Abstract
Safe and effective vaccines will provide essential medical countermeasures to tackle the COVID-19 pandemic. Here, we assessed the safety, immunogenicity and efficacy of the intradermal delivery of INO-4800, a synthetic DNA vaccine candidate encoding the SARS-CoV-2 spike protein in the rhesus macaque model. Single and 2 dose vaccination regimens were evaluated. Vaccination induced both binding and neutralizing antibodies, along with IFN-γ-producing T cells against SARS-CoV-2. Upon administration of a high viral dose (5 × 106 pfu) via the intranasal and intratracheal routes we observed significantly reduced virus load in the lung and throat, in the vaccinated animals compared to controls. 2 doses of INO-4800 was associated with more robust vaccine-induced immune responses and improved viral protection. Importantly, histopathological examination of lung tissue provided no indication of vaccine-enhanced disease following SARS-CoV-2 challenge in INO-4800 immunized animals. This vaccine candidate is currently under clinical evaluation as a 2 dose regimen.
Keywords: COVID-19, DNA vaccine, Nonhuman Primate Virus Challenge, SARS-CoV-2
1. Introduction
In March 2020, Coronavirus disease 2019 (COVID-19) was declared a pandemic [1]. As of, June 9, 2021 there have been>174,000,000 cases of COVID-19 and>3,7500,000 deaths worldwide (GISAID). The development of vaccines as medical countermeasures began in January 2020 upon the public release of the genetic sequence of the virus associated with an outbreak of pneumonia illness in Wuhan, China. The sequence revealed a beta coronavirus with 95% amino acid similarity to SARS-CoV-1 and was subsequently named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [2], [3]. Leveraging previous knowledge gained from vaccines developed against other beta coronaviruses, developers recognized the spike protein as a potential target antigen [4], [5], [6]. Currently the vast majority of vaccines in development have been designed based on the entire spike antigen, or a truncated form covering the receptor binding domain (RBD), with the goal to elicit host immune responses which will block interaction of this glycoprotein with its receptor angiotensin-converting enzyme receptor (ACE2) and protect against viral infection. Presently there are three vaccines approved for emergency use authorization (EUA) in the USA, two based on nucleic acid technology, BNT162b2, mRNA-1273 and the adenovirus-vectored Ad26.COV2.S. AZD1222, GAM-COVID-Vac and BBIBP-CorV COVID-19 vaccines have also received authorization outside of the USA. However, to immunize the global population billions of doses will be needed, which will require many more vaccine candidates to be approved for use.
Candidates which apply nucleic acid-based technology currently under clinical evaluation include mRNA-1273 [7], BNT162b2 [8], CVnCoV [9], ARCT-021 [10] and INO-4800 [11]. INO-4800 is a codon optimized plasmid DNA vaccine encoding the wildtype SARS-CoV-2 spike protein [11], [12]. Previous studies in animals have shown INO-4800 to be immunogenic, inducing T and B cell responses targeting SARS-CoV-2 [12], and mediating a reduction in viral replication upon challenge of human ACE2-transduced mice [13]. Furthermore, the induction of immunological memory by INO-4800 was shown to mediate a reduction of viral replication in a rhesus macaque SARS-CoV-2 challenge model [14]. Recently published Phase 1 clinical studies demonstrated INO-4800 vaccination was associated with a favorable safety and tolerability profile, and was immunogenic in 100% of recipients, inducing SARS-CoV-2 neutralizing antibodies and cellular immune responses [11]. INO-4800 is currently undergoing evaluation in a Phase 2 segment of a Phase 2/3 study which will further address safety, immunogenicity and efficacy (NCT04642638).
Here, we have evaluated the safety and efficacy of the synthetic DNA vaccine candidate, INO-4800 delivered as a 1 or 2 dose regimen, to the skin with CELLECTRA-ID electroporation technology in the rhesus macaque SARS-CoV-2 challenge model [15]. Nonhuman primates are a valuable model in the development of COVID-19 vaccines and therapeutics as they can be infected with wild-type SARS-CoV-2, and present with early infection that mimics aspects of mild human disease [16]. In contrast to previous NHP challenge studies with INO-4800, in this study we have used a high dose of SARS-CoV-2 Victoria01 strain (5X106 pfu) to specifically assess the impact of INO-4800 vaccination performed 4 weeks prior to challenge on lung disease burden to provide both vaccine safety and efficacy data. We have measured a broad range of lower respiratory tract disease parameters by applying histopathology, lung disease scoring metric system, in-situ hybridization, viral RNA RT-qPCR and computed tomography (CT) scans to provide an understanding of the impact of vaccine-induced immunity on protective efficacy and potential vaccine-enhanced disease (VED). To provide important translational information concerning this vaccine candidate, we have employed the method of administration of INO-4800 and a dosing regimen which match that currently undergoing clinical evaluation (NCT04642638).
2. Results
2.1. Immunogenicity of 1 and 2 dose regimens of INO-4800
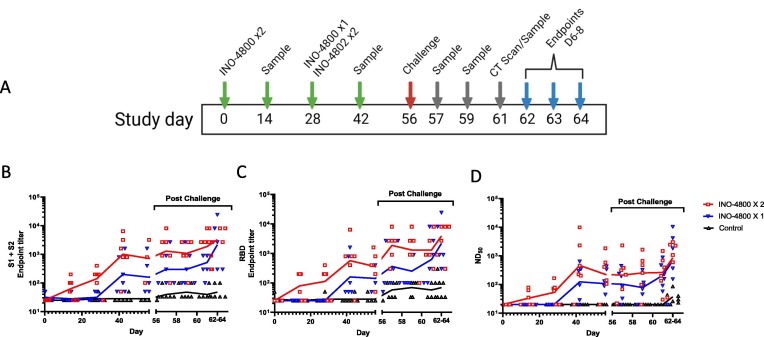
We vaccinated 12 (6 male and 6 female) rhesus macaques (Macaca mulatta). One group received one dose (6 animals) of INO-4800 administered on day 28. The second group (6 animals) received one dose of INO-4800 on day 0 and a second dose on day 28 (Fig. 1 A). For each treatment 1 mg INO-4800 was administered intradermally followed by CELLECTRA-ID EP. A further six age and sex matched animals were not vaccinated and provided the control group. Animals were observed and scored as alert and healthy for the duration of the study, and no adverse events or significant changes in clinical parameters were recorded in the animals (Supplemental Figure 1). The serum titers of SARS-CoV-2 spike antigen reactive IgG antibodies in all animals were measured biweekly between days 0 and 56. In the single dose group (INO-4800 X1) a mean endpoint binding titer of 467 against the SARS-CoV-2 spike antigen trimeric S1 + S2 ECD form and 442 against the RBD antigen, and a live virus (Victoria/01/2020 matched to the challenge strain) neutralization titer of 239 was measured 14 days after vaccination (Fig. 1B-D). In the 2- dose group (INO-4800 X2) a mean endpoint binding titer of 2,142 against the S1 + S2 ECD and 1,538 against the RBD antigen, and a live virus neutralization titer of 2,199 was measured 14 days after the 2nd vaccination (Fig. 1B-D). Vaccination with INO-4800 induced SARS-CoV-2 spike antigen-specific Th1 T cell responses in the PBMC population as measured by an IFN-γ ELISpot (Supplemental Figure 2A). In summary, intradermal delivery of INO-4800 induced a functional T cell response and SARS-CoV-2 spike protein directed humoral immunity which was boosted after a second dose. At the day of viral challenge (Day 56) the level of SARS-CoV-2 neutralizing antibodies in the serum was significantly higher in the vaccinated groups compared to the control group (P = 0.015). Following viral challenge there was a slight increase in SARS-CoV-2 spike binding and neutralizing antibody titers in all the groups between days 56 and 62–64 ( Fig. 1B-D). In the control group there was an increase in the cellular immune response to peptides spanning the SARS-CoV-2 spike antigen after viral challenge, but little change in the vaccinated groups, likely due to control of viral infection by the humoral arm of the host immune system (Supplemental Figure 2B).
