Abstract
Background
Visceral leishmaniasis (VL) is in elimination phase in India while cutaneous leishmaniasis (CL) is being reported from new foci. In Himachal Pradesh (HP), a foci of CL had been reported along Satluj River, but the causative agent poses a dilemma, hence the present study was undertaken in Shimla, Kullu and Kinnaur districts.
Methods
A total of 28 CL patients from Indira Gandhi Medical College and Hospital Shimla (IGMC) in 2018, were tested by rK39., Twelve fresh cases were subjected to microscopic detection of Leishmania parasite, PCR and sequencing. Skin biopsies of 3–4 mm diameter were cultured, as well as imprints were prepared for the detection of Leishmania amastigotes. Biopsy samples were inoculated into different culture media (M199, RPMI 1640, NNN) and were incubated at 22–24 °C. Polymerase chain reaction (PCR) was performed to characterize Leishmania parasite species.
Results
Of 28 patients, one was positive by rK39 dipstick test and one imprint was found positive for Leishmania amstigotes. Twelve biopsy DNA samples subjected to PCR for Leishmania kDNA, were found Lesihmania positive. Identification of Leishmania species was confirmed by PCR-RFLP and sequencing method. Of 12 Leishmania positive samples, six were identified as L. donovani, three L. tropica, two L.major and one remained unidentified.
Conclusions
This study revealed the existence of three species of parasites i.e., L. donovani, L. tropica and L. major indicating the existence of typical and atypical leishmaniasis in Himachal Pradesh. The occurrence of CL cases in HP, Kerala or elsewhere should not be ignored considering them just cases of CL alone. Further studies are warranted to confirm the existence of L.donovani zymodeme MON37 from cases of CL in HP or L.donovani zymodeme MON2 strain causing VL in Bihar. Elimination of CL should also be considered along with goal of Kala -Azar elimination.
Keywords: Leishmaniasis, Cutaneous leishmaniasis, Himachal Pradesh, India
Highlights
-
•
We studied the CL in Himachal Pradesh and found three species i.e L. donovani, L. tropica and L. major using PCR-RFLP and sequencing.
-
•
A study in three districts of HPwith existence of three parasite indicating the existence of typical leishmaniasis caused by L. donovani and L. tropica as well as atypical CL caused by L. donovani.
-
•
The present study as well as study undertaken by others, warrants that CL is widely distributed in India.
-
•
Further studies are warranted to confirm the existence of L.donovani zymodeme MON37 from cases of CL in HP or L. donovani zymodeme MON2 strain causing VL in Bihar.
-
•
Elimination of CL should also be considered along with goal of Kala -Azar elimination.
Leishmaniasis, Cutaneous Leishmaniasis, Himachal Pradesh, India.
1. Introduction
The leishmaniases are a group of diseases caused by the protozoan parasite, Leishmania. There are three main types of leishmaniasis: i) Visceral (VL), the most serious form of the disease often known as kala-azar; ii) Cutaneous (CL); and iii) Mucocutaneous [1]. The historical focus of CL in India, caused by L tropica and transmitted by Phlebotomus salehi, P. papatasi [2]. After 1973 a focus of CL has been reported from subalpine valley along the Satluj River in Himachal Pradesh. During 1988 to 2001, a total of 38 cases of CL were reported from the Satluj valley of Himachal Pradesh and 161 CL cases were also reported between 2001-2003 where L. tropica and L. donovani were identified as causative parasite and Phlebotomus longiductus as suspected vector [3, 4, 5, 18]. One patient was found positive with rK39 dipstick test [6]. A recent publication from Mahatma Gandhi Medical Service Complex, Rampur (Shimla) reported endemic foci of CL in Shimla and Kinnaur districts where 337 cases were clinically diagnosed and treated but identification of causing agent was not done [8, 9]. The present study was, therefore, carried out to identify and characterize the parasite species causing CL around Satluj river in Himachal Pradesh.
2. Material and methods
2.1. Epidemiological data and study area
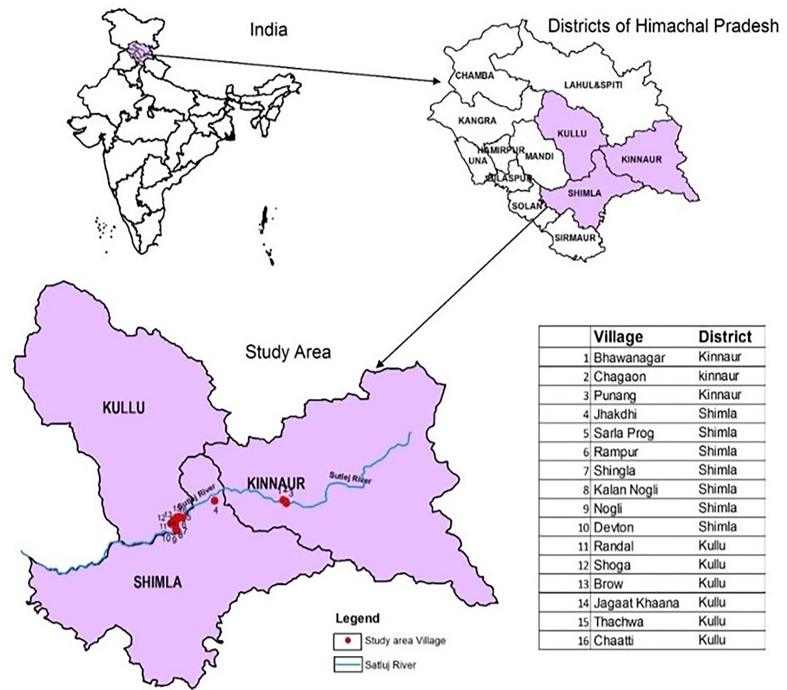
The present study was undertaken during 2018–19 in Shimla, Kullu and Kinnaur districts of Himachal Pradesh along the Sutlej river valley located between 31°05′0″N and 31°42′0″N latitude and 76°52′E and 78°24′0″E latitude. Based on the data recorded at Indira Gandhi Medical College (IGMC) Shimla, three blocks namely Rampur, Nirmand and Nichar were selected from Shimla, Kullu and Kinnaur districts respectively as almost all the cases were reported from these blocks. Distribution of villages reporting cases of CL is shown in Figure 1.
