Structured Abstract
Objectives:
1) To test the hypothesis that there would be proteomic differences in the composition of exosomes isolated from osteoclasts and odontoclasts and 2) to determine the clinical usefulness of these in vitro biomarker candidates.
Materials and Methods:
Mouse bone marrow-derived precursors were cultured on either dentin or bone slices and allowed to mature and begin resorption. Exosomes were isolated from cell culture media and characterized by mass spectrometry. The proteomic data obtained from this in vitro study were compared with the data obtained from human samples in our previous work.
Results:
There was a difference in the proteomic composition of exosomes from osteoclasts and odontoclasts. A total of 40 exosomal proteins were only present in osteoclast media, whereas six unique exosomal proteins were identified in odontoclast supernatants. Approximately 50% of exosomal proteins released by clastic cells in vitro can be found in oral fluids.
Conclusion:
Our data suggest that the mineralized matrix type plays a role in the final phenotypic characteristics of mouse clastic cells. Many in vitro biomarker candidates of bone and dentin resorption can also be found in human oral fluids, thus indicating that this approach may be a viable alternative in biomarker discovery.
Keywords: biomarker, exosomes, odontoclast, osteoclast
1 ∣. INTRODUCTION
The development of oral fluid-based tests as diagnostic tools or for monitoring of outcomes would be of tremendous benefit to both patients and dental care providers. Nevertheless, the field of “personalized dentistry” is moving slowly. The initial attempt to use biomarkers in gingival crevicular fluid (GCF) for discrimination of periodontal disease dates to the early 1970s.1 In orthodontics, the first attempts to detect biologically relevant GCF markers of bone remodelling and root resorption date back to the early 1990s2 and early 2000s,3 respectively. Despite these important initial attempts to identify biologically relevant markers, little progress has been made in developing point-of-care devices for testing oral fluids from patients.
Exploring oral fluids seems an attractive way to look at protein content; however, the broad range of concentrations of various molecules contained in these fluids is estimated to exceed ten orders of magnitude.4 Thus, it is possible that important biomarker candidates have not yet been identified because their concentration in oral fluids may be too low to be detected by conventional approaches. Recent research in the search for new biomarkers has revealed that many clinically relevant proteins are lodged in small extracellular vesicles called exosomes.5 Thus, by isolating exosomes, it could be possible to overcome the dynamic range challenge and facilitate characterization of markers that might more accurately represent ongoing biological processes. Moreover, the use of less complex samples, such as cell and/or tissue culture supernatants, may be helpful in the initial discovery phase.6,7
The major purpose of this article is to compare the protein profiles of exosomes derived from cells resorbing bone (“osteoclasts”) and dentin (“odontoclasts”). In addition, we compared this in vitro data with proteomic data derived from our previously published human GCF study8 to identify novel proteins that may be used as biomarkers of bone and dentin resorption in patients. We hypothesize that there will be a difference in the proteomic composition of exosomes from osteoclasts and odontoclasts; thus, this could be the first step in the discovery of biomarkers that would allow distinction between bone remodelling and dentin resorption.
2 ∣. MATERIAL AND METHODS
2.1 ∣. Cell culture
Adhering to all relevant Institutional Animal Care and Use Committee (IACUC) regulations at the University of Florida, progenitor cells were collected via extraction of bone marrow from the femurs of female C57BL/6 mice at 4-8 weeks of age by flushing with α-MEM (2.5 mL/bone) using a 10-mL syringe and 25-gauge needle. Mononuclear cells were plated at 0.5 x106 cells/well into treated 12-well cell culture plates containing either bone or dentin slices (Immunodiagnostic Systems, Gaithersburg, MD). Cells were cultured for 11 days in the presence of recombinant mouse M-CSF (20 ng/mL), recombinant soluble mouse RANKL (20 ng/mL), α-MEM (2 mL) and 10% exosome-depleted foetal bovine serum (Exo-FBS) (System Biosciences, Mountain View, CA). Conditioned media were replaced with fresh media on days 3 and 7. At each time point, visual inspection of clastic cell formation was carried out using a light microscope. On day 11, tartrate-resistant acid phosphatase (TRAP) activity was detected using the Leukocyte Acid Phosphatase Kit (Sigma-Aldrich; cat. no. 387AKT) following the instructions from the manufacturer. Resorbing activity of cells was confirmed by labelling the F-actin cytoskeleton with phalloidin as previously described.9 Experiments were performed in triplicate to reduce random error.
