Abstract
Over the past decade, critical, non-redundant roles of the ten-eleven translocation (TET) family of dioxygenase enzymes have been identified in the brain during developmental and postnatal stages. Specifically, TET-mediated active demethylation, involving the iterative oxidation of 5-methylcytosine to 5-hydroxymethylcytosine and subsequent oxidative derivatives, is dynamically regulated in response to environmental stimuli such as neuronal activity, learning and memory processes, and stressor exposure. Such changes may therefore perpetuate stable and dynamic transcriptional patterns within neuronal populations required for neuroplasticity and behavioural adaptation. In this review, we will highlight recent evidence supporting a role of TET protein function and active demethylation in stress-induced neuroepigenetic and behavioural adaptations. We further explore potential mechanisms by which TET proteins may mediate both the basal and pathological embedding of stressful life experiences within the brain of relevance to stress-related psychiatric disorders.
Keywords: Ten-eleven translocation proteins, 5-Hydroxymethylcytosine, Active demethylation, Stress, Epigenetics, DNA methylation
Abbreviations
- 5mC
5-methylcytosine
- 5hmC
5-hydroxymethylcytosine
- 5fC
5-formylmethylcytosine
- 5caC
5-carboxymethylcytosine
- CpG
cytosine-guanine dinucleotide
- CXXC
CXXC zinc finger domain
- mCH
non-CpG 5-methylcyotsine
- mCG
CpG 5-methylcytosine
- TET
Ten-eleven translocation
- TET1FL
TET1 full length isoform
- TET1s
TET1 short isoform
- TET3FL
TET3 full length isoform
- TET3s
TET3 short isoform
- TSS
Transcription start site
1. Introduction
The current aetiology of psychiatric disorders posits that the complex interaction of genetic predispositions and environmental factors confers disease risk. Stress is one of the major environmental risk factors for many psychiatric disorders including major depressive disorder, and post-traumatic stress disorder, which are both associated with maladaptive regulation of central stress response (Lu et al., 2008; Kessler et al., 1997; Kendler et al., 1999; Teicher et al., 2002; Mehta et al., 2013; Yehuda, 2009; Dean and Keshavan, 2017; Loman and Gunnar, 2010). This response is mediated via the autonomic nervous system and activation of the neuroendocrine stress system via the hypothalamic pituitary adrenal (HPA) axis. Activation of the HPA axis originates with corticotropin releasing hormone/factor (CRH/CRF) secretion from the parvocellular hypothalamic neurons of the paraventricular nucleus of the hypothalamus (PVN). CRF stimulates adrenocorticotropic hormone release from the anterior pituitary into the blood stream ultimately inducing the secretion of glucocorticoids from the adrenal cortex (Deussing and Chen, 2018). Within both the periphery and central nervous system, glucocorticoids act via the glucocorticoid receptor (GR) and mineralocorticoid receptor (MR), which provide negative feedback throughout the HPA axis and act as adaptive transcriptional regulators in response to stress exposure (De Kloet et al., 1998). The coordinated activation and regulation of the central stress response is required to enable adaptive behavioural responses in the face of stressful life experiences and are often disrupted in stress-related psychiatric disorders. Consequently, it is critical that we understand the molecular mechanisms by which stress mediates basal and pathological adaptations of the central stress response.
Clinical and pre-clinical evidence suggests that stressful life-events dynamically alter epigenetic mechanisms in both the periphery and central nervous system. Epigenetic mechanisms mediate the complex regulation of chromosomal regions without alteration of the DNA sequence itself and include DNA modifications, post-translational histone modifications, and non-coding RNA (e.g., microRNA), as well as a recently appreciated role of RNA modifications (Li et al., 2020a; Liu et al., 2020), which interact in a highly coordinated manner to establish cell type-specific epigenetic states and transcriptional profiles. Although known to be dynamically regulated throughout development, it is only in the past 10–15 years that epigenetics has become considered as a dynamic process within the brain throughout life. This has led to the term neuroepigenetics, which refers primarily to dynamic epigenetic processes within post-mitotic neurons of the brain. Neuroepigenetic mechanisms may therefore act as key mediators of the biological embedding of stressful life experiences at the molecular level (i.e., encoding of information related to stress exposure) within discrete neuronal populations in specific brain circuits, which may propagate aberrant brain function and behaviour associated with stress-related psychiatric disorders (Aristizabal et al., 2020). As a conduit for such change, DNA modifications are of particular interest due to their relative stability, especially in post-mitotic neurons, and the recent discovery of active demethylation pathways within the brain mediated via the Ten-eleven translocation (TET) family of dioxygenase enzymes. In this review, we will therefore focus on the role of TET proteins and dynamic DNA methylation processes in brain function and adaptation to stress.
The mammalian DNA methylome (i.e., genome-wide covalent DNA modifications) consists primarily of 5-methylcytosine (5mC) and its oxidative derivatives. 5mC is established by the de novo DNA methyltransferases DNMT3A and DNMT3B, whereas DNMT1 maintains methylation throughout genome replication in dividing cells (Jeltsch, 2006), with critical catalytic functions also in post-mitotic neurons (Hahn et al., 2020; Meadows et al., 2015) (Fig. 1A). Active DNA demethylation is catalysed by the evolutionary conserved TET proteins including TET1, TET2 and TET3, which mediate iterative oxidation of 5mC to 5-hydroxymethylcytosine (5hmC) and subsequently to 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC), which is then excised by the protein thymine DNA glycosylase (TDG) and base excision repair machinery (Fig. 1A) (Tahiliani et al., 2009; He et al., 2011; Ito et al., 2011; Shen et al., 2013; Zhang et al., 2012). Other DNA binding proteins such as the growth arrest and DNA damage-inducible protein (GADD45) family members are also implicated in active demethylation and brain function but will not be the focus of this review (Kohli and Zhang, 2013; Ma et al., 2009; Li et al., 2019; Labonte et al., 2019).
Fig. 1.
(A) Active DNA demethylation. Covalent addition of a methyl group to the 5th carbon of cytosine residues forming 5-mehtylcytosine (5mC) is catalysed by DNA methyltransferases (DNMTs), which can be further oxidised to 5-hydroxymethylcytosine (5hmC) by the ten-eleven translocation (TET) family of dioxygenases all of which are expressed within the post-mitotic neurons. 5hmC is then sequentially oxidised by TET enzymes to 5-formylcytosine (5fC) and 5-carboxyl-cytosine (5caC), which is then removed by Thymine DNA Glycosylase (TDG) and base excision repair (BER) machinery ultimately resulting in an unmodified cytosine. The dotted line indicates that TDG + BER-mediated excision of 5caC to C is yet to be empirically demonstrated in post-mitotic neurons. (B) Neuronal DNA methylome. Graphical representation of the major DNA modifications of the neuronal genome where 5-methylcytosine in the CpG (mCG) non-CpG context (mCH, H = A,C,T) accounts for ~50% and ~40–50% of all modified cytosines in neurons of the adult human and rodent brain, respectively. Enrichment of hydroxymethylation is found within the neuronal genome accounting for ~1% of modified cytosines and is primarily found in the CpG context (hmCG) in neurons of the adult human and rodent brain. A, adenine; C, cytosine; G, guanine; T, thymine. Figures reproduced with permission from (Dick and Chen, 2020).
Unlike the almost exclusive presence of 5mC at cytosine-guanine dinucleotides (CpGs) throughout the genome of most vertebrate tissues, 5mC is present in both the CpG (mCG) and CH (mCH) context (H = A, C, or T) in post-mitotic neurons with postnatal accumulation of mCH coinciding with synaptogenesis and synaptic pruning, reaching 40–50% of all modified cytosines in the adult rodent and human brain (Fig. 1B) (Lister et al., 2013; Mo et al., 2015; Lister and Mukamel, 2015; Guo et al., 2014). Notably, deposition of mCH is dependent on DNMT3A within neuronal populations in the mammalian brain (Guo et al., 2014; Stroud et al., 2017). Within both neuronal and non-neuronal cells, both mCG and mCH are anti-correlated with gene expression with mCH being the best predictor of gene expression in neuronal populations (Mo et al., 2015), although intragenic 5mC is positively correlated with transcription in certain contexts (Wu et al., 2010). Another unique feature of the neuronal methylome is the enrichment of 5hmC, which predominates in the CG context, and accounts for ~1% of all modified cytosines in the adult mammalian brain (Fig. 1B) (Lister et al., 2013; Mellen et al., 2017). Recent evidence also indicates a potential role of 5hmC within the human embryonic brain (with notable sex differences) of relevance to neurodevelopmental perturbations, such as prenatal stress (Spiers et al., 2017). Within neurons, 5hmC is generally enriched upstream of the transcription start site (TSS; although depleted at the TSS itself), as well as within gene bodies (where it is positively correlated with gene expression) and distal regulatory elements (e.g., enhancers) indicative of its regulatory function (Lister et al., 2013; Wu and Zhang, 2017). Moreover, both the stability of 5hmC and the binding of 5hmC “readers”, such as MECP2, within the brain indicate it is a functional epigenetic mark with discrete biological functions beyond being a transient modification throughout active demethylation (Mellen et al., 2012). These unique aspects of the neuronal DNA methylome are indicative of the increased molecular encoding capacity of the post-mitotic neurons and may mediate both stable and dynamic regulation of neuronal gene expression patterns required for experience-dependent plasticity and behavioural adaptation. Indeed, a role of dynamic regulation of the neuronal DNA methylome in brain function has been substantiated since the re-discovery of 5hmC (Tahiliani et al., 2009; Wyatt and Cohen, 1953; Penn et al., 1972) and the identification of TET-mediated active DNA demethylation.
