Abstract
Cancer stem cells (CSCs) are sparks for igniting tumor recurrence and the instigators of low response to immunotherapy and drug resistance. As one of the important components of tumor microenvironment, the tumor associated immune microenvironment (TAIM) is driving force for the heterogeneity, plasticity and evolution of CSCs. CSCs create the inhibitory TAIM (ITAIM) mainly through four stemness-related signals (SRSs), including Notch-nuclear factor-κB axis, Hedgehog, Wnt and signal transducer and activator of transcription. Ubiquitination and deubiquitination in proteins related to the specific stemness of the CSCs have a profound impact on the regulation of ITAIM. In regulating the balance between ubiquitination and deubiquitination, it is crucial for deubiquitinating enzymes (DUBs) to cleave ubiquitin chains from substrates. Ubiquitin-specific peptidases (USPs) comprise the largest family of DUBs. Growing evidence suggests that they play novel functions in contribution of ITAIM, including regulating tumor immunogenicity, activating stem cell factors, upregulating the SRSs, stabilizing anti-inflammatory receptors, and regulating anti-inflammatory cytokines. These overactive or abnormal signaling may dampen antitumor immune responses. The inhibition of USPs could play a regulatory role in SRSs and reversing ITAIM, and also have great potential in improving immune killing ability against tumor cells, including CSCs. In this review, we focus on the USPs involved in CSCs signaling pathways and regulating ITAIM, which are promising therapeutic targets in antitumor therapy.
Keywords: deubiquitylating enzymes, deubiquitination, cancer stem cells, stemness-related signals, inhibitory tumor-associated immune microenvironment
Introduction
Cancer cells are heterogeneous, and tumor cells in different states exhibit differences in expression products, immune interactions, tumor proliferation potential, and therapeutic responses (Castagnoli et al., 2020). There are many explanations for these differences, including the cancer stem cell (CSC) hypothesis, which is based on a set of cancer cells with self-renewal ability (Prager et al., 2019). Because CSCs are in an undifferentiated or poorly differentiated state, many stemness-related signals (SRSs) are key to their multiple functions. Some cancers heavily rely on SRSs to promote immunosuppression, metastasis, treatment resistance, and epithelial-mesenchymal transition (Lytle et al., 2018). CSCs can be compared to seeds with the surrounding tumor microenvironment considered the soil in which they grow. CSCs change the tumor microenvironment (TME) through SRSs.
Compared with regulating the entire TME, targeting one microenvironment seems to be a more effective method (Jin and Jin, 2020). As one of the important components of the TME, the tumor- associated immune microenvironment (TAIM) is driving force for the heterogeneity, plasticity and evolution of CSCs. CSCs can create inhibitory TAIMs (ITAIMs) by leveraging the immunomodulatory induction by SRSs (Clara et al., 2020), thereby regulating cytokines and chemokines to strengthen suppressive immune cells, upregulating suppressive immune checkpoints, and downregulating immunogenicity to reduce immune cell recognition and killing (Clara et al., 2020; Ho et al., 2020; Nowak and Klink, 2020). Breakthroughs in immunotherapy have yielded exceptional results in recent years. However, some tumors have a low response to immunotherapy, and the ITAIM is the chief culprit for these poor responses.
Ubiquitin can be linked by one of the seven Lys residues to form ubiquitin chains, including methionine1, K6, K11, K27, k29, k33, K48, and K63 (Wagner et al., 2011). These ubiquitin modifications perform different biological functions, among which K48 and k63 are the most thoroughly studied forms. The K48-linked chains act as a signal of proteasome degradation, and K63-linked chains play an important role in DNA repair (Kaushal and Ramakrishna, 2020). Several studies have documented that K48 and K63 play important roles in regulation of immune responses (Dzimianski et al., 2019). Similar to other post-translational modifications, ubiquitination is reversible. This reversal is achieved by deubiquitylating enzymes (DUBs) in a process called deubiquitination (Mevissen and Komander, 2017). DUBs protect the components of SRSs from degradation by deubiquitination, thus enhancing their reception of SRSs, and directly act on cytokines, inhibitory immune checkpoints or other stem cell factors in the TME. While DUBs activate and upregulate the SRSs, they can also potentially enhance the effect of CSCs on promoting ITAIM, thereby inducing tumor immunity escape, progression and drug resistance (Sun et al., 2020). Recently, DUBs inhibitors have been actively developed, and their therapeutic effects have been confirmed in preclinical trials. However, the space and potential for the exploration of DUBs between CSCs and TAIM regulation remain expansive.
This review summarizes the processes and mechanisms by which CSCs create ITAIMs through four main SRSs. In addition, it summarizes the unique roles of DUBs in regulating stem cell factors, SRSs and TAIMs. The research and application progress of potential target pathways and inhibitors are also reviewed. DUB inhibitors are expected to become CSC-targeted drugs and TAIM modulators in future clinical treatments (Fu et al., 2019; Huang et al., 2019; Dai et al., 2020; Li J. et al., 2020). They can be combined with immunotherapy to enhance the recognition and killing of CSCs to reduce tumor recurrence.
CSCs and ITAIMs
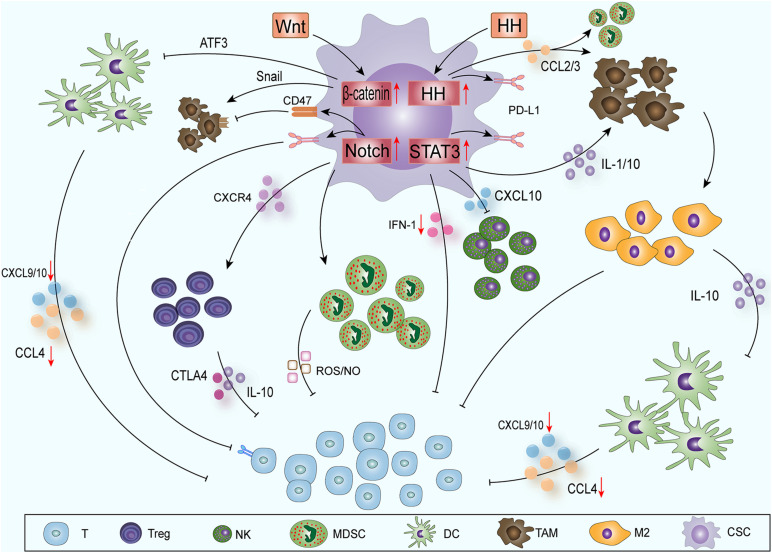
CSCs contain some molecular signaling pathways, such as Notch (Grazioli et al., 2017), nuclear factor κB (NF-κB) (Ferrandino et al., 2018), Hedgehog (HH) (Grund-Groschke et al., 2019), Wnt (Luke et al., 2019), signal transducer and activator of transcription (STAT3) (Fouse and Costello, 2013), phosphatase and tensin homolog deleted on chromosome ten (PTEN) (Li et al., 2017) and phosphatidylinositol 3-kinase (PI3K)/protein kinase B (also known as AKT) (Xia and Xu, 2015); these pathways can promote the epithelial-mesenchymal transition, immune cell migration, immunosuppression and treatment resistance. CSCs mainly influence the immune cells around the tumor through the Notch-NF-κB axis, HH, Wnt and STAT3 pathways to create ITAIMs (Figure 1).
FIGURE 1.
CSCs create ITAIMs mainly through four SRSs: The Notch-NF-κB axis, HH, Wnt and STAT3 pathways. Exogenous Wnt and HH proteins enter CSCs to activate corresponding SRSs. Wnt signaling acts on CD103+ DCs by inducing the transcription inhibitor ATF3. The reduction in CCL4, CXCL-9 and -10 reduces CD8+ T cell activation and infiltration. HH signaling recruits TAMs and immunosuppresses MDSCs by releasing CCL2 and CCL3. Notch-NF-κB axis signaling mainly regulates Tregs, MDSCs and T cells through a series of cytokines in the TAIM. Activated STAT3 signaling can promote the accumulation of immunosuppressive CD163+ TAMs through IL-1 and IL-10. It also downregulates the expression of CXCL-10 and reduces the cytotoxicity of NK cells. The activation of STAT3 inhibits the expression of type I IFN signaling and antitumor immune responses. The HH and Notch-NF-κB axis and STAT3 signaling pathways can also promote PD-L1 expression and suppress the immune response.
The Notch-NF-κB Axis
Notch is a classic and conservative signaling pathway. The target genes of Notch signaling mainly regulate the phenotype of CSCs, cell survival and differentiation and cell cycle and apoptosis (Andersson et al., 2011). At the same time, Notch is a regulator of CSCs and the TME, and its positive and negative effects have been confirmed by many studies (Weng et al., 2004; Venkatesh et al., 2018). Notch signaling mainly regulates three subgroups in the TAIM: regulatory T cells (Tregs), myeloid-derived suppressor cells (MDSCs) and T cells (Grazioli et al., 2017). In Tregs, Notch can also cascade with NF-κB transcription factors to play a regulatory role. In the TAIM, the classic NF-κB pathway is at the core of Treg functions regulated by Notch1 and Notch3 (Ferrandino et al., 2018). In leukemia, Notch1 can regulate the expression of NF-κB subunits directly to regulate T cells (Osipo et al., 2008) or can bind with the NF-κB subunit to regulate transcription indirectly in different environments and cells (Song L.L. et al., 2008). Notch can also enhance Treg activity through the C-X-C chemokine receptor type 4 (CXCR4) pathway and affect MDSCs through the interleukin (IL) -6-STAT3 axis, thereby promoting the formation of ITAIMs (Weng et al., 2004; Li et al., 2018).
Hedgehog
HH signaling plays an important role in embryonic development, but its abnormal activation has been shown to be related to the development of a variety of tumors (Katoh, 2019). In recent years, HH signaling has also been found to be related to ITAIM formation and immune escape. Hanna identified that HH signaling alters a critical kinomic signature that enables macrophages to assume the alternative M2 phenotype (Hanna et al., 2019). Chakrabarti’s study showed that HH signaling can induce the expression of programmed death-ligand 1 (PD-L1) in gastric tumor cells and can protect cancer cells from CTL-induced apoptosis (Chakrabarti et al., 2018). It has also been reported that in patients with advanced basal cell carcinoma, the application of HH inhibitors can improve the therapeutic effect of immune checkpoint inhibitors (Nikanjam et al., 2019). In glioma, HH signaling can recruit tumor-associated macrophages (TAMs) and can cause the immunosuppression MDSCs by releasing C-C motif chemokine ligand (CCL) 2/3 (Mi et al., 2020), thereby inducing PD-L1 expression, inducing stromal cells to produce IL10, and promoting the transcription factor forkhead box P3 (FOXP3) expression in Tregs (Grund-Groschke et al., 2019). HH signaling can also be activated by many other intracellular signals, including transforming growth factor β (TGF-β)-, kirsten rat sarcoma viral oncogene (KRAS)-mitogen activated protein kinase (MAPK)/extracellular regulated protein kinases (ERK)-, PI3K- AKT-, insulin-like growth factor—and tumor necrosis factor (TNF) -α induced mammalian target of rapamycin (mTOR)/ribosomal S6 kinase 1 (S6K1) activation (Takebe et al., 2015; Giroux-Leprieur et al., 2018).
Wnt
With the advancement of sequencing technology and the comprehensive understanding of cancer genomes, abnormal Wnt signaling has been observed in many cancer entities (Ruan et al., 2020; Yang et al., 2020). In the past few years, three types of Wnt cascade signaling have been described. Among these pathways, the typical Wnt signaling pathways related to tumors involves the activation of the T-cell-specific β-catenin–transcription factor–lymphoid enhancer-binding factor pathway, which plays a key role in immune regulation (Zhong and Virshup, 2020). The Wnt/β-catenin pathway has been identified as one of the important signaling pathways related to immune escape. In creating ITAIMs, Wnt signaling acts Wnt signaling on CD103+ dendritic cells (DCs) by inducing the expression of the transcription inhibitor ATF3, reducing the production of CCL4, C-X-C motif chemokine ligand (CXCL) -9 and -10 and inhibiting the activation and infiltration of CD8+ T cells (Fu et al., 2015). At the same time, in a pan-cancer analysis based on TCGA database information, Wnt/β-catenin pathway activation is usually associated with low T-cell infiltration and immune rejection (Luke et al., 2019). β-catenin signaling activation reduces the recruitment of DCs in hepatocellular carcinoma, thereby impairing T-cell activity (Ruiz de Galarreta et al., 2019). β-catenin signaling can also inhibit innate immunity by regulating interferon regulatory factor (IRF) 3, thereby downregulating the production of type I interferon (IFN) in colorectal cancer (Ding et al., 2018). Wnt signaling also affects TAMs through Snail to produce IL-1β and increase β-catenin activity (Spranger and Gajewski, 2015) to maintain Wnt signaling activation (Boulter et al., 2015).
