Abstract
Background
Laryngeal carcinoma is a primary malignant tumor originating from the laryngeal mucosa, and its pathogenesis is not fully understood. It is a rare type of cancer that shows a downward trend in the 5-year survival rate. In clinical practice, dysregulated microRNAs are often observed in patients with laryngeal cancer. In recent years, an increasing number of studies have confirmed that the strong biomarker potential of microRNAs. We conducted a systematic review and meta-analysis to identify and highlight multiple microRNAs as biomarkers for disease prognosis in patients with laryngeal cancer.
Methods
We actively searched the systematic reviews in PubMed, Embase, Web of Science and The Cochrane Library to select the studies that met the proposed guidelines. A total of 5307 patients with laryngeal cancer were included in this study to evaluate the association between microRNAs expression levels and patient outcomes. For overall survival in the clinical stage, a hazard ratio (HR) and corresponding 95% confidence interval (CI) are calculated to assess the effect of survival.
Results
A total of 36 studies on microRNAs and laryngeal cancer recovery were included in this meta-analysis. The selected endpoints for these studies included overall survival (OS) and disease-free survival (DFS).The comorbidities of overexpression and underexpression of microRNAs were 1.13 (95% CI 1.06–1.20, P < 0.05) and 1.10 (95% CI 1.00–1.20, P < 0.05), respectively.
Conclusion
MiRNA-100, miRNA-155, miRNA-21, miRNA-34a, miRNA-195 and miR-let-7 are expected to be potential noninvasive and simple markers for laryngeal cancer.
Keywords: Laryngeal cancer, Overall survival, Disease-free survival, Prognosis, MicroRNAs
Introduction
Laryngeal cancer is the second most common head and neck cancer (HNC), accounting for 1 to 5% of systemic malignancies. A recent study showed that the 5-year overall survival rate for all HNC sites was 51.4:50.3% for the oral cavity, 41.1% for the oropharynx, 35.0% for the hypopharynx and 63.9% for the larynx [1]. Fortunately, in the last decade, there have been major advances in the treatment of throat cancer [2]. Tobacco and alcohol are closely related factors of laryngeal cancer and lung cancer. These cancer types are more common among men because they are more exposed to these factors. Although tumors can develop anywhere in the throat, the glottis is the most common site, followed by the supraglottis and the subglottis [3]. Therefore, there is an urgent need to identify biomarkers for HNC to facilitate accurate diagnoses, provide information for patients and guide treatment.
MicroRNAs are small endogenous RNA molecules containing approximately 22 nucleotides that regulate gene expression by binding to specific messenger RNAs [4]. MicroRNAs are often located in genomic regions associated with cancer and are involved in the regulation of gene function [5]. It is believed that microRNAs can be used as tumor suppressor genes as well as oncogenes, and play an important role in the occurrence and development of cancer [6, 7]. MicroRNAs are short-stranded noncoding RNAs that play a very important role in malignant tumorigenesis, and they regulate the expression of tumor-related genes at the posttranscriptional level through specific binding to their target genes [8, 9]. MicroRNAs are highly stable and can be measured in biological fluids, including serum, plasma, and saliva, allowing for quick results and repeated analysis during and after treatment. This property strengthens the role of microRNAs as biomarkers and their great potential in the field of oncology [10]. MicroRNAs have been widely researched, and the results indicate that they may provide a sensitive method for the detection, monitoring and prognosis of laryngeal squamous cell carcinoma [11, 12]. In this study, we conducted a systematic review of the existing literature on this topic in PubMed, Embase, Web of Science and The Cochrane Library. Then, we conducted a meta-analysis of the survival rates (including overall survival [OS] and disease-free survival [DFS]) of patients expressing different levels of microRNAs.
Methods
Selection criteria
This meta-analysis was conducted based on an originally conceived protocol. To be qualified, studies had to meet all the following criteria: (1) the study discussed the expression of microRNAs in patients with laryngeal squamous cell carcinoma. (2) The study investigated the relationship between microRNAs expression levels and the survival status of cancer patients. (3) The study was published in full text and reported the risk ratio (HR) of DFS or OS based on microRNA status, with a confidence interval (CI) of 95%.
Studies that met one of the following criteria were excluded: (1) case reports, conference proceedings, letters, or reviews/meta-analyses; (2) thyroid and oral tumors; (3) animal studies; (4) laboratory studies; or (5) incomplete data (no NLR HR for OS/DSS). Incomplete study data (for example, studies that included only Kaplan–Meier curves or HRs that did not report a 95% confidence interval) were not initially ruled out. When this occurred, we contacted the authors and attempted to obtain the raw data.
Literature search strategy
A comprehensive search was performed in PubMed, EMBASE, and Web of Science databases from their inception through December 5, 2020. The search terms were “(Laryngeal Neoplasms OR Neoplasms, Laryngeal OR Laryngeal Neoplasm OR Neoplasm, Laryngeal OR Larynx Neoplasms OR Larynx Neoplasm OR Neoplasm, Larynx OR Neoplasms, Larynx OR Cancer of Larynx OR Larynx Cancers OR Laryngeal Cancer OR Cancer, Laryngeal OR Cancers, Laryngeal OR Laryngeal Cancers OR Larynx Cancer OR Cancer, Larynx OR Cancer of the Larynx) AND (MicroRNAs OR MicroRNA OR miRNAs OR Micro RNA OR miRNA OR Primary MicroRNA OR Primary miRNA OR pri-miRNA OR RNA, Small Temporal OR pre-miRNA OR pre-miRNA) AND (Prognosis OR Prognoses OR Prognostic Factors OR Factor, Prognostic OR Factors, Prognostic OR Prognostic Factor)”. In addition, we manually searched the selected review articles and reference lists of preliminary studies to ensure complete coverage.
Data extraction
To ensure the accuracy of data extraction, two authors extracted data according to the inclusion and exclusion criteria, and a third author assisted in making the final decision on the disputed information. Data were extracted, including author name, year of publication, country, tumor type, total number of patients and test method, follow-up time, microRNAs expression, HR and 95% CI. Meta-analyses were performed as required by the item statement of the systematic review and meta-analysis priority report.
Quality assessment
Methodological quality was systematically reviewed and meta-analyzed based on the quality assessment template of the National Heart, Lung and Blood Institute (NHLBI). Based on the opinions of the two reviewers, each study was subjectively rated as having a “high”, “moderate” or “low” risk of bias rating (Table 1).
Table 1.
Quality assessment of the selected studies for meta-analysis
| S. no | Criteria | High (0–55%) | Moderate (56–78%) | Low (79–100%) |
|---|---|---|---|---|
| 1 | Purpose of this study | 36 | – | – |
| 2 | Eligibility criteria | 25 | 6 | 5 |
| 3 | Sample size adjustment | 36 | – | – |
| 4 | Research group of people | 36 | – | – |
| 5 | Cut-off criteria (follow-up) | 33 | 3 | – |
| 6 | Range of anatomical parts | 36 | – | – |
| 7 | Definition of the measurement used | 36 | – | – |
| 8 | Outcome assessment (OS, DFS) | 31 | 5 | – |
| 9 | Outcome measures (HR, CI) | 26 | – | 10 |
| 10 | Follow-up rate | 27 | 9 | – |
| Total selected studies | 20 | 8 | 8 |
Statistical methods
Meta-analysis was performed on data from multiple included studies. Comprehensive meta-analysis software (RevMan5.3 and Stata 15.1) was used. A combined HR > 1 indicates that the prognosis of the group with elevated microRNA expression was poor. The red square shows a combined effect estimate of survival in patients with laryngeal cancer arranged by microRNA expression. We attempted to explore the heterogeneity of the results through subgroup analysis.
