Abstract
Identifying arrhythmia substrates and quantifying their heterogeneity has great potential to provide critical guidance for radio frequency ablation. However, quantitative analysis of heterogeneity on cardiac optical coherence tomography (OCT) images is lacking. In this paper, we conduct the first study on quantifying cardiac tissue heterogeneity from human OCT images. Our proposed method applies a dropout-based Monte Carlo sampling technique to measure the model uncertainty. The heterogeneity information is extracted by decoupling the intra/inter-tissue heterogeneity and tissue boundary uncertainty from the uncertainty measurement. We empirically demonstrate that our model can highlight the subtle features from OCT images, and the heterogeneity information extracted is positively correlated with the tissue heterogeneity information from corresponding histology images.
Keywords: Optical coherence tomography, Deep learning, Heterogeneity
1. Introduction
Arrhythmia is a major type of cardiovascular disease that afflicts millions of patients in the United States [17]. A standard intervention to treat arrhythmia is radio-frequency ablation (RFA), an intra-cardiac procedure that directs a catheter and delivers heat to areas where irregular rhythms are observed. Current clinical guidance of RFA is based on low-resolution imaging modalities with limited tissue composition information [26]. For many patients, an additional procedure is required to achieve a chronic successful termination of the arrhythmia. Thus, precise intervention is challenging. Knowledge of patients’ heart structure could help to optimize the intervention strategy by identifying arrhythmia substrates and avoiding critical structures. In addition, previous research has suggested that heterogeneity within the myocardium, such as fibrosis and adipose, are substrates that are potential mechanisms for the generation and maintenance of arrhythmias [1,11,28]. Thus, it is important to identify arrhythmia substrates and quantify their heterogeneity.
Optical coherence tomography (OCT) [14] is a depth-resolved imaging modality that can provide micron-level-resolution images in real time. It can detect micro-structures within cardiac tissues [4,8–10,21] and has great potential for precise guidance of RFA. Some forward-viewing OCT catheters have been designed to image cardiac tissues while the catheter is in contact with the cardiac surface [5,30]. As an interferometry imaging system which highlights the differences of reflective index of adjacent tissues, OCT can depict boundaries among tissue layers, opening a great possibility to segment various tissue compositions using either conventional machine learning or deep learning methods. However, quantitative analysis of heterogeneity within cardiac OCT images is still lacking.
Cardiac tissue heterogeneity refers to intermittent and spatially-varying structural distributions within the myocardium [11], or adipose-infiltrated fibrotic myocardium [28]. Histology heterogeneity assessment is used as a reference for cardiac fibrosis analysis, according to the conventional approach considering histology imaging as the gold standard [20]. OCT imaging provides an alternative non-invasive imaging solution, but quantification of heterogeneity in cardiac OCT images remains particularly challenging. First, unlike histology images, the cellular walls of cardiac muscle cells are not visible in OCT images. Second, there are subtle differences in pixel intensity and texture contrasts between normal myocardium and fibrotic myocardium. Third, OCT images are in grayscale, which have less tissue information than colored histology images. Other imaging modalities have a too low spatial resolution to identify cardiac fibrosis heterogeneity. These modalities rely upon indirect measures to assess cardiac tissue, such as mechanical deformation, strain, and wall thickness [2], which are only proxies to quantify cardiac fibrosis heterogeneity.
In this paper, we propose a novel deep learning framework for cardiac tissue heterogeneity measurement. We evaluate our method by comparing the heterogeneity information from OCT images with its corresponding histology images heterogeneity. In summary, this paper has following contributions:
This is the first paper, to the best of our knowledge, that attempts to extract the tissue heterogeneity information from OCT images via deep learning-based uncertainty measurement.
We empirically demonstrate that our model can highlight subtle features from OCT images, offering a way to extract the tissue heterogeneity information in a non-invasive way for real-time imaging and processing.
2. Methodology
Our goal is to measure the heterogeneity of human cardiac tissue on OCT images. As illustrated in Fig. 1, our proposed model consists of three major modules: 1) robust learning on region-based labels, 2) uncertainty measurement, and 3) heterogeneity information extraction. In the robust learning module, we use focal loss [19] to robustly learn the representative features from the region-based labels. Then, a novel dropout-based Monte Carlo sampling technique is applied for uncertainty measurement to get the uncertainty map of the prediction. Finally, the tissue heterogeneity information is extracted by decoupling the intra/inter-tissue heterogeneity and tissue boundary uncertainty from the uncertainty maps.
