Graphical abstract

Keywords: Quassinoids, Bruceine A, Phosphoproteomic, P38α MAPK activator, Binding affinity, Molecular simulations
Highlights
-
•
Bruceine A displays potent anti-pancreatic cancer activity in vitro and in vivo.
-
•
Phosphoproteomic analyses identify bruceine A induces phosphorylation of p38α MAPK.
-
•
Octet system and microscale thermophoresis reveal p38α MAPK has high affinity for bruceine A.
-
•
Molecular simulations illustrate determinants of bruceine A binding with p38α MAPK.
Abstract
Pancreatic cancer remains one of the cancers with the poorest prognosis bearing an overall 5-year survival rate of about 5%. Efficient new chemotherapic drugs are still highly desired. Here, bruceine A, a quassinoid identified from the dried fruits of Brucea javanica (L.) Merr., displayed the most potent anti-proliferation activity against pancreatic cancer in vitro and in vivo. Phosphoproteomic analysis revealed p38α MAPK phosphorylation was involved in bruceine A’s action in MIA PaCa-2 cells. Utilizing fortebio octet system and microscale thermophoresis, we found p38α MAPK had high affinity for bruceine A. Molecular docking and molecular dynamic simulations showed that bruceine A widely bound to residues (Leu171, Ala172, Met179, Thr180, Val183) in P-loop of p38α MAPK. Key determinants of bruceine A binding with P-loop of p38α MAPK were 19-C O, 22-CH3, 32-CH3, and 34-CH3. Taken together, our findings demonstrate that bruceine A binds directly to p38α MAPK, which can be used to probe the role of p38α MAPK phosphorylation in pancreatic cancer progression, and as a novel lead compound for pancreatic cancer therapy.
1. Introduction
Cancer is a debilitating disease known as one of the major health problems of global concern, and a substantial portion of the world population is currently affected by different types of cancers irrespective of age [1], [2]. Among the various types of cancers, pancreatic cancer remains one of the deadliest cancers with a 5% 5-year survival rate [3], only a slight improvement from 3% in 1970 over the past decades [4]. Pancreatic cancer is often diagnosed at advanced stages and responds poorly to chemotherapy, leading to low treatment success rates [3]. Both efficient new chemotherapic drugs and advanced diagnosis methods are thus highly desired.
Natural products and extracts derived from plants have been used traditionally for the survival and treatment of various types of ailments and diseases [5]. Quassinoids, a taxonomic marker of the Simaroubaceae family [6], have attracted long-term attention due to their unique structures and significant biological activities [7]. The ripe fruit of Brucea javanica (L.) Merr. (Simaroubaceae), a Chinese materia medica namely Fructus Bruceae recorded initially in Compendium of Materia Medica [8], is rich in quassionoids. Increasing attention has been paid to the remarkable antitumor activity of quassinoids of Fructus Bruceae since the discovery of bruceantin in 1973 [9].
Previous studies showed that several quassinoids of Fructus Bruceae possessed significant anticancer activity against pancreatic cancer cell lines (PANC-1, SW1990, CAPAN-1) [10], [11], [12], such as brusatol, bruceine D, and bruceine H. Among them, brusatol exhibited the most potent in vitro antipancreatic cancer cation, with half-maximal inhibitory concentration (IC50) values of 0.36 μM and 0.10 μM on PANC-1 and SW1990 cell lines, respectively [11]. Apparently, quassinoids as plant-derived natural products with unique skeletons and bioactivities drug leads and candidates, thereby constituting a hotspot in the field of drug discovery.
In this study, basic structural skeleton for quassinoids bearing potent antitumor activities was identified by comprehensive literatural survey. Quassinoids possessing identified structural skeleton were purified from the dried fruits of Brucea javanica (L.) Merr. via targeted isolation monitored by UPLC-MS/MS analysis. Bruceine A displayed the most potent anti-proliferation activity against MIA PaCa-2 cells. We observed that p38α MAPK phosphorylation was involved in bruceine A’s action by quantitative phosphoproteomic profiling together with a computational analysis. Thus, the molecular mechanism was further identified through western blot, molecular docking, fortebio octet system, and molecular dynamic (MD) simulations. Results showed the flexibility of the C13-side chain as well as the presence of 19-C O, 22-CH3, 32-CH3, and 34-CH3 were essential for the interaction of bruceine A with the P-loop of p38α MAPK. Taken together, this study revealed that bruceine A showed significant anti-pancreatic cancer activities both in vitro and in vivo by targeting p38α MAPK and it may be a potential therapeutic candidate for pancreatic cancer.
2. Materials and methods
2.1. Materials
Reagents: Dulbecco's modified eagle's medium (DMEM) (Sigma-Aldrich, MO, USA); fetal bovine serum (FBS) (Lonsera, UY, South America); p38 mitogen activated protein kinase (p38 MAPK) (D13E1) (8690, Cell Signaling Technology, MA, USA); phospho-p38 MAPK (P-p38 MAPK) (Thr180/Tyr182) (4631, Cell Signaling Technology, MA, USA); GAPDH (AF7021, Affinity Biosciences, Beijing, China); horseradish peroxidase (HRP)-conjugated anti-rabbit IgG (S0001, Affinity Biosciences, Beijing, China); phosSTOP phosphatase inhibitor (Roche, Basel, Switzerland); GC-rich PCR master mix (Sangon Biotech, Shanghai, China); acetonitrile (Fisher Chemical, Fairlawn, USA); triethylammonium bicarbonate (TEAB) (Sigma-Aldrich, MO, USA); Escherichia coli BL21(DE3) competent cells (Solarbio, Beijing, China); Ni NTA Beads 6FF (Smart-Lifesciences, Jiangsu, China); BCA protein assay kit (Beyotime, Shanghai, China); endofree plasmid midi kit (Cwbiotech, Beijing, China); fastpure gel DNA extraction mini kit (Vazyme Biotech, Jiangsu, China); Ni-NTA sensors (ForteBio, CA, USA); tandem mass tagging (TMT) Kit (Thermo, Waltham, USA). Ethylene diamine tetraacetic acid (EDTA), 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) and trypsin-EDTA (TE) were obtained from Beyotime Technology (Shanghai, China).
2.2. Cell culture
Human MIA PaCa-2 pancreatic cancer cells were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China) and cultured in DMEM supplemented with 10% FBS, 2.5% horse serum, 100 units/mL penicillin and 100 μg/mL streptomycin, and maintained at 37 °C in a fully humidified atmosphere with 5% CO2.
2.3. MTT assay
Cell viability for quassinoids was determined by the MTT assay [13]. Human MIA PaCa-2 pancreatic cancer cells (2.0 × 103 cells/well) were then cultured in 96-well plates with various concentrations of bruceantin, brusatol, bruceine A, bruceine B, and bruceantinol, respectively. After incubation for 24 h, 20 μL of MTT (5 mg/mL) was added to each well, and the cells were incubated for an additional 4 h at 37 °C. After the clear liquid was carefully aspirated, 150 µL of DMSO was added to each well, incubated at 37 °C for 30 min until the blue-violet crystals were completely dissolved. The absorbance of each well was measured at 570 nm via a microplate spectrophotometer (Thermo, Waltham, USA).
