Key Points
Question
Can major metabolite signatures be observed in gastric lesions that progress toward gastric cancer?
Findings
In this cohort study including 400 participants, distinct plasma metabolomic profiles were observed for precancerous gastric lesions and gastric cancer. Metabolite signatures, particularly α-linolenic acid, linoleic acid, and palmitic acid, were associated with progression of gastric lesions and risk of early gastric cancer.
Meaning
The findings of this study suggest that α-linolenic acid, linoleic acid, and palmitic acid may be meaningful biomarkers for assessing high-risk populations and early diagnosis of gastric cancer, benefiting targeted gastric cancer prevention and control in health care and general practice.
Abstract
Importance
Metabolic deregulation plays an important role in gastric cancer (GC) development. To date, no studies have comprehensively explored the metabolomic profiles along the cascade of gastric lesions toward GC.
Objective
To draw a metabolic landscape and define metabolomic signatures associated with the progression of gastric lesions and risk of early GC.
Design, Setting, and Participants
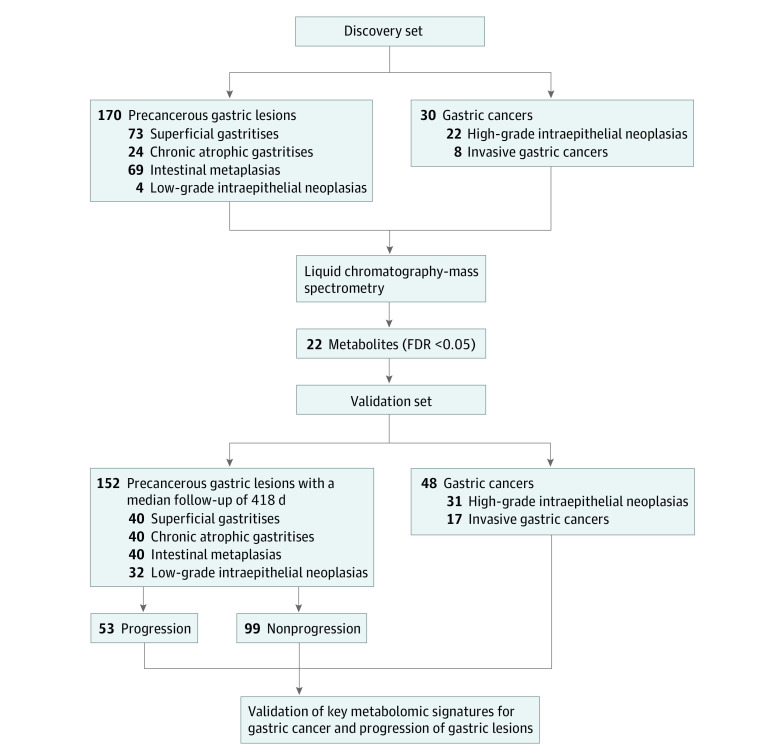
A 2-stage, population-based cohort study was initiated in 2017 in Linqu County, Shandong Province, China, a high-risk area for GC. Prospective follow-up was conducted during the validation stage (June 20, 2017, to May 27, 2020). A total of 400 individuals were included based on the National Upper Gastrointestinal Cancer Early Detection Program in China. The discovery stage involved 200 individuals with different gastric lesions or GC (high-grade intraepithelial neoplasia or invasive GC). The validation stage prospectively enrolled 152 individuals with gastric lesions who were followed up for 118 to 1063 days and 48 individuals with GC.
Exposures
Metabolomic profiles and metabolite signatures were examined based on untargeted plasma metabolomics assay.
Main Outcomes and Measures
The risk of GC overall and early GC (high-grade intraepithelial neoplasia), and progression of gastric lesions.
Results
Of the 400 participants, 124 of 200 (62.0%) in the discovery set were men; mean (SD) age was 56.8 (7.5) years. In the validation set, 136 of 200 (68.0%) were men; mean (SD) age was 57.5 (8.1) years. Distinct metabolomic profiles were noted for gastric lesions and GC. Six metabolites, including α-linolenic acid, linoleic acid, palmitic acid, arachidonic acid, sn-1 lysophosphatidylcholine (LysoPC)(18:3), and sn-2 LysoPC(20:3) were significantly inversely associated with risk of GC overall and early GC (high-grade intraepithelial neoplasia). Among these metabolites, the first 3 were significantly inversely associated with gastric lesion progression, especially for the progression of intestinal metaplasia (α-linolenic acid: OR, 0.42; 95% CI, 0.18-0.98; linoleic acid: OR, 0.43; 95% CI, 0.19-1.00; and palmitic acid: OR, 0.32; 95% CI, 0.13-0.78). Compared with models including only age, sex, Helicobacter pylori infection, and gastric histopathologic findings, integrating these metabolites significantly improved the performance for predicting the progression of gastric lesions (area under the curve [AUC], 0.86; 95% CI, 0.70-1.00 vs AUC, 0.69; 95% CI, 0.50-0.88; P = .02) and risk of early GC (AUC, 0.83; 95% CI, 0.58-1.00 vs AUC, 0.61; 95% CI, 0.31-0.91; P = .03).
Conclusions and Relevance
This study defined metabolite signatures that might serve as meaningful biomarkers for assessing high-risk populations and early diagnosis of GC, possibly advancing targeted GC prevention and control.
This cohort study examines plasma metabolomics in individuals with gastric lesions and gastric cancer to identify metabolite signatures that might serve as biomarkers for risk of progression of the lesions to cancer.
Introduction
Gastric cancer (GC) ranks as the fifth most commonly diagnosed cancer and the third leading cause of cancer death worldwide.1 In most patients, GC is diagnosed at a local advanced or advanced stage with unfavorable prognosis.2 The development of GC, particularly the intestinal type, involves a stepwise progression of a cascade of gastric lesions, from superficial gastritis (SG), chronic atrophic gastritis (CAG), intestinal metaplasia (IM), and low-grade intraepithelial neoplasia (LGIN) to high-grade intraepithelial neoplasia (HGIN) and invasive GC.3,4 Defining populations at a particularly high risk for the cascade progression of gastric lesions and identifying cases of GC at an early stage are necessary for implementing efficient prevention and management strategies. However, the cause of underlying gastric carcinogenesis remains poorly understood, even though Helicobacter pylori has been recognized as a major risk factor.5,6 In addition, biomarkers that can predict the progression of precancerous gastric lesions and development of GC are limited. Efforts are warranted to promote etiologic research and define novel molecular signatures behind gastric lesion progression and GC occurrence, considering challenges to GC prevention and control.
