Abstract
EGFR signaling promotes ovarian cancer tumorigenesis, and high EGFR expression correlates with poor prognosis. However, EGFR inhibitors alone have demonstrated limited clinical benefit for ovarian cancer patients, owing partly to tumor resistance and the lack of predictive biomarkers. Cotargeting EGFR and the PI3K pathway has been previously shown to yield synergistic antitumor effects in ovarian cancer. Therefore, we reasoned that PI3K may affect cellular response to EGFR inhibition. In this study, we revealed PI3K isoform-specific effects on the sensitivity of ovarian cancer cells to the EGFR inhibitor erlotinib. Gene silencing of PIK3CA (p110α) and PIK3CB (p110β) rendered cells more susceptible to erlotinib. In contrast, low expression of PIK3R2 (p85β) was associated with erlotinib resistance. Depletion of PIK3R2, but not PIK3CA or PIK3CB, led to increased DNA damage and reduced level of the nonhomologous end joining DNA repair protein BRD4. Intriguingly, these defects in DNA repair were reversed upon erlotinib treatment, which caused activation and nuclear import of p38 MAPK to promote DNA repair with increased protein levels of 53BP1 and BRD4 and foci formation of 53BP1. Remarkably, inhibition of p38 MAPK or BRD4 re-sensitized PIK3R2-depleted cells to erlotinib. Collectively, these data suggest that p38 MAPK activation and the subsequent DNA repair serve as a resistance mechanism to EGFR inhibitor. Combined inhibition of EGFR and p38 MAPK or DNA repair may maximize the therapeutic potential of EGFR inhibitor in ovarian cancer.
Keywords: Epidermal growth factor receptor, Erlotinib, PI3K, p85 regulatory subunit, DNA repair, p38 MAPK
Introduction
Epidermal growth factor receptor (EGFR) is a transmembrane receptor tyrosine kinase (RTK) that is overexpressed in >50% of ovarian cancer patients [[1], [2], [3], [4]]. EGFR signaling promotes ovarian tumorigenesis including proliferation, migration and angiogenesis [5,6]. Importantly, high EGFR expression is significantly associated with poor patient survival outcomes [7,8]. Together, these findings provide a rationale for the use of EGFR inhibitors in ovarian cancer therapy. However, studies to date have shown that ovarian cancer patients exhibited a partial or complete response rate of <10% to the EGFR inhibitor erlotinib or gefitinib [9], [10], [11]. The limited responses may be attributable to several reasons. First, the clinical trials conducted did not use biomarker stratification. In lung cancer, EGFR inhibitors were selectively given to patients with EGFR mutations that are associated with sensitivity (e.g., exon 19 deletion and the exon 21 L858R mutant) [12,13]. The objective response rate for the EGFR mutation-positive group was 71%, compared with 1% for the mutation-negative group [14]. The second reason, which is related to the first reason, is the lack of biomarkers that can be used to stratify patients who will likely respond. Most ovarian cancer patients do not have EGFR mutations that confer sensitivity to EGFR inhibitors [15,16]. Additionally, EGFR amplification or expression does not predict responsiveness [11,17]. Third, resistance of cancer cells to EGFR inhibition may occur, but the resistance mechanism remains understudied in ovarian cancer.
A major downstream signaling pathway of EGFR is the phosphoinositide 3-kinase (PI3K). The activating PIK3CA mutant p110α E545K has been shown to confer resistance of EGFR inhibitor in lung cancer [18]. Indeed, simultaneous inhibition of EGFR and the PI3K pathway has demonstrated synergistic antitumor effects in cancers, including ovarian cancer [19,20], implying an association between the PI3K pathway and sensitivity to EGFR inhibition. The class I PI3K is a heterodimeric lipid kinase composed of a catalytic p110 subunit and a regulatory subunit [21]. The heterodimer is activated upon RTK stimuli to catalyze phosphatidylinositol 3,4,5-trisphosphate production, thereby triggering downstream signaling such as the AKT/mTOR pathway. There are 4 catalytic subunit isoforms and 5 regulatory subunit isoforms. Gene aberrations, particularly copy number alterations of these 9 isoforms, are very frequent in ovarian cancer [15]. Intriguingly, accumulating evidence has demonstrated PI3K isoform-specific functions in cancers [22], [23], [24]. For example, p110α mediates RTK signaling, whereas p110β is downstream of G protein-coupled receptors [25]. We and others have shown that while p85α is a tumor suppressor [24,26,27], p85β is an oncogenic factor that promotes the acquisition of tumorigenic phenotypes[28,29]. Inhibition of p85β reduced the proliferation and metastasis of ovarian cancer cells [29]. We therefore explored the interaction between the PI3K isoforms and EGFR inhibitor in ovarian cancer. Strikingly, our findings revealed differential impact of the isoforms on the sensitivity to erlotinib. Inhibition of p110α (PIK3CA) and p110β (PIK3CB) potentiated the cytotoxicity of EGFR inhibitors. In contrast, low expression of p85β (PIK3R2) led to EGFR resistance, which was associated with p38 MAPK activation and enhanced DNA repair capacity. The combination of erlotinib with a p38 MAPK inhibitor induced DNA damage and apoptosis of ovarian cancer cells. These findings suggest that DNA repair efficiency is a determinant of the responsiveness to EGFR inhibition and have implications for the design of effective cancer therapies.
Materials and methods
Cell culture, siRNA and reagents
CAOV3, OVCAR3, SKOV3 were acquired from American Type Culture Collection (Manassas, VA). OVCAR4, OVCAR8, EFO21 and DOV13 were obtained from National Cancer Institute (Rockville, MD). OAW28 was obtained from the European Collection of Authenticated Cell Cultures (Salisbury, United Kingdom). FUOV1, HEYA8 and OVCAR5 were generously provided by Prof. Gordon Mills (Oregon Health Sciences University, Portland, OR). SKOV3, OVCAR3, OVCAR4, CAOV3, OVCAR5 and OVCAR8, DOV13, EFO21 FUOV1, HEYA8 were cultured in RPMI-1640 (Gibco, Carlsbad, CA) whereas OAW28 was grown in DMEM (Gibco) supplemented with 5% FBS (Gibco), 100 units/mL penicillin and 0.1 mg/mL streptomycin (Gibco). All cells were maintained at 37°C in a 5% CO2 incubator. All cell lines were authenticated using short tandem repeat DNA profiling and tested with negative mycoplasma contamination.
Pre-designed ON-TARGET plus siRNAs were purchased from Dharmacon (Lafayette, CO). Transfection of siRNA was carried out using Lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA) following the manufacturer's instructions. All siRNAs were used at a final concentration of 20 nM. Stable cells expressing PIK3R2 shRNA were established by lentiviral transduction and puromycin selection. The sequences of siRNAs and shRNA are provided in Supplementary Table 1. Human PIK3R2 expression plasmid was constructed by Gateway cloning into pLenti6.3-DEST vector (Invitrogen). Erlotinib and SB202190 were purchased from LC laboratories (Woburn, MA). Losmapimod and JQ1 were purchased from ApexBio Technology (Houston, TX). JNK-IN-8, BI-78D3, GDC00994 and trametinib were obtained from Selleck Chemicals (Houston, TX). ABT-888 was obtained from Santa Cruz Biotechnology (Santa Cruz, CA).
Western blotting
Cells were lysed with RIPA buffer (150 mM NaCl, 0.1% SDS, 1% NP-40, 1% sodium deoxycholate) supplemented with protease and phosphatase inhibitors (Thermo Fisher Scientific, Waltham, MA). Total protein concentration was quantified using PierceTM BCA Protein Assay (Thermo Fisher Scientific). Protein lysates were separated on 6-12% SDS-PAGE and transferred to PVDF membranes (GE Healthcare Life Sciences, Marlborough, MA). Membranes were blocked by 5% non-fat milk in TBST for 1 h and incubated overnight with primary antibodies at 4°C. After washing with TBST, membranes were incubated with HRP-conjugated secondary antibodies (1:5000, GE Healthcare Life Sciences) and signal was detected by chemiluminescence (Bio-Rad, Hercules, CA). All primary antibodies used in this study are listed in Supplementary Table 2. Image J was used for densitometric analysis.
Cell viability assay
Cells were seeded into 96-well plate at a density of 2,500 cells per well in triplicate. After 24 h, cells were treated with inhibitors for 72 h. For quantitation of viable cells, one tenth volume (10 μl per 100 μl) of 0.2 mg/ml Resazurin solution (Sigma-Aldrich, St. Louis, MO) was added to each well. Cells were incubated at 37°C for 1.5 h. Absorbance readings at 570 nm were obtained.
Mouse xenograft model
SKOV3 cells stably expressing PIK3R2 shRNA (7.5 × 106) or vector control (2 × 106) were injected intraperitoneally into 6-wk-old female nude mice (Charles River Lab, USA). Three weeks after cell inoculation, mice were randomly divided into subgroups (5 mice per group) and treated by oral gavage with drug suspension or vehicle 5 times a week for 2 consecutive weeks. Erlotinib was dissolved in 15% Captisol (CyDex Pharmaceuticals, Lawrence, KS) and losmapimod was dissolved in PBS with 50% PEG400 and 5% DMSO. Mice were euthanized for the collection of disseminated tumor nodules in the peritoneal cavity. Tumor burden based on the quantity and weight of tumor nodules was compared among subgroups. All animal procedures were carried out with the approval of the Ethics Committee (Committee on the Use of Live Animals in Teaching and Research) of the University of Hong Kong.
Cell cycle analysis
Cells (8 × 105) were serum starved overnight for synchronization prior to siRNA transfection and drug treatment for 72 h. On the day of harvest, cells were trypsinized and washed twice with cold PBS before fixation in cold 70% ethanol. Cells were then resuspended in RNase A solution (50 μg/mL; Sigma-Aldrich, St. Louis, MO) and incubated at 37°C for 30 min. Cells were stained with propidium iodide (PI; 20 μg/mL; Sigma-Aldrich) for 30 min in dark and analyzed using flow cytometer Canto II analyzer (BD Biosciences, San Jose, CA). Cell cycle distribution was analyzed by FlowJo software (Tree Star Inc, Ashland, OR).