1.1. TET proteins and their function within the brain
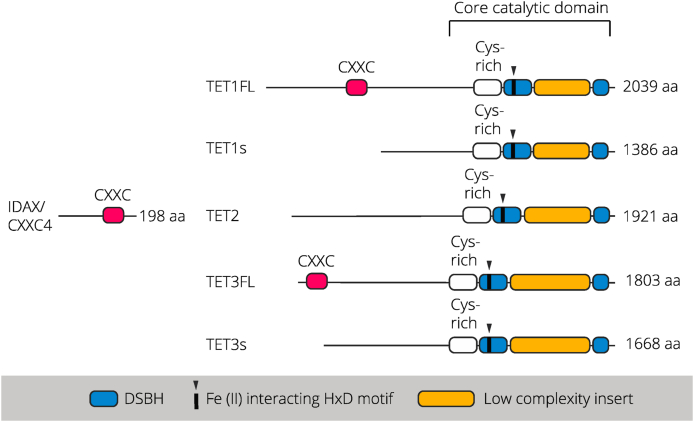
TET proteins catalyse the conversion of 5mC to 5hmC (and 5fC and 5caC) in a α-ketoglutarate (α-KG) and Fe(II)-dependent manner (He et al., 2011; Ito et al., 2010, 2011). This is mediated via a common C-terminal core catalytic domain, which consists of a double-stranded β-helix domain (mediating Fe(II), α-KG and 5mC interactions), a cysteine rich domain (stabilising TET-DNA interactions) and a low complexity insert for which the function remains elusive apart from demonstrated post-translational modification of this domain (Fig. 2) (Wu and Zhang, 2017) (Bauer et al., 2015). Both TET1 and TET3 have multiple cell-type specific isoforms of which the full-length isoform has an N-terminus CXXC domain that facilitates binding to CpG-rich sequences (Jin et al., 2016; Zhang et al., 2016). TET2 is present as a single isoform in mammalian cells with the putative TET2 CXXC domain encoded by the neighbouring gene Cxxc4/Idax, which was originally part of the ancestral Tet2 gene before evolutionary chromosomal rearrangement (Ko et al., 2013). Notably, the catalytic domain of each TET protein is sufficient for nuclear localisation and DNA demethylation indicative of the complex functional roles mediated via TET protein functional domains (Tahiliani et al., 2009; Ito et al., 2010).
Fig. 2.
Domain structure of TET protein isoforms expressed in the postnatal brain
TET protein isoforms share a conserved C-terminal core catalytic domain consisting of a cysteine rich (Cys-rich) domain mediating chromatin targeting, a double-stranded β-helix (DSBH) domain containing the key Fe (II) interacting HxD motif, and a low complexity insert for which the function is yet to be fully determined. TET1 full length (TET1FL) and TET3FL proteins have an N-terminal CXXC domain mediating DNA binding at CpG-rich sequences, which is absent in the truncated TET1 short (TET1s) and TET3 short (TET3s) isoforms abundantly expressed in the mature brain. TET2 lacks a CXXC domain yet interacts with the CXXC domain containing protein IDAX/CXXC4.
1.2. TET protein isoforms of the brain
Within the mouse and human cortex, single cell RNA-seq data (Allen Brain Atlas) indicates an abundance of Tet2 and Tet3 mRNA in neuronal populations with relatively lower levels of Tet1 mRNA present in the adult brain. In mammals, the TET1 full length (TET1FL) isoform containing the CXXC domain is primarily found within primordial germ cells, embryonic stem cells (ESCs) and the early embryo (Zhang et al., 2016). Interestingly, TET1FL also appears to be the predominant isoform within placental tissue (Yosefzon et al., 2017) suggesting likely CXXC-dependent functions in tissues sensitive to prenatal stress. The truncated TET1 short (TET1s) isoform lacking the N-terminus and CXXC domain, is the predominantly expressed TET1 isoform within most somatic tissues as well as within the brain (Zhang et al., 2016; Yosefzon et al., 2017) with Tet1s mRNA enriched within neuronal populations (Greer et al., 2020). Cell-type specific expression of TET1 isoforms is mediated via alternate promoter usage, with the TetFL promoter located upstream of the canonical TSS, and the Tet1s promoter located upstream of the canonical exon 2 (Zhang et al., 2016; Yosefzon et al., 2017; Greer et al., 2020). Within neurons, the Tet1FL promoter is marked by the repressive histone modification H3K27me3, which is absent from the Tet1s promoter resulting in the abundance of Tet1s mRNA within neuronal populations confirmed via quantitative real-time PCR (qRT-PCR) and RNA-seq (Zhang et al., 2016; Greer et al., 2020). Work from our own lab supports these findings as permissive chromatin accessibility (i.e., abundant ATAC-seq peak) is observed primarily at the Tet1s promoter within both excitatory (Neurod6 positive) and inhibitory forebrain neurons (Dlx5/6 positive) of the adult mouse brain (unpublished observations). Significantly, the lack of the CXXC domain confers TET1s with reduced global chromatin binding and reduced catalytic activity compared to TET1FL, although ChIP-seq data indicates similar genome wide binding patterns (i.e., enriched within CpG-rich regions and promoters) yet with decreased enrichment of TET1s in ESCs (Zhang et al., 2016). Functionally, TET1 isoforms appear to mediate distinct functions throughout development as well as in pathological states, with dysregulation of TET1s specifically associated with poor prognosis in several types of cancer (Zhang et al., 2016; Yosefzon et al., 2017; Good et al., 2017). Within the brain, a recent study also emphasizes the unique functions of TET1 isoforms (Greer et al., 2020). Neuronal activity and synaptic scaling induce regulation specifically of Tet1s mRNA with no changes in Tet1FL in primary hippocampal (HPC) neurons. Moreover, Transcription activator-like effector (TALE)-mediated knock-down (KD) of either Tet1s or Tet1FL results in disparate transcriptome changes and opposing modulation of glutamatergic transmission in HPC neurons in vitro (Greer et al., 2020). These changes likely mediate the divergent impairments in contextual fear memory observed following either Tet1FL or Tet1s KD within the dorsal HPC (dHPC) in this study. Thus, TET1 isoforms indeed appear to have unique functions within neuronal and non-neuronal cells, which must be considered in future research, particularly within the brain.
TET3 similarly has multiple cell-type specific isoforms in mammals although their distribution and function remain poorly understood. In oocytes and zygotes, a unique long Tet3 isoform lacking the CXXC domain is expressed (Tet3o) and is the most abundant TET protein in these tissues (Jin et al., 2016). Throughout neuronal differentiation, alternative splicing and alternate promoter usage mediates TET3 full length (TET3FL) and TET3 short (TET3s) expression (Fig. 2) with Tet3FL mRNA being more abundant within NPCs and the embryonic brain (Jin et al., 2016). In contrast, based on isoform-specific qRT-PCR analysis, we consistently observe that Tet3s opposed to Tet3FL mRNA is more abundant within both excitatory (Neurod6 positive) and inhibitory forebrain neurons (Dlx5/6 positive) of the adult mouse brain, which we also observe in nuclear RNA-seq from these neuronal populations (unpublished observations). Abundance of Tet3s mRNA is similarly observed in adult mouse retina (Perera et al., 2015) although the ratio of isoform expression within other cell types is presently not known. Unlike TET1 isoforms, TET3s has increased catalytic activity compared to TET3FL, with the TET3FL demonstrating unique preference for binding to 5caC over unmodified cytosines, 5mC, or 5hmC, which appears to mediate binding in proximity to the TSS of specific target genes in NPCs and embryonic brain (Jin et al., 2016). As for the TET1 isoforms, the TET3 isoform appears to mediate unique functions, likely due to differential chromatin binding and association with differential interacting protein partners (Perera et al., 2015). Thus, considering the abundance of truncated TET protein isoforms within the brain and their unique functions, further research is warranted to discern the contribution of different isoforms to the observed roles of TET family proteins within the brain.
1.3. TET function within the brain
Prior to the discovery of TET family proteins in 2009, the observed dynamic regulation of DNA methylation within the brain (albeit from bulk tissue analysis) repeatedly puzzled researchers as to how environmental stimuli, such as neuronal activity, learning and memory or stress could induce demethylation in post-mitotic cells. Thus, identification of TET-mediated active demethylation opened an exciting avenue of investigation to substantiate active processes underlying dynamic DNA methylation within the brain. To date, clear non-redundant roles of individual TET proteins have been identified in various aspects of brain function including: neuronal plasticity (Meadows et al., 2015; Kaas et al., 2013; Rudenko et al., 2013; Kumar et al., 2015; Yu et al., 2015) retention and extinction of conditioned fear memory (Kaas et al., 2013; Rudenko et al., 2013; Kumar et al., 2015; Li et al., 2014; Gontier et al., 2018), drug-induced plasticity (Feng et al., 2015), developmental (Ji et al., 2020; Hahn et al., 2013) and adult neurogenesis (Zhang et al., 2013; Montalban-Loro et al., 2019; Li et al., 2017; Choi et al., 2019), as well as age-related cognitive decline (Gontier et al., 2018) and neurodegeneration (Marshall et al., 2020; Li et al., 2020b). Clinical studies have also demonstrated a critical role for TET proteins in brain function as mutations and altered expression are associated with growth retardation, as well as cognitive and social deficits (Beck et al., 2020), neurodegenerative (Marshall et al., 2020) and psychiatric disorders (Dong et al., 2012). Moreover, altered 5hmC profiles are observed in multiple sclerosis (Kular et al., 2019; Chomyk et al., 2017), neurodegenerative diseases (Marshall et al., 2020), and psychiatric disorders such as autism spectrum disorder (Zhubi et al., 2014; Cheng et al., 2018a), psychosis (Dong et al., 2012), and major depressive disorder (Gross et al., 2017). Beyond these identified roles of TET function and 5hmC within the brain, recent evidence further implicates the TET proteins and active demethylation processes in modulation stress-induced molecular and behavioural adaptations (Table 1).