2.2 ∣. Exosome isolation
Conditioned media collected from osteoclasts and odontoclasts at day 7 were used for exosome characterization because this time point showed the higher number of multinuclear cells. Exosomes were isolated by differential centrifugation using ExoQuick-TC (System Biosciences, Mountain View, CA) as described previously.10 Aliquots of 10 μL were used for exosome quantitation using the manufacturer protocols in the EXOCET Quantitation Kit (System Biosciences, Mountain View, CA). Data were quantitated from the standard curve, and statistical analysis was determined by two-tailed t tests at 95% confidence interval. The remaining samples were centrifuged for two hours and the pellet frozen at −80°C for future analysis.
2.3 ∣. Sample digestion
Tryptic digests of exosome isolates were prepared by solubilizing exosome pellets in 200 μL of 1 M urea/0.2 M Tris/HCl buffer pH 7.6. Lysates were transferred into 0.5-mL Amicon filter units (10 kDa cut-off) and subjected to filter-aided sample preparation (FASP) digestion procedure with trypsin.11 Protein amounts were monitored using micro-BCA assay (Pierce, Rockford, IL). Resulting digest was acidified with trifluoroacetic acid (TFA) and purified by reversed-phase solid-phase extraction. Digested sample from each exosome isolate contained approximately 5-10 μg of the peptides (determined by NanoDrop 2000, Thermo Fisher). 1.5 μg of digested exosome protein extracts were subjected to one-dimensional liquid chromatography-tandem mass spectrometry (1D-LC-MS/MS) analysis.
2.4 ∣. LC-MS/MS settings
A splitless nano-flow LC system (Eksigent, Dublin, CA) was used with 500 nL/min flow rate and 100 μm×200 mm analytical column packed with 3-μm Luna C182 (Phenomenex, Torrance, CA). Samples (~1.5 μg of peptides in 10 μL of buffer A) were injected via a 300 μm×5 mm PepMap100 trap column. The gradient program included following steps: linear increase from 0.5 to 33% of buffer B (acetonitrile) in 108 minutes, 5-minutes column wash with 90% B and 7-min system equilibration using starting conditions of 0.5% B (120 minutes total analysis time). Both eluents A (water) and B (acetonitrile) contained 0.1% formic acid as ion-pairing modifier. Data-dependent acquisition TripleTOF 5600 mass spectrometer (Sciex, Concord, ON) was performed using following settings: 250 ms survey MS spectra (m/z 400-1500) followed by up to 20 MS/MS measurements on the most intense parent ions (300 counts/sec threshold, +2- +5 charge state, m/z 100-1500 mass range for MS/MS, 100 ms each, high sensitivity mode). Previously targeted parent ions were excluded from repetitive MS/MS acquisition for 12 seconds (100 mDa mass tolerance).
2.5 ∣. Data treatment and protein identification
Raw spectra files were converted into Mascot Generic File format (MGF) for protein identification by X!Tandem search algorithm (http://hs2.proteome.ca/tandem/thegpm_tandem_a.html). The following X!Tandem search parameters were used: 20 ppm and 50 ppm mass tolerance for parent and fragment ions, respectively; constant modification of Cys with iodoacetamide; default set post-translational modifications: oxidation of Met, Trp; N-terminal cyclization at Qln, Cys; N-terminal acetylation, phosphorylation (Ser, Thr, Tyr), deamidation (Asn and Gln); an expectation value cut-off of log(e)<–1 for both proteins and peptides. Protein quantitation was performed by calculating log(2) of the sum of intensity of all fragments from MS/MS spectra, which belong to a particular protein. Protein-level differences between osteoclast and odontoclast media were then mapped into Z-scores, that is the distance from the population mean in units of standard deviation. Proteins with Z-scores greater than 1.65 or smaller than −1.65 (the outermost 10% of the distribution) were considered upregulated in osteoclast or odontoclast media, respectively. As this experiment design did not include biological replicates, selecting the ∣Z∣>1.65 portion of the single difference distribution represented the most likely biologically interesting region.
