Abstract
Context-dependent trait exaggeration is a major contributor to phenotypic diversity. However, the genetic modifiers instructing development across multiple contexts remain largely unknown. We use the arthropod tibia, a hotspot for segmental differentiation, as a paradigm to assess the developmental mechanisms underlying the context-dependent structural exaggeration of size and shape through nutritional plasticity, sexual dimorphism and segmental differentiation. Using an RNAseq approach in the sexually dimorphic and male-polyphenic dung beetle Digitonthophagus gazella, we find that only a small portion (3.7%) of all transcripts covary positively in expression level with trait size across contexts. However, RNAi-mediated knockdown of the conserved sex-determination gene doublesex suggests that it functions as a context-dependent master mediator of trait exaggeration in D. gazella as well as the closely related dung beetle Onthophagus taurus. Taken together, our findings suggest (i) that the gene networks associated with trait exaggeration are highly dependent on the precise developmental context, (ii) that doublesex differentially shapes morphological exaggeration depending on developmental contexts and (iii) that this context-specificity of dsx-mediated trait exaggeration may diversify rapidly. This mechanism may contribute to the resolution of conflict arising from environment-dependent antagonistic selection among sexes and divergent developmental contexts in a wide range of animals.
Keywords: intralocus conflict, sexual dimorphism, nutritional plasticity, condition dependence, Onthophagus, geometric morphometrics
1. Background
Organismal growth and differentiation require developmental systems to respond to cues and signals originating from both outside and within the growing organism. Yet how organisms weave together various sources of intrinsic and extrinsic information into a cohesive developmental progression remains poorly understood. This applies in particular to traits subject to antagonistic selection depending on the genetic, developmental or environmental contexts within which they are expressed. A common feature of structural development associated with multiple such developmental contexts and various forms of conflict is trait exaggeration—a major component of organismal diversification [1]. This is especially true for male secondary sexual traits acting as ornaments and/or armaments [2,3] that are often nutritionally plastic. In these instances, trait exaggeration becomes a function of both nutritional conditions as well as sex. However, a third context is likely to be just as important: when traits possess serial homologues along the body axis. For example, the serially reiterated appendages of arthropods form through a shared and highly conserved set of developmental processes [4,5] yet often present dramatically different manifestations of the same homologous structure, such as the greatly elongated forelimbs of male harlequin beetles or the exaggerated hindlimbs of male leaf-footed bugs [6]. In these cases, trait exaggeration is contingent not only on sex or nutritional context, but also on the segmental identity of the serial homologue. Despite the importance of these interwoven contexts in driving trait exaggeration, the mechanisms that underlie and integrate context-dependent trait exaggeration in the same trait across multiple contrasts remain largely unexplored. This is despite the critical contribution of context-dependent development to the resolution of intralocus sexual conflict—a common phenomenon which arises when the optimal trait value of homologous structures differs between sexes [7]. Similar effects can be expected if the same trait is expressed in different external or internal environments, such as nutritional conditions or body regions. Using a combination of transcriptomic, functional genetic and geometric morphometric approaches, we sought to better understand the developmental underpinnings of trait exaggeration across diverse contexts, their integration within the formation of a singular structure—the dung beetle tibia— and the developmental resolution of potential conflicts arising when trait exaggeration in one context results in trait values inappropriate for another.
The evolution of trait exaggeration could be enabled by the deployment of taxon-restricted genes, recruitment and co-option of previously unrelated developmental processes, or modification of ancestrally plastic developmental processes via genetic accommodation [8,9]. Functional interrogation has identified several common developmental mechanisms underlying the evolution and diversification of exaggerated traits [3] including, among others, the homeobox genes Ubx/UbdA (in orthopteran hindlegs [10]), genes associated with insulin signalling (in beetle horns [11–13]) and genes ancestrally tasked with somatic sex determination (in dung and rhinoceros beetle horns and stag beetle mandibles [8,14–16]). Previous transcriptomic and quantitative genetic studies further suggest that sexual dimorphism and nutritional plasticity share developmental underpinnings. For example, sexual dimorphism has been shown to coincide with increased sex-bias in nutritional plasticity [17–21] and to co-evolve with sex-specific plasticity in traits under directional selection [22]. Together, these findings suggest that the same developmental processes may be recruited repeatedly during the convergent evolution of sex-biased trait exaggeration, hinting at the existence of a set of ‘core exaggeration genes'. Additive effects of environmental and intrinsic signals on such exaggeration genes could then mediate trait exaggeration that converges on optimal trait expression.
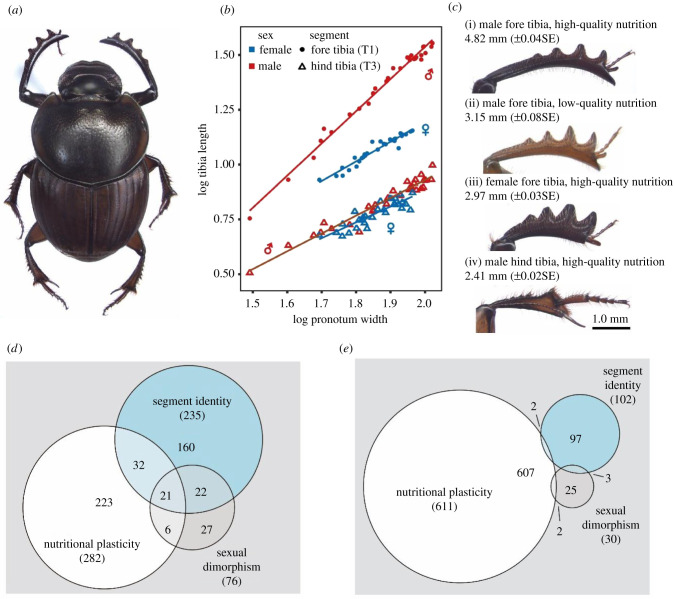
Here, we study the mechanisms underlying the multi-context-dependent exaggeration of insect tibia morphology. Tibiae—the fourth segments of all three pairs of insect legs—have an ancient evolutionary history, are exposed to selection in both sexes and, as integral components of critical locomotory appendages, are probably under strong functional constraints and selection. Yet, in insects, tibia exaggeration often varies between sexes, nutritional contexts and thoracic segments. For instance, male gazelle dung beetles (Digitonthophagus gazella; figure 1a) develop particularly long and elongated fore tibiae (figure 1b) used to hold on to the female elytra during copulation. This elongation is dependent on nutritional contexts in that fore tibia size increases disproportionately with larval nutrition and adult size (hyperallometry). Females on the other hand develop stout shovel-like fore tibiae used in the excavation of breeding tunnels and exhibit a largely isometric tibia length allometry in response to variation in larval nutrition (figure 1b,c). Furthermore, hind tibiae—which develop from serially homologous tissues in the third thoracic segment—are comparatively small and hypoallometric in both sexes (figure 1b,c). By comparing the developmental underpinnings of such sex-, nutrition- and segment-specific development of homologous tissues, we here aim to further our understanding of the mechanisms facilitating context-dependent trait exaggeration.