Fig. 1.
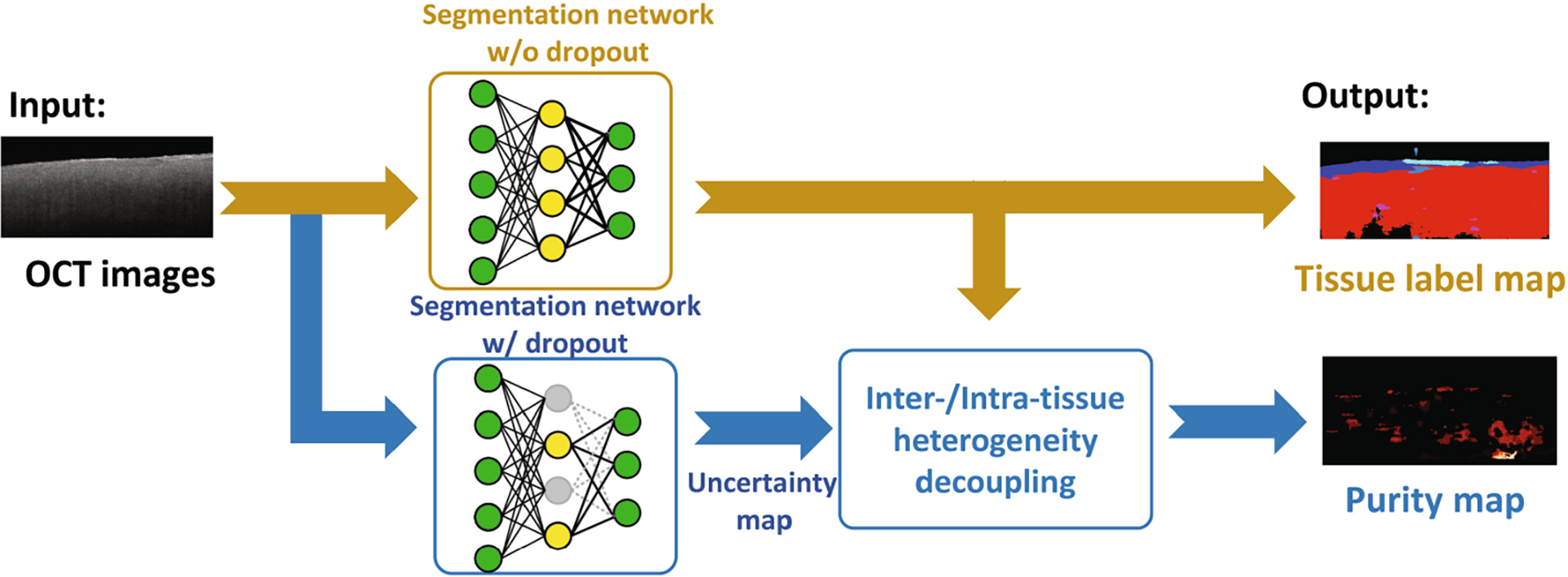
The framework of our proposed algorithm in testing. For each image, an uncertainty map is generated by the comparison of predictions from the network with dropout layers on or off. Based on the uncertainty map, a separate purity map is calculated to highlight the tissue heterogeneity information.
2.1. Segmentation Framework
We use the ReLayNet [24] architecture as the base of our network. ReLayNet is an end-to-end image segmentation framework that achieves state-of-the-art performance. Inspired by [16], we add dropout layers in the three inner encoder blocks in the training phase. The introduction of dropout addresses the overfitting issue for small datasets and also lays a foundation for uncertainty quantification in the next section. The network is jointly optimized by the following loss function:
| (1) |
where w is a trade-off parameter. FL is the focal loss [19] and DL is the Dice loss defined as:
| (2) |
where pk(x) is the predicted probability of class K at the pixel position x ∈ Ω with , qk(x) is the one-hot ground-truth label, and K is the number of classes. Based on Dice coefficient, Dice loss evaluates pixel-wise agreement between the prediction and the ground truth.