Apoptosis analysis
Apoptosis was detected by Human Cleaved Caspase-3 (Asp175) ELISA kit (Abcam, Cambridge, UK) following the manufacturer's instructions. Briefly, cells were washed with PBS and lysed with cell extraction buffer. Samples and cleaved caspase-3 (Asp175) standards were added to wells in microplate strips in triplicate. Then an antibody cocktail containing capture and detector antibodies was incubated with the samples for 1 h prior to the addition of TMB development solution and stop solution. Absorbance readings were measured at 450 nm.
Immunofluorescence and foci formation assay
SKOV3 cells (4 × 104) transfected with PIK3R2 siRNA were grown on sterilized glass coverslips for immunofluorescence (IF) microscopy. Twenty four hours after transfection, cells were treated as indicated for 3 d. Cells were fixed with 4% paraformaldehyde and permeabilized in 0.1% Triton X-100. Cells were then incubated with 3% BSA at room temperature for blocking non-specific signal. Samples were incubated with primary antibody overnight prior to Alexa Fluor 488-conjugated anti-rabbit secondary antibody or Alexa Fluor 594-conjugated anti-mouse secondary antibody (1:200, Thermo Fisher Scientific) for 1 hr in dark. After washing 3 times in PBS, slides were mounted on glass slide in mounting medium with DAPI. Images were captured under Carl Zeiss LSM 700 (Zeiss, Jena, Germany). Subnuclear foci formation of DNA damage repair proteins 53BP1 and RAD51 were analyzed by IF microscopy. Foci per cell were counted. For each data point, at least 80 cells were scored from 4-6 randomly captured images using an objective magnification of × 20.
Comet assay
PIK3R2 shRNA stable cells were treated with 10 μM erlotinib for 24 h or 72 h prior to assessment of DNA damages by alkaline comet assay using CometAssay Kit (Trevigen, Gaithersburg, MD) according to manufacturer's instructions. In brief, cells were suspended in cold PBS, combined with molten LMAgarose at a ratio of 1:10 (v/v) and were immediately transferred onto CometSlide. After overnight incubation in lysis solution at 4°C, slides were immersed in alkaline unwinding solution (200 mM NaOH, 1 mM EDTA, pH > 13) for 1 h in the dark. Electrophoresis was performed using alkaline unwinding solution at 17V for 35 min in the dark. Samples were then washed twice in water and 70% ethanol. Dried samples were stained in diluted SYBR Gold (1:30000). Pictures were taken using a Zeiss Axiophot fluorescence microscope. At least 50 comets were assessed per treatment condition. Percentage of DNA in comet tail (% tail DNA) were calculated by image processing software package Fiji using OpenComet plugin.
Copy number variations data from TCGA
Copy number status of each gene, which was determined by the GISTIC algorithm [30], was retrieved from the cBioPortal [15]. A GISTIC output of 0 indicates diploid; -2 indicates deep deletion (possibly a homozygous deletion); -1 indicates shallow deletion (possibly a heterozygous deletion); 1 indicates a gain; 2 indicates gene amplification.
Statistical analysis
The experiments were repeated 3 times. No statistical methods were used to predetermine sample size. No samples or animals were excluded from the analysis. The experiments were not randomized and were not carried out blind. No samples or animals were excluded from the analysis. All data were presented as means ± SD (or SEM for the in vivo data). Significance between 2 groups was determined by 2 sided Student's t test using GraphPad Prism. Analysis of variance (ANOVA) was used to determine statistical significance of difference among multiple groups. Differences were considered significant at P < 0.05.
Results
PI3K isoforms have distinct impacts on erlotinib sensitivity
Nine class I PI3K isoforms (PIK3CA, PIK3CB, PIK3CD, PIK3CG, PIK3R1, PIK3R2, PIK3R3, PIK3R5 and PIK3R6) in TCGA ovarian cancer samples were frequently aberrated at the genomic level (Supplementary Fig. 1A). Copy number alterations of these isoforms are more frequent than small-nucleotide mutations, consistent with the fact that ovarian cancer is primarily driven by copy number changes [31]. We first assessed the responses of 11 ovarian cancer cell lines to erlotinib, which is FDA-approved and a first-line treatment in EGFR-mutant non-small cell lung cancer. Total and phosphorylated protein levels of EGFR are ubiquitous in these cell lines, although lower EGFR expression was observed in DOV13, EFO21 and FUOV1 cells (Supplementary Fig. 1B). One cell line (CAOV3) carries EGFR mutation, while mutation in PIK3CA, PIK3CB or PIK3R1 are present in 3 cell lines (Supplementary Table 3). These cells were treated with increasing doses of erlotinib, from 3.2 nM to 50 µM. Six of the 11 cell lines were relatively resistant to erlotinib because the IC50 (the drug concentration that causes 50% growth inhibition) was not reached even at 50 µM (Supplementary Fig. 1C). Indeed, erlotinib at 10 µM and 50 µM caused similar effects on cell viability. The effectiveness of erlotinib in inhibiting EGFR and AKT activation was confirmed in 2 resistant cell lines and 2 sensitive cell lines (Supplementary Fig. 1D). The levels of phosphorylated EGFR and AKT were decreased upon erlotinib treatment in all these cell lines.
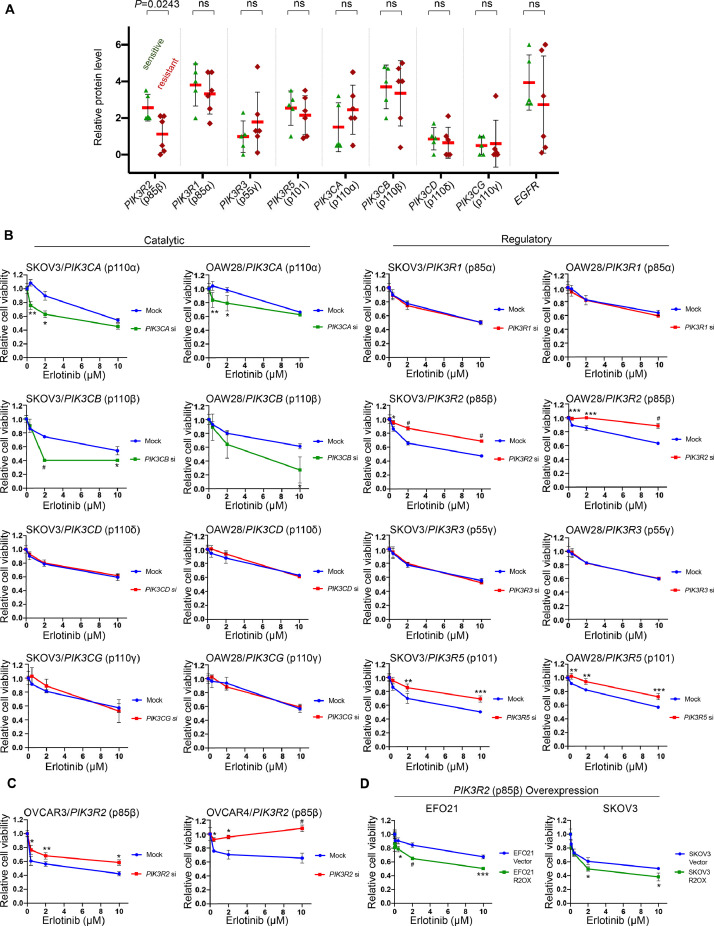
The protein levels of the PI3K isoforms in these cell lines were assessed (Supplementary Fig. 1B). PIK3R6 was excluded from the analysis due to the lack of an antibody that showed a specific band in our experiments. The correlation of the protein levels with erlotinib responsiveness was evaluated. Interestingly, cell lines that were relatively sensitive to erlotinib (those that exhibited ≥50% growth inhibition) had higher p85β protein levels than resistant cell lines (P < 0.05) (Fig. 1A). There was no significant difference in the levels of the other isoforms between erlotinib-sensitive and erlotinib-resistant cell lines (Fig. 1A).
Fig. 1.
PIK3R2 depletion confers erlotinib resistance in ovarian cancer.
(A) Correlation between the protein levels of EGFR or class 1A PI3K isoforms with cellular responses to erlotinib was evaluated. The protein levels are presented as the mean densitometry values of Western blotting data obtained from 3 independent experiments (representative blots are shown in Supplementary Figure 1B). Cell lines that showed ≥50% viability inhibition after 10 μM erlotinib treatment were defined as “sensitive”, whereas cell lines whose viability inhibition did not reach 50% were “resistant” (data are shown in Supplementary Figure 1C). P values were determined by 2-tailed t test; ns, not statistically significant. (B) SKOV3 or OAW28 cells were transfected with siRNA targeting individual PI3K isoforms for 24 h prior to treatment with erlotinib for another 72 h. The data presented show the cell viability relative to the corresponding untreated cells in 3 independent experiments performed in triplicate. (C-D) (C) PIK3R2 siRNA-transfected OVCAR3 and OVCAR4 cells or (D) EFO21 and SKOV3 cells overexpressing PIK3R2 were treated with erlotinib for 72 h before being subjected to a cell viability assay. The data presented show the cell viability relative to the corresponding untreated cells in 3 independent experiments performed in triplicate. *, P < 0.05; **, P < 0.005; ***, P < 0.001; #, P < 0.0001 by 2-way ANOVA with Sidak's multiple comparison test.
To further investigate the gene-drug interaction between erlotinib and these PI3K isoforms, the effect of individual gene knockdown on erlotinib sensitivity was examined in SKOV3 and OAW28 cells, which express EGFR and have moderate sensitivity to erlotinib among the cell lines. The efficiencies of the siRNAs were confirmed by Western blotting (Supplementary Fig. 2A). Fig. 1B shows the responses of mock-transfected and gene-specific siRNA-transfected cells to erlotinib, normalized to those of the corresponding transfected cells treated with DMSO. Depletion of the catalytic subunits p110α (PIK3CA) and p110β (PIK3CB) led to increased sensitivity, consistent with previous reports demonstrating that p110 inhibitor sensitized non-small cell lung cancer cells to EGFR inhibitor [20]. In contrast, cells with silenced regulatory subunits p85β (PIK3R2) or p101 (PIK3R5) became more resistant to erlotinib. Erlotinib sensitivity was independent of the other 4 isoforms (PIK3R1, PIK3R3, PIK3CD, and PIK3CG). Because the levels of p110α (PIK3CA) and p110δ (PIK3CD) are relatively low in SKOV3 cells, we confirmed the effects of these 2 genes in OVCAR3 cells, which express these isoforms. Consistent with the data obtained from SKOV3 and OAW28 cells, knocking down PIK3CA but not PIK3CD enhanced erlotinib sensitivity of OVCAR3 cells (Supplementary Fig. 2B-C).
