Graphical abstract
Abbreviations: DEG(s), differentially expressed gene(s); ER, endoplasmic reticulum; UPR, unfolded protein response. Genes names are given as shortened codes without preceding YALI0_; HSS(s), high-secretor strain(s); Pichia pastoris, traditional name for the renamed Komagataella phaffii; r(s)-Prot, recombinant (secretory) heterologous protein; SoA/TlG/scYFP/inYFP, strains overproducing a given heterologous protein (small and italicized letters); SOA/TLG/SCYFP/INYFP, genes encoding the target heterologous proteins (capitalized and italicized); SoA/TlG/scYFP/inYFP, heterologous proteins (small and non-italicized)
Keywords: Transcriptomics, Heterologous proteins, Secretory pathway, Yeast, Metabolic burden
Highlights
-
•
Secretion of a protein results in > 10-fold higher titer compared to its retention.
-
•
Overproduction of rs-Prots induces oxidative stress and detoxification response.
-
•
Excessive vacuolar protein degradation limits rs-Prot production in Y. lipolytica.
-
•
Non-classical export protein NCE102 is upregulated upon rs-Prot overloading.
-
•
Downregulation of cyclin CLN1 marks growth arrest in G1 under rs-Prot synthesis.
Abstract
Investigation of the yeast cell’s response to recombinant secretory protein (rs-Prot) overproduction is relevant for both basic and applied research. Imbalance, overloading or stress within this process impacts the whole cell. In the present study, by using steady-state cultures and transcriptomics, we investigated the cellular response of Yarrowia lipolytica challenged with high-level expression of genes encoding proteins with significantly different biochemical characteristics: a small protein retained within the cell i) or secreted ii), a medium size secretory protein with a high number of disulfide bonds iii), or glycosylation sites iv). Extensive analysis of omics data, supported by careful manual curation, led to some anticipated observations on oxidative and unfolded protein stress (CTT1, PXMP2/4, HAC1), glycosylation (ALGs, KTRs, MNTs, MNNs), folding and translocation (SSAs, SSEs) but also generated new exciting knowledge on non-conventional protein secretion (NCE102), transcriptional regulators (FLO11, MHY1, D01353 g, RSFA, E23925g or MAF1), vacuolar proteolysis targets in Y. lipolytica (ATGs, VPSs, HSE1, PRB1, PRC1, PEP4) or growth arrest (CLN1) upon rs-Prots overproduction.
1. Introduction
The investigation of the yeast cell’s response to recombinant secretory protein (rs-Prot) overproduction is relevant for both basic and applied research. It enables discovery of basic principles governing the cell, and identification of new targets for genetic engineering of the host to improve its performance. Synthesis of rs-Prot is a complex process taking place in multiple compartments of the cell. The be executed successfully, multiple processes must be coordinated, starting from transcription and translation, translocation into endoplasmic reticulum (ER) lumen, through folding, specific proteolytic cleavage, glycosylation, and disulfide bond formation, followed by a complex net of vesicular transit between compartments, all with an extensive system of quality control mechanisms and check points [1]. The cell must adjust the chaperone capacity, assure availability of resources, like amino acids, redox equivalents, ATP, membranes and glycans, as well as to adjust trafficking patterns, based on the collection of proteins that are synthesized at a given time. It has been estimated, that one-third of all the proteins is processed by the secretory pathway [2]. As such, substantial cellular resources are expended to maintain fluent operation of the secretory pathway, and consequently, imbalance, overloading or stress within this process severely impact the whole cellular metabolism. Due to high complexity of the pathway, dysfunctions may occur at many different levels, and dissecting the causative agent can be challenging [2]. Hence, the design of an efficient rs-Prot’s production platform requires an in-depth knowledge on its biology when challenged with this metabolically demanding task.
The world rs-Prot market constitutes one of the key branches of current industrial biotechnology, earning billion dollar (USD) revenues each year [3]. Within that field, yeast-based production platforms share the leadership with bacteria and fungi. Significant effort has gone into engineering yeast for increasing protein secretion (revised in [[4], [5], [6]]). Aside from engineering strategies targeting the r-Prot-encoding sequence itself, like shuffling with signal peptides or design of completely synthetic targeting elements [[7], [8], [9]], native mechanisms of the secretory pathway have been modified as well. The latter strategies targeted oxidative folding [[10], [11], [12]], elements of intracellular protein trafficking [13,14], the main transcription factor governing response to unfolded proteins (UPR) [[15], [16], [17]] or abolished excessive proteolysis [[18], [19], [20]].
Being aware of the complexity of the rs-Prot synthesis process, on a quest for new engineering targets the researchers harness omics analytical platforms. Global profiling of the metabolite, protein or transcript pools is used to scrutinize physiological response of a cell challenged with heterologous overproduction. Such a strategy was found very potent in identification of unobvious targets for cell engineering [[21], [22], [23]] or cultivation conditions favoring rs-Prot production [24,25]. The new targets were identified as the most responsive individual genes (differentially expressed genes; DEGs) or being a part of a highly responsive, up- or down-regulated biological process. In several studies, transcriptomics was used to comparatively study a global yeast cell’s response to overproduction of several different r(s)-Prots. In a series of papers on Saccharomyces cerevisiae perturbed with secretion of a small not glycosylated protein with even number of cysteines, and a larger protein with an odd number of Cys and a glycosylation site, numerous interesting observations were reported [2,5,[26], [27], [28]]. Recently, a comparative transcriptomics study on Yarrowia lipolytica challenged with overproduction of a small (55 kDa) and large (>100 kDa) beta-glycosidases has been reported [29]. All these studies provide substantial amounts of comprehensive, detailed and valuable knowledge, with high applicatory potential.
In the present study, we investigated cellular response of Y. lipolytica strains challenged with high-level expression of different r(s)-Prot-encoding genes. Y. lipolytica is one of the yeast strains considered an attractive protein production platform [4]. One of the biggest strengths of this species is an atypical, as for yeast, secretory pathway, resembling filamentous fungi [30]. We’ve been particularly interested in how Y. lipolytica cells respond to perturbations imposed by synthesis of proteins with significantly different biochemical characteristics. Hence, for our experimental set we’ve chosen: i) a small intracellular protein (inYFP), as a reference to its secretory counterpart (scYFP), ii) a small secretory protein with negligible posttranslational modifications (scYFP), iii) a medium size secretory protein with a high number of disulfide bonds (SoA;), iv) a medium size secretory protein with a high number of glycosylation sites (TlG; Table 1). The set of reporter r-Prots was composed in order to highlight cellular responses of Y. lipolytica to burdening the translational-secretory machinery at different points/processes. To get a thorough insight into the cell’s molecular response independently from the environmental conditions and the growth phase, the strains were cultured under the same conditions, in continuous mode to reach a steady-state. The following, global transcriptome profiling allowed to grasp expected, but here experimentally confirmed for Y. lipolytica phenomena, like an outburst of oxidative stress upon high rs-Prot synthesis; but also to observe many new, previously unreported for Y. lipolytica, responses. New targets for further cell engineering strategies have been identified and our knowledge on Y. lipolytica has been largely broadened. This knowledge can be used as a guidance for Y. lipolytica strains improvement towards more effective production of heterologous secretory proteins.
Table 1.
Biochemical properties of the r-Prot reporter proteins used in this study. Bioinformatic Tools used for determination of glycosylation sites and disulfide bonds are given in section 2.7.5.
| Protein | Molecular weight [Da]# | Number of aa# | Theoretical pI# | Predicted N-glycosylation sites* | Predicted O-glycosylation sites** | Number of Cys$ | Total different combinations S-S$ |
|---|---|---|---|---|---|---|---|
| YFP | 26991.54 | 239 | 5.58 | – | 1 | 2 | 2 |
| SoA | 5,214,040 | 474 | 4.88 | 1 | 8 | 11 | 35,696 |
| TlG | 65154.96 | 600 | 4.90 | 3 | 18 | 8 | 764 |
2. Materials and methods
2.1. Strains and culturing
All the yeast strains used in this study are derivatives of Y. lipolytica Po1h (Table S.1). The control strain (Po1h_Ura3) was transformed with a solo URA3 marker cassette to generate a prototroph. Recombinant Y. lipolytica strains were transformed with Golden Gate Assembly cassettes bearing a single transcription unit [TU] each with one of the target genes [31]. The target genes encoded either enzymatic (alpha-amylase – SoA, glucoamylase - TlG) or fluorescent (intracellular in- /secretory sc- form of YFP) proteins, expressed individually under the control of a synthetic hybrid promoter 4UASpTEF (4 direct repetitions of UAS and a minimal promoter of pTEF with CA environment). Secretory enzymatic reporters were transcriptionally fused with a signal peptide sequence native for Y. lipolytica exo‐1,3‐beta‐glucanase (B03564g) [7]. Yeast strains were routinely cultured in YPD medium (YPD [g L−1]: yeast extract (BTL, Lodz, Poland), 10; bactopeptone (BTL), 20; glucose, 20 (POCh, Gliwice, Poland)) liquid or solidified with agar at 15 [g L−1] (Biomaxima, Lublin, Poland), at 28 °C. For long-term storage, the strains were kept in 50 % glycerol stocks at −80 °C.
2.2. Chemostat cultivation
Continuous chemostat cultivations were performed according to previously described methodology [32] with some minor modifications. Briefly, the biomass from the glycerol stocks was transferred onto YPD agar plate and incubated at 30 °C for 24 h. The precultures were prepared by inoculating 30 mL of YPG20 medium ([g L−1]: yeast extract, 10; bactopeptone, 20; glycerol (POCh), 20) with a single colony, followed by cultivation at 28 °C for 22 h with agitation (250 rpm). Bioreactor cultures were conducted in fully instrumented and automatically controlled 0.5 L stirred tank bioreactors Multifors 2 (Infors HT, Bottmingen Basel, Switzerland) under the following conditions: temperature 28 °C, pO2 was kept at 20 % using cascade-control approach with stirring from 100 up to 1200 rpm, and constant aeration at 2.0 vvm. pH was maintained at 5.5 by automatic addition of 20 % NaOH and 10 % H2SO4. Foaming was minimized by automatic addition of a defoaming agent AntiFoam 204 (Sigma-Aldrich). The culturing medium YPG100 ([g L−1]: yeast extract, 10; bactopeptone, 20; glycerol, 100) in 300 mL volume was inoculated at 10 %. The culture was continued in a batch mode until glycerol was consumed, which was indicated by an increase in the pO2 parameter. Then the chemostat cultures were initiated at a dilution rate [D] of 0.20 [h−1]. Steady state was reached after at least 6 residence times [τ]. The experiments were performed in biological duplicate.
2.3. RNA extraction
Total RNA was extracted from the biomass samples collected from the steady-state culture. The RNA was isolated using Bead-Beat Total RNA Mini Kit (A&A Biotechnology, Gdynia, Poland) according to the manufacturer’s protocol. Extracts were obtained by mechanical disruption of the cells using Mixer Mill MM400 (Retsch, Germany) at 30 Hz for 30 s. The RNA extracts were verified for quantity using spectrophotometer (NanoDrop, Thermo Fisher Scientific) and for quality through agarose gel electrophoresis [33].
2.4. RTqPCR
The total RNA samples were reverse transcribed to the complementary DNA (cDNA) using SuperScript III Reverse Transcriptase and oligo(dT) primer, according to the manufacturer’s protocol (Thermo Fisher Scientific). The received cDNA samples were used as templates for RTqPCR reaction using RT HSPCR Mix SYBR® B (A&A Biotechnology) and gene-specific primers listed in Table S.2. The RTqPCR was carried out in an Applied Biosystems 7500 apparatus (Applied Biosystems, Foster City, USA) using the following thermal profile: 95 °C 3 min, (95 °C 15 s, 60 °C 30 s., 72 °C 30 s.) × 40, 72 °C 1 min, Melt Curve 94 °C 15 s, 60 °C 60 s, 95 °C 30 s., 60 °C 15 s.. The expression level of the genes was normalized to the expression level of actin- and translocation protein Sec62-encoding genes (ACT1; SEC62), used as internal calibrators, and to the reference strain (Y. lipolytica Po1h_Ura3+), used as the external calibrator, to which 1.0 expression level was assigned. All the samples were analyzed in biological duplicate and in technical triplicate. The results were presented as Log10ddCT calculated according to the ΔΔCT method [34].
2.5. RNA sequencing
Library construction and NGS sequencing were performed at Genomed S.A. (Warsaw, Poland) using HiSeq 4000 next-generation sequencer (Illumina) in a paired end mode. Reads with a length of 150 base pairs were generated. The raw sequencing reads are available on the NCBI Sequence Read Archive under the BioProject submission PRJNA701856 (see Data Availability section).
2.6. RNAseq data analysis
2.6.1. Raw data processing and data filtration
NGS adapters were removed using Cutadapt [35]. Initial standard filtering was done using q25 quality parameter and m15 the minimum length of the reads. Re-filtering was used to discard reads <20 bp. Quality control was conducted using FastQC [36]. Post-QC data were aligned and mapped to the Y. lipolytica CLIB122 reference genome (GenBank assembly identifier: GCF_000002525.2) using Hisat2 [37]. The number of readings pairs, mapped to an individual gene, was counted using the HTseq program [38]. Gene Ontology (GO) term annotations were assigned using the reference genome template (GCF_000002525.2). Comparative gene expression analysis was conducted using the DESeq2 package [39]. To compare the expression of different genes across the samples, raw read counts were converted into normalized RNAseq counts. Alternatively, relative gene expression level in reference to the control strain (Po1h_Ura3) was expressed as fold change. All Differentially Expressed Genes (DEGs) meet the requirement of FDR ≤ 0.05 (adjusted p value) tested via Wald test in DESeq2. Only DEGs meeting all those conditions were considered as responsive to introduced variable (overproduction of a specific heterologous protein) and subjected to further analyses. A complete list of all DEGs is attached as Table S.3.
2.6.2. Gene sets analysis
GO term statistical overrepresentation test was carried out by running DEGs list analysis with PANTHER v14.0-based online tool (rel. 20,200,728) [40]. First, GO terms were assigned to YALI DEGs according to GO Ontology database DOI: 10.5281/zenodo.4437524 Released 2021−01-01, specific for Y. lipolytica species. The test was carried out for biological process GO complete annotation data set, using Fisher’s exact test and False Discovery Rate for multiple testing. GSA was conducted on complete gene lists of all DEGs, as indicated by DESeq2. Only results with p < 0.05 are displayed.