STAT3
The activation of STAT3 signaling in CSCs is key to tumor progression because it promotes immunosuppressive factors and angiogenesis (Sasidharan Nair et al., 2018; Wang Y. et al., 2018). Many oncogenic signaling pathways, such as RAS/BRAF/MEK, can participate in the regulatory network of a TAIM. They cooperate with STAT3 and NF-κB signaling to regulate the expression of cytokines [IL-6, IL-1, IL-10, TNF, and vascular endothelial growth factors (VEGFs)] (Sasidharan Nair et al., 2018; Ji et al., 2019). Activated STAT3 signaling can promote the accumulation of immunosuppressive CD163+ TAMs and has an inhibitory effect on T cells. Depletion of CD163+ TAMs can enhance the infiltration of CD8+ T cells into melanoma and promote CD8+ T-cell-mediated tumor regression in mice (Etzerodt et al., 2019). T helper cell (Th) 2 and 17, Tregs and MDSCs produce inflammatory cytokines such as IL-1, IL-17, IL-10, TGF-β, and VEGFs through STAT3 signaling. These cytokines can promote the M2 polarization of macrophages and the proliferation of MDSCs, accelerating the formation of ITAIMs and the proliferation of tumor cells (Owen et al., 2019). The activation of STAT3 has been shown to inhibit the expression of STAT1, IRF7 and IRF 9 genes, type I IFN signaling and antitumor immune responses (Lu et al., 2018). Loannis confirmed that STAT3 signaling promotes PD-L1 expression and suppresses the immune response in breast cancer. In addition, STAT3 downregulates the expression of CXCL-10 and reduces the cytotoxicity induced by natural killer cells (NK) (Clark et al., 2019).
DUBs Involved in Stem Cell Factors, SRSs and ITAIM
Ubiquitination and deubiquitination constitute a whole, dynamic and specific biological process (Wang and Le, 2019). They are involved in the regulation of almost all life activities (Clague et al., 2019). Even in the progression and immune escape of a variety of cancer cells, DUBs also play novel functions, including regulating tumor immunogenicity, activating stem cell factors, upregulating the SRSs related to CSCs, stabilizing anti-inflammatory receptors, and regulating anti-inflammatory cytokines (Zhang et al., 2008; Qiu et al., 2017; Wang Y.C. et al., 2018; Huang et al., 2019; Lei et al., 2019; Li L. et al., 2019; Lai et al., 2020; Li J. et al., 2020). Different studies have emphasized the mechanisms of CSCs in the “re-editing” of the immune system to change the balance between antitumor immune cells and tumor-promoting immune cells. Research on DUBs that can regulate CSCs and ITAIMs has great potential for the use of synergistic immunotherapy. The therapeutic effects of DUB inhibitors such as USP1, USP4, USP7, USP14 and USP33 have been confirmed in prostate cancer, lung cancer, breast cancer and hematological malignancies (Ma et al., 2019; Xia et al., 2019a; Guo et al., 2020; Gutierrez-Diaz et al., 2020; Lai et al., 2020).
Ubiquitin-specific peptidases (USPs) comprise the largest family of deubiquitinases. Any abnormal immune response activation will cause pathological damage; therefore, the immune response is also strictly regulated. Among these regulators, DUBs in the ubiquitin-proteasome system play vital roles in regulating the immune response (Lopez-Castejon and Edelmann, 2016). Without exception, DUBs can directly affect the ITAIM by acting on cytokines and inhibitory immune checkpoints. In addition, nearly one-half of the USPs that have been identified can exhibit potential carcinogenic and cancer-promoting functions (Cheng et al., 2019). Among these USPs, USP4, USP7, USP14, USP15, USP22, and CYLD have been the most widely studied, and they could also regulate the stem cell factors and SRSs (Table 1 and Figure 2).
TABLE 1.
The DUBs associated with stem cell factors and the stemness-related signals.
| DUBs | Tumor | Substrates | Stemness-related signals | References |
| USP4 | Colon cancer | β-catenin | Wnt/β-catenin | Nguyen et al., 2019 |
| USP4 | Liver cancer | TGFβR-1 | HH | Qiu C. et al., 2018 |
| USP4 | Glioblastoma | TGFβR-1 | ERK˴HH | Zhou et al., 2019 |
| USP4 | Lung adenocarcinoma | β-catenin | Wnt/β-catenin | Hwang et al., 2016 |
| USP4 | IRF8 | Lin et al., 2017 | ||
| USP7 | T-Cell Leukemia | Notch1 | Notch | Jin et al., 2019 |
| USP7 | Colon cancer | β-catenin | Wnt/β-catenin | Novellasdemunt et al., 2017 |
| USP7 | Multiple myeloma | NEK2 | NF-κB˴Notch | Franqui-Machin et al., 2018 |
| USP7 | Promyelocytic leukemia protein | PTEN | PTEN/PI3K/AKT mTOR | Song M.S. et al., 2008 |
| USP7 | FOXP3 | FOXP3 | van Loosdregt et al., 2013 | |
| USP14 | Lung adenocarcinoma | β-catenin | Wnt/β-catenin | Wu et al., 2013 |
| USP14 | Colon cancer | DVL | Wnt | Jung et al., 2013 |
| USP15 | Glioblastoma | TGFβR-1 | HH | Eichhorn et al., 2012 |
| USP15 | TAB2 and TAB3 | Notch/NF-κB | Zhou Q. et al., 2020 | |
| USP15 | Glioblastoma | TGFβR-1 | HH | Eichhorn et al., 2012 |
| USP15 | MDM2 | MDM2 | Zou et al., 2014 | |
| USP15 | Malignant Hematopoiesis | MDM2 | MDM2 | Niederkorn et al., 2020 |
| USP15 | Melanoma | TET2 | Chen L.L. et al., 2020 | |
| USP15 | Breast cancer | BMI1 | BMI1 | Zhang L. et al., 2020 |
| USP22 | Non-small cell lung cancer | PD-L1 | Wang et al., 2020 | |
| USP22 | Liver cancer | PD-L1 | Huang X. et al., 2020 | |
| USP22 | Gastric cancer | SOS1 | PI3K/AKT | Lim et al., 2020 |
| USP22 | Glioblastoma | BMI1 | BMI1 | Qiu et al., 2020 |
| USP22 | Colon cancer | BMI1 | BMI1 | Yuan et al., 2019 |
| USP22 | Gastric cancer | MYC | c-Myc/NAMPT/SIRT1 | Liu H. et al., 2019 |
| USP22 | FOXP3 | FOXP3 | Cortez et al., 2020 | |
| USP22 | MED1 | Zhang Y. et al., 2020 | ||
| CYLD | Cylindroma skin tumors | DVL | Wnt/β-catenin | van Andel et al., 2017 |
| CYLD | Chronic myeloid leukemia | DVL | Wnt/β-catenin | Caliskan et al., 2017 |
| CYLD | Cervical carcinoma | TRAF˴Bcl-3 | Notch/NF-κB | Brummelkamp et al., 2003 |
| CYLD | Skin tumor | TRAF˴Bcl-3 | Notch/NF-κB | Massoumi et al., 2006 |
| CYLD | Salivary gland tumor | NF-κB | Notch/NF-κB | Fukuda et al., 2008 |
| CYLD | Breast cancer | NF-κB | Notch/NF-κB | Hayashi et al., 2014 |
| CYLD | Hepatocellular carcinoma | TRAIL | Notch/NF-κB | Chu et al., 2006 |
DUBs, deubiquitylating enzymes; CSCs, cancer stem cells; USP, ubiquitin-specific peptidase; TGFβR-1, TGF-β receptors; HH, hedgehog; ERK, extracellular regulated protein kinases; IRF, interferon regulatory factor; NF-κB, nuclear factor κB; PI3K, phosphatidylinositol 3-kinase; AKT, protein kinase B; mTOR, mammalian target of rapamycin; FOXP3, forkhead box P3; PTEN, phosphatase and tensin homolog deleted on chromosome ten; DVL, disheveled; PD-L1, programmed death-ligand 1; SOS1, son of sevenless 1; NAMPT, Nicotinamide phosphoribosyltransferase; TRAIL, TNF-related apoptosis-inducing ligand.
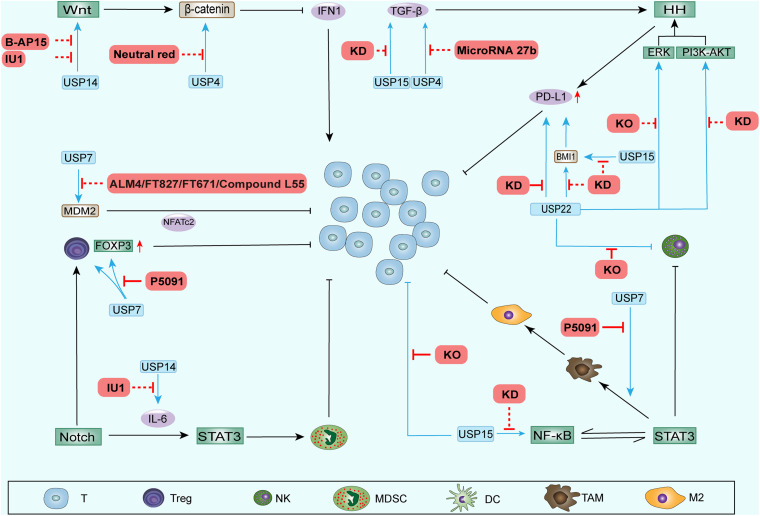
FIGURE 2.
DUBs involved in stem cell factors, SRSs and ITAIM. DUBs can regulate the stem cell factors and the components of SRSs, promoting stemness of CSCs. Additionally, DUBs can affect ITAIMs directly by acting on cytokines, inhibitory immune checkpoints or other oncogenes. The combination of DUBs with substrates leads to deubiquitination and protects substrates from degradation. Therefore, the protein level of the substrate and the signaling pathways involved are upregulated. The arrow at the end of the line in the figure represents upregulation, and the horizontal line represents downregulation.
USP4
USP4 not only plays a role in promoting cancer in a variety of tumor tissues but also indicates a poor prognosis when highly expressed (Yun et al., 2015; Cao et al., 2016; Qiu C. et al., 2018). USP4 can promote cancer in various ways, such as inhibiting SMAD4 monoubiquitination and promoting activin and BMP signaling (Zhou et al., 2017), promoting breast cancer metastasis via the p21-activated kinase 5-aspartyl aminopeptidase-USP4 axis (Geng et al., 2020) and promoting the invasion of breast cancer cells via the relaxin/TGF-β1/SMAD2/MMP-9 signaling pathway (Cao et al., 2016). In recent studies, we found that the USP4 are also involved in the regulation of the components in SRSs. USP4 promotes liver cancer metastasis by upregulating TGF-β signaling to induce the epithelial-mesenchymal transition (Qiu C. et al., 2018) and activate the ERK pathway by regulating TGF-β to promote the development of glioblastoma (Zhou et al., 2019). Both TGF-β and ERK signaling can activate HH signaling in CSCs. USP4 also stabilize β-catenin in brain metastasis lung adenocarcinoma to affect metastasis ability (Hwang et al., 2016). In colorectal cancer, USP4 upregulates Wnt/β-catenin signaling through deubiquitination and facilitates the nuclear localization of β-catenin (Yun et al., 2015; Nguyen et al., 2019). Recent studies have found that USP4 can deubiquitinate the transcriptional factor retinoic acid-related orphan receptor γt in an inflammatory environment and play a positive regulatory role in Th17 cell function (Yang et al., 2015). Th17 cells can produce inhibitory inflammatory cytokines through STAT3 signaling. In addition, USP4 stabilizes IRF8 through K48-linked deubiquitination and enhances the immunosuppressive function of Tregs (Lin et al., 2017). In summary, USP4 can strengthen the SRSs and directly act on immune cells to promote the ITAIM.
USP7
USP7 has been found to be highly expressed in a variety of tumors and is associated with poor prognosis (Zhang et al., 2016; Wang X. et al., 2018; Xia et al., 2019b; Zhang M.J. et al., 2019). The regulatory role of USP7 in TAIMs has attracted increasing attention, and it can also affect the SRSs. In T-cell leukemia, USP7 can regulate Notch1 at the transcriptional and post-translational levels and lead to increased expression of Notch1 target genes (Shan et al., 2018; Jin et al., 2019). After inhibiting USP7, Notch1 is ubiquitinated and degraded (Jin et al., 2019). In colorectal cancer, USP7 activates Wnt signaling through β-catenin deubiquitination to promote the growth of ectopic transplantation tumors (Novellasdemunt et al., 2017). It can also prevent the ubiquitination of NF-κB and its regulator NEK2 (Colleran et al., 2013; Franqui-Machin et al., 2018), activating NF-κB signaling. In addition, monoubiquitinated PTEN can be deubiquitinated by USP7, leading to the elimination of PTEN in the nucleus and activating the PI3K/AKT/mTOR signaling pathway. PTEN/PI3K/AKT, as one of the SRS pathways of CSCs, has been proven to maintain stemness and promote tumor growth in a variety of cancers (Li et al., 2017; Hu et al., 2019; Luongo et al., 2019). Classic PTEN/PI3K/AKT signaling plays a core role in maintaining the characteristics and stability of Tregs (Brandmaier et al., 2017). USP7 can also reduce and stabilize the ubiquitination of FOXP3 directly, enhancing the immunosuppressive ability of Tregs (van Loosdregt et al., 2013). Tregs suppress the immune response by expressing cytotoxic T-lymphocyte-associated protein 4 (CTLA-4), IL-10, TGFβ, etc. This immunosuppressive property promotes the immune escape of tumor cells (Li C. et al., 2020). In addition, immunosuppressive PD-1 signaling prevents CK2-mediated degradation of PTEN by reducing the expression of casein kinase 2 (CK2) (Patsoukis et al., 2013). In summary, USP7 can directly upregulate the SRSs of multiple cancers and enhance the effect of immunosuppressive cells.
