Graphical abstract
Molecular dynamics simulations have been performed on a complex in which clusters of boron in the form of molecules of the nanodiamond ortho-carborane have been inserted into the four large nonpolar cavities of a nanotube of the right-handed coiled-coil RHCC. The techniques of multi-configurational thermodynamic integration, steered molecular dynamics and umbrella sampling have been combined to investigate the energetics of storage of ortho-carborane in the cavities and to map out the free energy landscape of the complex along the central channel and along directions transverse to the central channel. The purpose of the study was to explore potential pathways for the diffusion of ortho-carborane between the cavities and the solvent and to assess the stability of the complex as a possible drug delivery system for boron neutron capture therapy (BNCT). The investigation reveals a complex free energy landscape with a multitude of peaks and valleys, all of which can be related to specific architectural elements of the , and the activation barriers for ortho-carborane capture and release support the requirements for rapid cargo uptake coupled with tight binding to the cavities.
Keywords: Right-handed coiled-coil tetrabrachion nanotube, Nonpolar cavities, Ortho-carborane, Molecular dynamics simulations, Free energy, Double-decoupling, Multi-configurational thermodynamic integration, Steered molecular dynamics, Umbrella sampling, Boron neutron capture therapy
Abstract
Molecular dynamics simulations have been performed on a complex in which clusters of boron in the form of molecules of the nanodiamond ortho-carborane have been inserted into the four large nonpolar cavities of a nanotube of the right-handed coiled-coil . The techniques of multi-configurational thermodynamic integration, steered molecular dynamics and umbrella sampling have been combined to investigate the energetics of storage of ortho-carborane in the cavities and to map out the free energy landscape of the complex along the central channel and along directions transverse to the central channel. The purpose of the study was to explore potential pathways for the diffusion of ortho-carborane between the cavities and the solvent and to assess the stability of the complex as a possible drug delivery system for boron neutron capture therapy (BNCT). The investigation reveals a complex free energy landscape with a multitude of peaks and valleys, all of which can be related to specific architectural elements of the , and the activation barriers for ortho-carborane capture and release support the requirements for rapid cargo uptake coupled with tight binding to the cavities.
1. Introduction
Boron neutron capture therapy (BNCT) is a next-generation non-invasive approach to cancer therapy. BNCT is a binary application requiring the administration of the nonradioactive isotope 10B followed by subsequent local irradiation by thermal neutrons (0.025 eV). Neutron capture by the 10B isotope, which features a high neutron capture cross section, results in the fission reaction [10B(n,α)7Li], with the production of high energy 7Li and α-particles that deliver a high linear energy transfer to the tumor tissue. The technique is very powerful and allows the selective and highly targeted elimination of malignant tumor cells while minimizing harm to healthy tissues. Due to the limited path lengths of these particles in tissues (5–9 μm), the destructive effects are limited to boron-containing cells. Carboranes are among the most promising boron compounds for BNCT [1], [2], [3]. However, the administration of free carborane molecules to target cells is complicated by the fact that boron compounds are usually absorbed by the intestines and rapidly cleared through the renal system and, as a consequence, the development of targeted carborane delivery agents remains an ongoing challenge. To fill the gap between technical and clinical development, the establishment of tumor selective targeting particles for BNCT has the highest priority.
Self-assembling coiled coil peptides are among the most commonly occurring structural motifs in biological systems, where they are associated with a range of functions. Coiled coil peptides have attracted considerable attention for their fundamental role in the rational design and engineering of self-assembling nanofibers, nanotubes and nanocages and for their biomedical applications in “drug free” macromolecular therapeutics and immunology, and as transport vehicles for the targeted delivery and controlled release of therapeutic drugs [4]. One source of such coiled coils is the tetrabrachion complex which constitutes the proteinaceous surface layer of the cell envelope of the hyperthermophilic archaebacterium Staphylothermus marinus. The complex is composed of subunits, each with a 70-nm stalk that branches into four arms, 24 nm in length [5], [6], [7]. X-ray diffraction measurements [5] performed on a polypeptide chain fragment of tetrabrachion show that the stalk is a tetrameric right-handed coiled-coil composed of four strands oriented in parallel in a right-handed fashion, forming a nanotube with a central channel and four large non-polar cavities which contain water clusters in the native conformation. The has been shown [5] to be stable at high salt concentrations, temperatures of over 100 ˚C, high pressures and extreme ranges of pH, and several studies have demonstrated that the hydrophobic cavities are able to uptake and store water clusters [5], elemental sulfur crowns [8], [9], atomic metals clusters [10], and polycyclic hydrocarbons [11].
Perhaps the most important equilibrium thermodynamic property of a biomolecular system is its free energy. The free energy is a state function that is independent of the process by which the equilibrium state is reached or prepared. By proceeding slowly and reversibly, it is possible to compute the free energy change for alchemically mutating one type of residue into another, or the absolute free energy to bind a ligand to variants of a given protein, thus providing a quantitative basis for rational protein engineering and improvements in drug design. The current investigation was designed to explore the energetics of storage of a specific carborane compound in in order to assess the viability of tetrabrachion as a carborane delivery agent for purposes of BNCT. Molecular dynamics simulations were performed on a complex of and molecules of ortho-carborane and binding free energies were computed using thermodynamic integration methodology, while steered molecular dynamics and umbrella sampling were employed to map out the free energy landscape of the along the central channel and transverse to the central channel. This in-depth free energy study reveals the potential pathways by which ortho-carborane can enter or leave the cavities of and provides information about the free energy barriers that prevent this motion. Our investigation reveals the existence of multiple locally stable configurations of ortho-carborane within the nanotube and demonstrates that provides an effective storage facility for ortho-carborane in which the up-take of ortho-carborane is highly favoured energetically over removal. These are ideal conditions for targeted Boron-Neutron capture therapy.
2. System and methods
This section surveys the molecular morphology of the system under investigation and identifies the source file for the atomic coordinates, describes the molecular dynamics simulation software and force field parameters, summarizes the basic MD simulation protocols, and reviews the analytical methods employed to compute absolute binding free energies and to map global free energy profiles along a specified reaction coordinate.
2.1. The system
All molecular dynamics simulations were performed on a complex in which all four of the non-polar cavities of identified in the crystallographic analysis as occupied by water molecules were instead simultaneously occupied by a single ortho-carborane molecule. For the purposes of the current investigation, a slightly unconventional notation is adopted by which these non-polar cavities determined by X-ray diffraction will be identified with labels , with cavity near the N-terminus and cavity near the C-terminus. The complex was formed by removing the water molecules from cavities through of the measured structure of an (PDB code 1FE6) [5] crystallized at in an aqueous environment and inserting a modeled structure of an ortho-carborane molecule into each cavity. Fig. 1 shows a cross section through a Pymol image of the in the van-der-Waals spheres/surface representation showing the location of the four ortho-carborane molecules in the four non-polar, water-bearing cavities identified in the original X-ray diffraction analysis.