Publication bias
We used Egger’s bias assessment graph test to construct funnel plots (scatter plots constructed using standard error [Y axis] and log (HR) [X axis]) for all included studies. The symmetry of the study distribution on the regression line is inversely proportional to the size of the publication bias in the meta-analysis. In the process of data selection and quality assessment, any differences were resolved through discussion among the reviewers.
Results
Study selection
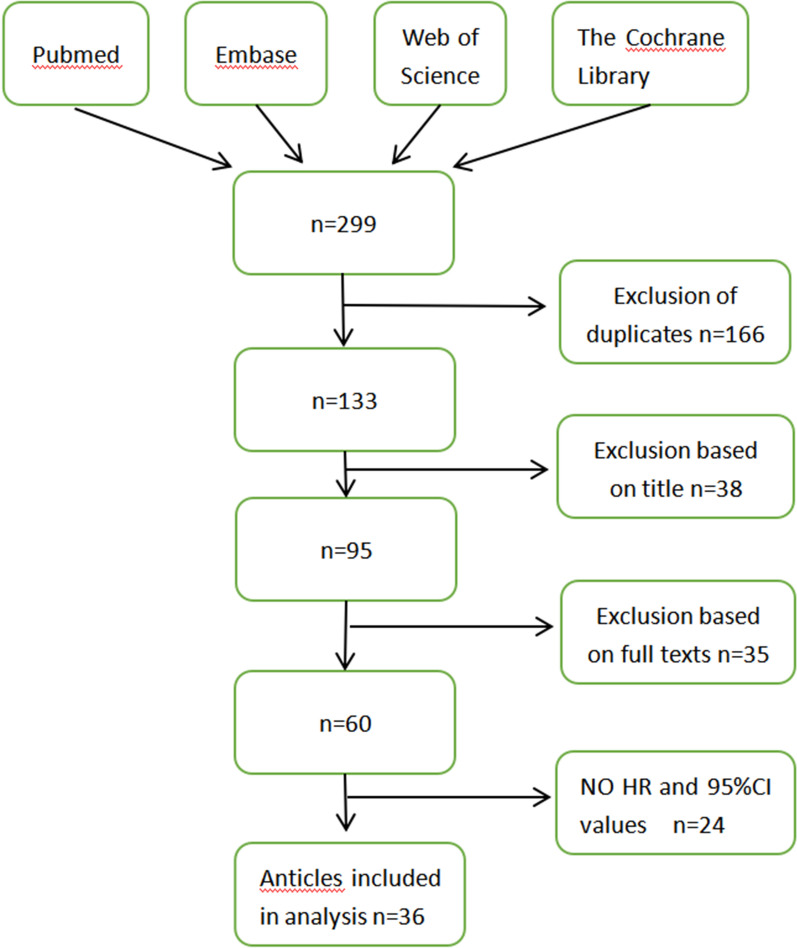
We found a total of 299 records and placed them into the EndNote Web reference manager to delete duplicate articles. A total of 36 studies were included in the meta-analysis. The details are shown in Fig. 1.
Fig. 1.
Flow chart describing search strategy
Study characteristics
The studies included in this systematic review included a sample of 36 research articles. The average patient age in most studies was approximately 60. We found that these studies originated from eight countries around the world, including The Netherlands (n = 1), Italy (n = 2), Germany (n = 1), Brazil (n = 2), U.S. A. (n = 6), Athens (n = 1), Japan (n = 1), and China (n = 22). MicroRNA expression was found in preserved tissue samples (23 studies), serum (10 studies) and plasma (3 studies). Eight studies used TaqMan low-density arrays to detect microRNAs expression profiles, and 27 studies used RT-PCR to detect microRNAs expression. The average period for follow-up studies was 3–7 years. In all the studies included in the systematic review and meta-analysis, a total of 24 microRNAs were reported. Of the 24 microRNAs included, 10 microRNAs were upregulated, and 14 microRNAs were downregulated. Table 2 describes the research design and patient information for all 36 studies (Table 2).
Table 2.
Characteristics of the included studies of the meta-analysis
| Author | Year | MiRNA | Case | Anatomic location | Assay Method | Country | Gender | Stage | Metastasis | Age | Outcomes | miRNA dysregulation |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Shuang et al. [13] | 2017 | miR-195 | 122 |
Glottis 61 Supraglottis 42 Subglottis 19 |
qRT-PCR | China | Male (65.5%) |
TI + T2 30 T3 + T4 92 |
N0 81 N + 41 |
53–69 y | OS | Down regulated |
| Zhao et al. [14] | 2018 | miR-145 | 132 |
Glottic 76 Supraglottic 56 |
qRT-PCR | China | Male (86.3%) |
T2 51 T3 + T4 81 |
N0 61 N+ 71 |
48–84 y | OS | Down regulated |
| Gao et al. [15] | 2019 | miR-145-5p | 188 |
Glottic 101 Supraglottic 83 Subglottic 4 |
TCGA | China | Male (88.8%) |
T1 + T2 111 T3 + T4 77 |
N0 142 N+ 46 |
≤ 60y 99 > 60y 89 |
OS | Down regulated |
| Li et al. [16] | 2015 | miR-101 | 80 | Larynx | qRT-PCR | China | Male (70%) |
T1 + T2 40 T3 + T4 40 |
N0 45 N + 35 |
≤ 60y 48 > 60y 32 |
OS | Down regulated |
| de Jong et al. [17] | 2015 | miR-203 | 34 | NA | qRT-PCR | The Netherlands | NA | NA | NA | NA | OS | Down regulated |
| Ding and Qi [18] | 2019 | miR-195 | 182 |
Supraglottic 50 Glottic 95 Subglottic 37 |
qRT-PCR | China | Male (65.3%) | NA |
N0 110 N+ 72 |
NA | OS | Down regulated |
| Fang et al. [19] | 2019 | miR-29c-3p | 66 |
Supraglottic 19 Glottic 45 Subglottic 2 |
qRT-PCR | China | Male (93.9%) |
T1 + T2 21 T3 + T4 45 |
N0 36 N+ 30 |
≤ 60y 26 > 60y 40 |
OS | Down regulated |
| He et al. [20] | 2017 | miR-300 | 133 | Larynx | qRT-PCR | China | Male (65.4%) |
T1 + T2 67 T3 + T4 66 |
N0 73 N+ 60 |
≤ 50y 47 > 50y 86 |
OS | Down regulated |
| Hui et al. [21] | 2019 |