Fig. 1.
Humoral responses in rhesus macaques vaccinated with INO-4800. Study outline (A). SARS-CoV-2 Spike-specific IgG (B), RBD-specific IgG (C) and live virus-neutralising antibodies (D) measured pre- and post-challenge in serum from rhesus macaques that received 1 or 2 doses of INO-4800 or were unvaccinated (Control). Lines represent the geometric means.
2.2. Viral loads in the upper and lower respiratory tracts after SARS-CoV-2 challenge
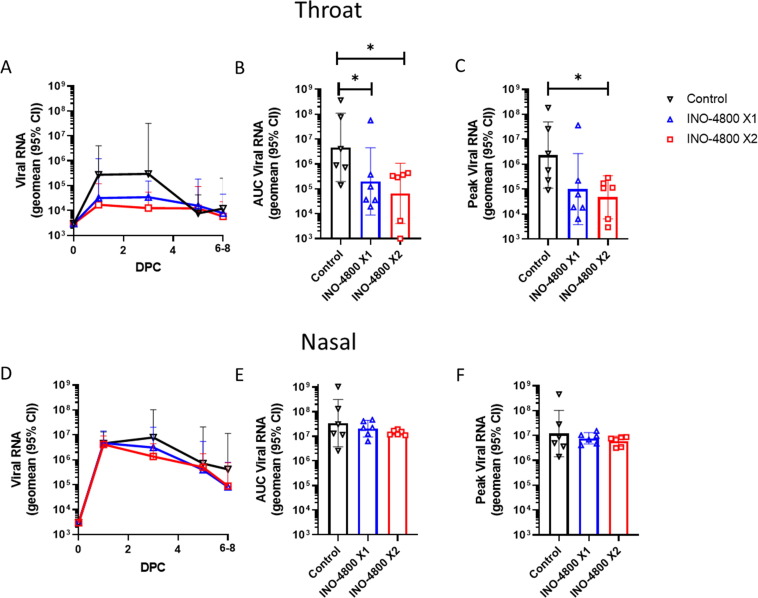
On day 56 all animals were challenged with a total of 5x106 pfu SARS-CoV-2 delivered to both the upper (IN) and lower (IT) respiratory tract. No overt clinical signs were observed throughout the duration (6–8 days) of the challenge in any of the animals (Supplemental Figure 1). At indicated timepoints nasal and throat swabs were collected from the animals. SARS-CoV-2 viral RNA and subgenomic (sgmRNA), which represents replicating virus were measured by RT-qPCR (Fig. 2 and Supplemental Figure 3). Analysis of viral RNA area under the curve (AUC) levels in the throat revealed significantly reduced levels in the vaccinated groups (Fig. 2B). Additionally, the peak viral load level measured in the INO-4800 X2 group was significantly reduced compared to the control group (Fig. 2C). Analysis revealed a significant negative correlation between throat viral loads and neutralizing and anti-RBD IgG titers (Supplemental Figure 4A-D). SARS-CoV-2 sgmRNA data revealed a similar trend of reduction of viral load in the vaccinated groups compared to controls (Supplemental Figure 3A-C). Analysis in the nasal compartment revealed a trend for reduction and accelerated clearance of viral RNA and sgmRNA in the vaccinated groups compared to control but did not reach a level of significance (Fig. 2D-F, and Supplemental Figure 3D-F). Analysis revealed a significant negative correlation between nasal viral loads and neutralizing and anti-RBD IgG titers on day 3, but not day 1 (Supplemental Figure 4E-H).
Fig. 2.
Upper respiratory tract viral loads detected by RT-qPCR following challenge with SARS-CoV-2. Animals received 1 (blue) or 2 (red) doses of INO-4800 or were unvaccinated (control - black). Viral load plotted as Log10 cDNA copies mL−1 for each animal in throat swabs (A-C) and nasal swabs (D-F). (A&D) Lines represent group geometric means with 95% CI. Area under the curve (AUC) of viral loads for (B) throat swabs and (E) nasal swabs for each experimental group. Peak viral loads measured in each animal during the challenge period for (C) throat swabs and (F) nasal swabs. LLOQ (lower limit of quantification, 3.80 log copies mL−1) and LLOD (lower limit of detection, 3.47 log copies mL−1). Positive samples detected below the LLOQ were assigned the value of 3.80 log copies mL−1. * P ≤ 0.05 with Mann-Whitney t test.
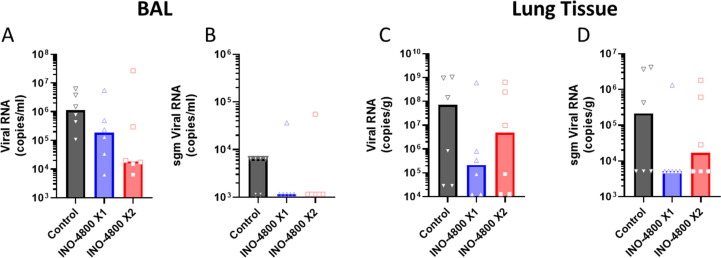
At the time of necropsy (6–8 days post challenge), bronchoalveolar lavage (BAL) fluid was collected from each animal. Measurement of the levels of SARS-CoV-2 viral RNA and sgmRNA revealed a reduction of the average virus in vaccinated groups, even though the levels were variable within each group dependent on the day of necropsy (Fig. 3 A&B). RT-qPCR was also performed on tissues collected at necropsy. At these timepoints post challenge the SARS-CoV-2 viral RNA levels detected were below limit of quantification in most tissues except the lungs (Supplemental Figure 5). Measurements of the level of SARS-CoV-2 viral mRNA and sgmRNA detected in the lung tissue samples indicated reduced average viral load in the vaccinated animals (Fig. 3C&D).
Fig. 3.
Lower respiratory tract viral loads detected by RT-qPCR following challenge with SARS-CoV-2. Animals received 1 (blue) or 2 (red) doses of INO-4800 or were unvaccinated (control - black). SARS-CoV-2 viral and subgenomic RNA were measured for individual animals in bronchoalveolar lavage (BAL (A&B)) and lung tissue (C&D) samples collected at necropsy (6–8 days post challenge). Bars represent group medians. Assay LLOQ’s and LLOD’s are provided in the methods section.
In summary data showed a significant reduction of viral load in the throat, and a trend for a reduction of viral loads in the lungs of the vaccinated groups. The collection of BAL and lung tissue samples at different timepoints (days 6, 7 or 8) after challenge likely added to the intragroup variability observed impacting statistical analysis. RT-qPCR viral load data indicated INO-4800 vaccination had a positive effect in reducing viral loads in rhesus macaques challenged with a high dose of SARS-CoV-2. In general, lower viral levels were measured in the 2-dose vaccine group compared to 1 dose vaccine group.