USP14
USP14 has been extensively studied because of its overexpression in lung cancer (Han et al., 2019), breast cancer (Xia et al., 2019a), ovarian cancer (Luo et al., 2019), and oesophageal squamous cell carcinoma (Sha et al., 2019). It may promote tumorigenesis in approximately 61% of cancers (Liu B. et al., 2019). USP14 inhibitors have also shown treatment efficacy in research settings (Xia et al., 2019a). We focus on the mechanism of USP14 involved in the regulation of SRS and ITAIM. In terms of WNT/β-catenin signaling, USP14 is overexpressed in lung adenocarcinoma and promotes cancer cell proliferation by upregulating β-catenin (Wu et al., 2013). Another study found that USP14 deubiquitinates the k63-linked polyubiquitin and dissociates from DVL. The DVL is subsequently phosphorylated and then activates and upregulates Wnt signaling (Jung et al., 2013). USP14 also promotes cell adhesion-mediated drug resistance by enhancing Wnt signaling in multiple myeloma cells (Xu et al., 2017). It has been reported that the inflammation regulation mediated by USP14 through lipopolysaccharides is related to the activation of NF-κB (Liu et al., 2017). Furthermore, USP14 also accelerates the degradation of IκBα mediated by the proteasome to promote the activation of NF-κB (Li M. et al., 2019). In addition, Xu found that USP14 cause resistance to cisplatin through the activation of AKT signaling in gastric cancer (Fu et al., 2018). In summary, the research on USP14 has made great progress in delineating its roles in the proliferation, migration and autophagy of different cancer cells. Although USP14 inhibitors have also been shown to be useful in cancer treatment, broad space for research on USP14 with respect to ITAIM regulation remains open because of its activation and upregulation of SRS.
USP22
USP22 was thought to have a cancer-promoting effect, and its gene was confirmed as one of 11 genes called “death-from-cancer” (Glinsky et al., 2005). However, in recent studies, the cancer-promoting or anticancer effects of USP22 were found to depend mainly on its environment (Jeusset and McManus, 2017). In most tumors, high expression of USP22 predicts a poor prognosis (Hu et al., 2015; Lim et al., 2020), and USP22 also been shown to promote tumor proliferation, invasion and drug resistance through a variety of substrates, including PD-L1 (Huang et al., 2019; Huang X. et al., 2020; Wang et al., 2020), RAS activator son of sevenless 1 (SOS1) (Lim et al., 2020), BMI1 (Ma et al., 2017; Qiu G.Z. et al., 2018; Yuan et al., 2019; Qiu et al., 2020), MYC (Liu H. et al., 2019; Zhang K. et al., 2019), and COX-2 (Xiao et al., 2015). These substrates include inhibitory immune checkpoints, transcriptional activators and oncogenes. In non-small cell lung cancer, USP22 can directly deubiquitinate PD-L1 and regulate the level of PD-L1 through the USP22-CSN5-PD-L1 axis, resulting in decreased T-cell proliferation and activation (Wang et al., 2020). Furthermore, studies have shown that knocking out USP22 inhibits the growth of liver cancer in an immune-dependent manner. Tumor immunogenicity and lymphocyte infiltration were enhanced, and the efficacy of PD-L1 immunotherapy and CDDP were also improved (Huang et al., 2019). SOS1, another substrate of USP22, was found to activate RAS signaling and upregulate the PI3K/AKT pathway (Lim et al., 2020). The PI3K/AKT pathway is a carrier of SRSs and promotes Treg activation (Brandmaier et al., 2017). The stem cell factors BMI1 has been proven to be a downstream target of USP22 in glioma, gastric and colorectal cancer. It is upregulated by USP22 deubiquitination (Ma et al., 2017; Qiu G.Z. et al., 2018; Yuan et al., 2019; Qiu et al., 2020). The latest research found that inhibiting BMI1 can lead to the recruitment and activation of CD8+ T cells by stimulating IRF3. Targeting BMI1 may eliminate CSCs by silencing PD-L1, and BMI1 inhibition can also attenuate tumor metastasis and recurrence by activating endogenous immunity (Jia et al., 2020). Therefore, USP22 may create ITAIMs through the BMI1 signaling pathway. USP22 can exert its cancer-promoting effect by upregulating the nicotinamide phosphoribosyltransferase (NAMPT)/SIRT1, AKT and ERK signaling pathways involved with c-Myc activation (Liu H. et al., 2019; Zhang K. et al., 2019). In a report, Myc upregulated the expression of CD47 and PD-L1 on the surface of CSCs. CD47 inhibited phagocytosis by binding to sirpα on macrophages (Park and Surh, 2017). In a study using CRISPR to screen the ubiquitination modulators of FOXP3 in Tregs, it was found that USP22 and Atxn713, members of the deubiquitination module of the SAGA stain complex, were positive regulators of FOXP3. In other words, USP22 may inhibit the immune response to tumor cells by directly acting on FOXP3 (Cortez et al., 2020). Furthermore, Han also proved that USP22 can promote tumor development through deubiquitination and immunosuppression in lung adenocarcinoma (Han et al., 2020). USP22 was found to activate MED1 transcriptionally to regulate NK cell development through histone H2A deubiquitination (Zhang Y. et al., 2020).
In summary, the regulation of USP22 in CSCs and TAIMs is abundant and has the potential to suppress immunity in many ways. In some preclinical experiments, USP22 used as a regulator achieved encouraging effects (Li J. et al., 2020).
USP15
USP15 has a high degree of homology with the abovementioned carcinogenic USP4, and its gene upregulation has been found in glioblastoma, breast cancer and ovarian cancer (Chou et al., 2017). However, USP15 is the same as USP22 in that its role in cancer depends on the environment (Cheng et al., 2019). USP15 acts as a regulator of the natural immune response in the anti-infection processes of the human body (Torre et al., 2017; Kusakabe et al., 2019; Huang L. et al., 2020). It has been reported that USP15 can participate in regulating multiple stem cell factors, including MDM2 (Zou et al., 2014; Niederkorn et al., 2020), TET2 (Chen L.L. et al., 2020), BMI1 (Zhang L. et al., 2020), NF-κB (Zhou Q. et al., 2020), and TGF-β (Eichhorn et al., 2012). The aforementioned signaling pathways activated by these factors can promote the production of IATIM. MDM2 not only can promote tumor progression by downregulating P53 (Niederkorn et al., 2020) but can also negatively regulate Tregs by acting on the transcription factor NFATc2 (Zou et al., 2014). In breast cancer, USP15 is found to deubiquitinate and stabilize BMI1 protein at lys-81 (Zhang L. et al., 2020). Recent studies have also found that USP15 exerts a deubiquitination effect at the K1299 site of TET2 and negatively regulates the activity of TET2. Therefore, USP15 weakens the TET2-mediated IFN/JAK/STAT pathway and reduces the expression of chemokines and lymphocytic infiltration. Knocking out USP15 improves the response of mice with transplanted melanoma to immunotherapy and prolongs survival (Chen L.L. et al., 2020). In summary, USP15 mainly acts on stem cell factors to affect CSCs and ITAIM, other potential pathways need to be explored and verified.
CYLD
CYLD plays a major role in cell development and proliferation, and it is also an important inflammation regulator (Hadian et al., 2011). CYLD inhibits the NF-κB pathway by deubiquitinating key factors, including NF-κB essential modulators IKKg and TRAF2 (Kovalenko et al., 2003). In contrast to other oncogenic DUBs of the USP family, CYLD is the DUB with the most typical tumor suppressor effects. Patients with high CYLD expression and multiple myeloma (van Andel et al., 2017), oral squamous cell carcinoma (Shinriki et al., 2018) or breast cancer (Hayashi et al., 2014) have a better prognosis. Several substrates of CYLD have been identified in recent studies, including DVL (Tauriello et al., 2010), TRAF (Brummelkamp et al., 2003), Bcl-3 (Massoumi et al., 2006), IKK (Chu et al., 2006), and p53 (Fernandez-Majada et al., 2016). Some of these substrates act as CSCs factors, which can activate and upregulate the SRSs. For example, the upregulation of CYLD causes DVL deubiquitination, significantly inhibiting the oncogenic Wnt pathway, thereby inhibiting the occurrence of tumors (Caliskan et al., 2017). The downregulation of CYLD promotes the accumulation of β-catenin in the nucleus through ubiquitination of DVL at Lys-63 (Tauriello et al., 2010). In addition, CYLD deubiquitinates TRAF and Bcl-3 proteins directly to downregulate NF-kB activity, inhibiting oncogenic transformation in keratinocytes (Brummelkamp et al., 2003; Massoumi et al., 2006). In salivary gland tumors, CYLD has a negative regulatory effect on NF-kB activity after TNF-α stimulation (Fukuda et al., 2008). Studies have also found that the downregulation of CYLD can promote breast cancer metastasis by activating NF-κB (Hayashi et al., 2014). In the inhibition process of TNF-related apoptosis-inducing ligand (TRAIL)-mediated NF-κB, the direct combination of CYLD and IKK enhances the effect of TRAIL (Chu et al., 2006).
The SRSs and CYLD influence each other. The upstream regulatory mechanism of CYLD transcription and translation has been extensively studied in tumors. A variety of oncogenic signaling pathways can regulate CYLD, and some of them belong to SRSs. Studies have shown that Hes-1, a downstream factor of Notch in T-cell leukemia, can reduce the expression of CYLD (D’Altri et al., 2011). STAT3 signaling can activate miR-21 (Iliopoulos et al., 2010) and miR-181b (Xu et al., 2016) to inhibit CYLD transcription. In chronic leukemia, lymphoid enhancer binding factor 1 (LEBF1) downregulates CYLD at the transcriptional level through the WNT/β-catenin signaling pathway (Liu et al., 2012). HH signaling in basal cell carcinoma downregulates the expression of CYLD through GLI1 under the action of the transcription repressor Snail (Kuphal et al., 2011).
In summary, CYLD, as a tumor-suppressing DUB, has a close relationship with CSCs. It potentially reverses the ITAIM by regulating the SRSs, and even improve the effect of immunotherapy.
Recurrence Caused by CSCs After Initial Treatment
Although surgery, radiotherapy, chemotherapy, and immunotherapy have been individualized and combined clinically, they still cannot completely solve the problems of tumor recurrence and treatment resistance (Das and Law, 2018). The initial comprehensive clinical treatment can eliminate most tumor cells, but CSCs can evade the killing of immune cells by creating ITAIMs, reducing their autoantigenicity, and upregulating inhibitory immune checkpoints. CSCs can also resist radiotherapy and chemotherapy through protective autophagy, efficient cell cycling, promotion of the epithelial-mesenchymal transition, removal of reactive oxygen species, drug efflux, DNA repair, and interactions with the TME (Najafi et al., 2019). These CSCs survive and may remain quiescent for a long time without losing their tumorigenic potential (Quayle et al., 2018; Brown et al., 2019). They are temporarily in balance with the immune system. However, due to aging, immunosuppressive therapy, disease or other factors, the immune system may lose its ability to suppress these CSCs, leading to refractory tumor recurrence (Bruttel and Wischhusen, 2014). As described above, CSCs act as tumor seeds. If they cannot be eradicated during the initial treatment, they will grow like weeds. A prairie fire cannot not destroy them; they shoot up again when the spring breezes blow.
In traditional treatment, most of the visible tumor tissue is removed by surgery, but this treatment creates a hypoxic environment that is beneficial to the maintenance of CSC stemness. In addition, lymph node dissection destroys the optimal microenvironment for T-cell activation (Najafi et al., 2020). Radiotherapy can only kill local tumor cells. Although chemotherapy can act on tumor cells in the blood, it cannot kill CSCs. Moreover, some cytotoxic drugs kill immune effector cells directly or indirectly, resulting in immune tolerance, suppression or incompetence (Shaked, 2016). Therefore, traditional treatment not only fails to prevent treatment failure and death caused by drug resistance and refractory recurrence but also destroys the immune system, which is not conducive to immunotherapy.
However, initial treatment can block the direct and indirect regulation pathways of ITAIM and reverse it. The purpose is to enhance the recognition and killing of CSCs by immunotherapy. Relying on autoimmune function to eliminate residual tumors, including CSCs, is the most promising way to prevent tumor recurrence.
Research on DUB Inhibitors and their Effects on SRSs and TAIM
Some DUB inhibitors have been actively developed and play inhibitory roles in tumor development. We summarize the recent progress in inhibitors of DUBs reported above (Table 2 and Figure 3).
TABLE 2.
Related research on inhibitors of DUBs that regulate TAIM.
| Regulate TAIM | DUBs inhibitors | DUBs | Selectivity | Function | References |
| Potential | Neutral red | USP4 | HS | Suppresses colorectal cancer by regulating β-Catenin signaling | Nguyen et al., 2019 |
| Potential | MicroRNA 27b | USP4 | HS | Protect Hepatocytes From TGF-β | Wu et al., 2019 |
| Potential | ALM4 | USP7 | HS | Increases p53 and decreases MDM2 total levels in cells | Gavory et al., 2018 |
| Potential | FT827 | USP7 | HS | Degradation of oncogenic E3 ligase MDM2 to inhibit tumor | Turnbull et al., 2017 |
| Potential | FT671 | USP7 | HS | Degradation of oncogenic E3 ligase MDM2 to inhibit tumor | Turnbull et al., 2017 |
| Detected | P5091 | USP7 | HS | Reduce the proportion of Treg cells | Fu et al., 2019 |
| Detected | P5091 | USP7 | HS | Impair Treg cell function and promotes antitumor immunity | Wang et al., 2016 |
| Detected | P5091 | USP7 | HS | Reprogram TAM to regulate antitumor immune response | Dai et al., 2020 |
| Potential | Compound L55 | USP7 | HS | Antitumor by inducing cell death and reduce the levels of MDM2 | Li M. et al., 2020 |
| Potential | IU1 | USP14 | PS | Decrease TNF-α, IL-1-β, IL-6 and IL-8, and increase I-κB | Kiprowska et al., 2017 |
| Potential | IU1 | USP14 | PS | Down regulation of Wnt/β- Catenin and PI3K/AKT pathway | Xia et al., 2019a |
| Potential | B-AP15 | USP14 | PS | Inhibition of Wnt /β- Catenin and TGFβ/ Smad pathway | Jiang et al., 2019 |
ND, not detected; HS, highly selective; PS poor selectivity.