3 ∣. RESULTS
3.1 ∣. Osteoclasts and odontoclasts secrete exosomes in vitro
Both cell types secrete exosomes in vitro. While not statistically significant (P>.05), there was a trend for higher numbers of exosomes in media from osteoclasts in the three rounds of experiment (Table 1).
TABLE 1.
Exosome quantity in cell culture media measured with EXOCET® Kit
| No. of Exosomes×109 Osteoclast (bone) media |
No. of Exosomes×109 Odontoclast (dentin) media |
|
|---|---|---|
| Round 1 | 5.9134 | 2.384 |
| Round 2 | 5.1291 | 3.2173 |
| Round 3 | 10.848 | 9.4428 |
| Average (SD) | 7.296 (3.10) | 5.014 (3.857) |
3.2 ∣. Proteomic composition of exosomes suggests biological differences between osteoclasts and odontoclasts
Table 2 shows the identification summary for the samples on the protein and peptide levels. The protein identification scores are expressed in the log(e) (expectation value) scale: the lower scores correspond to more confident identifications. We identified 457 native proteins in osteoclast supernatant vs 312 in odontoclast supernatant with log(e) scores <−10 and the presence of at least two peptides. Approximately 90% of these proteins are reported in the ExoCarta database as exosome-related (http://www.exocarta.org). Thus, the ratio of classified exosomes to overall observed protein count is much greater than for the overall proteome, suggesting there is indeed enrichment occurring in our samples.
TABLE 2.
Summary of protein/peptide identifications by 1D LC-MS analysis of cell culture supernatants from osteoclasts and odontoclasts that had been pre-enriched for exosomes
| Osteoclasts | Odontoclasts | |
|---|---|---|
| Number of MS/MS spectra | 51 811 | 35 659 |
| Total number of peptides | 6268 | 5058 |
| Number of non-redundant peptides | 2771 | 1930 |
| Proteins log(e) <−1 | 896 | 665 |
| Proteins log(e) <−3 | 656 | 450 |
| Proteins log(e) <−10 | 457 | 312 |
Figure 1 illustrates the log(2) protein expression values between odontoclasts and osteoclasts. The vast majority of proteins are present in similar amounts in both osteoclast and odontoclast exosomes, indicated by the dots within the diagonal ellipse (A) shown in Figure 1. Conversely, 40 exosomal proteins were only present in osteoclast media (B), whereas six unique exosomal proteins were identified uniquely in odontoclast supernatants (C). Some proteins are found in both samples but at higher expression values (∣Z∣>1.65) in either osteoclast (D) or odontoclast media (E). Table 3 shows the subset of exosomal proteins that were uniquely present in either osteoclast or odontoclast media (collections B, C in Figure 1); it included only proteins that were identified by four or more peptides as we consider these proteins the most confident for quantitation. Many interesting candidates for possible biomarkers were identified including syntenin-1 for dentin resorption (Figure 2) and vacuolar protein sorting-associated protein 35 for bone resorption (Figure 3).
FIGURE 1.
