Abstract
In recent years, the gut microbiota has been increasingly implicated in the development of many extraintestinal disorders, including neurodevelopmental and neurodegenerative disorders. Despite this growing connection, our understanding of the precise mechanisms behind these effects is currently lacking. Pattern recognition receptors (PRRs) are important innate immune proteins expressed on the surface and within the cytoplasm of a multitude of cells, both immune and otherwise, including epithelial, endothelial and neuronal. PRRs comprise four major subfamilies: the Toll-like receptors (TLRs), the nucleotide-binding oligomerization domain leucine rich repeats-containing receptors (NLRs), the retinoic acid inducible gene 1-like receptors and the C-type lectin receptors. Recognition of commensal bacteria by PRRs is critical for maintaining host–microbe interactions and homeostasis, including behaviour. The expression of PRRs on multiple cell types makes them a highly interesting and novel target for regulation of host–microbe signalling, which may lead to gut–brain signalling. Emerging evidence indicates that two of the four known families of PRRs (the NLRs and the TLRs) are involved in the pathogenesis of neurodevelopmental and neurodegenerative disorders via the gut–brain axis. Taken together, increasing evidence supports a role for these PRRs in the development of neurological disorders, including Alzheimer’s disease, Parkinson’s disease and multiple sclerosis, via the microbiota–gut–brain axis.
Keywords: gastrointestinal tract, microbiota, neurodegenerative, pattern recognition receptor
Graphical Abstract
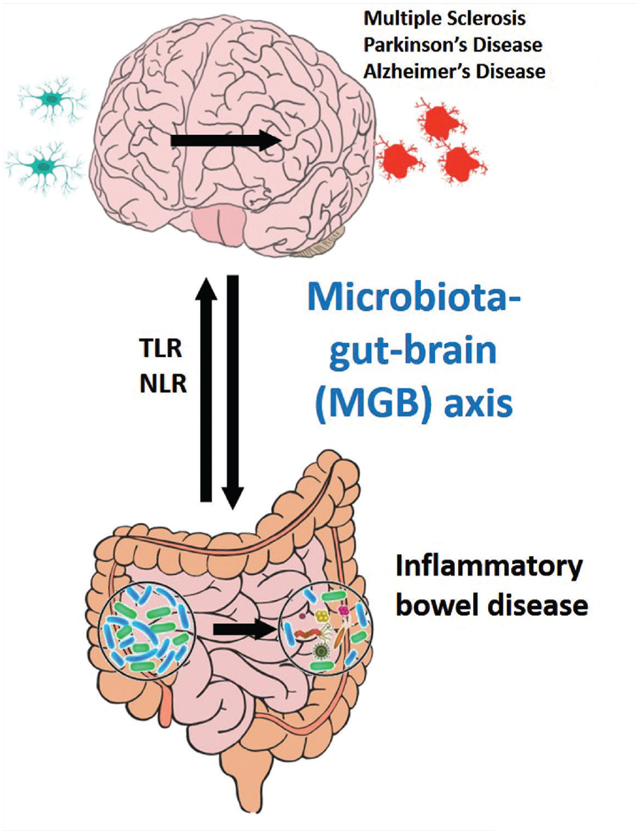
The microbiota-gut-brain (MGB) axis is involved in the pathogenesis of diseases both in the brain, including neurodevelopmental and neurodegenerative disorders, and diseases of the gut, including inflammatory bowel diseases. Pattern recognition receptors (PRRs) such as TLRs and NLRs are implicated in the development of these complex gut-brain disorders, in part via dysbiosis of the gut microbiota and alterations in the immune response.
Introduction
The understanding of the importance of the gut microbiota and its role in regulation of the physiology of the brain has grown exponentially in recent years. Humans are home to millions of microorganisms that reside both on the body (on the surface of the skin) and inside the body (gastrointestinal (GI) tract, nose and lungs). In fact, given this complex role, the gut microbiota is now considered to be a virtual organ in and of itself (Baquero & Nombela, 2012). Colonization of the microbiota begins at birth (Davis, 2016), with the early neonatal microbiome being dynamic and continuously modified as the child develops (Zhuang et al. 2019b). For example, breast-fed infants are host to species involved in the metabolism of colostrum present in breast milk, most notably Bifidobacteria infantis (Jiang et al. 2018). Upon the introduction of solid food, the infant microbiome shifts towards a more adult-like composition, increasing in its diversity and complexity (Ku et al. 2020). Additionally, studies have found that the gut microbiota is essential for the correct development of both the brain (Braniste et al. 2014; Lu et al. 2018) and the immune system (Schwarzer et al. 2019) beginning in early life. Therefore, recognition of commensal bacteria by the innate immune system is critical for maintaining host–microbe interactions and homeostasis, including behaviour.
Pattern recognition receptors
Pattern recognition receptors (PRRs) are part of the first line of innate immune defence following a pathological insult. They are expressed on multiple immune (leukocytes, macrophages, etc.) and non-immune cells (epithelial cells, endothelial cells and neurons) and respond to a variety of bacterial and viral ligands, including peptidoglycan (PGN), lipopolysaccharide (LPS), double stranded RNA, and CpG DNA, for example. PRRs comprise four major subfamilies: the Toll-like receptors (TLRs), the nucleotide-binding oligomerization domain leucin rich repeats-containing receptors (NLRs), the retinoic acid inducible gene 1-like receptors (RLRs), and the C-type lectin receptors (Walsh et al. 2013). In response to a pathological insult, the innate immune response is initiated by PRRs through binding of pathogen-associated molecular patterns (PAMPs), in turn triggering multiple intracellular signalling pathways, such as nuclear factor-κB (NF-κB), interferon regulatory factors and mitogen-activated protein kinase, resulting in production of cytokines and chemokines (Fawkner-Corbett et al. 2017). For the purposes of this review, we will focus on the TLR and the NLR families.
Despite continuous exposure to PAMPs in the lumen of the GI tract, intestinal epithelial cells (IECs) do not typically respond to commensal bacteria (Round & Mazmanian, 2009). This is due in part to PRR expression restricted to intracellular compartments, or basolateral expression in IECs, limiting their exposure to luminal PAMPs. In fact, commensal bacteria have been shown to be beneficial to the host (LeBlanc et al. 2017; Hiippala et al. 2018; Balakrishnan et al. 2019) by helping maintain immune surveillance. PRRs are crucial in maintaining these homeostatic interactions between the gut and the commensal microbiota, able to distinguish between pathogenic and commensal organisms. For example, distinct molecular signatures of the bacterial cell wall component PGN can elicit a variety of host immune gene patterns (Bersch et al. 2020). Commensal bacteria can drive myeloid differentiation primary response protein 88 (MyD88) signalling through TLR stimulation, inducing anti-microbial peptide production by Paneth cells that restrict bacterial colonization on the surface of the intestine, thereby limiting pro-inflammatory immune responses (Vaishnava et al. 2011). While other innate immune receptors also help maintain the balance between the host and the microbiota, these studies highlight a critical role for PRRs in this function.