Reprogramming cellular energy metabolism is one core hallmark of cancer cells and supports neoplastic proliferation most effectively and fosters carcinogenesis.7,8,9 Metabolic perturbation has been shown to play an important role in GC occurrence,10,11,12,13 corroborating the biological plausibility for characterizing metabolic profiles underlying GC development. The increased availability of high-throughput technologies of metabolomics has facilitated the identification and quantification of entire endogenous, low-molecular-weight metabolites14,15 that reflect the end product of the complex interaction of host, gene, and environment,16 therefore representing a potential tool for distinguishing molecular profiles and identifying novel biomarkers for GC.
A number of studies have examined metabolic profiles associated with GC previously, as summarized by a systematic review,12 but were often restricted by a modest sample size and lacking a replication data set. In addition, to our knowledge, no studies have comprehensively explored metabolomic profiles associated with the progression of precancerous gastric lesions to GC. In this study, plasma metabolomic profiling was conducted among a total of 400 individuals in 2 stages involving the cascade of gastric lesions and GC, with prospective endoscopic follow-up in the validation stage. We mapped the metabolomic landscape of precancerous gastric lesions and GC and comprehensively examined metabolic signatures associated with the progression of gastric lesions and GC occurrence.
Methods
Participants in this cohort study were enrolled based on the National Upper Gastrointestinal Cancer Early Detection (UGCED) Program from Linqu County in 2017 and 2018, an established high-risk area for GC in Shandong Province, China, where most GCs are of the intestinal type.4,6 Local residents aged 40 to 69 years attending the National UGCED program receive upper gastroendoscopy examination, and those diagnosed with HGIN, pathologically including high-grade dysplasia and GC in situ, would be classified as early GC and treated immediately. The discovery stage of our study enrolled 200 individuals, including 30 cases of GC (22 HGINs and 8 invasive GCs) and 170 individuals with a global diagnosis of different gastric lesions (73 SGs, 24 CAGs, 69 IMs, and 4 LGINs) (Figure 1). The validation stage enrolled another 200 independent individuals, including 48 with GC (31 HGINs and 17 invasive GCs) and 152 with gastric lesions of different stages (40 SGs, 40 CAGs, 40 IMs, and 32 LGINs). All 152 patients with gastric lesions were prospectively followed up for 118 to 1063 days (June 20, 2017, to May 27, 2020), with gastroendoscopy conducted again at the end point (Figure 1). The second endoscopy of each participant was scheduled considering baseline gastric histopathologic factors, family history of upper gastrointestinal cancers, presence of GC risk factors, and personal willingness, leading to a wide range of follow-up times.
Figure 1. Study Outline of Work Flow.
FDR indicates false discovery rate.
A 5-mL blood sample was collected from all participants. Helicobacter pylori infection status was determined by enzyme-linked immunosorbent assay for plasma IgG.17 The study was approved and informed consent was waived by the institutional review board of Peking University Cancer Hospital because study participants were selected within the framework of the National UGCED Program. This study followed the Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) reporting guideline for cohort studies.
Using video endoscopes (Olympus Corp), upper gastroendoscopy examinations were performed by 2 gastroenterologists. Formalin-fixed, paraffin-embedded tissue samples for biopsy were reviewed blindly by 2 pathologists (including Z.-W.L.) according to the criteria proposed by the Updated Sydney System18 and the Chinese Association of Gastric Cancer.19 The results of each biopsy were reported as normal, SG, CAG, IM, LGIN, HGIN, or invasive GC, and each participant was assigned a global diagnosis based on the most severe gastric histologic finding among any biopsy. Each participant was assigned a histologic severity score at baseline and end point. A participant was classified as having progression of gastric lesions if the end point score was higher than the baseline score.
Plasma samples were processed following a previously reported method with some modifications.20 Plasma metabolomic profiling was conducted using ultra high-performance liquid chromatography–mass spectrometry.21 Details on sample preparation, metabolomic profiling, and quality control are reported in the eMethods in the Supplement.
Bioinformatics and Statistical Analyses
All analyses were conducted using R, version 3.6.2 (R Foundation for Statistical Computing). Spearman correlation coefficients were calculated for the comparison of all quality control samples. Unsupervised principal component analysis and hierarchical clustering analysis were conducted to visualize metabolomic features across participants with mild gastric lesions (SG or CAG), advanced gastric lesions (IM or LGIN), and GC (HGIN or invasive GC). Trajectories of metabolomic profiles were also examined by locally estimated scatterplot smoothing regression, and hierarchical clustering was conducted to group metabolite clusters with similar trajectories.
Logistic regression analyses adjusted for age, sex, and H pylori infection were conducted to evaluate the associations of individual metabolites with the risk of advanced gastric lesions (IM or LGIN) and GC (HGIN or invasive GC), using mild gastric lesions (SG or CAG) as the reference. For each metabolite, odds ratios (ORs) and 95% CIs were calculated per 1-SD change of log-transformed metabolite level. The association with each metabolite was also examined for tertiles of plasma levels, with the lowest tertile as the reference. The Benjamini-Hochberg method was used to control for multiple comparisons, with false discovery rate–adjusted q values less than 0.05 considered statistically significant in the discovery set. For individual metabolites significantly associated with GC, metabolite set enrichment analyses were conducted to explore the significantly enriched pathways for GC, using R package (MetaboAnalystR), with P < .05 as the cutoff level. Metabolite-set enrichment analysis is a group-based enrichment analysis that identifies and interprets patterns of metabolite changes in a biologically meaningful context based on metabolite and metabolic pathway information from the literature and public databases.22
We further examined the associations between validated metabolites and the evolution of precancerous gastric lesions (progression vs nonprogression) using conditional logistic regression models stratified on baseline gastric histologic factors,6 with adjustment for age, sex, and H pylori infection. Because cohort follow-up completely relied on gastroendoscopy, we did not have information on the exact occurrence time of progression of gastric lesions, so use of the Cox proportional hazards regression model may not be optimal. Considering IM as a crucial preneoplastic lesion during gastric carcinogenesis, analyses were also conducted to examine the associations of key metabolites with the risk for progression of IM.
