SUMMARY
Motion vision is important in guiding animal behavior. Both the retina and the visual cortex process object motion in largely unbiased fashion: all directions are represented at all locations in the visual field. We investigate motion processing in the superior colliculus of the awake mouse, by optically recording neural responses across both hemispheres. Within the retinotopic map, one finds large regions of ~500 μm size where neurons prefer the same direction of motion. This preference is maintained in depth to ~350 μm. The scale of these patches, ~30 degrees of visual angle, is much coarser than the animal’s visual resolution (~2 degrees). A global map of motion direction shows approximate symmetry between the left and right hemispheres and a net bias for upward-nasal motion in the upper visual field.
eTOC Blurb
Li et al. investigate the functional architecture of visual motion direction in the superior colliculus of awake mice. Using two-photon and wide-field microscopy they explore most of the visual field, revealing a patchy organization: In small regions, measuring ~30 degrees across, neurons are all biased for the same direction of motion.
INTRODUCTION
The connection between the ecology of natural scenes and the neurobiology of vision has been a topic of enduring interest. To a first approximation, natural scenes are translation-invariant, meaning that any given visual feature or event is equally likely to happen anywhere in the image. This principle predicts that the neural representation of visual images should be approximately uniform everywhere in the visual field. Closer inspection suggests some departures from translation invariance. The upper visual field is often occupied by the sky, which differs in content from the lower visual field. Also animals direct their eyes to salient points in the scene, which selects for certain image features at the center of gaze.
The first station of the visual system seems to follow the principle of uniform coverage. In the mammalian retina, each of ~30 distinct types of retinal ganglion cells (RGCs) conveys information about a specific visual feature [1,2]. Each cell type tiles the entire retina in a “mosaic” arrangement, ensuring a representation of its special feature at every point in the visual space. The primary visual cortex (V1) continues this pattern. Individual neurons there are often tuned to one line orientation or motion direction, but all the possible orientations and directions are represented at every point, or at least within 1–4 degrees of visual angle [3–10].
An evolutionarily older visual pathway leads from the retina to the superior colliculus [11]. In the superior colliculus, the principle of uniform coverage appears to be broken. Some of the earliest recordings in cat and rabbit already suggested a preference for visual motion in specific directions depending on location in the visual field [12,13]. In mouse superior colliculus one finds large regions of the visual field, ~30 degrees across, where neurons prefer stimuli with a specific line orientation [14–17]. Whether the representation of visual motion is similarly biased in a coarse map remains debated [14,18,19]. Most of the studies to date were performed in anesthetized animals with an acute cortical lesion to uncover the underlying superior colliculus. Both of these perturbations are known to affect the direction-selectivity of neurons in the superior colliculus [17,20,21].
The goal of the present work is to reveal how the direction of visual motion is represented in the superior colliculus of awake mice at a large-scale population level. To this end, we applied two complementary approaches. Two-photon calcium imaging was used to record neuronal responses at single-cell resolution in the posterior-medial SC of behaving mice, leaving the cortex intact. To reveal the functional organization on a large scale over both hemispheres of the SC we applied a wide-field imaging method in a mutant mouse that lacks much of the overlying cortex.
RESULTS
Single-cell imaging reveals a coarse map for direction of motion
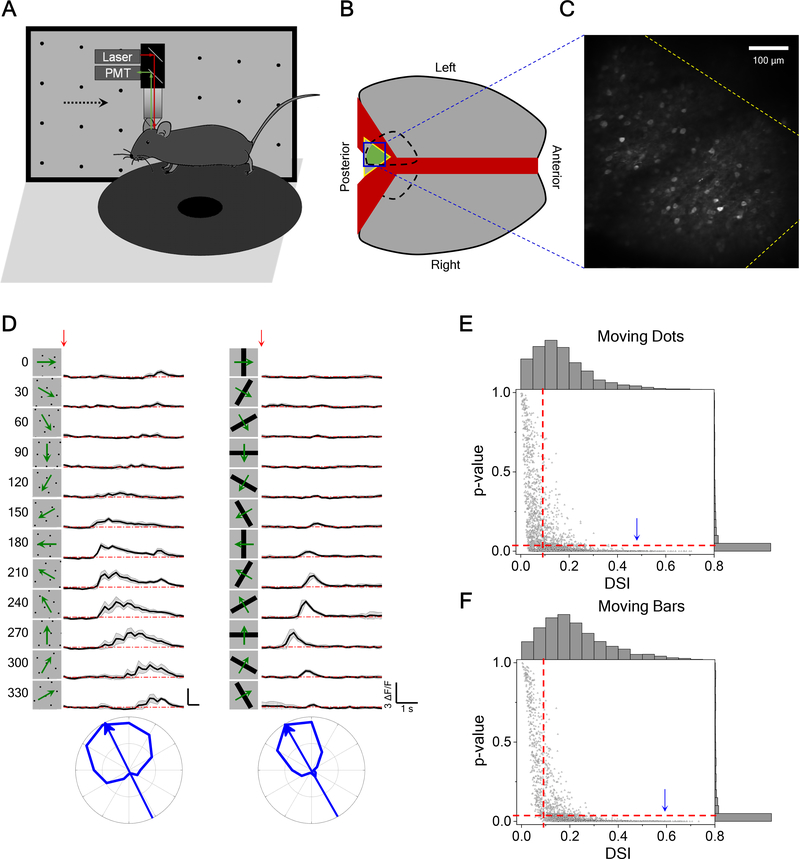
To investigate the tuning of SC neurons to the direction of motion, we imaged neuronal calcium responses to dots and bars moving in different directions using two-photon microscopy in head-fixed awake mice (Figure 1A). To maintain the integrity of the overlying cortex, one is limited to the posterior-medial sector of the mouse SC that corresponds to the upper lateral visual field [15] (Figures 1B and 1C). Direction-selective (DS) SC neurons in this region responded robustly to both moving dots and moving bars. Many neurons had a strong preference for one direction (Figure 1D). We defined an index of direction selectivity (DSI) and calculated its significance (see STAR Methods) and focused further analysis on neurons with DSI larger than 0.1 and p-value smaller than 0.05 (Figures 1E and 1F). Among these cells, the response to preferred direction motion was ~3 times larger on average than that to the null direction. Control experiments showed that the head-fixed animal performs only rare eye movements, and they are not systematically entrained by the moving stimuli (Figure S1A–B). Furthermore, when the animal runs on the circular treadmill the calcium responses increase in magnitude [22] but their direction tuning remains unchanged (Figure S1C–D).
Figure 1. Two-Photon Calcium Imaging Reveals Direction Tuning in Awake Mouse Superior Colliculus.
(A) Schematic of the experimental setup. Mice were head-fixed and free to run on a circular treadmill. Visual stimuli were presented on a screen. Neuronal calcium activity was imaged using two-photon microscopy. PMT, photomultiplier tube.
(B) Schematic of mouse brain anatomy after insertion of a triangular transparent plug to reveal the posterior-middle part of the SC underneath the posterior cortex.
(C) A standard deviation projection of calcium responses to moving dots.
(D) Calcium responses of a sample neuron to moving dots and moving bars at twelve directions. Red arrow indicates the stimulus onset. Gray shade indicates the standard deviation across identical trials. Small wiggles in the moving dot response reflect individual dots crossing the receptive field. Bottom, polar graphs of normalized peak responses with arrow pointing to the preferred direction.
(E) The p-value plotted against the direction selectivity index (DSI) of neurons responding to moving dots (3491 cells from 14 animals). Right and top show their histograms respectively. Dashed line indicates the threshold (DSI>0.1 and p < 0.05) applied for direction selectivity analysis. According to this criterion, 64% of recorded neurons are direction-selective. Arrowhead marks the cell in (D).
(F) The p-value and DSI of neurons responding to moving bars (2373 cells from 14 animals) displayed as in (E). 71% of recorded neurons are direction-selective.
See also Figure S1.
