Abstract
Purpose of Review
This review summarises the epidemiology of Candida auris infection and describes contemporary and emerging diagnostic methods for detection and identification of C. auris.
Recent Findings
A fifth C. auris clade has been described. Diagnostic accuracy has improved with development of selective/differential media for C. auris. Advances in spectral databases of matrix-associated laser desorption ionisation time-of-flight mass spectrometry (MALDI-TOF MS) systems have reduced misidentification. Direct detection of C. auris in clinical specimens using real time PCR is increasingly used, as is whole genome sequencing (WGS) to track nosocomial spread and to study phylogenetic relationships and drug resistance.
Summary
C. auris is an important transmissible, nosocomial pathogen. The microbiological laboratory diagnostic capacity has extended beyond culture-based methods to include PCR and WGS. Microbiological techniques on the horizon include the use of MALDI-TOF MS for early echinocandin antifungal susceptibility testing (AST) and expansion of the versatile and information-rich WGS methods for outbreak investigation.
Keywords: Candida auris, Candidaemia, Invasive candidiasis, Nosocomial infection, Multidrug resistance (MDR), Antifungal resistance, Matrix-associated laser desorption time of flight (MALDI-ToF), Antifungal susceptibility testing, Whole genome sequencing, Fungal outbreak, Future diagnostics
Introduction
Candida auris was first isolated in Japan in 2009 from the external auditory canal of a patient [1]. Since then, the rapid simultaneous global emergence of this pathogenic fungus has been described across all continents except Antarctica. Currently, C. auris is separated into four geographic clades which differ by > 10,000 single nucleotide polymorphisms (SNPs): the South Asian (clade I), East Asian (clade II), African (clade III) and South American (clade IV) clades. [2–4]. Recently, a 5th clade has been reported from Iran [5•]. The high number of SNPs between clades suggests the emergence of C. auris in multiple locations at once as opposed to a clonal source [4]. The drivers for this recent global co-emergence are unclear but may include increasing antifungal selection pressures within hospitals and agricultural practices, along with anthropogenic factors [4, 6•]. The pathogenicity of C. auris pertains to its multidrug resistance profile to antifungal agents, reported across all major classes [6•].
Although earlier reports of C. auris infection have been associated with predominantly nosocomial outbreaks [5•, 6•, 7, 17, 18], sporadic cases have also been described [19, 20•]. Furthermore, colonisation of patients with C. auris may occur without infection, and these patients can act as a reservoir for nosocomial spread [6•]. This highlights the need to understand local epidemiology and patterns of transmission. Further, it is essential that laboratories providing diagnostic services to healthcare institutions are capable of rapidly and accurately detecting C. auris in clinical samples.
This review aims to summarise the epidemiology of C. auris infections and describe contemporary diagnostic methods for detection and identification in microbiology laboratories.
Epidemiology and Clinical Features of C. auris Infection
The origin, transmission dynamics, epidemiology and environmental niches of C. auris remain incompletely understood. To date, there are no identified animal reservoirs [7, 8•].
C. auris infections have largely been healthcare-associated [9–11]. In the context of nosocomial candidaemia, C. auris is over-represented and has become endemic in South Africa [12] and India [13] where it accounts for 15% and 5–30% of the national reported candidaemia figures, respectively. Nosocomial spread causing protracted outbreaks involving critical and intensive care unit (ICU) settings and immunocompromised cohorts has occurred in Europe (Spain and the UK), the USA and Venezuela [3, 11, 14–17]. Other countries such as Norway, Germany and Australia [18, 19] have reported sporadic cases.
Candida auris infections have been documented in multiple (> 40) countries worldwide, affecting between 5 and 10% of C. auris-colonised patients [20•, 21]. The clinical spectrum of C. auris-related infections ranges from mild, superficial infections such as otitis media to invasive diseases similar in spectrum to invasive candidiasis due to other species [7, 14, 22]. The epidemiology for candidaemia is similar to that for other Candida spp. [7, 14, 22]. At-risk groups include those at extremes of age, ICU patients and patients with underlying immunosuppression or chronic diseases, especially following healthcare exposure. Crude mortality rates of C. auris invasive fungal disease remain high (30–60%) [9, 10, 14, 23].
Colonisation Due to C. auris
C. auris is unique among pathogenic fungi in its persistence within the clinical environment and inter-patient transmissibility. The limited efficacy of non-sporicidal disinfectants against this yeast contributes to the observed environmental persistence, for days to weeks, on inanimate surfaces, such as dry linen, reusable temperature probes and blood pressure cuffs [17, 25, 26•, 27, 28]. Contact with contaminated items is the most common method of colonisation, and the capacity to form high-burden biofilms has a key pathogenic role [14, 17, 22, 24–26•]. The highest prevalence of colonisation is reported in patients of ventilator-capable skilled nursing facilities (23-71%) [27]. Otherwise, close contacts of cases (current or past room contacts within the prior month) have a documented colonisation rate of 12–21% [14, 28].
Risk factors for colonisation and subsequent invasive disease vary between geographic surveys. Common identified risk factors include recent surgery (especially abdominal); severe concurrent medical conditions, often with ICU admission and/or mechanical ventilation; diabetes mellitus; immunosuppression; and the presence of invasive catheters and protracted antimicrobial administration, with nearly 50% of cases in one cohort receiving antifungal therapy at the time of, or immediately before diagnosis of C. auris infection [7]. The temporal dynamics of colonisation are uncertain and may be indefinite. Screening of healthcare workers is not recommended unless risk factors are identified, as they have not been linked to previous outbreaks [10, 11, 14, 24, 29].
Screening for C. auris Colonisation in Asymptomatic Patients
Screening for colonisation is recommended for patients with inpatient healthcare contact in settings where C. auris transmission has occurred, or close contacts of confirmed C. auris cases [7, 8•, 28, 29].
Public health laboratories commonly recommend superficial swabs of the axillae and groin for patient screening and environmental sponge samples for mycological culture. This is based on the ability of C. auris to colonise multiple body sites including nares, mouth, external ear canals, urine, wounds and rectum [15]. Swabs immersed in Amies transport medium are preferred over dry swabs as they promote the viability of the organism, and use of flocked swabs is likely to improve yield [26•, 29].
Diagnosis of C. auris Invasive Disease
As for other Candida spp., C. auris can be readily cultured from blood, urine, sputum and other bodily fluids. Prior to the description of C. auris, identification of yeasts from non-sterile clinical samples was ad hoc and limited. Yeasts such as Candida spp. are known colonisers and when isolated from non-sterile sites used to pique little interest in the microbiologist. Prompted by the emergence of this pathogenic fungus, major changes have been made to laboratory protocols in order to distinguish C. auris in clinical samples. Guidance and best practice documents from key professional bodies such as the Centers for Disease Control and Prevention (CDC) [29] and the Australasian Society for Infectious Diseases (ASID) [8•] recommend complete identification of yeasts to exclude C. auris from sterile sites and using clinical judgement regarding select non-sterile site specimens. Existing or prior cases of C. auris colonisation or infection in the same healthcare facility should heighten laboratory vigilance. In practice, it remains challenging to balance the need to exclude C. auris against the extra cost and time that is often applied to mixed growths of yeast with little clinical relevance. Anecdotally, whilst changes in recommendations have led to heightened awareness of C. auris among clinicians and increased complexity in the laboratory workup of yeasts, translational research that systematically compares the efficacy of this strategy as an adjunct to, or in comparison with, screening tests in non-endemic areas has not been reported.