Dysbiosis, or the disruption of the composition of the gut microbiota, has been implicated in numerous diseases not only those that impact the GI tract (e.g. inflammatory bowel diseases (IBD); Lupp et al. 2007; Kang et al. 2010) but also in diseases of the brain (e.g. neurodevelopmental and neurodegenerative diseases; Sampson et al. 2016; Hughes et al. 2018; Sun & Shen, 2018), lung (e.g. asthma; Liu et al. 2019; Zhuang et al. 2019a) and immune system (e.g. rheumatoid arthritis (Liu et al. 2013) and multiple sclerosis (MS; Cantarel et al. 2015)). While it remains unclear whether dysbiosis is causative or correlative in many cases, its impact on GI mucosal barrier function and host–microbe interactions can disrupt immune homeostasis in the rest of the body. Consequently, altered host–microbe interactions and subsequent GI pathophysiology could allow the commensal microbiota to gain access to the surrounding tissue, potentially leading to inflammation and damage (Garrett et al. 2010). Here we discuss the role of two PRR families, NLRs and TLRs, that are implicated in the development of neurological disorders and the intersection of the microbiota and the innate immune system (Fig. 1).
Figure 1. The role of NLRs and TLRs in gut–brain signaling.
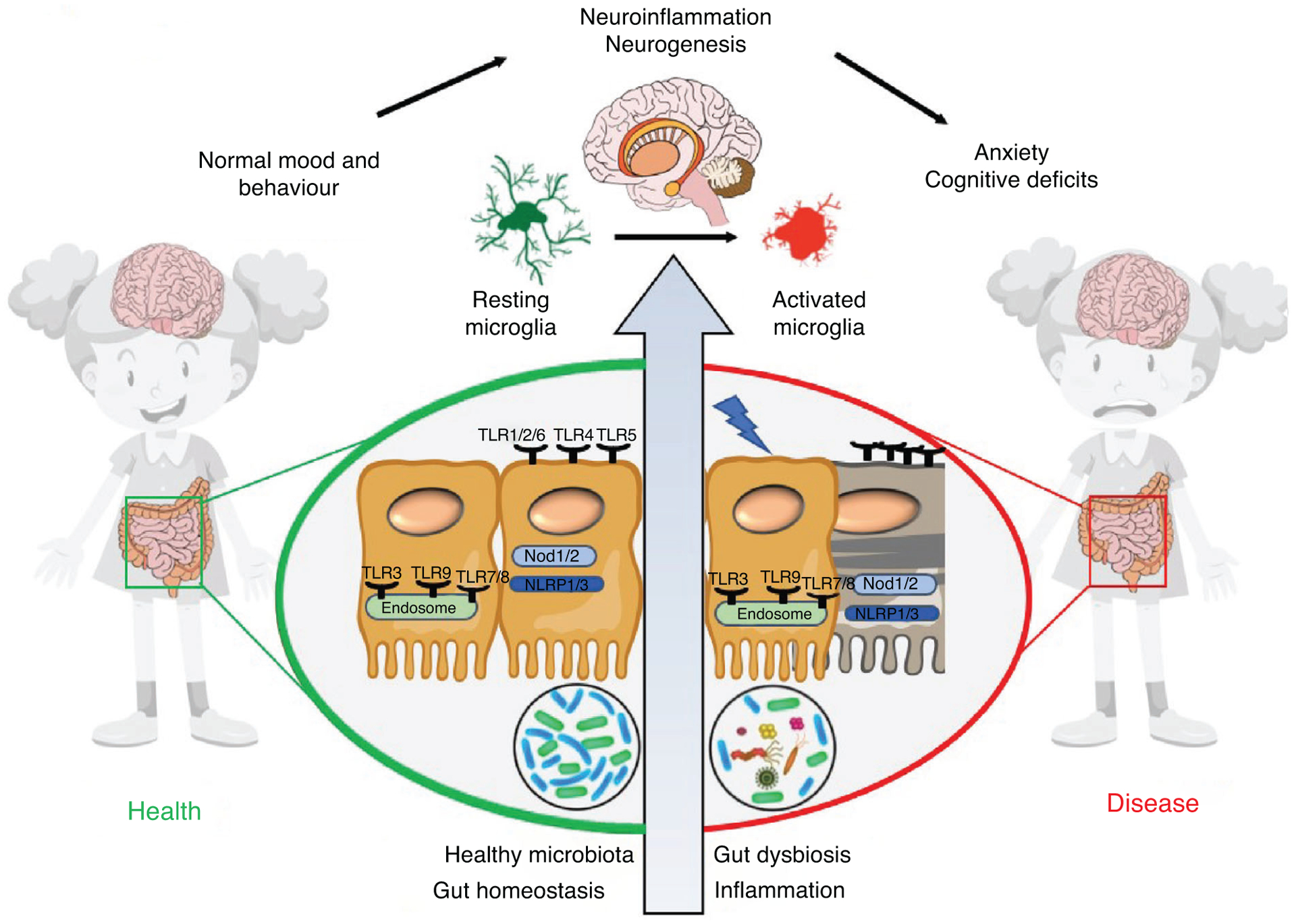
Under physiological conditions, host–microbe interactions maintain intestinal physiology via pattern recognition receptors (PRRs), including Nod-like receptors (NLRs) and Toll-like receptors (TLRs). In the brain, the PRRs regulate multiple cell types, including neurons, microglia and astrocytes. Dysbiosis, GI inflammation and neuroinflammation all contribute to diseases within the GI tract as well as in the brain. Understanding these pathways will provide new insight into disease pathogenesis and novel targets for restoration of physiology.
Nod-like receptors
The NLR receptor family can be divided into three different sub-groups: (1) inflammasome-forming NLRs (i.e. NLRP1, NLRP3), (2) positive regulatory NLRs (i.e. Nod1, Nod2) and (3) negative regulatory NLRs (i.e. NLRx1, NLRC3), each with a separate and distinct signalling pathway and downstream effect (Coutermarsh-Ott et al. 2016) (Fig. 2). The inflammasome forming group of NLRs consists of NLRP1, NLRP3, NLRP6, NLRP4 and NLRC5, which form multiprotein complexes. These NLR proteins multiplex with, for example, apoptosis-associated speck like protein and procaspase-1, to initiate proinflammatory cytokine expression.
Figure 2. The NLR signalling pathway.
