Abstract
Organoids have revolutionized cancer research as highly adaptable models that enable an array of experimental techniques to interrogate tissue morphology and function. Because they preserve the genetic, phenotypic, and behavioral traits of their source tissue, organoids have gained traction as the most relevant models for drug discovery, tracking therapeutic response and for personalized medicine. As organoids are indisputably becoming a mainstay of cancer research, this review specifically addresses how colon-derived organoids can be perfected as multidimensional, scalable, reproducible models of healthy, pre-neoplastic and neoplastic conditions of the colon and for use in high-throughput “Phase-0” human clinical trials-in-a-dish.
1. Introduction
Colorectal cancer (CRC) is the most frequently diagnosed heterogeneous disease in the digestive tract; it is the second most common cause of cancer-related death in the United States and the third leading cause worldwide (Siegel et al., 2020; Yan et al., 2020). Most deaths from CRCs can be attributed to cancer recurrence after initial treatment, which presents as distant metastases in secondary sites such as the liver or lung. Although the total global incidence of CRCs has dropped in the last decade, the incidence of early-onset colorectal cancer (EOCRC), defined as CRCs in patients aged 50 or younger, has been on the rise (Mauri et al., 2019). If left untreated, the 5-year survival rate of patients with metastatic CRC can be as low as 5% (Engel et al., 2020).
Like most cancers, CRCs are comprised of a group of molecularly heterogeneous subtypes, each characterized by a range of genomic and epigenomic alterations and distinct tumor drivers. Such heterogeneity makes the standard one-size-fits-all approach to CRC-treatment ineffective. Another major challenge is that the pathogenic mechanisms of some CRCs, such as EOCRCs, still remain poorly understood and insights into their molecular characterization remain scant. Without such insights, the development of relevant preclinical models also remains unfulfilled. Thus, the need for comprehensive studies that delineate the underlying molecular basis for disease pathogenesis using physiologically relevant human preclinical model systems is both urgent and unmet. Such studies could not only reveal disease mechanisms but also ensure that the translational potential of the discoveries remains high.
Since the ability to culture human intestinal organoids has been realized, several teams have established a library of patient-derived CRC organoid lines, encompassing different histological subtypes and clinical stages, as preclinical models for downstream genomic profiling (Sato et al., 2011; Schütte et al., 2017; van de Wetering et al., 2015; Weeber et al., 2015; Yan et al., 2020). Given the fact that sporadic CRCs arising through the classical adenoma-carcinoma sequence, with mutational activation of the WNT signaling pathway in ~80% of CRCs, the rationalized addition of WNT-3A and R-spondin1 in the culture medium can support normal organoid growth, and their withdrawal can prevent overgrowth. These insights have increased the success rate of tumor organoid establishment (van de Wetering et al., 2015). As organoids are indisputably becoming a mainstay of cancer research, we dedicate this review to not only highlight the challenges in modeling CRC initiation and progression and summarize what has been accomplished so far but also outline strategies that can help us realize the full potential of organoid-based CRC research by building multidimensional, scalable, reproducible models of healthy, pre-neoplastic and neoplastic conditions of the colon.
2. Challenges in modeling CRC initiation and progression
2.1. Complex etiopathology
While the etiopathology of CRC is not well known, it is assumed that CRCs are initiated by a cascade of DNA mutations within epithelial cells (Allen & Sears, 2019). It is also considered that some external stimuli like bacteria, and fungal toxins, incorporation of exoDNA into epithelial DNA and micro-environmental factors may alter the genetic and epigenetic signature of the cellular genome and may propel early to intermediate to late steps neoplastic progression of this disease (Allen & Sears, 2019; Bardhan & Liu, 2013; Chan & Giovannucci, 2010). Metastasis of CRCs, on the other hand, is fueled mostly by epigenetic mechanisms, believed to be of multifactorial triggers, the exact nature of which is less well understood (Bardhan & Liu, 2013). Epigenetic alterations that have been observed include DNA methylation, histone modifications, chromatin remodeling and non-coding RNAs, affect every aspect of tumor development from initiation to metastasis (Ju et al., 2011; Vaiopoulos, Athanasoula, & Papavassiliou, 2014). Without a complete understanding of the triggers or sequence of these events, their cause-effect relationships, or if they occur concomitantly and synergize, it is difficult to model such progressive events ex vivo. Informed models of tumor initiation as well as metastatic progression have not been realized, and whatever exists remains rudimentary at best.
2.2. Molecular subtypes
CRC is a heterogeneous disease; its subtypes carry distinct prognostic and predictive information that is associated with different patient outcomes (Inamura, 2018). CRCs are classified on the basis of their comprehensive molecular characterizations into The Cancer Genome Atlas (TCGA) and consensus molecular subtype (CMS). TCGA is further categorized into hypermutated and non-hypermutated CRC subtypes according to their DNA copy number, exome sequences, promoter methylation, messenger RNA as well as microRNA expression patterns. For CMS classification, gene expression profiles and TCGA data are used together to generate a consensus molecular subtypes of CRCs (Budinska et al., 2013; Marisa et al., 2013; Sadanandam et al., 2013). CRC subtypes include CMS1 (MSI immune, 14%), CMS2 (canonical, 37%), CMS3 (metabolic, 13%) and CMS4 (mesenchymal, 23%); they are categorized based on whether they are microsatellite proficient (i.e., stable; MSS) or deficient (i.e., unstable; MSI), and whether they have a CpG island methylator phenotype (CIMP) or somatic copy number alteration (SCNA) phenotype. CRCs are also categorized into distinct subtypes by considering their key molecular features and clinical outcomes. Based on their distinct clinicopathological features with regard to MSI, CIMP, BRAF and KRAS mutation status five distinct subtypes of CRCs have been recognized (Inamura, 2018; Phipps et al., 2015). Type 1 and 2 subtypes demonstrate a serrated morphology, type 3 reflects an alternate serrated pathway with origins in KRAS-mutated adenomas, type 4 subtype represents the origin of CRC from traditional adenoma-carcinoma sequence and type 5 subtype indicates possible Lynch syndrome with a high prevalence of CRC family history (Guinney et al., 2015). Patients with CRC subtypes 1 and 5 (MSI high) have the most favorable outcomes, whereas those with subtype 2 (MSS/MSI-low, CIMP-positive, BRAF-mutated, KRAS-mutation–negative) have the highest mortality (Jass, 2007; Phipps et al., 2015). The broad spectrum in genetic changes and mutations in various subtypes have created a gap between cancer genetics and patient treatment. These circumstances necessitate the creation of large genotyped phenotyped biobanks to generate reproducible findings. In this aspect, patient-derived organoids can help create a “living biobank,” which can minimize the gap between the disease and the experimental model, enhance the precision and personalization in the process of biomarker/drug discovery.
2.3. Lacking models for intermediate (pre-neoplastic) lesions
CRCs are the end results of a multistage process that is slow, and spans over years, as aberrant crypts eventually turn into malignant polyps. Because about 85% of CRCs begin as pre-cancerous polyps, the early detection and removal of polyps can notably reduce the risk of cancer (Gopalappa, Aydogan-Cremaschi, Das, & Orcun, 2011). But polyps are not all one and the same; several subtypes of polyps exist, e.g., hyperplastic polyps (HPs), sessile serrated adenoma/polyps (SSAPs) and traditional serrated adenoma (TSA) (Snover, Jass, Fenoglio-Preiser, & Batts, 2005). It is known that HPs polyps represent very low malignant potential where SSA/P and TSA both correspond as a precursor of cancer via the serrated neoplastic pathway (O’Brien et al., 2006; Spring et al., 2006; Uraoka et al., 2015). The SSA/P polyps are also recognized as precursors of sporadic colorectal cancer with findings of microsatellite instability and aberrant DNA hypermethylation (Bardhan & Liu, 2013; Goldstein, 2006; Noffsinger, 2009; Simon, 2016). However, the process of initiation and progression of polyps and the risk factors that propel such progression have not been comprehensively modeled to understand what constitutes such risk and why some polyps progress, and others do not (Harrington, Wei, Suriawinata, Mackenzie, & Hassanpour, 2020). Despite advances in the modeling and understanding of CRCs, precise preclinical ex-vivo models for investigating pre-neoplastic states (polyps) are lacking.
3. Conventional CRC models
3.1. 2D monolayers of CRC cell lines
Human 2D CRC cell line grown as monolayer is the most popular conventional CRC model that allows easy assessment of a variety of tumor cell phenotypes, e.g., cell viability, cell cycle, colony formation, migration, invasion, metastasis, and survival and apoptosis (Kannen et al., 2012; Nautiyal et al., 2011; Young, Ordonez, & Clarke, 2013). Moreover, the 2D monolayers have been used to study the efficacy of anti-cancer therapies, identification and validation of drug targets, and targeted drug delivery. For example, The role of JNK signaling in cell migration and invasion was studied in the COLO-205 cell line (Zhang et al., 2016), while the SW480 cell line was used to investigate β-catenin as a novel target for cancer therapy (Li, Zhou, Ji, & Luo, 2016). The popularity of the 2D monolayer model stems from multiple factors such as their low cost, high reproducibility, and their ease of creation and propagation (Joseph, Malindisa, & Ntwasa, 2018). However, this model has serious drawbacks, i.e., they do not recapitulate the tumor cell microenvironment and lack dimensionality (Porter, Murray, & McLean, 2020; Ronen, Hayat, & Akalin, 2019). Other disadvantages include (a) the molecular profiles of CRC cell lines often differ significantly from human tumors; (b) cancer cell lines are subjected to the risk of contamination and mislabeling, and genetic drifts during prolonged cultures; (c) lack of dimensionality leading to unnatural adhesion forces; and (d) lack the tumor-stroma and tumor-immune cell interactions (Domcke, Sinha, Levine, Sander, & Schultz, 2013; Huang, Liu, Zheng, & Shen, 2017).
3.2. 3D tumor spheroids of CRC cell lines
3D tumor spheroids, developed by either growing tumor cells within 3D scaffolds or as multicellular spheroids, recapitulate the in vivo tumor microenvironment and they have been developed to study the interactions between tumor and stromal cells as well as tumor cells and the extracellular matrix. This model mitigates some of the disadvantages of the 2D monolayer model, namely, as it is well documented to regain intrinsic properties and to better mimic the in vivo situation due to the added third dimension (Dolznig et al., 2011; Fischbach et al., 2009). Over the past few years, the 3D tumor spheroids have been preferred over 2D monolayers to study tumor cell metabolism, gene expression studies, and for tracking response to anti-tumor treatments (Desoize & Jardillier, 2000; Takagi et al., 2007). This model is advantaged by its simplicity, high degrees of reproducibility, and the ability to coculture with other cell types. Major disadvantages of this model are the poor structural organization and poor supply of oxygen and nutrients to the spheroid core (Porter et al., 2020). Moreover, the 2D and the 3D models have been shown to differentially activate signaling pathways, especially in the mTOR, AKT, S6K signaling in response to the therapeutic agents (Riedl et al., 2017). These apparent inconsistencies and major disadvantages require researchers to use pre-clinical animal models for human CRC research to confirm findings, forcing us one step backward (in terms of species).
3.3. Animal models
The main goal of modeling human CRC in animals is to understand the molecular etiology, pathology, and progression of CRC (Oliveira, Abrantes, Tralhão, & Botelho, 2020). This model has been used to translate and confirm the hypotheses derived from cell models (Johnson & Fleet, 2013). Moreover, it is used to study the precise molecular mechanisms as well as to test the possible potential preventative and therapeutic strategies for CRC (Oliveira et al., 2020). There five major strategies for the development of potential animal models for CRCs, including: (i) spontaneous intestinal cancers in various animal species including dogs, sheep and mice; (ii) chemically or environmentally induced cancers in rodents; (iii) mutagen-induced germline mutation models, including APCmin mouse and F344-Pirc Rat; (iv) genetically modified mice; and (v) implantation of tumor cells into mice, either syngeneic murine cells (Morikawa et al., 1988; Naito, von Eschenbach, Giavazzi, & Fidler, 1986) or xenotransplantation of human CRC tissues/cells (Johnson & Fleet, 2013; Oliveira et al., 2020). Although these animal models mitigate a lot of the disadvantages of the 2D and 3D models, there are still several weaknesses, for example, (i) the gut environment (diet, toxins, microbes) is non-human, and hence, the innate immune response in the gut lining will lack humanness; (ii) the ability to study both the initial and late stages (metastasis) of CRCs is limited because the models either allow studying initiation (e.g., spontaneous tumor models) or progression (syngeneic or xenograft models), but not both; (iii) The need for extensive resources, high costs related to animal care and breeding and ethical concerns (Johnson & Fleet, 2013; Porter et al., 2020). Furthermore, the human xenograft model, which requires the use of immune deficient mice has one major flaw that is unique to this model, in that, it lacks a key component in cancer progression, i.e., the host immune response to a growing tumor. Therefore, an optimal model that can be uniformly used to study both the early and late stages of CRCs is still lacking.
