Abstract
One year into the global COVID-19 pandemic, the focus of attention has shifted to the emergence and spread of SARS-CoV-2 variants of concern (VOCs). After nearly a year of the pandemic with little evolutionary change affecting human health, several variants have now been shown to have substantial detrimental effects on transmission and severity of the virus. Public health officials, medical practitioners, scientists, and the broader community have since been scrambling to understand what these variants mean for diagnosis, treatment, and the control of the pandemic through nonpharmaceutical interventions and vaccines. Here we explore the evolutionary processes that are involved in the emergence of new variants, what we can expect in terms of the future emergence of VOCs, and what we can do to minimise their impact.
Otto et al. review the evolutionary processes that have led to the emergence of variants of concern during the COVID-19 pandemic and describe efforts that could limit the future emergence of variants that spread faster, are more severe, or better escape immune responses.
Main text
Introduction
The December 2020 announcement by Public Health England1 that the variant B.1.1.7 exhibited a large number of mutations and a significant increase in transmission (50–100%2) rattled the world and served as a wake-up call on the importance of VOCs. Until then, there was little evidence that any mutations to the RNA genome of SARS-CoV-2 substantially increased viral fitness3. Within weeks, additional VOCs were reported with similar characteristics: greater than expected numbers of mutations and signatures of enhanced transmission (for example, B.1.351 in South Africa4 and P.1 in Brazil5).
The genomes of all viruses accumulate mutations over time. However, the pace of mutation accumulation and the consequences for transmission and disease in the host population depend on a range of factors, including the mutation rate and the impacts of mutation on viral dynamics within and between individual hosts. Together these factors determine the emergence and spread of viral variants and the evolution of pandemics. The genomes of RNA viruses are particularly prone to mutation6. But SARS-CoV-2, like related coronaviruses, encodes a proofreading domain (ExoN) that reduces its mutation rate relative to RNA viruses that do not (such as influenza, HIV, and hepatitis C viruses). The pace of change of the SARS-CoV-2 genome has thus been estimated at 1.87 × 10−6 nucleotide substitutions per site per day7, roughly 5-fold lower than influenza A/H3N2 (10.9 × 10−6 nucleotide substitutions per site per day; https://nextstrain.org 8; accessed 22 May 2021). Thus, across the ∼30,000 base-pair genome of SARS-CoV-2, approximately 20 genetic changes occur per year within a lineage.
Not all mutations that arise will persist long enough to be included in this estimate of the molecular clock of SARS-CoV-2. Assuming that synonymous mutations are largely neutral while non-synonymous mutations persist with probability , Wang et al. 9 used codon-based likelihood methods to estimate 0.56 among human SARS-CoV-2. Thus, roughly half of the mutations that alter amino acids are quickly lost. Given the proportion of sites subject to synonymous (p S = 0.22) and non-synonymous (p N = 0.78) mutations, we can thus back-calculate the rate at which mutations must have occurred, before selection acted, to be 2.84 × 10−6 (that is, 1.87 × 10−6/(p S + p N)). This correction is slight because selection has had little time to eliminate anything but the most deleterious mutations accumulated over the pandemic's short time frame. Over longer evolutionary time frames, such as that observed between SARS-CoV-2 and the most closely related bat sequences, Wang et al. 9 find that non-synonymous mutations are 14 times less likely to persist (0.039) than what we see today among the virus in humans, as selection has slowly eliminated weakly deleterious mutations.
The immense number of currently active cases — 12.1 million globally (https://www.worldometers.info/coronavirus; accessed 2 July 2021) — has greatly increased the opportunity for viruses with distinct characteristics to evolve. Several viral lineages have now been reported that have potential impacts (so called ‘variant of interest’, VOI) or demonstrated impacts (‘variant of concern’, VOC) on disease transmission and human health (see Box 1 for terminology). At the time they emerged, the first three VOCs (B.1.1.7, B.1.351 and P.1) exhibited almost twice as many changes to their genomes as other contemporaneous SARS-CoV-2 lineages, with 23 mutations characterizing B.1.1.710, 21 in B.1.3514, and 23 in P.15. The B.1.617.2 variant causing surging case numbers in India and recently designated a VOC (Table 1 ) also exhibits an excess of mutations, roughly similar to B.1.1.7 (Figure S1).
Box 1. Variant detection and terminology.
As viruses accumulate mutations, it can be challenging to know how to refer to the diverse virus forms that arise. A ‘variant’ is any virus with a different sequence from other viruses. A ‘lineage’ refers to viruses that are more closely related — cousins on the phylogenetic tree of the virus. A ‘strain’ is reserved for a broader grouping of viruses with different properties; for example, SARS-CoV-1 and SARS-CoV-2 represent different strains of coronavirus.
The World Health Organization71 has recently established guidelines for referring to viral variants for SARS-CoV-2. A ‘variant of interest’ (VOI) contains mutations thought likely to alter the phenotypic properties of the virus, with documented community transmission or international spread. Because VOIs might alter transmission rates or disease progression, impacting human health, they should be monitored closely.
In addition, a ‘variant of concern’ (VOC) has an established and detrimental effect on human health, with mutations that increase viral transmission rates, cause more severe health outcomes, and/or reduce the efficacy of public health measures such as vaccination. Systematic genomic surveillance is needed to allow the epidemiological analysis necessary to determine whether a VOI (that is, a variant of potential impact) should be designated a VOC (that is, one of demonstrated impact).
In many cases, variants have been observed to increase in frequency, but this alone is not sufficient information to document an increased transmission rate and justify designation of a VOC. Founder effects and transmission to a new segment of the population can lead, by chance, to the rapid growth of a lineage. Shifts in travel patterns over time can also cause changes in the frequency of viral lineages. Uneven sampling presents another challenge, especially if samples are more likely to be sequenced if they are suspected to be VOI or VOC. Particularly confounding is an immense statistical problem associated with multiple comparisons; with 2 million genomes sequenced and frequencies tracked in hundreds of regions across the globe, random changes in variant frequency may often be confused with a transmission advantage. Verifying a transmission advantage requires repeatedly showing that a variant increases in frequency, over multiple weeks and regions, as documented for B.1.1.7 in the UK1 , 2 , 29.
Table 1.
SARS-CoV-2 variants of concern and variants of interest.
| Pango lineage | Nextstrain clade8 | First detection location | VOI or VOC | WHO71 designation | Mutations of interest on S protein | Observed clinical effect |
||
|---|---|---|---|---|---|---|---|---|
| Transmissibility | Virulence | Antigenicity | ||||||
| B.1.1.7 | 20I/501Y.V1 | United Kingdom | VOC | Alpha | N501Y, E484K∗, P681H, D614G |
50–100% higher2 | 39–72% more lethal41 | No impact on NBA Minimal impact on NBSa |
| B.1.351 | 20H/501Y.V2 | South Africa | VOC | Beta | N501Y, K417N, E484K, D614G |
20–113% higher72 | – | Moderately reduced NBAa Reduced NBSa |
| P.1 | 20J/501Y.V3 | Brazil/Japan | VOC | Gamma | N501Y, K417T, E484K, D614G |
70–140% higher, evades immunity 21–46% more5 | 20–90% more lethal5 | Moderately reduced NBAa Reduced NBSa |
| B.1.427 and B.1.429 | 21C/S:452R | United States (California) | VOI | Epsilon | L452R, D614G |
18–22% higher73 | – | Moderately reduced NBAa Reduced NBSa |
| B.1.525 | 21D | United States (New York)/Nigeria | VOI | Eta | A67V, E484K, D614G, Q677H, F888L | – | – | Potentially reduced NBAa Potentially reduced NBSa |
| B.1.526 | 21F | United States (New York) | VOI | Iota | L5F∗, T95I, D253G, S477N∗, E484K, D614G, A701V∗ | – | – | Moderately reduced NBAa Reduced NBSa |
| B.1.617.1 | 21B/S:154K | India | VOI | Kappa | (T95I), G142D, E154K, L452R, E484Q, D614G, P681R, Q1071H | Secondary attack rates similar to B.1.1.774 | – | Potentially reduced NBAa Potentially reduced NBSa |
| B.1.617.2 | 21A/S:478K | India | VOC | Delta | T19R, G142D∗, 156del, 157del, R158G, L452R, T478K, D614G, P681R, D950N | Secondary attack rates higher than B.1.1.7; household transmission 64% higher than B.1.1.7 (26–113% higher)74 | – | Potentially reduced NBAa Potentially reduced NBSa |
| B.1.617.3 | 20A | India | VOI | T19R, G142D, L452R, E484Q, D614G, P681R, D950N | – | – | Potentially reduced NBAa Potentially reduced NBSa |
|
| P.2 | 20J | Brazil | VOI | Zeta | E484K, D614G, V1176F | – | – | Potentially reduced NBAa Reduced NBSa |
Here we have used the variant classification of the Centre for Disease Control of the United States and modified from the CDC data table, which lists additional mutations and information about clinical effectsa. Nextstrain clade names start with the year of origin and distinguish lineages that reach a global frequency of 20%, so the name of a lineage can change if it increases in frequency8. NBA: Neutralization by antibodies (monoclonal antibodies in therapeutic use); NBS: Neutralization by convalescent and/or post-vaccination sera (variant relative to non-variant). VOI and VOC designations vary over time (e.g. B.1.427 and B.1.429 were previously designated as VOC by the CDCa, accessed 13 June 2021) and by country (e.g. Canada designates the entire B.1.617 clade as a VOCb) because of different evaluations of the existing evidence.