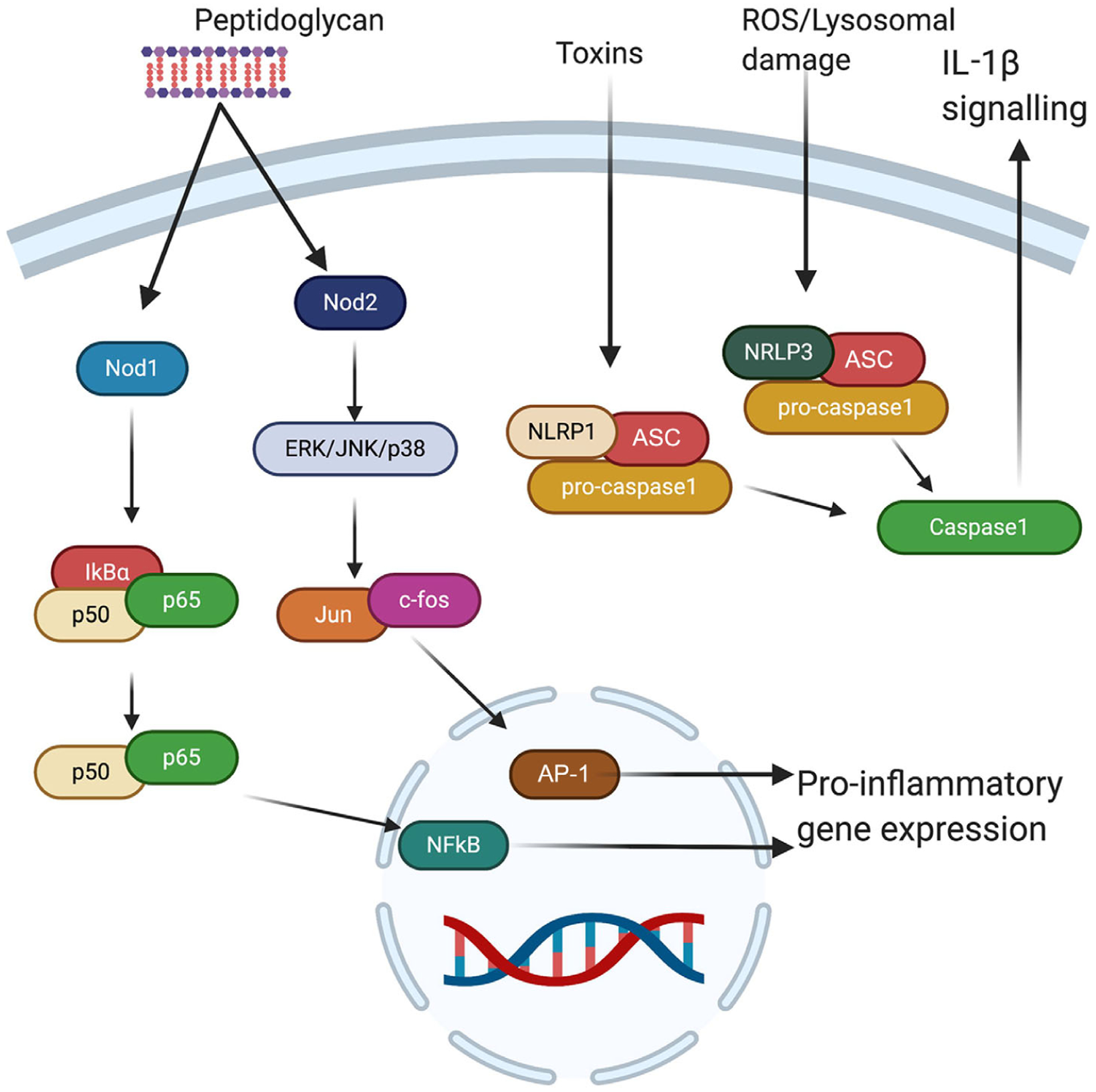
NLRs are predominantly present in the cytosol of cells, including immune, epithelial, endothelial and neuronal cells. Both Nod1 and Nod2 respond to bacterial peptidoglycan, with Nod1 initiating NF-κB-dependent pro-inflammatory gene expression and Nod2 initiating AP1 inflammatory genes via extracellular signal-regulated kinase (ERK)/c-Jun N-terminal kinase (JNK)/p38. NLRP1 activates caspase1 signalling in response to toxins and NLRP3 activates caspase1 in response to lysosomal damage and reactive oxygen species. AP-1, activator protein 1; ASC, apoptosis-associated speck-like protein containing a CARD; IκBα, inhibitor of nuclear factor-κB α; ROS, reactive oxygen species.
Inflammasome complexes, including NLRP1 and NLRP3, have been implicated in the development of many neurological disorders, for example, several single nucleotide polymorphisms of NLRP1 have been associated with Alzheimer’s disease (AD). Additionally, NLRP1 mRNA is upregulated in neurons of AD patients (Pontillo et al. 2012). Furthermore, amyloid-β plaques have been shown to stimulate purinergic receptors initiating activation of the inflammasome, in turn contributing to late-stage AD (Tan et al. 2014). In a mouse model of chronic constriction injury-induced neuropathic pain, the NLRP1 inflammasome was significantly activated in the hippocampus. Inhibition of the downstream product of NLRP1 attenuated the observed depression-like behaviour in these mice (Li et al. 2019). Dysbiosis has been observed in AD patients, suggesting a possible avenue of further research in order to fully elucidate the mechanisms and interconnectivity of NLRP1 in the brain with the host microbiota. In line with these findings, NLRP3 signalling has also been implicated in the development of major depressive disorder via the hypothalamic–pituitary–adrenal axis (Inserra et al. 2018). In Parkinson’s disease (PD) patients, NLRP3 levels were found to be upregulated in the serum, correlating with α-synuclein levels, a hallmark of disease severity (Chatterjee et al. 2020). Additionally, in mice it was found that α-synuclein activates NLRP3 via microglial endocytosis (Zhou et al. 2016). Deficiency in caspase-1, a member of the NLRP3 inflammasome complex, significantly reduced microglial activation indicating a possible role for the NLRP3 inflammasome in PD pathogenesis (Zhou et al. 2016; Gordon et al. 2018). Taken together, this suggests that intestinal dysbiosis may trigger altered NLRP signalling both in the gut and in the brain, leading to neurodegeneration in the brain.
The nucleotide-binding oligomerization domain (NOD) proteins are a family of positive regulatory NLRs that detect fragments within the cell wall of many bacteria, activating signalling pathways driving pro-inflammatory and anti-microbial responses. The two best characterized members of the NLR family are Nod1 and Nod2. They are unique in their function in that they sense bacterial PGN in the host cytosol as opposed to microbial ligands at the cell surface or within endosomes. Nod1 and Nod2 regulate activation of NF-κB transcription via a receptor-interacting serine/threonine-protein kinase 2-dependent mechanism in response to unique PGN fragments leading to the expression of pro-inflammatory cytokines (Caruso et al. 2014). Nod1 is ubiquitously expressed in many cell types, primarily in immune cells (Uhlen et al. 2015), neurons, endothelial cells and epithelial cells of many organs (Caruso et al. 2014). While Nod2 expression is slightly more restricted, it has been identified in lymphocytes, Paneth cells and IECs (Franchi et al. 2009). Importantly, studies have shown that both Nod1 and Nod2 receptors are also expressed in the brain, including within the hippocampus on multiple cell types such as neurons, astrocytes and microglia (Ogura et al. 2003; Arentsen et al. 2017) suggesting that they play an important role within the central nervous system.
Nod1 and Nod2 serve a critical role in responding to specific bacterial pathogens. For example, the enteric mouse pathogen Citrobacter rodentium induces an IL-17 response via a Nod1- and Nod2-dependent pathway (Rubino et al. 2013). Mice deficient in Nod1 and Nod2 are highly susceptible to infection with Listeria when they are first exposed to LPS or E. coli. The results of this study implicate that cells consistently exposed to microbial stimuli, such as in the GI tract, are characterized by low TLR expression and can become re-sensitized to commensal bacteria in the absence of Nod1 and Nod2 (Kim et al. 2008). Several studies have indicated that the recognition of pathogenic bacteria in intestinal cells lacking TLRs relies on Nod1 (Girardin et al. 2001; Zilbauer et al. 2007).
