Abstract
Interferon gamma (IFNγ) plays a context-dependent dual tumor-suppressor and pro-tumorigenic roles in cancer. IFNγ induces morphological changes in breast cancer (BC) cells with or without estrogen receptor alpha (ERα) expression. However, IFNγ-regulated genes in BC cells remain unexplored. Here, we performed a cDNA microarray analysis of MCF-7 (ERα+) and MDA-MB-231 (HER2-/PR-/ERα-) cells with and without IFNγ treatment. We identified specific IFNγ−modulated genes in each cell type, and a small group of genes regulated by IFNγ common in both cell types. IFNγ treatment for an extended time mainly repressed gene expression shared by both cell types. Nonetheless, some of these IFNγ-repressed genes were seemingly deregulated in human mammary tumor samples, along with decreased IFNGR1 (an IFNγ receptor) expression. Thus, IFNγ signaling-elicited anti-tumor activities may be mediated by the downregulation of main IFNγ target genes in BC; however, it may be deregulated by the tumor microenvironment in a tumor stage-dependent manner.
Keywords: IFNγ, Interferon-stimulated genes, Breast cancer, Estrogen receptor
Highlights
-
•
Identification of new potential genes regulated by IFNγ in breast cancer cells.
-
•
A small group of common genes is regulated by IFNγ in ERα- and ERα+ breast cancer cells.
-
•
IFNγ treatment for a long time mainly represses gene expression in breast cancer cells.
-
•
The tumor environment may lead to a decrease in IFNGR1 expression in mammary tumors.
1. Introduction
Interferon gamma (IFNγ) is the unique member of the type II interferon family [1,2]. The canonical signaling of IFNγ requires a specific heterotetrameric receptor complex and the JAK-STAT1 system. The receptor that recognizes IFNγ is a tetramer complex composed of two IFNGR1 and two IFNGR2 subunits. In this complex, the Janus activated kinases JAK1 and JAK2 are constitutively associated with IFNGR1 and IFNGR2. IFNγ is recognized by the ΙFNGRs, which undergoes a conformational change, activating and transphosphorylating JAK proteins [3]. STAT1 is phosphorylated by JAK1/2, forming a homodimer called gamma-activated factor, which is enriched in the nucleus and binds specific DNA sequences (TTNCNNNAA) called gamma-activated sites or GAS on the regulatory regions of target genes; thus, modulating IFNγ target gene transcription in a cell type-dependent manner [1]. One of the early IFNγ−induced genes is interferon regulatory factor 1 (IRF1), a transcription factor that induces other IFNγ target genes. In addition to the canonical IFNγ signaling, non-canonical pathways for IFNGRs and some genes are upregulated by IFNγ in a STAT1-independent manner [[4], [5], [6]]. Furthermore, the IFNγ signaling can activate NF-κB to regulate gene expression [[7], [8], [9]].
Breast cancer (BC) is a severe health problem worldwide. More than 70% of BC cases are estrogen receptor-α positive (ERα+), whereas “triple-negative BC” is highly metastatic and it does not express ERα, progesterone receptor (PR), and epidermal growth receptor (HER2) [10,11]. In this context, an IFN-dependent gene signature has been suggested as a marker for chemotherapy sensitivity in BC. However, some IFN-target genes have also been associated with chemotherapy resistance [12,13]. Particularly, IFNγ can induce apoptosis and cell cycle arrest. Furthermore, IFNγ autocrine signaling has been detected in BC cells [14,15]. Nevertheless, a limited number of studies have focused on molecular mechanisms underlying IFNγ regulation of BC biology. Some IFNγ-target genes are induced in specific cell types after short stimulation periods (0.5–2 h). However, the gene expression profile induced by IFNγ after extended periods of stimulation remains unexplored. In this study, we identified new IFNγ−regulated genes in both MCF-7 (ERα+) and MDA-MB-231 (triple-negative) BC cells using cDNA microarray analysis. We performed in silico analyses to understand the functional effects of the expression of genes modulated by IFNγ as part of molecular pathways shared by both BC cell types and the possible implications in mammary tumor samples.
2. Materials and methods
2.1. Reagents and cell lines
Recombinant human IFNγ (285-IF-00) was purchased from R&D Systems. The culture medium used for cell maintenance was obtained from Invitrogen. Cell staining reagents, calcein, and TRIzol were obtained from Invitrogen (Thermo Fisher Scientific). The MCF-7 (ERα+) and MDA-MB-231 (ERα–, triple-negative) BC cell lines were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% fetal bovine serum (FBS) and penicillin/streptomycin.
2.2. Calcein staining
A calcein-AM (green) assay was performed. Since viable cells possess an active metabolism, they present intracellular esterase enzymes capable of converting non-fluorescent calcein-AM to green fluorescent calcein. Hence, MCF-7 and MDA-MB-231 cells were treated with and without IFNγ for 24 h. Subsequently, cells were incubated with 1 μM calcein-AM at 37 °C for 30 min. After incubation, the morphology of calcein-stained cells was analyzed using fluorescence microscopy. The cell size was quantified using FIJI/ImageJ [16].