2.6.3. DEGs analysis through venn diagrams
Analysis of DEGs lists through Venn diagrams was carried out using InteractiVenn online tool [41]. Up to 6 sets of up or down regulated gene lists were analyzed. Generated Venn Diagrams were later adjusted using inkscape (https://inkscape.org/release/inkscape-1.0.2/), according to the InteractiVenn’s instruction. For the analysis using Venn Diagrams, DEGs predicted by DESeq2 were further filtered for 2-fold up- or down-regulation. Assignment of genes’ names and genes’ functions was manually curated by cross-referencing several databases – Panther, UniProt, GRYC and NCBI and blasting (blastp) the sequences against the databases’ collections. The most relevant matches were chosen to finally describe a given gene. All the identified DEGs sets, common or specific for a given strain, were subjected to Gene Sets Analysis (according to methodology in point 2.6.2.). Identified enriched biological processes were indicated, where found.
2.7. Analytical methods
2.7.1. Biomass concentration
Dry cellular weight (DCW; [g L−1]) was determined gravimetrically through drying the biomass in a laboratory dryer at 105 °C for 2–3 days, until a stable readout was reached. The dry biomass concentration was expressed in grams of the cell dry mass per liter [g L−1].
2.7.2. HPLC analysis
High-performance liquid chromatography was conducted using Agilent Technologies 1200 series chromatograph (Agilent Technologies, Santa Clara, CA, USA) equipped with a refractive-index detector (G1362A) and a Rezex ROA-Organic Acid H + column (Phenomenex, Torrance, CA, USA). Operating conditions were as follows: 0.005 N H2SO4 as eluent at a flow rate of 0.6 [mL min-1]; the column temperature was set at 40 °C.
2.7.3. Enzymatic assays
The activity of alpha-amylase (SoA) was measured in the culture supernatant samples using microSIT assay, according to [42]. One activity unit (alpha-amylase activity unit [AAU]) refers to the amount of enzyme that decomposes 1 mg of starch per 1 mL, during 1 min, at pH 5.0 and 40 °C, under the applied experimental conditions. The glucoamylase (TlG) activity was examined in the supernatant samples using microDNS (3,5-dinitrosalicylic acid) method, described previously [43]. One activity unit (glucoamylase activity unit [GAU]) was defined as the amount of enzyme that releases 1 μg of reducing sugar equivalents per 1 mL, during 1 h, at pH 5.0 and 40 °C, under the adopted assay conditions. The reference strain Po1h_Ura3+ was assayed in parallel to determine background level. All the readouts were normalized per blank reaction with distilled water. The colorimetric enzymatic assays were read using the Tecan Infinite M200 plate reader at 580 nm (microSIT) and 540 nm (microDNS) wavelength. Each sample was analyzed in technical duplicate.
2.7.4. Fluorescence determination
Fluorescence of the YFP reporter was measured in flat-bottomed MTP plates (Corning; Sigma-Aldrich) using Tecan Infinite M200 automatic plate reader. scYFP was assayed in 200 μl supernatant samples. inYFP was assayed in ddH2O-washed biomass re-suspended in the initial volume of ddH2O. The following wavelength settings were used: excitation 495 nm/emission 527 nm. The fluorescence was expressed as Relative Fluorescence Unit [RFU] defined as the sample median fluorescence value minus the background fluorescence value. scYFP was normalized vs fresh culture medium. inYFP was normalized vs Po1h_Ura3+ reference strain biomass. Each sample was analyzed in technical duplicate.
2.7.5. Basic bioinformatic tools
Nucleotide or protein sequences were withdrawn and analyzed using NCBI (https://www.ncbi.nlm.nih.gov/), GRYC (http://gryc.inra.fr/) and UniProt (https://www.uniprot.org/) platforms. Gene function classification and basic information on the identified DEGs or their homologs was obtained from either GRYC, SGD (https://www.yeastgenome.org/) or UniProt. The search for homologs was conducted using blastp tool using default settings. Basic biochemical properties of the target proteins were determined using ProtParam Tool from Expasy service (https://web.expasy.org/protparam/). Prediction of N- and O-glycosylation sites, and the disulfide bonds formation combinations was conducted using NetNGlyc and NetOGlyc tools at http://www.cbs.dtu.dk/services and NPSA tool at https://npsa-prabi.ibcp.fr, respectively.
3. Results and discussion
3.1. Basic characteristics of the strains and the reporter proteins
The experimental set of strains in this study covers four Y. lipolytica Po1h derivatives overproducing the following heterologous reporter proteins (r-Prots): i) small intracellular protein (inYFP), ii) small secretory protein with negligible posttranslational modifications (scYFP), iii) medium secretory protein with a high number of disulfide bonds (SoA), iv) medium secretory protein with a high number of glycosylation sites (TlG; Table 1). It was presumed that overproduction of such a set of varying proteins would highlight the cellular response of Y. lipolytica to burdening the translational-secretory machinery at different points.
To gain an insight into the cell’s molecular response, independently from the environmental conditions and the growth phase, the strains were cultured in continuous mode to reach a steady state (Fig. S.1). The amounts of the target proteins produced under steady states are given in Table 2. Interestingly, the amount of biomass-accumulated YFP in [kRFU/gDCW] was ∼10-fold higher for scYFP (intended for secretion) than inYFP (intracellular), even without accounting for the fraction secreted outside the scYFP cells. This observation suggests that due to the difference in the translation mechanism (on ER for scYFP, in cytoplasm for inYFP) and/or sequestration of the overproduced protein within the ER-Golgi-vesicle continuum, more of the protein could be synthesized when targeted for secretion. The practical significance of this observation is that targeting a protein for secretion allows more product to be obtained, at least in this particular case.
Table 2.
Amounts of r-Prots synthetized by Y. lipolytica strains under steady-state conditions in continuous cultures.
| Strain | Fraction | Amount of Protein in [AU/L] or [RFU/L] | Amount of Protein in [AU/g] or [RFU/gDCW] |
|---|---|---|---|
| SoA | Supernatant | 729.42 ± 11.20 [AAU/L] | 96.33 ± 6.55 [AAU/gDCW] |
| TlG | Supernatant | 1 069.83 ± 5.54 [GAU/L] | 75.43 ± 6.38 [GAU/gDCW] |
| inYFP | Biomass | 130 809.60 ± 5201.20 [kRFU/L] | 10 872.85 ± 810.41 [kRFU/gDCW] |
| scYFP | Biomass | 812 108.30 ± 5715.62[kRFU/L] | 109 519.42 ± 7 523.53 [kRFU/gDCW] |
| Supernatant | 24 785.28 ± 187.33 [kRFU/L] | 3 342.31 ± 53.80 [kRFU/gDCW] |
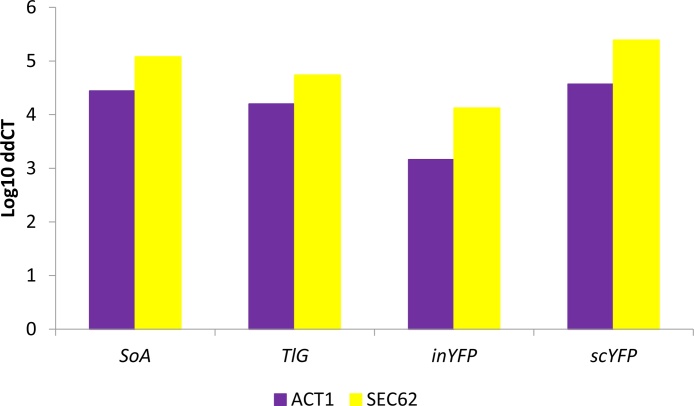
To determine how the amounts of the heterologous proteins correspond with gene expression levels, RTqPCR targeting the heterologous genes was carried out (Fig. 1). The expression level was comparable for the genes encoding secretory reporters – scYFP, SoA and TlG (in descending ranking). On the other hand, the inYFP-encoding gene’s expression was significantly lower (p < 0.05), even though all the genes were cloned under the same promoter. Such a trend was identical irrespective of the internal calibrator used (ACT1 or SEC62).
Fig. 1.
Relative expression level of the genes encoding heterologous r(s)-Prots in Y. lipolytica strains. X axis: Reporter gene (SOA, TLG, SCYFP and INYFP). Y axis: Log10 ddCt – relative quantitation value normalized to ACT1 (purple) or SEC62 (yellow) used as internal calibrators. (For interpretation of the references to colour in the Figure, the reader is referred to the web version of this article).
The conducted basic characterization indicated that our set of strains covers: i) scYFP – a highly expressing and efficiently producing strain, with no burden on disulfide bond formation or glycosylation (high-secretor strain; HSS), ii) inYFP – a moderately-producing strain, with significant limitations at the transcription level; the limitation cannot be caused by overloading the secretory pathway but some other mechanisms, iii) SoA – a strain with a high expression level and very high production and secretion (HSS) of the protein, possibly burdened at the oxidative folding level due to -S-S- bond formation; iv) TlG – a strain with decent expression and production levels; burdened at the glycosylation level due to the protein’s biochemical traits. How the perturbations introduced by the overproduction of the heterologous proteins impact the cellular response at the molecular level was further investigated through global transcriptional profiling. The analysis was conducted in both explorative and targeted mode, to study a general, global reaction of the cell, as well as to gain an insight into the processes taking place in the translational-secretory machinery.
3.2. Global transcriptome profiling – general observations
Quantification of raw counts for individual genes in the analyzed total RNA samples showed slightly different distributions, but none of the samples differed significantly from the rest (Fig. S.2). Moreover, biological replicates were characterized by high similarity in this regard. In terms of the overall similarity of the transcription profiles compared in terms of normalized read counts, strains expressing SCYFP and SOA were clustered together, while those expressing TLG and INYFP were more distant, generating a subcluster with some similarity to the control strain, located in the middle of the heatmap (Fig. S.3). The shape and local densities of volcano plots generated for the four strains (Fig. S.4) well illustrated those observations.
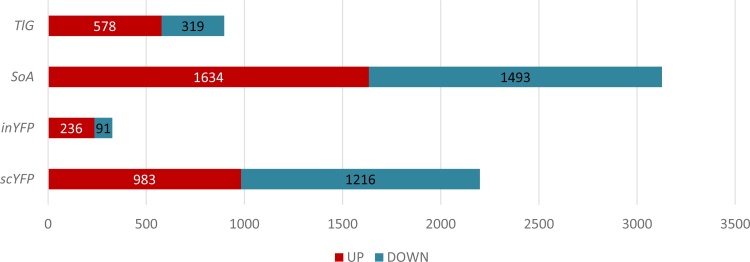
Primarily, the high dissimilarity of scYFP’s and inYFP’s transcriptomic profiles depicted in Fig. S.3 and Fig. S.4.a.b indicates that targeting a polypeptide for secretion induces massive rearrangements within the cell, exceeding the differences associated with biochemical properties of the proteins. Intracellular overproduction of YFP resulted in 327 DEGs vs the control strain, while its secretion resulted in 2199 DEGs, which is almost 7-fold more (Fig. 2). The high total number of DEGs in the SoA-producing strain (3127 DEGs; Fig. 2) suggests that high overproduction of a protein with a large number of Cys triggers substantial, multidirectional changes in the host cell. Expectedly, the number of DEGs was the lowest for inYFP-producing strains. The similarity of TlG’s transcriptional profile with that of inYFP may suggest that the maturation and secretion of such a polypeptide – highly glycosylated with low number of disulfide bonds – it less demanding. To gain more detailed insight into the specifics of transcriptional profiles generated by each of the strains, we aimed to test these hypotheses.
Fig. 2.
A total number of DEGs determined for each recombinant strain vs the control strain. X axis: number of DEGs: red fields – upregulated DEGs; blue fields – downregulated DEGs. Y axis: recombinant strains producing different r-Prots. (For interpretation of the references to colour in the Figure, the reader is referred to the web version of this article).