2.2. Uncertainty Measurement
We use dropout [27] based Monte Carlo sampling technique [7,13,26] for uncertainty measurement. For each segment, a pixel-wise uncertainty map is generated to present the posterior distribution for the tissue prediction map. During the training process, the dropout layers are opened to robustly learn the discriminative features. At the test time, as shown in Fig. 1, they are turned off to provide a baseline prediction pred. Then, served as proximal inference, these layers are turned on again Nit times to get the Monte Carlo sampling maps prediMC, i ∈ {1, 2, …, Nit}. For each pixel position x ∈ Ω, the uncertainty map UM is generated as:
| (3) |
where 1A is the indicator function of A. The value in the uncertainty map encodes the empirical confidence level of the network on its prediction. More specifically, a high uncertainty value on a pixel indicates a highly unreliable prediction, which corresponds to a low confidence.
2.3. Heterogeneity Measurement
In cardiac OCT images, the uncertainty of prediction comes from the intra/inter-tissue heterogeneity and the boundary effect. The boundary effect is caused by the subtle boundaries between two different tissue types. To remove this unwanted boundary effect, we generate a purity map by multiplying the uncertainty map with a boundary weight mask. This boundary weight mask is generated from the tissue prediction map via assigning smaller weights on the boundary. Similar to the weighting map in [24], lower weights are assigned to pixels at the boundary and higher weights are assigned to pixels in the central region of a tissue. Therefore, this boundary mask highlights the tissue boundary regions, decoupling the tissue heterogeneity and the boundary effect from the uncertainty map. Hence, the purity map is more informative to evaluate the tissue heterogeneity features. The whole process to generate a purity map is presented in Fig. 1.
Our tissue heterogeneity information is measured by homogeneity values [3]. Following previous research [3,6,15,29], the homogeneity values of both purity maps and tissue images (histology images) are calculated from gray-level co-occurrence matrix (GLCM). In particular, we average four directions (0°, 45°, 90° and 135°) to make the descriptor rotation invariant [15]. Then, we extract the homogeneity from the GLCM descriptors by using the following equation [3,6]:
| (4) |
where G(i, j) represents the (i, j) value of the GLCM. The homogeneity value can be used to measure the closeness of the distribution of pixel intensities in the GLCM [18], representing the amount of local variations in an image [3]. We compare the homogeneity values of purity maps with those of histology images. A high similarity value, which is positive correlation, between values from these two groups indicates that our purity maps can extract the tissue heterogeneity information.
3. Experimental Results
3.1. Dataset and Experimental Setup
We acquired an in-vitro cohort of 185 images taken from 15 human atria and ventricles from the Thorlabs OCT system. The samples were acquired through an National Disease Research Interchange (NDRI) approved protocol from Columbia University [8]. Upon imaging, sections of samples were stained with Masson’s Trichrome. We used white-light images acquired simultaneously with the OCT images to guarantee that histological slides and OCT volumes are from the same regions. This, along with inking, was used to match OCT with histology images. Each OCT image is of size 512 × 600 or 512 × 800 pixels with a field of view of 2.51 mm × 4 mm. Based on histology and the guidance from a pathologist, two investigators labeled the OCT images into the following tissue types: endocardium, myocardium, artifacts, adipose & fibrosis, and other tissue types. These investigators were blind to our algorithm results.
In order to fully utilize our dataset, we perform 6-fold cross-validation in evaluation. The images are randomized by donors. We have two pre-processing steps: first, we crop each OCT image into a depth of 360 pixels (~1.76 mm); next, we partition the images into a set of overlapping patches with a size of 360 × 64 pixels for data augmentation.
3.2. Quantitative Results
Learning from Region-Based Labels:
We empirically set the epoch = 500, learning rate = 0.001, and w = 0.5 (Eq. 1). Then, we use both Dice coefficient (DC) and accuracy (AC) to evaluate the segmentation performance of our proposed model. Table. 1 compares the experimental results averaged over the 6-fold cross-validation sets with the baseline algorithm, RelayNet [24], based on a similar network structure but without dropout and focal loss. However, these evaluation metrics cannot fully reflect the segmentation performance. The manual labels are region-based, which is not accurate enough for pixel-wise evaluation. Among six validation sets, the best accuracy is above 0.8. In addition, compared with the baseline algorithm, the use of focal loss and dropout layers improves both DC and AC. These results demonstrate that our network has the ability to robustly learn representative features from region-based labels.
Table 1.
Comparison of segmentation results
| Method | Accuracy (AC) | Dice coefficient (DC) |
|---|---|---|
| Proposed method | 0.726 ± 0.08 | 0.605 ± 0.04 |
| RelayNet architecture | 0.698 ± 0.10 | 0.596 ± 0.03 |
Visual results demonstrate the learning ability of our model on region-based labels. Figure 2 illustrates the predicted tissue maps, on two myocardium examples with some adipose and fibrosis regions. Our algorithm pinpoints isolated fibrosis in Fig. 2(c) and the large adipose region in Fig. 2(g). Overall, the prediction results are consistent with both OCT images and histology findings.
