Abstract
The molecular functions of the majority of RNA-binding proteins (RBPs) remain unclear, highlighting a major bottleneck to a full understanding of gene expression regulation. Here, we develop a plasmid resource of 690 human RBPs that we subject to luciferase-based 3′-untranslated-region tethered function assays to pinpoint RBPs that regulate RNA stability or translation. Enhanced UV-cross-linking and immunoprecipitation of these RBPs identifies thousands of endogenous mRNA targets that respond to changes in RBP level, recapitulating effects observed in tethered function assays. Among these RBPs, the ubiquitin-associated protein 2-like (UBAP2L) protein interacts with RNA via its RGG domain and cross-links to mRNA and rRNA. Fusion of UBAP2L to RNA-targeting CRISPR–Cas9 demonstrates programmable translational enhancement. Polysome profiling indicates that UBAP2L promotes translation of target mRNAs, particularly global regulators of translation. Our tethering survey allows rapid assignment of the molecular activity of proteins, such as UBAP2L, to specific steps of mRNA metabolism.
RBPs control the post-transcriptional processing of mRNA transcripts, thereby influencing the cellular transcriptome and thus the overall state of the cell. Following 5′-end capping, splicing, 3′-end cleavage and polyadenylation, mammalian mRNAs are exported to the cytoplasm, where RBPs control their turnover, subcellular localization and the efficiency with which they are translated. Dysfunction of RBPs is linked to dozens of multisystemic diseases, cancer and neurological disorders1-4. Despite their association with disease, and although the importance of regulating gene expression at these cytoplasmic stages of the mRNA life cycle is well appreciated, only a small fraction of the over 2,000 RBPs identified thus far have known RNA targets and molecular roles5-10. Cross-linking and immunoprecipitation (CLIP)-based approaches have enabled detailed studies of individual RBPs through the transcriptome-wide identification of their binding sites11-15. Molecular functions can then be inferred from integrative analyses of bound transcripts and region-level binding preferences in conjunction with transcriptome-wide changes in splicing levels or ribosome association of mRNAs upon RBP depletion or overexpression13,16-21. While powerful, these strategies are not easily scaled to thousands of RBPs.
Here, we turn to the well-described tethered function assay (TFA)22-24 as a complementary and orthogonal approach that is also scalable and efficient as a means to assign preliminary molecular functions to RBPs without requiring previous knowledge of their functional protein domains or natural RNA targets22. By developing an open reading frame (ORF) library of 690 RBPs (771 isoforms) fused to the bacteriophage MS2 coat protein (MCP) domain and using two reporter systems that direct RBP recruitment to luciferase mRNA via MS2 RNA stem-loop structures in the 3′ untranslated region (3′ UTR), we performed a large-scale tethering screen to assign functions in RNA stability and/or translation to known and predicted RBPs. Of the 50 candidate RBPs that scored positively in both reporter systems, we subjected 14 to enhanced CLIP (eCLIP) analyses11 to globally identify their endogenous RNA targets. Perturbation of the levels of these candidates revealed regulation of their natural substrates largely consistent with our reporter findings.
We further focused on the candidate ubiquitin-associated protein 2-like (UBAP2L) protein. We found that UBAP2L cross-links to rRNA and that its transcriptome-wide binding sites are enriched in coding mRNA regions. UBAP2L fused to RNA-targeting Cas9 (RCas9)25-27 conveys programmable enhancement of mRNA translation of a target mRNA. UBAP2L depletion followed by polysome profiling and sequencing reveals that UBAP2L positively regulates translation of genes that are themselves regulators of protein synthesis. Thus, we demonstrate that UBAP2L is a ribosome-associated RBP required for maintenance of global protein synthesis, exemplifying how rapid assignment of molecular function to annotated but poorly characterized or recently predicted RBPs enables their prioritization for detailed transcriptomic and mechanistic studies.
Results
Generation of RBP open reading frames fused to MS2 coat protein and tethered function assays.
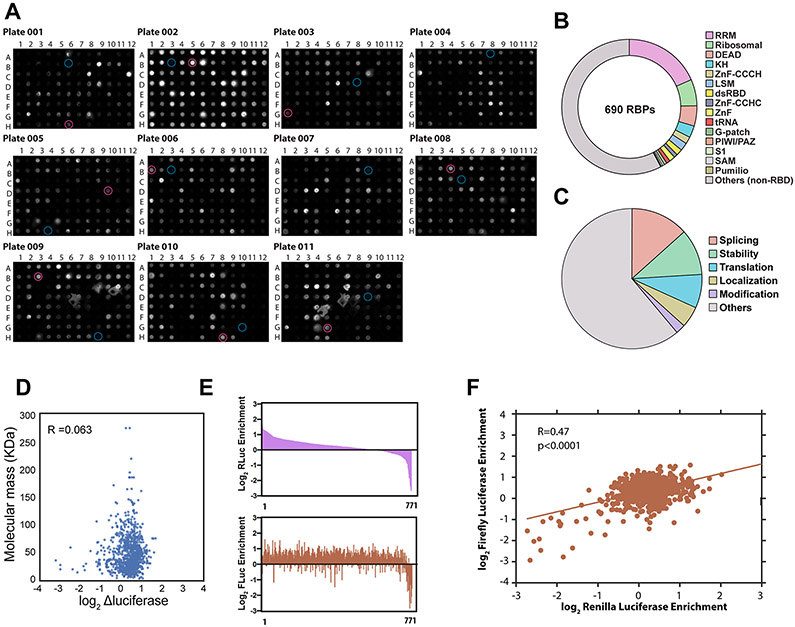
We identified putative RBPs using in-house bioinformatics tools to extract genes annotated to encode RNA-binding domains as predicted by PFAM28 and PRINTS29 and extended this set with mRNA-bound putative RBPs identified by UV-cross-linking and oligo(dT) capture followed by mass spectrometry2,3. We subcloned 881 RBP ORFs into a construct directing expression of the RBP with a V5 epitope tag and an MCP domain at the C terminus and ectopically expressed MCP–V5-tagged RBP ORFs in HeLa cells to assess the presence of the V5 epitope by western dot blot analysis (Extended Data Fig. 1a). We obtained a library of 690 unique RBPs from 771 expressible, full-length ORFs (Fig. 1a and Supplementary Table 1). Overall, ~40% of these RBPs contain known canonical RNA-binding motifs, while the remainder may associate with RNA through other interaction domains or binding modes (Extended Data Fig. 1b), similarly to previous reports2,3. Highlighting the need for assessing the roles of RBPs in RNA metabolism, Gene Ontology (GO) analysis showed that ~60% of these RBPs lack known RNA-related functions (Extended Data Fig. 1c). Thus, we have assembled a resource of representative ‘tethered’ and ‘untethered’ RBP expression libraries representing a comprehensive set of predicted and/or experimentally identified RBPs5-10.
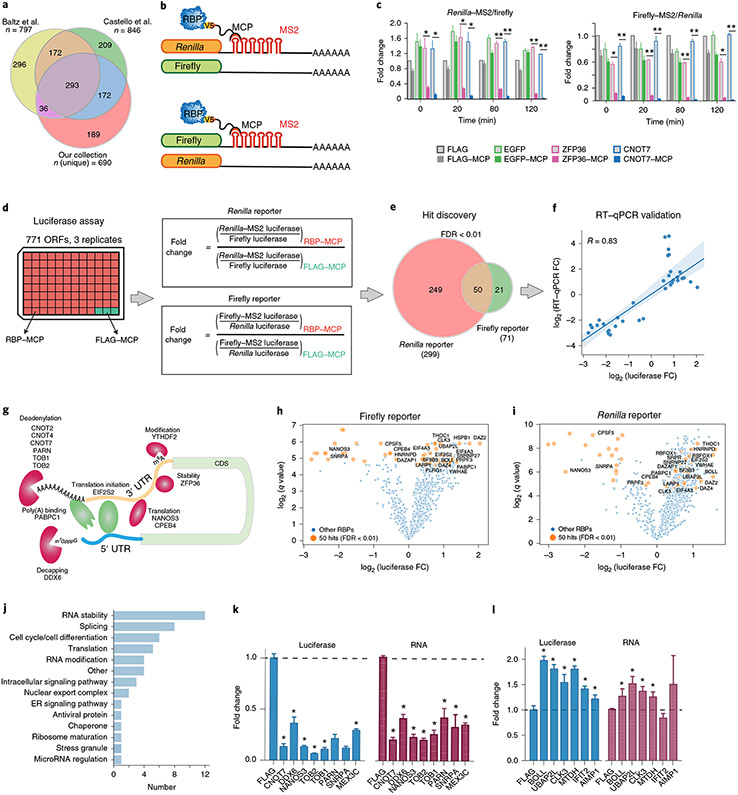
Fig. 1 ∣. A large-scale tethered function screen identifies RBPs regulating stability and translation.
a, Our collection of 771 ORFs for 690 unique RBPs and their overlap with those identified by Baltz et al.2 and Castello et al.3. b, Schematic of luciferase reporters with and without 3′-UTR MS2 stem–loops. c, Bar plots showing reporter activities for the indicated coexpressed known negative regulators of RNA stability (CNOT7, ZFP36) or negative controls (FLAG peptide, EGFP), without and with MCP fusion (light and dark shading, respectively), expressed as the ratios of the median luciferase activities with the indicated RBPs relative to those with FLAG controls at timepoint 0 (mean ± s.d., n = 3 replicate transfections; *P < 0.005, **P < 0.0005, two-tailed Student’s t-test). d, Experimental and analysis workflow. The effects of recruitment of 771 expressed MCP-tagged RBP ORFs were analyzed in both reporter contexts. e, Hit discovery. RBPs with effects at estimated FDR < 0.01 in both reporter assays were considered candidate regulators (n = 3 replicate transfections). f, RT–qPCR validation of reporter levels for 35 candidate RBP regulators. Means (n = 3 independent measurements) of log2-transformed fold changes (FC) of reporter mRNA levels, calculated analogously to those in d, are plotted against corresponding log2-transformed fold changes of reporter luciferase levels (line, least-squares linear regression fit; shaded area, 95% confidence interval; R, Pearson correlation coefficient). g, Examples of the 50 candidate RBP regulators known to affect RNA stability and/or translation. h,i, Volcano plots showing fold changes and q values for 50 RBP hits from the firefly (h) and Renilla (i) reporter assays. FDR < 0.01 in orange (n = 3 replicate transfections). j, GO classification of candidate RBP regulators by manual curation. k,l, Validation of the 9 negative (k) and 6 positive (l) candidate regulators by repeat luciferase (blue) and RT–qPCR (red) measurements. Values were calculated as in f (mean ± s.d., n = 4 replicate transfections; *P < 0.05 versus FLAG control, two-tailed Student’s t-test). Data for graphs in k and l are available as source data online.
Next, we constructed plasmid reporters that measure the effect of RBP recruitment to the 3′ UTR upon reporter expression: a construct encoding firefly luciferase followed by six MS2 hairpin sequences inserted into the 3′-UTR context of HBB (β-globin), a corresponding Renilla luciferase construct to evaluate potential reporter context dependencies and matched constructs lacking the MS2 sequences as negative controls (Fig. 1b). To validate our system, we introduced each reporter into HeLa cells along with constructs expressing MCP-fused and MCP-unfused versions of ZFP36, an RBP activator of AU-rich element-mediated RNA decay30, enhanced green fluorescent protein (EGFP) or FLAG peptide. As expected, ZFP36 but not EGFP or FLAG dramatically reduced luciferase levels in a manner that depended on the presence of the tether but not luciferase protein identity. This demonstrated that tethered ZFP36 can recruit functional CCR4–NOT deadenylase complexes to the reporter. Tethering of CNOT7 itself recapitulated this finding, indicating that productive recruitment is not limited to sequence-specific RBPs (such as ZFP36) but extends to effector RBPs (such as CNOT7) (Fig. 1c).
Large-scale tethered function screen reveals RBPs that affect reporter luciferase levels.
Next, we screened the 771 ORFs in triplicate using our two dual-luciferase reporter systems (Fig. 1d, left). We calculated the effect of RBP recruitment to the tethering reporter as the fold change in luciferase activity relative to the FLAG control, after normalization to the respective untethered reporters (Fig. 1d, right, and Supplementary Table 2). Supporting the validity of our screening approach, we confirmed that the effect was not correlated with RBP size (R = 0.063), indicating that steric hindrance is unlikely to account for these observations (Extended Data Fig. 1d). Although the magnitude of the effect on reporter transcript abundance generally depended on the reporter, fold changes in luciferase were significantly correlated (P < 0.0001) between the two reporters (Extended Data Fig. 1e,f).