A metabolic score was calculated by summing the weighted level of key metabolites associated with gastric lesion progression, calculated as the standardized expression value of each metabolite weighted by its regression coefficient from logistic regression. The association between metabolite score and risk of gastric lesion progression was examined using logistic regression models.
Random forest classifier was used to construct prediction models for the risk of gastric lesion progression as well as the occurrence of early GC integrating key metabolites with other variables. Random forest classifier is an ensemble of multiple decision trees such that each tree is constructed to predict the dependent variable based on a random bootstrap of samples and a random subset of variables proved to be effective in prediction.23 A receiver operating characteristic curve was plotted, with area under the curve (AUC) calculated. The Delong test was conducted to compare the predictive values of prediction models.
Results
The study involved a total of 400 individuals, including 200 in the discovery set (124 men [62.0%] and 76 women [38.0%]; mean [SD] age, 56.8 [7.5] years) and 200 in the validation set (136 [68.0%] men and 64 women [32.0%]; mean [SD] age, 57.5 [8.1] years) (eTable 1 in the Supplement). Assays of quality control samples showed Spearman correlation coefficients of 0.99 on average, indicating high reproducibility of metabolomics data generated from liquid chromatography–mass spectrometry analysis. All study samples and quality control samples showed good consistency in the quantification of plasma metabolite levels (eFigure 1 in the Supplement).
Distinct metabolomic profiles were revealed between participants with precancerous gastric lesions and GC (eFigure 2A,B in the Supplement). We examined alterations in global metabolite trajectories and defined 6 metabolite clusters that displayed dynamic changes in trajectories from precancerous lesions to GC. For example, metabolites of cluster 5 exhibited increased levels in participants with mild gastric lesions (SG or CAG) to advanced lesions (IM or LGIN) and then to HGIN or GC. In contrast, metabolites of cluster 1 displayed a decreasing trend with severity of gastric lesions but remained relatively steady from advanced gastric lesions to GC. The trajectories of clusters 2, 3, 4, and 6 metabolites appeared undulated, displaying J-shaped changes (eFigure 2C in the Supplement).
Compared with SG or CAG, 22 plasma metabolites were associated with GC (HGIN or invasive GC) in the discovery stage after correction for multiple comparisons (false discovery rate <0.05) (eTable 2 in the Supplement; Figure 2A). However, none of the metabolites was associated with IM or LGIN after multiple comparison adjustment (false discovery rate <0.05). Of the 22 metabolites, the association of 6 metabolites with GC, including α-linolenic acid (OR, 0.53; 95% CI, 0.36-0.78), linoleic acid (OR, 0.56; 95% CI, 0.38-0.81)), palmitic acid (OR, 0.56; 95% CI, 0.38-0.82), arachidonic acid (OR, 0.70; 95% CI, 0.50-0.99), sn-1 lysophosphatidylcholine (LysoPC)(18:3) (OR, 0.62; 95% CI, 0.43-0.88), and sn-2 LysoPC(20:3) (OR, 0.65; 95% CI, 0.45-0.94), were replicated in the validation stage (Table 1 and Figure 2A). Combining the discovery and validation sets, these 6 metabolites were inversely associated with the risk of GC in a dose-dependent manner (Figure 2B). Analyses restricted to early GC (HGIN) also found inverse associations with these 6 metabolites (Figure 2C).
Figure 2. Plasma Metabolites Associated With Advanced Gastric Lesions and Gastric Cancer (GC).
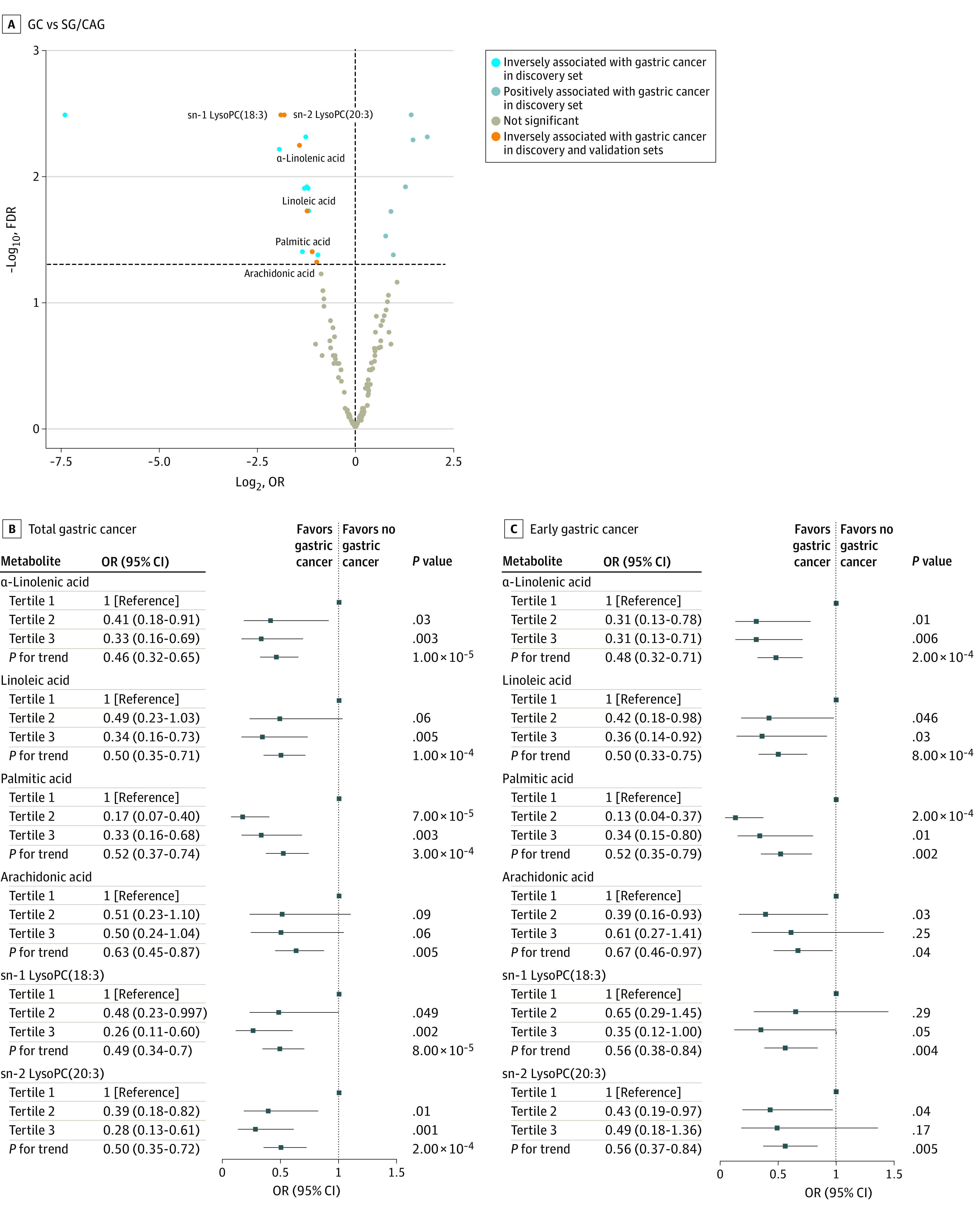
A, Metabolites associated with GC. Metabolites are significantly positively (false discovery rate [FDR], q <0.05 and odds ratio [OR], >1) or inversely (FDR, q <0.05 and OR, <1) associated with GC in the discovery set by logistic regression adjusting for age, sex, and Helicobacter pylori infection; orange color displaying metabolites successfully validated for GC. B, Six validated metabolites associated with GC. C, Six validated metabolites associated with early GC (high-grade intraepithelial neoplasia). Odds ratios (95% CIs) for the values in panels B and C were calculated by logistic regression adjusted for age, sex, and H pylori infection, combining the discovery and validation stages.