The data shown in Fig. 1A and 1B prompted us to confirm the effect of PIK3R2 on erlotinib sensitivity. Consistently, PIK3R2 siRNA conferred erlotinib resistance in additional ovarian cancer cell lines OVCAR3 and OVCAR4 (Fig. 1C). In reciprocal experiments, we overexpressed p85β in SKOV3 cells and in a cell line expressing low endogenous p85β level (EFO21). The overexpression of p85β was confirmed by Western blotting (Supplementary Fig. 3). Importantly, the overexpression sensitized these cells to erlotinib (Fig. 1D). Our data therefore indicate that PIK3R2 alters erlotinib response of ovarian cancer.
Cells with reduced sensitivity to erlotinib upon PIK3R2 depletion show activation of MAPK signaling
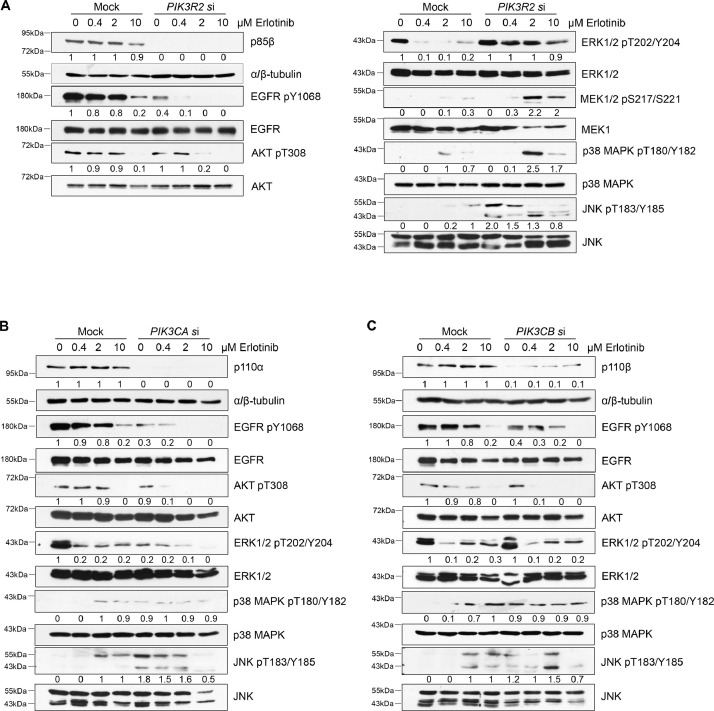
We attempted to elucidate the resistance mechanism conferred by PIK3R2 depletion. We also included cells transfected with PIK3CA and PIK3CB siRNA because silencing the 2 genes sensitized cells to erlotinib. Western blotting revealed that EGFR phosphorylation was reduced in untreated PIK3R2- or PIK3CA- or PIK3CB-depleted cells (Fig. 2A−C). Erlotinib treatment induced further reduction of EGFR phosphorylation in these cells, indicating the effectiveness of EGFR inhibition (Fig. 2A and 2B).
Fig. 2.
MAPK signaling is activated upon erlotinib treatment in cells with depleted PIK3R2 but not depleted PIK3CA or PIK3CB.
(A-C) SKOV3 cells were transfected with siRNA targeting (A) PIK3R2, (B) PIK3CA or (C) PIK3CB for 24 h prior to treatment with erlotinib for an additional 72 h. Protein lysates were collected for Western blotting with the indicated antibodies. Representative blots of 3 independent experiments are shown. The numbers below the blots indicate the mean densitometry values normalized to those of α/β-tubulin or to the corresponding total proteins (for phosphoproteins) of the 3 experiments.
The effects of erlotinib on the 2 major downstream signaling pathways of EGFR, the AKT and MAPK pathways, were then evaluated. First, consistent with the data in Supplementary Fig. 1D, phosphorylated AKT was decreased in all the transfected cells upon erlotinib treatment. Second, the phosphorylation of ERK1/2 was decreased by erlotinib in mock cells and similarly in PIK3CB-depleted cells. PIK3CA knockdown alone reduced ERK1/2 phosphorylation, and erlotinib caused a further reduction. In contrast, PIK3R2-depleted cells had sustained ERK1/2 phosphorylation in the presence of erlotinib. Activation of MEK1/2 by erlotinib was also observed in PIK3R2-depleted cells. Third, p38 MAPK was activated by PIK3CA or PIK3CB knockdown alone but was not further activated by erlotinib. p38 MAPK activation was, however, robustly activated by erlotinib in PIK3R2-depleted cells. Finally, the mock-transfected cells displayed increase in phosphorylated JNK upon erlotinib treatment. Silencing PIK3R2, PIK3CA or PIK3CB alone without erlotinib led to JNK activation, and the addition of erlotinib (10 μM) to these knockdown cells inhibited this activation. Together, these data demonstrated distinctive activation of ERK1/2 and p38 MAPK in PIK3R2-depleted cells upon erlotinib treatment.
Inhibition of p38 MAPK sensitizes PIK3R2-depleted ovarian cancer cells to erlotinib
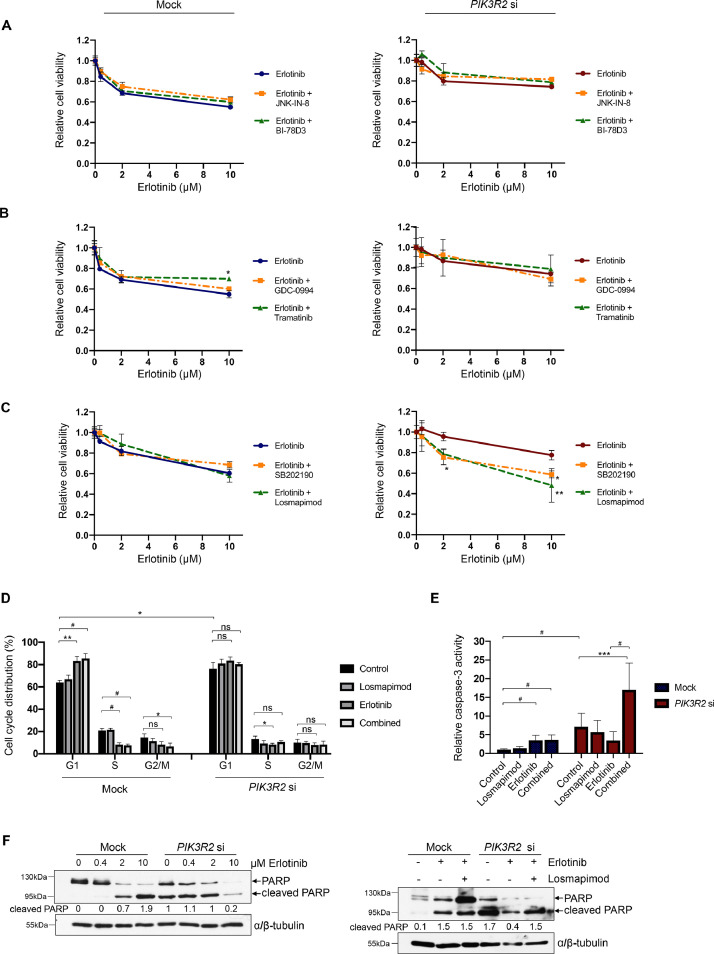
We next examined whether inhibition of the MAPK pathways restores the sensitivity of PIK3R2-depleted ovarian cancer cells to erlotinib. Our results showed that inhibitors of JNK (JNK-IN-8 [32] and BI-78D3 [33]), ERK1/2 (GDC-0994 [34]) and MEK1/2 (trametinib [35]) did not affect erlotinib sensitivity in PIK3R2-depleted cells (Fig. 3A and 3B). We observed an increase in the viability of the mock cells upon trametinib addition (Fig. 3B), and this finding warrants future investigation. Interestingly, p38 MAPK inhibitor treatment (SB202190 [36] and losmapimod [37]) conferred erlotinib sensitivity to PIK3R2-depleted cells, whereas the mock control cells were not sensitized (Fig. 3C).
Fig. 3.
Inhibitors of p38 MAPK render PIK3R2-silenced cells sensitive to erlotinib.
(A-C) SKOV3 cells transfected with or without PIK3R2 siRNA were treated with erlotinib alone or in combination with (A) JNK inhibitors (JNK-IN-8 and BI-78D3, 2 μM), (B) an ERK1/2 inhibitor (GDC-0994, 2 μM) or a MEK1/2 inhibitor (trametinib, 2 μM) or (C) p38 MAPK inhibitors (SB202190 and losmapimod, 10 μM) for 72 h. The data presented show the cell viability relative to the corresponding untreated cells in 3 independent experiments performed in triplicate. (D-E) SKOV3 cells transfected with or without PIK3R2 siRNA were treated with erlotinib alone (10 μM), the p38 MAPK inhibitor losmapimod alone (10 μM) or both in combination (10 μM each) for 72 h. The cells were harvested for (D) cell cycle analysis by flow cytometry or (E) active cleaved caspase-3 enzyme-linked immunosorbent assay. (F) SKOV3 cells transfected with or without PIK3R2 siRNA were treated with erlotinib alone or in combination with losmapimod for 72 h. These cells were harvested for Western blotting. α/β-tubulin was used as a loading control. The error bars represent the SD of 3 independent experiments performed in triplicate. *, P < 0.05; **, P < 0.005; ***, P < 0.001; #, P < 0.0001 by 2-way ANOVA test for cell viability data (Sidak's multiple comparison) and cell cycle data (Tukey's multiple comparison). Statistical analysis of the caspase-3 data was performed using ordinary one-way ANOVA for analysis within group (mock or PIK3R2 siRNA) and 2-way ANOVA for comparison between groups (mock vs PIK3R2 siRNA) with Sidak's multiple comparison test.