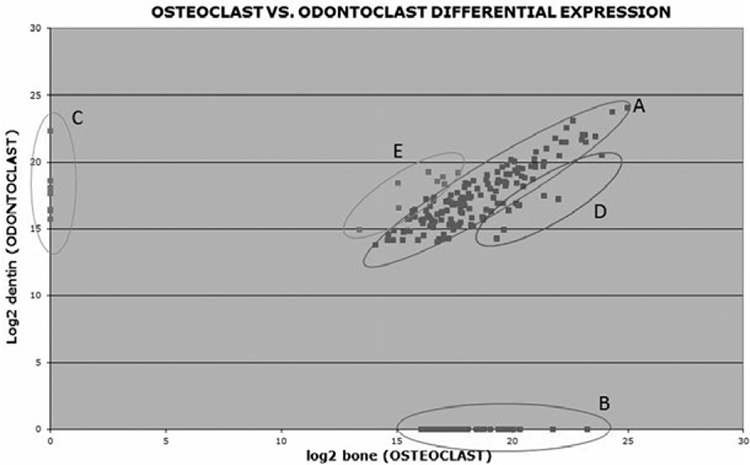
Differential expression between log2 values for exosomal proteins in osteoclast and odontoclast media. A, Proteins with similar values in osteoclasts and odontoclast media; B, proteins uniquely present in osteoclast media; C, proteins uniquely present in odontoclast media; D, proteins found in both samples but with higher number of peptides in osteoclast media (Z>1.65); E, proteins found in both samples but with higher abundance in odontoclast media (Z<−1.65)
TABLE 3.
Murine exosomal proteins uniquely present in either odontoclast or osteoclast media
| No. of Peptides | Log2 intensity | Gene Symbol | Protein name |
|---|---|---|---|
| ODONTOCLAST (DENTIN) MEDIA | |||
| 4 | 16.4 | ITGB2 | Integrin beta-2* |
| 4 | 16.43 | NAP1L1 | Nucleosome assembly protein 1-like 1 |
| 5 | 17.66 | SDCBP | Syntenin-1 |
| 4 | 18.08 | DSP | Desmoplakin* |
| 4 | 18.63 | C1QB | Complement C1q subcomponent subunit B* |
| 8 | 22.33 | HSPA1B | Heat-shock 70 kDa protein 1B* |
| OSTEOCLAST (BONE) MEDIA | |||
| 7 | 20.05 | SPP1 | Osteopontin |
| 13 | 20.05 | HEXB | Beta-hexosaminidase subunit beta |
| 13 | 19.8 | HEXA | Beta-hexosaminidase subunit alpha |
| 17 | 19.55 | ACTN4 | Alpha-actinin-4* |
| 14 | 19.4 | ATP6V1A | V-type proton ATPase catalytic subunit A |
| 7 | 19.03 | CALR | Calreticulin* |
| 10 | 18.78 | P4HB | Protein disulphide isomerase* |
| 9 | 18.65 | HSP90B1 | Endoplasmin |
| 5 | 18.44 | ARPC3 | Actin-related protein 2/3 complex subunit 3* |
| 9 | 18.06 | MSN | Moesin |
| 4 | 17.86 | ACTR2 | Actin-related protein 2* |
| 4 | 17.84 | RPLP2 | 60S acidic ribosomal protein P2 |
| 6 | 17.75 | ATP5B | ATP synthase subunit beta, mitochondrial* |
| 8 | 17.69 | VCL | Vinculin* |
| 4 | 17.66 | HSPA1A | Heat-shock 70 kDa protein 1A* |
| 6 | 17.47 | FTH1 | Ferritin heavy chain |
| 7 | 17.44 | PPP2R1A | Serine/threonine-protein phosphatase 2A 65 kDa (A alpha) |
| 4 | 17.41 | RAC2 | Ras-related C3 botulinum toxin substrate 2 |
| 5 | 17.36 | MDH2 | Malate dehydrogenase, mitochondrial* |
| 7 | 17.28 | ACO2 | Aconitate hydratase, mitochondrial |
| 5 | 17.24 | FSTL1 | Follistatin-related protein 1 |
| 7 | 17.08 | NCL | Nucleolin |
| 4 | 17 | CS | Citrate synthase, mitochondrial* |
| 5 | 16.83 | UAP1L1 | UDP-N-acetylhexosamine pyrophosphorylase-like protein 1 |
| 6 | 16.8 | TFRC | Transferrin receptor protein 1* |
| 6 | 16.78 | KPNB1 | Importin subunit beta-1* |
| 5 | 16.7 | EHD1 | EH domain-containing protein 1 |
