1. Introduction
Peto et al. [1] suggest that weekly testing of the entire UK population for SARS-CoV-2 combined with household quarantine could rapidly end the spread of COVID-19. The logistical challenges in conducting and processing 10 million tests per day over an extended period are formidable [2]. Even if these challenges could be overcome, it is not obvious that this strategy would reduce the effective reproduction number Reff sufficiently to control the epidemic. Peto et al. [1] support their claim with a statistical footnote. However, this does not actually calculate the proportion of transmissions that would be prevented, nor allow for high false-negative rates before symptom onset [3]. They also assume that one-third of transmissions are within households, which is likely to be an overestimate (see below).
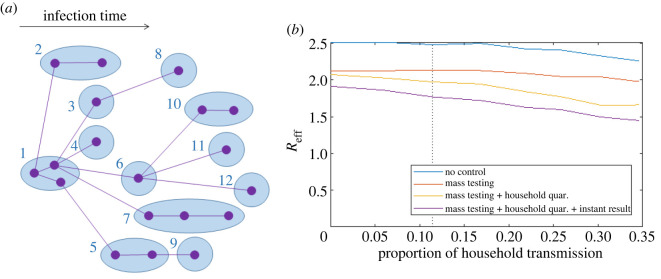
We use a simple household model of COVID-19 transmission (figure 1a), combined with data on reverse transcriptase polymerase chain reaction (RT-PCR) test sensitivity [3], to estimate the reduction in Reff from mass weekly testing and household quarantine. Even with optimistic assumptions about the test processing time (1 day) and the effectiveness of home isolation and quarantine (100% effective), this strategy would only reduce Reff by around 21%. If R0 = 2.5 in the absence of interventions, mass testing reduces Reff to 2.1 and the addition of household quarantine further reduces Reff to 2.0. Under more realistic parameter settings, such as partially effective quarantine [5] or incomplete testing coverage, Reff would be larger than this. Rapid antigen tests can provide faster turnaround of test results. While this may appear to be a solution to some of the problems outlined here, removing the assumed 1 day delay from test to result only reduces Reff to 1.8. In addition, these tests typically have lower sensitivity than RT-PCR tests [6], and relying solely on self-testing may compromise adherence to isolation and quarantine.
Figure 1.
(a) Household branching process model for COVID-19 transmission. Each household (blue ellipses) is randomly assigned a number of members between 1 and 8 according to the distribution of household sizes for England and Wales [4] (2011 census, all usual residents aged 18 and over in households). Households are numbered in order of the time of first infection; only infected individuals and their transmission routes are shown in the diagram. Once a household is placed in quarantine, all further onward transmission from all individuals in that household is prevented. (b) Effective reproduction number Reff with no control (blue), under mass testing (red) and mass testing plus household quarantine (yellow), and mass testing, household quarantine and instant return of test results (ttest = 0, purple) for varying proportions of household transmission. This was done by varying the household secondary attack rate SARhome between 0% (no household transmission) and 70% (33% of all transmission is within households) while fixing R0 = 2.5. Vertical dotted line shows the results when household secondary attack rate is 20%. Results are averaged over 200 independent realizations of the model, each initialized with one infected seed case and run for 30 days.
The results for Reff above assume that the household secondary attack rate (SARhome) is 20%, which means that around 12% of transmission is within households. This is at the upper end of empirically derived estimates [7–12] but lower than assumed by Peto et al. [1]. Since the proportion of household transmission may vary in different communities, we calculated Reff over a range of values of SARhome while keeping R0 fixed at 2.5 (figure 1b). Similar results could be obtained by varying the mean household size. This shows that, even when one-third of transmission is within households (SARhome = 70%), mass testing and quarantine reduce Reff to 1.6 at best.
