Abstract
Organoid technology has revolutionized the study of human organ development, disease and therapy response tailored to the individual. Although detailed protocols are available for the generation and long-term propagation of human organoids from various organs, such methods are lacking for breast tissue. Here we provide an optimized, highly versatile protocol for long-term culture of organoids derived from either normal human breast tissues or breast cancer (BC) tissues, as well as culturing conditions for a panel of 45 biobanked samples, including BC organoids covering all major disease subtypes (triple-negative, estrogen receptor-positive/progesterone receptor-positive and human epidermal growth receptor 2-positive). Additionally, we provide methods for genetic manipulation by Lipofectamine 2000, electroporation or lentivirus and subsequent organoid selection and clonal culture. Finally, we introduce an optimized method for orthotopic organoid transplantation in mice, which includes injection of organoids and estrogen pellets without the need for surgery. Organoid derivation from tissue fragments until the first split takes 7–21 d; generation of genetically manipulated clonal organoid cultures takes 14–21 d; and organoid expansion for xenotransplantation takes >4 weeks.
Introduction
BC remains the most widely diagnosed and second deadliest malignancy in women1. BC comprises multiple subtypes that arise through the sequential accumulation of mutations in resident epithelial cells and result in tumors with complex genomic and biological heterogeneity2–4. Standard therapeutic approaches are currently based on clinical and pathologic features, complemented by the expression status of estrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor 2 (HER2)5 and are, thereby, inadequately tailored to individual patients. An extensive understanding of the processes underlying normal breast development, carcinogenesis and disease progression is necessary to devise novel personalized therapies, but this is hampered by a lack of suitable and widely available in vitro models. Organoid technology—the long-term propagation of human adult stem cells in three-dimensional (3D) cultures6—has provided unprecedented and transformative ways to study healthy organ development and tumorigenesis in vitro. Here we describe optimized protocols for human normal breast and BC organoids to model breast development and cancer.
The described methods have been used to generate a biobank of BC organoids that typically recapitulated the original patient tumor in terms of histopathology, hormone receptor expression and HER2 status, as well as copy number variations and sequence changes, thereby capturing the full heterogeneity of BC (www.huborganoids.nl)7. Furthermore, normal organoid cultures contain basal/stem cells, luminal progenitors and mature luminal cells, thus preserving complex epithelial lineages8, and we previously engineered normal breast organoids using CRISPR–Cas9 to model the onset of BC9.
Overview of the procedures
A graphical overview of our protocol describing the efficient derivation and long-term culturing of human normal and BC organoids and their downstream processing is depicted in Fig. 1. First, we describe how organoids can be derived from both normal and tumor resections, outline their respective culture conditions and explain how to maintain and cryopreserve the cultures. Then, we discuss three different ways to genetically manipulate organoids and how to select and grow clonal organoids. Finally, we describe xenotransplantation of BC organoids.
Fig. 1 |. Schematic overview of the protocol describing human breast organoid derivation, culturing, genetic manipulation and xenotransplantation.
Organoids are established from resections of normal breast or tumor tissue, followed by organoid maintenance or freezing for long-term storage. Genetic manipulation by Lipofectamine-based transfection, electroporation-based transfection or lentiviral transduction is described, as well as (clonal) organoid selection. Orthotopic injection can be performed to grow organoid-derived tumors in vivo, with the stated time indicating time until first signs of tumor formation. Corresponding steps of the protocol and their timing are indicated in yellow boxes.
We provide information on morphology, growth rate and passaging conditions for 45 organoid cultures encompassing normal, primary tumor and metastatic cultures as a reference guide for organoid maintenance, taking into account inter-culture variation (Table 1). Establishing a breast organoid culture, from tissue isolation until the first passage, will take 7–21 d. Genetically manipulating an organoid culture and generating a selected clonal line at passage 1 will take 14–21 d. Expanding an organoid culture for transplantation usually requires more than 4 weeks and depends on the number of mice injected, as well as the organoid proliferation rate. To establish, maintain and manipulate breast organoids, expertise in 3D (organoid) culturing is useful but not essential. For in vivo work, appropriate facilities and expertise in mouse handling and maintenance are required.
Table 1 |.
Detailed information for 45 biobanked breast organoid cultures
| Organoid number | Organoid code | Origin | Tissuea |
Organoida |
Morphology | Long-term culture capacity | Passaging |
Reference | Available on request? | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ER | PR | HER2 | ER | PR | HER2 | Ratio | Frequency (d) | Method | Medium type | |||||||
| 1 | ORG24 | N | N/A | N/A | N/A | N/A | N/A | N/A | Mixed | >10 | 1:2 | 14 | Fragments | 1 | Ref. 8 | Yesb |
| 2 | ORG47 | N | N/A | N/A | N/A | N/A | N/A | N/A | Mixed | >10 | 1:2 | 14 | Fragments | 1 | Ref. 8 | Yesb |
| 3 | HUB-01-A2-033 | N | 95 | 100 | 0 | 5 | 80 | 0 | Mixed | >5 | 1:2 | 14 | Fragments | 2 | N/A | Yesc |
| 4 | HUB-01-A2-034 | N | 100 | 90 | 0 | 1 | 0 | 0 | Solid | >3 | 1:2 | 14 | Fragments | 2 | N/A | Yesc |
| 5 | HUB-01-A2-045 | N | 0 | 0 | 0 | 1 | 80 | 0 | Mixed | >10 | 1:1,5 | 14 | Fragments | 2 | N/A | Yesc |
| 6 | HUB-01-A2-056 | N | 60 | 100 | 0 | 0 | 30 | 0 | Mixed | >5 | 1:2 | 14 | Fragments | 2 | N/A | Yesc |
| 7 | HUB-01-A2-087 | N | 100 | 100 | 0 | 0 | 0 | 0 | Solid | >10 | 1:2 | 14 | Fragments | 2 | N/A | Yesc |
| 8 | HUB-01-A2-090 | N | 0 | 0 | 0 | 0 | 70 | 0 | Mixed | >5 | 1:1,5 | 14 | Fragments | 2 | N/A | Yesc |
| 9 | HUB-01-A2-095 | N | 90 | 10 | 0 | 1 | 1 | 0 | Mixed | >10 | 1:2 | 14 | Fragments | 2 | N/A | Yesc |
| 10 | HUB-01-A2-096 | N | 90 | 90 | 0 | N/A | N/A | N/A | Mixed | >5 | 1:2 | 14 | Fragments | 2 | N/A | Yesc |
| 11 | HUB-01-A2-134 | N | 100 | 90 | 0 | 95D | 80D | 0 | Solid | >20 | 1:2 | 14 | Fragments | 2 | N/A | Yesc |
| 12 | HUB-01-A2-150 | N | 50 | 30 | 0 | 0 | 2 | 0 | Solid | >5 | 1:2 | 14 | Fragments | 2 | N/A | Yesc |
| 13 | HUB-01-A2-186 | N | N/A | N/A | N/A | N/A | N/A | N/A | Solid | >5 | 1:2 | 14 | Fragments | 2 | N/A | Yesc |
| 14 | HUB-01-A2-187 | N | N/A | N/A | N/A | 5 | 1 | 0 | Mixed | >5 | 1:1,5 | 14 | Fragments | 2 | N/A | Yesc |
| 15 | HUB-01-B2-001 | P | 0 | 0 | +3 | 0 | 85 | 100 | Solid | >60 | 1:6 | 7 | Single cells | 1 | Ref. 7 | No |
| 16 | HUB-01-B2-003 | P | 0 | 0 | +1 | N/A | N/A | N/A | Grape-like | >80 | 1:6 | 7 | Fragments | 1 | Ref. 7 | No |
| 17 | HUB-01-B2-021 | P | 80 | 80 | +3 | N/A | N/A | N/A | Solid | >40 | 1:2 | 14 | Single cells | 1 | Ref. 7 | No |
| 18 | HUB-01-B2-004 | P | 10 | 0 | +1 | N/A | N/A | N/A | Solid | >50 | 1:4 | 7 | Single cells | 1 | Ref. 7 | No |
| 19 | HUB-01-B2-005 | P | 20 | 0 | +1 | N/A | N/A | N/A | Solid | >30 | 1:2 | 14 | Fragments | 1 | Ref. 7 | No |
| 20 | HUB-01-B2-011 | P | 20 | 10 | 0 | 30 | 30 | 0 | Solid | >80 | 1:6 | 7 | Single cells | 1 | Ref. 7 | No |
| 21 | HUB-01-B2-013 | P | 0 | 0 | +1 | N/A | N/A | N/A | Solid | >80 | 1:5 | 7 | Single cells | 1 | Ref. 7 | No |
| 22 | HUB-01-B2-060 | P | 0 | 0 | 0 | N/A | N/A | N/A | Mixed | >30 | 1:2 | 14 | Fragments | 1 | Ref. 7 | No |
| 23 | HUB-01-B2-061 | P | 0 | 0 | 0 | N/A | N/A | N/A | Solid | >30 | 1:2 | 14 | Fragments | 1 | Ref. 7 | No |
| 24 | HUB-01-B2-031 | P | 0 | 0 | +1 | N/A | N/A | N/A | Solid | >30 | 1:2 | 7 | Fragments | 1 | Ref. 7 | No |
| 25 | HUB-01-B2-033 | P | 95 | 100 | 0 | 50 | 5 | 0 | Mixed | >15 | 1:1,5 | 14 | Fragments | 1 | Ref. 7 | Yesc |
| 26 | HUB-01-B2-042 | P | 100 | 100 | 0 | 3 | 5 | 0 | Mixed | >5 | 1:1,5 | 14 | Fragments | 1 | Ref. 7 | Yesc |
| 27 | HUB-01-B2-043 | P | 80 | 10 | 0 | 80 | 80 | 0 | Grape-like | >10 | 1:2 | 14 | Fragments | 1 | Ref. 7 | No |
| 28 | HUB-01-B2-056 | P | 60 | 100 | 0 | 0 | 5 | 0 | Solid | > 10 | 1:2 | 14 | Fragments | 1 | Ref. 7 | Yesc |
| 29 | HUB-01-B2-058 | P | 0 | 0 | 0 | 0 | 0 | 0 | Solid | >10 | 1:2 | 14 | Fragments | 1 | Ref. 7 | Yesc |
| 30 | HUB-01-B2-066 | P | 0 | 0 | +3 | N/A | N/A | N/A | Solid | >40 | 1:2 | 14 | Single cells | 1 | Ref. 7 | No |
| 31 | HUB-01-B2-090 | P | 0 | 0 | +3 | N/A | N/A | N/A | Solid | >20 | 1:3 | 14 | Single cells | 1 | N/A | Yesc |
| 32 | HUB-01-B2-095 | P | 90 | 0 | 0 | 1 | 0 | 0 | Mixed | >10 | 1:2 | 14 | Fragments | 2 | N/A | Yesc |
| 33 | HUB-01-B2-125 | P | 100 | 60 | 0 | 20 | 30 | 0 | Solid | >20 | 1:1,5 | 14 | Fragments | 2 | Ref. 7 | Yesc |
| 34 | HUB-01-B2-135 | P | 100 | 100 | 0 | 1 | 0 | 0 | Cystic | >15 | 1:1,5 | 14 | Fragments | 1 | Ref. 7 | Yesc |
| 35 | HUB-01-B2-150 | P | 50 | 30 | 0 | 95 | 85 | 0 | Mixed | >5 | 1:2 | 14 | Fragments | 2 | Ref. 7 | Yesc |
| 36 | HUB-01-B2-186 | P | 0 | 0 | 0 | 0 | 5 | 0 | Mixed | >5 | 1:1,5 | 14 | Fragments | 2 | N/A | Yesc |
| 37 | HUB-01-B2-187 | P | 0 | 0 | 0 | 10 | 1 | 0 | Mixed | >5 | 1:1,5 | 14 | Fragments | 2 | N/A | Yesc |
| 38 | HUB-01-B2-190 | P | 0 | 0 | 0 | 1 | 20 | 0 | Mixed | >5 | 1:2 | 14 | Fragments | 2 | N/A | Yesc |
| 39 | HUB-01-C2-119 | M | 90 | 1 | 1 | N/A | N/A | N/A | Solid | >3 | 1:1,5 | 14 | Fragments | 2 | Ref. 7 | No |
| 40 | HUB-01-C2-120 | M | 95 | 80 | 1 | N/A | N/A | N/A | Solid | >5 | 1:2 | 14 | Fragments | 1 | Ref. 7 | No |
| 41 | HUB-01-C2-123 | M | pos | pos | 0 | 80 | 0 | 0 | Solid | >10 | 1:2 | 14 | Fragments | 1 | Ref. 7 | No |
| 42 | HUB-01-C2-152 | M | 0 | 0 | +3 | 0 | 10 | 0 | Grape-like | >40 | 1:4 | 7 | Fragments | 1 | Ref. 7 | Yesc |
| 43 | HUB-01-C2-156 | M | 60 | 5 | +1 | 90 | 80 | 0 | Solid | >40 | 1:4 | 7 | Fragments | 1 | Ref. 7 | Yesc |
| 44 | HUB-01-C2-189 | M | 0 | 0 | 0 | 0 | 10 | 10 | Grape-like | Stops -P20 | 1:2 | 14 | Fragments | 1 | N/A | Yesc |
| 45 | HUB-01-C2-140 | M | 0 | 0 | 0 | N/A | N/A | N/A | Solid | >12 | 1:2 | 14 | Fragments | 1 | Ref. 7 | Yesc |
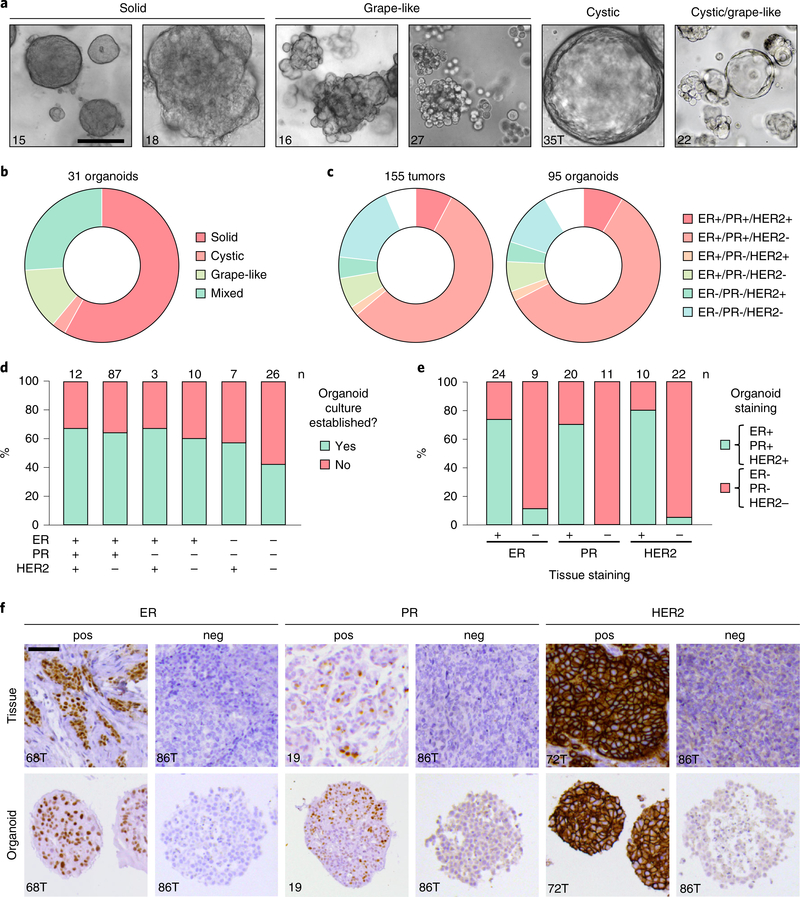
Expression status of the indicated receptors for tissue and organoid, indicated as percentage of positive cells (ER/PR) and immunohistochemical score (0–3; HER2). N/A indicates that the sample has not been tested or data are not available.