FIGURE 3.
DUB inhibitors and other inhibition methods performed to regulate the TAIM. Some inhibitors and other inhibition methods can block the upregulation of stemness-related signaling pathway components by DUBs. These processes weaken the function of CSCs during ITAIM creation through SRSs and possibly reverse the ITAIM. In addition, knocking out or knocking down some DUB genes can directly regulate the recruitment, quantity and function of immune cells in the TAIM. The red solid line represents the regulatory effect in the TAIM detected in preclinical trials, and the red dotted line represents the potential regulatory effect.
As one of the most extensively studied inhibitors, USP7 inhibitors have played an effective and highly specific role in many studies. They have a significant inhibitory effect on tumors (Lee et al., 2020; Leger et al., 2020; Ohol et al., 2020; Schauer et al., 2020a). P22077/50429 exerted anti-oncogene function in a variety of malignant tumors (Fan et al., 2013; Zhang W. et al., 2020) and improved chemoresistance in pancreatic cancer (Chen H. et al., 2020). FT827/671 inhibited tumors by degrading the oncogenic E3 ligase MDM2 (Turnbull et al., 2017). In the latest study, P5091 inhibitors showed robust tumor suppression effects in preclinical models of various tumors (An et al., 2017; Wang et al., 2017; Hayal et al., 2020; He et al., 2020; Lee et al., 2020). Among these cancers, in colorectal cancer, P5091 improves the antitumor immune response and reduces the proportion of Tregs in tumor xenografts in mice (Fu et al., 2019). P5091 has been reported to impair Tip60-dependent FOXP3+ Treg function and promote antitumor immunity in lung cancer (Wang et al., 2016). In addition, it modulated sensitivity to immunotherapy by reprogramming the TAM in lung cancer and has become a candidate drug for regulating immunotherapy (Dai et al., 2020).
Many other inhibitors have been found to inhibit USP14. It was found that VLX1570 can improve ibrutinib or bortezomib residence in Waldenstrom macroglobulinemia tumors by targeting USP14 and uchl5 and downregulating BCR-related components such as NF-κB and CXCR4 (Paulus et al., 2016). The Notch1 signaling pathway involves NF-κB and CXCR4 promoting the formation of ITAIMs. As a selective small-molecule inhibitor of USP14, IU1 can increase the expression of I-κB by reducing TNF-α, IL-1β, IL-6, and IL-8 levels (Kiprowska et al., 2017). TNF-α and IL-6 are stemness-related signaling pathway components in CSCs, and an increase in I-κB can inhibit NF-κB. In recent studies, the use of IU1 to inhibit USP14 enhanced the growth inhibition and apoptosis of breast cancer cells by enzalutamide. In addition, the combined application of enzalutamide and IU1 downregulated the Wnt/β-catenin and PI3K/AKT pathways associated with SRSs (Xia et al., 2019a). In diffuse large B-cell lymphoma, the small-molecule inhibitor of USP14 b-AP15 inhibited the spread of tumor cells by blocking the Wnt/β-catenin and TGFβ/Smad pathways (Jiang et al., 2019). b-AP15 and chemotherapeutic drugs (such as Adriamycin or VP-16) have a synergistic antitumor effect on neuroblastoma (Yu et al., 2019).
Thus, the widely studied DUB inhibitors can inhibit tumor progression and attenuate drug resistance by downregulating SRSs. The downregulation of these signaling pathways also reverses the negative impact of DUBs on TAIMs. Encouraging results have been seen in studies on other methods of DUB inhibition that regulate TAIMs (Table 3 and Figure 3). The focus has gradually shifted to how to regulate TAIMs to enhance the efficacy of immunotherapy.
TABLE 3.
Related research on other inhibition methods of DUBs that regulate TAIM.
| Regulate TAIM | Inhibition methods | DUBs | Function | References |
| Detected | KD | USP22 | Increase tumor immunogenicity and tumor-infiltrating lymphocytes and improve therapeutic efficacy of CD274-targeted immunotherapy and CDDP-based chemotherapy | Huang et al., 2019 |
| Detected | KD | USP22 | Inhibit tumorgenesis and promote T cell cytotoxicity | Wang et al., 2020 |
| Detected | KD | USP22 | Improve PD-L1-targeted for anti-liver cancer immunotherapy | Huang X. et al., 2020 |
| Potential | KD | USP22 | Reverse the effects of RAS signaling and the PI3K/AKT pathway | Lim et al., 2020 |
| Potential | KD | USP22 | Reduce proliferation through down-regulating BMI1 signaling in colon cancer cells | Yuan et al., 2019 |
| Potential | KD | USP22 | Reduce the stemness and proliferation of GSCs through down-regulating BMI1 signaling | Qiu et al., 2020 |
| Potential | KD | USP22 | Decrease proliferation, migration, and invasiveness of GC cells through c-Myc/NAMPT/SIRT1-dependent signaling | Liu H. et al., 2019 |
| Potential | KO | USP22 | Down regulate c-Myc signaling and AKT and ERK pathways | Zhang K. et al., 2019 |
| Detected | KO | USP22 | Down regulate FOXP3 signaling in Treg cells | Cortez et al., 2020 |
| Detected | KD | USP22 | Reverse tumor development caused by immunosuppression | Han et al., 2020 |
| Detected | KO | USP22 | Promote the infiltration of T cells and NK cells | Li J. et al., 2020 |
| Potential | KO | USP7 | Stabilize IκBα and blocking the NF-κB pathway | Yao et al., 2018 |
| Detected | KO | USP15 | Promote T cell activation in vitro and enhanced T cell responses to tumor challenge in vivo | Zou et al., 2014 |
| Potential | KD | USP15 | Down regulate TNFα- or IL-1β-triggered NF-κB activation | Zhou Q. et al., 2020 |
| Potential | KD | USP15 | Decrease TGF-β activity | Eichhorn et al., 2012 |
| Potential | KD | USP15 | Down regulate BMI1 expression | Zhang L. et al., 2020 |
| Detected | KO | USP15 | Improve the response of melanoma transplanted mice to immunotherapy | Chen L.L. et al., 2020 |
KD, gene knockdown; KO, gene knockout.
The Structure of USPs and the Binding Pattern and Development of Inhibitors
To date, about 100 DUBs have been identified and can be divided into different subclasses according to their protease domains. USPs, ubiquitin C-terminal hydrolases (UCHs), ovarian tumor proteases (OTUs), Machado Joseph disease proteases (MJDs), Mindys and ZUP1 are cysteine proteases, JAB1/MPN/Mov34 (JAMMs) are zinc-dependent metalloprotease (Clague et al., 2019).
Although DUB is an attractive target, the clinical development of small molecule inhibitors is limited by several obstacles. Most DUBs have an active site for catalytic cysteine, through which inhibitors can function. However, it is challenging to screen selective and effective small molecule inhibitors in DUBs with conserved catalytic pockets. In addition, the oxidative hydrolysis of cysteine at the active site will cause a high false positives in screening inhibitors (Harrigan et al., 2018). The mechanisms of DUB enzymatic activity are complex, including the use of allosteric effect or substrate-mediated catalysis, and the switching between active and inactive conformations, which make the development and selection of specific DUB inhibitors complex (D’Arcy et al., 2015). Finally, DUBs with larger molecular weight contain many domains in addition to the catalytic site. These domains can promote the recognition and connection of substrate and Ub, and regulate enzymatic kinetics. Different DUB families have divergent folds in their catalytic domain (Schauer et al., 2020b).
USPs, as the largest family in DUBs, contain three conserved domains (Cys, His, and ASP/ASN boxes), which are responsible for the reorganization of ubiquitin regulated molecules (Pfoh et al., 2015). These three domains form the binding pocket of ubiquitin and insert its C-terminal tail into the cleft between domains. At the end of the cleft, there is a cysteine triad which can catalyze the hydrolysis of peptide bond. Finally, there are two “blocking rings” (BL1 and BL2) near the binding sites of ubiquitin that can change conformation and orientation of active sites depending on substrate (Schauer et al., 2020b). Therefore, the development of most inhibitors utilizes the structural plasticity of BL1 and BL2 rings to induce the conformational changes in Ub bound DUB structures (Mevissen and Komander, 2017).
In addition, the DUBs interface in USP family is extensive, and there may be some allosteric sites in the enzyme to change the balance of ubiquitin binding. For example, the C-terminus of USP7 can directly bind to the USP domain and affect the activity (Pozhidaeva and Bezsonova, 2019). The catalytic domain of USP14 has the same structure as USP7 (Wertz and Murray, 2019). However, development of most other DUBs inhibitors focuses on compounds, which form reversible or irreversible covalent adducts with DUB catalytic cysteine. The highly reactive parts of some of these compounds may limit the drug selectivity and be toxic to patients (Kemp, 2016).
Conclusion
In recent years, with the deepening of research, CSCs have been found to evade being killed by immune cells by changing TAMs, especially TAIMs, which hinder immunotherapy. CSCs are the key components in refractory tumor recurrence after treatment. During initial treatment, immunotherapy that leverages the body’s immune cells is the most promising way to eliminate CSCs completely. Therefore, reversing ITAIMs to enhance the ability of immune cells to kill tumor cells, including CSCs, provides new ideas for antitumor therapy. The mechanisms and the efficacy of co-application with other drugs are valuable directions of research. However, many challenges still need to be addressed.
At present, researchers have separately studied the effects of DUBs and CSCs on the ITAIM, but there is no reliable research to establish a cascade among the three. Although DUBs have been clearly confirmed to affect CSCs, there is no research on DUBs regulate TAIM through CSCs. Obviously, the establishment of a connection between the three is a new and potential perspective for tumor immunotherapy. In this review, we summarized DUBs related to these SRSs and stem cell factors, focusing on their effects on tumor development by upregulating these signals. Therefore, we searched for all the potential DUBs that could affect the SRSs, including some articles that did not prove the direct effect of DUBs on CSCs. Nonetheless, cancer cells have the dynamic ability of bidirectional conversion from non-CSC state to CSC-state (Li and Laterra, 2012). Any modification or loss of controllable differentiation can result in cancer cells with stem cell-like characteristics, such as self-renewal capability, epithelial mesenchymal transition and chemotherapy resistance (Marjanovic et al., 2013). The process is also termed phenotypic switch of CSC. Among which the regulation of the ubiquitin–proteasome system plays a significant role (Okita et al., 2007). Therefore, CSCs and cancer cells are transformed into each other in TME. However, as a potential indirect mechanism, it is impossible to determine whether cells compensate through other pathways after inhibiting DUB. The complex interactions during DUB inhibition are currently unclear. Second, because of the complexity and variability of DUB domains, the development of DUB inhibitors is extremely difficult. Only a few inhibitors of DUBs have been developed, and inhibitors of DUBs such as USP22 and USP15 have not yet led to breakthrough treatments. Moreover, although the inhibitors developed to date have shown anticancer efficacy, they lack sufficient specificity. The third point is that the current studies involving the effects of DUBs and their inhibitors on tumors are limited to preclinical trials, and the effects in clinical treatment are not clear.
In recent years, with the development of technology, single-cell sequencing (Wen and Tang, 2016; Xu et al., 2020), spatial transcriptomics sequencing (Zhou Y. et al., 2020) and ubiquitin mass spectrometry technologies (Ohtake, 2020) have matured. Accurate verification can be performed at the single-cell level, RNA level, and protein level. These technologies will be of great help in our future discovery of antitumor and immune regulatory signal pathways of DUBs. New technologies such as X-ray crystallographic analysis, virtual screening and molecular dynamics simulations have also been used to develop specific DUBs inhibitors (Li M. et al., 2020). Targeted DUBs is expected to become a new strategy to regulate TAIMs, strengthen immunotherapy, and eliminate CSCs completely to reduce tumor recurrence.
Author Contributions
B-BC and W-LJ provided the idea and helped with the final revision of the article. J-NG, S-HD, and B-RX wrote the article. J-NG, CY, and Y-NP drew the figures and tables. All authors reviewed the manuscript and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
- CCL
C-C motif chemokine ligand
- CK2
Casein kinase 2
- CSCs
Cancer stem cells
- CTL
Cytotoxic T lymphocyte
- CTLA-4
Cytotoxic T-lymphocyte -associated protein 4
- CXCL
C-X -C motif chemokine ligand
- CXCR4
C-X -C chemokine receptor type 4
- DVL
Disheveled
- DCs
Dendritic cells
- DUBs
Deubiquitylating enzymes
- EGFR
Epidermal growth factor receptor
- ERK
Extracellular regulated protein kinases
- FOXP3
Forkhead box P3
- HH
Hedgehog
- KRAS
Kirsten rat sarcoma viral oncogene
- IFN
Interferon
- IL
Interleukin
- IRF
Interferon regulatory factor
- ITAIMs
Inhibitory tumor-associated immune microenvironment
- LEBF1
Lymphoid enhancer binding factor 1
- MAPK
Mitogen activated protein kinase
- MDSCs
Myeloid-derived suppressor cells
- MHC I
Major histocompatibility complex I
- mTOR
Mammalian target of rapamycin
- NAMPT
Nicotinamide phosphoribosyltransferase
- NF- κ B
Nuclear factor κ B
- NK
Natural killer cells
- PD-L1
Programmed death-ligand 1
- PI3K
Phosphatidylinositol 3-kinase
- PTEN
phosphatase and tensin homolog deleted on chromosome ten
- S6K1
Ribosomal S6 kinase 1
- SOS1
Son of sevenless 1
- SRSs
Stemness-related signals
- STAT3
signal transducer and activator of transcription
- TAIM
Tumor-associated immune microenvironment
- TAMs
Tumor-associated macrophages
- TAP
Transporter associated with antigen processing
- TGF- β
Transforming growth factor β
- Th
T helper cell
- TME
Tumor microenvironment
- TNF
Tumor necrosis factor
- TRAIL
TNF-related apoptosis-inducing ligand
- Tregs
Regulatory T cells
- Ub
Ubiquitin
- USPs
Ubiquitin-specific peptidases
- VEGFs
Vascular endothelial growth factors.