We prioritized candidates from each reporter system by using multiple t-tests at a threshold of P < 0.05 and calculated false discovery rates (FDRs) for each comparison using the Benjamini, Krieger and Yekutieli procedure31. We identified 299 and 71 RBPs at estimated FDR < 0.01 in the Renilla and firefly reporters, respectively, indicating that reporter contexts do factor in the regulatory impact of tethered RBPs. Fifty RBPs were recovered from both reporter contexts (Fig. 1e). As an independent metric, we measured luciferase transcript levels by reverse-transcription quantitative PCR (RT–qPCR) for 35 of the 50 RBPs. In general, reporter translation levels by luciferase assay were positively correlated (R = 0.83) with reporter transcript levels by RT–qPCR assay (Fig. 1f and Supplementary Tables 3 and 4). Among the strongest candidate negative regulators were RBP components of deadenylation-dependent and deadenylation-independent exonuclease decay pathways, including ZFP36, as well as members of the CCR4–NOT deadenylase complex (CNOT2, CNOT4, CNOT7, TOB1 and TOB2), the 3′-to-5′ exonuclease PARN and the decapping activator DDX6, which is recruited to the 5′ cap via interaction with the CCR4–NOT complex32. We also confirmed that YTHDF2, an N6-methyladenosine binding protein that recruits target RNAs to degradation bodies33, exerts a negative effect on target mRNA levels. The results of our screen also confirmed several known negative regulators of translation, such as NANOS3 specific to germ cells34 and CPEB4, which binds polyadenylation elements in the 3′ UTR and negatively regulates translation initiation by interacting with the translation initiation factor eIF3 (ref. 35). Interestingly, EIF2S2, with roles in promoting translation initiation, emerged as a positive regulator of translation when recruited to the 3′ UTR. We speculate that recruitment of this protein to the 3′ UTR brings it proximal to the mRNA cap and 5′ UTR, similarly to DDX6 and CPEB4 and consistent with the closed-loop model of translation36,37 (Fig. 1g). Nevertheless, while these proteins were previously proposed as translational regulators, we also saw changes in luciferase mRNA upon tethering, consistent with the intimate coupling between translation and mRNA stability38-40.
In summary, the screen revealed candidate regulators previously annotated to be linked to post-transcriptional gene regulation of metabolic processes, cell cycle and cell differentiation (DAZ family proteins BOLL, DAZ2 and DAZ4; DAZAP and NANOS3; refs. 41-43), stress granule-associated proteins (UBAP2L44-46), factors involved in translation (EIF2S2, LARP1, PABPC1 and CPEB4; refs. 35,47-49), endoplasmic reticulum proteins (SRPR50) and heat shock proteins (HSPB1). Our screen also identified eight annotated splicing factors (CLK3, CPSF5, PLRG1, PRPF3, RBFOX1, SF3B3, SNRNP27 and SNRPA51-53) and two nuclear export complex proteins (HNRNPD and THOC1; refs. 54,55) as candidates (Fig. 1h-j and Supplementary Table 5). We next reconfirmed 14 RBPs with significant effects (8 that enhanced and 6 that repressed luciferase mRNA and protein levels) for further investigation (Fig. 1k,l).
eCLIP identifies endogenous RNA targets of candidate regulators.
We focused on RBPs for which roles in RNA stability and/or translation are not known (UBAP2L, SNRPA, CLK3, MTDH, AIMP1 and IFIT2) and RBPs with known roles but where transcriptome-wide binding sites and preferences have not been described (CNOT7, DDX6, NANOS3, TOB1, TOB2, PARN, MEX3C and BOLL) (Fig. 2a). We investigated endogenous mRNA targets and transcriptome-wide binding sites using eCLIP followed by sequencing11. Briefly, HEK293T cells were UV-cross-linked and lysed, RNAs were fragmented and protein–RNA complexes were immunoprecipitated using validated antibodies56 (Extended Data Fig. 2a and Supplementary Table 6). We also transiently transfected cells with plasmids expressing V5-tagged fusions of those candidate RBPs that are not expressed in HEK293T cells or do not have RBP-specific antibodies (Extended Data Fig. 2b). In total, we generated duplicate eCLIP datasets for the 14 candidate proteins, with each replicate consisting of an RBP eCLIP (IP) library and a paired size-matched input (SMInput)11 library. Libraries were sequenced to >4 million reads (average, 27 million), of which >1 million (average, 7 million) mapped uniquely to the genome (Supplementary Table 7). All libraries passed our routine quality control metrics19 with average Pearson correlation coefficient >0.5 between replicates (Extended Data Fig. 2c). In summary, eCLIP libraries were successfully generated and yielded reproducible RBP-specific global binding profiles.
Fig. 2 ∣. eCLIP identifies endogenous RNA targets of candidate stabilizers and destabilizers.
a, Domain structures of 14 candidate RBPs with RNA-destabilizing (left) and RNA-stabilizing (right) effects in the tethering assay, with lengths of their polypeptide chains. b–d, Histograms showing region-based fold enrichment of read densities normalized to paired SMInput controls for BOLL and IFIT2, which show read density enrichment in 3′ UTRs (b); DDX6, TOB1 and PARN, which show read density enrichment in 5′ UTRs (c); and UBAP2L, which shows read density enrichment in CDS and 5′ UTRs (d). e, Bar graphs showing eCLIP binding cluster distribution across transcript regions for the eight destabilizers and six stabilizers. Peak assignment was performed using stringent enrichment criteria (≥4-fold enrichment and P ≤ 10−2 versus SMInput, Fisher’s exact test for read numbers <5 or χ2 test for read numbers ≥5). The region distribution of the entire transcriptome annotated in GENCODE v19 is indicated at the top. f–h, Example genome browser track views of eCLIP read densities (in reads per million, RPM) and corresponding SMInput read densities for BOLL and IFIT2, which show peak enrichment in 3′ UTRs (f); DDX6 and PARN, which show peak enrichment in 5′ UTRs (g); and UBAP2L, which shows peak enrichment across exons (h).
Next, we determined transcript binding region specificities using two distinct metrics, namely, read density and binding cluster enrichment. Read density enrichment within 5′ and 3′ UTRs and coding sequence (CDS) regions of annotated protein-coding genes was computed by the fold enrichment in the IP experiments normalized to their paired SMInput experiments for target transcripts. To illustrate, BOLL, a germ cell-specific RBP with documented roles in mRNA stabilization and translation enhancement, displayed a strong preference for 3′-UTR association. Surprisingly, IFIT2, which is known to inhibit expression of viral mRNAs, robustly displayed a strong 3′-UTR preference in human mRNAs (Fig. 2b). The helicase DDX6 was enriched for binding within 5′ UTRs and 3′ UTRs, consistent with its role in the assembly of the decapping complex and the closed-loop model of translation36,37. Unexpectedly, TOB family member TOB1, which recruits the catalytic subunits of the CCR4–NOT deadenylase complex to target mRNAs57, and PARN, which degrades poly(A) tails, showed preferences for 5′ UTRs in addition to 3′ UTRs and CDS, suggesting unexpected roles for these proteins (Fig. 2c). UBAP2L showed strong enrichment across CDS exons and 5′ UTRs (Fig. 2d).
To determine binding sites at higher resolution, we identified clusters58 and defined reproducible binding sites as clusters that overlapped between the two replicates. Cluster enrichment was computed by calculating the ratio of read densities between the IP and SMInput experiments within a reproducible cluster, and significant clusters were defined as those with P ≤ 10−2 (Fisher’s exact test for read numbers <5; χ2 test for read numbers ≥5) and ≥4-fold enrichment over SMInput11. Significant clusters were enriched for specific sequence motifs (Extended Data Fig. 2d), some of which reflected expected preferences. For example, the SNRPA motif GGUAAG resembles the consensus 5′ splice site (GGURAG), and the helicase DDX6 motif GGGGGG is consistent with its binding preferences to G-quadruplex RNA59. Interestingly, the binding motif identified for BOLL (AGUGUA) partially overlaps with the Pumilio response element UGUANAUA, consistent with complex formation of DAZ family proteins with PUM2 on RNA targets60,61. Binding cluster enrichment analyses of these RBPs that focused on the binding sites with high signal (Fig. 2e) generally agreed with read density enrichment analyses. Metagene plots further reinforced that BOLL and IFIT2 (Fig. 2f) as well as MEX3C, AIMP1 and CNOT7 were 3′-UTR-preferring binders (Extended Data Fig. 2e). DDX6 (Fig. 2g), TOB1, NANOS3 and TOB2 appeared to have 5′- and 3′-UTR, but lower CDS, preferences (Extended Data Fig. 2f). PARN (Fig. 2g) and CLK3 were enriched at the 5′ UTR, peaking near the start codon (Extended Data Fig. 2g). UBAP2L and MTDH binding clusters were predominantly in CDS (Extended Data Fig. 2h). Indeed, UBAP2L clusters were dispersed across exons (Fig. 2h). Overall, our analyses reveal not only previously unrecognized binding maps and preferences for RBPs known to affect mRNA stability and translation but also new RNA interactomes of candidate RBPs.
Integration of eCLIP and RNA-sequencing data defines regulatory classes of RBPs and transcripts.
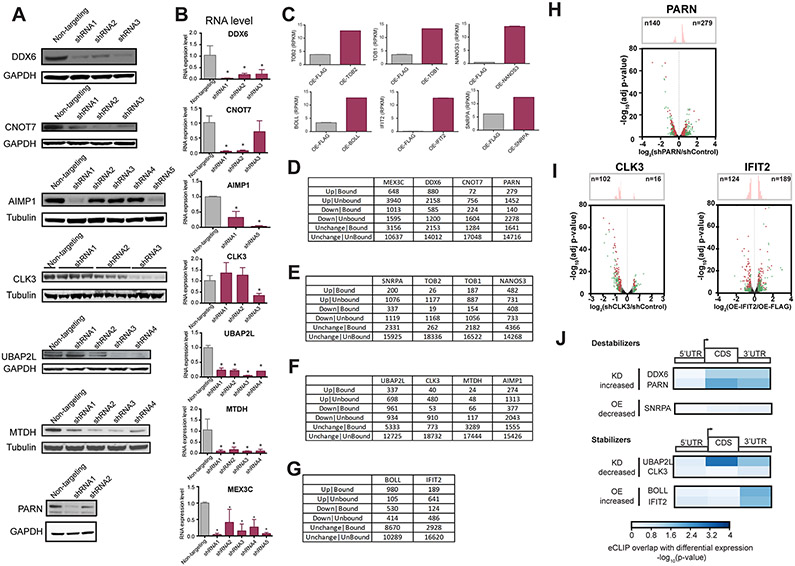
To gain insight into how our candidate RBPs affect transcriptome-wide mRNA levels, we depleted or exogenously expressed them in HEK293T cells and performed RNA-sequencing (RNA-seq) analysis. Specifically, we either depleted RBPs by lentiviral transduction of short hairpin RNAs (shRNAs) (Extended Data Fig. 3a,b and Supplementary Table 4) or ectopically expressed ORFs of those candidate RBPs that are not natively expressed in HEK293T cells or do not have RBP-specific shRNAs (Extended Data Fig. 3c). For each RBP, we either performed two independent transductions of two different targeting shRNAs and two nontargeting shRNAs or performed two independent transfections with a plasmid directing expression of the RBP as a V5-tagged fusion, with the FLAG construct as a control. We selected poly(A)+ RNA, prepared sequencing libraries and sequenced them to a depth of >32 million (>26 million uniquely mapped) reads (Supplementary Table 8).
To assess the effect of a candidate RBP on transcript levels, we measured the number of significantly up- or downregulated genes upon knockdown or overexpression (Extended Data Fig. 3d-g and Supplementary Table 9). In general, our manipulations of RBP levels resulted in a largely unperturbed population of transcripts, typically 80% at the threshold of statistical significance (≥1.23-fold, FDR-corrected P ≤ 0.05 versus nontargeting shRNA or FLAG control). This indicates that our candidate RBPs affect specific sets of target transcripts instead of having effects on global transcript stability. When we considered only those transcripts that were bound by the respective RBP, as measured by eCLIP (≥1 significantly enriched cluster per transcript), we observed higher numbers of targets that changed in the direction anticipated by the tethering assays than in the opposite direction for candidate destabilizers (negative regulators) DDX6 and SNRPA (Fig. 3a), as well as PARN (Extended Data Fig. 3h), and candidate stabilizers (positive regulators) UBAP2L and BOLL (Fig. 3b), as well as CLK3 and IFIT2 (Extended Data Fig. 3i). In other words, knockdown of specific predicted destabilizers led to more upregulated genes, whereas overexpression of destabilizers led to more downregulated genes, and reciprocal effects were observed in the alterations of stabilizing RBPs.