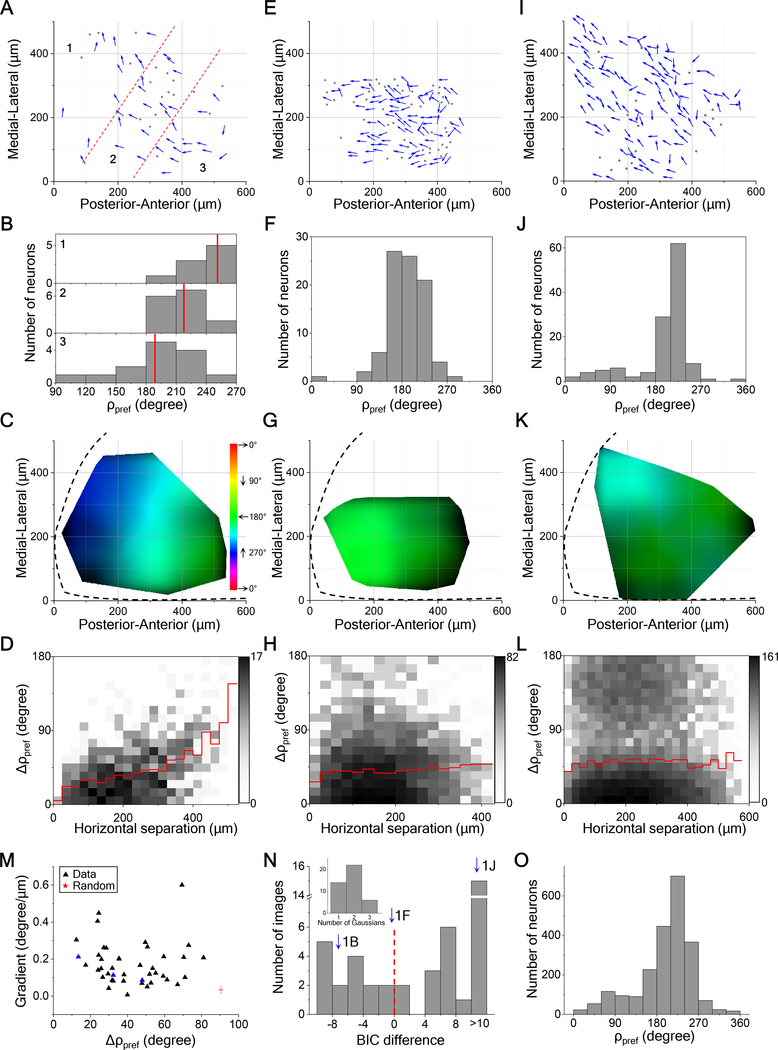
The moving-bar stimulus will drive feature detectors that are tuned to either motion direction or line orientation, leading to possible confounds, whereas a field of moving dots has no orientation signal. Therefore, we focused on results from moving dot stimuli. Figure 2 illustrates the direction tuning of neurons in the left posterior-medial SC of three different animals. In each case, neurons located near each other tended to have the same preferred direction of motion. That preferred direction varied only gradually across the surface of the SC (Figures 2A–2D), or remained constant over distances of several 100 μm (Figures 2E–2H, 2I–2L). At times, neurons that prefer two opposite directions were intermingled (Figures 2I–L). None of these arrangements of preferred direction resembles the salt-and-pepper organization in rodent V1 [10] (see more examples in Figure S2A–S2D). Instead, the preferred direction changes only slowly with the location on the SC, if at all. The regions of constant direction bias range over several hundred μm in size (Figures 2H and 2L), and indeed a larger field of view would be needed to assess their full extent. Furthermore, in all three animals, the preferred direction is the same, suggesting that the posterior-medial SC has a systematic bias for upward-nasal image motion.
Figure 2. Locally Uniform Direction Tuning.
(A) All direction-selective neurons in a sample field of view from left SC. Arrows are plotted at the anatomical location of the neurons and indicate the preferred direction in monitor coordinates. Dots are neurons with low direction-selectivity. Dashed lines divide the image into three regions.
(B) Histograms of preferred direction for the three regions. Red lines mark the mean.
(C) A smoothed version of the direction map in (A) where the hue encodes the preferred direction and the intensity varies inversely with the local variance of preferred directions. Note the systematic color gradient across the image. The boundary of the SC is indicated by the dashed line. Spatial scale as in (A).
(D) A 2-D histogram of the absolute difference in preferred directions of two neurons versus their horizontal distance (41 cells, 820 pairs). Red line indicates the mean.
(E, F, G, H) and (I, J, K, L) are single fields of view from two different mice represented in the same way as (A, B, C, D) with (88 cells, 382 pairs) and (124 cells, 7626 pairs) respectively. The dark regions in (K) indicate large local variance.
(M) The gradient of the map plotted against the absolute difference in preferred directions for pairs within 50 μm for all fields of view (42 images from 14 animals). Blue triangles are from the three sample images in (C, G, K). Red star indicates the mean of 1000 samples from a simulated random arrangement of preferred directions; error bar indicates SD.
(N) Histogram of the difference of Bayesian information criterion (BIC) between one-Gaussian and two-Gaussian fitting for all fields of view (42 images from 14 animals). Dashed line indicates the boundary between unimodal and bimodal. Arrows mark the three sample images. Inset, the number of Gaussians that gives the minimal BIC.
(O) Histograms of preferred direction for all direction-selective neurons (n = 2228 cells from 14 animals).
See also Figure S2.
To summarize the results across many recordings, we calculated for each field of neurons both a measure of the local scatter of preferred directions as well as the systematic gradient in the spatial map of preferred direction (Figure 2M, n = 42 images from 14 animals). The resulting distribution diverged dramatically from the null model in which each cell has an independent preferred direction (Figure 2M). In 26 of 42 cases, the preferred direction showed a bimodal distribution (Figure 2N). Across all samples, the preferred direction was strongly biased towards ~220 degrees (upward/nasal) and ~12% of DS neurons preferred the opposite direction (Figure 2O). This biased representation of motion direction was also observed in the responses to moving bars (Figure S2E–S2K).
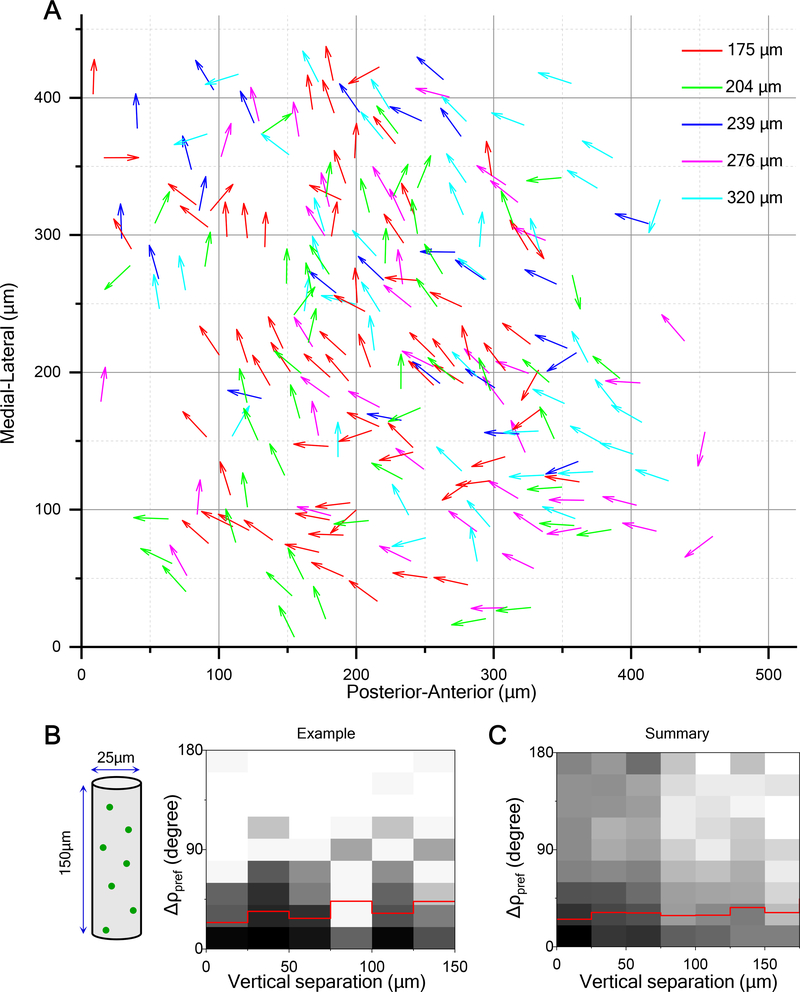
To explore the organization of preferred direction in the depth dimension, we imaged neuronal responses up to ~350 μm deep in steps of 20–30 μm. We projected the direction maps from different depths onto the same plane (Figure 3A). This revealed that neurons with short horizontal separation preferred a similar direction independent of depth (Figures 3A and 3B). Further analysis over 11 animals confirmed that preferred direction has a columnar organization, in that it varies little in the vertical dimension (Figure 3C). The average DSI did not change although the fraction of neurons with high DSI decreased somewhat at greater depth (Figure S3A) [19].
Figure 3. Direction Columns Extend in Depth.
(A) Horizontal plane projection of arrow plots (as in Figure 2A) at five different depths in the same field of view (229 cells).
(B) 2-D histogram of the absolute difference in the preferred direction versus vertical separation for this volume (236 pairs). Red line indicates mean.
(C) 2-D histogram of the absolute difference in the preferred direction versus vertical separation for all recorded neurons (2323 pairs from 11 animals).
See also Figure S3.