Available upon reasonable request from J.M.R.
Available upon reasonable request from C.S.V or S.F.B.
ER and PR levels are known to reduce at higher passage number.
M, metastasis; N, normal; P, primary tumor.
Applications
We anticipate that preclinical researchers in the BC field can use this protocol in a tailor-made fashion, by combining parts suited to their specific research needs. Normal organoids provide a valuable resource for studying development and function of the human mammary gland8. For understanding de novo oncogenesis, normal breast organoids can be subjected to CRISPR–Cas9 editing to efficiently knock out tumor suppressor genes9, or organoids from individuals with a hereditary cancer predisposition syndrome, such as BRCA1/BRCA2 mutations, can be used. Additionally, organoids can be used to assess the efficacy of novel drugs, both in vitro as well as upon engraftment in vivo. These organoid xenograft models can also be used to visualize tumor growth and cancer cell behavior in vivo through the introduction of a mammary imaging window10. Importantly, due to the retained genetic and histologic features of the original patient tumor7, BC organoids can be used as a clinical tool to aid personalized medicine, by assessing drug responses in a patient-specific manner. Although the methods described have been optimized for breast organoids, the sections on genetic manipulation and clonal selection can be applied to a wide variety of organoid cultures with minor adjustments, thus affecting a wide research field beyond BC.
Comparison with other methods and possible limitations
In comparison to commonly used BC cell lines11 or BC spheroids12, BC organoids can be grown long term, while recapitulating the complex genetic and phenotypic heterogeneity7 that is characteristic of BC13,14. Although several groups have reported 3D model systems for studying normal mammary development15,16, these do not allow the extended culture of all mammary lineages, in contrast to the normal breast organoids reported here8. Different from previously developed methods to generate patient-derived xenografts17, the injection-based method for estrogen pellet implantation and BC organoid xenotransplantation used here circumvents the need for complex surgical procedures. As compared to growth properties of human intestinal organoids, which can be passaged 1:6 every week with limited variation among samples6, the proliferation rate of breast organoids is relatively slow and can differ substantially between donors. The splitting ratio can vary from 1:6 weekly to 1:2 weekly, biweekly or even less frequently (Table 1). It should be noted that maintenance of 3D breast organoid cultures is relatively laborious and costly compared to most other in vitro breast cultures.
Experimental design
Expansion medium (reagent setup)
We describe the preparation of an expansion medium that was previously published7 (referred to as ‘Type 1’; Table 2). In addition, we introduce an alternative expansion medium, similar to that developed for long-term ovarian cancer organoid cultures18 (referred to as ‘Type 2’; Table 2), which can improve the growth properties of some organoid cultures (Fig. 2a and Table 1). We advise to grow newly established organoid cultures in both Type 1 and Type 2 expansion medium, to define the most optimal medium type per donor. We use homemade R-spondin-1, Noggin and Wnt3a conditioned media, for which detailed production protocols are available19–22. We strongly recommend using these homemade conditioned media, as we experienced superior performance compared to commercial compounds. However, we do provide commercial alternatives in the ‘Reagents’ section.
Table 2 |.
Overview of the components to add to adDMEM/F12+++ to generate Type 1 and Type 2 media, with the respective final concentrations per type of medium
| Component | Type 1 | Type 2 |
|---|---|---|
| Wnt3a | — | 20% conditioned medium |
| R-spondin1 | 10% conditioned medium | 10% conditioned medium |
| Noggin | 10% conditioned medium | 10% conditioned medium |
| B27 + VitA | 1× | 1× |
| Nicotinamide | 10 mM | 10 mM |
| N-acetylcysteine | 1.25 mM | 1.25 mM |
| Primocin | 100 μ/ml | 100 μg/ml |
| Hydrocortisone | — | 0.5 μg/ml |
| β-estradiol | — | 100 nM |
| Forskolin | — | 10 μM |
| Y-27632a | 5 μM | 5 μM |
| Heregulin B1 | 5 nM | 5 nM |
| FGF-7 | 5 ng/ml | — |
| FGF-10 | 20 ng/ml | 20 ng/ml |
| A83-01 | 0.5 μM | 0.5 μM |
| EGF | 5 ng/ml | 5 ng/ml |
| SB202190 | 1 μM | — |
Optional: Y-27632 can be removed from the medium 2–3 d after organoid establishment, passaging or thawing, as used in ref. 8.
VitA, vitamin A.
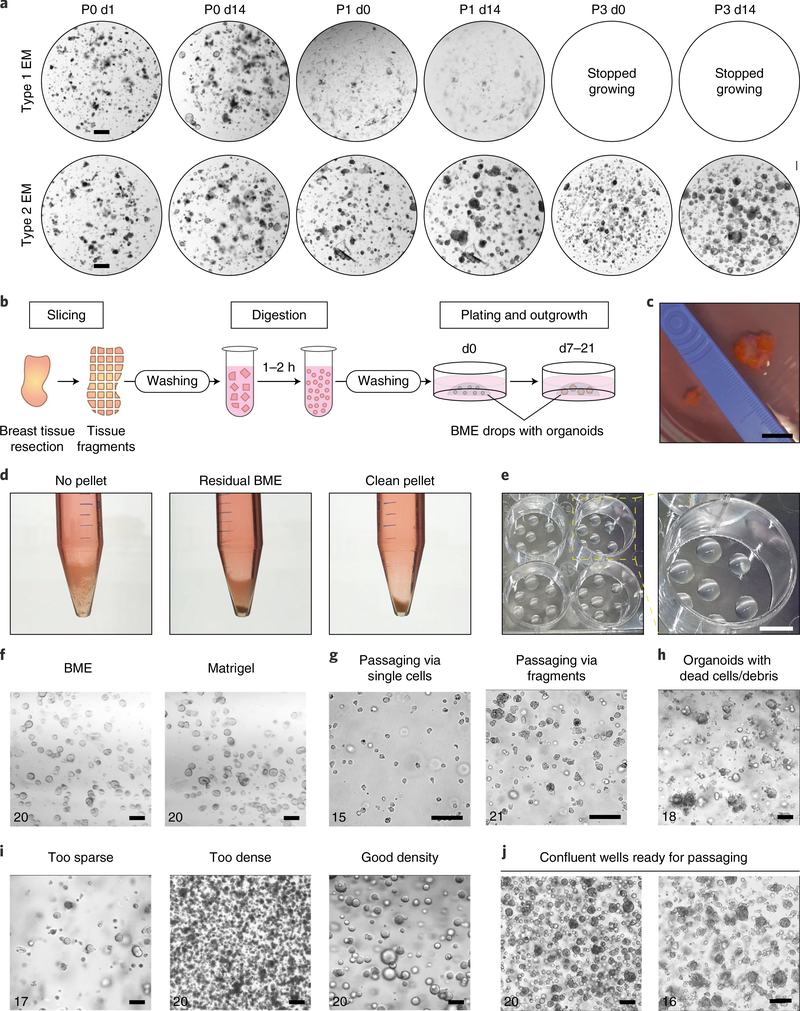
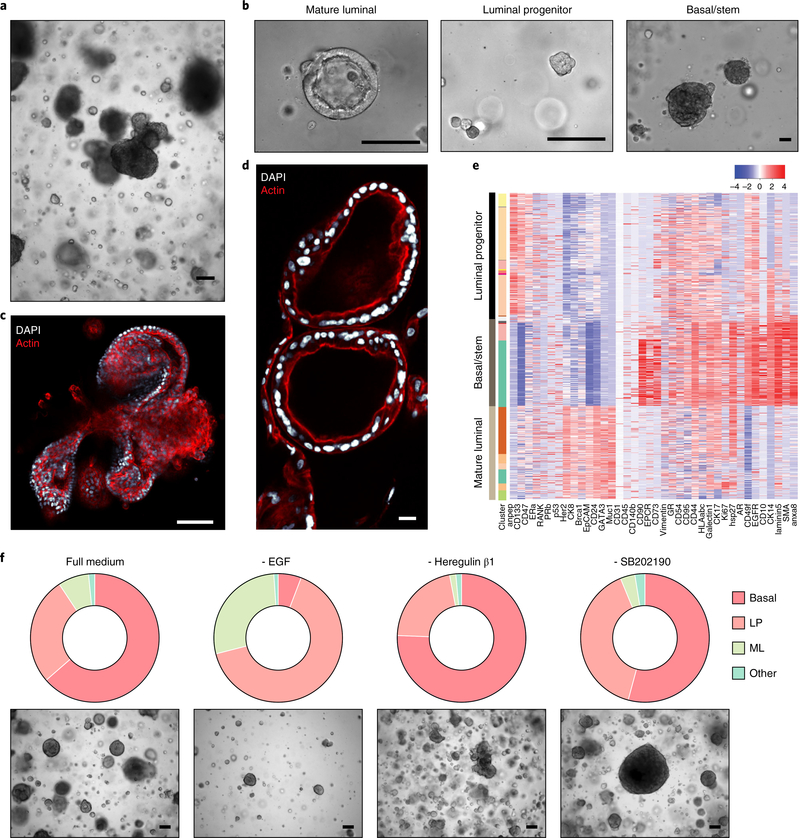
Fig. 2 |. Organoid derivation, culturing and passaging.
a, Bright-field images of BC organoids (Sample 36 of Table 1) grown in either Type 1 or Type 2 expansion medium (EM), demonstrating increased organoid outgrowth in Type 2 compared to Type 1 EM. P, passage number; d, number of days since last split. Scale bar, 400 μm. b, Schematic overview of the organoid derivation process. Biopsies are sliced into 0.5–1-mm3 pieces, digested with collagenase and plated in BME. c, Representative piece of healthy breast tissue. Scale bar, 1 cm. d, Representative images of the absence of a pellet (left), a pellet containing residual BME (middle) and a clean pellet (right), after organoid washing. e, Representative images of BME drops in a 12-well plate. Scale bar, 5 mm. f, Bright-field images of BC organoids grown in either Matrigel or BME, demonstrating similar organoid outgrowth. Scale bars, 100 μm. g, Representative images of BC organoid cultures passaged as single cells (left) or fragments of 5–20 cells (right). Scale bars, 100 μm. h, Representative image of an organoid culture with clear presence of cell debris. Scale bar, 100 μm. i, Representative bright-field images of BC organoids seeded too sparsely (left), too densely (middle) or at the proper density (right). Scale bars, 100 μm. j, Representative images of confluent organoid wells of solid (left) and grape-like (right) cultures. Scale bars, 100 μm. Numbers in f–j refer to sample numbers in Table 1.
Organoid derivation (Steps 1–40)
We describe the establishment of an organoid culture, as previously published7. Patient resection material is cut into small pieces, washed thoroughly and digested with collagenase. After additional washing and filtration, the digested tissue is plated in basement membrane extract (BME) and supplemented with expansion medium (Fig. 2b,c). BME, a gel-like substance that polymerizes at temperatures above 10 °C, is used to mimic the extracellular matrix and provide support for 3D organoids (Fig. 2d,e). Matrigel can be used as an alternative for BME with similar performance (Fig. 2f; see ‘Reagents’ section). Organoid derivation is optimal for tissues that contain a minimal amount of fat and necrotic tissue. The desired epithelial tissue for organoid generation usually appears as firm, pink-to-brownish tissue, whereas necrotic tissue is usually softer and darker. Fat tissue is soft and white to yellow and can be easily scraped or cut off.
Organoid maintenance (Steps 41–48)
The split ratio, passage interval and method of organoid dissociation differ considerably among donors and should be optimized for each newly established organoid culture (Table 1). Organoids can be passaged 7–21 d after organoid establishment (Steps 1–40) or after the previous split at a 1:2–1:8 ratio. Refer to Table 1 for a guideline. The time of passage and split ratio should be optimized for each newly established organoid culture based on confluency. The best moment to passage is just before organoids in the center of the BME drop start dying, characterized by shedding of debris or a darker appearance, or when organoids reach a diameter larger than 300 μm (further referred to as a ‘confluent well’). We describe both single-cell organoid passaging by trypsinization (Step 43A; Fig. 2g) and passaging via organoid fragments by mechanical disruption, referred to as shearing (Step 43B; Fig. 2g). The preferred way of passaging for most organoid cultures is via fragments rather than single cells (Table 1), because single cells are slower at growing into new organoids and have a higher chance of dying compared to fragments. Both methods can be tried for newly established cultures, especially for fast-growing cultures. In some occasions, for instance when cell counts are critical, single-cell passaging should be used. If you want to generate clonal organoid cultures after transfection, we recommend using an organoid culture that allows passaging via single cells. Using single cells, as opposed to fragments, will increase the likelihood that the organoids growing out after selection (e.g., by puromycin) indeed originate from single cells and are, thus, clonal.
