Abstract
Though Lake Michigan beaches in Chicago are not impacted by stormwater or wastewater outfalls, several of those beaches often exceed USEPA Beach Action Values (BAVs). We investigated the role of microbial source tracking (MST) as a complement to routine beach monitoring at Chicago beaches. In summer 2016, water samples from nine Chicago beaches were analyzed for E. coli by culture and enterococci by qPCR. A total of 195 archived samples were then tested for human (HF183/BacR287, HumM2), canine (DG3, DG37), and avian (GFD) microbial source tracking (MST) markers. Associations between MST and general fecal indicator bacteria (FIB) measures were evaluated and stratified based on wet and dry weather definitions. Among the 195 samples, HF183/BacR287 was quantifiable in 4%, HumM2 in 1%, DG3 in 6%, DG37 in 2%, and GFD in 23%. The one beach with a dog area was far more likely to have DG3 present in the quantifiable range than other beaches. Exceedance of general FIB BAVs increased the odds of human, dog and avian marker detection. MST marker weighted-average fecal scores for DG3 was 2.4 times, DG37 was 2.1 times, and GFD was 1.6 times higher during wet compared to dry weather conditions. HF183/BacR287 weighted-average fecal scores were not associated with precipitation. Associations between FIB BAV exceedance and MST marker detection were generally stronger in wet weather. Incorporating MST testing into routine beach water monitoring can provide information that beach managers can use when developing protection plans for beaches not impacted by point sources.
Keywords: Recreational water quality, fecal indicator bacteria, microbial source tracking, precipitation, non-point source pollution, qPCR
1. Introduction:
General fecal indicator bacteria (FIB), such as E. coli and enterococci, are widely used for routine beach water quality monitoring (United States Environmental Protection Agency, 2012). A limitation of this approach is that general FIB found in beach water can come from the fecal matter of a variety of host animals, such as humans, birds, dogs, ruminants and other animals (Korajkic, McMinn, & Harwood, 2018). Furthermore, general FIB identified through water testing does not necessarily indicate fecal pollution, as naturalized populations of general FIB can proliferate in beach wracks, submerged aquatic vegetation, sediments and sands of marine and freshwater beaches (Badgley et al., 2010; Byappanahalli & Fujioka, 2004; Byappanahalli et al., 2006; Imamura et al., 2011; Mathai et al., 2019; Nevers et al., 2016; Whitman & Nevers, 2003).
A clear understanding of the sources of general FIB in beach water can be useful in developing targeted mitigation strategies, particularly in settings without an obvious source of microbial pollution. In the past decade, microbial sources tracking (MST) methods have been developed to distinguish between sources of fecal waste by targeting DNA sequences or gene fragments of fecal indicator bacteria that vary among different animal hosts (at the genus or species level). Source-tracking technologies targeting human, canine, avian and other agricultural and wildlife wastes have been used to identify sources of fecal contamination within various water systems including fresh and marine recreational water around the United States (Harwood et al., 2014). MST methods have been useful in identifying avian fecal pollution as a problem at Great Lakes beaches, and in demonstrating benefits of interventions to reduce bird presence (and bird fecal pollution) at beaches (Converse et al., 2012; Kinzelman et al., 2008; Nevers et al., 2018). Characterizing the source(s) of general FIB might also be useful for estimating human health risk. Epidemiologic studies conducted at US marine and freshwater beaches have found that the risks of gastrointestinal illness among bathers increased in relation to general FIB levels (Wade et al., 2008, 2010). However, those studies were conducted at beaches thought to be impacted by wastewater discharges. Similar associations between general FIB and illness were not apparent at beaches with little human fecal pollution (Colford et al., 2007). At beaches with intermittent or seasonal human fecal pollution impacts, the FIB-health risk association has been observed only during periods in which human fecal pollution is thought to be present (Benjamin-Chung et al., 2017; Yau et al., 2014). Thus, sources of indicator bacteria may be important modifiers of associations between general FIB and human health risk. However, this is far from settled, as in a large epidemiologic study of swimming at beaches, markers of human fecal pollution were at best inconsistently (and in some cases, inversely) associated with human health risk (Napier et al., 2017, 2018).
Precipitation and the resulting flow of stormwater, either through outfalls or sheet flow across beaches, transport microbes to near-shore beach water. Precipitation is generally followed by elevated concentrations of E. coli (Ackerman & Weisberg, 2003; Dwight et al., 2011; Kirs et al., 2017; Kleinheinz et al., 2009; McLellan et al., 2007; Nevers & Whitman, 2011) and somewhat less consistently, of enterococci (Cordero et al., 2012; Jennings et al., 2018; Laureano-Rosario et al., 2017; Staley et al., 2013). Our understanding of the relationships between sources of fecal pollution, general FIB, and precipitation comes largely from beaches impacted by discharges from storm drains, combined sewer outfalls, stormwater channels, and/or wastewater treatment facilities at marine (Steele et al., 2018) and at Great Lakes beaches (Haack et al., 2013; Nevers et al., 2018; Staley & Edge, 2016). Other studies of pollution sources and precipitation have been conducted within the catchments of river systems (Brooks et al., 2019; Li et al., 2019; Riedel et al., 2015). In settings where fecal pollution is released from discrete points of discharge, localized control or treatment of those discharges could potentially improve water quality in wet weather. However, for beaches not impacted by storm or septic discharges, understanding the relationships between general FIB, MST markers, and precipitation should be helpful in estimating health risk and developing targeted wet weather and dry weather efforts to prevent fecal pollution from different sources from reaching bathing waters.
Despite progress made in identifying sensitive and specific MST methods, and the success of applying these to identify pollutant sources in particular locations, the role of MST testing as a complement to routine beach monitoring has yet to be evaluated. Lake Michigan beaches in Chicago present a simplified scenario to evaluate non-point source bacterial pollution sources at beaches. This is because stormwater and wastewater are discharged into the Chicago River, which has been engineered to flow away from Lake Michigan, diverting pollution away from the Lake and its beaches. As a result, Chicago beaches should not be impacted by wastewater discharges. Nevertheless, Beach Action Values (BAVs), for both enterococci qPCR and E. coli culture, are exceeded with some regularity at several of Chicago’s Lake Michigan beaches (Dorevitch, et al., 2017; Nevers & Whitman, 2011). Whether these exceedances reflect human fecal contamination (which would be unexpected) or other sources of fecal pollution, is still unknown. The incorporation of MST testing into routine beach monitoring in Chicago should be an opportunity to answer several questions:
Are host-associated genetic markers for human fecal pollution present at beaches thought to have no point sources of human fecal pollution?
Are the presence and/or concentration of human, dog, and bird MST markers associated with general FIB measurements generated for routine beach monitoring? If so, do the associations vary depending on the choice of general FIB measure?
Are the presence and/or concentration of human, dog, and bird MST markers associated with precipitation? Does precipitation modify associations between the detection of MST markers and general FIB measurements at these non-point source impacted beaches?
What is the potential value of incorporating MST testing into routine beach monitoring at non-point source impacted beaches?
2. Material and Methods:
2.1. Site description and sample collection
Chicago has 42 km of lakeshore with 27 public beaches (“Chicago Park District,” 2020). During the summer of 2016, beach monitoring using enterococci quantitative polymerase chain reaction (qPCR) was conducted five days a week (Wednesday-Sunday) at nine Chicago beaches (Table A.1 and Figure A.1). Water samples were collected in 1L polypropylene copolymer bottles at each of two transects, approximately 100 yards (roughly 92 meters) apart, at each beach. The sampling depth for samples was about knee deep (at least 6 inches below the water surface). At the same times and locations of sampling for qPCR analyses, water samples were collected for E. coli culture analyses (Wednesday-Friday). Samples were transported on ice to the University of Illinois at Chicago School of Public Health Water Microbiology Research Laboratory within approximately 1.5 hours of collection for immediate enterococci qPCR analysis as described in detail in Dorevitch et al., 2017. Water turbidity was measured in the laboratory using a HF Scientific MicroTPW (HF Scientific, Fort Myers, FL) turbidity meter, which was calibrated daily.
2.2. E. coli culture (cEC)
E. coli culture analyses were performed by STAT Analysis Corporation (STAT) laboratory (Chicago, IL) using the defined substrate test, Colilert® (IDEXX Laboratories, Westbrook, ME). The upper limit of quantification (uLOQ) for this method is 2,419 most probable number (MPN)/100 mL. Results > uLOQ were assigned the value of 2,420 MPN/ 100 mL for data analysis.