Table 1. Top Plasma Metabolites Associated With Advanced Gastric Lesions and GCa.
| Metabolite | Discovery set | Validation set | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IM/LGIN vs SG/CAG | GC vs SG/CAG | Early GCb vs SG/CAG | IM/LGIN vs SG/CAG | GC vs SG/CAG | Early GCb vs SG/CAG | |||||||||||||||||||||
| No./total No. | OR (95% CI) | P value | FDR | No./total No. | OR (95% CI) | P value | FDR | No./total No. | OR (95% CI) | P value | FDR | No./total No. | OR (95% CI) | P value | No./total No. | OR (95% CI) | P value | No./total No. | OR (95% CI) | P value | ||||||
| sn-2 LysoPC(20:3) | 73/97 | 1.31 (0.93-1.87) | .12 | 0.41 | 30/97 | 0.29 (0.14-0.51) | <.001 | 0.003 | 22/97 | 0.21 (0.08-0.43) | <.001 | 0.01 | 72/80 | 0.58 (1.19-1.21) | .001 | 48/80 | 0.65 (0.45-0.94) | .03 | 31/80 | 0.78 (0.53-1.16) | .15 | |||||
| sn-1 LysoPC(18:3) | 73/97 | 1.26 (0.90-1.77) | .18 | 0.47 | 30/97 | 0.27 (0.13-0.50) | <.001 | 0.003 | 22/97 | 0.26 (0.11-0.51) | <.001 | 0.01 | 72/80 | 0.68 (1.14-1.26) | .02 | 48/80 | 0.62 (0.43-0.88) | .01 | 31/80 | 0.76 (0.51-1.11) | .12 | |||||
| α-Linolenic acid | 73/97 | 1.09 (0.78-1.53) | .62 | 0.84 | 30/97 | 0.37 (0.21-0.62) | <.001 | 0.006 | 22/97 | 0.30 (0.14-0.55) | <.001 | 0.01 | 72/80 | 0.81 (0.80-1.84) | .13 | 48/80 | 0.53 (0.36-0.78) | .003 | 31/80 | 0.62 (0.41-0.92) | .02 | |||||
| Linoleic acid | 73/97 | 1.12 (0.80-1.56) | .51 | 0.76 | 30/97 | 0.43 (0.24-0.72) | .002 | 0.02 | 22/97 | 0.32 (0.15-0.60) | .001 | 0.02 | 72/80 | 0.80 (0.82-1.74) | .11 | 48/80 | 0.56 (0.38-0.81) | .01 | 31/80 | 0.63 (0.42-0.96) | .04 | |||||
| Palmitic acid | 73/97 | 1.23 (0.89-1.71) | .22 | 0.51 | 30/97 | 0.47 (0.26-0.79) | .007 | 0.04 | 22/97 | 0.32 (0.15-0.62) | .002 | 0.02 | 72/80 | 0.70 (1.11-1.30) | .02 | 48/80 | 0.56 (0.38-0.82) | .01 | 31/80 | 0.65 (0.43-0.98) | .04 | |||||
| Arachidonic acid | 73/97 | 1.35 (0.98-1.88) | .07 | 0.41 | 30/97 | 0.51 (0.28-0.85) | .01 | 0.05 | 22/97 | 0.42 (0.20-0.78) | .01 | 0.07 | 72/80 | 0.88 (0.58-2.44) | .22 | 48/80 | 0.70 (0.50-0.99) | .05 | 31/80 | 0.83 (0.57-1.21) | .21 | |||||
Abbreviations: CAG, chronic atrophic gastritis; FDR, false discovery rate; GC, gastric cancer; HGIN, high-grade intraepithelial neoplasia; IM, intestinal metaplasia; LGIN, low-grade intraepithelial neoplasia; LysoPC, lysophosphatidylcholine; OR, odds ratio; SG, superficial gastritis.
Individual metabolites associated with GC in the discovery stage (FDR <0.05) and validation stage (P < .05) are shown here. Logistic regression analyses were conducted adjusting for age, sex, and Helicobacter pylori infection. Metabolites are ranked by the P values for GC in the discovery set.
We defined participants with HGIN as early GC cases.
Metabolite-set enrichment analyses revealed that the pathways of glutamate metabolism, α-linolenic acid and linoleic acid metabolism (fold enrichment, 5.00; P = .006), and arachidonic acid metabolism (fold enrichment, 5.00; P = .04) were among the top enriched pathways associated with GC (eFigure 3 and eTable 3 in the Supplement).
Of the 6 validated metabolites associated with GC, α-linolenic acid, linoleic acid, and palmitic acid further demonstrated significantly inverse associations with the progression of gastric lesions during the follow-up period. Participants who had progression of gastric lesions during follow-up had significantly lower levels of α-linolenic acid (OR, 0.64; 95% CI, 0.45-0.91), linoleic acid (OR, 0.69; 95% CI, 0.48-0.99), and palmitic acid (OR, 0.63; 95% CI, 0.43-0.92) at baseline compared with those who did not have progression (Table 2 and Figure 3A). These 3 metabolites were also significantly inversely associated with progression of IM during follow-up (Table 2 and Figure 3B). Sensitivity analyses excluding the outliers (above or below 3 SDs from the mean level) did not materially change the results.