Fig. 3 ∣. Integration of eCLIP and RNA-seq data defines regulatory classes of RBPs and transcripts.
a,b, Volcano plots showing the distribution of fold changes in transcript levels upon modulation of destabilizers (a) and stabilizers (b), with distribution histograms shown at the top. a, Depletion of DDX6 (left) and overexpression of SNRPA (right). b, Depletion of UBAP2L (left) and overexpression of BOLL (right). Transcripts with log2 (fold change) ≥ 1.23 and FDR-corrected P ≤ 0.05 from two replicates are in color, with red and green denoting, respectively, transcripts with or without at least one significant RBP binding peak (≥4-fold enrichment and P ≤ 10−2 versus SMInput in two eCLIP replicates, Fisher’s exact test for read numbers <5 or χ2test for read numbers ≥5). c,d, Bar plots showing the percentage overlap between genes significantly up- or downregulated (log2 (fold change) ≥ 1.23 and FDR-corrected P ≤ 0.05 in two replicates) and significantly bound (≥4-fold enrichment and P ≤ 10−2 versus SMInput in two eCLIP replicates) upon knockdown (KD) or overexpression (OE) of candidate destabilizers (c) and stabilizers (d) (*P < 0.01, ***P < 10−3, ****P < 10−4, hypergeometric test versus nontargeting shRNA or FLAG overexpression, as appropriate). Data for graphs in c and d are available in Extended Data Fig. 3d-f. e–h, Cumulative distribution plots of transcript log2-transformed fold changes of overexpression versus vector control or shRNA-mediated knockdown versus nontargeting control, as indicated, for the destabilizers DDX6 (e) and SNRPA (f) and the stabilizers UBAP2L (g) and BOLL (h). Distributions are shown for transcripts with the indicated significant read enrichments over SMInput (P ≤ 10−2) from eCLIP analysis (4- to 8-fold, green; 8- to 16-fold, red) or transcripts that are not significantly bound (‘Not bound’, P > 10−2 or <4-fold enrichment, gray). n is the number of genes; P values are given versus ‘Not bound’, two-tailed Mann–Whitney U test from two replicates. i,j, Genome browser views from shRNA-mediated knockdowns showing RNA-seq reads (shRNA knockdown, maroon; nontargeting shRNA control, pink) and eCLIP reads (IP, blue; SMInput, gray) for PARN at the RPS21 locus (i) and CLK3 at the NELFCD locus (j). The y axes denote read density in RPM.
We also confirmed that the fraction of bound targets in the genes changing in the anticipated direction was statistically significantly enriched relative to unbound targets (Fig. 3c,d). In fact, we observed significant correlation between different eCLIP cluster fold enrichments of IP over SMInput and change in transcript levels for both candidate destabilizers (for example, DDX6 and SNRPA; Fig. 3e,f) and candidate stabilizers (for example, UBAP2L and BOLL; Fig. 3g,h). This indicates that our candidate RBPs directly engage hundreds of previously unknown target mRNAs to affect transcript levels in the predicted direction. For example, knockdown of the destabilizer PARN increased transcript levels of RPS21 mRNA, a PARN-bound transcript (Fig. 3i). Conversely, depletion of the stabilizer CLK3 reduced the abundance of its target NELFCD mRNA (Fig. 3j). Interestingly, UBAP2L binding within CDS emerged as the RBP-bound genic region most correlated with transcript levels (Extended Data Fig. 3j). In agreement with our tethering results, we conclude that the majority of our candidate RBPs affect the mRNA levels of their endogenous RNA targets.
UBAP2L increases mRNA polysome association and promotes translation.
Among our candidates, UBAP2L had the highest CDS read density enrichment, suggesting a role in translation. However, such a function for UBAP2L has not been described. We measured global protein synthesis in cells lacking UBAP2L by incorporation of puromycin (a structural analog of aminoacyl-tRNA) to label newly synthesized proteins (SUnSET assay62). HEK293T cells with UBAP2L deletion by CRISPR–Cas9-mediated genome editing (Fig. 4a) showed a ~40% reduction in protein synthesis (Fig. 4b and Extended Data Fig. 4a), indicating that UBAP2L promotes global translation. We next performed sucrose gradient centrifugation of HEK293T lysates to examine the ribosome association of UBAP2L. We detected enrichment of eukaryotic elongation factor 2 (EEF2) in the 40S, 60S and monosome but not the polysome fractions, as expected63. UBAP2L cofractionated with monosomes and polysomes, suggesting a role for UBAP2L in translation (Fig. 4c). To rule out the possibility that this observation is due to the presence of UBAP2L in non-ribosomal complexes of similar buoyant density, we treated cells with puromycin to release polysomes from transcripts. Puromycin treatment led to accumulation of 80S monosomes, as expected, and levels of UBAP2L in polysome fractions were strongly reduced (Extended Data Fig. 4b). We also treated cell lysates with EDTA to disassemble 80S monosomes into 40S and 60S ribosomal subunits and found that, similarly, UBAP2L was depleted from monosome fractions (Extended Data Fig. 4c). These results strongly suggest that UBAP2L directly interacts with translating ribosomes.
Fig. 4 ∣. UBAP2L is associated with translating ribosomes and promotes translation.
a,b, SUnSET translation assay. a, UBAP2L western blots from control (WT) and UBAP2L-knockout (KO) HEK293T lines. b, Left, representative puromycin western blot from puromycin-treated control and knockout cells. GAPDH, loading control. Right, densitometric quantification (mean ± s.d., n = 3 independent experiments; *P < 0.05 versus WT, two-tailed Student’s t-test). c, Polysome profile of UBAP2L. Top, absorbance plot of a HEK293T cell lysate fractionated through a sucrose gradient. Bottom, western blots of UBAP2L and EEF2 from corresponding fractions. d,e, Global polysome transcript association analysis. d, Scatterplots of log2-transformed transcript RPKM ratios (RPKM ≥ 1; averages from two replicates) of input transcript levels (x axis) and polysome transcript levels (y axis) between UBAP2L-knockout and control samples. e, Cumulative distribution plots of log2-transformed transcript levels (RPKM ≥ 1) in pooled polysome fractions of two UBAP2L-knockout and control HEK293T lines (n = 9,692 genes from two replicates; P values derived from two-sample Kolmogorov–Smirnov test versus respective input lysates). f, Bar graph showing log2-transformed ratios of input-normalized polysome transcript levels (n = 9,692 genes; RPKM ≥ 1 in all three samples; averaged for the two knockout lines) between the two UBAP2L-knockout lines and control lines. g, GO term analysis for UBAP2L exon target transcripts related to mRNA translation (n = 4,789 genes; Fisher’s exact test at FDR-corrected P < 0.01). h, Heat map showing the log2-transformed polysome association ratio between a UBAP2L-knockout line (KO2) and control (WT) for the indicated translation regulators. i, Left, representative western blots for the indicated proteins from UBAP2L-knockout cells. GAPDH, loading control. Right, densitometric quantification (mean ± s.d., n = 3 independent experiments; *P < 0.05 versus WT, two-tailed Student’s t-test). j–l, Quantitative flow cytometry reporter assay for mRNA translation using RCas9-fused UBAP2L. j, Transgene expression constructs. k, Bar graph showing mean YFP levels in RCas9–UBAP2L-expressing cells, normalized to cells expressing RCas9 only, for each gRNA (mean ± s.d., n = 2,000 RCas9–UBAP2L-expressing and n = 2,000 RCas9-expressing cells per gRNA; *P < 0.05, **P < 0.0001; NS, not significant at P = 0.7 versus nontargeting gRNA; two-tailed Student’s t-test). l, Bar graph showing YFP/RFP mRNA ratios with gRNA-2 in cells expressing RCas9–UBAP2L (mean ± s.d., n = 3 replicates; NS, not significant at P > 0.5 versus RCas9 only; two-tailed Student’s t-test). Uncropped images for a–c and i and data for graphs in b, i, k and l are available as source data online.
To identify transcripts translationally regulated by UBAP2L, we performed polysome profiling in cell lysates from two UBAP2L-knockout clonal isolates and from two control samples (Extended Data Fig. 4d). From two fractionations per sample, we isolated poly(A)+ mRNA from a portion of the input lysates and from pooled polysome fractions and prepared and sequenced RNA-seq libraries (Supplementary Table 10). We considered all transcripts with reads per kilobase per million mapped reads (RPKM) ≥ 1 in the input samples (Extended Data Fig. 4e). We observed that UBAP2L knockout resulted in a larger number of transcripts with changes in pooled polysome fractions compared to changes in input RNA abundance. Most of the transcripts (82%) were downregulated in pooled polysome fractions, but a similar fraction of transcripts was upregulated (55%) or downregulated (45%) in input RNA, indicating that the changes in pooled polysome fractions were independent of RNA abundance (Fig. 4d). In aggregate, these results suggest that UBAP2L predominantly acts at the translational level. As a measure of ribosome association, we computed the ratio of transcript RPKM in polysome pools to input for all transcripts. We found a significant decrease (P < 10−300; Mann–Whitney U test, two tailed) in mean transcript polysome enrichment in both UBAP2L-knockout lines compared to the controls (Fig. 4e). Replicate analyses showed excellent correlation (Extended Data Fig. 4f). When we isolated those transcripts that changed in the same direction in both knockout lines, we found that, overall, nearly tenfold more transcripts were reduced in translation (90.6%; n = 8,784) than enhanced (9.4%; n = 908) (Fig. 4f). Even more striking, 97% of the 4,789 UBAP2L exon target transcripts identified by eCLIP showed significant downregulation in polysome association upon UBAP2L knockout (Extended Data Fig. 4g). We confirmed these results for a subset of target transcripts by RT–qPCR assay from polysome pools (Extended Data Fig. 4h).
To investigate how depletion of UBAP2L affects global translation, we evaluated the gene function attributes of UBAP2L direct targets. We observed a significant enrichment in protein translation and ribosome biogenesis terms by GO analysis (Fig. 4g). We also found that UBAP2L depletion decreased polysome association on mRNAs encoding translation initiation factors, elongation factors and poly(A) binding proteins (Fig. 4h). Western blot analysis confirmed decreased protein levels of EIF4G1, EIF3B, DDX54 and EEF2 (Fig. 4i and Extended Data Fig. 4i). Taken together, these results suggest that UBAP2L enhances translation by directly binding mRNA substrates and by increasing translation of genes involved in global protein synthesis.
Programmable RNA-targeting CRISPR-mediated recruitment of UBAP2L promotes translation.
To assess the dependence of UBAP2L-mediated translational regulation on direct binding to its target mRNA, we employed a flow cytometry-based reporter assay using UBAP2L fused to RCas9 (refs. 25,26) (Fig. 4j). As a control, we performed our assay with RCas9-fused eukaryotic translation initiation factor 4E-binding protein 1 (EIF4EBP1, or 4EBP1), an inhibitor of translation initiation (Extended Data Fig. 4j). HEK293T cell lines expressing a fusion of RCas9 and UBAP2L, a fusion of RCas9 and 4EBP1, or Cas9 only were derived via transposase-mediated piggyBac genomic integration of plasmid constructs. We transfected a second, tripartite construct expressing a reporter that stably expresses red fluorescent protein (RFP) transcripts not regulated by RCas9, a guide RNA (gRNA) and a tetracycline-inducible yellow fluorescent protein (YFP) transcript harboring the gRNA target sequences. We designed seven different gRNAs targeting locations across the YFP transcript (5′ UTR, CDS and 3′ UTR) and a nontargeting gRNA. We then measured post-transcriptional regulation as changes in the normalized YFP/RFP fluorescence ratio between cells expressing Cas9 fusions and Cas9 only by flow cytometry. Because of the random nature of piggyBac-mediated integration in terms of construct integration sites and numbers, regulation for various RCas9 construct levels (cyan fluorescent protein, CFP) and reporter construct levels (RFP) can be quantified across thousands of data points (cells). With this highly sensitive and quantitative assay, we observed that the effect of UBAP2L on YFP reporter expression was dependent on UBAP2L being directed to targeting sites within the 3′ UTR and CDS (Fig. 4k). In contrast, significant 4EBP1-mediated reporter repression was observed only when 4EBP1 was targeted to the 5′ UTR, as expected (Extended Data Fig. 4k). Normalized YFP mRNA levels were not significantly different between RCas9–UBAP2L-expressing and RCas9-expressing cells transfected with gRNA-2 (which elicited the strongest increase with RCas9–UBAP2L), indicating that UBAP2L’s positive effect on reporter expression was not due to upregulation of reporter mRNA (Fig. 4l). Our UBAP2L–RCas9 results indicate a programmable means to enhance translation and further corroborate our observations from eCLIP and tethering.