The preferred direction is orthogonal to the preferred orientation
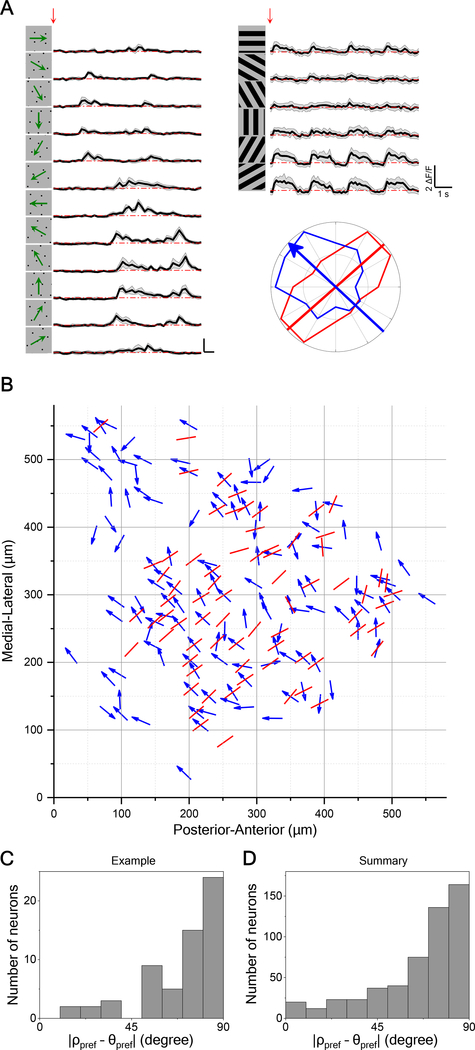
Early work on monkey and cat visual cortex discovered an orthogonal relationship between preferred orientation and preferred direction [6,23]. We examine whether this claim can be generalized to the mouse SC. To probe these two maps independently, we measured the direction preference with moving dots that carry no orientation signal and the orientation preference with flashed gratings that carry no motion signal (Figure 4A). Neurons with orientation selectivity index (OSI) > 0.1 and p < 0.05 were defined as orientation-selective neurons. About 24% of all direction-selective neurons were also orientation-selective (Figure S4I). Analysis of these doubly selective neurons in a window of cells spanning 580 μm revealed that the angle between their preferred direction and preferred orientation is about 90 degrees (Figures 4B and 4C). This orthogonal relationship was confirmed by histogramming the difference between preferred direction and orientation for neurons across many animals (Figure 4D).
Figure 4. Direction Maps Are Orthogonal to Orientation Maps.
(A) Calcium responses to moving dots and flashed gratings for a sample neuron exhibiting both direction- and orientation-selectivity. Red arrow indicates the stimulus onset. Gray shade indicates the standard deviation across identical trials. Polar plot shows peak response to dots (blue) and gratings (red). The blue arrow indicates the preferred direction and the red line indicate the preferred orientation. Here the preferred direction and orientation differ by 84 degrees.
(B) Overlay of maps for preferred direction (arrows) and orientation (lines) in a sample field of view, displayed as in Figure 2A.
(C) Histogram of the absolute difference between preferred direction and preferred orientation for the sample in (B) (60 cells).
(D) Same histogram for all direction- and orientation-selective neurons (530 cells from 14 animals).
See also Figure S4.
Wide-field imaging reveals the direction maps of two entire hemispheres
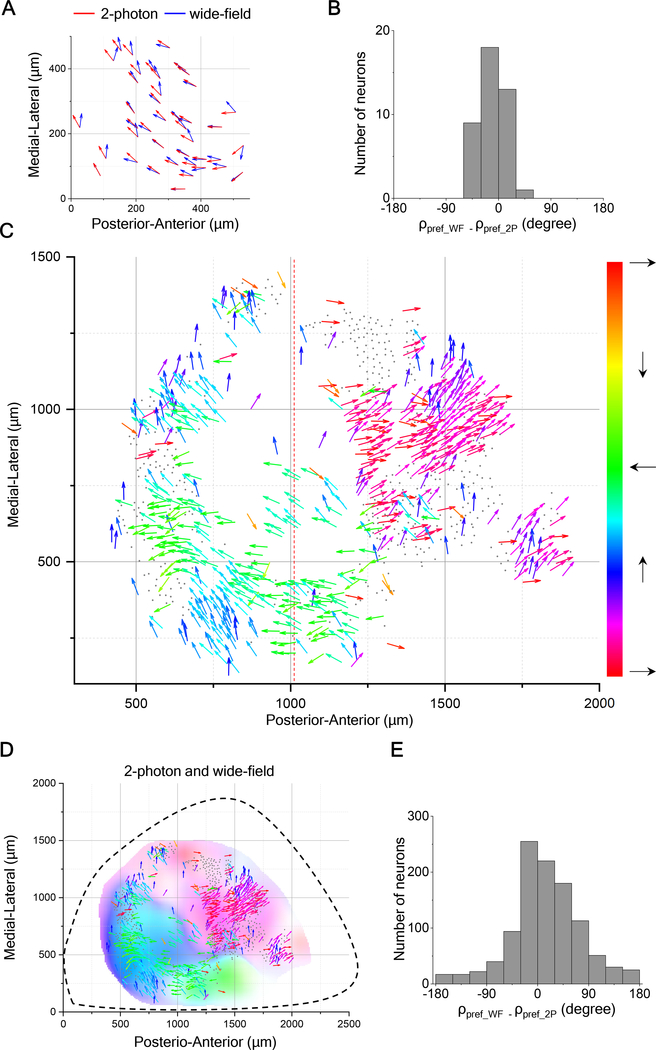
The bias in the preferred direction towards upward motion among neurons in the posterior-medial SC (see Figure 2O) motivated us to explore the functional organization of motion direction on a larger scale. To this end, we recorded from the entire SC using a wide-field imaging technique that sacrifices single-cell resolution but promises a global view of the gradual variations in direction tuning. We first validated the approach by imaging responses in the same brain using both two-photon and wide-field microscopy. Figures 5A and 5B illustrate the direction map in the posterior-medial sector of the SC obtained by both techniques, with very similar results (also see Figures 5SB and 5SC).
Figure 5. Direction Maps from Wide-Field Imaging And Two-Photon Imaging Agree.
(A) Arrow plots of preferred direction of single neurons measured by 2-photon imaging overlapped with the plots of preferred direction by wide-field imaging at the same anatomical location from the posterior-medial left superior colliculus of the same mouse.
(B) Histogram of difference of preferred directions for the data in (A).
(C) All direction-selective neurons in the SC of a mutant mouse with undeveloped cortex presented in the anatomical space with the preferred direction in monitor coordinates as in Figure 1A. Dashed line separates images that were acquired with the monitor at two different azimuth coordinates: centered on 95° for the left and 45° for the right.
(D) Overlay of arrow plots from 2-photon imaging data and direction map from wide-field imaging data. Hue encodes the preferred direction; saturation encodes the DSI.
(E) Histogram of difference of preferred directions for the data in (D).
See also Figure S5.
For broader coverage, we imaged the SC of mutant mice whose posterior cortex fails to develop [24], which leaves almost the entire SC directly accessible for optical recording (Figure S5A). Prior work on such partial-cortex mutants has shown that the visual response properties of collicular neurons are surprisingly normal [25,26]. For the present purposes we measured the visuotopic map on the surface of the SC (Figure S6F) and showed that it matches the features observed in normal mice [27–30]: elevation changes in the medio-lateral direction, azimuth in the anterior-posterior direction, and the magnification is ~25 μm/degree, varying somewhat with location and direction in the visual field. While this does not exclude the possibility of subtle developmental abnormalities, the close similarity to the normal mouse makes the mutant an attractive subject for wide-field imaging of subcortical structures.
We began by repeatedly moving the narrow window of the 2-photon microscope, and accumulated single-cell recordings of the preferred motion direction for ~600 neurons over much of the left SC (Figure 5C). A subsequent repeat of the same stimuli using the wide-field microscope yielded essentially the same map of preferred direction (Figures 5D and 5E). Note that the upward-nasal bias in the posterior-medial sector of the mutant is the same as in normal animals (Figure 2). These results show that wide-field imaging is a valid approach to reveal the functional organization of direction preference at a large scale (see Supplemental Videos 1–4 and Figure S6G).