miR-10a miR-16–2 |
22 | Larynx | TCGA | China | NA | NA | NA | NA | OS, RFS | Down regulated |
| Re et al. [22] | 2015 | miR-34c-5p | 90 |
Supraglottic 19 Transglottic 66 Subglottic 5 |
qRT-PCR | Italy | Male (96.6%) |
T3 60 T4 30 |
N0 29 N+ 61 |
Mean 66.51 | OS | Down regulated |
| Xu et al. [23] | 2016 | miR-149 | 97 | Larynx | qRT-PCR | China | NA |
T1 + T2 59 T3 + T4 38 |
N0 69 N+ 28 |
≤ 60y 46 > 60y 51 |
OS | Down regulated |
| Tian et al. [24] | 2014 | miR-203 | 56 |
Glottic 30 Supraglottic 26 |
qRT-PCR | China | Male (71.43%) |
T1 + T2 24 T3 + T4 32 |
N0 23 N+ 23 |
≤ 59y 32 > 59y 24 |
OS | Down regulated |
| Hess et al. [25] | 2017 |
miR-155 miR-146a miR-200b |
149 |
Oropharynx 77 Hypopharynx 72 |
qRT-PCR | Germany | NA | NA | NA | NA | OS, RFS | Down regulated |
| Zhao et al. [26] | 2018 | miR-181a | 127 |
Supraglottic 50 Glottic 77 |
qRT-PCR | China | Male (88.6%) |
T1 + T2 53 T3 + T4 74 |
N0 65 N+ 62 |
≤ 60y 79 > 60y 48 |
OS | Down regulated |
| Zhao et al. [27] | 2018 | miR-196b | 113 |
Supraglottic 43 Glottic 70 |
qRT-PCR | China | Male (85%) |
T1 + T2 47 T3 + T4 66 |
N0 65 N+ 48 |
≤ 60y 71 > 60y 42 |
OS | Upregulated |
| Zhao et al. [28] | 2018 | miR-155 | 120 |
Supraglottic 46 Glottic 74 |
qRT-PCR | China | Male (89.2%) |
T1 + T2 67 T3 + T4 53 |
N0 88 N+ 32 |
≤ 60y 63 > 60y 57 | OS | Upregulated |
| Guan et al. [29] | 2016 | miR-675 | 62 |
Larynx 46 Hypopharynx 14 Oropharynx 2 |
qRT-PCR | China | Male (94%) | NA | NA | Mean 64 | OS | Upregulated |
| Arantes et al. [30] | 2017 |
miR-21 miR-494 miR-720 |
71 |
Oropharynx 35 Larynx 28 Hypopharynx 8 |
TCGA | Brazil | Male (95.8%) |
T1 + T2 46 T3 + T4 25 |
N0 35 N+ 36 |
NA | OS | Upregulated |
| Avissar et al. [31] | 2009 | miR-21 | 169 | Larynx | qRT-PCR | USA | Male (68%) |
T1 + T2 46 T3 + T4 118 |
NA | Mean 61.5 | OS | Upregulated |
| Langevin et al. [32] | 2011 | miR-137 | 67 | Larynx | qRT-PCR | USA | Male (74.6%) |
T1 + T2 35 T3 + T4 32 |
N0 31 N+ 36 |
Mean 62.4 | OS | Upregulated |
| Qiang et al. [33] | 2019 | miR-31 | 55 | Larynx | qRT-PCR | China | Male (57.1%) |
T1 + T2 21 T3 + T4 35 |
NA | Mean 63.2 | OS | Upregulated |
| Wu et al. [34] | 2014 | miR-9 | 103 |
Supraglottic 66 Glottic 37 |
qRT-PCR | China | Male (47.5%) |
T1 + T2 55 T3 + T4 48 |
N0 31 N+ 74 |
≤ 60y 41 > 60y 62 |
OS | Upregulated |
| Wu et al. [35] | 2014 | miR-19a | 83 |
Supraglottic 35 Glottic 48 |
qRT-PCR | China | Male (68.6%) |
T1 + T2 52 T3 + T4 31 |
N0 54 N + 29 |
≤ 56y 42 > 56y 41 |
OS | Upregulated |
| Zhang et al. [36] | 2015 | miR-23a | 52 | Larynx | qRT-PCR | China | Male (86.5%) |
T1 + T2 24 T3 + T4 28 |
N0 34 N + 18 |
≤ 60y 22 > 60y 30 |
OS | Upregulated |
| Hu et al. [37] | 2014 | miR-21 | 46 |
Glottic 33 Supraglottic 11 Subglottic 2 |
qRT-PCR | China | Male (91.3%) |
T0 + T1 + T2 21 T3 + T4 25 |
NA | Mean 59.2 | OS | Upregulated |
| Saito et al. [38] | 2013 | miR-196a | 84 | Larynx | qRT-PCR | Japan | NA | NA | NA | NA | OS | Upregulated |
| Re and Magliulo [39] | 2017 | miR-34c-5p | 43 |
Supraglottic 8 Transglottic 33 Subglottic 2 |
qRT-PCR | Italy | Male (97.67%) |
T3 31 T4 12 |
N0 27 N+ 16 |
Mean 66.51 | DFS | Downregulated |
| Shen et al. [40] | 2012 | miR-34a | 69 | Larynx | qRT-PCR | China | NA |
T1 + T2 42 T3 + T4 27 |
N0 24 N+ 45 |
≤ 60y 33 > 60y 36 |
DFS | Downregulated |
| Danielle Maia [41] | 2015 | mIR-296-5P | 34 |
Supraglottic 7 Glottic 27 |
TCGA | Brazil | Male (88.2%) |
TI 16 T2 18 |
NA |
≤ 60y 16 > 60y 18 |
DFS | Downregulated |