3.3. Global response of the cell – explorative analysis
3.3.1. Biological processes and genes significantly responding to the reporter proteins’ overproduction
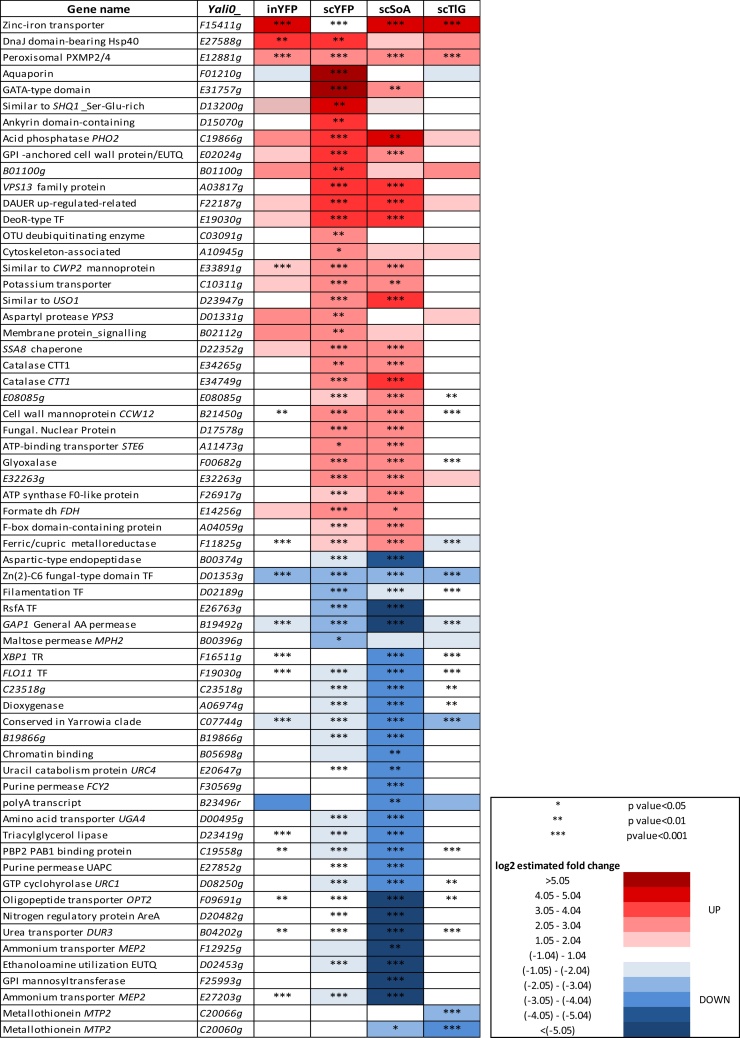
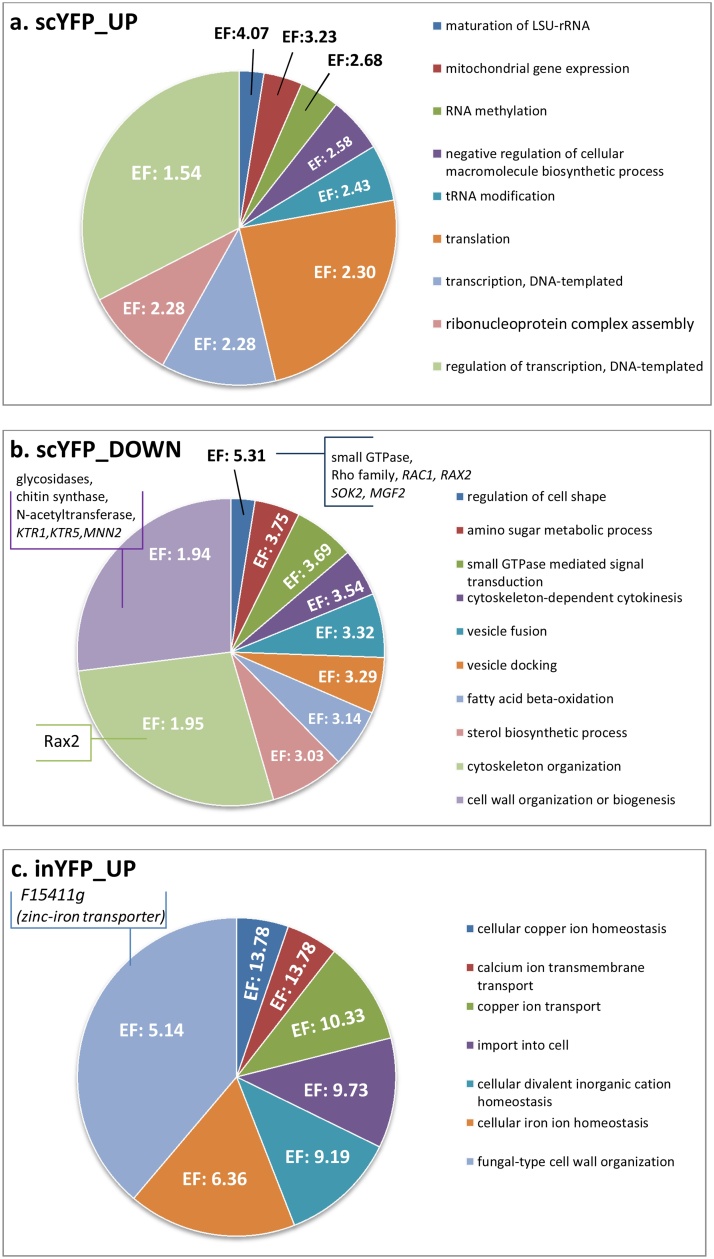
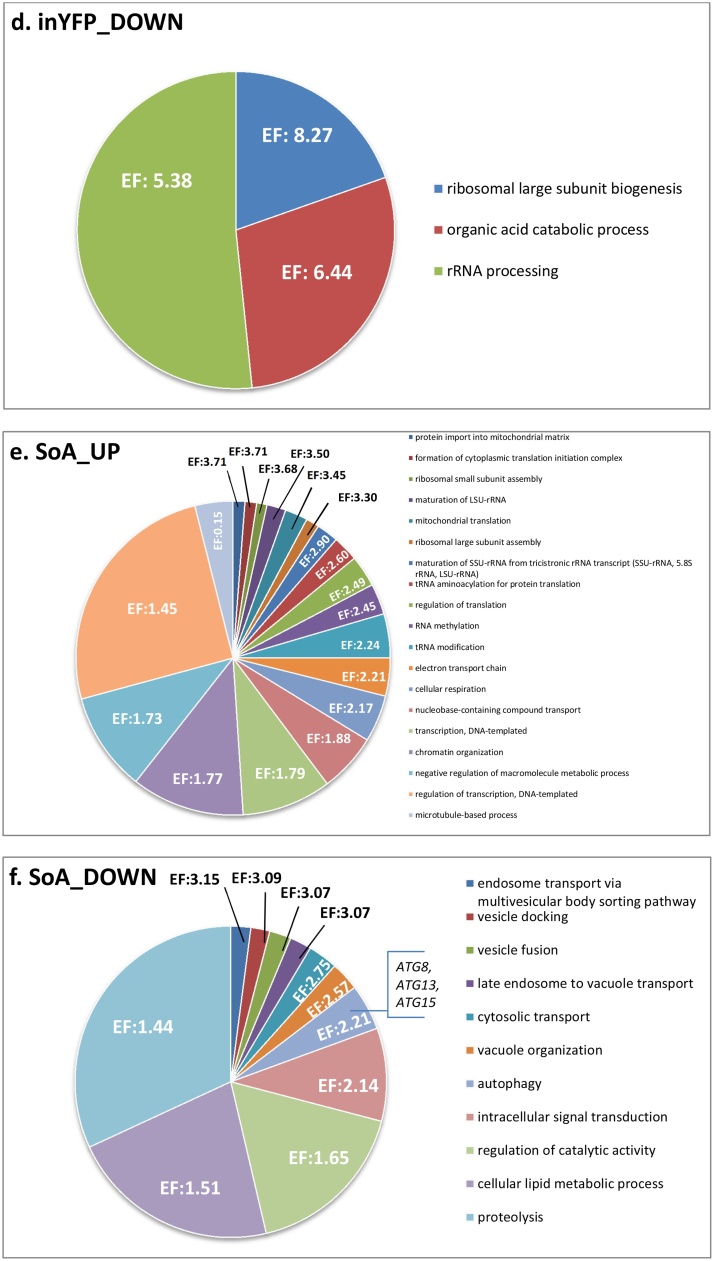
First we searched for biological processes that were up- and down-regulated upon overproduction of a specific protein type from our set. The transcriptional profiles of TlG- and inYFP-producing strains were enriched in upregulated genes involved in inorganic ion homeostasis (copper, calcium, zinc and iron; with the most up-regulated gene across the whole transcriptome – zinc-iron transporter F15411 g; Fig. 3), import into the cell, and fungal-type cell wall organization (Fig. 4.c.g). Microscopic observations demonstrated that the strains inYFP and TlG grew in the chemostat culture predominantly as ovoid-shaped cells, in contrast to the control and SoA strains, for which the dominant morphotype was filamentous and elongated ovoids (Fig. S.5). Since all the strains were maintained under the same conditions, it seems that the morphotype was dependent on the introduced variable – the type of overproduced protein and the resulting cellular response. No significant enrichment in the cell wall organization, morphogenesis, or cell shape control in SoA was recorded in the transcriptomic data, which indicates similarity in these biological processes for this and the control strain (corresponding to microscopic images). On the other hand, FLO11 (F19030g), involved in filamentous and invasive growth, was amongst the most downregulated genes in the SoA and scYFP strains’ transcriptome Fig. 3, which differentiates SoA and control strains in terms of the cell shape control. It thus was an open question whether this high downregulation of FLO11 is actually a hallmark of the onset of dimorphic transition to ovoid morphotype. In the second most active HSS (scYFP), which grew in both ovoid and very long filamentous forms (Fig. S.5), the cell shape regulation was the most enriched downregulated biological process (Enrichment fold 5.31; Fig. 4.b). This process was enriched in small GTPases, members of the Rho family (RHO1, RHO2, RHO3, RHO4), or RAC1, and RAX2, responsible for bud site selection. Interestingly, amongst the downregulated genes assigned to this biological process in the scYFP, we identified SOK2 (B19602g; similar to MGF1), which is a negative regulator of pseudohyphal growth, suggesting that at the moment of sample collection the cell population could be under dimorphic transition towards filaments. In addition, one of the most upregulated genes in scYFP and SoA codes for a GPI-anchored cell wall protein involved in cell wall organization and induction of hyphal growth (E02024g) (Fig. 3, Fig. 5.b). On the other hand, mycelial growth factors MGF2 (B19602g) and D02189g, mediators of RNA polymerase II involved in the regulation of the transition between yeast and filamentous forms, were downregulated in both SoA (-2.41/-1.91) and scYFP (-1.38/-2.12) (Fig. 5.c), which supports the hypothesis, conceived based on FLO11 downregulation and the gene set enrichment analysis, that the highly producing cells, representing both filamentous and ovoid morphotype, were under dimorphic transition towards ovoid shape. A high response of the “cell wall biogenesis” biological process was previously observed for S. cerevisiae overproducing r-Prot [2]; however, in that study, the researchers reported upregulation of invasive, filamentous growth, which was associated by the authors with increase in the traffic of secretory vesicles to the membrane. According to those authors’ argumentation, filamentous or pseudohyphal cells have a high surface-to-volume ratio and inherently require higher Golgi-to-cell membrane trafficking rates to supply cell membranes and cell wall components for growth. Our observations concerning Y. lipolytica suggest that upon high r-Prot overproduction, the cells reprogrammed the morphotype to ovoid. Additionally, in terms of combining Y. lipolytica morphotype and r-Prot overproduction, our recent study evidenced that ovoid cells are more active and efficient in secretory r-Prot synthesis compared to filamentous forms [44], but the molecular background of that observation was not known. Expression of SOK2 is regulated by RNA polymerase II, and MGF2, FLO11 and D02189g are the mediators of RNA polymerase II activity, which altogether provides a direct link between the gene overexpression and morphotype in Y. lipolytica. Those genes’ operation makes an important contribution to our understanding of the relationship between heterologous gene overexpression and morphology in Y. lipolytica.
Fig. 3.
Heatmap of the most responsive DEGs in the TlG-, inYFP-, scYFP- and SoA-overproducing strains. Shortened YALI_ code and a short description of a given gene are shown. Assigned names and functions were manually curated for all the ambiguous hits by blasting the sequence against UniProt and NCBI non-redundant protein database, and inferring the most accurate match. Red (up-regulated) and blue (down-regulated) squares were used to represent the transcription profiles of genes expressed as log2 Estimated fold change. Indicated genes meet the requirement of > 2.0 fold change in at least one of the overproducing strains. Abbreviations: dh - dehydrogenase, TF - Transcription Factor, TR – Transcription Repressor. (For interpretation of the references to colour in the Figure, the reader is referred to the web version of this article).
Fig. 4.
Pie charts showing significantly enriched biological processes determined via statistical overrepresentation test conducted using Panther tool on the complete DEGs lists. scYFP_UP (a.), scYFP_DOWN (b.), inYFP_UP (c.), inYFP_DOWN (d.), SoA_UP (e.), SoA_DOWN (f.), TlG_UP (g.) and TlG_DOWN (h.). Up- and down-regulated DEGs identified for each strain were analyzed separately to reveal up- and downregulated biological processes. Only biological processes enriched at significance level p < 0.05 are shown. The results were manually curated to reduce redundancy of several biological processes. EF – Enrichment fold. (For more convenient visualization of the Figure in color, the reader is referred to the web version of this article).
Fig. 5.
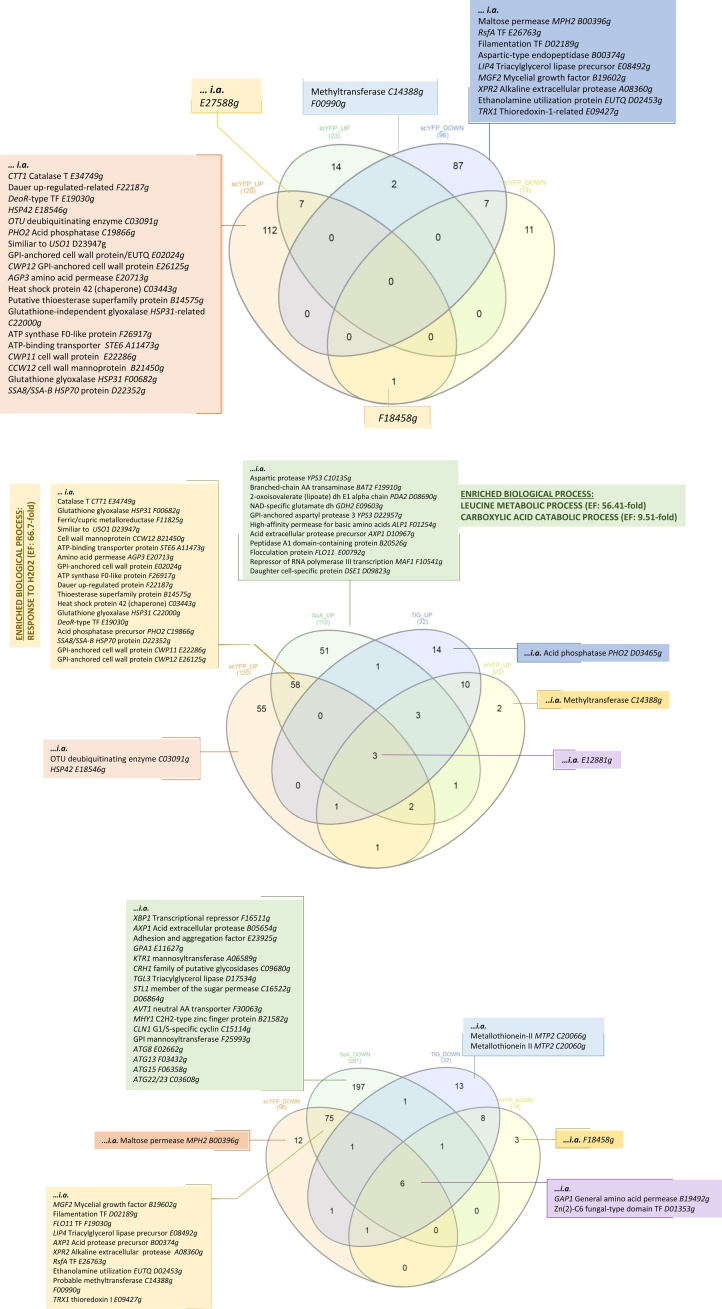
Venn Diagrams analysis illustrating shared and dissimilar genes/gene sets for individual strains. The numbers indicate the number of shared or strain-specific genes, up-/down-regulated in scYFP- and inYFP- producing strains (a.); up-regulated genes in SoA-, TlG-, scYFP- and inYFP-producing strains (b.); down-regulated genes in SoA-, TlG-, scYFP- and inYFP-producing strains (c.). Venn diagrams analysis was done with the DEGs lists restricted to 2- fold change. Shortened YALI_ code of the gene and a short description is given only for selected genes – full lists are given in Figure S.6. Significantly enriched biological processes within generated gene groups identified via GSA are given in BOLD and underlined (found only for two gene sets). Abbreviations: dh - dehydrogenase, TF - Transcription factor, TR – Transcription Repressor, AA- Amino Acid. (For more convenient visualization of the Figure in color, the reader is referred to the web version of this article).