We also provide guidelines for culturing at appropriate organoid densities and describe how to minimize cell death and recognize a confluent well of organoids (Step 49–53; Fig. 2h–j). Additionally, we describe procedures and conditions for long-term storage (Steps 54–57) and recovery after cryopreservation (Steps 58–64). We recommend cryopreserving at least six cryovials for each newly established organoid culture at an early passage (< passage 5).
Genetic manipulation (Steps 65A–C)
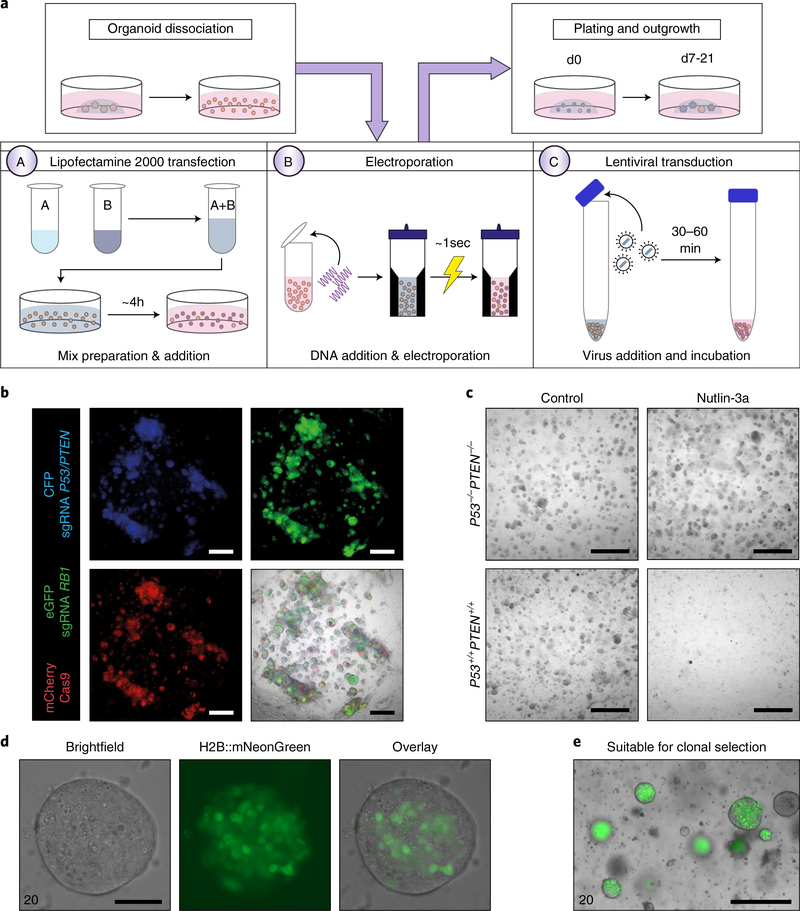
We discuss three different methods for genetically modifying breast organoids, including Lipofectamine 2000 for transient transfection with moderate-to-high efficiency (Step 65A), electroporation for transient transfection with high efficiency (Step 65B; used for Fig. S4 of ref. 7) or lentiviral infection for stable DNA transduction (Step 65C; used for Fig. 1 of ref. 9) (Fig. 3a–c). Specific conditions (e.g., DNA concentration, viral titer and number of organoid cells) or settings (e.g., voltage used for electroporation) require optimization for each experiment and culture. We recommend using control transfection or transduction vectors with a fluorescent label to assess efficiency of the procedure (Fig. 3b–e).
Fig. 3 |. Genetic manipulation of breast organoids.
a, Schematic overview of genetic manipulation of breast organoids. Dissociated organoids can be incubated with a Lipofectamine 2000-based transfection mix (A), electroporated (B) or incubated with high-titer lentivirus (C) and plated in BME. b, Representative fluorescent images of normal breast organoids 7 d in culture after transduction with lentivirus expressing fluorescent reporters and single guide RNAs or Cas9, as indicated. Scale bar, 500 μm. c, Representative bright-field images of normal breast organoids (control) or normal breast organoids knocked out for P53 and PTEN by CRISPR–Cas9 and treated with Nutlin-3a (10 μM) for 7 d. Scale bar, 200 μm. d,e, Representative images of a single organoid (d) or multipe organoids (e) (Sample 20 of Table 1) that were genetically edited to stably express H2B::mNeonGreen and a puromycin resistane gene after 14 d of puromycin selection. Scale bars, 100 μm (d) and 1,000 μm (e). Panels b and c were adapted from ref. 9.
Organoid selection and clonal cultures (Steps 66–77)
Genetically modified organoids can be selected by antibiotic addition if a resistance gene has been introduced or by addition or withdrawal of other medium components. For instance, addition of Nutlin-3a will kill all cells expressing wild-type P53 and can be used to select P53-mutated organoids (Fig. 3c)7,9. In addition, withdrawal of epidermal growth factor (EGF) can select for cells overexpressing KRAS23. We recommend defining the optimal concentration per selection agent and organoid culture, which is often the lowest concentration that kills 100% of untreated organoids. Selection and propagation of individual organoids can be used to generate a clonal culture (Fig. 3d,e). Non-manipulated organoids should be used as a control. Although sub-cloning for each organoid culture can be assessed, we recommend using organoid cultures that efficiently expand after passaging from single cells for better success rates.
Xenotransplantation (Steps 78–108)
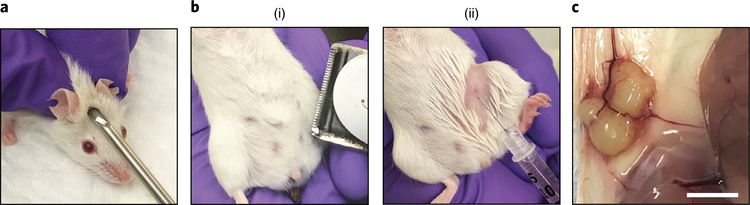
A large quantity of organoids is necessary for engraftment (Steps 78–89), usually varying between 0.25 × 106 and 1 × 106 cells per injected site in the form of intact organoids, for a better engraftment rate compared to single cells. These numbers require optimization per organoid culture. Organoid cultures with a fast growth rate in vitro tend to engraft better in vivo. We describe surgery-free estrogen pellet implantation (Steps 90–97; Fig. 4a) before orthotopic organoid injection (Steps 98–108; Fig. 4b) to provide an exogenous source of estrogen that facilitates tumor engraftment and growth24,25 (Fig. 4c). We recommend practicing organoid injection by injecting 30 μl of transplantation medium with 5% trypan blue, followed by dissection, to confirm localization in the mammary fad pad. It is important to be aware of potential bladder complications that might arise with estrogen pellets26. Of note, estrogen pellets either were9 or were not7 used in previous BC organoid transplantation studies. Mice should be monitored closely for signs of discomfort over the duration of the experiment.
Fig. 4 |. Estrogen pellet implantation and organoid xenotransplantation.
a, Subcutaneous estrogen pellet implantation. b, Shaving of the injection site (i) and orthotopic injection of organoids into the mammary fat pad (ii). c, Representative image of a tumor grown from orthotopically injected BC organoids. Scale bar, 5 mm.
Materials
Animals
Female NSG (NOD.Cg-Prkdcscid Il2rgtm1Wjl /SzJ) mice between 6 and 8 weeks of age !CAUTION Use of rodents for research purposes requires approval from the relevant national and institutional regulatory bodies. All experiments should be carried out in accordance with the local and national animal welfare guidelines and laws.
Biological material
This protocol can be applied to human resection or biopsy material of normal, pre-cancerous, cancerous and metastatic tissue. Fresh material is the preferred starting material, but tissue can be kept in adDMEM/F12+++ with 100 μg/ml of Primocin at 4 °C for up to 72 h before starting the procedure !CAUTION For the use of human material, approval of the relevant national and institutional regulatory bodies is necessary, and informed consent should be obtained from all donors. !CAUTION We recommend occasional profiling of the organoid cultures with a single-nucleotide polymorphism (SNP) array through commercial resources available (e.g., Thermo Fisher Scientific, cat. no. 4475394, following the company’s specific instructions for sample preparation) to rule out cross-contamination with a different culture and to regularly test for mycoplasma contamination.
Reagents
Bovine serum albumin (BSA), modified fraction V (Sigma-Aldrich, cat. no. A9418)
BSA, fatty acid free (Sigma-Aldrich, cat. no. A6003)
Cell Recovery Solution (Corning, cat. no. 354253)
Collagenase Type II (Thermo Fisher Scientific, cat. no. 17101–015)
Cultrex RGF BME, Type 2 (R&D Systems, cat. no. 3533–005-02); Matrigel (e.g., BD Biosciences, cat. no. 356231) can be used as an alternative with equal performance
Ethanol, 70% (vol/vol) (Klinipath, cat. no. 4070–9010) !CAUTION Ethanol is highly flammable.
Formalin, 4% solution (e.g., Sigma-Aldrich, cat. no. 1.00496)
DPBS (Dulbecco’s phosphate-buffered saline, no calcium, no magnesium, 1×; Thermo Fisher Scientific, cat. no. 14190–144)
Recovery cell culture freezing medium (Thermo Fisher Scientific, cat. no. 12648–010)
Trypan blue solution (Sigma-Aldrich, cat. no. T8154)
TrypLE Express (1×, Thermo Fisher Scientific, cat. no. 12605–010)
Fetal bovine serum (FBS), heat inactivated (Thermo Fisher Scientific, cat. no. 10500064)
Dimethyl sulfoxide (DMSO; Sigma-Aldrich, cat. no. D2650)
Reagents for organoid culture
A83–01 (Tocris Bioscience, cat. no. 2939)
Advanced DMEM/F12 (adDMEM/F12; Thermo Fisher Scientific, cat. no. 12634–028)
B27 Supplement (50×, Thermo Fisher Scientific, cat. no. 17504–44)
β-estradiol (Sigma-Aldrich, cat. no. E2257)
DMEM GlutaMAX (Thermo Fisher Scientific, cat. no. 31966–047)
Forskolin (Sigma-Aldrich, cat. no. F6886)
GlutaMAX (100×, Thermo Fisher Scientific, cat. no. 35050–061)
HEPES 1M (100×, Thermo Fisher Scientific, cat. no. 15630–080)
Heregulin β1 (PeproTech, cat. no. AF-100–03)
Hydrocortisone (Sigma-Aldrich, cat. no. H-0888)
N-acetyl-L-cysteine (Sigma-Aldrich, cat. no. A9165–5g)
Nicotinamide (Sigma-Aldrich, cat. no. N0636)
Noggin-conditioned medium, produced by a Noggin-producing cell line, which can be obtained from the laboratory of Prof. H. Clevers (Hubrecht Institute). Preparation instructions can be found in Box 2 of ref. 19 ▲CRITICAL Commercially available Noggin (e.g., U-Protein Express BV, cat. no. N002, at a final concentration of 1–2% (vol/vol)) or recombinant Noggin (PeproTech, cat. no. 120–10C, at a final concentration of 100 ng/ml) can be used as alternatives, but we strongly recommend using in-house-produced conditioned medium.
Nutlin-3a (Merck, cat. no. SML0580)
Penicillin–streptomycin (Thermo Fisher Scientific, cat. no. 15140–122)
Primocin (Invivogen, cat. no. Ant-pm-1)
Recombinant human epidermal growth factor (EGF) (PeproTech, cat. no. AF-100–15)
Recombinant human fibroblast growth factor (FGF)-7 (PeproTech, cat. no. AF-100–19)
Recombinant human FGF-10 (PeproTech, cat. no. AF-100–26)
Red blood cell lysis buffer (Sigma-Aldrich, cat. no. 11814389001)
R-spondin-1-conditioned medium, produced by a R-spondin-1-producing cell line, which can be obtained from the laboratory of Prof. C. Kuo (Stanford University). Preparation instructions can be found in Box 2 of refs. 20–22 ▲CRITICAL Commercially available recombinant R-spondin-3 (eg, R&D Systems, cat. no. 3500-RS/CF, at a final concentration of 250 ng/ml) can be used as an alternative, but we strongly recommend using in-house-produced conditioned medium.
SB 202190 (Sigma-Aldrich, cat. no. S7067)
Wnt3a-conditioned medium, produced by an L Wnt3A-producing cell line, which can be obtained from the laboratory of Prof. H. Clevers (Hubrecht Institute). Preparation instructions can be found in Box 2 of refs. 21,22 ▲CRITICAL Commercially available Wnt surrogate (U-Protein Express BV, cat. no. N001, at a final concentration of 0.2 nM) can be used as an alternative, but we strongly recommend using in-house-produced conditioned medium.
Y-27632 dihydrochloride (ROCK inhibitor; AbMole Bioscience, cat. no. M1817)
Reagents for genetic manipulation
Lipofectamine 2000 (Thermo Fisher Scientific, cat. no. 11668–019)
Opti-MEM, reduced serum medium (Thermo Fisher Scientific, cat. no. 11058–021)
Reagents for xenotransplantation
17β-estradiol pellet, 0.36 mg/pellet, 60-day release (Innovative Research of America, cat. no. SE-121)
Isoflurane (100% (wt/wt); inhalation anesthetic (e.g., Isoflutek; Laboratorios Karizoo) !CAUTION Isoflurane can irritate skin, eyes and lungs and is a suspected teratogen. Work in a biosafety cabinet.
