471. doi: 10.1007/s11030-020-10140-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar D., Chauhan G., Kalra S., Kumar B., Gill M.S. A perspective on potential target proteins of COVID-19: Comparison with SARS-CoV for designing new small molecules. Bioorg. Chem. 2020;104 doi: 10.1016/j.bioorg.2020.104326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loschwitz J., Jaeckering A., Keutmann M., Olagunju M., Eberle R.J., Coronado M.A., Olubiyi O.O., Strodel B. Novel inhibitors of the main protease enzyme of SARS-CoV-2 identified via molecular dynamics simulation-guided in vitro assay. Bioorg. Chem. 2020;111 doi: 10.1016/j.bioorg.2021.104862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo S., Xie H., Lei Y., Liu B., Zhang L., Xu Y., Zuo Z. Discovery of novel inhibitors against main protease (Mpro) of SARS-CoV-2 via virtual screening and biochemical evaluation. Bioorg. Chem. 2020;110 doi: 10.1016/j.bioorg.2021.104767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun L.-Y., Chen C., Su J., Li J.-Q., Gao H., Chigan J.-Z., Zhai H.-H.L., Yang K.-W. Ebsulfur and Ebselen as highly potent scaffolds for the development of potential SARS-CoV-2 antivirals. Bioorg. Chem. 2020;112:104889. doi: 10.1016/j.bioorg.2021.104889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dominguez-Villa F.X., Duran-Iturbide N.A., Avila-Zarraga J.G. Synthesis, molecular docking, and in silico ADME/Tox profiling studies of new 1-aryl-5-(3-azidopropyl)indol-4-ones: Potential inhibitors of SARS CoV-2 main protease. Bioorg. Chem. 2020;106 doi: 10.1016/j.bioorg.2020.104497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Panda Siva S, Girgis Adel S, Honkanadavar Hitesh H, George Riham F, Srour Aladdin M. Synthesis of new ibuprofen hybrid conjugates as potential anti-inflammatory and analgesic agents. Future Med. Chem. 2020;12(15):1369–1386. doi: 10.4155/fmc-2020-0109. [DOI] [PubMed] [Google Scholar]
- 22.Panda Siva S., Girgis Adel S., Thomas Sean J., Capito Jason E., George Riham F., Salman Asmaa, El-Manawaty May A., Samir Ahmed. Synthesis, pharmacological profile and 2D-QSAR studies of curcumin-amino acid conjugates as potential drug candidates. Eur. J. Med. Chem. 2020;196:112293. doi: 10.1016/j.ejmech.2020.112293. [DOI] [PubMed] [Google Scholar]
- 23.Seliem Israa A., Panda Siva S., Girgis Adel S., Nagy Yosra I., George Riham F., Fayad Walid, Fawzy Nehmedo G., Ibrahim Tarek S., Al‐Mahmoudy Amany M.M., Sakhuja Rajeev, Abdel‐samii Zakaria K.M. Design, synthesis, antimicrobial, and DNA gyrase inhibitory properties of fluoroquinolone-dichloroacetic acid hybrids. Chem. Biol. Drug Des. 2020;95(2):248–259. doi: 10.1111/cbdd.13638. [DOI] [PubMed] [Google Scholar]
- 24.Panda S.S., Girgis A.S., Mishra B.B., Elagawany M., Devarapalli V., Littlefield W.F., Samir A., Fawzy N.G., Srour A.M. Novel pyrazinoic acid-isoniazid conjugates with amino acid linker: Microwave assisted synthesis, anti-infective properties, and molecular modeling studies. RSC Adv. 2019;9(35):20450–20462. doi: 10.1039/c9ra03380g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Faidallah Hassan M., Girgis Adel S., Tiwari Anand D., Honkanadavar Hitesh H., Thomas Sean J., Samir Ahmed, Kalmouch Atef, Alamry Khalid A., Khan Khalid A., Ibrahim Tarek S., AL-Mahmoudy Amany M.M., Asiri Abdullah M., Panda Siva S. Synthesis, antibacterial properties and 2D-QSAR studies of quinolone-triazole conjugates. Eur. J. Med. Chem. 2018;143:1524–1534. doi: 10.1016/j.ejmech.2017.10.042. [DOI] [PubMed] [Google Scholar]
- 26.Faidallah Hassan M., Panda Siva S., Serrano Juan C., Girgis Adel S., Khan Khalid A., Alamry Khalid A., Therathanakorn Tanya, Meyers Marvin J., Sverdrup Francis M., Eickhoff Christopher S., Getchell Stephen G., Katritzky Alan R. Synthesis, antimalarial properties and 2D-QSAR studies of novel triazole-quinine conjugates. Bioorg. Med. Chem. 2016;24(16):3527–3539. doi: 10.1016/j.bmc.2016.05.060. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Bo. Comprehensive review on the anti-bacterial activity of 1,2,3-triazole hybrids. Eur. J. Med. Chem. 2019;168:357–372. doi: 10.1016/j.ejmech.2019.02.055. [DOI] [PubMed] [Google Scholar]