Fig. 2.
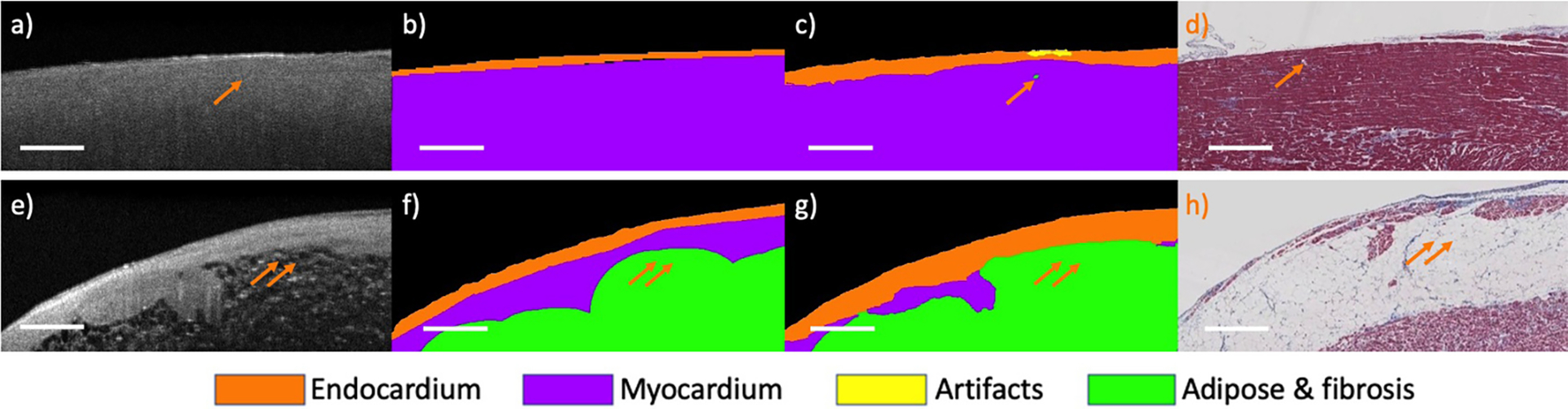
Segmentation results from the proposed method on two myocardium regions. (a, e) original OCT images; (b, f) manual labels; (c, g) prediction results; (d, h) corresponding histology images. Arrows highlight locations of excellent agreement between our tissue prediction results and histology images. Scale bar: 500 μm
Uncertainty Measurement:
We set Nit to 10, which is the smallest number leading to a stable uncertainty map. In Fig. 3, we compare an uncertainty map with its histology to show the effectiveness of our uncertainty measurement. In concordance with the results in [16], we observe that the uncertainty map has a high value at the tissue boundaries and subtle features that are hard to visualize. Therefore, the homogeneous regions appear low uncertainty to the model. These results visually demonstrate that our proposed algorithm can provide accurate localization of model uncertainty and this uncertainty is related to the tissue heterogeneity.
Fig. 3.
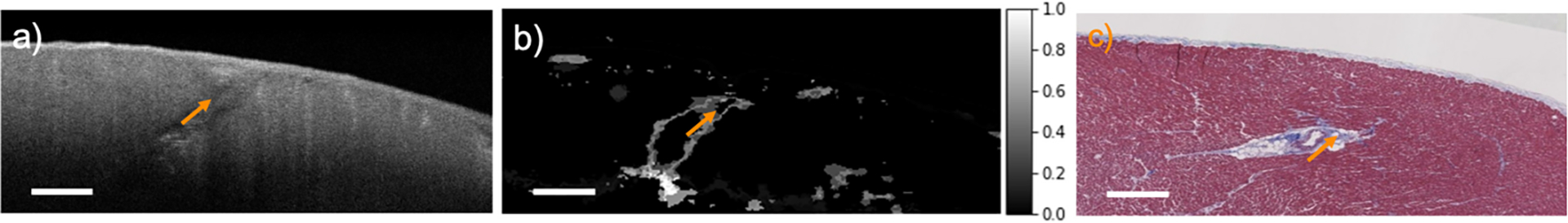
An uncertainty map from a human myocardium region. (a) the original OCT image; (b) the uncertainty map; (c) the corresponding histology image. The uncertainty map highlights heterogeneous regions caused by fibrosis and adipose. Scale bar: 500 μm.