4. The development of patient-derived colorectal organoids as 3D models for studying CRCs
Recent technological innovation in stem cell research has had a significant impact specifically on CRC research, enabling the development of 3D colorectal organoid models and their optimized growth conditions which bypass some of the disadvantages of the conventional models (Du et al., 2020; Ghosh et al., 2020; Pleguezuelos-Manzano et al., 2020; Sayed, Chakraborty, et al., 2020; Sayed, Sahan, et al., 2020; Sayed, Suarez, et al., 2020; Yuki, Cheng, Nakano, & Kuo, 2020). Although most of the published papers used the murine APCmin colon-derived tissue to grow organoids, others (Sato et al., 2011) and we Ghosh et al. (2020), Sayed, Chakraborty, et al. (2020), Sayed, Sahan, et al. (2020), Sayed, Suarez, et al. (2020) have shown that the long-term culture of colorectal organoids from healthy subjects and patients with inflammatory bowel disease (IBD) (d’Aldebert et al., 2020; Sayed, Chakraborty, et al., 2020; Sayed, Sahan, et al., 2020; Sayed, Suarez, et al., 2020), adenoma and colorectal cancers (Bruun et al., 2020; Sato et al., 2011). These organoids spurred an unprecedented growth of associated technologies, all of which have highlighted the limitless potential of this human pre-clinical model system (see Fig. 1).
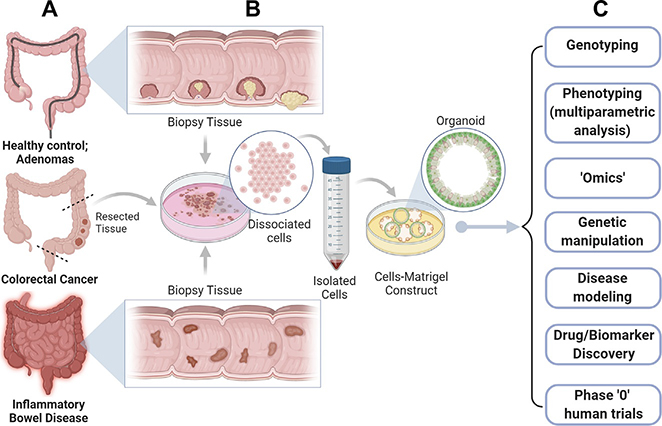
Fig. 1.
The limitless potential of colorectal organoids. Organoids can be generated from virtually all colon tissues representing diverse pathophysiologic states of the colon (A), e.g., normal healthy mucosa and pre-neoplastic states such as diverse types of colon polyps (e.g., tubular, villous and serrated adenomas) (top), colorectal cancers (middle), and finally, inflammatory conditions, e.g., inflammatory bowel diseases (IBD), checkpoint colitis, etc., as long as the tissue can be preserved to serve as a source of adult stem cells. (B) Schematic outlines key steps in the process of organoid generation. Briefly, fresh tissue or viable frozen tissue is dissociated via enzymatic and mechanical methods to release the stem cells. Dissociated cells are subsequently embedded within the extracellular matrix (Matrigel). Under the right growth conditions, which includes a delicate ratio of growth factors (WNT, R-spondin and Noggin), 3D organoids are formed, which contain all cell types normally found in the colonic epithelium. These adult stem-cell-derived organoids can be passaged, expanded and biobanked. (C) The organoid model system can be exploited for several applications such as multiparametric analysis, genetic manipulation, drug and biomarker discovery and for carrying out phase “0” clinical trials in humans to assess not just drug toxicity/safety but also efficacy.
4.1. Standardized methodologies for culturing 3D organoids, enhancing reproducibility
The optimization of the 3D organoids’ culture media has had an enormous impact on CRC research. Sato et al. had successfully established mouse intestinal crypt-villus organoids from single sorted LGR5+ stem cells using WNT 3a, R-spondin and Noggin (WRN) containing culture media and 3D serum-free extracellular matrix such as Matrigel (Sato et al., 2009). The composition of WRN media and other constituents required for organoid culture are listed in Table 1. This media supports the growth of LGR5+ stem cells into crypt-villus-forming epithelial domains containing all the differentiated cell types that are present in the colon epithelium in vivo. It is notable that these organoids can be maintained for long-term in culture despite the absence of a mesenchymal niche (more than 8 months) (Sato et al., 2009).
Table 1.
Composition of the culture media required from the growth of healthy and CRC colon organoid.
| Component required for organoid culture | Function | References |
|---|---|---|
| Wnt | • Important for crypt proliferation | Kim et al. (2005) and Pinto, Gregorieff, Begthel, and Clevers (2003) |
| R-spondins | • Upregulate the WNT signaling pathway through a specific effect on the Lgr receptors driving the crypt hyperplasia in vivo | de Lau et al. (2011) |
| Noggin | • Inhibitor for bone morphogenetic protein (BMP) signaling pathway • Helps in the expansion of crypt numbers |
Haramis et al. (2004) |
| Epidermal growth factor (EGF) | • Important for stem cell proliferation and inhibit apoptosis | Dignass and Sturm (2001) |
| Rho-kinase inhibitor Y-27632 | • Inhibits anoikis in isolated embryonic stem cells • Decreases cell death |
Watanabe et al. (2007) |
| Notch-agonistic peptide | • Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells | Li et al. (1998) and van Es et al. (2005) |
| Extracellular Matrix (Matrigel) | • Rich with laminin (αl and α2) • Support the intestinal epithelial growth through laminin enrichment at the crypt base |
Sasaki, Giltay, Talts, Timpl, and Talts (2002) |
| Nicotinamide | • Inhibitors to ROCK kinase • Promotes Cell Survival and Differentiation • Required for long-term growth of human derived organoids and organoids from CRC patients, mouse APC-deficient adenomas, and Barrett’s esophagus |
Jung et al. (2011), Meng et al. (2018), and Sato et al. (2011) |
| A83–01 | • Potent inhibitor of activin receptor-like kinase (ALK) including ALK5 (type I transforming growth factor-β receptor), ALK4 (type IB activin receptor), and ALK7 (type I NODAL receptor) • Inhibits TGF-β-induced epithelial-to-mesenchymal transition • Allow long-term culture of organoid from CRC patients and APC deficient adenoma |
Jung et al. (2011), Sato et al. (2011), and Wu et al. (2017) |
| SB202190 | • Inhibits p38 of mitogen-activated protein kinase (MAPK) • Decreases the degradation of EGF receptor • Increase plating efficiency and allows long-term culture of organoid from CRC patients and APC deficient adenoma |
Frey, Dise, Edelblum, and Polk, (2006), Jung et al. (2011), and Sato et al. (2011) |
| Gastrin | • Not always required • Increase the culture efficiency |
Jung et al. (2011) and Sato etal. (2011) |
The addition of nicotinamide, A83–01 (an ACTIVIN/NODAL/TGF-β pathway inhibitor), and SB202190 (a p38 MAPK inhibitor) to the WRN media allows the long-term culture of organoids derived from the human small intestine and colon tissues and mouse APC-deficient adenomas, but also from human colorectal cancer cells and metaplastic epithelia from regions of Barrett’s esophagus (Jung et al., 2011; Sato et al., 2011) Table 1. The use of such conditioned media (instead of custom media with cocktails) has been shown to improve reproducibility (VanDussen, Sonnek, & Stappenbeck, 2019). With the optimization of R-spondin-based culture conditions, organoid technology has been expanded to enable several other human carcinomas, including gastric (Nanki et al., 2018), breast (Sachs et al., 2018), ovarian (Kopper et al., 2019), lung (Sachs et al., 2019), and liver cancers (Broutier et al., 2017); spurring the notion that if there are LGR5+ adult tissue stem cells, establishment of organoids is feasible. Importantly, the organoids derived from human CRC samples, especially tissues that represent the adenoma-carcinoma transition stage, can grow without the niche factors WNT, R-spondin, EGF, and Noggin; this is because, the mutational background of these samples ensure that the associated downstream signaling pathways are constitutively activated (Drost et al., 2015; Fujii et al., 2016; Matano et al., 2015). Recently, it has been shown that Fibroblast growth factor 2 (FGF2) is essential and necessary for the self-renewal of colon cancer organoids (Otte et al., 2019) especially to grow organoids from tumor metastases.
Thus, the past decade has seen a concerted push toward the standardization and simplification of protocols for organoid isolation and expansion, and most groups converge on the fundamentals (i.e., what to add). However, like most external manipulations in cell biology, we emphasize that “less is more” and that such a restrained approach which avoids the addition of too many/too much external growth factors might be best in order to preserve the epigenetic and phenotypic properties of the organoid closest to its tissue of origin.
4.2. Generation of a living organoid biobank from CRC patients
The importance of building patient tumor-derived organoid biobank has been documented before with other cancers, e.g., primary pancreatic ductal adenocarcinoma (Huang, Holtzinger, et al., 2015; Seino et al., 2018; Tiriac et al., 2018), breast cancer (Sachs et al., 2018), prostate cancer (Gao et al., 2014), and primary liver cancer (Broutier et al., 2017). These efforts not only spurred the discovery of novel pathways and actionable targets, but also personalize anti-cancer therapeutics to match the molecular phenotype. In the case of CRCs, efforts to build similar biobank(s) have been reported previously (van de Wetering et al., 2015; Weeber et al., 2015); van de Wetering and colleagues established patient-derived organoids from 20 patients using tumor tissues that represented major CRC molecular subtypes and matching adjacent healthy regions (van de Wetering et al., 2015). The tumor organoids recapitulated the mutation spectra found in the parent CRC tissue (van de Wetering et al., 2015), making them ideal for use in high-throughput drug screening assays to not just facilitate personalized therapy but also exploit for genomic and functional studies on the individual patient (van de Wetering et al., 2015). Weeber and colleagues showed that organoids reflect the mutation load of original metastases from which they were derived, capturing ~90% of the somatic mutations seen in the patient, as determined by sequencing of 14 patient-derived organoids (PDOs) for 1977 cancer-relevant genes (Weeber et al., 2015). Similarly, Fujii et al. demonstrated that organoids (n = 55 PDOs) preserve the histopathological grade and differentiation status of their parental tumors in vitro and develop tumors after implantation to the immunodeficient mice (Fujii et al., 2016). Moreover, the OncoTrack consortium generated a large biobank of 106 tumors, 35 organoids, and 59 xenografts from CRC patients (stages I–IV) (n = 106 PDOs), and developed a preclinical platform generating a compendium of drug sensitivity data more than 4000 assays for evaluation of 16 anti-cancer drugs (Schütte et al., 2017). By combining the molecular profiles and drug sensitivity patterns, they could identify biomarkers to predict sensitivity to the EGFR inhibitor cetuximab (Schütte et al., 2017).
These key proof-of-concept studies demonstrate that a well-annotated organoid biobank accurately reflecting the patient’s tumor and its unique biology are useful for drug and biomarker discovery, for strategizing personalized treatment plans, and could even serve as platforms for carrying out phase “0” pre-clinical human clinical trials for novel targets/compounds (Fig. 1). The key, however, is to have proper metadata collection, as complete as possible, which not just includes information such as age, gender, source (biopsy or surgical resection), clinical staging, prior treatment history (chemotherapy or targeted), location and histology (moderately differentiated or poorly differentiated), but also mutation load estimates, MSI/MSS status, and molecular makeup. In other words, biobanks would benefit from diligent collection of PDO-associated metadata, which is any and all information that can aid in the selection of therapeutics and formulation of treatment plans.