For simplicity, we ignored individual heterogeneity in non-household transmission. To test this assumption, we simulated the model with a negative binomial distribution for the number of non-household secondary cases, with values of the overdispersion parameter k = 1, 0.5 and 0.1 [13]. These values are in the range of estimates for COVID-19 transmission [14–16]. Individual heterogeneity modelled in this way affects the dynamics of small outbreaks and increases the probability of extinction for an outbreak from a single seed case at a given reproduction number [13]. However, our focus here is on the effect of a particular control measure on a large epidemic, in which case the effective reproduction number is the most important measure. Although heterogeneity increases variability in individual reproduction numbers, we found that Reff was insensitive to introducing heterogeneity and varying the value of k (table 1) [13]. Including age-specific contact patterns would allow the spread of the outbreak through different age groups to be modelled, but is unlikely to dramatically change the impact of the interventions examined on Reff.
Table 1.
Effective reproduction number Reff under different control measures and with differing levels of individual heterogeneity in transmission rates (Poisson distribution has no heterogeneity, negative binomial distributions with smaller values of k have more heterogeneity). Results are for SARhome = 0.2 and are averaged over 500 independent realizations of the model, each initialized with one infected seed case and run for 30 days.
| no control | mass testing | mass testing + HHQ | mess testing + HHQ + instant | |
|---|---|---|---|---|
| Poisson (Rout) | 2.49 | 2.12 | 2.01 | 1.78 |
| NegBin (Rout, k = 1) | 2.50 | 2.11 | 1.99 | 1.77 |
| NegBin (Rout, k = 0.5) | 2.51 | 2.13 | 2.01 | 1.78 |
| NegBin (Rout, k = 0.1) | 2.49 | 2.09 | 2.05 | 1.82 |
Rapid and widely available testing is undoubtedly a critical part of the response to COVID-19. However, it is dangerous to assert that particular interventions will be sufficient to control the spread of the virus without supporting evidence. A simple model shows that mass weekly testing and household quarantine, even if it were perfectly achievable, would not be sufficient to control the spread of COVID-19. This is due to a combination of significant pre-symptomatic transmission [22,23], low test sensitivity prior to symptom onset [3], delays in processing tests and returning results, and substantial non-household transmission. This implies that other measures would also need to be added. Given the logistical barriers to a programme of population-wide weekly testing, focusing resources on more targeted controls, such as symptom-based testing combined with rapid tracing and quarantining of contacts, is preferable to an indiscriminate mass testing approach.
2. Methods
Table 2 shows data sources, parameter values and references. We assume that in the absence of any interventions R0 = 2.5. This is split into non-household transmission Rout and household transmission where SARhome is the household secondary attack rate and is the average number of household contacts [4]. Each household is randomly assigned a size according to the proportion of the population living in a household of size 1 to 8 in England and Wales [4] (2011 census, all usual residents aged 18 and over in households). In the absence of interventions, each clinical case has a probability SARhome of infecting each other member of the household. Each clinical case also infects an independent Poisson distributed number of people outside the household with mean Rout. To model individual heterogeneity in transmission rates, we ran simulations where the number of secondary cases outside the household was instead drawn from a negative binomial distribution with mean Rout and overdispersion parameter k. We assume no individual can be re-infected, so each household transmission reduces the susceptible pool within that household. New cases infected outside the household are all assigned to new households, which are assumed to be fully susceptible initially. Times of secondary infections are distributed according to a Weibull distribution, time-shifted such that 35% of onward transmission occurs before symptom onset. We assume 33% of infections are subclinical and these are 50% as infectious as clinical cases and have 65% test sensitivity.
Table 2.
Model parameter values, data sources and references.