2.4. Sample preparation for quantitative phosphoproteomic analysis and data acquisition
Three biological replicas of control (C1 ~ C3) and bruceine A treated (B1 ~ B3) groups were analyzed. Then, these cell samples were sonicated three times on ice using a high intensity ultrasonic processor in lysis buffer. The remaining debris was removed by centrifugation at 12,000 g at 4 °C for 10 min. The supernatant was collected, and the protein concentration was determined with BCA kit according to the manufacturer’s instructions. After trypsin digestion, peptide was desalted by strata X C18 SPE column and vacuum dried. Peptide was reconstituted in 0.5 M TEAB and processed according to the manufacturer’s protocol for TMT kit/iTRAQ kit. The tryptic peptides were fractionated by high pH reverse-phase HPLC using Thermo Betasil C18 column (5 μm particles, 10 mm ID, 250 mm length). Briefly, peptides were separated with a 60 min linear gradient from 8% to 32% acetonitrile (pH = 9.0). Phosphopeptides elute were automatically collected and combined into six fractions. Finally, each fraction was desalted for LC-MS/MS analysis.
MS/MS data were processed using Maxquant search engine (v.1.5.2.8). We also used Soft MoMo (motif-x algorithm) to assess the model of sequences constituted with amino acids in specific positions of modify-21-mers (10 amino acids upstream and downstream of the site, but phosphorylation with modify-13-mers that 6 amino acids upstream and downstream of the site) in all protein sequences. Gene Ontology (GO) annotation proteome was derived from the UniProt-GOA database, Kyoto Encyclopedia of Genes and Genomes (KEGG) and InterProScan (v.5.14–53.0). All differentially expressed modified protein database accession or sequence were searched against the STRING database version 10.1 for protein–protein interactions (PPI).
2.5. Western blot analysis
The protein was extracted from tumor tissues and cells using RIPA buffer supplemented with protease inhibitor cocktail and phosSTOP phosphatase inhibitor. Total proteins were then separated by 10% sodium dodecyl sulfate polyacrylamide electrophoresis (SDS-PAGE) gels and transferred to PVDF membrane. The membranes were blocked with 5% BSA and then incubated overnight at 4 °C with primary antibodies against P-p38 MAPK (1:1000), p38 MAPK (1:1000), or GAPDH (1:3000), respectively. The next day, after incubation with secondary antibodies (HRP-conjugated anti-rabbit IgG, 1:3000) at room temperature for 1 h, detection was performed using chemiluminescence imaging system (Tanon, Shanghai, China).
2.6. Expression and purification of p38α MAPK
The human p38α MAPK sequence was synthesized using published sequence data (NM_001315.3) from the NCBI database and constructed in the pcDNA3.1 vector. The p38α MAPK gene were separated on 1% agarose gel and recovered by gel extraction kit. The pET-28a-H-p38α MAPK prokaryotic expression plasmid was constructed and extracted according to the protocol recommended by the manufacturer. Then, the pET-28a-H-p38α MAPK plasmid was transformed into the Escherichia coli BL21(DE3) competent cells [14], [15]. Briefly, they were plated on a LB agar containing kanamycin. Positive clones were identified by colony-PCR, sequence analysis or by restriction endonucleases. The bacterial growth was induced by IPTG to a final OD600 0.4 ~ 0.6 followed by an ultrasonication. The supernatant was collected by centrifugation for 20 min at 4 ℃, 12000 rpm. Recombinant His-tagged p38α MAPK protein was purified by chromatography on a Ni2+-NTA agarose resin column. Subsequently, the elution, wash, flow-through buffer was collected for 12% SDS-PAGE analysis then stained by coomassie blue, respectively. As a result, the purified p38α MAPK protein was obtained from the E1, E2, E3 elution buffer and determined by western blot.
2.7. Fortebio octet system assay for affinity of bruceine A or brusatol to p38α MAPK
The binding affinities of bruceine A or brusatol to p38α MAPK were tested by the fortebio octet system (ForteBio, CA, USA) [16]. The Ni-NTA sensors were wetted in Phosphate buffered saline Tween 20 (PBST) for 10 min before detection. The p38α MAPK was loaded onto the Ni-NTA sensors for 15 min. Then the processes of adsorption and desorption of the bruceine A or brusatol molecule were monitored in parallel. After equilibrated with PBST for 3 min, the sensors were transferred into bruceine A or brusatol solution at the 2-fold serial dilutions concentrations of 31.3–500 μM. The data were analyzed using the system software data analysis 11.0 (ForteBio, CA, USA). A graph of binding curves was drawn using ORIGIN 9.5 software (OriginLab, Northampton, MA).
2.8. Microscale Thermophoresis (MST)
The interaction between bruceine A and p38α MAPK was measured by microscale thermophoresis using the NanoTemper Monolith NT.115 instrument set at 2% Pico-RED and Medium MST power. Each measurement consists of 16 reaction mixtures where fluorescent-labeled p38α MAPK concentration was set constant at 20 nM and two-fold diluted bruceine A ranging from 16 μM to 0.488 nM was used. The MO.affinity Analysis v2.3 software was used to fit the data and to determine the Kd.
2.9. Molecular docking
The molecular docking calculations were performed with the Discovery Studio 3.0 software (Accelrys, San Diego, USA). The three-dimensional (3D) structures of the ligand was constructed and optimized via Sketching protocol in DS3.0. The crystal structure of p38α MAPK (ID: 5UOJ) was downloaded from protein data bank (https://www.rcsb.org/), which was preprocessed separately by deleting water molecules and adding hydrogen atoms for molecular docking studies.
The LibDock protocol of DS3.0 was employed to conduct the semi-flexible docking between ligand (bruceine A, bruceine B, bruceantin, brusatol, bruceantinol) and receptor (p38α MAPK) in the CHARMm force field [17]. Firstly, the hotspot plot was calculated for active sites of the receptor p38α MAPK. Then, the ligands of different conformations were rigidly superimposed to the hotspot plot to form a more suitable binding mode. Energy optimization was performed to obtain the most stable configurations.
All assigned atom numbers in the quassinoid structures were generated by molecular simulation software randomly rather than following the rules of natural product nomenclature.
2.10. MD simulations
We performed MD simulations on the complexes of bruceine A-p38α MAPK and brusatol-p38α MAPK with DS3.0 software [18]. The solvation protocol was performed for the addition of Na+ and Cl– counter ions to mimic an in vivo environment. A binding constraint of 5.0 kcal·mol−1·Å−2 was applied to limit all atoms to optimize solvent and ion molecules using the setup constraints protocol. The five steps of the standard dynamics cascade process involved minimization 1, minimization 2, heating, equilibration, and production. The energy minimization of the system was divided into two steps. The first 2000 steps used the steepest descent algorithm and the last 500 steps used conjugate gradient algorithm. Then, the system was gradually heated from 50 to 300 K and the heating progress was maintained for 2000 steps with the force constant of 2.0 kcal·mol−1·Å−2 in the constant-temperature, constant-volume ensemble. The equilibrium MD simulations with the force constant of 2.0 kcal·mol−1·Å−2 were carried out in the constant-temperature, constant-pressure ensemble to ensure the stability of the molecular systems. Finally, the MD simulations at 300 K in production protocol. Periodic boundary conditions were applied to avoid edge effects. The time step size of 2 fs was adopted for all MD simulations. For all produced trajectories, persists for records coordinate trajectories every 2 ps.
2.11. Xenograft pancreatic cancer model
30 male BALB/c nude mice (18–20 g) were purchased from GemPharmatech Co., Ltd (Jiangsu, China) and maintained in an animal facility under standard laboratory conditions for 7 days prior to experiments. 1 × 106 of MIA PaCa-2 cells were suspended in 100 µL of phosphate-buffered saline (PBS) and injected subcutaneously into the right flank regions of each mouse. Then the mice were divided into 5 respective groups: vehicle (PBS, i.p.), positive (gemcitabine, 100 mg/kg, i.p.), bruceine A low (1 mg/kg, i.p.), bruceine A medium (2 mg/kg, i.p.), and bruceine A high (4 mg/kg, i.p.). During the treatment, the tumor volumes and body weight were determined every 2 days. On the 42nd day, all mice were sacrificed, the organs and tumor tissues were harvested.