2.3. Enterococci qPCR (qENT) analysis
The procedures outlined in USEPA Method 1609.1 (USEPA, 2015) were followed for the filtration, extraction, amplification, and quantification of enterococci DNA as described in Dorevitch et al., 2017. The procedure is briefly described below. From each beach water sample, 100 mL of water was filtered through 0.4 mm pore size 47 mm diameter polycarbonate filters (MilliporeSigma, Burlington, MA). Filters were folded and placed in a 2-mL extraction tubes containing 0.3 g of acid-washed glass beads (Generite, LLC, North Brunswick, NJ). A total of 600 mL 0.2 mg/L single stranded salmon testes DNA (SSDNA) (Sigma-Aldrich, St. Louis, MO) was added to each extraction tube. Genomic DNA was extracted by bead beating for 60s at 5000 rpm. Tubes were subsequently centrifuged at 12,000 × g for 1 min. Supernatants were transferred to sterile 1.5 mL low-retention microcentrifuge tubes (Sarstedt, Inc., Newton, NC), which were centrifuged at 12,000 × g for 5 min. Genomic DNA (in the supernatant) was transferred to a sterile 1.5 mL low- retention microcentrifuge tube and analyzed immediately. In addition to the filters that were analyzed immediately, additional sets of filters from each beach water samples were archived in sterile, pre-loaded glass bead tubes (GeneRite, LLC, North Brunswick, NJ) at −80°C for future MST analyses (<6 months).
Undiluted 5 μL of final genomic DNA extracts were added to 20 μL of reagents, in duplicate. All reactions were performed on the Applied Biosystems StepOnePlus Real-Time PCR platform (Applied Biosystems, Foster City, CA) specified elsewhere (USEPA, 2015). Additional details can be found in Table A.3. For the quantification of enterococci DNA, the comparative cycle threshold (ΔΔCt) method as described in Method 1609.1 was used. Results for qENT were reported as calibrator cell equivalents (CCE)/ 100 mL.
2.4. Sample selection for MST marker analysis
A total of 195 samples from the set of archived filters described above were selected for MST testing based on their general FIB levels (cEC and qENT). The primary goal of sample selection for MST analysis was to efficiently contrast the likelihood of marker presence at beach-days (number of beaches multiplied by the number of days of testing) when general FIB levels were relatively high versus relatively low. For that reason, rather than analyzing a random sample of archived water samples (filters), we stratified sample selection so that A) approximately 50% were from beach-days in which qENT CCE >320/100mL (50% of the USEPA’s BAV meant to limit recreational waterborne illness to 32 cases/1000 bathers); B) approximately 20% were from beach-days in which cEC MPN>160/100mL (50% of the USEPA’s statistical threshold value (STV) meant to limit recreational waterborne illness to 32 cases/1000 bathers) and qENT was <320 CCE/100mL; and C) approximately 30% of beach-days with the lowest qENT (<320 CCE/100mL) and cEC values (<160 MPN/100mL). No samples from two beaches, North Avenue and South Shore, were used in the MST analysis as no general FIB BAV exceedances occurred at these two sites. Of the 195 selected samples analyzed for MST markers, 170 samples had corresponding cEC results (qPCR analysis was done Wednesday-Sunday while E. coli culture testing was done Wednesday-Friday).
2.5. qPCR analysis for MST markers
Two canine markers, DG3 and DG37 (Green, White, et al., 2014), one general avian marker, GFD (Green et al., 2012), and two human-associated markers, HF183/BacR287 (Green et al., 2014; USEPA, 2019a) and HumM2 (Shanks et al., 2009; USEPA, 2019b), were used. Genomic DNA extracted from the archived samples was analyzed using methods described previously (Kelty et al., 2012; Shanks et al., 2016). Three method extraction blanks (MeBs), with purified water substituted for test sample, were performed with each sample processing batch (38 samples/batch). DNA extracts from filters were stored at 4°C in 1.5 mL low-retention microcentrifuge tubes (Sarstedt, Inc., Newton, NC) until the time of analysis (<24 hours).
Two microliters of purified genomic DNA extracted from the archived samples was added to 23 μL of PCR master mix (25 μL total volume). The qPCR master mix included 1X TaqMan Environmental Master Mix (Version 2.0; Thermo Fisher Scientific, Grand Island, NY), 0.1X SYBR Green I dye (GFD assay only; Thermo Fisher Scientific, Grand Island, NY), 1 μM each primer, 80 nM 6-carboxyfluorescein (FAM)-labeled probe, and 80 nM VIC-labeled probe (HF183/BacR287 and HumM2 multiplex reactions only), and 0.2 mg/mL bovine serum albumin (Sigma-Aldrich, St. Louis, MO).
Moreover, the PCR master mix for HF183/BacR287 and HumM2 also included 2 μL of internal amplification control (IAC) plasmid to test for amplification inhibition (Li et al., 2019). MST analyses were performed at the USEPA Center for Environmental Measurement and Modeling (Cincinnati, OH) on the Applied Biosystems QuantStudio 3 Real-Time PCR System (Thermo Fisher Scientific, Waltham, MA). Information on primers, probes and thermal cycling settings for all MST markers are summarized in Table A.2 and Table A.3. All reactions were performed in triplicate. Six replicate reactions of no template control (NTC) were included in every MST qPCR instrument run to screen for potential contamination. The MST marker concentrations were estimated using data acceptance metrics previously described in Shanks et al., 2016. Briefly, data acceptance metrics included using multiplex IAC procedures for human-associated genetic markers in order to check qPCR amplification inhibition. Poor DNA recovery from water sample filters was monitored by means of a sample processing control (SPC) protocol using Sketa22 assay as described in Li et al., 2019. However, we made one modification: the Sketa22 qPCR SPC proficiency acceptance threshold was changed to ≤ 0.71 quantification cycle (Cq) standard deviation rather than ≤.0.62 Cq in order to include data from all instrument runs. The lower limit of quantification (LLOQ) threshold for each MST assay was defined as the upper bound of the 95% credible interval corresponding to the respective master standard curve model at 10 copies/reaction. Each standard curve included triplicate reactions of serially diluted plasmid standards in the following range of concentrations: 101, 102, 103, 104 and 105 copies/reaction. Standard curves generated from six independent instrument runs were used to calculate master calibration models for each host-associated qPCR assay. The master calibration curves and estimates of sample concentration were determined using a Bayesian Monte Carlo Markov Chain approach that incorporates within and between run variability (Sivaganesan et al., 2008).
2.6. Precipitation data
Precipitation data were downloaded from the National Weather Service (Midway Airport, Station 14819) and applied to all beaches (which are between 16–27 km from the weather station). Based on the hourly rainfall data, the total precipitation amount (mm) during the 24 hours preceding sample collection was calculated, assuming each sample was taken at 8:00 a.m. every day. For assessing the effect of precipitation, samples were categorized into three categories: wet, intermediate and dry weather samples. Two or more millimeter of rainfall within the 24-hours prior to sample collection was classified as “wet” weather. Rainfall between 0–2 mm in preceding 24 hours of sample collection was categorized as “intermediate” weather. Finally, no precipitation in prior 24 hours of sample collection was categorized as “dry” weather.
2.7. Reference fecal sample analysis
In order to verify host-association of genetic markers in reference samples from the Chicago area, dog (N=21), gull (N=5) and goose (N=5) fecal samples were collected as previously described (Shanks et al., 2010). Primary effluent sewage sample from a local wastewater treatment plant (N=1) that uses secondary treatment but no disinfection was used as a surrogate for human fecal reference. A standardized concentration (1 ng/reaction) was tested in triplicates for each sample and assay combination as previously reported (Kelty et al., 2012). Reference sample DNA extractions were performed using a DNA-EZ RW02 kit (GeneRite, LLC, North Brunswick, NJ) according to the manufacturer’s instructions as previously described (Green et al., 2014; Kelty et al., 2012).
2.8. Data analysis
Data analyses were conducted using SAS software for Windows, version 9.4 (SAS Institute, Cary, NC) unless noted otherwise. For each MST qPCR assay, sample results were categorized as non-detect (ND) and detect (D). The detect results were further characterized as detect but non-quantifiable (DNQ) or quantifiable (Q). ND for a given assay was defined as two or more Cq values (among three triplicates) were not detected after 40 cycles of amplification. A DNQ occurred when two or more replicate reactions had Cq values <LLOQ. If all replicate reactions had Cq values ≤ LLOQ were categorized as Q. Quantifiable MST results were reported as log10 copies per reaction. Descriptive statistics of the MST results were summarized as percentage of ND, DNQ and Q for each MST marker tested. The normality of distribution of turbidity, cEC, and qENT results were determined by Kolmogorov-Smirnov tests. Since none of those measures were normally distributed, data were log10 transformed. All results and relationships were considered significant at alpha (α) <0.05.