UBAP2L binds to RNA via the RGG domain and cross-links to the expansion segments of the ribosome.
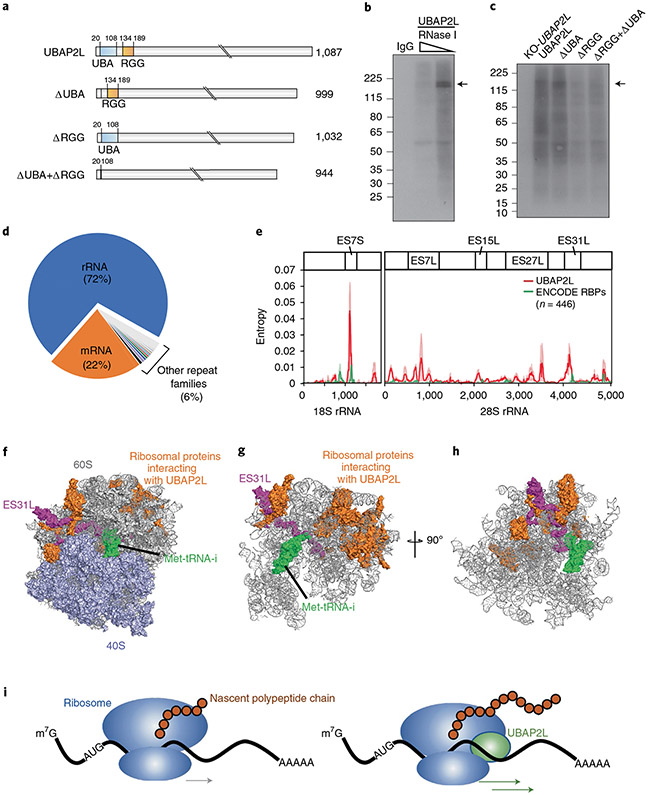
To gain deeper insight into the mechanisms by which UBAP2L enhances mRNA translation, we determined which protein domains mediate UBAP2L’s interaction with RNA. UBAP2L is predicted to contain a ubiquitin-associated (UBA) domain and an RGG domain, a common RNA-binding domain. Using inducible lentiviral vectors, we expressed UBAP2L or truncated versions lacking the UBA domain, the RGG domain or both (Fig. 5a) in UBAP2L-knockout HEK293T cells. We then performed UV-cross-linking, IP, RNA fragmentation and radiolabeling to visualize RNA bound to UBAP2L (Fig. 5b). Deletion of the RGG domain resulted in dramatically reduced recovery of RNA, indicating that the interaction between UBAP2L and RNA is mainly mediated by the RGG domain (Fig. 5c).
Fig. 5 ∣. UBAP2L binds directly to the ribosome.
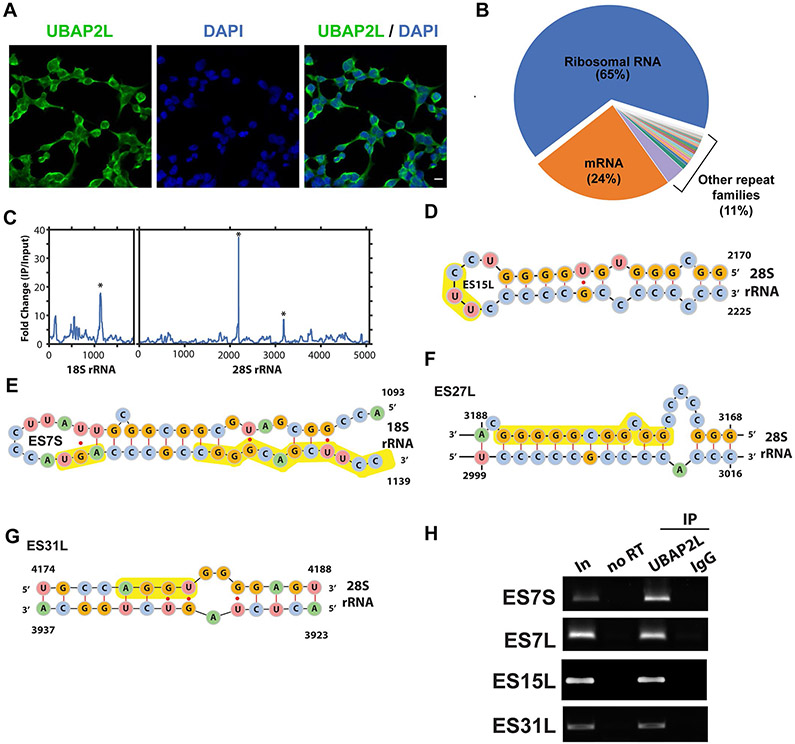
a, Domain structures of the UBAP2L constructs inducibly expressed in UBAP2L-knockout HEK293T cells. The UBA domain (blue) and RGG region (orange) are indicated. b,c, Autoradiographs of UBAP2L–RNA complexes immunoprecipitated from lysates of UV-cross-linked HEK293T cells treated with RNase I, radiolabeled and separated on an SDS–polyacrylamide gel. Arrows indicate the expected molecular weight of UBAP2L. b, Lysates from wild-type HEK293T cells treated with increasing RNase concentrations. c, Lysates from UBAP2L-knockout cells (KO-UBAP2L) expressing the indicated constructs. d, Pie chart showing fractions of UBAP2L eCLIP reads from HEK293T cells unambiguously mapping to mRNAs, rRNAs and other repeat families. e, Locations of UBAP2L binding sites on rRNAs. Line plots show the Kullback–Leibler divergence (relative entropy) for UBAP2L in HEK293T cells (red) and the mean of 446 other RBPs analyzed by the ENCODE consortium19 (green; https://www.encodeproject.org/, accession code ENCSR456FVU) on 18S and 28S rRNAs (mean of relative entropy; lightly shaded areas indicate 10–90% confidence intervals). f–h, Model of the interactions of UBAP2L on the human ribosome structure (PDB 6EK0)66. f, Surface view with 60S ribosomal subunits in gray and lavender, respectively, with Met-tRNA (green), ribosomal proteins previously identified as UBAP2L interactors by IP and mass spectrometry65 (orange) and expansion segment ES31L (magenta). g, View as in f with the non-highlighted proteins removed. h, View as in g rotated 90° around the z axis. i, UBAP2L (green) is proposed to enhance translation by interacting with the ribosome (blue) during nascent polypeptide synthesis (brown). Uncropped images for b and c are available as source data online.
Given that UBAP2L cofractionated with monosomes and polysomes in sucrose gradients, we reasoned that UBAP2L may interact directly with functional ribosomes. We first confirmed that UBAP2L is indeed localized to the cytoplasm. UBAP2L showed non-punctate staining throughout the cytoplasm (Extended Data Fig. 5a). We next examined two UBAP2L eCLIP datasets using a repeat-family-centric mapping strategy, which maps reads to consensus transcripts from repetitive and recurrent genomic loci, including rRNA genes. Remarkably, rRNA reads constituted the largest fraction, with 72% in replicate 1 and 65% in replicate 2, while mRNA reads totaled 22% and 24%, respectively (Fig. 5d and Extended Data Fig. 5b). Closer inspection showed that reads were most highly enriched over SMInput at expansion segments ES15L and ES27L of 28S rRNA and ES7S of 18S rRNA (Extended Data Fig. 5c-g), which are located at the solvent-exposed surface of ribosomes and are thought to engage with RBPs to modulate translation64. As a further confidence measure, we utilized an information-theoretic metric, relative entropy, which scales enrichment with the strength of evidence (that is, read depth) at each peak19. We confirmed that the peaks at ES15L, ES27L and ES7S (and an additional peak at ES31L) had high information content (Fig. 5e). In contrast, the mean of 446 other RBPs19 showed very limited information content, reflecting their limited rRNA specificity. As further confirmation, we performed RNA IP (RIP) followed by RT–PCR on ES7S, ES7L, ES15L and ES31L. Indeed, UBAP2L immunoprecipitated rRNA targets, indicating that UBAP2L interacts with ribosomes on expansion segments (Extended Data Fig. 5h). This is consistent with a recent UBAP2L IP–mass spectrometry study that recovered peptides from 15 ribosomal proteins65, further supporting a UBAP2L–ribosome interaction.
To assess the spatial arrangement of UBAP2L and the ribosome, we mapped these interactions onto the cryo-EM structure of the mammalian ribosome66. The top ribosomal proteins that co-immunoprecipitated with UBAP2L65 cluster in the 60S subunit (Fig. 5f). In addition, ES31L, highly enriched for UBAP2L binding, lies close to the region of the 60S subunit occupied by tRNA in the exit site during protein synthesis (Fig. 5g,h). Collectively, these data support a model in which UBAP2L’s function is associated with its interactions with the ribosome.
Discussion
Tethering of RBPs reveals new candidate regulators and rules.
To assign molecular functions to the growing number of predicted RBPs, we have developed a comprehensive resource enabling large-scale RBP TFA analyses. Using two pairs of 3′-UTR reporters interrogating nearly 700 RBPs, we discovered at least 50 RBPs with significant positive and negative effects on both reporters. Extrapolating to the over 2,000 RBPs encoded in the human genome5-10, we speculate that over 100 may have yet unrecognized roles in RNA metabolism by regulating mRNA stability and/or translation.
Unappreciated roles for several RBPs.
SNRPA (or U1A) is a dual-function protein functioning as a component of the spliceosomal U1 snRNP important for 5′ splice site recognition and, in a snRNP-free form, couples splicing to polyadenylation67. SNRPA interacts with stem–loop structures in 3′ UTRs called polyadenylation inhibitory elements to inhibit poly(A) polymerase68. In our assay, SNRPA emerged as an RNA destabilizer, its overexpression led to significant downregulation of its targets, and its binding was enriched, expectedly, in intronic regions (59% of significant peaks) and also in 3′ UTRs (7.3%), including the polyadenylation inhibitory element in its own transcript. Previously, only a handful of SNRPA 3′-UTR targets were known. We identified 344 SNRPA 3′-UTR targets, suggesting that this mode of regulation is widespread.
IFIT2 is an interferon-induced protein that blocks translation via sequestration of the eIF3 factor essential for cap-dependent translational initiation. Together with other IFIT family members, IFIT2 binds viral cap structures and sequesters viral proteins and RNAs; however, its role for modulating the host response is unclear69. We find that IFIT2 is highly enriched for binding the 3′ UTRs of cellular genes and regulates the stability of a small fraction of them (14%). Based on our tethering assay, we speculate that IFIT2 may also act at the translational level to regulate the host antiviral response by increasing translation of relevant endogenous RNAs.
AIMP1 is a multifunctional protein acting as an auxiliary factor of aminoacyl-tRNA synthetase complexes and as an intracellular and extracellular signaling molecule promoting inflammation and suppressing tumorigenesis. However, a role for this protein in mRNA stability and translation has not been described70. Surprisingly, we find that AIMP1 binds a large number of endogenous mRNAs.
UBAP2L as a global translation factor.
Recent reports have demonstrated a role for UBAP2L in the assembly and disassembly of cytoplasmic stress granules, a function that is mediated by its intrinsically disordered C-terminal domain44-46. These dynamic membraneless organelles coalesce in response to cellular stress and contain mRNAs that are translationally stalled at the initiation step71. In this study, we demonstrate a role for UABP2L in regulating protein homeostasis in the absence of conditions that induce stress granule formation (Extended Data Fig. 5a). Specifically, we find that UBAP2L acts by increasing the expression of target transcripts at the translational level but also stabilizes mRNAs, reflecting the coupling between translation efficiency and mRNA stability72. We present evidence that UBAP2L is an RBP that interacts with RNA via its RGG domain. We observe a limited number of sites on rRNA to which UBAP2L cross-links, supporting a specific set of interactions. Intriguingly, all interactions map to the eukaryote-specific expansion segments, flexible RNA structures protruding from the surface of the ribosome. A recent report showed that binding of methionine aminopeptidase to ES27L controls translational fidelity64. Association of UBAP2L with the ribosome is consistent with our eCLIP observations that UBAP2L binds to the coding regions of thousands of transcripts. Notably, mRNAs targeted by UBAP2L are themselves enriched for central regulators of translation and protein synthesis. Thus, our working model proposes that UBAP2L is dynamically recruited to translating ribosome–mRNA complexes to enhance translation on many targets (Fig. 5i), including translational regulators, to affect global protein synthesis. UBAP2L joins a growing list of stress granule-associated RBPs with documented additional cytosolic roles16,73.