Animal welfare and experimental procedures were conducted in accordance with the Provision and General Recommendation of Chinese Experimental Animals Administration Legislation and were approved by Institutional Animal Ethical Committee, Nanjing University of Chinese Medicine (No.201910A005).
2.12. Statistical analysis
All data were represented mean ± standard deviation (SD) of three individual experiments. The statistical analysis was assessed using GraphPad Prism v.8 software. We used one-way ANOVA followed by Dunnett's post hoc test when comparing more than two groups of data and one-way ANOVA, non-parametric Kruskal–Wallis test, followed by Dunn's post hoc test when comparing multiple independent groups. p < 0.05 was considered as statistically significant.
3. Results
3.1. Effects of quassinoids on the viability of MIA PaCa-2 cells
Quassinoids could be roughly divided into several classes according to their anti-cancer potential (Fig. S1). It was found that quassinoids bearing similar skeleton to that of brusatol usually exhibited potent anti-cancer activities with IC50 values as low as 13 nM [19]. Thus, it was reasonable to seek potential anti-cancer lead compounds from quassinoids bearing the specific skeleton as given in Fig. S1D. With aid of UPLC-MS/MS analysis, we finally identified and purified five of these quassinoids from the EtOAc-soluble extract of Fructus Bruceae by repeated column chromatography and HPLC separation (Fig. 1A-H). Based on interpretation of HRMS and NMR data, these targeted quassionoids were identified to be bruceine B, brusatol, bruceine A, bruceantinol, and bruceantin, respectively. To confirm the effects of quassinoids on pancreatic cancer cell viability, we treated MIA PaCa-2 cells with various concentrations of quassinoids for 24 h. The MTT assay revealed the IC50 values of quassinoids on MIA PaCa-2 cells, bruceine A (0.029 μM), brusatol (0.034 μM), bruceine B (0.065 μM), bruceantinol (0.669 μM), and bruceantin (0.781 μM).
Fig. 1.
The identification of quassinoids by UPLC-ESI-Q-TOF-MS/MS. (A) The BPI chromatograms of the fragment ions in EtOAc-soluble extract of Fructus Bruceae. Peaks labeled 1–5 were characterized with daughter ion at m/z 421.1493. (B-H) were MS/MS spectral information of five isolates, bruceine B, brusatol, bruceine A, bruceantin, bruceantinol, and detailed information, respectively.
3.2. Bruceine A activates p38α MAPK signaling pathway in MIA PaCa-2 cells
Protein phosphorylation in cells is one of the most active posttranslational modifications in response to various stresses and phosphorylation status of all the proteins in cells subject to a particular treatment [20]. Considering that bruceine A exhibited the highest anti-proliferation activity on MIA PaCa-2 cells, we used quantitative phosphoproteomic approaches to find the key signaling components regulated by bruceine A. A total of 12,284 phosphorylated sites in 3862 phosphoproteins were quantitatively analyzed (Fig. S2A). Compared with the control group, 2112 phosphoproteins significantly changed after bruceine A treatment, including 1066 proteins up-regulated and 1046 down-regulated (Fig. S2B). As shown in Fig. 2A, 3557 phosphorylation sites were detected with significant changes between the treated and control groups, consisting of 1788 sites up-regulated and 1769 down-regulated. The hierarchical clustering of the differentially expressed phosphoproteins indicated that the two groups were separated from each other in both sample sets, and three replicates in each group proved to be reproducible (Fig. 2B).
Fig. 2.
Bruceine A activates p38α MAPK signaling pathway in MIA PaCa-2 cells. (A) Volcano plots showing up-, non-, and downregulated protein sites among bruceine A and control groups. (B) Heat map and cluster analysis of all differentially expressed proteins in two groups (bruceine A vs. control). Protein expression levels are color-coded, showing higher and lower expression in red and blue, respectively. (C) PPI network analysis of P-p38α MAPK. The regulations were connected to the P-p38α MAPK by various shapes and the log2 ratio of bruceine A vs control was represented by color. (D) Significant biological processes affected by bruceine A. Top-22 biological processes derived from GO enrichment analysis were shown. The x-axis displayed the significance represented as -log10 (p value). (E) Significant pathways affected by bruceine A. Top-22 pathways derived from KEGG were shown, the significance level was represented as -log10 (p value). The red bar indicated pathway associated with p38α MAPK. (F) Western blot results showed the expression level of p38α MAPK in MIA PaCa-2 cells following a 24 h treatment with bruceine A at various concentration, GAPDH was used as control. Data were presented as the mean ± SD; n = 4, ⁎p < 0.05 vs the control group. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
To clarify protein kinases that were potentially involved in bruceine A induced anti-proliferation, all differentially expressed modified protein database accession or sequence were searched against the STRING database version 10.1 for protein–protein interactions. Interestingly, we noticed that P-p38α MAPK (ratio of bruceine A: control = 1.281, p value = 0.00026084) was one of the differentially expressed between the treated and control groups (Table S1). The p38α MAPK signaling pathway is critical for human cancer cell survival, dissemination, and resistance to drug therapy, which makes the components of the signaling cascade interesting targets for therapeutic intervention [21], [22]. Additionally, to understand the overall biological effects, interactions between p38α MAPK and the other proteins were displayed as PPI networks in Fig. 2C, the phosphor-MAPK14 (also known as phospho-p38α MAPK, P-p38α MAPK), as an nodal protein, exhibited varying degrees of interactions with 38 proteins, including 17 proteins up-regulated, 20 down-regulated and 1 up/down-regulated.
Moreover, the results of GO enrichment analysis were displayed as biological processes. The top-22 biological processes were listed in Fig. 2D and p38α MAPK was implicated in the mRNA stabilization, which was a highly important process in control of gene expression [23] (Fig. 2D, red color indicated). Furthermore, the KEGG pathway analysis was used to obtain information about “pathways” for better exploration of the internal changes correlated with bruceine A treatment. We observed that multiple signal pathways were regulated, such as spliceosome, RNA transport, and AMPK signaling pathway. Specifically, in the top-22 pathways, we observed that p38α MAPK involved in four signaling pathway, including fluid shear and atherosclerosis, GnRH signaling pathway, platelet activation and FoxO signaling pathway (Fig. 2E, red color indicated). Thus, the regulation of p38α MAPK may represent a worth exploring mode in the anticancer effect of bruceine A. Based on the identification of the phosphorylation sites, it assumed that the Threonine 180 (Thr180) residue was a potential bruceine A-induced p38α MAPK phosphorylation site (Table S1). To validate our phosphoproteomic analysis, we further used western blot to examine the expression level of P-p38α MAPK in bruceine A-treated MIA PaCa-2 cells. Consistently, we found that bruceine A induced the p38α MAPK phosphorylation in a dose- and time-dependent manner (Fig. 2F, Fig. S3). Moreover, we also observed that the phosphorylation of p38α MAPK wasn’t caused by upstream kinases (MKK3/6, MKK4 and JNK) in bruceine A-treated MIA PaCa-2 cells (Fig. S4). These results verify the notion that bruceine A activates p38α MAPK signaling pathway in MIA PaCa-2 cells.