Figure 1.
(a) Digitonthophagus gazella males have generally (b,c) longer and more nutritionally plastic fore tibiae compared to females (β♂ = 1.47 [1.40,1.53] (95%CI), n = 30; β♀ = 0.90 [0.80, 0.99], n = 30; sex × log pronotum width interaction: p < 0.001, n = 60). By contrast, the hind tibia is much smaller and scales hypoallometrically in both sexes (β♂ = 0.81 [0.74, 0.89], n = 30; β♀ = 0.72 [0.57, 0.87], n = 30; sex × log pronotum width interaction: p = 0.265, n = 60). (d,e) Venn diagrams depicting the number of differentially expressed genes (p adj < 0.05) that correlate positively (d) and negatively (e) in expression levels with trait size. (Online version in colour.)
2. Material and methods
(a) . Animal husbandry
Digitonthophagus gazella (Fabricius, 1787) was collected in Santa Fe, Florida, in spring 2019 and shipped to Bloomington, Indiana, where a laboratory colony was established following standard procedures [23].
(b) . Sex-specific static allometries in Digitonthophagus gazella
To quantify variation in tibia size due to sexual dimorphism, nutritional plasticity and segment identity, we first reared F1 offspring under various amounts of food (1.0 to 3.0 g of homogenized cow dung) in 12-well tissue culture plates at 29°C (as described previously [23]). Upon adult eclosion, individuals were sacrificed and stored in 70% ethanol. As an estimate of overall body size, we measured pronotum width (a common measure in this group of organisms that shows a multivariate allometric slope close to isometry in this species). Body size and fore and hind tibia length (see electronic supplementary material) were measured for 30 individuals per sex spanning the entire body size range. Sex-specific allometric coefficients were calculated using ordinary least-squares regression of log trait size against log pronotum width.
(c) . Sample generation, sequencing and sequence analysis
To study variation in gene expression, we again reared newly hatched (F1) offspring in 12-well plates. This time, half or all animals received a full well (3.2 g) of homogenized cow dung, while the other half received only 50% as much food (1.6 g). Upon reaching the third larval stage, individuals were sexed based on gonad morphology [24], weighed using a Mettler Toledo (AL54 Ohio, USA, d = 0.1 mg) scale and dissected (see electronic supplementary material, figures S6 and S7). Following [25], we first rinsed all prepupae with RNAse-free water and carefully removed the larval cuticle in 0.05% Triton-X in phosphate-buffered saline, removed the prepupal fore- and hindleg primordia of all individuals, and dissected only the tissue that gives rise to the adult fore and hind tibia (see electronic supplementary material, figure S6). We collected fore- and hindleg tissue for six males reared under high food conditions, foreleg tissue of six males reared under poor food conditions, and foreleg tissue of six females reared under high food conditions (24 samples of 18 individuals total). Tissue was stored immediately in RNAlater and kept at −80°C. After thawing, tissues were homogenized with disposable polypropylene RNase-free pestles and RNA was extracted using a RNeasy Plus Mini Kit (Qiagen) following the manufacturer's instructions. Total RNA quality was checked using an RNA ScreenTape TapeStation System (Agilent). Libraries were quantified using a Quant-iT DNA Assay Kit (Thermo Fisher), pooled in equal molar amounts and sequenced as single-end reads using a 75-cycle High kit on the NextSeq500 platform (Illumina, San Diego, CA). Samples were all pooled to control for batch effects.
The quality of raw reads was checked using FastQC [26]. Illumina adapter sequences were removed using trimmomatic [27] (electronic supplementary material, table S1). The transcripts of all 24 tibia samples were used to generate a de novo transcriptome assembly using Trinity v2.4.0 [28] (only considering longest isoforms using get_longest_isoform_seq_per_trinity_gene.pl; electronic supplementary material, table S2). BUSCO [29] was used to assess the completeness of the transcriptome and Trinotate (https://trinotate.github.io/) was used for gene annotation (electronic supplementary material, table S2). The trimmed reads were mapped against the de novo transcriptome using bowtie2 (v2.3.2) [30] and quantified with rsem (v1.3.0) [31] using Trinity's built in ‘align_and_estimate_abundance’ and ‘abundance_estimates_to_matrix’ tool. We performed pairwise comparisons of normalized gene expression levels across all contrasts as implemented in the R-package DESEq2 [32]. For all contrasts, we used a critical false discovery rate threshold of 0.05 with Benjamini–Hochberg adjustment.
(d) . RNA interference: dsRNA synthesis and injection
To assess the function of dsx, we revisited individuals subjected to RNAi-mediated knockdown of dsx expression (dsxRNAi) generated in a previous study [8]. dsRNA synthesis and injection followed methods described elsewhere [14,33]. In brief, dsx template DNA was amplified by PCR using dsx-specific primers attached to a T7 promoter sequence. MEGAscript T7 transcription and MEGAclear kits (Invitrogen) were used to synthesize and purify dsRNA. dsRNA was then diluted in injection buffer to reach a concentration of 1.0 µg µl−1 dsRNA. Using a hand-held syringe, 3 µg dsRNA were consequently injected into the thorax of early L3 larvae. Control injections were performed by injecting buffer solution only. The sex of larvae was determined according to gonad morphology [24]. After complete sclerotization, emerging adults were sacrificed and stored in 70% ethanol.