Mutation detected in some sequences within the lineage. Ranges give 95% confidence intervals or credible intervals, except for the transmission rate of B.1.1.7 (a consensus estimate across models2) and for P.1 (50% Bayesian credible intervals).
https://www.cdc.gov/coronavirus/2019-ncov/cases-updates/variant-surveillance/variant-info.html; ‘moderate’ includes modest decreases and/or cases where alternative antibody treatments remain available.
Factors at play in the emergence and spread of VOCs
The establishment and spread of such variants depend on two main factors: what happens within the individuals in whom the mutations arose and what happens afterwards as the virus transmits among individuals.
Processes occurring within individuals
Mutations arise as viruses replicate within an infected individual, and thus new variants initially face selective forces within that individual. For SARS-CoV-2, these within-individual evolutionary processes have been best documented in immunocompromised patients11, 12, 13, 14. These patients maintain high viral loads over prolonged periods of time, allowing more opportunities for viral replication and selection and leading to elevated substitution rates (see elevated branch lengths in, for example, references11 , 13, which reflect a larger than expected number of substitutions). By sequencing the virus at multiple time points, these studies have documented rapid changes to the composition of the viral population within a patient, over the course of days. These rapid changes are faster than expected by drift in a large population and point to natural selection for the virus to better replicate or to evade a weakened immune system, as well as the intensive antibody therapies received by these patients11, 12, 13. A disproportionate fraction of mutations in many of these patients are clustered in the gene encoding Spike, the protein that juts out of the virus and binds to the ACE2 receptor that allows entry into host cells: for example, 57% of 15 changes in the final sample reported by Choi et al. 11 and 33% of 9 changes in the final nose and throat sample reported by Kemp et al. 13, compared to an expectation of only 13% based on the length of Spike. Many of these mutations were found in the receptor binding domain, which is essential for host entry. This concentration in Spike may reflect, in part, relaxed selective constraints within immunocompromised individuals, but many of these mutations are non-synonymous (for example, all eight mutations in Spike reported by Choi et al. 11), consistent with selection favouring changes to the Spike protein. Furthermore, several mutations have arisen in parallel in different patients, suggesting selection for those changes15. For example, McCarthy et al. 14 report four independent cases of patients bearing deletions in the same amino-terminal domain of Spike, which is important for host entry and antibody evasion. These non-random patterns of genomic changes suggest that selective pressures, alongside mutation, strongly shape viral evolution within immunocompromised individuals. Immunocompromised patients should be protected from COVID-19 by prioritizing their contacts for vaccination, and any patient with a prolonged infection should be treated with great care as a potential source of new variants.
The changes observed in the three main VOCs to date (B.1.1.7, P.1, B.1.351) echo the evolutionary changes observed in immunocompromised individuals, with a larger than expected number of mutations that are non-randomly distributed across the genome. This pattern led Public Health England1 to suggest that B.1.1.7 might have originated in an immunocompromised individual. Many mutations in these VOCs are also concentrated in Spike (35% of the 23 mutations in B.1.1.710). All three contain a specific deletion (Orf1ab:3675–3677del) affecting NSP6, a protein involved in intracellular processes that alter the balance between viral replication and clearance. All three lineages also contain S:N501Y, and two VOCs (B.1.351 and P.1) bear mutations in common at three other sites in spike (S:L18F, S:K417N/T, S:E484K; where this notation indicates, for example, ‘S’ for the spike gene, ‘L’ for the original amino acid leucine, ‘18’ for the amino acid position, and ‘F’ for the mutant amino acid phenylalanine). Given the number of characteristic mutations in these three VOCs and the genome size, there is a <5% probability that we would see the same site mutated in any of the variants just by chance. These parallel changes are thus a strong signal that selection played a role in the initial emergence of the VOCs, especially given the weak constraints generally observed9.
We have focused above on immunocompromised individuals because there is limited opportunity for evolution within typical infections (that is, those within immunocompentent hosts). With exponential growth of the population of viruses within a host, most mutations at peak infectivity would have occurred in the last round or two of replication, leaving little time for selection within that individual (this is the principle underlying the classic experiment of Luria and Delbrück16). Furthermore, estimates of the bottleneck size between transmission events suggest that few viruses found subsequent infections; for example, only 1–8 virions17. As a result, there is little genetic diversity, allowing little opportunity for evolutionary change within typical infections17 , 18. Instead, drift is likely to dominate which viruses cause new infections, with one main exception: the elimination of strongly deleterious mutations. Indeed, Lythgoe et al. 17 report a similar reduction in the number of nonsynonymous mutations observed within individuals ( = 0.55) as estimated from different individuals ( = 0.56)9, suggesting that the elimination of strongly deleterious mutations primarily occurs within individual hosts.
Elevated mutation rates can also be caused by genetic changes in SARS-CoV-2 that increase the error rate during viral replication19. Takada et al. 19 showed that mutation P203L in NSP14 alters the ExoN proofreading domain and nearly doubles the mutation rate. Such ‘mutator’ lineages could also generate variants with rare combinations of new mutations, but to date, these mutators have not risen in frequency, possibly because of the higher numbers of deleterious mutations that result19.
Another potential source of novel variants within individual infections is recombination. There is debate about the extent of recombination in SARS-CoV-220 , 21, in part because detecting recombination is challenging when there is relatively little genomic variation22. All circulating SARS-CoV-2 viruses are similar and closely related by descent from the virus that first infected humans in late 2019. Furthermore, artefacts generated during genomic sequencing and assembly can mimic recombination and may explain some of the apparent recombination events21. That said, a recent analysis provides compelling evidence that sampled genomes from the UK arose by recombination during a period when B.1.1.7 and non-variant lineages were both prevalent23. Recombination is a concern because it generates new combinations, potentially bringing together components of different lineages in a way that benefits the virus, although no fitness advantage has been detected for the recombinants observed to date23.
Processes occurring among individuals
For those SARS-CoV-2 mutations that do make it out of the body and cause new infections, the probability that the variant becomes established depends on its transmissibility in the population in which it emerges, as well as the nature and extent of contacts among individuals and chance events. For many infectious diseases, a small fraction of individuals tends to be responsible for a large fraction of the transmission events (the so-called ‘80/20 rule’24, in which 80% of new infections are thought to be caused by 20% of cases). This pattern has also been observed for SARS-COV-225 , 26 and is sometimes referred to as ‘overdispersion’, meaning that the variation in the number of new infections generated by different individuals is larger than expected (typically with reference to a Poisson distribution).
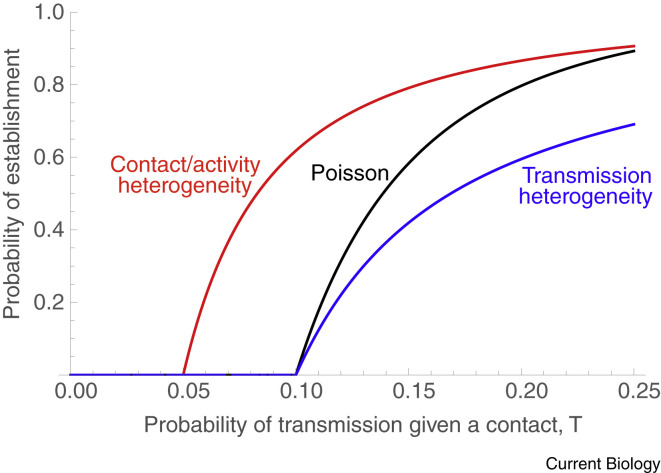
Overdispersion can arise through various processes, and the details have a strong effect on the probability that a new variant establishes within a population (by established, we mean that the variant is present in enough active cases that it is very unlikely to be lost by chance). If overdispersion arises because individuals vary in infectivity, with some cases transmitting much less often and others much more often than expected (for example, ‘superspreaders’), then this variability tends to decrease the probability of establishment of a variant27 (Figure 1 , compare black and blue curves). On the other hand, if the overdispersion occurs because some individuals have many fewer contacts or much lower activity levels and others many more, this tends to increase the probability of establishment (Figure 1, red curve compared to a Poisson distributed number of contacts in black).
Figure 1.
The role of heterogeneity in the probability that a variant establishes within a population.