Table 1.
Summary of stress-related behavioural phenotypes and molecular adaptations following modulation of TET family proteins within the rodent brain. 5mC 5-methylcytosine, 5hmC 5-hydroxymethylcytosine, AMG Amygdala, BNST Bed nucleus of the stria terminalis, CD Catalytic domain, CDm Catalytic domain mutant, CER Cerebellum, cKO conditional knock-out, CRS Chronic restraint stress, CSDS Chronic social defeat stress, CTX cortex, DaLi Dark light test, DEG Differentially expressed gene, DG Dentate gyrus, dHPC dorsal hippocampus, EPM Elevated plus maze, EZM Elevated zero maze, FST Forced swim test, HPC hippocampus, IEG Immediate early gene, KD Knockdown, KO knockout, LTD Long-term depression, LTP Long-term potentiation, mEPSC miniature excitatory post-synaptic currents, mIPSC miniature inhibitory post-synaptic currents, mESCs mouse embryonic stem cells, MWM Morris water maze, NAc Nucleus accumbens, NOR Novel object recognition, OE overexpression, OF open field, OLM Object location memory, RAWM Radial arm water maze, shRNA short hairpin RNA, SPT Sucrose preference test, TALE Transcription Activator Like Effectors, TST Tail suspension test, vHPC ventral hippocampus.
| Manipulation | Brain region | Molecular effects | Physiological/Behavioural effects | Reference |
|---|---|---|---|---|
| Tet1-CD OE | dHPC | ↓ global 5mC and ↑ global 5hmC; ↑ IEG (Arc, Egr1, Fos) and Bdnf mRNA expression (both TET1-CD and Tet1-CDm OE) ↑ Tdg, Mbd4, Smug1 mRNA expression (only Tet1-CD OE) |
OF: No change Context fear: Short term (1hr) no change; Long term (24hr): ↓ freezing (both TET1-CD and Tet1-CDm OE) |
Kaas et al. (2013) |
| Tet1KO (exon 4 deletion) | Constitutive | ↓ global 5hmC (cortex and HPC) ↓ IEG mRNA expression (Npas4, Fos, Arc, Egr2) ↑ 5 mC at Npas4 locus |
OF, EPM, FST, cued fear (24hr): No change Context fear (1 × 0.8 mA): Long term (24hr) no change; Extinction: ↑ freezing MWM: Impaired reversal learning dHPC CA1 LTP and basal neurophysiology properties: No change dHPC CA1 LTD: Enhanced |
Rudenko et al. (2013) |
| Tet1KO (exon 4 deletion) | Constitutive | 5hmC decrease in DG, HPC, CTX Dysregulation of IEGs and methylation machinery |
OF, EPM, 3-chamber social interaction: No change Context fear (1 × 0.5 mA or 1 × 0.8 mA) ↑ freezing at 24hr, 15d and 30d Cued fear (1 × 0.5 mA or 1 × 0.8 mA) ↑ freezing at 24hr dHPC CA1 LTP and basal neurophysiological properties: No change (also observed upon Tet1-CD and Tet1-CDm OE in dHPC CA1) |
Kumar et al. (2015) |
| Tet1KO (exon 3 deletion) | Constitutive | Decreased body weight and increased lethality at birth; Compensatory ↑ Tet2 mRNA in mESCs but not adult HPC Decreased Oxtr and Npas4 mRNA associated with increased 5 mC around TSS |
OF: Decreased locomotion and centre time No Change DaLi, NOR, CTX and cued fear, MWM Resident intruder: females more aggressive with modest shift in threatening postures in both sexes Maternal care impaired HPC: No change basal neurophysiological properties, LTP or sIPSCs. |
Towers et al. (2018) |
| Tet1KO (exon 13 deletion) | Constitutive | Epigenetic regulation of neurogenic genes | Impaired adult HPC neurogenesis and spatial memory (MWM) | Zhang et al. (2013) |
| Tet1KO (exon 4 deletion) | Constitutive | Altered 5hmC profiles in PFC largely overlapping with stress-induced hypermethylated loci in wild-type PFC | FST: ↓ immobility at baseline and after 1 and 2 weeks CRS TST: No change at baseline; ↓ immobility 1 and 2 weeks CRS |
Cheng et al. (2018b) |
| Tet2KO (exon 3 deletion) | Constitutive | Altered 5hmC profiles in PFC with modest overlap with stress-induced hypomethylated loci in wild-type PFC | FST: ↑ immobility at baseline and after 1 and 2 weeks CRS TST: No change at baseline; ↑ immobility after 2 weeks CRS |
|
| Tet1 OE | Constitutive | ↑ Tet1 mRNA and TET1 protein in PFC, AMG, HPC, CER | FST and rotarod: No change OF: ↓ locomotor activity and centre time EPM: ↓ open arm time Y maze deficit Passive avoidance: ↑ latency at 1d and 7d ↑ HPC adult neurogenesis |
Kwon et al. (2018) |
| Tet1cKO (exon 4 deletion) | NAc | Altered NAc gene expression profiles with ~250 DEGs following CSDS vs WT controls enriched for gene networks mediating immune response. | Social interaction and FST: No change at baseline SPT: ↑ preference at baseline OF: ↑ centre time at baseline EPM: ↑ open arm time at baseline CSDS: ↑ proportion of resilient Tet1cKO after 10d CSDS |
Feng et al. (2017) |
| Tet1FL KD (TALE) | HPC primary neurons | Altered gene expression enriched for immune response gene networks | ↑ mEPSC frequency and amplitude | Greer et al. (2020) |
| Tet1FL KD (TALE) | dHPC CA1 | No analysis conducted | OF and EZM: No change Context fear: ↓ freezing (24h) |
|
| Tet1s KD (TALE) | HPC primary neurons | Altered gene expression enriched for neuronal function gene networks | ↓ mEPSC frequency with no change in amplitude | |
| Tet1s KD (TALE) | dHPC CA1 | No analysis conducted | OF and EZM: No change Context fear: ↑ freezing (24h) |
|
| Tet2 KD (shRNA) | dHPC | No analysis conducted | Impaired HPC neurogenesis RAWM: Impaired short (24h) and long term (5d) memory Contextual fear: ↓ freezing (24h); Cued fear: No change |
Gontier et al. (2018) |
| Tet2cKO (exon 3 deletion) | Adult neural progenitor cells (Tet2cKO-Nestin) | No analysis conducted | Impaired HPC neurogenesis RAWM: Impaired long term (5d) memory Contextual fear: ↓ freezing (24h); Cued fear: No change |
|
| Tet2 OE | dHPC | Altered gene expression and 5hmC profiles associated with pro neurogenesis pathways | Rescues age-related neurogenesis and cognitive impairments | |
| Tet3cKO (exon 7 deletion) | Forebrain excitatory neurons (Tet3cKO-Camk2a) | Reduced Tet3 mRNA in AMG, PFC, HPC; No change in BNST No change in global 5mC Altered dHPC gene expression with 20 DEGs including ↑ Fos and Npas4 mRNA Altered vHPC gene expression with 143 DEGs including ↑ Npas4 and ↓ Crhr2 mRNA |
↑ basal am and pm circulating corticosterone levels TST, FST, NOR: No change OF: ↓ centre time EPM: ↓ open arm head dips and ↓ latency to enter open arms MWM: No change in acquisition and retention; Altered search strategy vHPC dendritic spine maturation altered with no change in density (no change dHPC) |
Antunes et al. (2020) |
1.4. Role of TET proteins and demethylation in the central stress response
Prior to the discovery of TET-mediated active demethylation, clinical and pre-clinical studies had identified stress-induced regulation of DNA methylation at candidate genes associated primarily with HPA axis function (e.g., Nr3c1 encoding GR) following prenatal, early-life and adult stress exposures (Provencal and Binder, 2015; Dick and Chen, 2020; Dirven et al., 2017). GR binding upon stimulation with dexamethasone (DEX, synthetic GR agonist) induced stable DNA demethylation of glucocorticoid response elements (GREs) in vitro (Thomassin et al., 2001; Makkonen et al., 2009), as well as within stress responsive genes such as intronic demethylation of GREs within the Fkbp5 locus (encoding the GR co-chaperone FKBP51) in the mouse hypothalamus (HYP) following chronic corticosterone (CORT) treatment (Lee et al., 2010). Similarly, demethylation of intronic Fkbp5 GREs was observed following DEX stimulation in a human HPC cell line, subsequently demonstrated also within peripheral blood cells as a consequence of child trauma (i.e., early life stress) (Klengel et al., 2013).