5. The coculture models with CRC organoids and its benefits in CRC research
5.1. Coculture of CRC organoids with the immune cells
Somatic mutations in the cancer cells have the potential to encode immunogenic antigens, named neoantigens (Le et al., 2015). The host immune response produced against the neoantigens may be insufficient to neutralize them, leading to immune escape and survival of cancer cells. To overcome this challenge, patient’s immune cells could be expanded in vitro to generate large quantities of activated immune cells for in vivo use, and this is the principle of cancer immunotherapy (Chen & Mellman, 2017; Sharma & Allison, 2015). Immune cells can be expanded in vitro in coculture with organoids (see Fig. 2). Nozaki and colleagues successfully established a coculture model between mouse intestinal epithelial organoids and intraepithelial lymphocytes (IELs) that allows efficient expansion and motility analysis of IELs in the presence of IL-2, IL-7, and IL-15 (Nozaki et al., 2016). IELs showed a fourfold increase in both αβT and γδT IELs for 2 weeks and were efficiently maintained within and outside of organoids (Nozaki et al., 2016). Finnberg and colleagues generated 3D-tumoroids, organoids derived from CRC patients, cocultured them with the patient’s peripherally and tumor-derived immune cells in vitro to mimic the tumor microenvironment and assess the tumor response to chemotherapy such as 5-fluorouracil (Finnberg et al., 2017). Similarly, Neal and colleagues used the air-liquid interface method to develop a coculture of organoids, derived from more than 100 human biopsies and mouse tumors, and native embedded immune cells (T, B, and NK cells and macrophages) to recapitulate the tumor microenvironment (Neal et al., 2018). They also found that human and murine derived organoids successfully modeled immune checkpoint blockade with anti-PD-1- and/or anti-PD-L1 expanding and activating tumor antigen-specific CD8+ T lymphocytes and eliciting tumor cytotoxicity (Neal et al., 2018). Furthermore, Dijkstra and colleagues showed that cocultures of autologous tumor organoids, derived from mismatch repair-deficient CRC patients and non-small-cell lung cancer, and peripheral blood lymphocytes were efficient in enrichment and induction the tumor activity of the circulating T cells that specifically kill autologous tumor organoids (Dijkstra et al., 2018). Moreover, analysis of immune cells from human mammary ductal epithelial organoids revealed the presence of T lymphocytes subset, Vδ2+ T cells, which produced antitumor cytokine IFNγ and efficiently killed breast carcinoma cells in the presence of FDA—approved drugs bisphosphonate drugs (Zumwalde et al., 2016). In addition, the development of reverse-engineered thymus-derived organoids can be applied in regenerative medicine and solid organ transplantation as they provide a physiological thymic microenvironment that can be a source for tumor-specific lymphocytes (Tajima, Pradhan, Trucco, & Fan, 2016).
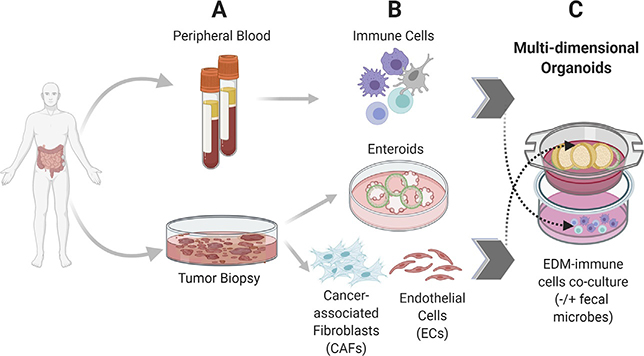
Fig. 2.
Building multi-dimensional organoids that emulate the complexity of the gut. Schematic displays the isogenic cell components derived from the same patient that can be used to build multidimensional organoid-based CRC models. Blood and tumor tissue collected from the same patient (A) can be used as a source of immune cells (B, top) or tumor organoids and tumor-associated stromal cells (B, bottom). Coculture models can then be assembled to preserve the physiologic orientation of tissue (i.e., basolateral side of the tumor epithelium facing stroma and immune cells; see C). Complexity can be further increased with the addition of fecal microbes from the same patient. Although not shown here, components of the enteric nervous system can also be incorporated into the coculture where desired. Such an understanding-by-modeling approach is critical to creating wholesome models for testing drug efficacy or for testing fundamental research hypotheses in the setting of complex cell types.
The tumor immune microenvironment (TME) can be generated using CRC organoids by three approaches: (a) reconstituted models, in which immune cells can be cocultured with organoids; (b) native TME models, in which the intrinsic immune microenvironment of tumors are preserved when establishing tumor organoids; (c) air–liquid interface (ALI) culture models, in which tissue fragments containing both tumor cells and immune components are embedded in collagen gels (Yuki et al., 2020). All these models are promising platforms for use in interrogating immunotherapeutic strategies.
5.2. Organoid models to assess the contribution of microbes in cancer initiation and progression
Microbes have been implicated in both initiation and progression of cancers, as exemplified by the oncopathogen that increases the risk of gastric cancers, Helicobacter pylori. Cocultures of microbes and organoids are useful to assess the mechanisms and risk factors for cancer development. Bartfeld and colleagues first developed a 3D organoid culture of human gastric stem cells and challenged with H. pylori by microinjecting the bacteria into the lumen; they noted a strong inflammatory response that was characterized by an induction of IL-8 and activation of NF-κB (Bartfeld et al., 2015). Since then, several groups used similar coculture models of human gastric organoids with H. pylori to understand the host epithelial cell responses to the microbe and the microbial response to host metabolites and host signaling pathways (Bartfeld et al., 2015; Holokai et al., 2019; Huang, Sweeney, et al., 2015; Sebrell et al., 2019). We recently used gastric organoid-derived monolayers and challenged them with H. pylori and observed altered DNA repair pathways, specifically, the base excision repair process, followed by induction of inflammatory response and oxidative DNA damage (Sayed, Sahan, et al., 2020); the latter are key cellular events that have been implicated in gastric cancer initiation and progression.
Besides cocultures of H. pylori and gastric organoids, murine gallbladder organoids have been cocultured with the enteric pathogen, Salmonella (Scanu et al., 2015); this study showed that the microbe can induce host signaling pathways that culminate in the amplification of c-MYC through the activation of MAPK and AKT signaling pathways, and that these events were permissive to cellular transformation associated with gall bladder carcinoma.
In the case of CRCs, it is now widely accepted that gut microbes contribute to CRC progression through various mechanisms such as the induction of chronic inflammatory state and the production of toxic metabolites, which in turn affects stem cell precursors (Tsilimigras, Fodor, & Jobin, 2017). The well-known CRC associated microbes are Fusobacterium nucleatum (Fn), enterotoxigenic Bacteroides fragilis (ETBF), and colibactin producing E. coli (Alhinai, Walton, & Commane, 2019). The anaerobe F. nucleatum resides in the oral cavity and plays a role in CRC progression (Kostic et al., 2013, 2012), possibly by activating β-catenin and Wnt signaling and upregulates inflammatory responses (Rubinstein et al., 2013). In addition, high levels of F. nucleatum in CRC patients’ tissues was associated with poor patient outcome and recurrence of post-chemotherapy (Yu et al., 2017). Much like H. pylori, F. nucleatum also induces DNA damage through the effect on the DNA repair pathway (Geng, Zhang, Lu, Zhang, & Pan, 2020). Recently, using 3D colon-derived organoids and polarized 2D epithelial cells derived from healthy humans, we have shown that F. nucleatum induces DNA damage that can influence CRC initiation and/or progression (Sayed, Chakraborty, et al., 2020).
The enterotoxigenic Bacteroides fragilis (ETBF) is another example of a CRC-associated microbe that secretes a metalloprotease toxin, which causes human inflammatory diarrhea (Sears et al., 2008). ETBF has been shown to enhance CRC progression in APC min mice through immunologic mechanisms via selective activation to the colonic signal transducer and activator of transcription-3 (Stat3) and T-helper 17 (Th17), causing inflammation-induced cancer (Wu et al., 2009). In addition, purified Bacteriodes fragilis toxin (BFT) upregulates the enzyme spermine oxidase (SMO) in the colon epithelial cells resulting in the accumulation of reactive oxygen species (ROS) and the induction of γ-H2AX, a marker of DNA damage (Goodwin et al., 2011).
E. coli with the polyketide synthase genomic island (pks+ E. coli) are also associated with CRC and responsible for the synthesis of colibactin (genotoxin). Colibactin producing E. coli NC101 strain induces colon carcinogenesis through induction of DNA double-strand breaks triggering genomic instability (Arthur et al., 2012; Cuevas-Ramos et al., 2010; Nougayrède et al., 2006). A recent study done by Dejea and colleagues reported that patients with familial adenomatous polyposis (FAP) harbor colonic biofilms containing tumorigenic bacteria composed of colibactin producing E. coli and ETBF (Dejea, Fathi, & Craig, 2018). Adherent invasive E. coli, associated with Crohn’s disease, increases the risk of CRC, especially in IBD patients through induction of chronic inflammation and accumulation of DNA damage (Martin et al., 2004).
Taken together, these studies have converged on a common overall mechanism, one that involves infection, chronic inflammation, leading to DNA damage coupled with altered core DNA repair mechanisms in the epithelium, which in turn triggers aberrant cell behavior and immune responses (Alhinai et al., 2019). Despite these insights, whether CRC-associated microbes can “cause” CRCs has been met with a healthy dose of criticism. One plausible hypothesis is that they may simply preferentially colonize cancerous lesions because of growth advantage (Coleman & Nunes, 2016; Wassenaar, 2018).
6. The advantage of the organoid model in the CRC research
6.1. Models to interrogate the precise contributions of host genetics and microbes in CRC initiation and propagation
It is conceivable that the 3D colon organoid and polarized epithelial cell model that we already developed for use in coculture assays (Sayed, Chakraborty, et al., 2020; Sayed, Sahan, et al., 2020; Sayed, Suarez, et al., 2020) could be systemically infected with each strain to assess the impact of microbes on CRC initiation. Colon organoids isolated from patients with hereditary polyposis syndromes (derived from uninvolved mucosal tissue that is at risk, as well as from adenomatous tissue) or those from patients with sporadic adenomas and/or CRCs (Fig. 3A) could be cocultured with the aforementioned CRC-associated bacterial strains (in the preceding paragraphs) or even fecal microbiota isolated from the same patient (Fig. 3C).
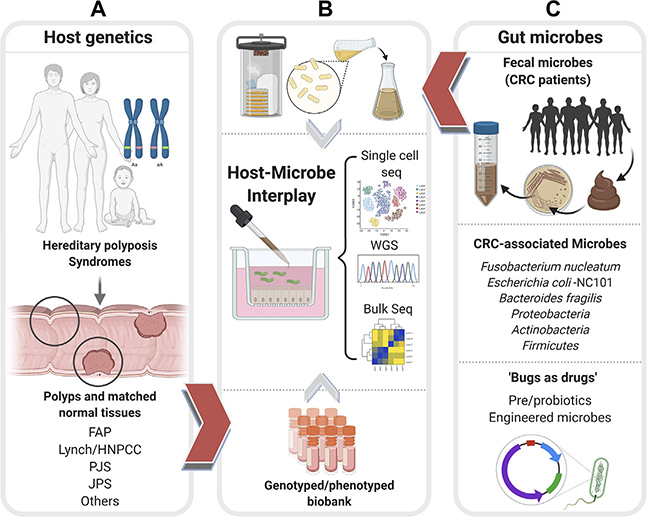
Fig. 3.
Patient-derived organoids can enable an objective assessment of the relative contributions of host genetics and microbes toward CRC initiation, reveal the potential use of microbes as therapeutics. Schematic displays how patient-derived organoids (PDOs) from uninvolved normal mucosal biopsies and involved adenomatous regions of colons representing diverse CRC syndromes (A), e.g., Familial adenomatous polyposis (FAP), Lynch or Hereditary nonpolyposis colorectal cancer (HNPCC), Peutz-Jeghers syndrome (PJS), Juvenile polyposis syndrome (JPS) of known genotypes can be used to represent host contributions in organoid-microbe coculture models in panel (B). PDOs can also help interrogate and assess the impact of both harmful and beneficial gut microbes (C). Fecal microbes collected from high-risk patients with CRCs/excessive polyposis (C, top) can be cultured using aerobic or anaerobic conditions (B, top) prior to use in coculture studies with normal PDOs. Bacterial strains associated with CRCs (C, middle) can similarly be used in cocultures with PDOs with known genotypes. Pre- and probiotics and engineered microbes (C, bottom) can be similarly tested for their beneficial effects on PDOs. In each coculture model, the PDOs can be objectively assessed in an unbiased way using “omics”- and computational approaches (B, middle) to measure the impact of host genetics, microbes, individually and when combined.
Such a study model, that is systematically designed to interrogate the 2-way interactions between the host genetics and the gut microbe (either fecal slurry or specific strains) could help objectively assess the exact nature of and the relative contributions of the complex interplay between the microbes and the host in CRC initiation and progression (Fig. 3B).
Such a powerful well-controlled “reductionistic” model could be paired with unbiased “omics”-based analyses (e.g., whole genome or bulk or single-cell sequencing of the transcriptome; Fig. 3B) to ask numerous interesting and exciting questions. For example, what sequence of events may be triggered by microbes in the healthy vs the vulnerable mucosa, and which microbes may be able to “shift” the transcriptome toward CRCs. On the flip side, this very same coculture model could be exploited to detect, interrogate and harness for therapeutic purposes the beneficial effect of probiotics and prebiotics that may shift the cellular processes and transcriptomics away from CRCs.