Illustrated here is a predominantly susceptible population, with an average number of ten contacts per case and no competition for susceptible hosts between the variant and non-variant. If the number of contacts per individual is Poisson distributed and there is a constant chance of infection per contact, the probability of establishment rises with the chance of infection per contact as shown by the black curve. Here, variants are not expected to persist unless the transmission probability is above 10%, as only then are cases expected to give rise to at least one new case (Rt > 1). If cases vary in their infectiousness, then variants are less likely to establish because more cases fail to have any onward transmission (blue curve, where we assume that half of the cases are three times as infective as the other half). If, however, there is variability in contact number or activity level, variants are more likely to establish because individuals with more contacts are more likely to get infected and then more likely to pass on the variant (red curve, assuming the contact distribution is negative binomial with a dispersion parameter of k = 3). Because the disease spreads more easily among the subset of active people, heterogeneity in contacts also reduces the critical transmission probability above which establishment is possible (red curve rises above zero earlier, causing Rt > 1). (Based on methods in reference75.)
The key difference between the two cases is that when overdispersion is due to chance events affecting the course of infection, then there is no relationship between the likelihood of an individual acquiring a variant infection and that individual’s propensity to transmit to others. As a result, this simply introduces additional noise into the system, decreasing the probability of establishment (blue curve). By contrast, with overdispersion due to contact or activity heterogeneity (red curve), variants are more likely to infect highly connected and active individuals who also have a higher propensity to transmit by virtue of their high connectivity. This process, whereby the most active individuals contribute disproportionately to transmission, also explains why targeting vaccines and nonpharmaceutical interventions to essential workers is particularly effective at reducing cases28.
After a favoured variant becomes established within a population, its spread through a population rises in a more deterministic fashion. Tracing the week-after-week spread of B.1.1.7 across many regions of the UK in great detail was made possible because of a deletion in the spike gene of B.1.1.7 (S:H69–V70del) that caused one of three molecular probes to ‘drop out’ in the PCR test for COVID-19 used in the UK (referred to as S-gene target failure’, or SGTF). By analysing data on the proportion of PCR tests in which this probe dropped out, B.1.1.7 was estimated to be 50–100% more transmissible than previously circulating variants, depending on the model2 , 29. The higher probability of transmission increases the likelihood of B.1.1.7 establishing and spreading in new areas (Figure 1). Within three months of the Public Health England announcement, over 100 countries had reported B.1.1.730, despite increased restrictions on travel from the UK.
The way that selection is expected to act on new SARS-CoV-2 variants is affected by the particular features of COVID-19, including the extreme variability in disease presentation. About 17% of cases never exhibit symptoms (‘asymptomatic’ cases31), and yet they can still infect others, although less often (42% lower transmission, on average31). Even for those who do have symptoms, roughly half of the transmission events are estimated to occur before individuals exhibit symptoms (‘pre-symptomatic’ transmission), at least in areas where individuals are encouraged and able to self-isolate32. Most symptomatic individuals recover from COVID-19, but even so the infection fatality rate is high at about 0.68% (95% CI: 0.53–0.82%) globally33. In many infectious diseases, direct selection is expected to reduce virulence34. However, because severe symptoms and mortality from COVID-19 typically occur weeks after peak viral load, direct selection against virulence is expected to be extremely weak35, 36, 37. By contrast, selection strongly favours an increased transmission rate in the early stages of a pandemic, when the abundance of susceptible hosts is high35 , 38, 39, 40. These general predictions are also borne out in SARS-CoV-2-specific models, which additionally show that selection favours shorter latent periods prior to infectivity, higher infectiousness of asymptomatic individuals, and prolonged infectious periods37.
Although the death rate caused by SARS-CoV-2 may be under relatively weak direct selection, mutations that affect transmission rate or other disease attributes can have correlated effects on mortality. Variants with even a moderately enhanced transmission rate can readily spread during a pandemic, whether they increase or decrease death rates. Thus, the evolution of virulence depends strongly on the nature of the mutations that arise. For example, if increased transmission rate is due to a higher viral load, more severe illness may result. Alternatively, if mutations cause milder symptoms and so raise activity levels of infected individuals, they may increase transmissibility but reduce severity and death rates.
Unfortunately, of these possibilities, B.1.1.7 is both more transmissible and more virulent41 , 42. Based on structural modelling, the changes in the Spike protein in B.1.1.7 are predicted to cause conformational changes that allow the viruses to bind more easily to cells in the respiratory tract of an uninfected person43 (and so a lower infectious dose might be needed). Data also suggest that infected individuals have a slightly higher viral load and longer infectious period44. However, for B.1.1.7 at least, a shorter serial interval does not explain the observed spread in the UK2. The increased transmission has been accompanied by increased virulence, with an estimated 64% higher mortality rate (95% CI: 32–104%)41 , 42. Currently unknown is which of the mutations in B.1.1.7 are responsible for the observed changes in transmission and virulence, which are selectively favoured, and which have risen by hitchhiking despite being neutral or even deleterious.
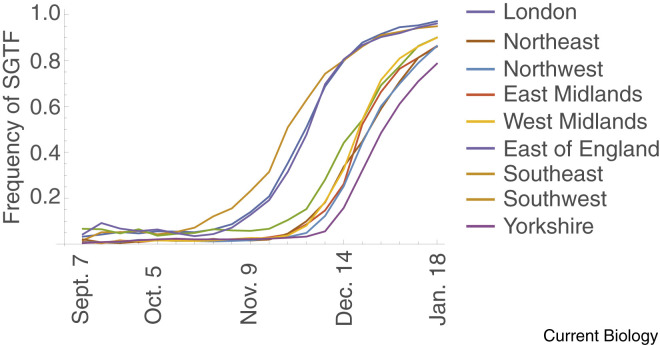
While public health measures have reduced the reproductive number (R t) of non-variant SARS-CoV-2 to below 1 in many countries, these measures were not initially strong enough to control the spread of B.1.1.7, allowing it to become the predominant variant in many countries within months. Where B.1.1.7 has established (say at 5–10% frequency), the subsequent rise in frequency has been fast. In the UK, the odds that a case was caused by B.1.1.7 (that is, the frequency of B.1.1.7 divided by the frequency of non-variants) doubled every 1–2 weeks depending on the region (Figure 2 ). The average reproductive number is expected to increase alongside the rise in variant frequency, requiring even stronger public health measures to control the disease. This recent history is now being repeated with B.1.617.2, which is estimated to be 64% more transmissible within households than B.1.1.7 (Table 1).
Figure 2.
The rise in frequency of B.1.1.7 in nine regions of England.
Data are weekly measures of the fraction of Pillar 2 tests that showed SGTF, taken from Public Health England Technical Briefing 5 (http://www.gov.uk/government/publications/investigation-of-novel-sars-cov-2-variant-variant-of-concern-20201201).
Processes occurring among species
Predicting the course of SARS-CoV-2 evolution is further complicated by the broad potential host range and known transmissibility to animals45. Were transmission in domesticated and/or wild animals to become an important reservoir for SARS-CoV-2, then selection for greater transmission in these animals could lead to different disease attributes. For example, evolution of transmissibility of West Nile virus in American robins (a key reservoir host) resulted in increased virulence to crows46. For SARS-CoV-2, a host jump in Denmark from humans to farmed mink and back to humans led to a highly mutated variant (‘Cluster 5’), which was less easily neutralized by antibodies from convalescent sera47. Cluster 5 initially reached a high frequency (30%) in communities near affected farms, but through massive control efforts, including the culling of 3–4 million mink, appears to have been eradicated47. As concluded by Larsen et al. 47, keeping susceptible animals in such large and dense populations poses a threat to human health by presenting an opportunity for viral adaptation.
Future evolution
What types of variants are we likely to see over the next year? Although it is impossible to answer this question precisely, evolutionary models can help us explore the possibilities.
As long as circulating strains cannot infect recovered or immunized individuals, then models predict that, as the number of susceptible individuals falls, selection for increased transmissibility will become weaker, with selection increasingly favouring prolonged infectious periods35 , 37, 38, 39, 40.
However, as the proportion of the population that is immunologically naïve shrinks through infection and immunization, selection will also increasingly favour variants that are partially or wholly able to overcome the immunity of previously protected individuals (Box 2 ). In some countries, particularly with limited medical and public-health resources, estimates suggest that most of the population may now have been infected, becoming naturally immune and increasing the relative fitness of such immune-escape variants48.
Box 2. Escape mutations.