2.3. Disease burden in the lungs after SARS-CoV-2 challenge
The pulmonary disease burden was assessed on harvested lung tissues collected at necropsy 6 to 8 days after challenge. Analysis was performed on all animals in the study in a double blinded manner. Histopathological analysis was performed on multiple organ tissues, but only the lungs showed remarkable lesions, compatible with SARS-CoV-2 infection. Pulmonary lesions consistent with infection with SARS-CoV-2 were observed in the lungs of animals from the unvaccinated control and at a reduced level in vaccinated groups. Representative histopathology images are provided in Supplemental Figure 6. Briefly, the lung parenchyma was comprised of multifocal to coalescing areas of pneumonia surrounded by unaffected parenchyma. Alveolar damage, with necrosis of pneumocytes was a prominent feature in the affected areas. Alveolar spaces and interalveolar septa contained mixed inflammatory cells (including macrophages, lymphocytes, viable and degenerated neutrophils, and occasional eosinophils), and oedema. Type II pneumocyte hyperplasia was also observed in bronchiolo-alveolar junctions. In the larger airways occasional, focal, epithelial degeneration and sloughing was observed in the respiratory epithelium. Low numbers of mixed inflammatory cells, comprising neutrophils, lymphoid cells, and occasional eosinophils, infiltrated bronchial and bronchiolar walls. In the lumen of some airways, mucus admixed with degenerative cells, mainly neutrophils and epithelial cells, was seen. Within the parenchyma, perivascular and peribronchiolar cuffing was also observed, being mostly lymphoid cells comprising the infiltrates.
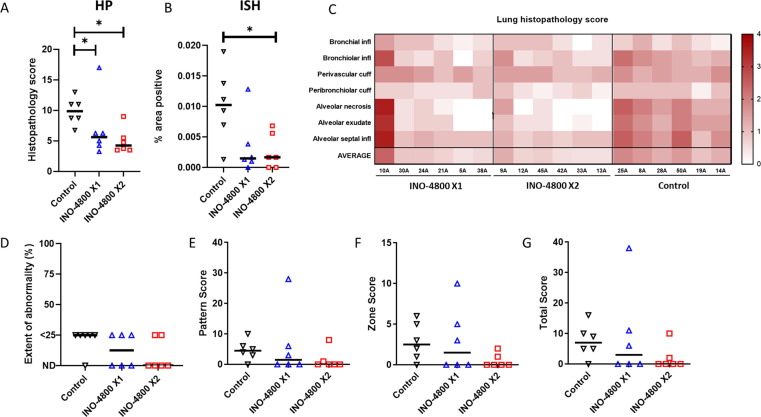
The histopathology score and percent tissue area of SARS-CoV-2 RNA positivity were applied to quantify the disease burden. The unvaccinated group showed the highest histopathological scores in the lung when compared with the vaccinated groups (Fig. 4 A&C). Animals from vaccinated groups showed reduced pathology when compared with unvaccinated animals, except for animal #10A from INO-4800 X1 group, which showed histopathological scores similar to the unvaccinated animals. In line with this observation, the antibody levels in animal 10A, comparable to the unvaccinated animals, remained at unchanged throughout the pre-challenge phase of the study. To detect the presence of SARS-CoV-2 RNA in the lung tissue, in situ hybridization (ISH) was performed. Viral RNA was observed in pneumocytes and inflammatory cells within the histopathological lesions with reduced frequency in the vaccinated animals (Fig. 4B).
Fig. 4.
Lung disease burden measured by histopathology and CT scan following challenge with SARS-CoV-2. Total histopathology score (A), and image analysis of positively stained area in ISH RNAScope labelled sections for viral RNA (B). (C) Heat map illustration of histopathology scoring for each parameter for individual animals. CT radiology scores for individual animals (D-G). (D) The extent of abnormality as a percentage of the lung affected. (E) COVID-19 disease pattern with scoring based on presence of nodules, ground glass opacity, and consolidation. (F) Zone classification (lung is divided into 12 zones and each zone showing abnormalities is attributed 1 point). (G) Total cumulative CT score (Pattern + Zone scores). Line on graphs represent median value of group. * P ≤ 0.05 with Mann-Whitney’s U test.
CT scans were performed to provide an in-life, unbiased, and quantifiable metric of lung disease. Results from lung CT imaging performed 5 days after challenge with SARS-CoV-2 were evaluated for the presence of COVID-19 disease features: ground glass opacity (GGO), consolidation, crazy paving, nodules, peri-lobular consolidation; distribution - upper, middle, lower, central 2/3, peripheral, bronchocentric, and for pulmonary embolus. The medical radiologist was blinded to the animal’s treatment and clinical status. The extent of lung involvement was evaluated and quantified using a scoring system developed for COVID disease. The score system parameters are provided in the materials and methods section. Pulmonary abnormalities characteristic of COVID-19 disease where observed in 3 out of 6 and 2 out of 6 animals in the INO-4800 1 dose or 2 dose groups, respectively, and in 5 out of 6 unvaccinated animals in the control group (representative CT scan images are provided in Supplemental Figure 7). The extent of lung involvement in the animals with disease involvement was<25% and considered low level disease (Fig. 4D). There was a trend for disease scores to be highest in the unvaccinated control group with a reduction in the INO-4800 1 and 2 dose groups (Fig. 4E-G). The comparison of scores between groups did not reach statistical difference (P = 0.0584 between INO-4800 2-dose group and no vaccine group, Mann Whitney’s U test). One outlier animal (10A) in the INO-4800 X1 group scored higher than other animals. However, the level of disease was still considered low and comparable disease burden had been observed in other NHP SARS-CoV-2 challenge studies performed under the same conditions. In summary, CT scanning provides a useful measure of SARS-CoV-2-induced disease in rhesus macaques. Day 5 post SARS-CoV-2 infection, abnormalities where present were reported at low levels (<25% of lung involved). Evidence from CT scans suggested trends for differences in pulmonary disease burden between groups, with disease burden highest in the nonvaccinated control group.
In summary, after high dose SARS-CoV-2 challenge of nonhuman primates the disease burden was reduced in the animals receiving a 1 or 2 dose regimen of INO-4800 vaccine. There was no indication of VED, even in animals receiving a 1 dose vaccination regimen.
3. Discussion
This report describes the safety, immunogenicity and efficacy assessments of the SARS-CoV-2 synthetic DNA vaccine INO-4800 in a stringent high dose nonhuman primate challenge model. We report intradermal delivery of 1 mg of INO-4800 to rhesus macaques induced both humoral and T cell responses against the SARS-CoV-2 spike antigen in both a 2 dose regimen and a 1 dose regimen. Throughout the study no overt clinical events were recorded in the animals, which along with our clinical data with INO-4800 supports the safety of the vaccine [11]. After a high dose SARS-CoV-2 challenge we observed a reduction in viral loads and lung disease burden in both the 1 and 2 dose vaccine groups supporting the efficacy of INO-4800. Importantly, vaccine enhanced disease was not observed, even in the 1 dose group.