The effect of erlotinib on the cell cycle distribution of these cells was analyzed by flow cytometry. Previous studies have shown that erlotinib induces G1 phase arrest and apoptosis of erlotinib-sensitive cancer cells [38,39]. As expected, erlotinib significantly increased the proportion of control cells in the G1 phase (from 64% to 83%), with decreases in the S phase (from 21 to 9%) and G2 phase (15 to 8%) (Fig. 3D). In line with decrease in EGFR activation (Fig. 2A), a greater proportion of PIK3R2-depleted cells were in the G1 stage than control cells (64% to 77%), indicating that PIK3R2 depletion causes G1 phase cell cycle arrest. In contrast to its effect on the control cells, erlotinib had no prominent effect on the cell cycle phases of PIK3R2-depleted cells. Erlotinib only slightly increased the proportion in G1 phase (from 77% to 84%), with modest decreases in the proportion in S phase (14% to 8%) and G2 phase (from 10 to 8%). The proportion of cells in G1 phase due to PIK3R2 depletion may lead to a compromised inhibitory effect of erlotinib on cell cycle progression. Moreover, losmapimod treatment had minimal effect on cell distribution in the cell cycle. Therefore, the viability of PIK3R2-depleted cells in response to erlotinib is cell cycle-independent.
Apoptosis is a cell death pathway contributing to cytotoxicity. Erlotinib triggered apoptosis in mock control cells, but combined treatment with losmapimod did not increase apoptosis of these cells (Fig. 3E). A greater number of untreated PIK3R2-depleted cells underwent apoptosis compared with their wild-type counterparts, and this effect was reduced upon erlotinib treatment. Concurrent with our finding that inhibiting p38 MAPK sensitized PIK3R2-depleted cells to erlotinib, combination treatment of erlotinib and losmapimod led to a marked increase in apoptosis (a 2.4-fold and 4.9-fold increase compared with untreated and erlotinib-treated PIK3R2-depleted cells, respectively). We also observed corresponding changes in the protein levels of cleaved PARP, which is an apoptosis marker (Fig. 3F). Collectively, these data indicated that erlotinib induces G1 phase arrest and apoptosis of erlotinib-sensitive mock control cells. In contrast, reduction in apoptosis underlies the resistance of PIK3R2-depleted cells to erlotinib.
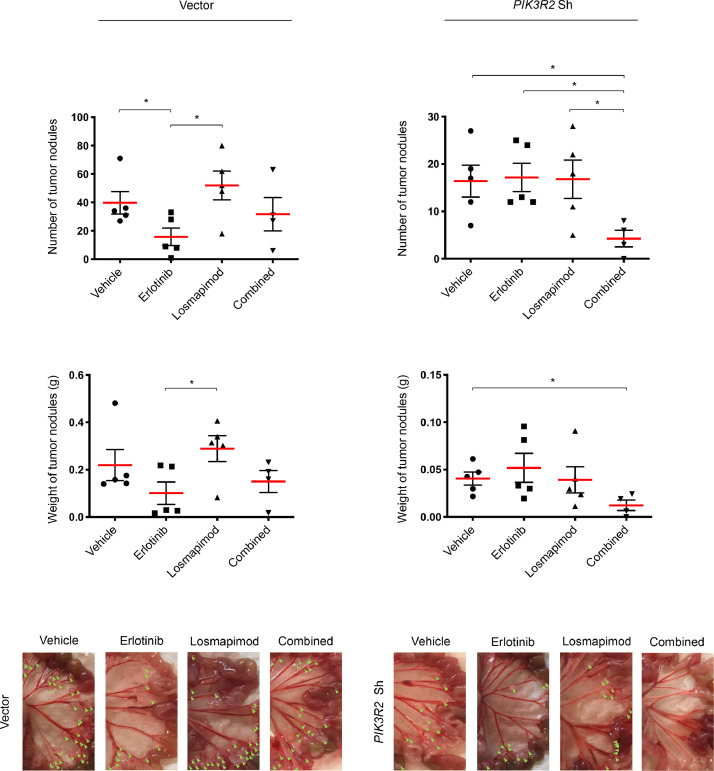
We next investigated whether the in vitro drug responses are reproducible in mouse xenograft models established from cells with stable shRNA-mediated PIK3R2 knockdown. Consistent with the behavior of the PIK3R2 siRNA-transfected cells, the cells displayed reduced sensitivity to erlotinib in vitro (Supplementary Fig. 4A) and increased phosphorylation of p38 MAPK upon erlotinib treatment (Supplementary Fig. 4B). Knocking down PIK3R2 reduced tumor growth in vivo, as previously reported (Fig. 4) [29]. Single treatment with erlotinib, but not single treatment with losmapimod, reduced tumor formation by the vector control xenografts (Fig. 4). Xenograft tumors formed by PIK3R2-depleted cells were not responsive to erlotinib, concordant with the observed resistance in vitro. Strikingly, the combination of erlotinib and losmapimod significantly reduced both nodule numbers and tumor weight compared with the vehicle group. Notably, these effects upon the combination treatment were not observed in the vector control xenografts. These findings indicated that p38 MAPK inhibition can overcome erlotinib insensitivity in PIK3R2-depleted cells. Future in vivo investigations with larger sample size are warranted to further explore the antitumor effects of the drug combination.
Fig. 4.
p38 MAPK inhibitor losmapimod re-sensitizes PIK3R2-depleted tumors to erlotinib in vivo.
SKOV3 cells stably expressing vector control or PIK3R2 shRNA were injected i.p. into nude mice. Tumor-bearing mice (n=5) were randomly selected for treatment with vehicles, erlotinib alone (50 mg/kg/d), the p38 MAPK inhibitor losmapimod alone (12 mg/kg/d) or both drugs in combination for 2 wk. Bar graphs of tumor nodule number and tumor weight are presented. Representative images show peritoneal disseminated tumor nodules as indicated by green arrows. The error bars represent SEM. *, P < 0.05 based on 2-tailed t test.
DNA repair capability is reduced in PIK3R2-depleted cells but is enhanced upon erlotinib treatment
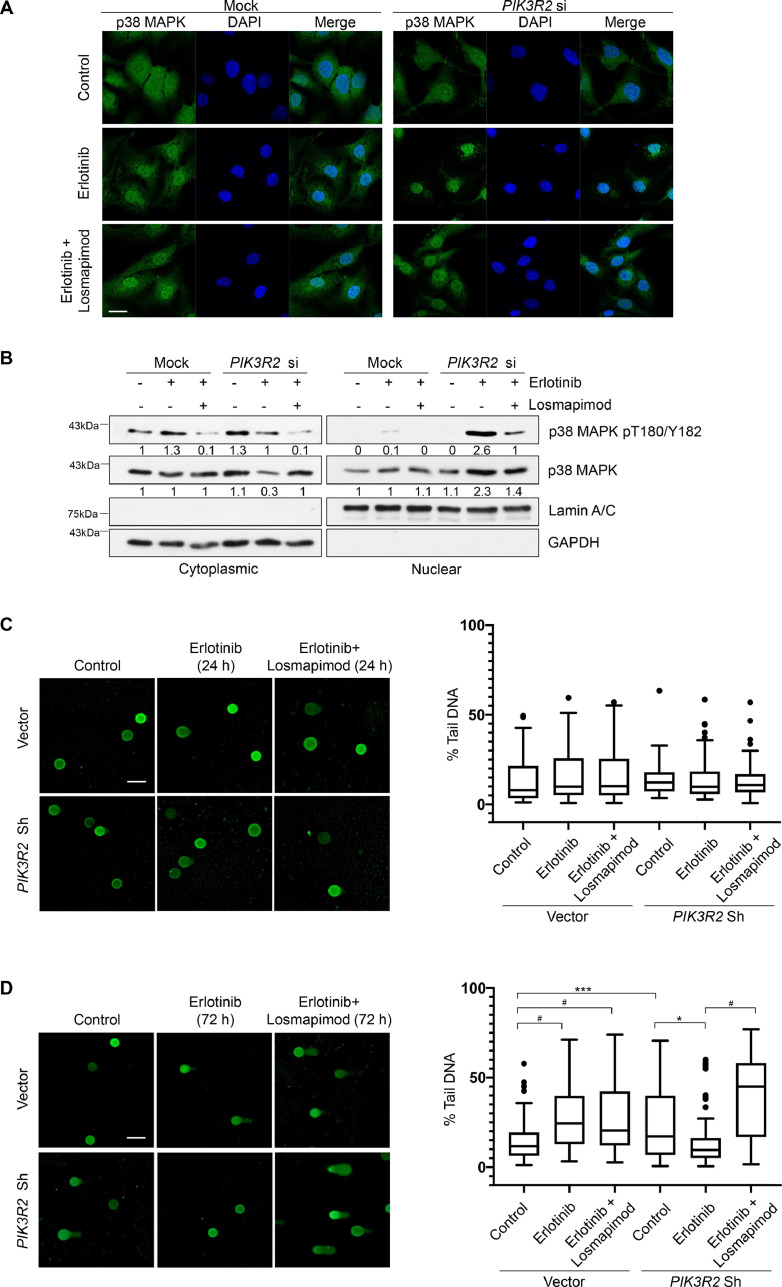
p38 MAPK signaling involves in a number of cellular processes, including cancer cell behaviors, genomic stability and inflammation [40,41]. It has been suggested that the subcellular localization of p38 MAPK associates with the type of stimulating signal, with its nuclear translocation being predominantly induced upon DNA damage [42]. By immunofluorescence staining, we detected cytoplasmic and nuclear p38 MAPK in both control and PIK3R2-depleted cells (Fig. 5A). Strikingly, treatment of PIK3R2-depleted cells with erlotinib led to prominent nuclear localization of p38 MAPK, which was reversed by the addition of losmapimod. The localization of p38 MAPK in control cells was not altered by erlotinib or losmapimod. These observations were further confirmed by subcellular fractionation (Fig. 5B). Level of phosphorylated p38 MAPK in the nucleus was markedly increased in PIK3R2-depleted cells upon erlotinib treatment.
Fig. 5.
Insensitivity to erlotinib in PIK3R2-depleted cells correlates with p38 MAPK nuclear translocation and p38 MAPK-mediated DNA repair.