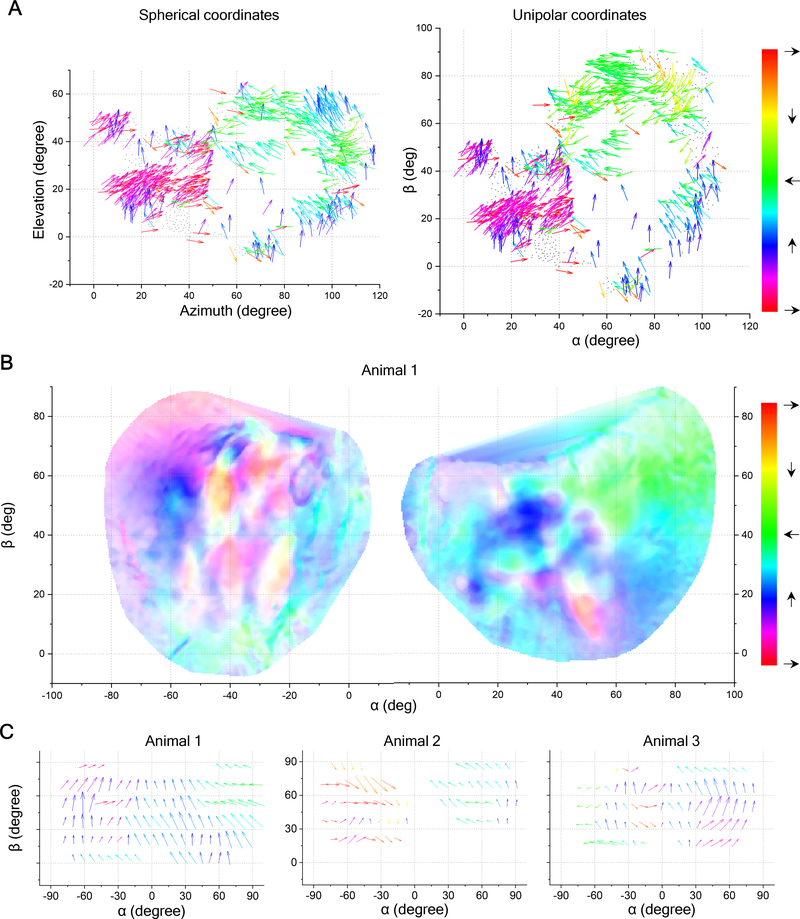
The patchy anatomy of preferred direction on the SC suggests that large parts of the visual field experience a bias for certain motion directions. To investigate this functional map, one needs to represent each neuron at the location of its receptive field, along with its direction preference in visual field coordinates. The need to represent motion directions throughout the entire visual field brings up an interesting challenge for graphic display. The conventional spherical coordinate system for visual space has singularities at the “north” and “south” poles (elevation = +90° and −90°) where the direction of motion cannot be expressed with respect to the coordinates (at the North Pole all directions point south). But the north pole of the spherical coordinate system is well within the visual field of the mouse [31]. To eliminate these pathologies, we chose to represent the sphere with a “unipolar” coordinate system that has only one singularity. That special point can be placed dead behind the animal, outside of the visual field (Figures S6A–S6D, see STAR Methods). In the following, we will use these unipolar coordinates for large-scale maps of the mouse visual field.
For the sample recording of Figure 5C we mapped the receptive fields and preferred directions of all neurons into visual space (Figure 6A). As predicted, the preferred direction varies with location in the visual field. Patches of approximately constant bias, ranging from 10 degrees to 40 degrees in size, are separated by regions with sharp changes. The gradient of preferred direction varies across the visual space. Wide-field imaging performed across both sides of the brain confirmed the patchy organization and the net bias for upward motion. Furthermore, regions representing the left and right visual fields appear to prefer directions that are mirror-symmetric about the midline (Figure 6B). Additional examples (Figure 6C) illustrate that the patchy organization varies among individuals, while the net upward bias is prevalent in almost all cases.
Figure 6. Large-Scale Direction Maps in Visual Space.
(A) All direction-selective neurons in the SC of a partial-cortex mouse presented in visual space using spherical and unipolar coordinates. Same two-photon recording as Figure 5C, but the arrows are plotted at receptive field locations. Note that the coordinate change transforms both locations and directions, and hence the colors of a given arrow differ in the two systems. See Videos S1–S4.
(B) Direction map obtained from wide-field imaging of both left and right SC of a different mouse, plotted in visual space with unipolar coordinates. Hue encodes the preferred direction; saturation encodes the DSI. Note the temporal direction is green on the right and red on the left.
(C) Arrow plots of the preferred direction in the SC of 3 animals across the visual field in unipolar coordinates.
See also Figure S6.
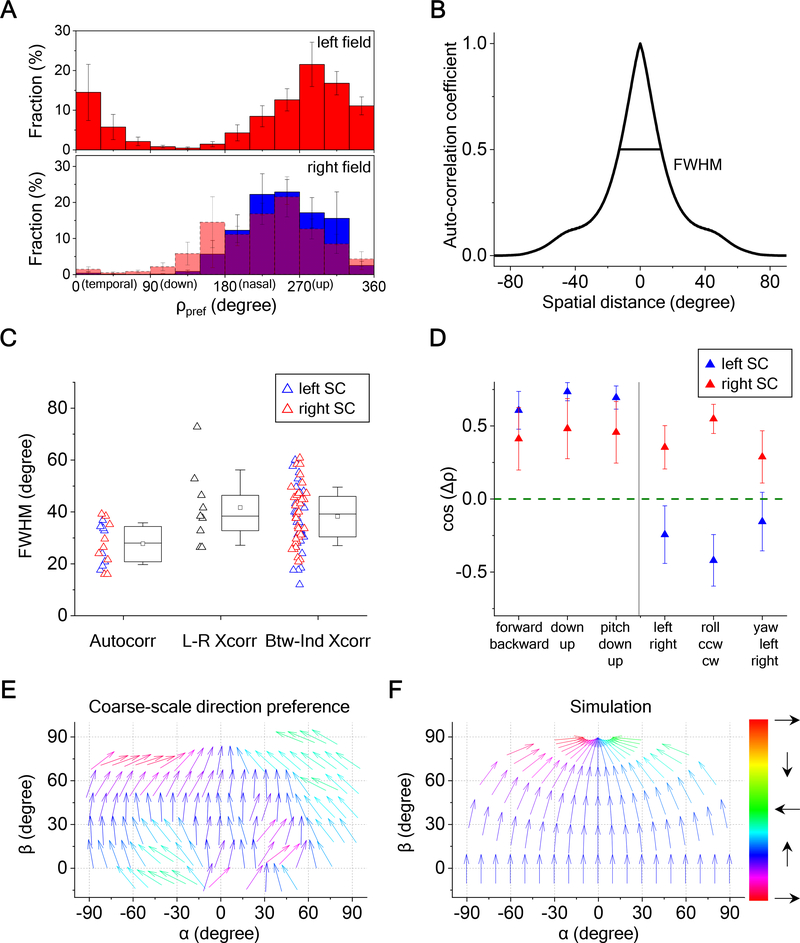
To identify the systematic aspects of direction-sensitivity, we analyzed these global maps across nine mutant mice. The simplest overarching feature is the overall bias for certain directions of motion, slightly nasal of upward, which is mirror-symmetric across the two sides of the visual field (Figure 7A). To quantify the size of the patches of constant direction, we calculated the autocorrelation function for each map, which yielded a full width at half maximum of 30 degrees (Figures 7B and S7A). As might be expected this average patch size is the same in the left and right half of the SC (Figure 7C). The direction patterns in left and right visual fields do not align perfectly: A cross-correlation between the two sides has a somewhat wider peak of ~40 degrees (Figure 7C). This concordance is similar to that observed across different animals (Figures 7C, S7B and S7C).
Figure 7. Bias at Various Scales in the Direction Map.
(A) Distribution of preferred directions encountered in both visual hemifields (9 animals). Error bars represent one standard error across animals. Bottom: The histogram for the left hemifield was flipped about the vertical and overlaid on the histogram for the right hemifield.
(B) The radial profile of autocorrelation for a sample direction map. Full-width at half maximum (FWHM) is indicated.
(C) Box plots of the FWHM for the autocorrelation, the cross-correlation between left and right SCs, and the cross-correlation between individuals for the same side of the SC (9 animals). The box plots 25th to 75th percentiles, the band inside the box is the median, and the square inside the box is the mean. The ends of the whisker indicate one standard deviation above and below the mean.
(D) Projections of preferred-direction maps from experimental data onto motion maps representing different optic flows (9 animals). Error bars represent one standard error. Wilcoxon signed-rank test or one-sample t-test were applied to test the significance of the difference from 0; the p-values are 4.3e-4, 5.1e-7, 5.0e-6, 0.17, 0.019, and 0.37 for the left SC; 0.098, 0.039, 0.027, 0.019, 1.5e-4, and 0.08 for the right SC. Mann-Whitney U-test or two-sample t-test were applied to test the significance of the difference between left and right SCs, and the p-values are 9.1e-3, 2.5e-5, and 0.061 for the right half figure.
(E) Arrow plots of the preferred direction in the SC, averaged over 9 animals, and plotted across the visual field in unipolar coordinates.