Morphological transition to a filamentous form is a known marker of stress in Y. lipolytica cells [45,46]. In the present study, filamentation of the control strain is not clearly understood, as the cells were maintained at high carbon/nutrient provision and at a pH that should not promote filamentation (pH 5.5; filament formation is induced at pH close to 7.0; Gorczyca, Kaźmierczak et al., to be published). On the other hand, both HSSs were overexpressing CTT1 (E34749g) encoding a peroxisomal catalase, which is a known, ubiquitous marker of an ongoing cellular stress response (Fig. 3, Fig. 5.b). Its expression was over 13- and 6-fold lower in a filamenting control strain compared to HSSs. Hence, it suggests that filamentation in the control strain was not associated with the general stress response but was probably caused by some other factors. Formation of filaments by the SoA-overproducing strain and the above discussed dimorphic transition ongoing in scYFP are more straightforward to explain. In this regard, CTT1, which plays a key role in the oxidative stress response and in protecting proteins against oxidative inactivation, was one of the most responsive, overexpressed DEGs in HSSs. It is known that overproduction of rs-Prots is inherently associated with oxidative stress due to energy- and material-consuming oxidative folding taking place in the ER [30]. Additionally, transcriptomic profiles of SoA and scYFP were characterized by high overexpression of glutathione-independent glyoxalase Hsp31-related (F00682g; Fig. 3; Fig. 5.b), which plays a role in detoxifying endogenously produced glyoxals [47,48] and is involved in protection against reactive oxygen species (ROS), similarly to CTT1. This further supports the statement on induction of oxidative stress in the HSSs. Notably, it has been demonstrated that oxidative stress is induced in the overproducing strains, even if the overproduced protein bears only a limited number of Cys residues [2,12,49], which relates to the scYFP case.
In contrast, upon TlG overproduction, the most downregulated genes were metallothioneins (C20060g and C20066g; Fig. 3; Fig. 5.c), which are cysteine-rich heavy-metal-binding proteins, localized to the membrane of the Golgi apparatus and cytoplasm, serving as the primary regulators of trace metal homeostasis in yeast [50]. Their principal role is protection against metal toxicity and oxidative stress (as CTT1), and regulation of zinc and copper pools. Biosynthesis of metallothioneins in yeast increases by several-fold throughout oxidative stress. Their downregulation is in high contrast to the most upregulated zinc-iron transporter (F15411g); and such a co-expression pattern is not fully understood. Likewise, one of the most upregulated genes in SoA (F11825 g; Fig. 3) encodes a ferric/cupric metalloreductase responsible for reducing extracellular iron and copper prior to import. In each of the studied transcriptomes, genes involved in “metal ion homeostasis and import” head the list of upregulated DEGs, highlighting the significance of this biological process upon enhanced protein production.
The Rax2 protein mentioned above, responsible for bud site selection, was also assigned to several significantly enriched downregulated biological processes in scYFP (Fig. 4.b), including cytoskeleton organization (Enrichment fold: 1.95), and cytoskeleton-dependent cytokinesis (Enrichment fold: 3.54), which was composed of cytoskeletal proteins, actin, tubulins, actin binding motor proteins, GTPase-activating protein and tropomyosins. Their downregulation in the intensively secreting strain is not currently clear, except for the hypothesis on the HSSs’ transition to ovoid morphotype. On the other hand, high upregulation of USO1 (D23947g) in both HSSs suggests enhancement of the intracellular protein transport (Fig. 3; Fig. 5.b) and USO1’s dominant role in that biological process. Uso1 is an essential protein involved in vesicle-mediated ER to Golgi transport; it binds membranes, acts during vesicle docking to the Golgi, and is required for assembly of the ER-to-Golgi SNARE complex.
The cell wall organization process (Enrichment fold: 1.94) was significantly downregulated in the scYFP strain. Manual inspection of the genes assigned to this process revealed several glycosidases, chitin synthase, phosphoacetylglucosamine mutase, glucosamine-6-phosphate isomerase and N-acetyl transferase. These genes were clustered together with mannose-6-phosphate isomerase, cell wall mannoprotein cis3-related (note that fungal-type cell wall organization was upregulated in the ovoid-shaped inYFP and TlG strains) and mannosyltransferases, including KTR1, KTR5, and MNN2 (Fig. 4.b). Although the mannosyltransferases were assigned to a cell wall organization process, it is known that apart from being involved in cell wall macromolecule biosynthesis, they play a role in glycosylation of proteins. Likewise, the individual gene (not a part of an enriched biological process) F25993g, encoding Golgi membrane-bound mannosyltransferase 2, involved in protein glycosylation and vesicular trafficking, was one of the most downregulated DEGs in the SoA strain (Fig. 3; Fig. 5.c). Downregulation of the secretory pathway constituents in the HSSs requires additional comment. Both the proteins are scarcely glycosylated (Table 1). It may be speculated that the prevalence of the non-glycosylated scYFP/SoA as cargos in the secretory pathway resulted in relative downregulation of mannosyltransferases versus the control strain, which were involved in the native polypeptide maturation in the latter. Indeed, manual inspection of the gene sets revealed that all the main secretory proteases and lipases, including TGL3 (D17534g), LIP4 (E08492g), AXP1 (B00374g, B05654g) and XPR2 (A08360g), were downregulated in SoA, and three of them – XPR2, AXP1, LIP4 – were downregulated in scYFP (Fig. 5.a.c). These proteins differ in terms of number of glycosylation sites, but they bear on average 3 N-glycosylations and 8 O-glycosylations. Their downregulation improves the capacity of the secretory pathway, required for SoA/scYFP synthesis, but at the same time decreases the requirement for glycosylating enzymes. Furthermore, in relevance to the cell wall restructuring in HSSs, a gene encoding cell wall mannoprotein, CCW12 (B21450g), was identified amongst the most upregulated genes, together with STE6 (A11473g), which is an ABC-type peptide-transporting ATPase involved in the export of methylated and prenylated a-factor pheromone [51] (for scYFP – only upregulated; not “the most responsive”; Fig. 3). While it has been postulated that Ste6 is specific towards a-pheromone, its closest mammalian homologue encodes a multidrug resistance (MDR1) channel, involved in excretion of numerous toxic compounds. It can be speculated that upon high oxidative stress in the HSS overproducing cells, many toxic compounds are generated and are pumped out of the cell via the Mdr1/Ste6 membrane channel. More details on cell wall biogenesis-related genes are given hereafter.
Another significantly downregulated biological process in TlG-/inYFP-producing strains was associated with ribosome biogenesis, rRNA processing and organic acid metabolic (catabolic) processes (Fig. 4.d.h), which was again rather surprising, considering the ongoing overproduction of heterologous proteins. Still, those biological processes were only slightly down-regulated, since when conducting the enrichment analysis with the gene sets filtered for 2-fold change, they were not identified as significant. In contrast, transcriptional profiles generated upon overproduction of SoA and scYFP were significantly enriched in upregulated genes involved in ribosome biogenesis and assembly, transcription, translation, regulation of DNA-templated transcription, chromatin modification, histones synthesis and modification, which is straightforward to explain by intensive ongoing transcription and translation (Fig. 4.a.e). High downregulation of E26763g (RSFA) in scYFP (−2.14) and SoA (−5.39) strains (Fig. 3; Fig. 5.c), which is a ribosomal silencing factor [52], well corresponds with those observations. RsfA helps cells to adapt to slow-growth/stationary phase conditions by down-regulating protein synthesis, which is one of the most energy-consuming processes. Its downregulation releases ribosome assembly from inhibitory action of RsfA, which is of key importance for the overproduction of heterologous proteins. Intriguingly, RSFA was not significantly downregulated in TlG and inYFP, which were also overproducing r-Prot. In this regard, it was previously observed that overproduction of a large r-Prot resulted in repression of overall transcription and translation [2]. That observation contrasts with our results on RSFA and high enrichment in ribosome assembly and biogenesis process upregulation in HSSs. The authors explained that the repression is a general, broad-spectrum response used to adjust the expression rates to reduced capacity of folding and maturation. It was further explained that if the processes upstream oxidative folding are not downregulated, the risk of severe oxidative stress and excessive consumption of reducing equivalents is high [2]. Considering the high upregulation of oxidative stress response genes in HSSs studied here, it is possible that unrestricted high transcription and translation led to imbalanced flow through the secretory pathway. Results on limited expression level of INYFP and TLG (Fig. 1) together with lack of downregulation of RSFA and no oxidative stress response outburst in these strains collectively support that statement. However, it remains unclear why the defensive processes were not implemented in HSSs; and it requires further deepened studies.
Within this subject, amongst significantly upregulated biological processes in HSSs, we identified “negative regulation of cellular macromolecule biosynthesis” (Fig. 4.a.e). In our previous work we postulated that upon high overproduction of a rs-Prot, transcription of its gene is repressed to avoid overloading the secretory pathway [32], as was previously reported for S. cerevisiae [2]. In the present research, we observed that the biological process “transcription” is upregulated in the HSSs, but one of the key genes assigned to that process is specific towards polymerase III transcribing small RNAs required for translation, cell growth and the cell cycle (and not mRNAs). The responsible gene (F10541g; MAF1) is a conserved repressor of RNA polymerase III, so its upregulation results in decreased transcription of the governed genes (Fig. 5.b); which, in this case, corresponds to previous observations [2] made for S. cerevisiae.
Vacuole organization, autophagy and proteolysis (Enrichment fold: 2.57, 2.21, 1.44, respectively) were significantly downregulated biological processes in SoA (Fig. 4.f; only a tendency observed for scYFP); including such genes as ATG8, ATG13, and ATG15 (autophagy-related proteins). Autophagy, through the sequestration and delivery of cargo to the lysosomes, is the major route for degrading long-lived proteins and cytoplasmic organelles [[53], [54], [55]]. Formation of the autophagic vesicles requires the recruitment of Atg8 (downregulated in HSSs), a ubiquitin-like protein, to the membrane of the nascent autophagosomes. Their downregulation suggests limited protein degradation in comparison to the control strain. Additionally, one of the most upregulated genes in scYFP was C03091g, associated with protein degradation. It bears high similarity to OTU domain-containing protein acting as thiol-dependent deubiquitinating enzyme [56] (Fig. 3; Fig. 5.a). The transcriptome of SoA-overproducing strain was enriched in an upregulated panel of peptidase A1 domain-containing proteins, encoded by B20526g, C10135g, D10967g, and D22957 g (Fig. 5.b). The former three peptidases are extracellular, membrane-unbound, aspartic-type endopeptidases that play a role in extracellular proteolysis. The latter peptidase is Yps3, a GPI-anchored aspartyl protease which is involved in fungal-type cell wall organization and may also play a role in the maturation of GPI-mannoproteins associated with the cell wall. YPS3 was also one of the most upregulated DEGs in scYFP (Fig. 3). Yps3 belongs to the yapsin family of known, negative impact on the stability of secretory proteins [57]. Disruption of yapsin-encoding genes [58] by combined knock-out of YPS1/PEP4 increased the titer of the non-degraded product from ∼40 up to 80 % [59]. In Y. lipolytica, the most popular expression platform strains, belonging to the Po1-series, typically bear disruption of one or both major extracellular proteases – XPR2 and AXP1, but yapsins have not been considered for disruption to date. It would be interesting to test a similar strategy in Y. lipolytica, since now it is clear that the genes are highly expressed upon r-Prot production.
Additionally, within the scope of protein and amino acid turnover and transportation, we observed common significant downregulation of GAP1 encoding a general amino acid permease (B19492g; Fig. 3; Fig. 5.c), which was surprising considering the high demand for the protein building blocks. In contrast, some other amino acid permeases were identified as upregulated within the strains’ transcriptomes, including the amino acid permease AGP3 and the high-affinity permease for basic amino acid import ALP1. On the other hand, genes involved in amino acid turnover and transformation were significantly upregulated in HSSs. BAT2 (branched chain amino acid aminotransferase), PDA2 (2-oxoisovalerate dehydrogenase), 3-hydroxy-3-methylglutarate-CoA lyase, and GDH2 (glutamate dehydrogenase) constitute the central branch point in the amino acid turnover net [60,61]. They were all upregulated in the SoA-overproducing strain, indicating intensive amino acid metabolism in the cell (Fig. 5.b). Taken together, these results suggest that upon limited autophagy in the HSSs, extracellular proteolysis of peptone and YE components is catalyzed by the upregulated set of extracellular/membrane-bound proteases and results in high amino acid provision in the medium. Amino acids are taken up by the upregulated amino acid transporters (e.g. AGP3 and ALP1) and the imported moieties are subjected to transformations by the amino acid turnover net, according to the requirements of the ongoing overproduction.
Finally, in terms of the general cellular response to a specific protein’s overproduction, beta-oxidation and cellular lipid metabolic processes were downregulated in scYFP and SoA (Enrichment fold: 3.14 and 1.51), respectively (Fig. 4.b.f). Modulation of the lipid metabolism under heterologous secretory protein’s overproduction could be caused by increased requirement for phospholipids for the intracellular membrane trafficking. It has been postulated that limitation of membrane availability may be the key limiting factor for efficient operation of the secretory pathway [62]. High upregulation of the EUTQ gene (E02024g; ethanolamine utilization protein) in the SoA producer and downregulation of the putative ethanolamine utilization protein D02453g (also in scYFP) are direct indications of intensive turnover of phospholipids ongoing in the cell (Fig. 3). Manual inspection of the biological processes’ representatives showed that the downregulated lipid metabolism gene set in SoA consists mainly of numerous ER-located membrane-bound proteins, including ceramide very long chain fatty acid hydroxylase SCS7, diacylglycerol O-acyltransferase, GPI mannosyltransferase 4, GPI anchor transamidase, dolichyl glycosyltransferase (N-glycosylation), GlcNAc-1-P transferase (playing a role in N-linked glycosylation), dehydrodolichyl diphosphate synthase (which is a prenyltransferase involved in protein glycosylation), or diacylglycerol O-acyltransferase 1. Although being assigned to the downregulated lipid metabolism, these genes encode ER-/Golgi-membrane proteins involved in protein maturation. The low number of predicted glycosylation sites and the presumed abundance of SoA/scYFP as cargos in the secretory pathway may account for such observations. Still, some fatty acid metabolic process-related genes were included in that downregulated group (in SoA), including peroxisomal acyl-CoA oxidase, and enoyl-CoA isomerase 2 (Fig. 4.f). Likewise, the scYFP-producing strain showed downregulation in lipid metabolism, including peroxisomal multifunctional enzyme type 2, acyl-coenzyme A oxidase 2 and 3, enoyl-CoA hydratase and isomerase (Fig. 4.b). Furthermore, the cells overproducing SoA (and scYFP, but to lesser extent) were also particularly active in cellular respiration and generation of energy associated with upregulated protein import into the mitochondrion, and mitochondrial gene expression (Fig. 4.a.e). In this regard, amongst the most responsive upregulated DEGs we identified F26917g, which encodes ATP synthase F0-like protein. Increased energy demand for the r-Prot overproduction easily explains that observation (Fig. 3). As demonstrated previously [2], the secretory stress shifts metabolism to increased oxygen and ATP requirements.
3.3.2. Similarities and dissimilarities in the global gene response profiles of Y. lipolytica to different proteins’ overproduction
Venn diagram analysis is useful for identification of common and distinct genes and processes characterizing a given transcriptomic profile. Depending on the input gene sets, it is possible to identify molecular objects “hallmarking” a given biological process. In this research we aimed to identify genes specifically responding to protein secretion (comparison of inYFP vs scYFP; Fig. 5.a) and marking intensified oxidative folding (SoA vs other) or glycosylation (TlG vs other) (Fig. 5.b.c). A complete list of genes (accompanied by a short description and YALI name) used for Venn diagrams analysis is available in the Supplementary material (Fig. S.6).