As Nod1 and Nod2 are activated by PGN, they are also important in maintaining gut homeostasis by priming the immune system in the absence of infection, employing the gut microbiota as its stimulus (Clarke et al. 2010; Claes et al. 2015). Mice deficient in both Nod1 and Nod2 (NodDKO) display stress-induced anxiety-like behaviour, cognitive impairment and depression (Pusceddu et al. 2019). In the hippocampus, NodDKO mice displayed decreased 5-HT at baseline and following acute stress. In particular, Nod1 expression on IECs was identified as a specific factor in regulating the stress response and serotonergic signalling, but the precise signalling mechanisms behind this effect have yet to be fully elucidated (Pusceddu et al. 2019). Numerous studies have implicated the NLR family as important PRRs in the development of neurological diseases mediated by the gut microbiota. However, much is still unclear about the mechanism of action of these effects. Given these findings, the NLR family, in particular Nod1 and Nod2, remain attractive targets for the development of therapeutics for treatment of many of the aforementioned neurological disorders.
PGN derived from commensal gut bacteria can cross the blood–brain barrier into the central nervous system (CNS), with levels of PGN within the brain increasing with age (Arentsen et al. 2017). Several PGN-sensing molecules, such as the peptidoglycan recognition protein (PGRP) PRRs and NLRs, are highly expressed in the neonatal brain during early development and are highly susceptible to changes in the gut microbiota (Arentsen et al. 2017). Knockout of PGN recognition molecule 2 (Pglyrp2) induces behavioural changes and alterations in the autism spectrum disorder risk gene c-Met in a sex-specific manner (Arentsen et al. 2017). Taken together, these findings highlight a novel role for PRRs in maintaining behaviour and CNS function.
Toll-like receptors
TLRs are a highly expressed family of transmembrane PRRs responsible for initiating downstream signal transduction in response to PAMPs and tissue damage. Localization of each TLR allows classification into two groups, those expressed on the plasma membrane (TLR1, TLR2, TLR4, TLR5, TLR6, TLR11) and those expressed within the cytoplasm and organelles (TLR3, TLR7, TLR8, TLR9) (Fig. 3). To date, 11 human and 13 mouse TLRs have been characterized (Akira & Takeda, 2004). Activation of each TLR, following specific PAMP recognition, causes a conformational change in the receptor allowing recruitment of the appropriate downstream signalling adaptor, in turn activating specific transcription factors, and subsequent innate immune responses (Takeuchi & Akira, 2001). Four adaptor proteins have been identified, each one responsible for a specific immune response. For example, the universal adaptor protein MyD88 is known to induce activation of NF-κB and activator protein 1 (AP-1) triggering the expression of inflammatory cytokines such as tumour necrosis factor-α (TNFα). Alternatively, TLR3 and TLR4 can signal through the adaptor protein Toll/IL-1 receptor domain-containing adapter inducing interferon-β (TRIF) in order to activate type-I interferon (IFN) (Fitzgerald et al. 2003). The regulation of these responses is tightly controlled via post-translational modifications such as glycosylation (Weber et al. 2004; Sun et al. 2006; Abdulkhalek et al. 2011; Iavarone et al. 2011) and ubiquitination (Boone et al. 2004; Chuang & Ulevitch, 2004; Shembade et al. 2010; Guedes et al. 2014; Kinsella et al. 2018) and through negative feedback (Scott et al. 1993; Renner & Schmitz, 2009). Disruption of TLR activation or maturation can lead to dysregulation of the immune response (Barrat et al. 2005; Reynolds et al. 2010; Ziegler et al. 2011; Suarez-Farinas et al. 2013; Cavalcante et al. 2018). Despite continuous exposure to TLR ligands in the gut lumen, IECs express low levels of TLRs. Introduction of pathogenic bacteria causes upregulation of some TLRs, namely TLR2, TLR4, TLR5 and TLR9 (Muzio et al. 2000; Gewirtz et al. 2001; Ewaschuk et al. 2007), while others are differentially expressed in response to pathogenic bacteria. TLRs play an important role in maintaining host–microbe interactions and mucosal immunity in the GI tract.
Figure 3. The TLR signalling pathway.
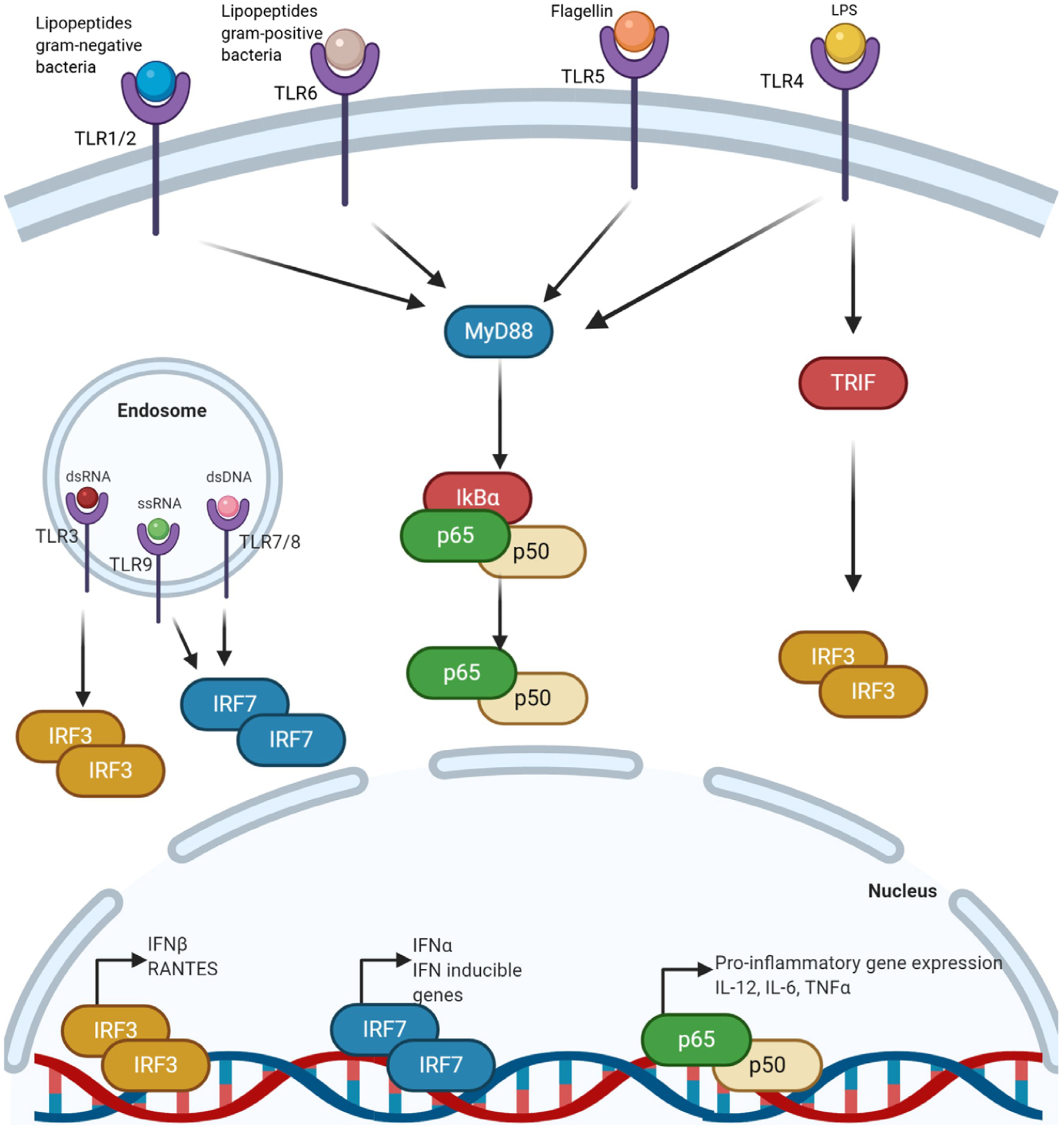
TLRs are membrane bound or endosomally bound. Membrane bound TLRs (TLR1, 2, 6, 4 and 5) signal through MyD88 in response to a variety of stimuli, such as lipopeptides from gram-negative or gram-positive bacteria, flagellin or lipopolysaccharide, and activate NF-κB-dependent pro-inflammatory genes. TLR4 can also signal through a MyD88 independent pathway and activate interferon (IFN) signalling. Endosomally bound TLRs (TLR3, 9, 7 and 8) are activated by dsRNA, ssRNA and dsDNA and activate IFN signalling in response to these stimuli. IRF, interferon regulatory factor; TRIF, Toll/IL-1 receptor domain-containing adaptor-inducing IFN-β.