Fig. 1.
A cross section through the showing the locations of the four ortho-carborane molecules in the four non-polar cavities identified in the original X-ray diffraction analysis. The ortho-carborane molecules are shown as clusters of van-der-Waals spheres (in teal). Three of the protein coils are shown in the van-der-Waals spheres/surface representation and the fourth coil is shown in the ribbon representation (in green). The terminus of the tetramer is on the left and the terminus is on the right, and the cavities are identified by black ovals and labels and . (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.2. Molecular dynamics setup
Molecular dynamics simulations were performed with the GROMACS molecular dynamics simulation package [12] using the AMBER force field parm94 [13] and the TIP3P water model. The force field parameters for ortho-carborane appropriate for use with Amber 99 were obtained from Sarosi et al [14] which were adapted from the bond stretching and torsion parameters developed from HF/6–31 + G* optimization and GAFF values by Timofeeva et al [15] and from the bonded terms developed by Gamba et al [16]. The partial charges on boron, carbon, and hydrogen were obtained from closo-carboranyl antifolate models [17] generated using electrostatic potential fitting in Gaussian, and calculated at the HF/6–31 + G* level.
The complex was solvated in a rectangular box with dimensions containing 8560 solvent water molecules and a set of 16 charge neutralizing Na + ions. The closest distance between any point on the complex and the surface of the simulation box was 1.2 nm. Long-range electrostatic interactions were treated with the particle mesh Ewald method. The cut-off radius for all non-bonded interactions was taken to be 1.0 nm and the neighbour list for all non-bonded interactions was updated every 10 time steps. The measured structure was initially energy minimized to using the method of steepest descent and then re-minimized for decreasing values of the energy until convergence was achieved typically around .
The energy minimized system was first heated gradually from 0 K to 50 K over a time interval of 20 ps while position-restraining the non-hydrogen atoms, using velocities chosen randomly from a Maxwell-Boltzmann distribution. Heating and stabilization was repeated at 150 K and 300 K over the same time interval of 20 ps, with initial velocities at each new temperature chosen from the final frame of the equilibrated trajectory at the previous temperature. During the production run, the entire system ( tetrabrachion plus four ortho-carboranes plus solvent) was maintained at a pressure of 1 atm and a temperature of 300 K using a Berendsen barostat [18] and a velocity-rescaling thermostat [19], respectively, with time constants of 0.1 ps. Production runs were performed for 2 ns in time steps of 1 fs using temperature and pressure scaling [20].
2.3. Transfer free energies
The free energy for transferring one molecule from the solvent into one of the cavities through of was calculated by the method of double-decoupling [21], [22]:
is the free energy to remove a single, unrestrained, fully-interacting carborane molecule from solvent to the gas phase. is the free energy to remove a single carborane molecule from one of the cavities through of to the gas phase in the presence of a flat bottom harmonic well (FBHW) restraint potential [8] that restricts the carborane molecule to the volume of the cavity, where and . is the free energy cost of removing the constraint from the carborane molecule, which includes a correction for the standard concentration.
and are computed using the method of multi-configurational thermodynamic integration (MCTI) [23], [24], [25], [26] in a single topology format, which employs a hybrid potential energy function for the non-bonded electrostatic and van der Waals interactions with a coupling parameter which allows to be varied incrementally and reversibly from an initial state in which the target molecule to be decoupled is fully interacting with the rest of the system to a final state in which the target molecule is free and, in the case of the cavity, interacts only with the restraint potential. The current investigation employed approximately 20 values of the non-bonded coupling parameters and . The restraint free energy was calculated analytically [8] using where is the effective FBHW simulation volume and is the standard state volume corresponding to a concentration of .
The energy minimization and thermal equilibration procedures were repeated for each value of λ before generating production runs for 2 ns. Structures were saved for analysis every 0.5 ps during the production runs and the resulting set of 4000 structures was used to determine for each value of λ. The mean value was calculated using g_analyze of the Gromacs MD simulation package and the errors were computed by block averaging. The statistical error in was determined from the autocorrelation of the data over the 2 ns trajectory. Soft-core interactions were incorporated into all simulations presented in this paper in order to remove the discontinuity in the non-bonded parameters as the interactions are turned off and to allow for proper convergence. A soft-core parameter was found to produce the smoothest free energy curves and the best convergence. The sampling of configuration space during end-state removals (in the non-interacting limit ) is believed to be sufficiently extensive based on the low statistical error in , which is typically between 0.5% and 3%. For all λ steps, the values of converged to a time-independent constant within the first 2 ps of each 2 ns trajectory and the derivative varied smoothly with , both of which are indicative of convergence.
Re-solvation of the cavity following the extraction of ortho-carborane can be accomplished in a similar way by tagging an appropriate number of water molecules and transferring them in sequence, one at a time, from the solvent into the cavity by reversibly turning off the interactions between each tagged water molecule and the solvent molecules, relocating that ghost water molecule into the cavity and then reversibly turning the interactions back on between that water molecule, the protein host and the water molecules previously transferred into the cavity. The configuration of cavity waters matched the configuration obtained by X-ray diffraction. Further details may be obtained by consulting reference [9].
2.4. Exit pathways for ortho-carborane
Potential pathways for the movement of ortho-carborane between the cavities through of and the solvent were identified using the method of steered molecular dynamics (SMD) [27], [28], [29], [30], [31]. A moving harmonic steering force was applied to the centre of mass of the molecule and the molecule was pulled at constant speed along a nominally linear path starting from its initial position in the cavity and terminating in the solvent. The harmonic steering force has the form where is the reaction coordinate along the pulling direction, is the point of application of the steering force, is the coordinate of the centre of mass of the carborane molecule along the pulling direction and . The steering force constant was chosen so that extraction could be completed within a time window while also allowing the molecule freedom to respond to the local environment and deviate from the nominal pulling direction. All positions were measured relative to a coordinate origin which coincided with one of the backbone atoms of a specific reference residue located on coil A, close to the pulling axis. Throughout all of the steering processes, coil A of was held immobile using a stationary harmonic constraint with a large force constant. The remaining three coils B, C and D were unrestrained. Longitudinal trajectories where the ortho-carborane was steered along the central channel of were investigated as well as trajectories that were transverse to the central channel of and penetrated the walls of between two neighbouring coils.