(A-B) SKOV3 cells transfected with or without PIK3R2 siRNA were treated with erlotinib alone (10 μM) or in combination with losmapimod (10 μM) for 72 h. (A) An anti-p38 MAPK antibody (green) and DAPI (blue) were used for immunofluorescence staining. Scale bar, 20 μm. (B) Cells were subjected to subcellular fractionation. Representative blots of 3 independent experiments are shown. The numbers below the blots indicate the mean densitometry values normalized to those of GAPDH (cytoplasmic marker) or lamin A/C (nuclear marker). (C-D) SKOV3 cells stably expressing PIK3R2 shRNA or vector control were treated with erlotinib alone (10 μM) or in combination with losmapimod (10 μM) for (C) 24 h or (D) 72 h. DNA damage was visualized by comet assay (Left). Scale bar, 100 μm. The amount of damaged DNA was expressed as percentage of DNA in comet tail (% tail DNA) (right). The box and whisker plot was plotted by Graphpad Prism software using the Tukey method. *, P < 0.05; **, P < 0.005; ***, P < 0.001; #, P < 0.0001 using ordinary one-way ANOVA for analysis within group (vector or PIK3R2 shRNA) and 2-way ANOVA for comparison between groups (vector vs PIK3R2 shRNA) with Sidak's multiple comparison test.
The reported role of p38 MAPK in DNA damage response (DDR) and the nuclear staining observed prompted us to assess cellular DNA repair indirectly using comet assays. As shown by data obtained from stable PIK3R2 knockdown cells at an early time point (24 h), the amount of DNA damage was not significantly different between untreated cells and cells with the indicated treatments (Fig. 5C). At the late time point (72 h), the untreated stable PIK3R2 knockdown cells displayed remarkably more damaged DNA, as reflected by higher percent DNA in comet tail, suggesting inefficient DNA damage repair (Fig. 5D). Strikingly, erlotinib treatment led to a significant reduction in unrepaired DNA in these cells that correlated with the nuclear shuffling of p38 MAPK. This reduction was reversed in the presence of the p38 MAPK inhibitor losmapimod (Fig. 5D) or SB202190 (Supplementary Fig. 5), confirming that the DNA repair is mediated by p38 MAPK. Remarkably, these responses were not observed in the control cells, which indeed showed increased DNA damage upon erlotinib treatment, consistent with previous reports [43]. It has been shown that DNA damage induces apoptosis [44]. Intriguingly, the extent of DNA damage demonstrated in the cells echoed that of apoptosis (Fig. 3E).
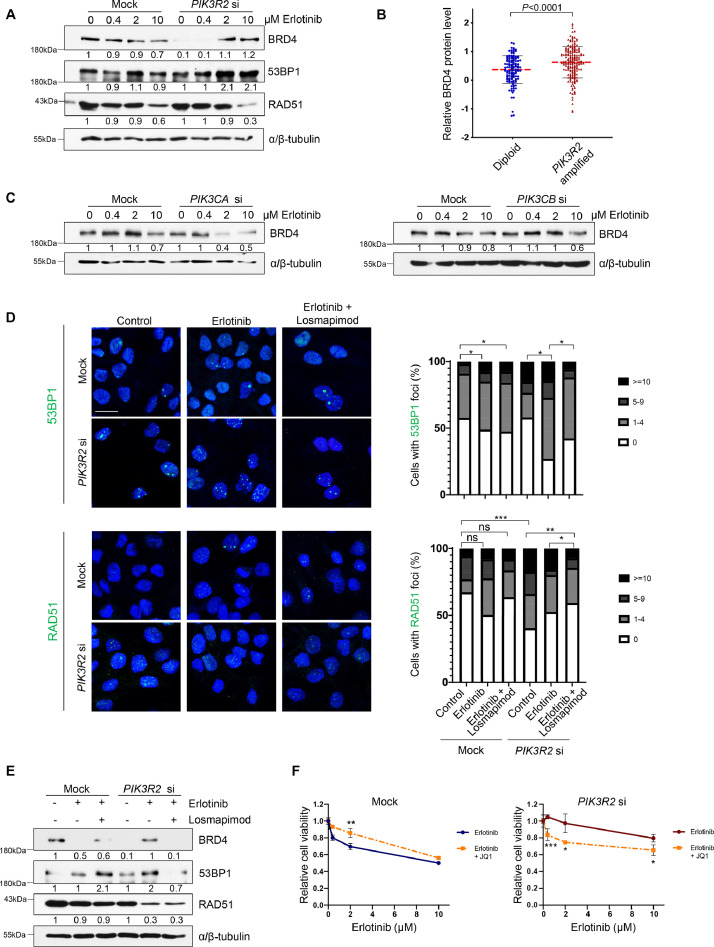
We next identified the potential DNA repair pathway involved in the observed DNA repair response. We examined the protein levels of 53BP1 and BRD4, which are key DDR factors promoting nonhomologous end joining (NHEJ) repair [45], and RAD51 which mediates homologous recombination (HR) [46]. Interestingly, untreated PIK3R2-depleted cells had reduced level of BRD4 compared with the control cells (Fig. 6A). This suggested that the accumulation of DNA breaks in PIK3R2-depleted cells is likely due to a deficient NHEJ repair pathway. The regulation of BRD4 protein level by PIK3R2 was further supported by the observation that PIK3R2-amplified ovarian tumor samples from the TCGA database displayed significantly higher BRD4 protein levels (Fig. 6B). Intriguingly, the levels of BRD4 and 53BP1 were then gradually increased by increasing doses of erlotinib (Fig. 6A). These increases aligned with the activation of p38 MAPK, as shown in Fig. 2A. We also examined the effects of PIK3CA and PIK3CB on BRD4 levels. The results showed that, in contrast to PIK3R2 siRNA, neither PIK3CA nor PIK3CB siRNA reduced BRD4 levels in the untreated cells (Fig. 6C). Further, erlotinib treatment of these PIK3CA- or PIK3CB-silenced cells did not enhance BRD4 protein levels (Fig. 6C). Indeed, BRD4 level were markedly decreased in PIK3CA-depleted cells upon erlotinib treatment.
Fig. 6.
p38 MAPK promotes NHEJ repair in PIK3R2-depleted cells treated with erlotinib.
(A) SKOV3 cells were transfected with or without PIK3R2 siRNA for 24 h prior to erlotinib treatment for 72 h. Protein lysates were collected for Western blotting with the indicated antibodies. (B) Relative BRD4 protein levels were compared between TCGA serous ovarian cancer tumor samples (n=304) categorized according to PIK3R2 copy number. The P value was calculated by the nonparametric Mann-Whitney test. (C) SKOV3 cells were transfected with or without siRNA targeting PIK3CA or PIK3CB for 24 h prior to erlotinib treatment for 72 h. Protein lysates were collected for Western blotting with the indicated antibodies. (D-E) SKOV3 cells transfected with or without PIK3R2 siRNA were treated with erlotinib alone (10 μM) or in combination with losmapimod (10 μM) for 72 h. (D) Representative immunofluorescence images of 53BP1 or RAD51 foci (left) and the quantification of the cells with foci (right) are shown. Scale bar, 20 μm. (E) Western blotting with collected protein lysates was performed using the indicated antibodies. (F) SKOV3 cells transfected with or without PIK3R2 siRNA were treated with erlotinib alone or in combination with BRD4 inhibitor (JQ1; 1 μM) for 72 h. The data presented show the relative cell viability relative to the corresponding untreated cells. The error bars represent the SD of 3 independent experiments performed in triplicate. *, P < 0.05; **, P < 0.005; #, P < 0.0001 using ordinary one-way ANOVA for analysis within group (mock or PIK3R2 siRNA) and 2-way ANOVA for comparison between groups (mock vs PIK3R2 siRNA) with Sidak's multiple comparison test. In the Western blot analysis, α/β-tubulin was used as a loading control. Representative blots of 3 independent experiments are shown. The numbers below the blots indicate the mean densitometry values normalized to those of α/β-tubulin.
Our data suggest that p38 MAPK signaling mediates these alterations in DNA repair machinery. Interestingly, the formation of 53BP1 nuclear foci was induced by erlotinib in PIK3R2-depleted cells (Fig. 6D). The induction was reversed by losmapimod (Fig. 6D). Similarly, erlotinib increased the protein levels of BRD4 and 53BP1 in PIK3R2-depleted cells, and this effect was abolished by losmapimod (Fig. 6E). Consistent with the ability of 53BP1 to antagonize HR [47], protein expression and nuclear foci formation of the HR repair protein RAD51 were indeed decreased in the PIK3R2-depleted cells upon erlotinib treatment (Fig. 6D and 6E). Building on the findings that BRD4 is upregulated by erlotinib in PIK3R2-depleted cells, we reasoned that inhibiting BRD4 would confer sensitivity to erlotinib. When PIK3R2-depleted cells were cotreated with JQ1 (BRD4 inhibitor) and erlotinib, cell viability was significantly reduced compared with erlotinib treatment alone (P < 0.05) (Fig. 6F).
Discussion
Reported resistance mechanisms to EGFR inhibition in cancer include the acquisition of the EGFR gatekeeper T790M mutation, activation of bypass signaling (such as by HER2, c-Met, AXL or IGF-1R) and aberrant activation of downstream Ras/MAPK and PI3K/AKT pathways [48]. While new generation EGFR inhibitors are designed to enhance treatment outcomes by targeting the gatekeeper mutant, overcoming non-EGFR-centric adaptation may rely on simultaneous inhibition of compensatory pathways. Several combination strategies have been evaluated to increase the sensitivity of ovarian cancer cells to EGFR inhibition. For example, EGFR inhibitors can be combined with frontline therapies, including chemotherapy [49,50] or antiangiogenic agent [51]. However, these combination regimens showed only modest clinical benefit. More investigations are warranted to understand the underlying mechanism of insensitivity towards EGFR inhibition in ovarian cancer and to derive rational combinational treatments accordingly. In this study, our data suggested that insensitivity to erlotinib upon PIK3R2 depletion is a result of p38 MAPK activation, which initiates DNA repair and thereby protects cells from DNA damage-induced apoptosis. Importantly, inhibition of p38 MAPK leads to accumulated DNA lesions and sensitizes ovarian cancer cells to erlotinib (Fig. 7).