Figure 1.
Map of villages reporting CL cases in the study area of Himachal Pradesh.
2.2. Patients’ sample collection
The cases of CL were diagnosed clinically by the appearance of plaque, erythematous nodules or ulcerated lesions by a dermatologist. Of 28 cases, registered and admitted in the ward of IGMC, Shimla, samples were collected only from 12 cases, being fresh. The skin biopsies of 3–4 mm in diameter were taken under anesthetic and sterile conditions from the active site of the lesions. A portion of biopsy sample was transferred to culture vial, another was used for making imprint on slide as well preserved in formalin for PCR. The culture media were M199, RPMI supplemented with 25 mmol/L HEPES (pH 7.5) and 10% fetal bovine serum (FCS) and NNN medium and incubated at 22–24 °C). Cultures were examined microscopically for the growth of promastigotes up to four weeks. All the 28 patients clinically diagnosed for CL were also screened by rK39 dipstick test. Venous blood from 28 clinically diagnosed CL patients were collected in buffered citrate. These were centrifuged for 30 min at 3000 rpm and serum was separated with sterile pipettes for rapid rK39 immunochromatographic dipstick test. The dipstick test was performed according to the manufacturers' (InBios International, Inc. Seattle, WA 98104) instructions. Briefly, 20 μl each of serum was added to the sample wells of the strip, followed by 100–150 μl of buffer solution provided with the test kit. The results were read after 10 min and were considered positive when two distinct red lines, one in the test region and another in the control region, appeared [6]. The study was approved by Ethical committee of ICMR -National Institute of Malaria Research, Delhi (committee's reference number NIMR/ECR/2015/369). Consent form was filled and signed by the patients' prior to taking the samples.
The CL patients were treated at IGMC Shimla with Sodium stibogluconate (SSG) injected intra-lesional, three injections in a month and repeated until healing was complete. In most of the patients, complete resolution of lesions was seen after 3–6 months. Intralesional SSG was found consistently effective without any major side effects except painful injections.
2.3. DNA extraction
Total DNA was extracted from the biopsy samples collected and preserved in formalin buffer using the Qiagen DNA mini kit (Qiagen,Valencia, CA), as per manufacturer's instructions and eluted in 20ul elution buffer. DNA extracts were stored at -20 °C for further use. Genomic DNA was extracted from in-vitro cultures of L. donovani Ag83 and was used as positive control. L. tropica (MHOM/AF/87/RUP), and L. major (MHOM/SN/74/SD) positive controls were procured from BEI, Resource USA.
2.4. Polymerase chain reaction- Restriction Fragment Length Polymorphism (PCR –RFLP)
The polymerase chain reaction was carried out on DNA isolates from biopsy samples.The Leishmania specific JW11/12 primer set amplifies a 120bp fragment of KDNA of genus Leishmania [10] and the LITSR/L5.8S set amplifies a 320 bp fragment of ITS1 region of Leishmania genus-specific [11, 12]. Primers sequences and relevant PCR conditions are given in Table 1. A volume of 2μl of extracted DNA from patients and L. donovani, L. tropica and L. major positive controls were amplified for each forward and reverse primer in the presence of 2 X Kappa master mix in a final volume of 20 μl in Bio Rad PCR Thermocycler. The amplified PCR products were electrophoresed on 1.5–2% (w/v) agarose gel stained with ethidium bromide (EtBr) (Sigma Aldrich) in 1X tris-acetate-EDTA buffer at 100 V for 45 min, Gel images were captured on a computerized gel documentation unit.
Table 1.
Amplification conditions of Thermocycler Polymerase Chain Reaction.
| PCR | Primer sequence | Amplification programme |
|---|---|---|
| KDNA genus Leishmania specific PCR (Rodgers et al. 1990) [10] |
JW11 (forward): 5′-CCTATTTTACACCAACCCCCAGT-3′ JW12 (reverse): 5′-GGGTAGGGGCGTTCTGCGAAA-3′ |
Initial denaturation at 94 °C for 1 min followed by 34 cycles of denaturation at 94 °C for 30s,annealing at 58 °C for 30 s, and extension at 72 °C for 30 s |
| Internal transcribed spacer 1 PCR genus Leishmania specific (El Tai et al. 2000) [11] |
LITSR (forward): 5′-CTGGATCATTTTCCGATG-3′ L 5.8S (reverse): 5′-TGATACCACTTATCGCACTT-3′ |
Initial denaturation at 95 °C for 2 min, followed by 34 cycles of denaturation at 95 °C for 20 s, annealing at 53 °C for 30 s, and extension at 72 °C for 1 min, with a final extension of 72 °C for 6 min |
For Leishmania species identification, PCR-Restriction Fragment Length Polymorphism was carried out, the relevant PCR conditions are given in Table 1. Ten microlitre of LITSR/L5.8S amplified PCR products were digested with HaeIII enzyme (Thermoscientific) over night at 37 °C, according to the manufacturer's instructions. The restriction fragments were analysed by gel electrophoresis in 3% metaphor gel [12].
2.5. DNA sequencing
DNA sequencing was got done commercially from Apical Scientific Selangor, Malaysia for LITSR/L5.8S amplified Leishmania positive PCR products using LITSR/L5.8S Leishmania specific primer as mentioned in Table 1. The sequences obtained were cleaned, reverse complemented and submitted to GenBank under accession No. MT423519, MT423520, MT423521, MT423522 and MT423523.