Systematic comparison of DEGs sets of inYFP and scYFP was expected to reveal markers of high overproduction of intracellular and secretory r-Prot in Y. lipolytica. Such genes were initially searched within the groups of strain-specific and differential expression direction-specific genes (n = 112, n = 14, n = 87, n = 11; Fig. 5.a), but no enrichment in any specific biological process could be found there. Interestingly, transcriptomes of inYFP and scYFP shared 7 common upregulated genes, including E27588g, which at the same time is the second most upregulated gene in inYFP (Fig. 3, Fig. 5.a). The gene encodes a DnaJ-domain-bearing Hsp40 chaperone regulating the ATPase activity of Hsp70, involved in folding of nascent proteins, translocation of polypeptides across organelle membranes, coordinating responses to stress, and targeting selected proteins for degradation. Its specific, high upregulation in both YFP-producing strains may indicate its specific role in folding of non-post-translationally modified proteins. Then we sought for markers of high overproduction of the intracellular and secretory r-Prot in the common gene groups but of opposite regulation in in-/sc-YFP. A single gene, F18458g, was downregulated in inYFP and upregulated in scYFP. The F18458g gene after the blastp search showed weak similarity to the bifunctional transcriptional activator/DNA repair enzyme Ada, bearing an assigned function of methylguanine-DNA cysteine methyltransferase in Clavispora lusitaniae. Due to the lack of a more specific functional annotation it is impossible to infer its role in the studied processes. Amongst the genes of the opposite pattern of regulation (up in inYFP, down in scYFP) we identified a methyltransferase (C14388g) and a GPI-anchored cell wall protein (F00990g) involved in cell wall organization and hyphal growth that is induced specifically in response to hyphal growth (Fig. 5.a). The latter again confirms our hypothesis that the HSSs were undergoing a dimorphic transition to ovoid morphotype.
The conducted Venn diagram analysis revealed that the only significantly enriched biological process common for upregulated genes in HSSs (n = 58; Fig. 5.b) was the “response to hydrogen peroxide” (Enrichment fold: 66.7). Aside from that, manual inspection of the list of common genes allowed us to identify the uncharacterized gene F22187 g, which was amongst the most upregulated genes in both HSSs (Fig. 3). Basic search demonstrated its similarity to another uncharacterized gene (NCU09057) from Neurospora crassa. Establishing its putative function was difficult, indicating lack of interest in this protein to date. A blastp search against the GRYC database revealed F22187g’s similarity with (depending on the region of similarity): 1) glutaredoxin (thioltransferase; belonging to the LEA type 4 family; 28 % within a range of 18–634 amino acids) from Planoprotostelium fungivorum, 2) protein LEA-1, isoform j from P. fungivorum, 3) LEA protein DR_1172 from Deinococcus radiodurans, or 4) CsbD domain-containing protein, being an integral membrane component in Williamsia spp. On the other hand, in the Panther database, F22187g was assigned to a dauer upregulated-related protein family. The term “dauer” denotes a growth arrest stage in a model nematode (C. elegans) intensively studied in relation to longevity and aging processes. The dauer (growth arrest) gene family includes a number of different heat shock proteins (e.g. Hsp20, smHsps) and oxidoreductases (e.g. cytochrome P450, glutathionyl-, glucuronosyl/glucosyl-, short-chain- oxidoreductases), as well as UDP-glucuronosyltransferases and glutathione S-transferases [63]. It was proposed that broad spectrum detoxification and repair of damaged proteins constitute a molecular basis of the growth arrest and the consequent prolonged viability. Interestingly, the smooth ER was proposed to be a “longevity organelle”, where detoxification and repair take place. In relation to the current findings and the blast search results, it could be postulated that F22187g is an ER-resident thioltransferase, upregulated in SoA- and scYFP-overproducing strains, supporting homeostasis maintenance; however, it must be kept in mind that the function was assigned only based on the blast similarity search. In reference to the current study, growth arrest and upregulation of detoxification processes may be the fundamental biological processes accompanying intensive r-Prot overproduction, with the key role of ER. The other upregulated genes supporting this statement, and shared by scYFP and SoA, were thioesterase (B14575g), HSP42 (C03443g) and glyoxalase HSP31 (F00682g, C22000g) (Fig. 5.b).
Within the scope of the proposed mechanism of growth arrest associated with r-Prot overproduction, we observed intriguing, high downregulation of F16511g (−302; aka YALI2_F00325g; Fig. 3) specific for SoA (Fig. 5.c). F16511g/F00325g has an assigned role as an XBP1 transcriptional repressor involved in G1/S transition of the mitotic cell cycle. In S. cerevisiae (YIL101C) Xbp1 binds to RNA polymerase II-specific genes’ promoter sequences of the cyclin genes. Manual inspection of SoA’s DEGs showed significant downregulation of CLN1 G1/S-specific cyclin (C15114 g; promoting G1 to S transition), indicating growth arrest (G1→G0) (Fig. 5.c). That observation contradicts the possible involvement of XBP1 in CLN1’s downregulation, at least, according to the known mechanism (its downregulation should increase expression of CLN1, and promote G1 → S transition). On the other hand, it is known that CLN1 expression is also controlled by other transcriptional regulators – Swi4/Swi6 and Mbp1/Swi6. However, Y. lipolytica’s homologs were not found within SoA-induced DEGs. Nevertheless, CLN1’s downregulation is a hallmark of cell transition to quiescence, maintaining G1 arrest. XBP1’s downregulation upon SoA overproduction is also interesting due to its other interactions, namely, its role in the unfolded protein response (UPR). Upon ER stress, XBP1 mRNA is spliced by ER-resident Ire1 (like HAC1, discussed below), thereby generating functional spliced XBP1, translocating into the nucleus to initiate transcriptional programs that regulate a subset of UPR- and non-UPR-associated genes. It is also known that XBP1 expression is induced by different stress conditions. Its high downregulation in SoA is unclear, considering the high enrichment fold in cellular “response to hydrogen peroxide” in both HSSs. Taken together, downregulation of XBP1 accompanied by no transcriptional response from HAC1 (discussed below), and downregulation of CLN1, could collectively suggest that HSSs entered the quiescence state rather than UPR. Upregulation of another transcription factor, DEOR (E19030g), in both HSSs (Fig. 3; Fig. 5.b) further supports that statement. DeoR-type transcription factors bearing a sugar-binding domain, which is also present in the enzymes of the phosphosugar isomerase family, are involved in regulation of sugar metabolism in bacteria [64], e.g. as a repressor of sugar import [65]. It can be speculated that significant E19030g upregulation is associated with downregulation of maltose permease MPH2 (B00396 g) in scYFP and sugar permease STL1 (C16522g) in SoA (Fig. 5.c). Reduced transcription of some less important genes in order to supply enough energy and substrates for synthesis of r-Prot has also been suggested based on transcriptomics data from continuous cultivation of P. pastoris overproducing r-Prot [66].
No scYFP-specific up- or downregulated biological process was identified. On the other hand, SoA-specific upregulated genes (n = 51; Fig. 5.b) accounted for enrichment of the “leucine metabolic process” (Enrichment fold: 56.41) and carboxylic acid catabolic process (Enrichment fold: 9.51). Manual inspection demonstrated that the central metabolism of amino acid conversions and transformations was grouped in those enriched processes (including BAT2, PDA2, MVA1, GDH2, discussed above). While no particular biological process was enriched by specific (n = 12/n = 197) or common (n = 75) downregulated genes in SoA and scYFP (Fig. 5.c), what attracted our attention was a common downregulation MGF2 transcription factor, backed up with SoA-specific downregulation of MHY1, FLO11, “adhesion and aggregation factor” (E23925g) involved in filamentation. Intriguing downregulation of the cytoplasmic thioredoxin TRX1 (E09427g) (within a group n = 75), shared by HSSs, is discussed below.
While no TlG-specific biological process was enriched, we observed common upregulation of the PHO2 acid phosphatase-encoding gene in all the strains overproducing rs-Prots (scYFP, SoA, TlG). PHO2 is involved in scavenging of inorganic phosphorus required for energy storage and transfer. Detailed analysis of Fig. 5.b (and Fig. S.6) showed however that the same PHO2 activity was encoded by different genes: 1) C19866g (highly similar to D03465g) in HSSs, and 2) D03465 g in TlG. Importantly, PHO2 was amongst the top most responsive upregulated DEGs in both HSSs (Fig. 3), which is logical in the face of high-energy demanding r-Prot overproduction. The difference between the two isogenes, and the reason for strain-specific upregulation, is not understood. It would be interesting to know whether enhanced burden on the secretory pathway is the actual causative agent for PHO2 upregulation.
Interestingly, all the transcriptomes studied here were characterized by uniform high upregulation of an uncharacterized protein E12881g (SoA log2 fold change: 2.6, scYFP: 2.6, TlG: 2.82, inYFP: 2.74) (Fig. 3). Blasting the polypeptide sequence of E12881g against the protein database indicated its ∼30 % similarity to a putative membrane protein belonging to the peroxisomal PXMP2/4 family in Metschnikowia aff. pulcherrima, Ogataea parapolymorpha and Komagataella phaffii. PXMP2/4 s are putatively involved in pore-forming activity and may contribute to the unspecific permeability of the peroxisomal membrane. Their uniform upregulation makes them an interesting target in further studies on DEGs significantly responding to heterologous protein overproduction, in general. On the other hand, all the strains studied here bear a common pattern of high, significant downregulation of the uncharacterized gene D01353g, encoding Zn(2)C6 fungal-type domain-containing protein (Fig. 3). The gene was downregulated to a very high degree in all tested strains: SoA (log2 fold change: -273) scYFP (-215), TlG (-279), inYFP (-244). While the primary search in the GRYC database showed that D01353g is an uncharacterized zinc-binding transcription factor, blasting it against the UniProt database showed some similarities with a ZCF32 transcription factor from C. albicans, which is involved in negative regulation of transcription by RNA polymerase II (specifically). Hence, its uniform downregulation in all the overproducing strains suggests release from transcription inhibition by the “ZCF32-like” homolog under ongoing overexpression of inYFP, scYFP, SoA and TlG proteins in Y. lipolytica. Such a mechanism, of downregulation of the RNA polymerase II repressor, is additionally enhanced by the above-discussed E26763g (RSFA) (Fig. 3; Fig. 5.c) in scYFP (-2.14) and SoA (-5.39) strains.
3.4. Specific response of the secretory pathway to the heterologous secretory proteins’ overproduction
The targeted analysis was conducted on a set of pre-selected genes of known involvement in protein maturation and the secretory pathway. The list of genes was developed based on previous evidence of their role in those biological processes in yeast [1,22,23]. We distinguished 27 categories, according to which the genes were classified (Table S.3). The normalized expression level of selected genes from that set is given in Fig. 6, whereas the relative expression ratio vs the control strain for all the genes assigned to this biological process (∼ 700 genes) is shown in Table S.4. Such focused analysis allowed us to gain a deeper insight into fine changes in the secretory machinery triggered by overproduction of different rs-Prots. As previously discussed, the changes in mRNA levels are subtle and specific when induced solely by overproduced r-Prots [17].
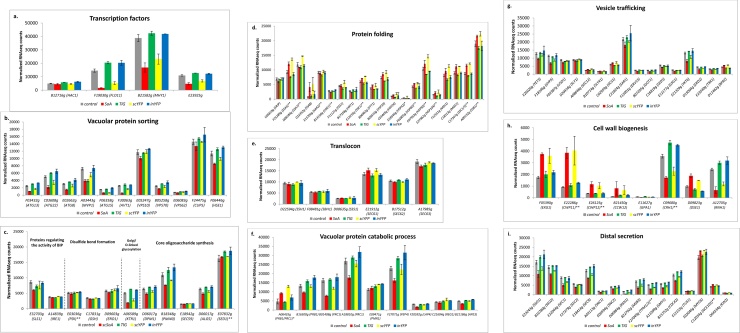
Fig. 6.
Expression level of selected genes involved in the r-Prot folding, maturation and secretion. Normalized transcript counts were used for quantification of the transcript levels. Transcription factors (a), Vacuolar protein sorting (b), disulfide bonds formation, O- glycosylation, core oligosaccharides proteins (c), Protein folding (d), Translocon’s elements (e), Vacuolar protein catabolic process (f), Vesicle trafficking (g), Cell wall biogenesis (h), Distal secretion (i). Targeted transcriptome analysis. X axis: shortened YALI_ code of a gene with a three letter name. Y axis: Normalized Counts of transcript. For clarity of the Figure some quantification values were * multiplied by 10, ** divided by 10. (For more convenient visualization of the Figure in color, the reader is referred to the web version of this article).
Primarily, we saw that the key transcription factor HAC1 (B12716g), which initiates UPR in yeast [[15], [16], [17]], was upregulated by 23–27 % solely in TlG and inYFP, without any significant changes in the expression level in SoA/scYFP vs the control strain (Fig. 6.a). Its expression was relatively low, but the gene was continuously expressed, either when challenged with r-Prot or not. The same was observed in P. pastoris continuous cultures [67]. However, in that study, continuous expression of HAC1 was interpreted as a hallmark of continuous activation of UPR and ERAD. In S. cerevisiae, HAC1 was continuously upregulated, together with its downstream targets, when the cells were used as r-Prot production platforms [27]. On one hand, our observation was highly surprising, as we expected to observe signs of high UPR in the HSSs. Moreover, it is known that Hac1 regulates transcription of several hundred genes, so its upregulation should induce substantial transcriptomic changes as well [16]; this was limited for inYFP, having the largest increase in HAC1 expression. Likewise, the typical downstream processes responding to UPR (including protein folding and proteasomal degradation, ERAD; Table S.4) were not activated in TlG/inYFP strains, except for several genes involved in vacuolar protein sorting: ATG8 (E02662g), ATG13 (F03432g), VPS70 (B05258g) and CUP5 (F24475g); and vacuolar proteases PEP4 (F27071g), PRC1 (A18810g), PRB1 (B16500g) (Fig. 6.b.f). It was impossible to determine whether upregulation of those vacuolar protein sorting/degradation genes already signifies activation of UPR in TlG/inYFP. In contrast, the expression level of the UPR’s regulators, including IRE1 (A14839g), KAR2 (BIP; E13706g), ERJ (C05819g), and the key protein disulfide isomerase PDI1 (E03036g), together with its reactivator ERO1 (D09603g) and glutathione synthetase GSH (C17831g), all remained at the control level in all the recombinant strains studied here (Fig. 6.c.d). Such an observation was particularly surprising for SoA, rich in disulfide bonds. Lack of their upregulation was previously interpreted as lack of an actual HAC1-triggered response [2]. In relation to oxidative folding, disulfide bond formation and, associated with it, oxidative stress, we observed downregulation of the cytoplasmic thioredoxin TRX1 (E09427g) shared by HSSs (within a group with n = 75; Fig. 5.c). Thioredoxin activity is intrinsically associated with the process of disulfide bond formation, mediated by Pdi1 and Ero1, and relies on stochastic oxidation-reduction of cysteine side chains, which consumes considerable amounts of oxidizing and reducing agents (O2 and glutathione, respectively). Although oxidative folding takes place within the ER lumen, neither thioredoxin nor glutathione redox systems were identified within the ER [49]. According to the current model, a cytoplasmic thioredoxin (e.g. TRX1; E09427g) shuttles electrons into the ER to reduce oxidized Pdi [68], while glutathione is actively transported from the cytoplasm by specific transporters [69]. In the face of that, the downregulation of TRX1 in HSSs is not clear, and contrasted by 17 % upregulation of SEC61 (E21912g) in SoA (Fig. 6.e), which is known to transport glutathione, required for oxidative folding, into the ER lumen [69]. That slight upregulation of the SEC61 channel is the only, faint marker of enhanced oxidative folding and disulfide bond formation in SoA.