Studies are emerging highlighting a novel connection between TLRs and neurodegenerative diseases, including AD, PD and MS. A link between gut microbiota-associated inflammation and brain amyloidosis in AD has been shown, with bacterial amyloids able to initiate expression of inflammatory cytokines (Nishimori et al. 2012). In AD brains, a higher bacterial LPS load was observed (Zhan et al. 2016) and administration of LPS in mice led to prolonged elevation of amyloid-β and cognitive deficits (Kahn et al. 2012). Further studies have observed increases in amyloid-β in mouse brains alongside alterations in the gut microbiota (Kaji et al. 2010). LPS-dependent TLR4 signalling is reduced in AD mice, suggesting TLR4 may play a role in disease manifestation (Go et al. 2016). In PD patients, misfolded α-synuclein protein activates microglia via TLR2, activating MyD88-dependent NF-κB signalling, in turn increasing the expression of TLRs. TLR4 also has an observable interaction with α-synuclein and a genetic knockout of TLR4 protected mice from neurodegeneration (Stefanova et al. 2011). Increased gut permeability and bacterial translocation leading to TLR4 activation in the pre-frontal cortex has been observed in mice with depressive behaviour (Martin-Hernandez et al. 2016). Finally, TLR2 expression is upregulated in both MS patients and, in the mouse model, experimental autoimmune encephalomyelitis (EAE) (Fujiwara et al. 2018). While the signalling pathways involved are not well understood, TLR2 knock-out mice develop attenuated EAE, suggesting a role for TLR2 (Fujiwara et al. 2018). Studies have found that the microbiota in MS patients is directly responsible for the dysregulation of TLR2 and its subsequent role in pathology (Wasko et al. 2020). Taken together, these studies indicate the importance of TLR signalling in development of multiple neurological disorders and present novel therapeutic strategies with which to treat them.
Our understanding of neurological disorders and the importance of the gut microbiota in their development is continually expanding, with new targets for treatment being identified. PRRs in particular can make attractive targets due to their function as the first line of defence against pathogens and their dysregulation in many disease pathologies (Mullen et al. 2015). Targeting of TLR2 signalling by increased TLR2 tolerance in a mouse model of MS significantly enhanced CNS remyelination (Wasko et al. 2019). A loss-of-function mutation in the TLR4 gene was shown to suppress the activation of microglia and monocytes by Alzheimer’s amyloid peptides in in vitro studies (Walter et al. 2007). Melatonin treatment was beneficial in attenuating NLRP3 inflammasome activation following LPS-induced depressive-like behaviours, in part via reduced microglia activation (Arioz et al. 2019). Further studies of the mechanisms these PRRs regulate are needed to identify more targets in the treatment and prevention of many neurodegenerative and neurodevelopmental disorders.
PRRs in other diseases
As well as their role in neurological disorders, PRRs have been implicated in the development of other disorders, including autoimmune diseases. Polymorphisms of both Nod1 and Nod2 have been associated with an increased susceptibility to Guillain–Barre syndrome, an autoimmune disorder that attacks the peripheral nervous system (Kharwar et al. 2016). Furthermore, increased expression of Nod1 and Nod2 has been noted in the pathogenesis of Vogt–Koyanagi–Harada syndrome, a rare autoimmune disorder (Deng et al. 2016). Additionally, polymorphisms in the NLR family increase the risk of developing IBD, with Nod1 and Nod2 deficient mice displaying an increase in DSS-induced colitis severity (Natividad et al. 2012) and Nod2 mutations correlating with dysbiosis in IBD patients (Aschard et al. 2019). A single nucleotide polymorphism (SNP) located on chromosome 1q44 downstream of NLRP3 has previously been implicated in increased susceptibility to Crohn’s disease (CD) (Villani et al. 2009), but more recent studies in a Chinese Han patient population (Zhang et al. 2014) and a panel of patients in the UK (Lewis et al. 2011) indicate that SNPs in the NLRP3 gene are more closely associated with ulcerative colitis (UC) than with CD. Despite this, loss-of-function CARD8 mutation in CD patients leads to increased NLRP3 activation, indicating that there may be a role for NRLP3 in CD pathogenesis (Schoultz et al. 2009; Mao et al. 2018). NLRP3−/− mice displayed higher sensitivity to oxazolone-induced colitis, indicating a protective role for the inflammasome in UC (Itani et al. 2016).
In the case of the TLRs, receptor activation and subsequent NF-κB signalling in the gut is important for the survival of enteric neurons responsible for gut motility. Knockout mouse models indicate that TLR4 is important for gut motility, with delayed GI motility associated with decreased numbers of nitrergic neurons (Anitha et al. 2012). Preliminary studies suggest a role for TLR4 in the development of multiple system atrophy (MSA), where, similar to PD, MSA patients were found to have disrupted tight junction proteins and a higher expression of TLR4 in their colonic sigmoid mucosa when compared to healthy controls (Engen et al. 2017). Taken together, these findings demonstrate the critical role NLRs play in multiple disease pathways, highlighting their potential in maintaining normal physiology.
Conclusions
PRRs, in particular the NLR and TLR families, have been implicated as novel signalling mechanisms in the development of many complex neurological disorders, likely acting in concert with numerous other signalling pathways. Their high levels of expression in many tissues, in particular the GI tract, make them an attractive target for further study of the gut microbiota and its impact on human health and development of gut–brain function.
Funding
This research was supported by the NIH 1R01AT009365-01 (to M.G.G.).