The homologous gene sequences for ITS1 Leishmania species- and region-specific L.donovani isolates were retrieved from the NCBI GenBank database. ITS1 nucleotide query sequences from 31 specimens were analyzed using NCBI BLASTN ((https://blast.ncbi.nlm.nih.gov/Blast.cgi), multiple alignment software and Bio edit sequence alignment editor (version 7.2.5) [24]. The maximum parsimony method was used to create tree from the aligned sequences obtained with default parameters using the MEGAX program [25]. In order to find out the genetic and geographic relatedness of identified isolates among themselves and with standard reference strains, we performed ITS1 microsatellite repeat analysis and phylogenetic classification.
3. Results
3.1. Clinical features of patients
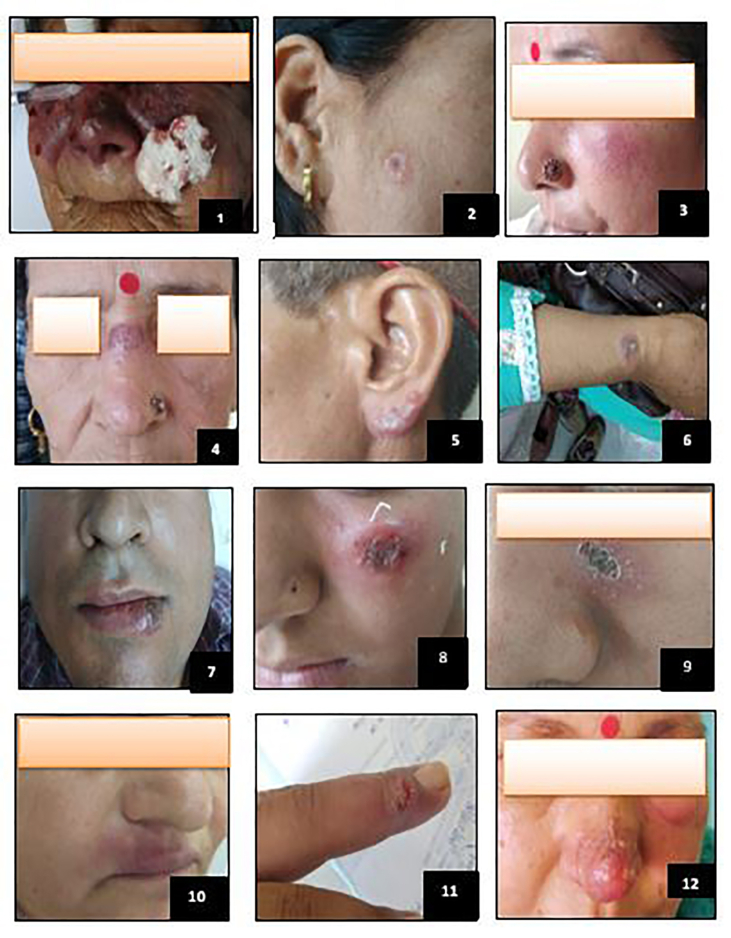
The age of CL patients ranged from 1-72 years of which 10 were male and 18 female. Mostly female students were affected. Duration of the lesions ranged from three weeks to two years. In most of the cases, lesions were mainly found on face while in three cases (Figure 2 ID-1,7,12) the lesions were of mucocutaneous type involving upper and lower lip and angle of mouth in three cases. The number of lesions per patient ranged from one to two and in majority of the cases single lesion was found. Size of the lesions ranged from 0.5 × 1cm to 5 × 5cm, the lesions were mostly erythematous, papule, and nodule-ulcerative plaques with or without crusting (Figure 2). None of the patients exhibited typical symptoms of VL. As all the 28 patients were native of villages along the Satluj River valley (HP) endemic for CL, the diagnosis of CL was reliable as it was done by trained dermatologist.
Figure 2.
Clinical presentation of cutaneous leishmaniasis patients reported at IGMC Shimla.
3.1.1. Confirmation of diagnosis by smear, rK39 and culture
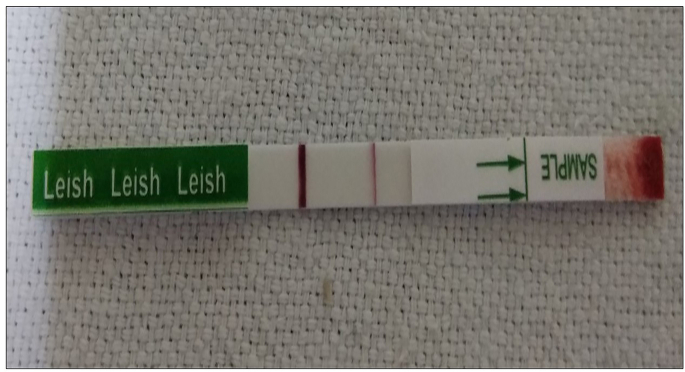
Of 12 patients from which imprint smear was prepared, only one was found positive for Leishmania amastigotes. The culture was not grown because of bacterial contamination as some Leishmania strains are difficult to grown [19, 20, 21]. None of the skin biopsy samples (12) was found positive for promastigotes. As regards screening through rK39 rapid diagnostic kit, only one out of 28 patients showed positivity (Figure 3).The rK39 positive patient was detected as having Leishmania donovani (Figure 2 Table 2-ID-1).
Figure 3.
Rapid immunochromtographic rK39 dipstick test results. The single band represents a negative control, while the double band shows positive result.
Table 2.