Despite the established regulation of DNA methylation following stress, initial studies into TET function within the brain did not directly assess their role in stress-related behaviours and/or adaptations to stress exposure. It was demonstrated that constitutive knock out (KO) of Tet1 (Tet1KO) in mice does not alter anxiety-like behaviours as assessed in the open field (OF) and elevated plus maze (EPM), with no changes in depressive-like or social behaviours assessed in the forced swim test (FST) and 3-chamber social interaction test, respectively (Rudenko et al., 2013; Kumar et al., 2015). Overexpression (OE) of the catalytic domain of TET1 (TET1-CD) or the TET1-CD mutant (TET1-CDm) within the mouse dHPC also had no effects on anxiety-like behaviour in the OF (Kaas et al., 2013) although further characterisation of stress-related behaviours were not conducted in these studies. However, direct assessment of depressive-like behaviours indicates that TET1 and TET2 may differentially modulate susceptibility to chronic stress in adult male mice (Cheng et al., 2018b). Specifically, chronic restraint stress (CRS) induced decreases in global 5hmC and Tet1 mRNA levels as well as altered genome-wide 5hmC profiles specifically within the prefrontal cortex (PFC) following CRS. Interestingly, depressive-like behaviours assessed in the FST and tail suspension test (TST), were decreased in Tet1KO (same strain as mentioned above) and increased in constitutive Tet2KO mice. Moreover, Tet1KO mice had a blunted response to CRS as assessed in the FST and TST in contrast to an apparent increased sensitivity to CRS in Tet2KO mice compared to wild-type control mice, which the authors suggest as stress resiliency and susceptibility phenotypes in Tet1KO and Tet2KO, respectively. However, no further investigation of stress-related physiological parameters (e.g., adrenal hypertrophy), HPA axis function, or other behavioural measures was conducted such that the claim of stress resiliency vs. susceptibility must be considered with caution. Mechanistically, stress-induced differentially hydroxymethylated regions (DhMRs) of the genome were predominantly hypermethylated following CRS (despite decreased global levels) with an enrichment of the transcription factor motif of the hypoxia inducible factor (HIF) family (Cheng et al., 2018b). Co-immunoprecipitation (Co-IP) experiments revealed a direct TET1-HIF1α protein-protein interaction (PPI) within the mouse PFC only following CRS with HIF1α ChIP-seq analysis confirming a small but significant enrichment of HIF1α binding sites within a small subset of DhMRs within the PFC following CRS. Thus, authors suggested that HIF1α may recruit TET1 to a small subset of candidate loci in response to chronic stress within the rodent PFC. Unfortunately, the lack of RNA expression analysis and further characterisation of physiological or behavioural parameters limits the interpretation of this study. However, it should be noted that a direct TET1-HIF1α PPI has also been observed in zebrafish and mice in response to hypoxia (Wang et al., 2017).
Generation of a novel constitutive Tet1KO mouse (deletion of Tet1 exon 3 leading to premature stop codon in exon 6 disrupting catalytic domain) also identified aberrant social behaviour and maternal care in female Tet1KO mice associated with hypermethylation and decreased expression of the Oxtr locus (encoding the oxytocin receptor) and the IEG Npas4 in the HPC with no other stress-related behavioural or cognitive deficits observed (Towers et al., 2018). In contrast, transgenic constitutive OE of the full length Tet1 gene (Tet1OE) in mice induces an anxiogenic phenotype (i.e., increased anxiety) in adult males with no differences in depressive-like behaviours (Kwon et al., 2018). Stress-related learning is also augmented as Tet1OE mice display increased latency to enter a conditioned aversive chamber in the passive avoidance task up to 7 days after conditioning. Increased adult neurogenesis and TET1-mediated increases in Egr1 expression within the HPC of Tet1OE mice were suggested as potential mechanisms for anxiogenic phenotype and aberrant retention of stress-related memory in the passive avoidance task (Kwon et al., 2018). Such studies must be considered with caution due to the developmental confounds of TET modulation and the lack of tissue specificity, particularly considering the roles TET proteins in neurodevelopment and hippocampal neurogenesis. Yet, it appears that decreased or increased levels of TET1 throughout life modulate depressive- and anxiety-like behaviours in opposing directions, with decreased TET2 levels promoting depressive-like behaviours in adult male mice.
Interestingly, the depressive-like phenotype observed in Wilms tumour 1 (Wt1) conditional KO (cKO) mice, is mediated in part via embryonic perturbation of a WT1-TET2 complex that in wild-type mice binds to the promoter of the important neurodevelopmental gene Erythropoietin (Epo), driving TET2-mediated demethylation and increasing Epo mRNA expression for functional embryonic neurogenesis (Ji et al., 2020). Such findings are indicative of a potential mechanism of depressive-like phenotypes observed in Tet2KO mice (Cheng et al., 2018b). Conversely, support for the role of reduced TET1 levels mediating antidepressant-like phenotypes was demonstrated in Tet1cKO mice (Tet1 exon 4 flanked by LoxP sites such that cre-dependent KO results in an unstable truncated protein lacking the catalytic domain), which display anxiolytic and antidepressant-like phenotypes following targeted Tet1 KD within the nucleus accumbens (NAc) in adult male mice (Feng et al., 2017). Viral-mediated KD of Tet1 within the NAc (~80% Tet1 mRNA of controls) increased sucrose preference (a measure of hedonic reward processing) and induced anxiolytic-like effects in the OF and EPM with no difference in social interaction in adult male mice. Utilising chronic social defeat stress (CSDS), the authors further demonstrated that NAc-specific Tet1 KD modestly increased the proportion of stress resilient mice compared to wild-type controls, although overall Tet1 KD mice still displayed social avoidance, an indicator of stress susceptibility in this model (Golden et al., 2011). This finding contradicted the reduced Tet1 mRNA levels observed in the NAc of mice susceptible to CSDS, which is in line with decreased PFC Tet1 mRNA and TET1 protein levels in a genetic rat model of depressive-like behaviour (Wei et al., 2014) emphasizing the necessity for refined cell-type specific molecular analysis and TET modulation within the brain. Moreover, opposed to the baseline characterisation of Tet1 KD mice, only the social interaction test (commonly employed to assess resiliency/susceptibility to CSDS), was employed to analyse the effects of CSDS so no conclusions regarding anxiety-like and depressive-like behaviours or HPA axis function following chronic stress can be drawn from this study (Feng et al., 2017).
Collectively, these studies indicate that both developmental and targeted reduction in TET1 levels within the brain may result in a pro stress resilience phenotype in male mice, whereas developmental increases in TET1 levels appears to be anxiogenic with developmental reduction of TET2 levels and perturbation of TET2 interacting proteins inducing depressive-like behaviours. As to how these behavioural phenotypes manifest following TET modulation remains to be seen due the confounds of developmental transgenic manipulations and the limited molecular characterisation conducted. Moreover, the lack of thorough analysis of both behavioural and physiological parameters before and after stress exposure, restrict the conclusions that can be drawn in relation to TET function in stress adaptation. Despite this, considering the observed phenotypes following modulation of TET1 and TET2, this begs the question; what is the role of TET3 in adaptive behaviour in response to stress?
Unlike the viability of Tet1KO and Tet2KO mice, Tet3 deletion results in neonatal lethality (Guo et al., 2011) and thus genetic manipulations of Tet3 remain relatively scarce in adult animals. However, TET3 appears to be critical for basal and stimulus-induced brain functions with it being most sensitive to homeostatic synaptic plasticity and modulating glutamatergic transmission to a greater extent than TET1 or TET2 in HPC neurons (Yu et al., 2015). A role of TET3 within the mouse medial PFC (mPFC) has also been demonstrated for behavioural adaptation upon the extinction of cued-fear memory (Li et al., 2014). Moreover, TET3 appears to be sensitive to GR stimulation as DEX treatment induces a specific upregulation of Tet3 mRNA in rat neural stem cells (NSCs) in vitro, although prenatal DEX exposure increased the expression of Tet1, Tet2, and Tet3 mRNA within the postnatal pup cortex in this study (Bose et al., 2015). Further evidence for the interaction of TET3 and stress-induced signalling pathways in the brain was demonstrated via epigenetic priming (i.e., latent epigenetic changes modulating gene inducibility) of the Crh locus in the PVN of male chicks in response to early-life heat stress (Cramer et al., 2019). Heat shock stress at postnatal day (PND) 3 induces coordinated regulation of chromatin state and DNA methylation within an intronic repressor element of the Crh locus, with increased Tet3 mRNA and TET activity suggested to mediate active demethylation of this Crh repressor element, decreasing Crh transcription and CRH levels in resilient chicks. However, in a complex interplay of epigenetic factors, a TET3-REST complex binds to this Crh repressor specifically in vulnerable chicks so that it does not undergo active demethylation and therefore results in increased Crh transcription and CRH levels upon a subsequent stress challenge at PND 10. This is a good example of stress-induced epigenetic metaplasticity or epigenetic priming and demonstrates that TET3-mediated active demethylation does indeed function in stress adaptation in the brain. Notably, thorough investigation of DNA methylation and gene expression changes upon DEX stimulation in a human HPC neuronal cell line identified stable changes in DNA methylation that did not alter basal expression of proximal genes but rather primed them to a subsequent DEX challenge indicative of a role of DNA methylation in GR-mediated epigenetic priming in neurons (Provencal et al., 2020). Upregulation of TET1 and UHRF1 mRNA (DNMT1-interacting protein) was also observed in this study upon DEX stimulation although this was not stably maintained following washout of DEX from cell culture medium yet could indicate a transient role of GR-induced TET1 activity in this system. Thus, TET proteins may mediate stress-induced epigenetic priming/metaplasticity of relevance to the molecular embedding of stressful life experiences in neurons.