As culture requires significant time and may not detect all cases, the use of biomarkers to assist in identifying invasive C. auris infections is attractive. 1, 3-Beta-D-glucan (BDG) is a biomarker with reasonable sensitivity for the diagnosis of invasive candidiasis. However, in one South African study, BDG performed poorly in the detection of C. auris candidaemia, with a sensitivity of 71% which was the lowest of all Candida spp. Given this, biomarker use with BDG for C. auris diagnosis is not currently recommended [30].
Laboratory Diagnosis of C. auris
Whether screening or clinical specimens are submitted, culture-based diagnostics are the current stalwart of the clinical mycology laboratory. Turn-around time and diagnostic sensitivity are improved by the use of molecular technologies, particularly for specimens from individuals with a high pre-test probability of C. auris colonisation or infection. Molecular technologies are increasingly being used in most laboratories as an adjunct to culture-based techniques.
Culture-Based Approaches for C. auris Isolation
Mycological culture remains the cornerstone of the laboratory diagnosis of C. auris. Distinguishing features to ensure an accurate identification and minimise misidentifications are outlined in Tables 1, 2 and Fig. 1.
Table 1.
Reported misidentifications of Candida auris from some commercial biochemical and MALDI-TOF-MS identification systems. Table adapted from [3, 29]
| Identification method | Candida auris can be misidentified as | Comments |
|---|---|---|
| Biochemical method | ||
| Vitek 2 YST |
Candida haemulonii Candida duobushaemulonii Candida spp. not identified |
Updated software version 8.01 will identify C. auris. |
| API 20C AUX |
Rhodotorula glutinis (characteristic red colour not present) Candida sake |
Reflex to MALDI-TOF or sequencing |
| API ID 32C |
Candida intermedia Candida sake Saccharomyces kluyveri Candida spp. not identified |
Reflex to MALDI-TOF or sequencing |
| BD Phoenix yeast identification system |
Candida haemulonii Candida catenulate Candida spp. not identified |
Reflex to MALDI-TOF or sequencing |
| MicroScan |
Candida famata/Debaryomyces hansenii Candida guilliermondii*/Meyerozyma guilliermondii Candida lusitaniae*/Clavispora lusitaniae Candida parapsilosis* Also rarely misidentifies as C. albicans and C. tropicalis Candida spp. not identified |
Reflex to MALDI-TOF or sequencing |
| RapID Yeast Plus |
Candida parapsilosis* Candida spp. not identified |
Reflex to MALDI-TOF or sequencing |
| MALDI-TOF MS | ||
| VITEK MS (bioMerieux) |
C. albicans C. haemulonii C. lusitaniae/Clavispora lusitaniae Candida spp. not identified |
Reliable updated databases include IVD library v3.2 onwards RUO library with Saramis Version 4.14 database and Saccharomycetaceae update |
| MALDI Biotyper (BRUKER Daltronics) |
Neisseria meningitidis serogroup A Pseudomonas rhizosphaerae |
Reliable updated databases include CA System library Version Claim 4 RUO libraries versions 2014 (5627) and more recent |
Abbreviations: C. auris, Candida auris; MALDI-TOF matrix-assisted laser desorption ionisation time-of-flight, RUO research use only
*Yeast isolates which identify as Candida guilliermondii/Meyerozyma guilliermondii, Candida lusitaniae/Clavispora lusitaniae or Candida parapsilosis can be further evaluated using cornmeal agar. If no hyphae or pseudohyphae are present on cornmeal agar, these can be ruled out; the isolate will require further workup for C. auris. If hyphae or pseudohyphae are present, the isolate is likely one of the above. However, some C. auris strains have had hyphae or pseudohyphae; thus, this distinguishing feature is for guidance rather than confirmation.
Table 2.
| Characteristics | C. auris | C. haemulonii | C. duobushaemulonii | C. haemulonii var. vulnera | C. pseudohaemulonii |
|---|---|---|---|---|---|
| Fermentation of: | |||||
| Raffinose | - | - | + | - | - |
| Sucrose | + | + | + | + | - |
| Growth at: | |||||
| 37 oC | + | - | + | + | + |
| 40 oC | + | - | - | - | - |
| Growth on SAB1: | |||||
| Dextrose | + | - | - | - | - |
| Dulcitol | + | - | - | - | - |
| Mannitol | + | - | - | - | - |
| Growth in 60% glucose | - | - | + | - | ND |
| Vitamin-free medium | + | - | - | - | ND |
1SAB, Sabouraud broth incubated at 40 oC with dextrose, dulcitol, and mannitol as carbon sources
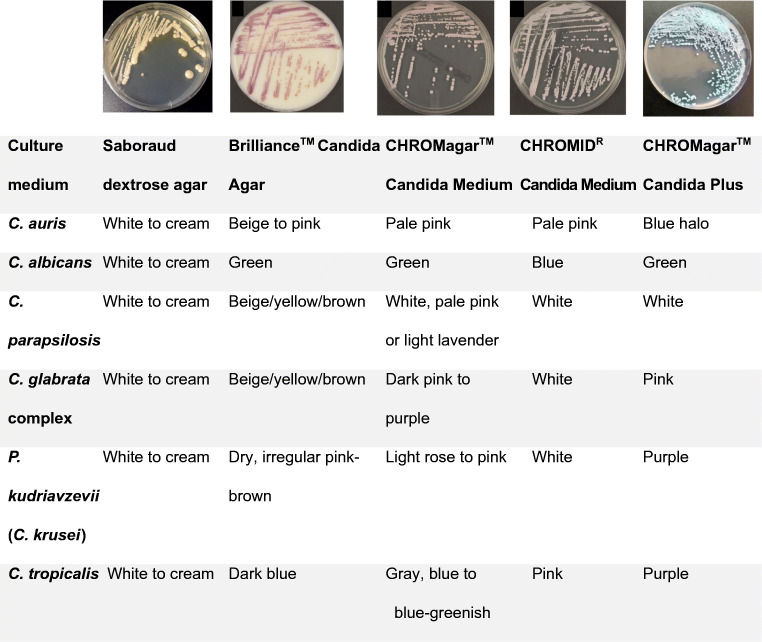
Fig. 1.