Characteristics of lesions and result of diagnosis of 12 patients of CL.
| Patient's Id | Smear | rK39 | Culture | Leishmania species -specific identification by PCR- RFLP | Leishmania species -specific identification by sequencing method | Type,size and site of lesion | Village/block/district |
|---|---|---|---|---|---|---|---|
| 1 | Negative | Positive | Negative | L. donovani | L. donovani | An well-defined Single erythematous plaque over the dorsum of nose and adjacent cheeks of size 15 cm × 4 cm approx., yellowish crusting over surface, discharge of pus surrounding erythematous border. | Chagon/Nichar/kinnuar |
| 2 | Negative | Negative | Negative | L. donovani | L. donovani | An well-defined Single shining plaque on left cheek near the ear of size 1.5 × 1cm approx. | Anni/Anni/Kullu |
| 3 | Positive | Negative | Negative | L. donovani | L. donovani | An well-defined shining plaque over the left cheek of size 3 × 2cm approx. | Jagaat khaana/Nirmand/Kullu |
| 4 | Negative | Negative | Negative | L. major | L. major | A single well defined nodular plaque over the dorsum of nose of size 3 × 3cm approx. | Gasso/Rampur/Shimla |
| 5 | Negative | Negative | Negative | L. major | L. major | A single well defined plaque with two satellite lesion in vicinity on ear lobule of size 3 × 1.5cm approx. | Chaura/Nichar/Kinnaur |
| 6 | Negative | Negative | Negative | L. donovani | L. donovani | A single well defined erythematous plaque of size 1.5 × 1.5cm approx. on right forearm with central crusting. | Anni/Kullu |
| 7 | Negative | Negative | Negative | L. tropica | L. tropica | A single well defined ulcerated plaque with damage over lower and upper lip of size 2 × 2cm approx. | Yangpa/Nichar/Kinnaur |
| 8 | Negative | Negative | Negative | L. donovani | L. donovani | An well-defined single shining papule with no secondary changes over left cheek of size 4 × 4cm approx. | Randal/Brow/Kullu |
| 9 | Negative | Negative | Negative | Not identified | Sequences not readable | A single well defined crusted plaque present below left eye of size 1 × 1cm approx. | Jagaat khaana/Nirmand/Kullu |
| 10 | Negative | Negative | Negative | L. tropica | L. tropica | An well-defined shining plaque over left upper lip of size 4 × 4 cm approx. | Ranjan/theog/Shimla |
| 11 | Negative | Negative | Negative | L. donovani | L. donovani | An well Ulcerated plaque over distil pharynx of middle finger of size 2 × 2cm approx. | Brow/Nirmand/kullu |
| 12 | Negative | Negative | Negative | L. tropica | L. tropica | An well defined erythematous nodule plaque of size 3 × 3cm over the tip of the nose, adjacent cheeks and on wrist. | Kimcha/Kushwa/Kullu |
3.2. Analysis of biopsy specimens by PCR –RFLP
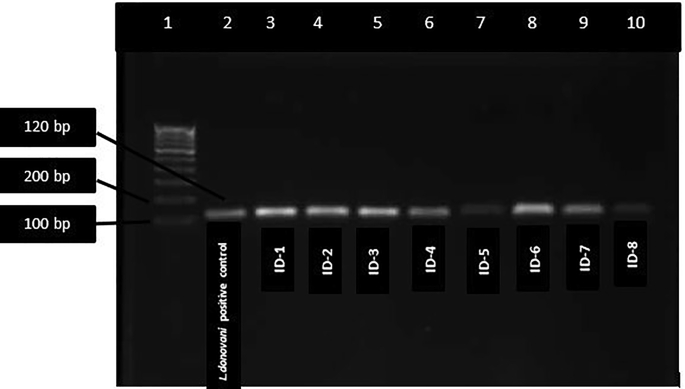
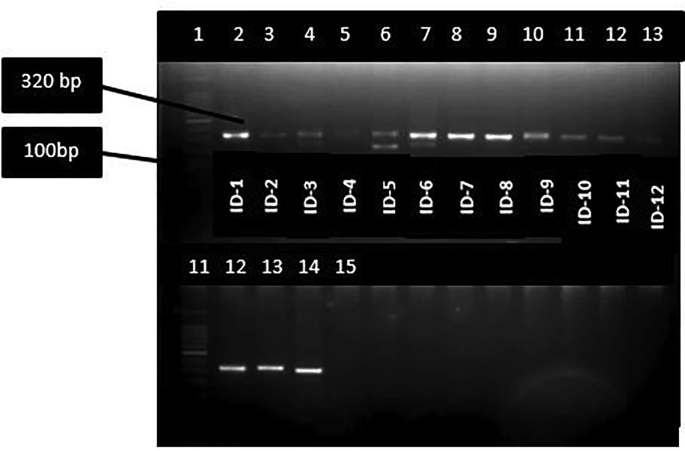
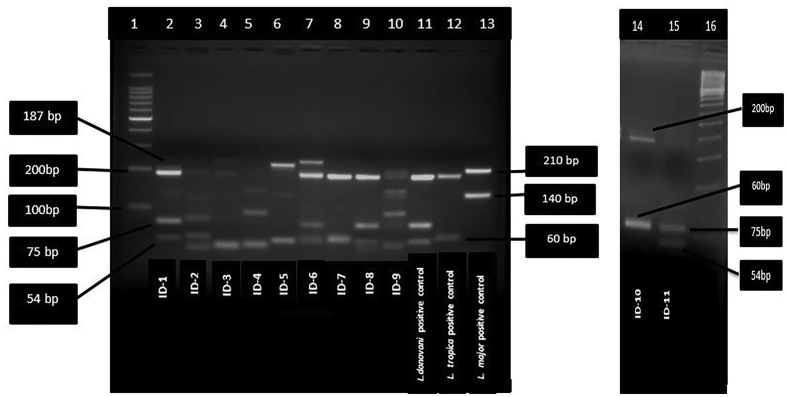
Of 12 biopsy samples, eight were found positive for Leishmania with JW11/12 set of primers (Table 2 and Figure 4) while all the 12 samples were found positive with ITS1 Leishmania specific set of primers (Figure 5). Of twelve Leishmania positive samples, six were identified as L. donovani with band size 187bp, 75bp and 54bp, 3 were identified as L. tropica with band size of 200 bp and 60bp, and two were identified as L. major with band size of 210 and 140 by PCR- RFLP (Figure 6). One sample remained unidentified (Table 2,ID-9) showing multiple bands in gel image and unreadable sequences by sequencing, however, the patient was found positive for Leishmania by PCR. The L. major identified cases were autochthonous and native of Himachal Pradesh only. Patient with ID-4, belongs to Jhakhadi village of Rampur (Shimla) and patient ID-5 belongs to Chaura village of Nichar (Kinnaur).