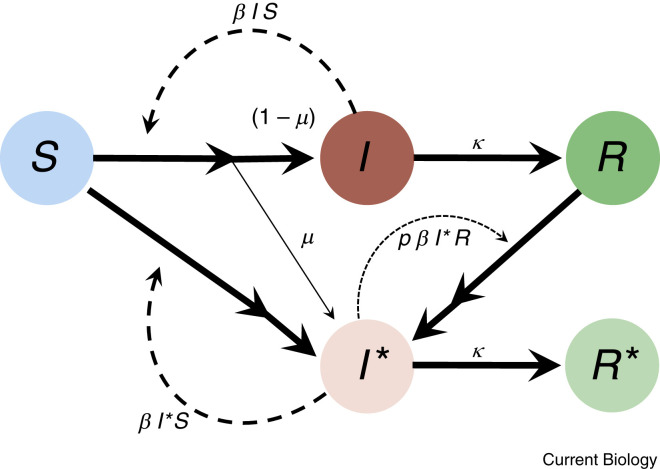
Each infection carries the risk of giving rise to a mutation that evades resistance in individuals who have specific immunity due to either prior infection or vaccination. To illustrate the main factors determining this risk, we simplify SARS-CoV-2 dynamics and consider a population consisting only of S susceptible individuals, I infected, and R resistant individuals, ignoring case heterogeneity (presymptomatic, asymptomatic, vaccination type and dose, etc.; see reference67 for a more detailed model focused on within-individual adaptation). Although these variables all change over time, we focus on a region with a small current fraction of active infections, so that S and R can be treated as constant over the short term, allowing us to focus on the dynamics of the infectious class and escape variants .
Before escape variants are present, new cases arise at a rate per day and are cleared at rate (Box 2 figure, below). Each new infection has a chance of giving rise to an escape variant by mutation. Because we are considering the first such mutation, the ability of the variant to infect resistant individuals is only partial, reducing the transmission rate to resistant individuals from to .
At this snapshot in the epidemic, the number of standard cases and the number of escape-variant cases are expected to change at rates:
| (1) |
| (2) |
The chance that an escape mutation appears on any particular day is thus proportional to . While cases remain roughly constant in number, the time until the first escape mutation appears is approximately exponentially distributed with a mean of days. The waiting time is thus shorter in locations with ineffective control measures (higher ), many circulating cases (higher ), and many remaining susceptibles (higher ).
Once an escape mutant has appeared, equation (2) indicates that the mutant strain will increase in frequency at a faster rate than the original strain, raising the reproductive number for the new variant from to . Preventing the selective accumulation of partially resistant mutations thus requires reducing contacts between cases and resistant individuals , boosting resistance where possible by vaccinating naturally infected individuals and completing recommended vaccine doses (reducing p), and persisting with public health measures that reduce transmission in general (reducing ).
This basic model also clarifies the challenges inherent in deciding whether to spread available vaccines to more people in single doses or to complete two-dose regimes. Vaccinating more people with a single dose can decrease the number of infections circulating in a region (reducing and ) and reduce the reproductive number of all variants (by reducing S), but it also has the potential to increase the probability of onward transmission to partially resistant individuals (increasing p and the selective advantage of escape variants). The right policy thus depends on resistance after one versus two doses and the impact on the number of severe cases.
A simplified model of the evolution of the first step escape mutant (I∗) using an SIR model framework.
The transmission rate to susceptible individuals and the clearance rate are assumed to be equal for both variants. Only the escape mutant is partially able to infect resistant individuals (at a rate reduced by a factor p relative to the rate of infecting susceptible individuals). The model considers only the first step in the accumulation of mutations that increase the ability to reinfect otherwise resistant individuals but could be extended to model subsequent steps by mutation.
The evolution of resistance to vaccines typically occurs more slowly and is observed less often than the evolution of resistance to therapeutic agents like antimicrobials49. Both natural immunity and vaccine-induced immunity generally result in a broad range of antibodies and T cells that recognize distinct parts of the virus, particularly Spike and nucleoproteins50 , 51. Even though current mRNA vaccines only encode Spike proteins, a range of antibody and T-cell responses are produced, recognizing different parts (‘epitopes’) of the protein52, making it less likely that single mutational changes will substantially reduce protection. Even with the multiple mutations observed in the currently circulating VOCs, substantial natural and vaccine-induced antibody responses, and especially T-cell responses, are observed against the variants tested (B.1.1.7, B.1.351, and P.152 , 53). Importantly, a mutant virus that escapes the immune response of one individual may not escape the immune response of another.
Although full escape from vaccine-induced immunity in SARS-CoV-2 has not yet been documented, variants displaying partial escape are already here. P.1, B.1.617.2, and especially B.1.351 evade monoclonal antibody therapies, at least for some antibodies (Table 1). Many also exhibit weaker inhibition by neutralizing antibodies in serum samples from vaccinated individuals54, requiring 6.4-fold higher titers of antibodies to neutralize B.1.351 and 3.5-fold to neutralize P.1 after two doses of Moderna (mRNA-1273). Real-world estimates also find slightly lower vaccine effectiveness against known infections by variants B.1.351 (Pfizer BNT162b2)55 and B.1.617.2 (Pfizer BNT162b2 and AstraZeneca ChAdOx1)56. Nevertheless, vaccine effectiveness increases with the second dose and/or over time, reaching high levels of protection against infection (>75%) and almost complete protection against severe disease55.
Over the long term, the accumulation of multiple mutations in SARS-CoV-2 will eventually reduce the efficacy of immune responses in a large proportion of vaccinated and naturally immune people (Box 2). How long might we expect immunity to last? In another coronavirus (229E), antibodies within blood collected in the 1980s were shown to decline in neutralizing ability after a decade of evolutionary changes (‘antigenic drift’57), suggesting years of immunity. But with so many active COVID-19 cases, further bursts of mutations, as seen in immunocompromised patients and via passage through animals, should be expected, accelerating evolutionary change. Altogether, it seems plausible that boosters may be needed within the next year or two, with longer gaps between boosters once case numbers fall globally. Fortunately, updating mRNA vaccines to target new variants is relatively straightforward, and clinical studies are already underway for variant-specific vaccines. For example, Moderna recently reported on phase 2 trials with mRNA-1273.351, a vaccine that matches changes in Spike observed in the B.1.351 variant58. That said, the results showed very modest increases in antibodies targeting B.1.351 following two doses of the standard Moderna vaccine with mRNA-1273.351, suggesting that variant-specific vaccines may primarily boost previous immune responses rather than convey new immune responses to the variant epitopes. Targeting escape variants with vaccines that target other proteins, like the nucleocapsid, might be an effective alternative strategy.
Even without processes that make multiple changes more likely, selection can favour the accumulation of mutations one at a time that together reduce detection by the immune system and increase the risk of reinfection. The gradual build-up of escape mutants is particularly likely in regions where there are both high case numbers and high numbers of immunized individuals (Box 2).
In summary, the emergence of new variants should be expected, initially driven by selection for greater transmissibility and a longer duration of infectivity and then increasingly shaped by selection to evade immune responses, enabling transmission to vaccinated and naturally immunized people. Although there currently appears to be only weak selection on virulence, virulence may increase (or decrease) if coupled by mutation with these other features (as a pleiotropic side effect or through linkage). The further emergence of variants with multiple genetic changes, due to infections in immunocompromised individuals, mutators, and/or recombination, is expected and should be closely monitored.
What are the implications of ongoing evolution for control strategies during the pandemic?
The emergence of VOCs and the ongoing evolution of SARS-CoV-2 have potentially important implications for how best to control the pandemic. We first consider implications for detecting VOCs and then turn to different potential control measures that can help slow their emergence and spread.
Detection and surveillance of VOCs
Identifying VOCs as they emerge allows for the earlier implementation of control measures. Whole-genome sequencing followed by bioinformatic analysis can identify lineages that are increasing in frequency or that can infect previously immune individuals. Coupled with structural modelling of potential effects on viral function and the ability to evade antibodies, variants of interest (VOIs) can be identified (Box 1). Systematic surveillance and contact tracing are then needed to track a VOI to determine the mechanisms by which it is increasing in frequency, as well as whether it causes altered disease characteristics, such as increased transmission rates, longer periods of infectiousness, greater virulence, or the capacity to infect previously immune individuals. Assessing changes to the infectious period of a variant is, for example, critical for setting effective quarantine intervals.
Once a variant of interest is elevated to the status of VOC (Box 1), reducing its spread also requires effective genetic surveillance. Importantly, the higher the fraction of cases that are genetically typed, the easier it will be to contain a VOC59, because the variant is more likely to be detected early when there are fewer VOC infections. For this reason, rapid PCR-based methods that detect a VOC (for example the SGTF that allowed detection of B.1.1.7 in the UK1) are an important tool, alongside whole genome sequencing, to detect VOCs early enough to isolate cases and prevent establishment.