Logistic regression assessed the relationship between the D versus ND of the MST markers in selected sample filters with the general FIB concentrations (expressed as log10 E. coli MPN/100 mL and log10 enterococci CCE/100 mL). Additionally, agreement between exceedance of BAVs (cEC or qENT) and detection of MST markers were characterized using Cohen’s Kappa statistic which accounts for the expected agreement due to chance alone. The Kappa statistic for agreement were interpreted as following: 0–0.20, none; 0.21–0.39, minimal; 0.40–0.59, weak; 0.60–0.79, moderate; 0.80–0.89, strong; ≥0.90, near perfect (McHugh, 2012).
A qPCR censored-data method as described in Cao et al., 2018 was utilized to generate weighted-average fecal scores (log10 copies per 100 mL) from a series of samples grouped by either FIB BAV definition or cumulative precipitation 24 hours prior to sampling for each eligible MST marker and beach combination. Briefly, prior to calculating a weighted-average fecal score, the mean Cq of the MST markers for each sample (no amplification was set to 40 Cq) was classified into two groups: a range of quantification (ROQ) group if mean MST Cq ≤ LLOQ or an MST MPN group (if respective mean MST Cq > LLOQ). After the samples were classified into either the ROQ group or the MST MPN group, weighted-average fecal scores (which utilizes all data, including non-detects) for each MST assays were calculated as described elsewhere (Cao et al., 2018). The weighted-average fecal scores for the MST assays can be considered as an estimate of the level of fecal contamination from a particular source (dog, bird or human) present at a given FIB BAV grouping or precipitation category based on the average concentration of the source-specific gene observed in all the water samples in the study. For BAV grouping, we evaluated whether fecal scores differed among ordinal categories of FIB concentrations: Group 1: FIB <10% percent of the BAV, Group 2: FIB ≥ 10% of the BAV to FIB < BAV, and Group 3: FIB ≥ BAV. These comparisons were made for each FIB BAV definition (for cEC BAV= 235 MPN/100mL and for qENT BAV= 1000 CC-E/ 100mL). For precipitation grouping, fecal scores were determined based on two categories including: Group 1: ≥ 2 mm cumulative rainfall in past 24 hours (wet) and Group 2: No precipitation in past 24 hours (dry).
Since there was only one “intermediate” weather day with 1.02 mm of rainfall in the past 24 hours, it was combined with “dry” weather day for analysis purposes. Only sample groupings composed of ≥ 10 samples with a minimum of at least one sample in the ROQ group and at least one D across all MST MPN group samples were eligible for fecal score determination.
For categorical data analyses, such as the association between the D/ND of MST markers in samples collected under dry and wet weather chi-square tests were performed, generating odds ratios (OR). Finally, analyses of association were also conducted for the dataset overall, stratified by precipitation category. Evaluation of modification of general FIB BAV exceedance and MST marker presence stratified by weather conditions were performed using Cochran-Mantel-Haenszel (CMH) tests (weather-condition specific ORs and interaction terms).
3. Results:
3.1. Data quality
All quality control (QC) requirements for the USEPA Method 1609.1 were met or exceeded. The linearity of the standard curves for enterococci and all the five MST markers tested was high (R2 > 0.991 for all assays). Amplification efficiencies for MST assays ranged from 92.3% to 96.5% and LLOQ values ranged from 35.09 to 37.35 Cq based on repeated measures from six instrument runs. Calibration model performance parameter information from the pooled standard curves for individual assays are summarized in Table A.4. Out of the 780 NTC and MeB total reactions, 99.5% were DNA-free (N = 4 false positives) suggesting minimal DNA contamination (Table A.5). A total of 71 sets of enterococci calibrator samples (positive controls) were analyzed throughout the 2016 beach season for enterococci qPCR. Precision was high based on the low coefficients of variation (CV) for the SPC and for the enterococci calibrator cells (Table A.6). No amplification inhibition was observed in any of the 195 archived filters’ DNA extracts (Table A.7).
3.2. Reference fecal sample results
Human-associated (HF183/BacR287 and HumM2) genetic markers were detected in all replicates of the sewage sample but were not detected in any dog or goose fecal samples. Bird-associated (GFD) genetic marker was detected in 50% of goose sample reactions, with no false positives observed in dog fecal or sewage samples. Poor DNA recovery from gull fecal samples prevented the testing of local reference samples for the GFD marker, though previous studies report the presence of the GFD marker in 90% to 100% of gull fecal samples from the US Midwest region (Green et al., 2012). Dog-associated genetic markers were detected in 41.3% (DG3) to 76.2% (DG37) of the reactions of dog fecal samples, with no false positives observed in goose samples. Both DG3 (66.7%) and DG37 (33.3%) were detected in the primary effluent sample reactions.
3.3. MST results and their associations with general FIB measures
Among the 195 samples, 95 (48.7%) were in the relatively high qENT category (qENT CCE >320/100mL), 42 samples (21.5 %) had high cEC (cEC MPN>160/100mL) but low qENT (qENT CCE <320/100mL), and 58 samples (29.7%) had both cEC <160 MPN/100mL and qENT<320 CCE/100mL. The MST marker detected most frequently was GFD (bird), in 40% of samples. Dog marker (DG3) was detected in 14% of samples, while human marker (HF183/BacR287) was detected in 10% of samples (Table 1). Host-associated genetic markers were in the quantifiable (Q) range in 4% (n=8) of samples for HF183/BacR287, 1% (n=3) for HumM2, 6% (n=12) for DG3, 2% (n=4) for DG37 and 23% (n=45) of samples for GFD. Among the human markers, HF183/BacR287 was detected more frequently, though in two samples HumM2 was detected when HF183/BacR287 was not. Considerable variability was observed in marker detection by beach (Table 1). The human markers were detected at six of the seven beaches and they were detected and quantifiable on ten different sampling dates, with no more than one of the seven beaches testing positive on any single day. The dog marker, DG3, was quantifiable exclusively at one beach, Montrose (MN), which is the only beach with a designated “dog beach” area, where dogs are allowed at the beach and into the water. At that beach, mean concentrations of DG3 and DG37 among quantifiable samples ranged from 1.3 to 2.7 log10 copies per reaction and 1.1 to 1.4 log10 copies per reaction respectively.
Table 1:
Results of MST qPCR analysis, by beach (N=195)
| Beach | N | DG3 N (%) | DG37 N (%) | GFD N (%) | HF183/BacR287 N (%) | HumM2 N (%) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ND | DNQ | Q | ND | DNQ | Q | ND | DNQ | Q | ND | DNQ | Q | ND | DNQ | Q | ||
| MN | 28 | 6 (21) | 10 (36) | 12 (43) | 22 (78) | 3 (11) | 3 (11) | 15 (53) | 5 (18) | 8 (29) | 22 (78) | 3 (11) | 3 (11) | 25 (89) | 2 (7) | 1 (4) |
| OS | 30 | 29 (97) | 1 (3) | 0 (0) | 29 (97) | 0 (0) | 1 (3) | 19 (63) | 6 (20) | 5 (17) | 25 (83) | 3 (10) | 2 (7) | 29 (97) | 0 (0) | 1 (3) |
| TS | 22 | 22 (100) | 0 (0) | 0 (0) | 22 (100) | 0 (0) | 0 (0) | 11 (50) | 8 (36) | 3 (14) | 21 (95) | 0 (0) | 1 (5) | 22 (100) | 0 (0) | 0 (0) |
| MB | 28 | 26 (93) | 2 (7) | 0 (0) | 28 (100) | 0 (0) | 0 (0) | 16 (57) | 5 (18) | 7 (25) | 28 (100) | 0 (0) | 0 (0) | 28 (100) | 0 (0) | 0 (0) |
| ST | 31 | 30 (97) | 1 (3) | 0 (0) | 31 (100) | 0 (0) | 0 (0) | 18 (58) | 4 (13) | 9 (29) | 30 (97) | 1 (3) | 0 (0) | 29 (94) | 1 (3) | 1 (3) |
| RB | 27 | 26 (96) | 1 (4) | 0 (0) | 27 (100) | 0 (0) | 0 (0) | 20 (74) | 4 (15) | 3 (11) | 26 (96) | 1 (4) | 0 (0) | 27 (100) | 0 (0) | 0 (0) |
| CL | 29 | 29 (100) | 0 (0) | 0 (0) | 29 (100) | 0 (0) | 0 (0) | 17 (59) | 2 (7) | 10 (34) | 24 (83) | 3 (10) | 2 (7) | 27 (93) | 2 (7) | 0 (0) |
| All Sites | 195 | 168 (86) | 15 (8) | 12 (6) | 188 (96) | 3 (2) | 4 (2) | 116 (60) | 34 (17) | 45 (23) | 176 (90) | 11 (6) | 8 (4) | 187 (96) | 5 (3) | 3 (1) |
Note: ND = Non-detect.; DNQ = Detect but not quantifiable; Q = Quantifiable.