The MD steering simulation was used to extract approximately 20 equally spaced positions of the CM of the molecule along the exit trajectory, and the free energy along the exit path as a function of the position of the CM was constructed by the method of Umbrella Sampling [32], [33], [34]. At each of the chosen locations, an initial equilibration was performed for , during which the motion of the molecule was constrained to the vicinity of the reference point by a harmonic bias potential applied to the CM of the molecule. This was followed by an MD production run for which yielded a biased probability distribution for finding the molecule in sampling window at a displacement from a selected atom on the stationary reference residue, from which the free energy in each window was computed. For each cavity, multiple umbrella force constants were used to generate each distribution. The umbrella sampling technique produced distributions which overlapped with those in neighbouring windows and the individual results were combined using the weighted histogram analysis method (WHAM) [33], [35] to generate a global free energy . Errors were estimated using the bootstrapping technique [36], [37].
3. Results
3.1. Transfer free energies
Table 1 summarizes the calculated values of all three contributions to the free energy for transferring one carborane molecule from the solvent into each of the four cavities through of . The calculated solvation free energy for agreed within computational error with other calculations of the solvation free energy [38] for several other closely related substituted 3D-aromatic neutral carboranes (typically ). As the table shows, all four cavities are energetically favourable for carborane relative to the aqueous solvent.
Table 1.
standard free energy in to transfer from solvent to the four cavities of RHCC-tetrabrachion.
3.2. Exit pathways for ortho-carborane
Table 2 summarizes the details of the various steering trajectories, including the carborane being steered (identified by the cavity of origin through ), the steering direction (longitudinal or transverse to the central channel), the steering force constant, the steering reference residue and the umbrella force constants used to generate the histograms and free energy profiles. The designation included with the longitudinal trajectories indicates steering towards the channel exit at the C-terminus and the designation indicates steering in the opposite direction towards the N-terminus. Before performing the longitudinal trajectories out of cavity , the ortho-carborane molecules in cavities and were converted into ghosts by removing the non-bonded interactions. A restraint force constant was applied to all of the backbone atoms on coil A of to ensure that the reference residues remained stationary throughout the steering process. As mentioned earlier, smaller steering force constants are preferred because they allow the ortho-carborane molecule more freedom to respond to the local environment and more time to explore shallow traps bounded by low free energy barriers, thus exposing more of the fine structure corrugations in the free energy hypersurface. Larger force constants will reduce wandering by rapidly extinguishing the smaller energy barriers, yielding smoother (more featureless) and less informative free energy profiles. While calculated free energy profiles are thus expected to depend not only on the specific direction of steering due to variations in the local environments encountered by the ortho-carborane molecule along the different extraction pathways but also on the magnitude of the steering force constant, all of the profiles presented here were generated with the smallest force constant that allowed complete extraction of the ortho-carborane molecule from the cavity of origin with minimal penetration into the solvent within the time constraint of the simulations.
Table 2.
Steering and Umbrella Sampling Parameters.
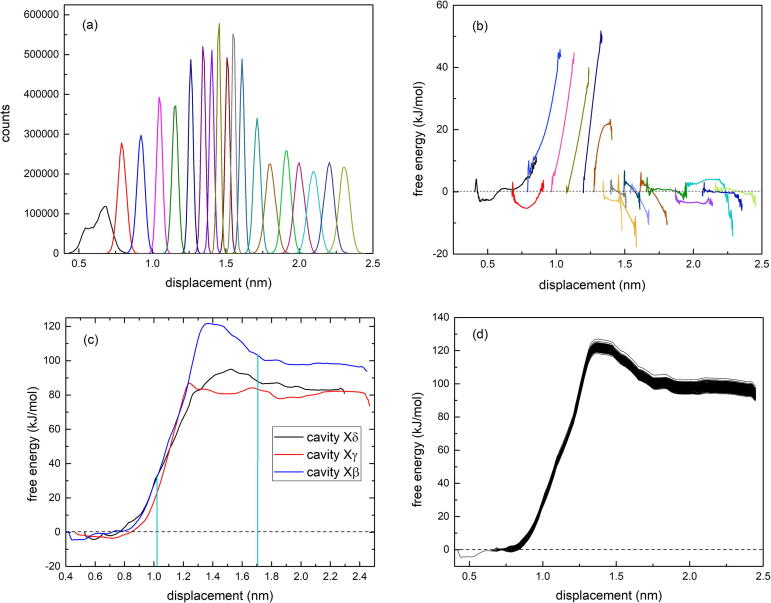
Fig. 2(a) shows a typical set of biased probability distributions generated by umbrella sampling for a complete set of sampling windows along the transverse steering trajectory out of cavity , plotted as a function of the displacement of the centre-of-mass of the ortho-carborane molecule measured from the location of the backbone atom on the reference residue , and Fig. 2(b) shows the complete set of corresponding free energy profiles for the individual sampling windows. Fig. 2(c) shows the global free energy profile as a function of the displacement for all three transverse pathways linking cavities and with the solvent generated, by a single application of WHAM, from the corresponding set of distributions and free energy profiles for each pathway, and Fig. 2(d) shows a typical WHAM free energy profile for cavity generated with bootstrapping. The solid vertical lines in Fig. 2(c) mark the approximate locations of the inner and outer surfaces of the nanotube wall. Fig. 3 shows the global free energy profiles for the single longitudinal channel trajectory out of cavity 1 (Fig. 3(a)) and the two longitudinal channel trajectories out of cavity (Fig. 3(b) and (c)). The schematic diagram at the top of each figure shows the location of the ortho-carborane molecule in the cavity of origin with a black dot and the direction of steering with an arrow. The horizontal axis in the plot is the displacement of the ortho-carborane molecule during steering measured relative to a reference residue. For all channel steering directed to the right in Fig. 3, the reference residue is located to the left of the starting point in the cavity and thus corresponds to a positive displacement of ortho-carborane down the channel, while for all channel steering directed to the left in Fig. 3, the reference residue is located to the right of the starting point in the cavity and thus corresponds to a negative displacement of ortho-carborane.
Fig. 2.
(a) A complete set of biased probability distributions generated by umbrella sampling for all of the selected sampling windows along the transverse exit trajectory of the ortho-carborane molecule out of cavity through the walls of as a function of the displacement of the CM of relative to the location of the atom of the harmonically restrained reference residue . (b) The corresponding set of free energy profiles for all of the sampling windows. (c) A plot of the global free energy generated by a single application of WHAM from the set of biased distributions for the three transverse exit trajectories of ortho-carborane out of cavities and . The vertical lines mark the approximate locations of the interior and exterior surfaces of the protein wall. (d) WHAM free energy profiles for the cavity trajectory generated with bootstrapping.