The study was designed to match the dosing regimen and vaccine administration protocol currently undergoing evaluation in clinical trials. Specifically, this included the use of intradermal CELLECTRA EP (ID-EP) technology. This technology has been widely studied in the preclinical setting and shown to enhance delivery of DNA vaccines to professional antigen presenting cells in the skin [17], [18]. In contrast to intramuscular delivery, skin administration has an improved tolerability profile, can be dose sparing as it targets a tissue rich in APCs, and the smaller portable EP devices may be favorable for mass vaccination [19], [20], [21]. ID-EP has been used in multiple clinical trials to deliver vaccines (including INO-4800) to the dermis [11], [22]. This immunization protocol has been shown to induce a balanced T and B cell response [11], [22]. Together with the favorable stability profile of the pDNA vaccine formulations this platform technology may have an important global role in bringing vaccines to resource poor settings. Global Phase 3 trials are planned for INO-4800 employing this EP technology.
The rhesus macaque model has become a widely employed model for assessing medical countermeasures against SARS-CoV-2. Importantly, wildtype non-adapted SARS-CoV-2 replicates in the respiratory tract of rhesus macaques, and the animal presents with some of the characteristics observed in humans with mild COVID-19 symptoms [15], [16]. Here, we focused on the lung disease burden in SARS-CoV-2 challenged rhesus macaques which had been vaccinated with INO-4800. While the level of lung disease burden measured in the animals was mild, we observed a significant reduction in histopathological and viral detection scores in the lungs of vaccinated animals (Fig. 4). This was a very encouraging signal as it suggests the potential for a positive impact on the LRT disease which is observed in COVID-19 patients who progress to severe disease. Interestingly, we also observed a significant reduction in viral loads in the throat compartment in the upper respiratory tract, but only a trend for reduction in the nasal compartment. Currently we cannot give a definitive answer to why this was observed, but we can speculate. It may be that differential induction of mucosal immunity exists between the throat and nasal compartment. Interestingly, we observed a significant negative correlation between the RBD targeting and neutralizing antibodies in the serum with viral loads in the throat, but not the nasal compartment at day 1 post challenge (Supplemental Figure 4). However, we did not measure the antibody level in either of these URT compartments, which could have provided further evidence of the presence of increased levels of functional antibodies in the throat compared to nasal passage. Another possibility could be that viral control in the nasal compartment where the extensive (5x106 pfu) SARS-CoV-2 challenge dose was directly instilled may be a higher bar than in other mucosal compartments. In support of this, data in the control animals showed nasal swabs yielded higher viral titers than throat swabs, with similar observations being reported in COVID-19 subjects [23].
Importantly, the data indicated that enhanced respiratory disease (ERD) was not associated with INO-4800 immunization in either the 1 dose or 2 dose regimen. In the INO-4800 1 dose group, one animal (10A) did present with the highest lung histopathology score and CT scan score. However, the multifocal lesions in animal 10A showed a similar histopathological pattern as those observed in the animals from the nonvaccinated group, with no apparent influx of different inflammatory cell subpopulations in the infiltrates. A potential hallmark of VED is the increased influx of inflammatory cells such as eosinophils [24], [25]. We believe the CT scan and histopathology data for animal 10A to not be associated with ERD, but rather a disease score and pattern similar to that of nonvaccinated animals. Similar lung histopathology inflammation scores ranging from minimal-mild to mild-moderate were reported in samples analyzed 7 or 8 days after challenge in rhesus macaques receiving other vaccine candidates [26]. Currently, VED remains a theoretical concern with SARS-CoV-2 vaccination and attempts to induce enhanced disease using a formalin inactivated whole virus preparation of SARS-CoV-2 have failed to repeat the lung pathology previously reported for other inactivated respiratory viral vaccines [27].
This data complements an earlier NHP SARS-CoV-2 challenge study which demonstrated reduction in LRT viral loads several months after INO-4800 immunization [14]. However, there are distinct differences between the studies, including different doses and variants used for the challenge stock, and the timing of the challenge. In the study described in this manuscript we challenged the animal four weeks after the last vaccination, at a timepoint when high levels of circulating neutralizing antibodies were present. In the other study, the level of serum SARS-CoV-2 neutralizing antibody was low at the time of challenge, protection appeared to be dependent on the recall of a memory response, with a strong humoral and cellular response against SARS-CoV-2 spike antigen detected in the animals [14]. Here, we did not observe an anamnestic response of a similar magnitude, suggesting protection may have been mediated by the antibodies present in circulation at time of challenge which is supported by the correlation between serum SARS-CoV-2 targeting antibody levels and reductions in viral loads (Supplemental Figure 4). This is in line with reports from other vaccines [26], [28], [29].
Recently, a number of SARS-CoV-2 variants have demonstrated increased transmissibility, potential for increased pathogenicity and escape of vaccine-induced neutralizing antibody responses [30], [31], [32], [33]. We have tested INO-4800-induced antibody and T cell responses against a panel of SARS-CoV-2 variants of concern. Data generated from samples collected from INO-4800 vaccinees indicated the vaccine-induced T cell responses were impervious to epitope changes in the VOC spike antigens, and neutralizing antibody activity was maintained against B.1.1.7 (Alpha) and P.1 (Gamma), but were reduced against the B.1.351 (Beta) variant [34]. In response to the rising threat of VOCs nucleic acid vaccines are in a strong position to respond rapidly. Vaccines matched to the Beta variant are already in clinical testing [35], and in a proactive approach to the likelihood of many more VOCs arising, vaccines designed to provide pan-SARS-CoV-2 coverage are undergoing testing [36], [37]. For example, we have employed SynCon® technology to create a synthetic DNA vaccine based on a single spike immunogen which induces a host immune response which broadly neutralizes a panel of major VOCs [36]. Phase I/II clinical testing is planned to start imminently.
In conclusion, the results here in a stringent preclinical SARS-CoV-2 animal model provide further support for the efficacy and safety of the synthetic DNA vaccine INO-4800 as a prophylactic countermeasure against COVID-19. Importantly, tested as a single dose immunization we observed a positive impact on the lung disease burden and no VED. Together with the clinical data, we believe INO-4800 has many attributes in terms of safety, efficacy and logistical feasibility due its high stability, potentially removing the need for challenging cold chain distribution requirements for global access. Furthermore, synthetic DNA vaccine technology is amenable to highly accelerated developmental timelines, permitting rapid design and testing of candidates against new SARS-CoV-2 variants which display potential for immune escape [38], [39] .
4. Methods
4.1. Vaccine
The optimized DNA sequence encoding SARS-CoV-2 spike was created using Inovio’s proprietary in silico Gene Optimization Algorithm to enhance expression and immunogenicity [12]. INO-4800 spike antigen is homologous to the Wuhan strain. The optimized DNA sequence was synthesized, digested with BamHI and XhoI, and cloned into the expression vector pGX0001 under the control of the human cytomegalovirus immediate-early promoter and a bovine growth hormone polyadenylation signal.