| Ogawa et al. [42] | 2012 | miR-34a | 24 | Larynx | TCGA | Japan | Male (66.6%) |
T3 10 T4 14 |
NA |
≤ 60y 10 > 60y 14 |
DFS | Downregulated |
| Pantazis et al. [43] | 2020 | miR-20b-5p | 105 | Larynx | qRT-PCR | Athens | Male (63.1%) |
T1 + T2 31 T3 + T4 74 |
NA | NA | DFS | Downregulated |
| Wilkins [44] | 2019 | miR-100 | 136 | Larynx | TCGA | USA | NA | NA | NA | NA | DFS | Downregulated |
| Li et al. [45] | 2019 | miR-424-5p | 106 |
Glottic 55 Supraglottic 40 Subglottic 3 Transglottic 8 |
TCGA | China | Male (93.3%) |
T1 + T2 58 T3 + T4 48 |
N0 80 N+ 26 |
≤ 60y 47 > 60y 59 |
DFS | Upregulated |
| Childs et al. [46] | 2009 | miR-let-7d | 73 |
Oropharynx 32 Hypopharynx 9 Larynx 32 |
qRT-PCR | USA | Male (68%) |
T1 + T2 17 T3 + T4 56 |
NA |
≤ 60y 30 > 60y 43 |
OS | Downregulated |
| Liu et al. [47] | 2017 | miR-let-7a | 131 | Larynx | qRT-PCR | China | Male (33%) | T1 + T2 51 T3 + T4 80 | NA |
≤ 60y 88 > 60y 43 |
OS | Downregulated |
| Wilkins et al. [48] | 2018 | miR-let-7a | 2083 |
Pharynx 1458 Larynx 625 |
TCGA | USA | Male (24.5%) |
T1 + T2 541 T3 + T4 1542 |
NA | NA | OS | Downregulated |
Meta-analysis
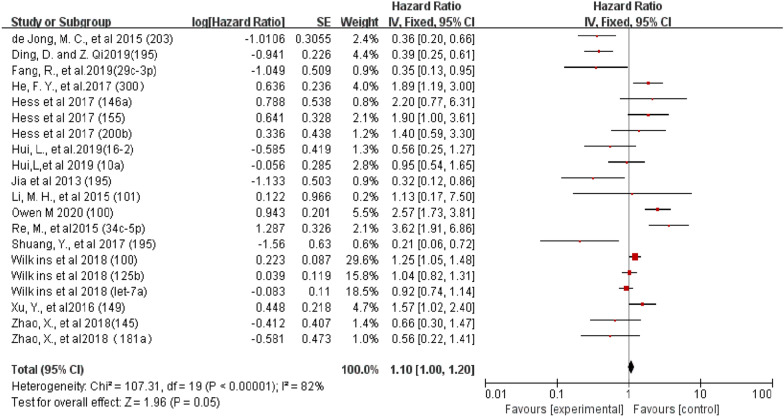
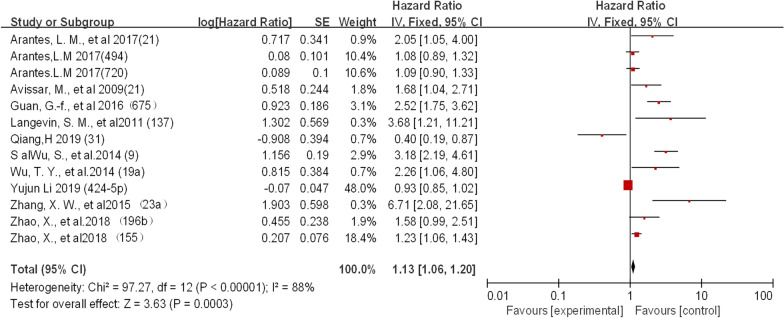
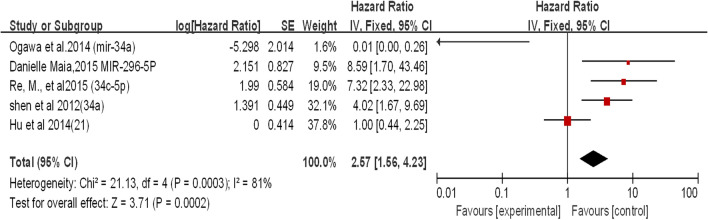
The association of OS with at least one microRNA in laryngeal squamous cell carcinoma (LSCC) was reported in 36 studies. The expression of these molecules was detected primarily at the RNA level, and the upregulated expression was compared with that in normal tissues. The meta-analysis pooled HR and 95% CI (n = 36) from studies with OS as the endpoint, providing an overall effect of a downregulated microRNA size estimate (HR) of 1.10 (95% CI 1.00–1.20), while the estimated pooling effect size (HR) of upregulated microRNAs was 1.13 (95% CI 1.06–1.20). The effect size estimate (HR) for studies using DFS as their survival endpoint was 2.57 (95% CI 1.56–4.23). When OS was used as a survival endpoint, there was no significant difference between the upregulated and downregulated expression of these microRNAs. This finding suggests that any change in microRNA expression is associated with a lower survival rate in patients with LSCC. The apparent overall heterogeneity between these studies using OS as a survival endpoint was found to be high (I2 = 88.05;). Similar results were found in DFS (Figs. 2, 3 and 4).
Fig. 2.
Meta-analysis of downregulated miRNA expression for overall survival in LSCC
Fig. 3.
Meta-analysis of upregulated miRNA expression for OS in LSCC
Fig. 4.
Meta-analysis of upregulated and downregulated miRNA expression for disease-free survival in LSCC
Subgroup analysis
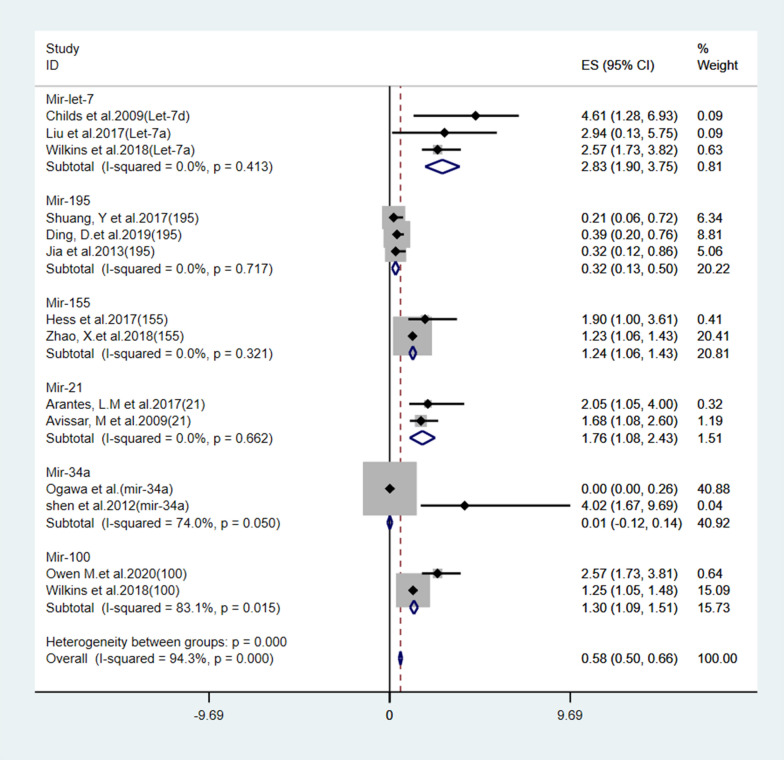
MicroRNAs of six subgroups were included in Stata 15.1 to obtain the combined forest plot (Fig. 5). The heterogeneity between groups was large (I2 = 94.3%), so we conducted a subgroup analysis. We divided microRNA subgroups according to the survival end points (OS and DFS). The microRNA subgroups included miR-195, miR-100, miR-21, miR-155 and miR-let-7 for the OS group (Figs. 6, 7, 8, 9 and 10) and miR-34a for the DFS group (Fig. 11).
Fig. 5.
The total Forest plot of the 6 miRNA subgroups in LSCC
Fig. 6.
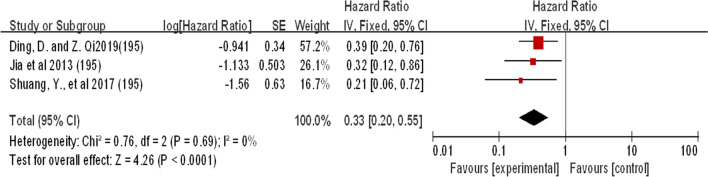
Subgroup analysis of miRNA-195 expression for OS in LSCC
Fig. 7.
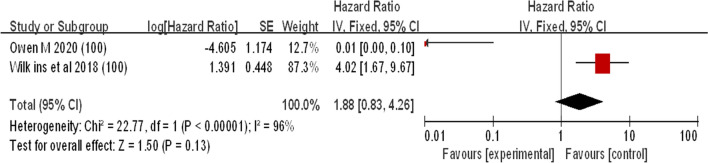
Subgroup analysis of miRNA-100 expression for OS in LSCC
Fig. 8.
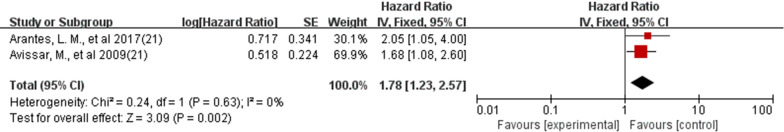
Subgroup analysis of miRNA-21 expression for OS in LSCC
Fig. 9.