Detection of the two human markers and the dog marker, DG37, were not frequent enough to model by beach. Beach-specific logistic regression models demonstrated that general FIB concentrations (on a log10 scale) predicted the presence of avian (GFD) and dog (DG3) markers (Table 2). The odds of detecting the GFD marker more than doubled for a log10 increase in the concentrations of enterococci CCE [OR (95% CI) =2.3 (1.5, 3.4)] and E. coli MPN [OR (95% CI) =2.2 (1.5, 3.3)]. The odds of detecting the DG3 marker also increased with log10 concentrations of enterococci CCE [OR (95% CI) =2.3 (1.3, 4.1)]. Beach-specific regression analysis showed that the relationships between general FIB and the odds of MST marker detection varied by beach. Both log10 concentrations of qENT and cEC results showed similar associations with MST marker presence (Table 2), with one notable exception: at one beach, MN, GFD was strongly associated with qENT but not with cEC [OR (95% CI) = 92.9 (2.5- >999) and 4.8 (0.6–36.2), respectively]. After accounting for chance alone, the agreement between DG3 detection and qENT BAV exceedance was minimal (Kappa=0.22); for all other combinations of maker detection and FIB BAV exceedance (either qENT or cEC), there was no agreement beyond what would be expected due to chance alone (Kappa <0.2). Associations between water turbidity and MST marker detection did not reach statistical significance, and are summarized in Supplementary Information Table A.8.
Table 2:
Logistic regression: Association between MST presence and log10 FIB concentrations
| Beach | General FIB- qENT | General FIB- cEC | ||||
|---|---|---|---|---|---|---|
| N | GFD-OR (95 % CI) | DG3-OR (95 % CI) | N | GFD-OR (95 % CI) | DG3-OR (95 % CI) | |
| MN | 28 | 92.9* (2.5- >999) | 2.4 (0.7–8.5) | 22 | 4.8 (0.6–36.2) | 0.4 (0.03–3.9) |
| OS | 30 | 2.8* (1.2–6.7) | 8.7 (0.1–866.4) | 26 | 7.7* (1.5–40.4) | 7.8 (0.4–159.6) |
| TS | 22 | 2.7 (0.7–10.5) | NA | 19 | 2.7 (0.8–9.7) | NA |
| MB | 28 | 1.2 (0.5–2.9) | 3.0 (0.3–28.4) | 24 | 2.6 (0.95–7.2) | 2.4 (0.4–16.6) |
| ST | 31 | 1.4 (0.5–4.3) | 0.5 (0.03–8.6) | 27 | 1.6 (0.4–6.5) | 1.7 (0.03–86.9) |
| RB | 27 | 1.3 (0.4–4.3) | 0.2 (0.01–3.5) | 26 | 1.3 (0.5–3.5) | NA |
| CL | 29 | 4.9* (1.1–22.9) | NA | 26 | 4.7* (1.0–20.8) | NA |
| All Sites | 195 | 2.3* (1.5–3.4) | 2.3* (1.3–4.1) | 170 | 2.2* (1.5–3.3) | 1.98* (1.1–3.6) |
Statistically significant (p<0.05).
Note: FIB = Fecal Indicator Bacteria; qENT= Enterococci qPCR; cEC= E. coli culture; OR = Odds Ratio; CI = Confidence Interval; NA = Not Applicable.
3.4. Beach-specific weighted-average fecal scores and FIB levels
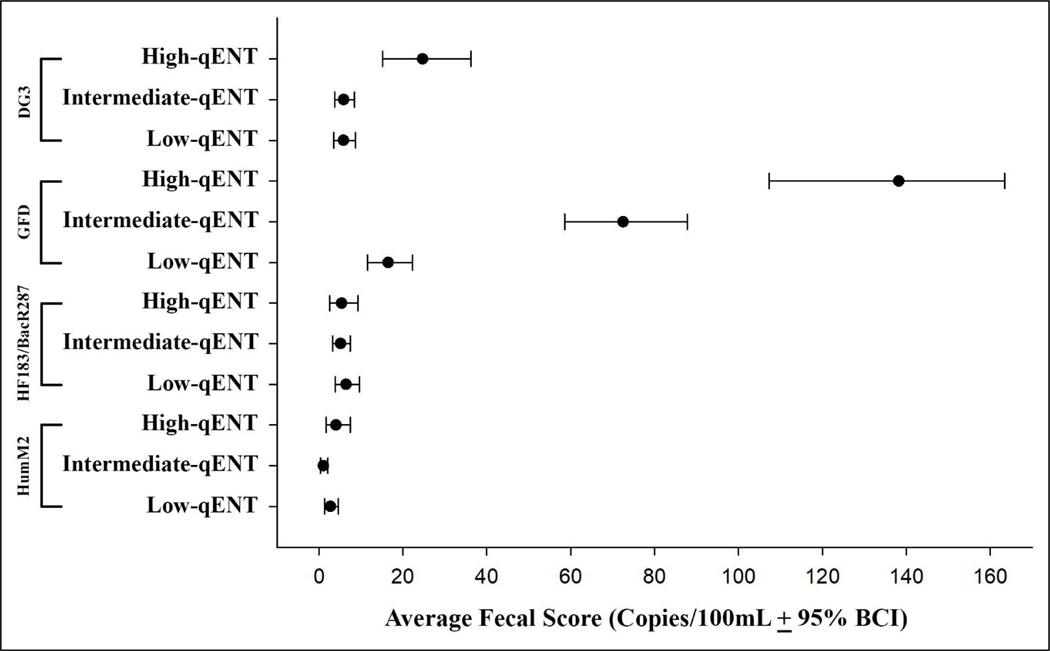
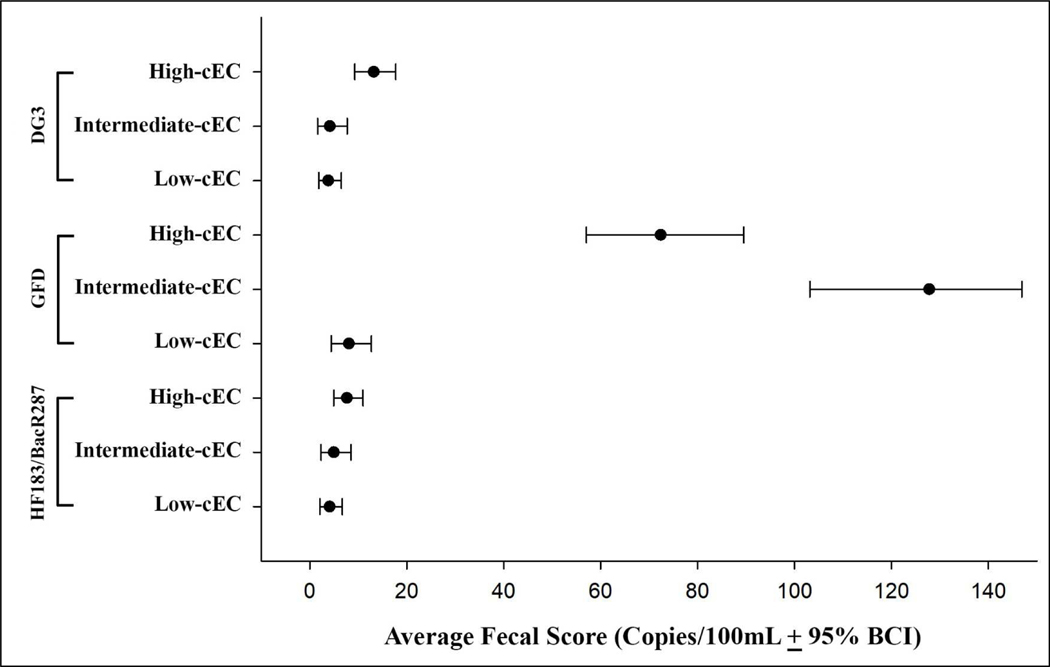
Weighted-average fecal scores with 95% Bayesian confidence interval (BCI) for each marker by beach, and by general FIB BAV exceedance category (enterococci qPCR BAV exceedance and E. coli culture BAV exceedance) are presented in Table A.9 and Table A.10. While variability of fecal scores for a given marker was relatively small among beaches, the mean scores differed substantially among markers, with much higher scores for GFD than the other MST markers. An exception to this is the very high weighted-average fecal score for DG3 at the beach with an area for dogs to swim (MN). The weighted-average fecal scores for some MST markers were much higher on beach-days when the qENT BAV was exceeded relative to beach-days when the qENT values was <10% of the BAV. Weighted-average fecal score for GFD was 8.4 times higher and for DG3 was 4.2 times higher in samples that exceeded the qENT BAV compared to samples that were <10% of qENT BAV as shown in Figure 1. A similar pattern was observed in samples that exceeded the E. coli culture-based BAV compared to samples < 10% of the cEC BAV. For GFD the weighted-average fecal scores were 9 times higher and for DG3 3.5 times higher as seen in Figure 2. However, weighted- average fecal scores for GFD for the intermediate category of cEC (between 10% of the BAV and the BAV) were considerably higher than the low and high E. coli groups. In contrast, both human-associated marker average concentrations were not different, regardless of general FIB BAV sample groupings (Figure 1 and Figure 2).