(e) . (Geometric) morphometric analysis in Digitonthophagus gazella
Using a digital camera (Scion, Frederick, MD, USA) mounted on a Leica MZ-16 stereomicroscope (Bannockburn, IL, USA), we took calibrated pictures of the pronotum as well as the fore and hind tibiae. Pronotum width and tibia length were measured using tpsDig2 [34]. To test for an effect of dsx knockdown on logarithmized fore and hind tibia length, we used ANOVAs (type II sums of squares) with injection treatment (buffer or dsxRNAi) and sex as fixed effects and logarithmized pronotum width as covariate.
To quantify the effect of dsx knockdown on tibia shape we again used tpsDig2 [34] to acquire nine two-dimensional coordinates to quantify fore tibia shape and five landmarks to quantify hind tibia shape (see electronic supplementary material, figure S8). Landmark coordinates were subjected to a full Procrustes analysis using the function gpagen() of the R-package geomorph v. 3.1.1 [35]. Centroid size, a shape-independent measure of structural size [36], was retrieved. To test for an effect of dsx knockdown and sex differences in tibia shape, we used Procrustes ANOVAs with residual randomization permutation procedure (as implemented in geomorph). Logarithmized centroid size was included to control for (sex-specific) shape variation due to allometry. To illustrate the variation in tibia shape, and to quantify how strongly sexes and RNAi treatments were differentiated, we performed a canonical variate analysis (CVA) with jack-knife cross-validation (as implemented in Morpho [37]). To explore whether dsx causes differences in the direction or magnitude (vector norm) of shape change between sexes and treatment groups, we used trajectory analysis (TA) [38] with a randomized residual permutation procedure (as implemented in the geomorph function trajectory.analysis())
(f) . Evolutionary diversification of the function of doublesex
To assess whether the function of doublesex in tibia development is unique to D. gazella or whether it is conserved among species, we revisited a previous study that used dsx knockdown in Onthophagus taurus, a related species of dung beetle [14]. We haphazardly chose several dsx dsRNA-injected and control individuals per sex and again measured pronotum width, as well as fore and hind tibia length. We then used (Procrustes) ANOVAs to test for species differences in the function of dsx in the sex-specific development of size and shape (electronic supplementary material, tables S5 and S6).
(g) . Sex-specific nutritional plasticity in horns, legs and thorax
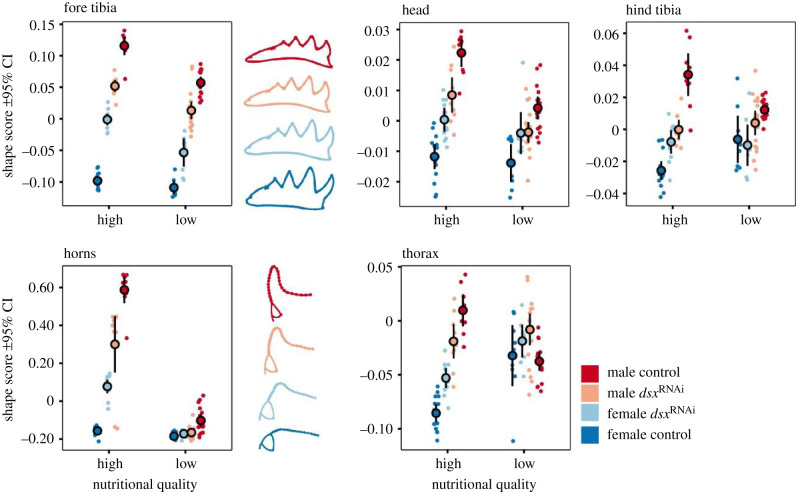
To investigate the effect of dsxRNAi on sex-specific nutritional plasticity, we again reared 144 newly hatched (L1) larvae in 12-well plates. As in the RNAseq experiment described above, half of all individuals were provided with 3.2 g of homogenized cow dung, while the other half received only half as much food (1.6 g). This nutritional manipulation was crossed with an RNAi treatment (dsRNA or buffer injection). After adult emergence and sclerotization, we quantified the shape of fore and hind tibiae (as described above), heads (6 full- and 8 semi-landmarks), horns (4 full- and 26 semi-landmarks) and thoraces (3 full- and 18 semi-landmarks) as indicated in electronic supplementary material, figure S8. The positioning of semi-landmarks was optimized by minimizing bending energy. Procrustes ANOVAs were used to test for the effect of dsxRNAi and nutritional manipulation on sex-specific shape. To visualize the effect of nutrition and dsxRNAi on sexual shape dimorphism, we extracted vectors of coefficients for the sex effect in high-quality nutrition (β) and projected the shape data (Y) onto this vector as [39]. This renders an individual ‘shape score’ (s) for each trait (figure 4). Small values indicate a more female-like shape while larger values indicate more masculinized morphology. Sex-specific vectors of the effect of nutritional manipulation on shape were computed for buffer and dsx dsRNA-injected individuals using multivariate linear models. Vector correlations between male- and female-specific vectors of regression coefficients (β) were computed as (figure 5). 95% CIs were generated using non-parametric bootstrapping. See electronic supplementary material, table S10 for vector correlation between sexual shape dimorphism vectors (vectors of regression coefficients for the effect of sex) of animals injected with buffer and individuals injected with dsx dsRNA.
Figure 4.
Combined effect of dsxRNAi and nutritional quality on shape in D. gazella. Shape scores [42] represent projections of the shape data onto the vector defined by sexual shape dimorphism of animals reared on a high-quality diet. Small values indicate a more ‘female-like’ shape while larger scores indicate more masculinized morphology. Sketches correspond to the sex-specific mean shapes of control and dsxRNAi individuals for horn and fore tibia morphology (see electronic supplementary material, figure S3 for a principal component analysis). (Online version in colour.)
Figure 5.