In contrast to a constant level of KAR2 and IRE1 expression, the expression of SLS1 (E32703g), encoding a mediator of interaction between Ire1 and Kar2 [70], essential for co-translation translocation in Y. lipolytica [71,72], was downregulated upon overproduction of all the rs-Prots (SoA, TlG and scYFP), but not for inYFP (Fig. 6.c). Sls1 operates as an ADP to ATP exchange factor for Kar2 [72]. In an ATP-bound form, Kar2 is in an Ire1-interaction-favoring state. In that state, Ire1 cannot execute its endonucleolytic activity towards HAC1/XBP1 mRNAs, which is required for their translation. Hence, it could be speculated that by downregulation of SLS1, the interaction of Kar2-Ire1 is limited, and consequently, Ire1 is released for processing HAC1 mRNA to a translationally active form. In that way, although not upregulated in HSSs, HAC1 could be translated and initiate massive rearrangements in its transcriptomes (Fig. 2) even from its basic transcription level (Fig. 6.a). Moreover, in the case of HSSs, systematic changes in protein folding, vesicular transportation and vacuolar protein catabolic processes were observed, which could support the proposed model. Definitely, those hypotheses require further deepened biochemical investigations. Additionally, in view of the lack of significant change of HAC1 expression in HSSs, a comment on the high CTT1 upregulation is required. While indeed HAC1 may induce expression of genes involved in the oxidative stress response, which is important for managing the secretory pathway, it was previously demonstrated that the oxidative stress response is not directly managed through the UPR [2], which explains the current observations made for HSSs.
Furthermore, the postulated limited rate of Kar2-Ire1 interaction would suggest that the translocation takes place via the post-translational translocation mechanism, i.e. the less efficient mode in Y. lipolytica [1], or via both pathways. This hypothesis is supported by the highly elevated expression of cytosolic chaperones of the Ssa family, in particular SSA5 (F25289g) and SSA8 (D22352g), in HSSs (Fig. 6.d; Table S.4). While the expression of the other Ssa-family members, SSA5 and SSA7 (D08184g) was very high in all the analyzed strains, including the control (Fig. 6.d), the expression level was most changed for SSA8 (5.5- and 6.2-fold higher expression in HSSs vs the control) (Table S.4). The Ssa chaperones belong to the Hsp70 family with the primary role in securing the nascent polypeptide in a translocation competent state, when the polypeptide is to be translocated via a post-translational mechanism. A similar expression pattern (significant upregulation in HSSs) was observed for another member of the cytosolic chaperones, SSE1 (E13255g) (Fig. 6.d). SSE1 (and SSE2 in P. pastoris; a single gene in Y. lipolytica) assists in protein folding by binding to the nascent peptide and maintaining it in a folding-competent state, but cannot actively promote folding reaction. Co-expression of a heterologous gene with SSE1 as a helper protein allowed the target protein amount to be increased by 2.3-fold [21]. Additionally, we did not observe any substantial changes in the expression of the genes encoding the co-translational translocation mechanism-associated activities, SRP receptor or SRP itself. Again, this supports the hypothesis that the rs-Prots studied here were translocated via a post-translational mechanism, or, possibly, via both in SoA.
With respect to r-Prot-specific cellular responses, we observed systematic, substantial upregulation of the genes involved in synthesis of core oligosaccharides (DPM1: D06017g, PMI40: B18348g, SEC59: E18942g, ALG5: D06017g) and in glycoprotein processing in TlG (Fig. 6.c; Table S.4). Seven in ten genes belonging to the Alg family, involved in glycoprotein processing, were downregulated in HSSs, with concomitant lack of change for TlG and inYFP. A similar expression profile was also observed for many genes assigned to the GPI-anchoring process (Table S.4). Only a single gene involved in that process, SED1 (E07832g), was upregulated by 20 %, and that upregulation was observed solely in TlG. All these data indicate that the selection of TlG as a model of highly glycosylated protein was relevant.
Likewise, none of the genes involved in glycoproteins’ maturation or processing was significantly up-/downregulated in inYFP. The same observation relates to protein folding, disulfide bond formation, protein degradation, ERAD, and vesicular transportation, with only four exceptions in ∼250 genes, assigned to those processes; these were: downregulation of FES1 (F11121g) and BFR2 (D11462g), upregulation of HSP26 (C03465g) and HRT1 (B17358g) (Fig. 6.d.g). HSP26 encodes a chaperone suppressing unfolded proteins’ aggregation, also possessing mRNA-binding activity. The gene was also upregulated in SoA (1.47-fold change) and scYFP (1.84-fold change), and was significantly downregulated in TlG (0.66-fold change). While differential expression of HSP26 in all the studied strains demonstrates its importance for r-Prot synthesis, the pattern of expression is unclear. Likewise, the expression level of another cytoplasmic chaperone, HSP42 (E18546g), having unfolded protein-binding and protein-sequestering activity, was significantly elevated, by 2.26- and 4.48-fold, for SoA and scYFP, respectively. It is plausible that, except for Ssas, it assists nascent polypeptides to the ER surface for translocation.
Several other interesting observations were made based on the results of the targeted analysis (Fig. 6; Table S.4), namely: 1) vacuolar protein sorting and protein degradation-associated genes’ expression was on average 2-fold lower in HSSs; in contrast 2) several above-mentioned vacuolar protein sorting-involved genes (Fig. 6.b), ATG13, ATG8, CUP5, VPS70, but also HSE1 (F06446g) and VPS62 (E06083g), were significantly upregulated in TlG- and inYFP-producing strains, suggesting increased protein degradation occurring in these strains. Whether it is a result of HAC1 upregulation and induction of UPR remains an open question; 3) in all the strains, expression level of the genes involved in ERAD either remained unchanged or was decreased (five genes in SoA, three genes in scYFP and a single gene in TlG); 4) within the protein folding biological process we identified several genes significantly upregulated in HSSs, that were all localized to mitochondria, including MDJ1 (F12551g), MGE1 (C18513g), HSP60 (F02805g), and HSP10 (B05610g) (Fig. 6.d), and SoA-specific upregulation of PRX1 (A19426g), with high similarity to mitochondrial peroxiredoxin with thioredoxin peroxidase activity, reactivated by glutathione (involved in cell redox homeostasis and the cellular response to oxidative stress). Such high overrepresentation of upregulated mitochondrial genes corresponds well with the significantly enriched biological process “mitochondrial gene expression and translation” as well as with “cellular respiration”, identified for HSSs (Fig. 4.a.e).
According to the conducted biological process enrichment analysis (Fig. 4), protein degradation, autophagy and vacuolar protein sorting were significantly downregulated in HSSs, which was a consequence of many minor downregulations of specific DEGs assigned to those processes, as shown in Table S.4. In approximately 80 genes assigned to the vacuolar protein sorting process, 18 and 30 remained at an unchanged expression level, 2 and 1 were upregulated and the remaining ∼60 and ∼50 were significantly downregulated in SoA and scYFP strains, with concomitant lack of changes in the expression level for TlG and inYFP (with few exceptions). Amongst the most downregulated DEGs, we found ATG13, ATG22/23 (C03608g), ATG15 (F06358g), and AVT1 (F30063g), and the level of their downregulation was correlated for the two HSSs (Fig. 6.b; Table S.4). Functionally associated with “vacuolar protein sorting”, the “vacuolar protein catabolic process” was the most DEG-enriched category out of the 27 distinguished. All the genes assigned to that process were somehow affected by the introduced variable. Several genes showed prevailing upregulation, e.g. PMR1 (E09471g), significantly upregulated in all the strains, or PRB1/PRC1 (A06435g) and REG1 (C21604g), upregulated in three strains and unchanged in the fourth strain (Fig. 6.f; Table S.4). Several genes showed a differential pattern of expression depending on the strain, such as PEP4 (F27071g), PRC1 (A18810g), PRB1 (B16500g) and APE3 (B21186g), which were downregulated in HSSs and upregulated in TlG/inYFP. The remaining genes assigned to that process were either downregulated or at an unchanged expression level, depending on the strain (Table S.4). The uniformly upregulated PMR1 encodes Ca2+-ATPase, responsible for transport of Ca2+ and Mn2+ inside the Golgi lumen, which are required for glycosylation, sorting and ERAD. Due to the latter implication, PMR1 is assigned to a vacuolar protein catabolic process, although it localizes to the Golgi membrane. Its disruption was shown to increase secretion of heterologous proteins in S. cerevisiae, Kluyveromyces lactis and Hansenula polymorpha [[73], [74], [75]]. Its upregulation observed in the present study and the previous results obtained for the other yeast species could be interpreted as undesired targeting and degradation of some portion of correctly folded proteins which could be successfully secreted. It was previously pointed out that mis-sorting to the vacuole causes severe losses of rs-Prots in P. pastoris [20]. Likewise, it was shown that significant amounts of r-Prots accumulate in the vacuole in S. cerevisiae, and material and energy expenses for their production and degradation were incurred [76]. In relevance to this, we observed high (and undesired) upregulation of the key, vacuolar serine protease PRB1/PRC1 (A06435g) in SoA (2.03-fold change), scYFP (2.58-fold change) and inYFP (1.46-fold change) (Fig. 6.f), indicating increased protein degradation processes in these strains. Deletion of this gene was a prerequisite for successful modification (Δvps21) of the early steps of the vacuolar sorting and degradation pathway targeting improved heterologous protein secretion in P. pastoris [20]. In the view of its currently documented response to r-Prot overproduction in Y. lipolytica, and the previous data for P. pastoris, it would be relevant and interesting to investigate the impact of Δprb1 genotype (accompanied by another modification) on the secretory capacity of Y. lipolytica. Likewise, PEP4, PRC1 (A18810g) and PRB1 (B16500g) genes, encoding vacuolar aspartyl protease and broad-specificity C-terminal exopeptidase involved in non-specific protein degradation in the vacuole, were upregulated in TlG and inYFP. Together with the previously discussed upregulation of vacuolar protein sorting markers, including ATG8, ATG13, CUP5, and VPS70, they represent interesting targets for a genetic engineering strategy aiming at decreased degradation of heterologous protein production in Y. lipolytica (at least for strains overproducing TlG and inYFP). Downregulation of the above-mentioned vacuolar protein sorting markers as well as PEP4, PRC1 (A18810g), and PRB1 (B16500g) could account for, at least part of, the predominance of HSSs over TlG/inYFP in terms of the produced protein amounts (Table 2). It would well align with the previous observations and conclusions [27,67,77] that even though UPR activates foldases, chaperones and other folding and secretory helpers, its continuous activation reduces the final yields of r-Prots.
Cell wall biogenesis- and distal secretion-related biological processes were enriched in the genes responsive to the introduced variable. However, no general trend could be observed depending on the overproduced protein (as could be done in the case of protein folding or glycoprotein processing). This means that the final set of significant DEGs was built by many up-/and down-regulated genes in a given strain. In nearly 170 genes assigned to cell wall biogenesis, only a few were highly responsive, while the majority were up-/down-regulated by not more than 20–30 %. As mentioned above, regulation of cell wall biogenesis was conducted by several highly responsive transcription factors, including FLO11, MHY1, and E23925g, that apparently fine-tuned the repertoire of the downstream genes, as required for adequate cell wall modifications (Fig. 6.a). Amongst the most responsive genes in HSSs (for which the cell wall biogenesis was a significantly changed biological process; Fig. 3; Fig. 4; Fig. 6.h), we identified highly upregulated CCW12 (B21450g) (3.32/2.68-fold change in SoA/scYFP), encoding a cell wall mannoprotein that plays a role in maintenance of newly synthesized areas of the cell wall; DSE1 (D09823g; 2.01/1.14-fold change in SoA/scYFP), involved in cell separation after cytokinesis; EXG1 (F05390g; 2.23/1.93-fold change in SoA/scYFP), the major exo-1,3-beta-glucanase of the cell wall, involved in cell wall beta-glucan assembly; and two structural cell wall proteins CWP11/CWP12 (E22286g/E26125g). Amongst the most responsive downregulated genes in HSSs we identified GPA1 (E11627g), a plasma membrane/endosome G-protein involved in signaling, and CRH1 (C09680g), encoding chitin transglycosylase, transferring chitin to beta-glucans, both being slightly upregulated in TlG/inYFP strains and downregulated in HSSs.
Within the vesicular transportation and membrane fusion processes (Fig. 6.g), we identified several up- and downregulated genes, but mainly for HSSs. A common expression pattern in both HSSs was observed for: 1) upregulated SEC12 (A08646g), BFR2 (D11462g), SLY41 (B10714g), GCS1 (C02959g), and downregulated SAR1 (C21824g), ERV15 (F12001g), GOT1 (B07205g), and USO1 (F02387g) involved in COPII anterograde transport; 2) as well as SED5 (C16819g), several representatives of Sec and Trs families, GSG1 (E12177g), YKT6 (E21329g), playing a role in membrane fusion; 3) and also ERD2 (D15004g) and TRX1 (E23540g), involved in retrograde Golgi to ER transport. It is not possible to discuss the specifics of all the abovementioned genes here, but the responsive genes may be potentially considered as targets in genetic engineering strategies enhancing rs-Prot production. Such a strategy was successfully implemented [14]. Interestingly, BFR1 (D11462g), upregulated here, was previously shown to be a useful secretory helper in P. pastoris, increasing the target r-Prot production by 1.5-fold [21], which demonstrates plausible similarities in its operation in the two yeast species.