Footnotes
Funding. This work was supported by the Natural Science Foundation of Heilongjiang Province of China (No. ZD2017019), the Natural Science Foundation of China (No. 81872430), the Beijing Medical Award Foundation (No. YXJL-2019-0072-0023), the Nn10 Program of Harbin Medical University Cancer Hospital (No. Nn102017-02), and the No.12 Special Fund in China Postdoctoral Science Foundation (No. 2019T120281).
References
- An T., Gong Y., Li X., Kong L., Ma P., Gong L., et al. (2017). USP7 inhibitor P5091 inhibits Wnt signaling and colorectal tumor growth. Biochem. Pharmacol. 131 29–39. 10.1016/j.bcp.2017.02.011 [DOI] [PubMed] [Google Scholar]
- Andersson E. R., Sandberg R., Lendahl U. (2011). Notch signaling: simplicity in design, versatility in function. Development 138 3593–3612. 10.1242/dev.063610 [DOI] [PubMed] [Google Scholar]
- Boulter L., Guest R. V., Kendall T. J., Wilson D. H., Wojtacha D., Robson A. J., et al. (2015). WNT signaling drives cholangiocarcinoma growth and can be pharmacologically inhibited. J. Clin. Invest. 125 1269–1285. 10.1172/JCI76452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandmaier A., Hou S. Q., Demaria S., Formenti S. C., Shen W. H. (2017). PTEN at the interface of immune tolerance and tumor suppression. Front. Biol. (Beijing) 12:163–174. 10.1007/s11515-017-1443-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown Y., Hua S., Tanwar P. S. (2019). Extracellular matrix-mediated regulation of cancer stem cells and chemoresistance. Int. J. Biochem. Cell Biol. 109 90–104. 10.1016/j.biocel.2019.02.002 [DOI] [PubMed] [Google Scholar]
- Brummelkamp T. R., Nijman S. M., Dirac A. M., Bernards R. (2003). Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-kappaB. Nature 424 797–801. 10.1038/nature01811 [DOI] [PubMed] [Google Scholar]
- Bruttel V. S., Wischhusen J. (2014). Cancer stem cell immunology: key to understanding tumorigenesis and tumor immune escape? Front. Immunol. 5:360. 10.3389/fimmu.2014.00360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caliskan C., Pehlivan M., Yuce Z., Sercan O. (2017). Dishevelled proteins and CYLD reciprocally regulate each other in CML cell lines. Mol. Biol. Rep. 44 391–397. 10.1007/s11033-017-4122-3 [DOI] [PubMed] [Google Scholar]
- Cao W. H., Liu X. P., Meng S. L., Gao Y. W., Wang Y., Ma Z. L., et al. (2016). USP4 promotes invasion of breast cancer cells via Relaxin/TGF-beta1/Smad2/MMP-9 signal. Eur. Rev. Med. Pharmacol. Sci. 20 1115–1122. [PubMed] [Google Scholar]
- Castagnoli L., De Santis F., Volpari T., Vernieri C., Tagliabue E., Di Nicola M., et al. (2020). Cancer stem cells: devil or savior-looking behind the scenes of immunotherapy failure. Cells 9:555. 10.3390/cells9030555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti J., Holokai L., Syu L., Steele N. G., Chang J., Wang J., et al. (2018). Hedgehog signaling induces PD-L1 expression and tumor cell proliferation in gastric cancer. Oncotarget 9 37439–37457. 10.18632/oncotarget.26473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Zhu X., Sun R., Ma P., Zhang E., Wang Z., et al. (2020). Ubiquitin-specific protease 7 is a druggable target that is essential for pancreatic cancer growth and chemoresistance. Invest. New Drugs 38 1707–1716. 10.1007/s10637-020-00951-0 [DOI] [PubMed] [Google Scholar]
- Chen L. L., Smith M. D., Lv L., Nakagawa T., Li Z., Sun S. C., et al. (2020). USP15 suppresses tumor immunity via deubiquitylation and inactivation of TET2. Sci. Adv. 6:eabc9730. 10.1126/sciadv.abc9730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J., Guo J., North B. J., Wang B., Cui C. P., Li H., et al. (2019). Functional analysis of deubiquitylating enzymes in tumorigenesis and development. Biochim. Biophys. Acta Rev. Cancer 1872:188312. 10.1016/j.bbcan.2019.188312 [DOI] [PubMed] [Google Scholar]
- Chou C. K., Chang Y. T., Korinek M., Chen Y. T., Yang Y. T., Leu S., et al. (2017). The regulations of deubiquitinase USP15 and its pathophysiological mechanisms in diseases. Int. J. Mol. Sci. 18:483. 10.3390/ijms18030483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu L., Gu J., He Z., Xiao T., Liu X. (2006). Adenoviral vector expressing CYLD augments antitumor activity of TRAIL by suppression of NF-kappaB survival signaling in hepatocellular carcinoma. Cancer Biol. Ther. 5 615–622. 10.4161/cbt.5.6.2662 [DOI] [PubMed] [Google Scholar]
- Clague M. J., Urbe S., Komander D. (2019). Breaking the chains: deubiquitylating enzyme specificity begets function. Nat. Rev. Mol. Cell Biol. 20 338–352. 10.1038/s41580-019-0099-1 [DOI] [PubMed] [Google Scholar]
- Clara J. A., Monge C., Yang Y., Takebe N. (2020). Targeting signalling pathways and the immune microenvironment of cancer stem cells – a clinical update. Nat. Rev. Clin. Oncol. 17 204–232. 10.1038/s41571-019-0293-2 [DOI] [PubMed] [Google Scholar]
- Clark S. E., Burrack K. S., Jameson S. C., Hamilton S. E., Lenz L. L. (2019). NK Cell IL-10 production requires IL-15 and IL-10 driven STAT3 activation. Front. Immunol. 10:2087. 10.3389/fimmu.2019.02087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colleran A., Collins P. E., O’Carroll C., Ahmed A., Mao X., McManus B., et al. (2013). Deubiquitination of NF-kappaB by Ubiquitin-Specific Protease-7 promotes transcription. Proc. Natl. Acad. Sci. U.S.A. 110 618–623. 10.1073/pnas.1208446110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez J. T., Montauti E., Shifrut E., Gatchalian J., Zhang Y., Shaked O., et al. (2020). CRISPR screen in regulatory T cells reveals modulators of Foxp3. Nature 582 416–420. 10.1038/s41586-020-2246-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X., Lu L., Deng S., Meng J., Wan C., Huang J., et al. (2020). USP7 targeting modulates anti-tumor immune response by reprogramming Tumor-associated Macrophages in Lung Cancer. Theranostics 10 9332–9347. 10.7150/thno.47137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Altri T., Gonzalez J., Aifantis I., Espinosa L., Bigas A. (2011). Hes1 expression and CYLD repression are essential events downstream of Notch1 in T-cell leukemia. Cell Cycle 10 1031–1036. 10.4161/cc.10.7.15067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Arcy P., Wang X., Linder S. (2015). Deubiquitinase inhibition as a cancer therapeutic strategy. Pharmacol. Ther. 147 32–54. 10.1016/j.pharmthera.2014.11.002 [DOI] [PubMed] [Google Scholar]
- Das M., Law S. (2018). Role of tumor microenvironment in cancer stem cell chemoresistance and recurrence. Int. J. Biochem. Cell Biol. 103 115–124. 10.1016/j.biocel.2018.08.011 [DOI] [PubMed] [Google Scholar]
- Ding C., He J., Zhao J., Li J., Chen J., Liao W., et al. (2018). beta-catenin regulates IRF3-mediated innate immune signalling in colorectal cancer. Cell Prolif. 51:e12464. 10.1111/cpr.12464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzimianski J. V., Beldon B. S., Daczkowski C. M., Goodwin O. Y., Scholte F. E. M., Bergeron E., et al. (2019). Probing the impact of nairovirus genomic diversity on viral ovarian tumor domain protease (vOTU) structure and deubiquitinase activity. PLoS Pathog. 15:e1007515. 10.1371/journal.ppat.1007515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichhorn P. J., Rodon L., Gonzalez-Junca A., Dirac A., Gili M., Martinez-Saez E., et al. (2012). USP15 stabilizes TGF-beta receptor I and promotes oncogenesis through the activation of TGF-beta signaling in glioblastoma. Nat. Med. 18 429–435. 10.1038/nm.2619 [DOI] [PubMed] [Google Scholar]
- Etzerodt A., Tsalkitzi K., Maniecki M., Damsky W., Delfini M., Baudoin E., et al. (2019). Specific targeting of CD163(+) TAMs mobilizes inflammatory monocytes and promotes T cell-mediated tumor regression. J. Exp. Med. 216 2394–2411. 10.1084/jem.20182124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y. H., Cheng J., Vasudevan S. A., Dou J., Zhang H., Patel R. H., et al. (2013). USP7 inhibitor P22077 inhibits neuroblastoma growth via inducing p53-mediated apoptosis. Cell Death Dis. 4:e867. 10.1038/cddis.2013.400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Majada V., Welz P. S., Ermolaeva M. A., Schell M., Adam A., Dietlein F., et al. (2016). The tumour suppressor CYLD regulates the p53 DNA damage response. Nat. Commun. 7:12508. 10.1038/ncomms12508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrandino F., Grazioli P., Bellavia D., Campese A. F., Screpanti I., Felli M. P. (2018). Notch and NF-kappaB: coach and players of regulatory T-cell response in cancer. Front. Immunol. 9:2165. 10.3389/fimmu.2018.02165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouse S. D., Costello J. F. (2013). Cancer stem cells activate STAT3 the EZ way. Cancer Cell 23 711–713. 10.1016/j.ccr.2013.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franqui-Machin R., Hao M., Bai H., Gu Z., Zhan X., Habelhah H., et al. (2018). Destabilizing NEK2 overcomes resistance to proteasome inhibition in multiple myeloma. J. Clin. Invest. 128 2877–2893. 10.1172/JCI98765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu C., Liang X., Cui W., Ober-Blobaum J. L., Vazzana J., Shrikant P. A., et al. (2015). beta-Catenin in dendritic cells exerts opposite functions in cross-priming and maintenance of CD8+ T cells through regulation of IL-10. Proc. Natl. Acad. Sci. U.S.A. 112 2823–2828. 10.1073/pnas.1414167112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu C., Zhu X., Xu P., Li Y. (2019). Pharmacological inhibition of USP7 promotes antitumor immunity and contributes to colon cancer therapy. Onco Targets Ther. 12 609–617. 10.2147/OTT.S182806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Ma G., Liu G., Li B., Li H., Hao X., et al. (2018). USP14 as a novel prognostic marker promotes cisplatin resistance via Akt/ERK signaling pathways in gastric cancer. Cancer Med. 7 5577–5588. 10.1002/cam4.1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda M., Hiroi M., Suzuki S., Ohmori Y., Sakashita H. (2008). Loss of CYLD might be associated with development of salivary gland tumors. Oncol. Rep. 19 1421–1427. [PubMed] [Google Scholar]
- Gavory G., O’Dowd C. R., Helm M. D., Flasz J., Arkoudis E., Dossang A., et al. (2018). Discovery and characterization of highly potent and selective allosteric USP7 inhibitors. Nat. Chem. Biol. 14 118–125. 10.1038/nchembio.2528 [DOI] [PubMed] [Google Scholar]
- Geng N., Li Y., Zhang W., Wang F., Wang X., Jin Z., et al. (2020). A PAK5-DNPEP-USP4 axis dictates breast cancer growth and metastasis. Int. J. Cancer 146 1139–1151. 10.1002/ijc.32523 [DOI] [PubMed] [Google Scholar]
- Giroux-Leprieur E., Costantini A., Ding V. W., He B. (2018). Hedgehog signaling in lung cancer: from oncogenesis to cancer treatment resistance. Int. J. Mol. Sci. 19:2835. 10.3390/ijms19092835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glinsky G. V., Berezovska O., Glinskii A. B. (2005). Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J. Clin. Invest. 115 1503–1521. 10.1172/JCI23412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grazioli P., Felli M. P., Screpanti I., Campese A. F. (2017). The mazy case of Notch and immunoregulatory cells. J. Leukoc. Biol. 102 361–368. 10.1189/jlb.1VMR1216-505R [DOI] [PubMed] [Google Scholar]