| parameter |
value |
Source | |||||||
|---|---|---|---|---|---|---|---|---|---|
| distribution of generation times (days) | 1.3 + Weibull (scale = 5.67, shape = 2.83) | [17,18] | |||||||
| basic reproduction number | R0 = 2.5 | assumed | |||||||
| relative infectiousness of subclinical cases | csub = 0.5 | [19,20] | |||||||
| relative onward transmission from isolated or quarantined cases | cisol = 0 | perfect isolation and quarantine assumed | |||||||
| proportion of subclinical infections | psub = 0.33 | [20] | |||||||
| relative test sensitivity for subclinical infections | ptestsubclin = 0.65 | [21] | |||||||
| household secondary attack rate | SARhome = 0.2 (other values simulated in figure 1b) | [12] | |||||||
| time from testing to return of test result | ttest = 1 day | estimated | |||||||
| test sensitivity (probability of an infected individual testing positive) as a function of the number of days (0–38) since infection. Sensitivity is assumed to be zero more than 38 days after infection. | 0 | 1 | 2 | 3 | 4 | 5 | 6 | [3] | |
| 0.00 | 0.00 | 0.01 | 0.04 | 0.33 | 0.62 | 0.75 | |||
| 7 | 8 | 9 | 10 | 11 | 12 | 13 | |||
| 0.79 | 0.80 | 0.79 | 0.76 | 0.72 | 0.68 | 0.64 | |||
| 14 | 15 | 16 | 17 | 18 | 19 | 20 | |||
| 0.60 | 0.56 | 0.52 | 0.48 | 0.44 | 0.40 | 0.37 | |||
| 21 | 22 | 23 | 24 | 25 | 26 | 27 | |||
| 0.34 | 0.32 | 0.30 | 0.28 | 0.26 | 0.24 | 0.22 | |||
| 28 | 29 | 30 | 31 | 32 | 33 | 34 | |||
| 0.20 | 0.18 | 0.16 | 0.14 | 0.12 | 0.10 | 0.08 | |||
| 35 | 36 | 37 | 38 | ||||||
| 0.06 | 0.04 | 0.02 | 0.00 | ||||||
| proportion of people living in a household of size 1 to 8 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | [4] |
| 17% | 36% | 19% | 17% | 7% | 3% | 0.7% | 0.6% | ||
To simulate mass testing, we assume that everyone is tested weekly and has a positive test probability that depends on the time since infection [3]. This probability is very low in the first 3 days after infection and increases to a peak of 80% in 8 days after infection. We assume that more than 21 days after infection (the longest time with available data), the positive test probability continues to decrease by 2 percentage points per day (table 2). We assume that it takes one day for the test result to be returned and, following a positive test result, all subsequent transmission from the positive case and, in the household quarantine scenario, all their household members is prevented. If the test is a false negative, the individual is retested 7 days later with an independent probability of testing positive.
Supplementary Material
Data accessibility
This article has no additional data.
Competing interests
We declare we have no competing interests.
Funding
The authors were funded by Te Pūnaha Matatini, New Zealand's Centre of Research Excellence in Complex Systems, and the New Zealand Ministry of Business, Innovation and Employment.
References
- 1.Peto J, et al. 2020. Weekly COVID-19 testing with household quarantine and contact tracing is feasible and would probably end the epidemic. R. Soc. Open Sci. 7, 200915. ( 10.1098/rsos.200915) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smeeth L. 2020. Should the UK really test 10 million people per day for COVID-19? London School of Hygiene & Tropical Medicine, Expert Opinion. Retrieved from: https://www.lshtm.ac.uk/newsevents/expert-opinion/should-uk-really-test-10-million-people-day-covid-19. [Google Scholar]
- 3.Kucirka LM, Lauer SA, Laeyendecker O, Boon D, Lessler J. 2020. Variation in false-negative rate of reverse transcriptase polymerase chain reaction–based SARS-CoV-2 tests by time since exposure. Ann. Intern. Med. 173, 262-267. ( 10.7326/M20-1495) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.CT0412_2011 Census – Age by household size by household composition by family status [Internet]. 2015 [cited 11 August 2020]. See https://www.ons.gov.uk/peoplepopulationandcommunity/housing/adhocs/004118ct04122011censusagebyhouseholdsizebyhouseholdcompositionbyfamilystatusnationaltoregion.
- 5.James A, et al. 2020. Successful contact tracing systems for COVID-19 rely on effective quarantine and isolation. medRxiv. https://www.medrxiv.org/content/10.1101/2020.06.10.20125013v.
- 6.Centers for Disease Control and Prevention. 2020. Interim Guidance for Rapid Antigen Testing for SARS-CoV-2. https://www.cdc.gov/coronavirus/2019-ncov/lab/resources/antigen-tests-guidelines.html.