Figure 4.
DNA amplification of different parasite isolates of Leishmania from CL patients. JW11/JW12 primer set amplifies 120 bp fragment of KDNA of genus Leishmania.lane 1:100 bp ladder, lane 2: L.donovani positive control, lane 3–10: DNA isolated from patients.Negative control run in separate gel.
Figure 5.
DNA amplification of different parasite and DNA isolates from CL patients. LITSR/L5.8S set amplifies a 320 bp fragment of ITS1 region of Leishmania genus-specific DNA. Lane 1:100bp Ladder, lane 2–12 DNA isolate from patient, lane 11: 100 bp ladder,lane 12: L.donovani positive control, lane 13: L.tropica positive control, lane 14: L.major control, lane 15: Negative control.
Figure 6.
Leishmania species-specific restriction fragment length polymorphism –polymerase chain reaction assay of different parasite strain and isolates from patients. lane 1 : 100 bp DNA ladder; lane 2–10 patients samples (remaining sample and negative control run separately); lane 11: L.donovani positive control; lane 12; L.tropica positive control; lane 13: L.major positive control.lane 14 patient isolate (identified as L. tropica)-15 patient isolate (identified as L.donovani); lane 16: 50 bp ladder.
3.3. Sequence analysis
3.3.1. In silico analysis
The BLAST-N analysis of identified sequences (MT423519-23) revealed a total of 204 unique accession/locus numbers showing >40% similarity with the identified sequences. Only MT423523 sequence was found having less nucleotides i.e. 180 bp showing high similarity with the sequence of L. major from Turkey/Syria (MH347926).
Three accessions (MT423519-21) showed close similarity 98.19–100% with the L. donovani sequences. Among these three, only MT423520 sequence showed a single base ‘C’ addition at 271 nt (nucleotide) position.
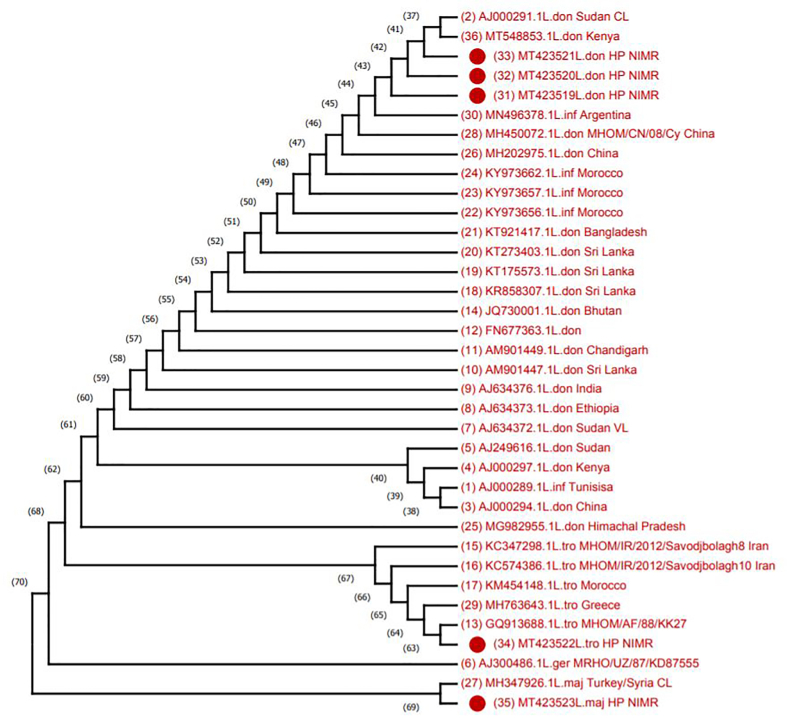
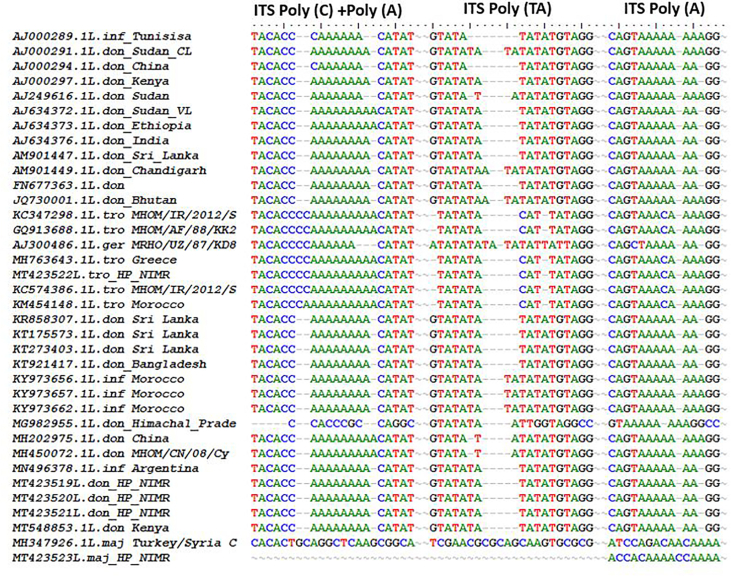
To make a phylogenetic proximity analysis, search was fixed to 36 sequences (including 5 sequences from the present study) where we retained all the accessions retrieved from NCBI database. In the tree obtained by maximum parsimony analysis, it has been found that three sequences with accession numbers MT423519-21 were 98.20–100% similar to L. donovani sequences from Kenya (MT548853) and 98.19–100% similar to Sudan (AJ00029) as well as 98.2–100% similarity with AM901449 and AJ634376 from India (Figure 7). Although these sequences showed ~100% similarity to L. donovani but also showed similarity to L. infantum with accession no. Argentina (MN496378) and Morocoo (KY973662, KY973657, KY973656). However,the ITS1 polymorphic microsatellite repeat analysis indicated that the isolates in the present study were different from L. infantum and closest to L. donovani isolates from Kenya (Table 3 and Figure 8).