- Grund-Groschke S., Stockmaier G., Aberger F. (2019). Hedgehog/GLI signaling in tumor immunity - new therapeutic opportunities and clinical implications. Cell Commun. Signal. 17:172. 10.1186/s12964-019-0459-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F., Zhang C., Wang F., Zhang W., Shi X., Zhu Y., et al. (2020). Deubiquitinating enzyme USP33 restrains docetaxel-induced apoptosis via stabilising the phosphatase DUSP1 in prostate cancer. Cell Death Differ. 27 1938–1951. 10.1038/s41418-019-0473-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Diaz B. T., Gu W., Ntziachristos P. (2020). Deubiquitinases: pro-oncogenic activity and therapeutic targeting in blood malignancies. Trends Immunol. 41 327–340. 10.1016/j.it.2020.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadian K., Griesbach R. A., Dornauer S., Wanger T. M., Nagel D., Metlitzky M., et al. (2011). NF-kappaB essential modulator (NEMO) interaction with linear and lys-63 ubiquitin chains contributes to NF-kappaB activation. J. Biol. Chem. 286 26107–26117. 10.1074/jbc.M111.233163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han B., Sun Y., Yang D., Zhang H., Mo S., Chen X., et al. (2020). USP22 promotes development of lung adenocarcinoma through ubiquitination and immunosuppression. Aging (Albany NY) 12 6990–7005. 10.18632/aging.103056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K. H., Kwak M., Lee T. H., Park M. S., Jeong I. H., Kim M. J., et al. (2019). USP14 inhibition regulates tumorigenesis by inducing autophagy in lung cancer in vitro. Int. J. Mol. Sci. 20:5300. 10.3390/ijms20215300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna A., Metge B. J., Bailey S. K., Chen D., Chandrashekar D. S., Varambally S., et al. (2019). Inhibition of Hedgehog signaling reprograms the dysfunctional immune microenvironment in breast cancer. Oncoimmunology 8:1548241. 10.1080/2162402X.2018.1548241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrigan J. A., Jacq X., Martin N. M., Jackson S. P. (2018). Deubiquitylating enzymes and drug discovery: emerging opportunities. Nat. Rev. Drug Discov. 17 57–78. 10.1038/nrd.2017.152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayal T. B., DoGan A., Hb S. I., Kiratli B., SahIn F. (2020). Ubiquitin-specific protease 7 downregulation suppresses breast cancer in vitro. Turk. J. Biol. 44 145–157. 10.3906/biy-1912-83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M., Jono H., Shinriki S., Nakamura T., Guo J., Sueta A., et al. (2014). Clinical significance of CYLD downregulation in breast cancer. Breast Cancer Res. Treat. 143 447–457. 10.1007/s10549-013-2824-3 [DOI] [PubMed] [Google Scholar]
- He Y., Wang S., Tong J., Jiang S., Yang Y., Zhang Z., et al. (2020). The deubiquitinase USP7 stabilizes Maf proteins to promote myeloma cell survival. J. Biol. Chem. 295 2084–2096. 10.1074/jbc.RA119.010724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho W. J., Jaffee E. M., Zheng L. (2020). The tumour microenvironment in pancreatic cancer - clinical challenges and opportunities. Nat. Rev. Clin. Oncol. 17 527–540. 10.1038/s41571-020-0363-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Yang D., Zhang H., Liu W., Zhao Y., Lu H., et al. (2015). USP22 promotes tumor progression and induces epithelial-mesenchymal transition in lung adenocarcinoma. Lung Cancer 88 239–245. 10.1016/j.lungcan.2015.02.019 [DOI] [PubMed] [Google Scholar]
- Hu M., Zhu S., Xiong S., Xue X., Zhou X. (2019). MicroRNAs and the PTEN/PI3K/Akt pathway in gastric cancer (Review). Oncol. Rep. 41 1439–1454. 10.3892/or.2019.6962 [DOI] [PubMed] [Google Scholar]
- Huang L., Liu H., Zhang K., Meng Q., Hu L., Zhang Y., et al. (2020). Ubiquitin-conjugating enzyme 2S enhances viral replication by inhibiting type I IFN production through recruiting USP15 to deubiquitinate TBK1. Cell Rep. 32:108044. 10.1016/j.celrep.2020.108044 [DOI] [PubMed] [Google Scholar]
- Huang X., Zhang Q., Lou Y., Wang J., Zhao X., Wang L., et al. (2019). USP22 Deubiquitinates CD274 to suppress anticancer immunity. Cancer Immunol. Res. 7 1580–1590. 10.1158/2326-6066.CIR-18-0910 [DOI] [PubMed] [Google Scholar]
- Huang X., Zhang X., Bai X., Liang T. (2020). Blocking PD-L1 for anti-liver cancer immunity: USP22 represents a critical cotarget. Cell Mol. Immunol. 17 677–679. 10.1038/s41423-019-0348-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang S. J., Lee H. W., Kim H. R., Lee H., Shin C. H., Yun S. I., et al. (2016). Ubiquitin-specific protease 4 controls metastatic potential through beta-catenin stabilization in brain metastatic lung adenocarcinoma. Sci. Rep. 6:21596. 10.1038/srep21596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliopoulos D., Jaeger S. A., Hirsch H. A., Bulyk M. L., Struhl K. (2010). STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol. Cell 39 493–506. 10.1016/j.molcel.2010.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeusset L. M., McManus K. J. (2017). Ubiquitin specific peptidase 22 regulates histone H2B mono-ubiquitination and exhibits both oncogenic and tumor suppressor roles in cancer. Cancers (Basel) 9:167. 10.3390/cancers9120167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z., He L., Regev A., Struhl K. (2019). Inflammatory regulatory network mediated by the joint action of NF-kB, STAT3, and AP-1 factors is involved in many human cancers. Proc. Natl. Acad. Sci. U.S.A. 116 9453–9462. 10.1073/pnas.1821068116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia L., Zhang W., Wang C. Y. (2020). BMI1 inhibition eliminates residual cancer stem cells after PD1 blockade and activates antitumor immunity to prevent metastasis and relapse. Cell Stem Cell 27 238–253.e6. 10.1016/j.stem.2020.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Sun Y., Wang J., He Q., Chen X., Lan X., et al. (2019). Proteasomal cysteine deubiquitinase inhibitor b-AP15 suppresses migration and induces apoptosis in diffuse large B cell lymphoma. J. Exp. Clin. Cancer Res. 38:453. 10.1186/s13046-019-1446-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin M. Z., Jin W. L. (2020). The updated landscape of tumor microenvironment and drug repurposing. Signal. Transduct. Target Ther. 5 166. 10.1038/s41392-020-00280-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Q., Martinez C. A., Arcipowski K. M., Zhu Y., Gutierrez-Diaz B. T., Wang K. K., et al. (2019). USP7 cooperates with NOTCH1 to drive the oncogenic transcriptional program in T-cell leukemia. Clin. Cancer Res. 25 222–239. 10.1158/1078-0432.CCR-18-1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H., Kim B. G., Han W. H., Lee J. H., Cho J. Y., Park W. S., et al. (2013). Deubiquitination of Dishevelled by Usp14 is required for Wnt signaling. Oncogenesis 2:e64. 10.1038/oncsis.2013.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh M. (2019). Genomic testing, tumor microenvironment and targeted therapy of Hedgehog-related human cancers. Clin. Sci. (Lond.) 133 953–970. 10.1042/CS20180845 [DOI] [PubMed] [Google Scholar]
- Kaushal K., Ramakrishna S. (2020). Deubiquitinating enzyme-mediated signaling networks in cancer stem cells. Cancers (Basel) 12:3253. 10.3390/cancers12113253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp M. (2016). Recent advances in the discovery of Deubiquitinating enzyme inhibitors. Prog. Med. Chem. 55 149–192. 10.1016/bs.pmch.2015.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiprowska M. J., Stepanova A., Todaro D. R., Galkin A., Haas A., Wilson S. M., et al. (2017). Neurotoxic mechanisms by which the USP14 inhibitor IU1 depletes ubiquitinated proteins and Tau in rat cerebral cortical neurons: relevance to Alzheimer’s disease. Biochim. Biophys. Acta Mol. Basis Dis. 1863 1157–1170. 10.1016/j.bbadis.2017.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalenko A., Chable-Bessia C., Cantarella G., Israel A., Wallach D., Courtois G. (2003). The tumour suppressor CYLD negatively regulates NF-kappaB signalling by deubiquitination. Nature 424 801–805. 10.1038/nature01802 [DOI] [PubMed] [Google Scholar]
- Kuphal S., Shaw-Hallgren G., Eberl M., Karrer S., Aberger F., Bosserhoff A. K., et al. (2011). GLI1-dependent transcriptional repression of CYLD in basal cell carcinoma. Oncogene 30 4523–4530. 10.1038/onc.2011.163 [DOI] [PubMed] [Google Scholar]
- Kusakabe S., Suzuki T., Sugiyama Y., Haga S., Horike K., Tokunaga M., et al. (2019). USP15 participates in hepatitis C virus propagation through regulation of viral RNA translation and lipid droplet formation. J. Virol. 93:e01708-18. 10.1128/JVI.01708-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C. Y., Yeh D. W., Lu C. H., Liu Y. L., Chuang Y. C., Ruan J. W., et al. (2020). Epigenetic silencing of ubiquitin specific protease 4 by snail1 contributes to macrophage-dependent inflammation and therapeutic resistance in lung cancer. Cancers (Basel) 12:148. 10.3390/cancers12010148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. E., Park C. M., Kim J. H. (2020). USP7 deubiquitinates and stabilizes EZH2 in prostate cancer cells. Genet. Mol. Biol. 43:e20190338. 10.1590/1678-4685-GMB-2019-0338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leger P. R., Hu D. X., Biannic B., Bui M., Han X., Karbarz E., et al. (2020). Discovery of potent, selective, and orally bioavailable inhibitors of USP7 with in vivo antitumor activity. J. Med. Chem. 63 5398–5420. 10.1021/acs.jmedchem.0c00245 [DOI] [PubMed] [Google Scholar]
- Lei C. Q., Wu X., Zhong X., Jiang L., Zhong B., Shu H. B. (2019). USP19 inhibits TNF-alpha- and IL-1beta-triggered NF-kappaB activation by Deubiquitinating TAK1. J. Immunol. 203 259–268. 10.4049/jimmunol.1900083 [DOI] [PubMed] [Google Scholar]
- Li B., Lu Y., Yu L., Han X., Wang H., Mao J., et al. (2017). miR-221/222 promote cancer stem-like cell properties and tumor growth of breast cancer via targeting PTEN and sustained Akt/NF-kappaB/COX-2 activation. Chem. Biol. Interact. 277 33–42. 10.1016/j.cbi.2017.08.014 [DOI] [PubMed] [Google Scholar]
- Li B., Zeng Y., Reeves P. M., Ran C., Liu Q., Qu X., et al. (2018). AMD3100 augments the efficacy of mesothelin-targeted, immune-activating VIC-008 in mesothelioma by modulating intratumoral immunosuppression. Cancer Immunol. Res. 6 539–551. 10.1158/2326-6066.CIR-17-0530 [DOI] [PubMed] [Google Scholar]
- Li C., Jiang P., Wei S., Xu X., Wang J. (2020). Regulatory T cells in tumor microenvironment: new mechanisms, potential therapeutic strategies and future prospects. Mol. Cancer 19:116. 10.1186/s12943-020-01234-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Yuan S., Norgard R. J., Yan F., Yamazoe T., Blanco A., et al. (2020). Tumor cell-intrinsic USP22 suppresses antitumor immunity in pancreatic cancer. Cancer Immunol. Res. 8 282–291. 10.1158/2326-6066.CIR-19-0661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Wei J., Li S., Jacko A. M., Weathington N. M., Mallampalli R. K., et al. (2019). The deubiquitinase USP13 stabilizes the anti-inflammatory receptor IL-1R8/Sigirr to suppress lung inflammation. EBioMedicine 45 553–562. 10.1016/j.ebiom.2019.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Liu S., Chen H., Zhou X., Zhou J., Zhou S., et al. (2020). N-benzylpiperidinol derivatives as novel USP7 inhibitors: structure-activity relationships and X-ray crystallographic studies. Eur. J. Med. Chem. 199:112279. 10.1016/j.ejmech.2020.112279 [DOI] [PubMed] [Google Scholar]
- Li M., Zhao J., Jia L. (2019). USP14-mediated IkappaBalpha degradation exacerbates NF-kappaB activation and IL-1beta-stimulated chondrocyte dedifferentiation. Life Sci. 218 147–152. 10.1016/j.lfs.2018.12.014 [DOI] [PubMed] [Google Scholar]
- Li Y., Laterra J. (2012). Cancer stem cells: distinct entities or dynamically regulated phenotypes? Cancer Res. 72, 576–580. 10.1158/0008-5472.CAN-11-3070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C., Xu J. C., Chen T. Y., Xu J. X., Chen W. F., Hu J. W., et al. (2020). Ubiquitin-specific peptide 22 acts as an oncogene in gastric cancer in a son of sevenless 1-dependent manner. Cancer Cell Int. 20:45. 10.1186/s12935-020-1137-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R., Nie J., Ren J., Liang R., Li D., Wang P., et al. (2017). USP4 interacts and positively regulates IRF8 function via K48-linked deubiquitination in regulatory T cells. FEBS Lett. 591 1677–1686. 10.1002/1873-3468.12668 [DOI] [PubMed] [Google Scholar]
- Liu B., Chen J., Zhang S. (2019). Emerging role of ubiquitin-specific protease 14 in oncogenesis and development of tumor: therapeutic implication. Life Sci. 239:116875. 10.1016/j.lfs.2019.116875 [DOI] [PubMed] [Google Scholar]
- Liu H., Liu N., Zhao Y., Zhu X., Wang C., Liu Q., et al. (2019). Oncogenic USP22 supports gastric cancer growth and metastasis by activating c-Myc/NAMPT/SIRT1-dependent FOXO1 and YAP signaling. Aging (Albany NY) 11 9643–9660. 10.18632/aging.102410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N., Kong T., Chen X., Hu H., Gu H., Liu S., et al. (2017). Ubiquitin-specific protease 14 regulates LPS-induced inflammation by increasing ERK1/2 phosphorylation and NF-kappaB activation. Mol. Cell Biochem. 431 87–96. 10.1007/s11010-017-2978-0 [DOI] [PubMed] [Google Scholar]