3.3. Anti-pancreatic cancer effect of quassinoids based on the nodal protein p38α MAPK
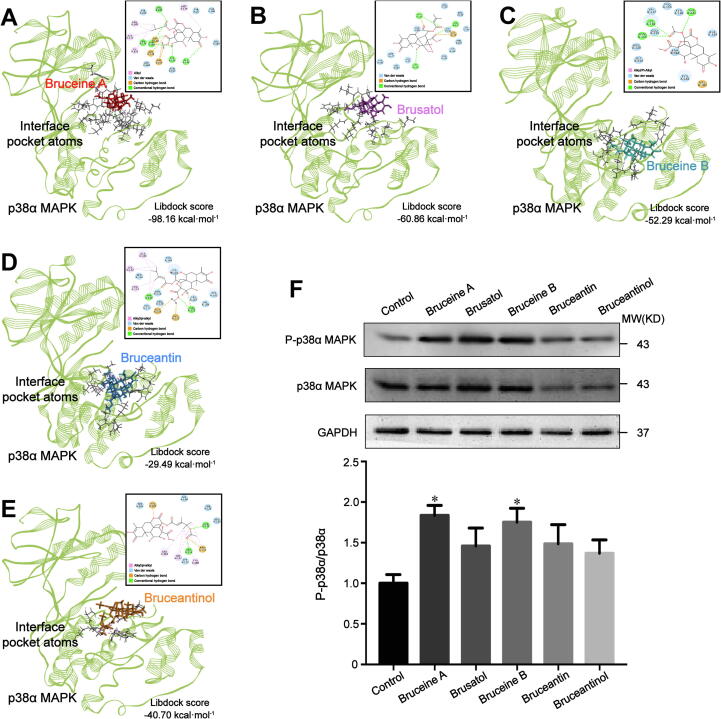
Since the p38α MAPK signaling pathway has been identified as a promising therapeutic target for the modern cancer treatment [22], we speculated that p38α MAPK may be an important target of bruceine A. First, we employed molecular docking to identify the binding mode of p38α MAPK with bruceine A, bruceine B, brusatol, bruceantin, and bruceantinol, respectively (Fig. 3A-E). The docking score was applied for evaluating the receptor-ligand binding ability. The absolute value of the docking score represents the strength of the binding ability. Among them, the p38α MAPK-bruceine A complex was the potent conformation with a libdock score of −98.158 kcal·mol−1. Also, we noticed that five quassinoids interacted with different residues in the phosphorylation activation loop (P-loop, that extends from Leu171 to Val183) of p38α MAPK. Then, the receptor-ligand interactions tool was used to analyze the hydrogen bonding and hydrophobic interaction presented in the p38α MAPK-quassinoid complexes. All the detailed data of amino acids involved in quassinoid-p38α MAPK interactions were depicted in Table S2.
Fig. 3.
Anti-pancreatic cancer effect of quassinoids based on the nodal protein p38α MAPK. (A-E) Interactions of p38α MAPK with bruceine A, brusatol, bruceine B, bruceantin, and bruceantinol. The interaction on a 2D diagram was in the upper-right corner of each panel. (F) Western blot analysis of p38α MAPK expression in MIA PaCa-2 cells untreated (control) or treated with quassinoids at the IC50. GAPDH was analyzed as an internal control. Data were presented as the mean ± SD; n = 3, ⁎p < 0.05 vs the control group.
Subsequently, we performed western blot to assess the effect of five quassinoids on p38α MAPK signaling pathway in MIA PaCa-2 cells. As shown in Fig. 3F, bruceine A-treatment group demonstrated the most potent activation on p38α MAPK signaling pathway compared with the control group (p < 0.05). Overall, our data indicate that bruceine A exerts more potent activation on the p38α MAPK than the other four quassinoids which may be through direct interaction.
3.4. P38α MAPK binds with higher apparent affinity to bruceine A than brusatol
Both bruceine A and brusatol are composed of an α, β-unsaturated cyclohexan ring (A), two cyclohexane rings (B and C), a six-membered lactone ring (D), and a tetrahydrofuran ring (E) [24], [25]. The only structural difference is that the C13-side chain of the brusatol is O-isobutenyl while that of bruceine A is O-isobutyl. To date, brusatol potentiated gemcitabine-induced apoptosis and growth inhibition in pancreatic cancer cells, which has been widely explored [11], [26]. Given that brusatol is structurally similar to bruceine A, we firstly focused on the underlying mechanism of the structural difference in activation p38α MAPK.
To determine the binding affinity of bruceine A or brusatol to p38α MAPK, we performed binding kinetics experiments using fortebio octet system. At first, we conducted the expression and purification of p38α MAPK protein (Fig. S5). The interaction analysis revealed that the adsorption capacity of bruceine A to p38α MAPK rapidly increased at the beginning and then reached equilibrium at 30 s (Fig. 4A), whereas brusatol reached equilibrium at 8 s (Fig. 5A). Obviously, the adsorption and dissociation equilibrium of brusatol was achieved more quickly than bruceine A. The affinity (KD = 0.96 M) of p38α MAPK for brusatol was also lower than that of p38α MAPK for bruceine A (KD = 0. 221 mM). Moreover, the binding affinity of bruceine A to p38α MAPK has been verified through microscale thermophoresis (Kd = 0.031 μM) (Fig. 4B). Collectively, these results support the notion that bruceine A can directly bind to p38α MAPK with higher affinity than brusatol.
Fig. 4.
Bruceine A interacts with p38α MAPK. (A) The binding affinity of bruceine A with p38α MAPK were determined by fortebio octet system assay. (B) The binding affinity of bruceine A with p38α MAPK were determined by microscale thermophoresis. (C) The stereo view of the binding mode. (D) The 2D diagram of the interaction. (E) The 2D structure of bruceine A, the element number was in accordance with the software default system number. (F) The superimposition of bruceine A structure before and after docking. (G) Variation of the angles and dihedral angles of bruceine A before and after docking. (H) The hydrophobic groove of the interaction. (I) The hydrophilic groove of the interaction. (J) The superimposition of the p38α MAPK crystal structure.
Fig. 5.
Brusatol interacts with p38α MAPK. (A) Determination of brusatol binding to p38α MAPK by fortebio octet system assay. (B) The stereo view of the binding mode. (C) The 2D diagram of the interaction. (D) The 2D structure of brusatol, the element number was in accordance with the software default system number. (E) The superimposition of brusatol structure before and after docking. (F) Variation of the angles and dihedral angles of brusatol before and after docking. (G) The hydrophobic groove of the interaction. (H) The hydrophilic groove of the interaction. (I) The superimposition of the p38α MAPK crystal structure.
Like most kinases, p38α MAPK is composed of two lobes: the smaller N-terminal lobe (N-lobe), consisting mostly of β-sheets, and the α-helical C-terminal lobe (C-lobe)[27]. The lobes are linked by a flexible hinge and the red region is the P-loop in p38α MAPK consists of Leu171–Val183 [27], [28]. Although structure for p38α MAPK have been determined, how they interact with bruceine A or brusatol are poorly understood. Here, we initially performed bruceine A-p38α MAPK docking using libdock protocol to understand its binding mode. As shown in the Fig. 4C and D, bruceine A had wider non-covalent interactions with the C-lobe and P-loop of p38α MAPK, including nine hydrogen bonds (H-bonds), six alkyl hydrophobic interactions, and seven van der waals interactions (Table S3).