Figure 7.
ITS1-based molecular analysis of clinical isolates from CL patients, Himachal Pradesh, India. Phylogenetic tree of ITS1 sequences from test isolates (designated as HP NIMR) and standard WHO Leishmania strains. Tree constructed by using maximum-parsimony tree method using the phylogeny program of MEGAX builder (10.1.8). GenBank accession numbers are indicated. ITS1, internal transcribed spacer 1.
Table 3.
Leishmania species used in ITS -based microstatillite polymorphism and phylognteic analysis of Leishmania isolates identified.
| GenBank accession | Standard Leishmania strains | Place of origin | Poly C (24–39) | Poly A (24–39) | Poly TA (61–76) | Poly A (124–134) |
|---|---|---|---|---|---|---|
| AJ000289.1 | L.infantum | Tunisisa | 3 | 6 | 4 | 8 |
| AJ000291.1 | L.donovani | Sudan CL | 2 | 8 | 6 | 8 |
| AJ000294.1 | L.donovani | China | 5 | 6 | 4 | 7 |
| AJ000297.1 | L.donovani | Kenya | 2 | 8 | 5 | 7 |
| AJ249616.1 | L.donnovani | Sudan | 2 | 7 | 5 | 8 |
| AJ300486.1 | L. gerbilli | MRHO/UZ/87/KD87555 | 4 | 6 | 6, 2TTA | 6 |
| AJ634372.1 | L.donovani | Sudan VL | 2 | 9 | 5 | 7 |
| AJ634373.1 | L.donovani | Ethiopia | 2 | 9 | 5 | 7 |
| AJ634376.1 | L.donovani | India | 2 | 8 | 5 | 7 |
| AM901447.1 | L.donovani | Sri Lanka | 2 | 8 | 5 | 7 |
| AM901449.1 | L.donovani | Chandigarh | 2 | 8 | 2,TAA,3 | 7 |
| FN677363.1 | L.donovani | 2 | 8 | 5 | 7 | |
| GQ913688.1 | L.tropica | MHOM/AF/88/KK27 | 4 | 9 | 3, CAT,2 | 3, C, 4 |
| JQ730001.1 | L.donovani | Bhutan | 2 | 8 | 2,TAA,3 | 8 |
| KC347298.1 | L.tropica | MHOM/IR/2012/Savodjbolagh8 | 4 | 9 | 3, CAT,2 | 3, C, 4 |
| KC574386.1 | L.tropica | MHOM/IR/2012/Savodjbolagh10 | 4 | 9 | 3, CAT,2 | 3, C, 4 |
| KM454148.1 | L.tropica | Morocco | 3 | 10 | 3, CAT,2 | 3, C, 4 |
| KR858307.1 | L.donovani | Sri Lanka | 2 | 8 | 5 | 7 |
| KT175573.1 | L.donovani | Sri Lanka | 2 | 8 | 5 | 7 |
| KT273403.1 | L.donovani | Sri Lanka | 2 | 8 | 5 | 7 |
| KT921417.1 | L.donovani | Bangladesh | 2 | 8 | 5 | 7 |
| KY973656.1 | L.infantum | Morocco | 2 | 8 | 6 | 8 |
| KY973657.1 | L.infantum | Morocco | 2 | 8 | 6 | 8 |
| KY973662.1 | L.infantum | Morocco | 2 | 8 | 6 | 8 |
| MG982955.1 | L.donovani | Himachal Pradesh | Heterogenous | 2,TAA,3 | 8 | |
| MH202975.1 | L.donovani | China | 2 | 9 | 5 | 8 |
| MH347926.1 | L.major | Turkey/Syria CL | Heterogenous | |||
| MH450072.1 | L.donovani | China | 2 | 9 | 5 | 8 |
| MH763643.1 | L.tropica | Greece | 4 | 9 | 3, CAT,2 | 3, C, 4 |
| MN496378.1 | L.infantum | Argentina | 2 | 8 | 5 | 7 |
| MT423519 | L.donovani | HP NIMR | 2 | 8 | 5 | 7 |
| MT423520 | L.donovani | HP NIMR | 2 | 8 | 5 | 7 |
| MT423521 | L.donovani | HP NIMR | 2 | 8 | 5 | 7 |
| MT423522 | L.tropica | HP NIMR | 4 | 9 | 3, CAT,2 | 3, C, 4 |
| MT423523 | L.major | HP NIMR | NA | 4, CC 4 | ||
| MT548853.1 | L.donovani | Kenya | 2 | 8 | 5 | 7 |
Figure 8.
Multiple sequence alignment of ITS1 microsatellite repeat sequences of representative parasite isolates from patients with those of L. donovani complex reference strains from different geographic regions. Sequences were aligned by using BioEdit sequence alignment program.
4. Discussion
Leishmaniasis is thought to be a disease of low altitude, with no occurrence of cases above 2000 feet (600 m) [28], However, considerable number of cases have been reported from subalpine valley of Himachal Pradesh. Before 1988, CL was not known to occur in Himachal Pradesh but thereafter, sporadic cases are being reported continuously. In conratst to detection of L. tropica from CL cases from Rajasthan, L. donovani and L. tropica have also been reported from cases of CL in Himachal Pradesh.
In our study we found three parasites i.e. L donovani, L. tropica and L. major from cases of CL from Rampur (Shimla), Kinnaur and Kullu districts. L donovani has also been reported from CL cases from Kerala, India [13]. LITSR/L5.8S set of primers were found better than JW11/12 as the former could detect Leishmania in all the 12 samples. PCR-RFLP and sequencing methods were found equally good in species identification of Leishmania. The other gene region that can differentiate L.donovani complex from L. infantum better than ITS1 region, (like Hsp70) may also be explored.