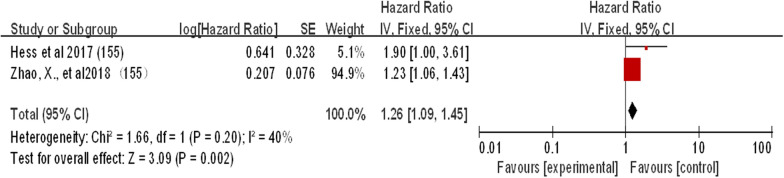
Subgroup analysis of miRNA-155 expression for OS in LSCC
Fig. 10.
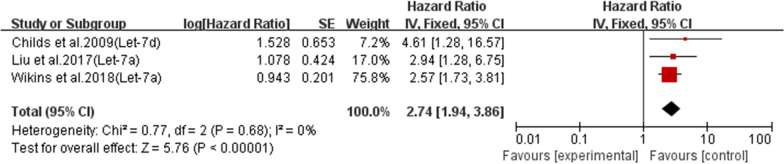
Subgroup analysis of miRNA-let-7 expression for OS in LSCC
Fig. 11.
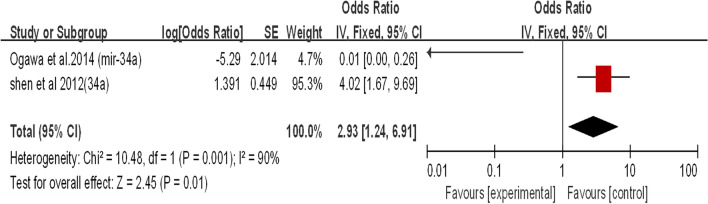
Subgroup analysis of miRNA-34a expression in LSCC (DFS)
miRNA-195
Three studies revealed the expression of miR-195 in patients with laryngeal cancer (Fig. 6). All three studies showed that the expression of the miR-195 gene was downregulated in patients with LSCC. The combined effect size estimate (HR) was 0.33 (0.20–0.55; P < 0.05). All three studies showed that downregulation of miR-195 expression resulted in higher survival rates. The heterogeneity of the studies was 0 (I2 = 0%), indicating that the three studies supported the same finding (Fig. 6).
miRNA-100
Two studies investigated miR-100 expression in LSCC patients (Fig. 7). Both studies showed that miRNA-100 expression was upregulated in patients with laryngeal squamous cell carcinoma. The combined effect size estimate (HR) was 1.88 (95% CI 0.83–4.26). As shown in Fig. 7, there was significant heterogeneity between the two groups (I2 = 96%). The work of Owen et al. [43] contradicts the work of another author. To identify the source of the difference, we searched existing databases and found that high miR-100 expression levels were associated with lower survival rates in oral squamous cell carcinoma (46 cases) and esophageal carcinoma (47 cases). By removing this study, the merger heterogeneity effect of the upregulated group will be reduced (Fig. 7).
miRNA-21
Two studies reviewed the expression of miR-21 in patients with laryngeal squamous cell carcinoma (Fig. 8). The two studies demonstrated similar results regarding miR-21 expression in laryngeal squamous cell carcinoma (I2 = 0%). This similarity in results indicated that the two studies agreed that overexpression of miR-21 in laryngeal cancer tissue leads to poor survival. The combined effect estimate (HR) was statistically significant, with a value of 1.78 (95% CI 1.23–2.57; P < 0.05) (Fig. 8).
miRNA-155
Two studies reviewed miR-155 expression in patients with laryngeal squamous cell carcinoma (Fig. 9). The combined heterogeneity of the two studies was very low (I2 = 40%) and thus could be ignored. Both studies showed that overexpression of miR-155 in laryngeal squamous cell carcinoma resulted in reduced survival. The combined effect estimate (HR) value was 1.26 (95% CI 1.09–1.45; P < 0.05) (Fig. 9).
miRNA-let-7
Three studies reviewed the expression of miR-let-7 in patients with laryngeal squamous cell carcinoma (Fig. 10). The three studies demonstrated similar results regarding miR-let-7 expression in laryngeal squamous cell carcinoma (I2 = 0%). This similarity in results indicated that the three studies agreed that overexpression of miR-let-7 in laryngeal cancer tissue leads to poor survival. The combined effect estimate (HR) was statistically significant, with a value of 2.74 (95% CI 1.94–3.86; P < 0.05) (Fig. 10).
Disease-free survival group
miRNA-34a
Two studies reviewed the expression of miR-34a in LSCC patients (Fig. 11). Both studies showed that miR-34a was downregulated in laryngeal squamous cell carcinoma. The combined effect size (HR) was 2.93 (95% CI 1.24–6.91; P < 0.05). In Fig. 11, we can clearly see that when the two studies were combined, the overall heterogeneity was high (I2 = 90%). Based on a review of published studies, it has been demonstrated that downregulation of miR-34a promotes laryngeal cancer cell proliferation and migration by targeting cyclin D1 [49]. The prognostic results obtained by Ogawa et al. contradicted the experimental phenomenon. Therefore, we consider that the study of Ogawa et al. led to a high degree of heterogeneity (Fig. 11).
Publication bias
Funnel plot
Figures 12 and 13 indicate that in terms of survival results, the funnel chart shows slight asymmetry between different studies. In the studies included in the analysis, visual observation of the funnel shape was the first step we used to assess publication bias. The evidence of asymmetry revealed the potential publication bias, and the publication bias of OS and DFS was detected by Begg and Egger methods. This suggests that a few studies may have been missing from our search, including results related to increased survival (Figs. 12, 13).
Fig. 12.
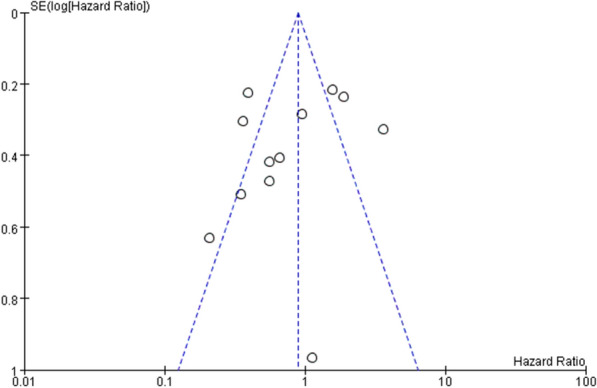
Funnel plot of studies correlating overall patient survival and downregulated miRNA expression
Fig. 13.