To further verify the conformational changes of bruceine A, we superimposed the conformation before and after docking. We observed that the methyl formate side chain at the C13- on the C ring of bruceine A was inverted by 180°, while the side chain at the C15- was reversed by 180° (Fig. 4E-G). Indeed, the considerable reverse is favorable for the decrease of steric hindrance of the bruceine A-p38α MAPK interaction. This may contribute to the formation of non-covalent interactions with the surrounding amino acid residues. Subsequently, the unsaturated carbonyl group at the C1- and the free hydroxyl group at the C16- of bruceine A formed classical H-bonds with the surrounding amino acid residues, promoting the binding of bruceine A with p38α MAPK. Additionally, bruceine A bound to the nonpolar residues in the hydrophobic groove of p38α MAPK non-covalently, including Leu171, Ala184, Phe169, Gly170, Ala172, Met179, Val183, and Phe169 (Fig. 4H). In parallel, bruceine A primary established H-bonds and van der waals interactions with the polar residues in the hydrophilic groove of p38α MAPK, consisting of Lys152, Thr180, Thr185, Arg189, Asp168, Asp150, Asn155, Arg149, Tyr200, and Tyr35 (Fig. 4I). Upon bruceine A binding, the flexibility of the interaction pocket was remarkably reduced for p38α MAPK, as stabilized by the binding of bruceine A (Fig. 4J). The environment of the active pocket in p38α MAPK was composed of hydrophilic and hydrophobic grooves and the presence of multiple non-covalent interactions enhanced the binding affinity. Together, our results clearly show that bruceine A widely interacts with residues (Leu171, Ala172, Met179, Thr180, Val183) in the P-loop of p38α MAPK.
In comparison, we also identified the binding mode of brusatol with p38α MAPK using molecular docking. Unlike bruceine A, p38α MAPK primarily interacted with the side chains of brusatol (Fig. 5B). Also, the interaction diagrams indicated that the side chains of brusatol bind to the pocket atoms of p38α MAPK by non-covalent bonds comprised of six H-bonds and twelve van der waals interactions (Fig. 5B, C and Table S4). Upon the conformational changes of brusatol before and after docking, we observed that the side chain at the C13-of brusatol converted from stretched to folded state and only had one H-bond with Gly170 of p38α MAPK (Fig. 5E and F). Similar to brucine A, the side chain at C15- of brusatol was inverted by 180° and extended into the active pocket of p38α MAPK to form H-bonds with the surrounding residues. However, the branch at the C13-side chain could not rotate freely, probably owing to the present of the double bond at the C31-, ultimately resulting in the free range of brusatol less than that of bruceine A. Furthermore, we noticed that carbonyl groups at the C13- and the C15-side chain of brusatol competed with the residue Gly170 and Ala172 of p38α MAPK respectively, leading to its affinity for p38α MAPK lower than that of bruceine A. The primary interactive region was in the hydrophilic groove of p38α MAPK, which contained Lys152, Gly170, Thr180, Tyr35, Asn155, Asp168, Arg189, and Arg149 (Fig. 5G). Also, brusatol displayed several non-covalent interactions with the residues of hydrophobic groove, including Leu171, Ala172, Tyr35, Met179, Val183, Phe169, and Ile193 (Fig. 5H). Simultaneously, the flexibility of the interaction pocket of p38α MAPK showed a remarkable increase, indicating the high fluctuations of p38α MAPK upon brusatol binding (Fig. 5I). Like bruceine A, brusatol interacted with residues in the P-loop of p38α MAPK, including Leu171, Ala172, Met179, Thr180, and Val183. However, the folding C13-side chain of brusatol caused the increased steric hindrance, primarily resulting in the lower binding than that of bruceine A. Overall, the data indicate that p38α MAPK binds with higher apparent affinity to bruceine A than does brusatol.
3.5. Bruceine A possesses more stable binding to p38α MAPK than brusatol
Having verified the stronger affinity of bruceine A to p38α MAPK than brusatol, we used MD simulations to investigate the molecular stability of the complexes and dynamic trajectories of interactions. Root mean square deviation (RMSD) is the most commonly used quantitative measure of the similarity between two superimposed atomic coordinates [29]. The RMSD analysis of backbone atoms indicated that bruceine A-p38α MAPK complex reached relatively stable states with a trajectory of maximal average of 1.44 ± 0.07 Å and that of brusatol-p38α MAPK complex was 1.65 ± 0.08 Å (Fig. 6A). It was possible to summarize the RMSD value of brusatol-p38α MAPK was higher than that of bruceine A-p38α MAPK complex during the simulation. Besides, the RMSD value of brusatol-p38α MAPK complex increased from 1.50 Å until 1.90 Å and fluctuated over the simulation time. Conversely, the RMSD value of bruceine A-p38α MAPK complex fluctuated throughout the first 80 conformations of simulation, but was then stable with an average of 1.41 Å until the end of simulation. The increasing value of RMSD in brusatol-p38α MAPK complex was due to enhanced motions between the atoms. Our results clearly showed that bruceine A-p38α MAPK complex had better conformational relative stability in comparison with brusatol-p38α MAPK complex.
Fig. 6.
Bruceine A possesses more stable binding to p38α MAPK than brusatol. (A) RMSD plot of the complexes versus the conformations. (B) RMSF for each Cα atoms of each residue in complexes. (C, D) The change of the energy of bruceine A-p38α MAPK complex and brusatol-p38α MAPK complex during the MD simulations.
Additionally, the functions of proteins which are inherently flexible molecules depends critically on changes of different molecular conformations [30]. To better understand structural variations and conformational flexibility of proteins in complexes, we conducted the root mean-square fluctuation (RMSF) of Cα on the protein backbone to explore fluctuations of each residue in the simulation procedure. The enhancement of flexibility could be observed in terms of the average interatomic distances between the atoms [31]. The 3D structure of amino acid residues in regions of Leu13-Glu19, Val52-Arg70, Ala93-Val102, and Asp168-Thr185 of bruceine A-p38α MAPK complex were fluctuated significantly, while that of brusatol-p38α MAPK complex were Gln11-Arg23, Gly31-Ser37, Val50-Leu75, Ala93-Phe99, and Asp168-Trp187 (Fig. 6B). Compared to brusatol, bruceine A binding site on p38α MAPK exhibits less conformation fluctuations, suggesting that bruceine A binding stabilizes the local conformation.
Furthermore, we analyzed the total energy, kinetic energy, and potential energy of complexes. As shown in Fig. 6C and D, for bruceine A-p38α MAPK complex, the mean total energy, kinetic energy, and potential energy were −8.70 × 104 KJ·mol−1, 2.00 × 104 KJ·mol−1, and −10.68 × 104 KJ·mol−1, respectively. Correspondingly, for brusatol-p38α MAPK complex, the average of total energy, kinetic energy, and potential energy were −8.06 × 104 KJ·mol−1, 1.85 × 104 KJ·mol−1, and −9.91 × 104 KJ·mol−1, respectively. Obviously, bruceine A-p38α MAPK complex possesses stronger interaction energy than that of brusatol, providing its more substantial stability.
3.6. Bruceine A suppresses the growth of pancreatic cancer in vivo
To explore whether bruceine A can be used for in vivo pancreatic cancer targeting p38α MAPK, we used xenografts tumor model in nude mice. As indicated in Fig. 7A and B, mice were divided into five groups randomly: vehicle, positive control with gemcitabine, 1 mg/kg of bruceine A, 2 mg/kg of bruceine A, and 4 mg/kg of bruceine A. All mice were sacrificed after 42 days, and the tumors were obtained. Tumor volume (Fig. 7C) and tumor weight (Fig. 7D) were dose dependently reduced in mice treated with bruceine A compared with vehicle group. Importantly, the body weights of mice were stable in bruceine A-treated groups with no obvious distinctions to vehicle group (Fig. 7E). Meanwhile, the heart, liver, and lung weight indexes of mice were slightly increased in parts of the groups. While kidney and spleen weight indexes were not affected in all dose groups (Fig. S6). Compared with the vehicle group, the expression of P-p38α MAPK protein was up-regulated by bruceine A in a dose-dependent manner (p < 0.05) (Fig. 7F and G). Taken together, the results suggest that bruceine A suppresses tumor growth and activated p38α MAPK in vivo.
Fig. 7.