Equipment
1.5-ml Safe-lock tubes (Eppendorf, cat. no. 0030120086)
2.0-ml Safe-lock tubes (Eppendorf, cat. no. 0030120094)
2.0-ml Cryovials (Greiner Bio-One, cat. no. 122263)
Biosafety cabinet
−80 °C freezer (e.g., Eppendorf CryoCube F740hi, cat. no. 740320011)
CoolCell LX Freezing Container (Corning, cat. no. CLS432001)
Counting chamber (e.g., Bürker-Türk, Merck, cat. no. BR719520)
Micro-dissecting forceps (Merck, cat. no. F3767)
Cell culture dishes, 60 mm × 15 mm (Greiner Bio-One, cat. no. 664160)
Cell strainer, 70 μm (Greiner Bio-One, cat. no. 542070)
Cell strainer, 100 μm (Greiner, cat. no. 542000)
Centrifuge for 15/50 ml Falcon Bio-One tubes (e.g., Eppendorf, cat. no. 5804R)
CO2 incubator, 37 °C and 5% CO2 (e.g., Thermo Fisher Scientific, model no. 4141)
Disposable scalpels (Swann-Morton, code 0501)
Electroporation chambers (2 mm), Cuvettes Plus (Thermo Fisher Scientific, cat. no. BTX620)
Falcon tubes, 15 ml, conical bottom (Greiner Bio-One, cat. no. 188271)
Falcon tubes, 50 ml, conical bottom (Greiner Bio-One, cat. nos. 227261)
Gene electroporator (NEPA GENE, model no. NEPA21 Super Electroporator)
Heated incubator oven (e.g., Panasonic, model no. MIR-H163-PE)
Petri dish, glass, 100 mm × 20 mm (VMR, cat. no. 391–0579)
Inverted bright-field microscope, objectives ×5, ×10 and ×20 (e.g., Leica, model no. DMi1)
Microcentrifuge (e.g., Eppendorf, cat. no. 5424R)
Millex-GS Syringe Filter Unit, 0.22 μm (Merck, cat. no. SLGS033SS)
Multi-well suspension plates, 12-, 24- and 48-well (Greiner Bio-One, cat. nos. 665102, 662102 and 677102)
Orbital shaker (e.g., Panasonic, model no. MIR-S100-PE)
Pipetman L, P20L, P200L, P1000L (Gilson, cat. nos. FA10003M, FA10005M and FA10006M)
Pipette tips with filter (e.g., Gilson Pipetman, Diamond tips)
Serological pipettes, 5-, 10- and 25-ml (Greiner Bio-One, cat. nos. 606180, 607160 and 760160)
Equipment for xenotransplantation
MicroFine+ Insulin Syringes, 29-gauge (Becton Dickinson, cat. no. 324825)
Precision trochar, 10-gauge (Innovative Research of America, cat. no. MP-182)
Trimmer, Exacta GT416 (Aesculap, cat. no. AEX415)
Equipment setup
For organoid electroporation, set the NEPA21 electroporator to the following settings:
| Poring pulse | Transfer pulse | |
|---|---|---|
| Voltage | 200 V | 20 V |
| Pulse length | 5 ms | 50 ms |
| Pulse interval | 50 ms | 50 ms |
| Number of pulses | 2 | 5 |
| Decay rate | 10% | 40% |
| Polarity | + | +/− |
Reagent setup
Collagenase Type II
To prepare a 20 mg/ml stock solution, dissolve 1 g of powder in 50 ml of adDMEM/F12+++ (room temperature (RT), 18–22 °C) and filter through a 0.22-μm filter. Store 1-ml aliquots, protected from light, at −20 °C for up to 3 months.
DMEM with 0.1% (wt/vol) BSA (D-BSA)
Supplement 500 ml of DMEM GlutaMAX with 5 ml of penicillin–streptomycin (1% vol/vol) and 5 ml of 10% BSA (fatty acid free, wt/vol in DPBS) solution (final concentration: 0.1% BSA). Store at 4 °C for up to 4 weeks.
adDMEM/F12+++
Supplement 500 ml of Advanced DMEM/F12 with 5 ml of penicillin–streptomycin (1% vol/vol), 5 ml of GlutaMAX (1% vol/vol) and 5 ml of HEPES (1% vol/vol). Store at 4 °C for up to 6 months.
DPBS with 0.1% (wt/vol) BSA (DPBS-B)
Used for diluting specific growth factors. Dissolve 0.1 g of BSA (Modified Fraction V) per 100 ml of DPBS (RT) and filter-sterilize over a 0.22-μm filter. Store 1-ml aliquots at −20 °C.
Type 1 expansion medium
The medium components and final concentrations are listed in Table 2. To make 200 ml of Type 1 expansion medium, add to 153 ml of adDMEM/F12+++: 20 ml of R-spondin-1-conditioned medium, 20 ml of Noggin-conditioned medium, 4 ml of 50× B27 supplement, 2 ml of nicotinamide (1 M in DPBS), 500 μl of N-acetyl-L-cysteine (500 mM in water), 400 μl of Primocin (50 mg/ml), 10 μl of Y-27632 (100 mM in DMSO), 100 μl of Heregulin β1 (10 μM in DPBS-B), 100 μl of human FGF-7 (10 μg/ml in DPBS-B), 100 μl of human FGF-10 (40 μg/ml in 0.1% DPBS-B), 20 μl of A83–01 (5 mM in DMSO), 2 μl of human EGF (500 μg/ml in 0.1% DPBS-B) and 6.8 μl of SB202190 (30 mM in DMSO). This medium can be stored for 1 week at 4 °C.
Type 2 expansion medium
The medium components and final concentrations are listed in Table 2. To make 200 ml of Type 2 expansion medium, add to 112 ml of adDMEM/F12+++: 40 ml of Wnt3a-conditioned medium, 20 ml of R-spondin-1-conditioned medium, 20 ml of Noggin-conditioned medium, 4 ml of 50× B27 supplement, 2 ml of nicotinamide (1 M in DPBS), 500 μl of N-acetyl-L-cysteine (500 mM in water), 400 μl of hydrocortisone (250 μg/ml), 200 μl of β-estradiol (100 μM), 200 μl of Forskolin (10 mM in DMSO), 400 μl of Primocin (50 mg/ml), 10 μl of Y-27632 (100 mM in DMSO), 100 μl of Heregulin β1 (10 μM in DPBS-B), 100 μl of human FGF-10 (40 μg/ml in DPBS-B), 20 μl of A83–01 (5 mM in DMSO) and 2.0 μl of human EGF (500 μg/ml in 0.1% DPBS-B). This medium can be stored for 1 week at 4 °C.
Wnt3a-conditioned medium
The procedure of Wnt3a-conditioned medium production is described in Box 1 of refs. 21,22. Wnt3a-conditioned medium can be stored at 4 °C for 6 months.
R-spondin-1-conditioned medium
The procedure of R-spondin-1-conditioned medium production is described in Box 1 of refs. 20–22. R-spondin-1-conditioned medium can be stored at 4 °C for 6 months.
Noggin-conditioned medium
The procedure of Noggin-conditioned medium production is described in Box 2 of ref. 19. Noggin-conditioned medium can be stored at 4 °C for 6 months.
A83–01
To prepare a 5 mM stock solution (10,000×), dissolve 10 mg of powder in 5 ml of DMSO (RT). Store 50-μl aliquots at −20 °C for up to 1 year.
β-estradiol
To make a 100 μM stock solution (1,000×), dissolve 1 mg of powder in 1 ml of 100% EtOH (RT). Add 35.7 ml of adDMEM/F12+++ and sterilize over a 0.22-μm filter. Store 100-μl aliquots at −20 °C for up to 1 year, avoiding freeze–thaw cycles.
Forskolin
To prepare a 10 mM stock solution (1,000×), dissolve 50 mg of powder in 12.2 ml of DMSO (RT). Store 100-μl aliquots at −20 °C for up to 1 year.
Nicotinamide
To prepare a 1 M stock solution (100×), dissolve 6 g of powder in 50 ml of DPBS (RT) and sterilize over a 0.22-μm filter. Store 1- and 5-ml aliquots at −20 °C for up to 1 year.
N-acetyl-L-cysteine
To prepare a 500 mM stock solution (400×), dissolve 815 mg of powder in 10 ml of water (RT) and sterilize over a 0.22-μm filter. Store 500-μl aliquots at −20 °C for up to 1 year.
Nutlin-3a
To prepare a 1 mM stock solution, dissolve 5 mg of powder in 860 μl of DMSO (RT). Store 50-μl aliquots at −20 °C for up to 1 year, avoiding freeze–thaw cycles.
Heregulin β1
To prepare a 10 μM stock solution (2,000×), dissolve 100 μg of powder in 1,333 μl of DPBS (RT). Store 100-μl aliquots at −20 °C for up to 1 year, avoiding freeze–thaw cycles.
Human EGF
To prepare a 500 μg/ml stock solution (100,000×), dissolve 500 μg of powder in 1 ml of DPBS-B (RT). Store 100-μl aliquots at −20 °C for up to 1 year, avoiding freeze–thaw cycles.
Human FGF7
To prepare a 100 μg/ml master stock solution (20,000×), dissolve 100 μg of powder in 1 ml of DPBS-B (RT). Store 100-μl aliquots at −80 °C for up to 1 year. To prepare a 10 μg/ml stock solution (2,000×), dilute the master stock ten times in DPBS-B (add 100 μl of master stock to 900 μl of DPBS-B). Store 100-μl aliquots at −20 °C for up to 1 year, avoiding freeze–thaw cycles.
Human FGF10
To prepare a 40 μg/ml stock solution (2,000×), dissolve 100 μg of powder in 2.5 ml of DPBS-B (RT). Store 100-μl aliquots at −20 °C for up to 1 year, avoiding freeze–thaw cycles.
Hydrocortisone
To make a 250 μg/ml stock solution (500×), dissolve 10 mg of powder in 500 μl of 100% EtOH (RT). Add 39.5 ml of adDMEM/F12+++ (RT) and sterilize over a 0.22-μm filter. Store 200-μl aliquots at −20 °C for up to 1 year, avoiding freeze–thaw cycles.
SB 202190
To prepare a 30 mM stock solution (30,000×), dissolve 5 mg of powder in 500 μl of DMSO (RT). Store 10-μl aliquots at −20 °C for up to 1 year.
Y-27632 dihydrochloride (ROCK inhibitor)
To prepare a 100 mM stock solution (20,000×), dissolve 10 mg of powder in 312 μl of DMSO (RT). Store 100-μl aliquots at −20 °C for up to 1 year. ▲CRITICAL Be aware that the published half-life of Y-27632 is shorter than the time we advise to store Type 1 and Type 2 expansion medium (1 week). In general, 4 °C-stored expansion medium can be used at any moment for organoid propagation with good performance within a week from preparation. However, for organoid establishment from tissue (Steps 11–40), after genetic manipulation (Steps 65–89) or when passaging single organoids (Steps 90–101), we recommend using expansion medium that is as fresh as possible for optimal recovery of single cells.
Transplantation medium
Prepare this medium while harvesting organoids for transplantation (Steps 105–107). Account for 40 μl of transplantation buffer per injection, plus at least 10% extra. Mix equal volumes of ice-cold Type 1 or Type 2 medium (donor dependent) and BME, avoiding bubbles. Store on ice until organoid pellet is ready for resuspension.
Procedure
Establishment of organoids from breast tissue resections ● Timing 4.5–6.5 h
▲CRITICAL This section describes the isolation of epithelial cells from breast tissue resections for the establishment of organoid cultures (Fig. 2b). When pipetting disrupted tissue, always pre-wet the 5- or 10-ml sterile serological pipettes with D-BSA medium for preventing tissue sticking to the plastic (Steps 12, 13, 19, 21, 24 and 32). When receiving resected material from two different locations (ie, normal and tumor), start processing the normal tissue and, while this is digesting, process the tumor tissue. The derivation success of normal organoids depends on the viability of the stem cells in the normal tissue, which are usually present in lower numbers than cancer (stem) cells in tumor tissue. Normal tissue should, thus, be processed as soon as possible upon arrival. Resected tissue should be placed in a Falcon tube containing 14 ml of adDMEM/F12+++ with Primocin (100 μg/ml) and be transferred to the lab as soon as possible for isolation.
!CAUTION As the derivation process is performed with primary human material, which could potentially carry human pathogens, all steps should be performed wearing appropriate personal protective equipment (PPE), while working in a biosafety cabinet, and with the tissue in closed tubes or Petri dishes when checking it under the microscope. Working in a biosafety cabinet also reduces the risk of contaminating the tissue and isolated organoids with microorganisms.
▲CRITICAL Store the Falcon tube containing tissue on wet ice in case it takes up to 72 h to reach the lab for isolation.
▲CRITICAL The procedures are described for working with 12-well plates. Refer to Table 3 in case a culture plate with a different number of wells is desired.
Table 3 |.
Standard number of BME drops and volumes of BME and culture medium per type of culture plate
| Culture plate | BME (μl) | BME drops | Medium (μl) |
|---|---|---|---|
| 96-well | 10 | 1 | 100 |
| 48-well | 25 | 1 | 200 |
| 24-well | 50 | 4 | 500 |
| 12-well | 100 | 5–8 | 750–1,000 |
| 6-well | 200 | 10–16 | 1,500–2,000 |
Tissue preparation ● Timing 30–50 min
-
1
Transfer each tissue sample (<3–4 cm3) into a separate 50-ml Falcon tube filled with D-BSA. Keep tissues on ice until starting the isolation.
-
2
Register the new sample in the corresponding lab management system.
-
3
(Optional) Generate labels for collecting pieces of tissue for different purposes (DNA, RNA, protein and/or histology).
-
4
Collect the 50-ml Falcon tube containing the normal or tumor tissue and transfer the tissue to a 10-cm glass Petri dish by decanting the 50-ml Falcon tube (Fig. 2c).
-
5
Take a microscopic picture of the tissue, including a ruler in the picture (Fig. 2c). Make a note of how the tissue looks (ie, size, fatty, vascularized, necrotic, etc) in the corresponding lab note.