In the first study employing cell-type specific Tet3 deletion within the brain, Antunes et al., 2020 found that early adult deletion of Tet3 within excitatory forebrain neurons induces an anxiogenic phenotype associated with increased basal CORT levels in adult male mice. Employing inducible calcium/calmodulin-dependent protein kinase 2α (Camk2a)-CreERT2 mice crossed with Tet3cKO mice (Tet3cKO-Camk2a; Tet3 exon 7 encoding catalytic domain flanked by loxP sites), deletion of Tet3 in forebrain excitatory neurons reduced bulk levels (i.e., not cell-type specific) of Tet3 mRNA within the PFC, amygdala, dHPC and ventral (vHPC) with no changes in the bed nucleus of the stria terminalis (BNST) (Antunes et al., 2020). Tet3cKO-Camk2a mice had a moderate anxiogenic phenotype assessed in the OF and EPM with no changes in depressive-like behaviours observed in the FST and TST or cognitive performance in the novel object recognition or Morris water maze (despite altered strategy). Significantly, Tet3cKO-Camk2a mice had increased basal circulating CORT levels at both a.m. and p.m. time points compared to wild-type controls, although stress-induced modulation of HPA axis function was not further investigated. Morphological abnormalities of neurons in vHPC but not dHPC were observed in Tet3CKO−Camk2a mice and associated with modest alterations in bulk gene expression in which several stress-associated candidate genes were dysregulated, such as Crhr2 and IEGs Fos and Npas4 (Antunes et al., 2020). Due to the forebrain-wide deletion of Tet3 within excitatory neurons in this study, it cannot be concluded that the anxiogenic phenotype and/or increased CORT levels observed are due to Tet3 modulation within the HPC alone. Around 1/3 of Crh neurons within both the anterior BNST and central amygdala also co-express Camk2a and were identified as long-range GABAergic projection neurons targeting regions such as the ventral tegmental area of the midbrain (Dedic et al., 2018). Interestingly, Crh KO within these Camk2a positive anterior BNST and central amygdala neurons employing CrhcKO-Camk2a mice, similarly induces an anxiogenic phenotype as in the Tet3cKO-Camk2a although unlike Tet3 KO, no differences in basal or stress-induced CORT levels were observed following Crh KO (Dedic et al., 2018). Considering these findings and the evidence of TET3 modulation of Crh expression within the HYP (Cramer et al., 2019), one can posit that Tet3cKO-Camk2a mice will also delete Tet3 in these Camk2a-Crh co-expressing neurons of the anterior BNST and central amygdala, potentially altering Crh levels within these regions contributing to the anxiogenic phenotype and altered CORT levels reported in Antunes et al., 2020). Clearly, further investigation into the role of TET3 upon HPA axis function and stress adaptation is required. Moreover, direct comparison of targeted cell-type specific TET modulation should be addressed within the context of stress.
Although our understanding remains limited, there is an increasing appreciation for the non-redundant role of TET family proteins as well as demethylation and 5hmC in adaptive and maladaptive responses to stress. Generally, reduced TET1 levels within the brain appears to confer pro stress resiliency and anti-depressant-like effects opposed to the depressive-like or anxiogenic phenotypes observed upon reduce levels of TET2 and TET3, respectively, within the male rodent brain. Yet, many open questions remain as to mechanisms by which TET proteins function in the brain, particularly within the context of stress. For example; are there sex-specific functions of TET proteins in stress adaptation? Are catalytic and catalytic-independent functions of TET proteins, both of which are observed in the brain (Montalban-Loro et al., 2019), involved in their interaction with the central stress response? Are TET-mediated dynamic methylation processes simply permissive or necessary for the molecular embedding and perpetuation of stress-induced transcriptional activity within the brain? Finally, one prominent question pertains to how specific targeting of dynamic DNA methylation processes occurs at discrete genomic regions in response to environmental stimuli, such as stress. As such, do TET interacting partners mediate TET genome targeting and/or modulate TET activity within the brain to mediate transcriptional and behavioural adaptions to stress?
2. TET interacting proteins in the brain and potential role in the central stress response
Due to the predominance of truncated TET protein isoforms lacking the CXXC domain within the adult brain, an important question remains as to the identity and role of TET interacting proteins for TET-mediated targeting and function in response to stressful stimuli. For example, several studies have identified REST as a direct interacting partner of TET3 within the brain (Cramer et al., 2019), main olfactory epithelium (Yang et al., 2020), and retina (Perera et al., 2015). These studies suggest that REST may recruit TET3 to candidate loci inducing local demethylation, as for the Crh locus within the chick HYP upon heat shock stress (Cramer et al., 2019). Although within the main olfactory epithelium, it appears that TET3 plays mainly a non-catalytic role stabilising the REST protein (Yang et al., 2020). Moreover, Cheng et al., 2018 identified a direct stress-induced interaction of HIF1α and TET1 within the mouse PFC suggesting that, at least for a subset of candidate loci, HIF1α may guide TET1 to stress-sensitive genomic loci to modulate local DNA methylation and transcription (Cheng et al., 2018b). Another promising TET interaction partner is early growth response protein 1 (EGR1) also known as nerve growth factor-inducible protein A, which has a clear role in stress-induced transcription in the brain (Duclot and Kabbaj, 2017). Recently, a direct interaction of EGR1 and TET1s (via their C-terminals) was demonstrated within the mouse frontal cortex with a suggested role of EGR1 in recruiting TET1s to modulate the neuronal DNA methylome throughout postnatal neurodevelopment and in response to neuronal activity (Sun et al., 2019). However, whether stress-induced modulation of EGR1 may also coordinate genomic targeting and/or activity of TET1 (and potentially TET2/3) within neurons to mediate stress-induced transcription remains an open question. A potential example of such an effect has, however, previously been described in a prominent series of studies employing a rat maternal care model (low licking and grooming model) (Buschdorf and Meaney, 2015). EGR1 was demonstrated to bind a single CpG within an EGR1 response element in the Nr3c1 exon 1 (Dean and Keshavan, 2017) promoter resulting in induction of a permissive chromatin state enabling DNA demethylation and increased HPC Nr3c1 mRNA in response to increased maternal care in rats (Weaver et al., 2004a, 2004b, 2007). As a consequence of low maternal care (i.e., early life stress), this EGR1 binding and subsequent demethylation process is lost resulting hypermethylation at this CpG and stable downregulation of Nr3c1 mRNA within the rat HPC associated with increased HPA axis responsivity in adulthood (Buschdorf and Meaney, 2015; Weaver et al., 2004a, 2004b, 2007; Hellstrom et al., 2012). Hypermethylation of the orthologous human promoter of this locus (NR3C1 1F) within the HPC of suicide victims exposed to child abuse (and other NR3C1 promoters) (Labonte et al., 2012a, 2012b) further suggested that similar mechanisms may be involved within the human brain following early life stress (McGowan et al., 2009). The mechanism by which active demethylation occurs at this locus has not been subsequently pursued. However, in light of the recently identified TET1-EGR1 interaction within the brain and the proposed EGR1 targeting model it is tempting to hypothesise that EGR1indeed recruits TET1 to this locus to induce the observed demethylation, however this requires experimental validation.
Notably, recent evidence in HEK293T cells also indicates that TET proteins are direct interacting partners of several steroid hormone receptors including the thyroid hormone receptor α1 (ΤRα1), TRβ1, oestrogen receptor alpha (ERRα) and androgen receptor (AR) (Guan et al., 2017). This interaction appears to be mediated via the catalytic domain and was strongest for TET3. Importantly, direct TET3-CD interactions were shown to increase chromatin binding of ΤRα1, TRβ1, ERRα and AR indicative of the coordinated action of TET3 and these receptors for appropriate chromatin localisation. Moreover, TET3 stabilises ΤRα1 likely via non-catalytic limitation of polyubiquitination, such that Tet3 KD or KO results in reduced ΤRα1 protein but not mRNA (Guan et al., 2017). Further evidence of TET-steroid receptor interactions has been observed in various non-neuronal cell types. For example, within prostate cells, hormone-induced stimulation of AR activity also appears to recruit TET1 and TDG to stress-sensitive loci such as the Sgk1 locus modulating its transcription (Dhiman et al., 2015). TET1, specifically TET1s, has also been shown to be involved in the differentiation of gonadotrope cells of the pituitary gland with gonadotropin-releasing hormone inducing AR and ESR1 binding to the Tet1 locus regulating Tet1s mRNA (Yosefzon et al., 2017). Thus, HPA axis function at level of pituitary may also involve TET proteins as well as at the level of the adrenal gland as Tet3 mRNA is increased in cortisol-producing adenoma (Zhong et al., 2019). Considering the degree of overlap between the interactome of steroid receptors, such as GR and AR (Lempiainen et al., 2017), it is intriguing as to whether such TET/steroid receptor interactions would extend to GR and MR, in which contexts, and whether these interactions also occur within the brain. Moreover, do such interactions contribute the molecular encoding of stressful experiences as suggested for the TET1-EGR1 interaction?