- Liu P., Xu B., Shen W., Zhu H., Wu W., Fu Y., et al. (2012). Dysregulation of TNFalpha-induced necroptotic signaling in chronic lymphocytic leukemia: suppression of CYLD gene by LEF1. Leukemia 26 1293–1300. 10.1038/leu.2011.357 [DOI] [PubMed] [Google Scholar]
- Lopez-Castejon G., Edelmann M. J. (2016). Deubiquitinases: novel therapeutic targets in immune surveillance? Mediat. Inflamm. 2016:3481371. 10.1155/2016/3481371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Zhu F., Zhang M., Li Y., Drennan A. C., Kimpara S., et al. (2018). Gene regulation and suppression of type I interferon signaling by STAT3 in diffuse large B cell lymphoma. Proc. Natl. Acad. Sci. U.S.A. 115 E498–E505. 10.1073/pnas.1715118115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke J. J., Bao R., Sweis R. F., Spranger S., Gajewski T. F. (2019). WNT/beta-catenin pathway activation correlates with immune exclusion across human cancers. Clin. Cancer Res. 25 3074–3083. 10.1158/1078-0432.CCR-18-1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H., Wang X., Ge H., Zheng N., Peng F., Fu Y., et al. (2019). Inhibition of ubiquitinspecific protease 14 promotes connexin 32 internalization and counteracts cisplatin cytotoxicity in human ovarian cancer cells. Oncol. Rep. 42 1237–1247. 10.3892/or.2019.7232 [DOI] [PubMed] [Google Scholar]
- Luongo F., Colonna F., Calapa F., Vitale S., Fiori M. E., De Maria R. (2019). PTEN tumor-suppressor: the dam of stemness in cancer. Cancers (Basel) 11:1076. 10.3390/cancers11081076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lytle N. K., Barber A. G., Reya T. (2018). Stem cell fate in cancer growth, progression and therapy resistance. Nat. Rev. Cancer 18 669–680. 10.1038/s41568-018-0056-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma A., Tang M., Zhang L., Wang B., Yang Z., Liu Y., et al. (2019). USP1 inhibition destabilizes KPNA2 and suppresses breast cancer metastasis. Oncogene 38 2405–2419. 10.1038/s41388-018-0590-8 [DOI] [PubMed] [Google Scholar]
- Ma Y., Fu H. L., Wang Z., Huang H., Ni J., Song J., et al. (2017). USP22 maintains gastric cancer stem cell stemness and promotes gastric cancer progression by stabilizing BMI1 protein. Oncotarget 8 33329–33342. 10.18632/oncotarget.16445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marjanovic N. D., Weinberg R. A., Chaffer C. L. (2013). Cell plasticity and heterogeneity in cancer. Clin. Chem. 59, 168–179. 10.1373/clinchem.2012.184655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massoumi R., Chmielarska K., Hennecke K., Pfeifer A., Fassler R. (2006). Cyld inhibits tumor cell proliferation by blocking Bcl-3-dependent NF-kappaB signaling. Cell 125 665–677. 10.1016/j.cell.2006.03.041 [DOI] [PubMed] [Google Scholar]
- Mevissen T. E. T., Komander D. (2017). Mechanisms of deubiquitinase specificity and regulation. Annu. Rev. Biochem. 86 159–192. 10.1146/annurev-biochem-061516-044916 [DOI] [PubMed] [Google Scholar]
- Mi Y., Guo N., Luan J., Cheng J., Hu Z., Jiang P., et al. (2020). The emerging role of myeloid-derived suppressor cells in the glioma immune suppressive microenvironment. Front. Immunol. 11:737. 10.3389/fimmu.2020.00737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najafi M., Farhood B., Mortezaee K., Kharazinejad E., Majidpoor J., Ahadi R. (2020). Hypoxia in solid tumors: a key promoter of cancer stem cell (CSC) resistance. J. Cancer Res. Clin. Oncol. 146 19–31. 10.1007/s00432-019-03080-1 [DOI] [PubMed] [Google Scholar]
- Najafi M., Mortezaee K., Majidpoor J. (2019). Cancer stem cell (CSC) resistance drivers. Life Sci. 234:116781. 10.1016/j.lfs.2019.116781 [DOI] [PubMed] [Google Scholar]
- Nguyen H. H., Kim T., Nguyen T., Hahn M. J., Yun S. I., Kim K. K. (2019). A selective inhibitor of ubiquitin-specific protease 4 suppresses colorectal cancer progression by regulating beta-catenin signaling. Cell Physiol. Biochem. 53 157–171. 10.33594/000000127 [DOI] [PubMed] [Google Scholar]
- Niederkorn M., Hueneman K., Choi K., Varney M. E., Romano L., Pujato M. A., et al. (2020). TIFAB regulates USP15-mediated p53 signaling during stressed and malignant hematopoiesis. Cell Rep. 30 2776–2790.e6. 10.1016/j.celrep.2020.01.093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikanjam M., Cohen P. R., Kato S., Sicklick J. K., Kurzrock R. (2019). Advanced basal cell cancer: concise review of molecular characteristics and novel targeted and immune therapeutics. Ann. Oncol. 30:1675. 10.1093/annonc/mdz213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novellasdemunt L., Foglizzo V., Cuadrado L., Antas P., Kucharska A., Encheva V., et al. (2017). USP7 is a tumor-specific WNT activator for APC-mutated colorectal cancer by mediating beta-catenin Deubiquitination. Cell Rep. 21 612–627. 10.1016/j.celrep.2017.09.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak M., Klink M. (2020). The role of tumor-associated macrophages in the progression and chemoresistance of ovarian cancer. Cells 9:1299. 10.3390/cells9051299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohol Y. M., Sun M. T., Cutler G., Leger P. R., Hu D. X., Biannic B., et al. (2020). Novel, selective inhibitors of USP7 uncover multiple mechanisms of antitumor activity in vitro and in vivo. Mol. Cancer Ther. 19 1970–1980. 10.1158/1535-7163.MCT-20-0184 [DOI] [PubMed] [Google Scholar]
- Ohtake F. (2020). Mass spectrometry technologies for deciphering the ubiquitin code. Trends Biochem. Sci. 45 820–821. 10.1016/j.tibs.2020.04.008 [DOI] [PubMed] [Google Scholar]
- Okita K., Ichisaka T., Yamanaka S. (2007). Generation of germline-competent induced pluripotent stem cells. Nature 448 313–317. 10.1038/nature05934 [DOI] [PubMed] [Google Scholar]
- Osipo C., Golde T. E., Osborne B. A., Miele L. A. (2008). Off the beaten pathway: the complex cross talk between Notch and NF-kappaB. Lab. Invest. 88 11–17. 10.1038/labinvest.3700700 [DOI] [PubMed] [Google Scholar]
- Owen K. L., Brockwell N. K., Parker B. S. (2019). JAK-STAT signaling: a double-edged sword of immune regulation and cancer progression. Cancers (Basel) 11:2002. 10.3390/cancers11122002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S. A., Surh Y. J. (2017). Modulation of tumor microenvironment by chemopreventive natural products. Ann. N. Y. Acad. Sci. 1401 65–74. 10.1111/nyas.13395 [DOI] [PubMed] [Google Scholar]
- Patsoukis N., Li L., Sari D., Petkova V., Boussiotis V. A. (2013). PD-1 increases PTEN phosphatase activity while decreasing PTEN protein stability by inhibiting casein kinase 2. Mol. Cell Biol. 33 3091–3098. 10.1128/MCB.00319-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulus A., Akhtar S., Caulfield T. R., Samuel K., Yousaf H., Bashir Y., et al. (2016). Coinhibition of the deubiquitinating enzymes, USP14 and UCHL5, with VLX1570 is lethal to ibrutinib- or bortezomib-resistant Waldenstrom macroglobulinemia tumor cells. Blood Cancer J. 6:e492. 10.1038/bcj.2016.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfoh R., Lacdao I. K., Saridakis V. (2015). Deubiquitinases and the new therapeutic opportunities offered to cancer. Endocr. Relat. Cancer 22 T35–T54. 10.1530/ERC-14-0516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozhidaeva A., Bezsonova I. (2019). USP7: structure, substrate specificity, and inhibition. DNA Repair (Amst) 76 30–39. 10.1016/j.dnarep.2019.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prager B. C., Xie Q., Bao S., Rich J. N. (2019). Cancer stem cells: the architects of the tumor ecosystem. Cell Stem Cell 24 41–53. 10.1016/j.stem.2018.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu C., Liu Y., Mei Y., Zou M., Zhao Z., Ye M., et al. (2018). Ubiquitin-speci fi c protease 4 promotes metastasis of hepatocellular carcinoma by increasing TGF-beta signaling-induced epithelial-mesenchymal transition. Aging (Albany NY) 10 2783–2799. 10.18632/aging.101587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu G. Z., Liu Q., Wang X. G., Xu G. Z., Zhao T., Lou M. Q. (2020). Hypoxia-induced USP22-BMI1 axis promotes the stemness and malignancy of glioma stem cells via regulation of HIF-1alpha. Life Sci. 247:117438. 10.1016/j.lfs.2020.117438 [DOI] [PubMed] [Google Scholar]
- Qiu G. Z., Mao X. Y., Ma Y., Gao X. C., Wang Z., Jin M. Z., et al. (2018). Ubiquitin-specific protease 22 acts as an oncoprotein to maintain glioma malignancy through deubiquitinating B cell-specific Moloney murine leukemia virus integration site 1 for stabilization. Cancer Sci. 109 2199–2210. 10.1111/cas.13646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu G. Z., Sun W., Jin M. Z., Lin J., Lu P. G., Jin W. L. (2017). The bad seed gardener: Deubiquitinases in the cancer stem-cell signaling network and therapeutic resistance. Pharmacol. Ther. 172 127–138. 10.1016/j.pharmthera.2016.12.003 [DOI] [PubMed] [Google Scholar]
- Quayle L. A., Ottewell P. D., Holen I. (2018). Chemotherapy resistance and stemness in mitotically quiescent human breast cancer cells identified by fluorescent dye retention. Clin. Exp. Metastasis 35 831–846. 10.1007/s10585-018-9946-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Z., Liang M., Lai M., Shang L., Deng X., Su X. (2020). KYA1797K down-regulates PD-L1 in colon cancer stem cells to block immune evasion by suppressing the beta-catenin/STT3 signaling pathway. Int. Immunopharmacol. 78:106003. 10.1016/j.intimp.2019.106003 [DOI] [PubMed] [Google Scholar]
- Ruiz de Galarreta M., Bresnahan E., Molina-Sanchez P., Lindblad K. E., Maier B., Sia D., et al. (2019). beta-Catenin activation promotes immune escape and resistance to anti-PD-1 therapy in hepatocellular carcinoma. Cancer Discov. 9 1124–1141. 10.1158/2159-8290.CD-19-0074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasidharan Nair V., Toor S. M., Ali B. R., Elkord E. (2018). Dual inhibition of STAT1 and STAT3 activation downregulates expression of PD-L1 in human breast cancer cells. Expert Opin. Ther. Targets 22 547–557. 10.1080/14728222.2018.1471137 [DOI] [PubMed] [Google Scholar]
- Schauer N. J., Liu X., Magin R. S., Doherty L. M., Chan W. C., Ficarro S. B., et al. (2020a). Selective USP7 inhibition elicits cancer cell killing through a p53-dependent mechanism. Sci. Rep. 10:5324. 10.1038/s41598-020-62076-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer N. J., Magin R. S., Liu X., Doherty L. M., Buhrlage S. J. (2020b). Advances in discovering Deubiquitinating enzyme (DUB) inhibitors. J. Med. Chem. 63 2731–2750. 10.1021/acs.jmedchem.9b01138 [DOI] [PubMed] [Google Scholar]
- Sha B., Chen X., Wu H., Li M., Shi J., Wang L., et al. (2019). Deubiquitylatinase inhibitor b-AP15 induces c-Myc-Noxa-mediated apoptosis in esophageal squamous cell carcinoma. Apoptosis 24 826–836. 10.1007/s10495-019-01561-9 [DOI] [PubMed] [Google Scholar]
- Shaked Y. (2016). Balancing efficacy of and host immune responses to cancer therapy: the yin and yang effects. Nat. Rev. Clin. Oncol. 13 611–626. 10.1038/nrclinonc.2016.57 [DOI] [PubMed] [Google Scholar]
- Shan H., Li X., Xiao X., Dai Y., Huang J., Song J., et al. (2018). USP7 deubiquitinates and stabilizes NOTCH1 in T-cell acute lymphoblastic leukemia. Signal Transduct. Target Ther. 3:29. 10.1038/s41392-018-0028-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinriki S., Jono H., Maeshiro M., Nakamura T., Guo J., Li J. D., et al. (2018). Loss of CYLD promotes cell invasion via ALK5 stabilization in oral squamous cell carcinoma. J. Pathol. 244 367–379. 10.1002/path.5019 [DOI] [PubMed] [Google Scholar]
- Song L. L., Peng Y., Yun J., Rizzo P., Chaturvedi V., Weijzen S., et al. (2008). Notch-1 associates with IKKalpha and regulates IKK activity in cervical cancer cells. Oncogene 27 5833–5844. 10.1038/onc.2008.190 [DOI] [PubMed] [Google Scholar]