Fig. 3.
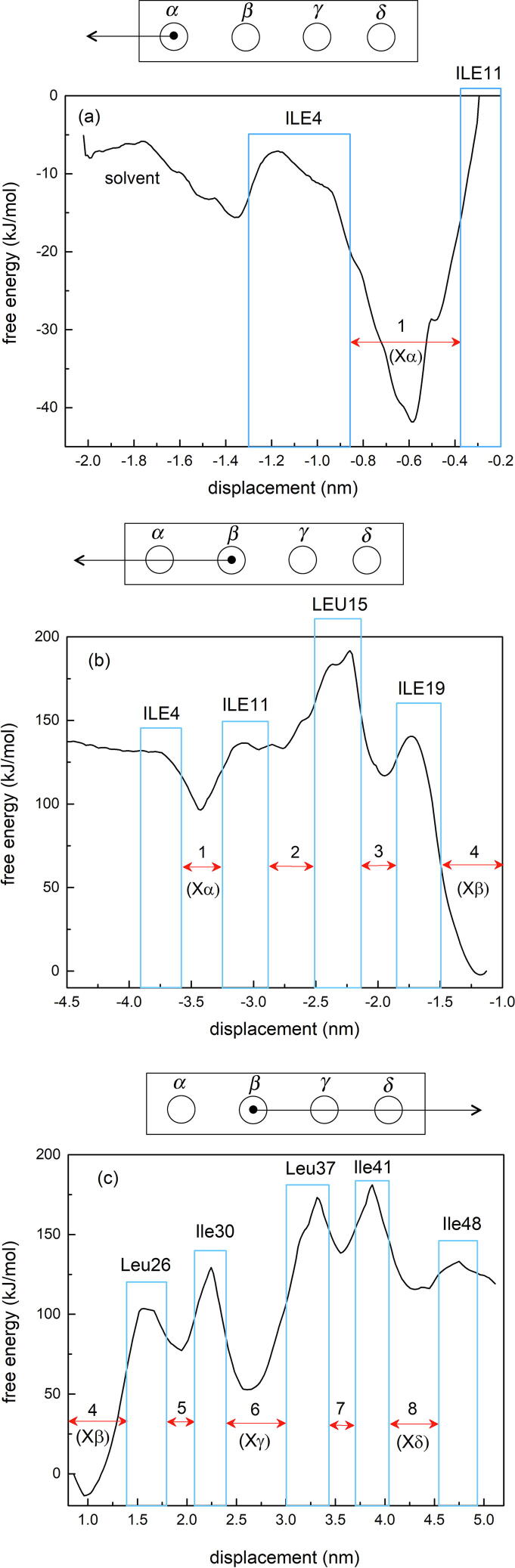
The global free energy profiles for (a) the single longitudinal channel trajectory out of cavity and, (b) and (c) the two longitudinal channel trajectories out of cavity . The horizontal red arrows show the locations of the major and minor channel cavities of as determined by the free energy analysis and the blue rectangles show the free energy barriers between the cavities labelled by the residue rings that are responsible for the barriers. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
It is clear from the profiles in Fig. 2(c) that occupancy of cavities and is significantly favoured over the solvent by and that the thermally-induced shuttling of ortho-carborane in and out of these cavities transversely through the walls of is controlled by a system of energy barriers that exhibits a significant asymmetry with respect to capture from the solvent and release from the cavity, with and .
Fig. 4 shows a sequence of five snapshots of the complex at five different times and as the ortho-carborane molecule is steered transversely out of cavity through a gap between coils C and D of . The image on the left of each snapshot is a view from outside transverse to the central channel and looking inward along the exit trajectory towards cavity 2, while the image on the right is a complementary view along the central channel and transverse to the exit trajectory which shows the location of the ortho-carborane molecule as it passes between coils C and D. In snapshot (a), the ortho-carborane molecule is still inside the cavity and close to the inner wall. In snapshots (b), (c) and (d), the ortho-carborane molecule passes between coils C and D, distorting the helical backbone structure and bending the residue sidechains. In snapshot (e), the ortho-carborane is entering the solvent and the coils are relaxing into their original configurations.
Fig. 4.
A series of snapshots of the complex at five different times and as the ortho-carborane molecule is steered transversely out of cavity through a gap between coils C and D of . The ortho-carborane molecule is shown in blue as non-bonded spheres and dots and the five residues which suffer the greatest distortion due to the passage of ortho-carborane are shown in orange as sticks. The image on the left of each snapshot is a view from outside transverse to the central channel and looking inward along the exit trajectory towards cavity 2, while the image on the right is a complementary view along the central channel and transverse to the exit trajectory which shows the location of the ortho-carborane molecule as it passes between coils C and D. In snapshot (a), the ortho-carborane molecule is still inside the cavity and close to the inner wall. In snapshots (b), (c) and (d), the ortho-carborane molecule passes between coils C and D, distorting the helical backbone structure and bending the residue sidechains. In snapshot (e), the ortho-carborane is entering the solvent and the coils are relaxing into their original configurations. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
By contrast with the transverse free energy profiles in Fig. 2, the longitudinal free energy profiles in Fig. 3 exhibit complicated structure with multiple peaks and valleys and even a single inflection point bounded by a peak on one side and a shoulder on the other. However, as the notations in the diagrams suggest, in spite of this complexity it is possible to relate each of the structural features of the free energy profiles in Fig. 3(a), (b) and (c) to specific architectural elements of the nanotube. The identification begins with a visual inspection of a Pymol movie of the steering process which shows that each longitudinal trajectory consists of short segments of varying lengths in which the ortho-carborane molecule translates more or less freely down the channel under the influence of the steering force. These segments terminate in a clearly visible acceleration of the ortho-carborane molecule during which it is attracted into a “stationary” configuration where it oscillates and tumbles within a small volume on the order of the size of the molecule before it slowly emerges and continues to propagate down the channel. By inspecting the movie frames corresponding to each of the “stationary” configurations, it was possible to determine the position of the centre-of-mass of the ortho-carborane molecule in each of the eight “stationary” configurations observed in the steering movie and each was found to be a precise match with either one of the seven local minima or with the solitary inflection point in the free energy landscape, identified by horizontal red arrows with numerical or alphabetical labels in Fig. 3. Since there is little or no discernible change in the rigidity of the tetrameric structure of for temperatures up to at least , all of the free energy profiles presented here are expected to be valid under the physiological conditions which prevail during therapy.