Figure 1:
Weighted-average fecal scores for MST markers for low, intermediate and high enterococci CCE levels
Note: qENT = Enterococci qPCR; BCI = Bayesian Confidence Interval.
Figure 2:
Weighted-average fecal scores for MST markers for low, intermediate and high E. coli MPN level
Note: cEC= E. coli culture; BCI = Bayesian Confidence Interval.
3.5. Precipitation and MST markers
Generally, log odds associations were not suggested between MST marker detection and wet weather (Table 3). An association between detection of DG3 and wet weather was suggested by the odds ratio of 2.20, but did not reach statistical significance at the 0.05 level, as evidenced by the fact that the 95% confidence interval (0.94, 5.18) includes 1.0. This association was driven by beaches without dog areas [OR (95% CI) =4.42 (0.48, 40.37)]; at the beach with a dog area, the odds of detecting DG3 were comparable in wet and dry weather [OR (95% CI) =1.75 (0.28, 10.81)]. The weighted-average fecal scores of MST markers differed in dry and wet weather (Table 4). Weighted-average fecal scores for HumM2, could not be calculated for different weather conditions as that marker was only detected in dry weather. DG3 (dog) weighted-average fecal scores were 2.4 times higher, DG37 (dog) were 2.1 times and GFD (bird) were 1.6 times higher in wet weather samples compared to dry weather samples (Table 4). Conversely, overall HF183/BacR287 (human) concentrations were slightly higher in samples during dry weather than wet weather, though this marker was detected much less frequently than the bird or dog markers.
Table 3:
Detection of MST markers under different weather conditions (N=195)
| MST Marker | Detect N (%) | Non-Detect N (%) | OR (95% CI) | ||
|---|---|---|---|---|---|
| Dry weather | Wet weather | Dry weather | Wet weather | ||
| DG3 | 9 (33) | 18 (67) | 88 (52) | 80 (48) | 2.20 (0.94, 5.18) |
| DG37 | 2 (29) | 5 (71) | 95 (51) | 93 (49) | 2.55 (0.48, 13.49) |
| GFD | 37 (47) | 42 (53) | 60 (52) | 56 (48) | 1.22 (0.69, 2.16) |
| HF183/BacR287 | 11 (58) | 8 (42) | 86 (49) | 90 (51) | 0.70 (0.27, 1.81) |
| HumM2 | 6 (75) | 2 (25) | 91 (49) | 96 (51) | 0.32 (0.06, 1.61) |
Note: OR = Odds Ratio (odds of detection under wet versus dry conditions); CI = Confidence Interval.
Table 4:
Beach-specific enterococci qPCR BAV exceedance under different weather conditions and weighted-average fecal scores with 95% BCI for the MST assays (N=195)
| Beach | Weather | N | qENT BAV Exceedance N (%) | Weighted-Average Fecal Score in copies/ 100 mL (95 % BCI) | |||
|---|---|---|---|---|---|---|---|
| DG3 | DG37 | GFD | HF183/ BacR287 | ||||
| MN | Dry | 11 | 2 (7.1) | 204.7 (145.2–321) | NA | 61.81 (36.05–82.92) | 6.15 (0.93–15.63) |
| Wet | 17 | 7 (25.0) | 385.5 (274.1–505.3) | NA | 73.17 (44.79–101.3) | 24.68 (13.07–39.35) | |
| OS | Dry | 17 | 4 (13.3) | NA | NA | 25.33 (15.84–33.32) | 10.97 (4.74–19.76) |
| Wet | 13 | 2 (6.7) | NA | NA | 43.14 (14.92–82.27) | 6.92 (1.67–15.62) | |
| TS | Dry | 8 | 0 (0.0) | NA | NA | NA | NA |
| Wet | 14 | 1 (4.6) | NA | NA | NA | NA | |
| MB | Dry | 13 | 4 (14.3) | NA | NA | 42.25 (23.81–59.19) | NA |
| Wet | 15 | 4 (14.3) | NA | NA | 94.66 (52.44–141.1) | NA | |
| ST | Dry | 17 | 3 (9.7) | NA | NA | 74.80 (40.24–117.7) | NA |
| Wet | 14 | 4 (12.9) | NA | NA | 58.56 (32.93–84.67) | NA | |
| RB | Dry | 17 | 2 (7.4) | NA | NA | 25.52 (14.50–38.05) | NA |
| Wet | 10 | 2 (7.4) | NA | NA | 30.85 (12.85–53.35) | NA | |
| CL | Dry | 14 | 1 (3.5) | NA | NA | 30.65 (10.18–59.50) | NA |
| Wet | 15 | 1 (3.5) | NA | NA | 116.2 (68.88–163.5) | NA | |
| All | Dry | 97 | 16 (8.2) | 4.71 (2.86–6.99) | 0.95 (0.31–1.95) | 39.53 (31.61–48.40) | 6.11 (4.02–8.68) |
| Wet | 98 | 21 (10.8) | 11.37 (8.17–15.06) | 1.95 (0.91–3.38) | 63.89 (51.30–77.56) | 4.95 (3.16–7.12) | |
Note: BAV = Beach Action Value; BCI = Bayesian Confidence Interval; qENT= Enterococci qPCR; NA = Not Applicable.
Beach-specific DG3 fecal scores were very high at the beach with an area for dogs to swim, with weighted-average fecal scores 1.9 times higher in wet weather than dry weather. The beach-specific GFD values and their associations with precipitation also varied among beaches. Weighted-average fecal scores were 3.8 to 1.2 times higher during wet weather when compared to dry weather at six of the beaches, while 63rd Street (ST) beach samples showed an inverse pattern (Table 4). The two beaches that had eligible data for beach-specific HF183/BacR287 weighted-average fecal scores Montrose (MN) and Ohio Street (OS) suggest very different associations with precipitation. HF183/BacR287 concentrations in wet weather samples from MN were 4 times higher than in dry weather samples, with little overlap in the Bayesian confidence interval (BCIs). HF183/BacR287 concentrations in samples from OS were comparable in wet and dry weather conditions.
3.6. Precipitation as a modifier of associations between general FIB BAV exceedances and MST markers
Associations between general FIB BAV exceedances and MST marker presence varied by weather conditions (Table 5). In wet weather conditions, the odds of DG3 presence and (separately) GFD presence were higher when qENT BAV was exceeded (Table 5). However, this was not the case with cEC BAV exceedances. Under dry weather conditions the detection of HumM2 was positively associated with exceedance of cEC BAV but not with exceedance of qENT BAV.
Table 5:
Association between BAV exceedance and MST marker detection stratified by weather condition
| Exceedance of qENT BAV OR (95% CI) | Exceedance of cEC BAV OR (95% CI) | |||||||
|---|---|---|---|---|---|---|---|---|
| Wet | Dry | M-H adjusted | p-value† | Wet | Dry | M-H adjusted | p- value† | |
| DG3 | 5.67 (1.87– 17.18) | 1.51 (0.29– 8.04) | 3.68 (1.52– 8.89) | 0.20 | 2.75 (0.80–9.43) | 3.86 (0.85–17.60) | 3.12 (1.19– 8.15) | 0.73 |
| DG37 | 6.25 (0.97–40.22) | 0.96* (0.04–21.02) | 3.29 (0.69–15.70) | 0.98 | 8.39* (0.44–160.71) | 0.65* (0.03–16.45) | 5.53 (0.47–64.68) | 0.96 |
| GFD | 3.50 (1.26– 9.70) | 1.79 (0.61– 5.28) | 2.57 (1.24– 5.35) | 0.38 | 1.50 (0.97–3.46) | 2.34 (0.91– 6.05) | 1.83 (0.97– 3.46) | 0.50 |
| HF183/BacR287 | 4.29 (0.98– 18.92) | 0.19* (0.01–3.32) | 1.19 (0.37– 3.79) | 0.96 | 6.83 (0.80–58.05) | 1.70 (0.42–6.95) | 2.96 (0.98– 8.96) | 0.29 |
| HumM2 | 19.87* (0.92–430.95) | 1.01 (0.11–9.31) | 3.05 (0.67– 13.90) | 0.98 | 4.46* (0.21–95.66) | 12.05 (1.33–109.13) | 15.44 (1.65– 144.78) | 0.99 |
Note:
0.5 was added to all cells to calculate this OR because of a zero cell in the MST marker detection and precipitation 2×2 table
p-value for the interaction term qENT= Enterococci qPCR; cEC= E. coli culture; BAV= Beach Action Value; OR= Odds Ratio; M-H: Mantel-Haenszel.