Bruceine A suppresses the growth of pancreatic cancer in vivo. (A) A schematic of the treatment plan for nude mice bearing pancreatic cancer MIA PaCa-2 cells. (B) Representative pictures of tumors dissected from the mice per group. (C-E) Tumor volume, tumor weight, and body weight of each group during the treatment period. (F, G) Protein expression of p38α MAPK after treatment determined by western blot analysis. GAPDH was analyzed as an internal control. Data were presented as the mean ± SD; n = 6, compared with vehicle group, ⁎p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
4. Discussion
Pancreatic cancer remains one of the cancers with the poorest prognosis, with an overall 5-year survival rate of about 5%, without much difference between high-income countries and low-income and middle-income countries [32]. Hence, this urgent medical problem demands a comprehensive solution. Many quassinoids has been reported to display outstanding antitumor activity in different potencies, such as bruceantin and glaucarubinone [33].
Basic structural skeleton for quassinoids bearing potent antitumor activities was identified by comprehensive literature survey. Currently, quassinoids possess fierce anticancer activities against multiple cancers, including colorectal [34], [35], hepatoma [34], [35], gastric [34], [35], [36], pancreatic [37], [38], [39], breast cancer [38], [40] and so on. Particularly, a previous study reported bruceine A showed anticancer activities against human breast cancer MCF-7 and MDA-MB-231cell lines after 72 h, having IC50 values of (0.182 ± 0.048) μM and (0.228 ± 0.020) μM, respectively [41]. We previously reported that, among seven quassinoids of Fructus Bruceae including brusatol, bruceine D, bruceine H, yadanzioside A, yadanzioside G, javanicoside C, and bruceantinoside A, brusatol exhibited the most potent in vitro anti-pancreatic cancer action on PANC-1 and SW1990 cell lines, respectively [11]. Presently, for the first time, we reported that bruceine A, a quassinoid of Fructus Bruceae, possessed higher anti-proliferation activity on human MIA PaCa-2 pancreatic cancer cells than brusatol and the other four quassinoids, with an IC50 value of 29 nM. More importantly, we observed that bruceine A showed no significant organ (heart, lung, liver, kidney, spleen) toxicities in vivo. Thus, our findings provide further support for the notion that bruceine A may become a novel anti-pancreatic cancer drug candidate.
P38α MAPK, a highly conserved member of the MAPKs, play critical roles in cellular responses, proliferation, survival, cell cycle, and migration in cancer [42]. Therefore, p38α MAPK is emerging as a bona fide drug target for pancreatic cancer therapy [43]. As the highest anti-proliferation activity of bruceine A on MIA PaCa-2 cells, a potential mechanism indicated by phosphoproteomic data was the regulation of p38α MAPK signaling pathway. Concordantly, our western blot results showed that bruceine A exhibited dose-dependent up-regulation of the P-p38α MAPK protein in vitro and in vivo. Molecular docking results were consisted with western blot analysis, suggesting that bruceine A exerted more potent activation on p38α MAPK than the other four quassinoids. In previous work, activation of p38α MAPK is necessary for gemcitabine-induced cytotoxicity in human pancreatic cancer cells [44], [45]. Intriguingly, a discovery set of patients’ pancreatic cancer tissues suggests that activated p38α MAPK correlated positively with the patient survival rates, leading support to the notion that the functional p38α MAPK signaling in pancreatic cancer is a predictive marker of outcome in resected pancreatic cancer [46], [47]. In this work, combining our phosphoproteomic analysis, the identification of phosphorylation sites assumed that the Thr180 residue was a potential bruceine A-induced p38α MAPK phosphorylation site. In support of this view, our molecular docking results suggested that bruceine A interacted with residues in the P-loop of p38α MAPK, including Leu171, Ala172, Met179, Thr180, and Val183.
Both bruceine A and brusatol are composed of an α, β-unsaturated cyclohexan (A), two cyclohexane rings (B and C), a six-membered lactone ring (D), and a tetrahydrofuran ring (E) [24], [25]. The only structural difference is that the C13-side chain of the brusatol is O-isobutenyl while that of bruceine A is O-isobutyl. Recent years, brusatol potentiated gemcitabine-induced apoptosis and growth inhibition in pancreatic cancer cells, which has been widely explored [11], [26]. Notably, brusatol induced apoptosis by activating p38α MAPK pathways in PANC-1 and PATU-8988 cell lines [12]. Here, our fortebio octet system assay identified bruceine A could directly bind to p38α MAPK with higher affinity than brusatol. Moreover, the binding affinity of bruceine A to p38α MAPK has been verified through microscale thermophoresis (Kd = 0.031 μM). Combined with molecular docking results, p38α MAPK bound with higher apparent affinity to bruceine A than did to brusatol. In a further step, MD simulations indicated bruceine A possessed more stable binding to p38α MAPK than that of brusatol. Surprisingly, brusatol also interacted with the residues (Leu171, Ala172, Met179, Thr180, Val183) in the P-loop of p38α MAPK, as bruceine A did. By comparison, similarly to bruceine A, the side chain at C15- of brusatol was inverted by 180° and extended into the active pocket of p38α MAPK to form H-bonds with the surrounding residues. However, the present of the double bond at the C31- in brusatol, the branch at the C13-side chain could not rotate freely, leading to the free range of brusatol was less than that of bruceine A. Impressively, after interaction with p38α MAPK, the side chains at C13- and C15- of bruceine A exhibited certain degrees of reverse, generally resulting in the decreased steric hindrance. Moreover, the folding C13-side chain of brusatol caused the increased steric hindrance, inducing its lower binding affinity. This intriguing behavior suggested that the decreased steric hindrance and free range at C13-side chain of bruceine A to be key for a tighter binding affinity with p38α MAPK, in spite of binding to the same binding site. Ultimately, we speculated that key determinants for bruceine A binding of P-loop of p38α MAPK were the 19-C O, 22-CH3, 32-CH3, and 34-CH3, while that of brusatol were the 12-C O, 19-C O, and 27-C O.
As mentioned before [33], some general structural requirements for optimal cytotoxic and solid tumor selectivity included: a) a four ring skeleton with a lactone D-ring; b) an α,β-unsaturated ketone and an α-hydroxyl group next to the carbonyl in the A-ring; c) two free hydroxyl groups and an oxygen-methylene bridge in the C-ring; and d) an ester group at either C-15 or C-6. Furthermore, we concluded evidences of critical binding determinants for quassinoids binding with the P-loop of p38α MAPK in the present work including the decreased steric hindrance at the C13-side chain and an ester group at C19. Accordingly, the binding of quassinoid with p38α MAPK provides efforts to their potent anti-pancreatic cancer activity.
Moreover, small molecules that specifically interact with protein recognition are becoming increasingly important in antitumor drug development. However, the characteristics and properties of the protein complex interaction interface remains unknown, introducing difficulties to the design for targeted agents. In this study, some properties and characteristics of the p38α MAPK complex binding pocket were analyzed and summarized (Fig. S7, 8 and Table S5, 6). Firstly, compared with brusatol, the more stable interaction conformation revealed that bruceine A formed a variety of H-bonds while interacting with p38α MAPK accounted for its higher stability. Secondly, both Phe and Tyr are aromatic amino acids and the side chain has the phenyl ring [48]. The hydrophobic interaction of Phe is stronger than that of Tyr. Next, the Phe169 forms classical H-bonds with the weak hydrophilic ester at 34-CH3 in bruceine A, while the phenolic hydroxyl group of Tyr (Tyr35 and Tyr200) was easily dissociated to provoke van der waals interactions with bruceine A. By contrast, the folding C13-side chain of brusatol caused the increase of steric hindrance, the Phe169 only formed a van der waals interaction with the hydrophobic of C15-side chain of brusatol, explaining its lower affinity. More importantly, the solvent accessible surface (SAS) [49] accessibility (349 Å) of the interaction between brusatol and p38α MAPK was lower than that of bruceine A (388.5 Å), suggesting p38α MAPK binds with higher apparent affinity to bruceine A than does to brusatol. In total, we hypothesize that hydrophobic anticancer molecules with negative charges may give rise to its excellent selectivity and affinity for targeting the P-loop of p38α MAPK protein, which may make efforts to design p38α MAPK targeting agents toward more efficient and specific targeted anti-pancreatic cancer.