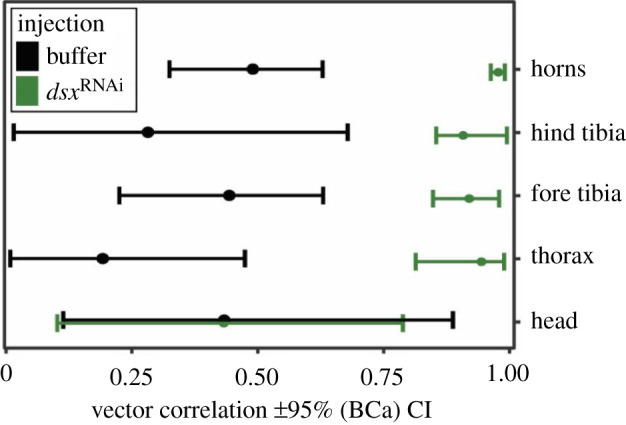
In control animals, shape changes caused by nutritional plasticity strongly differ between the sexes (as indicated by moderate vector correlations). However, with the exception of anterior head shape, these sex differences in nutritional plasticity are largely eliminated when dsx expression is knocked down (vector correlations close to 1). This indicates that dsx function is required for the sexes to follow independent nutrition-sensitive development. (Online version in colour.)
3. Results and discussion
(a) . Transcriptional underpinnings of context-dependent tibia exaggeration
We sought to investigate the developmental mechanisms mediating context-dependent exaggeration of homologous traits. To do so, we assessed the transcriptomic and functional genetic underpinnings of sex-, nutrition- and segment-specific development of tibia size and shape in dung beetles. To explore whether trait enlargement driven by sexual dimorphism, nutritional plasticity or segment identity is governed by a set of ‘core exaggeration genes' or, alternatively, by discrete developmental processes, we compared gene expression patterns across homologous prepupal tibia tissues at the time point during which the prepupal tibia is reshaped into the adult leg. Of the 17 776 longest isoforms detected overall, 1220 (6.8%) showed differential expression (DE) in one or more of the three contrasts investigated. Nutritional plasticity caused the weakest change in tibia length (figure 1c) but affected the largest number of transcripts (n = 893; figure 1d,e). By contrast, the stronger morphological differences between fore and hind tibiae and male and female fore tibiae (figure 1c) were associated with much fewer DE transcripts (n = 337 and 106, respectively; figure 1d,e). This is in contrast with previous reports of a stronger effect of sexual dimorphism compared to nutritional plasticity across various types of tissues in beetles [17,20,40] and highlights that differences in the morphological exaggeration of (serially) homologous tissues do not necessarily mirror transcriptional disparity.
To assess whether trait exaggeration in different contexts relies on a shared set of developmental processes, we were particularly interested in differentially expressed transcripts overlapping across various contexts that give rise to an exaggerated morphology as well as those that overlap in contexts that generate less exaggerated morphology. We thus compared transcripts differentially expressed uniquely in conditions that lead to a larger or smaller structure (figure 1d,e, respectively). If a common set of ‘exaggeration genes’ is responsible for the exaggerated growth of homologous tissues, we expected candidate genes directly linked to leg growth, tibia development and nutritional plasticity to be shared in all pairwise comparisons. In contrast with our expectation, only a small portion (n = 21, 3.7%; figure 1d) of all transcripts that covaried positively in expression level with trait size were shared across all three contrasts and, surprisingly, none of these transcripts had detectable orthologues in the genomes of Drosophila melanogaster, Tribolium castaneum or the close relative Onthophagus taurus. This indicates that exaggeration due to nutritional plasticity, sexual dimorphism and segment identity are likely underlain by disparate transcriptomic landscapes and, by extension, largely non-overlapping gene networks and developmental pathways. This is further reflected in that a majority of differentially expressed genes were entirely unique to a given contrast—that is, only 12.2% of all upregulated transcripts were contained in one of the three possible pairwise overlaps. However, 64.4% (49/76) of all genes that were overexpressed in males relative to females (sexual dimorphism) exhibited the same direction in one of the other two contrasts (figure 1d). This adds to previous studies that suggest nutritional plasticity and sexual dimorphism share developmental underpinnings [19] but indicates that the degree of transcriptional overlap may depend on the specific contrasts in question. Lastly, when considering only genes that were upregulated in tissues giving rise to smaller adult structures, the overlap between contrasts was minimal (0.01%; figure 1e). Taken together, these findings indicate that some exaggeration contrasts recruit a similar set of putative ‘exaggeration genes’; yet, overall, the overlap in DE genes between contrasts was surprisingly small. This implies that the genes, gene networks and developmental pathways involved in tibia exaggeration are strongly dependent on the developmental context—a finding inconsistent with the ‘core exaggeration genes' hypothesis. By extension, these results raise the possibility that differential exaggeration is not caused by the accumulation of additive effects in multiple contrasts, but possibly by context-dependent expression of a developmental master mediator gene able to integrate various sources of information in a sex-, segment- and nutrition-dependent manner.
(b) . Doublesex acts as functional integrator of sex-, segment- and environment-specific exaggeration
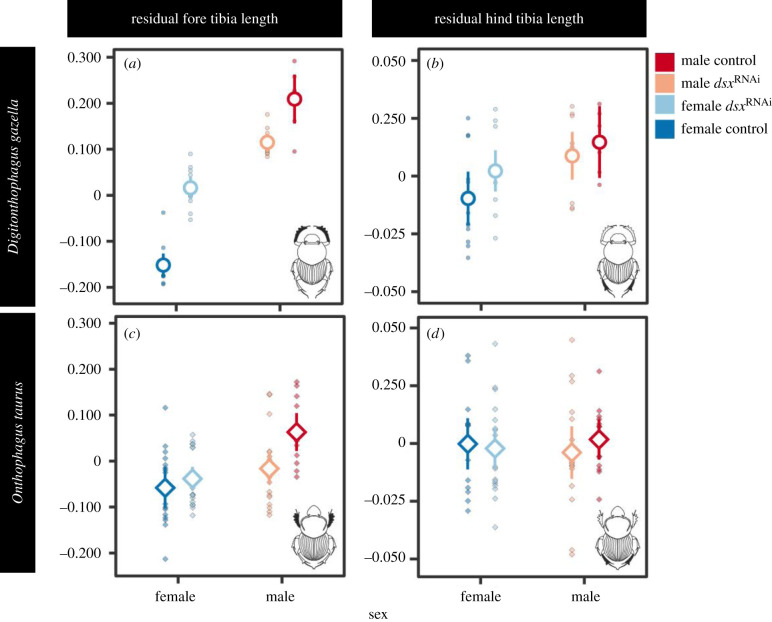
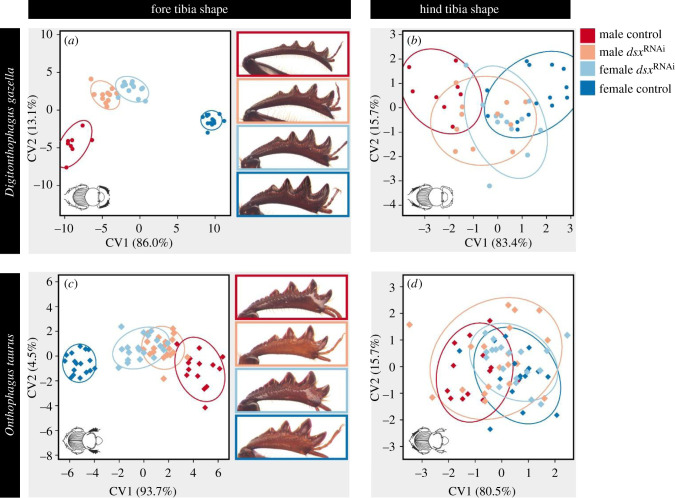