2.3. RNA extraction and cDNA microarray
TRIzol was used to isolate total RNA from MCF-7 and MDA-MB-231 BC cells treated with and without IFNγ (100 ng/mL) for 24 h. RNA concentration and purity were determined using a NanoDrop, ND-1000. RNA integrity was evaluated using agarose gel electrophoresis. Total RNA (2 μg) was used for cDNA synthesis for the microarray.
The cDNA from MCF-7 control cells was marked with Alexa 555, and IFNγ-treated MCF-7 cells were marked with Alexa 647, mixed, and hybridized at GeneChip Human Mapping 10 K Array (CHIP H10KA_07_20). MDA-MB-231 cells with and without IFNγ were marked similarly to MCF-7 cells and hybridized with another GeneChip Human Mapping 10 K Array (CHIP H10KA_07_21). The results obtained from the quantification of cDNA microarray images were analyzed using the GenArise software. Microarray service from the Instituto de Fisiología Celular (IFC), Unidad de Microarreglos de DNA was used, and downregulated genes with a Z-score > 2 were selected to study expression changes. cDNA microarray results were analyzed and represented using heatmaps generated with MATLAB.
2.4. In silico analysis
We used the Gene Ontology resource to define the functions of IFNγ-modulated genes (http://geneontology.org/) [17,18]. We also used Genemania to analyze the functions and predict the pathways associated with the identified IFNγ-modulated genes (https://genemania.org/) [19].
Functional interactions between the products from identified IFNγ-modulated genes were analyzed using STRING 10.5 (https://string-db.org/cgi/network.pl) [20]. Additionally, we analyzed functional nodes for the products of IFNγ target genes using Chemical-Protein Interaction Networks “STITCH” (http://stitch.embl.de/) and integrated the information on interactions from metabolic pathways [21].
To identify the putative sites for transcription factors activated by IFNγ in the promoter regions of genes, we used Interferome v2.0 (http://www.interferome.org/interferome/home.jspx) [22].
2.5. Gene expression analysis in mammary tumor samples
We used Curtis databases from the cancer microarray database OncomineTM (www.oncomine.org) to analyze the gene expression in patient-derived mammary tumors and normal mammary tissues. The analysis of IFNGR1 in mammary tumor samples was performed using UALCAN using the Cancer Genome Atlas (TCGA) data set (http://ualcan.path.uab.edu/) [23].
2.6. Statistical analysis
Unpaired Student's t-tests were performed using GraphPad Prism 5 software (GraphPad Software, Inc.); p < 0.05 (*), p < 0.01 (**), and p < 0.001 (***) were considered statistically significant vs. control condition. ### means p < 0.001 between indicated groups.
3. Results
3.1. IFNγ induces morphological changes in MCF-7 and MDA-MB-231 cells, modulating a small group of genes
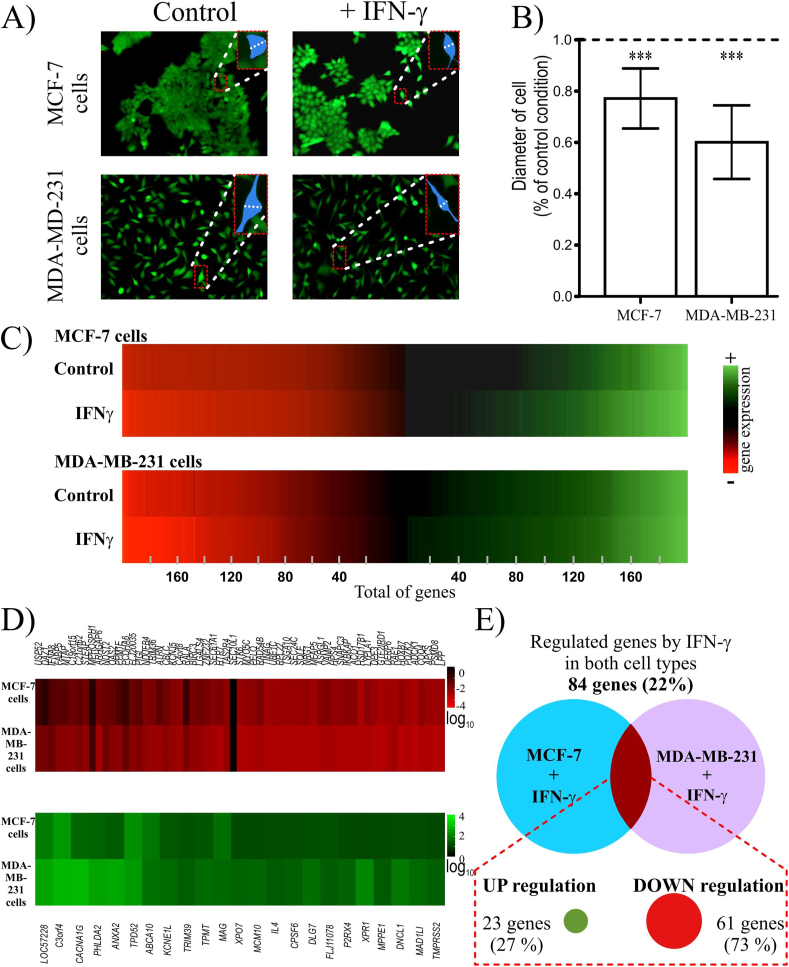
Morphological changes of MCF-7 and MDA-MB-231 cells were observed after 24 h of IFNγ treatment without an effect on their viability (Fig. 1A). We detected that in response to IFNγ, the diameter of MDA-MB-231 cells reduced compared to control cells (Fig. 1A and B). Moreover, control MCF-7 cells showed an epithelial morphology; however, upon IFNγ treatment, the cellular diameter was similarly reduced (Fig. 1A and B). Therefore, we set to perform a cDNA microarray and analyze common molecular pathways triggered by IFNγ treatment for 24 h, which may drive the morphological changes of both cell types. As a result, we identified 374 and 393 IFNγ−modulated genes in MCF-7 and MDA-MB-231 cells, respectively. In both cases, approximately 50% of genes were upregulated and 50% downregulated (Fig. 1C).