| 4 | 16.69 | ATP6V1F | V-type proton ATPase subunit F |
| 5 | 16.65 | VPS35 | Vacuolar protein sorting-associated protein 35 * |
| 5 | 16.65 | MVP | Major vault protein |
| 7 | 16.64 | COPS4 | COP9 signalosome complex subunit 4 |
| 4 | 16.64 | TGFBI | Transforming growth factor-beta-induced protein ig-h3* |
| 5 | 16.58 | CORO1C | Coronin-1C* |
| 5 | 16.58 | TUBB6 | Tubulin beta-6 chain |
| 4 | 16.56 | EFEMP1 | EGF-containing fibulin-like extracellular matrix protein 1 |
| 4 | 16.5 | RAB7A | Ras-related protein Rab-7a |
| 5 | 16.37 | LCP1 | Plastin-2 |
| 4 | 16.15 | PSMB5 | Proteasome subunit beta type-5 |
| 5 | 16.07 | ITGB3 | Integrin beta-3 |
| 4 | 16.05 | HSPA4 | Heat-shock 70 kDa protein 4* |
Proteins in bold were also identified in human gingival crevicular fluid (GCF) in a previous study 8
FIGURE 2.
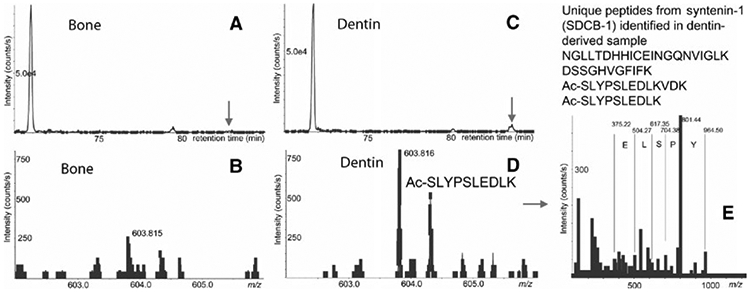
Identification of syntenin-1 (SDCB-1) peptides in odontoclast-derived exosomes by mass spectrometry (MS). A, B: Extracted ion chromatogram (XIC) for m/z 603.81 and MS spectra at 82.9 min from LC-MS of bone-derived exosome digest; C, D: respective XIC and MS spectra from dentin-derived sample. Note that respective peak for syntenin-1 in panel B (bone) is lower than a threshold for MS/MS acquisition (300 counts per second). E: MS/MS spectra of 603.816 peak with assignment of selected y-ions
FIGURE 3.
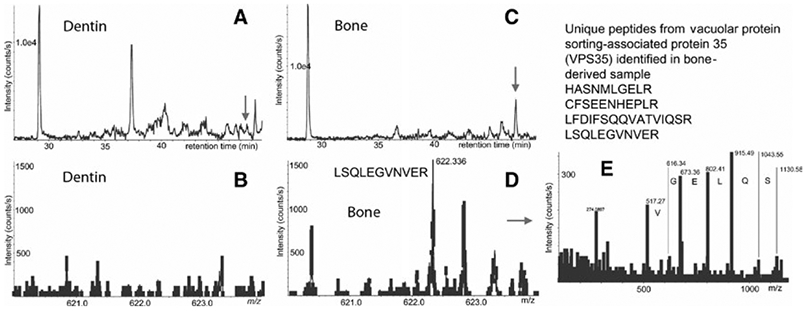
Identification of vacuolar protein sorting-associated protein 35 (VPS35) in osteoclast-derived exosomes by mass spectrometry (MS). A, B, Extracted ion chromatogram (XIC) for m/z 622.36 and MS spectra at 50 min from LC-MS of dentin-derived exosome digest; C, D, respective XIC and MS spectra from bone-derived sample. Note that respective peak for VPS35 in panel B (dentin) is lower than a threshold for MS/MS acquisition (300 counts per second). E, MS/MS spectra of 622.36 peak with assignment of selected y-ions
3.3 ∣. Expression in human GCF samples
To determine the clinical usefulness of these biomarker candidates, we compared the list in Table 3 with our human GCF proteomic data extracted from resorbing and non-resorbing primary teeth that is published elsewhere.8 Our preliminary results demonstrate that approximately 50% of exosomal proteins released by clastic cells in vitro can be found in oral fluids (Table 3, proteins in bold/asterisk), thus supporting the use of cell culture platforms in the identification of novel biomarkers.