Similarly, in 65 genes assigned to the distal secretion process (Fig. 6.i), only one (and only in the SoA strain) met the requirement of fold change >2 for DEG (RSN1; D06864g; -2.25), while the remaining genes’ expression was changed to a smaller degree. We paid attention to the genes that were previously indicated as interesting targets for improving r-Prot overproduction in yeast, e.g. SSO1/2 (E23243g, B10780g), encoding a syntaxin homolog, acting in late stages of secretion post-Golgi t-SNARE fusion [14,78], SNC1/2 (A03113g, E00594g), vesicle membrane receptor protein (v-SNARE); involved in the fusion between Golgi-derived secretory vesicles with the plasma membrane, and SEC1/2/9 (E22044g, F27379g, E18414g), encoding an SM-like protein involved in docking and fusion of exocytic vesicles, which binds to assembled SNARE complexes at the membrane (reviewed by Celińska et al. [30]). They all shared a common expression pattern of downregulation in HSSs and slight upregulation or no change in TlG/inYFP. Their downregulation upon very high overproduction and secretion, with no evident upregulation of any redundant activities, is unclear. The same relates to KIN2 (D22770g), which was previously used as a very efficient secretion helper, increasing the target protein production by 2.2-fold upon co-expression [21]. However, in the present research, production of either of the studied r-Prots did not trigger changes in its expression profile. Moreover, exactly the same expression pattern was observed for many other genes assigned to the distal secretion process, but exerting different functions, including the kinesin-like myosin passenger-protein SMY1 (A11099g), tropomyosin TPM1/2 (F27049g), Rab GTPase-activating protein MSB3 (B22792g) important for polarized growth, and rho-like GTPase CDC42 (B15752g), having a key role in establishment and maintenance of cell polarity and regulating actin assembly (Fig. 6.i; Table S.4). Their downregulation could result from the action of the filamentation factors, and previously postulated transition into ovoid forms, and not be related to r-Prot transportation and secretion. Only a few genes assigned to the distal secretion process escaped that general expression pattern. These were, for example, the rho1-GDP exchange factor TUS1 (E12155g), required for signaling of cell wall defects to Rho1, and the type-I myosin MYO5 (E02046g) that promotes actin assembly, which were upregulated in HSSs. Interestingly, another gene, NCE102 (C12034g), was upregulated in all the secretory protein-overproducing strains (no change for inYFP solely). Nce102 stands for non-classical export protein 2, which is involved in secretion of proteins that lack classical secretory signal sequences. While its upregulation upon overproduction of the secretory heterologous proteins is logical, it was surprising considering the fact that all the heterologous genes were transcriptionally fused to a signal peptide of typical structure and function (exo‐1,3‐beta‐glucanase; B03564g) [7]. Nce102 is implicated in export of proteins which lack a cleavable signal sequence. However, the non-classical export pathway is known to operate as an alternative clearance/detoxification pathway to eliminate damaged material, when the basic pathway is not sufficient. Thus, we postulate that upon the very high secretion of SoA and scYFP, NCE102 works as an alternative route of secretion of correctly folded, functional proteins, as we observed in the activity/fluorescence tests (Table 2).
4. Conclusions
The conducted research improved our understanding of phenomena taking place inside Y. lipolytica cells upon r-Prot overproduction. First of all, based on global comparison of the expression profiles generated by SoA, scYFP, TlG and inYFP, we conclude that targeting a protein for secretion (as opposed to its retention within a cell) induces massive transcriptional changes, greatly exceeding those resulting from secreting a protein with different biochemical characteristics. Interestingly, if the same protein (YFP) was targeted for secretion, its yield was > 10-fold higher. Current data, confirmed for Y. lipolytica, as is already known for P. pastoris and S. cerevisiae, show that high overproduction of r-Prot is inherently associated with oxidative stress. Here it was indicated by high upregulation of a “response to hydrogen peroxide” biological process, and upregulation of catalases, as the most responsive DEGs. Importantly, the phenomenon does not rely on the number of disulfide bonds within the r-Prot’s secondary structure, at least for the two HSSs studied here. Overproduction of r-Prots triggered massive rearrangements within the cell by acting through transcriptional regulators, such as D01353g, downregulated upon overproduction of any r-Prot, irrespective of its biochemical characteristics, or specific for HSSs – RSFA or MAF1. Based on their putative function, their downregulation in HSSs led to release of RNA polymerase II and III, respectively, from their inhibitory activity. On the other hand, the key transcription factor, typically associated with the response to overproduction of r-Prots, HAC1, was upregulated solely in the strains producing lower yields of r-Prots (inYFP/TlG). Those strains were also characterized by upregulation in vacuolar protein sorting and protein degradation. Whether it was caused by the actual onset of UPR could not be definitely established, as the other processes, typically upregulated under UPR, remained at the control level. Likewise, whether lower yields of TlG/inYFP were caused by the UPR remains to be experimentally verified.
The global approach undertaken here allowed us to perceive unobvious relationships, such as overproduction of r-Prot, with given characteristics (or at a given rate) and the cell’s morphotype. Genes putatively linking these phenomena were indicated here. The unexpected downregulation of SLS1 (an interaction mediator of IRE1 and KAR2), shared by all the secretory r-Prot producers, led us to form conclusions on the mechanism of the nascent polypeptides’ translocation and to speculations on its regulation. Based on cytosolic chaperones’ expression pattern, we postulate that the polypeptides were in the majority of cases translocated into the ER lumen via a post-translational translocation mechanism. Traditionally, the interaction between a polypeptide’s signal peptide and SRP was considered the decision-making point [79]. Based on the current data, we speculate that the ER occupancy also participates in the decision making. However, this hypothesis requires experimental testing. We also report implication of NCE102 in rs-Prot secretion, which was highly upregulated in all the rs-Prot overproducers. While its assigned function is secretion of proteins that lack typical signal peptides (not true for SoA, TlG, or scYFP), we postulate its significant role in secretion of functional r-Prots, equipped with a typical signal peptide, under overproduction conditions. And finally, we hypothesize that the cells enter into a growth arrest phase (G1 phase) upon high overproduction of r-Prots, as a strategy undertaken by the cells to withstand the high burden imposed on them by genetic engineering. The hypothesis is supported by downregulation of a G1/S-specific cyclin gene, activation of detoxification processes (common upregulation of peroxisomal PXMP2/4), and transition into ovoid morphotype in HSSs.
Author statement
PKW designed the experiments, performed experimental work, analyzed omics data and wrote the manuscript. EC conceived the study, analyzed omics data and wrote the manuscript.
Funding
This study was financially supported by the Ministry of Science and Higher Education (currently, Ministry of Education and Science) project number: DI 2017 000947.
Data availability
All data accompanying this research are presented directly in the manuscript, supplementary materials or are available in Sequence Read Archive, NCBI database under BioProject number: PRJNA701856.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The Authors would like to thank Dr Piotr Kubiak for guidance and assistance in chemostat cultures. DBFM’s chromatography team is acknowledged for great help in HPLC analyses.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.btre.2021.e00646.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- 1.Delic M., Valli M., Graf A.B., Pfeffer M., Mattanovich D., Gasser B. The secretory pathway: exploring yeast diversity. FEMS Microbiol. Rev. 2013;37:872–914. doi: 10.1111/1574-6976.12020. [DOI] [PubMed] [Google Scholar]
- 2.Tyo K.E.J., Liu Z., Petranovic D., Nielsen J. Imbalance of heterologous protein folding and disulfide bond formation rates yields runaway oxidative stress. BMC Biol. 2012;10 doi: 10.1186/1741-7007-10-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Çelik E., Çalık P. Production of recombinant proteins by yeast cells. Biotechnol. Adv. 2012;30:1108–1118. doi: 10.1016/j.biotechadv.2011.09.011. [DOI] [PubMed] [Google Scholar]
- 4.Madzak C. Engineering Yarrowia lipolytica for use in biotechnological applications: a review of major achievements and recent innovations. Mol. Biotechnol. 2018;60:621–635. doi: 10.1007/s12033-018-0093-4. [DOI] [PubMed] [Google Scholar]
- 5.Hou J., Tyo K.E.J., Liu Z., Petranovic D., Nielsen J. Metabolic engineering of recombinant protein secretion by Saccharomyces cerevisiae. FEMS Yeast Res. 2012;12:491–510. doi: 10.1111/j.1567-1364.2012.00810.x. [DOI] [PubMed] [Google Scholar]
- 6.Zahrl R.J., Peña D.A., Mattanovich D., Gasser B. Systems biotechnology for protein production in Pichia pastoris. FEMS Yeast Res. 2017;17:1–15. doi: 10.1093/femsyr/fox068. [DOI] [PubMed] [Google Scholar]
- 7.Celińska E., Borkowska M., Białas W., Korpys P., Nicaud J.M. Robust signal peptides for protein secretion in Yarrowia lipolytica: identification and characterization of novel secretory tags. Appl. Microbiol. Biotechnol. 2018;102:5221–5233. doi: 10.1007/s00253-018-8966-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duan G., Ding L., Wei D., Zhou H., Chu J., Zhang S., Qian J. Screening endogenous signal peptides and protein folding factors to promote the secretory expression of heterologous proteins in Pichia pastoris. J. Biotechnol. 2019 doi: 10.1016/j.jbiotec.2019.06.297. [DOI] [PubMed] [Google Scholar]
- 9.Yarimizu T., Nakamura M., Hoshida H., Akada R. Synthetic signal sequences that enable efficient secretory protein production in the yeast Kluyveromyces marxianus. Microb. Cell Fact. 2015;14 doi: 10.1186/s12934-015-0203-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith J.D., Tang B.C., Robinson A.S. Protein disulfide isomerase, but not binding protein, overexpression enhances secretion of a non-disulfide-bonded protein in yeast. Biotechnol. Bioeng. 2004;85:340–350. doi: 10.1002/bit.10853. [DOI] [PubMed] [Google Scholar]
- 11.Tang H., Bao X., Shen Y., Song M., Wang S., Wang C., Hou J. Engineering protein folding and translocation improves heterologous protein secretion in Saccharomyces cerevisiae. Biotechnol. Bioeng. 2015;112:1872–1882. doi: 10.1002/bit.25596. [DOI] [PubMed] [Google Scholar]
- 12.Hou J., Tang H., Liu Z., Österlund T., Nielsen J., Petranovic D. Management of the endoplasmic reticulum stress by activation of the heat shock response in yeast. FEMS Yeast Res. 2014;14:481–494. doi: 10.1111/1567-1364.12125. [DOI] [PubMed] [Google Scholar]
- 13.Bao J., Huang M., Petranovic D., Nielsen J. Moderate expression of SEC16 increases protein secretion by Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2017;83 doi: 10.1128/AEM.03400-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Zyl J.H.D., Den Haan R., Van Zyl W.H. Overexpression of native Saccharomyces cerevisiae ER-to-Golgi SNARE genes increased heterologous cellulase secretion. Appl. Microbiol. Biotechnol. 2016;100:505–518. doi: 10.1007/s00253-015-7022-2. [DOI] [PubMed] [Google Scholar]
- 15.Graf A., Gasser B., Dragosits M., Sauer M., Leparc G.G., Tüchler T., Kreil D.P., Mattanovich D. Novel insights into the unfolded protein response using Pichia pastoris specific DNA microarrays. BMC Genomics. 2008;9:1–13. doi: 10.1186/1471-2164-9-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guerfal M., Ryckaert S., Jacobs P.P., Ameloot P., Van Craenenbroeck K., Derycke R., Callewaert N. The HAC1 gene from Pichia pastoris: characterization and effect of its overexpression on the production of secreted, surface displayed and membrane proteins. Microb. Cell Fact. 2010;9:1–12. doi: 10.1186/1475-2859-9-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gasser B., Maurer M., Rautio J., Sauer M., Bhattacharyya A., Saloheimo M., Penttilä M., Mattanovich D. Monitoring of transcriptional regulation in Pichia pastoris under protein production conditions. BMC Genomics. 2007;8:1–18. doi: 10.1186/1471-2164-8-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodríguez-Limas W.A., Tannenbaum V., Tyo K.E.J. Blocking endocytotic mechanisms to improve heterologous protein titers in Saccharomyces cerevisiae. Biotechnol. Bioeng. 2015;112:376–385. doi: 10.1002/bit.25360. [DOI] [PubMed] [Google Scholar]
- 19.Idiris A., Tohda H., Sasaki M., Okada K., Kumagai H., Giga-Hama Y., Takegawa K. Enhanced protein secretion from multiprotease-deficient fission yeast by modification of its vacuolar protein sorting pathway. Appl. Microbiol. Biotechnol. 2010;85:667–677. doi: 10.1007/s00253-009-2151-0. [DOI] [PubMed] [Google Scholar]
- 20.Marsalek L., Gruber C., Altmann F., Aleschko M., Mattanovich D., Gasser B., Puxbaum V. Disruption of genes involved in CORVET complex leads to enhanced secretion of heterologous carboxylesterase only in protease deficient Pichia pastoris. Biotechnol. J. 2017;12 doi: 10.1002/biot.201600584. [DOI] [PubMed] [Google Scholar]
- 21.Gasser B., Sauer M., Maurer M., Stadlmayr G., Mattanovich D. Transcriptomics-based identification of novel factors enhancing heterologous protein secretion in yeasts. Appl. Environ. Microbiol. 2007;73:6499–6507. doi: 10.1128/AEM.01196-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swennen D., Beckerich J.M. Yarrowia lipolytica vesicle-mediated protein transport pathways. BMC Evol. Biol. 2007;7:219. doi: 10.1186/1471-2148-7-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swennen D., Henry C., Beckerich J.M. Folding proteome of Yarrowia lipolytica targeting with uracil permease mutants. J. Proteome Res. 2010;9:6169–6179. doi: 10.1021/pr100340p. [DOI] [PubMed] [Google Scholar]
- 24.Matthews C.B., Kuo A., Love K.R., Love J.C. Development of a general defined medium for Pichia pastoris. Biotechnol. Bioeng. 2018;115:103–113. doi: 10.1002/bit.26440. [DOI] [PubMed] [Google Scholar]