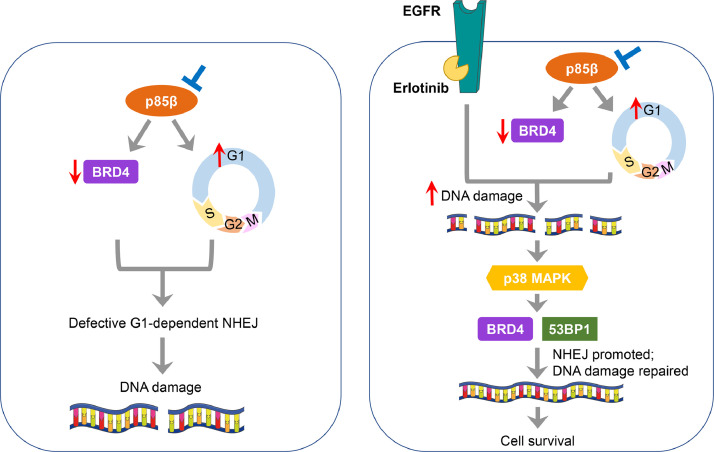
Fig. 7.
The proposed model of erlotinib insensitivity of ovarian cancer cells with low PIK3R2 expression.
Left, Depletion of PIK3R2 (p85β) leads to G1 cell cycle arrest and a reduction in BRD4 protein level. These 2 events collectively result in defective nonhomologous end joining (NHEJ) repair and DNA damage. Right, DNA damage is further exacerbated in these PIK3R2-depleted cells upon treatment with erlotinib because erlotinib itself also inhibits DNA repair. The accumulated genotoxic stress activates p38 MAPK signaling, which in turn increases the expression levels of BRD4 and 53BP1, thereby repairing damaged DNA and sparing cells from apoptosis. p38 MAPK activation and induced DNA repair confer enhanced viability of PIK3R2-depleted cells treated with erlotinib and represent resistance mechanisms to EGFR inhibition.
EGFR inhibition suppresses tumor development by modulating cell proliferation, apoptosis and angiogenesis [38,52,53]. Erlotinib has also been shown to inhibit tumorigenesis by inducing DNA double-strand breaks (DSBs) [43], providing an additional level of antitumoral action. Mechanistically, erlotinib downregulates homology-directed recombinational repair by inhibiting nuclear localization of BRCA1 [43]. Indeed, regulation of DNA repair pathways by EGFR has been reported. In addition to that of HR, EGFR can modulate other DNA repair pathways, including NHEJ [54] and nucleotide excision repair pathways [55]. Nuclear EGFR physically binds to DNA-dependent protein kinase (DNA-PK) [55,56], which is a component of the NHEJ machinery that promotes DNA repair [57]. Collectively, these studies indicate DNA repair signaling as a downstream pathway of EGFR. Aberrant DNA damage signaling may be critical for EGFR inhibitor responsiveness, particularly in tumor cells under DNA damage stress.
Our data showed that depleting p85β reduced BRD4 protein level, suggesting that p85β involves in the NHEJ DNA repair pathway. p85 has not been shown to modulate DNA damage repair. Evidence of DNA repair pathway regulation by the PI3K pathway so far has been focused on the p110 catalytic subunit and its downstream signaling. While p110β promotes the binding of the HR repair protein NBS1 to damaged DNA [58,59], AKT promotes NHEJ repair by regulating DNA-PK [60] or inhibits HR by impairing BRCA1 and RAD51 foci formation [61]. We postulate that the increase in DNA damage upon PIK3R2 depletion is at least partially due to the inhibition of NHEJ repair and an accumulation of cells in the G1 phase, in which the NHEJ pathway predominates [62]. Whether the effect of p85β on DDR is mediated through the classical p110 or AKT signaling remains to be examined. However, notably, in contrast to the depletion of p85β, the depletion of p110α or p110β in the untreated cells did not alter BRD4 protein levels. Moreover, erlotinib caused opposing effect on BRD4 level in PIK3CA-depleted cells compared with PIK3R2-depleted cells.
In response to DNA damage, p38 MAPK regulates DNA repair at multiple levels. p38 MAPK initiates signaling cascades that regulate cell cycle checkpoints prior to cell division. This process allows the coordination of the DDR machinery to repair damaged DNA before entry into the next cell cycle [63]. Phosphorylated p38 MAPK can also be translocated into the nucleus by the induction of DSBs. Nuclear p38 MAPK binds to components of the DDR machinery including histone 2A variant X (H2AX) and C-terminal-binding protein interacting protein (CtIP) [64], which interact with DNA repair proteins such as the MRE11-RAD50-NBS1 complex to promote HR and NHEJ repair [65], [66], [67]. Nuclear p38 MAPK phosphorylates H2AX at Ser139 [68] and CtIP at multiple amino acid residues [40], thereby promoting the activities of H2AX and CtIP. p38 MAPK may also regulate the recruitment of 53BP1 to DSBs [69]. Inhibiting p38 MAPK thus reduces DNA repair activity, along with increased replication fork stalking and DNA damage [40]. Aligning with the role of p38 MAPK in DNA repair, erlotinib treatment of the PIK3R2-depleted cells in this study led to increases in p38 MAPK phosphorylation and nuclear import with a concomitant decrease in the amount of damaged DNA. Intriguingly, p38 MAPK was activated as a consequence of p85β depletion and erlotinib treatment but not upon either event alone. This raises an interesting possibility that the DNA damage resulting from the 2 events together induces a sufficiently strong signal to activate p38 MAPK and thereby the DDR machinery. The DNA damage caused by either event alone might not reach the threshold level required for DDR activation. In accordance with this proposition, distinct DNA damage thresholds associated with different cellular outcomes and cell fates have been postulated [70,71]. Further, it has indeed been shown that increased unrepaired DSBs in cells with compromised NHEJ proficiency induced sustained activation of p38 MAPK [72]. Therefore, the impact of EGFR inhibitors on DNA damage and treatment outcomes may be dependent on the intrinsic DNA repair ability.
Conclusion
In summary, our data suggest that the copy number status or expression levels of PIK3R2, PIK3CA and PIK3CB may affect treatment response to EGFR inhibitors. The identification of response biomarkers and our proposed model of resistance mechanism may help unlock the potential of EGFR inhibitors in treating ovarian cancer.
Data availability
The data generated in this study are available from the corresponding author upon reasonable request.
Authors’ contribution
Victor CY Mak: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing.
Xinran Li: Investigation, Methodology.
Ling Rao: Investigation.
Yuan Zhou: Investigation.
Sai-Wah Tsao: Formal analysis, Writing – review & editing.
Lydia WT Cheung: Conceptualization, Formal analysis, Funding acquisition, Project administration, supervision, Writing – original draft, Writing – review & editing.
Acknowledgments
The authors thank the Core Facility of the LKS Faculty of Medicine HKU for the technical support with flow cytometer and confocal microscope.
Footnotes
✩Funding: This work was supported by the National Natural Science Foundation of China (82022078) and the Hong Kong Research Grants Council (17111618) to LWC.
✩✩Conflicts of interest: The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neo.2021.05.009.
Appendix. Supplementary materials
References
- 1.Ilekis JV, Connor JP, Prins GS, Ferrer K, Niederberger C, Scoccia B. Expression of epidermal growth factor and androgen receptors in ovarian cancer. Gynecol Oncol. 1997;66(2):250–254. doi: 10.1006/gyno.1997.4764. [DOI] [PubMed] [Google Scholar]
- 2.Nielsen JS, Jakobsen E, Holund B, Bertelsen K, Jakobsen A. Prognostic significance of p53, Her-2, and EGFR overexpression in borderline and epithelial ovarian cancer. Int J Gynecol Cancer. 2004;14(6):1086–1096. doi: 10.1111/j.1048-891X.2004.14606.x. [DOI] [PubMed] [Google Scholar]
- 3.Noske A, Schwabe M, Weichert W, Darb-Esfahani S, Buckendahl AC, Sehouli J, Braicu EI, Budczies J, Dietel M, Denkert C. An intracellular targeted antibody detects EGFR as an independent prognostic factor in ovarian carcinomas. BMC Cancer. 2011;11:294. doi: 10.1186/1471-2407-11-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Skirnisdottir I, Sorbe B, Seidal T. The growth factor receptors HER-2/neu and EGFR, their relationship, and their effects on the prognosis in early stage (FIGO I-II) epithelial ovarian carcinoma. Int J Gynecol Cancer. 2001;11(2):119–129. doi: 10.1046/j.1525-1438.2001.011002119.x. [DOI] [PubMed] [Google Scholar]
- 5.Alper O, Bergmann-Leitner ES, Bennett TA, Hacker NF, Stromberg K, Stetler-Stevenson WG. Epidermal growth factor receptor signaling and the invasive phenotype of ovarian carcinoma cells. J Natl Cancer Inst. 2001;93(18):1375–1384. doi: 10.1093/jnci/93.18.1375. [DOI] [PubMed] [Google Scholar]
- 6.Siwak DR, Carey M, Hennessy BT, Nguyen CT, McGahren Murray MJ, Nolden L, Mills GB. Targeting the epidermal growth factor receptor in epithelial ovarian cancer: current knowledge and future challenges. J Oncol. 2010;2010 doi: 10.1155/2010/568938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Psyrri A, Kassar M, Yu Z, Bamias A, Weinberger PM, Markakis S, Kowalski D, Camp RL, Rimm DL, Dimopoulos MA. Effect of epidermal growth factor receptor expression level on survival in patients with epithelial ovarian cancer. Clin Cancer Res. 2005;11:8637–8643. doi: 10.1158/1078-0432.CCR-05-1436. 24 Pt 1. [DOI] [PubMed] [Google Scholar]
- 8.Tas F, Karabulut S, Serilmez M, Ciftci R, Duranyildiz D. Increased serum level of epidermal growth factor receptor (EGFR) is associated with poor progression-free survival in patients with epithelial ovarian cancer. Cancer Chemother Pharmacol. 2014;73(3):631–637. doi: 10.1007/s00280-014-2396-x. [DOI] [PubMed] [Google Scholar]