Table 2. Association of Top Metabolites With the Progression of Gastric Lesions During Follow-up of the Validation Set Participantsa.
| Metabolite | Progression vs nonprogression for all participants with prospective follow-up | Progression vs nonprogression for IM cases only | ||||
|---|---|---|---|---|---|---|
| No./total No. | OR (95% CI) | P value | No./total No. | OR (95% CI) | P value | |
| sn-2 LysoPC(20:3) | 53/99 | 1.13 (0.80-1.59) | .27 | 9/31 | 0.72 (0.35-1.48) | .23 |
| sn-1 LysoPC(18:3) | 53/99 | 0.82 (0.59-1.15) | .17 | 9/31 | 0.58 (0.27-1.25) | .12 |
| α-Linolenic acid | 53/99 | 0.64 (0.45-0.91) | .02 | 9/31 | 0.42 (0.18-0.98) | .05 |
| Linoleic acid | 53/99 | 0.69 (0.48-0.99) | .05 | 9/31 | 0.43 (0.19-1.00) | .05 |
| Palmitic acid | 53/99 | 0.63 (0.43-0.92) | .02 | 9/31 | 0.32 (0.13-0.78) | .02 |
| Arachidonic acid | 53/99 | 0.90 (0.65-1.25) | .30 | 9/31 | 0.47 (0.21-1.07) | .07 |
Abbreviations: IM, intestinal metaplasia; LysoPC, lysophosphatidylcholine; OR, odds ratio.
Individual metabolites validated to be associated with gastric cancer are shown here, ranked in the same order as given in Table 1. Conditional logistic regression analyses stratified on baseline gastric histopathologic factors were conducted, with adjustment for age, sex, and Helicobacter pylori infection.
Figure 3. Validated Metabolites and Prediction Models for Gastric Lesion Progression and Risk of Early Gastric Cancer (GC).
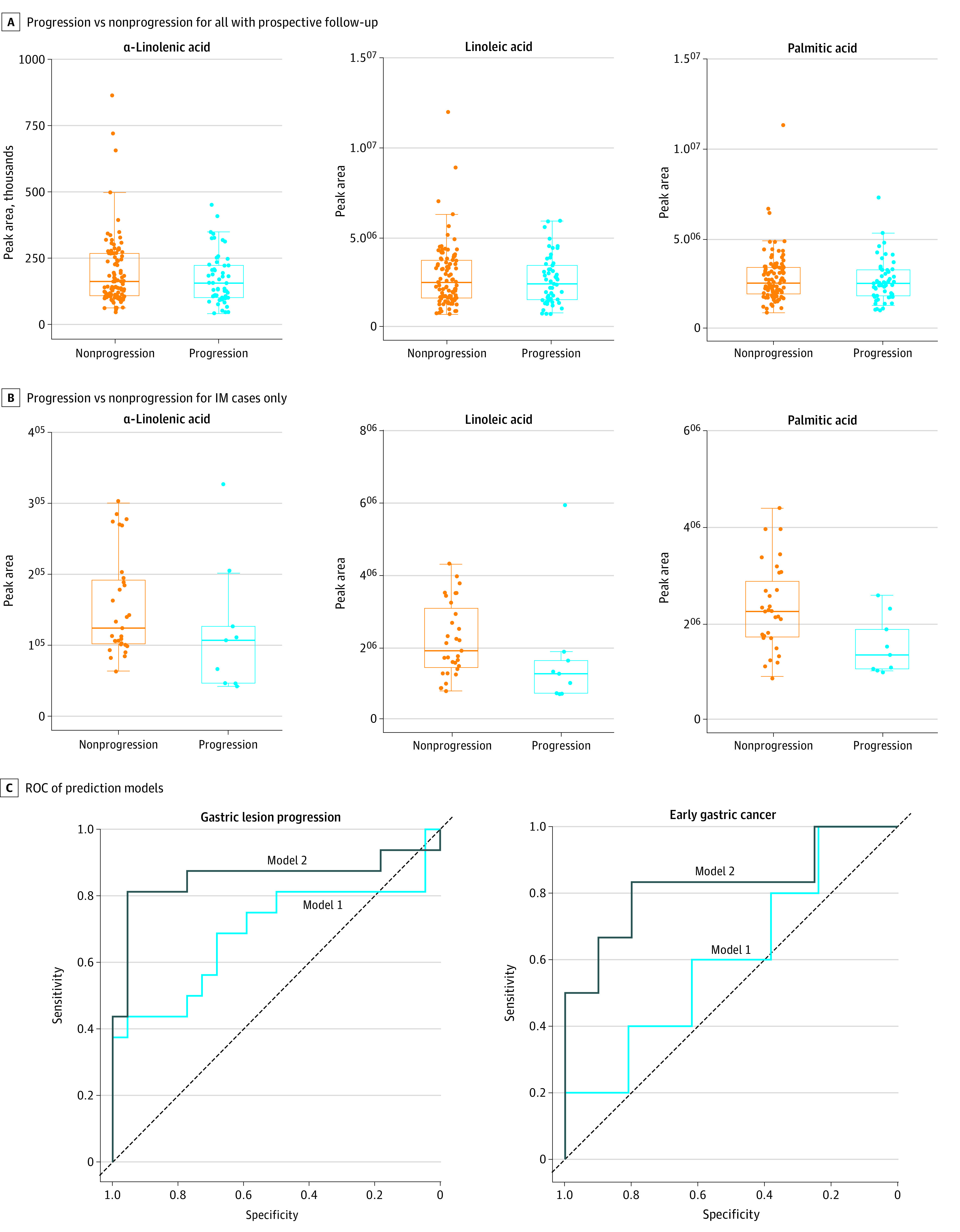
A-B, P values are available in eTable 2 in the Supplement. C, Model 1: age, sex, H pylori infection, and baseline histologic factors; model 2: metabolic scores of the highlighted 3 metabolites. For gastric lesion progression, model 1: area under the curve (AUC), 0.69 (95% CI, 0.50-0.88); P = .05; model 2: AUC, 0.86 (95% CI, 0.70-1.00); P = 7.50 × 10−6. For early GC, model 1: AUC, 0.61 (95% CI, 0.31-0.91); P = .47; model 2: AUC, 0.83 (95% CI, 0.58-1.00); P = .009. IM indicates intestinal metaplasia; ROC, receiver operating characteristic.