- Song M. S., Salmena L., Carracedo A., Egia A., Lo-Coco F., Teruya-Feldstein J., et al. (2008). The deubiquitinylation and localization of PTEN are regulated by a HAUSP-PML network. Nature 455 813–817. 10.1038/nature07290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spranger S., Gajewski T. F. (2015). A new paradigm for tumor immune escape: beta-catenin-driven immune exclusion. J. Immunother. Cancer 3:43. 10.1186/s40425-015-0089-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T., Liu Z., Yang Q. (2020). The role of ubiquitination and deubiquitination in cancer metabolism. Mol. Cancer 19:146. 10.1186/s12943-020-01262-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takebe N., Miele L., Harris P. J., Jeong W., Bando H., Kahn M., et al. (2015). Targeting Notch, Hedgehog, and Wnt pathways in cancer stem cells: clinical update. Nat. Rev. Clin. Oncol. 12 445–464. 10.1038/nrclinonc.2015.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauriello D. V., Haegebarth A., Kuper I., Edelmann M. J., Henraat M., Canninga-van Dijk M. R., et al. (2010). Loss of the tumor suppressor CYLD enhances Wnt/beta-catenin signaling through K63-linked ubiquitination of Dvl. Mol. Cell 37 607–619. 10.1016/j.molcel.2010.01.035 [DOI] [PubMed] [Google Scholar]
- Torre S., Polyak M. J., Langlais D., Fodil N., Kennedy J. M., Radovanovic I., et al. (2017). USP15 regulates type I interferon response and is required for pathogenesis of neuroinflammation. Nat. Immunol. 18 54–63. 10.1038/ni.3581 [DOI] [PubMed] [Google Scholar]
- Turnbull A. P., Ioannidis S., Krajewski W. W., Pinto-Fernandez A., Heride C., Martin A. C. L., et al. (2017). Molecular basis of USP7 inhibition by selective small-molecule inhibitors. Nature 550 481–486. 10.1038/nature24451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Andel H., Kocemba K. A., de Haan-Kramer A., Mellink C. H., Piwowar M., Broijl A., et al. (2017). Loss of CYLD expression unleashes Wnt signaling in multiple myeloma and is associated with aggressive disease. Oncogene 36 2105–2115. 10.1038/onc.2016.368 [DOI] [PubMed] [Google Scholar]
- van Loosdregt J., Fleskens V., Fu J., Brenkman A. B., Bekker C. P., Pals C. E., et al. (2013). Stabilization of the transcription factor Foxp3 by the deubiquitinase USP7 increases Treg-cell-suppressive capacity. Immunity 39 259–271. 10.1016/j.immuni.2013.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh V., Nataraj R., Thangaraj G. S., Karthikeyan M., Gnanasekaran A., Kaginelli S. B., et al. (2018). Targeting Notch signalling pathway of cancer stem cells. Stem Cell Investig. 5:5. 10.21037/sci.2018.02.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S. A., Beli P., Weinert B. T., Nielsen M. L., Cox J., Mann M., et al. (2011). A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol. Cell Proteomics 10:M111.013284. 10.1074/mcp.M111.013284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Kumar S., Dahiya S., Wang F., Wu J., Newick K., et al. (2016). Ubiquitin-specific Protease-7 inhibition impairs Tip60-dependent Foxp3+ T-regulatory cell function and promotes antitumor immunity. EBioMedicine 13 99–112. 10.1016/j.ebiom.2016.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Zhang Y., Wang T., Zhang J., Zhou Z., Sun Y., et al. (2017). The USP7 inhibitor P5091 induces cell death in ovarian cancers with different P53 status. Cell Physiol. Biochem. 43 1755–1766. 10.1159/000484062 [DOI] [PubMed] [Google Scholar]
- Wang X., Zhang Q., Wang Y., Zhuang H., Chen B. (2018). Clinical significance of ubiquitin specific protease 7 (USP7) in predicting prognosis of hepatocellular carcinoma and its functional mechanisms. Med. Sci. Monit. 24 1742–1750. 10.12659/msm.909368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Le W. D. (2019). Autophagy and ubiquitin-proteasome system. Adv. Exp. Med. Biol. 1206 527–550. 10.1007/978-981-15-0602-4_25 [DOI] [PubMed] [Google Scholar]
- Wang Y., Shen Y., Wang S., Shen Q., Zhou X. (2018). The role of STAT3 in leading the crosstalk between human cancers and the immune system. Cancer Lett. 415 117–128. 10.1016/j.canlet.2017.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Sun Q., Mu N., Sun X., Wang Y., Fan S., et al. (2020). The deubiquitinase USP22 regulates PD-L1 degradation in human cancer cells. Cell Commun. Signal. 18:112. 10.1186/s12964-020-00612-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. C., Wu Y. S., Hung C. Y., Wang S. A., Young M. J., Hsu T. I., et al. (2018). USP24 induces IL-6 in tumor-associated microenvironment by stabilizing p300 and beta-TrCP and promotes cancer malignancy. Nat. Commun. 9:3996. 10.1038/s41467-018-06178-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L., Tang F. (2016). Single-cell sequencing in stem cell biology. Genome Biol. 17:71. 10.1186/s13059-016-0941-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng A. P., Ferrando A. A., Lee W., Morris J. P. T., Silverman L. B., Sanchez-Irizarry C., et al. (2004). Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 306 269–271. 10.1126/science.1102160 [DOI] [PubMed] [Google Scholar]
- Wertz I. E., Murray J. M. (2019). Structurally-defined deubiquitinase inhibitors provide opportunities to investigate disease mechanisms. Drug Discov. Today Technol. 31 109–123. 10.1016/j.ddtec.2019.02.003 [DOI] [PubMed] [Google Scholar]
- Wu H. M., Kim T. H., Kim A., Koo J. H., Joo M. S., Kim S. G. (2019). Liver X receptor alpha-induced cannabinoid receptor 2 inhibits ubiquitin-specific peptidase 4 through miR-27b, protecting hepatocytes from TGF-beta. Hepatol. Commun. 3 1373–1387. 10.1002/hep4.1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N., Liu C., Bai C., Han Y. P., Cho W. C., Li Q. (2013). Over-expression of deubiquitinating enzyme USP14 in lung adenocarcinoma promotes proliferation through the accumulation of beta-catenin. Int. J. Mol. Sci. 14 10749–10760. 10.3390/ijms140610749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia P., Xu X. Y. (2015). PI3K/Akt/mTOR signaling pathway in cancer stem cells: from basic research to clinical application. Am. J. Cancer Res. 5 1602–1609. [PMC free article] [PubMed] [Google Scholar]
- Xia X., Huang C., Liao Y., Liu Y., He J., Guo Z., et al. (2019a). Inhibition of USP14 enhances the sensitivity of breast cancer to enzalutamide. J. Exp. Clin. Cancer Res. 38:220. 10.1186/s13046-019-1227-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X., Liao Y., Huang C., Liu Y., He J., Shao Z., et al. (2019b). Deubiquitination and stabilization of estrogen receptor alpha by ubiquitin-specific protease 7 promotes breast tumorigenesis. Cancer Lett. 465 118–128. 10.1016/j.canlet.2019.09.003 [DOI] [PubMed] [Google Scholar]
- Xiao H., Tian Y., Yang Y., Hu F., Xie X., Mei J., et al. (2015). USP22 acts as an oncogene by regulating the stability of cyclooxygenase-2 in non-small cell lung cancer. Biochem. Biophys. Res. Commun. 460 703–708. 10.1016/j.bbrc.2015.03.093 [DOI] [PubMed] [Google Scholar]
- Xu D. D., Zhou P. J., Wang Y., Zhang L., Fu W. Y., Ruan B. B., et al. (2016). Reciprocal activation between STAT3 and miR-181b regulates the proliferation of esophageal cancer stem-like cells via the CYLD pathway. Mol. Cancer 15:40. 10.1186/s12943-016-0521-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G., Liu Y., Li H., Liu L., Zhang S., Zhang Z. (2020). Dissecting the human immune system with single cell RNA sequencing technology. J. Leukoc. Biol. 107 613–623. 10.1002/JLB.5MR1019-179R [DOI] [PubMed] [Google Scholar]
- Xu X., Liu J., Shen C., Ding L., Zhong F., Ouyang Y., et al. (2017). The role of ubiquitin-specific protease 14 (USP14) in cell adhesion-mediated drug resistance (CAM-DR) of multiple myeloma cells. Eur. J. Haematol. 98 4–12. 10.1111/ejh.12729 [DOI] [PubMed] [Google Scholar]
- Yang J., Xu P., Han L., Guo Z., Wang X., Chen Z., et al. (2015). Cutting edge: ubiquitin-specific protease 4 promotes Th17 cell function under inflammation by deubiquitinating and stabilizing RORgammat. J. Immunol. 194 4094–4097. 10.4049/jimmunol.1401451 [DOI] [PubMed] [Google Scholar]
- Yang W., Li Y., Gao R., Xiu Z., Sun T. (2020). MHC class I dysfunction of glioma stem cells escapes from CTL-mediated immune response via activation of Wnt/beta-catenin signaling pathway. Oncogene 39 1098–1111. 10.1038/s41388-019-1045-6 [DOI] [PubMed] [Google Scholar]
- Yao Y., Zhang Y., Shi M., Sun Y., Chen C., Niu M., et al. (2018). Blockade of deubiquitinase USP7 overcomes bortezomib resistance by suppressing NF-kappaB signaling pathway in multiple myeloma. J. Leukoc. Biol. 104 1105–1115. 10.1002/JLB.2A1017-420RR [DOI] [PubMed] [Google Scholar]
- Yu Y., Zhao Y., Fan Y., Chen Z., Li H., Lu J., et al. (2019). Inhibition of ubiquitin-specific protease 14 suppresses cell proliferation and synergizes with chemotherapeutic agents in neuroblastoma. Mol. Cancer Ther. 18 1045–1056. 10.1158/1535-7163.MCT-18-0146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X., Wang H., Xu A., Zhu X., Zhan Y., Wang W. (2019). Ubiquitin-specific peptidase 22 promotes proliferation and metastasis in human colon cancer. Oncol. Lett. 18 5567–5576. 10.3892/ol.2019.10872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun S. I., Kim H. H., Yoon J. H., Park W. S., Hahn M. J., Kim H. C., et al. (2015). Ubiquitin specific protease 4 positively regulates the WNT/beta-catenin signaling in colorectal cancer. Mol. Oncol. 9 1834–1851. 10.1016/j.molonc.2015.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Yang L., Wang J., Sun T., Guo Y., Nelson R., et al. (2019). Ubiquitin-specific protease 22 is critical to in vivo angiogenesis, growth and metastasis of non-small cell lung cancer. Cell Commun. Signal. 17:167. 10.1186/s12964-019-0480-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Qiang J., Yang X., Wang D., Rehman A. U., He X., et al. (2020). IL1R2 blockade suppresses breast tumorigenesis and progression by impairing USP15-dependent BMI1 stability. Adv. Sci. (Weinh) 7:1901728. 10.1002/advs.201901728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Wang H., Tian L., Li H. (2016). Expression of USP7 and MARCH7 is correlated with poor prognosis in epithelial ovarian cancer. Tohoku J. Exp. Med. 239 165–175. 10.1620/tjem.239.165 [DOI] [PubMed] [Google Scholar]
- Zhang M. J., Chen D. S., Li H., Liu W. W., Han G. Y., Han Y. F. (2019). Clinical significance of USP7 and EZH2 in predicting prognosis of laryngeal squamous cell carcinoma and their possible functional mechanism. Int. J. Clin. Exp. Pathol. 12 2184–2194. [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Zhang J., Xu C., Zhang S., Bian S., Jiang F., et al. (2020). Ubiquitin-specific protease 7 is a drug-able target that promotes hepatocellular carcinoma and chemoresistance. Cancer Cell Int. 20:28. 10.1186/s12935-020-1109-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. Y., Varthi M., Sykes S. M., Phillips C., Warzecha C., Zhu W., et al. (2008). The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Mol. Cell 29 102–111. 10.1016/j.molcel.2007.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wang Y., Gao B., Sun Y., Cao L., Genardi S. M., et al. (2020). USP22 controls iNKT immunity through MED1 suppression of histone H2A monoubiquitination. J. Exp. Med. 217:e20182218. 10.1084/jem.20182218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Z., Virshup D. M. (2020). Wnt signaling and drug resistance in cancer. Mol. Pharmacol. 97 72–89. 10.1124/mol.119.117978 [DOI] [PubMed] [Google Scholar]
- Zhou F., Xie F., Jin K., Zhang Z., Clerici M., Gao R., et al. (2017). USP4 inhibits SMAD4 monoubiquitination and promotes activin and BMP signaling. EMBO J. 36 1623–1639. 10.15252/embj.201695372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Cheng C., Wei Y., Yang J., Zhou W., Song Q., et al. (2020). USP15 potentiates NF-kappaB activation by differentially stabilizing TAB2 and TAB3. FEBS J. 287 3165–3183. 10.1111/febs.15202 [DOI] [PubMed] [Google Scholar]
- Zhou Y., Jia E., Pan M., Zhao X., Ge Q. (2020). Encoding method of single-cell spatial transcriptomics sequencing. Int. J. Biol. Sci. 16 2663–2674. 10.7150/ijbs.43887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Liang P., Ji W., Yu Z., Chen H., Jiang L. (2019). Ubiquitin-specific protease 4 promotes glioblastoma multiforme via activating ERK pathway. Onco Targets Ther. 12 1825–1839. 10.2147/OTT.S176582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Q., Jin J., Hu H., Li H. S., Romano S., Xiao Y., et al. (2014). USP15 stabilizes MDM2 to mediate cancer-cell survival and inhibit antitumor T cell responses. Nat. Immunol. 15 562–570. 10.1038/ni.2885 [DOI] [PMC free article] [PubMed] [Google Scholar]