Interestingly, UBAP2L’s translational enhancement activity is transferable to a target mRNA via RCas9 fused to UBAP2L, indicating that UBAP2L recruitment to an mRNA substrate is sufficient to increase its translation. Indeed, when UBAP2L is recruited to the CDS of the reporter via RCas9, UBAP2L increases reporter translation by ~35-50%, depending on the location within the coding region (or 3′ UTR). While moderate, this magnitude is congruent with the size of the decrease in protein synthesis (~40%) and in polysome association of UBAP2L-targeted transcripts upon UBAP2L knockdown (median, 31%). Incidentally, this orthogonal approach also further expands in vivo applications of RCas9 (refs. 25,26,74).
In conclusion, our results provide proof of principle for the utility of large-scale 3′-UTR tethering assays for identification of candidate RBPs involved in affecting mRNA and protein levels. While these assays are not without caveats (discussed further in the Supplementary Note), we anticipate that our RBP–MCP fusion library and screening methods will enable massively parallel assays aimed at elucidating the roles of RBPs in other RNA metabolic processes. In light of new experimental and computational approaches that have unearthed hundreds of candidate novel RBPs2,3,75 and noncanonical RNA-binding domains76, we speculate that high-throughput approaches such as ours will be of increasing utility to assign molecular functions.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41594-020-0477-6.
Methods
Generation of expression plasmids for MCP-tagged RBPs.
The majority of ORF clones were obtained in pENTR vectors from the CCSB human ORFeome collection77 (Dana-Farber Cancer Institute) or the DNASU Plasmid Repository (Arizona State University). Some ORFs were purchased in standard expression clones, amplified by PCR (Phusion polymerase, NEB) with oligonucleotide primers containing attB recombination sites and recombined into pDONR221 using BP clonase II (Thermo Fisher). ORFs were then recombined into a custom pEF DEST51 destination vector (Thermo Fisher) engineered to direct expression of the ORFs as fusion proteins with a V5 epitope tag and MCP appended C terminally and under the control of the EF1-alpha promoter to create ORF–V5–MS2BP constructs. The identity of all cDNA clones was verified by Sanger sequencing. Plasmid libraries are available on Addgene (155390–156159). Supplementary Table 1 lists all ORFs and relevant information.
Gene Ontology analysis.
Panther was used for GO analysis78,79. For library RBPs, the following GO terms related to RNA processing were used: splicing (‘RNA splicing’), stability (‘RNA stabilize’, ‘RNA stability’, ‘RNA stabilization’, ‘RNA decay’, ‘RNA turnover’, ‘RNA deadenylation’, ‘RNA cleavage’, ‘RNA cleaving’, ‘RNA degradation’), translation (‘translation’), localization (‘RNA localization’, ‘RNA transport’, ‘RNA localize’, ‘RNA export’) and modification (‘RNA methylation’, ‘RNA modification’). Significant GO terms were determined by Fisher’s exact test after FDR correction at P < 0.01 and sorted by fold enrichment. For GO analysis of UBAP2L-regulated genes, significantly enriched GO terms were determined by Fisher’s exact test after FDR correction at P < 0.01 and sorted by fold enrichment.
Cell lines.
HEK293T, HEK293XT and HeLa cells were purchased from the American Type Culture Collection and were not further authenticated. Cells were routinely tested for mycoplasma contamination with a MycoAlert mycoplasma test kit (Lonza) and were found negative for mycoplasma.
Dot blots.
HeLa cells were grown in antibiotic-free DMEM (Life Technologies) with 10% FBS. Plasmid (50 ng) was prepared for transfection using Lipofectamine 3000 reagent (Thermo Fisher). After a 15-min incubation, plasmid was transferred to 96-well tissue culture plates coated with poly(d-lysine) hydrobromide (Sigma-Aldrich). Cells (3.5 × 105) were subsequently plated in each well. After 48 h, cells were washed with PBS and lysed in lysis buffer (50 mM Tris-HCl, 100 mM NaCl, 1% NP-40, 0.1% SDS, 0.5% sodium deoxycholate; pH 7.4) with Protease Inhibitor Cocktail Set III (EMD Millipore). Lysate was transferred to nitrocellulose membrane prewetted with PBS using the Bio-Dot Microfiltration Apparatus (Bio-Rad), following the manufacturer’s instructions. Membranes were blocked in blocking buffer (TBS containing 5% (wt/vol) dry milk powder) for 30 min and then probed with rabbit anti-V5 tag antibody (Bethyl) in blocking buffer for 16 h at 4 °C. Membranes were washed three times with TBS and probed with secondary HRP-conjugated antibody in blocking buffer for 1 h at room temperature. Signal was detected by Pierce ECL substrate (Thermo Fisher) and collected using the Azure c600 imager (Azure Biosystems). Dot intensity was calculated using the ImageJ gel analyzer, with each column of the blot treated as a gel lane. Fold change was calculated for each sample over the highest-intensity negative-control well from the membrane containing the sample. Positive detection was called for fold changes >1.0.
Generation of luciferase reporter assay constructs.
Standard restriction enzyme cloning was used to generate reporter constructs directing expression of firefly (FLuc) or Renilla (RLuc) luciferase fused to protein destabilizing domains CP1 and PEST, with a 3′ UTR consisting of that from the HBB 3′ UTR, under the control of the tetracycline response element promoter (pTET2). Six MS2 hairpin structures80 were inserted into the 3′ UTR to generate FLuc–6MS and RLuc–6MS by standard restriction enzyme cloning. All constructs were sequence verified.
Luciferase reporter screen.
For time course analyses, Tet-Off Advanced HeLa cells (Clontech) were grown in DMEM (Thermo Fisher) with tetracycline-free FBS (10%; Clontech), penicillin-streptomycin (1×; Thermo Fisher) and G418 (100 μg ml−1; Corning). Before transfection, G418 was removed. A 6:1:1 mix of RBP–MCP, firefly–MS2 (or Renilla–MS2) and Renilla (or firefly) luciferase reporter (transfection control) constructs was diluted in 150 mM NaCl and mixed for transfection with polyethyleneimine (PEI; Polysciences) at a ratio of 1 μg DNA to 4 μg PEI. Cells were transfected at 50-60% cellular confluency, with a total of 125 ng and 250 ng DNA for 48-well plates and 24-well plates, respectively, and grown in the absence of G418. Reporter transcription was suppressed by the addition of tetracycline (1 μg ml−1; Sigma) 48 h after transfection. Cells were lysed after 20, 80 and 120 min, and luciferase activity was measured with the Dual-Luciferase Reporter Assay System (Promega), following the manufacturer’s directions, in a microplate reader. Values were expressed as the ratio of the mean luciferase activity of MS2-tagged over MS2-untagged reporters from three replicates. For the screen and validations, transfections were done as for the time course assay, and luciferase activities were measured 48 h after transfection. Supplementary Table 2 lists the results of the luciferase assays.
RT–qPCR.
Total RNA was isolated by lysing cells in TRIzol (Thermo Fisher) and purification with Direct-zol RNA kits (Zymo), following the manufacturers’ protocols. Reverse transcription of 0.5–1 μg total RNA was performed using SuperScript III with oligo(dT)12–18 primers (Thermo Fisher). cDNA was diluted 20-fold in water, and target transcripts were quantified with Power SYBR Green Master Mix (Thermo Fisher) using the primer pairs listed in Supplementary Table 4. Three biological replicate samples were analyzed, and RT–qPCR was carried out in three technical triplicates. Mean Ct values were calculated from each triplicate set. Biological replicates were averaged to generate mean fold changes, and values expressed as fold differences to control samples were calculated using the ΔΔCt method. Significance was assessed by a two-tailed Student’s t-test.
eCLIP library preparation and sequencing.
eCLIP was performed essentially as described11. Briefly, for each RBP, 3 × 107 HEK293T cells were UV-cross-linked (400 mJ cm−2, 254 nm) and lysed. Lysates were sonicated and treated with RNase I to fragment RNA. Two percent of each lysate sample was stored for preparation of a parallel SMInput library. The remaining lysates were immunoprecipitated using RBP-specific antibodies (Supplementary Table 6). Bound RNA fragments in the immunoprecipitates were dephosphorylated and 3′-end ligated to an RNA adaptor. Protein–RNA complexes from SMInputs and immunoprecipitates were run on an SDS–polyacrylamide gel and transferred to nitrocellulose membrane. Membrane regions comprising the exact RBP sizes to 75 kDa above were excised, and RNA was released from the complexes with proteinase K. SMInput samples were dephosphorylated and 3′-end ligated to an RNA adaptor. All RNA samples (immunoprecipitates and SMInputs) were reverse transcribed with AffinityScript (Agilent). cDNAs were 5′-end ligated to a DNA adaptor. cDNA yields were quantified by qPCR, and 100–500 fmol of library was generated with Q5 PCR mix (NEB).
Computational analysis of eCLIP sequencing data.
Reads were processed essentially as described11. Briefly, reads were adaptor trimmed and mapped to human-specific repetitive elements from RepBase (version 18.05) by STAR81. Repeat-mapping reads were removed, and remaining reads were mapped to human genome assembly hg19 with STAR. PCR duplicate reads were removed using the unique molecular identifier sequences in the 5′ adaptor, and remaining reads were retained as ‘usable reads’. Peaks were called on the usable reads by CLIPper58 and assigned to gene regions annotated in GENCODE v19 with the following order of descending priority: CDS, 5′ UTR, 3′ UTR, proximal intron and distal intron. Proximal intron regions are defined as extending up to 500 bp from an exon–intron junction. Each peak was normalized to the SMInput by calculating the fraction of the number of usable reads from the IP sample relative to the usable reads from the SMInput sample. Peaks were deemed significant at ≥4-fold enrichment and P ≤ 10−5 (χ2 test, or Fisher’s exact test if the observed or expected read number in eCLIP or SMInput was below 5). Reproducible clusters were defined as clusters that overlapped in both replicates. Target transcripts were defined as transcripts that contained at least one significant reproducible cluster. Sequencing and processing statistics are listed in Supplementary Table 7. Code is available on GitHub (https://github.com/YeoLab/eclip).
eCLIP region-based fold enrichment analyses.
Region-based fold enrichment was calculated as described previously82. Briefly, usable reads were counted across regions for all annotated transcripts in GENCODE v19 (comprehensive). Possible regions included CDS, 5′ UTR and 3′ UTR. For each gene, a read was first queried for overlap with CDS regions and then with 5′-UTR or 3′-UTR regions. Reads were then summed across all exons for the gene to obtain final region counts, and a pseudocount of 1 was added to classes for which no reads were observed. Read counts were normalized by the total number of usable reads (RPM normalization). Only regions with at least ten reads in one IP or SMInput sample, and where at least ten reads would be expected in the opposite dataset given the total number of usable reads, were considered. The fold enrichment was calculated as the ratio of normalized read counts in IP over SMInput.
Repeat-family-centric mapping.
Binding to rRNA was quantified using a family-aware repeat element mapping pipeline19. Briefly, reads were mapped to a database of 7,419 multicopy element transcripts, including the 5S, 5.8S, 18S and 28S rRNAs as well as tRNAs, retrotransposable elements and numerous other RNAs. Reads mapping to multiple element families were not considered for further analysis. To summarize relative enrichment between IP and input, relative information was defined as the Kullback–Leibler divergence (relative entropy): , where pi is the fraction of total reads in the IP sample that map to a queried repetitive element i and qi is the fraction of total reads in input for the same element. Code is available on GitHub (https://github.com/YeoLab/repetitive-element-mapping).
Metagene mapping analyses.
Metagene plots were created using the intersection of eCLIP peaks and a set of mRNA regions. To generate the list of each CDS, 5′ UTR and 3′ UTR, non-overlapping gene annotations from GENCODE v19 were used. First, low-expression transcripts (TPM < 1) were removed. Then, transcripts with the highest TPM were selected, resulting in a single transcript per gene in the CDS. For each 5′ UTR, CDS and 3′ UTR in a gene, the entire set of exons making up the region was concatenated and overlapped with eCLIP peaks, resulting in a vector of positions across the spliced transcript containing values of 1 if a peak was found at a given position or 0 otherwise. Plotted lines represent the number of total peaks found at each position divided by the total number of unique transcripts. The length of each region within the metagene was then scaled to 8%, 62% and 30%, corresponding to the average length of regions from the most highly expressed transcripts in ENCODE HepG2 RNA-seq control datasets19. The peak density was calculated as the percentage of peaks at a given position (https://github.com/YeoLab/rbp-maps).