Characteristics of C. auris on SAB and on chromogenic media. BrillianceTM Candida Agar (Oxoid, Basingstoke, UK), CHROMagarTM Candida Medium (Becton Dickinson, Heidelberg, Germany), CHROMIDR Candida medium (bioMérieux, Marcy l’Étoile, France), CHROMagarTM Candida Plus (CHROMagar, France). Images adapted from [63–66]
C. auris grows on routine laboratory and mycological media such as Sabouraud dextrose agar (SDA) and chromogenic media and has an optimum growth temperature of 37–40 oC [1]. A prolonged incubation time of up to 10 days may be required for screening of primary clinical samples [8•]. In practice, 48 h incubation after enrichment is probably sufficient [10, 31•].
Standard Candida chromogenic agar does not permit differentiation of C. auris from other common Candida spp. as it lacks a distinctive colony colour to Pichia kudriavzevii (formerly C. krusei), C. glabrata complex or Meyerozyma guilliermondii (formerly C. guilliermondii) [29]. A recently described novel chromogenic agar, CHROMagarTM Candida Plus, has promising utility for rapid identification and differentiation of C. auris from other Candida species [32•]. On CHROMagarTM Candida Plus (CHROMagar, France), all four lineages of C. auris have pale cream colonies with a distinctive blue halo (Fig. 1). In an evaluation of more than 50 Candida species, only the clinically very rare C. diddensiae was noted to have a similar appearance [32•]. Growth may be faster on CHROMagarTM Candida Plus, with detectable colony growth as early as 36 h [32•].
The use of selective enrichment broth media optimised to C. auris growth characteristics achieves a faster recovery time in clinical specimens, along with improved sensitivity and specificity [14, 26•]. The CDC currently recommends use of 10% salt Sabouraud Dulcitol (SDD) broth medium with chloramphenicol and gentamicin, inoculated then shaken and subsequently incubated at 37–40 oC [26•]. This utilises C. auris’ unique ability to grow at elevated temperatures and in the presence of saline conditions (10% weight/volume) [26•]. C. haemulonii and C. duobushaemulonii may also grow in such conditions but require glucose as a carbon source, in contrast to C. auris which can use dulcitol or mannitol as a carbon source [26•].
Local practice varies according to in-house laboratory processes, with some sites following CDC recommendations by incubating for 5 days in an enrichment broth and then plate culture for 48 h, whilst others opt to plate directly and incubate on selective agar for 10 days with the goal of reducing turn-around time as most grow within 48 h. The aforementioned CHROMagarTM Candida Plus may also be used as a primary isolation medium with clinical screening samples, though whether the sensitivity of this approach will be equivalent to using an enrichment step remains to be seen [32•].
On microscopy, C. auris demonstrates oval-elongated, budding single or aggregates of yeast, approximately 2.0–3.0 x 2.5–5.0 μm [1, 33], with smooth, white-cream colonies on SDA [1, 31, 33], beige to pink colonies on the BrillianceTM Candida Agar (Oxoid, UK) [1, 31, 33], pale pink colonies on Candida CHROMagarTM (Becton Dickinson, Heidelberg, Germany) [31] and pale pink colonies on CHROMIDR Candida (BioMerieux, France) (Fig. 1) [1, 31, 33]. Clade-specific phenotypic characteristics have been observed. Pseudohyphae are rare for isolates of the South African and Japanese/Korean clades but may be present in isolates of the Southern Asian clade [31, 34]. In contrast, cell aggregate formation is more frequently observed in isolates of the South African and Southern Asian clades [34]. C. auris does not form germ tubes [31, 34].
Phenotypic and Biochemical Identification of C. auris
Reliable identification through traditional phenotypic and biochemical methods is challenging, and these have largely been superseded by proteomics and molecular methods. The conventional assimilation methods are unable to differentiate between C. auris and phylogenetically related C. haemulonii complex [35]. The main phenotypic and growth characteristic differences between the two are described in Table 2. Cyclohexamide inhibits the growth of C. auris. C. auris ferments glucose, sucrose and trehalose and assimilates glucose, sucrose, maltose, D-trehalose, D-raffinose, D-melezitose, D-mannitol, sorbitol, citrate, inulin, starch, ribitol, galactitol, N-acetyl glucosamine, succinate and gluconate [33]. A notable exception, however, are Japanese and Korean reference isolates which do not assimilate N-acetyl glucosamine [36].
The Emerging Role of Mass Spectrometry
In contrast to biochemical methods, matrix-assisted laser desorption ionisation time-of-flight mass spectrometry (MALDI-TOF MS) has proven to be an accurate, discriminatory, high throughput method for pathogen identification [3, 5, 11, 13, 15, 36]. Successful, reliable and reproducible identification of C. auris by MALDI-TOF has been demonstrated on isolates from a variety of agars and also directly from blood culture bottles [3, 5•, 11, 13, 15, 36].
Extraction Methods for MALDI-TOF MS
Some studies report reliable log scores using ‘direct smear’ spotting of tiles [3, 5•, 11, 13, 15, 36]; most, however, note higher identification log scores with full tube extraction methods and slightly lower (but not statistically significant) log scores with partial extraction techniques [37–39]. Full tube extraction requires vortexing a volume of fresh Candida culture in sterile double distilled or Milli-Q® water, followed by addition of ethanol then a further centrifugation step to create a pellet. The supernatant is then discarded; the pellet air dried and resuspended in equal volumes of 70% formic acid and acetonitrile. The resuspension undergoes a final vortex step, and 1 μL is pipetted onto the MALDI target plate and overlaid with the Bruker MatrixTM solution after air drying [35, 38]. In contrast, described partial extraction methods emulsify a loop of fresh culture in sterile water (Fraser et al. standardised the suspension at 2 McFarland density), with either the addition of ethanol or concentrating to a pellet and repeating the emulsification and then spotting 1 μL of this solution to the MALDI plate and overlaying with 70% formic acid and Bruker MatrixTM solution after air drying at each step [37, 38].
MALDI-TOF MS Database Requirements
Accurate spectral databases are pivotal to diagnostic accuracy (Table 1). The CDC diagnostic algorithm for C. auris includes the Bruker research use only (RUO) libraries from 2014 onwards, or the United States Food and Drug Administration approved Bruker CA System library Version 4, BioMerieux RUO library 4.14 with Saccharomycetaceae update or BioMerieux IVD library version 3.2 databases [29]. One study has observed superior performance of the aforementioned Bruker database compared to BioMerieux 3.2 for the identification of C. auris [40]. The BioMerieux 3.2 database has been updated, and recent Australian quality assurance activity has demonstrated excellent performance for identification of C. auris [41]. Many C. auris studies utilise customised in-house or commercial databases, highlighting the value of supplementary in-house databases in achieving accurate comparative spectra [35, 37, 38, 42]. Additionally, a CDC-curated database, MicrobeNet, includes C. auris spectra and is freely available for Bruker database supplementation (https://www.cdc.gov/microbenet/index.html) (last accessed 22 March, 2021).