4.2. Animals
Eighteen rhesus macaques of Indian origin (Macaca mulatta) from a UK breeding colony were used in this study. Study groups comprised three males and three females of each species and all were adults aged between 2.5 and 3.5 years of age and weighing > 4 Kg at time of challenge. Prior to the start of the experiment, socially compatible animals were randomly assigned to challenge groups, to minimize bias. Animals were housed in compatible social groups, in cages in accordance with the UK Home Office Code of Practice for the Housing and Care of Animals Bred, Supplied or Used for Scientific Procedures (2014) and National Committee for Refinement, Reduction and Replacement (NC3Rs) Guidelines on Primate Accommodation, Care and Use, August 2006. Housing prior and for the duration of challenge is described in [15]. All experimental work was conducted under the authority of a UK Home Office approved project license that had been subject to local ethical review at PHE Porton Down by the Animal Welfare and Ethical Review Body (AWERB) and approved as required by the Home Office Animals (Scientific Procedures) Act 1986. Animals were sedated by intramuscular (IM) injection with ketamine hydrochloride (Ketaset, 100 mg mL−1, Fort Dodge Animal Health Ltd, Southampton, UK; 10 mg kg−1) for procedures requiring removal from their housing. None of the animals had been used previously for experimental procedures.
Animals received 1 mg of SARS-CoV-2 DNA vaccine, INO-4800, by intradermal injection at day 28 only (1 dose group) or 0 and 28 (2 dose group) followed by an EP treatment using the CELLECTRA 2000® Adaptive Constant Current Electroporation Device with a 3P array (Inovio Pharmaceuticals).
Serum and heparinised whole blood were collected whilst animals were sedated at bi-weekly intervals during the vaccination phase. Nasal and throat swabs were also collected on the day of challenge on D56. After challenge, nasal swabs, throat swabs and serum were collected at 1, 3, 5 dpc and at cull (6, 7 or 8 dpc – staggered due to the high level of labour involved in procedures), with heparinised whole blood collected at 3 dpc and at cull. Nasal and throat swabs were obtained as described [15].
4.3. Clinical observations
Animals were monitored multiple times per day for behavioral and clinical changes. Behavior was evaluated for contra-indicators including depression, withdrawal from the group, aggression, changes in feeding patterns, breathing pattern, respiration rate and cough. Animals were observed and scored as follows for activity and health throughout the study. Key: Activity Level: A0 = Active & Alert; A1 = Only active when stimulated by operator; A2 = Inactive even when stimulated/Immobile; H = Healthy; S = Sneeze, C = Cough, Nd = Nasal Discharge, Od = Ocular Discharge, Rn = Respiratory Noises, Lb = Laboured breathing, L = Lethargy, Di = Diarrhoea, Ax = Loss of Appetite, Dx = Dehydration, RD = Respiratory Distress. Animal body weight, temperature and haemoglobin levels were measured and recorded throughout the study.
4.4. Viruses and cells
SARS-CoV-2 Victoria/01/2020[40] was generously provided by The Doherty Institute, Melbourne, Australia at P1 after primary growth in Vero/hSLAM cells and subsequently passaged twice at PHE Porton Down in Vero/hSLAM cells [ECACC 04091501]. Infection of cells was with ~ 0.0005 MOI of virus and harvested at day 4 by dissociation of the remaining attached cells by gentle rocking with sterile 5 mm borosilicate beads followed by clarification by centrifugation at 1,000 × g for 10 mins. Whole genome sequencing was performed, on the P3 challenge stock, using both Nanopore and Illumina as described previously [41]. Virus titre of the challenge stocks was determined by plaque assay on Vero/E6 cells [ECACC 85020206]. Cell lines were obtained from the European Collection of Authenticated Cell Cultures (ECACC) PHE, Porton Down, UK. Cell cultures were maintained at 37 °C in Minimum essential medium (MEM) (Life Technologies, California, USA) supplemented with 10% fetal bovine serum (FBS) (Sigma, Dorset, UK) and 25 mM HEPES (Life Technologies, California, USA). In addition, Vero/hSLAM cultures were supplemented with 0.4 mg mL−1 of geneticin (Invitrogen) to maintain the expression plasmid. Challenge substance dilutions were conducted in phosphate buffer saline (PBS). Inoculum (5 × 106 PFU) was delivered by intratracheal route (2 ml) and intranasal instillation (1.0 ml total, 0.5 ml per nostril).
4.5. Clinical signs and in-life imaging by computerised tomography
CT scans were performed two weeks before and five days after challenge with SARS-CoV2. CT imaging was performed on sedated animals using a 16 slice Lightspeed CT scanner (General Electric Healthcare, Milwaukee, WI, USA) in both the prone and supine position and scans evaluated by a medical radiologist expert in respiratory diseases (as described previously [15]). To provide the power to discriminate differences between individual NHP’s with low disease volume (i.e. < 25% lung involvement), a refined score system was designed in which scores were attributed for possession of abnormal features characteristic of COVID in human patients (COVID pattern score) and for the distribution of features through the lung (Zone score). The COVID pattern score was calculated as sum of scores assigned for the number of nodules identified, and the possession and extent of GGO and consolidation according to the following system: Nodule(s): Score 1 for 1, 2 for 2 or 3, 3 for 4 or more; GGO: each affected area was attributed with a score according to the following: Score 1 if area measured < 1 cm, 2 if 1 to 2 cm, 3 if 2–3 cm, 4 if > 3 cm and scores for each area of GGO were summed to provide a total GGO score; Consolidation: each affected area was attributed with a score according to the following: 1 if area measured < 1 cm, 2 if 1 to 2 cm, 3 if 2–3 cm, 4 if > 3 cm. Scores for each area of consolidation are summed to provide a total consolidation score. To account for estimated additional disease impact on the host of consolidation compared to GGO, the score system was weighted by doubling the score assigned for consolidation. To determine the zone score, the lung was divided into 12 zones and each side of the lung divided (from top to bottom) into three zones: the upper zone (above the carina), the middle zone (from the carina to the inferior pulmonary vein), and the lower zone (below the inferior pulmonary vein). Each zone was further divided into two areas: the anterior area (the area before the vertical line of the midpoint of the diaphragm in the sagittal position) and the posterior area (the area after the vertical line of the mid-point of the diaphragm in the sagittal position). This results in 12 zones in total where a score of one is attributed to each zone containing structural changes. The COVID pattern score and the zone are summed to provide the Total CT score.
4.6. Post-mortem examination and histopathology
Animals were euthanized at 3 different time-points, in groups of six (one male and one female from each group) at 6, 7 and 8 dpc. The bronchial alveolar lavage fluid (BAL) was collected at necropsy from the right lung. The left lung was dissected prior to BAL collection and used for subsequent histopathology and virology procedures. At necropsy nasal and throat swabs, heparinised whole blood and serum were taken alongside tissue samples for histopathology. Samples from the left cranial and left caudal lung lobe together with spleen, kidney, liver, mediastinal and axillary lymph nodes, small intestine (duodenum), large intestine (colon), trachea, larynx inoculation site and draining lymph node, were fixed by immersion in 10% neutral-buffered formalin and processed routinely into paraffin wax. Four µm sections were cut and stained with hematoxylin and eosin (H&E) and examined microscopically. A lung histopathology scoring system [15] was used to evaluate lesions affecting the airways and the parenchyma. Three tissue sections from each left lung lobe were used to evaluate the lung histopathology. In addition, samples were stained using the RNAscope technique to identify the SARS-CoV-2 virus RNA in lung tissue sections. Briefly, tissues were pre-treated with hydrogen peroxide for 10 mins (RT), target retrieval for 15 mins (98–102 °C) and protease plus for 30 mins (40 °C) (Advanced Cell Diagnostics). A V-nCoV2019-S probe (SARS-CoV-2 Spike gene specific) was incubated on the tissues for two hours at 40 °C. In addition, samples were stained using the RNAscope technique to identify the SARS-CoV-2 virus RNA. Amplification of the signal was carried out following the RNAscope protocol using the RNAscope 2.5 HD Detection kit – Red (Advanced Cell Diagnostics, Biotechne). All H&E and ISH stained slides were digitally scanned using a Panoramic 3D-Histech scanner and viewed using CaseViewer v2.4 software. The presence of viral RNA by ISH was evaluated using the whole lung tissue section slides. Digital image analysis was performed in RNAscope labelled slides to ascertain the percentage of stained cells within the lesions, by using the Nikon-NIS-Ar software package.