De novo motif analysis.
HOMER was used to identify de novo motifs using the command ‘findMotifsGenome.pl <foreground> hg19 <output location> -rna -S 20 -len 6 -p 4 -bg <background>’. The foreground was a bed file of significant peaks; the background was randomly defined peaks within the same annotated region as the foreground peaks. Code is available on GitHub (https://github.com/YeoLab/clip_analysis_legacy).
eCLIP correlation analysis.
We utilized Pearson correlation statistics to measure the reproducibility between pairwise comparisons of replicate eCLIP experiments. The read density in peaks was normalized to the SMInput by calculating the fraction of the number of usable reads from the IP sample relative to the usable reads from the SMInput sample. The correlation was the comparison of fold enrichment in both datasets for all peaks.
Lentiviral shRNA knockdowns and transient plasmid transfections.
To generate lentiviral particles for RBP knockdown, we seeded 3.8 × 106 HEK293XT cells in 10-cm plates in antibiotic-free DMEM (Gibco) with 10% FBS. After 24 h, cells were transfected with sequence-verified shRNA plasmids (pLKO.1; Supplementary Table 9) and packaging plasmids (pMD2.G: Addgene 12259; psPAX2: Addgene 12260; both gifts from D. Trono, École polytechnique fédérale de Lausanne) using Lipofectamine 3000 (Thermo Fisher). Virus-containing medium was collected, replaced with 15 ml of medium with 20% FBS, and collected again a further 24 h later. Virus-containing media were pooled. For lentiviral transduction of HEK293T cells, cells were seeded in six-well plates at 8 × 105 cells per well and grown for 16 h in DMEM with 10% FBS. Cells were transduced with virus-containing medium diluted 1:1 in fresh medium. After 24 h, medium was replaced with fresh medium containing 2 μg μl−1 puromycin. After a further 72 h, cells were collected and analyzed for shRNA knockdown efficiency by western blot and RT–qPCR and for RNA-seq analysis.
For RBP overexpression, 3.8 × 106 HEK293XT cells were seeded in 10-cm plates in antibiotic-free DMEM (Gibco) with 10% FBS at 37 °C. After 24 h, cells were transfected with 24 μg RBP plasmids using Lipofectamine 3000 (Thermo Fisher). After a further 48 h, cells were collected and analyzed by western blot for successful overexpression, by RNA-seq and by eCLIP analysis as indicated.
RNA-seq library preparation and analysis.
RNA was extracted from cells with TRIzol (Invitrogen). Strand-specific RNA-seq libraries were prepared from 0.5–3 μg total RNA using the TruSeq Stranded mRNA Sample Preparation kit (Illumina). Libraries were sequenced on the Illumina HiSeq 4000 platform at a depth of at least 12 × 106 reads per sample in SE50 mode. RNA-seq reads were trimmed of adaptor sequences using cutadapt (v1.4.0) and mapped to repetitive elements (RepBase v18.04) using STAR (v2.4.0i). Reads that did not map to repetitive elements were then mapped to the human genome (hg19). GENCODE v19 gene annotations and featureCounts (v.1.5.0) were used to create read count matrices. Differential expression was calculated using DESeq2 version 1.10.1 (ref. 83), individually pairing each knockdown or overexpression experiment with its respective controls. Genes with RPKM < 1 were not used. Sequencing and processing statistics are described in Supplementary Table 8.
Generation of UBAP2L-knockout HEK293T cells.
Annealed pairs of gRNA sequences targeting three sites within the UBAP2L coding sequence (CCTCAAAGTCAGCATCATTA and TAGACTTGCACAGATGATTT, both targeting the second coding exon, and AAGCAATCACACATTCATCC, targeting the third coding exon) were inserted into plasmid pX330-U6-Chimeric_BB-CBh-hSpCas9 (Addgene 42230; a gift from F. Zhang (McGovern Institute) by restriction cloning. An equimolar mix of the three plasmids was transfected into HEK293T cells using Lipofectamine 3000 (Thermo Fisher). Cells were replated at clonal density 48 h after transfection. Clonal isolates were expanded and assayed for absence of UBAP2L protein expression by western blotting. Cell lines are available upon request.
Polysome profiling.
For lysate preparation, 4 × 107 HEK293T cells were prepared. Before collection, cells were treated with cycloheximide (CHX) at 100 μg ml−1 for 5 min at 37 °C. Culture medium was removed, and cells were washed two times with cold PBS containing 100 μg ml−1 CHX (PBS-CHX), resuspended in PBS-CHX by centrifugation at 200g at 4 °C for 5 min, collected in PBS-CHX and snap-frozen in liquid nitrogen. Cells were lysed by trituration through a 27-gauge needle in 400 μl polysome lysis buffer (20 mM Tris-HCl (pH 7.4), 150 mM NaCl, 5 mM MgCl2) with 1× protease inhibitor cocktail (EMD Millipore), 100 μg ml−1 CHX, 1 mM DTT, 25 U ml−1 DNase (TURBO DNase; Thermo Fisher) and 20 U ml−1 RNase inhibitor (RNaseOUT; Thermo Fisher) and incubation on ice for 30 min. Lysates were clarified by centrifugation at 17,500g at 4 °C for 5 min; 50 μl was reserved for inputs, and the remainder was used for fractionation.
For puromycin treatment to release polysomes, cells were treated with puromycin at 0.5 mM for 40 min and then with CHX at 100 μg ml−1 for 5 min. For disassembly of monosomes into 40S and 60S ribosomal subunits, the polysome lysis buffer was supplemented with 30 mM EDTA.
For fractionation, a 14-ml 10-50% (wt/vol) sucrose gradient was prepared in polysome buffer. Samples were loaded on the sucrose gradient and centrifuged in a swinging bucket rotor at 35,000g at 4 °C for 3 h. Fractions were collected from the top, and UV absorbance was monitored using a Gradient Station (BioComp) equipped with an Econo UV monitor (Bio-Rad). Fractions (500 μl each) were collected using an FC 203B (Gilson) fraction collector. Fractions containing polysomes were pooled. Total RNA from the inputs and polysome pools was extracted in TRIzol LS (Thermo Fisher) and purified with Direct-zol RNA kits (Zymo). RNA-seq libraries were generated and sequenced and reads were processed as described above.
For analysis of fractions by western blotting, 2 μg BSA was added to each fraction and protein was precipitated by addition of trichloroacetic acid to 20% (vol/vol). Protein was precipitated for 16 h at 4 °C and collected by centrifugation at 15,000g for 20 min at 4 °C. Protein pellets were washed twice with 1 ml ice-cold acetone, centrifuged at 15,000g for 15 min at 4 °C, dried at room temperature, neutralized, resuspended and denatured by incubation in 50 mM Tris-HCl, pH 7.4, at 65 °C for 30 min and 98 °C for 15 min. Western blotting was performed as described above.
Polysome association analysis.
The transcript RPKM of input and polysome fractions was calculated from the read count matrices. Only genes with RPKM ≥ 1 were considered. Polysome association was measured by calculating the RPKM ratio of transcript levels in polysomes over input. Polysome association ratios between depletion samples and their respective controls were calculated, log2 transformed, sorted and used to calculate cumulative probabilities. P values were calculated using a two-sided Kolmogorov–Smirnov test. Sequencing and processing statistics are listed in Supplementary Table 10.
Western blot.
Cells were washed with PBS and lysed in lysis buffer (50 mM Tris-HCl, 100 mM NaCl, 1% NP-40, 0.1% SDS, 0.5% sodium deoxycholate; pH 7.4) with Protease Inhibitor Cocktail Set III (EMD Millipore). Lysates were sonicated in a water bath sonicator (Diagenode) at 4 °C for 5 min with 30-s on/off pulses at the low setting. Protein extracts were denatured at 75 °C for 20 min and run at 150 V for 1.5 h on 4-12% NuPAGE Bis-Tris gels in NuPAGE MOPS running buffer (Thermo Fisher). Proteins were transferred to polyvinylidene difluoride membrane using NuPAGE transfer buffer (Thermo Fisher) with 10% methanol. Membranes were blocked in blocking buffer (TBS containing 5% (wt/vol) dry milk powder) for 30 min and probed with primary antibodies in blocking buffer for 16 h at 4 °C. Primary antibodies are described in Supplementary Table 6. Membranes were washed three times with TBS and probed with secondary HRP-conjugated antibodies in blocking buffer for 1 h at room temperature. Signal was detected by Pierce ECL substrate (Thermo Fisher) and exposure to film.
SUnSET assay.
De novo protein synthesis was measured by the SUnSET method62. Control HEK293T cells and those with homozygous deletion of UBAP2L were treated with puromycin (10 μg ml−1) for 10 min and then harvested on ice by lysing cells in eCLIP lysis buffer. Protein concentration was determined with the Pierce BCA protein assay kit (Thermo Scientific). Equal amounts of protein were analyzed by western blotting, as described above. Newly synthesized proteins were detected with an anti-puromycin antibody (1:20,000). Membranes were stripped and reblotted with an anti-GAPDH antibody (1:8,000) as a loading control. Immunoblots were quantified by densitometric analysis in ImageJ to obtain levels of protein synthesis for each sample.
RCas9–UBAP2L tethered translation assay.
HEK293T cells were grown in antibiotic-free DMEM (Life Technologies) with 10% FBS at 37 °C. Cells were transfected at 50-60% cellular confluency with a 4:1 mix of a piggyBac transposon vector coexpressing CFP, a piggyBac transposase vector and either RCas9–UBAP2L or RCas9–4EBP1 (or RCas9 only) using FuGENE HD transfection reagent (Promega). CFP-positive cells (integrants) were collected by fluorescence-activated cell sorting (FACS), expanded and transfected again with a piggyBac transposon vector constitutively expressing RFP, YFP under the control of a Tet-inducible promoter and a gRNA targeting the YFP reporter. RFP-positive cells were collected by FACS and expanded. Cells were induced with doxycycline (10 ng ml−1) for 36 h and quantified by FACS. For each cell, the YFP/RFP fluorescence ratio was quantified as a metric of RCas9–UBAP2L-mediated or RCas9–4EBP1-mediated post-transcriptional regulation of the target transcript, and CFP fluorescence was used to quantify expression levels of RCas9–UBAP2L, RCas9–4EBP1 or RCas9 only.
Immunocytochemistry.
HEK293T cells were fixed with 4% paraformaldehyde for 10 min at room temperature. Cells were permeabilized with PBS with 0.1% Triton X-100 (PBST) and blocked with blocking buffer (5% goat serum in PBST) for 1 h at room temperature. Cells were then incubated with anti-UBAP2L antibody (1:1,500) in blocking buffer for 16 h at 4 °C, washed with PBST three times for 5 min each at room temperature, and then incubated with secondary antibody (goat anti-rabbit secondary IgG (H+L) Superclonal Recombinant Secondary Antibody, Alexa Fluor 488 (Invitrogen)) in blocking buffer for 1 h. After staining, cells were washed again in PBST three times for 5 min each at room temperature. Staining of nuclei with 4′,6-diamidino-2-phenylindole (DAPI) was performed with mounting solution. Images were captured on a Zeiss LSM 710 confocal microscope.
Radiolabeling of RBP-bound RNA fragments.
UV-cross-linked HEK293T cells (20 × 106) were lysed in 550 μl lysis buffer (50 mM Tris-HCl (pH 7.4), 100 mM NaCl, 1% NP-40, 0.1% SDS, 0.5% sodium deoxycholate) with protease inhibitor (Roche). Lysates were sonicated for 5 min (Bioruptor; low setting, 30-s on/off) in an ice-cold water bath. After addition of 2.2 μl TURBO DNase (2 U μl−1; Thermo Fisher) and undiluted RNase I stock (100 U μl−1) or RNase I stock diluted 1:3 (high RNase) or 1:25 (low RNase) in low-stringency wash buffer (20 mM Tris-HCl (pH 7.4), 10 mM MgCl2, 0.2% Tween-20), samples were incubated at 37 °C for 5 min with shaking. RNase digestion was stopped with 11 μl murine RNase inhibitor (40 U μl−1; NEB), and insoluble material was removed by centrifugation (15 min, 15,000g, 4 °C). Protein–RNA complexes were immunoprecipitated for 16 h at 4 °C with anti-UBAP2L antibody or normal rabbit IgG (Thermo Fisher) precoupled to magnetic beads (Dynabeads M-280 Sheep Anti-Rabbit IgG; Thermo Fisher). A series of wash steps was employed to ensure stringency, as follows: two washes with low-stringency wash buffer (see above), two washes with high-stringency buffer (15 mM Tris-HCl (pH 7.4), 5 mM EDTA, 2.5 mM EGTA, 1% Triton-X 100, 1% sodium deoxycholate, 0.1% SDS, 120 mM NaCl, 25 mM KCl), two washes with high-salt wash buffer (50 mM Tris-HCl (pH 7.4), 1 M NaCl, 1 mM EDTA, 1% NP-40, 0.1% SDS, 0.5% sodium deoxycholate), two washes with low-stringency wash buffer and two washes with no-salt wash buffer (50 mM Tris-HCl (pH 7.4), 10 mM MgCl2, 0.5% NP-40). Protein–RNA complexes were radiolabeled on beads in 40-μl reactions with T4 polynucleotide kinase (NEB) and 2 μl [γ-32P]ATP (6,000 Ci mmol−1, 10 mCi ml−1) for 10 min at 37 °C. Beads were washed three times in low-salt wash buffer and resuspended in NuPAGE LDS sample buffer (Thermo Fisher) containing 0.1 M DTT. Protein–RNA complexes were denatured at 75 °C for 15 min, run on 4-12% NuPAGE Bis-Tris gels in NuPAGE MOPS running buffer (all Thermo Fisher) at 150 V for 1.5 h and wet-transferred to nitrocellulose membrane using NuPAGE transfer buffer (Thermo Fisher) with 10% methanol for 3 h at 200 mA. The membrane was exposed to film for 20 min at room temperature, and the film was developed.