Heterogeneity Measurement:
The positive correlation between purity map heterogeneity and the tissue heterogeneity can be observed in the scatter diagram. Figure 4 shows the normalized heterogeneity results on a cross-validation set with 95% confidence interval. The homogeneity from the purity map has the strongest correlation relationship with tissue homogeneity. The correlation coefficient obtained on these testing images is 0.7117, which achieves the highest value among other methods (OCT images: 0.3501, Uncertainty maps: 0.5959). From the interpreting table in [22,25], the homogeneity values obtained from the purity maps have a high positive correlation with the homogeneity values obtained from the histology images. Therefore, tissue boundary removal is essential to highlight purity map information within tissue regions. In addition, from the results of 95% confidence interval, using a purity map for heterogeneity measurement has less outliers. Hence, the purity map highlights the tissue heterogeneity information and it has the ability for tissue heterogeneity measurement.
Fig. 4.
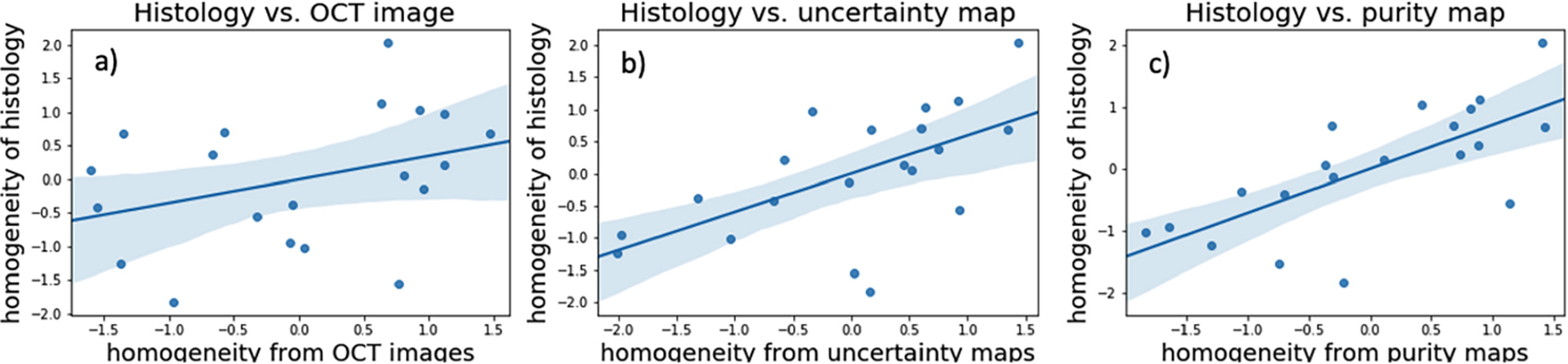
Scatter diagram for normalized homogeneity values. (a) homogeneity values of OCT images vs. histology images; (b) homogeneity values of uncertainty maps vs. histology images; (c) homogeneity values of purity maps vs. histology images. Compared with the OCT images and uncertainty maps, homogeneity obtained from purity maps has the highest correlation with tissue homogeneity.
Our algorithm shows good performance on quantifying both inter-tissue heterogeneity and intra-tissue heterogeneity. Figure 5 shows two purity maps from different human hearts, demonstrating two scenarios of intra/inter-tissue heterogeneity. A high value in the purity map indicates a high heterogeneity and a low value indicates a high homogeneity. In Fig. 5 (a)–(c), the purity map has low values (high homogeneity) all through the homogeneous cardiac regions. Meanwhile, it highlights heart fibers that are not well aligned due to the dilation of the myocardium. In Fig. 5 (d)–(f), two clusters with high heterogeneity are enhanced (arrow and double arrow). The purity map successfully highlights these subtle features in the OCT images. This intra/inter-tissue heterogeneity information is entirely determined by our purity maps, which cannot be observed directly from the labels since all manual labels are region-based.
Fig. 5.