(F) The motion vectors in the mouse’s visual field produced by an aerial predator approaching at constant height from various directions.
See also Figure S7.
One possible interpretation of the global inhomogeneity is that it relates somehow to the optic flow pattern created by self-motion of the animal. To evaluate this notion, we compared the experimental preferred-direction maps to optic flow maps predicted from the six basic modes of self-motion (Figures S7D). By projecting the optic flow map onto the direction map one gains an estimate of how strongly that optic flow stimulus activates the superior colliculus. For those self-motions without a lateral component, the two sides of the SC are tuned in the same way. In particular, they are more strongly activated by forward translation, down translation, and downward pitch (Figure 7D). For those motions with a lateral component, the two sides are tuned in the opposite way. In particular, the left SC prefers translation to the right, clockwise roll, and yaw to the right; vice versa for the right SC. Most of these biases are highly significant and consistent across animals.
Alternatively one can relate the motion bias of the superior colliculus to movements in the visual scene outside of the animal. In the upper visual field, there is an intriguing match between the direction tuning of the SC and the motion vectors produced by an aerial predator approaching at a constant height from the horizon: Such a bird’s trajectory describes an upward path in the visual field that eventually bends in a nasal direction towards the north pole (Figures 7E and 7F).
DISCUSSION
This study extends our understanding of the functional architecture of visual features in the brain. We showed that the mouse superior colliculus represents visual motion with a bias that depends on location in the visual field. Nearby neurons prefer a similar direction of motion even over distances of ~500 μm on the collicular surface (Figure 2), with an average patch size of ~30 degrees of visual angle (Figure 7). The direction bias extends vertically over several 100 μm to form columns of preferred direction (Figure 3). For neurons that are selective both for direction and orientation, the preferred direction is orthogonal to the preferred orientation (Figure 4). Large-scale wide-field imaging revealed a global map of preferred direction with coarse symmetry between two sides of the superior colliculus and a strong bias for upward-nasal motion in the upper visual field (Figures 5–7). Here, we briefly discuss these conclusions and their implications.
Relation to prior work
Some of the features reported here have been described before: For example, de Malmazet et al [18] report that nearby neurons in certain regions of the retinotopic map tend to prefer the same direction of motion, and that this local preference also extends in depth. There has been controversy on those points that may be attributed to the effects of anesthesia, variations in imaging depth, or image size [17,19], but our results reliably confirm the observation of a local direction bias. More specifically de Malmazet et al [18] focused on the border of the binocular visual field, at ~30° azimuth and ±15° elevation, and reported that the direction bias switches from upward-nasal to upward-temporal across that line. This is broadly consistent with our consensus map for orientation bias (Figure S7F). Others have commented on the general prevalence of neurons with an upward [16] or upward-nasal [14] direction bias; this is a prominent feature shared across animals in our results as well (Figures 6C, 7E).
As regards the relationship between orientation-selectivity and direction-selectivity, our results differ somewhat from those in prior work. De Malmazet et al [18] measured the orientation-preference and the direction preference in different sets of neurons, and concluded that the two bias maps are unrelated to each other. By contrast we focused explicitly on individual neurons that were both well-tuned to direction and orientation (24% of the DS neurons) and we used two entirely different stimuli to establish orientation- and direction-tuning. At this single-cell level the orthogonal relationship between orientation- and direction-preference is unambiguous (Figures 4, S4).
Given this orthogonal relationship, one can also compare the present work to prior results on orientation-selectivity (OS). Again, these are available in only small portions of the visual field. Feinberg and Meister [15] reported a patchy OS-map in the posterior-medial sector of the SC, corresponding to high azimuth and elevation. The scale of these patches accords with the DS-patches in the present report. Furthermore the reported variability in OS-patches across individuals is consistent with the individual DS-maps reported here (Figure S7E). Ahmadlou and Heimel [14] reported that near the origin (0 degrees azimuth and elevation) the preferred orientation is perpendicular to the radius from the origin. Under the assumption of orthogonality the preferred direction should therefore point radially in that region. Indeed our consensus map of direction bias has such a region with an expanding vector field near the origin (Figure S7G).
Going beyond this prior work, we present here a global analysis of direction coding in the SC. We extend the measurements to a larger visual field, especially in the superior elevations ignored in earlier studies. We also record from both sides of the brain, covering almost the entire surface of the SC accessible from above. We established that the global map consists of multiple iso-direction patches and measured their size. Furthermore, we assessed the symmetry of these patches across the two sides of the SC and evaluated how the global direction map interacts with the global optic flow fields that result from locomotion during natural behavior.
Functional anatomy in different species
The functional organization of visual processing has been reported in the brains of many species. The visual cortex of cat [10], monkey [6], and ferret [5] shows a functional map, where a neuron’s response features – such as preferred orientation and direction – depend systematically on its location in the brain. In the rodent cortex there is less organization, and neurons preferring different orientations are intermingled in a “salt-and-pepper” pattern (but see [32]). Despite this difference, the common point is that neurons of all preferred orientations (or directions) are represented within a visual region smaller than the receptive field of a single neuron. In the V1 of the monkey, the orientation columns measure about 0.1 degrees on average, only one-tenth of the receptive field size [33]. In V1 of the tree shrew, all orientations are represented within a 4 degrees visual angle, again smaller than the average receptive field size [9]. By contrast, in the mouse superior colliculus, the patches with uniform preferred orientation span ~30 degrees, which greatly exceeds the RF size of ~10 degrees [17], or the visual acuity of 2 degrees [34]. The results reported here show that the representation of object motion in the superior colliculus is similarly dominated by such local biases. An intriguing new feature is that the iso-direction patches contain a minority of neurons tuned to the diametrically opposite direction (Figures 2I–2L). It will be important to understand the synaptic relationship of these two neural populations within the local circuitry, in particular whether they inhibit each other.
How does the patchy direction map in the SC arise?
Several explanations can be contemplated: (1) The local direction bias may exist already in the neural projections from the retina. (2) It may result from selective connectivity between the retinal inputs and the target neurons in the SC. (3) Or it may be constructed by postsynaptic circuitry within the SC. It has been shown that much of the direction selectivity in the mouse SC arises already in the excitatory synaptic inputs from retinal ganglion cells [35], which argues for a diminished role of collicular interneurons in the phenomenon. The mouse retina contains 8 types of direction-selective ganglion cells (DSGCs), and their global distribution across the retina has been analyzed. Certain DSGC types do indeed favor the forward-translation optic flow [36], as does the SC (Figure 7D). However, there is no indication of local inhomogeneities in the retina that could explain the patchy organization in the direction map of the SC [36–38]. Furthermore, the projections of DSGCs to the superficial SC appear to form a layer of uniform density, with no indication that one subtype dominates over the others in any local region [39,40]. This prior work suggests that each retinotopic location on the SC receives an equally balanced input of direction-selective signals from the retina, quite unlike what occurs in the optic tectum of the zebrafish [41]. If so, then the local bias observed in iso-direction patches must result from selective connectivity between SC neurons and specific subtypes of DSGCs, and that connectivity bias must somehow be coordinated among nearby neurons. Conceivably the local circuitry of interneurons contributes to this lateral coordination.
Behavioral relevance
Is there an intelligible purpose to the direction architecture in the mouse superior colliculus? On a coarse scale, cells in the upper visual field systematically prefer motion in the upward and nasal direction. Because activity in the upper visual field is generally associated with defensive reactions [42,43], one may speculate that the direction tuning is adapted to those functions. Indeed, we found that the global pattern of preferred direction across the visual field aligns with the motion vectors that would be produced by an aerial predator approaching from the horizon.
In addition, the global bias in the upper visual field also predicts a selectivity for certain optic flows produced by self-motion. For example, the superior colliculus as a whole should respond more strongly to forward motion of the animal than to backward motion, whereas horizontal turns left or right should produce excess activity in the right and left colliculus respectively. Such strong global signals may play a role in rapid feedback during the control of locomotion.
The other characteristic scale of the direction architecture is the size of smaller patches that deviate from the average tuning, measuring ~30 degrees of visual angle. The meaning of these specializations is harder to discern. The effects are substantial: For a direction-selective neuron in such a patch, the median ratio between preferred and null direction response is about 2.2. Thus, differently tuned patches may differ by a factor of 4 in their relative response to two moving objects. This should produce noticeable regional gradients in the animal’s perceptual sensitivity to motion, a prediction to be tested in future experiments.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests should be directed to and will be fulfilled by the Lead Contact: Markus Meister at meister@caltech.edu.
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
Data and code used in generating figures are available on CaltechDATA: https://doi.org/10.22002/D1.1441.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animal
Laboratory mice of both sexes were used at age 2–4 months. The strains were C57BL/6J (wild-type) and Emx1-Cre:Pals1flox/wt (partial-cortex[24]). All animal procedures were performed according to relevant guidelines and approved by the Caltech IACUC.