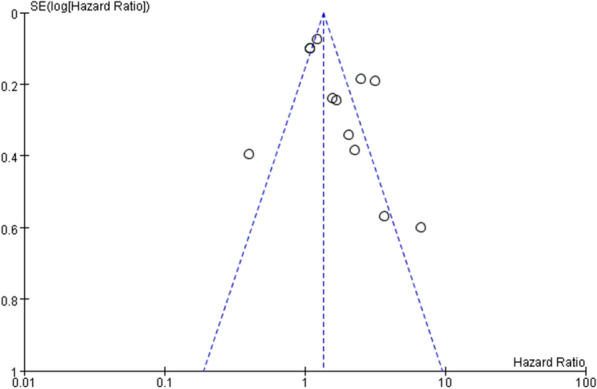
Funnel plot of studies correlating overall patient survival and upregulated miRNA expression
Discussion
MicroRNAs are a class of nonprotein coding small RNAs 21–25 nucleotides in length which play a key role in gene regulation [50]. Circulating biomarkers are often used by clinicians to diagnose the early stages of LSCC. In contrast, invasive examination is not conducive to observing the progress of LSCC, and patients often reject invasive procedures because the examination may cause them pain. MicroRNAs are very stable, and clinical studies investigating their role have provided reproducible and consistent results [51]. In addition, we can easily detect microRNAs by RT qPCR for LSCC diagnosis. Circulating microRNAs can resist RNase A digestion and other adverse conditions, including repeated freeze-thaw cycles and long-term storage under unsuitable pH conditions [52]. MicroRNAs are promising biomarkers for early diagnosis and prognosis [53].
Our meta-analysis examined 36 articles including a patient population of 5307 individuals. In the different studies, there were 10 upregulated microRNAs and 14 downregulated microRNAs. Then, according to the expression level, they were divided into two groups, which were put into the tool to obtain the forest map. It is generally believed that if I2 is less than 50% this means that the heterogeneity between studies is small, and if it is greater than 50% then the heterogeneity is large. It does not make sense to combine HR of all microRNAs because their heterogeneity is too high. For prognostic biomarkers, it is more appropriate to focus on the prognostic efficiency of each microRNA.
In this study, six microRNAs (miR-34a, miR-195, miR-100, miR-21, miR-155 and miR-let-7) had high prognostic value. The remaining microRNAs were of low value. The two groups that showed great heterogeneity were miRNA-34a and miRNA-100. We tried to find the source of the heterogeneity by exploring the basic biomolecular experimental findings. We conclude that high expression of five microRNAs (miR-34a, miR-100, miR-21, miR-155 and miR-let-7) indicates poor overall survival, while high expression of miRNA-195 indicates improved overall survival. DFS is only reported only in miR-34a.
In 2008, Thian-Sze et al. conducted the first study to detect the expression level of miR-195 in head and neck tumors. Real-time quantitative PCR was used to evaluate the upregulation of miR-195 expression in four types of tongue cancer cells [54]. In 2019, Tian et al. studied two human laryngeal squamous cell carcinoma cell lines with different in vitro radiosensitivity. It was found that miR-195 is a direct downstream target of DGCR5. Moreover, the overexpression of miR-195 enhanced the radiosensitivity of Hep-2R cells [55].
Lidia et al. isolated extracellular vesicles from the plasma of 60 patients with thyroid cancer in 2020 and analyzed vesicle microRNAs. The area under the curve (AUC) value of miR-let-7 was 0.814 [56]. This result shows that miR-let-7 has a high diagnostic ability.
Yao et al. confirmed that the miR-34a-5p/Axl axis plays an aggressive role in oral cancer cells through the Akt/GSK-3 and β/β-catenin/Snail signal transduction pathways and may be a therapeutic target for oral squamous cell carcinoma [57].
The latest study by Weina et al. found that in the epithelial-mesenchymal transition of LSCC cells induced by TGF-β, miR-155HG can regulate epithelial–mesenchymal transition markers through the miR-155/SOX10 axis [58]. The author concluded that the miR-155HG/miR-155-5p/SOX10 axis is a key part of the process in promoting LSCC.
The studies in this analysis were drawn from all currently available LSCC studies that investigate microRNA expression and prognosis. Compared with other studies, the results of this study show certain advantages in accuracy. One of the shortcomings of our analysis is that the sample size of this study is relatively small. Because our study was retrospective, we were limited by the existing literature. In addition, we only chose the study endpoints of OS and DFS because of the lack of other survival outcomes, such as relapse-free survival. In addition, many risk factors contribute to the development of laryngeal squamous cell carcinoma. In the future, we can also collect data on early glottic diseases that suggest the occurrence of laryngeal cancer.
Conclusions
Overall, despite the limitations of our review, our data still provide evidence suggesting that miRNA-100, miR-155, miR-21, miR-34a, miR-195 and miR-let-7 are potential noninvasive and simple humoral tumor markers for laryngeal cancer.
Acknowledgements
This work is supported by Scientific Projects of Jiangsu Province (BK20191157), Scientific Program of Changzhou (CE20195048) and Funding from Young Talent Development Plan of Changzhou Health Commission (2020-233).
Abbreviations
- miRNA
MicroRNA
- LSCC
Laryngeal squamous cell carcinoma
- NA
Not available
- DFS
Disease-free survival
- OS
Overall survival
- y
Years
- N0
No lymph node metastasis
- N+
Lymph node metastasis
- Mean
Mean years
- HNSCC
Head and neck cancer
- HR
Hazard ratio
- OSCC
Oral squamous cell carcinoma
- RFS
Relapse free survival
- DGCR5
DiGeorge syndrome critical region gene 5
- TGF-β
Transforming growth factor-β
Authors’ contributions
Conceptualization: YH, MG. Data curation: MG, YT. Formal analysis: YT, ZS. Funding acquisition: JL. Investigation: YT, MG. Project administration: ZS. Software: YH, YT. Supervision: YT, ZL. Writing—original draft: YH. Writing—review and editing: YH. All authors read and approved the final manuscript.
Availability of data and materials
All data are included in this article.
Declarations
Ethics approval and consent to participate
The study was approved by the Human Research Ethics Committees of The Affiliated Changzhou No.2 People’s Hospital of Nanjing Medical University, Changzhou, China.
Consent for publication
All authors agree to publish.
Competing interests
All authors have no conflict of interest in this meta-analysis.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yan Huan and Min Gu contributed equally to this study.
Contributor Information
Judong Luo, Email: judongluo@suda.edu.cn.
Zhe Li, Email: lizhezsh@126.com.