Fig. 1.
IFNγ induces morphological changes in breast cancer cells and modulates their transcriptome.
A) The morphology of MDA-MB-231 and MCF-7 cells changed after IFNγ treatment for 24 h. Cells were stained with calcein. Magnified view of a representative cell marked in blue, indicating the quantified diameter. B) Cell diameters were quantified to demonstrate the morphological changes in breast cancer cells in response to IFNγ. C) Heat maps of IFNγ-regulated genes in both breast cancer cells. D) Heat maps of the commonly regulated genes in both MCF-7 and MDA-MB-231 cells by IFNγ. IFNγ down- and upregulated genes are displayed in red and green color, respectively. E) Venn diagram showing the percentage of genes regulated in both breast cancer cell lines.
Although our analysis indicated that approximately 78% of the identified IFNγ-regulated genes differed and were specific for each BC cell line (MCF-7 vs. MDA-MB-231 cells), a small group of 84 genes (22% of the total genes) were regulated in both cell types after IFNγ treatment (Fig. 1D and E). Notably, 73% of the IFNγ−regulated genes in both cell lines were downregulated (61 genes), whereas 27% of them (23 genes) were upregulated (Fig. 1D and E). In this study, we exclusively analyzed genes regulated in both cell types to understand the IFNγ-mediated molecular mechanisms shared by both cell lines independently of the ERα/PR/HER2 status.
We analyzed the promoter regions (−1500/+500 bp) of these 84 genes regulated by IFNγ using Interferome, detecting one or more putative sites, for the transcription factors, namely STAT1, STAT3, IRF1, IRF7, IRF8, and NF-κB, and predicting their possible regulation by canonical and non-canonical IFNγ signaling pathways.
Gene ontology analysis indicated that the IFNγ-upregulated genes are related to the modulation of subcellular localization and cellular processes, whereas IFNγ-downregulated genes are involved in catalytic activities, biological regulation, cellular and metabolic processes. Using the Genemania program, we found that some upregulated genes were involved in cell cycle checkpoints, cell cycle, and mitotic pathways, whereas the IFNγ-downregulated genes were related to molecular transport, immune system, proteolysis, and RNA metabolism.
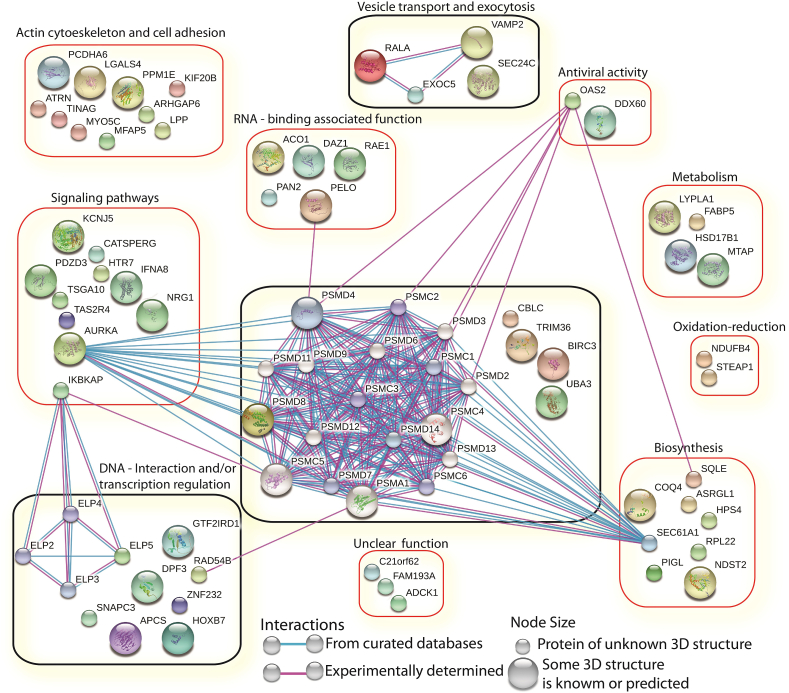
3.2. IFNγ downregulates genes associated mainly with transcription elongation, proteolysis, and vesicular transport in BC cells
Our analysis via STITCH displayed three groups of genes with interaction nodes (Fig. 2). One group of genes was related to proteolysis (CBLC, TRIM36, BIRC3, UBA3, and PSMD8). PSMD8 was associated with AURKA, PELO, OAS2, RAD54B, SEC61A1, and PSMD proteasome subunits. Another group comprised IKBKAP and the transcription elongation factor ELP5, which was predicted to be related to other elongation factors such as ELP2, 3, and 4. The last group contained genes such as RALA, VAMP2, EXOC5, and SEC24C, associated with vesicle and membrane transport and exocytosis. On the STITCH interactome, we grouped genes according to their reported activities. We defined eight groups of genes associated with 1) actin cytoskeleton and cell adhesion, 2) signaling pathways, 3) DNA-interaction and/or transcription regulation, 4) vesicle transport and exocytosis, 5) RNA-binding associated functions, 6) antiviral activity, 7) metabolism, 8) oxide reduction and biosynthesis, and 9) function not completely known (Fig. 2). These data suggest that IFNγ signaling can specifically inhibit the expression of target genes in MCF-7 and MDA-MB-231 BC cell lines, independently of the ERα/PR/HER2 status.