The Future of MALDI-TOF MS for C. auris
Future uses of MALDI-TOF MS profiles for laboratories identifying C. auris include rapid clade assessment to aid epidemiological investigations and antifungal resistance testing (namely for the echinocandins: elaborated on below). Spectral variation has been reported between clades, with reliable inter-clade clustering, providing quicker assessment of isolate relatedness than WGS might otherwise allow [43•]. Whilst ClinProToolsTM had shown promise for sub-speciation of Candida spp. as an add-on to the Bruker MALDI-TOF, this software is no longer supported (personal communication with the manufacturer).
Molecular Laboratory Methods for C. auris Diagnosis and Epidemiological Studies
Significant advances have been made in the area of molecular diagnostics for C. auris. Conventional phenotypic methods are limited in that they may misidentify C. auris or only identify the pathogen to genus level only. Conventional PCR methods that amplify the internal transcribed spacer (ITS) and/or D1/D2 regions of the 28S ribosomal DNA (rDNA) gene followed by DNA sequencing or WGS provide options for accurate species identification [4, 8•, 44–47]. This requires the isolate to be cultured, is expensive and requires expertise [31, 48, 49]. Although MALDI-TOF MS improves timeliness of identification, the isolate must still be cultured, taking up to 10 days. This can become onerous if applied to multiple colonies from screening swabs [32•, 48].
Nucleic Acid Amplification Testing (NAAT)
To facilitate a rapid turn-around time, many commercial and academic groups have designed real time PCR assays to identify C. auris nucleic acid directly from patient samples, enabling results within hours [8•]. These assays are exquisitely sensitive (with limits of detection in the range of 1–10 C. auris colony forming units (CFU)/PCR reaction) and specific, demonstrated by testing large panels of pathogens including closely related yeasts and other fungi on both clinical and screening samples [48, 50]. Adams et al. demonstrated the increased sensitivity of PCR when compared to culture on environmental screening isolates across New York healthcare facilities, where an additional 19 of 781 screening samples were found to be PCR positive, having been previously negative by culture [14].
Sexton et al. have recently developed a SYBR Green (TaKaRa Bio, Japan)-based quantitative PCR assay with melt curve analysis for use on skin swabs (groin and axilla) [51]. Compared to culture-based methods, this assay demonstrated a sensitivity of 93% and a specificity of 96% and was able to provide same day results. Lima et al. developed a Taqman probe-based assay performed on the BD Max system (Becton Dickinson Diagnostics, Sparks, MD) using primers and probes targeting the partial sequence of ITS1 and 28S rRNA gene and complete sequence of 5.8S rRNA gene and ITS2 [49]. Other rapid diagnostic assays include T2Candida aurisTM (T2 Biosytems®) Panel RUO assay and loop-mediated isothermal amplification assays that have been validated for use on screening swabs [51, 52].
Other than having a role in species identification of C. auris, sequencing of ITS or D1/D2 regions may be used for phylogenetic analysis to assess for clonality with other isolates [47].
Whole Genome Sequencing
WGS is increasingly being utilised for phylogenetic analysis to investigate nosocomial outbreaks. It offers superior discriminative power and shorter turn-around time (8–72 h) than older typing methods such as amplified fragment length polymorphism and multilocus sequence typing; however, WGS requires significant bioinformatics expertise and is expensive [47, 53]. In the non-outbreak setting, WGS allows the differentiation of new importation events from recent transmission events [2]. Lockhart et al. carried out WGS on isolates from 54 patients, clearly demonstrating low genetic diversity within clades in contrast to isolates from different regions [4]. A challenge presented by the exceptionally low diversity within some clades makes it difficult to establish a specific cut-off for the number of SNPs required to differentiate an institutional outbreak from circulating environmental isolates in endemic areas. Isolates from within hospital outbreaks are reported to be highly genomically related, with as little as <3 SNP differences [2, 4]. Eyre et al. used greater than 5 SNP differences as a threshold for identifying distinct genomes in an ICU outbreak involving 70 patients in the UK; sequencing of patient and environmental isolates implicated axillary temperature probes as the source [17]. In contrast, WGS on clinical and screening isolates obtained from 4 different patients in Australia found that although all isolates belonged to clade III, they arose from independent importation events [2].
In addition to its role in molecular epidemiology, WGS has played a pivotal role in the understanding of virulence and resistance (see below)
Assessment for Antifungal Drug Resistance
Reference standards for antifungal susceptbitlity testing for yeasts are published by the Clinical Laboratory Standards Institute (CLSI) and the European Committee for Antimicrobial Susceptibility Testing (EUCAST). These two standards differ in the methodology of broth microdilution (BMD) and interpretative clinical breakpoints (CBPs), which for C. auris have yet to be set by either group. However, CBPs that have been established in other Candida species may (or may not) be applicable to infections with C. auris based on pharmacokinetic/pharmacodynamic data [8•, 54]. Such tentative breakpoints for antifungal resistance have been offered by the CDC for use with CLSI methodology; these are as follows: fluconazole, ≥ 32mg/L (with the suggestion to use susceptibility as a surrogate for other second generation triazoles and voriconazole); amphotericin B, ≥ 2mg/L; anidulafungin; and micafungin, ≥ 4mg/L.
The high proportion of isolates with likely acquired resistance has complicated the establishment of breakpoints. Assessment of 123 C. auris isolates from India by Arendrup et al. resulted in MIC distributions that did not fulfil EUCAST criteria for epidemiological cut off (ECOFF) determinations [18]. The amphotericin B tentative breakpoint of 1–2 mg/L was consistent with that proposed by the CDC. However, for the triazoles, more than one peak was observed in the minimum inhibitory concentration (MIC), likely due to a large proportion of isolates included that were non-wild type. It is worth noting that MIC distribution varies with geography and specifically by clade [2, 34]. The echinocandin tentative breakpoints were 0.25–1mg/L, lower than the CDC proffered breakpoints and for anidulafungin, lacked reproducibility in MIC determination due to partial inhibition over several dilutions [18].
Antifungal susceptibility interpretation is further complicated by the need for expertise in interpretation. Sensititre YeastOne YO10 (ThermoScientific, USA), E-test (BioMerieux, France) and Vitek 2 YST AST (BioMerieux, France) offer greater ease of use and decreased cost compared with reference methods. However, a higher MIC for amphotericin B was noted with Vitek 2 when compared with E-test, and poor agreement of echinocandins on Vitek 2 and E-test with BMD has been observed [8•, 33, 48]. Echinocandin MIC determination may be impacted by paradoxical growth effect, most significantly with caspofungin for which testing displays poor reproducibility, and azoles may exhibit trailing growth [46•]. Thus, MIC values obtained for C. auris should be confirmed in a reference laboratory.