4.7. Viral load quantification by RT-qPCR
RNA was isolated from nasal swabs and throat swabs. Samples were inactivated in AVL (Qiagen) and ethanol. Downstream extraction was then performed using the BioSprint™96 One-For-All vet kit (Indical) and Kingfisher Flex platform as per manufacturer’s instructions. Tissues were homogenized in Buffer RLT + betamercaptoethanol (Qiagen). Tissue homogenate was then centrifuged through a QIAshredder homogenizer (Qiagen) and supplemented with ethanol as per manufacturer’s instructions. Downstream extraction from tissue samples was then performed using the BioSprint™96 One-For-All vet kit (Indical) and Kingfisher Flex platform as per manufacturer’s instructions.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) targeting a region of the SARS-CoV-2 nucleocapsid (N) gene was used to determine viral loads and was performed using TaqPath™ 1-Step RT-qPCR Master Mix, CG (Applied Biosystems™), 2019-nCoV CDC RUO Kit (Integrated DNA Technologies) and QuantStudio™ 7 Flex Real-Time PCR System. Sequences of the N1 primers and probe were: 2019-nCoV_N1-forward, 5′ GACCCCAAAATCAGCGAAAT 3′; 2019-nCoV_N1-reverse, 5′ TCTGGTTACTGCCAGTTGAATCTG 3′; 2019-nCoV_N1-probe, 5′ FAM-ACCCCGCATTACGTTTGGTGGACC-BHQ1 3′. The cycling conditions were: 25 °C for 2 min, 50 °C for 15 min, 95 °C for 2 min, followed by 45 cycles of 95 °C for 3 s, 55 °C for 30 s. The quantification standard was in vitro transcribed RNA of the SARS-CoV-2 N ORF (accession number NC_045512.2) with quantification between 1 and 6 log copies µL−1. Positive swab and fluid samples detected below the limit of quantification (LoQ) of 4.11 log copies mL−1, were assigned the value of 5 copies µL−1, this equates to 3.81 log copies mL−1, whilst undetected samples were assigned the value of < 2.3 copies µL−1, equivalent to the assay’s lower limit of detection (LoD) which equates to 3.47 log copies mL−1. Positive tissue samples detected below the limit of quantification (LoQ) of 4.76 log copies mL−1 were assigned the value of 5 copies µL−1, this equates to 4.46 log copies g−1, whilst undetected samples were assigned the value of < 2.3 copies µL−1, equivalent to the assay’s lower limit of detection (LoD) which equates to 4.76 log copies g−1.
Subgenomic RT-qPCR was performed on the QuantStudio™ 7 Flex Real-Time PCR System using TaqMan™ Fast Virus 1-Step Master Mix (Thermo Fisher Scientific) and oligonucleotides as specified by Wolfel et al.[42], with forward primer, probe and reverse primer at a final concentration of 250 nM, 125 nM and 500 nM respectively. Sequences of the sgE primers and probe were: 2019-nCoV_sgE-forward, 5′ CGATCTCTTGTAGATCTGTTCTC 3′; 2019-nCoV_sgE-reverse, 5′ ATATTGCAGCAGTACGCACACA 3′; 2019-nCoV_sgE-probe, 5′ FAM- ACACTAGCCATCCTTACTGCGCTTCG-BHQ1 3′. Cycling conditions were 50 °C for 10 min, 95 °C for 2 min, followed by 45 cycles of 95 °C for 10 s and 60 °C for 30 s. RT-qPCR amplicons were quantified against an in vitro transcribed RNA standard of the full length SARS-CoV-2 E ORF (accession number NC_045512.2) preceded by the UTR leader sequence and putative E gene transcription regulatory sequence described by Wolfel et al [42]. Positive samples detected below the lower limit of quantification (LLOQ) were assigned the value of 5 copies µL−1, whilst undetected samples were assigned the value of ≤ 0.9 copies µL−1, equivalent to the assays lower limit of detection (LLOD). For nasal swab, throat swab and BAL samples extracted samples this equates to an LLOQ of 4.11 log copies mL−1 and LLOD of 3.06 log copies mL−1. For tissue samples this equates to an LLOQ of 4.76 log copies g−1 and LLOD of 3.71 log copies g−1.
4.8. Plaque reduction neutralisation test
Neutralising virus titres were measured in heat-inactivated (56 °C for 30 min) serum samples. SARS-CoV-2 was diluted to a concentration of 1.4 × 103 pfu mL−1 (70 pfu in 50 µl) and mixed 50:50 in 1% FCS/MEM with doubling serum dilutions from 1:10 to 1:320 in a 96-well V-bottomed plate. The plate was incubated at 37 °C in a humidified box for one hour to allow the antibody in the serum samples to neutralise the virus. The neutralised virus was transferred into the wells of a washed 96-well plate containing virus-susceptible VERO/E6 cells, allowed to adsorb at 37 °C for a further hour, and overlaid with 1% CMC in 10% FCS/MEM. After a 24-hour incubation at 37 °C in a humified box, the plates were formaldehyde-fixed before infected cell foci were detected by immunostaining with anti-SARS-CoV-2 RBD spike protein, a polyclonal rabbit horseradish peroxidase conjugate and TrueBlue Substrate. The percentage reduction of foci in serum compared to virus only control was calculated using SoftMax Pro v7.0.3GxP and neutralization titres (ND50) are reported as the serum dilution that neutralized 50% of the virus foci.
4.9. Antigen binding ELISA
Recombinant SARS-CoV-2 Spike- and RBD-specific IgG responses were determined by ELISA. A full-length trimeric and stabilised version of the SARS-CoV-2 Spike protein was supplied by Lake Pharma (#46328). Recombinant SARS-CoV-2 Receptor-Binding-Domain (319–541) Myc-His was developed and kindly provided by MassBiologics.
High-binding 96-well plates (Nunc Maxisorp, 442404) were coated with 50 µl per well of 2 µg mL−1 Spike trimer (S1 + S2) or RBD in 1x PBS (Gibco) and incubated overnight at 4 °C. The ELISA plates were washed and blocked with 5% Foetal Bovine Serum (FBS, Sigma, F9665) in 1x PBS/0.1% Tween 20 for 1 h at room temperature. Serum collected from animals after vaccination had a starting dilution of 1:50 followed by 8 two-fold serial dilutions. Post-challenge samples were inactivated in 0.5% triton and had a starting dilution of 1:100 followed by 8 three-fold serial dilutions. Serial dilutions were performed in 10% FBS in 1x PBS/0.1% Tween 20. After washing the plates, 50 µl per well of each serum dilution was added to the antigen-coated plate in duplicate and incubated for 2 h at room temperature. Following washing, anti-monkey IgG conjugated to HRP (Invitrogen, PA1-84631) was diluted (1: 10,000) in 10% FBS in 1X PBS containing 0.1% Tween 20 and 100 µl per well was added to the plate. Plates were then incubated for 1 h at room temperature. After washing, 1 mg mL-1O-Phenylenediamine dihydrochloride solution (Sigma P9187) was prepared and 100 µl per well were added. The development was stopped with 50 µl per well 1 M Hydrochloric acid (Fisher Chemical, J/4320/15) and the absorbance at 490 nm was read on a Molecular Devices versamax plate reader using Softmax (version 7.0). Titers were determined using the endpoint titer determination method. For each sample, an endpoint titer is defined as the reciprocal of the highest sample dilution that gives a reading (OD) above the cut-off. The cut-off was determined for each experimental group as the mean OD + 3SD of naïve samples.