Fig. 2.
The genes downregulated by IFNγ in MCF-7 and MDA-MB-231 cells are related to different molecular pathways. The analysis of genes downregulated by IFNγ was performed using STITCH.
3.3. Decreased IFNGR1 gene expression in BC
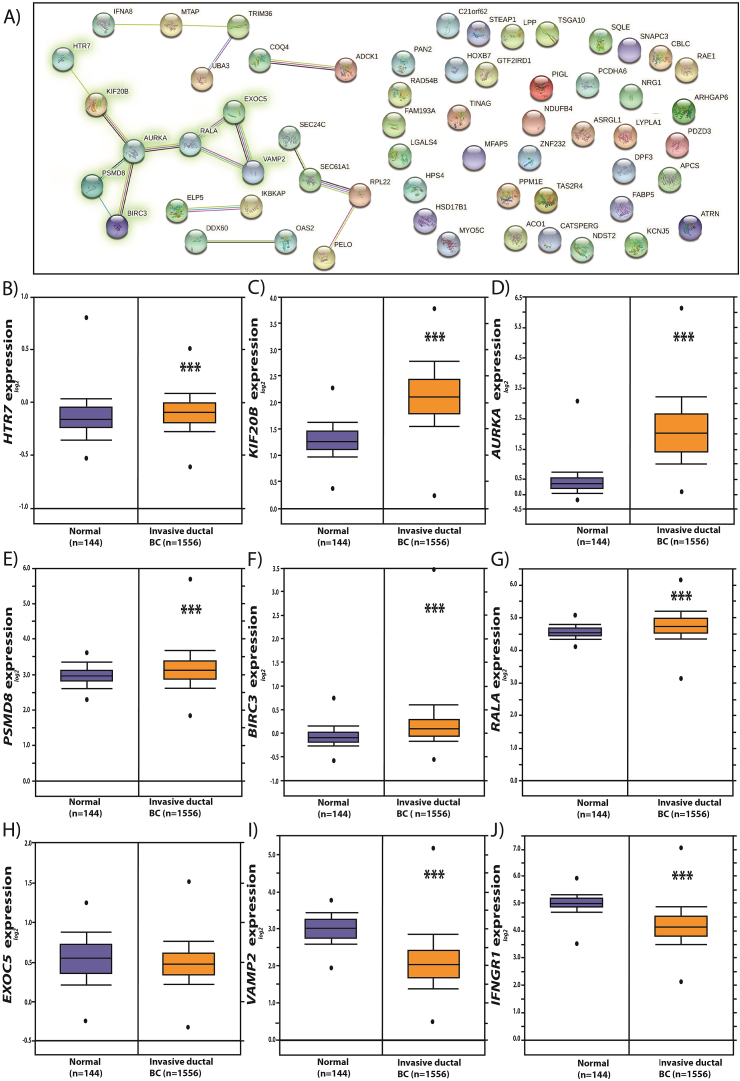
To explore the relevance of the expression of IFNγ-downregulated genes in mammary tumors, we performed an analysis using the Curtis data set from Oncomine™. Of the 22 IFNγ−downregulated genes, we selected a group of eight genes (HTR7, KIF20B, AURKA, PSMD8, BIRC3, RALA, EXOC5, and VAMP2), which constituted the major predicted node of interaction or experimentally tested according to our analysis from STRING (Fig. 3A). Our results revealed that the expression of these genes was significantly higher in mammary tumors than in normal mammary tissue, except for EXOC5, which did not significantly change, and VAMP2, which was decreased in mammary tumors (Fig. 3B–I). Since the IFNGR1 gene encodes one of the receptor subunits that recognizes IFNγ and transduces the signal into cells, we evaluated its expression in mammary tumors. We detected that IFNGR1 expression was lower in mammary tumors than in normal mammary tissues (Fig. 3J).
Fig. 3.
The expression of some genes downregulated by IFNγ in MCF-7 and MDA-MB-231 cells is increased in mammary tumor samples. A) IFNγ-downregulated genes analyzed using the STRING software. B) The expression of HTR7, KIF20B, PSMD8, AURKA, BIRC3, RALA, EXOC5, VAMP2, and IFNGR1 was evaluated using the Curtis dataset from Oncomine (C–K).