6.2. Genome editing and genetic cancer modeling in organoids
Colon-derived organoids have proven to be the perfect models for controlled and sequential genetic manipulation to mimic what is currently known to occur during CRC initiation and progression. CRISPR/Cas9 genome-editing system has enabled researchers to introduce mutations in the tumor suppressor genes APC, SMAD4, and TP53, and/or the oncogenes KRAS/BRAF and/or PIK3CA/PTEN, either alone, or in combination, either sequentially and/or randomly into the normal human intestinal epithelium. Using such an approach, Matano et al. reported that the organoids harbored all five mutations, i.e., APC, SMAD4, TP53, KRAS and PIK3CA, could grow independently of niche factors (Matano et al., 2015). These isogenic genetically manipulated organoids formed tumors after implantation under the kidney in mice, and they formed micrometastases containing tumor initiating cells after injection in the mice spleen, but neither progressed to form distant metastases in the liver. These findings suggested that the mutations may be sufficient for stem cell maintenance in the hostile tumor microenvironment, but not for metastatic progression (Matano et al., 2015). In addition, Drost et al. reported that loss of APC and TP53 are key drivers of chromosomal instability and aneuploidy that lead to CRC (Drost et al., 2015). Using the CRISPR/Cas9 technique for the generation of quadruple mutant organoids (mutant for APC, P53, KRAS, and SMAD4), they showed that the mutant organoids could grow independently of all stem-cell-niche factors and tolerate the presence of the P53 stabilizer nutlin-3 (Drost et al., 2015). Once again, these mutant organoids grew independently of niche growth factors from the culture medium, and they could grow as tumors with features of invasive carcinoma upon xenotransplantation into mice (Drost et al., 2015). Furthermore, Fumagalli et al. studied the genetic dissection of CRC progression by orthotopic transplantation of human colon organoids engineered to harbor different combinations of CRC-associated mutations (Fumagalli et al., 2017). They showed that the sequential accumulation of oncogenic mutations in critical signaling pathways, e.g., the WNT, EGFR, P53, and TGF-β/SMAD mediates efficient tumor growth, migration, and metastatic colonization (Fumagalli et al., 2017). Besides, Verissimo and colleagues developed normal organoids and tumor organoids with oncogenic KRAS mutation introduced via CRISPR and used them to evaluate the therapeutic potential of RAS-pathway inhibitors and for use in drug screening (Verissimo et al., 2016). These studies revealed that the presence of mutant RAS correlated with resistance to the targeted therapies and that such resistance stemmed from a drug-induced transient cell-cycle arrest rather than cell death (Verissimo et al., 2016). In another study aimed at exploring the origin of cancer-associated mutational signatures (Drost & van Boxtel, 2017), the authors deleted essential DNA repair genes in human colon organoids via CRISPR/Cas9 and found that mutation accumulation in organoids deficient in the mismatch repair gene MLH1 resembles the mutation profiles recorded in mismatch repair-deficient CRCs. By contrast, mutation accumulation in organoids deficient in the base excision repair protein NTHL1 resembles mutation previously reported in a breast cancer cohort (Drost & van Boxtel, 2017). Moreover, genome editing enabled the characterization of cancer-associated genes and identified the entire spectrum of tumor progression and metastasis (Roper et al., 2017). Using gene-editing of the APC and TP53 tumor suppressor genes in colon epithelial cells, followed by orthotopic transplantation of APC-edited colon organoids, Roper and colleagues reported that APCΔ/Δ;KrasG12D/+;TP53Δ/Δ (AKP) mouse colon organoids and human CRC organoids engrafted in the distal colon and metastasized to the liver suggesting that a high-grade dysplasia developed with two driver mutation combinations (Roper et al., 2017).
6.3. CRC organoids as platforms for drug screening
Tumor cell lines grown as monolayers in 2D have been used as models for tumor research and drug screening for a long time (Barretina et al., 2012; Sharma, Haber, & Settleman, 2010). However, gathering evidence suggests that the response of 2D models to drugs often do not mirror the drug response in clinical patients (Liu et al., 2020; Xia, Li, He, Aji, & Gao, 2019). Patient-derived xenograft (PDX) is another model used in cancer research for drug screening and it has advantages over 2D cell lines of being preserving the molecular and morphological characteristics and revealing the heterogenicity of the primary tumor (DeRose et al., 2011; Liu et al., 2017; Wang et al., 2019). However, the PDX model lacks the immune system, and tumor evolution occurred in PDX (Liu et al., 2020). Cancer organoid provided an alternative strategy for drug screening which has advantages over 2D models and PDX models in terms of stability, fidelity, and the possibility of coculture with the immune cells, and hence the cancer organoid is a promising tool for preclinical drug screening and personalized therapy (Kondo & Inoue, 2019; Yang et al., 2019). In addition, the development of organoid biobanks from CRC tissues and adjacent healthy tissue enables high-throughput drug screening and detection of gene-drug associations (van de Wetering et al., 2015). Using CRC organoids, van de Wetering and colleagues screened 83 compound library including drugs in clinical use (n = 25), chemotherapeutics (n = 10), drugs in clinical trials (n = 29), and experimental anti-cancer compounds (n = 29). They showed that a single organoid culture showed high sensitivity to WNT secretion inhibitors (porcupine) was mutated for ring finger protein 43 (RNF43), the negative WNT feedback regulator, rather than in APC (van de Wetering et al., 2015). Vlachogiannis and colleagues compared the relationship of drug responses between patients and their metastatic gastrointestinal tumor organoids. They assessed 55 drugs that are in phase I–III studies or currently approved for clinical use and concluded the beneficial application of tumor organoids in precision cancer medicine (Vlachogiannis et al., 2018). Moreover, Schütte et al. developed a preclinical platform with more than 4000 assays testing the drug sensitivity of 16 clinical drugs using CRC patient organoids (Schütte et al., 2017). Analysis of drug sensitivity patterns identified biomarkers such as a signature outperforming RAS/RAF mutations that could predict the sensitivity to the EGFR inhibitor cetuximab (Schütte et al., 2017).
6.4. Organoids aid in personalizing anti-cancer therapies
Because patient-derived tumor organoids recapitulate the original biopsies that they were derived from (Weeber et al., 2015), the development of living organoid biobanks that represent all CRC subtypes could be ideal models for testing markers of treatment resistance (Schütte etal., 2017) or for optimizing personalized anti-cancer therapies. For example, using organoids as mainstay models, it was established that CRCs harboring KRAS/BRAF mutations are resistant to targeted therapies (Karapetis et al., 2008; Loupakis et al., 2009).
Reversal of the mutation overcomes the resistance and reinstates sensitivity to anti-cancer therapies (Zhai et al., 2017). Mechanistically, resistance was attributed to the ability of KRAS mutations to allow tumor cells to arrest cell cycle when exposed to combination therapies (EGFR/MEK inhibitors and/or EGRF/ERK inhibitors), and persist (Verissimo et al., 2016); these therapy-resisting persistors rapidly restarted tumor growth when the treatment was stopped (Verissimo et al., 2016). Interestingly, the addition of a low dose of navitoclax, a clinically tested BCL2/BCLXL inhibitor, to the EGFR/MEK/ERK pathway through the targeted combinational therapies primed the RAS mutant CRC organoids into apoptosis, indicating the strong potential of patient-derived CRC organoid libraries in evaluating pathway inhibitors and drug combinations in a preclinical setting (Verissimo et al., 2016). Another prospective clinical study demonstrated that PDOs could predict response to chemotherapy in metastatic CRC patients (Ooft & Weeber, 2019). PDOs were used for evaluation of sensitivity to a standard of care chemotherapy in CRC, and they predicted response of the biopsied lesion in more than 80% of patients treated with irinotecan-based therapies without misclassifying patients who could have treatment benefits (Ooft & Weeber, 2019). However, the PDOs failed to predict the outcome for treatment with 5-fluorouracil plus oxaliplatin, suggesting that the prediction was specific to irinotecan-based chemotherapy (Ooft & Weeber, 2019). Moreover, a rectal cancer organoid platform was used to study individual responses to chemoradiation (Ganesh, Wu, O’Rourke, & Szeglin, 2019); the latter were generated from primary, metastatic, and recurrent tumors (n = 65) and subsequently used to assess the responses of individuals to chemoradiation. The response of tumoroids to clinically relevant chemotherapy and radiation treatment were heterogeneous but correlated with the clinical responses noted in individual patients’ tumors (Ganesh et al., 2019). Taken together, all these studies generally agree that PDOs recapitulate the patient tumor phenotype and genotype, and therefore, they are ideal models for enhancing both precision and personalization in CRC treatment.
6.5. Impact of CRC organoids on clinical trials
Vlachogiannis and colleagues carried out phase I/II clinical trials to evaluate the clinical value of PDOs collected from metastatic, heavily pretreated CRC and gastroesophageal cancer patients (n = 71) in the evaluation of the treatment response (Vlachogiannis et al., 2018). Using 3D screening assays and 55 agents under assessment in phase I–III clinical trials or already clinically acceptable standard of care, the drug responses in cancer organoids mirrored the clinical response in the corresponding patients with 100% sensitivity and 93% specificity. These results provide proof-of-concept that PDOs can recapitulate patient responses in the clinic and may become crucial pre-clinical models for early phase clinical trials and for personalizing therapeutics in the clinic (Vlachogiannis et al., 2018). Another study done by Yao and colleagues used PDOs to predict chemoradiation responses in locally advanced rectal cancer (LARC) patients treated with neoadjuvant chemoradiation who were enrolled in a concomitant phase III clinical trial (Yao et al., 2020). The chemoradiation responses in patients showed 84.43% accuracy, 78.01% sensitivity, and 91.97% specificity, compared to the responses in PDOs, indicating that the PDOs predict LARC patient responses in the clinic and could be a helpful prognostic tool in the personalization of management for rectal cancer (Yao et al., 2020).
7. Current challenges in the organoid model
Although the organoid model is a relevant system that physiologically mimics the in vivo environment, some challenges are still associated with this system. Standalone, the organoid model lacks the multicellular nature of the tumor microenvironment, such as the tumor-stromal fibroblasts, vascular endothelial cells, immune cells, and microbial factors. To overcome this challenge, coculture techniques have been developed, featuring immune cells (Finnberg et al., 2017; Neal et al., 2018), and/or fibroblasts (Öhlund, Handly-Santana, Biffi, Elyada, & Almeida, 2017). However, the coculture of the organoids with multiple elements at the same time to mimic the tumor microenvironment remains challenging. Also, the success rate for organoid development from metastatic CRC or from patients undergoing therapy is low, probably due to the low amount of live stem cells found in these biopsies. Besides, fresh biopsies are sometimes not available. Even when tissues are available, organoid biobanking may not be as cost effective in most laboratories unless they have the necessary infrastructure, dedicated funding and expertise that is found only in a handful research laboratories around the world (Drost & Clevers, 2018; Ji & Wu, 2020). In addition, some components in the organoid media such as matrigel and fetal calf serum may affect the experimental readouts, for example, in the setting of drug screening (Drost & Clevers, 2018; Ji & Wu, 2020). Researchers have tried to solve this problem by replacing matrigel with synthetic matrices that support the growth of mouse and human intestinal organoids without interference in the experiment results (Gjorevski et al., 2016). Also, researchers have tried to use serum-free WNT media by replacing fetal calf serum with water-soluble ligands for Frizzled/low-density lipoprotein receptor-related protein 5 (LRP5) or LRP6 heterodimerizes that act as surrogate WNT agonists and can support the growth of many humanoid organoids (Janda et al., 2017). Alternatively, stabilization of WNT protein, using phospholipids and cholesterol, enables the serum-free culture of human organ stem cells (Tüysüz et al., 2017). Also, adequate characterization of each PDO, including whole-exome sequencing, copy number assessment, RNA sequencing, and drug sensitivity tests, is highly recommended for achieving assay specificity, sensitivity, and reproducibility (Ji & Wu, 2020).
8. The next-generation CRC organoids
The traditional 3D culture method for growing 3D organoids depends on the addition of growth factors and extracellular matrix. However, the existing culture systems have reduced the capacity to recapitulate the complex and dynamic microenvironment of a developing organ and organogenesis. Therefore, stem cell researchers, bioengineers, and physicists have collaboratively developed advanced in vitro technologies for organoid research such as organ-on-a-chip and 3D bioprinting of organoid culture system that combine dimensionality and multicellularity with physical forces (Clevers, 2017; Fan, Demirci, & Chen, 2019; Park, Georgescu, & Huh, 2019).
8.1. Organ-on-a-chip
Organs-on-a-chip is microfabricated cell culture device designed to model the functional units of human organs in vitro (Bhatia & Ingber, 2014; Esch, Bahinski, & Huh, 2015). This chip can be used for the development of organoid models because it overcomes the challenges associated with the traditional 3D cultures, such as microenvironmental control (both, physical and chemical), modeling tissue–tissue and multiorgan interactions, and reducing the variability (Park et al., 2019). These advantages are expanded below:
First, organs-on-a-chip mimics tissues perfused by blood vessels, simulated through the use of a microfluidic platform, which mimics the vascularization of organoids and recapitulates the in vivo tumor microenvironment with regard to nutrient supply, hypoxia, etc. (Hasan, Paul, Memic, & Khademhosseini, 2015; Kim, Lee, Chung, & Jeon, 2013; Wang et al., 2016).
Second, these chips are associated with mechanically actuatable microengineered platforms that can generate and apply in vivo–like mechanical forces to organoids such as rhythmic contraction of the stomach (Lee et al., 2018).