4. Discussion:
Water samples collected several times per week from seven urban Lake Michigan beaches thought to be free of discharges from wastewater treatment plants or combined sewer overflows identified evidence of human fecal pollution sporadically, without any clear spatial or temporal patterns. Evidence of bird fecal pollution was common, and evidence of dog fecal pollution was common at the one location with a ‘dog’ beach, and rare elsewhere. On wet weather days, an increased odds of dog (DG3) marker detection was suggested (Table 3), but not human or avian makers. Dog and bird MST marker estimated concentrations (weighted-average fecal scores) were increased approximately 3- and 2-fold, respectively (Table 4). Exceedance of general FIB BAVs increased the odds of dog and bird marker detection; for human markers this reached borderline statistical significance.
Associations between MST marker detection and the exceedance of BAVs differed in wet and dry weather. Dog marker (DG3) was 5.7 times as likely to be present with qENT BAV exceedance in wet weather; there was no increased likelihood of detecting this marker with BAV exceedances in dry weather. Similar observations were noted for avian and human markers, though only in relation to exceedance of the qENT BAV and not for the cEC BAV.
Human markers, HF183/BacR287 and HumM2, were detected in 10 and 4% of samples, respectively. This is lower than rates of detecting these markers in 2,330 samples from point-source impacted marine and freshwater beaches at which the NEEAR study was conducted (28% and 10%, respectively) (Napier et al., 2017), and lower than rates of detection at a marine beach in Southern California (Riedel et al., 2015) that may have been impacted by domestic and other septic systems. The lower rate of human marker detection in Chicago is consistent with expectations based on the absence of point-source and septic system discharges in Chicago. The HF183/BacR287 marker detection frequency was greater than that observed at Great Lakes beaches in Indiana that were not down-current of wastewater treatment discharge (5/448 samples, or 1.1%), but comparable to that at a beach which was down-current of a canal that released treated wastewater into the Lake (15/112 or 13.3%). (Nevers et al., 2018). The sources of human fecal pollution at Chicago’s beaches are not known, but might include bathers themselves, dirty diapers, and illicit discharges from boats. The fact that wet weather was not associated with human fecal marker detection (Table 3) suggests that sewer overflows or stormwater discharges are unlikely the problem. It must be noted that methods were not standardized across laboratories in the studies cited above. As a result, subtle differences in detection rates should not be assumed to be meaningful.
In 43% of water samples at the beach with an adjacent “dog beach”, DG3 was in the quantifiable range, while none of the 167 samples from other beaches were. This suggests that the beach management practice of banning dogs from beaches may be effective at preventing dog fecal pollution in nearshore waters. This observed rate of dog marker detection at beaches without dog areas is lower than that observed at Great Lakes beaches in Toronto impacted by riverine flow (Staley & Edge, 2016).
The marker for avian fecal pollution was by far the one most commonly detected (40% of samples) and most widely found across beaches. This is similar to findings at other Great Lakes beaches (Converse et al., 2012; Haack et al., 2013; Nevers et al., 2018). Given that strategies to reduce avian populations at bathing beaches has been followed by reductions in general FIB and avian marker detection rates (Converse et al., 2012; Goodwin et al., 2016; Haack et al., 2013; Nevers et al., 2018). Efforts to protect natural habitats of birds, while making constructed, intensively managed urban beaches less hospitable to them, would be expected to be reduce bird fecal pollution at Chicago’s bathing beaches.
The log10 general FIB concentrations increased the odds of GFD marker and (separately) of DG3 marker detection by about two-fold, however, this was beach-specific (Table 2). This, may be due to local spatial factors, such as embayment of some locations that have been shown to impact general FIB levels (Whitman & Nevers, 2008). Concentrations of human-associated markers were similar across categories of BAV exceedance frequency (Figures 1 and 2). The detection of HumM2 suggested an increased odd of qENT BAV exceedance in wet weather samples [OR (95% CI) = 19.87 (0.92, 430.95)]; this was not the case with cEC BAV exceedance.
The role of precipitation on BAV exceedance and on host-associated marker presence is important in developing strategies to improve water quality. We found that precipitation is associated with an approximate doubling of the odds of dog marker presence, and a 1.9-fold increase in the dog fecal score at the “dog beach”. At beaches overall, no association between precipitation and avian marker presence was observed. However, a 4-fold increase in the avian marker was observed at one beach (CL) with precipitation, pointing to an opportunity to develop targeted efforts to reduce bird presence or sheet flow of rainwater across the beach to the nearshore area. The fact that the strong association between dog, avian, and human marker presence and qENT BAV exceedance were limited to wet weather suggests that engineering solutions to reduce flow across or near beaches, as well as investigations to identify unrecognized outfalls, may reduce general FIB BAV exceedance frequency.
Two different general FIB, qENT and cEC, were measured and the associations between the general FIB and MST as well as the associations between the general FIB and precipitation were comparable. Exceptions include the monotonic increase in GFD weighted-average fecal score across ordinal categories of qENT (Figure 1 and 2), which was not observed with cEC (though the GFD weighted-average fecal scores were clearly higher on days that cEC was greater than 10% of the BAV vs. <10% of the BAV). Associations between qENT BAV exceedance and host-associated marker detection seemed much more dependent on precipitation status than those between cEC-BAV exceedance and host marker detection. On the other hand, in dry weather, the human marker HumM2 was strongly associated with exceedance of cEC BAV, but not with exceedance of qENT BAV. If this is observed in other settings, human MST markers specific for wet weather and dry weather pollutant sources might be identified. Others have shown that the bacterial DNA targets of qPCR assays persist longer in water environments than do culturable bacteria themselves (Anderson et al., 2005; Korajkic et al., 2014; Yamahara et al, 2012). This may explain the observation that in wet weather (but not dry weather), the association between qENT and MST marker detection was stronger than the association between cEC and MST marker detection (runoff from shore may add MST targets and non-viable enterococci to a greater extent than any addition of culturable E. coli to nearshore waters).
The present study points to several potential benefits of incorporating MST testing in the context of monitoring beaches not thought to be impacted by human fecal pollution. First, human fecal pollution presence appears to occur sporadically, both temporally and spatially. This would not be known if a small number of samples had been collected; thus, sampling strategies for human source markers at non-point source impact sites should be comprehensive. Likewise, the ability to characterize source presence and weighted-average fecal score in relation to precipitation could only have occurred with regular sampling over several months. Our observations regarding widespread avian marker presence and the localized presence of dog markers are entirely consistent with visual inspections of beaches. This supports the role of sanitary surveys in identifying fixed and variable sources of fecal pollution at beaches. Presently, MST testing of archived samples collected over the course of a beach season may be most useful as an adjunct to the annual sanitary survey; real-time testing may be more useful for promptly investigating and potentially mitigating sources of fecal pollution that were not apparent on visual inspections of beaches. Because the present study was conducted over a single summer, the additional benefit of routine testing over multiple seasons is not known. Further research that would assess human health risk in relation to the presence, concentrations and weighted-average fecal scores of host-associated markers will help shed light on the health risk information contained in MST data.
5. Conclusions:
Non-human fecal pollution sources including dogs and birds may influence recreational water quality at Chicago beaches. Corrective water management actions targeting canine and avian non-point fecal pollution sources may be helpful in improving water quality at these non-point source impacted beaches.
Infrequent detection and low concentrations of human fecal markers in the samples tested indicate that human waste generally does not contaminate Chicago beaches, likely due to the effectiveness of Chicago’s engineered system of wastewater and stormwater drainage infrastructure which protects Lake Michigan from urban discharges.
The role of precipitation on BAV exceedance and on host-associated marker presence is important in developing strategies to improve water quality. This information may be critical for remediation and management of recreational waters for better public health protection.
MST findings coupled with precipitation information can provide a better picture of different sources of fecal pollution and their pathways.
Finally, our study highlights various benefits of incorporating MST testing in the context of monitoring beaches not thought to be impacted by human fecal pollution.
Simultaneous use of precipitation data, MST results and general FIB used for routine beach monitoring and notification may be useful for remediation and management of recreational waters to better heath protection of beachgoers.
Supplementary Material
Highlights.
Exceedance of BAVs is associated with higher odds of dog and bird marker detection.
Dog markers were more likely to be detected following precipitation.
MST findings coupled with precipitation information can guide beach management.
MST can provide actionable information as a supplement to routine beach monitoring.