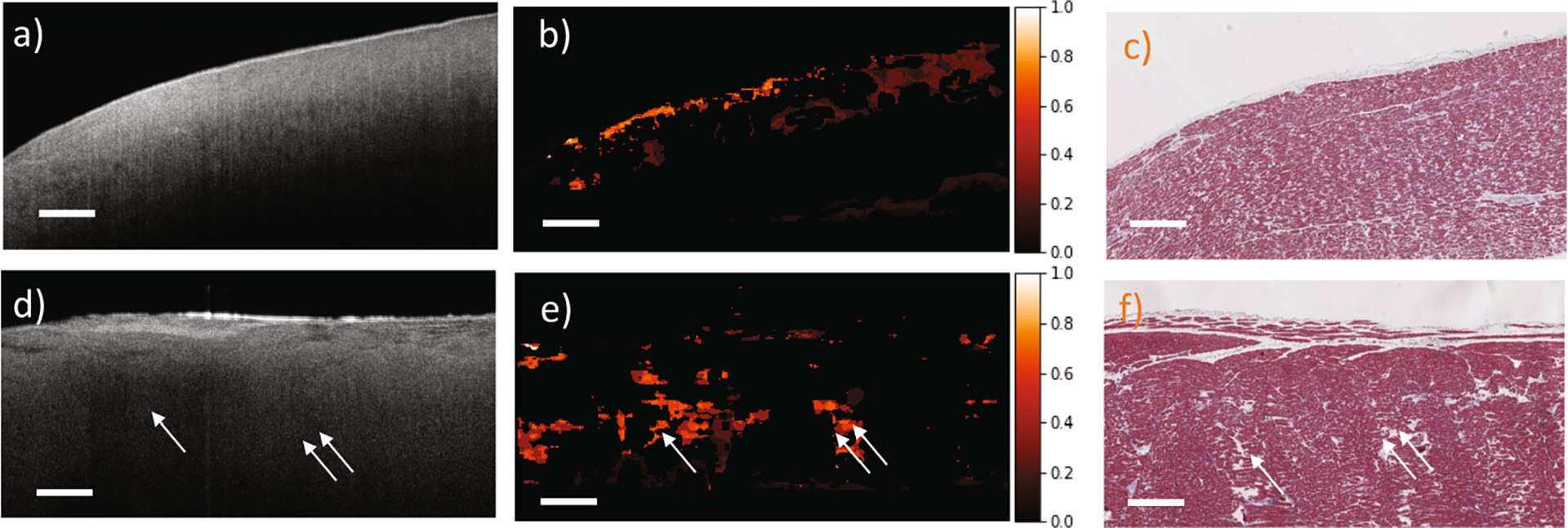
Purity maps from human myocardium regions. (a, d) original OCT images; (b, e) purity maps; (c, f) corresponding histology images. The purity maps highlight the heterogeneity within the human myocardium regions. Scale bar: 500 μm.
Cardiac heterogeneity measurement from OCT images is very challenging and complicated. Figure 6 shows two representative outlier cases from Fig. 4 (c). In Fig. 6 (a)–(c), the adipose regions in the purity map are more heterogeneous than the histology images. It is caused by the fixation and dehydration in the histology process. During the histology process, scattering changes and tissue architectural distortion are inevitable due to the shrinkage of epithelial, muscle, and connective tissue layers [12]. Structural information may get lost after the shrinkage, especially in highly heterogeneous regions. Therefore, a lower value of heterogeneity is observed, leading to a low correlation value. In addition, stain variation in histology could also result in a lower correlation value. In Fig. 6, the loose and dense collagen regions in (a)-(c) are purple and blue, while in (d)-(f), they are pink and pale with isolated light blue strands. This is due to the concentration of the stains and the timings of staining [23]. This issue may be mitigated by improving the histology protocol with better control on the amount of stain used in processing.
Fig. 6.