5. Conclusions
We demonstrate here that bruceine A is a novel p38α MAPK activator. It strongly inhibits pancreatic cancer cell growth in vitro and in vivo by interacting with residues (Leu171, Ala172, Met179, Thr180, Val183) in P-loop of p38α MAPK. Critical binding determinants of bruceine A are found to be 19-C O, 22-CH3, 32-CH3, and 34-CH3. Our data suggest that bruceine A could be used as a novel lead compound for pancreatic cancer therapy.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The research was financially supported by the National Natural Science Foundation of China (No. 81773876), Postgraduate Research & Practice Innovation Program of Jiangsu Province (KYCX20_1495), Natural Science Foundation of Jiangsu Province, China (BK20191413) and the Fund for the Specially Appointed Professor of Jiangsu Province.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.06.011.
Contributor Information
Jin-Ao Duan, Email: dja@njucm.edu.cn.
Ming Zhao, Email: mingzhao@njucm.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Fitzmaurice C., Akinyemiju T.F., Al Lami F.H., Alam T., Alizadeh-Navaei R., Allen C. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 29 cancer groups, 1990 to 2016: A systematic analysis for the global burden of disease study. JAMA Oncol. 2018;4(11):1553. doi: 10.1001/jamaoncol.2018.2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.David A.R., Zimmerman M.R. Cancer: an old disease, a new disease or something in between? Nat Rev Cancer. 2010;10(10):728–733. doi: 10.1038/nrc2914. [DOI] [PubMed] [Google Scholar]
- 3.Ilic M., Ilic I. Epidemiology of pancreatic cancer. World J Gastroenterol. 2016;22(44):9694. doi: 10.3748/wjg.v22.i44.9694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jemal A, Siegel R, Xu JQ, Ward E. Cancer statistics. 2010. CA Cancer J Clin 2010;60:277-300. [DOI] [PubMed]
- 5.Dias D.A., Urban S., Roessner U. A historical overview of natural products in drug discovery. Metabolites. 2012;2(2):303–336. doi: 10.3390/metabo2020303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Almeida M.M.B., Arriaga Â.M.C., Santos A.K.L., Lemos T.L., Braz-Filho R., Vieira I.J.C. Occurrence and biological activity of quassinoids in the last decade. Qumica Nova. 2007;30:935–951. [Google Scholar]
- 7.Vieira C., Ivo J. Quassinoids: structural diversity, biological activity and synthetic studies. Stud Nat Prod Chem. 2006;33:433–492. [Google Scholar]
- 8.Wei Y.-J., Qi L.-W., Li P., Luo H.-W., Yi L., Sheng L.-H. Improved quality control method for Fufang Danshen preparations through simultaneous determination of phenolic acids, saponins and diterpenoid quinones by HPLC coupled with diode array and evaporative light scattering detectors. J Pharm Biomed Anal. 2007;45(5):775–784. doi: 10.1016/j.jpba.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 9.Kupchan S.M., Britton R.W., Ziegler M.F., Sigel C.W. Bruceantin, a new potent antileukemic simaroubolide from Brucea antidysenterica. J Org Chem. 1973;38(1):178–179. doi: 10.1021/jo00941a049. [DOI] [PubMed] [Google Scholar]
- 10.Lau S.T., Lin Z.X., Liao Y. Bruceine D induces apoptosis in pancreatic adenocarcinoma cell line PANC-1 through the activation of p38-mitogen activated protein kinase. Cancer Lett. 2009;281:42–52. doi: 10.1016/j.canlet.2009.02.017. [DOI] [PubMed] [Google Scholar]
- 11.Zhao M., Lau S.T., Leung P.S., Che C.T., Lin Z.X. Seven quassinoids from Fructus Bruceae with cytotoxic effects on pancreatic adenocarcinoma cell lines. Phytother Res. 2011;25(12):1796–1800. doi: 10.1002/ptr.3477. [DOI] [PubMed] [Google Scholar]
- 12.Xiang Y., Ye W., Huang C., Lou B., Zhang J., Yu D. Brusatol inhibits growth and induces apoptosis in pancreatic cancer cells via JNK/p38 MAPK/NF-κb/Stat3/Bcl-2 signaling pathway. Biochem Biophys Res Commun. 2017;487(4):820–826. doi: 10.1016/j.bbrc.2017.04.133. [DOI] [PubMed] [Google Scholar]
- 13.Wu J., Liu J., Wei X.Q., Niu X.H., Tang S. A feature-based analysis identifies COL1A2 as a regulator in pancreatic cancer. J Enzyme Inhib Med Chem. 2019;34(1):420–428. doi: 10.1080/14756366.2018.1484734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bukhtiyarova M., Northrop K., Chai X.M., Casper D., Karpusas M., Springman E. Improved expression, purification, and crystallization of p38alpha MAP kinase. Protein Expr Purif. 2004;37:154–161. doi: 10.1016/j.pep.2004.05.017. [DOI] [PubMed] [Google Scholar]
- 15.Özcan D., SİpahİoĞlu H.M. Simultaneous production of alpha and beta amylase enzymes using separate gene bearing recombinant vectors in the same Escherichia coli cells. Turk. J Biol. 2020;44:201–207. doi: 10.3906/biy-2001-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petersen R.L. Strategies using biolayer interferometry biosensor technology for vaccine research and development. Biosensors-Basel. 2017;7(4):49. doi: 10.3390/bios7040049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lv D., Cao Y., Chen L.D., Zhu Z.Y., Chen X.F., Li D. Simulation strategies for characterizing phosphodiesterase-5 inhibitors in botanical dietary supplements. Anal Chem. 2018;90(18):10765–10770. doi: 10.1021/acs.analchem.8b01609. [DOI] [PubMed] [Google Scholar]
- 18.Kufareva I., Abagyan R. Methods of protein structure comparison. Methods Mol Biol. 2011;857:231–257. doi: 10.1007/978-1-61779-588-6_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cuendet M., Christov K., Lantvit D.D., Deng Y.F., Hedayat S., Helson L. Multiple myeloma regression mediated by bruceantin. Clin Cancer Res. 2004;10(3):1170–1179. doi: 10.1158/1078-0432.ccr-0362-3. [DOI] [PubMed] [Google Scholar]
- 20.Pi E., Qu L., Hu J. Mechanisms of soybean roots' tolerances to salinity revealed by proteomic and phosphoproteomic comparisons between two cultivars. Mol Cell Proteomics. 2016;15(1):266–288. doi: 10.1074/mcp.M115.051961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luca A.D., Maiello M.R., D'Alessio A., Pergameno M., Nicola N. The RAS/RAF/MEK/ERK and the PI3K/AKT signalling pathways: role in cancer pathogenesis and implications for therapeutic approaches. Expert Opin Ther Targets. 2012;16(sup2):S17–S27. doi: 10.1517/14728222.2011.639361. [DOI] [PubMed] [Google Scholar]
- 22.Burotto M., Chiou V.L., Lee J.M., Kohn E.C. The MAPK pathway across different malignancies: a new perspective. Cancer. 2014;120(22):3446–3456. doi: 10.1002/cncr.28864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shim J., Karin M. The control of mRNA stability in response to extracellular stimuli. Mol Cells. 2002;14:323–331. [PubMed] [Google Scholar]