Third, the application of continuous flow in the chip leads to the expansion of endothelial progenitors within the organoids; such expansion is critical for the structural and functional development of several organoid models such as kidney, pancreas, and the intestines (Homan, Gupta, & Kroll, 2019; Tao et al., 2019; Workman et al., 2018).
Fourth, organs-on-a-chip provides an automated digital microfluidic platform with an array of electrodes that enables the automated culture of 3D organoid in media droplets, which provides a producible and precise method and reduces the variability compared to the traditional 3D culture system (Au, Chamberlain, Mahesh, Sefton, & Wheeler, 2014; Yin et al., 2016). Moreover, some culture platforms, especially for multiorgan culture, include biosensing elements to permit continuous screening of organoids behavior that will minimize the variability. For example, a multiorgan-on-a-chip device integrated with multi-sensors for long-term monitoring of cardiac, liver organoids, and primary hepatic spheroids during drug treatment (Zhang, Aleman, Shin, & Kilic, 2017).
Fifth, the organs-on-a-chip platform also enables the coculture of different cell/tissue types and organs, e.g., a vascularized liver organoid-on-a-chip is a multicompartment microdevice in which hepatic cells are cocultured with human endothelial cells in an ECM hydrogel (Jin et al., 2018). Jin and colleagues used a microfluidic array to coculture stem cell-derived liver, intestinal, and stomach organoids that were maintained in separate compartments but were communicated via rocker-induced media flow between culture chambers (Jin et al., 2018). This multi-organ model was used to study the complex biology and enterohepatic circulation of bile acids (Jin et al., 2018). Similarly, Skardal and colleagues developed microengineered heart-lung-liver model to reveal how cardiotoxicity of bleomycin is mediated due to the cytokine-associated cross-talk between the lung and the heart tissues (Skardal et al., 2017). In another study, a multiorgan microfluidic chip comprised of four organs, lung, brain, liver, and bone was developed to study the metastasis of primary lung cancer (Xu et al., 2016).
Due to the previously mentioned advantages, brain and/or liver-on-a-chip are promising tools as a preclinical model for drug screening and discovery (Wang et al., 2018; Zhu et al., 2017). Also, organoids-on-a-chip can be used for the generation of patient- and population-specific disease models, which mimics the disease phenotypes in vivo and hence an excellent tool for personalized medicine (Clevers, 2017). Besides, organoids-on-a-chip can be used in regenerative medicine through the transplanting in vitro expanded organoids into animals to repair damaged organs (Nakamura & Sato, 2018).
8.2. 3D-bioprinting of organoid culture
3D bioprinting is a biofabrication process that generates biological components, including ECM scaffolds, cells, and growth factors in 3D (Horváth et al., 2015; Wu et al., 2016). This technology closely mimics the human in vivo microenvironment and has the advantages of being high-precision, high-accuracy, and high-throughput (Knowlton, Yenilmez, & Tasoglu, 2016; Peng, Unutmaz, & Ozbolat, 2016). The standard 3D organoid culture method depends on the manual mixing of cells with ECM substrates in certain ratios before gelling or manually laying the cells on the preformed ECM gel, which could lead to a different size, morphology, distribution, and cell types within the formed organoids (Reid et al., 2016; Yonemura, 2014). Such variation could affect the results of experiments and/or make experiments uninterpretable, or findings irreproducible (Reid, Mollica, Bruno, & Sachs, 2018; Reid et al., 2016; Yonemura, 2014). 3D bioprinting technology overcomes this limitation in the traditional 3D culture method through the use of a computer-aided process that precisely allows the deposition of the cells layer-by-layer in the organoids to confirm the experimental consistency (Zhou et al., 2013). Reid and colleagues successfully adapted a low-cost 3D-printer for precise cell placement and used this system for printing human induced pluripotent stem cells (hiPSCs) within 3D hydrogels (Reid et al., 2016). This system was subsequently used to generate large 3D mammary organoids in hydrogels (Reid et al., 2018). This printer was later used as a platform for mechanistic analysis of tumoroids and chimeric mammary organoids to study the microenvironmental control of breast cancer (Reid & Palmer, 2019). In the context of CRCs, Rios de la Rosa and colleagues developed and validated a biofabrication method to model the early-stage CRC tumor monitoring two biomarkers (CD44 and HIF-1α) (Rios de la Rosa, Wubetu, Tirelli, & Tirella, 2018). Using alginate hydrogels and HCT-116 cells, they successfully biofabricated the 3D model and monitored two biomarkers (CD44 and HIF-1α) during tumor development. This model mimics the tumor environment and closely represents the heterogeneous tumor mass and has distinct advantages over the traditional 3D culture system such as spheroid and monolayer. Therefore, this high throughput 3D system is a promising tool for preclinical drug testing in CRC (Rios de la Rosa et al., 2018).
9. Conclusion
Patient-derived organoids have immense applications in CRC research as they retain the molecular, cellular, and histological complexity and genetic heterogeneity of the individual patients after long-term culture and can be used for the personalizing optimal treatment strategies. Technological advancement in organoid-based studies, especially those that allow their use in the high throughput format, with higher reproducibility, and quality control is expected to spur more accurate modeling of disease, organ replacement and drug testing. Approximately 15% of oncology drugs proceed from Phase I to FDA approval (DiMasi, Reichert, Feldman, & Malins, 2013; Hay, Thomas, Craighead, Economides, & Rosenthal, 2014). The success rate may improve if we can include pre-clinical human models, e.g., patient-specific organoids with cutting edge organ-on-a-chip and 3D bioprinting models before the candidate drugs enter Phase I. The addition of all the necessary cell types and the microbiome population of the same patient will open new avenues in the detection, measurement, tracking of and tackling (through therapeutics) the risk of CRC initiation and progression. Breakthroughs in cancer research in the coming years are expected to include the development of novel treatment strategies that program either the tumor cell, or its environment, or the host immune system and/or some combination of them all; organoids are bound to be big players in that journey. Their use is expected to help maintain our focus on what is relevant and real in humans, and thereby improve precision in patient care.
Acknowledgment
This work was supported, partly by National Institute of Health Grants: R01 AI141630, CA238042, and CA100768 (to P.G.); DK107585, AG069689, C3 Padre Pedal MCC pilot grant (to S.D.); and UG3TR002968 (to P.G. and S.D.). A.A.A. is supported by an NIH-funded Cancer Therapeutics Training Program (CT2, T32 CA121938).
References
- Alhinai EA, Walton GE, & Commane DM (2019). The role of the gut microbiota in colorectal cancer causation. International Journal of Molecular Sciences, 20(21), 5295. 10.3390/ijms20215295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen J, & Sears CL (2019). Impact of the gut microbiome on the genome and epigenome of colon epithelial cells: Contributions to colorectal cancer development. Genome Medicine, 11(1), 11. 10.1186/s13073-019-0621-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur JC, Perez-Chanona E, Mühlbauer M, Tomkovich S, Uronis JM, Fan TJ, et al. (2012). Intestinal inflammation targets cancer-inducing activity of the microbiota. Science, 338(6103), 120–123. 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au SH, Chamberlain MD, Mahesh S, Sefton MV, & Wheeler AR (2014). Hepatic organoids for microfluidic drug screening. Lab on a Chip, 14(17), 3290–3299. 10.1039/c4lc00531g. [DOI] [PubMed] [Google Scholar]
- Bardhan K, & Liu K (2013). Epigenetics and colorectal cancer pathogenesis. Cancers (Basel), 5(2), 676–713. 10.3390/cancers5020676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. (2012). The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature, 483(7391), 603–607. 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartfeld S, Bayram T, van de Wetering M, Huch M, Begthel H, Kujala P, et al. (2015). In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology, 148(1), 126–136. e126. 10.1053/j.gastro.2014.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia SN, & Ingber DE (2014). Microfluidic organs-on-chips. Nature Biotechnology, 32(8), 760–772. 10.1038/nbt.2989. [DOI] [PubMed] [Google Scholar]
- Broutier L, Mastrogiovanni G, Verstegen MM, Francies HE, Gavarró LM, Bradshaw CR, et al. (2017). Human primary liver cancer-derived organoid cultures for disease modeling and drug screening. Nature Medicine, 23(12), 1424–1435. 10.1038/nm.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruun J, Kryeziu K, Eide PW, Moosavi SH, Eilertsen IA, Langerud J, et al. (2020). Patient-derived organoids from multiple colorectal cancer liver metastases reveal moderate intra-patient pharmacotranscriptomic heterogeneity. Clinical Cancer Research, 26(15), 4107–4119. 10.1158/1078-0432.CCR-19-3637. [DOI] [PubMed] [Google Scholar]
- Budinska E, Popovici V, Tejpar S, D’Ario G, Lapique N, Sikora KO, et al. (2013). Gene expression patterns unveil a new level of molecular heterogeneity in colorectal cancer. The Journal of Pathology, 231(1), 63–76. 10.1002/path.4212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan AT, & Giovannucci EL (2010). Primary prevention of colorectal cancer. Gastroenterology, 138(6), 2029–2043. e2010. 10.1053/j.gastro.2010.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen DS, & Mellman I (2017). Elements of cancer immunity and the cancer-immune set point. Nature, 541(7637), 321–330. 10.1038/nature21349. [DOI] [PubMed] [Google Scholar]
- Clevers H (2017). Advances in organoid technology: Hans Clevers, Madeline Lancaster, and Takanori Takebe. Cell Stem Cell, 20, 759–762. [Google Scholar]
- Coleman OI, & Nunes T (2016). Role of the microbiota in colorectal cancer: Updates on microbial associations and therapeutic implications. Biores Open Access, 5(1), 279–288. 10.1089/biores.2016.0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuevas-Ramos G, Petit CR, Marcq I, Boury M, Oswald E, & Nougayrède JP (2010). Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America, 107(25), 11537–11542. 10.1073/pnas.1001261107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Aldebert E, Quaranta M, Sebert M, Bonnet D, Kirzin S, Portier G, et al. (2020). Characterization of human colon organoids from inflammatory bowel disease patients. Frontiers in Cell and Development Biology, 8, 363. 10.3389/fcell.2020.00363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lau W, Barker N, Low TY, Koo BK, Li VS, Teunissen H, et al. (2011). Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature, 476(7360), 293–297. 10.1038/nature10337. [DOI] [PubMed] [Google Scholar]
- Dejea CM, Fathi P, & Craig JM (2018). Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science, 359(6375), 592–597. 10.1126/science.aah3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRose YS, Wang G, Lin YC, Bernard PS, Buys SS, Ebbert MT, et al. (2011). Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nature Medicine, 17(11), 1514–1520. 10.1038/nm.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desoize B, & Jardillier J-C (2000). Multicellular resistance: a paradigm for clinical resistance? Critical Reviews in Oncology/Hematology, 36(2–3), 193–207. 10.1016/s1040-8428(00)00086-x. [DOI] [PubMed] [Google Scholar]
- Dignass AU, & Sturm A (2001). Peptide growth factors in the intestine. European Journal of Gastroenterology & Hepatology, 13(7), 763–770. 10.1097/00042737200107000-00002. [DOI] [PubMed] [Google Scholar]
- Dijkstra KK, Cattaneo CM, Weeber F, Chalabi M, van de Haar J, Fanchi LF, et al. (2018). Generation of tumor-reactive T cells by co-culture of peripheral blood lymphocytes and tumor organoids. Cell, 174(6), 1586–1598. e1512. 10.1016/j.cell.2018.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMasi JA, Reichert JM, Feldman L, & Malins A (2013). Clinical approval success rates for investigational cancer drugs. Clinical Pharmacology and Therapeutics, 94(3), 329–335. 10.1038/clpt.2013.117. [DOI] [PubMed] [Google Scholar]