Biography

Ciara Keogh received her BSc in Genetics (Hons) from Dublin City University in Dublin, Ireland in 2018. During her PhD in University College Dublin, under the supervision of Dr Eoin Cummins, she worked on the effects of carbon dioxide on inflammatory signalling. She then moved to UC Davis, School of Veterinary Medicine, where she studies the role of antibiotics in the microbiota–gut–brain axis signalling in neonatal mice under the supervision of Dr Melanie Gareau.
Footnotes
Competing interests
The authors declare no competing interests.
References
- Abdulkhalek S, Amith SR, Franchuk SL, Jayanth P, Guo M, Finlay T, Gilmour A, Guzzo C, Gee K, Beyaert R & Szewczuk MR (2011). Neu1 sialidase and matrix metalloproteinase-9 cross-talk is essential for Toll-like receptor activation and cellular signaling. J Biol Chem 286, 36532–36549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akira S & Takeda K (2004). Toll-like receptor signalling. Nat Rev Immunol 4, 499–511. [DOI] [PubMed] [Google Scholar]
- Anitha M, Vijay-Kumar M, Sitaraman SV, Gewirtz AT & Srinivasan S (2012). Gut microbial products regulate murine gastrointestinal motility via Toll-like receptor 4 signaling. Gastroenterology 143, 1006–1016.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arentsen T, Qian Y, Gkotzis S, Femenia T, Wang T, Udekwu K, Forssberg H & Diaz Heijtz R (2017). The bacterial peptidoglycan-sensing molecule Pglyrp2 modulates brain development and behavior. Mol Psychiatry 22, 257–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arioz BI, Tastan B, Tarakcioglu E, Tufekci KU, Olcum M, Ersoy N, Bagriyanik A, Genc K & Genc S (2019). Melatonin attenuates LPS-induced acute depressive-like behaviors and microglial NLRP3 inflammasome activation through the SIRT1/Nrf2 pathway. Front Immunol 10, 1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschard H, Laville V, Tchetgen ET, Knights D, Imhann F, Seksik P, Zaitlen N, Silverberg MS, Cosnes J, Weersma RK, Xavier R, Beaugerie L, Skurnik D & Sokol H (2019). Genetic effects on the commensal microbiota in inflammatory bowel disease patients. PLoS Genet 15, e1008018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakrishnan B, Luckey D & Taneja V (2019). Autoimmunity-associated gut commensals modulate gut permeability and immunity in humanized mice. Mil Med 184, 529–536. [DOI] [PubMed] [Google Scholar]
- Baquero F & Nombela C (2012). The microbiome as a human organ. Clin Microbiol Infect 18 2–4. [DOI] [PubMed] [Google Scholar]
- Barrat FJ, Meeker T, Gregorio J, Chan JH, Uematsu S, Akira S, Chang B, Duramad O & Coffman RL (2005). Nucleic acids of mammalian origin can act as endogenous ligands for Toll-like receptors and may promote systemic lupus erythematosus. J Exp Med 202, 1131–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bersch K, DeMeester K, Zagani R, Wodzanowski K, Reinecker H-C & Grimes C (2020). Bacterial peptidoglycan fragments differentially regulate innate immune signaling. bioRxiv, 10.1101/2020.09.03.278705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone DL, Turer EE, Lee EG, Ahmad R-C, Wheeler MT, Tsui C, Hurley P, Chien M, Chai S, Hitotsumatsu O, McNally E, Pickart C & Ma A (2004). The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat Immunol 5, 1052–1060. [DOI] [PubMed] [Google Scholar]
- Braniste V, Al-Asmakh M, Kowal C, Anuar F, Abbaspour A, Toth M, Korecka A, Bakocevic N, Ng LG, Kundu P, Gulyas B, Halldin C, Hultenby K, Nilsson H, Hebert H, Volpe BT, Diamond B & Pettersson S (2014). The gut microbiota influences blood-brain barrier permeability in mice. Sci Transl Med 6, 263ra158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel BL, Waubant E, Chehoud C, Kuczynski J, DeSantis TZ, Warrington J, Venkatesan A, Fraser CM & Mowry EM (2015). Gut microbiota in multiple sclerosis: possible influence of immunomodulators. J Investig Med 63, 729–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruso R, Warner N, Inohara N & Nunez G (2014). NOD1 and NOD2: signaling, host defense, and inflammatory disease. Immunity 41, 898–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalcante P, Barzago C, Baggi F, Antozzi C, Maggi L, Mantegazza R & Bernasconi P (2018). Toll-like receptors 7 and 9 in myasthenia gravis thymus: amplifiers of autoimmunity? Ann N Y Acad Sci 1413, 11–24. [DOI] [PubMed] [Google Scholar]
- Chatterjee K, Roy A, Banerjee R, Choudhury S, Mondal B, Halder S, Basu P, Shubham S, Dey S & Kumar H (2020). Inflammasome and alpha-synuclein in Parkinson’s disease: A cross-sectional study. J Neuroimmunol 338, 577089. [DOI] [PubMed] [Google Scholar]
- Chuang TH & Ulevitch RJ (2004). Triad3A, an E3 ubiquitin-protein ligase regulating Toll-like receptors. Nat Immunol 5, 495–502. [DOI] [PubMed] [Google Scholar]
- Claes AK, Zhou JY & Philpott DJ (2015). NOD-like receptors: guardians of intestinal mucosal barriers. Physiology 30, 241–250. [DOI] [PubMed] [Google Scholar]
- Clarke TB, Davis KM, Lysenko ES, Zhou AY, Yu Y & Weiser JN (2010). Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat Med 16, 228–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutermarsh-Ott S, Eden K & Allen IC (2016). Beyond the inflammasome: regulatory NOD-like receptor modulation of the host immune response following virus exposure. J Gen Virol 97, 825–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CD (2016). The gut microbiome and its role in obesity. Nutr Today 51, 167–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng B, Ye Z, Li L, Zhang D, Zhu Y, He Y, Wang C, Wu L, Kijlstra A & Yang P (2016). Higher expression of NOD1 and NOD2 is associated with Vogt-Koyanagi-Harada (VKH) Syndrome but not Behcet’s Disease (BD). Curr Mol Med 16, 424–435. [DOI] [PubMed] [Google Scholar]
- Engen PA, Dodiya HB, Naqib A, Forsyth CB, Green SJ, Voigt RM, Kordower JH, Mutlu EA, Shannon KM & Keshavarzian A (2017). The potential role of gut-derived inflammation in multiple system atrophy. J Parkinsons Dis 7, 331–346. [DOI] [PubMed] [Google Scholar]
- Ewaschuk JB, Backer JL, Churchill TA, Obermeier F, Krause DO & Madsen KL (2007). Surface expression of Toll-like receptor 9 is upregulated on intestinal epithelial cells in response to pathogenic bacterial DNA. Infect Immun 75, 2572–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawkner-Corbett D, Simmons A & Parikh K (2017). Microbiome, pattern recognition receptor function in health and inflammation. Best Pract Res Clin Gastroenterol 31, 683–691. [DOI] [PubMed] [Google Scholar]