- 25.Nieto-Taype M.A., Garrigós-Martínez J., Sánchez-Farrando M., Valero F., Garcia-Ortega X., Montesinos-Seguí J.L. Rationale-based selection of optimal operating strategies and gene dosage impact on recombinant protein production in Komagataella phaffii (Pichia pastoris) Microb. Biotechnol. 2020;13:315–327. doi: 10.1111/1751-7915.13498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hou J., Österlund T., Liu Z., Petranovic D., Nielsen J., Osterlund T., Liu Z., Petranovic D., Nielsen J. Heat shock response improves heterologous protein secretion in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2013;97:3559–3568. doi: 10.1007/s00253-012-4596-9. [DOI] [PubMed] [Google Scholar]
- 27.Liu Z., Hou J., Martínez J.L., Petranovic D., Nielsen J. Correlation of cell growth and heterologous protein production by Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2013;97:8955–8962. doi: 10.1007/s00253-013-4715-2. [DOI] [PubMed] [Google Scholar]
- 28.Martínez J.L., Meza E., Petranovic D., Nielsen J. The impact of respiration and oxidative stress response on recombinant α-amylase production by Saccharomyces cerevisiae. Metab. Eng. Commun. 2016;3:205–210. doi: 10.1016/j.meteno.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swietalski P., Hetzel F., Seitl I., Fischer L. Secretion of a low and high molecular weight β-glycosidase by Yarrowia lipolytica. Microb. Cell Fact. 2020;19:1–13. doi: 10.1186/s12934-020-01358-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Celińska E., Nicaud J.-M. Filamentous fungi-like secretory pathway strayed in a yeast system: peculiarities of Yarrowia lipolytica secretory pathway underlying its extraordinary performance. Appl. Microbiol. Biotechnol. 2019;103:39–52. doi: 10.1007/s00253-018-9450-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Celińska E., Ledesma-Amaro R., Larroude M., Rossignol T., Pauthenier C., Nicaud J.M. Golden gate assembly system dedicated to complex pathway manipulation in Yarrowia lipolytica. Microb. Biotechnol. 2017;10:450–455. doi: 10.1111/1751-7915.12605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Korpys-Woźniak P., Kubiak P., Białas W., Celińska E. Impact of overproduced heterologous protein characteristics on physiological response in Yarrowia lipolytica steady-state-maintained continuous cultures. Appl. Microbiol. Biotechnol. 2020 doi: 10.1007/s00253-020-10937-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook J., Russell D. 3rd ed. Cold Spring Harbor Laboratory Press; New York: 2001. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 34.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 35.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011;17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 36.Andrews S. 2010. A Quality Control Tool for High Throughput Sequence Data.https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ [Google Scholar]
- 37.Kim D., Paggi J., Park C., Bennett C., Salzberg S. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4.Graph-Based. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anders S., Pyl P.T., Huber W. HTSeq-A python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:1–21. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mi H., Muruganujan A., Huang X., Ebert D., Mills C., Guo X., Thomas P. Protocol update for large-scale genome and gene function analysis with PANTHER classification system (v.14.0) Nat. Protoc. 2019;14:703–721. doi: 10.1038/s41596-019-0128-8.Protocol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heberle H., Meirelles V.G., da Silva F.R., Telles G.P., Minghim R. InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams. BMC Bioinformatics. 2015;16:1–7. doi: 10.1186/s12859-015-0611-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Borkowska M., Białas W., Kubiak M., Celińska E. Rapid micro-assays for amylolytic activities determination: customization and validation of the tests. Appl. Microbiol. Biotechnol. 2019;103:2367–2379. doi: 10.1007/s00253-018-09610-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goncalves C., Rodriguez-Jasso R.M., Gomes N., Teixeira J.A., Belo I. Adaptation of dinitrosalicylic acid method to microtiter plates. Anal. Methods. 2010;2:2046–2048. doi: 10.1039/c0ay00525h. [DOI] [Google Scholar]
- 44.Gorczyca M., Kaźmierczak J., Steels S., Fickers P., Celińska E. Impact of oxygen availability on heterologous gene expression and polypeptide secretion dynamics in Yarrowia lipolytica-based protein production platforms. YEAST. 2020;37:559–568. doi: 10.1002/yea.3499. [DOI] [PubMed] [Google Scholar]
- 45.Ruiz-Herrera J., Sentandreu R. Different effectors of dimorphism in Yarrowia lipolytica. Arch. Microbiol. 2002;178:477–483. doi: 10.1007/s00203-002-0478-3. [DOI] [PubMed] [Google Scholar]
- 46.Kawasse F.M., Amaral P.F., Rocha-Leão M.H.M., Amaral A.L., Ferreira E.C., Coelho M.A.Z. Morphological analysis of Yarrowia lipolytica under stress conditions through image processing. Bioprocess Biosyst. Eng. 2003;25:371–375. doi: 10.1007/s00449-003-0319-z. [DOI] [PubMed] [Google Scholar]
- 47.Zhao Q., Su Y., Wang Z., Chen C., Wu T., Huang Y. Identification of glutathione (GSH)-independent glyoxalase III from Schizosaccharomyces pombe. BMC Evol. Biol. 2014;14:1–18. doi: 10.1186/1471-2148-14-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hasim S., Hussin N.A., Alomar F., Bidasee K.R., Nickerson K.W., Wilson M.A. A glutathione-independent glyoxalase of the DJ-1 superfamily plays an important role in managing metabolically generated methylglyoxal in Candida albicans. J. Biol. Chem. 2014;289:1662–1674. doi: 10.1074/jbc.M113.505784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guerrero-Gómez D., Mora-Lorca J.A., Sáenz-Narciso B., Naranjo-Galindo F.J., Muñoz-Lobato F., Parrado-Fernández C., Goikolea J., Cedazo-Minguez Á., Link C.D., Neri C., Sequedo M.D., Vázquez-Manrique R.P., Fernández-Suárez E., Goder V., Pané R., Cabiscol E., Askjaer P., Cabello J., Miranda-Vizuete A. Loss of glutathione redox homeostasis impairs proteostasis by inhibiting autophagy-dependent protein degradation. Cell Death Differ. 2019;26:1545–1565. doi: 10.1038/s41418-018-0270-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Buchman C., Skroch P., Welch J., Fogel S., Karin M. The CUP2 gene product, regulator of yeast metallothionein expression, is a copper-activated DNA-binding protein. Mol. Cell. Biol. 1989;9:4091–4095. doi: 10.1128/mcb.9.9.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Michaelis S., Barrowman J. Biogenesis of the Saccharomyces cerevisiae pheromone a-Factor, from yeast mating to human disease. Microbiol. Mol. Biol. Rev. 2012;76:626–651. doi: 10.1128/mmbr.00010-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Häuser R., Pech M., Kijek J., Yamamoto H., Titz B., Naeve F., Tovchigrechko A., Yamamoto K., Szaflarski W., Takeuchi N., Stellberger T., Diefenbacher M.E., Nierhaus K.H., Uetz P. RsFA (YbeB) proteins are conserved ribosomal silencing factors. PLoS Genet. 2012;8:1–12. doi: 10.1371/journal.pgen.1002815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fabrizio P., Hoon S., Shamalnasab M., Galbani A., Wei M., Giaever G., Nislow C., Longo V.D. Genome-wide screen in Saccharomyces cerevisiae identifies vacuolar protein sorting, autophagy, biosynthetic, and tRNA methylation genes involved in life span regulation. PLoS Genet. 2010;6:1–14. doi: 10.1371/journal.pgen.1001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alers S., Wesselborg S., Stork B. ATG13: Just a companion, or an executor of the autophagic program? Autophagy. 2014;10:1481. doi: 10.4161/auto.29717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ramya V., Rajasekharan R. ATG15 encodes a phospholipase and is transcriptionally regulated by YAP1 in Saccharomyces cerevisiae. FEBS Lett. 2016;590:3155–3167. doi: 10.1002/1873-3468.12369. [DOI] [PubMed] [Google Scholar]
- 56.Reyes Turcu F.E., Ventii K.H., Wilkinson K.D. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu. Rev. Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Puxbaum V., Mattanovich D., Gasser B. Quo vadis? The challenges of recombinant protein folding and secretion in Pichia pastoris. Appl. Microbiol. Biotechnol. 2015;99:2925–2938. doi: 10.1007/s00253-015-6470-z. [DOI] [PubMed] [Google Scholar]
- 58.Silva C.I.F., Teles H., Moers A.P.H.A., Eggink G., de Wolf F.A., Werten M.W.T. Secreted production of collagen-inspired gel-forming polymers with high thermal stability in Pichia pastoris. Biotechnol. Bioeng. 2011;108:2517–2525. doi: 10.1002/bit.23228. [DOI] [PubMed] [Google Scholar]
- 59.Wu M., Shen Q., Yang Y., Zhang S., Qu W., Chen J., Sun H., Chen S. Disruption of YPS1 and PEP4 genes reduces proteolytic degradation of secreted HSA/PTH in Pichia pastoris GS115. J. Ind. Microbiol. Biotechnol. 2013;40:589–599. doi: 10.1007/s10295-013-1264-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Celińska E., Olkowicz M., Grajek W. L-Phenylalanine catabolism and 2-phenylethano synthesis in Yarrowia lipolytica-mapping molecular identities through whole-proteome quantitative mass spectrometry analysis. FEMS Yeast Res. 2015;15 doi: 10.1093/femsyr/fov041. [DOI] [PubMed] [Google Scholar]
- 61.Celińska E., Borkowska M., Białas W., Kubiak M., Korpys P., Archacka M., Ledesma-Amaro R., Nicaud J.J.-M. Genetic engineering of Ehrlich pathway modulates production of higher alcohols in engineered Yarrowia lipolytica. FEMS Yeast Res. 2019;19:1–13. doi: 10.1093/femsyr/foy122. [DOI] [PubMed] [Google Scholar]
- 62.Idiris A., Tohda H., Kumagai H., Takegawa K. Engineering of protein secretion in yeast: strategies and impact on protein production. Appl. Microbiol. Biotechnol. 2010;86:403–417. doi: 10.1007/s00253-010-2447-0. [DOI] [PubMed] [Google Scholar]
- 63.Gems D., McElwee J.J. Broad spectrum detoxification: the major longevity assurance process regulated by insulin/IGF-1 signaling? Mech. Ageing Dev. 2005;126:381–387. doi: 10.1016/j.mad.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 64.Aravind L., Anantharaman V., Balaji S., Babu M.M., Iyer L.M. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol. Rev. 2005;29:231–262. doi: 10.1016/j.femsre.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 65.Gaigalat L., Schlüter J.P., Hartmann M., Mormann S., Tauch A., Pühler A., Kalinowski J. The DeoR-type transcriptional regulator SugR acts as a repressor for genes encoding the phosphoenolpyruvate: sugar phosphotransferase system (PTS) in Corynebacterium glutamicum. BMC Mol. Biol. 2007;8:1–20. doi: 10.1186/1471-2199-8-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liang S., Wang B., Pan L., Ye Y., He M., Han S., Zheng S., Wang X., Lin Y. Comprehensive structural annotation of Pichia pastoris transcriptome and the response to various carbon sources using deep paired-end RNA sequencing. BMC Genomics. 2012;13 doi: 10.1186/1471-2164-13-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hesketh A.R., Castrillo J.I., Sawyer T., Archer D.B., Oliver S.G. Investigating the physiological response of Pichia (Komagataella) pastoris GS115 to the heterologous expression of misfolded proteins using chemostat cultures. Appl. Microbiol. Biotechnol. 2013;97:9747–9762. doi: 10.1007/s00253-013-5186-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Poet G.J., Oka O.B., Lith M., Cao Z., Robinson P.J., Pringle M.A., Arnér E.S., Bulleid N.J. Cytosolic thioredoxin reductase 1 is required for correct disulfide formation in the ER. EMBO J. 2017;36:693–702. doi: 10.15252/embj.201695336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ponsero A.J., Igbaria A., Darch M.A., Miled S., Outten C.E., Winther J.R., Palais G., D’Autreaux B., Delaunay-Moisan A.A., Toledano M.B., D’Autréaux B., Delaunay-Moisan A.A., Toledano M.B. Endoplasmic reticulum transport of glutathione by Sec61 is regulated by Ero1 and bip. Mol. Cell. 2017;67:962–973. doi: 10.1016/j.molcel.2017.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Babour A., Kabani M., Boisramé A., Beckerich J.M. Characterization of Ire1 in the yeast Yarrowia lipolytica reveals an important role for the Sls1 nucleotide exchange factor in unfolded protein response regulation. Curr. Genet. 2008;53:337–346. doi: 10.1007/s00294-008-0190-1. [DOI] [PubMed] [Google Scholar]
- 71.Boisramé A., Kabani M., Beckerich J.M., Hartmann E., Gaillardin C. Interaction of Kar2p and Sls1p is required for efficient co- translational translocation of secreted proteins in the yeast Yarrowia lipolytica. J. Biol. Chem. 1998;273:30903–30908. doi: 10.1074/jbc.273.47.30903. [DOI] [PubMed] [Google Scholar]
- 72.Boisramé A., Beckerich J.M., Gaillardin C. Sls1p, an endoplasmic reticulum component, is involved in the protein translocation process in the yeast Yarrowia lipolytica. J. Biol. Chem. 1996;271:11668–11675. doi: 10.1074/jbc.271.20.11668. [DOI] [PubMed] [Google Scholar]
- 73.Zhao H.L., Xue C., Wang Y., Yao X.Q., Liu Z.M. Increasing the cell viability and heterologous protein expression of Pichia pastoris mutant deficient in PMR1 gene by culture condition optimization. Appl. Microbiol. Biotechnol. 2008;81:235–241. doi: 10.1007/s00253-008-1666-0. [DOI] [PubMed] [Google Scholar]
- 74.Harmsen M.M., Bruyne M.I., Raué H.A., Maat J. Overexpression of binding protein and disruption of the PMR1 gene synergistically stimulate secretion of bovine prochymosin but not plant Thaumatin in yeast. Appl. Microbiol. Biotechnol. 1996;46:365–370. doi: 10.1007/BF00166231. [DOI] [PubMed] [Google Scholar]
- 75.Sohn Y.S., Park C.S., Lee S.B., Ryu D.D.Y. Disruption of PMR1, encoding a Ca2+-ATPase homolog in Yarrowia lipolytica, affects secretion and processing of homologous and heterologous proteins. J. Bacteriol. 1998;180:6736–6742. doi: 10.1128/jb.180.24.6736-6742.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tyo K.E.J., Liu Z., Magnusson Y., Petranovic D., Nielsen J. Impact of protein uptake and degradation on recombinant protein secretion in yeast. Appl. Microbiol. Biotechnol. 2014;98:7149–7159. doi: 10.1007/s00253-014-5783-7. [DOI] [PubMed] [Google Scholar]
- 77.Kauffman K.J., Pridgen E.M., Doyle F.J., Dhurjati P.S., Robinson A.S. Decreased protein expression and intermittent recoveries in BiP levels result from cellular stress during heterologous protein expression in Saccharomyces cerevisiae. Biotechnol. Prog. 2002;18:942–950. doi: 10.1021/bp025518g. [DOI] [PubMed] [Google Scholar]
- 78.Toikkanen J.H., Sundqvist L., Keränen S. Kluyveromyces lactis SSO1 and SEB1 genes are functional in Saccharomyces cerevisiae and enhance production of secreted proteins when overexpressed. Yeast. 2004;21:1045–1055. doi: 10.1002/yea.1151. [DOI] [PubMed] [Google Scholar]
- 79.Ng D.T.W., Brown J.D., Walter P. Signal sequences specify the targeting route to the endoplasmic reticulum membrane. J. Cell Biol. 1996;134:269–278. doi: 10.1083/jcb.134.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data accompanying this research are presented directly in the manuscript, supplementary materials or are available in Sequence Read Archive, NCBI database under BioProject number: PRJNA701856.