One other promising TET interacting partner likely to mediate such effects, although not directly in the brain, is the O-linked N-acetylglucosamine transferase (OGT), which stably interacts with all TET proteins and can be recruited by them in a non-catalytic manner to mediate transcriptional regulation via post-translational O-GlcNAcylation of predominantly chromatin modifiers (Hrit et al., 2018; Chen et al., 2013; Deplus et al., 2013; Vella et al., 2013). Interestingly, placental OGT is significantly lower in males, as opposed to females, in humans and mice (due to its X-linkage). Prenatal stress-induced or genetic reductions in placental OGT result in transcriptional and epigenetic regulation in placental tissue and the embryonic HYP associated with maladaptive stress responses in adulthood in a sex-specific manner (Howerton et al., 2013; Howerton and Bale, 2014). Moreover, work from our own lab suggests that modulation of Tet and Dnmt mRNA levels are modulated in a sex-specific manner within placental tissue following prenatal stress in mice (Schroeder et al., 2018). Thus, considering the widespread epigenetic and transcriptional changes upon placental OGT perturbations, the abundant expression of TET proteins within the placenta (Yosefzon et al., 2017) and stress-sensitivity of TETs in placental tissue, it is likely that TET-OGT interactions play a role in the response of placental tissue and thus subsequent developmental trajectories following prenatal stress exposure. TET protein interacting partners may therefore act as a functional link between stress-induced signalling cascades and TET-mediated transcriptional regulation within the brain in both healthy and pathological states warranting much further investigation.
3. Future directions
The described studies identify non-redundant roles of TET proteins in modulating the neuroepigenetic and behavioural adaptation in response to stress exposure by yet undefined mechanisms. To date, the role of TET proteins in the central stress response has focused primarily on developmental genetic manipulations with only a few studies investigating the molecular mechanisms underlying TET-mediated behavioural dysfunction. Future studies should focus on developmental stage, region- and cell-type specific TET modulation employing sophisticated viral-mediated and transgenic approaches. Due to the region- and cell-type specific nature of the neuronal DNA methylome (Luo et al., 2017), it is critical that future studies employ modern cell-type specific analysis of both the transcriptome and epigenome changes in response to stress or TET modulations via the use of single cell omics, transgenic mouse models (Mo et al., 2015), or fluorescent activated cell sorting. Moreover, considering the inability of classical bisulfite sequencing to differentiate between 5mC and 5hmC, future studies should employ genome-wide, base resolution methylation profiling techniques assessing both 5mC and 5hmC (e.g., oxidative bisulfite sequencing) in CG and non-CG contexts to enable the identification of novel loci of relevance to long-term embedding of stress exposure at the molecular level. As discussed, further investigation of TET interacting partners in the brain must also be conducted, with the aim of furthering our understanding of how targeted regulation of DNA methylation can occur at discrete genomic loci in neuronal populations in response to environmental stimuli, such as stress. Novel Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/dCas9 tools such as CRISPR activation (Savell et al., 2019) and CRISPR interference (Zheng et al., 2018) should also be employed to modulate endogenous TET levels in a more physiological range as well as isoform specific manner as recently demonstrated with TALE-mediated repression of Tet1 isoforms in the brain (Greer et al., 2020). Moreover, as the functional utility of epigenome editing tools, such as dCas9-TET1 and dCas9-DNMT3A, have been demonstrated within the rodent brain (Liu et al., 2016, 2018), the causal interaction of stress-induced DNA methylation changes must be investigated to further our mechanistic understanding of how TET proteins and dynamic regulation of the DNA methylome may be a conduit for the biological embedding of stressful life experiences within neurons of the brain.
4. Conclusions
Since the re-discovery of 5hmC and the identification of the TET family dioxygenases over 10 years ago, a significant body of research has identified critical, non-redundant roles of the TET proteins and dynamic modulation of the DNA methylome within the brain in developmental and postnatal stages. TET protein function and the active demethylation also appear to be involved in stress-induced neuroepigenetic and behavioural adaptations through as yet largely undefined mechanisms. Thus, although still relatively underexplored, TET proteins may play a key role in the biological embedding of stressful life experiences within the brain ultimately perpetuating stable and dynamic transcriptional patterns underlying adaptive and/or maladaptive behavioural states of relevance to stress-related psychiatric disorders.
CRediT authorship contribution statement
Alec Dick: Writing – original draft, Writing – review & editing. Alon Chen: Writing – original draft, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors would like to acknowledge Jessica Keverne for proofing of the manuscript. A.C. is the incumbent of the Vera and John Schwartz Professorial Chair in Neurobiology at the Weizmann Institute and the head of the Max Planck Society–Weizmann Institute of Science Laboratory for Experimental Neuropsychiatry and Behavioural Neurogenetics gratefully funded by the Max Planck Foundation. This work is supported by Ruhman Family Laboratory for Research in the Neurobiology of Stress (A.C.); research support from Bruno and Simone Licht; the Perlman Family Foundation, founded by Louis L. and Anita M. Perlman (A.C.); the Adelis Foundation (A.C.); and Sonia T. Marschak (A.C.).
Data availability
No data was used for the research described in the article.
References
- Antunes C. Tet3 ablation in adult brain neurons increases anxiety-like behavior and regulates cognitive function in mice. Mol. Psychiatr. 2020 doi: 10.1038/s41380-020-0695-7. [DOI] [PubMed] [Google Scholar]
- Aristizabal M.J. Biological embedding of experience: a primer on epigenetics. Proc. Natl. Acad. Sci. U. S. A. 2020;117:23261–23269. doi: 10.1073/pnas.1820838116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer C. Phosphorylation of TET proteins is regulated via O-GlcNAcylation by the O-linked N-acetylglucosamine transferase (OGT) J. Biol. Chem. 2015;290:4801–4812. doi: 10.1074/jbc.M114.605881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck D.B. Delineation of a human mendelian disorder of the DNA demethylation machinery: TET3 deficiency. Am. J. Hum. Genet. 2020;106:234–245. doi: 10.1016/j.ajhg.2019.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose R. Tet3 mediates stable glucocorticoid-induced alterations in DNA methylation and Dnmt3a/Dkk1 expression in neural progenitors. Cell Death Dis. 2015;6 doi: 10.1038/cddis.2015.159. e1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschdorf J.P., Meaney M.J. Epigenetics/Programming in the HPA Axis. Comprehensive Physiology. 2015;6:87–110. doi: 10.1002/cphy.c140027. [DOI] [PubMed] [Google Scholar]
- Chen Q., Chen Y., Bian C., Fujiki R., Yu X. TET2 promotes histone O-GlcNAcylation during gene transcription. Nature. 2013;493:561–564. doi: 10.1038/nature11742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y. 5-Hydroxymethylcytosine alterations in the human postmortem brains of autism spectrum disorder. Hum. Mol. Genet. 2018;27:2955–2964. doi: 10.1093/hmg/ddy193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y. Ten-eleven translocation proteins modulate the response to environmental stress in mice. Cell Rep. 2018;25:3194–3203. doi: 10.1016/j.celrep.2018.11.061. e3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C., Kim T., Chang K.T., Min K.T. DSCR1-mediated TET1 splicing regulates miR-124 expression to control adult hippocampal neurogenesis. EMBO J. 2019;38 doi: 10.15252/embj.2018101293. e101293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomyk A.M. DNA methylation in demyelinated multiple sclerosis hippocampus. Sci. Rep. 2017;7:8696. doi: 10.1038/s41598-017-08623-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer T., Rosenberg T., Kisliouk T., Meiri N. Early-life epigenetic changes along the corticotropin-releasing hormone (CRH) gene influence resilience or vulnerability to heat stress later in life. Mol. Psychiatr. 2019;24:1013–1026. doi: 10.1038/s41380-018-0280-5. [DOI] [PubMed] [Google Scholar]
- De Kloet E.R., Vreugdenhil E., Oitzl M.S., Joels M. Brain corticosteroid receptor balance in health and disease. Endocr. Rev. 1998;19:269–301. doi: 10.1210/edrv.19.3.0331. [DOI] [PubMed] [Google Scholar]
- Dean J., Keshavan M. The neurobiology of depression: an integrated view. Asian journal of psychiatry. 2017;27:101–111. doi: 10.1016/j.ajp.2017.01.025. [DOI] [PubMed] [Google Scholar]
- Dedic N. Chronic CRH depletion from GABAergic, long-range projection neurons in the extended amygdala reduces dopamine release and increases anxiety. Nat. Neurosci. 2018;21:803–807. doi: 10.1038/s41593-018-0151-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deplus R. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. 2013;32:645–655. doi: 10.1038/emboj.2012.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deussing J.M., Chen A. The corticotropin-releasing factor family: physiology of the stress response. Physiol. Rev. 2018;98:2225–2286. doi: 10.1152/physrev.00042.2017. [DOI] [PubMed] [Google Scholar]
- Dhiman V.K., Attwood K., Campbell M.J., Smiraglia D.J. Hormone stimulation of androgen receptor mediates dynamic changes in DNA methylation patterns at regulatory elements. Oncotarget. 2015;6:42575–42589. doi: 10.18632/oncotarget.6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick A.L.W., Chen A. vols. 37–47. Elsevier; 2020. (Stress: Genetics, Epigenetics and Genomics). [Google Scholar]