References
- 1.Giraldi L, Leoncini E, Pastorino R, Wünsch-Filho V, de Carvalho M, Lopez R, et al. Alcohol and cigarette consumption predict mortality in patients with head and neck cancer: a pooled analysis within the International Head and Neck Cancer Epidemiology (INHANCE) Consortium. Ann Oncol. 2017;28(11):2843–2851. doi: 10.1093/annonc/mdx486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Steuer CE, El-Deiry M, Parks JR, Higgins KA, Saba NF. An update on larynx cancer. CA Cancer J Clin. 2017;67(1):31–50. doi: 10.3322/caac.21386. [DOI] [PubMed] [Google Scholar]
- 3.Salvador-Coloma C, Cohen E. Multidisciplinary care of laryngeal cancer. J Oncol Pract. 2016;12(8):717–724. doi: 10.1200/JOP.2016.014225. [DOI] [PubMed] [Google Scholar]
- 4.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–540. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 5.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 6.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10(10):704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allegra A, Alonci A, Campo S, Penna G, Petrungaro A, Gerace D, et al. Circulating microRNAs: new biomarkers in diagnosis, prognosis and treatment of cancer (review) Int J Oncol. 2012;41(6):1897–1912. doi: 10.3892/ijo.2012.1647. [DOI] [PubMed] [Google Scholar]
- 8.Felekkis K, Touvana E, Stefanou C, Deltas C. microRNAs: a newly described class of encoded molecules that play a role in health and disease. Hippokratia. 2010;14(4):236–240. [PMC free article] [PubMed] [Google Scholar]
- 9.Wahid F, Shehzad A, Khan T, Kim YY. MicroRNAs: synthesis, mechanism, function, and recent clinical trials. Biochem Biophys Acta. 2010;1803(11):1231–1243. doi: 10.1016/j.bbamcr.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 10.Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, et al. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56(11):1733–1741. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105(30):10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abd-El-Fattah AA, Sadik NA, Shaker OG, Aboulftouh ML. Differential microRNAs expression in serum of patients with lung cancer, pulmonary tuberculosis, and pneumonia. Cell Biochem Biophys. 2013;67(3):875–884. doi: 10.1007/s12013-013-9575-y. [DOI] [PubMed] [Google Scholar]
- 13.Shuang Y, Li C, Zhou X, Huang Y, Zhang L. MicroRNA-195 inhibits growth and invasion of laryngeal carcinoma cells by directly targeting DCUN1D1. Oncol Rep. 2017;38(4):2155–2165. doi: 10.3892/or.2017.5875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao X, Zhang W, Ji W. MYO5A inhibition by miR-145 acts as a predictive marker of occult neck lymph node metastasis in human laryngeal squamous cell carcinoma. Onco Targets Ther. 2018;11:3619–3635. doi: 10.2147/OTT.S164597. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Gao W, Zhang C, Li W, Li H, Sang J, Zhao Q, et al. Promoter methylation-regulated miR-145-5p inhibits laryngeal squamous cell carcinoma progression by targeting FSCN1. Mol Ther J Am Soc Gene Ther. 2019;27(2):365–379. doi: 10.1016/j.ymthe.2018.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li M, Tian L, Ren H, Chen X, Wang Y, Ge J, et al. MicroRNA-101 is a potential prognostic indicator of laryngeal squamous cell carcinoma and modulates CDK8. J Transl Med. 2015;13:271. doi: 10.1186/s12967-015-0626-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Jong MC, Ten Hoeve JJ, Grénman R, Wessels LF, Kerkhoven R, Te Riele H, et al. Pretreatment microRNA expression impacting on epithelial-to-mesenchymal transition predicts intrinsic radiosensitivity in head and neck cancer cell lines and patients. Clin Cancer Res. 2015;21(24):5630–5638. doi: 10.1158/1078-0432.CCR-15-0454. [DOI] [PubMed] [Google Scholar]
- 18.Ding D, Qi Z. Clinical significance of miRNA-195 expression in patients with laryngeal carcinoma. J BUON. 2019;24(1):315–322. [PubMed] [Google Scholar]
- 19.Fang R, Huang Y, Xie J, Zhang J, Ji X. Downregulation of miR-29c-3p is associated with a poor prognosis in patients with laryngeal squamous cell carcinoma. Diagn Pathol. 2019;14(1):109. doi: 10.1186/s13000-019-0893-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He FY, Liu HJ, Guo Q, Sheng JL. Reduced miR-300 expression predicts poor prognosis in patients with laryngeal squamous cell carcinoma. Eur Rev Med Pharmacol Sci. 2017;21(4):760–764. [PubMed] [Google Scholar]
- 21.Hui L, Wang J, Zhang J, Long J. lncRNA TMEM51-AS1 and RUSC1-AS1 function as ceRNAs for induction of laryngeal squamous cell carcinoma and prediction of prognosis. PeerJ. 2019;7:e7456. doi: 10.7717/peerj.7456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Re M, Çeka A, Rubini C, Ferrante L, Zizzi A, Gioacchini FM, et al. MicroRNA-34c-5p is related to recurrence in laryngeal squamous cell carcinoma. Laryngoscope. 2015;125(9):E306–E312. doi: 10.1002/lary.25475. [DOI] [PubMed] [Google Scholar]
- 23.Xu Y, Lin YP, Yang D, Zhang G, Zhou HF. Clinical significance of miR-149 in the survival of patients with laryngeal squamous cell carcinoma. Biomed Res Int. 2016;2016:8561251. doi: 10.1155/2016/8561251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tian L, Li M, Ge J, Guo Y, Sun Y, Liu M, et al. MiR-203 is downregulated in laryngeal squamous cell carcinoma and can suppress proliferation and induce apoptosis of tumours. Tumour Biol J Int Soc Oncodev Biol Med. 2014;35(6):5953–5963. doi: 10.1007/s13277-014-1790-7. [DOI] [PubMed] [Google Scholar]
- 25.Hess AK, Müer A, Mairinger FD, Weichert W, Stenzinger A, Hummel M, et al. MiR-200b and miR-155 as predictive biomarkers for the efficacy of chemoradiation in locally advanced head and neck squamous cell carcinoma. Eur J Cancer (Oxford, Engl: 1990) 2017;77:3–12. doi: 10.1016/j.ejca.2017.02.018. [DOI] [PubMed] [Google Scholar]
- 26.Zhao X, Zhang W, Ji W. miR-181a targets GATA6 to inhibit the progression of human laryngeal squamous cell carcinoma. Future Oncol (Lond, Engl) 2018;14(17):1741–1753. doi: 10.2217/fon-2018-0064. [DOI] [PubMed] [Google Scholar]
- 27.Zhao X, Zhang W, Ji W. miR-196b is a prognostic factor of human laryngeal squamous cell carcinoma and promotes tumor progression by targeting SOCS2. Biochem Biophys Res Commun. 2018;501(2):584–592. doi: 10.1016/j.bbrc.2018.05.052. [DOI] [PubMed] [Google Scholar]
- 28.Zhao X, Zhang W, Ji W. YB-1 promotes laryngeal squamous cell carcinoma progression by inducing miR-155 expression via c-Myb. Future Oncol (Lond, Engl) 2018;14(16):1579–1589. doi: 10.2217/fon-2018-0058. [DOI] [PubMed] [Google Scholar]
- 29.Guan GF, Zhang DJ, Wen LJ, Xin D, Liu Y, Yu DJ, et al. Overexpression of lncRNA H19/miR-675 promotes tumorigenesis in head and neck squamous cell carcinoma. Int J Med Sci. 2016;13(12):914–922. doi: 10.7150/ijms.16571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arantes LM, Laus AC, Melendez ME, de Carvalho AC, Sorroche BP, De Marchi PR, et al. MiR-21 as prognostic biomarker in head and neck squamous cell carcinoma patients undergoing an organ preservation protocol. Oncotarget. 2017;8(6):9911–9921. doi: 10.18632/oncotarget.14253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Avissar M, McClean MD, Kelsey KT, Marsit CJ. MicroRNA expression in head and neck cancer associates with alcohol consumption and survival. Carcinogenesis. 2009;30(12):2059–2063. doi: 10.1093/carcin/bgp277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Langevin SM, Stone RA, Bunker CH, Lyons-Weiler MA, LaFramboise WA, Kelly L, et al. MicroRNA-137 promoter methylation is associated with poorer overall survival in patients with squamous cell carcinoma of the head and neck. Cancer. 2011;117(7):1454–1462. doi: 10.1002/cncr.25689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qiang H, Zhan X, Wang W, Cheng Z, Ma S, Jiang C. A study on the correlations of the miR-31 expression with the pathogenesis and prognosis of head and neck squamous cell carcinoma. Cancer Biother Radiopharm. 2019;34(3):189–195. doi: 10.1089/cbr.2018.2621. [DOI] [PubMed] [Google Scholar]