- 24.Hu S.Z., Jin L., Yu T., Tian H.Y., Jiang R.W. Brusatol. Acta Crystallogr Sect E Struct Rep. 2012;68(6):o1592–o1593. doi: 10.1107/S1600536812018582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feng X.H., Zhang Y.N., He W.Z., Zhang L., Jiang H.Y. Bruceine A. Acta Crystallogr Sect E Struct Rep. 2010;66(4):o854–o855. doi: 10.1107/S1600536810007646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiang Y.K., Ye W., Huang C.H., Yu D.L., Chen H., Deng T. Brusatol enhances the chemotherapy efficacy of gemcitabine in pancreatic cancer via the Nrf2 signalling pathway. Oxi Med Cell Longev. 2018;2018:1–10. doi: 10.1155/2018/2360427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar G.S., Clarkson M.W., Kunze M.B., Granata D., Wand A.J., Lindorff-Larsen K. Dynamic activation and regulation of the mitogen-activated protein kinase p38. Proc. Natl Acad Sci U S A. 2018;115(18):4655–4660. doi: 10.1073/pnas.1721441115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y.Y., Wu J.W., Wang Z.X. Mitogen-activated protein kinase (MAPK) phosphatase 3-mediated cross-talk between MAPKs ERK2 and p38α. J Biol Chem. 2011;286(18):16150–16162. doi: 10.1074/jbc.M110.203786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sutthibutpong T., Rattanarojpong T., Khunrae P. Effects of helix and fingertip mutations on the thermostability of xyn11A investigated by molecular dynamics simulations and enzyme activity assays. J Biomol Struct Dyn. 2018;36(15):3978–3992. doi: 10.1080/07391102.2017.1404934. [DOI] [PubMed] [Google Scholar]
- 30.Liu J.F., Wei B.B., Che C.H., Gong Z.J., Jiang Y.S., Si M.R. Enhanced stability of manganese superoxide dismutase by amino acid replacement designed via molecular dynamics simulation. Int J Biol Macromol. 2019;128:297–303. doi: 10.1016/j.ijbiomac.2019.01.126. [DOI] [PubMed] [Google Scholar]
- 31.Gu J.F., Bai F., Li H.L., Wang X.C. A generic force field for protein coarse-grained molecular dynamics simulation. Int J Mol Sci. 2012;13(12):14451–14469. doi: 10.3390/ijms131114451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McGuigan A., Kelly P., Turkington R.C., Jones C., Coleman H.G., McCain R.S. Pancreatic cancer: a review of clinical diagnosis, epidemiology, treatment and outcomes. World J Gastroenterol. 2018;24(43):4846–4861. doi: 10.3748/wjg.v24.i43.4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo Z., Vangapandu S., Sindelar R.W., Walker L.A., Sindelar R.D. Biologically active quassinoids and their chemistry: potential leads for drug design. Curr Med Chem. 2005;12:173–190. doi: 10.2174/0929867053363351. [DOI] [PubMed] [Google Scholar]
- 34.Liu J.H., Zhao N., Zhang G.J., Yu S.S., Wu L.J., Qu J. Bioactive quassinoids from the seeds of Brucea javanica. J Nat Prod. 2012;75(4):683–688. doi: 10.1021/np200920c. [DOI] [PubMed] [Google Scholar]
- 35.Chen H., Bai J., Fang Z.F., Yu S.S., Ma S.G., Xu S. Indole alkaloids and quassinoids from the stems of Brucea mollis. J Nat Prod. 2011;74(11):2438–2445. doi: 10.1021/np200712y. [DOI] [PubMed] [Google Scholar]
- 36.Chen Y, Zhu L, Yang X, Wei C, Chen CR, He Y, et al. Ailanthone induces G2/M cell cycle arrest and apoptosis of SGC‑7901 human gastric cancer cells. Mol Med Rep 2017;16:6821-7. [DOI] [PMC free article] [PubMed]
- 37.Yeo D., Huynh N., Beutler J.A., Christophi C., Shulkes A., Baldwin G.S. Glaucarubinone and gemcitabine synergistically reduce pancreatic cancer growth via down-regulation of P21-activated kinases. Cancer Lett. 2014;346(2):264–272. doi: 10.1016/j.canlet.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Win N.N., Ito T., Ismail Kodama T., Win Y.Y., Tanaka M. Picrajavanicins A-G, quassinoids from Picrasma javanica collected in myanmar. J Nat Prod. 2015;78(12):3024–3030. doi: 10.1021/acs.jnatprod.5b00824. [DOI] [PubMed] [Google Scholar]
- 39.Win N.N., Ito T., Ismail Tondra I., Kodama T. Picrajavanicins H-M, new quassinoids from Picrasma javanica collected in myanmar and their antiproliferative activities. Tetrahedron. 2016;72(5):746–752. [Google Scholar]
- 40.Prema, Wong C.P., Nugroho A.E., Awouafack M.D., Win Y.Y., Win N.N. Two new quassinoids and other constituents from Picrasma javanica wood, and their biological activities. J Nat Med. 2019;73(3):589–596. doi: 10.1007/s11418-018-01279-z. [DOI] [PubMed] [Google Scholar]
- 41.Ye Q.M., Bai L.L., Hu S.Z., Tian H.Y., Ruan L.J., Tan Y.F. Isolation, chemotaxonomic significance and cytotoxic effects of quassinoids from Brucea javanica. Fitoterapia. 2015;105:66–72. doi: 10.1016/j.fitote.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 42.Robinson M.J., Cobb M.H. Mitogen-activated protein kinase pathways. Curr Opin Cell Biol. 1997;9(2):180–186. doi: 10.1016/s0955-0674(97)80061-0. [DOI] [PubMed] [Google Scholar]
- 43.Yang L., Sun X.T., Ye Y., Lu Y.T., Zuo J., Liu W. P38α mitogen-activated protein kinase is a druggable target in pancreatic adenocarcinoma. Front Oncol. 2019;9:1294. doi: 10.3389/fonc.2019.01294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kiozumi K., Tanno S., Nakano Y., Habiro A., Izama T., Mizukami Y. Activation of p38 mitogen-activated protein kinase is necessary for gemcitabine-induced cytotoxicity in human pancreatic cancer cells. Anticancer Res. 2005;25:3347–3353. [PubMed] [Google Scholar]
- 45.Burris H.A., Moore M.J., Andersen J., Green M.R., Rothenberg M.L., Modiano M.R. Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: a randomized trial. J Clin Oncol. 1997;15(6):2403–2413. doi: 10.1200/JCO.1997.15.6.2403. [DOI] [PubMed] [Google Scholar]
- 46.Korc M. P38 MAPK in pancreatic cancer: finding a protective needle in the haystack. Clin Cancer Res. 2014;20(23):5866–5868. doi: 10.1158/1078-0432.CCR-14-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhong Y., Naito Y., Cope L., Naranjo-Suarez S., Saunders T., Hong S.M. Functional p38 MAPK identified by biomarker profiling of pancreatic cancer restrains growth through JNK inhibition and correlates with improved survival. Clin Cancer Res. 2014;20(23):6200–6211. doi: 10.1158/1078-0432.CCR-13-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Neidhardt M.M., Schmitt K., Baro A., Schneider C., Bilitewski U., Laschat S. Self-assembly and biological activities of ionic liquid crystals derived from aromatic amino acids. Phys Chem Chem Phys. 2018;20(31):20371–20381. doi: 10.1039/c8cp03404d. [DOI] [PubMed] [Google Scholar]
- 49.Lee B., Richards F.M. The interpretation of protein structures: estimation of static accessibility. J Mol Biol. 1971;55(3):379–IN4. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.