METHOD DETAILS
Visual Stimulation
For 2-photon imaging experiments, an LED screen (22”) was placed 18 cm away from the mouse’s right eye. The center of the monitor was at 95° azimuth and 25° elevation in the laboratory frame, and the monitor covered a visual field of 106° × 79°. The monitor’s LED illuminator was strobed for 12 μs at the end of each laser scan line to minimize interference of the stimulus with fluorescence detection. The monitor was gamma-corrected. For measuring neuronal tuning to orientation and motion direction we applied three types of stimuli. (1) A 2-d array of moving dots with a diameter of 5° spaced at 54°. The array moved in 12 possible directions at 50 degrees/s. This stimulus contains no orientation signal. (2) Square gratings with a spatial frequency of 0.08 cycles per degree were flashed at 6 orientations and 4 phases. The gratings stayed on for 2 s with an interstimulus interval of 4 sec. This stimulus contains no motion signal. (3) A moving dark bar with 5° width moving in 12 possible directions at 50 degrees/s. The sequence of directions or orientations was pseudo-randomized.
For the wide-field imaging setup, a large LED screen (43”) was placed 15 cm away from the mouse’s eye covering a visual field of 145° × 121°. The center of the monitor was at 25° elevation and either 0° azimuth (covering both eyes), 45° azimuth (mostly right eye) or −45° azimuth (mostly left eye). To avoid excessive fisheye distortion in these wide-field experiments, we applied a spherical correction to the images on the monitor [44].
Viral Injection
We injected adeno-associated virus (AAV) expressing non-floxed GCaMP6 (AAV2/1.hSyn1.GCaMP6f.WPRE.SV40) into the SC of wild-type mice or the SC of partial cortex mutant mice. After 2–3 weeks, we implanted a cranial window coupled to a transparent silicone plug that rested on the surface of the SC and exposed its caudomedial portion. This portion of the SC corresponds to the up-temporal part of the visual field. The optics remained clear for several months, which enabled long-term monitoring of the same neurons. Two-photon microscopy was applied to image calcium signals in the SC of head-fixed awake mice 3 weeks to 2 months after viral injection.
In Vivo Two-Photon Calcium Imaging
For imaging experiments, the animal was fitted with a head bar, and head-fixed while resting on a rotating treadmill. The animal was awake and free to move on the treadmill, but not engaged in any conditioned behavior. Two-photon imaging was performed on a custom-built microscope with a 16×, 0.8 NA, 3 mm WD objective (Nikon) and controlled by custom software written in Labview (National Instruments). A Ti:Sapphire laser with mode-locking technique (Spectra-Physics Mai Tai HP DeepSee) was scanned by galvanometers (Cambridge). GCaMP6f was excited at 920 nm and laser power at the sample plane was typically 20–80 mW. A 600 μm × 600 μm field of view was scanned at 4.8 Hz as a series of 250 pixel × 250 pixel images and the imaging depth was up to 350 μm. Emitted light was collected with a T600/200dcrb dichroic (Chroma), passed through a HQ575/250m-2p bandpass filter (Chroma), and detected by a photomultiplier tube (R3896, Hamamatsu). Artifacts of the strobed stimulus were eliminated by discarding 8 pixels on either end of each line. The animal’s locomotion on the treadmill and its pupil positions were recorded and synchronized to the image acquisition.
In Vivo Wide-Field Imaging
The wide-field microscope was built with a 50-mm / 105-mm tandem lens [45]. Illumination was from a blue LED with an excitation filter (469 nm ± 35 nm) and the camera used an emission filter (525 nm ± 39 nm). Images were acquired using a CMOS camera (Basler ace acA2000–165μm NIR USB 3.0 Camera) at an acquisition rate of 10 Hz. The camera covers an area of 5.36 mm × 2.85 mm at a spatial resolution of 2.63 μm.
Visual Field Coordinate Systems
During experiments with visual stimulation, the animal was head-fixed with the lambda-bregma plane horizontal in the laboratory frame. To report locations in the visual field, we use either spherical coordinates (azimuth,elevation)=(φ,θ) or unipolar coordinates (α,β). The lambda-bregma plane is at θ =0° and β= 0°.
Unipolar Coordinates
We introduce this coordinate system to avoid the singularities occurring at the poles of spherical coordinates. Suppose the observer sits at the origin of a Cartesian system facing in the z-direction (Figure 6A). The horizontal plane is (x, z) and the y-axis points up. A sphere of radius r is centered on the origin. For a given point on the sphere, spherical coordinates (azimuth,elevation)=( φ,θ) are defined as
| (1) |
and unipolar coordinates (α,β) as
| (2) |
Note the only singular point of the unipolar system is directly behind the observer at . The transformation from spherical coordinates to unipolar coordinates is:
| (3) |
The transformation from unipolar coordinates to spherical coordinates is:
| (4) |
The direction of motion on the sphere in unipolar coordinates is defined as
| (5) |
where dα and dβ are increments of motion along α and β respectively.
Analysis of Calcium Responses Measured by Two-Photon Imaging
Brain motion during imaging was corrected using SIMA [46] or NoRMCorre [47]. Regions of interest (ROIs) were drawn manually using Cell Magic Wand Tool (ImageJ) and fitted with an ellipse in Matlab. Fluorescence traces of each ROI were extracted after estimating and removing contamination from surrounding neuropil signals as described previously [15,48,49]. The true fluorescence signal of a neuron is Ftrue =Fraw -(r×Fneuropil), where r is the out-of-focus neuropil contamination factor and the estimated value for our setup is ~0.7. Slow baseline fluctuations were removed by subtracting the eighth percentile value from a 15-s window centered on each frame [50].
Two criteria were applied to interpret ROIs as neurons: 1) The size of ROI was limited to 10 – 20 μm to match the size of a neuron; 2) ROIs with a signal-to-noise ratio (SNR) smaller than 0.4 were discarded from further analysis [51].
| (6) |
Where C is the T×R response matrix (time samples × stimulus repetitions), 〈〉 is the mean and Var[] is the variance.
For any given stimulus, the response of a neuron was defined by the fluorescence trace in its ROI during the stimulus period
| (7) |
where the baseline is derived from a 0.5-s period prior to stimulation.
To quantify the tuning of a neuron to motion direction, we computed the preferred direction as the argument of the response-weighted vector sum of all directions:
| (8) |
where R(ρk) is the response amplitude in the kth direction ρk. The direction 615 selectivity index (DSI) was calculated as the normalized amplitude of the sum:
| (9) |
Similarly, for orientation tuning, the preferred orientation was calculated as the argument of the response-weighted vector sum of all orientations:
| (10) |
where R(θk) is the response amplitude at the kth orientation θk. The orientation selectivity index (OSI) was calculated as the normalized amplitude of the sum:
| (11) |
We assessed the statistical significance of DSI and OSI using a permutation test [52]: Surrogate trials were generated by randomly shuffling the trial labels and recomputing the DSI or OSI. This procedure was repeated 1000 times to generate a null distribution. Neurons were classified as direction-selective if DSI > 0.1 and p< 0.05, and as orientation-selective if OSI > 0.1 and p< 0.05.
The direction map was generated by linearly interpolating the single-cell-resolution data and then applying a mean filter over a 200-μm square region. To calculate the gradient of the map, the map of the preferred direction was fitted by:
| (12) |
where x and y are the anatomical coordinates. The gradient was calculated as:
| (13) |
The artificial random arrangement of preferred direction (Figure 2M) was simulated as 26 × 26 neurons, each of which prefers a random direction, evenly distributed on a 500 μm × 500 μm anatomical space.
To find the optimal number of Gaussians to fit the preferred direction distribution (Figure 2N), we evaluated the Bayesian information criterion (BIC) [53]:
| (14) |
where L is the maximized likelihood, M is the number of parameters in the model, and N is the number of neurons.
The center of the receptive field (RF) of each neuron was calculated based on the moving-bar responses. For each direction of motion, the time of peak response depends on the location of the RF and the neuron’s response latency. By evaluating responses to all directions (Figure 1D), one can extract the response latency and determine the best-fit location of the RF (Figure S6E).
Analysis of Calcium Responses Measured by Wide-Field Imaging
Each acquired image was convolved with a normalized box filter and down-sampled to 1/10 of the original size. An ROI was drawn manually over the SC to exclude signals from the surrounding areas. For each pixel in the image, the response was defined as described above for ROIs in 2-photon experiments. The RF center of each pixel was calculated from moving bar responses as described above, such that each pixel is assigned a location in visual space (Figure S6E–F).