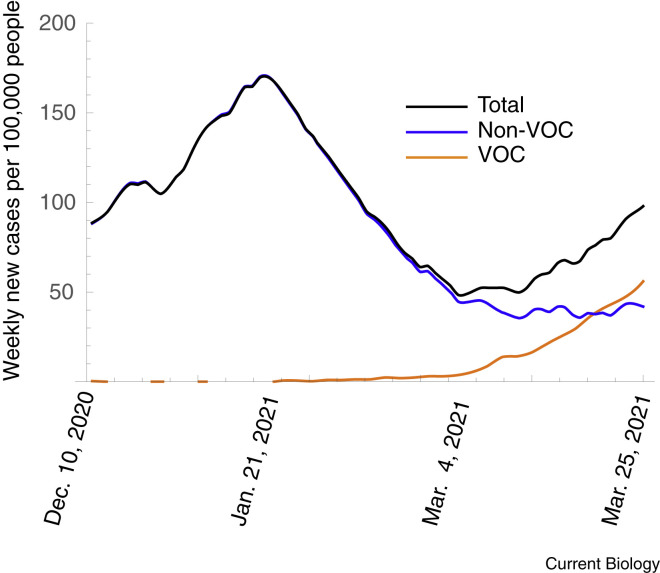
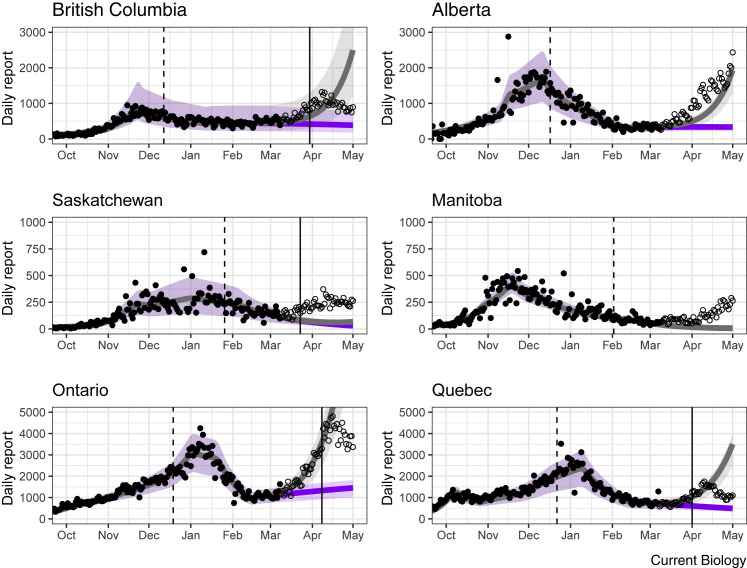
Effective ongoing surveillance also provides information about evolutionary change during the pandemic. Understanding the impact of these evolutionary changes can be important for guiding control measures and for solidifying public support for these measures. For example, as in many regions, B.1.1.7 recently increased in prevalence in Ontario Canada (Figure 3 ). During the initial spread (from January–February 2021), however, the total new daily case count steadily decreased, leading many people to believe that the pandemic was under control. But this steady decrease in total case count masked the fact that control measures at the time were insufficient to control B.1.1.7. As a result, once the prevalence of B.1.1.7 reached a high enough value, it began to dominate the total case count, causing the overall daily case count to start rising once again (Figure 3). Although models predicted this evolutionary dynamic and the subsequent spike in cases based on the selective differences observed in the UK (Figure 4 ), accurate surveillance data for B.1.1.7 were not readily available to demonstrate unequivocally what was happening. Consequently, it was challenging to galvanize support for enhancing control measures proactively until the total daily case count began to climb once again.
Figure 3.
Weekly new cases per 100,000 people for Ontario, Canada.
Data displayed as total count, those due to non-VOC, and those due to VOC (primarily B.1.1.7, as measured by a PCR test for the N501Y mutation). Plots are moving seven-day averages, using data from https://covid19-sciencetable.ca/ontario-dashboard/. The decreasing total case count between 21 January, 2021 and early March, 2021 masked an underlying increase in the case count due to VOCs.
Figure 4.
The spike in case numbers in the spring of 2021 was predicted by models of VOC dynamics.
Case numbers in Canadian provinces (black circles; data up to 8 March, 2021) were fit using a dynamic modeling approach, either ignoring VOC (purple) or allowing the spread of VOC with a transmission advantage of 50% (grey). In each panel, the VOC is introduced a week before the date of the first publicly reported case in each province (vertical dashed line) with initial numbers set to match the observed VOC numbers in early March. Subsequent case numbers, which were not used in the model fits, are shown as hollow circles. The spike in cases led to various emergency restrictions (vertical solid lines), which subsequently brought cases down over the next couple of weeks. Poor model predictions in a couple of provinces are likely due to migration among provinces and/or a low sampling rate for VOC (for example, genomics now indicates that B.1.1.7 was in Manitoba at least 19 days earlier than the first reported case). (Based on model fits using the Public Health Agency of Canada/McMaster model76.)
Since the Public Health England announcement1, countries worldwide have also increased mandatory testing before and/or after travel, as well as quarantine periods following travel, which can slow the spread of VOCs into regions where they have yet to establish. Enhanced measures to reduce transmission, including travel restrictions and lockdown measures that prohibit various activities, have subsequently reversed the rise in case numbers in many regions (Figure 4).
Detection of immune-escape VOCs will pose specific challenges. Preliminary data suggest that vaccinated individuals who become infected are less likely to feel symptoms, making breakthrough infections less likely to be detected, although severe infections continue to occur60. To detect and break transmission chains involving immune-escape variants, backwards contact tracing61, as well as regular testing of asymptomatic individuals, may be essential tools, followed by enhanced control measures to prevent onward transmissions. Distinguishing escape mutations from chance breakthrough cases will also be a challenge. Flagging transmission chains that occur between vaccinated individuals, as well as whole-genome sequencing of breakthrough cases, will help detect new immune-escape VOCs earlier. Hampering this effort, however, is the fact that vaccination status, reason for sequencing, contact history, and case information (for example, severity) are typically not shared alongside genomic data, limiting the power to detect escape variants and prevent their global spread.
Finally, internationally coordinated genomic surveillance will be important for identifying which variants to include in future booster vaccines, with an eye to prioritizing immune-escape VOCs and highly divergent SARS-CoV-2 lineages.
Control measures to slow the evolution of new variants
Although we cannot generally reduce the rate of mutation, we may be able to reduce the risk that multiple mutations arise. In particular, we can provide a halo of protection around immunocompromised individuals by prioritizing vaccination of their households and caregivers, reducing their risk of exposure. If long-lasting infections do occur, enhanced surveillance and contact tracing should be considered to prevent onward transmissions. Heightened measures to reduce onward transmission may also be warranted for lineages associated with higher-than-expected mutation rates (for example, mutants that alter proofreading capacity19).
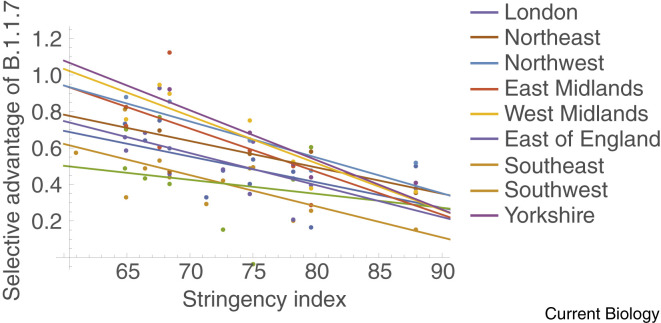
More broadly, we can reduce the rate of emergence of new VOCs and slow the spread of existing ones by reducing overall case numbers through vaccination at a global scale and by maintaining or enhancing the nonpharmaceutical interventions that have contributed to controlling the pandemic (case detection and isolation, contact tracing and quarantine, masking and personal distancing, and improved ventilation)62 , 63. Having low case numbers makes it easier to test and genotype a high fraction of cases and increases the efficacy of contact tracing measures to stop onward transmission64. Furthermore, mathematical models predict that measures that reduce contact rates with susceptible individuals will not only slow the spread of SARS-CoV-2 overall but will also reduce the relative advantage of variants that have a transmission advantage37. Thus, the measures taken to reduce contacts and limit the number of COVID-19 cases may have the added benefit of slowing the rate at which VOCs with a transmission advantage overtake the wild type. This predicted pattern, with selection weakening as stringency measures are increased, appears to be borne out in data for B.1.1.7 from England (Figure 5 ) and British Columbia, Canada (Figure S2).
Figure 5.
Selective advantage of B.1.1.7 declines with increasing social restrictions in the UK.
The same data as in Figure 2 but now the selective advantage of B.1.1.7, as measured by the weekly change in log(frequency SGTF/frequency of non-SGTF), is plotted against the stringency index of restrictions during that week in the UK (taken from http://www.bsg.ox.ac.uk/research/research-projects/covid-19-government-response-tracker). All data points corresponding to a frequency of less than 10% were excluded to ensure SGTF data predominantly reflect the presence of B.1.1.7.