- 34.Wu S, Jia S, Xu P. MicroRNA-9 as a novel prognostic biomarker in human laryngeal squamous cell carcinoma. Int J Clin Exp Med. 2014;7(12):5523–5528. [PMC free article] [PubMed] [Google Scholar]
- 35.Wu TY, Zhang TH, Qu LM, Feng JP, Tian LL, Zhang BH, et al. MiR-19a is correlated with prognosis and apoptosis of laryngeal squamous cell carcinoma by regulating TIMP-2 expression. Int J Clin Exp Pathol. 2014;7(1):56–63. [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang XW, Liu N, Chen S, Wang Y, Zhang ZX, Sun YY, et al. High microRNA-23a expression in laryngeal squamous cell carcinoma is associated with poor patient prognosis. Diagn Pathol. 2015;10:22. doi: 10.1186/s13000-015-0256-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu A, Huang JJ, Xu WH, Jin XJ, Li JP, Tang YJ, et al. MiR-21/miR-375 ratio is an independent prognostic factor in patients with laryngeal squamous cell carcinoma. Am J Cancer Res. 2015;5(5):1775–1785. [PMC free article] [PubMed] [Google Scholar]
- 38.Saito K, Inagaki K, Kamimoto T, Ito Y, Sugita T, Nakajo S, et al. MicroRNA-196a is a putative diagnostic biomarker and therapeutic target for laryngeal cancer. PLoS ONE. 2013;8(8):e71480. doi: 10.1371/journal.pone.0071480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Re M, Magliulo G, Gioacchini FM, Bajraktari A, Bertini A, Çeka A, et al. Expression levels and clinical significance of miR-21-5p, miR-let-7a, and miR-34c-5p in laryngeal squamous cell carcinoma. Biomed Res Int. 2017;2017:3921258. doi: 10.1155/2017/3921258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shen Z, Zhan G, Ye D, Ren Y, Cheng L, Wu Z, et al. MicroRNA-34a affects the occurrence of laryngeal squamous cell carcinoma by targeting the antiapoptotic gene survivin. Med Oncol (Northwood, Lond, Engl) 2012;29(4):2473–2480. doi: 10.1007/s12032-011-0156-x. [DOI] [PubMed] [Google Scholar]
- 41.Maia D, de Carvalho AC, Horst MA, Carvalho AL, Scapulatempo-Neto C, Vettore AL. Expression of miR-296-5p as predictive marker for radiotherapy resistance in early-stage laryngeal carcinoma. J Transl Med. 2015;13:262. doi: 10.1186/s12967-015-0621-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ogawa T, Saiki Y, Shiga K, Chen N, Fukushige S, Sunamura M, et al. miR-34a is downregulated in cis-diamminedichloroplatinum treated sinonasal squamous cell carcinoma patients with poor prognosis. Cancer Sci. 2012;103(9):1737–1743. doi: 10.1111/j.1349-7006.2012.02338.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pantazis TL, Giotakis AI, Karamagkiolas S, Giotakis I, Konstantoulakis M, Liakea A, et al. Low expression of miR-20b-5p indicates favorable prognosis in laryngeal squamous cell carcinoma, especially in patients with non-infiltrated regional lymph nodes. Am J Otolaryngol. 2020;41(5):102563. doi: 10.1016/j.amjoto.2020.102563. [DOI] [PubMed] [Google Scholar]
- 44.Wilkins OM, Titus AJ, Salas LA, Gui J, Eliot M, Butler RA, et al. MicroRNA-related genetic variants associated with survival of head and neck squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2019;28(1):127–136. doi: 10.1158/1055-9965.EPI-18-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Y, Liu J, Hu W, Zhang Y, Sang J, Li H, et al. miR-424-5p promotes proliferation, migration and invasion of laryngeal squamous cell carcinoma. Onco Targets Ther. 2019;12:10441–10453. doi: 10.2147/OTT.S224325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Childs G, Fazzari M, Kung G, Kawachi N, Brandwein-Gensler M, McLemore M, Chen Q, Burk RD, Smith RV, Prystowsky MB, et al. Low-level expression of microRNAs let-7d and miR-205 are prognostic markers of head and neck squamous cell carcinoma. Am J Pathol. 2009;174:736–745. doi: 10.2353/ajpath.2009.080731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu N, Zhou Q, Qi YH, Wang H, Yang L, Fan QY. Effects of long non-coding RNA H19 and microRNA let7a expression on thyroid cancer prognosis. Exp Mol Pathol. 2017;103:71–77. doi: 10.1016/j.yexmp.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 48.Wilkins OM, Titus AJ, Salas LA, Gui J, Eliot M, Butler RA, Sturgis EM, Li G, Kelsey KT, Christensen BC. MicroRNA-related genetic variants associated with survival of head and neck squamous cell carcinoma. Cancer Epidemiol Biomark Prev. 2019;28:127–136. doi: 10.1158/1055-9965.EPI-18-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ye J, Li L, Feng P, Wan J, Li J. Downregulation of miR-34a contributes to the proliferation and migration of laryngeal carcinoma cells by targeting cyclin D1. Oncol Rep. 2016;36(1):390–398. doi: 10.3892/or.2016.4823. [DOI] [PubMed] [Google Scholar]
- 50.Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer. 2006;6(4):259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 51.Kosaka N, Iguchi H, Yoshioka Y, Takeshita F, Matsuki Y, Ochiya T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem. 2010;285:17442–17452. doi: 10.1074/jbc.M110.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 53.Mahmood N, Hanif M, Ahmed A, Jamal Q, Mushtaq S, Khan A, Saqib M. Circulating miR-21 as a prognostic and predictive biomarker in oral squamous cell carcinoma. Pak J Med Sci. 2019;35:1408–1412. doi: 10.12669/pjms.35.5.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP, Wei WI. Mature miR-184 as potential oncogenic microRNA of squamous cell carcinoma of tongue. Clin Cancer Res. 2008;14(9):2588–2592. doi: 10.1158/1078-0432.CCR-07-0666. [DOI] [PubMed] [Google Scholar]
- 55.Tang T, Shan G, Zeng F. Knockdown of DGCR5 enhances the radiosensitivity of human laryngeal carcinoma cells via inducing miR-195. J Cell Physiol. 2019;234(8):12918–12925. doi: 10.1002/jcp.27958. [DOI] [PubMed] [Google Scholar]
- 56.Zabegina L, Nazarova I, Knyazeva M, Nikiforova N, Slyusarenko M, Titov S, et al. MiRNA let-7 from TPO(+) extracellular vesicles is a potential marker for a differential diagnosis of follicular thyroid nodules. Cells. 2020;9(8):1917. doi: 10.3390/cells9081917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li YY, Tao YW, Gao S, Li P, Zheng JM, Zhang SE, et al. Cancer-associated fibroblasts contribute to oral cancer cells proliferation and metastasis via exosome-mediated paracrine miR-34a-5p. EBioMedicine. 2018;36:209–220. doi: 10.1016/j.ebiom.2018.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cui W, Meng W, Zhao L, Cao H, Chi W, Wang B. TGF-β-induced long non-coding RNA MIR155HG promotes the progression and EMT of laryngeal squamous cell carcinoma by regulating the miR-155-5p/SOX10 axis. Int J Oncol. 2019;54(6):2005–2018. doi: 10.3892/ijo.2019.4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are included in this article.