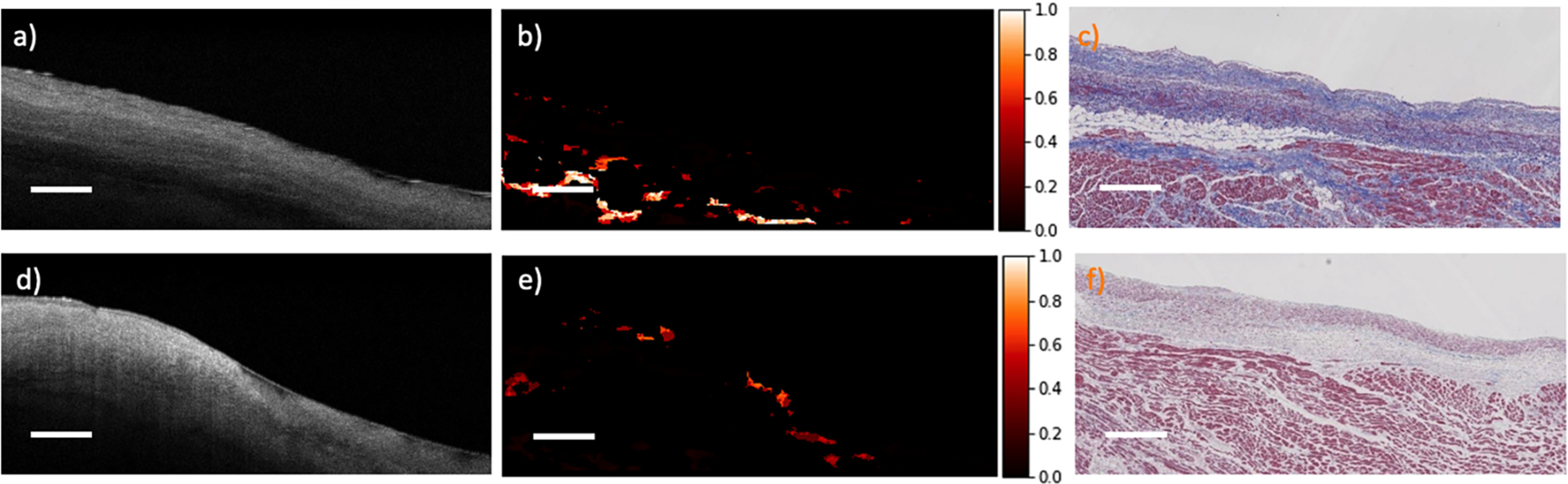
Examples of two outliers from Fig. 4 (c). (a, d) original OCT images; (b, e) purity maps; (c, f) corresponding histology images. Scale bar: 500 μm.
4. Conclusion
In this paper, we propose the first deep learning framework for cardiac tissue heterogeneity measurement on OCT images. Our proposed algorithm consists of three powerful modules: robust learning on region-based labels, uncertainty measurement, and heterogeneity information extraction. We show that these modules are necessary and benefit from each other. Our results indicate that the purity maps can successfully highlight the subtle features from the OCT images and they are highly consistent with the corresponding histology images. Combining with the tissue prediction map, the purity map could be used for the guidance of RFA intervention by identifying regions that have diffusive adipose-fibrosis or heterogeneous myofiber alignment. In the future, we will evaluate our method on a larger dataset and further improve our model in 3D.
Acknowledgements.
The study was funded in part by the National Institute of Health (4DP2HL127776-02 and 1R01HL149369-01, CPH), the National Science Foundation Career Award (1454365, CPH).
References
- 1.Aslanidi OV, Boyett MR, Dobrzynski H, Li J, Zhang H: Mechanisms of transition from normal to reentrant electrical activity in a model of rabbit atrial tissue: interaction of tissue heterogeneity and anisotropy. Biophysical J 96(3), 798–817 (2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baues M, et al. : Fibrosis imaging: current concepts and future directions. Adv. Drug Deliv. Rev 121, 9–26 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buch K, et al. : Using texture analysis to determine human papillomavirus status of oropharyngeal squamous cell carcinomas on CT. AJNR Am. J. Neuroradiol 36(7), 1343–1348 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cua M, et al. : Morphological phenotyping of mouse hearts using optical coherence tomography. J. Biomed. Opt 19(11), 116007 (2014) [DOI] [PubMed] [Google Scholar]
- 5.Fleming CP, Rosenthal N, Rollins AM, Arruda M: First in vivo real-time imaging of endocardial RF ablation by optical coherence tomography. J. Innov. Card. Rhythm Manag 2, 199–201 (2011) [Google Scholar]
- 6.Fujima N, et al. : The utility of MRI histogram and texture analysis for the prediction of histological diagnosis in head and neck malignancies. Cancer Imaging 19(1), 5 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gal Y, Ghahramani Z: Dropout as a bayesian approximation: representing model uncertainty in deep learning. In: International Conference on Machine Learning pp. 1050–1059 (2016) [Google Scholar]
- 8.Gan Y, Lye TH, Marboe CC, Hendon CP: Characterization of the human myocardium by optical coherence tomography. J. Biophotonics 12(12), e201900094 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gan Y, Tsay D, Amir SB, Marboe CC, Hendon CP: Automated classification of optical coherence tomography images of human atrial tissue. J. Biomed. Opt 21(10), 101407 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goergen CJ, et al. : Optical coherence tractography using intrinsic contrast. Opt. Lett 37(18), 3882–3884 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haissaguerre M, et al. : Intermittent drivers anchoring to structural heterogeneities as a major pathophysiological mechanism of human persistent atrial fibrillation. J. Physiol 594(9), 2387–2398 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsiung PL, Nambiar PR, Fujimoto JG: Effect of tissue preservation on imaging using ultrahigh resolution optical coherence tomography. J. Biomed. Opt 10(6), 064033 (2005) [DOI] [PubMed] [Google Scholar]