4.10. Peripheral blood mononuclear cell isolation and resuscitation
PBMCs were isolated from whole blood anticoagulated with heparin (132 Units per 8 ml blood) (BD Biosciences, Oxford, UK) using standard methods. PBMCs isolated from tissues were stored at − 180 °C. For resuscitation PBMCs were thawed, washed in R10 medium (consisting of RPMI 1640 supplemented with 2 mM L-glutamine, 50 U/ml penicillin- 50 μg mL−1 streptomycin, and 10% heat-inactivated FBS) with 1 U mL−1 of DNase (Sigma), and resuspended in R10 medium and incubated at 37 °C 5% CO2 overnight.
5. Elispot
An IFNγ ELISpot assay was used to estimate the frequency and IFNγ production capacity of SARS-CoV-2-specific T cells in PBMCs using a human/simian IFNγ kit (MabTech, Nacka. Sweden), as described previously [43]. The cells were assayed at 2 × 105 cells per well. Cells were stimulated overnight with SARS-CoV-2 peptide pools spanning the ECD spike protein. Five peptide pools were used, comprising of 15mer peptides, overlapping by 9 amino acids. Phorbol 12-myristate (Sigma) (100 ng mL−1) and ionomycin (CN Biosciences, Nottingham, UK) (1 mg mL−1) were used as a positive control. Results were calculated and reported as spot forming units (SFU) per million cells. All SARS-CoV-2 peptides were assayed in duplicate and media only wells subtracted to give the antigen-specific SFU. ELISPOT plates were analysed using a CTL scanner and software (CTL, Germany) and further analysis carried out using GraphPad Prism (GraphPad Software, USA).
6. Statistics
All statistical analyses were performed using GraphPad Prism 7 or 8 software (La Jolla, CA). These data were considered significant if p < 0.05. The type of statistical analysis performed is detailed in the figure legend. No samples or animals were excluded from the analysis.
CRediT authorship contribution statement
Karen E. Gooch: Data curation, Formal analysis, Writing - original draft, Writing - review & editing. Trevor R.F. Smith: Conceptualization, Writing - original draft, Writing - review & editing. Francisco J. Salguero: Data curation, Formal analysis, Writing - original draft, Writing - review & editing. Susan A. Fotheringham: Data curation, Formal analysis, Writing - review & editing. Robert J. Watson: Data curation, Formal analysis, Writing - review & editing. Mike J. Dennis: Data curation, Formal analysis, Writing - review & editing. Alastair Handley: Data curation Formal analysis, Writing - review & editing. Holly E. Humphries: Data curation, Formal analysis, Writing - review & editing. Stephanie Longet: Data curation, Formal analysis, Writing - review & editing. Tom Tipton: Data curation, Formal analysis, Writing - review & editing. Charlotte Sarfas: Data curation, Formal analysis, Writing - review & editing. Laura Sibley: Data curation, Formal analysis, Writing - review & editing. Gillian S. Slack: Data curation, Formal analysis, Writing - review & editing. Emma Rayner: Data curation, Formal analysis, Writing - review & editing. Kathryn A. Ryan: Data curation, Formal analysis, Writing - review & editing. Katherine Schultheis: Data curation, Formal analysis, Writing - review & editing. Stephanie J. Ramos: Data curation, Formal analysis, Writing - review & editing. Andrew White: Data curation, Formal analysis, Writing - review & editing. Sue Charlton: Conceptualization, Writing - review & editing. Sally A. Sharpe: Data curation, Formal analysis, Writing - original draft, Writing - review & editing. Fergus Gleeson: Data curation, Formal analysis, Writing - review & editing. Laurent M. Humeau: Conceptualization, Writing - review & editing. Yper Hall: Conceptualization, Writing - review & editing. Kate E. Broderick: Conceptualization, Writing - review & editing. Miles W. Carroll: Conceptualization, Writing - review & editing.
Declaration of Competing Interest
TRFS, LMH, SJR, KS and KEB are employees of Inovio Pharmaceuticals and as such receive salary and benefits, including ownership of stock and stock options, from the company. All other authors report there are no conflicting interests.
Acknowledgments
Acknowledgements
The studies described in this manuscript were funded by a grant from the Coalition for Epidemic Preparedness Innovations (CEPI). The authors would like to thank Amy Shurtleff for her expert advice throughout this study. We would also like to thank the David Weiner laboratory at the Wistar Institute for their valuable support of the INO-4800 vaccine program.
We would like to thank Maria Yang and the Inovio plasmid production team for generating the DNA constructs and formulation, Matthew Coultas and the Inovio logistics team for overseeing shipment of the biologics and drug delivery equipment.
Thank you to Lauren Allen, Emily Brunt, Breeze E Cavell, Rebecca Cobb, Konstantinos Gkolfinos, Kerry J Godwin, Jade Gouriet, Richard Hesp, Charlotte Hind, Catherine M. K. Ho, Stephanie Leung, Jennifer Logue, Vanessa Lucas, Adam Mabbutt, Alexandra Morrison, Jemma Patterson, Elizabeth J. Penn, Stephen Thomas, Nadina Wand for assistance with sample processing and analysis, Laura Hunter and Chelsea Kennard for histology analysis and Debbie Harris, Nathan Wiblin and all Biological Investigations Group staff for animal husbandry and assistance with in vivo procedures.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.vaccine.2021.06.057.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.World Health, O., Coronavirus disease 2019 (COVID-19): situation report, 51. 2020, World Health Organization: Geneva.
- 2.Zhu N., et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu F., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pallesen J., et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc Natl Acad Sci. 2017;114(35):E7348–E7357. doi: 10.1073/pnas.1707304114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kirchdoerfer R.N., et al. Pre-fusion structure of a human coronavirus spike protein. Nature. 2016;531(7592):118–121. doi: 10.1038/nature17200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muthumani, K., et al., A synthetic consensus anti–spike protein DNA vaccine induces protective immunity against Middle East respiratory syndrome coronavirus in nonhuman primates. Sci Translational Med 2015. 7(301): p. 301ra132–301ra132. [DOI] [PMC free article] [PubMed]
- 7.Baden L.R., et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N Engl J Med. 2020 doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Polack F.P., et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. New England J Med. 2020;383(27):2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kremsner, P., et al., Phase 1 Assessment of the Safety and Immunogenicity of an mRNA- Lipid Nanoparticle Vaccine Candidate Against SARS-CoV-2 in Human Volunteers. medRxiv, 2020: p. 2020.11.09.20228551.