One of the genes that showed differential expression in two of our three focal contrasts was the somatic sex-determination gene doublesex (dsx). Specifically, dsx was upregulated in the fore relative to the hind tibia of D. gazella and exhibited higher expression in the fore tibia of males with access to high-quality nutrition compared to low nutrition males. Previous work showed that strictly sex-specific splice variants underpin sexual dimorphism in several species, including the close relatives Onthophagus taurus and O. sagittarius [14,33]. When re-analysing differential expression to differentiate individual isoforms (electronic supplementary material, figure S1; note that by restricting our initial analysis to the longest isoforms male- and female-specific splicing variants are masked in figure 1d,e) we, too, are able to document sex-specific DE in 3 out of the 5 dsx isoforms detected in D. gazella. dsx therefore emerged as a likely candidate simultaneously mediating tissue exaggeration associated with sexual dimorphism (via the expression of sex-specific isoforms), nutritional plasticity (via upregulation in the fore tibia of high nutrition males) and segment identity (via upregulation in fore compared to hind tibiae of the same individual). To begin assessing this possibility experimentally, we perturbed dsx function through RNAi-mediated transcript depletion. Systemic dsx expression knockdown of all known isoforms in males and females reared on standardized laboratory conditions [8] revealed that dsx expression results in the exaggeration of fore tibia length in males but the reduction of fore tibia length in females (sex × injection interaction: F1,39 = 184.88, p < 0.001, n = 45; electronic supplementary material, table S3; figure 2a). dsxRNAi further revealed strong sex-specific effects on fore tibia shape (Procrustes ANOVA, sex × injection interaction: F1,39 = 41.65, p < 0.001, n = 45; electronic supplementary material, table S4; CVA cross-validation success: 91.1%; figure 3a), reducing sexual dimorphism as measured in unit Procrustes distance in fore tibia shape from 0.20 to 0.05 (TA: p < 0.001). By contrast, even though dsx knockdown also reduced sexual shape dimorphism in the hind tibia (Procrustes ANOVA, sex × injection interaction: F1,40 = 3.21, p = 0.010, n = 45; electronic supplementary material, table S4; figure 3b), compared to the fore tibia, this effect was much weaker (CVA cross-validation success: 53.3%). These results suggest that dsx regulates both sex- and segment-specific exaggeration of tibial size and shape.
Figure 2.
RNAi-mediated systemic knockdowns of doublesex reduce fore (a) and hind tibia (b) length in male D. gazella, but has the opposite effect in females. These sexually antagonistic phenotypic effects are also evident in the fore tibia of the closely related O. taurus (c), but there was no effect on hind tibiae (d). Residuals were extracted from a linear regression of log tibia length against log pronotum width. Bars represent 95% CIs. (Online version in colour.)
Figure 3.
(a) Doublesex expression knockdown in D. gazella drastically reduces sexual shape dimorphism in fore tibiae (pictures indicate phenotypes of each treatment group). (b) Similar yet much weaker effects were found for hind tibia morphology. (c) dsxRNAi also strongly affected sex-specific fore tibia shape in O. taurus, while having (d) no effect on the shape of hind tibiae. (Online version in colour.)
Next, we sought to test whether dsx also mediates nutrition-dependent exaggeration of tibial development. To do so, we reared larvae in a strictly controlled, fully factorial design crossing nutritional manipulation with the RNAi-mediated downregulation of dsx expression [41]. This additional round of dsxRNAi treatments confirmed results reported above regarding sexual dimorphism in fore tibia morphology, and in addition, demonstrated that the dsxRNAi-mediated reduction of sexual dimorphism in fore tibia size was particularly pronounced among individuals that had access to a high-quality diet (electronic supplementary material, figure S2). This was driven by a reduction of allometric slopes in males by dsxRNAi (from 1.55 to 1.46) and an increase in the slope in females from 1.03 to 1.37 (sex × log body size × injection interaction: F1,93.4 = 10.36, p = 0.002, n = 102). Correspondingly, the dsxRNAi-mediated reduction of sexual shape dimorphism was stronger among individuals with access to high-quality nutrition (figure 4; electronic supplementary material, figure S3; Procrustes ANOVA: sex × nutrition × injection interaction: F1,93 = 3.81, p = 0.009, n = 102). Furthermore, dsxRNAi largely eliminated sex differences in nutritional plasticity of fore tibia shape (increasing vector correlations between sexes from 0.43 to 0.97; figure 4). These findings indicate that dsx expression mediates the exaggeration of fore tibia size and shape across all three focal contexts: sexual dimorphism, segment-specific differentiation, as well as nutrition-responsive growth. Previous studies have shown that dsx's target repertoires are highly sex- and tissue-specific [33], thus, our results support the hypothesis that dsx functions as a master mediator able to differentially employ discrete sets of target genes in different exaggeration contexts via the nutrition- and tissue-specific expression of sex-specific isoforms. Next, we sought to test whether changes in doublesex function may contribute to evolutionary divergences among species.