- 13.Hu S, et al. : Supervised uncertainty quantification for segmentation with multiple annotations. In: Shen D, et al. (eds.) MICCAI 2019 LNCS, vol. 11765, pp. 137–145. Springer, Cham: (2019). 10.1007/978-3-030-32245-816 [DOI] [Google Scholar]
- 14.Braunmühl T: Optical coherence tomography. Der Hautarzt 66(7), 499–503 (2015). 10.1007/s00105-015-3607-z [DOI] [PubMed] [Google Scholar]
- 15.Kather JN, et al. : Multi-class texture analysis in colorectal cancer histology. Scientific Reports 6, 27988 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kendall A, Badrinarayanan V, Cipolla R: Bayesian segnet: Model uncertainty in deep convolutional encoder-decoder architectures for scene understanding arXiv preprint arXiv:1511.02680 (2015) [DOI] [PubMed]
- 17.Khurshid S, et al. : Frequency of cardiac rhythm abnormalities in a half million adults. Circ. Arrhythm Electrophysiol 11(7), e006273 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laplante P: Encyclopedia of Image Processing CRC Press, United States: (2018) [Google Scholar]
- 19.Lin TY, Goyal P, Girshick R, He K, Dollár P: Focal loss for dense object detection. In: IEEE International Conference on Computer Vision. pp. 2980–2988 (2017) [Google Scholar]
- 20.López B, et al. : Circulating biomarkers of myocardial fibrosis: the need for a reappraisal. J. Am. Coll. Cardiol 65(22), 2449–2456 (2015) [DOI] [PubMed] [Google Scholar]
- 21.Lye TH, Iyer V, Marboe CC, Hendon CP: Mapping the human pulmonary venoatrial junction with optical coherence tomography. Biomed. Opt. Express 10(2), 434–448 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mukaka M: A guide to appropriate use of correlation coefficient in medical research. Malawi Med. J 24(3), 69–71 (2012) [PMC free article] [PubMed] [Google Scholar]
- 23.Rotimi O, Cairns A, Gray S, Moayyedi P, Dixon M: Histological identification of helicobacter pylori: comparison of staining methods. J. Clin. Pathol 53(10), 756–759 (2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roy AG, et al. : Relaynet: retinal layer and fluid segmentation of macular optical coherence tomography using fully convolutional networks. Biomed. Opt. Express 8(8), 3627–3642 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schober P, Boer C, Schwarte LA: Correlation coefficients: appropriate use and interpretation. Anesthesia & Analgesia 126(5), 1763–1768 (2018) [DOI] [PubMed] [Google Scholar]
- 26.Sedai S, Antony B, Mahapatra D, Garnavi R: Joint segmentation and uncertainty visualization of retinal layers in optical coherence tomography images using bayesian deep learning. In: Stoyanov D, et al. (eds.) OMIA/COMPAY −2018 LNCS, vol. 11039, pp. 219–227. Springer, Cham: (2018). 10.1007/978-3-030-00949-626 [DOI] [Google Scholar]
- 27.Srivastava N, Hinton G, Krizhevsky A, Sutskever I, Salakhutdinov R: Dropout: a simple way to prevent neural networks from overfitting. J. Mach. Learn. Res 15(1), 1929–1958 (2014) [Google Scholar]
- 28.Tereshchenko LG, et al. : Infiltrated atrial fat characterizes underlying atrial fibrillation substrate in patients at risk as defined by the aric atrial fibrillation risk score. Int. J. Cardiol 172(1), 196–201 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei L, Gan Q, Ji T: Cervical cancer histology image identification method based on texture and lesion area features. Comput. Assist. Surg 22(sup1), 186–199 (2017) [DOI] [PubMed] [Google Scholar]
- 30.Zhao X, et al. : Integrated RFA/PSOCT catheter for real-time guidance of cardiac radio-frequency ablation. Biomed. Opt. Express 9(12), 6400–6411 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]