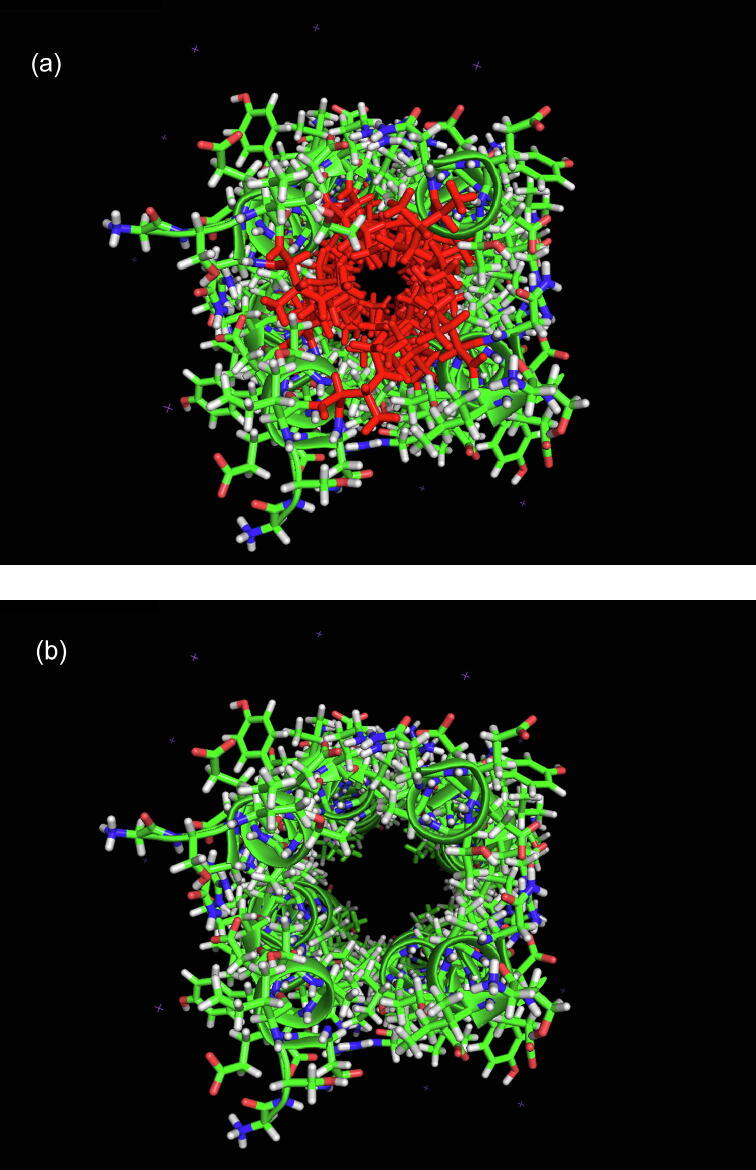
In order to establish the physical origin of the energy barriers that separate the free energy minima, an investigation of the architecture of the bare nanotube (all cavities empty) was conducted on Pymol and nine key rings of residues were discovered distributed along the length of the nanotube. Each ring consisted of four identical Ile or Leu residues, one contributed by each of the four coils of and all nine rings shared a common structural feature that was not observed in any of the other rings that constituted the molecule, namely, that at least two or three of the four residues possessed side chains that extended directly inward, partially blocking the central channel. In order to illustrate the extent of the channel blockage, Fig. 5 shows a view down the channel of (all carboranes removed) with (a) all nine rings of Ile and Leu in place and highlighted in red and (b) with all Ile and Leu rings removed. It is clear that the cross-sectional area of the central channel is significantly diminished by the presence of the Ile and Leu side chains by a factor of between 4 and 8. Fig. 6 shows two Pymol reconstructions of the complex created by superposing eight movie frames corresponding to the eight “stationary” configurations of the ortho-carborane molecules, shown as clusters of white spheres. The nine residue rings are highlighted in different colours in a stick representation. Fig. 6(a) shows the channel trajectory in the direction of the channel exit closest to cavity 4 and Fig. 6(b) shows the channel trajectory in the opposite direction. An inspection of these figures shows that the nine residue rings define the physical boundaries of the eight “stationary” configurations and that the location of each ring coincides with either the location of one of the free energy barriers or with the solitary free energy shoulder in Fig. 3. The nine residue rings are shown as blue rectangles in Fig. 3 and are labelled with the residue names.
Fig. 5.
A view down the channel of (all carboranes removed) with (a) all nine rings of Ile and Leu in place and highlighted in red and (b) with all Ile and Leu rings removed. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 6.
Pymol reconstructions of the complex showing the nine residue rings highlighted in different colours in a stick representation and individually labeled as isoleucine or leucine, and the “stationary” ortho-carborane configurations, shown as clusters of white spheres, for (a) the positive channel trajectory in the direction of the channel exit closest to the terminus and (b) the negative channel trajectory in the direction of the channel exit closest to the terminus.
4. Discussion
The comprehensive free energy analysis of the complex presented here shows that the provides an energetically stabilizing environment for ortho-carborane molecules relative to immersion in an aqueous bath. As mentioned previously, X-ray crystallography measurements performed on the native structure reveal the existence of four large hydrophobic cavities and also show [5] that all cavities were filled with water molecules with occupancies of 2, 9, 5, and 1 in cavities through , respectively, with the nine water molecules in cavity arranged in a non-planar clathrate-like ring conformation. Molecular dynamics simulations performed on the [5], [39] show that the water clusters in the two largest cavities and are marginally favourable due to intra-cluster hydrogen bonds between the cavity waters as well as to the formation of temporary hydrogen bonds with the carbonyl oxygens of four isoleucine residues in the walls of the cavities.
By contrast, in the current investigation, the free energy profiles derived from a steering and umbrella analysis of potential diffusion pathways of ortho-carborane between the central channel of the and the aqueous bath reveal a much richer free energy landscape than suggested by crystallographic analysis and show that ortho-carborane is trapped in eight different locations along the axis of the nanotube by local energy barriers. The traps are labelled sequentially from 1 through 8 in Fig. 3 starting from the terminus (with and ) and consist of seven metastable local free energy minima plus a single marginally stable inflection point (trap 2), bounded by local activation barriers ranging from a minimum height of for the inflection point shoulder to a maximum height of . As expected, four of these “stationary” configurations coincide with the four non-polar storage cavities through that were identified in the original structural analysis of the RHCC tetramer [1] as containing clusters of water molecules and these correspond to the free energy minima labelled 1, 4, 6 and 8 in Fig. 3. Of these, cavities 4 and 6 provide the most attractive environment for the ortho-carborane molecule relative to the solvent with and have the highest activation barriers . Of the remaining set of four traps which emerge from the free energy analysis labelled 2, 3, 5 and 7 in Fig. 3, cavities 3, 5 and 7 are comparable in depth to cavities 1 and 8 identified in the original crystallographic analysis and cavities 3 and 5 have free energies which lie below the solvent free energy by an amount comparable to cavity 1, that is, between . Furthermore, all eight cavities (including the inflection point) share the common feature that they are bounded on either end by rings of Isoleucine and Leucine residues whose side chains penetrate into the central channel and provide the local stabilizing energy barriers in the form of steric hindrance that block the migration of the ortho-carborane molecule down the central channel during steering.