(c) . Doublesex-mediated context-dependent trait exaggeration is evolutionarily labile
To assess if and to what extent dsx-mediated trait exaggeration contributes to species divergences, we compared dsx knockdown phenotypes in D. gazella to those in Onthophagus taurus, a species of dung beetle belonging to the same tribe (Onthophagini) which possesses much stouter and less sexually dimorphic fore tibiae. As in D. gazella, dsx knockdown led to a reduction in fore tibia length in O. taurus males while having the opposite effect in females (sex-by-treatment interaction: F1,62 = 73.41, p < 0.001, n = 69; electronic supplementary material, table S5). dsx knockdown also affected sex-specific shape exaggeration in the fore tibia (sex-by-injection interaction: F1,61 = 25.20, p < 0.001, n = 69; electronic supplementary material, table S6; CVA cross-validation success: 85.5%), but there was little to no effect on hind tibia shape (sex-by-injection interaction: F1,64 = 0.84, p = 0.496; electronic supplementary material, table S6; CVA cross-validation success: 39.1%). While the directionality of dsxRNAi effects were similar in both species, knockdown phenotypes were considerably stronger in D. gazella (sex × injection × species interaction for tibia length: F1,102 = 62.05, p < 0.001, n = 114; electronic supplementary material, table S7, and shape: F1,100 = 2.20, p = 0.028, n = 114; TA: p < 0.001; electronic supplementary material, table S8). This is opposite to dsxRNAi effects in cephalic horns [8], suggesting that species differences in RNAi penetrance cannot explain the observed differences between the two species. Furthermore, dsxRNAi did not show species-specific effects for hind tibia length (F1,102 = 0.85, p = 0.347, n = 114; electronic supplementary material, table S7) or shape (F1,102 = 0.95, p = 0.150, n = 114; electronic supplementary material, table S8). Collectively, these findings suggest that interspecific divergences in sexual dimorphism in fore tibia exaggeration between two closely related dung beetle species are largely driven by diversification in sex- and segment-specific dsx function during tibia ontogeny. Whether this functional shift is enabled by divergences in the regulation of dsx expression or changes in how DSX interacts with downstream targets, however, requires further scrutiny.
(d) . Dsx mediates sex-specific nutritional plasticity in legs, heads, horns and thorax
In addition to dsx's function in instructing context-dependent development in tibia shape and size, our data also allowed us to document its effect on sexual shape dimorphism in diverse other traits, in particular horns, heads and the dorsal prothorax (figure 4a–d; electronic supplementary material, figures S3–S5). Paralleling our results for fore tibia development detailed above, this reduction was especially pronounced in horn and thorax shape of individuals that had access to a high-quality diet (sex × injection × nutritional quality interactions: p < 0.005). Comparing the vectors of shape change associated with nutritional plasticity between sexes revealed that dsx knockdown largely eliminates sex differences in nutritional plasticity in the shape of tibiae, thorax and horns, but not of the shape of the anterior head (figure 5). Taken together, doublesex therefore emerges as a ubiquitous mediator of tissue-specific sexual dimorphism by modulating sex-specific nutritional plasticity in the shape of diverse traits. More generally, these findings highlight the significance of developmental plasticity in generating sexual dimorphism in overall size and shape.
(e) . Condition-dependent sexual dimorphism and the resolution of sexual conflict
The evolution of sexual dimorphism requires the resolution of intralocus sexual conflict generated by sexually antagonistic selection acting on a genome largely shared between sexes [7]. Intralocus conflict is best understood in horns of O. taurus where long horns benefit large males but are exceedingly costly in females and small males due to manoeuvrability costs [43]. Resolving this conflict requires horn exaggeration to be nutrition-sensitive, sex-limited, as well as trait-specific. Our findings support the hypothesis that the sex-specific splice variants of a promiscuous transcription factor, dsx, generate the sex-limited environment-sensitive mechanism necessary to resolve sexual conflict across nutritional contexts in a trait-specific manner as predicted by theory [19]. That is, by virtue of the nutrition-dependent expression of sex-specific isoforms, males and females are enabled to develop divergent phenotypes depending on the environment they experience. Although the sex-specific fitness consequences of tibia and thorax shape remain to be fully determined in our study species (but see [44,45]), trait exaggeration in insects is generally costly [46,47], underscoring dsx's potential to facilitate conflict resolution. As dsx is highly conserved [48,49], including the expression of strictly sex-specific isoforms (e.g. Diptera [50], Lepidoptera [51,52], Hymenoptera [41] and Coleoptera [14,16]) and commonly exhibits a high degree of tissue specificity (e.g. [53]), dsx is likely to contribute to the resolution of trait-specific sexual conflict in a broad range of arthropods.
4. Conclusion
Optimal trait expression often depends on environmental and genetic factors. Developmental systems thus need to integrate various sources of intrinsic and extrinsic information. We found that exaggeration in tibia size and shape in different contexts is unlikely to be driven by a shared set of ‘exaggeration genes’. Instead, our findings are consistent with the hypothesis that dsx expression integrates tissue-, sex- and environment-dependent information and thereby instructs context-dependent exaggeration to varying degrees throughout the organism. In doing so, it serves as a mechanism generating sex-specific interactions that aid in resolving trait- and environment-specific sexual conflict. These interactions evolve across species and are probably common in arthropods.
Supplementary Material
Acknowledgements
We thank Erik Parker for collecting beetles in the wild and Anna Macagno, Kayla Copper and Levi Burdine with help in maintaining beetle colonies.
Data accessibility
Data underlying the analysis presented in this manuscript are provided as electronic supplementary material. Raw reads are deposited at https://www.ncbi.nlm.nih.gov/sra/PRJNA718544.
The data are provided in the electronic supplementary material [54].
Authors' contributions
P.T.R.: conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology, visualization, writing-original draft, writing-review and editing; D.M.L.: conceptualization, formal analysis, investigation, methodology, writing-original draft, writing-review and editing; A.P.M.: conceptualization, project administration, resources, writing-review and editing
All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Competing interests
We declare we have no competing interests.
Funding
This work was supported by an Early Postdoc.Mobility fellowship by the Swiss National Science Foundation (grant no. P2ZHP3_184003 and P400PB_199257 to P.T.R.). Additional support was provided by National Science Foundation (grant nos. IOS 1256689 and 1901680) as well as grant 61369 from the John Templeton Foundation. The opinions, interpretations, conclusions and recommendations are those of the authors and are not necessarily endorsed by the National Science Foundation, or the John Templeton Foundation.