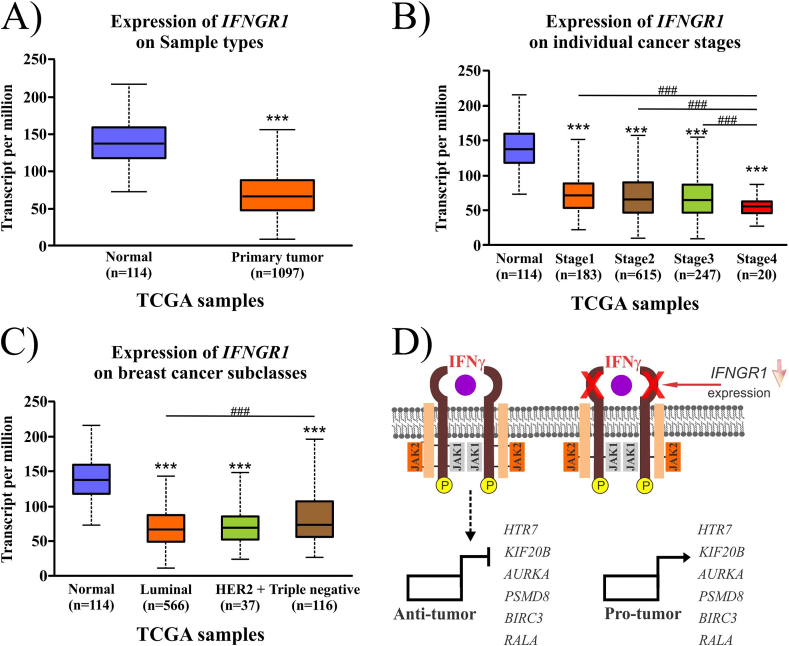
As IFNGR1 gene expression is central for modulating IFNγ-dependent gene expression. We used the TCGA dataset from UALCAN to analyze the IFNGR1 expression profile in BC. First, we observed that IFNGR1 expression was lower in mammary tumors than in normal mammary tissue, corroborating the previous result from the Curtis dataset (Fig. 4A). Second, we detected that IFNGR1 expression was significantly decreased, mainly in stage 4 compared to stages 1–3 of mammary tumors (Fig. 4B). Third, IFNGR1 downregulation was shared in different BC types (MCF-7 and MDA-MB-231). Moreover, a significant decrease in IFNGR1 gene expression was observed in luminal BC tumors compared to triple-negative tumors (Fig. 4C). Thus, IFNγ/IFNGR1 signaling pathways are seemingly affected in BC tumors, partly explaining why some identified IFNγ-downregulated genes can be de-repressed by IFNGR1 downregulation in mammary tumors while other genes co-regulated by collateral signaling pathways may not be directly affected.
Fig. 4.
The expression of IFNGR1 is altered in mammary tumors from patients. The analysis of IFNGR1 expression in breast cancer with respect to normal mammary tissues (A), tumor stage (B), and breast cancer type (C) was evaluated using UALCAN. The proposed model based on our analysis is presented in D.
4. Discussion
IFNγ stimulation for 24 h led to morphological changes in MCF-7 and MD-MB-231 BC cells. Hence, we considered that: 1) IFNγ may modulate the gene expression patterns after prolonged treatment as compared to those usually reported (short exposure: 1–2 h). 2) Some of these IFNγ−regulated genes in both cell types may partly explain the observed morphological changes.
Our study identified new upregulated and downregulated genes by IFNγ in BC cells. Only a small percentage of all genes (22%, 84 genes) were regulated in both cell types (ERα+ and triple-negative). Many of these genes were downregulated (73%, 61 genes). This is a novel result because IFNγ-dependent gene inhibition has been limitedly studied compared to genes modulated by IFNγ/STAT1 canonical signaling in cancer cells. Moreover, these results suggest that IFNγ signaling may promote morphological changes through a common pathway that mainly involves gene repression in both BC cell types in an ERα/PR/HER2 status-independent manner.
Gene induction (MAD1L1, MCM10, and DYNLL1) by IFNγ may be associated with cytoskeletal organization and plays a central role in controlling the cell cycle. For instance, DYNLL1 can regulate checkpoint activation [24], and MAD1L1 is a checkpoint gene whose mutations can affect its functions in several cancer types, including BC [25]. Our data also indicated that IFNγ might negatively regulate the expression of genes involved in transport, and vesicle pathways, which are central in BC pathophysiology [26]. Furthermore, IFNγ probably requires the modulation of several subsets of genes to promote morphological changes in BC cells.