RNA immunoprecipitation and RT–PCR.
HEK293T cells (1.5 × 107) were washed with PBS and lysed in lysis buffer (50 mM Tris-HCl, 100 mM NaCl, 1% NP-40, 0.1% SDS, 0.5% sodium deoxycholate; pH 7.4) with Protease Inhibitor Cocktail Set III (EMD Millipore). Five percent of each lysate sample was stored for preparation of the input RNA sample. The remaining lysates were split into two aliquots, immunoprecipitated using 10 μg anti-UBAP2L antibody or control IgG and incubated at 4 °C for 8 h. Bound RNA fragments in the IPs were washed six times in wash buffer (5 mM Tris-HCl, 150 mM NaCl, 0.1% Triton X-100). Immunoprecipitated RNA was isolated in TRIzol and purified with Direct-zol RNA kits, following the manufacturers’ protocols. Ten percent of RNA was saved for the no–reverse-transcriptase control. The remaining RNA was reverse-transcribed using SuperScript III with random primer mix (Thermo Fisher). cDNA was diluted tenfold in water, and target transcript cDNAs were amplified by PCR. Primer sequences are listed in Supplementary Table 4.
Reporting Summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Extended Data
Extended Data Fig. 1 ∣. Source of RBP ORFs, correlation between luciferase levels and RBP sizes, concordance between two luciferase systems, and correlation of reporter RNA and luciferase levels.
a, Western dot blot analysis of transiently expressed MCP-V5-tagged RBP ORFs in HeLa cells using a V5 antibody. Blue circles denote negative controls (no plasmid), red circles denote positive controls (CNOT7-V5-MCP). The order of wells and fold changes over negative controls are listed in Extended Data Table 1. b, Distribution of known classical and nonclassical RNA-binding domains in RBPs represented in our library. c, Distribution of molecular categories for RNA-related functions of RBPs represented in our library. d, Scatter plot of RBP size and luciferase effect. R, Pearson correlation coefficient. e, Luciferase activities from two different reporter constructs. Bar graphs showing log2-fold changes of the activity of Renilla (top) or firefly (bottom) luciferase reporters in presence of the MCP-fusion ORFs over FLAG control. Each vertical line represents a tethered ORF. f, Scatter plot of luciferase activities from the two reporter constructs. Values are expressed as log2-fold changes of the mean luciferase activity in the presence of MCP-fusion ORFs over FLAG controls. R, Pearson correlation coefficient.
Extended Data Fig. 2 ∣. IP validation from eCLIP experiments, correlation between eCLIP libraries, and de novo sequence motifs and metagene maps for candidate RBPs.
a,b, In-line western blots of eCLIP IPs of candidate RBPs. Extracts from HEK293T cells (a) or HEK293T cells transfected with the indicated V5-tagged RBP ORFs (b) immunoprecipitated with nonimmune (IgG) control antibodies, and western blot analysis using either RBP-specific (a) or anti-V5 (b) antibodies. The molecular weights (in kilodaltons) of standards are indicated on the right. Arrowheads indicate the calculated molecular weight for each RBP or RBP-V5 fusion protein. c, Heatmap of the Pearson correlation coefficients of fold enrichment of eCLIP peaks for the indicated 14 RBPs analyzed in duplicate. d, De novo sequence motifs in significant eCLIP peaks of the indicated RBP candidates enriched above background, with associated binomial P value. e–h, Metagene maps showing the distribution of eCLIP peak densities at target transcripts. The x axis indicates the relative length of each region. Dark red lines indicate the average number of significantly enriched peaks (≥4-fold enriched and P ≤ 10−2 versus SMInput) of eCLIP peak densities at all transcripts for BOLL, IFIT2, MEX3C, AIMP1 and CNOT7 (e), which show peak enrichment in 3′ UTR; DDX6, TOB1, NANOS3 and TOB2 (f), which show peak enrichment in 5′ UTR/3′ UTR; PARN and CLK3 (g), which show peak enrichment in 5′ UTR; and UBAP2L and MTDH (h), which show peak enrichment in CDS. Light shaded areas denote the 95% confidence interval.
Extended Data Fig. 3 ∣. Confirmation of RBP knockdown/overexpression, numbers of genes differentially regulated/unaffected by RBP perturbation, and region-level analysis of bound transcripts.
a,b, shRNA-mediated depletion of RBPs in HEK293T cells using 3-5 distinct shRNAs for each RBP, as indicated, compared to nontargeting shRNA control. a, Western blots with GAPDH or tubulin serving as loading controls, as indicated. b, Bar graphs indicating RBP transcript levels determined by RT–qPCR, normalized to levels of 18S rRNA. Data are shown as mean ± SD (n = 3 replicates). Asterisks denote significance at P < 0.05 determined by two-sided Student’s t-test. c, Overexpression of RBPs in HEK293T cells. Bar plots showing transcript levels (RPKM) for each RBP following transfection of RBP expression constructs or FLAG vector control. Data are shown as mean ± s.d. (n = 2 replicates). d–g, Numbers of up- or downregulated (log2-fold change ≥ 1.23 and FDR-corrected P ≤ 0.05) or unchanged genes for transcripts bound (≥4-fold enriched and P ≤ 10−2 versus eCLIP SMInput) or not bound by the indicated RBP for knockdown (d) and overexpression (e) of destabilizing RBPs and for knockdown (f) and overexpression (g) of stabilizing RBPs. h,i, Volcano plots showing the distribution of fold changes in transcript levels, with distribution histograms at the top, upon depletion of the destabilizer PARN (h) and depletion of the stabilizer CLK3 (left) and overexpression of the destabilizer IFIT2 (right) (i). Transcripts with log2(fold change) ≥1.23 and FDR-corrected P ≤ 0.05 are in color, with red and green denoting transcripts with or without at least one significant RBP binding peak (≥4-fold enriched and P ≤ 10−2 versus SMInput in eCLIP), respectively. j, Heatmap showing significance in differential expression of genes significantly differentially expressed (log2(fold change) ≥1.23 and FDR-corrected P ≤ 0.05) and significantly bound (≥4-fold enriched and P ≤ 10−2 versus SMInput in eCLIP) versus all unbound genes upon knockdown (KD) or overexpression (OE) of candidate RBPs in each region. Significance was calculated using a two-tailed Mann-Whitney U test. Uncropped images for a are available as source data online, and data for graphs in b, c are available as source data online.
Extended Data Fig. 4 ∣. Translation monitoring in a UBAP2L knockout replicate line, replicate concordance, and validation of polysome analyses in UBAP2L knockouts.
a, Translation monitoring using puromycin incorporation. Anti-puromycin western blot of extracts from puromycin-treated UBAP2L knockout (KO1) and parental (WT) HEK293T cell lines. GAPDH served as loading control. b,c, Polysome profile of UBAP2L after treatment of cells with 0.5 mM puromycin (b) and treatment of lysates with 30 mM EDTA (c). Top, absorbance (at 260 nm) plot of a HEK293T cell lysate fractionated through a 10-50% sucrose gradient. Bottom, western blots of UBAP2L from the corresponding fractions. d, Polysome profiles of HEK293T cells (WT, n = 2) and UBAP2L knockout HEK293T cells (KO, n = 4) fractionated through 10-50% sucrose gradients. Light-colored lines indicate means from each set (WT, light blue; KO, pink), and darkly shaded areas denote s.d. (WT, blue; KO, red). e, Bar graphs showing percentages of transcripts with RPKM ≥ 1 of all transcripts with ≥10 reads per transcript for two UBAP2L knockout lines (KO, 2 replicates each) and control samples (WT, two replicates). f, Scatter plots showing correlation of log2-transformed ratios of input-normalized polysome transcript levels (RPKM) between the two UBAP2L knockout HEK293T lines. R, Pearson correlation coefficient. g, Bar graph showing the percentage of regulated transcripts in UBAP2L targets and nontargets. *P < 0.0001 (χ2 test with Yates’s correction). h, RT–qPCR validation of reduced polysome association for the indicated transcripts. Transcript levels in inputs and polysome fractions were measured for KO and WT samples. KO/WT ratios of input-normalized polysome association of transcripts were then calculated. i, Western blots of DDX54, EIF4G1, EIF3B, and EEF2 in UBAP2L knockout cells (KO1, KO2). GAPDH served as a loading control. j,k, Quantitative flow cytometry reporter assay for mRNA translation using RCas9-fused 4EBP1. j, Plasmid design for the RCas9-4EBP1 experiment. k, Bar graph showing mean YFP levels in RCas9-4EBP1-expressing cells, normalized to RCas9-expressing cells, on each targeting site. Error bars denote s.d. from n > 5,000 RCas9-4EBP1-expressing and n > 5,000 rCas9-expressing cells per site. *P < 0.005; n.s., not significant (P > 0.5); two-tailed Student’s t-test. Uncropped images for a–c and i and data for graphs in h and k are available as source data online.
Extended Data Fig. 5 ∣. Repetitive element analysis of UBAP2L eCLIP data.
a, Immunofluorescence images showing UBAP2L (green) in HEK293T cells. DAPI (blue) marks nuclei. Scale bar, 10 μm. b, Pie chart showing fractions of UBAP2L replicate 2 eCLIP reads unambiguously mapping to repeat families in HEK293T cells. c, Line plot of UBAP2L binding sites on rRNAs. Fold enrichment of reads for IP over SMInput is plotted against the nucleotide positions of 18S and 28S rRNAs. Asterisk (*) denotes relative entropy ≥0.01. d–g, Location of UBAP2L binding sites on rRNA. d, ES15L; e, ES7S; f, ES27L; g, ES31L. Nucleotides with significant binding are highlighted in yellow. h, RIP of UBAP2L-RIP and RT–PCR in HEK293T cell lysates. The RIP assay was performed using anti-UBAP2L antibody or rabbit nonimmune IgG. RT–PCR was performed using primer sets within UBAP2L target regions ES7S, ES7L, ES15L, and ES31L. Uncropped images for h are available as source data online.
Supplementary Material
Acknowledgements
We thank members of the Yeo lab, in particular E. Wheeler and F. Krach, for helpful discussions and A. Palazzo for advice on polysome fractionation. E.-C.L. was partly supported by a study-abroad graduate student fellowship from the Taiwanese government. J.C.S. was partially supported by a Natural Sciences and Engineering Research Council of Canada Postgraduate Scholarships–Doctoral (PGS D-532649-2019). Y.H. was supported by the UCSD Frontiers of Innovation Scholars Program. G.A.P. was supported by a graduate fellowship from the National Science Foundation. F.E.T. was supported by a postdoctoral fellowship from the American Cancer Society (129547-PF-16-060-01-RMC). S.M. was supported by a postdoctoral fellowship from the Larry L. Hillblom Foundation (2014-A-027-FEL). This work was supported by grants from the NIH (R01HG004659, U19MH107367, R01NS103172 and U41HG009889) to G.W.Y.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Code availability
All code described in the Methods is publicly available and can be found at https://github.com/YeoLab/.
Competing interests
G.W.Y. is a cofounder and a member of the board of directors, is on the scientific advisory board and is an equity holder and paid consultant for Locana and Eclipse BioInnovations and a visiting professor at the National University of Singapore. The interests of G.W.Y. have been reviewed and approved by the University of California, San Diego in accordance with its conflict-of-interest policies. The authors declare no other competing financial interests.