- Dirven B.C.J., Homberg J.R., Kozicz T., Henckens M. Epigenetic programming of the neuroendocrine stress response by adult life stress. J. Mol. Endocrinol. 2017;59:R11–R31. doi: 10.1530/JME-17-0019. [DOI] [PubMed] [Google Scholar]
- Dong E., Gavin D.P., Chen Y., Davis J. Upregulation of TET1 and downregulation of APOBEC3A and APOBEC3C in the parietal cortex of psychotic patients. Transl. Psychiatry. 2012;2:e159. doi: 10.1038/tp.2012.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duclot F., Kabbaj M. The role of early growth response 1 (EGR1) in brain plasticity and neuropsychiatric disorders. Front. Behav. Neurosci. 2017;11:35. doi: 10.3389/fnbeh.2017.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J. Role of Tet1 and 5-hydroxymethylcytosine in cocaine action. Nat. Neurosci. 2015;18:536–544. doi: 10.1038/nn.3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J. Tet1 in nucleus accumbens opposes depression- and anxiety-like behaviors. Neuropsychopharmacology. 2017;42:1657–1669. doi: 10.1038/npp.2017.6. official publication of the American College of Neuropsychopharmacology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden S.A., Covington H.E., 3rd, Berton O., Russo S.J. A standardized protocol for repeated social defeat stress in mice. Nat. Protoc. 2011;6:1183–1191. doi: 10.1038/nprot.2011.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gontier G. Tet2 rescues age-related regenerative decline and enhances cognitive function in the adult mouse brain. Cell Rep. 2018;22:1974–1981. doi: 10.1016/j.celrep.2018.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good C.R. A novel isoform of TET1 that lacks a CXXC domain is overexpressed in cancer. Nucleic Acids Res. 2017;45:8269–8281. doi: 10.1093/nar/gkx435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer C.B. Tet1 isoforms differentially regulate gene expression, synaptic transmission and memory in the mammalian brain. J. Neurosci. : the official journal of the Society for Neuroscience. 2020 doi: 10.1523/JNEUROSCI.1821-20.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross J.A. Gene-body 5-hydroxymethylation is associated with gene expression changes in the prefrontal cortex of depressed individuals. Transl. Psychiatry. 2017;7 doi: 10.1038/tp.2017.93. e1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W. Methylcytosine dioxygenase TET3 interacts with thyroid hormone nuclear receptors and stabilizes their association to chromatin. Proc. Natl. Acad. Sci. U. S. A. 2017;114:8229–8234. doi: 10.1073/pnas.1702192114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J.U., Su Y., Zhong C., Ming G.L., Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. S0092-8674(11)00299-6 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J.U. Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat. Neurosci. 2014;17:215–222. doi: 10.1038/nn.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn M.A. Dynamics of 5-hydroxymethylcytosine and chromatin marks in Mammalian neurogenesis. Cell Rep. 2013;3:291–300. doi: 10.1016/j.celrep.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn A. DNA methyltransferase 1 (DNMT1) function is implicated in the age-related loss of cortical interneurons. Front Cell Dev Biol. 2020;8:639. doi: 10.3389/fcell.2020.00639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y.F. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellstrom I.C., Dhir S.K., Diorio J.C., Meaney M.J. Maternal licking regulates hippocampal glucocorticoid receptor transcription through a thyroid hormone-serotonin-NGFI-A signalling cascade. Phil. Trans. Roy. Soc. Lond. B Biol. Sci. 2012;367:2495–2510. doi: 10.1098/rstb.2012.0223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howerton C.L., Bale T.L. Targeted placental deletion of OGT recapitulates the prenatal stress phenotype including hypothalamic mitochondrial dysfunction. Proc. Natl. Acad. Sci. U. S. A. 2014;111:9639–9644. doi: 10.1073/pnas.1401203111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howerton C.L., Morgan C.P., Fischer D.B., Bale T.L. O-GlcNAc transferase (OGT) as a placental biomarker of maternal stress and reprogramming of CNS gene transcription in development. Proc. Natl. Acad. Sci. U. S. A. 2013;110:5169–5174. doi: 10.1073/pnas.1300065110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrit J. OGT binds a conserved C-terminal domain of TET1 to regulate TET1 activity and function in development. Elife. 2018;7 doi: 10.7554/eLife.34870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeltsch A. On the enzymatic properties of Dnmt1: specificity, processivity, mechanism of linear diffusion and allosteric regulation of the enzyme. Epigenetics. 2006;1:63–66. doi: 10.4161/epi.1.2.2767. [DOI] [PubMed] [Google Scholar]
- Ji F., Wang W., Feng C., Gao F., Jiao J. Brain-specific Wt1 deletion leads to depressive-like behaviors in mice via the recruitment of Tet2 to modulate Epo expression. Mol. Psychiatr. 2020 doi: 10.1038/s41380-020-0759-8. [DOI] [PubMed] [Google Scholar]
- Jin S.G. Tet3 reads 5-carboxylcytosine through its CXXC domain and is a potential guardian against neurodegeneration. Cell Rep. 2016;14:493–505. doi: 10.1016/j.celrep.2015.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaas G.A. TET1 controls CNS 5-methylcytosine hydroxylation, active DNA demethylation, gene transcription, and memory formation. Neuron. 2013;79:1086–1093. doi: 10.1016/j.neuron.2013.08.032. S0896-6273(13)00791-5 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler K.S., Karkowski L.M., Prescott C.A. Causal relationship between stressful life events and the onset of major depression. Am. J. Psychiatr. 1999;156:837–841. doi: 10.1176/ajp.156.6.837. [DOI] [PubMed] [Google Scholar]
- Kessler R.C., Davis C.G., Kendler K.S. Childhood adversity and adult psychiatric disorder in the US National Comorbidity Survey. Psychol. Med. 1997;27:1101–1119. doi: 10.1017/s0033291797005588. [DOI] [PubMed] [Google Scholar]
- Klengel T. Allele-specific FKBP5 DNA demethylation mediates gene-childhood trauma interactions. Nat. Neurosci. 2013;16:33–41. doi: 10.1038/nn.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M. Modulation of TET2 expression and 5-methylcytosine oxidation by the CXXC domain protein IDAX. Nature. 2013;497:122–126. doi: 10.1038/nature12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli R.M., Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013;502:472–479. doi: 10.1038/nature12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kular L. Neuronal methylome reveals CREB-associated neuro-axonal impairment in multiple sclerosis. Clin. Epigenet. 2019;11:86. doi: 10.1186/s13148-019-0678-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar D. Tet1 oxidase regulates neuronal gene transcription, active DNA hydroxy-methylation, object location memory, and threat recognition memory. Neuroepigenetics. 2015;4:12–27. doi: 10.1016/j.nepig.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon W. Tet1 overexpression leads to anxiety-like behavior and enhanced fear memories via the activation of calcium-dependent cascade through Egr1 expression in mice. Faseb. J. 2018;32:390–403. doi: 10.1096/fj.201601340RR. [DOI] [PubMed] [Google Scholar]
- Labonte B. Genome-wide epigenetic regulation by early-life trauma. Arch. Gen. Psychiatr. 2012;69:722–731. doi: 10.1001/archgenpsychiatry.2011.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labonte B. Differential glucocorticoid receptor exon 1(B), 1(C), and 1(H) expression and methylation in suicide completers with a history of childhood abuse. Biol. Psychiatr. 2012;72:41–48. doi: 10.1016/j.biopsych.2012.01.034. [DOI] [PubMed] [Google Scholar]
- Labonte B. Gadd45b mediates depressive-like role through DNA demethylation. Sci. Rep. 2019;9:4615. doi: 10.1038/s41598-019-40844-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R.S. Chronic corticosterone exposure increases expression and decreases deoxyribonucleic acid methylation of Fkbp5 in mice. Endocrinology. 2010;151:4332–4343. doi: 10.1210/en.2010-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lempiainen J.K. Agonist-specific protein interactomes of glucocorticoid and androgen receptor as revealed by proximity mapping. Mol. Cell. Proteomics. 2017;16:1462–1474. doi: 10.1074/mcp.M117.067488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. Neocortical Tet3-mediated accumulation of 5-hydroxymethylcytosine promotes rapid behavioral adaptation. Proc. Natl. Acad. Sci. U. S. A. 2014;111:7120–7125. doi: 10.1073/pnas.1318906111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. Ten-eleven translocation 2 interacts with forkhead box O3 and regulates adult neurogenesis. Nat. Commun. 2017;8 doi: 10.1038/ncomms15903. 15903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. The DNA repair-associated protein Gadd45gamma regulates the temporal coding of immediate early gene expression within the prelimbic prefrontal cortex and is required for the consolidation of associative fear memory. J. Neurosci. : the official journal of the Society for Neuroscience. 2019;39:970–983. doi: 10.1523/JNEUROSCI.2024-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. N(6)-Methyladenosine co-transcriptionally directs the demethylation of histone H3K9me2. Nat. Genet. 2020;52:870–877. doi: 10.1038/s41588-020-0677-3. [DOI] [PubMed] [Google Scholar]
- Li L. Reduction of Tet2 exacerbates early stage Alzheimer's pathology and cognitive impairments in 2xTg-AD mice. Hum. Mol. Genet. 2020;29:1833–1852. doi: 10.1093/hmg/ddz282. [DOI] [PubMed] [Google Scholar]
- Lister R., Mukamel E.A. Turning over DNA methylation in the mind. Front. Neurosci. 2015;9:252. doi: 10.3389/fnins.2015.00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341 doi: 10.1126/science.1237905. 1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.S. Editing DNA methylation in the mammalian genome. Cell. 2016;167:233–247. doi: 10.1016/j.cell.2016.08.056. e217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.S. Rescue of fragile X syndrome neurons by DNA methylation editing of the FMR1 gene. Cell. 2018;172:979–992. doi: 10.1016/j.cell.2018.01.012. e976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. N (6)-methyladenosine of chromosome-associated regulatory RNA regulates chromatin state and transcription. Science. 2020;367:580–586. doi: 10.1126/science.aay6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loman M.M., Gunnar M.R. Early Experience, S. & Neurobehavioral Development, C. Early experience and the development of stress reactivity and regulation in children. Neurosci. Biobehav. Rev. 2010;34:867–876. doi: 10.1016/j.neubiorev.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W., Mueser K.T., Rosenberg S.D., Jankowski M.K. Correlates of adverse childhood experiences among adults with severe mood disorders. Psychiatr. Serv. 2008;59:1018–1026. doi: 10.1176/appi.ps.59.9.101810.1176/ps.2008.59.9.1018. [DOI] [PubMed] [Google Scholar]