-
6
Aspirate the excess D-BSA and cut the tissue into small pieces (2–4 mm3) using two scalpels by holding one scalpel in each hand and slicing the tissue while pressing the blades of both scalpels against each other.
-
7
(Optional) Use a micro-dissecting forceps cleaned with 70% EtOH to transfer 1–2 pieces of 2–4-mm3 tissue into a 2-ml Eppendorf tube labeled for DNA analysis, snap freeze on dry ice and transfer to −20 °C for storage.
▲CRITICAL This tissue sample can be used as the reference DNA sample for SNP analysis (see ‘Biological material’ section).
-
8
(Optional) Fix two pieces of 2–4-mm3 tissue for histology by transferring them into a 15-ml Falcon tube labeled with histology label and containing 5 ml of 4% formalin.
!CAUTION Perform this step in a fume hood, as formalin is toxic.
-
9
(Optional) When future transcriptomic analyses are required, transfer 1–2 pieces of 2–4-mm3 tissue into a 2-ml microtube labeled with RNA label, snap freeze on dry ice and transfer to a −80 °C freezer.
-
10
(Optional) When future proteomic analyses are required, transfer 1–2 representative pieces of tissue into a 2-ml microtube labeled with protein label, snap freeze on dry ice and transfer to a −80 °C freezer.
▲CRITICAL Be aware that optional Steps 7–10 will reduce tissue volume for organoid generation, which might result in a longer organoid propagation phase before frozen stocks can be generated. On average, 8–10 mm3 would be enough for one well of a 12-well plate, but this is highly variable depending on the cellularity of the tissue sample.
▲CRITICAL For Steps 7–10, make sure to select tissue pieces that represent different colors or regions of the resected tissue to better capture different cell populations present.
Organoid isolation from tissue ● Timing 1.5–5 h
-
11
Mince the remaining tissue into smaller pieces (0.5–1 mm3) with two scalpels as stated in Step 6, until the tissue mass looks uniform and appears somewhat viscous.
-
12
Add 5 ml of D-BSA into the glass Petri dish and transfer the minced tissue to a 50-ml Falcon tube using a 5-ml sterile serological pipette pre-wetted with D-BSA. Wash the same glass Petri dish with 5 ml of D-BSA and collect residual minced tissue into the same 50-ml Falcon tube.
-
13
Add 35 ml of D-BSA to the 50-ml Falcon tube and use a 10-ml sterile serological pipette pre-wetted with D-BSA to resuspend the minced tissue by pipetting up and down five times.
-
14
Let the tissue sediment by gravity for 2–3 min, until all tissue pieces are at the bottom of the tube, before aspirating all but 10 ml of D-BSA.
-
15
Repeat Steps 13 and 14 two more times to wash away potential microorganisms, cell types not contributing to the establishment of organoids, such as blood cells, and dead cells and debris.
-
16
Aspirate all supernatant carefully, so that only the sedimented tissue pellet remains, and add 10 ml of Type 1 medium.
-
17
Add 500 μl of collagenase (20 mg/ml stock, final concentration 1 mg/ml) and 10 μl of ROCK inhibitor (10 mM stock, final concentration 10 μM) to the 50-ml Falcon tube containing the minced tissue.
-
18
Wrap the 50-ml Falcon tube with Parafilm and place the tube at a slight angle (~15°) on top of an orbital shaker located inside a heated incubator oven at 37 °C. Set the orbital shaker at 140 r.p.m. and digest the tissue for 1–2 h. At this moment, the processing of the next tissue can be initiated.
▲CRITICAL Placing the tube at a slight angle facilitates the distribution of minced tissue along the tube for proper digestion, as it increases the surface area of tissue exposed to the digestion reagent. The angle can be achieved by placing the top of the tube on a 0.5–1-cm platform and resting the end of the tube on the shaker.
-
19
Pipette up and down 5–10 times every 30 min using a 5-ml sterile serological pipette pre-wetted with D-BSA medium to aid digestion. Monitor the digestion of the tissue at least every 30 min, using an inverted bright-field microscope (×5, ×10 or ×20 objective). When clusters of 5–10 cells are observed (Fig. 2g), digestion is complete.
? TROUBLESHOOTING
▲CRITICAL Normal tissue digestion typically takes 2 h, whereas tumor tissue digestion typically takes 0.5–2 h. The time of digestion depends on several variables, such as the time from tissue isolation to digestion, tissue size and consistency and the quality and activity of the digestion reagent. Prolonged tissue storage before processing or freshly made digestion reagent reduces the digestion time, whereas solid, firm tissue or an older digestion reagent increases the digestion time.
-
20
After incubation, take the tube from the heated incubator oven and clean with 70% EtOH. Add 1 ml of FBS directly to the 50-ml Falcon tube to stop the collagenase digestion.
-
21
Homogenize the digested tissue by pipetting it up and down vigorously with a 10-ml sterile serological pipette pre-wetted with D-BSA.
-
22
Pre-wet a 100-μm strainer by pipetting 10 ml of D-BSA on the strainer into a new 50-ml Falcon tube. Swirl the 50-ml tube to wet the sides of the tube and discard the D-BSA. Then, strain the digested tissue over the 100-μm strainer into the 50-ml Falcon tube (both pre-wetted with D-BSA).
-
23
Transfer the unfiltered tissue pieces back into the initial 50-ml Falcon tube by reversing the filter and flushing with 10 ml of D-BSA.
-
24
Pipette the digested tissue up and down vigorously, using a 10-ml sterile serological pipette pre-wetted with D-BSA.
-
25
Reverse the 100-μm filter and strain the tissue again, collecting the cell suspension in the same 50-ml Falcon tube from Step 22.
-
26
Repeat Steps 23–25 two more times, using the same filter to avoid wasting precious tissue.
-
27
Centrifuge the flow-through from Steps 22–26 at 450g for 5 min at 8 °C.
-
28
Aspirate the supernatant, leaving the last 4 ml to avoid disturbing the pellet. Resuspend the tissue pellet in the remaining 4 ml with a sterile serological pipette and transfer it to a 15-ml Falcon tube pre-wetted with D-BSA.
-
29
Add 10 ml of D-BSA with a sterile serological pipette and homogenize by pipetting up and down several times.
-
30
Centrifuge the 15-ml Falcon tube at 450g for 5 min at 8 °C. Aspirate the supernatant very carefully with a vacuum system and clean tip until 1 ml is left. Let the tube rest for 1 min to allow the D-BSA from the side of the tube to settle at the bottom and remove the residual D-BSA with a P1000 pipette.
-
31
(Optional) If the pellet is partially red, add 1–2 ml of red blood cell lysis buffer with a P1000 pipette, resuspend the pellet by pipetting up and down several times and incubate for 2 min at RT before continuing with Step 32.
▲CRITICAL For pellets <100 μl in size, 1 ml of red blood cell lysis buffer is sufficient. For pellets >100 μl, use 2 ml of red blood cell lysis buffer.
-
32
Add 10 ml D-BSA to the 15-ml Falcon tube with a sterile serological pipette and pipette up and down several times using the same pipette.
-
33
Centrifuge the cells and aspirate the medium as described in Step 30.
■PAUSE POINT At this point, cells can be frozen with Recovery Cell Freezing Medium by adding 1 ml, resuspending the cell pellet and transferring to a cryovial. Preserving part of the isolation yield will ensure a backup of the sample in case the culture is lost at an early passage before cryopreservation (e.g., due to bacterial or fungal contamination or improper handling by an inexperienced user).
-
34
Resuspend the pellet in an appropriate amount of BME. As a guideline, 200 μl is sufficient for a pellet of ~50 μl. The amount of BME to add scales linearly with the pellet volume.
▲CRITICAL After filtering the digested tissue, the cell pellet should be easy to resuspend in BME. However, if the pellet is very sticky, cut the tip of the 200-μl low-retention filter tip.
▲CRITICAL BME must be kept on ice to prevent solidification. Work quickly to prevent the BME from solidifying but carefully to prevent bubble formation.
-
35
Plate 100 μl of BME-containing cells in multiple small drops (<20 μl each) per well in a 12-well plate (Fig. 2e). When tissue yield is limited, plating can be scaled down to 24-well format by adapting all volumes accordingly (Table 3).
-
36
Turn the plate upside down and leave in the biosafety cabinet for 5 min.
-
37
Transfer the plate, upside down, into a 37 °C incubator and leave to solidify for 30 min.
▲CRITICAL Solidifying the BME upside down prevents the tissue fragments from sinking and attaching to the bottom of the plate.
-
38
Add 750 μl of pre-warmed (RT to 37 °C) Type 1 or Type 2 medium to each well and transfer to an incubator at 37 °C and 5% CO2. Refer to Table 3 for appropriate media volumes per type of culture plate.
▲CRITICAL For each culture, test both Type 1 and Type 2 expansion medium (Fig. 2a; Table 2). Occasionally monitor organoid growth with an inverted bright-field microscope (×5, ×10 or ×20 objective) and continue with the type of medium that results in the highest organoid confluency, thus yielding the highest outgrowth. Type 1 is typically better for BC organoids; Type 2 is typically better for normal organoids (Table 1).
-
39
(Optional) Take microscopic pictures of representative drops for documentation of organoid density and morphology (×2.5 and ×10 magnifications).
-
40
Refresh culture medium every 2–4 d by following Steps 41 and 42. After isolation, organoids can be split for the first time after 7–21 d by following Steps 43–48.
? TROUBLESHOOTING
▲CRITICAL After 3–4 d, it should be possible to identify organoids with an inverted bright-field microscope (×5, ×10 or ×20 objective). Monitor organoid growth closely and ensure that most have reached a size of at least 100 μm before passaging.
▲CRITICAL At early passages, it is possible to observe highly elongated fibroblast growing on the bottom of the culture plate. Avoid scratching the bottom of the plate while passaging to prevent transfer of the fibroblast to the next passage.
Refreshing medium ● Timing 5 min
-
41
Tilt the plate at a 45° angle and aspirate all expansion medium from the well containing cultured organoids by placing the aspiration tip to the side of the well. Avoid getting close to the BME drops.
▲CRITICAL Use a clean tip for each different organoid culture and remove the lid from one culture plate at a time to avoid cross-contamination.
-
42
Tilt the plate at a 45° angle and gently add dropwise 750 μl of expansion medium to each well using a P1000 pipette when refreshing a few wells or a 5-ml or 100-ml sterile serological pipette when refreshing many wells. Aim at the bottom corner of the well to avoid disturbing the BME drops.
▲CRITICAL Do not touch the well while adding expansion medium to avoid cross-contamination.
Organoid maintenance and cryopreservation ● Timing 75 min–2 h
Organoid dissociation ● Timing 30–60 min
▲CRITICAL This section describes the culture of normal breast and BC organoids in a 12-well plate. Scale the volumes of reagents needed using Table 3 as a reference if using a different well size.
▲CRITICAL To prevent cross-contamination of different organoid cultures, work cautiously when handling different cultures simultaneously.
-
43
Dissociate organoids into either single cells (Option A) or fragments (Option B).
(A). Dissociating organoids into single cells by TrypLE digestion ● Timing 10–40 min
Aspirate all expansion medium from a well containing organoids by holding the plate at a 45° angle and placing the aspiration tip to the side of the well, not getting close to the BME drops. When passaging multiple wells of organoids of the same culture, all medium of those wells can be aspirated at once with the same tip.
Add 1 ml of TrypLE to the well and dissociate the BME by forcefully pipetting up and down 6–10 times per well with a P1000 pipette, aiming at the BME drops.
(Optional) Repeat Step (ii) with other wells of the same organoid culture by transferring the 1 ml of organoids in TrypLE to the next well with organoids without expansion medium and repeating the pipetting process. Up to three wells of organoids can be combined in 1 ml of TrypLE.
Directly after completing Step (ii) or (iii), hold the plate at a 45° angle and forcefully pipette the BME-TrypLE mixture with organoids up and down ten times. This will dissociate most of the BME, but the organoids will remain intact. Bubble formation is not harmful but can accelerate the dissociation.
Incubate the plate at RT for 5–10 min.
Hold the plate at a 45° angle and forcefully pipette the BME-TrypLE mixture with organoids up and down ten times to mechanically dissociate the organoids.
Determine if a single-cell solution has been achieved by checking the well with an inverted bright-field microscope (×5, ×10 or ×20 objective) (Fig. 2g). If partially dissociated organoids are still visible, repeat Steps (v) and (vi) until a single-cell solution has been achieved. Continue with Step 44.
▲CRITICAL Incubation time varies among donors, but the total incubation time should, ideally, take no more than 20–25 min. For cultures that are hard to dissociate, the plates can be incubated at 37 °C instead of RT in Step (v).
(B). Dissociating organoids to fragments by shearing with TrypLE ● Timing 10–20 min
Remove 500 μl of expansion medium from the well containing organoids, leaving ~250 μl in the well, by holding the plate at a 45° angle and placing the aspiration tip to the side of the well, not getting close to the BME drops.
Add TrypLE to the well, to a final volume of 1 ml, and dissociate the BME by forcefully pipetting up and down 6–10 times per well with a P1000 pipette, aiming at the BME drops.
(Optional) Repeat Step (ii) with other wells of the same organoid culture by transferring the 1 ml of organoids in TrypLE to the next well with organoids without expansion medium and repeating the pipetting process. Up to three wells of organoids can be combined in 1 ml of TrypLE.
Directly after Step (iii), hold the plate at a 45° angle and forcefully pipette the organoids in TrypLE up and down ten times. This will dissociate most of the BME, but the organoids will still be intact.
To shear the organoids, hold the plate at a 45° angle and forcefully pipette up and down 10–20 times, firmly pressing the 1-ml tip to the bottom corner of the well to create a small opening for organoid disruption.
Check the organoids with an inverted bright-field microscope (×5, ×10 or ×20 objective). Repeat Step (v) until the desired fragment size has been achieved (Fig. 2g). Continue with Step 44.
▲CRITICAL Organoid shearing will result in a suspension of single cells and small-to-large organoid fragments. Shearing is optimal when most cells are present in the form of small organoid fragments (usually containing 5–20 cells or 20–50 μm in diameter).