We selected IFNγ−repressed genes with functional interactions in the major nodes of the STRING analysis, such as HTR7, KIF20B, PSMD8, AURKA, BIRC3, RALA, EXOC5, and VAMP2. Interestingly, KIF20B, RALA, PSMD8, and AURKA upregulation is reportedly associated with BC progression. For example, KIF20B overexpression in BC correlates with poor prognoses [27]. Similarly, RALA upregulation is a marker of poor prognosis in BC patients [28]. Notably, our analysis suggested that proteasome-related pathways may be affected by IFNγ signaling, implying that: 1) some proteasome inhibitors could be tested as pharmacological tools for different cancer types, including BC [29,30]; 2) proteasome subunit (as PSMD8) deregulation can result in cancer progression, and resistance to proteasome inhibitors [31,32]; 3) IFNγ increases ISGylation, a protein modification associated with protein stability by competing with the ubiquitination pathway in BC [[33], [34], [35]]; and 4) the IFNγ repression of proteolysis-associated genes may be helpful in the development of novel strategies to treat this pathology.
On the other hand, changes in cell morphology are reportedly implicated in migration and invasion and the inhibition of growth and reduction of invasiveness [36,37]. Although the effects of IFNγ-induced morphological changes in BC cells remain unclear, our results suggested that IFNγ may trigger secondary molecular pathways modulating gene expression (mainly by repression) after prolonged treatment, inhibiting the pro-tumorigenic characteristics of BC cells.
Nevertheless, the mammary tumors showed a decrease in IFNGR1 expression. These tumors developed into a microenvironment enriched by secreted factors from the same cancer cells, stroma, and immune cells. As T lymphocytes and natural killer cells secrete IFNγ, constant signaling of this interferon may negatively affect IFNGR1 expression in BC cells. Furthermore, the expression of some IFNγ-downregulated genes identified in this study were higher in mammary tumor samples than in normal mammary tissue. These results indicated that the IFNγ/IFNGR1 signaling pathways are affected in mammary tumors from BC patients, leading to the de-repression of specific target genes such as HTR7, KIF20B, PSMD8, AURKA, BIRC3, and RALA. For instance, AURKA, a kinase-encoding gene, is associated with BC progression, and its inhibitors are being investigated as promising therapies for this disease [38]. The downregulation of AURKA by IFNγ suggests that the anti-tumor effects of this interferon may be mediated by the transcriptional inhibition of genes critical in BC. However, in advanced mammary tumor stage 4, IFNGR1 expression was reduced. Consequently, IFNγ signaling may decrease, affecting the regulation of its target genes and compromising its anti-tumor activity (Fig. 4D).
Nevertheless, the mechanisms underlying the regulation of IFNγ-mediated repression need to be further elucidated. Moreover, in this study, we focused on common genes regulated by IFNγ in BC cells; however, it is essential to consider that approximately 78% of the total identified genes are specific for each BC cell type, ERα+ and triple-negative. It is noteworthy that the overall gene modulation by IFNγ may contribute to the final cellular behavior in BC.
In summary, we identified new IFNγ−modulated genes in triple-negative and ERα+ BC cells, MDA-MB-231 and MCF-7, respectively. IFNγ modulated the transcriptome of both BC cell types, but only a small group of genes was commonly regulated. IFNγ-dependent repression appears to be a central mechanism that modulates proteolysis, vesicle trafficking pathways, and transcriptional regulation, which may decrease BC cell tumorigenicity. Some of these putative IFNγ−downregulated genes are important in BC, such as AURKA. The tumor environment may lead to a decrease in IFNGR1 expression in mammary tumors, stimulating the de-repression of some IFNγ-inhibited genes and affecting BC progression.
Author contributions
A.C. T.-C. designed the research, and participated in the analysis, organization, and writing of the manuscript. M. M.-S. participated in the research and the improvement of this article. J.O. R.-J. and B. M.-A. participated in the analysis and figure preparation.
Declaration of competing interest
The authors declare that they have no conflict of interest.
Acknowledgments
We thank Dr. Jorge Ramírez and members of the Unidad de Microarreglos de ADN (Instituto de Fisiología Celular, Universidad Nacional Autónoma de México). We also thank Dr. Claudia Rivera Cerecedo for her valuable comments. We thank the Universidad Autónoma de la Ciudad de México (UACM) for the support provided.