To measure the size of direction-tuned patches in a direction map, we calculated the normalized 2-D autocorrelation of the map. Similarly to evaluate the correspondence between maps on the left and right SC or across different mice, we calculated the normalized 2-D cross-correlation of two maps. Suppose a direction map is given by , where at each location x in the visual field is a vector that points in the preferred direction ρ with length proportional to the DSI. For two such direction maps and we define the cosine correlation as
| (15) |
For every possible shift vector y, this computes the normalized dot product of one map with the shifted version of the other map. We first identified the shift vector ym that maximizes the correlation; in the case of an autocorrelation ym=0. Then we averaged the correlation map around that peak over all shift vectors of the same distance |y − ym| = r to obtain a radial profile C(r). The full width at half maximum (FWHM) of the correlation map (Figure 7B) was defined as twice the radius where this profile dropped to half of its maximum.
To produce the arrow plots at the global scale, the map was mean-filtered with a 16-degree window and down-sampled to an 8-degree resolution.
QUANTIFICATION AND STATISTICAL ANALYSIS
No statistical method was used to predetermine the sample size. A permutation test was used to assess the statistical significance of DSI and OSI as described above. The Shapiro-Wilk test was applied to assess whether data were normally distributed. In the case of a normal distribution, a t-test was applied; otherwise, a nonparametric test (Mann–Whitney U-test or One-sample Wilcoxon signed-rank test) was applied. The statistical details of experiments can be found in the results and figure legends.
Supplementary Material
Video S1. Calcium response of both sides of superior colliculus of a partial cortex mutant mouse to moving dots in 12 directions at a monitor that covers mostly left eye. Related to Figure 6. The monitor was at the 25° elevation and −45° azimuth and covered a visual field of 145° × 121°. The videos are recorded at 10 Hz and displayed at a 5x speed.
Video S2. Calcium response of both sides of superior colliculus of a partial cortex mutant mouse to moving dots in 12 directions at a monitor that covers mostly right eye. Related to Figure 6. The monitor was at the 25° elevation and 45° azimuth and covered a visual field of 145° × 121°. The videos are recorded at 10 Hz and displayed at a 5x speed.
Video S3. Calcium response of both sides of superior colliculus of a partial cortex mutant mouse to moving bars in 12 directions at a monitor that covers mostly left eye. Related to Figure 6. The monitor was at the 25° elevation and −45° azimuth and covered a visual field of 145° × 121°. The videos are recorded at 10 Hz and displayed at a 5x speed.
Video S4. Calcium response of both sides of superior colliculus of a partial cortex mutant mouse to moving bars in 12 directions at a monitor that covers mostly right eye. Related to Figure 6. The monitor was at the 25° elevation and 45° azimuth and covered a visual field of 145° × 121°. The videos are recorded at 10 Hz and displayed at a 5x speed.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| AAV1.Syn.GCaMP6f.WPRE.SV40 | University of Pennsylvania Vector Core | AV-1-PV2822 |
| Experimental Models: Organisms/Strains | ||
| C57BL/6J | The Jackson laboratory | JAX:000664 |
| Software and Algorithms | ||
| MATLAB 2019b | Mathworks | N/A |
| Python | Python Software Foundation | N/A |
| NoRMCorre: Non-Rigid Motion Correction | [47] | https://github.com/flatironinstitute/NoRMCorre#ref |
| SIMA: Motion correction | [46] | http://www.losonczylab.org/sima/1.3/api/motion.html |
| ImageJ 1.52g | NIH Image | https://imagej.nih.gov/ij/index.html |
| ReadImageJROI | Dylan Muir | https://www.mathworks.com/matlabcentral/fileexchange/32479-readimagejroi |
| swtest | Ahmed BenSaïda | https://www.mathworks.com/matlabcentral/fileexchange/13964-shapiro-wilk-and-shapiro-francia-normality-tests |
| Data repository including custom Python and MATLAB code | This paper | https://doi.org/10.22002/D1.1441 |
Highlights.
Mouse superior colliculus is biased towards certain directions of visual motion.
In patches of the visual field ~30 deg across neurons prefer the same direction.
Many neurons are tuned to oriented lines perpendicular to the preferred direction.
The global map is bilaterally symmetric with a bias for upward-nasal motion.
ACKNOWLEDGMENTS
M.M was supported by a grant from NIH (R01 NS111477). Y.L. was supported by NEI K99EY028640 and a Helen Hay Whitney Postdoctoral Fellowship.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Roska B, and Meister M (2014). The retina dissects the visual scene into distinct features. In The new visual neurosciences, Werner JS and Chalupa LM, eds. (Cambridge, MA: MIT Press; ), pp. 163–82. [Google Scholar]
- 2.Sanes JR, and Masland RH (2015). The Types of Retinal Ganglion Cells: Current Status and Implications for Neuronal Classification. Annu. Rev. Neurosci. 38, 221–246. [DOI] [PubMed] [Google Scholar]
- 3.Blasdel GG, and Salama G (1986). Voltage-sensitive dyes reveal a modular organization in monkey striate cortex. Nature 321, 579–585. [DOI] [PubMed] [Google Scholar]
- 4.Bonhoeffer T, and Grinvald A (1991). Iso-orientation domains in cat visual cortex are arranged in pinwheel-like patterns. Nature 353, 429–431. [DOI] [PubMed] [Google Scholar]
- 5.Weliky M, Bosking WH, and Fitzpatrick D (1996). A systematic map of direction preference in primary visual cortex. Nature 379, 725–728. [DOI] [PubMed] [Google Scholar]
- 6.Malonek D, Tootell RBH, and Grinvald A (1994). Optical imaging reveals the functional architecture of neurons processing shape and motion in owl monkey area MT. Proc R Soc Lond B 258, 109–119. [DOI] [PubMed] [Google Scholar]
- 7.Tusa RJ, Rosenquist AC, and Palmer LA (1979). Retinotopic organization of areas 18 and 19 in the cat. J. Comp. Neurol. 185, 657–678. [DOI] [PubMed] [Google Scholar]
- 8.Law MI, Zahs KR, and Stryker MP (1988). Organization of primary visual cortex (area 17) in the ferret. J. Comp. Neurol. 278, 157–180. [DOI] [PubMed] [Google Scholar]
- 9.Bosking WH, Crowley JC, and Fitzpatrick D (2002). Spatial coding of position and orientation in primary visual cortex. Nat. Neurosci. 5, 874–882. [DOI] [PubMed] [Google Scholar]
- 10.Ohki K, Chung S, Ch’ng YH, Kara P, and Reid RC (2005). Functional imaging with cellular resolution reveals precise micro-architecture in visual cortex. Nature 433, 597. [DOI] [PubMed] [Google Scholar]
- 11.Basso MA, and May PJ (2017). Circuits for Action and Cognition: A View from the Superior Colliculus. Annu. Rev. Vis. Sci. 3, 197–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schaefer K-P (1966). Mikroableitungen im Tectum opticum des frei beweglichen Kaninchens. Arch. Für Psychiatr. Nervenkrankh. 208, 120–146. [DOI] [PubMed] [Google Scholar]
- 13.Straschill M, and Hoffmann KP (1969). Functional aspects of localization in the cat’s tectum opticum. Brain Res. 13, 274–283. [DOI] [PubMed] [Google Scholar]
- 14.Ahmadlou M, and Heimel JA (2015). Preference for concentric orientations in the mouse superior colliculus. Nat. Commun. 6, 6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feinberg EH, and Meister M (2015). Orientation columns in the mouse superior colliculus. Nature 519, 229–232. [DOI] [PubMed] [Google Scholar]
- 16.Dräger UC, and Hubel DH (1975). Responses to visual stimulation and relationship between visual, auditory, and somatosensory inputs in mouse superior colliculus. J. Neurophysiol. 38, 690–713. [DOI] [PubMed] [Google Scholar]
- 17.Wang L, Sarnaik R, Rangarajan K, Liu X, and Cang J (2010). Visual Receptive Field Properties of Neurons in the Superficial Superior Colliculus of the Mouse. J. Neurosci. 30, 16573–16584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Malmazet D, Kühn NK, and Farrow K (2018). Retinotopic Separation of Nasal and Temporal Motion Selectivity in the Mouse Superior Colliculus. Curr. Biol. 28, 2961–2969.e4. [DOI] [PubMed] [Google Scholar]
- 19.Inayat S, Barchini J, Chen H, Feng L, Liu X, and Cang J (2015). Neurons in the Most Superficial Lamina of the Mouse Superior Colliculus Are Highly Selective for Stimulus Direction. J. Neurosci. 35, 7992–8003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosenquist AC, and Palmer LA (1971). Visual receptive field properties of cells of the superior colliculus after cortical lesions in the cat. Exp. Neurol. 33, 629–652. [DOI] [PubMed] [Google Scholar]
- 21.Wickelgren BG, and Sterling P (1969). Influence of visual cortex on receptive fields in the superior colliculus of the cat. J. Neurophysiol. 32, 16–23. [DOI] [PubMed] [Google Scholar]