Acknowledgements:
The information in this paper has been subjected to U.S. EPA peer and administrative review and has been approved for external publication. Any opinions expressed in this paper are those of the authors and do not necessarily reflect the official positions and policies of the U.S. EPA. Any mention of trade names or commercial products does not constitute endorsement or recommendation for use. The HumM2 qPCR assay is patented by U.S. Environmental Protection Agency (U.S. Patent No. 7572584).
Funding:
Funding for the Summer 2016 beach monitoring project was provided by the Chicago Park District. This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Footnotes
Declaration of interests
☐ The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
The authors have no financial stake in the products described or in this research. The employer of two of the authors (AS and SD), the University of Illinois at Chicago, is contracted by the Chicago Park District to conduct beach monitoring, and that monitoring is managed by those authors. The beaches managed by the Chicago Park District are the subject of the manuscript.
References:
- Ackerman D, & Weisberg SB (2003). Relationship between rainfall and beach bacterial concentrations on Santa Monica Bay beaches. Journal of Water and Health, 1(2), 85–87. Retrieved from http://citeseerx.ist.psu.edu/viewdoc/download;jsessionid=60B8A14FB5E17F3BA5DC4EC7 A8AEE865?doi=10.1.1.495.79&rep=rep1&type=pdf [PubMed] [Google Scholar]
- Anderson KL, Whitlock JE, & Harwood VJ (2005). Persistence and Differential Survival of Fecal Indicator Bacteria in Subtropical Waters and Sediments. Applied and Environmental Microbiology, 71(6), 3041–3048. 10.1128/AEM.71.6.3041-3048.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badgley BD, Nayak BS, & Harwood VJ (2010). The importance of sediment and submerged aquatic vegetation as potential habitats for persistent strains of enterococci in a subtropical watershed. Water Research, 44(20), 5857–5866. 10.1016/j.watres.2010.07.005 [DOI] [PubMed] [Google Scholar]
- Benjamin-Chung J, Arnold BF, Wade TJ, Schiff K, Griffith JF, Dufour AP, … Colford JM (2017). Coliphages and Gastrointestinal Illness in Recreational Waters: Pooled Analysis of Six Coastal Beach Cohorts. Epidemiology, 28(5), 644–652. 10.1097/EDE.0000000000000681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks YM, Spirito CM, Bae JS, Hong A, Mosier EM, Sausele DJ, … Richardson RE (2019). Fecal indicator bacteria, fecal source tracking markers, and pathogens detected in two Hudson River tributaries. Water Research, 115342. 10.1016/j.watres.2019.115342 [DOI] [PubMed] [Google Scholar]
- Byappanahalli M, & Fujioka R. (2004). Indigenous soil bacteria and low moisture may limit but allow faecal bacteria to multiply and become a minor population in tropical soils. Water Science and Technology, 50(1), 27–32. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/15318482 [PubMed] [Google Scholar]
- Byappanahalli MN, Whitman RL, Shively DA, Ting WTE, Tseng CC, & Nevers MB (2006). Seasonal persistence and population characteristics of Escherichia coli and entercocci in deep backshore sand of two freshwater beaches. Journal of Water and Health, 4(3), 313–320. 10.2166/wh.2006.018 [DOI] [PubMed] [Google Scholar]
- Cao Y, Sivaganesan M, Kelty CA, Wang D, Boehm AB, Griffith JF, … Shanks OC (2018). A human fecal contamination score for ranking recreational sites using the HF183/BacR287 quantitative real-time PCR method. Water Research, 128, 148–156. 10.1016/j.watres.2017.10.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicago Park District. (2020). Retrieved June 26, 2018, from https://www.chicagoparkdistrict.com/parks-facilities/beaches
- Colford JM, Wade TJ, Schiff KC, Wright CC, Griffith JF, Sandhu SK, … Weisberg SB (2007). Water quality indicators and the risk of illness at beaches with nonpoint sources of fecal contamination. Epidemiology (Cambridge, Mass.), 18(1), 27–35. 10.1097/01.ede.0000249425.32990.b9 [DOI] [PubMed] [Google Scholar]
- Converse RR, Kinzelman JL, Sams EA, Hudgens E, Dufour AP, Ryu H, … Wade TJ (2012). Dramatic improvements in beach water quality following gull removal. Environmental Science and Technology, 46(18), 10206–10213. 10.1021/es302306b [DOI] [PubMed] [Google Scholar]
- Cordero L, Norat J, Mattei H, & Nazario C. (2012). Seasonal variations in the risk of gastrointestinal illness on a tropical recreational beach. Journal of Water and Health, 10(4), 579–593. 10.2166/wh.2012.076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorevitch S, Shrestha A, DeFlorio-Barker S, Breitenbach C, & Heimler I. (2017). Monitoring urban beaches with qPCR vs. culture measures of fecal indicator bacteria: Implications for public notification. Environmental Health : A Global Access Science Source, 16(1), 45. 10.1186/s12940-017-0256-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwight RH, Caplan JS, Brinks MV, Catlin SN, Buescher G, & Semenza JC (2011). Influence of Variable Precipitation on Coastal Water Quality in Southern California. Water Environment Research, 83(December), 2121–2130. 10.2175/106143011X12928814444574 [DOI] [PubMed] [Google Scholar]
- Goodwin KD, Gruber S, Vondrak M, & Crumpacker A. (2016). Watershed Assessment with Beach Microbial Source Tracking and Outcomes of Resulting Gull Management. Environmental Science and Technology, 50(18), 9900–9906. 10.1021/acs.est.6b02564 [DOI] [PubMed] [Google Scholar]
- Green HC, Dick LK, Gilpin B, Samadpour M, & Field KG (2012). Genetic markers for rapid PCR-based identification of gull, Canada goose, duck, and chicken fecal contamination in water. Applied and Environmental Microbiology, 78(2), 503–510. 10.1128/AEM.05734-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green HC, Haugland RA, Varma M, Millen HT, Borchardt MA, Field KG, … Shanks OC (2014). Improved HF183 quantitative real-time PCR assay for characterization of human fecal pollution in ambient surface water samples. Appl. Environ. Microbiol, 80(10), 3086–3094. 10.1128/AEM.04137-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green HC, White KM, Kelty CA, & Shanks OC (2014). Development of rapid canine fecal source identification PCR-based assays. Environmental Science and Technology, 48(19), 11453–11461. 10.1021/es502637b [DOI] [PubMed] [Google Scholar]
- Haack SK, Fogarty LR, Stelzer EA, Fuller LM, Brennan AK, Isaacs NM, & Johnson HE (2013). Geographic setting influences Great Lakes beach microbiological water quality. Environmental Science and Technology, 47(21), 12054–12063. 10.1021/es402299a [DOI] [PubMed] [Google Scholar]
- Harwood VJ, Staley C, Badgley BD, Borges K, & Korajkic A. (2014, January). Microbial source tracking markers for detection of fecal contamination in environmental waters: Relationships between pathogens and human health outcomes. FEMS Microbiology Reviews. 10.1111/1574-6976.12031 [DOI] [PubMed] [Google Scholar]
- Imamura GJ, Thompson RS, Boehm AB, & Jay JA (2011). Wrack promotes the persistence of fecal indicator bacteria in marine sands and seawater. FEMS Microbiology Ecology, 77(1), 40–49. 10.1111/j.1574-6941.2011.01082.x [DOI] [PubMed] [Google Scholar]
- Jennings WC, Chern EC, O ‘donohue D, Kellogg MG, & Boehm AB (2018). Frequent detection of a human fecal indicator in the urban ocean: environmental drivers and covariation with enterococci. Environmental Science: Processes & Impacts. 10.1039/c7em00594f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelty CA, Varma M, Sivaganesan M, Haugland RA, & Shanks OC (2012). Distribution of genetic marker concentrations for fecal indicator bacteria in sewage and animal feces. Applied and Environmental Microbiology, 78(12), 4225–4232. 10.1128/AEM.07819-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinzelman J, McLellan SL, Amick A, Preedit J, Scopel CO, Olapade O, … Sedmak G. (2008). Identification of human enteric pathogens in gull feces at Southwestern Lake Michigan bathing beaches. Canadian Journal of Microbiology, 54(12), 1006–1015. 10.1139/W08-096 [DOI] [PubMed] [Google Scholar]
- Kirs M, Kisand V, Wong M, Caffaro-Filho RA, Moravcik P, Harwood VJ, … Fujioka RS (2017). Multiple lines of evidence to identify sewage as the cause of water quality impairment in an urbanized tropical watershed. Water Research, 116, 23–33. 10.1016/j.watres.2017.03.024 [DOI] [PubMed] [Google Scholar]