-
44
Transfer single cells or organoid fragments into a 15-ml Falcon tube with 10 ml of ice-cold adDMEM/F12+++, using a P1000 pipette. Rinse the well with 1 ml of adDMEM/F12+++ to ensure that no dissociated organoids remain in the well. A maximum of six wells with organoids from the same culture can be pooled in one 15-ml Falcon tube.
■PAUSE POINT The 15-ml Falcon tube with processed organoids can be stored on ice for 1 h to first harvest multiple cultures before continuing with Step 45, with minimal effect on organoid recovery after passaging.
-
45
Centrifuge the 15-ml Falcon tube with organoids at 300g for 5 min at 4 °C and confirm presence of a BME-free pellet by eye (Fig. 2d). Presence of residual BME in the pellet should be avoided as this layer might contain dead cells and debris that can negatively influence organoid growth.
? TROUBLESHOOTING
-
46
Remove the supernatant by decanting the fluid into a waste bottle and put the pellet on ice for 1 min. Remove the remaining supernatant carefully with a P1000 pipette.
-
47
(Optional) If you want to count the cells at this stage, resuspend the cell pellet in 250–500 μl of expansion medium per harvested well, taking 10 μl of cells and mixing it with 10 μl trypan blue solution and count using a counting chamber and an inverted bright-field microscope (×10 or ×20 objective). One confluent well of a 12-well plate commonly contains 200,000–500,000 cells. Before continuing with Step 48, spin down the non-counted cells at 300g for 5 min and remove the supernatant.
-
48
To directly plate organoids for passaging, continue to Step 49. To genetically manipulate organoids, continue to Step 65A for Lipofectamine 2000-based transfection, to Step 65B for electroporation-based transfection and to Step 65C for lentiviral transduction.
(Optional) Plating organoids for passaging ● Timing 15–20 min
-
49
Use a P200 or P1000 pipette to resuspend the pellet of dissociated organoids in the 15-ml tube in ice-cold BME by carefully pipetting up and down approximately ten times. Plate in multiple small drops (<20 μl each) per well (Fig. 2e). Refer to Table 3 for appropriate number of BME drops and total BME volume for different culture plates.
▲CRITICAL In addition to using a new tip, we recommend cleaning the P200 or P1000 pipette with 70% EtOH before using it to resuspend the pellet of the next culture, to prevent cross-contamination.
-
50
Swiftly but carefully flip the plate upside down before placing in the incubator to prevent organoids from sinking to the bottom and attaching to the plate.
-
51
Place the plate in an incubator (5% CO2, 37 °C) for 10–20 min to allow the BME to solidify.
-
52
Supplement the well with freshly passaged organoids with 750 μl of pre-warmed (RT, 37 °C) Type 1 or Type 2 medium in a dropwise manner. Refer to Table 3 for the appropriate media volumes when using a 6, 24, 48 or 96-well culture plate.
-
53
Refresh expansion medium every 2–4 d by following Steps 41 and 42. Occasionally check organoid density and morphology with an inverted bright-field microscope (×5, ×10 or ×20 objective).
▲CRITICAL Organoids secrete factors that stimulate cell proliferation. Therefore, culturing with an organoid density that is not too sparse (risk of slow growth and cell death) or too dense (risk of overgrowth) is a critical factor for a well-growing, viable culture (Fig. 2h-j).
? TROUBLESHOOTING
(Optional) Freezing intact organoids ● Timing 10–15 min
▲CRITICAL Make sure to freeze organoids 2–7 d before they reach optimal confluency for splitting, usually when they reach a diameter of 100–150 μm. This ensures a recovery efficiency after thawing of close to 90%.
-
54
Follow Steps 43A, (i)–(iv) to remove intact organoids from the BME.
-
55
Follow Steps 44–46 to wash and pellet the intact organoids.
-
56
Resuspend the pelleted organoids in Recovery Cell Freezing Medium. Combine up to four wells of a 12-well plate of the same donor in 1 ml of freezing medium. Transfer the organoids in freezing medium to a 2-ml cryovial, storing 1 ml per vial.
-
57
Transfer the cryovials to a −80 °C cell-freezing container. Store at −80 °C at least overnight before transferring to a longer-term storage method.
■PAUSE POINT Organoids can be stored at −80 °C for up to 1 month or in liquid nitrogen indefinitely.
(Optional) Thawing organoids ● Timing 20 min
-
58
Transport the cryovial containing organoids from the −80 °C or liquid nitrogen storage to the cell culture laboratory on dry ice.
-
59
Quickly thaw the cryovial containing organoids in a 37 °C water bath, clean the vial using 70% EtOH and dropwise add 1 ml of pre-warmed (37 °C) adDMEM/F12 +++ supplemented with 10 μM ROCK inhibitor with a P1000 pipette while tapping the bottom of the vial.
▲CRITICAL ROCK inhibitor is added to enhance recovery from freeze–thawing.
-
60
Gently resuspend two times, transfer the content of the vial to a 15-ml Falcon tube and dropwise add 10 ml of pre-warmed (37 °C) adDMEM/F12 +++ supplemented with 10 μM ROCK inhibitor.
-
61
Centrifuge the 15-ml Falcon tube with organoids at 300g for 5 min at 4 °C
-
62
Remove the supernatant and put the pellet on ice for a few minutes to allow the residual supernatant to reach the bottom of the tube. Remove the remaining supernatant carefully with a P200 pipette.
-
63
Follow Steps 49–53 to plate the organoids.
-
64
Refresh the expansion medium every 2–4 d by following Steps 41 and 42.
(Optional) Genetic manipulation
-
65
If you want to genetically manipulate the organoids (Fig. 3a), follow Option A (Lipofectamine 2000), Option B (electroporation) or Option C (lentiviral transduction (Fig. 3b).
▲CRITICAL Use 20–50% of one confluent well of organoids (12-well plate) or 100,000–200,000 single organoid cells for each transfection or transduction condition.
▲CRITICAL Plate the manipulated organoids at least twice as densely as compared to standard passaging.
▲CRITICAL For optimal efficiency and organoid recovery, make sure to use an organoid culture with minimal presence of cell debris and dying organoids as starting material (Fig. 2i,j) and process organoids into small fragments containing several cells (Fig. 2g).
▲CRITICAL After genetic manipulation of organoids, expression of transgenes can disappear due to silencing or genetic rearrangements, especially in tumor organoids. It is, therefore, important to supplement the expansion medium with the appropriate selection agent from the manipulation onward to maintain transgene expression in close to 100% of cells.
(A). Transfection of organoids using Lipofectamine 2000 ● Timing 5 h
Dissociate and pellet the organoids as described in Steps 43A/43B–46. After aspirating the supernatant from the organoid pellet as described in Step 46, continue with Step (ii).
-
Resuspend the pellet of dissociated organoids in 450 μl of Type 1 or Type 2 expansion medium per condition using a P1000 pipette, by pipetting five times up and down, and transfer 450 μl to each well of a 24-well suspension plate.
■PAUSE POINT Place the plate at 37 °C in 5% CO2 while the transfection mix is being prepared.
Prepare two 1.5-ml Eppendorf tubes. To the first, add Lipofectamine 2000 (4 μl + 10% per transfection) and Opti-MEM (21 μl + 10% per transfection). To the second, add plasmid DNA (4 μl of a 1 μg/μl stock + 10% per transfection) and Opti-MEM (21 μl + 10% per transfection).
Vortex both Eppendorf tubes briefly at maximum speed and incubate at RT for 5 min.
Transfer the content of the first to the second Eppendorf tube, using a P200 pipette, vortex the Eppendorf tube briefly and incubate at RT for 20 min.
Add 50 μl of transfection mix to 450 μl of expansion medium with organoids, using a P200 pipette.
Mix the transfection mix with the organoids by gently pipetting up and down three times, using a P1000 pipette, preventing bubble formation.
Incubate at 37 °C in 5% CO2 for 4 h.
Transfer the transfected organoids to a 15-ml Falcon tube containing 3 ml of adDMEM/F12+++, using a P1000 pipette.
Centrifuge the Falcon tube with transfected organoids for 5 min at 300g and plate as described in Steps 49–53.
When the transfected DNA contains a gene encoding a fluorescent protein (Fig. 3d,e), expression can be detected within 1 d after transfection. For selection of organoids using a selection agent, follow Steps 66–68. The transfection efficiency is usually between 5% and 50%.
? TROUBLESHOOTING
(B). Transfection of organoids by electroporation ● Timing 50–60 min
-
i
Dissociate and pellet the organoids as described in Steps 43A/43B–46. After aspirating the supernatant from the organoid pellet as described in Step 46, continue with Step (ii). Place the Falcon tube with dissociated organoids on ice while preparing the transfection mix.
-
ii
Prepare the transfection mix. Per condition, add 10 μg of plasmid (in max 10 μl) to a 1.5-ml Eppendorf tube and top up with Opti-MEM to a final volume of 100 μl. Vortex the Eppendorf tube briefly and place on ice for 1 min.
-
iii
Resuspend the pellet of dissociated organoids in the transfection mix by gently pipetting up and down five times and transfer the sample (~100 μl) into a 2-mm electroporation cuvette, using a P200 pipette. Make sure the sample reaches the bottom and prevent bubble formation.
-
iv
Place the cuvette in the NEPA21 electroporator and check the resistance (ohm).
▲CRITICAL The transfection is most optimal between 0.030 ohm and 0.055 ohm. The sample should be diluted in transfection mix (prepared in Step (ii)) if the resistance is too high.
-
v
Electroporate using the settings as described in the ‘Equipment setup’. Vary in V (100–300) and pulse length (2–8 ms) to optimize settings for each organoid culture.
? TROUBLESHOOTING
-
vi
Add 400 μl of ice-cold expansion medium to the cuvettes and transfer the sample into a clean 1.5-ml Eppendorf tube using a P200 pipette.
-
vii
Centrifuge the Eppendorf tube with electroporated organoids for 5 min at 300g and plate transfected cells as described in Steps 49–53.
-
viii
When the transfected DNA contains a gene encoding a fluorescent protein (Fig. 3d,e), expression can be detected within 1 d after transfection. For selection of organoids using a selection agent, follow Steps 66–68. The transfection efficiency is usually between 20% and 80%.
? TROUBLESHOOTING
(C). Lentiviral transduction of organoids ● Timing 1–1.5 h
!CAUTION Follow institutional biosafety guidelines for working with infectious material while working with lentivirus.
Dissociate and pellet the organoids as described in Steps 43A/43B–46. After aspirating the supernatant from the organoid pellet as described in Step 46, continue with Step (ii). When performing a cell count is desired, perform Step 47 before continuing with Step (ii). We describe transduction using lentivirus concentrated to a titer of 50 × 106 infectious viral particles per ml (pfu/ml).
Resuspend the pellet of dissociated organoids in 10 μl of virus (500,000 infective virus particles) + 90 μl of Type 1 or Type 2 expansion medium by gently pipetting up and down five times with a P200 pipette. When 100,000 organoid cells are used (often similar to ~1/3 of a confluent well of a 12-well plate), the multiplicity of infection (MOI) is 5.
Incubate the 15-ml Falcon tube at 37 °C in 5% CO2 for 30–60 min with a loose lid to allow diffusion of gas to the organoids.
Add 10 ml of cold adDMEM/F12+++ and centrifuge the Eppendorf tube with transduced organoids for 5 min at 300g and plate as described in Steps 49–53.
When the transduced DNA contains a gene encoding a fluorescent protein (Fig. 3d,e), expression can be detected within 2–3 d after transduction. For selection of organoids using a selection agent, follow Steps 66–68. The transduction efficiency is usually between 20% and 90%.
? TROUBLESHOOTING
Organoid selection ● Timing 10 min
▲CRITICAL Organoid selection (Fig. 3c) can be started 3 d after transfection or transduction, by adding the selection agent to the expansion medium. It is advised that the concentration of the selection agent is first optimized by culturing organoids using a titration of the selection agent. We advise to use the lowest concentration that kills 100% of untreated organoids after 3–10 d of culture.
-
66
Three days after transfection (Step 65A or 65B) or transduction (Step 65C), aspirate all medium from a well containing organoids by holding the plate at a 45° angle and placing the aspiration tip to the side of the well, avoiding getting close to the BME drops.
-
67
Gently add 750 μl of expansion medium (with the appropriate concentration of selection agent) in a dropwise manner to each well.
-
68
Refresh the expansion medium every 2–4 d following Steps 41 and 42 until organoids have reached a size sufficient for further passaging (diameter of <300 μm; Steps 43–53) or clone picking (diameter of 200–500 μm; Steps 69–77).
Clone picking ● Timing 1.5–2 h
▲CRITICAL To ensure that individual organoids used for clone picking originated from a single cell, it is best to generate a culture with sparsely growing organoids after selection (Fig. 3e). If no selection is applied, this can also be achieved by passaging at low density.
-
69
Prepare 1.5-ml Eppendorf tubes, one per organoid, with 100 μl of TrypLE.
-
70
Disrupt the BME drops with cultured organoids by pipetting the 750-μl expansion medium up and down 8–10 times with a P1000 pipette, aiming at the top of the BME drops, and transfer the organoids to a 10-cm culturing dish with 10 ml of adDMEM/F12+++ at RT.
-
71
Use an inverted bright-field microscope (×20 objective) and pick individual organoids with a P20 pipette, thereby aspirating as little volume as possible (<10 μl). Transfer each individual organoid to a separate 1.5-ml Eppendorf tube with TrypLE. When working outside of a biosafety cabinet with primary human cells, it is essential to work fast to avoid contamination and wear appropriate PPE.
▲CRITICAL Pre-wet the 20-μl tip with Type 1 or Type 2 expansion medium to prevent organoids sticking to the tip. Alternatively, D-BSA or adDMEM/F12+++ can be used.
-
72
Incubate the Eppendorf tubes with a single organoid each for 5 min at RT. Using a P200 pipette, add 100 μl of FBS at RT to the Eppendorf tube and directly resuspend the clone by pipetting 3–10 times up and down.