Combining 3 validated metabolites (α-linolenic acid, linoleic acid, and palmitic acid) for the risk of gastric lesion progression and early GC, a metabolite score was calculated for each individual, which was significantly associated with the progression of gastric lesions, with an OR of 5.25 (95% CI, 1.01-30.72) per unit increase of score (eFigure 4 in the Supplement).
We sought to integrate metabolite score as a potential predictor for the risk of gastric lesion progression and early GC based on prospectively enrolled participants in the validation set (Figure 3C). Compared with the model including age, sex, H pylori infection, and baseline histologic findings, the additional combination of metabolite score significantly improved the performance in predicting the progression of gastric lesions (AUC, 0.86; 95% CI, 0.70-1.00 vs AUC, 0.69; 95% CI, 0.50-0.88; Delong test P = .02). In addition, the combination of the score also significantly strengthened the ability to predict the risk of early GC (AUC, 0.83; 95% CI, 0.58-1.00 vs AUC, 0.61; 95% CI, 0.31-0.91; Delong test P = .03).
Discussion
Based on populations from a high-risk area for GC, we noted distinct plasma metabolomic profiles for precancerous gastric lesions and GC and defined metabolomic signatures associated with the progression of gastric lesions and risk of early GC (HGIN). Six plasma metabolites were significantly inversely associated with the risk of GC overall and early GC, with 3 of these metabolites—α-linolenic acid, linoleic acid, and palmitic acid—significantly inversely associated with the risk of gastric lesion progression, especially for the progression of IM. Integrating these 3 metabolites significantly improved the ability to predict the progression of gastric lesions and the risk of early GC.
Altered metabolism is a primary feature for cancer cells during tumorigenesis.7,8,9 Metabolic alterations have been increasingly reported in GC.10,11,12,13 Accumulated evidence has shown the alterations of glucose, amino acid, lipid, and nucleotide metabolism in GC but often with discrepant findings.10,11,12,13 Most earlier studies were limited by cross-sectional or case-control design in modest sample sizes and lacking a replication set addressing concerns of false-positive probability. In our study with a reasonable sample size involving 2 stages, we found differential metabolomic profiles for precancerous gastric lesions and GC, supporting the application of metabolomic profiling in clarifying GC etiologic factors and identifying GC-related biomarkers. Six metabolite clusters were aggregated with similar changing trajectories from mild to advanced gastric lesions and GC, indicating the complexity of metabolite networks underlying gastric carcinogenesis.
Our study validated 3 plasma metabolites, including α-linolenic acid, linoleic acid, and palmitic acid, inversely associated with the risk of gastric lesion progression, GC overall, and early GC. Previous evidence suggested that linoleic acid could induce the formation of lipid peroxides that further promote apoptosis of cancer cells by enhancing reactive oxygen species generation and inducing mitochondrial dysfunction.24,25 The anticancer action of α-linolenic acid to suppress the growth and induce the apoptosis of cancer cells has also been reported by increasing reactive oxygen species generation in tumor cells.26,27 As essential fatty acids, linoleic acid and α-linolenic acid may inhibit the growth of gastric tumor cells,27 therefore suppressing gastric lesion progression. A previous study also reported an inverse association of serum linoleic acid with GC (n = 30 GCs vs n = 30 non-GCs).28 However, a small-scale study reported upregulated serum α-linolenic acid in GCs (n = 24 pairs of GCs and non-GCs in the discovery stage and 14 pairs in the validation stage).29 Palmitic acid is one major saturated fatty acid in human blood and could suppress cancer cell proliferation, viability, invasion, and migration of gastric and hepatocellular cancer cells both in vitro and in vivo.30,31,32 A recent study reported downregulated palmitic acid in GC (n = 29 GCs vs n = 20 non-GCs).31 In addition, decreased tissue palmitoleic acid levels, a product of palmitic acid, has been associated with GC (n = 30 GCs vs n = 30 adjacent healthy tissues).33 However, another study reported discrepant findings of elevated serum palmitic acid levels in GC (n = 17 GCs and 15 postoperation GCs vs n = 20 SGs).34
In our study combining α-linolenic acid, linoleic acid, and palmitic acid, a metabolite score was independently associated with the progression of gastric lesions, with a higher score indicating an increased risk of progression. Integrating these metabolites, a prediction model materially strengthened the ability to predict the progression of gastric lesions. We placed special interest in IM, considering IM as a crucial preneoplastic lesion during gastric carcinogenesis. Notably, these 3 metabolites were also inversely associated with the progression of IM. Pathway analyses revealed the enrichment of an α-linolenic acid and linoleic acid metabolism pathway in GC. Collectively, these results support the possible role of downregulation of these metabolites in early gastric carcinogenesis, but laboratory studies are needed to clarify underlying mechanisms for the observed findings.
Our study also reported significantly lower plasma arachidonic acid, sn-1 LysoPC(18:3), and sn-2 LysoPC(20:3) levels for GC overall and early GC. Arachidonic acid is also an essential fatty acid and has been involved in the inhibition of malignant cell proliferation and apoptosis, cell survival, tumor metastasis, inflammation, and neoangiogenesis.35 Altered arachidonic acid metabolism in the tumor microenvironment may have a substantial effect on tumorigenesis.35 Small-scale studies have found decreased tissue arachidonic acid levels in GC.33,36 Our study also noted an enriched arachidonic acid metabolism pathway in GC. LysoPC is a class of lipid biomolecules derived by the cleaving of PC and has been linked to tumor cell proliferation, apoptosis, migration, invasion, and inflammation in tumorigenesis.37,38 To our knowledge, no studies have reported an association of sn-1 LysoPC(18:3) and sn-2 LysoPC(20:3) with GC, but earlier studies showed decreased serum LysoPC and PC,39 plasma LysoPC, and PC levels40 as well as tissue PC levels41 in GC, consistent with our finding of downregulated plasma LysoPC in GC.
In our study, all highlighted metabolites were negative markers, displaying decreased levels with progression of gastric lesions and risk of GC. Even so, these metabolites still had meaningful abundance in GC. Instead of focusing on GCs at local advanced or advanced stages for potential therapeutic targets, the study was designed as part of our ongoing efforts to advance risk assessment and early detection of GC, concentrating on different gastric lesions and early GC (HGIN).