Importantly, the existence of VOCs means that scientists and public health officials cannot group all cases together when inferring the dynamics of SARS-CoV-2 and predicting future case loads. Each VOC will have its own reproductive number (or growth rate) and, consequently, different requisite measures for effective control. Additional restrictions (for example, further closures of schools, businesses, and leisure venues65) can slow the spread and reduce the impact of VOCs, despite their higher transmission advantage29. By flattening both non-VOC and VOC COVID-19 curves, more people can be vaccinated and protected before peak case numbers are reached.
Over the near future, epidemic control by nonpharmaceutical interventions will be replaced by control via vaccination, increasing selection in favour of immune escape. Reducing case numbers during this time will continue to be important. In particular, reducing exposure of immunized individuals to active cases should slow the evolution of immune-escape variants (Box 2) by reducing their selective advantage. Vaccinating whole communities or workplaces at once, coupling vaccination campaigns with temporary stay-at-home orders to reduce community transmission, and communicating the risks of exposure to vaccinated individuals are all potential actions that could reduce the evolution of immune escape variants, by reducing contact rates between infectious and immunized individuals. Having effective public health measures also reduces the likelihood that recombinant variants arise by driving down the chance of coinfection.
Dosing strategies
Data suggest that individuals who have received only a single dose of the Pfizer BNT162b2 vaccine or who have natural immunity elicit a weaker antibody response to VOCs than those who have received two vaccine doses, although T-cell responses were similar for variants and non-variants52. Recent reports also indicate lower real-world effectiveness of vaccines against VOCs after one dose (Pfizer: 29.5% [95% CI: 22.9–35.5] for B.1.351 versus 16.9% [95% CI: 10.4–23.0] for B.1.1.755; Pfizer: 33.2% [95% CI: 8.3–51.4] for B.1.617.2 versus 49.2% [95% CI: 42.6–55.0] for B.1.1.756; AstraZeneca 32.9% [95% CI: 19.3–44.3] for B.1.617.2 versus 51.4% [95% CI: 47.3–55.2] for B.1.1.756). These studies, though preliminary, also find that efficacy rises for all variants following the second dose. At this time, however, it is unknown how much of the rise in efficacy against VOCs after the second dose is due to the booster shot itself and how much is due to the maturation of the immune response given more time since the first dose. At face value, these data indicate that completing the recommended number of doses for two-dose vaccines reduces the risk of transmission of partial-escape variants.
Over the short-term, however, the reduction in case numbers made possible by administering one dose to more people is likely to both save lives and reduce the chance that more resistance mutants arise in the first place66 (Box 2). As long as one dose provides sufficient immunity, reducing the burden of disease by vaccinating as many people as possible with one dose and delaying the second dose better prevents the evolution of immune escape variants than more slowly vaccinating the population with two doses at a short inter-dose interval66 , 67.
Another important factor to consider when determining the dosing regime comes from recent studies showing that delaying the second dose enhances immunity, with elevated antibody levels following a longer dosing interval (12 weeks) than a shorter interval (<6 weeks) for both the Pfizer68 and the AstraZeneca69 vaccines. Delayed second doses might thus increase the selective advantage of partial escape mutations over the short term but reduce it over the longer term given the more robust immune responses observed with a delayed booster68 , 69.
Reopening
As we move to reopen societies, rebuild economies, and resume social activities, contact rates will increase. This is expected to increase the selective advantage of VOCs that are more transmissible and/or more able to infect immunized individuals (Figure 5). Increased activity will also make contact tracing more difficult, as will the potential rise in asymptomatic cases among vaccinated individuals60. Coupling reopening with expanded use of rapid testing and environmental sampling would allow cases, and particularly asymptomatic breakthrough cases, to be identified and controlled. Border measures, with testing before, during, and after travel, along with quarantine measures, will continue to be important while cases remain high globally to slow the spread of immune-escape variants and other VOCs that emerge. Ultimately, though, variants will continue to emerge until cases are globally controlled through, for example, equitable access to vaccines. In the words of Ursula von der Leyen (President of the European Commission), “none of us will be safe until everyone is safe.”
Conclusions
As COVID-19 transitions from a pandemic to an endemic disease, VOCs present new global challenges to health by virtue of increased transmissibility and virulence and evasion of natural and vaccine-induced immunity. In this article we have explored the selective forces that shape how VOCs emerge and become established. We also identify possible steps that we can take to limit their emergence and, when they do arise, their impact. Moving forward, we must also consider how SARS-CoV-2 transmits to and amongst other animal species, placing both them and us at further risk. It will therefore be important to adopt a multidisciplinary One Health approach70 for future pandemic management that accounts for the interrelated nature of human, animal, and ecosystem health.
Acknowledgements
The authors would like to thank Art Poon for providing the deviations from the molecular clock used in Figure S1B, Hongru Wang and Rasmus Nielsen for providing estimates of p S to estimate the mutation rate, and two referees for helpful suggestions. Funding was provided by the Natural Sciences and Engineering Research Council of Canada to S.P.O. (RGPIN-2016-03711) and T.D. (RGPIN-2013-217636), by Canada 150 Research Chair support for C.C., by the Public Health Agency of Canada to M.L., S.M., G.V.D., and N.H.O., by the CIHR 2019 Novel Coronavirus (COVID-19) rapid research program to J.W., and by the M.G. DeGroote Institute for Infectious Disease Research at McMaster University to J.D. and D.J.D.E.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.cub.2021.06.049.
Supplemental information
References
- 1.Public Health England . 2020. Investigation of novel SARS-CoV-2 variant: Variant of Concern 202012/01. Version 1, release date 21/12/2020.https://www.gov.uk/government/publications/investigation-of-novel-sars-cov-2-variant-variant-of-concern-20201201 [Google Scholar]
- 2.Volz E., Mishra S., Chand M., Barrett J.C., Johnson R., Geidelberg L., Hinsley W.R., Laydon D.J., Dabrera G., O'Toole A., et al. Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature. 2021;593:266–269. doi: 10.1038/s41586-021-03470-x. [DOI] [PubMed] [Google Scholar]
- 3.van Dorp L., Richard D., Tan C.C.S., Shaw L.P., Acman M., Balloux F. No evidence for increased transmissibility from recurrent mutations in SARS-CoV-2. Nat. Commun. 2020;11:5986–5988. doi: 10.1038/s41467-020-19818-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., Doolabh D., Pillay S., San E.J., Msomi N., et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature. 2021;592:438–443. doi: 10.1038/s41586-021-03402-9. [DOI] [PubMed] [Google Scholar]
- 5.Faria N.R., Mellan T.A., Whittaker C., Claro I.M., Candido D.D.S., Mishra S., Crispim M.A.E., Sales F.C.S., Hawryluk I., McCrone J.T., et al. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science. 2021;372:815–821. doi: 10.1126/science.abh2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holmes E.C. Oxford University Press; Oxford: 2009. The Evolution and Emergence of RNA Viruses. [Google Scholar]
- 7.Vasilarou M., Alachiotis N., Garefalaki J., Beloukas A., Pavlidis P. Population genomics insights into the first wave of COVID-19. Life. 2021;11:129. doi: 10.3390/life11020129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hadfield J., Megill C., Bell S.M., Huddleston J., Potter B., Callender C., Sagulenko P., Bedford T., Neher R.A. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang H., Pipes L., Nielsen R. Synonymous mutations and the molecular evolution of SARS-CoV-2 origins. Virus Evol. 2020;7:veaa098. doi: 10.1093/ve/veaa098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rambaut A., Loman N., Pybus O., Barclay W., Barrett J., Carabelli A., Connor T., Peacock T., Robertson D.L., Volz E., on behalf of the COVID-19 Genomics Consortium UK . 2020. Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations.https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563 virological.org. [Google Scholar]