-
73
Use an inverted bright-field microscope (×10 or ×20 objective) to check the dissociation. Repeat Step 72 until the organoid is dissociated into small fragments and/or single cells (see Fig. 2g as reference).
▲CRITICAL The incubation time (usually 5–20 min total) and number of times to resuspend (usually 5–30 times total) need to be optimized per organoid culture. The digestion time of a clonal organoid is often similar to the digestion time of the parental culture when digesting for passaging.
▲CRITICAL Make sure not to over-digest the organoid, as this will lead to a decreased viability. Do not digest the single organoid for longer than 25 min.
-
74
Centrifuge the Eppendorf tubes at 300g for 5 min. Carefully aspirate the supernatant with a P200 pipette and resuspend the pellet in 30 μl of ice-cold BME by gently pipetting up and down five times.
-
75
Plate the dissociated organoid in a 48-well plate in one drop of 30-μl BME, in the center of the well, and incubate for 10–20 min at 37 °C in 5% CO2.
-
76
Tilt the plate at a 45° angle and gently add 250 μl of expansion medium to each well in a dropwise manner, using a P1000 pipette. Aim at the bottom corner of the well, avoiding the BME drops.
▲CRITICAL Continue to supplement the expansion medium with the selection agent to prevent potential outgrowth of organoids in which the transgene is silenced.
-
77
Refresh the expansion medium every 3–4 d. After ~7–21 d, when organoids reach a diameter of 200–500 μm, they can be passaged as described in Steps 43A/43B and Steps 44–53 to an appropriate culture plate (Table 3). The expected outgrowth efficiency is 20–80% and depends on the organoid culture.
? TROUBLESHOOTING
Xenotransplantation in mouse mammary fat pad
▲CRITICAL Before scaling up organoid cultures for transplantation, we recommend measuring the number of cells harvested on average per well, using Steps 78–84 below, and using this number to estimate the required number of wells to expand for transplantation.
▲CRITICAL Use organoids for transplantation when they are still in their proliferative state, 2–5 d before they would normally be split, as this will improve engraftment efficiency. As a guideline, we recommend harvesting organoids passaged at a weekly interval at day 5 and organoids passaged biweekly at day 10, counted from the day they were last split. Avoid growing organoids larger than 70 μm, because they will be filtered out during organoid harvesting to avoid clogging the injection needle.
Defining the number of wells required for transplantation ● Timing 30–60 min
-
78
Place a 70-μm cell strainer on a 50-ml Falcon tube and pre-wash the strainer with 5 ml of ice-cold adDMEM/F12+++.
-
79
Harvest intact organoids from two representative wells of a 12-well plate as described in Steps 43A (i)–(iv). Pipette this solution over the 70-μm strainer with a P1000 pipette and rinse the strainer with another 5 ml of adDMEM/F12+++. Transfer the filtrated solution to a 15-ml Falcon tube.
-
80
Centrifuge the 15-ml Falcon tube for 5 min at 300g at 4 °C. Remove the supernatant by decanting into a waste bottle; place the Falcon tube at RT for 1 min; remove the remaining supernatant with a P1000 pipette; and resuspend the organoid pellet in 1 ml of TrypLE.
-
81
Incubate for 5 min at 37 °C and thoroughly pipette up and down ten times with a P1000 pipette.
-
82
Use an inverted bright-field microscope (×5, ×10 or ×20 objective) to assess digestion. If required, repeat Step 81 until a single-cell suspension has been achieved.
-
83
Add 9 ml of adDMEM/F12+++ and centrifuge the 15-ml Falcon tube for 5 min at 300g at 4 °C. Leave 1 ml of supernatant in the Falcon tube and resuspend the cells.
-
84
Mix 10 μl of the single-cell suspension with 10 μl of trypan blue solution and count using a counting chamber and an inverted bright-field microscope (×10 or ×20 objective). To calculate the average number of cells per well, take the mean of the two wells counted. From this, extrapolate the number of wells necessary for the desired number of injections.
▲CRITICAL Account for at least 10–20% excess. For example, when injecting ten mammary fat pads with 1 × 106 cells, prepare 10 × 106 plus 20% = 12 × 106 cells.
Organoid harvesting for transplantation ● Timing Option A 60–90 min; Timing Option B 30–60 min
-
85
To prepare the cells for transplantation, two different methods can be used to take the organoids out of the BME. For using Cell Recovery Solution, follow Option A; for using TrypLE, follow Option B.
▲CRITICAL Compared to organoid harvesting by TrypLE, harvesting by Cell Recovery Solution requires less hands-on time and dissolves all BME more efficiently, usually resulting in a tight, BME-free organoid pellet. However, Option A might occasionally result in organoid dissociation, which is more likely to happen for organoid lines that dissociate fast into single cells upon TrypLE digestion (Step 43A). If this happens, dissociated cells can be propagated again in BME by following Steps 44–53. However, we advise to test one well of organoid culture by following Option A before the day of transplantation, to test if they remain intact throughout the process. If not, follow Option B.
(A). Removing organoids from BME with Cell Recovery Solution
Remove the expansion medium from the number of wells needed, as calculated in Steps78–84, by holding the plate at a 45° angle and aspirating with a vacuum system and clean tip.
-
Add 1 ml of Cell Recovery Solution per well. Incubate for 30–60 min on a horizontal orbital shaker at 20 r.p.m. at 4 °C.
▲CRITICAL The incubation is ready when the BME drops are breaking up and detaching from the plate.
Place a 70-μm cell strainer on a 50-ml Falcon tube and pre-wash the strainer with 2 ml of ice-cold adDMEM/F12+++.
Resuspend the organoid suspension by pipetting up and down 5–10 times with a P1000 pipette, holding the plate at a 45° angle. Pipette the suspension with intact organoids over the 70-μm strainer into the 50-ml Falcon tube, combining up to six wells of the same culture. Rinse the strainer with 6 ml of adDMEM/F12+++ and transfer the filtrated solution to a 15-ml Falcon tube.
Continue to Step 86.
(B). Removing organoids from BME with TrypLE
Place a 70-μm cell strainer on a 50-ml Falcon tube and pre-wash the strainer with 5 ml of ice-cold adDMEM/F12+++.
Remove the expansion medium from the number of wells needed, calculated in Steps78–84, by holding the plate at a 45° angle and aspirating with a vacuum system and clean tip.
Resuspend up to three wells of the same culture in 1 ml of TrypLE, as described in Steps43A (ii)–(iv), to break down the BME but leave the organoids intact.
- Pipette the intact organoid solution over the 70-μm strainer into the 50-ml Falcon tube, combining up to six wells of the same culture. Rinse the strainer with 5 ml of adDMEM/F12+++ and transfer the filtrated solution to a 15-ml Falcon tube.
-
86Centrifuge the 15-ml Falcon tube at 300g for 5 min at 4 °C. Confirm by eye that a tight BME-free pellet is present (Fig. 2d).
-
86
? TROUBLESHOOTING
-
87
Remove the supernatant by decanting the fluid into a waste bottle. Let the tube rest for 1 min to allow the adDMEM/F12+++ from the side of the tube to settle at the bottom and remove the residual adDMEM/F12+++ with a P1000 pipette.
-
88
Pool the organoids from the same culture together in 1 ml of Type 1 or Type 2 expansion medium and transfer to a 1.5-ml Eppendorf tube. Centrifuge the tube at 300g for 5 min at 4 °C.
-
89
Remove all supernatant carefully with a P1000 pipette, estimate the volume of the pellet and add ice-cold transplantation buffer up to a total volume of 40 ul × the number of injection sites (eg, for ten injections, the total volume of organoid pellet + transplantation buffer = 400 ul). Mix well by pipetting up and down ten times with a P1000 pipette. Prevent bubble formation.
▲CRITICAL Example: for ten injections, make at least enough for 12 injections.
■PAUSE POINT Store and transport the tubes on ice until injection.
Implantation of the estrogen pellet ● Timing ~ 5 min per mouse
!CAUTION All animal experiments should conform to local and national animal welfare regulations and guidelines.
-
90
Implant the estrogen pellet while the organoids are stored on ice until injection, or implant up to 1 d before organoid injection to minimize the time the organoids are stored on ice.
-
91
Prepare the implantation by placing the estrogen pellet in the trocar.
▲CRITICAL Make sure the trocar has been cleaned and sterilized with 70% alcohol and is completely dry before inserting the estrogen pellet.
-
92
Anesthetize the mouse with 3–4% (vol/vol) isoflurane in an induction chamber.
-
93
Carefully take the mouse out of the induction chamber and place in front of you with its back facing up and toward you from the anterior–lateral side.
▲CRITICAL The mouse will no longer receive anaesthesia at this point, so it is important to proceed swiftly and carefully.
-
94
Fix the dorsal skin of the neck between the thumb and index finger. Position the trocar with estrogen pellet flat on the head of the mouse (Fig. 4a) and inject the needle subcutaneously, with the tip facing down, into the skin of the neck.
-
95
Insert the estrogen pellet by pushing the inner cylinder forward, releasing the pellet into the skin pocket.
-
96
Carefully take out the trocar while keeping the pellet steady in the skin between the thumb and index finger. When organoid injection directly follows estrogen pellet injection, continue with Step 102; when organoids are injected the next day, continue with Step 97.
-
97
Gently place the mouse back into the cage and monitor until recovery from anesthesia.
Xenotransplantation of BC organoids ● Timing ~5–10 min per mouse
-
98
Pre-cool insulin syringes with a 29-gauge needle on ice. Use one syringe for up to five injections.
-
99
Perform the procedures described in Steps 100–106 for one mouse at a time.
-
100
Anesthetize the mouse with 3–4% (vol/vol) isoflurane in an induction chamber.
-
101
Take the mouse out of the induction chamber and carefully place it on a foam board wrapped with aluminum foil.
-
102
Place the mouse with its back down and nose inserted into the nose piece that supplies isoflurane. Lower the anesthesia to 1.5–2% (vol/vol) isoflurane. Tape down all four legs of the mouse for fixation.
▲CRITICAL Continuously monitor breathing and movement reflexes to ensure that the anesthesia is not too high or too low.
-
103
Shave the area around the 4th nipple using a trimmer and disinfect with 70% EtOH (Fig. 4b).
▲CRITICAL Shaving the injection area is required to better visualise the nipple.
-
104
Shortly resuspend the Eppendorf tube with organoids, prepared in Steps 85–89, by flicking the sides a few times. Fill the pre-cooled insulin syringes with 40 μl of organoid suspension per injection for up to five injections per needle.
▲CRITICAL Work swiftly to prevent premature BME polymerization, which might clog the syringe. Place the syringe on ice in between the two injections.
-
105
Position the syringe flat on the mouse and insert the needle ~3 mm below the 4th nipple and move the tip of the needle right under the nipple, ~1 mm deep into the fat pad (Fig. 4b). Steadily inject 40 μl of organoid suspension into the fat pad.
-
106
Take the mouse off anesthesia and gently place it into a clean cage, placed on a heating pad, to recover from anesthesia.
-
107
Monitor the mouse until awake and mobile.
-
108
Monitor tumor growth by palpation of the injection site at least twice per week. The tumor onset depends on the organoid culture and typically ranges from 2 weeks to 4 months (Fig. 4c). The proportion of organoid cultures that can engraft in vivo is ~50% for cultures that can be passaged at least 1:2 every 2 weeks.
Troubleshooting
Troubleshooting advice can be found in Table 4.
Table 4 |.
Troubleshooting table
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 19 | The tissue is not noticeably digested after 2 h | Digestion was not suffcient | Centrifuge the 15-ml Falcon tube at 450g for 5 min at 8 °C and repeat Steps 16–19 |
| 40 | Expansion medium appears very yellow (and dead cells are present in the culture) | Plating density is too high | Passage organoids at lower density |
| Insufficient expansion medium | Refresh expansion medium more often and/or use 1 ml of expansion medium per well | ||
| BME drops are floating | The drops were not properly attached to the plate | Skip Step 51 and/or incubate for 30 min in Step 50 | |
| 45, 86 | The organoid pellet after centrifugation is fuzzy and contains undissolved BME | The TrypLE treatment was insufficient to dissolve the BME, or the amount of adDMEM/F12+++ was insufficient to wash away the BME, or the adDMEM/F12+++ was not cold enough to aid dissolving the BME | (1) Resuspend the pellet in 1 ml of TrypLE, incubate for 5 min at RT and wash with adDMEM/F12+++; or (2) wash once more in >10 ml of adDMEM/F12+++ following Steps 44–46; or (3) place the tube on ice for 10 min, resuspend the well using a 10-ml pipette and centrifuge at 300g for 5 min at 4 °C |
| The organoid pellet after centrifugation is smaller than expected | Single organoid cells or fragments are stuck to the side of the tube | Add 2% (vol/vol) FBS to the tube, resuspend the well using a 10-ml pipette and centrifuge at 300g for 5 min 4 °C | |
| 53 | The culture is growing slowly or reduces growth rate | Organoids are growing at too low density | Increase plating density by passaging with low split ratio (sometimes lower than 1:1, thus plating in a smaller total volume of BME compared to the previous passage) |
| The organoid culture contains a lot of dead cells/cell debris | Passaging conditions are too harsh | Passage with milder conditions by reducing TrypLE concentration, incubation time or times of resuspension | |
| Culture is changing in growth speed or morphology | The culture is cross-contaminated with a different culture | Avoid cross-contamination by following the critical steps at 41–43 and 49 Perform a SNP analysis (see ‘Human material’ section) to check for culture purity | |
| Discard the culture and continue with an early passage | |||
| 65A(x) | Lipofectamine 2000 transfection efficiency is low | The specific organoid culture is difficult to transfect | Try electroporation or select a different organoid culture |
| Try electroporation | |||
| The plasmid is difficult to transfect (e.g., too large) | |||
| 65B(v), (ix) | Organoid survival after transfection by electroporation is low | The settings are too harsh for the cells | Use lower voltage and pulse length settings |
| The specific organoid culture is sensitive to electroporation | Use a different organoid culture or try Lipofectamine 2000-based transfection | ||
| Transfection efficiency after transfection by electroporation is low | The settings are too mild | Use higher voltage and pulse length settings | |
| 65C(iv) | Organoid survival after lentiviral transduction is low | MOI is too high | Lower the MOI by adding less virus or by increasing the number of organoid cells (up to 300,000) |
| The specific organoid culture is sensitive to lentiviral transduction | Use a different organoid culture | ||
| Transduction efficiency is low | MOI or incubation time is too low | Increase the MOI (up to 10) by adding more virus or by decreasing the number of organoid cells (>50,000) or increasing incubation time | |
| 77 | Low efficiency of clone outgrowth | Passaging conditions were too harsh | Reduce TrypLE incubation time or times of resuspending |
| Organoids were small while picking | Grow larger organoids (e.g., >300 μm in diameter) before picking | ||
Timing
The times stated below are estimated based on experienced users carrying out the procedures.