WGS and targeted sequencing have highlighted mechanisms of drug resistance in C. auris. A limited number have been linked to clinical failure. Echinocandin resistance has been shown in relation to amino acid substitutions FKS1 at position S639, which is the homologue of hot spot 1 position S645 in C. albicans [2, 46•, 48, 55–57]. Azole resistance has been demonstrated in association with amino acid substitutions in ERG11 including F126L, Y132F and K143R [2, 4, 58, 59]; most mutations are clade-specific, with clade I isolates from India tending to display the Y132F and K143R mutations in ERG11, whilst the F126L mutation is observed in clade III isolates from South Africa. The links between mutations and amphotericin B resistance are yet to be proven [46•]. The roles of transporters, regulators and additional elements such as increased copy number variation [45•] call for further research to clarify their role in drug resistance [45•, 46•, 57].
Molecular markers of antifungal drug resistance offer promise. Targeting recognised mutations associated with resistance to echinocandin or azole therapy makes an approach akin to that used for rifampicin use in Mycobacterium tuberculosis feasible [46]. This would enable information about likely non-susceptibility much quicker than standard laboratory methods, or indeed WGS, especially if applied directly to clinical specimens to enable refinement of empiric therapeutic regimens. A duplex assay designed to detect the more common mutations in ERG11 and FKS1 for C. auris has been developed [60]. This approach currently carries more potential for detection of likely echinocandin resistance than for azole resistance, as few isolates with azole resistance had detectable mutations in the ERG11 gene, reflecting the multifaceted nature of the mechanisms of azole resistance [46•, 61]. Given the high prevalence of raised fluconazole MIC in C. auris [18, 43•], azole therapy should not form part of the primary empiric treatment regimen.
Further, a technology that is increasingly accessible that may provide rapid assessment of antifungal drug resistance is proteomics [43•]. MALDI-TOF MS is now the standard method of identification of fungal isolates, and recent data suggested excellent concordance for assessment of echinocandin non-susceptibility [43•]. Detection of echinocandin resistance using the minimal profile change concentration (the minimal drug concentration needed to detect changes in MALDI-TOF MS spectra after incubation with an antifungal drug for 6 h) was found to correlate > 95% of the time with echinocandin MICs determined by (CLSI) methods, apart from caspofungin [43•, 62]. This will potentially allow earlier optimisation of antifungal therapy and improve clinical outcomes.
Conclusion
Candida auris is an important transmissible, nosocomial pathogen. As awareness increases, directing paradigm shifts in screening and infection control practices is of great importance. Whilst the focus of diagnosis remains culture-based, molecular techniques for screening are increasingly used for rapid results. Commercial MALDI-TOF MS identification systems have incorporated reliable spectra into their databases. Clinical breakpoints from CLSI and EUCAST for this MDR-fungus may be available in the near future. Future microbiological techniques include use of MALDI-TOF-MS for early echinocandin AST and clade typing and expansion of WGS methods for outbreak investigation.
Declarations
Conflict of Interest
Keighley C, Garnham K, Harch S A J, Robertson M, Chaw K and Teng JC declare that they have no conflict of interest. Chen SC-A reports untied educational grants from MSD Australia and F2G Ltd outside the submitted work.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Footnotes
This article is part of the Topical Collection on Advances in Diagnosis of Invasive Fungal Infections
Keighley C. and Garnham K. are first author
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance
- 1.Satoh K, Makimura K, Hasumi Y, Nishiyama Y, Uchida K, Yamaguchi H. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol. 2009;53(1):41–44. doi: 10.1111/j.1348-0421.2008.00083.x. [DOI] [PubMed] [Google Scholar]
- 2.Biswas C, Wang Q, van Hal SJ, Eyre DW, Hudson B, Halliday CL, et al. Genetic heterogeneity of Australian Candida auris isolates: insights from a non-outbreak setting using whole-genome sequencing. Open Forum Infect Dis. 2020;7(5):ofaa158. doi: 10.1093/ofid/ofaa158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jeffery-Smith A, Taori SK, Schelenz S, Jeffery K, Johnson EM, Borman A, et al. Candida auris: a review of the literature. Clin Microbiol Rev. 2018;31(1). 10.1128/CMR.00029-17. [DOI] [PMC free article] [PubMed]
- 4.Lockhart SR, Etienne KA, Vallabhaneni S, Farooqi J, Chowdhary A, Govender NP, Colombo AL, Calvo B, Cuomo CA, Desjardins CA, Berkow EL, Castanheira M, Magobo RE, Jabeen K, Asghar RJ, Meis JF, Jackson B, Chiller T, Litvintseva AP. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis. 2017;64(2):134–140. doi: 10.1093/cid/ciw691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chow NA, de Groot T, Badali H, Abastabar M, Chiller TM, Meis JF. Potential fifth clade of Candida auris, Iran, 2018. Emerg Infect Dis. 2019;25(9):1780–1781. doi: 10.3201/eid2509.190686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.• Kean R, Brown J, Gulmez D, Ware A, Ramage G. Candida auris: a decade of understanding of an enigmatic pathogenic yeast. J Fungi. 2020;6(1). 10.3390/jof6010030Excellent review including epidemiology. [DOI] [PMC free article] [PubMed]
- 7.Forsberg K, Woodworth K, Walters M, Berkow EL, Jackson B, Chiller T, Vallabhaneni S. Candida auris: the recent emergence of a multidrug-resistant fungal pathogen. Medical mycology. 2019;57(1):1–12. doi: 10.1093/mmy/myy054. [DOI] [PubMed] [Google Scholar]
- 8.Ong CW, Chen SC, Clark JE, Halliday CL, Kidd SE, Marriott DJ, et al. Diagnosis, management and prevention of Candida auris in hospitals: position statement of the Australasian Society for Infectious Diseases. Intern Med J. 2019;49(10):1229–1243. doi: 10.1111/imj.14612. [DOI] [PubMed] [Google Scholar]
- 9.Al Maani A, Paul H, Al-Rashdi A, Wahaibi AA, Al-Jardani A, Al Abri AMA, et al. Ongoing challenges with healthcare-associated Candida auris outbreaks in Oman. J Fungi (Basel). 2019;5(4). 10.3390/jof5040101. [DOI] [PMC free article] [PubMed]
- 10.Ruiz-Gaitán A, Moret AM, Tasias-Pitarch M, Aleixandre-López AI, Martínez-Morel H, Calabuig E, Salavert-Lletí M, Ramírez P, López-Hontangas JL, Hagen F, Meis JF, Mollar-Maseres J, Pemán J. An outbreak due to Candida auris with prolonged colonisation and candidaemia in a tertiary care European hospital. Mycoses. 2018;61(7):498–505. doi: 10.1111/myc.12781. [DOI] [PubMed] [Google Scholar]