- 11.Choi B., Choudhary M.C., Regan J., Sparks J.A., Padera R.F., Qiu X., Solomon I.H., Kuo H.-H., Boucau J., Bowman K., et al. Persistence and evolution of SARS-CoV-2 in an immunocompromised host. New Engl. J. Med. 2020;383:2291–2293. doi: 10.1056/NEJMc2031364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Avanzato V.A., Matson M.J., Seifert S.N., Pryce R., Williamson B.N., Anzick S.L., Barbian K., Judson S.D., Fischer E.R., Martens C., et al. Case study: prolonged infectious SARS-CoV-2 shedding from an asymptomatic immunocompromised individual with cancer. Cell. 2020;183:1901–1912. doi: 10.1016/j.cell.2020.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kemp S.A., Collier D.A., Datir R.P., Ferreira I.A.T.M., Gayed S., Jahun A., Hosmillo M., Rees-Spear C., Mlcochova P., Lumb I.U., et al. SARS-CoV-2 evolution during treatment of chronic infection. Nature. 2021;592:277–282. doi: 10.1038/s41586-021-03291-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCarthy K.R., Rennick L.J., Nambulli S., Robinson-McCarthy L.R., Bain W.G., Haidar G., Duprex W.P. Recurrent deletions in the SARS-CoV-2 spike glycoprotein drive antibody escape. Science. 2021;371:1139–1142. doi: 10.1126/science.abf6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCormick K.D., Jacobs J.L., Mellors J.W. The emerging plasticity of SARS-CoV-2. Science. 2021;371:1306–1308. doi: 10.1126/science.abg4493. [DOI] [PubMed] [Google Scholar]
- 16.Luria S., Delbruck M. Mutations of bacteria from virus sensitivity to virus resistance. Genetics. 1943;28:491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lythgoe K.A., Hall M., Ferretti L., de Cesare M., MacIntyre-Cockett G., Trebes A., Andersson M., Otecko N., Wise E.L., Moore N., et al. SARS-CoV-2 within-host diversity and transmission. Science. 2021;372:eabg0821. doi: 10.1126/science.abg0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Braun K., Moreno G.K., Wagner C., Accola M.A., Rehrauer W.M., Baker D.A., Koelle K., O’Connor D.H., Bedford T., Friedrich T.C., Moncla L.H. Limited within-host diversity and tight transmission bottlenecks limit SARS-CoV-2 evolution in acutely infected individuals. bioRxiv. 2021 doi: 10.1101/2021.04.30.440988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takada K., Ueda M.T., Watanabe T., Nakagawa S. Genomic diversity of SARS-CoV-2 can be accelerated by a mutation in the nsp14 gene. bioRxiv. 2020 doi: 10.1101/2020.12.23.424231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richard D., Owen C.J., van Dorp L., Balloux F. No detectable signal for ongoing genetic recombination in SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.12.15.422866. [DOI] [Google Scholar]
- 21.Ignatieva A., Hein J., Jenkins P.A. Ongoing recombination in SARS-CoV-2 revealed through genealogical reconstruction. bioRxiv. 2021 doi: 10.1101/2021.01.21.427579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.VanInsberghe D., Neish A.S., Lowen A.C., Koelle K. Recombinant SARS-CoV-2 genomes are currently circulating at low levels. bioRxiv. 2021 doi: 10.1101/2020.08.05.238386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jackson B., Rambaut A., Pybus O.G., Robertson D.L., Connor T., Loman N.J., on behalf of the COVID-19 Genomics UK Consortium . 2021. Recombinant SARS-CoV-2 genomes involving lineage B.1.1.7 in the UK.https://virological.org/t/recombinant-sars-cov-2-genomes-involving-lineage-b-1-1-7-in-the-uk/658 virological.org. [Google Scholar]
- 24.Woolhouse M.E., Dye C., Etard J.-F., Smith T., Charlwood J.D., Garnett G.P., Hagan P., Hii J.L., Ndhlovu P.D., Quinnell R.J., et al. Heterogeneities in the transmission of infectious agents: implications for the design of control programs. Proc. Natl. Acad. Sci. USA. 1997;94:338–342. doi: 10.1073/pnas.94.1.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Endo A., Abbott S., Kucharski A.J., Funk S. Estimating the overdispersion in COVID-19 transmission using outbreak sizes outside China. Wellcome Open Res. 2020;5:67. doi: 10.12688/wellcomeopenres.15842.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Althouse B.M., Wenger E.A., Miller J.C., Scarpino S.V., Allard A., Hébert-Dufresne L., Hu H. Superspreading events in the transmission dynamics of SARS-CoV-2: Opportunities for interventions and control. PLoS Biol. 2020;18:e3000897. doi: 10.1371/journal.pbio.3000897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lloyd-Smith J.O., Schreiber S.J., Kopp P.E., Getz W.M. Superspreading and the effect of individual variation on disease emergence. Nature. 2005;438:355–359. doi: 10.1038/nature04153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mulberry N., Tupper P., Kirwin E., McCabe C., Colijn C. Vaccine rollout strategies: The case for vaccinating essential workers early. medRxiv. 2021 doi: 10.1101/2021.02.23.21252309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davies N.G., Abbott S., Barnard R.C., Jarvis C.I., Kucharski A.J., Munday J.D., Pearson C.A.B., Russell T.W., Tully D.C., Washburne A.D., et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021;372:eabg3055. doi: 10.1126/science.abg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O’Toole Á., Hill V. 2021. Global report investigating novel coronavirus haplotypes: B.1.1.7.https://cov-lineages.org/global_report_B.1.1.7.html [Google Scholar]
- 31.Byambasuren O., Cardona M., Bell K., Clark J., McLaws M.-L., Glasziou P. Estimating the extent of asymptomatic COVID-19 and its potential for community transmission: systematic review and meta-analysis. JAMMI. 2020;5:223–234. doi: 10.3138/jammi-2020-0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tindale L.C., Stockdale J.E., Coombe M., Garlock E.S., Lau W.Y.V., Saraswat M., Zhang L., Chen D., Wallinga J., Colijn C. Evidence for transmission of COVID-19 prior to symptom onset. eLife. 2020;9:e57149. doi: 10.7554/eLife.57149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meyerowitz-Katz G., Merone L. A systematic review and meta-analysis of published research data on COVID-19 infection fatality rates. Int. J. Infect. Dis. 2020;101:138–148. doi: 10.1016/j.ijid.2020.09.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bull J.J. Virulence. Evolution. 1994;48:1423–1437. doi: 10.1111/j.1558-5646.1994.tb02185.x. [DOI] [PubMed] [Google Scholar]
- 35.Frank S.A. Models of parasite virulence. Q. Rev. Biol. 1996;71:37–78. doi: 10.1086/419267. [DOI] [PubMed] [Google Scholar]
- 36.Day T. Virulence evolution and the timing of disease life-history events. Trends Ecol. Evol. 2003;18:113–118. [Google Scholar]
- 37.Day T., Gandon S., Lion S., Otto S.P. On the evolutionary epidemiology of SARS-CoV-2. Curr. Biol. 2020;30:R849–R857. doi: 10.1016/j.cub.2020.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lenski R.E., May R.M. The evolution of virulence in parasites and pathogens: reconciliation between two competing hypotheses. J. Theor. Biol. 1994;169:253–265. doi: 10.1006/jtbi.1994.1146. [DOI] [PubMed] [Google Scholar]
- 39.Day T., Gandon S. Applying population-genetic models in theoretical evolutionary epidemiology. Ecol. Lett. 2007;10:876–888. doi: 10.1111/j.1461-0248.2007.01091.x. [DOI] [PubMed] [Google Scholar]
- 40.Bull J.J., Ebert D. Invasion thresholds and the evolution of nonequilibrium virulence. Evol. Appl. 2008;1:172–182. doi: 10.1111/j.1752-4571.2007.00003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davies N.G., Jarvis C.I., CMMID COVID-19 Working Group. Edmunds W.J., Jewell N.P., Diaz-Ordaz K., Keogh R.H. Increased mortality in community-tested cases of SARS-CoV-2 lineage B.1.1.7. Nature. 2021;593:270–274. doi: 10.1038/s41586-021-03426-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Challen R., Brooks-Pollock E., Read J.M., Dyson L., Tsaneva-Atanasova K., Danon L. Risk of mortality in patients infected with SARS-CoV-2 variant of concern 202012/1: matched cohort study. BMJ. 2021;372:n579. doi: 10.1136/bmj.n579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Teruel N., Mailhot O., Najmanovich R. Modelling conformational state dynamics and its role on infection for SARS-CoV-2 spike protein variants. bioRxiv. 2020 doi: 10.1021/2020.12.16.423118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Calistri P., Amato L., Puglia I., Cito F., Di Giuseppe A., Danzetta M.L., Morelli D., Di Domenico M., Caporale M., Scialabba S., et al. Infection sustained by lineage B.1.1.7 of SARS-CoV-2 is characterised by longer persistence and higher viral RNA loads in nasopharyngeal swabs. Int. J. Infect. Dis. 2021;105:753–755. doi: 10.1016/j.ijid.2021.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Damas J., Hughes G.M., Keough K.C., Painter C.A., Persky N.S., Corbo M., Hiller M., Koepfli K.-P., Pfenning A.R., Zhao H., et al. Broad host range of SARS-CoV-2 predicted by comparative and structural analysis of ACE2 in vertebrates. Proc. Natl. Acad. Sci. USA. 2020;117:22311–22322. doi: 10.1073/pnas.2010146117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brault A.C., Huang C.Y., Langevin S.A., Kinney R.M., Bowen R.A., Ramey W.N., Panella N.A., Holmes E.C., Powers A.M., Miller B.R. A single positively selected West Nile viral mutation confers increased virogenesis in American crows. Nat. Genet. 2007;39:1162–1166. doi: 10.1038/ng2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Larsen H.D., Fonager J., Lomholt F.K., Dalby T., Benedetti G., Kristensen B., Urth T.R., Rasmussen M., Lassaunière R., Rasmussen T.B., et al. Preliminary report of an outbreak of SARS-CoV-2 in mink and mink farmers associated with community spread, Denmark, June to November 2020. Euro. Surveill. 2021;26:2100009. doi: 10.2807/1560-7917.ES.2021.26.5.210009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Watson O.J., Abdelmagid N., Ahmed A., Elhameed A.E.A.A., Whittaker C., Brazeau N., Hamlet A., Walker P., Hay J., Ghani A., et al. Imperial College London; London: 2020. Characterising COVID-19 epidemic dynamics and mortality underascertainment in Khartoum, Sudan. [DOI] [Google Scholar]