- Dolznig H, Rupp C, Puri C, Haslinger C, Schweifer N, Wieser E, et al. (2011). Modeling colon adenocarcinomas in vitro: A 3D co-culture system induces cancer-relevant pathways upon tumor cell and stromal fibroblast interaction. The American Journal of Pathology, 179(1), 487–501. 10.1016/j.ajpath.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domcke S, Sinha R, Levine DA, Sander C, & Schultz N (2013). Evaluating cell lines as tumour models by comparison of genomic profiles. Nature Communications, 4(2126). 10.1038/ncomms3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drost J, & Clevers H (2018). Organoids in cancer research. Nature Reviews Cancer, 18(7), 407–418. 10.1038/s41568-018-0007-6. [DOI] [PubMed] [Google Scholar]
- Drost J, & van Boxtel R (2017). Use of CRISPR-modified human stem cell organoids to study the origin of mutational signatures in cancer. Science, 358(6360), 234–238. 10.1126/science.aao3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drost J, van Jaarsveld RH, Ponsioen B, Zimberlin C, van Boxtel R, Buijs A, et al. (2015). Sequential cancer mutations in cultured human intestinal stem cells. Nature, 521(7550), 43–47. 10.1038/nature14415. [DOI] [PubMed] [Google Scholar]
- Du Y, Li X, Niu Q, Mo X, Qui M, Ma T, et al. (2020). Development of a miniaturized 3D organoid culture platform for ultra-high-throughput screening. Journal of Molecular Cell Biology, 12(8), 630–643. 10.1093/jmcb/mjaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel RM, Chan WH, Nickless D, Hlavca S, Richards E, Kerr G, et al. (2020). Patient-derived colorectal cancer organoids upregulate revival stem cell marker genes following chemotherapeutic treatment. Journal of Clinical Medicine, 9(1), 128. 10.3390/jcm9010128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esch EW, Bahinski A, & Huh D (2015). Organs-on-chips at the frontiers of drug discovery. Nature Reviews. Drug Discovery, 14(4), 248–260. 10.1038/nrd4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H, Demirci U, & Chen P (2019). Emerging organoid models: Leaping forward in cancer research. Journal of Hematology & Oncology, 12(1), 142. 10.1186/s13045-019-0832-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnberg NK, Gokare P, Lev A, Grivennikov SI, MacFarlane AW, Campbell KS, et al. (2017). Application of 3D tumoroid systems to define immune and cytotoxic therapeutic responses based on tumoroid and tissue slice culture molecular signatures. Oncotarget, 8(40), 66747–66757. 10.18632/oncotarget.19965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach C, Kong HJ, Hsiong SX, Evangelista MB, Yuen W, & Mooney DJ (2009). Cancer cell angiogenic capability is regulated by 3D culture and integrin engagement. Proceedings of the National Academy of Sciences, 106(2), 399–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey MR, Dise RS, Edelblum KL, & Polk DB (2006). p38 kinase regulates epidermal growth factor receptor downregulation and cellular migration. The EMBO Journal, 25(24), 5683–5692. 10.1038/sj.emboj.7601457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii M, Shimokawa M, Date S, Takano A, Matano M, Nanki K, et al. (2016). A colorectal tumor organoid library demonstrates progressive loss of niche factor requirements during tumorigenesis. Cell Stem Cell, 18(6), 827–838. 10.1016/j.stem.2016.04.003. [DOI] [PubMed] [Google Scholar]
- Fumagalli A, Drost J, Suijkerbuijk SJ, van Boxtel R, de Ligt J, Offerhaus GJ, et al. (2017). Genetic dissection of colorectal cancer progression by orthotopic transplantation of engineered cancer organoids. Proceedings of the National Academy of Sciences, 114(12), E2357–e2364. 10.1073/pnas.1701219114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesh K, Wu C, O’Rourke KP, & Szeglin BC (2019). A rectal cancer organoid platform to study individual responses to chemoradiation. Nature Medicine, 25(10), 1607–1614. 10.1038/s41591-019-0584-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D, Vela I, Sboner A, Iaquinta PJ, Karthaus WR, Gopalan A, et al. (2014). Organoid cultures derived from patients with advanced prostate cancer. Cell, 159(1), 176–187. 10.1016/j.cell.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng F, Zhang Y, Lu Z, Zhang S, & Pan Y (2020). Fusobacterium nucleatum caused DNA damage and promoted cell proliferation by the Ku70/p53 pathway in oral cancer cells. DNA and Cell Biology, 39(1), 144–151. 10.1089/dna.2019.5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh P, Swanson L, Sayed IM, Mittal Y, Lim BB, Ibeawuchi SR, et al. (2020). The stress polarity signaling (SPS) pathway serves as a marker and a target in the leaky gut barrier: Implications in aging and cancer. Life Science Alliance, 3(3), e201900481. 10.26508/lsa.201900481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjorevski N, Sachs N, Manfrin A, Giger S, Bragina ME, Ordóñez-Morán P, et al. (2016). Designer matrices for intestinal stem cell and organoid culture. Nature, 539(7630), 560–564. 10.1038/nature20168. [DOI] [PubMed] [Google Scholar]
- Goldstein NS (2006). Serrated pathway and APC (conventional)-type colorectal polyps: Molecular-morphologic correlations, genetic pathways, and implications for classification. American Journal of Clinical Pathology, 125(1), 146–153. [PubMed] [Google Scholar]
- Goodwin AC, Destefano Shields CE, Wu S, Huso DL, Wu X, Murray-Stewart TR, et al. (2011). Polyamine catabolism contributes to enterotoxigenic Bacteroides fragilis-induced colon tumorigenesis. Proceedings of the National Academy of Sciences of the United States of America, 108(37), 15354–15359. 10.1073/pnas.1010203108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalappa C, Aydogan-Cremaschi S, Das TK, & Orcun S (2011). Probability model for estimating colorectal polyp progression rates. Health Care Management Science, 14(1), 1–21. 10.1007/s10729-010-9138-3. [DOI] [PubMed] [Google Scholar]
- Guinney J, Dienstmann R, Wang X, de Reynies A, Schlicker A, Soneson C, et al. (2015). The consensus molecular subtypes of colorectal cancer. Nature Medicine, 21(11), 1350–1356. 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haramis AP, Begthel H, van den Born M, van Es J, Jonkheer S, Offerhaus GJ, et al. (2004). De novo crypt formation and juvenile polyposis on BMP inhibition in mouse intestine. Science, 303(5664), 1684–1686. 10.1126/science.1093587. [DOI] [PubMed] [Google Scholar]
- Harrington LX, Wei JW, Suriawinata AA, Mackenzie TA, & Hassanpour S (2020). Predicting colorectal polyp recurrence using time-to-event analysis of medical records. AMIA Joint Summits on Translational Science Proceedings, 2020, 211–220. [PMC free article] [PubMed] [Google Scholar]
- Hasan A, Paul A, Memic A, & Khademhosseini A (2015). A multilayered microfluidic blood vessel-like structure. Biomedical Microdevices, 17(5), 88. 10.1007/s10544-015-9993-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay M, Thomas DW, Craighead JL, Economides C, & Rosenthal J (2014). Clinical development success rates for investigational drugs. Nature Biotechnology, 32(1), 40–51. 10.1038/nbt.2786. [DOI] [PubMed] [Google Scholar]
- Holokai L, Chakrabarti J, Broda T, Chang J, Hawkins JA, Sundaram N, et al. (2019). Increased programmed death-ligand 1 is an early epithelial cell response to helicobacter pylori infection. PLoS Pathogens, 15(1), e1007468. 10.1371/journal.ppat.1007468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homan KA, Gupta N, & Kroll KT (2019). Flow-enhanced vascularization and maturation of kidney organoids in vitro. Nature Methods, 16(3), 255–262. 10.1038/s41592-019-0325-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horváth L, Umehara Y, Jud C, Blank F, Petri-Fink A, & Rothen-Rutishauser B (2015). Engineering an in vitro air-blood barrier by 3D bioprinting. Scientific Reports, 5, 7974. 10.1038/srep07974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Holtzinger A, Jagan I, BeGora M, Lohse I, Ngai N, et al. (2015). Ductal pancreatic cancer modeling and drug screening using human pluripotent stem cell- and patient-derived tumor organoids. Nature Medicine, 21(11), 1364–1371. 10.1038/nm.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Liu Y, Zheng C, & Shen C (2017). Investigation of cross-contamination and misidentification of 278 widely used tumor cell lines. PLoS One, 12(1), e0170384. 10.1371/journal.pone.0170384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JY, Sweeney EG, Sigal M, Zhang HC, Remington SJ, Cantrell MA, et al. (2015). Chemodetection and destruction of host urea allows helicobacter pylori to locate the epithelium. Cell Host & Microbe, 18(2), 147–156. 10.1016/j.chom.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inamura K (2018). Colorectal cancers: An update on their molecular pathology. Cancers (Basel), 10(1), 26. 10.3390/cancers10010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda CY, Dang LT, You C, Chang J, de Lau W, Zhong ZA, et al. (2017). Surrogate Wnt agonists that phenocopy canonical Wnt and β-catenin signalling. Nature, 545(7653), 234–237. 10.1038/nature22306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jass JR (2007). Classification of colorectal cancer based on correlation of clinical, morphological and molecular features. Histopathology, 50(1), 113–130. 10.1111/j.1365-2559.2006.02549.x. [DOI] [PubMed] [Google Scholar]
- Ji D-B, & Wu A-W (2020). Organoid in colorectal cancer: Progress and challenges. Chinese Medical Journal, 133(16), 1971–1977. 10.1097/cm9.0000000000000882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Kim J, Lee JS, Min S, Kim S, Ahn D-H, et al. (2018). Vascularized liver organoids generated using induced hepatic tissue and dynamic liver-specific microenvironment as a drug testing platform. Advanced Functional Materials, 28(37), 1801954. 10.1002/adfm.201801954. [DOI] [Google Scholar]
- Johnson RL, & Fleet JC (2013). Animal models of colorectal cancer. Cancer and Metastasis Reviews, 32(1–2), 39–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph JS, Malindisa ST, & Ntwasa M (2018). Two-dimensional (2D) and three-dimensional (3D) cell culturing in drug discovery. IntechOpen. [Google Scholar]
- Ju HX, An B, Okamoto Y, Shinjo K, Kanemitsu Y, Komori K, et al. (2011). Distinct profiles of epigenetic evolution between colorectal cancers with and without metastasis. The American Journal of Pathology, 178(4), 1835–1846. 10.1016/j.ajpath.2010.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung P, Sato T, Merlos-Suárez A, Barriga FM, Iglesias M, Rossell D, et al. (2011). Isolation and in vitro expansion of human colonic stem cells. Nature Medicine, 17(10), 1225–1227. 10.1038/nm.2470. [DOI] [PubMed] [Google Scholar]
- Kannen V, Hintzsche H, Zanette DL, Silva WA Jr., Garcia SB, Waaga-Gasser AM, et al. (2012). Antiproliferative effects of fluoxetine on colon cancer cells and in a colonic carcinogen mouse model. PLoS One, 7(11), e50043. 10.1371/journal.pone.0050043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karapetis CS, Khambata-Ford S, Jonker DJ, O’Callaghan CJ, Tu D, Tebbutt NC, et al. (2008). K-ras mutations and benefit from cetuximab in advanced colorectal cancer. The New England Journal of Medicine, 359(17), 1757–1765. 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- Kim KA, Kakitani M, Zhao J, Oshima T, Tang T, Binnerts M, et al. (2005). Mitogenic influence of human R-spondin1 on the intestinal epithelium. Science, 309(5738), 1256–1259. 10.1126/science.1112521. [DOI] [PubMed] [Google Scholar]
- Kim S, Lee H, Chung M, & Jeon NL (2013). Engineering of functional, perfusable 3D microvascular networks on a chip. Lab on a Chip, 13(8), 1489–1500. 10.1039/c3lc41320a. [DOI] [PubMed] [Google Scholar]
- Knowlton S, Yenilmez B, & Tasoglu S (2016). Towards single-step biofabrication of organs on a chip via 3D printing. Trends in Biotechnology, 34(9), 685–688. 10.1016/j.tibtech.2016.06.005. [DOI] [PubMed] [Google Scholar]
- Kondo J, & Inoue M (2019). Application of cancer organoid model for drug screening and personalized therapy. Cell, 8(5). 10.3390/cells8050470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopper O, de Witte CJ, Lõhmussaar K, Valle-Inclan JE, Hami N, Kester L, et al. (2019). An organoid platform for ovarian cancer captures intra- and interpatient heterogeneity. Nature Medicine, 25(5), 838–849. 10.1038/s41591-019-0422-6. [DOI] [PubMed] [Google Scholar]
- Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, et al. (2013). Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host & Microbe, 14(2), 207–215. 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, et al. (2012). Genomic analysis identifies association of fusobacterium with colorectal carcinoma. Genome Research, 22(2), 292–298. 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. (2015). PD-1 blockade in tumors with mismatch-repair deficiency. The New England Journal of Medicine, 372(26), 2509–2520. 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KK, McCauley HA, Broda TR, Kofron MJ, Wells JM, & Hong CI (2018). Human stomach-on-a-chip with luminal flow and peristaltic-like motility. Lab on a Chip, 18(20), 3079–3085. 10.1039/c8lc00910d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Milner LA, Deng Y, Iwata M, Banta A, Graf L, et al. (1998). The human homolog of rat Jagged1 expressed by marrow stroma inhibits differentiation of 32D cells through interaction with Notch1. Immunity, 8(1), 43–55. 10.1016/s1074-7613(00)80457-4. [DOI] [PubMed] [Google Scholar]