- 28.Kim Dae-Kee, Kim Joonseop, Park Hyun-Ju. Synthesis and biological evaluation of novel 2-pyridinyl[1,2,3]triazoles as inhibitors of transforming growth factor β1 type 1 receptor. Bioorg. Med. Chem. Lett. 2004;14(10):2401–2405. doi: 10.1016/j.bmcl.2004.03.024. [DOI] [PubMed] [Google Scholar]
- 29.Whiting Matthew, Tripp Jonathan C., Lin Ying-Chuan, Lindstrom William, Olson Arthur J., Elder John H., Sharpless K. Barry, Fokin Valery V. Rapid discovery and structure-activity profiling of novel inhibitors of human immunodeficiency virus type 1 protease enabled by the copper(I)-catalyzed synthesis of 1,2,3-triazoles and their further functionalization. J. Med. Chem. 2006;49(26):7697–7710. doi: 10.1021/jm060754+. [DOI] [PubMed] [Google Scholar]
- 30.Cheng Z.-Y., Li W.-J., He F., Zhou J.-M., Zhu X.-F. Synthesis and biological evaluation of 4-aryl-5-cyano-2H-1,2,3-triazoles as inhibitor of HER2 tyrosine kinase. Bioorg. Med. Chem. 2007;15(3):1533–1538. doi: 10.1016/j.bmc.2006.09.041. [DOI] [PubMed] [Google Scholar]
- 31.da Silva Fernando de C., de Souza Maria Cecilia B.V., Frugulhetti Izabel I.P., Castro Helena C., Souza Silmara L. de O., de Souza Thiago Moreno L., Rodrigues Diego Q., Souza Alessandra M.T., Abreu Paula A., Passamani Fabiana. Synthesis, HIV-RT inhibitory activity and SAR of 1-benzyl-1H-1,2,3-triazole derivatives of carbohydrates. Eur. J. Med. Chem. 2009;44(1):373–383. doi: 10.1016/j.ejmech.2008.02.047. [DOI] [PubMed] [Google Scholar]
- 32.Meanwell N.A. Fluorine and fluorinated motifs in the design and application of bioisosteres for drug design. J. Med. Chem. 2018;61(14):5822–5880. doi: 10.1021/acs.jmedchem.7b01788. [DOI] [PubMed] [Google Scholar]
- 33.Panda Siva S., Jain Subhash C. New trifluoromethyl quinolone derivatives: Synthesis and investigation of antimicrobial properties. Bioorg. Med. Chem. Lett. 2013;23(11):3225–3229. doi: 10.1016/j.bmcl.2013.03.120. [DOI] [PubMed] [Google Scholar]
- 34.Maracic S., Lapic J., Djakovic S., Opacak-Bernardi T., Glavas-Obrovac L., Vrcek V., Raic-Malic S. Quinoline and ferrocene conjugates: Synthesis, computational study and biological evaluations. Appl. Organometal Chem. 2019;33(1) [Google Scholar]
- 35.Feoktistova Maria, Geserick Peter, Leverkus Martin. Crystal violet assay for determining viability of cultured cells. Cold Spring Harb Protoc. 2016;2016(4) doi: 10.1101/pdb.prot087379. [DOI] [PubMed] [Google Scholar]
- 36.Mostafa A., Kandeil A., Elshaier Y.A.M.M., Kutkat O., Moatasim Y., Rashad A.A., Shehata M., Gomaa M.R., Mahrous N., Mahmoud S.H., GabAllah M., Abbas H., El Taweel A., Kayed A.E., Kamel M.N., El Sayes M., Mahmoud D.B., El-Shesheny R., Kayali G., Ali M.A. FDA-approved drugs with potent in vitro antiviral activity against severe acute respiratory syndrome coronavirus 2. Pharmaceuticals. 2020;13(12):443. doi: 10.3390/ph13120443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alnajjar Radwan, Mostafa Ahmed, Kandeil Ahmed, Al-Karmalawy Ahmed A. Molecular docking, molecular dynamics, and in vitro studies reveal the potential of angiotensin II receptor blockers to inhibit the COVID-19 main protease. Heliyon. 2020;6(12):e05641. doi: 10.1016/j.heliyon.2020.e05641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.http://www.rcsb.org/structure/6LU7.
- 39.Jin Zhenming, Du Xiaoyu, Xu Yechun, Deng Yongqiang, Liu Meiqin, Zhao Yao, Zhang Bing, Li Xiaofeng, Zhang Leike, Peng Chao, Duan Yinkai, Yu Jing, Wang Lin, Yang Kailin, Liu Fengjiang, Jiang Rendi, Yang Xinglou, You Tian, Liu Xiaoce, Yang Xiuna, Bai Fang, Liu Hong, Liu Xiang, Guddat Luke W., Xu Wenqing, Xiao Gengfu, Qin Chengfeng, Shi Zhengli, Jiang Hualiang, Rao Zihe, Yang Haitao. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582(7811):289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.