- 49.Kennedy D.A., Read A.F. Why does drug resistance readily evolve but vaccine resistance does not? Proc. R. Soc. B. 2017;284:20162562. doi: 10.1098/rspb.2016.2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Le Bert N., Tan A.T., Kunasegaran K., Tham C.Y., Hafezi M., Chia A., Chng M.H.Y., Lin M., Tan N., Linster M., et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 51.Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Skelly D.T., Harding A.C., Gilbert-Jaramillo J., Knight M.L., Longet S., Brown A., Adele S., Adland E., Brown H., et al. Vaccine-induced immunity provides more robust heterotypic immunity than natural infection to emerging SARS-CoV-2 variants of concern. Research Square. 2021 doi: 10.21203/rs.3.rs-226857/v1. [DOI] [Google Scholar]
- 53.Tarke A., Sidney J., Methot N., Zhang Y., Dan J.M., Goodwin B., Rubiro P., Sutherland A., da Silva Antunes R., Frazier A., et al. Negligible impact of SARS-CoV-2 variants on CD4+ and CD8+ T cell reactivity in COVID-19 exposed donors and vaccinees. bioRxiv. 2021 doi: 10.1101/2021.02.27.433180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu K., Werner A.P., Koch M., Choi A., Narayanan E., Stewart-Jones G.B., Colpitts T., Bennett H., Boyoglu-Barnum S., Shi W., et al. Serum neutralizing activity elicited by mRNA-1273 vaccine. New Engl. J. Med. 2021;384:1468–1470. doi: 10.1056/NEJMc2102179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Abu-Raddad L.J., Chemaitelly H., Butt A.A., on behalf of the National Study Group for COVID-19 Vaccination Effectiveness of the BNT162b2 COVID-19 vaccine against the B.1.1.7 and B.1.351 variants. New Engl. J. Med. 2021;385:187–189. doi: 10.1056/NEJMc2104974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lopez Bernal J., Andrews N., Gower C., Gallagher E., Simmons R., Thelwall S., Stowe J., Tessier E., Groves N., Dabrera G., et al. Effectiveness of COVID-19 vaccines against the B.1.617.2 variant. medRxiv. 2021 doi: 10.1101/2021.05.22.21257658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eguia R.T., Crawford K.H.D., Stevens-Ayers T., Kelnhofer-Millevolte L., Greninger A.L., Englund J.A., Boeckh M.J., Bloom J.D. A human coronavirus evolves antigenically to escape antibody immunity. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu K., Choi A., Koch M., Ma L., Hill A., Nunna N., Huang W., Oestreicher J., Colpitts T., Bennett H., et al. Preliminary analysis of safety and immunogenicity of a SARS-CoV-2 variant vaccine booster. medRxiv. 2021 doi: 10.1101/2021.05.05.21256716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Otto S.P., Phillips B., Colijn C. 2021. Are there COVID variants amongst us. BC COVID-19 Modelling Group blog.https://bccovid-19group.ca/post/covid-variants-amongst-us/ [Google Scholar]
- 60.Teran, R.A., Walblay, K.A., Shane, E.L., Xydis, S., Gretsch, S., Gagner, A., Samala, U., Choi, H., Zelinski, C., and Black, S.R. (2021). Postvaccination SARS-CoV-2 infections among skilled nursing facility residents and staff members – Chicago, Illinois, December 2020-March 2021. Morb. Mortal. Wkly. Rep. 70, 632–638. [DOI] [PMC free article] [PubMed]
- 61.Kojaku, S., Hébert-Dufresne, L., Mones, E., Lehmann, S., and Ahn, Y.-Y. The effectiveness of backward contact tracing in networks. Nat. Physics 17, 652–658. [DOI] [PMC free article] [PubMed]
- 62.Ludwig A., Berthiaume P., Orpana H., Nadeau C., Diasparra M., Barnes J., Hennessy D., Otten A., Ogden N. Assessing the impact of varying levels of case detection and contact tracing on COVID-19 transmission in Canada during lifting of restrictive closures using a dynamic compartmental model. Can. Commun. Dis. Rep. 2020;46:409–421. doi: 10.14745/ccdr.v46i1112a08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tang B., Scarabel F., Bragazzi N.L., McCarthy Z., Glazer M., Xiao Y., Heffernan J.M., Asgary A., Ogden N.H., Wu J. De-escalation by reversing the escalation with a stronger synergistic package of contact tracing, quarantine, isolation and personal protection: feasibility of preventing a COVID-19 rebound in Ontario, Canada, as a case study. Biology. 2020;9:100. doi: 10.3390/biology9050100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tupper P., Otto S., Colijn C. Fundamental limitations of contact tracing for COVID-19. medRxiv. 2020 doi: 10.1101/2020.12.15.20248299. [DOI] [Google Scholar]
- 65.Ng V., Fazil A., Waddell L., Turgeon P., Otten A., Ogden N.H. Modelling the impact of shutdowns on resurging SARS-CoV-2 transmission in Canada. R. Soc. Open Sci. 2021;8:210233. doi: 10.1098/rsos.210233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cobey S., Larremore D.B., Grad Y.H., Lipsitch M. Concerns about SARS-CoV-2 evolution should not hold back efforts to expand vaccination. Nat. Rev. Immunol. 2021;21:330–335. doi: 10.1038/s41577-021-00544-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Saad-Roy C.M., Morris S.E., Metcalf C.J.E., Mina M.J., Baker R.E., Farrar J., Holmes E.C., Pybus O.G., Graham A.L., Levin S.A., et al. Epidemiological and evolutionary considerations of SARS-CoV-2 vaccine dosing regimes. Science. 2021;372:363–370. doi: 10.1126/science.abg8663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Parry H., Bruton R., Stephens C., Brown K., Amirthalingam G., Hallis B., Otter A., Zuo J., Moss P. Extended interval BNT162b2 vaccination enhances peak antibody generation in older people. medRxiv. 2021 doi: 10.1101/2021.05.15.21257017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Voysey M., Clemens S.A.C., Madhi S.A., Weckx L.Y., Folegatti P.M., Aley P.K., Angus B., Baillie V.L., Barnabas S.L., Bhorat Q.E., et al. Single-dose administration and the influence of the timing of the booster dose on immunogenicity and efficacy of ChAdOx1 nCoV-19 (AZD1222) vaccine: a pooled analysis of four randomised trials. Lancet. 2021;397:881–891. doi: 10.1016/S0140-6736(21)00432-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ogden N.H., Wilson J.R., Richardson D.M., Hui C., Davies S.J., Kumschick S., Le Roux J.J., Measey J., Saul W.C., Pulliam J.R.C. Emerging infectious diseases and biological invasions: a call for a One Health collaboration in science and management. R. Soc. Open Sci. 2019;6:181577. doi: 10.1098/rsos.181577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.World Health Organization . 2021. COVID-19 Weekly Epidemiological Update (25 February 2021) https://apps.who.int/iris/handle/10665/339859. [Google Scholar]
- 72.Pearson C., Russell T.W., Davies N.G., Kucharski A.J., CMMID COVID-19 working group. Edmunds W.J., Eggo R.M. Centre for Mathematical Modelling of Infectious Diseases Repository; 2021. Estimates of severity and transmissibility of novel SARS-CoV-2 variant 501Y.V2 in South Africa.https://cmmid.github.io/topics/covid19/sa-novel-variant.html [Google Scholar]
- 73.Deng X., Garcia-Knight M.A., Khalid M.M., Servellita V., Wang C., Morris M.K., Sotomayor-González A., Glasner D.R., Reyes K.R., Gliwa A.S., et al. Transmission, infectivity, and neutralization of a spike L452R SARS-CoV-2 variant. Cell. 2021;184:3426–3437. doi: 10.1016/j.cell.2021.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Public Health England . Public Health England; London: 2021. SARS-CoV-2 variants of concern and variants under investigation in England: Technical briefing 15.https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/993198/Variants_of_Concern_VOC_Technical_Briefing.pdf [Google Scholar]
- 75.Alexander H.K., Day T. Risk factors for the evolutionary emergence of pathogens. J. R. Soc. Interface. 2010;7:1455–1474. doi: 10.1098/rsif.2010.0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bolker B., Li M., Dushoff J., Earn D., So M., Levine Z., Gharouni A., Papst I., Walker S. Initial release for Zenodo; 2021. McMasterPandemic. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.