The possible existence of other cavities beyond those identified in the original crystallographic analysis has been inferred previously through a geometrical analysis of the molecule [40] but the simulations described here represent the first rigorous confirmation of the existence of eight cavities by free energy mapping, which not only allows a quantitative energy-based comparison between the cavities but which also reveals the presence of one marginally stable cavity not apparent from the geometrical analysis, in the form of an inflection point.
The recognition that coiled-coil systems like possess physical and chemical properties that are particularly well suited for purposes of selectively targeted drug delivery has existed for some time [40]. The drug can be uploaded as cargo into the storage cavities and the encapsulating walls overcome complications related to the hydrophobicity of the cargo and prevent damage to healthy cells while themselves exerting no cytotoxic effect on the cells. Furthermore, the and terminals can be easily conjugated to numerous epitopes that bind to specific cell markers, thus enabling targeted delivery. A study by Erriksson et al. [41] demonstrated the successful binding of the anticancer drug cisplatin to the with, on average, one molecule of cisplatin bound per cavity, and that the complex was stable for up to twelve hours in solution and taken up into intracellular vesicles. The fusion of coiled-coil domains to epitopes may also enable the production of therapeutic vaccines. Schroeder et al. [42] and Pimentel et al. [43] demonstrated that the repetitive arrangement of epitopes on the surface of a peptide nanoparticle was able to elicit a specific immunogenic response which could be exploited in the design of vaccines.
In a similar manner, it is possible to contemplate a potentially promising treatment for malign tumors in which clusters of the nonradioactive boron isotope 10B are uploaded into the cavities of and delivered either in close proximity or even into the interior of tumor cells and then activated by neutron bombardment, producing a 7Li particle and an particle as the destructive agent. Since the cargo is, by design, intended to remain with the carrier throughout the delivery and activation process, such a delivery system demands a tightly bound complex, and the free energy barriers in Fig. 2(c) which control the diffusion of ortho-carborane through the walls of clearly satisfy this requirement. The energy barriers for capture are on the order of the thermal energy with at while the energy barriers for release are on the order of . According to transition state theory, the rate constant for release at would be and, using the rate equation for a unimolecular reaction , this would correspond to a half-life of for ortho-carborane in cavities 4, 6 and 8. Similar considerations apply to the free energy barriers along the central channel of shown in Fig. 3. The diffusion of the ortho-carboranes in cavities 4 and 6 down the central channel is tightly constrained by free energy barriers with activation energies similar in magnitude to those which characterize the walls. The ortho-carboranes in cavities 1 and 8 are similarly constrained with respect to diffusion inward along the central channel, towards cavities 4 and 6, but appear to be much less constrained with respect to motion in and out of the channel exits at the and terminals, suggesting that these are most likely the routes by which they are uploaded into the from the solvent.
It is was pointed out earlier in connection with Fig. 4 that all steering trajectories, whether they are directed transversely through gaps between the coiled-coils or down the central channel, are observed to produce distortions of the helices, typically on the order of , with transverse distortions appearing to relax more quickly than longitudinal distortions. This immediately raises the question of whether ortho-carborane is capable of generating the necessary distortions and creating a pathway into the on its own, solely through the mechanism of thermal fluctuations or whether migration is assisted by spontaneous fluctuations of the protein in the form of dynamic breathing modes which are intrinsic to the protein. Fully atomistic, room temperature molecular dynamics simulations of complete transit trajectories of small nonpolar ligands between the solvent and the heme-binding pocket of myoglobin [44], [45] coupled with a free volume fluctuation analysis reveal the existence of identifiable entrance and exit portals on the surface of the protein and of intrinsic dynamic pathways where expansion and contraction of internal cavities regulated by the opening and closing of gates controlled by dihedral rotations in specific amino acids, and large structural fluctuations in the spaces between the helices of the myoglobin scaffolding may facilitate internal migration of ligands through the protein matrix. The ligand movements along the pathways consist of a sequence of rapid hops between transient docking sites where the ligand dwells for relatively long periods of time [45], a description which matches that of down-channel steering in the current study. Such breathing modes may well play a role not only in the uptake and transport of ortho-carborane by but also in substantiating one of the biological functions proposed for the anaerobic hyperthermophilic archeon S. marinus, namely, the capture and storage of cyclooctasulfur rings from its native sulfur-rich environment in order to make elemental sulfur available as a terminal electron acceptor for energy producing redox-reactions [9]. Future work will focus on searching for evidence of the necessary dynamic structural flexibility in .
Finally, it is important to call attention to other approaches that have been developed for mapping entire networks of entry/exit channels in biomolecules based on a detailed reconstruction of the landscape of local metastable free energy traps (pockets) which characterize such complex systems [46], [47], [48]. These approaches are computationally much more intensive than the current approach and employ a variety of enhanced sampling techniques to analyze active site binding and unbinding events of ligands that might adversely affect the biological functionality, for the purpose of formulating novel drug design strategies. By contrast, In the current investigation, where the emphasis is on ligand diffusion for the purposes of simple storage rather than chemical transformation, and where the pathways between the solvent and the cavities tend to lack the diversity and complexity of those in other systems, a more heuristic approach has been adopted which depends on direct steering through gaps in the protein walls.
5. Conclusion
The use of nanoparticles such as Albumin to carry drugs of interest [49], [50] has allowed for an expansion of molecular libraries used for medical treatments. Nevertheless, these nanoparticle-ligand complexes must be examined on an individual basis in order to establish their specific physicochemical properties, mechanisms of action and delivery capabilities. The helical coiled-coil is among the most prevalent oligomerization motifs encountered in biological systems and plays a significant role in many key biological processes such as transcription, membrane fusion, intercellular communication and molecular motors. Its simple, robust and highly adaptable design and its ability to bind biologically relevant molecules have suggested its potential application to medical nanotechnology. Molecular dynamics simulations performed on a complex of and ortho-carborane show that the tetrameric host has at least seven and possibly as many as eight storage cavities of varying sizes distributed along its central channel, each capable of accommodating a single ortho-carborane molecule. The asymmetric nature of the free energy activation barriers identified in this MD-based analysis permit rapid up take of ortho-carborane from the solvent coupled with slow release through gaps between the coiled-coil walls and also through the channel exits at the terminus and terminus of the nanotube. These conditions position the complex as an ideal candidate vehicle for boron-neutron capture therapy.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The presented work was funded by the Natural Science and Engineering Council of Canada (NSERC). JS is a Canada Research Chair in Structural Biology and Biophysics. There is no conflict of interest to declare.