- Luo C. Single-cell methylomes identify neuronal subtypes and regulatory elements in mammalian cortex. Science. 2017;357:600–604. doi: 10.1126/science.aan3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D.K., Guo J.U., Ming G.L., Song H. DNA excision repair proteins and Gadd45 as molecular players for active DNA demethylation. Cell Cycle. 2009;8:1526–1531. doi: 10.4161/cc.8.10.8500. 8500 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makkonen H., Kauhanen M., Paakinaho V., Jaaskelainen T., Palvimo J.J. Long-range activation of FKBP51 transcription by the androgen receptor via distal intronic enhancers. Nucleic Acids Res. 2009;37:4135–4148. doi: 10.1093/nar/gkp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall L.L. Epigenomic analysis of Parkinson's disease neurons identifies Tet2 loss as neuroprotective. Nat. Neurosci. 2020;23:1203–1214. doi: 10.1038/s41593-020-0690-y. [DOI] [PubMed] [Google Scholar]
- McGowan P.O. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat. Neurosci. 2009;12:342–348. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meadows J.P. DNA methylation regulates neuronal glutamatergic synaptic scaling. Sci. Signal. 2015;8:ra61. doi: 10.1126/scisignal.aab0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta D. Childhood maltreatment is associated with distinct genomic and epigenetic profiles in posttraumatic stress disorder. Proc. Natl. Acad. Sci. U. S. A. 2013;110:8302–8307. doi: 10.1073/pnas.1217750110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellen M., Ayata P., Dewell S., Kriaucionis S., Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151:1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellen M., Ayata P., Heintz N. 5-hydroxymethylcytosine accumulation in postmitotic neurons results in functional demethylation of expressed genes. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E7812–E7821. doi: 10.1073/pnas.1708044114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo A. Epigenomic signatures of neuronal diversity in the mammalian brain. Neuron. 2015;86:1369–1384. doi: 10.1016/j.neuron.2015.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montalban-Loro R. TET3 prevents terminal differentiation of adult NSCs by a non-catalytic action at Snrpn. Nat. Commun. 2019;10:1726. doi: 10.1038/s41467-019-09665-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn N.W., Suwalski R., O'Riley C., Bojanowski K., Yura R. The presence of 5-hydroxymethylcytosine in animal deoxyribonucleic acid. Biochem. J. 1972;126:781–790. doi: 10.1042/bj1260781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera A. TET3 is recruited by REST for context-specific hydroxymethylation and induction of gene expression. Cell Rep. 2015;11:283–294. doi: 10.1016/j.celrep.2015.03.020. [DOI] [PubMed] [Google Scholar]
- Provencal N., Binder E.B. The effects of early life stress on the epigenome: from the womb to adulthood and even before. Exp. Neurol. 2015;268:10–20. doi: 10.1016/j.expneurol.2014.09.001. [DOI] [PubMed] [Google Scholar]
- Provencal N. Glucocorticoid exposure during hippocampal neurogenesis primes future stress response by inducing changes in DNA methylation. Proc. Natl. Acad. Sci. U. S. A. 2020;117:23280–23285. doi: 10.1073/pnas.1820842116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudenko A. Tet1 is critical for neuronal activity-regulated gene expression and memory extinction. Neuron. 2013;79:1109–1122. doi: 10.1016/j.neuron.2013.08.003. S0896-6273(13)00714-9 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savell K.E. A neuron-optimized CRISPR/dCas9 activation system for robust and specific gene regulation. eNeuro. 2019;6 doi: 10.1523/ENEURO.0495-18.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder M. Sex dependent impact of gestational stress on predisposition to eating disorders and metabolic disease. Mol Metab. 2018;17:1–16. doi: 10.1016/j.molmet.2018.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L. Genome-wide analysis reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics. Cell. 2013;153:692–706. doi: 10.1016/j.cell.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiers H., Hannon E., Schalkwyk L.C., Bray N.J., Mill J. 5-hydroxymethylcytosine is highly dynamic across human fetal brain development. BMC Genom. 2017;18:738. doi: 10.1186/s12864-017-4091-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H. Early-life gene expression in neurons modulates lasting epigenetic states. Cell. 2017;171:1151–1164. doi: 10.1016/j.cell.2017.09.047. e1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z. EGR1 recruits TET1 to shape the brain methylome during development and upon neuronal activity. Nat. Commun. 2019;10:3892. doi: 10.1038/s41467-019-11905-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teicher, M. H., Andersen, S. L., Polcari, A., Anderson, C. M. & Navalta, C. P. Developmental neurobiology of childhood stress and trauma. Psychiatr. Clin. 25, 397-426, vii-viii (2002). [DOI] [PubMed]
- Thomassin H., Flavin M., Espinas M.L., Grange T. Glucocorticoid-induced DNA demethylation and gene memory during development. EMBO J. 2001;20:1974–1983. doi: 10.1093/emboj/20.8.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towers A.J. Epigenetic dysregulation of Oxtr in Tet1-deficient mice has implications for neuropsychiatric disorders. JCI Insight. 2018;3 doi: 10.1172/jci.insight.120592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vella P. Tet proteins connect the O-linked N-acetylglucosamine transferase Ogt to chromatin in embryonic stem cells. Mol. Cell. 2013;49:645–656. doi: 10.1016/j.molcel.2012.12.019. [DOI] [PubMed] [Google Scholar]
- Wang J. Tet1 facilitates hypoxia tolerance by stabilizing the HIF-alpha proteins independent of its methylcytosine dioxygenase activity. Nucleic Acids Res. 2017;45:12700–12714. doi: 10.1093/nar/gkx869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver I.C. Epigenetic programming by maternal behavior. Nat. Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- Weaver I.C., Diorio J., Seckl J.R., Szyf M., Meaney M.J. Early environmental regulation of hippocampal glucocorticoid receptor gene expression: characterization of intracellular mediators and potential genomic target sites. Ann. N. Y. Acad. Sci. 2004;1024:182–212. doi: 10.1196/annals.1321.099. [DOI] [PubMed] [Google Scholar]
- Weaver I.C. The transcription factor nerve growth factor-inducible protein a mediates epigenetic programming: altering epigenetic marks by immediate-early genes. J. Neurosci. : the official journal of the Society for Neuroscience. 2007;27:1756–1768. doi: 10.1523/JNEUROSCI.4164-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y., Melas P.A., Wegener G., Mathe A.A., Lavebratt C. Antidepressant-like effect of sodium butyrate is associated with an increase in TET1 and in 5-hydroxymethylation levels in the Bdnf gene. Int. J. Neuropsychopharmacol. 2014;18 doi: 10.1093/ijnp/pyu032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Zhang Y. TET-mediated active DNA demethylation: mechanism, function and beyond. Nat. Rev. Genet. 2017;18:517–534. doi: 10.1038/nrg.2017.33. [DOI] [PubMed] [Google Scholar]
- Wu H. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science. 2010;329:444–448. doi: 10.1126/science.1190485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt G.R., Cohen S.S. The bases of the nucleic acids of some bacterial and animal viruses: the occurrence of 5-hydroxymethylcytosine. Biochem. J. 1953;55:774–782. doi: 10.1042/bj0550774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D., Wu X., Zhou Y., Wang W., Wang Z. The microRNA/TET3/REST axis is required for olfactory globose basal cell proliferation and male behavior. EMBO Rep. 2020;21 doi: 10.15252/embr.201949431. e49431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehuda R. Status of glucocorticoid alterations in post-traumatic stress disorder. Ann. N. Y. Acad. Sci. 2009;1179:56–69. doi: 10.1111/j.1749-6632.2009.04979.x. [DOI] [PubMed] [Google Scholar]
- Yosefzon Y. An epigenetic switch repressing Tet1 in gonadotropes activates the reproductive axis. Proc. Natl. Acad. Sci. U. S. A. 2017;114:10131–10136. doi: 10.1073/pnas.1704393114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H. Tet3 regulates synaptic transmission and homeostatic plasticity via DNA oxidation and repair. Nat. Neurosci. 2015;18:836–843. doi: 10.1038/nn.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L. Thymine DNA glycosylase specifically recognizes 5-carboxylcytosine-modified DNA. Nat. Chem. Biol. 2012;8:328–330. doi: 10.1038/nchembio.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R.R. Tet1 regulates adult hippocampal neurogenesis and cognition. Cell Stem Cell. 2013;13:237–245. doi: 10.1016/j.stem.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W. Isoform switch of TET1 regulates DNA demethylation and mouse development. Mol. Cell. 2016;64:1062–1073. doi: 10.1016/j.molcel.2016.10.030. [DOI] [PubMed] [Google Scholar]
- Zheng Y. CRISPR interference-based specific and efficient gene inactivation in the brain. Nat. Neurosci. 2018;21:447–454. doi: 10.1038/s41593-018-0077-5. [DOI] [PubMed] [Google Scholar]
- Zhong J.Y. Aberrant DNA methylation of synaptophysin is involved in adrenal cortisol-producing adenoma. Aging (Albany NY) 2019;11:5232–5245. doi: 10.18632/aging.102119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhubi A. Increased binding of MeCP2 to the GAD1 and RELN promoters may be mediated by an enrichment of 5-hmC in autism spectrum disorder (ASD) cerebellum. Transl. Psychiatry. 2014;4:e349. doi: 10.1038/tp.2013.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.