- 11.Schelenz S, Hagen F, Rhodes JL, Abdolrasouli A, Chowdhary A, Hall A, Ryan L, Shackleton J, Trimlett R, Meis JF, Armstrong-James D, Fisher MC. First hospital outbreak of the globally emerging. Antimicrob Resist Infect Control. 2016;5:35. doi: 10.1186/s13756-016-0132-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Schalkwyk E, Mpembe RS, Thomas J, Shuping L, Ismail H, Lowman W, Karstaedt AS, Chibabhai V, Wadula J, Avenant T, Messina A, Govind CN, Moodley K, Dawood H, Ramjathan P, Govender NP, for GERMS-SA Epidemiologic shift in Candidemia driven by Candida auris, South Africa, 2016-2017. Emerg Infect Dis. 2019;25(9):1698–1707. doi: 10.3201/eid2509.190040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chakrabarti A, Sood P, Rudramurthy SM, Chen S, Kaur H, Capoor M, Chhina D, Rao R, Eshwara VK, Xess I, Kindo AJ, Umabala P, Savio J, Patel A, Ray U, Mohan S, Iyer R, Chander J, Arora A, Sardana R, Roy I, Appalaraju B, Sharma A, Shetty A, Khanna N, Marak R, Biswas S, Das S, Harish BN, Joshi S, Mendiratta D. Incidence, characteristics and outcome of ICU-acquired candidemia in India. Intensive Care Med. 2015;41(2):285–295. doi: 10.1007/s00134-014-3603-2. [DOI] [PubMed] [Google Scholar]
- 14.Adams E, Quinn M, Tsay S, Poirot E, Chaturvedi S, Southwick K, Greenko J, Fernandez R, Kallen A, Vallabhaneni S, Haley V, Hutton B, Blog D, Lutterloh E, Zucker H, Workgroup CI. Candida auris in Healthcare Facilities, New York, USA, 2013-2017. Emerg Infect Dis. 2018;24(10):1816–1824. doi: 10.3201/eid2410.180649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chowdhary A, Sharma C, Meis JF. Candida auris: A rapidly emerging cause of hospital-acquired multidrug-resistant fungal infections globally. PLoS Pathog. 2017;13(5):e1006290. doi: 10.1371/journal.ppat.1006290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vallabhaneni S, Kallen A, Tsay S, Chow N, Welsh R, Kerins J, Kemble SK, Pacilli M, Black SR, Landon E, Ridgway J, Palmore TN, Zelzany A, Adams EH, Quinn M, Chaturvedi S, Greenko J, Fernandez R, Southwick K, Furuya EY, Calfee DP, Hamula C, Patel G, Barrett P, Lafaro P, Berkow EL, Moulton-Meissner H, Noble-Wang J, Fagan RP, Jackson BR, Lockhart SR, Litvintseva AP, Chiller TM. Investigation of the first seven reported cases of Candida auris, a globally emerging invasive, multidrug-resistant fungus-United States, May 2013-August 2016. Am J Transplant. 2017;17(1):296–299. doi: 10.1111/ajt.14121. [DOI] [PubMed] [Google Scholar]
- 17.Eyre DW, Sheppard AE, Madder H, Moir I, Moroney R, Quan TP, Griffiths D, George S, Butcher L, Morgan M, Newnham R, Sunderland M, Clarke T, Foster D, Hoffman P, Borman AM, Johnson EM, Moore G, Brown CS, Walker AS, Peto TEA, Crook DW, Jeffery KJM. A Candida auris outbreak and its control in an intensive care setting. N Engl J Med. 2018;379(14):1322–1331. doi: 10.1056/NEJMoa1714373. [DOI] [PubMed] [Google Scholar]
- 18.Arendrup MC, Prakash A, Meletiadis J, Sharma C, Chowdhary A. Comparison of EUCAST and CLSI reference microdilution MICs of eight antifungal compounds for Candida auris and associated tentative epidemiological cutoff values. Antimicrob Agents Chemother. 2017;61(6). 10.1128/aac.00485-17. [DOI] [PMC free article] [PubMed]
- 19.Worth LJ, Harrison SJ, Dickinson M, van Diemen A, Breen J, Harper S, et al. Candida auris in an Australian health care facility: importance of screening high risk patients. Med J Aust. 2020;212(11):510-1 e1. 10.5694/mja2.50612. [DOI] [PubMed]
- 20.• Centers for Disease Control and Prevention. Tracking Candida auris. (https://www.cdc.gov/fungal/candida-auris/tracking-c-auris.html#world). Accessed 1st August 2020. An informative, user friendly guide to laboratory diagnostic methods, the CDC algorithm is useful to those working in similarly resourced laboratories.
- 21.Karmarkar E, O'Donnell K, Prestel C, Forsberg K, Schan D, Chow N, et al. Regional assessment and containment of Candida auris transmission in post-acute care settings - Orange County, California, 2019. Open Forum Infectious Diseases. 2019;6:S993. doi: 10.1093/ofid/ofz415.2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kohlenberg A, Struelens M, Monnet D. Candida auris: epidemiological situation, laboratory capacity and preparedness in European Union and European Economic Area countries, 2013-2017. D Pla, Eurosurveillance. 2018;23(13). 10.2807/1560-7917.ES.2018.23.13.18-00136. [DOI] [PMC free article] [PubMed]
- 23.Lee WG, Shin JH, Uh Y, Kang MG, Kim SH, Park KH, Jang HC. First three reported cases of nosocomial fungemia caused by Candida auris. J Clin Microbiol. 2011;49(9):3139–3142. doi: 10.1128/JCM.00319-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Biswal M, Rudramurthy SM, Jain N, Shamanth AS, Sharma D, Jain K, Yaddanapudi LN, Chakrabarti A. Controlling a possible outbreak of Candida auris infection: lessons learnt from multiple interventions. J Hosp Infect. 2017;97(4):363–370. doi: 10.1016/j.jhin.2017.09.009. [DOI] [PubMed] [Google Scholar]
- 25.Piedrahita CT, Cadnum JL, Jencson AL, Shaikh AA, Ghannoum MA, Donskey CJ. Environmental surfaces in healthcare facilities are a potential source for transmission of Candida auris and other Candida species. Infect Control Hosp Epidemiol. 2017;38(9):1107–1109. doi: 10.1017/ice.2017.127. [DOI] [PubMed] [Google Scholar]
- 26.Welsh RM, Bentz ML, Shams A, Houston H, Lyons A, Rose LJ, et al. Survival, persistence, and isolation of the emerging multidrug-resistant pathogenic yeast Candida auris on a plastic health care surface. J Clin Microbiol. 2017;55(10):2996–3005. doi: 10.1128/JCM.00921-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pacilli M, Kerins JL, Clegg WJ, Walblay KA, Adil H, Kemble SK, Xydis S, McPherson TD, Lin MY, Hayden MK, Froilan MC, Soda E, Tang AS, Valley A, Forsberg K, Gable P, Moulton-Meissner H, Sexton DJ, Jacobs Slifka KM, Vallabhaneni S, Walters MS, Black SR. Regional emergence of Candida auris in Chicago and lessons learned from intensive follow-up at one ventilator-capable skilled nursing facility. Clin Infect Dis. 2020;71:e718–e725. doi: 10.1093/cid/ciaa435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsay S, Welsh RM, Adams EH, Chow NA, Gade L, Berkow EL, et al. Notes from the field: ongoing transmission of Candida auris in health care facilities - United States, June 2016-May 2017. MMWR Morbidity and mortality weekly report. 2017;66(19):514–515. doi: 10.15585/mmwr.mm6619a7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Centers for Disease Control and Prevention. Identification of Candida auris. https://www.cdc.gov/fungal/candida-auris/identification.html? CDC_AA_refVal=https%3A%2F%2Fwww.cdc.gov%2Ffungal%2Fcandida-auris%2Frecommendations.html. Accessed 1st August 2020.