References
- 1.Hoppenz P., Els-Heindl S., Kellert M., Kuhnert R., Saretz S., Lerchen H.-G. A selective carborane-functionalized gastrin-releasing peptide receptor agonist as boron delivery agent for boron neutron capture therapy. J. Org. Chem. 2020;85(3):1446–1457. doi: 10.1021/acs.joc.9b02406. [DOI] [PubMed] [Google Scholar]
- 2.Assaf KI, Wilińska J, Gabel D. 2018. “Ionic Boron Clusters as Superchaotropic Anions: Implications for Drug Design” from “Boron-Based Compounds: Potential and Emerging Applications in Medicine, John Wiley & Sons First Edition. p. 109–125.
- 3.Lutz Jr et al. Carborane-containing matrix metalloprotease (MMP) ligands as candidates for boron neutron-capture therapy, ChemMedChem 15 (2020): 1–13, 10.1002/cmdc.202000470. [DOI] [PubMed]
- 4.Wu Y., Collier J.H. α-Helical coiled coil peptide materials for biomedical applications. Wiley Interdisciplinary Rev Nanomed Nanobiotechnol. 2017;9(2):e1424. doi: 10.1002/wnan.1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stetefeld J., Jenny M., Schulthess T., Landwehr R., Engel J., Kammerer R.A. Crystal structure of a naturally occurring parallel right-handed coiled coil tetramer. Nat Struct Biol. 2000;7:772–776. doi: 10.1038/79006. [DOI] [PubMed] [Google Scholar]
- 6.Peters J, Baumeister W, Lupas A. J Mol Biol 257 (1996): 1031–41. [DOI] [PubMed]
- 7.Peters J, Nitsch M, Kuḧlmorgen B, Golbik R, Lupas A, Kellermann J, et al. J Mol Biol 245 (1995): 385–401. [DOI] [PubMed]
- 8.McDougall Matthew, Francisco Olga, Harder-Viddal Candice, Roshko Roy, Meier Markus, Stetefeld Jörg. Archaea S-Layer Nanotube from a “ Black Smoker ” in Complex with Cyclo-Octasulfur (S8) Rings. Proteins. 2017;85(12):2209–2216. doi: 10.1002/prot.v85.1210.1002/prot.25385. [DOI] [PubMed] [Google Scholar]
- 9.Harder-Viddal C., McDougall M., Roshko R.M., Stetefeld J. Energetics of storage and diffusion of water and cyclo-octasulfur for a nonpolar cavity of RHCC tetrabrachion by molecular dynamics simulations. Comput Struct Biotechnol J. 2019;17:675–683. doi: 10.1016/j.csbj.2019.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McDougall Matthew, McEleney Kevin, Francisco Olga, Trieu Benchmen, Ogbomo Efehi Kelly, Tomy Gregg. Reductive power of the archaea right-handed coiled coil nanotube (RHCC-NT) and incorporation of mercury clusters inside protein cages. J Struct Biol. 2018;203(3):281–287. doi: 10.1016/j.jsb:2018.05.013. [DOI] [PubMed] [Google Scholar]
- 11.McDougall Matthew, Francisco Olga, Harder-Viddal Candice, Roshko Roy, Heide Fabian, Sidhu Shubleen. Proteinaceous nano container encapsulate polycyclic aromatic hydrocarbons. Sci Rep. 2019;9(1) doi: 10.1038/s41598-018-37323-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A.E., Berendsen H.J. GROMACS: Fast, flexible, and free. J Comput Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 13.Wang J, et al. Comp. Chem. 21 (2000): 1049–1074.
- 14.Sarosi and Lybrand J, Chem Inf Model 58 (2018): 1990−1999. [DOI] [PubMed]
- 15.Timofeeva Tatjana V., Suponitsky Kyrill Yu., Yanovsky Alexander I., Allinger Norman L. The MM3 force field for 12-vertex boranes and carboranes. J Organomet Chem. 1997;536–537:481–488. [Google Scholar]
- 16.Gamba Z., Powell B.M. The condensed phases of carboranes. J Chem Phys. 1996;105:2436–2440. [Google Scholar]
- 17.Mahasenan K, Pavlovicz R, Li C, and Tjarks W, Carborane clusters in computational drug design: A comparative docking evaluation using Autodock, Flexx, Glideand Surflex Rohit Tiwari, J Chem Inf Model 49(6) (2009): 1581–1589. 10.1021/ci900031y. [DOI] [PMC free article] [PubMed]
- 18.Berendsen H.J.C., Postma J.P.M., van Gunsteren W.F., DiNola A., Haak J.R. Molecular dynamics with coupling to an external bath. J Chem Phys. 1984;81(8):3684–3690. [Google Scholar]
- 19.Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J Chem Phys. 2007;126 doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 20.All simulations were performed on a MacPro Workstation with Mac OS, 12 core Intel Xeon E5 processor; for each nanosecond of simulation time, the core computing time was 70h, and the wall time was 3h.