Researchers new to the procedures might find that they take longer to complete. Steps 1–10, Tissue cataloging and preparation for isolation: 30–50 min
Steps 11–40, Organoid isolation from normal or tumor resection material: 1.5–5 h
Steps 41 and 42, Medium exchange: 5 min
Step 43, Organoid dissociation to single cells (A): 10–40 min; to fragments (B): 10–20 min
Steps 44–53, Plating organoids: 15–20 min
Steps 54–57, Freezing organoids: 10–15 min
Steps 58–64, Thawing organoids: 20 min
Step 65A, Genetic manipulation by Lipofectamine 2000 transfection, excluding organoid dissociation: 5 h
Step 65B, Genetic manipulation by electroporation, excluding organoid dissociation: 50–60 min
Step 65C, Genetic manipulation by lentiviral transduction, excluding organoid dissociation: 1–1.5 h
Steps 66–68, Selection of organoids with selection agent: 10 min
Steps 69–77, Picking individual organoids for clonal culture: 1.5–2 h
Steps 78–84, Estimating cell numbers for xenotransplantation, excluding organoid dissociation: 30–60 min, and then up to 4 weeks to grow sufficient organoids
Steps 85–89, Harvesting organoids for xenotransplantation using (A) Cell Recovery Solution: 60–90 min; using (B) TrypLE: 30–60 min
Steps 90–97, Estrogen pellet implantation: 5 min per mouse
Steps 98–107, Orthotopic injection of organoids in the mammary fat pad: 5–10 min per mouse
Step 108, Monitoring for tumor growth: 5 min per mouse
Anticipated results
Organoids derived from BC tissue can display solid (~60% of cultures), cystic (<5%), grape-like (~20%) or mixed morphologies (~20%) (Fig. 5a,b). BC organoids vary substantially in their growth rate but can reach high expansion speeds for some donors (1:6 every week; Table 1). Once established (e.g., > passage 5 in vitro), BC organoids rarely stop growing or reduce growth speed. Organoid establishment efficiency ranges between 55% and 70% for most BC subtypes and is ~40% for triple-negative breast cancer (TNBC; Fig. 5c,d), suggesting that it is challenging for the often aggressive and genetically unstable TNBC cells to adapt to in vitro culture conditions. However, because these data were collected while we were optimizing methods9, success rates will likely be substantially higher by following the guidelines provided in this protocol (e.g., by comparing both the published Type 1 medium7 with the Type 2 expansion medium introduced here).
Fig. 5 |. Characterization of BC organoid cultures.
a, Representative bright-field images of different morphologies of BC organoids. Scale bars, as indicated in the bottom right corner of the first image, 100 μm. b, Donut chart depicting the distribution of the major morphologies of 31 BC organoid cultures. c, Donut chart showing the distribution of BC subtypes in 155 tissue samples and 95 related organoid cultures. d, Stacked bar chart demonstrating the success rate of organoid establishment, grouped by receptor expression status. ‘Yes’ indicates successful establishment (>5 in vitro passages); ‘No’ indicates unsuccessful establishment. e, Stacked bar charts showing the percentage of organoid cultures with positive (green) or negative (pink) receptor status, as compared to the original tissue receptor status. f, Comparative immunohistochemical images of BC tissue and derived organoids. Shown are representative images of receptor positive (pos) and receptor negative (neg) samples. Scale bar, as indicated in the top left corner of the first image, 100 μm. Panels a, c, e and f were adapted from ref. 7. In panels a and f, numbers 15, 18, 16, 27, 22 and 19 refer to sample numbers in Table 1, and 35T, 68T, 86T and 72T refer to sample numbers as published in ref. 7.
Immunohistochemical comparison to the original tumor specimen indicated that organoids sometimes lose expression of ER (~25% of samples), PR (~25% of samples) or HER2 (~20% of samples) during culturing, and few samples gain ER (~10%) or HER2 (<5%) expression (Fig. 5e,f and Table 1). The time in culture at which receptor expression reduces or increases can vary and should be assessed for each culture. Reasons for these changes are currently unclear but could be influenced by propagating organoids in Type 1 or Type 2 expansion medium, culture-induced gene silencing or outgrowth of certain subclones. Researchers should be aware that organoids generated from tumor resections can be contaminated with normal cells, which is expected to be the case for <10% of tumor samples. However, as normal organoids generally grow slowly (Table 1) and ultimately stop growing9, normal cells will probably be lost during culture over time. It is not possible to distinguish normal cells from tumor cells by eye using light microscopy, but pathologists are often able to confirm normal or tumorous origin of a culture by immunohistochemical analysis. When the tumor harbors p53 mutations, Nutlin-3a can be added to the expansion medium to select for tumor cells, ensuring death of all normal cells that are potentially present (Fig. 3c). Confirmation of the cancerous nature of the sample can be achieved by assessing genomic alterations, either by whole-genome or whole-exome sequencing or by comparative genomic hybridization4. We have used BC organoids to illustrate the potential of targeting BCL2 in combination with CDK4/6 inhibitors in ER+ BC organoids in vitro7 and have, furthermore, shown that BC cultures can retain responsiveness or resistance to HER2 blocking by afatinib in vitro after xenotransplantation in vivo7.
Regarding normal breast organoid cultures, the rate of establishment is almost 100%8,9. However, their growth rate is generally low (split ratio of 1:2 every 2 weeks at best; Table 1) and declines over time, and cultures have limited growth potential (up to in vitro passage ~20)9. Normal breast organoids display different morphologies (Fig. 6a), including mature luminal-type cystic structures, luminal progenitor-type solid, budding structures and basal/stem cell-type dense, solid structures (Fig. 6b)8. Normal organoids can display as branching or cystic structures containing a central lumen (Fig. 6c,d). CyTOF analysis confirms the presence of basal, luminal progenitor and mature luminal cells in cultured normal organoids (Fig. 6e).
Fig. 6 |. Characteristics of breast organoid cultures derived from histologically normal breast tissues.
a, Diverse structure types can be seen in a single normal breast organoid culture at passage 5. b, Representative examples of organoid cultures showing structures of the mature luminal type (acinar structures with a large lumen), luminal progenitor type (smaller structures with a small inner lumen that can exhibit budding) and basal/stem cell type (larger structure types that can exhibit budding or branching). c, Fluorescent confocal image of a representative branching organoid at passage 3 of culture labeled for DAPI (white) and F-actin (red). d, Fluorescent confocal image of a luminal structure at passage 5 of culture labeled for DAPI (white) and F-actin (red). e, CyTOF analysis using a breast-specific antibody panel demonstrates the presence of all of the major mammary epithelial cell types in a single culture. Heat map showing normalized protein expression levels for the indicated markers (x axis) for single cells in a representative organoid culture (y axis). f, The indicated medium components were omitted from Type 1 expansion medium, and breast organoid cultures were generated. The distribution of mammary lineages as measured by CyTOF is shown in the donut plots (upper panel), and representative bright-field images of the cultures are shown (bottom panel). Basal: basal/stem cell; LP, luminal progenitor; ML, mature luminal. Other: cells that fall outside of the CD49f and EpCAM gating parameters, so cannot be defined as basal, LP or ML. In all panels, organoids were maintained in Type 1 expansion medium from which Y-27632 was removed 2–3 d after organoid establishment, passaging or thawing. Scale bars, 100 μm (a–c) and 20 μm (d). Panels b, e and f were adapted from ref. 8.
We offer different options to alter the ratio of basal, luminal progenitor and mature luminal cells (Fig. 6f). These manipulations can be useful for short-term study of specific cell types but can affect the morphology and long-term growth of the culture. Furthermore, we have genetically edited normal breast organoids using CRISPR–Cas9 (Fig. 3b,c), showing that mutating P53, PTEN and RB1 in normal organoids is sufficient to transform them into organoids that form ER+ tumors upon xenotransplantation in vivo (Fig. 4c)9. This illustrates the potential utility of this model to increase understanding of the early molecular events that culminate in specific subtypes of BC.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
All data described in this protocol are included in the article figures or were published previously in the supporting primary research papers (https://doi.org/10.1016/j.cell.2017.11.010, https://doi.org/10.1038/s41467–020-15548- and https://doi.org/10.1038/s41596–019-0160-).
Supplementary Material
Acknowledgements
We are very grateful for the technical support of the Princess Máxima Center for Pediatric Oncology, the Hubrecht Institute, the Foundation Hubrecht Organoid Technology (HUB), the Dana-Farber Cancer Institute and the Walter and Eliza Hall Institute of Medical Research for generation of the described methods and the HUB for managing the breast organoid biobank. This work was financially supported by the Princess Máxima Center for Pediatric Oncology. J.F.D. was supported by a Marie Curie Global Fellowship and a VENI grant from the Dutch Organization for Scientific Research. J.M.R. and J.S.B. were supported by an R35 grant (CA242428) from the National Cancer Institute and acknowledge the support of the Susan G. Komen Breast Cancer Foundation. J.M.R. was supported by the American Cancer Society. J.E.V. was supported by the Australian National Health and Medical Research Council. A.C.R. was supported by a Starting Grant 2019 (project 804412) from the European Research Council.
Footnotes
Competing interests
J.F.D., N.S., S.F.B., H.C. and A.C.R. are inventors on patents related to the organoid technology (WO201309812-A3/WO2016083612-Al/P309013GB).
Additional information
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41596-020-00474-1.
Peer review information Nature Protocols thanks the anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.DeSantis CE et al. Breast cancer statistics, 2019. CA Cancer J. Clin 69, 438–451 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Nik-Zainal S et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 534, 47–54 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Curtis C et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486, 346–352 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rios AC et al. Intraclonal plasticity in mammary tumors revealed through large-scale single-cell resolution 3D imaging. Cancer Cell 35, 618–632 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Hoon Tan P et al. The 2019 World Health Organization classi!cation of tumours of the breast. Histopathology 77, 181–185 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Sato T et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Sachs N et al. A living biobank of breast cancer organoids captures disease heterogeneity. Cell 172, 373–386 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Rosenbluth JM et al. Organoid cultures from normal and cancer-prone human breast tissues preserve complex epithelial lineages. Nat. Commun 11, 1711 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dekkers JF et al. Modeling breast cancer using CRISPR–Cas9-mediated engineering of human breast organoids. J. Natl. Cancer Inst 112, 540–544 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kedrin D et al. Intravital imaging of metastatic behavior through a mammary imaging window. Nat. Methods 5, 1019–1021 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holliday DL & Speirs V Choosing the right cell line for breast cancer research. Breast Cancer Res. 13, 76–99 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kapałczyńska M et al. 2D and 3D cell cultures—a comparison of different types of cancer cell cultures. Arch. Med. Sci. 14, 910–919 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vargo-Gogola T & Rosen JM Modelling breast cancer: one size does not fit all. Nat. Rev. Cancer 7, 659–672 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Neve RM et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 10, 515–527 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller DH, Sokol ES & Gupta PB 3D primary culture model to study human mammary development. Methods Mol. Biol 1612, 139–147 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Linnemann JR, Meixner LK, Miura H & Scheel CH An organotypic 3D assay for primaryhuman mammary epithelial cells that recapitulates branching morphogenesis. Methods Mol. Biol. 1612, 125–137 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Lim E et al. Aberrant luminal progenitors as the candidate target population for basal tumor development inBRCA1 mutation carriers. Nat. Med 15, 907–913 (2009). [DOI] [PubMed] [Google Scholar]
- 18.Kopper O et al. An organoid platform for ovarian cancer captures intra- and interpatient heterogeneity. Nat. Med 25, 838–849 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Cattaneo CM et al. Tumor organoid–T-cell coculture systems. Nat. Protoc 15, 15–39 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drost J et al. Organoid culture systems for prostate epithelial and cancer tissue. Nat. Protoc 11, 347–358 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fujii M, Matano M, Nanki K & Sato T Efficient genetic engineering of human intestinal organoids using electroporation. Nat. Protoc 10, 1474–1485 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Broutier L et al. Culture and establishment of self-renewing human and mouse adult liver and pancreas 3Dorganoids and their genetic manipulation. Nat. Protoc 11, 1724–1743 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Drost J et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature 521, 43–47 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Dall G, Vieusseux J, Unsworth A, Anderson R & Britt K Low dose, low cost estradiol pellets can supportMCF-7 tumour growth in nude mice without bladder symptoms. J. Cancer 6, 1331–1336 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dobrolecki LE et al. Patient-derived xenograft (PDX) models in basic and translational breast cancer research. Cancer Metastasis Rev 35, 547–573 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gakhar G, Wight-Carter M, Andrews G, Olson S & Nguyen TA Hydronephrosis and urine retention in estrogen-implanted athymic nude mice. Vet. Pathol 46, 505–508 (2009). [DOI] [PubMed] [Google Scholar]
- 27.Whittle JR et al. Dual targeting of CDK4/6 and BCL2 pathways augments tumor response in estrogen receptor positive breast cancer. Clin. Cancer Res 26, 4120–4134 (2020). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data described in this protocol are included in the article figures or were published previously in the supporting primary research papers (https://doi.org/10.1016/j.cell.2017.11.010, https://doi.org/10.1038/s41467–020-15548- and https://doi.org/10.1038/s41596–019-0160-).