- 30.Chibabhai V, Fadana V, Bosman N, Nana T. Comparative sensitivity of 1,3 beta-D-glucan for common causes of candidaemia in South Africa. Mycoses. 2019;62(11):1023–1028. doi: 10.1111/myc.12982. [DOI] [PubMed] [Google Scholar]
- 31.Chew SM, Sweeney N, Kidd SE, Reed C. Candida auris arriving on our shores: an Australian microbiology laboratory's experience. Pathology. 2019;51(4):431–433. doi: 10.1016/j.pathol.2019.01.009. [DOI] [PubMed] [Google Scholar]
- 32.• Borman AM, Fraser M, Johnson EM. CHROMagarTM Candida Plus: a novel chromogenic agar that permits the rapid identification of Candida auris. Medical mycology. 2020. 10.1093/mmy/myaa049Report of a novel chromogenic agar. [DOI] [PubMed]
- 33.Kathuria S, Singh PK, Sharma C, Prakash A, Masih A, Kumar A, Meis JF, Chowdhary A. Multidrug-resistant Candida auris misidentified as Candida haemulonii: characterization by matrix-assisted laser desorption ionization-time of flight mass spectrometry and DNA sequencing and its antifungal susceptibility profile variability by Vitek 2, CLSI broth microdilution, and Etest method. J Clin Microbiol. 2015;53(6):1823–1830. doi: 10.1128/jcm.00367-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Szekely A, Borman AM, Johnson EM. Candida auris isolates of the Southern Asian and South African lineages exhibit different phenotypic and antifungal susceptibility profiles in vitro. J Clin Microbiol. 2019;57(5). 10.1128/jcm.02055-18. [DOI] [PMC free article] [PubMed]
- 35.Cendejas-Bueno E, Kolecka A, Alastruey-Izquierdo A, Theelen B, Groenewald M, Kostrzewa M, Cuenca-Estrella M, Gómez-López A, Boekhout T. Reclassification of the Candida haemulonii complex as Candida haemulonii (C. haemulonii group I), C. duobushaemulonii sp. nov. (C. haemulonii group II), and C. haemulonii var. vulnera var. nov.: three multiresistant human pathogenic yeasts. J Clin Microbiol. 2012;50(11):3641–3651. doi: 10.1128/JCM.02248-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chowdhary A, Sharma C, Duggal S, Agarwal K, Prakash A, Singh PK, Jain S, Kathuria S, Randhawa HS, Hagen F, Meis JF. New clonal strain of Candida auris, Delhi. India. Emerg Infect Dis. 2013;19(10):1670–1673. doi: 10.3201/eid1910.130393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fraser M, Brown Z, Houldsworth M, Borman AM, Johnson EM. Rapid identification of 6328 isolates of pathogenic yeasts using MALDI-ToF MS and a simplified, rapid extraction procedure that is compatible with the Bruker Biotyper platform and database. Medical mycology. 2016;54(1):80–88. doi: 10.1093/mmy/myv085. [DOI] [PubMed] [Google Scholar]
- 38.Ghosh AK, Paul S, Sood P, Rudramurthy SM, Rajbanshi A, Jillwin TJ, Chakrabarti A. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry for the rapid identification of yeasts causing bloodstream infections. Clin Microbiol Infect. 2015;21(4):372–378. doi: 10.1016/j.cmi.2014.11.009. [DOI] [PubMed] [Google Scholar]
- 39.Mizusawa M, Miller H, Green R, Lee R, Durante M, Perkins R, Hewitt C, Simner PJ, Carroll KC, Hayden RT, Zhang SX. Can multidrug-resistant Candida auris be reliably identified in clinical microbiology laboratories? J Clin Microbiol. 2017;55(2):638–640. doi: 10.1128/JCM.02202-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grenfell RC, da Silva Junior AR, Del Negro GM, Munhoz RB, Gimenes VM, Assis DM, et al. Identification of Candida haemulonii Complex Species: Use of ClinProTools(TM) to Overcome Limitations of the Bruker Biotyper(TM), VITEK MS(TM) IVD, and VITEK MS(TM) RUO Databases. Front Microbiol. 2016;7:940. doi: 10.3389/fmicb.2016.00940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.RCPAQAP. Mycology Generic Report. Sydney, Australia: the Royal College of Pathologists of Australasia Quality Assurance Program (RCPAQAP)2018 25/10/2018.