References
- 1.Thompson DAW. 1917. On growth and form. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 2.Andersson B. 1994. Sexual selection. Princeton, NJ: Princeton Universty Press. [Google Scholar]
- 3.Lavine L, Gotoh H, Brent CS, Dworkin I, Emlen DJ. 2015. Exaggerated trait growth in insects. Annu. Rev. Entomol. 60, 453-472. ( 10.1146/annurev-ento-010814-021045) [DOI] [PubMed] [Google Scholar]
- 4.Kojima T. 2017. Developmental mechanism of the tarsus in insect legs. Curr. Opin. Insect Sci. 19, 36-42. ( 10.1016/j.cois.2016.11.002) [DOI] [PubMed] [Google Scholar]
- 5.Casares F, Mann RS. 1998. Control of antennal versus leg development in Drosophila. Nature 392, 723-726. ( 10.1038/33706) [DOI] [PubMed] [Google Scholar]
- 6.Emlen DJ. 2014. Reproductive contests and the evolution of extreme weaponry. In The evolution of insect mating systems (eds Shuker D, Simmons L), pp. 92-105. Oxford, UK: OUP. [Google Scholar]
- 7.Arnqvist G, Rowe GAL, Rowe L. 2005. Sexual conflict. Princeton, NJ: Princeton University Press. [Google Scholar]
- 8.Casasa S, Zattara EE, Moczek AP. 2020. Nutrition-responsive gene expression and the developmental evolution of insect polyphenism. Nat. Ecol. Evol. 4, 970-978. ( 10.1038/s41559-020-1202-x) [DOI] [PubMed] [Google Scholar]
- 9.Moczek AP, et al. 2011. The role of developmental plasticity in evolutionary innovation. Proc. R. Soc. B 278, 2705-2713. ( 10.1098/rspb.2011.0971) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mahfooz NS, Li H, Popadic A. 2004. Differential expression patterns of the hox gene are associated with differential growth of insect hind legs. PNAS 101, 4877-4882. ( 10.1073/pnas.0401216101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emlen DJ, Szafran Q, Corley LS, Dworkin I. 2006. Insulin signaling and limb-patterning: candidate pathways for the origin and evolutionary diversification of beetle ‘horns’. Heredity 97, 179-191. ( 10.1038/sj.hdy.6800868) [DOI] [PubMed] [Google Scholar]
- 12.Warren IA, Gotoh H, Dworkin IM, Emlen DJ, Lavine LC. 2013. A general mechanism for conditional expression of exaggerated sexually-selected traits. Bioessays 35, 889-899. ( 10.1002/bies.201300031) [DOI] [PubMed] [Google Scholar]
- 13.Casasa S, Moczek AP. 2018. Insulin signalling's role in mediating tissue-specific nutritional plasticity and robustness in the horn-polyphenic beetle Onthophagus taurus. Proc. R. Soc. B 285, 20181631. ( 10.1098/rspb.2018.1631) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kijimoto T, Moczek AP, Andrews J. 2012. Diversification of doublesex function underlies morph-, sex-, and species-specific development of beetle horns. PNAS 109, 20 526-20 531. ( 10.1073/pnas.1118589109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gotoh H, Miyakawa H, Ishikawa A, Ishikawa Y, Sugime Y, Emlen DJ, Lavine LC, Miura T. 2014. Developmental link between sex and nutrition; doublesex regulates sex-specific mandible growth via juvenile hormone signaling in stag beetles. PLoS Genet. 10, e1004098. ( 10.1371/journal.pgen.1004098) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ito Y, et al. 2013. The role of doublesex in the evolution of exaggerated horns in the Japanese rhinoceros beetle. EMBO Rep. 14, 561-567. ( 10.1038/embor.2013.50) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ledon-Rettig CC, Moczek AP. 2016. The transcriptomic basis of tissue- and nutrition-dependent sexual dimorphism in the beetle Onthophagus taurus. Ecol. Evol. 6, 1601-1613. ( 10.1002/ece3.1933) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wyman MJ, Agrawal AF, Rowe L. 2010. Condition-dependence of the sexually dimorphic transcriptome in Drosophila melanogaster. Evolution 64, 1836-1848. ( 10.1111/j.1558-5646.2009.00938.x) [DOI] [PubMed] [Google Scholar]
- 19.Bonduriansky R. 2007. The genetic architecture of sexual dimorphism: the potential roles of genomic imprinting and condition-dependence. In Sex, size and gender roles (eds Fairbairn DJ, Blanckenhorn WU, Székely T), pp. 176-184. Oxford, UK: Oxford University Press. [Google Scholar]
- 20.Zinna R, Emlen D, Lavine LC, Johns A, Gotoh H, Niimi T, Dworkin I. 2018. Sexual dimorphism and heightened conditional expression in a sexually selected weapon in the Asian rhinoceros beetle. Mol. Ecol. 27, 5049-5072. ( 10.1111/mec.14907) [DOI] [PubMed] [Google Scholar]
- 21.Rohner PT, Teder T, Esperk T, Lüpold S, Blanckenhorn WU. 2018. The evolution of male-biased sexual size dimorphism is associated with increased body size plasticity in males. Functional Ecol. 32, 581-591. [Google Scholar]
- 22.Rohner PT, Blanckenhorn WU. 2018. A comparative study of the role of sex-specific condition dependence in the evolution of sexually dimorphic traits. Am. Nat. 192, E202-E215. ( 10.1086/700096) [DOI] [PubMed] [Google Scholar]
- 23.Shafiei M, Moczek AP, Nijhout HF. 2001. Food availability controls the onset of metamorphosis in the dung beetle Onthophagus taurus (Coleoptera : Scarabaeidae). Physiol. Entomol. 26, 173-180. ( 10.1046/j.1365-3032.2001.00231.x) [DOI] [Google Scholar]
- 24.Moczek AP, Nijhout HF. 2002. A method for sexing final instar larvae of the genus Onthophagus Latreille (Coleoptera: Scarabaeidae). Coleopterists Bullet. 56, 279-284. ( 10.1649/0010-065X(2002)056[0279:AMFSFI]2.0.CO;2) [DOI] [Google Scholar]
- 25.Linz DM, Moczek AP. 2020. Integrating evolutionarily novel horns within the deeply conserved insect head. BMC Biol. 18, 41. ( 10.1186/s12915-020-00773-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. See https://github.com/s-andrews/FastQC.