- Fitzgerald KA, Rowe DC, Barnes BJ, Caffrey DR, Visintin A, Latz E, Monks B, Pitha PM & Golenbock DT (2003). LPS-TLR4 signaling to IRF-3/7 and NF-κB involves the toll adapters TRAM and TRIF. J Exp Med 198, 1043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchi L, Warner N, Viani K & Nunez G (2009). Function of Nod-like receptors in microbial recognition and host defense. Immunol Rev 227, 106–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara M, Anstadt EJ, Flynn B, Morse K, Ng C, Paczkowski P, Zhou J, Mackay S, Wasko N, Nichols F & Clark RB (2018). Enhanced TLR2 responses in multiple sclerosis. Clin Exp Immunol 193, 313–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett WS, Gordon JI & Glimcher LH (2010). Homeostasis and inflammation in the intestine. Cell 140, 859–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gewirtz AT, Navas TA, Lyons S, Godowski PJ & Madara JL (2001). Cutting edge: bacterial flagellin activates basolaterally expressed TLR5 to induce epithelial proinflammatory gene expression. J Immunol 167, 1882–1885. [DOI] [PubMed] [Google Scholar]
- Girardin SE, Tournebize R, Mavris M, Page AL, Li X, Stark GR, Bertin J, DiStefano PS, Yaniv M, Sansonetti PJ & Philpott DJ (2001). CARD4/Nod1 mediates NF-κB and JNK activation by invasive Shigella flexneri. EMBO Rep 2, 736–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go M, Kou J, Lim JE, Yang J & Fukuchi KI (2016). Microglial response to LPS increases in wild-type mice during aging but diminishes in an Alzheimer’s mouse model: Implication of TLR4 signaling in disease progression. Biochem Biophys Res Commun 479, 331–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon R, Albornoz EA, Christie DC, Langley MR, Kumar V, Mantovani S, Robertson AAB, Butler MS, Rowe DB, O’Neill LA, Kanthasamy AG, Schroder K, Cooper MA & Woodruff TM (2018). Inflammasome inhibition prevents α-synuclein pathology and dopaminergic neurodegeneration in mice. Sci Transl Med 10, eaah4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guedes RP, Csizmadia E, Moll HP, Ma A, Ferran C & da Silva CG (2014). A20 deficiency causes spontaneous neuroinflammation in mice. J Neuroinflammation 11, 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiippala K, Jouhten H, Ronkainen A, Hartikainen A, Kainulainen V, Jalanka J & Satokari R (2018). The potential of gut commensals in reinforcing intestinal barrier function and alleviating inflammation. Nutrients 10, 988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes HK, Rose D & Ashwood P (2018). The gut microbiota and dysbiosis in autism spectrum disorders. Curr Neurol Neurosci Rep 18, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iavarone C, Ramsauer K, Kubarenko AV, Debasitis JC, Leykin I, Weber AN, Siggs OM, Beutler B, Zhang P, Otten G, D’Oro U, Valiante NM, Mbow ML & Visintin A (2011). A point mutation in the amino terminus of TLR7 abolishes signaling without affecting ligand binding. J Immunol 186, 4213–4222. [DOI] [PubMed] [Google Scholar]
- Inserra A, Rogers GB, Licinio J & Wong ML (2018). The microbiota-inflammasome hypothesis of major depression. Bioessays 40, 1800027. [DOI] [PubMed] [Google Scholar]
- Itani S, Watanabe T, Nadatani Y, Sugimura N, Shimada S, Takeda S, Otani K, Hosomi S, Nagami Y, Tanaka F, Kamata N, Yamagami H, Tanigawa T, Shiba M, Tominaga K, Fujiwara Y & Arakawa T (2016). NLRP3 inflammasome has a protective effect against oxazolone-induced colitis: a possible role in ulcerative colitis. Sci Rep 6, 39075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang T, Liu B, Li J, Dong X, Lin M, Zhang M, Zhao J, Dai Y & Chen L (2018). Association between sn-2 fatty acid profiles of breast milk and development of the infant intestinal microbiome. Food Funct 9, 1028–1037. [DOI] [PubMed] [Google Scholar]
- Kahn MS, Kranjac D, Alonzo CA, Haase JH, Cedillos RO, McLinden KA, Boehm GW & Chumley MJ (2012). Prolonged elevation in hippocampal Aβ and cognitive deficits following repeated endotoxin exposure in the mouse. Behav Brain Res 229, 176–184. [DOI] [PubMed] [Google Scholar]
- Kaji R, Kiyoshima-Shibata J, Nagaoka M, Nanno M & Shida K (2010). Bacterial teichoic acids reverse predominant IL-12 production induced by certain lactobacillus strains into predominant IL-10 production via TLR2-dependent ERK activation in macrophages. J Immunol 184, 3505–3513. [DOI] [PubMed] [Google Scholar]
- Kang S, Denman SE, Morrison M, Yu Z, Dore J, Leclerc M & McSweeney CS (2010). Dysbiosis of fecal microbiota in Crohn’s disease patients as revealed by a custom phylogenetic microarray. Inflamm Bowel Dis 16, 2034–2042. [DOI] [PubMed] [Google Scholar]
- Kharwar NK, Prasad KN, Paliwal VK & Modi DR (2016). Association of NOD1 and NOD2 polymorphisms with Guillain-Barre syndrome in Northern Indian population. J Neurol Sci 363, 57–62. [DOI] [PubMed] [Google Scholar]
- Kim YG, Park JH, Shaw MH, Franchi L, Inohara N & Nunez G (2008). The cytosolic sensors Nod1 and Nod2 are critical for bacterial recognition and host defense after exposure to Toll-like receptor ligands. Immunity 28, 246–257. [DOI] [PubMed] [Google Scholar]
- Kinsella S, Fichtner M, Watters O, Konig HG & Prehn JHM (2018). Increased A20-E3 ubiquitin ligase interactions in bid-deficient glia attenuate TLR3- and TLR4-induced inflammation. J Neuroinflammation 15, 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku HJ, Kim YT & Lee JH (2020). Microbiome study of initial gut microbiota from newborn infants to children reveals that diet determines its compositional development. J Microbiol Biotechnol 30, 1067–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc JG, Chain F, Martin R, Bermudez-Humaran LG, Courau S & Langella P (2017). Beneficial effects on host energy metabolism of short-chain fatty acids and vitamins produced by commensal and probiotic bacteria. Microb Cell Fact 16, 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis GJ, Massey DC, Zhang H, Bredin F, Tremelling M, Lee JC, Berzuini C & Parkes M (2011). Genetic association between NLRP3 variants and Crohn’s disease does not replicate in a large UK panel. Inflamm Bowel Dis 17, 1387–1391. [DOI] [PubMed] [Google Scholar]
- Li Q, Liu S, Zhu X, Mi W, Maoying Q, Wang J, Yu J & Wang Y (2019). Hippocampal PKR/NLRP1 inflammasome pathway is required for the depression-like behaviors in rats with neuropathic pain. Neuroscience 412, 16–28. [DOI] [PubMed] [Google Scholar]