L. donovani is the established causative agent of VL in Bihar (India) but the strain is L. donovani zymodeme MON2 [22, 23, 27]. A recent study undertaken in Himachal Pradesh provided evidence that the L. donovani parasite isolated from the cases of CL is similar to L. donovani zymodeme MON37 but distinct from L. donovani zymodeme MON2 causing VL [26] and termed it as atypical CL. However, further studies on whole genome sequencing are required to confirm the preponderance of exclusively L. donovani zymodeme MON 37 strain from CL cases in Himachal Pradesh. The countries like Kenya, Iraq, Sri Lanka, Yemen [14, 15, 16, 17] have also reported L. donovani as causative parasite for CL. In our study we found three species of Leishmania, (with L. donovani as predominant), suggesting that in Himachal Pradesh there is existence of typical as well as atypical CL and the problem is quite wide spread in five districts (Shimla, Kinnaur, Kullu, Mandi and Solan) [26]. The L. donovani -infantum complex suspected by Sharma et al (2005) has been confirmed as L. donovani by the present study as well as by Thakur et al. al (2020). In view of findings of the present study, it is suggested that state wide point surveillance studies be undertaken in Himachal Pradesh so that foci in other districts can be unraveled. A latest review on atypical leishmaniasis in India has mapped the distribution of CL from the states of Punjab, Assam, Haryana, Delhi, Jammu & Kashmir and Varanasi with atypical leishmaniasis from Kerala [7] suggesting that CL has spreaded in several parts of India. Since the study area is close to China border,where P.longiductus is the vector for VL, it is likely that there are cases of VL also in Himachal Pradesh.Therefore detailed studies in the wider geographic area bordering districts of Himachal Pradesh with China are required. In the wake of achieving the goal of elimination of kala-azar from India, elimination of CL should also be considered.
5. Conclusions
Detection of three species of Leishmania, i.e. L. donovani, L. tropica and L. major from the patients of CL from Himachal Pradesh confirms the existence of typical as well as atypical CL. While India is close to elimination of VL, the occurrence of CL cases in HP, Kerala or elsewhere should not be ignored considering them just cases of CL alone. Further studies are warranted to confirm the existence of L. donovani zymodeme MON37 from cases of CL in Himachal Pradesh or L. donovani zymodeme MON2 strain causing VL in Bihar. The goal of elimination of VL should be considerd comprehensively as elimination of leishmaniasis encompassing CL.
Declarations
Author contribution statement
Suman Lata: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Sandhya Kumari: Performed the experiments; Analyzed and interpreted the data.
Ram Das: Analyzed and interpreted the data.
Shweta Pasi: Contributed reagents, materials, analysis tools or data.
Ramesh C. Dhiman: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This work was supported by the Indian Council of Medical Research New Delhi, India (Grant No. 68/10/2014-NCD-1).
Data availability statement
Data associated with this study has been deposited at NCBI under the accession numbers MT423519, MT423520, MT423521, MT423522, MT423523.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
We acknowledge the Indian Council of Medical Research for funding the project. Thanks are also due to the State Programme Officer, Himachal Pradesh and staff of Dermatology department, IGMC, Shimla.
References
- 1.World Health Organization Global Health Observatory . 2017. Leishmaniasis.http://www.who.int/gho/neglected_diseases/leishmaniasis/en/ cited 20th June 2020. Available from: [Google Scholar]
- 2.World Health Organization . March 2010. Control of the Leishmaniasis: Report of a Meeting of the WHO Expert Committee on the Control of Leishmaniases, Geneva; pp. 22–26. (WHO technical report series; no. 949) [Google Scholar]
- 3.Sharma N.L., Mahajan V.K., Kanga A., Sood A., Katoch V.M., Mauricio I. Localized Cutaneous leishmaniasis due to Leishmania donovani and Leishmania tropica: preliminary findings of the study of 161 new cases from a new endemic focus in Himachal Pradesh, India. Am. J. Trop. Med. Hyg. 2005;72:819–824. PMID: 15964970. [PubMed] [Google Scholar]
- 4.Sharma R.C., Mahajan V.K., Sharma N.L., Sharma A. A new focus of cutaneous leishmaniasis in Himachal Pradesh (India) Indian J. Dermatol. Venereol. Leprol. 2003;69(2):170–172. [PubMed] [Google Scholar]
- 5.Sharma N.L., Mahajan V.K., Ranjan N., Verma G.K., Negi A.K., Mehta K.I.S. The sandflies of the Satluj river valley, Himachal Pradesh (India): some possible vectors of the parasite causing human cutaneous and visceral leishmaniases in this endemic focus. J. Vector Borne Dis. 2009;46:136–140. PMID: 19502693. [PubMed] [Google Scholar]
- 6.Sharma N.L., Mahajan V.K., Negi A.K., Verma G.K. The rK39 immunochromatic dipstick testing: a study for rK39 seroprevalence in dogs and human leishmaniasis patients for possible animal reservoir of cutaneous and visceral leishmaniasis in endemic focus of Satluj river valley of Himachal Pradesh (India Indian. J Dermatol Venereol Leprol. 2009;75(1):52–55. doi: 10.4103/0378-6323.45221. [DOI] [PubMed] [Google Scholar]