Limitations
This study has limitations. First, we have not yet thoroughly longitudinally followed up the discovery stage participants by endoscopy. Second, we were not able to evaluate biological functions and underlying mechanisms for the highlighted metabolomic profiles and significant metabolites. Third, all participants were enrolled from the census-registered population in Linqu County. The homogeneity of participants may have enhanced the standardized sample collection and minimized possible residual confounding from host genetic background, but the extrapolation of findings requires caution.
Conclusions
In this study, we set metabolomic profiles and identified metabolomic signatures for precancerous gastric lesions progressing to cancer in a 2-stage metabolomics investigation. Six plasma metabolites appear to be inversely associated with the risk of GC overall and early GC (HGIN), with 3 of these metabolites (α-linolenic acid, linoleic acid, and palmitic acid) potentially predicting the risk of gastric lesion progression and early GC. Pending further validation from large-scale, multicenter prospective studies, our findings may have translational implications in preventive and clinical practice. Decreased levels of these plasma metabolites may serve as noninvasive biomarkers distinguishing populations at a particularly high-risk for GC so that appropriate primary intervention strategies can be implemented. Such individuals may also benefit from targeted frequent GC screening, aiding early detection and management of GC.
eMethods. Detailed Methods
eFigure 1. Quality Control During LC-MS Analysis
eFigure 2. Metabolomic Profiles for Precancerous Gastric Lesions and GC
eFigure 3. Metabolite Set Enrichment Analysis for the Enriched Metabolite Pathways Associated With GC
eFigure 4. Metabolite Scores Associated With the Progression of Gastric Lesions During the Follow-up
eTable 1. Characteristics of Study Subjects in the Discovery and Validation Sets
eTable 2. Plasma Metabolites Associated With Advanced Gastric Lesions and Gastric Cancer
eTable 3. Metabolite Set Enrichment Analysis for Plasma Metabolites Associated With Gastric Cancer
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394-424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Zong L, Abe M, Seto Y, Ji J. The challenge of screening for early gastric cancer in China. Lancet. 2016;388(10060):2606. doi: 10.1016/S0140-6736(16)32226-7 [DOI] [PubMed] [Google Scholar]
- 3.Correa P. Human gastric carcinogenesis: a multistep and multifactorial process—First American Cancer Society Award Lecture on Cancer Epidemiology and Prevention. Cancer Res. 1992;52(24):6735-6740. [PubMed] [Google Scholar]
- 4.You WC, Li JY, Blot WJ, et al. Evolution of precancerous lesions in a rural Chinese population at high risk of gastric cancer. Int J Cancer. 1999;83(5):615-619. doi: [DOI] [PubMed] [Google Scholar]
- 5.Amieva M, Peek RM Jr. Pathobiology of Helicobacter pylori–induced gastric cancer. Gastroenterology. 2016;150(1):64-78. doi: 10.1053/j.gastro.2015.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li WQ, Zhang JY, Ma JL, et al. Effects of Helicobacter pylori treatment and vitamin and garlic supplementation on gastric cancer incidence and mortality: follow-up of a randomized intervention trial. BMJ. 2019;366:l5016. doi: 10.1136/bmj.l5016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646-674. doi: 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 8.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even Warburg did not anticipate. Cancer Cell. 2012;21(3):297-308. doi: 10.1016/j.ccr.2012.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pearce EL, Poffenberger MC, Chang CH, Jones RG. Fueling immunity: insights into metabolism and lymphocyte function. Science. 2013;342(6155):1242454. doi: 10.1126/science.1242454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiao S, Zhou L. Gastric cancer: metabolic and metabolomics perspectives. Int J Oncol. 2017;51(1):5-17. doi: 10.3892/ijo.2017.4000 [DOI] [PubMed] [Google Scholar]
- 11.Abbassi-Ghadi N, Kumar S, Huang J, Goldin R, Takats Z, Hanna GB. Metabolomic profiling of oesophago-gastric cancer: a systematic review. Eur J Cancer. 2013;49(17):3625-3637. doi: 10.1016/j.ejca.2013.07.004 [DOI] [PubMed] [Google Scholar]
- 12.Huang S, Guo Y, Li Z, et al. A systematic review of metabolomic profiling of gastric cancer and esophageal cancer. Cancer Biol Med. 2020;17(1):181-198. doi: 10.20892/j.issn.2095-3941.2019.0348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jayavelu ND, Bar NS. Metabolomic studies of human gastric cancer: review. World J Gastroenterol. 2014;20(25):8092-8101. doi: 10.3748/wjg.v20.i25.8092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nicholson JK, Lindon JC. Systems biology: metabonomics. Nature. 2008;455(7216):1054-1056. doi: 10.1038/4551054a [DOI] [PubMed] [Google Scholar]
- 15.Holmes E, Wilson ID, Nicholson JK. Metabolic phenotyping in health and disease. Cell. 2008;134(5):714-717. doi: 10.1016/j.cell.2008.08.026 [DOI] [PubMed] [Google Scholar]
- 16.Rochfort S. Metabolomics reviewed: a new “omics” platform technology for systems biology and implications for natural products research. J Nat Prod. 2005;68(12):1813-1820. doi: 10.1021/np050255w [DOI] [PubMed] [Google Scholar]
- 17.Zhang L, Blot WJ, You WC, et al. Helicobacter pylori antibodies in relation to precancerous gastric lesions in a high-risk Chinese population. Cancer Epidemiol Biomarkers Prev. 1996;5(8):627-630. [PubMed] [Google Scholar]
- 18.Dixon MF, Genta RM, Yardley JH, Correa P. Classification and grading of gastritis: the Updated Sydney System; International Workshop on the Histopathology of Gastritis, Houston 1994. Am J Surg Pathol. 1996;20(10):1161-1181. doi: 10.1097/00000478-199610000-00001 [DOI] [PubMed] [Google Scholar]
- 19.You WC, Blot WJ, Li JY, et al. Precancerous gastric lesions in a population at high risk of stomach cancer. Cancer Res. 1993;53(6):1317-1321. [PubMed] [Google Scholar]