- 42.Girard V, Mailler S, Chetry M, Vidal C, Durand G, van Belkum A, Colombo AL, Hagen F, Meis JF, Chowdhary A. Identification and typing of the emerging pathogen Candida auris by matrix-assisted laser desorption ionisation time of flight mass spectrometry. Mycoses. 2016;59(8):535–538. doi: 10.1111/myc.12519. [DOI] [PubMed] [Google Scholar]
- 43.Vatanshenassan M, Boekhout T, Meis JF, Berman J, Chowdhary A, Ben-Ami R, et al. Candida auris identification and rapid antifungal susceptibility testing against Echinocandins by MALDI-TOF MS. Front Cell Infect Microbiol. 2019;9:20. doi: 10.3389/fcimb.2019.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Almaghrabi RS, Albalawi R, Mutabagani M, Atienza E, Aljumaah S, Gade L, Forsberg K, Litvintseva A, Althawadi S. Molecular characterisation and clinical outcomes of Candida auris infection: single-centre experience in Saudi Arabia. Mycoses. 2020;63(5):452–460. doi: 10.1111/myc.13065. [DOI] [PubMed] [Google Scholar]
- 45.• ElBaradei A. A decade after the emergence of Candida auris: what do we know? Eur J Clin Microbiol Infect Dis. 2020. doi:10.1007/s10096-020-03886-9. Excellent review including epidemiology. [DOI] [PubMed]
- 46.• Kordalewska M, Perlin DS. Molecular diagnostics in the times of surveillance for Candida auris. J Fungi (Basel). 2019;5(3). 10.3390/jof5030077Excellent review focusing on molecular diagnostics and mechanisms of resistance inC. auris. [DOI] [PMC free article] [PubMed]
- 47.Osei SJ. Candida auris: A systematic review and meta-analysis of current updates on an emerging multidrug-resistant pathogen. Microbiologyopen. 2018;7(4):e00578. doi: 10.1002/mbo3.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kordalewska M, Lee A, Park S, Berrio I, Chowdhary A, Zhao Y, et al. Understanding echinocandin resistance in the emerging pathogen Candida auris. Antimicrob Agents Chemother. 2018;62(6). 10.1128/aac.00238-18. [DOI] [PMC free article] [PubMed]
- 49.Lima A, Widen R, Vestal G, Uy D, Silbert S. A TaqMan probe-based real-time PCR assay for the rapid identification of the emerging multidrug-resistant pathogen Candida auris on the BD max system. Journal of Clinical Microbiology. 2019;57(7):e01604–e01618. doi: 10.1128/JCM.01604-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leach L, Russell A, Zhu Y, Chaturvedi S, Chaturvedi V. A rapid and automated sample-to-result Candida auris real-time PCR assay for high-throughput testing of surveillance samples with the BD max open system. J Clin Microbiol. 2019;57(10). 10.1128/JCM.00630-19. [DOI] [PMC free article] [PubMed]
- 51.Sexton DJ, Kordalewska M, Bentz ML, Welsh RM, Perlin DS, Litvintseva AP. Direct detection of emergent fungal pathogen Candida auris in clinical skin swabs by SYBR green-based quantitative PCR assay. Journal of Clinical Microbiology. 2018;56(12):e01337–e01318. doi: 10.1128/JCM.01337-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamamoto M, Alshahni MM, Tamura T, Satoh K, Iguchi S, Kikuchi K, Mimaki M, Makimura K. Rapid detection of Candida auris based on loop-mediated isothermal amplification (LAMP) Journal of Clinical Microbiology. 2018;56(9):e00591–e00518. doi: 10.1128/JCM.00591-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oh BJ, Shin JH, Kim MN, Sung H, Lee K, Joo MY, Shin MG, Suh SP, Ryang DW. Biofilm formation and genotyping of Candida haemulonii, Candida pseudohaemulonii, and a proposed new species (Candida auris) isolates from Korea. Medical mycology. 2011;49(1):98–102. doi: 10.3109/13693786.2010.493563. [DOI] [PubMed] [Google Scholar]
- 54.Lepak AJ, Zhao M, Berkow EL, Lockhart SR, Andes DR. Pharmacodynamic optimization for treatment of invasive Candida auris infection. Antimicrob Agents Chemother. 2017;61(8). 10.1128/aac.00791-17. [DOI] [PMC free article] [PubMed]
- 55.Chowdhary A, Prakash A, Sharma C, Kordalewska M, Kumar A, Sarma S, Tarai B, Singh A, Upadhyaya G, Upadhyay S, Yadav P, Singh PK, Khillan V, Sachdeva N, Perlin DS, Meis JF. A multicentre study of antifungal susceptibility patterns among 350 Candida auris isolates (2009-17) in India: role of the ERG11 and FKS1 genes in azole and echinocandin resistance. J Antimicrob Chemother. 2018;73(4):891–899. doi: 10.1093/jac/dkx480. [DOI] [PubMed] [Google Scholar]
- 56.Berkow EL, Lockhart SR. Activity of CD101, a long-acting echinocandin, against clinical isolates of Candida auris. Diagn Microbiol Infect Dis. 2018;90(3):196–197. doi: 10.1016/j.diagmicrobio.2017.10.021. [DOI] [PubMed] [Google Scholar]
- 57.Munoz JF, Gade L, Chow NA, Loparev VN, Juieng P, Berkow EL, et al. Genomic insights into multidrug-resistance, mating and virulence in Candida auris and related emerging species. Nat Commun. 2018;9(1):5346. doi: 10.1038/s41467-018-07779-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Healey KR, Kordalewska M, Jiménez Ortigosa C, Singh A, Berrío I, Chowdhary A et al. Limited ERG11 mutations identified in isolates of Candida auris directly contribute to reduced azole susceptibility. Antimicrob Agents Chemother. 2018;62(10). doi:10.1128/aac.01427-18. [DOI] [PMC free article] [PubMed]
- 59.Rhodes J, Abdolrasouli A, Farrer RA, Cuomo CA, Aanensen DM, Armstrong-James D, Fisher MC, Schelenz S. Genomic epidemiology of the UK outbreak of the emerging human fungal pathogen Candida auris. Emerg Microbes Infect. 2018;7(1):43–12. doi: 10.1038/s41426-018-0045-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hou X, Lee A, Jiménez-Ortigosa C, Kordalewska M, Perlin DS, Zhao Y. Rapid detection of ERG11-Associated azole resistance and FKS-associated echinocandin resistance in Candida auris. Antimicrobial Agents and Chemotherapy. 2019;63(1):e01811–e01818. doi: 10.1128/AAC.01811-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kwon YJ, Shin JH, Byun SA, Choi MJ, Won EJ, Lee D, et al. Candida auris clinical isolates from South Korea: identification, antifungal susceptibility, and genotyping. J Clin Microbiol. 2019;57(4). 10.1128/jcm.01624-18. [DOI] [PMC free article] [PubMed]
- 62.Delavy M, Dos Santos AR, Heiman CM, Coste AT. Investigating antifungal susceptibility in Candida Species with MALDI-TOF MS-based assays. Front Cell Infect Microbiol. 2019;9:19. doi: 10.3389/fcimb.2019.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hata DJ, Humphries R, Lockhart SR. Candida auris: an emerging yeast pathogen posing distinct challenges for laboratory diagnostics, treatment, and infection prevention. Archives of pathology & laboratory medicine. 2020;144(1):107–114. doi: 10.5858/arpa.2018-0508-RA. [DOI] [PubMed] [Google Scholar]
- 64.Pekard-Amenitsch S, Schriebl A, Posawetz W, Willinger B, Kölli B, Buzina W. Isolation of Candida auris from ear of otherwise healthy patient, Austria, 2018. Emerging Infectious Disease journal. 2018;24(8):1596–1597. doi: 10.3201/eid2408.180495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Letscher-Bru V, Meyer M-H, Galoisy A-C, Waller J, Candolfi E. Prospective evaluation of the new chromogenic medium Candida ID, in comparison with Candiselect, for isolation of molds and isolation and presumptive identification of yeast species. Journal of Clinical Microbiology. 2002;40(4):1508–1510. doi: 10.1128/JCM.40.4.1508-1510.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mulet Bayona JV, García CS, Palop NT, Cardona CG. Evaluation of a novel chromogenic medium for Candida spp. identification and comparison with CHROMagar™ Candida for the detection of Candida auris in surveillance samples. Diagnostic Microbiology and Infectious Disease. 2020. 10.1016/j.diagmicrobio.2020.115168. [DOI] [PubMed]