References
- 1.Alspach E., Lussier D.M., Schreiber R.D. Interferon gamma and its important roles in promoting and inhibiting spontaneous and therapeutic cancer immunity. Cold Spring Harb Perspect Biol. 2019;11 doi: 10.1101/cshperspect.a028480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zaidi M.R. The interferon-gamma paradox in cancer. J. Interferon Cytokine Res. 2019;39:30–38. doi: 10.1089/jir.2018.0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jorgovanovic D., Song M., Wang L., Zhang Y. Roles of IFN-gamma in tumor progression and regression: a review. Biomark Res. 2020;8:49. doi: 10.1186/s40364-020-00228-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gil M.P., Bohn E., O'Guin A.K., Ramana C.V., Levine B., Stark G.R., Virgin H.W., Schreiber R.D. Biologic consequences of Stat1-independent IFN signaling. Proc. Natl. Acad. Sci. U. S. A. 2001;98:6680–6685. doi: 10.1073/pnas.111163898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramana C.V., Gil M.P., Schreiber R.D., Stark G.R. Stat1-dependent and -independent pathways in IFN-gamma-dependent signaling. Trends Immunol. 2002;23:96–101. doi: 10.1016/s1471-4906(01)02118-4. [DOI] [PubMed] [Google Scholar]
- 6.Roy S.K., Wachira S.J., Weihua X., Hu J., Kalvakolanu D.V. CCAAT/enhancer-binding protein-beta regulates interferon-induced transcription through a novel element. J. Biol. Chem. 2000;275:12626–12632. doi: 10.1074/jbc.275.17.12626. [DOI] [PubMed] [Google Scholar]
- 7.Lee W.H., Chung M.H., Tsai Y.H., Chang J.L., Huang H.M. Interferon-gamma suppresses activin A/NF-E2 induction of erythroid gene expression through the NF-kappaB/c-Jun pathway. Am. J. Physiol. Cell Physiol. 2014;306:C407–C414. doi: 10.1152/ajpcell.00312.2013. [DOI] [PubMed] [Google Scholar]
- 8.Tang M., Tian L., Luo G., Yu X. Interferon-gamma-mediated osteoimmunology. Front. Immunol. 2018;9:1508. doi: 10.3389/fimmu.2018.01508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang S., Yu M., Sun L., Xiao W., Yang X., Sun L., Zhang C., Ma Y., Yang H., Liu Y., Lu D., Teitelbaum D.H., Yang H. Interferon-gamma-induced intestinal epithelial barrier dysfunction by NF-kappaB/HIF-1alpha pathway. J. Interferon Cytokine Res. 2014;34:195–203. doi: 10.1089/jir.2013.0044. [DOI] [PubMed] [Google Scholar]
- 10.Jitariu A.A., Cimpean A.M., Ribatti D., Raica M. Triple negative breast cancer: the kiss of death. Oncotarget. 2017;8:46652–46662. doi: 10.18632/oncotarget.16938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tecalco-Cruz A.C., Ramirez-Jarquin J.O., Cruz-Ramos E. Estrogen receptor alpha and its ubiquitination in breast cancer cells. Curr. Drug Targets. 2019;20:690–704. doi: 10.2174/1389450119666181015114041. [DOI] [PubMed] [Google Scholar]
- 12.Legrier M.E., Bieche I., Gaston J., Beurdeley A., Yvonnet V., Deas O., Thuleau A., Chateau-Joubert S., Servely J.L., Vacher S., Lassalle M., Depil S., Tucker G.C., Fontaine J.J., Poupon M.F., Roman-Roman S., Judde J.G., Decaudin D., Cairo S., Marangoni E. Activation of IFN/STAT1 signalling predicts response to chemotherapy in oestrogen receptor-negative breast cancer. Br. J. Canc. 2016;114:177–187. doi: 10.1038/bjc.2015.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ning Y., Riggins R.B., Mulla J.E., Chung H., Zwart A., Clarke R. IFNgamma restores breast cancer sensitivity to fulvestrant by regulating STAT1, IFN regulatory factor 1, NF-kappaB, BCL2 family members, and signaling to caspase-dependent apoptosis. Mol. Canc. Therapeut. 2010;9:1274–1285. doi: 10.1158/1535-7163.MCT-09-1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gooch J.L., Herrera R.E., Yee D. The role of p21 in interferon gamma-mediated growth inhibition of human breast cancer cells. Cell Growth Differ. 2000;11:335–342. [PubMed] [Google Scholar]
- 15.Niu X.L., Wang Y., Yao Z., Duan H., Li Z., Liu W., Zhang H., Deng W.M. Autocrine interferon-gamma may affect malignant behavior and sensitivity to tamoxifen of MCF-7 via estrogen receptor beta subtype. Oncol. Rep. 2015;34:3120–3130. doi: 10.3892/or.2015.4294. [DOI] [PubMed] [Google Scholar]
- 16.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.Y., White D.J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Harris M.A., Hill D.P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J.C., Richardson J.E., Ringwald M., Rubin G.M., Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gene C. Ontology, the Gene Ontology resource: enriching a GOld mine. Nucleic Acids Res. 2021;49:D325–D334. doi: 10.1093/nar/gkaa1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Warde-Farley D., Donaldson S.L., Comes O., Zuberi K., Badrawi R., Chao P., Franz M., Grouios C., Kazi F., Lopes C.T., Maitland A., Mostafavi S., Montojo J., Shao Q., Wright G., Bader G.D., Morris Q. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–W220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szklarczyk D., Morris J.H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N.T., Roth A., Bork P., Jensen L.J., von Mering C. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuhn M., von Mering C., Campillos M., Jensen L.J., Bork P. STITCH: interaction networks of chemicals and proteins. Nucleic Acids Res. 2008;36:D684–D688. doi: 10.1093/nar/gkm795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rusinova I., Forster S., Yu S., Kannan A., Masse M., Cumming H., Chapman R., Hertzog P.J. Interferome v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res. 2013;41:D1040–D1046. doi: 10.1093/nar/gks1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chandrashekar D.S., Bashel B., Balasubramanya S.A.H., Creighton C.J., Ponce-Rodriguez I., Chakravarthi B., Varambally S. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.West K.L., Kelliher J.L., Xu Z., An L., Reed M.R., Eoff R.L., Wang J., Huen M.S.Y., Leung J.W.C. LC8/DYNLL1 is a 53BP1 effector and regulates checkpoint activation. Nucleic Acids Res. 2019;47:6236–6249. doi: 10.1093/nar/gkz263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsukasaki K., Miller C.W., Greenspun E., Eshaghian S., Kawabata H., Fujimoto T., Tomonaga M., Sawyers C., Said J.W., Koeffler H.P. Mutations in the mitotic check point gene, MAD1L1, in human cancers. Oncogene. 2001;20:3301–3305. doi: 10.1038/sj.onc.1204421. [DOI] [PubMed] [Google Scholar]
- 26.Barbosa A.M., Martel F. Targeting glucose transporters for breast cancer therapy: the effect of natural and synthetic compounds. Cancers. 2020;12 doi: 10.3390/cancers12010154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li T.F., Zeng H.J., Shan Z., Ye R.Y., Cheang T.Y., Zhang Y.J., Lu S.H., Zhang Q., Shao N., Lin Y. Overexpression of kinesin superfamily members as prognostic biomarkers of breast cancer. Canc. Cell Int. 2020;20:123. doi: 10.1186/s12935-020-01191-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghoroghi S., Mary B., Larnicol A., Asokan N., Klein A., Osmani N., Busnelli I., Delalande F., Paul N., Halary S., Gros F., Fouillen L., Haeberle A.M., Royer C., Spiegelhalter C., Andre-Gregoire G., Mittelheisser V., Detappe A., Murphy K., Timpson P., Carapito R., Blot-Chabaud M., Gavard J., Carapito C., Vitale N., Lefebvre O., Goetz J.G., Hyenne V. Ral GTPases promote breast cancer metastasis by controlling biogenesis and organ targeting of exosomes. Elife. 2021;10 doi: 10.7554/eLife.61539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frankland-Searby S., Bhaumik S.R. The 26S proteasome complex: an attractive target for cancer therapy. Biochim. Biophys. Acta. 2012;1825:64–76. doi: 10.1016/j.bbcan.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soave C.L., Guerin T., Liu J., Dou Q.P. Targeting the ubiquitin-proteasome system for cancer treatment: discovering novel inhibitors from nature and drug repurposing. Canc. Metastasis Rev. 2017;36:717–736. doi: 10.1007/s10555-017-9705-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deng S., Zhou H., Xiong R., Lu Y., Yan D., Xing T., Dong L., Tang E., Yang H. Over-expression of genes and proteins of ubiquitin specific peptidases (USPs) and proteasome subunits (PSs) in breast cancer tissue observed by the methods of RFDD-PCR and proteomics. Breast Canc. Res. Treat. 2007;104:21–30. doi: 10.1007/s10549-006-9393-7. [DOI] [PubMed] [Google Scholar]
- 32.Tsvetkov P., Sokol E., Jin D., Brune Z., Thiru P., Ghandi M., Garraway L.A., Gupta P.B., Santagata S., Whitesell L., Lindquist S. Suppression of 19S proteasome subunits marks emergence of an altered cell state in diverse cancers. Proc. Natl. Acad. Sci. U. S. A. 2017;114:382–387. doi: 10.1073/pnas.1619067114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Desai S.D., Haas A.L., Wood L.M., Tsai Y.C., Pestka S., Rubin E.H., Saleem A., Nur E.K.A., Liu L.F. Elevated expression of ISG15 in tumor cells interferes with the ubiquitin/26S proteasome pathway. Canc. Res. 2006;66:921–928. doi: 10.1158/0008-5472.CAN-05-1123. [DOI] [PubMed] [Google Scholar]
- 34.Tecalco-Cruz A.C., Cruz-Ramos E. Protein ISGylation and free ISG15 levels are increased by interferon gamma in breast cancer cells. Biochem. Biophys. Res. Commun. 2018;499:973–978. doi: 10.1016/j.bbrc.2018.04.030. [DOI] [PubMed] [Google Scholar]
- 35.Tecalco-Cruz A.C., Ramirez-Jarquin J.O., Cruz-Ramos E. Regulation and action of interferon-stimulated gene 15 in breast cancer cells. Hum. Cell. 2020;33:954–962. doi: 10.1007/s13577-020-00414-x. [DOI] [PubMed] [Google Scholar]
- 36.Brandhagen B.N., Tieszen C.R., Ulmer T.M., Tracy M.S., Goyeneche A.A., Telleria C.M. Cytostasis and morphological changes induced by mifepristone in human metastatic cancer cells involve cytoskeletal filamentous actin reorganization and impairment of cell adhesion dynamics. BMC Canc. 2013;13:35. doi: 10.1186/1471-2407-13-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tarhan Y.E., Kato T., Jang M., Haga Y., Ueda K., Nakamura Y., Park J.H. Morphological changes, cadherin switching, and growth suppression in pancreatic cancer by GALNT6 knockdown. Neoplasia. 2016;18:265–272. doi: 10.1016/j.neo.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Du R., Huang C., Liu K., Li X., Dong Z. Targeting AURKA in Cancer: molecular mechanisms and opportunities for Cancer therapy. Mol. Canc. 2021;20:15. doi: 10.1186/s12943-020-01305-3. [DOI] [PMC free article] [PubMed] [Google Scholar]