- 7.Bi Q, et al. 2020. Epidemiology and transmission of COVID-19 in 391 cases and 1286 of their close contacts in Shenzhen, China: a retrospective cohort study. Lancet Infect. Dis. 20, 911-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng H-Y, Jian S-W, Liu D-P, Ng T-C, Huang W-T, Lin H-H. 2020. Contact tracing assessment of COVID-19 transmission dynamics in Taiwan and risk at different exposure periods before and after symptom onset. JAMA Intern. Med. 180, 1156-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jing Q-L, et al. 2020. Household secondary attack rate of COVID-19 and associated determinants in Guangzhou, China: a retrospective cohort study. Lancet Infect. Dis. 20, 1141-1150. ( 10.1016/S1473-3099(20)30471-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Y, et al. 2020. The epidemiological characteristics of infection in close contacts of COVID-19 in Ningbo city. Chin. J. Epidemiol. 41, 668-672. [DOI] [PubMed] [Google Scholar]
- 11.Luo L, et al. 2020. Modes of contact and risk of transmission in COVID-19 among close contacts. medRxiv. https://www.medrxiv.org/content/10.1101/2020.03.24.20042606v1.
- 12.Kucharski AJ, et al. 2020. Effectiveness of isolation, testing, contact tracing and physical distancing on reducing transmission of SARS-CoV-2 in different settings: a mathematical modelling study. Lancet Infect. Dis. 20, 1151-1160. ( 10.1016/S1473-3099(20)30457-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lloyd-Smith JO, Schreiber SJ, Kopp PE, Getz WM. 2005. Superspreading and the effect of individual variation on disease emergence. Nature 438, 355-359. ( 10.1038/nature04153) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Endo A, Abbott S, Kucharski AJ, Funk S. 2020. Estimating the overdispersion in COVID-19 transmission using outbreak sizes outside China. Wellcome Open Res. 5, 67. ( 10.12688/wellcomeopenres.15842.3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Riou J, Althaus CL. 2020. Pattern of early human-to-human transmission of Wuhan 2019 novel coronavirus (2019-nCoV), December 2019 to January 2020. Eurosurveillance 25, 2000058. ( 10.2807/1560-7917.ES.2020.25.4.2000058) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang L, et al. 2020. U1 snRNP regulates cancer cell migration and invasion in vitro. Nat. Commun. 11, 1-6. ( 10.1038/s41467-019-13993-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferretti L, et al. 2020. Quantifying SARS-CoV-2 transmission suggests epidemic control with digital contact tracing. Science 368, eabb6936. ( 10.1126/science.abb6936) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lauer SA, et al. 2020. The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann. Intern. Med. 172, 577-582. ( 10.7326/M20-0504) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davies NG, et al. 2020. Age-dependent effects in the transmission and control of COVID-19 epidemics. Nat. Med. 26, 1205-1211. ( 10.1038/s41591-020-0962-9) [DOI] [PubMed] [Google Scholar]
- 20.Davies NG, et al. 2020. Effects of non-pharmaceutical interventions on COVID-19 cases, deaths, and demand for hospital services in the UK: a modelling study. Lancet Public Health 5, E375-E385. ( 10.1016/S2468-2667(20)30133-X) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clifford S, et al. 2020. Strategies to reduce the risk of SARS-CoV-2 re-introduction from international travellers. medRxiv. https://www.medrxiv.org/content/10.1101/2020.07.24.20161281v2.
- 22.Ganyani T, Kremer C, Chen D, Torneri A, Faes C, Wallinga J, Hens N. 2020. Estimating the generation interval for coronavirus disease (COVID-19) based on symptom onset data, March 2020. Eurosurveillance 25, 2000257. ( 10.2807/1560-7917.ES.2020.25.17.2000257) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tindale L, et al. 2020. Transmission interval estimates suggest pre-symptomatic spread of COVID-19. eLife 9, e57149.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This article has no additional data.