- 22.Ito S, Feldheim DA, and Litke AM (2017). Segregation of Visual Response Properties in the Mouse Superior Colliculus and Their Modulation during Locomotion. J. Neurosci. 37, 8428–8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shmuel A, and Grinvald A (1996). Functional Organization for Direction of Motion and Its Relationship to Orientation Maps in Cat Area 18. J. Neurosci. 16, 6945–6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim S, Lehtinen MK, Sessa A, Zappaterra MW, Cho S-H, Gonzalez D, Boggan B, Austin CA, Wijnholds J, Gambello MJ, et al. (2010). The Apical Complex Couples Cell Fate and Cell Survival to Cerebral Cortical Development. Neuron 66, 69–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shanks JA, Ito S, Schaevitz L, Yamada J, Chen B, Litke A, and Feldheim DA (2016). Corticothalamic Axons Are Essential for Retinal Ganglion Cell Axon Targeting to the Mouse Dorsal Lateral Geniculate Nucleus. J. Neurosci. 36, 5252–5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee KH, Tran A, Turan Z, and Meister M (2020). The sifting of visual information in the superior colliculus. eLife 9, e50678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dräger UC, and Hubel DH (1976). Topography of visual and somatosensory projections to mouse superior colliculus. J. Neurophysiol. 39, 91–101. [DOI] [PubMed] [Google Scholar]
- 28.Mrsic-Flogel TD, Hofer SB, Creutzfeldt C, Cloëz-Tayarani I, Changeux J-P, Bonhoeffer T, and Hübener M (2005). Altered Map of Visual Space in the Superior Colliculus of Mice Lacking Early Retinal Waves. J. Neurosci. 25, 6921–6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cang J, Wang L, Stryker MP, and Feldheim DA (2008). Roles of Ephrin-As and Structured Activity in the Development of Functional Maps in the Superior Colliculus. J. Neurosci. 28, 11015–11023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cang J, and Feldheim DA (2013). Developmental Mechanisms of Topographic Map Formation and Alignment. Annu. Rev. Neurosci. 36, 51–77. [DOI] [PubMed] [Google Scholar]
- 31.Sterratt DC, Lyngholm D, Willshaw DJ, and Thompson ID (2013). Standard Anatomical and Visual Space for the Mouse Retina: Computational Reconstruction and Transformation of Flattened Retinae with the Retistruct Package. PLoS Comput. Biol. 9, e1002921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fahey PG, Muhammad T, Smith C, Froudarakis E, Cobos E, Fu J, Walker EY, Yatsenko D, Sinz FH, Reimer J, et al. (2019). A global map of orientation tuning in mouse visual cortex. bioRxiv, 745323. [Google Scholar]
- 33.Blasdel G, and Campbell D (2001). Functional Retinotopy of Monkey Visual Cortex. J. Neurosci. 21, 8286–8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prusky GT, and Douglas RM (2004). Characterization of mouse cortical spatial vision. Vision Res. 44, 3411–3418. [DOI] [PubMed] [Google Scholar]
- 35.Shi X, Barchini J, Ledesma HA, Koren D, Jin Y, Liu X, Wei W, and Cang J (2017). Retinal origin of direction selectivity in the superior colliculus. Nat. Neurosci. 20, 550–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sabbah S, Gemmer JA, Bhatia-Lin A, Manoff G, Castro G, Siegel JK, Jeffery N, and Berson DM (2017). A retinal code for motion along the gravitational and body axes. Nature 546, 492–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim I-J, Zhang Y, Yamagata M, Meister M, and Sanes JR (2008). Molecular identification of a retinal cell type that responds to upward motion. Nature 452, 478–482. [DOI] [PubMed] [Google Scholar]
- 38.Yonehara K, Ishikane H, Sakuta H, Shintani T, Nakamura-Yonehara K, Kamiji NL, Usui S, and Noda M (2009). Identification of Retinal Ganglion Cells and Their Projections Involved in Central Transmission of Information about Upward and Downward Image Motion. PLOS ONE 4, e4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huberman AD, Wei W, Elstrott J, Stafford BK, Feller MB, and Barres BA (2009). Genetic Identification of an On-Off Direction- Selective Retinal Ganglion Cell Subtype Reveals a Layer-Specific Subcortical Map of Posterior Motion. Neuron 62, 327–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kay JN, Huerta I.D. la, Kim I-J, Zhang Y, Yamagata M, Chu MW, Meister M, and Sanes JR (2011). Retinal Ganglion Cells with Distinct Directional Preferences Differ in Molecular Identity, Structure, and Central Projections. J. Neurosci. 31, 7753–7762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nikolaou N, Lowe AS, Walker AS, Abbas F, Hunter PR, Thompson ID, and Meyer MP (2012). Parametric Functional Maps of Visual Inputs to the Tectum. Neuron 76, 317–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dean P, Mitchell IJ, and Redgrave P (1988). Responses resembling defensive behaviour produced by microinjection of glutamate into superior colliculus of rats. Neuroscience 24, 501–510. [DOI] [PubMed] [Google Scholar]
- 43.Sahibzada N, Dean P, and Redgrave P (1986). Movements resembling orientation or avoidance elicited by electrical stimulation of the superior colliculus in rats. J. Neurosci. 6, 723–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marshel JH, Garrett ME, Nauhaus I, and Callaway EM (2011). Functional Specialization of Seven Mouse Visual Cortical Areas. Neuron 72, 1040–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ratzlaff EH, and Grinvald A (1991). A tandem-lens epifluorescence macroscope: Hundred-fold brightness advantage for wide-field imaging. J. Neurosci. Methods 36, 127–137. [DOI] [PubMed] [Google Scholar]
- 46.Kaifosh P, Zaremba JD, Danielson NB, and Losonczy A (2014). SIMA: Python software for analysis of dynamic fluorescence imaging data. Front. Neuroinformatics 8, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pnevmatikakis EA, and Giovannucci A (2017). NoRMCorre: An online algorithm for piecewise rigid motion correction of calcium imaging data. J. Neurosci. Methods 291, 83–94. [DOI] [PubMed] [Google Scholar]
- 48.Göbel W, and Helmchen F (2007). In Vivo Calcium Imaging of Neural Network Function. Physiology 22, 358–365. [DOI] [PubMed] [Google Scholar]
- 49.Kerlin AM, Andermann ML, Berezovskii VK, and Reid RC (2010). Broadly Tuned Response Properties of Diverse Inhibitory Neuron Subtypes in Mouse Visual Cortex. Neuron 67, 858–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dombeck DA, Khabbaz AN, Collman F, Adelman TL, and Tank DW (2007). Imaging Large-Scale Neural Activity with Cellular Resolution in Awake, Mobile Mice. Neuron 56, 43–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baden T, Berens P, Franke K, Román Rosón M, Bethge M, and Euler T (2016). The functional diversity of retinal ganglion cells in the mouse. Nature 529, 345–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ecker AS, Berens P, Cotton RJ, Subramaniyan M, Denfield GH, Cadwell CR, Smirnakis SM, Bethge M, and Tolias AS (2014). State Dependence of Noise Correlations in Macaque Primary Visual Cortex. Neuron 82, 235–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kass RE, and Raftery AE (1995). Bayes Factors. J. Am. Stat. Assoc. 90, 773–795. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1. Calcium response of both sides of superior colliculus of a partial cortex mutant mouse to moving dots in 12 directions at a monitor that covers mostly left eye. Related to Figure 6. The monitor was at the 25° elevation and −45° azimuth and covered a visual field of 145° × 121°. The videos are recorded at 10 Hz and displayed at a 5x speed.
Video S2. Calcium response of both sides of superior colliculus of a partial cortex mutant mouse to moving dots in 12 directions at a monitor that covers mostly right eye. Related to Figure 6. The monitor was at the 25° elevation and 45° azimuth and covered a visual field of 145° × 121°. The videos are recorded at 10 Hz and displayed at a 5x speed.
Video S3. Calcium response of both sides of superior colliculus of a partial cortex mutant mouse to moving bars in 12 directions at a monitor that covers mostly left eye. Related to Figure 6. The monitor was at the 25° elevation and −45° azimuth and covered a visual field of 145° × 121°. The videos are recorded at 10 Hz and displayed at a 5x speed.
Video S4. Calcium response of both sides of superior colliculus of a partial cortex mutant mouse to moving bars in 12 directions at a monitor that covers mostly right eye. Related to Figure 6. The monitor was at the 25° elevation and 45° azimuth and covered a visual field of 145° × 121°. The videos are recorded at 10 Hz and displayed at a 5x speed.
Data Availability Statement
Data and code used in generating figures are available on CaltechDATA: https://doi.org/10.22002/D1.1441.