- 21.Gilson M.K., Given J.A., Bush B.L., McCammon J.A. The statistical-thermodynamic basis for computation of binding affinities: a critical review. Biophys J. 1997;72(3):1047–1069. doi: 10.1016/S0006-3495(97)78756-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hamelberg D., McCammon J.A. Standard free energy of releasing a water molecule from the binding pocket of proteins: Double-decoupling method. J Am Chem Soc. 2004;126:7683–7689. doi: 10.1021/ja0377908. [DOI] [PubMed] [Google Scholar]
- 23.Gilson M.K., Given J.A., Bush B.L., McCammon J.A. The statistical- thermodynamic basis for computation of binding affinities: a critical review. Biophys J. 1997;72(3):1047–1069. doi: 10.1016/S0006-3495(97)78756-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamelberg D., McCammon J.A. Standard free energy of releasing a water molecule from the binding pocket of proteins: double- decoupling method. J Am Chem Soc. 2004;126:7683–7689. doi: 10.1021/ja0377908. [DOI] [PubMed] [Google Scholar]
- 25.Straatsma T.P., McCammon J.A. Theoretical calculations of relative affinities of binding. Methods Enzymol. 1991;202:497–511. doi: 10.1016/0076-6879(91)02025-5. [DOI] [PubMed] [Google Scholar]
- 26.Boresch S., Karplus M. The meaning of component analysis: decomposition of the free energy in terms of specific interactions. J Mol Biol. 1995;254(5):801–807. doi: 10.1006/jmbi.1995.0656. [DOI] [PubMed] [Google Scholar]
- 27.Grubmu ¨ller H, Heymann B, Tavan P, Ligand binding and molecular mechanics calculation of the streptavidin-biotin rupture force. Science 271 (1996): 997–999. [DOI] [PubMed]
- 28.Izrailev S., Stepaniants S., Balsera M., Oono Y., Schulten K. Molecular dynamics study of unbinding of the avidin-biotin complex. Biophys J. 1997;72(4):1568–1581. doi: 10.1016/S0006-3495(97)78804-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Evans E., Ritchie K. Dynamic strength of molecular adhesion bonds. Biophys J. 1997;72(4):1541–1555. doi: 10.1016/S0006-3495(97)78802-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Balsera M., Stepaniants S., Izrailev S., Oono Y., Schulten K. Reconstructing potential energy functions from simulated force-induced unbinding processes. Biophys J. 1997;73(3):1281–1287. doi: 10.1016/S0006-3495(97)78161-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Isralewitz B., Gao M., Schulten K. Steered molecular dynamics and mechanical functions of proteins. Curr Opin Struct Biol. 2001;11:224–230. doi: 10.1016/s0959-440x(00)00194-9. [DOI] [PubMed] [Google Scholar]
- 32.Torrie G.M., Valleau J.P. Monte-Carlo free-energy estimates using non-Boltzmann sampling: Application to subcritical Lennard-Jones fluid. Chem Phys Lett. 1974;28:578–581. [Google Scholar]
- 33.Torrie G.M., Valleau J.P. Nonphysical sampling distributions in Monte Carlo free-energy estimation: Umbrella sampling. J Comput Phys. 1977;23(2):187–199. [Google Scholar]
- 34.Kästner Johannes. Umbrella sampling. WIREs Comput Mol Sci. 2011;1(6):932–942. [Google Scholar]
- 35.Kumar S., Bouzida D., Swendsen R.H., Kollman P.A., Rosenberg J.M. The weighted histogram analysis method for free-energy calculations on biomolecules. 1. The method. J Comput Chem. 1992;13:1011–1021. [Google Scholar]
- 36.Hub Jochen S., de Groot Bert L., van der Spoel David. G_Wham - A free weighted histogram analysis implementation including robust error and autocorrelation estimates. J Chem Theory Comput. 2010;6(12):3713–3720. [Google Scholar]
- 37.Hub J.S., de Groot B.L. Mechanism of selectivity in aquaporins and aquaglyceroporins. PNAS. 2008;105(4):1198–1203. doi: 10.1073/pnas.0707662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rabindranath Lo R., Fanfrlık J., Lepsik M., Hobza P. The properties of substituted 3D-aromatic neutral carboranes: the potential for r-hole bonding. Phys Chem Chem Phys. 2015;17:20814–20821. doi: 10.1039/c5cp03617h. [DOI] [PubMed] [Google Scholar]
- 39.Yin Hao, Hummer Gerhard, Rasaiah Jayendran C. Metastable water clusters in the nonpolar cavities of the thermostable protein tetrabrachion. J Am Chem Soc. 2007;129(23):7369–7377. doi: 10.1021/ja070456h. [DOI] [PubMed] [Google Scholar]
- 40.McFarlane Ainsley A., Orriss George L., Stetefeld Jörg. The use of coiled-coil proteins in drug delivery systems. Eur J Pharmacol. 2009;625(1–3):101–107. doi: 10.1016/j.ejphar.2009.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eriksson M., Hassan S., Larsson R., Linder S., Ramqvist T., Lövborg H. Utilization of a right-handed coiled-coil protein from archaebacteria Staphylothermus marinus as a carrier for cisplatin. Anticancer Res. 2009;29:11–18. [PubMed] [Google Scholar]
- 42.Schroeder U., Graff A., Buchmeier S., Rigler P., Silvan U., Tropel D. Peptide nanoparticles serve as a powerful platform for the immunogenic display of poorly antigenic actin determinants. J Mol Biol. 2009;386:1368–1381. doi: 10.1016/j.jmb.2008.11.023. [DOI] [PubMed] [Google Scholar]
- 43.Burkhard P, Peptide nanoparticles as novel immunogens: design and analysis of a prototypic severe acute respiratory syndrome vaccine. Chem Biol Drug Des 73 (2009): 53–61. [DOI] [PMC free article] [PubMed]
- 44.Scorciapino M.A., Robertazzi A., Casu M., Ruggerone P., Ceccarelli M. Breathing motions of a respiratory protein revealed by molecular dynamics simulations. J Am Chem Soc. 2009;131:11825–11832. doi: 10.1021/ja9028473. [DOI] [PubMed] [Google Scholar]
- 45.Ruscio J.Z., Deept Kumar D., Shukla M., Prisant M.G., Murali T.M., Onufriev A.V. Atomic level computational identification of ligand migration pathways between solvent and binding site in myoglobin. Proc Nat Acad Sci. 2008;105:9204–9209. doi: 10.1073/pnas.0710825105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dickson Alex, Lotz Samuel D. Multiple ligand unbinding pathways and ligand-induced destabilization revealed by WExplore. Biophys J. 2017;112(4):620–629. doi: 10.1016/j.bpj.2017.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu Yong, Mohammadi Mohammadjavad, Vashisth Harish. Diffusion network of CO in FeFe-hydrogenase. J Chem Phys. 2018;149(20):204108. doi: 10.1063/1.5054877. [DOI] [PubMed] [Google Scholar]
- 48.Schuetz Doris A., Bernetti Mattia, Bertazzo Martina, Musil Djordje, Eggenweiler Hans-Michael, Recanatini Maurizio. Predicting residence time and drug unbinding pathway through scaled molecular dynamics. J Chem Inf Model. 2019;59(1):535–549. doi: 10.1021/acs.jcim.8b00614. [DOI] [PubMed] [Google Scholar]
- 49.Paál K., Müller J., Hegedûs L. High affinity binding of paclitaxel to human serum albumin. Eur J Biochem. 2001;268:2187–2191. doi: 10.1046/j.1432-1327.2001.02107.x. [DOI] [PubMed] [Google Scholar]
- 50.Siri M, Grasselli M, Alonso S del V, Correlation between assembly structure of a gamma irradiated albumin nanoparticle and its function as a drug delivery system. Colloids Surfaces A Physicochem Eng Asp 603 (2020): 125176.