4 ∣. DISCUSSION
Our data suggest that the exosome proteome profiles of odontoclasts and osteoclasts differ from each other, thus endorsing the principle that mineralized matrix type plays a role in the terminal differentiation of clastic cells. It is well established that the interactions between cells and matrix components strongly influence cell differentiation and affect cell phenotype.12 Osteoclasts and odontoclasts are defined as multinuclear cells that break down bone and dentin, respectively.13 Several biological features are shared by these two types of “clastic” cells; however, many differences have been identified as well that likely result from differences in cell’s interactions with the different mineralized matrices.13 We hypothesize that different matrices induce unique phenotypes in “clastic” cells resulting in very specific markers, which in turn can be used for the development a non-invasive diagnostic assays using oral fluids.
Recently published articles have proposed that osteoclasts secrete exosomes in vitro;10 however, the molecular repertoire of exosomes released by “clastic” cells and how this varies during mineralized tissue disorders remain largely unexplored. GCF is a rich source of biomarkers; however, it contains more than 2500 proteins with range of concentrations that exceed ten orders of magnitude.4,8 This may pose a challenge to the discovery of novel biomarkers using mass spectrometry, as many low-abundant proteins fail to be ionized in the presence of peptides from high-abundant proteins. Thus, our group chose to develop cell culture platforms for biomarker discovery, one of which is described in this article. While one could argue that our screen in mouse cells may not accurately reflect the human clastic cells we wish to detect, it is noteworthy to point out that recent studies demonstrate common patterns in mammalian proteomes.14 Furthermore, the fact that approximately 50% of the proteins were common between murine cell culture media and human GCF is encouraging.
Another important challenge is the selection of meaningful candidates from this preliminary in vitro data, which is still an ongoing endeavour for our group. At first, we chose to focus on proteins that are only present in one media vs another (Table 3, Figure 1 B,C). Syntenin-1, a PDZ scaffolding protein which is a known controller of the biogenesis of exosomes,15 was enriched in exosomes from odontoclasts. Syntenin-1 is also able to link transmembrane proteins to the actin cytoskeleton; thus, it is involved in a plethora of biological processes, such as cell migration and cell adhesion.15 Work by Baietti et al.16 has demonstrated that the interaction between syntenin-1 and small proteoglycans called syndecans is key for membrane trafficking in pathological processes. Although the role of these two molecules in cancer cells has been extensively studied, their ability to regulate mineralized tissue resorption has not been fully investigated yet. Using cell culture platforms, Benad-Mehner et al.17 observed that syndecans play a role in osteoclast biology as it modulates osteoprotegerin (OPG) expression. Thus, it seems worthwhile to investigate whether syntenin-1 can be detected by specific antibodies in human oral fluids that had been pre-enriched for exosomes. The exosomal protein VPS35 was found to be present only in conditioned media from osteoclasts. One of the known molecular functions associated with this protein is to regulate the amount of RANKL on the surface of osteoclasts to control bone remodelling.18 Thus, the fact that VPS35 was not identified in conditioned media from odontoclast merits further investigation as a biomarker to discriminate between osteoclast and odontoclast function.
5 ∣. CONCLUSIONS
The proteome profiles of exosomes isolated from osteoclasts and odontoclasts are not completely identical, indicating that the matrix type plays a role in the final phenotypic characteristics of clastic cells. Exosomal proteins detected in cell culture media were also found in human samples, thus suggesting that this approach may be a viable alternative in biomarker discovery.
ACKNOWLEDGEMENTS
This work was supported by the American Association of Orthodontists Foundation (AAOF).