- Li K, Zhou ZY, Ji PP, & Luo HS (2016). Knockdown of β-catenin by siRNA influences proliferation, apoptosis and invasion of the colon cancer cell line SW480. Oncology Letters, 11(6), 3896–3900. 10.3892/ol.2016.4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JF, Palakurthi S, Zeng Q, Zhou S, Ivanova E, Huang W, et al. (2017). Establishment of patient-derived tumor xenograft models of epithelial ovarian cancer for preclinical evaluation of novel therapeutics. Clinical Cancer Research, 23(5), 1263–1273. 10.1158/1078-0432.ccr-16-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Qin T, Huang Y, Li Y, Chen G, & Sun C (2020). Drug screening model meets cancer organoid technology. Translational Oncology, 13(11), 100840. 10.1016/j.tranon.2020.100840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loupakis F, Ruzzo A, Cremolini C, Vincenzi B, Salvatore L, Santini D, et al. (2009). KRAS codon 61, 146 and BRAF mutations predict resistance to cetuximab plus irinotecan in KRAS codon 12 and 13 wild-type metastatic colorectal cancer. British Journal of Cancer, 101(4), 715–721. 10.1038/sj.bjc.6605177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marisa L, de Reyniès A, Duval A, Selves J, Gaub MP, Vescovo L, et al. (2013). Gene expression classification of colon cancer into molecular subtypes: Characterization, validation, and prognostic value. PLoS Medicine, 10(5), e1001453. 10.1371/journal.pmed.1001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin HM, Campbell BJ, Hart CA, Mpofu C, Nayar M, Singh R, et al. (2004). Enhanced Escherichia coli adherence and invasion in Crohn’s disease and colon cancer. Gastroenterology, 127(1), 80–93. 10.1053/j.gastro.2004.03.054. [DOI] [PubMed] [Google Scholar]
- Matano M, Date S, Shimokawa M, Takano A, Fujii M, Ohta Y, et al. (2015). Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nature Medicine, 21(3), 256–262. 10.1038/nm.3802. [DOI] [PubMed] [Google Scholar]
- Mauri G, Sartore-Bianchi A, Russo AG, Marsoni S, Bardelli A, & Siena S (2019). Early-onset colorectal cancer in young individuals. Molecular Oncology, 13(2), 109–131. 10.1002/1878-0261.12417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Y, Ren Z, Xu F, Zhou X, Song C, Wang VY, et al. (2018). Nicotinamide promotes cell survival and differentiation as kinase inhibitor in human pluripotent stem cells. Stem Cell Reports, 11(6), 1347–1356. 10.1016/j.stemcr.2018.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa K, Walker SM, Nakajima M, Pathak S, Jessup JM, & Fidler IJ (1988). Influence of organ environment on the growth, selection, and metastasis of human colon carcinoma cells in nude mice. Cancer Research, 48(23), 6863–6871. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/2846163. [PubMed] [Google Scholar]
- Naito S, von Eschenbach AC, Giavazzi R, & Fidler IJ (1986). Growth and metastasis of tumor cells isolated from a human renal cell carcinoma implanted into different organs of nude mice. Cancer Research, 46(8), 4109–4115. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/3731078. [PubMed] [Google Scholar]
- Nakamura T, & Sato T (2018). Advancing intestinal organoid technology toward regenerative medicine. Cellular and Molecular Gastroenterology and Hepatology, 5(1), 51–60. 10.1016/j.jcmgh.2017.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanki K, Toshimitsu K, Takano A, Fujii M, Shimokawa M, Ohta Y, et al. (2018). Divergent routes toward Wnt and R-spondin niche independency during human gastric carcinogenesis. Cell, 174(4), 856–869. e817. 10.1016/j.cell.2018.07.027. [DOI] [PubMed] [Google Scholar]
- Nautiyal J, Banerjee S, Kanwar SS, Yu Y, Patel BB, Sarkar FH, et al. (2011). Curcumin enhances dasatinib-induced inhibition of growth and transformation of colon cancer cells. International Journal of Cancer, 128(4), 951–961. 10.1002/ijc.25410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neal JT, Li X, Zhu J, Giangarra V, Grzeskowiak CL, Ju J, et al. (2018). Organoid modeling of the tumor immune microenvironment. Cell, 175(7), 1972–1988. e1916. 10.1016/j.cell.2018.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noffsinger AE (2009). Serrated polyps and colorectal cancer: New pathway to malignancy. Annual Review of Pathology, 4, 343–364. 10.1146/annurev.pathol.4.110807.092317. [DOI] [PubMed] [Google Scholar]
- Nougayrède JP, Homburg S, Taieb F, Boury M, Brzuszkiewicz E, Gottschalk G, et al. (2006). Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science, 313(5788), 848–851. 10.1126/science.1127059. [DOI] [PubMed] [Google Scholar]
- Nozaki K, Mochizuki W, Matsumoto Y, Matsumoto T, Fukuda M, Mizutani T, et al. (2016). Co-culture with intestinal epithelial organoids allows efficient expansion and motility analysis of intraepithelial lymphocytes. Journal of Gastroenterology, 51(3), 206–213. 10.1007/s00535-016-1170-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien MJ, Yang S, Mack C, Xu H, Huang CS, Mulcahy E, et al. (2006). Comparison of microsatellite instability, CpG island methylation phenotype, BRAF and KRAS status in serrated polyps and traditional adenomas indicates separate pathways to distinct colorectal carcinoma end points. The American Journal of Surgical Pathology, 30(12), 1491–1501. 10.1097/01.pas.0000213313.36306.85. [DOI] [PubMed] [Google Scholar]
- Öhlund D, Handly-Santana A, Biffi G, Elyada E, & Almeida AS (2017). Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. The Journal of Experimental Medicine, 214(3), 579–596. 10.1084/jem.20162024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira RC, Abrantes AM, Tralhão JG, & Botelho MF (2020). The role of mouse models in colorectal cancer research—The need and the importance of the orthotopic models. Animal Models and Experimental Medicine, 3(1), 1–8. 10.1002/ame2.12102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooft SN, & Weeber F (2019). Patient-derived organoids can predict response to chemotherapy in metastatic colorectal cancer patients. Science Translational Medicine, 11(513), eaay2574. 10.1126/scitranslmed.aay2574. [DOI] [PubMed] [Google Scholar]
- Otte J, Dizdar L, Behrens B, Goering W, Knoefel WT, Wruck W, et al. (2019). FGF signalling in the self-renewal of colon cancer organoids. Scientific Reports, 9(1), 17365. 10.1038/s41598-019-53907-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SE, Georgescu A, & Huh D (2019). Organoids-on-a-chip. Science, 364(6444), 960–965. 10.1126/science.aaw7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng W, Unutmaz D, & Ozbolat IT (2016). Bioprinting towards physiologically relevant tissue models for pharmaceutics. Trends in Biotechnology, 34(9), 722–732. 10.1016/j.tibtech.2016.05.013. [DOI] [PubMed] [Google Scholar]
- Phipps AI, Limburg PJ, Baron JA, Burnett-Hartman AN, Weisenberger DJ, Laird PW, et al. (2015). Association between molecular subtypes of colorectal cancer and patient survival. Gastroenterology, 148(1), 77–87. e72. 10.1053/j.gastro.2014.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Gregorieff A, Begthel H, & Clevers H (2003). Canonical Wnt signals are essential for homeostasis of the intestinal epithelium. Genes & Development, 17(14), 1709–1713. 10.1101/gad.267103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleguezuelos-Manzano C, Puschhof J, van den Brink S, Geurts V, Beumer J, & Clevers H (2020). Establishment and culture of human intestinal organoids derived from adult stem cells. Current Protocols in Immunology, 130(1), e106. 10.1002/cpim.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter RJ, Murray GI, & McLean MH (2020). Current concepts in tumour-derived organoids. British Journal of Cancer, 123(8), 1209–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid JA, Mollica PA, Bruno RD, & Sachs PC (2018). Consistent and reproducible cultures of large-scale 3D mammary epithelial structures using an accessible bioprinting platform. Breast Cancer Research, 20(1), 122. 10.1186/s13058018-1045-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid JA, Mollica PA, Johnson GD, Ogle RC, Bruno RD, & Sachs PC (2016). Accessible bioprinting: Adaptation of a low-cost 3D-printer for precise cell placement and stem cell differentiation. Biofabrication, 8(2), 025017. 10.1088/17585090/8/2/025017. [DOI] [PubMed] [Google Scholar]
- Reid JA, & Palmer XL (2019). A 3D bioprinter platform for mechanistic analysis of tumoroids and chimeric mammary organoids. Scientific Reports, 9(1), 7466. 10.1038/s41598-019-43922-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl A, Schlederer M, Pudelko K, Stadler M, Walter S, Unterleuthner D, et al. (2017). Comparison of cancer cells in 2D vs 3D culture reveals differences in AKT–mTOR–S6K signaling and drug responses. Journal of Cell Science, 130(1), 203–218. 10.1242/jcs.188102. [DOI] [PubMed] [Google Scholar]
- Rios de la Rosa JM, Wubetu J, Tirelli N, & Tirella A (2018). Colorectal tumor 3D in vitro models: Advantages of biofabrication for the recapitulation of early stages of tumour development. Biomedical Physics & Engineering Express, 4(4), 045010. 10.1088/2057-1976/aac1c9. [DOI] [PubMed] [Google Scholar]
- Ronen J, Hayat S, & Akalin A (2019). Evaluation of colorectal cancer subtypes and cell lines using deep learning. Life Science Alliance, 2(6), e201900517. 10.26508/lsa.201900517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roper J, Tammela T, Cetinbas NM, Akkad A, Roghanian A, Rickelt S, et al. (2017). In vivo genome editing and organoid transplantation models of colorectal cancer and metastasis. Nature Biotechnology, 35(6), 569–576. 10.1038/nbt.3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, & Han YW (2013). Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA Adhesin. Cell Host & Microbe, 14(2), 195–206. 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs N, de Ligt J, Kopper O, Gogola E, Bounova G, Weeber F, et al. (2018). A living biobank of breast cancer organoids captures disease heterogeneity. Cell, 172(1–2), 373–386. e310. 10.1016/j.cell.2017.11.010. [DOI] [PubMed] [Google Scholar]
- Sachs N, Papaspyropoulos A, Zomer-van Ommen DD, Heo I, Böttinger L, Klay D, et al. (2019). Long-term expanding human airway organoids for disease modeling. EMBO Journal, 38(4). 10.15252/embj.2018100300, e100300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadanandam A, Lyssiotis CA, Homicsko K, Collisson EA, Gibb WJ, Wullschleger S, et al. (2013). A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nature Medicine, 19(5), 619–625. 10.1038/nm.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Giltay R, Talts U, Timpl R, & Talts JF (2002). Expression and distribution of laminin alpha1 and alpha2 chains in embryonic and adult mouse tissues: An immunochemical approach. Experimental Cell Research, 275(2), 185–199. 10.1006/excr.2002.5499. [DOI] [PubMed] [Google Scholar]
- Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van den Brink S, et al. (2011). Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology, 141(5), 1762–1772. 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, et al. (2009). Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature, 459(7244), 262–265. 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- Sayed IM, Chakraborty A, Abd El-Hafeez AA, Sharma A, Sahan AZ, Huang WJM, et al. (2020). The DNA glycosylase NEIL2 suppresses fusobacterium-infection-induced inflammation and DNA damage in colonic epithelial cells. Cells, 9(9), 1980. 10.3390/cells9091980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayed IM, Sahan AZ, Venkova T, Chakraborty A, Mukhopadhyay D, Bimczok D, et al. (2020). Helicobacter pylori infection down-regulates the DNA glycosylase NEIL2, resulting in increased genome damage and inflammation in gastric epithelial cells. The Journal of Biological Chemistry, 295(32), 11082–11098. 10.1074/jbc.RA119.009981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayed IM, Suarez K, Lim E, Singh S, Pereira M, Ibeawuchi SR, et al. (2020). Host engulfment pathway controls inflammation in inflammatory bowel disease. The FEBS Journal, 287(18), 3967–3988. 10.1111/febs.15236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanu T, Spaapen RM, Bakker JM, Pratap CB, Wu LE, Hofland I, et al. (2015). Salmonella manipulation of host signaling pathways provokes cellular transformation associated with gallbladder carcinoma. Cell Host & Microbe, 17(6), 763–774. 10.1016/j.chom.2015.05.002. [DOI] [PubMed] [Google Scholar]