- 20.Dunn WB, Broadhurst D, Begley P, et al. ; Human Serum Metabolome (HUSERMET) Consortium . Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat Protoc. 2011;6(7):1060-1083. doi: 10.1038/nprot.2011.335 [DOI] [PubMed] [Google Scholar]
- 21.Song JW, Lam SM, Fan X, et al. Omics-driven systems interrogation of metabolic dysregulation in COVID-19 pathogenesis. Cell Metab. 2020;32(2):188-202.e5. doi: 10.1016/j.cmet.2020.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xia J, Wishart DS. MSEA: a web-based tool to identify biologically meaningful patterns in quantitative metabolomic data. Nucleic Acids Res. 2010;38(web server issue):W71-W77. doi: 10.1093/nar/gkq329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Breiman L. Random forests. Machine Learning. 2001;45(1):5-32. doi: 10.1023/A:1010933404324 [DOI] [Google Scholar]
- 24.Lu X, Yu H, Ma Q, Shen S, Das UN. Linoleic acid suppresses colorectal cancer cell growth by inducing oxidant stress and mitochondrial dysfunction. Lipids Health Dis. 2010;9:106. doi: 10.1186/1476-511X-9-106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu F, Zuo X, Liu Y, et al. Suppression of membranous LRP5 recycling, Wnt/β-catenin signaling, and colon carcinogenesis by 15-LOX-1 peroxidation of linoleic acid in PI3P. Cell Rep. 2020;32(7):108049. doi: 10.1016/j.celrep.2020.108049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chamberland JP, Moon HS. Down-regulation of malignant potential by alpha linolenic acid in human and mouse colon cancer cells. Fam Cancer. 2015;14(1):25-30. doi: 10.1007/s10689-014-9762-z [DOI] [PubMed] [Google Scholar]
- 27.Dai J, Shen J, Pan W, Shen S, Das UN. Effects of polyunsaturated fatty acids on the growth of gastric cancer cells in vitro. Lipids Health Dis. 2013;12:71. doi: 10.1186/1476-511X-12-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song H, Peng JS, Dong-Sheng Y, et al. Serum metabolic profiling of human gastric cancer based on gas chromatography/mass spectrometry. Braz J Med Biol Res. 2012;45(1):78-85. doi. doi: 10.1590/S0100-879X2011007500158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang D, Li W, Zou Q, et al. Serum metabolomic profiling of human gastric cancer and its relationship with the prognosis. Oncotarget. 2017;8(66):110000-110015. doi: 10.18632/oncotarget.21314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin L, Ding Y, Wang Y, et al. Functional lipidomics: palmitic acid impairs hepatocellular carcinoma development by modulating membrane fluidity and glucose metabolism. Hepatology. 2017;66(2):432-448. doi: 10.1002/hep.29033 [DOI] [PubMed] [Google Scholar]
- 31.Sun Q, Yu X, Peng C, et al. Activation of SREBP-1c alters lipogenesis and promotes tumor growth and metastasis in gastric cancer. Biomed Pharmacother. 2020;128:110274. doi: 10.1016/j.biopha.2020.110274 [DOI] [PubMed] [Google Scholar]
- 32.Pan J, Fan Z, Wang Z, et al. CD36 mediates palmitate acid–induced metastasis of gastric cancer via AKT/GSK-3β/β-catenin pathway. J Exp Clin Cancer Res. 2019;38(1):52. doi: 10.1186/s13046-019-1049-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song H, Wang L, Liu HL, et al. Tissue metabolomic fingerprinting reveals metabolic disorders associated with human gastric cancer morbidity. Oncol Rep. 2011;26(2):431-438. doi: 10.3892/or.2011.1302 [DOI] [PubMed] [Google Scholar]
- 34.Aa J, Yu L, Sun M, et al. Metabolic features of the tumor microenvironment of gastric cancer and the link to the systemic macroenvironment. Metabolomics. 2012;8(1):164-173. doi: 10.1007/s11306-011-0297-0 [DOI] [Google Scholar]
- 35.Hyde CA, Missailidis S. Inhibition of arachidonic acid metabolism and its implication on cell proliferation and tumour-angiogenesis. Int Immunopharmacol. 2009;9(6):701-715. doi: 10.1016/j.intimp.2009.02.003 [DOI] [PubMed] [Google Scholar]
- 36.Song H, Peng JS, Yao DS, et al. Metabolic disorders of fatty acids and fatty acid amides associated with human gastric cancer morbidity. Chin Med J (Engl). 2012;125(5):757-763. [PubMed] [Google Scholar]
- 37.Law SH, Chan ML, Marathe GK, Parveen F, Chen CH, Ke LY. An updated review of lysophosphatidylcholine metabolism in human diseases. Int J Mol Sci. 2019;20(5):E1149. doi: 10.3390/ijms20051149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uehara T, Kikuchi H, Miyazaki S, et al. Overexpression of lysophosphatidylcholine acyltransferase 1 and concomitant lipid alterations in gastric cancer. Ann Surg Oncol. 2016;23(suppl 2):S206-S213. doi: 10.1245/s10434-015-4459-6 [DOI] [PubMed] [Google Scholar]
- 39.Yang T, Luo P, Li Y, Hua R, Yin P, Xu G. A serum metabolomics study of gastric cancer based on pseudotargeted liquid chromatography-mass spectrometry approach [Chinese]. Se Pu. 2014;32(2):126-132. doi: 10.3724/sp.j.1123.2013.11050 [DOI] [PubMed] [Google Scholar]
- 40.Lee GB, Lee JC, Moon MH. Plasma lipid profile comparison of five different cancers by nanoflow ultrahigh performance liquid chromatography-tandem mass spectrometry. Anal Chim Acta. 2019;1063:117-126. doi: 10.1016/j.aca.2019.02.021 [DOI] [PubMed] [Google Scholar]
- 41.Kwon SY, Choi SH, Park YS, et al. Lipid MALDI MS profiles of gastric cancer. Open Proteomics J. 2014;7(1). doi: 10.2174/1875039701407010001 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eMethods. Detailed Methods
eFigure 1. Quality Control During LC-MS Analysis
eFigure 2. Metabolomic Profiles for Precancerous Gastric Lesions and GC
eFigure 3. Metabolite Set Enrichment Analysis for the Enriched Metabolite Pathways Associated With GC
eFigure 4. Metabolite Scores Associated With the Progression of Gastric Lesions During the Follow-up
eTable 1. Characteristics of Study Subjects in the Discovery and Validation Sets
eTable 2. Plasma Metabolites Associated With Advanced Gastric Lesions and Gastric Cancer
eTable 3. Metabolite Set Enrichment Analysis for Plasma Metabolites Associated With Gastric Cancer