- Kleinheinz GT, McDermott CM, Hughes S, & Brown A. (2009). Effects of Rainfall on E. coli Concentrations at Door County, Wisconsin Beaches. International Journal of Microbiology, 2009, 1–9. 10.1155/2009/876050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korajkic A, McMinn B, & Harwood V. (2018). Relationships between Microbial Indicators and Pathogens in Recreational Water Settings. International Journal of Environmental Research and Public Health, 15(12), 2842. 10.3390/ijerph15122842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korajkic A, McMinn BR, Shanks OC, Sivaganesan M, Fout GS, & Ashbolt NJ (2014). Biotic interactions and sunlight affect persistence of fecal indicator bacteria and microbial source tracking genetic markers in the upper mississippi river. Applied and Environmental Microbiology, 80(13), 3952–3961. 10.1128/AEM.00388-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laureano-Rosario AE, Symonds EM, Rueda-Roa D, Otis D, & Muller-Karger FE (2017). Environmental factors correlated with culturable enterococci concentrations in tropical recreational waters: A case study in escambron beach, San Juan, Puerto Rico. International Journal of Environmental Research and Public Health, 14(12), 1602. 10.3390/ijerph14121602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Sivaganesan M, Kelty CA, Zimmer-Faust A, Clinton P, Reichman JR, … Shanks OC (2019). Large-scale implementation of standardized quantitative real-time PCR fecal source identification procedures in the Tillamook Bay Watershed. PLoS ONE, 14(6), e0216827. 10.1371/journal.pone.0216827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathai PP, Dunn HM, Magnone P, Zhang Q, Ishii S, Chun CL, & Sadowsky MJ (2019). Association between submerged aquatic vegetation and elevated levels of Escherichia coli and potential bacterial pathogens in freshwater lakes. Science of the Total Environment, 657, 319–324. 10.1016/j.scitotenv.2018.11.484 [DOI] [PubMed] [Google Scholar]
- McHugh ML (2012). Interrater reliability: the kappa statistic. Biochemia Medica, 22(3), 276–282. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/23092060 [PMC free article] [PubMed] [Google Scholar]
- McLellan SL, Hollis EJ, Depas MM, van Dyke M, Harris J, & Scopel CO (2007). Distribution and fate of Escherichia coli in Lake Michigan following contamination with urban stormwater and combined sewer overflows. Journal of Great Lakes Research, 33, 566–580. 10.3394/0380-1330(2007)33 [DOI] [Google Scholar]
- Napier MD, Haugland R, Poole C, Dufour AP, Stewart JR, Weber DJ, … Wade TJ (2017). Exposure to human-associated fecal indicators and self-reported illness among swimmers at recreational beaches: A cohort study. Environmental Health: A Global Access Science Source, 16(1), 103. 10.1186/s12940-017-0308-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napier MD, Poole C, Stewart JR, Weber DJ, Glassmeyer ST, Kolpin DW, … Wade TJ (2018). Exposure to Human-Associated Chemical Markers of Fecal Contamination and Self-Reported Illness among Swimmers at Recreational Beaches. Environmental Science & Technology. 10.1021/acs.est.8b00639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevers MB, Byappanahalli MN, Shively D, Buszka PM, Jackson PR, & Phanikumar MS (2018). Identifying and eliminating sources of recreational water quality degradation along an urban coast. Journal of Environmental Quality, 47(5), 1042–1050. 10.2134/jeq2017.11.0461 [DOI] [PubMed] [Google Scholar]
- Nevers MB, Przybyla-Kelly K, Spoljaric A, Shively D, Whitman RL, & Byappanahalli MN (2016). Freshwater wrack along Great Lakes coasts harbors Escherichia coli: Potential for bacterial transfer between watershed environments. Journal of Great Lakes Research, 42(4), 760–767. 10.1016/j.jglr.2016.04.011 [DOI] [Google Scholar]
- Nevers MB, & Whitman RL (2011). Efficacy of monitoring and empirical predictive modeling at improving public health protection at Chicago beaches. Water Research, 45(4), 1659–1668. 10.1016/j.watres.2010.12.010 [DOI] [PubMed] [Google Scholar]
- Riedel TE, Thulsiraj V, Zimmer-Faust AG, Dagit R, Krug J, Hanley KT, … Jay JA (2015). Long-term monitoring of molecular markers can distinguish different seasonal patterns of fecal indicating bacteria sources. Water Research, 71, 227–243. 10.1016/j.watres.2014.12.037 [DOI] [PubMed] [Google Scholar]
- Shanks OC, Kelty CA, Oshiro R, Haugland RA, Madi T, Brooks L, … Sivaganesan M. (2016). Data acceptance criteria for standardized human-associated fecal source identification quantitative real-time PCR methods. Applied and Environmental Microbiology, 82(9), 2773–2782. 10.1128/AEM.03661-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanks OC, Kelty CA, Sivaganesan M, Varma M, & Haugland RA (2009). Quantitative PCR for genetic markers of human fecal pollution. Applied and Environmental Microbiology, 75(17), 5507–5513. 10.1128/AEM.00305-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanks OC, White K, Kelty CA, Sivaganesan M, Blannon J, Meckes M, & Varma M. (2010). Performance of PCR Based Assays Targeting Bacteriodales Genetic Markers of Human Fecal Pollution in Sewage and Fecal Samples. Environmental Science and Technology, 44(16), 6281–6288. 10.1021/es100311n [DOI] [PubMed] [Google Scholar]
- Sivaganesan M, Seifring S, Varma M, Haugland RA, & Shanks OC (2008). A Bayesian method for calculating real-time quantitative PCR calibration curves using absolute plasmid DNA standards. BMC Bioinformatics, 9. 10.1186/1471-2105-9-120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley ZR, Chase E, Mitraki C, Crisman TL, & Harwood VJ (2013). Microbial water quality in freshwater lakes with different land use. Journal of Applied Microbiology, 115(5), 1240–1250. 10.1111/jam.12312 [DOI] [PubMed] [Google Scholar]
- Staley ZR, & Edge TA (2016). Comparative microbial source tracking methods for identification of fecal contamination sources at Sunnyside Beach in the Toronto region area of concern. Journal of Water and Health, 14(5), 839–850. 10.2166/wh.2016.296 [DOI] [PubMed] [Google Scholar]
- Steele JA, Blackwood AD, Griffith JF, Noble RT, & Schiff KC (2018). Quantification of pathogens and markers of fecal contamination during storm events along popular surfing beaches in San Diego California. Water Research, 136, 137–149. 10.1016/j.watres.2018.01.056 [DOI] [PubMed] [Google Scholar]
- United States Environmental Protection Agency. (2012). Recreational Water Quality Criteria. [Google Scholar]
- USEPA. (2015). Method 1609.1: Enterococci in Water by Taqman Quantitative Polymerase Chain Reaction (qPCR) with Internal Amplification Control (IAC) Assay. 10.1017/CBO9781107415324.004 [DOI]
- USEPA. Method 1696: Characterization of Human Fecal Pollution in Water by HF183/BacR287 TaqMan® Quantitative Polymerase Chain Reaction (qPCR) Assay (2019). Retrieved from www.epa.gov
- USEPA. (2019b). Method 1697: Characterization of Human Fecal Pollution in Water by HumM2 TaqMan® Quantitative Polymerase Chain Reaction (qPCR) Assay®. Retrieved from www.epa.gov
- Wade TJ, Calderon RL, Brenner KP, Sams E, Beach M, Haugland R, … Dufour AP (2008). High Sensitivity of Children to Swimming-Associated Gastrointestinal Illness. Epidemiology, 19(3), 375–383. 10.1097/EDE.0b013e318169cc87 [DOI] [PubMed] [Google Scholar]
- Wade TJ, Sams E, Brenner KP, Haugland R, Chern E, Beach M, … Dufour AP (2010). Rapidly measured indicators of recreational water quality and swimming-associated illness at marine beaches: a prospective cohort study. Environmental Health : A Global Access Science Source, 9(1), 66. 10.1186/1476-069X-9-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman RL, & Nevers MB (2003). Foreshore Sand as a Source of Escherichia coli in Nearshore Water of a Lake Michigan Beach. Applied and Environmental Microbiology, 69(9), 5555–5562. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/12957945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman RL, & Nevers MB (2008). Summer E. coli patterns and responses along 23 Chicago beaches. Environmental Science and Technology, 42(24), 9217–9224. 10.1021/es8019758 [DOI] [PubMed] [Google Scholar]
- Yamahara KM, Sassoubre LM, Goodwin KD, & Boehm AB (2012). Occurrence and persistence of bacterial pathogens and indicator organisms in beach sand along the California coast. Applied and Environmental Microbiology, 78(6), 1733–1745. 10.1128/AEM.06185-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yau VM, Schiff KC, Arnold BF, Griffith JF, Gruber JS, Wright CC, … Colford JM (2014). Effect of submarine groundwater discharge on bacterial indicators and swimmer health at Avalon Beach, CA, USA. Water Research, 59, 23–36. 10.1016/j.watres.2014.03.050 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.