- 9.Gordon AN, Finkler N, Edwards RP, Garcia AA, Crozier M, Irwin DH, Barrett E. Efficacy and safety of erlotinib HCl, an epidermal growth factor receptor (HER1/EGFR) tyrosine kinase inhibitor, in patients with advanced ovarian carcinoma: results from a phase II multicenter study. Int J Gynecol Cancer. 2005;15(5):785–792. doi: 10.1111/j.1525-1438.2005.00137.x. [DOI] [PubMed] [Google Scholar]
- 10.Schilder RJ, Sill MW, Chen X, Darcy KM, Decesare SL, Lewandowski G, Lee RB, Arciero CA, Wu H, Godwin AK. Phase II study of gefitinib in patients with relapsed or persistent ovarian or primary peritoneal carcinoma and evaluation of epidermal growth factor receptor mutations and immunohistochemical expression: a Gynecologic Oncology Group Study. Clin Cancer Res. 2005;11(15):5539–5548. doi: 10.1158/1078-0432.CCR-05-0462. [DOI] [PubMed] [Google Scholar]
- 11.Vergote IB, Jimeno A, Joly F, Katsaros D, Coens C, Despierre E, Marth C, Hall M, Steer CB, Colombo N. Randomized phase III study of erlotinib versus observation in patients with no evidence of disease progression after first-line platin-based chemotherapy for ovarian carcinoma: a European Organisation for Research and Treatment of Cancer-Gynaecological Cancer Group, and Gynecologic Cancer Intergroup study. J Clin Oncol. 2014;32(4):320–326. doi: 10.1200/JCO.2013.50.5669. [DOI] [PubMed] [Google Scholar]
- 12.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350(21):2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 13.Sholl LM, Xiao Y, Joshi V, Yeap BY, Cioffredi LA, Jackman DM, Lee C, Janne PA, Lindeman NI. EGFR mutation is a better predictor of response to tyrosine kinase inhibitors in non-small cell lung carcinoma than FISH, CISH, and immunohistochemistry. Am J Clin Pathol. 2010;133(6):922–934. doi: 10.1309/AJCPST1CTHZS3PSZ. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu YL, Fukuoka M, Mok TS, Saijo N, Thongprasert S, Yang JC, Chu DT, Yang JJ, Rukazenkov Y. Tumor response and health-related quality of life in clinically selected patients from Asia with advanced non-small-cell lung cancer treated with first-line gefitinib: post hoc analyses from the IPASS study. Lung Cancer. 2013;81(2):280–287. doi: 10.1016/j.lungcan.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 15.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wheler JJ, Falchook GS, Tsimberidou AM, Hong DS, Naing A, Piha-Paul SA, Chen SS, Fu S, Stephen B, Fok JY. Aberrations in the epidermal growth factor receptor gene in 958 patients with diverse advanced tumors: implications for therapy. Ann Oncol. 2013;24(3):838–842. doi: 10.1093/annonc/mds524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mehner C, Oberg AL, Goergen KM, Kalli KR, Maurer MJ, Nassar A, Goode EL, Keeney GL, Jatoi A, Radisky DC. EGFR as a prognostic biomarker and therapeutic target in ovarian cancer: evaluation of patient cohort and literature review. Genes Cancer. 2017;8(5-6):589–599. doi: 10.18632/genesandcancer.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Engelman JA, Mukohara T, Zejnullahu K, Lifshits E, Borras AM, Gale CM, Naumov GN, Yeap BY, Jarrell E, Sun J. Allelic dilution obscures detection of a biologically significant resistance mutation in EGFR-amplified lung cancer. J Clin Invest. 2006;116(10):2695–2706. doi: 10.1172/JCI28656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glaysher S, Bolton LM, Johnson P, Atkey N, Dyson M, Torrance C, Cree IA. Targeting EGFR and PI3K pathways in ovarian cancer. Br J Cancer. 2013;109(7):1786–1794. doi: 10.1038/bjc.2013.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ihle NT, Paine-Murrieta G, Berggren MI, Baker A, Tate WR, Wipf P, Abraham RT, Kirkpatrick DL, Powis G. The phosphatidylinositol-3-kinase inhibitor PX-866 overcomes resistance to the epidermal growth factor receptor inhibitor gefitinib in A-549 human non-small cell lung cancer xenografts. Mol Cancer Ther. 2005;4(9):1349–1357. doi: 10.1158/1535-7163.MCT-05-0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wong KK, Engelman JA, Cantley LC. Targeting the PI3K signaling pathway in cancer. Curr Opin Genet Dev. 2010;20(1):87–90. doi: 10.1016/j.gde.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Utermark T, Rao T, Cheng H, Wang Q, Lee SH, Wang ZC, Iglehart JD, Roberts TM, Muller WJ, Zhao JJ. The p110alpha and p110beta isoforms of PI3K play divergent roles in mammary gland development and tumorigenesis. Genes Dev. 2012;26(14):1573–1586. doi: 10.1101/gad.191973.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thorpe LM, Yuzugullu H, Zhao JJ. PI3K in cancer: divergent roles of isoforms, modes of activation and therapeutic targeting. Nat Rev Cancer. 2015;15(1):7–24. doi: 10.1038/nrc3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheung LW, Hennessy BT, Li J, Yu S, Myers AP, Djordjevic B, Lu Y, Stemke-Hale K, Dyer MD, Zhang F. High frequency of PIK3R1 and PIK3R2 mutations in endometrial cancer elucidates a novel mechanism for regulation of PTEN protein stability. Cancer Discov. 2011;1(2):170–185. doi: 10.1158/2159-8290.CD-11-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murga C, Fukuhara S, Gutkind JS. A novel role for phosphatidylinositol 3-kinase beta in signaling from G protein-coupled receptors to Akt. J Biol Chem. 2000;275(16):12069–12073. doi: 10.1074/jbc.275.16.12069. [DOI] [PubMed] [Google Scholar]
- 26.Thorpe LM, Spangle JM, Ohlson CE, Cheng H, Roberts TM, Cantley LC, Zhao JJ. PI3K-p110alpha mediates the oncogenic activity induced by loss of the novel tumor suppressor PI3K-p85alpha. Proc Natl Acad Sci U S A. 2017;114(27):7095–7100. doi: 10.1073/pnas.1704706114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li X, Mak VCY, Zhou Y, Wang C, Wong ESY, Sharma R, Lu Y, Cheung ANY, Mills GB, Cheung LWT. Deregulated Gab2 phosphorylation mediates aberrant AKT and STAT3 signaling upon PIK3R1 loss in ovarian cancer. Nat Commun. 2019;10(1):716. doi: 10.1038/s41467-019-08574-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gonzalez-Garcia A, Carrera AC. p85beta increases phosphoinositide 3-kinase activity and accelerates tumor progression. Cell Cycle. 2012;11(19):3523–3524. doi: 10.4161/cc.21961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rao L, Mak VCY, Zhou Y, Zhang D, Li X, Fung CCY, Sharma R, Gu C, Lu Y, Tipoe GL. p85beta regulates autophagic degradation of AXL to activate oncogenic signaling. Nat Commun. 2020;11(1):2291. doi: 10.1038/s41467-020-16061-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beroukhim R, Getz G, Nghiemphu L, Barretina J, Hsueh T, Linhart D, Vivanco I, Lee JC, Huang JH, Alexander S. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc Natl Acad Sci U S A. 2007;104(50):20007–20012. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ciriello G, Miller ML, Aksoy BA, Senbabaoglu Y, Schultz N, Sander C. Emerging landscape of oncogenic signatures across human cancers. Nat Genet. 2013;45(10):1127–1133. doi: 10.1038/ng.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang T, Inesta-Vaquera F, Niepel M, Zhang J, Ficarro SB, Machleidt T, Xie T, Marto JA, Kim N, Sim T. Discovery of potent and selective covalent inhibitors of JNK. Chem Biol. 2012;19(1):140–154. doi: 10.1016/j.chembiol.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stebbins JL, De SK, Machleidt T, Becattini B, Vazquez J, Kuntzen C, Chen LH, Cellitti JF, Riel-Mehan M, Emdadi A. Identification of a new JNK inhibitor targeting the JNK-JIP interaction site. Proc Natl Acad Sci U S A. 2008;105(43):16809–16813. doi: 10.1073/pnas.0805677105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blake JF, Burkard M, Chan J, Chen H, Chou KJ, Diaz D, Dudley DA, Gaudino JJ, Gould SE, Grina J. Discovery of (S)-1-(1-(4-Chloro-3-fluorophenyl)-2-hydroxyethyl)-4-(2-((1-methyl-1H-pyrazol-5-y l)amino)pyrimidin-4-yl)pyridin-2(1H)-one (GDC-0994), an Extracellular Signal-Regulated Kinase 1/2 (ERK1/2) Inhibitor in Early Clinical Development. J Med Chem. 2016;59(12):5650–5660. doi: 10.1021/acs.jmedchem.6b00389. [DOI] [PubMed] [Google Scholar]
- 35.Yamaguchi T, Kakefuda R, Tajima N, Sowa Y, Sakai T. Antitumor activities of JTP-74057 (GSK1120212), a novel MEK1/2 inhibitor, on colorectal cancer cell lines in vitro and in vivo. Int J Oncol. 2011;39(1):23–31. doi: 10.3892/ijo.2011.1015. [DOI] [PubMed] [Google Scholar]
- 36.Wilson KP, McCaffrey PG, Hsiao K, Pazhanisamy S, Galullo V, Bemis GW, Fitzgibbon MJ, Caron PR, Murcko MA, Su MS. The structural basis for the specificity of pyridinylimidazole inhibitors of p38 MAP kinase. Chem Biol. 1997;4(6):423–431. doi: 10.1016/s1074-5521(97)90194-0. [DOI] [PubMed] [Google Scholar]
- 37.Willette RN, Eybye ME, Olzinski AR, Behm DJ, Aiyar N, Maniscalco K, Bentley RG, Coatney RW, Zhao S, Westfall TD. Differential effects of p38 mitogen-activated protein kinase and cyclooxygenase 2 inhibitors in a model of cardiovascular disease. J Pharmacol Exp Ther. 2009;330(3):964–970. doi: 10.1124/jpet.109.154443. [DOI] [PubMed] [Google Scholar]