- Schütte M, Risch T, Abdavi-Azar N, Boehnke K, Schumacher D, Keil M, et al. (2017). Molecular dissection of colorectal cancer in pre-clinical models identifies biomarkers predicting sensitivity to EGFR inhibitors. Nature Communications, 8, 14262. 10.1038/ncomms14262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears CL, Islam S, Saha A, Arjumand M, Alam NH, Faruque AS, et al. (2008). Association of enterotoxigenic Bacteroides fragilis infection with inflammatory diarrhea. Clinical Infectious Diseases, 47(6), 797–803. 10.1086/591130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebrell TA, Hashimi M, Sidar B, Wilkinson RA, Kirpotina L, Quinn MT, et al. (2019). A novel gastric spheroid co-culture model reveals chemokine-dependent recruitment of human dendritic cells to the gastric epithelium. Cellular and Molecular Gastroenterology and Hepatology, 8(1), 157–171. e153. 10.1016/j.jcmgh.2019.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seino T, Kawasaki S, Shimokawa M, Tamagawa H, Toshimitsu K, Fujii M, et al. (2018). Human pancreatic tumor organoids reveal loss of stem cell niche factor dependence during disease progression. Cell Stem Cell, 22(3), 454–467. e456. 10.1016/j.stem.2017.12.009. [DOI] [PubMed] [Google Scholar]
- Sharma P, & Allison JP (2015). The future of immune checkpoint therapy. Science, 348(6230), 56–61. 10.1126/science.aaa8172. [DOI] [PubMed] [Google Scholar]
- Sharma SV, Haber DA, & Settleman J (2010). Cell line-based platforms to evaluate the therapeutic efficacy of candidate anticancer agents. Nature Reviews. Cancer, 10(4), 241–253. 10.1038/nrc2820. [DOI] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Goding Sauer A, Fedewa SA, Butterly LF, Anderson JC, et al. (2020). Colorectal cancer statistics, 2020. CA: A Cancer Journal for Clinicians, 70(3), 145–164. 10.3322/caac.21601. [DOI] [PubMed] [Google Scholar]
- Simon K (2016). Colorectal cancer development and advances in screening. Clinical Interventions in Aging, 11, 967–976. 10.2147/cia.s109285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skardal A, Murphy SV, Devarasetty M, Mead I, Kang HW, Seol YJ, et al. (2017). Multi-tissue interactions in an integrated three-tissue organ-on-a-chip platform. Scientific Reports, 7(1), 8837. 10.1038/s41598-017-08879-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snover DC, Jass JR, Fenoglio-Preiser C, & Batts KP (2005). Serrated polyps of the large intestine: A morphologic and molecular review of an evolving concept. American Journal of Clinical Pathology, 124(3), 380–391. 10.1309/v2ep-tplj-rb3f-ghjl. [DOI] [PubMed] [Google Scholar]
- Spring KJ, Zhao ZZ, Karamatic R, Walsh MD, Whitehall VL, Pike T, et al. (2006). High prevalence of sessile serrated adenomas with BRAF mutations: A prospective study of patients undergoing colonoscopy. Gastroenterology, 131(5), 1400–1407. 10.1053/j.gastro.2006.08.038. [DOI] [PubMed] [Google Scholar]
- Tajima A, Pradhan I, Trucco M, & Fan Y (2016). Restoration of thymus function with bioengineered thymus organoids. Current Stem Cell Reports, 2(2), 128–139. 10.1007/s40778-016-0040-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi A, Watanabe M, Ishii Y, Morita J, Hirokawa Y, Matsuzaki T, et al. (2007). Three-dimensional cellular spheroid formation provides human prostate tumor cells with tissue-like features. Anticancer Research, 27(1A), 45–53. [PubMed] [Google Scholar]
- Tao T, Wang Y, Chen W, Li Z, Su W, Guo Y, et al. (2019). Engineering human islet organoids from iPSCs using an organ-on-chip platform. Lab on a Chip, 19(6), 948–958. 10.1039/c8lc01298a. [DOI] [PubMed] [Google Scholar]
- Tiriac H, Belleau P, Engle DD, Plenker D, Deschenes A, Somerville TDD, et al. (2018). Organoid profiling identifies common responders to chemotherapy in pancreatic cancer. Cancer Discovery, 8(9), 1112–1129. 10.1158/2159-8290.CD-18-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsilimigras MC, Fodor A, & Jobin C (2017). Carcinogenesis and therapeutics: The microbiota perspective. Nature Microbiology, 2, 17008. 10.1038/nmicrobiol.2017.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tüysüz N, van Bloois L, van den Brink S, Begthel H, Verstegen MM, Cruz LJ, et al. (2017). Lipid-mediated Wnt protein stabilization enables serum-free culture of human organ stem cells. Nature Communications, 8, 14578. 10.1038/ncomms14578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uraoka T, Higashi R, Horii J, Harada K, Hori K, Okada H, et al. (2015). Prospective evaluation of endoscopic criteria characteristic of sessile serrated adenomas/polyps. Journal of Gastroenterology, 50(5), 555–563. 10.1007/s00535-014-0999-y. [DOI] [PubMed] [Google Scholar]
- Vaiopoulos AG, Athanasoula K, & Papavassiliou AG (2014). Epigenetic modifications in colorectal cancer: Molecular insights and therapeutic challenges. Biochimica et Biophysica Acta, 1842(7), 971–980. 10.1016/j.bbadis.2014.02.006. [DOI] [PubMed] [Google Scholar]
- van de Wetering M, Francies HE, Francis JM, Bounova G, Iorio F, Pronk A, et al. (2015). Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell, 161(4), 933–945. 10.1016/j.cell.2015.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Es JH, van Gijn ME, Riccio O, van den Born M, Vooijs M, Begthel H, et al. (2005). Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature, 435(7044), 959–963. 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
- VanDussen KL, Sonnek NM, & Stappenbeck TS (2019). L-WRN conditioned medium for gastrointestinal epithelial stem cell culture shows replicable batch-to-batch activity levels across multiple research teams. Stem Cell Research, 37, 101430. 10.1016/j.scr.2019.101430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verissimo CS, Overmeer RM, Ponsioen B, Drost J, Mertens S, Verlaan-Klink I, et al. (2016). Targeting mutant RAS in patient-derived colorectal cancer organoids by combinatorial drug screening. eLife, 5, e18489. 10.7554/eLife.18489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachogiannis G, Hedayat S, Vatsiou A, Jamin Y, Fernández-Mateos J, Khan K, et al. (2018). Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science, 359(6378), 920–926. 10.1126/science.aao2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Fu S, Zhao J, Zhao W, Shen Z, Wang D, et al. (2019). Transbronchoscopic patient biopsy-derived xenografts as a preclinical model to explore chemorefractory-associated pathways and biomarkers for small-cell lung cancer. Cancer Letters, 440–441, 180–188. 10.1016/j.canlet.2018.10.014. [DOI] [PubMed] [Google Scholar]
- Wang X, Phan DT, Sobrino A, George SC, Hughes CC, & Lee AP (2016). Engineering anastomosis between living capillary networks and endothelial cell-lined microfluidic channels. Lab on a Chip, 16(2), 282–290. 10.1039/c5lc01050k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang H, Deng P, Chen W, Guo Y, Tao T, et al. (2018). In situ differentiation and generation of functional liver organoids from human iPSCs in a 3D perfusable chip system. Lab on a Chip, 18(23), 3606–3616. 10.1039/c8lc00869h. [DOI] [PubMed] [Google Scholar]
- Wassenaar TM (2018). E. coli and colorectal cancer: A complex relationship that deserves a critical mindset. Critical Reviews in Microbiology, 44(5), 619–632. 10.1080/1040841X.2018.1481013. [DOI] [PubMed] [Google Scholar]
- Watanabe K, Ueno M, Kamiya D, Nishiyama A, Matsumura M, Wataya T, et al. (2007). A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nature Biotechnology, 25(6), 681–686. 10.1038/nbt1310. [DOI] [PubMed] [Google Scholar]
- Weeber F, van de Wetering M, Hoogstraat M, Dijkstra KK, Krijgsman O, Kuilman T, et al. (2015). Preserved genetic diversity in organoids cultured from biopsies of human colorectal cancer metastases. Proceedings of the National Academy of Sciences of the United States of America, 112(43), 13308–13311. 10.1073/pnas.1516689112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman MJ, Gleeson JP, Troisi EJ, Estrada HQ, Kerns SJ, Hinojosa CD, et al. (2018). Enhanced utilization of induced pluripotent stem cell-derived human intestinal organoids using microengineered chips. Cellular and Molecular Gastroenterology and Hepatology, 5(4), 669–677. e662. 10.1016/j.jcmgh.2017.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Rhee KJ, Albesiano E, Rabizadeh S, Wu X, Yen HR, et al. (2009). A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nature Medicine, 15(9), 1016–1022. 10.1038/nm.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Su X, Xu Y, Kong B, Sun W, & Mi S (2016). Bioprinting three-dimensional cell-laden tissue constructs with controllable degradation. Scientific Reports, 6, 24474. 10.1038/srep24474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Tran T, Dwabe S, Sarkissyan M, Kim J, Nava M, et al. (2017). A83–01 inhibits TGF-β-induced upregulation of Wnt3 and epithelial to mesenchymal transition in HER2-overexpressing breast cancer cells. Breast Cancer Research and Treatment, 163(3), 449–460. 10.1007/s10549-017-4211-y. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Xia X, Li F, He J, Aji R, & Gao D (2019). Organoid technology in cancer precision medicine. Cancer Letters, 457, 20–27. 10.1016/j.canlet.2019.04.039. [DOI] [PubMed] [Google Scholar]
- Xu Z, Li E, Guo Z, Yu R, Hao H, Xu Y, et al. (2016). Design and construction of a multi-organ microfluidic chip mimicking the in vivo microenvironment of lung cancer metastasis. ACS Applied Materials & Interfaces, 8(39), 25840–25847. 10.1021/acsami.6b08746. [DOI] [PubMed] [Google Scholar]
- Yan HHN, Siu HC, Ho SL, Yue SSK, Gao Y, Tsui WY, et al. (2020). Organoid cultures of early-onset colorectal cancers reveal distinct and rare genetic profiles. Gut, 69(12), 2165–2179. 10.1136/gutjnl-2019-320019. [DOI] [PubMed] [Google Scholar]
- Yang L, Yang S, Li X, Li B, Li Y, Zhang X, et al. (2019). Tumor organoids: From inception to future in cancer research. Cancer Letters, 454, 120–133. 10.1016/j.canlet.2019.04.005. [DOI] [PubMed] [Google Scholar]
- Yao Y, Xu X, Yang L, Zhu J, Wan J, Shen L, et al. (2020). Patient-derived organoids predict chemoradiation responses of locally advanced rectal cancer. Cell Stem Cell, 26(1), 17–26. e16. 10.1016/j.stem.2019.10.010. [DOI] [PubMed] [Google Scholar]
- Yin X, Mead BE, Safaee H, Langer R, Karp JM, & Levy O (2016). Engineering stem cell organoids. Cell Stem Cell, 18(1), 25–38. 10.1016/j.stem.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonemura S (2014). Differential sensitivity of epithelial cells to extracellular matrix in polarity establishment. PLoS One, 9(11), e112922. 10.1371/journal.pone.0112922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M, Ordonez L, & Clarke AR (2013). What are the best routes to effectively model human colorectal cancer? Molecular Oncology, 7(2), 178–189. 10.1016/j.molonc.2013.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, et al. (2017). Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell, 170(3), 548–563. e516. 10.1016/j.cell.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuki K, Cheng N, Nakano M, & Kuo CJ (2020). Organoid models of tumor immunology. Trends in Immunology, 41(8), 652–664. 10.1016/j.it.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Z, Yu X, Yang B, Zhang Y, Zhang L, Li X, et al. (2017). Colorectal cancer heterogeneity and targeted therapy: Clinical implications, challenges and solutions for treatment resistance. Seminars in Cell & Developmental Biology, 64, 107–115. 10.1016/j.semcdb.2016.08.033. [DOI] [PubMed] [Google Scholar]
- Zhang YS, Aleman J, Shin SR, & Kilic T (2017). Multisensor-integrated organs-on-chips platform for automated and continual in situ monitoring of organoid behaviors. Proceedings of the National Academy of Sciences of the United States of America, 114(12), E2293–e2302. 10.1073/pnas.1612906114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Lin L, Jin Y, Lin Y, Cao Y, & Zheng C (2016). Overexpression of WNT5B promotes COLO 205 cell migration and invasion through the JNK signaling pathway. Oncology Reports, 36(1), 23–30. 10.3892/or.2016.4772. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Arai T, Horiguchi Y, Ino K, Matsue T, & Shiku H (2013). Multiparameter analyses of three-dimensionally cultured tumor spheroids based on respiratory activity and comprehensive gene expression profiles. Analytical Biochemistry, 439(2), 187–193. 10.1016/j.ab.2013.04.020. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Wang L., Yu H., Yin F., Wang Y., Liu H., et al. (2017). In situ generation of human brain organoids on a micropillar array. Lab on a Chip, 17(17), 2941–2950. 10.1039/c7lc00682a. [DOI] [PubMed] [Google Scholar]
- Zumwalde NA, Haag JD, Sharma D, Mirrielees JA, Wilke LG, Gould MN, et al. (2016). Analysis of immune cells from human mammary ductal epithelial organoids reveals Vδ2+ T cells that efficiently target breast carcinoma cells in the presence of bisphosphonate. Cancer Prevention Research (Philadelphia, Pa.), 9(4), 305–316. 10.1158/1940-6207.capr-15-0370-t. [DOI] [PMC free article] [PubMed] [Google Scholar]