- 10.de Alwis, R., et al., A Single Dose of Self-Transcribing and Replicating RNA Based SARS-CoV-2 Vaccine Produces Protective Adaptive Immunity In Mice. bioRxiv, 2020: p. 2020.09.03.280446. [DOI] [PMC free article] [PubMed]
- 11.Tebas P., et al. Safety and immunogenicity of INO-4800 DNA vaccine against SARS-CoV-2: A preliminary report of an open-label, Phase 1 clinical trial. EClinicalMedicine. 2021;31 doi: 10.1016/j.eclinm.2020.100689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith T.R.F., et al. Immunogenicity of a DNA vaccine candidate for COVID-19. Nat Commun. 2020;11(1):2601. doi: 10.1038/s41467-020-16505-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gary, E.N., et al., A novel mouse AAV6 hACE2 transduction model of wild-type SARS-CoV-2 infection studied using synDNA immunogensGary, Ebony N. et al. iScience, 2021. Volume 24, Issue 7, 102699. [DOI] [PMC free article] [PubMed]
- 14.Patel, A., et al., Intradermal-delivered DNA vaccine provides anamnestic protection in a rhesus macaque SARS-CoV-2 challenge model. bioRxiv, 2020: p. 2020.07.28.225649.
- 15.Salguero, F.J., White, A.D., Slack, G.S. et al. Comparison of rhesus and cynomolgus macaques as an infection model for COVID-19. Nat Commun 12, 1260 (2021). [DOI] [PMC free article] [PubMed]
- 16.Muñoz-Fontela, C., et al., Animal models for COVID-19. Nature, 2020. 586(7830): p. 509–515. [DOI] [PMC free article] [PubMed]
- 17.Amante D.H., et al. Direct transfection of dendritic cells in the epidermis after plasmid delivery enhanced by surface electroporation. Hum Gene Ther Methods. 2014;25(6):315–316. doi: 10.1089/hgtb.2014.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amante D.H., et al. Skin Transfection Patterns and Expression Kinetics of Electroporation-Enhanced Plasmid Delivery Using the CELLECTRA-3P, a Portable Next-Generation Dermal Electroporation Device. Hum Gene Ther Methods. 2015;26(4):134–146. doi: 10.1089/hgtb.2015.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diehl, M.C., et al., Tolerability of intramuscular and intradermal delivery by CELLECTRA((R)) adaptive constant current electroporation device in healthy volunteers. Hum Vaccin Immunother, 2013. 9(10): p. 2246–52. [DOI] [PMC free article] [PubMed]
- 20.Tebas P., et al. Intradermal SynCon(R) Ebola GP DNA Vaccine Is Temperature Stable and Safely Demonstrates Cellular and Humoral Immunogenicity Advantages in Healthy Volunteers. J Infect Dis. 2019;220(3):400–410. doi: 10.1093/infdis/jiz132. [DOI] [PubMed] [Google Scholar]
- 21.De Rosa, S.C., et al., Robust antibody and cellular responses induced by DNA-only vaccination for HIV. JCI Insight, 2020. 5(13). [DOI] [PMC free article] [PubMed]
- 22.Mammen, M.P., et al., Safety and immunogenicity of INO-4800 DNA vaccine against SARS-CoV-2: a preliminary report of a randomized, blinded, placebo-controlled, Phase 2 clinical trial in adults at high risk of viral exposure. 2021: p. 2021.05.07.21256652.
- 23.Mohammadi A., et al. SARS-CoV-2 detection in different respiratory sites: A systematic review and meta-analysis. EBioMedicine. 2020;59 doi: 10.1016/j.ebiom.2020.102903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bolles M., et al. A double-inactivated severe acute respiratory syndrome coronavirus vaccine provides incomplete protection in mice and induces increased eosinophilic proinflammatory pulmonary response upon challenge. J Virol. 2011;85(23):12201–12215. doi: 10.1128/JVI.06048-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yasui F., et al. Prior Immunization with Severe Acute Respiratory Syndrome (SARS)-Associated Coronavirus (SARS-CoV) Nucleocapsid Protein Causes Severe Pneumonia in Mice Infected with SARS-CoV. J Immunol. 2008;181(9):6337–6348. doi: 10.4049/jimmunol.181.9.6337. [DOI] [PubMed] [Google Scholar]
- 26.Corbett K.S., et al. Evaluation of the mRNA-1273 Vaccine against SARS-CoV-2 in Nonhuman Primates. N Engl J Med. 2020;383(16):1544–1555. doi: 10.1056/NEJMoa2024671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bewley, K.R., et al., Immunological and pathological outcomes of SARS-CoV-2 challenge after formalin-inactivated vaccine immunisation of ferrets and rhesus macaques. 2020: p. 2020.12.21.423746. [DOI] [PMC free article] [PubMed]
- 28.Rauch, S., et al., mRNA vaccine CVnCoV protects non-human primates from SARS-CoV-2 challenge infection. bioRxiv, 2020: p. 2020.12.23.424138.
- 29.Vogel, A., et al., A prefusion SARS-CoV-2 spike RNA vaccine is highly immunogenic and prevents lung infection in non-human primates. bioRxiv, 2020.
- 30.Chen R.E., et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat Med. 2021;27(4):717–726. doi: 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davies N.G., et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021;372(6538) doi: 10.1126/science.abg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davies N.G., et al. Increased mortality in community-tested cases of SARS-CoV-2 lineage B.1.1.7. Nature. 2021;593(7858):270–274. doi: 10.1038/s41586-021-03426-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garcia-Beltran W.F., et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell. 2021;184(9):2523. doi: 10.1016/j.cell.2021.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andrade, V.M., et al., INO-4800 DNA Vaccine Induces Neutralizing Antibodies and T cell Activity Against Global SARS-CoV-2 Variants. 2021: p. 2021.04.14.439719. [DOI] [PMC free article] [PubMed]
- 35.Wu, K., et al., Preliminary Analysis of Safety and Immunogenicity of a SARS-CoV-2 Variant Vaccine Booster. 2021: p. 2021.05.05.21256716.
- 36.Reed, C.C., et al., Design and immunogenicity of a Pan-SARS-CoV-2 synthetic DNA vaccine. 2021: p. 2021.05.11.443592.
- 37.Saunders K.O., et al. Neutralizing antibody vaccine for pandemic and pre-emergent coronaviruses. Nature. 2021 doi: 10.1038/s41586-021-03594-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wibmer, C.K., et al., SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. 2021: p. 2021.01.18.427166. [DOI] [PubMed]
- 39.Moore, J.P, P.A. Offit. SARS-CoV-2 Vaccines and the Growing Threat of Viral Variants. JAMA, 2021. [DOI] [PubMed]
- 40.Caly L., et al. Isolation and rapid sharing of the 2019 novel coronavirus (SARS-CoV-2) from the first patient diagnosed with COVID-19 in Australia. Med J Aust. 2020;212(10):459–462. doi: 10.5694/mja2.50569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lewandowski K., et al. Metagenomic Nanopore Sequencing of Influenza Virus Direct from Clinical Respiratory Samples. J Clin Microbiol. 2019;58(1) doi: 10.1128/JCM.00963-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wolfel R., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(7809):465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 43.Sibley L.S., et al. ELISPOT Refinement Using Spot Morphology for Assessing Host Responses to Tuberculosis. Cells. 2012;1(1):5–14. doi: 10.3390/cells1010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.