Extended data is available for this paper at https://doi.org/10.1038/s41594-020-0477-6.
Supplementary information is available for this paper at https://doi.org/10.1038/s41594-020-0477-6.
Peer review information Anke Sparmann was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Reprints and permissions information is available at www.nature.com/reprints.
Data availability
Sequencing data are available at NCBI GEO (accession number GSE117294). Source data are provided with this paper.
References
- 1.Lukong KE, Chang KW, Khandjian EW & Richard S RNA-binding proteins in human genetic disease. Trends Genet. 24, 416–425 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Baltz AG et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell 46, 674–690 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Castello A et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149, 1393–1406 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Gerstberger S, Hafner M, Ascano M & Tuschl T Evolutionary conservation and expression of human RNA-binding proteins and their role in human genetic disease. Adv. Exp. Med. Biol 825, 1–55 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerstberger S, Hafner M & Tuschl T A census of human RNA-binding proteins. Nat. Rev. Genet 15, 829–845 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trendel J et al. The human RNA-binding proteome and its dynamics during translational arrest. Cell 176, 391–403 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Queiroz RML et al. Comprehensive identification of RNA–protein interactions in any organism using orthogonal organic phase separation (OOPS). Nat. Biotechnol 37, 169–178 (2019); erratum 37, 692 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Urdaneta EC et al. Purification of cross-linked RNA–protein complexes by phenol-toluol extraction. Nat. Commun 10, 990 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graindorge A et al. In-cell identification and measurement of RNA–protein interactions. Nat. Commun 10, 5317 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beckmann BM et al. The RNA-binding proteomes from yeast to man harbour conserved enigmRBPs. Nat. Commun 6, 10127 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Nostrand EL et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat. Methods 13, 508–514 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wheeler EC, Van Nostrand EL & Yeo GW Advances and challenges in the detection of transcriptome-wide protein–RNA interactions. Wiley Interdiscip. Rev. RNA 9, e1436 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Licatalosi DD et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature 456, 464–469 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hafner M et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141, 129–141 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Konig J et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat. Struct. Mol. Biol 17, 909–915 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kapeli K et al. Distinct and shared functions of ALS-associated proteins TDP-43, FUS and TAF15 revealed by multisystem analyses. Nat. Commun 7, 12143 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martinez FJ et al. Protein–RNA networks regulated by normal and ALS-associated mutant HNRNPA2B1 in the nervous system. Neuron 92, 780–795 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Nostrand EL, Huelga SC & Yeo GW Experimental and computational considerations in the study of RNA-binding protein–RNA interactions. Adv. Exp. Med. Biol 907, 1–28 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Nostrand EL et al. A large-scale binding and functional map of human RNA binding proteins. Nature 10.1038/s41586-020-2077-3 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ule J et al. CLIP identifies Nova-regulated RNA networks in the brain. Science 302, 1212–1215 (2003). [DOI] [PubMed] [Google Scholar]
- 21.Ule J et al. An RNA map predicting Nova-dependent splicing regulation. Nature 444, 580–586 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Coller JM, Gray NK & Wickens MP mRNA stabilization by poly(A) binding protein is independent of poly(A) and requires translation. Genes Dev. 12, 3226–3235 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coller J & Wickens M Tethered function assays: an adaptable approach to study RNA regulatory proteins. Methods Enzymol. 429, 299–321 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Bos TJ, Nussbacher JK, Aigner S & Yeo GW Tethered function assays as tools to elucidate the molecular roles of RNA-binding proteins. Adv. Exp. Med. Biol 907, 61–88 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Batra R et al. Elimination of toxic microsatellite repeat expansion RNA by RNA-targeting Cas9. Cell 170, 899–912 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nelles DA et al. Programmable RNA tracking in live cells with CRISPR/Cas9. Cell 165, 488–496 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Connell MR et al. Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature 516, 263–266 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Punta M et al. The Pfam protein families database. Nucleic Acids Res. 40, D290–D301 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Attwood TK et al. PRINTS and its automatic supplement, prePRINTS. Nucleic Acids Res. 31, 400–402 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lykke-Andersen J & Wagner E Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev. 19, 351–361 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benjamini Y, Krieger AM & Yekutieli D Adaptive linear step-up procedures that control the false discovery rate. Biometrika 93, 491–507 (2006). [Google Scholar]
- 32.Nishimura T et al. The eIF4E-binding protein 4E-T is a component of the mRNA decay machinery that bridges the 5′ and 3′ termini of target mRNAs. Cell Rep. 11, 1425–1436 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Wang X et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garneau NL, Wilusz J & Wilusz CJ The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol 8, 113–126 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Hu W, Yuan B & Lodish HF Cpeb4-mediated translational regulatory circuitry controls terminal erythroid differentiation. Dev. Cell 30, 660–672 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vicens Q, Kieft JS & Rissland OS Revisiting the closed-loop model and the nature of mRNA 5′–3′ communication. Mol. Cell 72, 805–812 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rissland OS The organization and regulation of mRNA–protein complexes. Wiley Interdiscip. Rev. RNA 8, e1369 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bicknell AA & Ricci EP When mRNA translation meets decay. Biochem. Soc. Trans 45, 339–351 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Radhakrishnan A & Green R Connections underlying translation and mRNA stability. J. Mol. Biol 428, 3558–3564 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Roy B & Jacobson A The intimate relationships of mRNA decay and translation. Trends Genet. 29, 691–699 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fu XF et al. DAZ family proteins, key players for germ cell development. Int. J. Biol. Sci 11, 1226–1235 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosario R, Childs AJ & Anderson RA RNA-binding proteins in human oogenesis: balancing differentiation and self-renewal in the female fetal germline. Stem Cell Res. 21, 193–201 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Julaton VT & Reijo Pera RA NANOS3 function in human germ cell development. Hum. Mol. Genet 20, 2238–2250 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cirillo L et al. UBAP2L forms distinct cores that act in nucleating stress granules upstream of G3BP1. Curr. Biol 30, 698–707 (2020). [DOI] [PubMed] [Google Scholar]
- 45.Markmiller S et al. Context-dependent and disease-specific diversity in protein interactions within stress granules. Cell 172, 590–604 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Youn JY et al. High-density proximity mapping reveals the subcellular organization of mRNA-associated granules and bodies. Mol. Cell 69, 517–532. e11 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Deragon JM & Bousquet-Antonelli C The role of LARP1 in translation and beyond. Wiley Interdiscip. Rev. RNA 6, 399–417 (2015). [DOI] [PubMed] [Google Scholar]
- 48.Gray GA & Gray NK A tail of translational regulation. Elife 6, e29104 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Andreev DE et al. Translation control of mRNAs encoding mammalian translation initiation factors. Gene 651, 174–182 (2018). [DOI] [PubMed] [Google Scholar]
- 50.Hortsch M, Griffiths G & Meyer DI Restriction of docking protein to the rough endoplasmic reticulum: immunocytochemical localization in rat liver. Eur. J. Cell Biol 38, 271–279 (1985). [PubMed] [Google Scholar]
- 51.Gehman LT et al. The splicing regulator Rbfox1 (A2BP1) controls neuronal excitation in the mammalian brain. Nat. Genet 43, 706–711 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Duncan PI, Stojdl DF, Marius RM, Scheit KH & Bell JC The Clk2 and Clk3 dual-specificity protein kinases regulate the intranuclear distribution of SR proteins and influence pre-mRNA splicing. Exp. Cell Res 241, 300–308 (1998). [DOI] [PubMed] [Google Scholar]
- 53.Scotti MM & Swanson MS RNA mis-splicing in disease. Nat. Rev. Genet 17, 19–32 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen CY, Xu N, Zhu W & Shyu AB Functional dissection of hnRNP D suggests that nuclear import is required before hnRNP D can modulate mRNA turnover in the cytoplasm. RNA 10, 669–680 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luna R, Rondon AG & Aguilera A New clues to understand the role of THO and other functionally related factors in mRNP biogenesis. Biochim. Biophys. Acta 1819, 514–520 (2012). [DOI] [PubMed] [Google Scholar]
- 56.Sundararaman B et al. Resources for the comprehensive discovery of functional RNA elements. Mol. Cell 61, 903–913 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miyasaka T et al. Interaction of antiproliferative protein Tob with the CCR4–NOT deadenylase complex. Cancer Sci. 99, 755–761 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lovci MT et al. Rbfox proteins regulate alternative mRNA splicing through evolutionarily conserved RNA bridges. Nat. Struct. Mol. Biol 20, 1434–1442 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee DSM, Ghanem LR & Barash Y Integrative analysis reveals RNA G-quadruplexes in UTRs are selectively constrained and enriched for functional associations. Nat. Commun 11, 527 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Urano J, Fox MS & Reijo Pera RA Interaction of the conserved meiotic regulators, BOULE (BOL) and PUMILIO-2 (PUM2). Mol. Reprod. Dev 71, 290–298 (2005). [DOI] [PubMed] [Google Scholar]
- 61.Moore FL et al. Human Pumilio-2 is expressed in embryonic stem cells and germ cells and interacts with DAZ (Deleted in AZoospermia) and DAZ-like proteins. Proc. Natl Acad. Sci. USA 100, 538–543 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schmidt EK, Clavarino G, Ceppi M & Pierre P SUnSET, a nonradioactive method to monitor protein synthesis. Nat. Methods 6, 275–277 (2009). [DOI] [PubMed] [Google Scholar]
- 63.Liu B & Qian SB Characterizing inactive ribosomes in translational profiling. Translation 4, e1138018 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fujii K, Susanto TT, Saurabh S & Barna M Decoding the function of expansion segments in ribosomes. Mol. Cell 72, 1013–1020 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Maeda M et al. Arginine methylation of ubiquitin-associated protein 2-like is required for the accurate distribution of chromosomes. FASEB J. 30, 312–323 (2016). [DOI] [PubMed] [Google Scholar]
- 66.Natchiar SK, Myasnikov AG, Kratzat H, Hazemann I & Klaholz BP Visualization of chemical modifications in the human 80S ribosome structure. Nature 551, 472–477 (2017). [DOI] [PubMed] [Google Scholar]
- 67.Lutz CS, Cooke C, O’Connor JP, Kobayashi R & Alwine JC The snRNP-free U1A (SF-A) complex(es): identification of the largest subunit as PSF, the polypyrimidine-tract binding protein-associated splicing factor. RNA 4, 1493–1499 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Boelens WC et al. The human U1 snRNP-specific U1A protein inhibits polyadenylation of its own pre-mRNA. Cell 72, 881–892 (1993). [DOI] [PubMed] [Google Scholar]
- 69.Fensterl V & Sen GC Interferon-induced Ifit proteins: their role in viral pathogenesis. J. Virol 89, 2462–2468 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liang D, Halpert MM, Konduri V & Decker WK Stepping out of the cytosol: AIMp1/p43 potentiates the link between innate and adaptive immunity. Int. Rev. Immunol 34, 367–381 (2015). [DOI] [PubMed] [Google Scholar]
- 71.Protter DSW & Parker R Principles and properties of stress granules. Trends Cell Biol. 26, 668–679 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hanson G, Alhusaini N, Morris N, Sweet T & Coller J Translation elongation and mRNA stability are coupled through the ribosomal A-site. RNA 24, 1377–1389 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fischer JW, Busa VF, Shao Y & Leung AKL Structure-mediated RNA decay by UPF1 and G3BP1. Mol. Cell 78, 70–84 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cox DBT et al. RNA editing with CRISPR–Cas13. Science 358, 1019–1027 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brannan KW et al. SONAR discovers RNA-binding proteins from analysis of large-scale protein–protein interactomes. Mol. Cell 64, 282–293 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Castello A et al. Comprehensive identification of RNA-binding domains in human cells. Mol. Cell 63, 696–710 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rual JF et al. Human ORFeome version 1.1: a platform for reverse proteomics. Genome Res. 14, 2128–2135 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.The Gene Ontology Consortium. Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res. 45, D331–D338 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ashburner M et al. Gene Ontology: tool for the unification of biology. Nat. Genet 25, 25–29 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Clement SL & Lykke-Andersen J A tethering approach to study proteins that activate mRNA turnover in human cells. Methods Mol. Biol 419, 121–133 (2008). [DOI] [PubMed] [Google Scholar]
- 81.Dobin A et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Conway AE et al. Enhanced CLIP uncovers IMP protein–RNA targets in human pluripotent stem cells important for cell adhesion and survival. Cell Rep. 15, 666–679 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data are available at NCBI GEO (accession number GSE117294). Source data are provided with this paper.