- 27.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114-2120. ( 10.1093/bioinformatics/btu170) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grabherr MG, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644-652. ( 10.1038/nbt.1883) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210-3212. ( 10.1093/bioinformatics/btv351) [DOI] [PubMed] [Google Scholar]
- 30.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357-359. ( 10.1038/nmeth.1923) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 12, 323. ( 10.1186/1471-2105-12-323) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. ( 10.1186/s13059-014-0550-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ledón-Rettig CC, Zattara EE, Moczek AP. 2017. Asymmetric interactions between doublesex and tissue- and sex-specific target genes mediate sexual dimorphism in beetles. Nat. Commun. 8, 14593. ( 10.1038/ncomms14593) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rohlf FJ. 2009. Tpsdig. Version 2.14. Stony Brook, NY: Department of Ecology and Evolution, State University of New York at Stony Brook. [Google Scholar]
- 35.Adams DC, Collyer ML, Kaliontzopoulou A. 2020. Geomorph:Software for geometric morphometric analyses. R package version 3.2.1.
- 36.Klingenberg CP. 2016. Size, shape, and form: concepts of allometry in geometric morphometrics. Dev. Genes Evol. 226, 113-137. ( 10.1007/s00427-016-0539-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schlager S. 2017. Morpho and Rvcg - Shape Analysis in R. In Statistical shape and deformation analysis: methods, implementation and applications (eds Zheng G, Li S, Szekely G), pp. 217-256. San Diego, CA: Elsevier Science. [Google Scholar]
- 38.Adams DC, Collyer ML. 2009. A general framework for the analysis of phenotypic trajectories in evolutionary studies. Evolution 63, 1143-1154. ( 10.1111/j.1558-5646.2009.00649.x) [DOI] [PubMed] [Google Scholar]
- 39.Drake AG, Klingenberg CP. 2008. The pace of morphological change: historical transformation of skull shape in St Bernard dogs. Proc. R. Soc. B 275, 71-76. ( 10.1098/rspb.2007.1169) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kijimoto T, Snell-Rood EC, Pespeni MH, Rocha G, Kafadar K, Moczek AP. 2014. The nutritionally responsive transcriptome of the polyphenic beetle Onthophagus taurus and the importance of sexual dimorphism and body region. Proc. R. Soc. B 281, 20142084. ( 10.1098/rspb.2014.2084) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rohner PT. In press. A role for sex-determination genes in life history evolution? Doublesex mediates sexual size dimorphism in the gazelle dung beetle. J. Evol. Biol. ( 10.1111/jeb.13877) [DOI] [PubMed] [Google Scholar]
- 42.Cho S, Huang ZY, Zhang J. 2007. Sex-specific splicing of the honeybee doublesex gene reveals 300 million years of evolution at the bottom of the insect sex-determination pathway. Genetics 177, 1733-1741. ( 10.1534/genetics.107.078980) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moczek AP, Emlen DJ. 2000. Male horn dimorphism in the scarab beetle, Onthophagus taurus: do alternative reproductive tactics favour alternative phenotypes? Anim. Behav. 59, 459-466. ( 10.1006/anbe.1999.1342) [DOI] [PubMed] [Google Scholar]
- 44.Macagno AL, Moczek AP, Pizzo A. 2016. Rapid divergence of nesting depth and digging appendages among tunneling dung beetle populations and species. Am. Nat. 187, E143-E151. ( 10.1086/685776) [DOI] [PubMed] [Google Scholar]
- 45.Linz DM, Hu Y, Moczek AP. 2019. The origins of novelty from within the confines of homology: the developmental evolution of the digging tibia of dung beetles. Proc. R. Soc. B 286, 20182427. ( 10.1098/rspb.2018.2427) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O'Brien DM, et al. 2019. Muscle mass drives cost in sexually selected arthropod weapons. Proc. R. Soc. B 286, 20191063. ( 10.1098/rspb.2019.1063) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Emlen DJ. 2001. Costs and the diversification of exaggerated animal structures. Science 291, 1534-1536. ( 10.1126/science.1056607) [DOI] [PubMed] [Google Scholar]
- 48.Shukla JN, Nagaraju J. 2010. Doublesex: a conserved downstream gene controlled by diverse upstream regulators. J. Genet. 89, 341-356. ( 10.1007/s12041-010-0046-6) [DOI] [PubMed] [Google Scholar]
- 49.Price DC, Egizi A, Fonseca DM. 2015. The ubiquity and ancestry of insect doublesex. Sci. Rep. 5, 13068. ( 10.1038/srep13068) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baker BS, Wolfner MF. 1988. A molecular analysis of doublesex, a bifunctional gene that controls both male and female sexual differentiation in Drosophila melanogaster. Genes Dev. 2, 477-489. ( 10.1101/gad.2.4.477) [DOI] [PubMed] [Google Scholar]
- 51.Ohbayashi F, Suzuki MG, Mita K, Okano K, Shimada T. 2001. A homologue of the Drosophila doublesex gene is transcribed into sex-specific mRNA isoforms in the silkworm, Bombyx mori. Comp. Biochem. Phys. B 128, 145-158. ( 10.1016/S1096-4959(00)00304-3) [DOI] [PubMed] [Google Scholar]
- 52.Prakash A, Monteiro A. 2020. Doublesex mediates the development of sex-specific pheromone organs in Bicyclus butterflies via multiple mechanisms. Mol. Biol. Evol. 37, 1694-1707. ( 10.1093/molbev/msaa039) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rice GR, Barmina O, Luecke D, Hu K, Arbeitman M, Kopp A. 2019. Modular tissue-specific regulation of doublesex underpins sexually dimorphic development in Drosophila. Development 146, dev178285. ( 10.1242/dev.178285) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rohner PT, Linz DM, Moczek AP. 2021. Data from: Doublesex mediates species-, sex-, environment- and trait-specific exaggeration of size and shape. Figshare. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Rohner PT, Linz DM, Moczek AP. 2021. Data from: Doublesex mediates species-, sex-, environment- and trait-specific exaggeration of size and shape. Figshare. [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
Data underlying the analysis presented in this manuscript are provided as electronic supplementary material. Raw reads are deposited at https://www.ncbi.nlm.nih.gov/sra/PRJNA718544.
The data are provided in the electronic supplementary material [54].