- 38.Gong Y, Somwar R, Politi K, Balak M, Chmielecki J, Jiang X, Pao W. Induction of BIM is essential for apoptosis triggered by EGFR kinase inhibitors in mutant EGFR-dependent lung adenocarcinomas. PLoS Med. 2007;4(10):e294. doi: 10.1371/journal.pmed.0040294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huether A, Hopfner M, Sutter AP, Schuppan D, Scherubl H. Erlotinib induces cell cycle arrest and apoptosis in hepatocellular cancer cells and enhances chemosensitivity towards cytostatics. J Hepatol. 2005;43(4):661–669. doi: 10.1016/j.jhep.2005.02.040. [DOI] [PubMed] [Google Scholar]
- 40.Canovas B, Igea A, Sartori AA, Gomis RR, Paull TT, Isoda M, Perez-Montoyo H, Serra V, Gonzalez-Suarez E, Stracker TH. Targeting p38alpha Increases DNA Damage, Chromosome Instability, and the Anti-tumoral Response to Taxanes in Breast Cancer Cells. Cancer Cell. 2018;33(6):e8. doi: 10.1016/j.ccell.2018.04.010. 1094-110. [DOI] [PubMed] [Google Scholar]
- 41.Koul HK, Pal M, Koul S. Role of p38 MAP Kinase Signal Transduction in Solid Tumors. Genes Cancer. 2013;4(9-10):342–359. doi: 10.1177/1947601913507951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wood CD, Thornton TM, Sabio G, Davis RA, Rincon M. Nuclear localization of p38 MAPK in response to DNA damage. Int J Biol Sci. 2009;5(5):428–437. doi: 10.7150/ijbs.5.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li L, Wang H, Yang ES, Arteaga CL, Xia F. Erlotinib attenuates homologous recombinational repair of chromosomal breaks in human breast cancer cells. Cancer Res. 2008;68(22):9141–9146. doi: 10.1158/0008-5472.CAN-08-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Norbury CJ, Zhivotovsky B. DNA damage-induced apoptosis. Oncogene. 2004;23(16):2797–2808. doi: 10.1038/sj.onc.1207532. [DOI] [PubMed] [Google Scholar]
- 45.Zimmermann M, de Lange T. 53BP1: pro choice in DNA repair. Trends Cell Biol. 2014;24(2):108–117. doi: 10.1016/j.tcb.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tashiro S, Walter J, Shinohara A, Kamada N, Cremer T. Rad51 accumulation at sites of DNA damage and in postreplicative chromatin. J Cell Biol. 2000;150(2):283–291. doi: 10.1083/jcb.150.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Panier S, Boulton SJ. Double-strand break repair: 53BP1 comes into focus. Nat Rev Mol Cell Biol. 2014;15(1):7–18. doi: 10.1038/nrm3719. [DOI] [PubMed] [Google Scholar]
- 48.Nagano T, Tachihara M, Nishimura Y. Mechanism of Resistance to Epidermal Growth Factor Receptor-Tyrosine Kinase Inhibitors and a. Potential Treatment Strategy. Cells. 2018;7(11) doi: 10.3390/cells7110212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Blank SV, Christos P, Curtin JP, Goldman N, Runowicz CD, Sparano JA, Liebes L, Chen HX, Muggia FM. Erlotinib added to carboplatin and paclitaxel as first-line treatment of ovarian cancer: a phase II study based on surgical reassessment. Gynecol Oncol. 2010;119(3):451–456. doi: 10.1016/j.ygyno.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Secord AA, Blessing JA, Armstrong DK, Rodgers WH, Miner Z, Barnes MN, Lewandowski G, Mannel RS. Gynecologic Oncology G. Phase II trial of cetuximab and carboplatin in relapsed platinum-sensitive ovarian cancer and evaluation of epidermal growth factor receptor expression: a Gynecologic Oncology Group study. Gynecol Oncol. 2008;108(3):493–499. doi: 10.1016/j.ygyno.2007.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nimeiri HS, Oza AM, Morgan RJ, Friberg G, Kasza K, Faoro L, Salgia R, Stadler WM, Vokes EE, Fleming GF. Efficacy and safety of bevacizumab plus erlotinib for patients with recurrent ovarian, primary peritoneal, and fallopian tube cancer: a trial of the Chicago, PMH, and California Phase II Consortia. Gynecol Oncol. 2008;110(1):49–55. doi: 10.1016/j.ygyno.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen X, Yang S, Pan Y, Li X, Ma S. Mitochondrial pathway-mediated apoptosis is associated with erlotinib-induced cytotoxicity in hepatic cells. Oncol Lett. 2018;15(1):783–788. doi: 10.3892/ol.2017.7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang SM, Li J, Armstrong EA, Harari PM. Modulation of radiation response and tumor-induced angiogenesis after epidermal growth factor receptor inhibition by ZD1839 (Iressa) Cancer Res. 2002;62(15):4300–4306. [PubMed] [Google Scholar]
- 54.Kriegs M, Kasten-Pisula U, Rieckmann T, Holst K, Saker J, Dahm-Daphi J, Dikomey E. The epidermal growth factor receptor modulates DNA double-strand break repair by regulating non-homologous end-joining. DNA Repair (Amst) 2010;9(8):889–897. doi: 10.1016/j.dnarep.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 55.Liccardi G, Hartley JA, Hochhauser D. EGFR nuclear translocation modulates DNA repair following cisplatin and ionizing radiation treatment. Cancer Res. 2011;71(3):1103–1114. doi: 10.1158/0008-5472.CAN-10-2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang SC, Hung MC. Nuclear translocation of the epidermal growth factor receptor family membrane tyrosine kinase receptors. Clin Cancer Res. 2009;15(21):6484–6489. doi: 10.1158/1078-0432.CCR-08-2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Davis AJ, Chen BP, Chen DJ. DNA-PK: a dynamic enzyme in a versatile DSB repair pathway. DNA Repair (Amst) 2014;17:21–29. doi: 10.1016/j.dnarep.2014.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kumar A, Fernandez-Capetillo O, Carrera AC. Nuclear phosphoinositide 3-kinase beta controls double-strand break DNA repair. Proc Natl Acad Sci U S A. 2010;107(16):7491–7496. doi: 10.1073/pnas.0914242107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tauchi H, Kobayashi J, Morishima K, van Gent DC, Shiraishi T, Verkaik NS, vanHeems D, Ito E, Nakamura A, Sonoda E. Nbs1 is essential for DNA repair by homologous recombination in higher vertebrate cells. Nature. 2002;420(6911):93–98. doi: 10.1038/nature01125. [DOI] [PubMed] [Google Scholar]
- 60.Toulany M, Lee KJ, Fattah KR, Lin YF, Fehrenbacher B, Schaller M, Chen BP, Chen DJ, Rodemann HP. Akt promotes post-irradiation survival of human tumor cells through initiation, progression, and termination of DNA-PKcs-dependent DNA double-strand break repair. Mol Cancer Res. 2012;10(7):945–957. doi: 10.1158/1541-7786.MCR-11-0592. [DOI] [PubMed] [Google Scholar]
- 61.Plo I, Laulier C, Gauthier L, Lebrun F, Calvo F, Lopez BS. AKT1 inhibits homologous recombination by inducing cytoplasmic retention of BRCA1 and RAD51. Cancer Res. 2008;68(22):9404–9412. doi: 10.1158/0008-5472.CAN-08-0861. [DOI] [PubMed] [Google Scholar]
- 62.Mao Z, Bozzella M, Seluanov A, Gorbunova V. DNA repair by nonhomologous end joining and homologous recombination during cell cycle in human cells. Cell Cycle. 2008;7(18):2902–2906. doi: 10.4161/cc.7.18.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thornton TM, Rincon M. Non-classical p38 map kinase functions: cell cycle checkpoints and survival. Int J Biol Sci. 2009;5(1):44–51. doi: 10.7150/ijbs.5.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gong X, Ming X, Deng P, Jiang Y. Mechanisms regulating the nuclear translocation of p38 MAP kinase. J Cell Biochem. 2010;110(6):1420–1429. doi: 10.1002/jcb.22675. [DOI] [PubMed] [Google Scholar]
- 65.Davies OR, Forment JV, Sun M, Belotserkovskaya R, Coates J, Galanty Y, Demir M, Morton CR, Rzechorzek NJ, Jackson SP. CtIP tetramer assembly is required for DNA-end resection and repair. Nat Struct Mol Biol. 2015;22(2):150–157. doi: 10.1038/nsmb.2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nakamura AJ, Rao VA, Pommier Y, Bonner WM. The complexity of phosphorylated H2AX foci formation and DNA repair assembly at DNA double-strand breaks. Cell Cycle. 2010;9(2):389–397. doi: 10.4161/cc.9.2.10475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Quennet V, Beucher A, Barton O, Takeda S, Lobrich M. CtIP and MRN promote non-homologous end-joining of etoposide-induced DNA double-strand breaks in G1. Nucleic Acids Res. 2011;39(6):2144–2152. doi: 10.1093/nar/gkq1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lu C, Shi Y, Wang Z, Song Z, Zhu M, Cai Q, Chen T. Serum starvation induces H2AX phosphorylation to regulate apoptosis via p38 MAPK pathway. FEBS Lett. 2008;582(18):2703–2708. doi: 10.1016/j.febslet.2008.06.051. [DOI] [PubMed] [Google Scholar]
- 69.Lee DH, Acharya SS, Kwon M, Drane P, Guan Y, Adelmant G, Kalev P, Shah J, Pellman D, Marto JA. Dephosphorylation enables the recruitment of 53BP1 to double-strand DNA breaks. Mol Cell. 2014;54(3):512–525. doi: 10.1016/j.molcel.2014.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bartek J, Lukas J, Bartkova J. DNA damage response as an anti-cancer barrier: damage threshold and the concept of 'conditional haploinsufficiency. Cell Cycle. 2007;6(19):2344–2347. doi: 10.4161/cc.6.19.4754. [DOI] [PubMed] [Google Scholar]
- 71.Khanna A. DNA damage in cancer therapeutics: a boon or a curse? Cancer Res. 2015;75(11):2133–2138. doi: 10.1158/0008-5472.CAN-14-3247. [DOI] [PubMed] [Google Scholar]
- 72.Kanungo J.DNA-PK, P38 MAPK. A Kinase Collusion in Alzheimer's Disease? Brain Disord Ther. 2017;6(2):232. doi: 10.4172/2168-975X.1000232. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are available from the corresponding author upon reasonable request.