Footnotes
CONFLICT OF INTERESTS
The authors have stated that there is no conflict of interests.
REFERENCES
- 1.Golub LM, Borden SM, Kleinberg I. Urea content of gingival crevicular fluid and its relation to periodontal diseases in humans. J Periodontal Res. 1971;6:243–251. [DOI] [PubMed] [Google Scholar]
- 2.Miyajima K, Ohno Y, Iwata T, Tanida K, Iizuka T. The lactic acid and citric acid content in the gingival fluid of orthodontic patients. Aichi Gakuin Dent Sci. 1991;4:75–82. [PubMed] [Google Scholar]
- 3.Mah J, Prasad N. Dentine phosphoproteins in gingival crevicular fluid during root resorption. Eur J Orthod. 2004;26:25–30. [DOI] [PubMed] [Google Scholar]
- 4.Corthals GL, Wasinger VC, Hochstrasser DF, Sanchez JC. The dynamic range of protein expression: a challenge for proteomic research. Electrophoresis. 2000;21:1104–1115. [DOI] [PubMed] [Google Scholar]
- 5.Johnstone RM. Exosomes biological significance: a concise review. Blood Cells Mol Dis. 2006;36:315–321. [DOI] [PubMed] [Google Scholar]
- 6.Kulasingam V, Diamandis EP. Tissue culture-based breast cancer biomarker discovery platform. Int J Cancer. 2008;123:2007–2012. [DOI] [PubMed] [Google Scholar]
- 7.Leung F, Bernardini MQ, Clarke B, Rouzbahman M, Diamandis EP, Kulasingam V. Discovery of novel subtype-specific ovarian cancer biomarkers via integrated tissue proteomics. Clin Cancer Res. 2016;22(Supplement 2):B13. [Google Scholar]
- 8.Rody WJ Jr, Holliday LS, McHugh KP, Wallet SM, Spicer V, Krokhin O. Mass spectrometry analysis of gingival crevicular fluid in the presence of external root resorption. Am J Orthod Dentofacial Orthop. 2014;145:787–798. [DOI] [PubMed] [Google Scholar]
- 9.Crotti TN, O’Sullivan RP, Shen Z, et al. Bone matrix regulates osteoclast differentiation and annexin A8 gene expression. J Cell Physiol. 2011;226:3413–3421. [DOI] [PubMed] [Google Scholar]
- 10.Huynh N, VonMoss L, Smith D, et al. Characterization of Regulatory Extracellular Vesicles from Osteoclasts. J Dent Res. 2016;95:673–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–362. [DOI] [PubMed] [Google Scholar]
- 12.Lin CQ, Bissell MJ. Multi-faceted regulation of cell differentiation by extracellular matrix. FASEB J. 1993;7:737–743. [DOI] [PubMed] [Google Scholar]
- 13.Wang Z, McCauley LK. Osteoclasts and odontoclasts: signaling pathways to development and disease. Oral Dis. 2011;17:129–142. [DOI] [PubMed] [Google Scholar]
- 14.Kanapin A, Batalov S, Davis MJ, et al. Mouse proteome analysis. Genome Res. 2003;13:1335–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friand V, David G, Zimmermann P. Syntenin and syndecan in the biogenesis of exosomes. Biol Cell. 2015;107:331–341. [DOI] [PubMed] [Google Scholar]
- 16.Baietti MF, Zhang Z, Mortier E, et al. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat Cell Biol. 2012;14:677–685. [DOI] [PubMed] [Google Scholar]
- 17.Benad-Mehner P, Thiele S, Rachner TD, Gobel A, Rauner M, Hofbauer LC. Targeting syndecan-1 in breast cancer inhibits osteoclast functions through up-regulation of osteoprotegerin. J Bone Oncol. 2014;3:18–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xia WF, Tang FL, Xiong L, et al. Vps35 loss promotes hyperresorptive osteoclastogenesis and osteoporosis via sustained RANKL signaling. J Cell Biol. 2013;200:821–837. [DOI] [PMC free article] [PubMed] [Google Scholar]