- 7.Thakur L., Singh K.K., Shanker V., Negi A., Jain A., Matlashewski G. Atypical leishmaniasis: a global perspective with emphasis on the Indian subcontinent. PLoS Neglected Trop. Dis. 2018;12(9) doi: 10.1371/journal.pntd.0006659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sandhya Negi A., Garg A., Thakur K., Lal P., Ahluwalia A.K. Clinico-epidemiological trends of cutaneous leishmaniasis along the Satluj valley of Himachal Pradesh-A new focus with emerging infection. J Med Sci Clin Res. 2017;5 [Google Scholar]
- 9.Kumari S., Garg A. Lip leishmaniasis: a new emerging clinical form of cutaneous leishmaniasis from sub himalayan region. J Med Sci Clin Res. 2018;6 [Google Scholar]
- 10.Rodgers M.R., Popper S.J., Wirth D.F. Amplification of kinetoplast DNA as a tool in the detection and diagnosis of Leishmania. Exp. Parasitol. 1990;71(3):267–275. doi: 10.1016/0014-4894(90)90031-7. [DOI] [PubMed] [Google Scholar]
- 11.El Tai N.O., Osman O.F., el Fari M., Presber W., Schönian G. Genetic heterogeneity of ribosomal internal transcribed spacer in clinical samples of Leishmania donovani spotted on filter paper as revealed by single-strand conformation polymorphisms and sequencing. Trans. R. Soc. Trop. Med. Hyg. 2000;94(5):575–579. doi: 10.1016/s0035-9203(00)90093-2. [DOI] [PubMed] [Google Scholar]
- 12.Ranasinghe S., Wickremasinghe R., Hulangamuwa S. Polymerase chain reaction detection of Leishmania DNA in skin biopsy samples in Sri Lanka where the causative agent of cutaneous leishmaniasis is Leishmania donovani. Mem. Inst. Oswaldo Cruz. 2015;110(8):1017–1023. doi: 10.1590/0074-02760150286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar N.P., Srinivasan R., Anish T.S.G., Nandakumar G., Jambulingam P. Cutaneous leishmaniasis caused by Leishmania donovani in the tribal population of the agasthyamala biosphere reserve forest, Western Ghats, Kerala, India. J. Med. Microbiol. 2015;64:157–163. doi: 10.1099/jmm.0.076695-0. 2015. [DOI] [PubMed] [Google Scholar]
- 14.Mebrahtu Y.B., van Eys G., Guizani I., Lawyer P.G., Pamba H., Koech D. Human cutaneous leishmaniasis caused by Leishmania donovani. in Kenya. Trans. R. Soc. Trop. Med. Hyg. 1993;87:598–601. doi: 10.1016/0035-9203(93)90101-u. [DOI] [PubMed] [Google Scholar]
- 15.Al-Diwany L.J., al-Awkati N.A., Atia M., Rassam M.B. Concomitant natural infection with L. donovani and L. major: a case report from Iraq. J. Trop. Med. 2014;2014:8pp. doi: 10.1007/BF01354478. [DOI] [PubMed] [Google Scholar]
- 16.Karunaweera N.D., Pratlong F., Siriwardane H.V., Ihalamulla R.L., Dedet J.P. Sri Lankan cutaneous leishmaniasis is caused by Leishmania donovani zymodeme Mon-37. Trans. R. Soc. Trop. Med. Hyg. 2003;97:380–381. doi: 10.1016/s0035-9203(03)90061-7. [DOI] [PubMed] [Google Scholar]
- 17.Pratlong F., Bastien P., Perello R., Lami P., Dedet J.P. Human cutaneous leishmaniasis caused by Leishmania donovani sensu stricto in Yemen. Trans. R. Soc. Trop. Med. Hyg. 1995;89(4):398–399. doi: 10.1016/0035-9203(95)90025-x. [DOI] [PubMed] [Google Scholar]
- 18.Sharma N., Sood A., Arora S., Kanga A., Mahajan V.K., Negi A. Characteristics of Leishmania spp. isolated from a mixed focus of cutaneous and visceral Leishmaniasis in Himachal Pradesh (India) Internet J. Caribb. Third World Med. 2008;7(2):1–7. [Google Scholar]
- 19.Schuster F.L., Sullivan J.J. Cultivation of clinically significant hemoflagellates. Clin. Microbiol. Rev. 2002;15(3):374–389. doi: 10.1128/CMR.15.3.374-389.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mathis A., Deplazes P. PCR and in vitro cultivation for detection of Leishmania spp. in diagnostic samples from humans and dogs. J. Clin. Microbiol. 1995;33(5):1145–1149. doi: 10.1128/jcm.33.5.1145-1149.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marsden P.D., Jones T.C. Clinical manifestations, diagnosis and treatment of Leishmaniasis. In: Chang K.P., Bray R.S., editors. Human Parasitic Diseases: Leishmaniasis. Elsevier Science Publishers; Oxford: 1985. pp. 183–198. 1985. [Google Scholar]
- 22.Brahmachari U.N. A new form of cutaneous leishmaniasis-dermal leishmanoid. Indian Med. Gaz. 1922;57(4):125–127. PMID: 29008368. [PMC free article] [PubMed] [Google Scholar]
- 23.Desjeux P., Ghosh R.S., Dhalaria P., Strub-Wourgaft N., Zijlstra E.E. 2013. Report of the Post Kala-Azar Dermal Leishmaniasis (PKDL) Consortium Meeting, New Delhi, India, 27-29 June 2012. Parasit Vectors; p. 196. 6. Published 2013 Jul 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hall T.A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- 25.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. Mega X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thakur L., Singh K.K., Kushwaha H.R., Sharma S.K., Shankar V., Negi A. Leishmania donovani infection with atypical cutaneous manifestations, Himachal Pradesh, India, 2014–2018. Emerg. Infect. Dis. 2020;26(8):1864–1869. doi: 10.3201/eid2608.191761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thakur C.P., Dedet J.P., Narain S., Pratlong F. Leishmania species, drug unresponsiveness and visceral leishmaniasis in Bihar, India. Trans. R. Soc. Trop. Med. Hyg. March-April 2001;95(2):187–189. doi: 10.1016/s0035-9203(01)90160-9. [DOI] [PubMed] [Google Scholar]
- 28.Park K. In: Park's Text Book of Preventive and Social Medicine. sixth ed. Park K., editor. Jabalpur; India: 2007. pp. 256–258. Banarsidas Bhanot. (Leishmaniasis) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data associated with this study has been deposited at NCBI under the accession numbers MT423519, MT423520, MT423521, MT423522, MT423523.