- Liu F, Li J, Guan Y, Lou Y, Chen H, Xu M, Deng D, Chen J, Ni B, Zhao L, Li H, Sang H & Cai X (2019). Dysbiosis of the gut microbiome is associated with tumor biomarkers in lung cancer. Int J Biol Sci 15, 2381–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zou Q, Zeng B, Fang Y & Wei H (2013). Analysis of fecal Lactobacillus community structure in patients with early rheumatoid arthritis. Curr Microbiol 67, 170–176. [DOI] [PubMed] [Google Scholar]
- Lu J, Synowiec S, Lu L, Yu Y, Bretherick T, Takada S, Yarnykh V, Caplan J, Caplan M, Claud EC & Drobyshevsky A (2018). Microbiota influence the development of the brain and behaviors in C57BL/6J mice. PLoS One 13, e0201829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC & Finlay BB (2007). Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe 2, 119–129. [DOI] [PubMed] [Google Scholar]
- Mao L, Kitani A, Similuk M, Oler AJ, Albenberg L, Kelsen J, Aktay A, Quezado M, Yao M, Montgomery-Recht K, Fuss IJ & Strober W (2018). Loss-of-function CARD8 mutation causes NLRP3 inflammasome activation and Crohn’s disease. J Clin Invest 128, 1793–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Hernandez D, Caso JR, Bris AG, Maus SR, Madrigal JL, Garcia-Bueno B, MacDowell KS, Alou L, Gomez-Lus ML & Leza JC (2016). Bacterial translocation affects intracellular neuroinflammatory pathways in a depression-like model in rats. Neuropharmacology 103, 122–133. [DOI] [PubMed] [Google Scholar]
- Mullen LM, Chamberlain G & Sacre S (2015). Pattern recognition receptors as potential therapeutic targets in inflammatory rheumatic disease. Arthritis Res Ther 17, 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzio M, Bosisio D, Polentarutti N, D’Amico G, Stoppacciaro A, Mancinelli R, van’t Veer C, Penton-Rol G, Ruco LP, Allavena P & Mantovani A (2000). Differential expression and regulation of toll-like receptors (TLR) in human leukocytes: selective expression of TLR3 in dendritic cells. J Immunol 164, 5998–6004. [DOI] [PubMed] [Google Scholar]
- Natividad JM, Petit V, Huang X, de Palma G, Jury J, Sanz Y, Philpott D, Garcia Rodenas CL, McCoy KD & Verdu EF (2012). Commensal and probiotic bacteria influence intestinal barrier function and susceptibility to colitis in Nod1−/−;Nod2−/− mice. Inflamm Bowel Dis 18, 1434–1446. [DOI] [PubMed] [Google Scholar]
- Nishimori JH, Newman TN, Oppong GO, Rapsinski GJ, Yen JH, Biesecker SG, Wilson RP, Butler BP, Winter MG, Tsolis RM, Ganea D & Tukel C (2012). Microbial amyloids induce interleukin 17A (IL-17A) and IL-22 responses via Toll-like receptor 2 activation in the intestinal mucosa. Infect Immun 80, 4398–4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura Y, Lala S, Xin W, Smith E, Dowds TA, Chen FF, Zimmermann E, Tretiakova M, Cho JH, Hart J, Greenson JK, Keshav S & Nunez G (2003). Expression of NOD2 in Paneth cells: a possible link to Crohn’s ileitis. Gut 52, 1591–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontillo A, Catamo E, Arosio B, Mari D & Crovella S (2012). NALP1/NLRP1 genetic variants are associated with Alzheimer disease. Alzheimer Dis Assoc Disord 26, 277–281. [DOI] [PubMed] [Google Scholar]
- Pusceddu MM, Barboza M, Keogh CE, Schneider M, Stokes P, Sladek JA, Kim HJD, Torres-Fuentes C, Goldfild LR, Gillis SE, Brust-Mascher I, Rabasa G, Wong KA, Lebrilla C, Byndloss MX, Maisonneuve C, Baumler AJ, Philpott DJ, Ferrero RL, Barrett KE, Reardon C & Gareau MG (2019). Nod-like receptors are critical for gut-brain axis signalling in mice. J Physiol 597, 5777–5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner F & Schmitz ML (2009). Autoregulatory feedback loops terminating the NF-κB response. Trends Biochem Sci 34, 128–135. [DOI] [PubMed] [Google Scholar]
- Reynolds JM, Pappu BP, Peng J, Martinez GJ, Zhang Y, Chung Y, Ma L, Yang XO, Nurieva RI, Tian Q & Dong C (2010). Toll-like receptor 2 signaling in CD4+ T lymphocytes promotes T helper 17 responses and regulates the pathogenesis of autoimmune disease. Immunity 32, 692–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round JL & Mazmanian SK (2009). The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol 9, 313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubino SJ, Geddes K, Magalhaes JG, Streutker C, Philpott DJ & Girardin SE (2013). Constitutive induction of intestinal Tc17 cells in the absence of hematopoietic cell-specific MHC class II expression. Eur J Immunol 43, 2896–2906. [DOI] [PubMed] [Google Scholar]
- Sampson TR, Debelius JW, Thron T, Janssen S, Shastri GG, Ilhan ZE, Challis C, Schretter CE, Rocha S, Gradinaru V, Chesselet MF, Keshavarzian A, Shannon KM, Krajmalnik-Brown R, Wittung-Stafshede P, Knight R & Mazmanian SK (2016). Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson’s Disease. Cell 167, 1469–1480.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoultz I, Verma D, Halfvarsson J, Torkvist L, Fredrikson M, Sjoqvist U, Lordal M, Tysk C, Lerm M, Soderkvist P & Soderholm JD (2009). Combined polymorphisms in genes encoding the inflammasome components NALP3 and CARD8 confer susceptibility to Crohn’s disease in Swedish men. Am J Gastroenterol 104, 1180–1188. [DOI] [PubMed] [Google Scholar]
- Schwarzer M, Hermanova P, Srutkova D, Golias J, Hudcovic T, Zwicker C, Sinkora M, Akgun J, Wiedermann U, Tuckova L, Kozakova H & Schabussova I (2019). Germ-free mice exhibit mast cells with impaired functionality and gut homing and do not develop food allergy. Front Immunol 10, 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott ML, Fujita T, Liou HC, Nolan GP & Baltimore D (1993). The p65 subunit of NF-κB regulates IκB by two distinct mechanisms. Genes Dev 7, 1266–1276. [DOI] [PubMed] [Google Scholar]
- Shembade N, Ma A & Harhaj EW (2010). Inhibition of NF-κB signaling by A20 through disruption of ubiquitin enzyme complexes. Science 327, 1135–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanova N, Fellner L, Reindl M, Masliah E, Poewe W & Wenning GK (2011). Toll-like receptor 4 promotes α-synuclein clearance and survival of nigral dopaminergic neurons. Am J Pathol 179, 954–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez-Farinas M, Arbeit R, Jiang W, Ortenzio FS, Sullivan T & Krueger JG (2013). Suppression of molecular inflammatory pathways by Toll-like receptor 7, 8, and 9 antagonists in a model of IL-23-induced skin inflammation. PLoS One 8, e84634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Duffy KE, Ranjith-Kumar CT, Xiong J, Lamb RJ, Santos J, Masarapu H, Cunningham M, Holzenburg A, Sarisky RT, Mbow ML & Kao C (2006). Structural and functional analyses of the human Toll-like receptor 3. Role of glycosylation. J Biol Chem 281, 11144–11151. [DOI] [PubMed] [Google Scholar]
- Sun MF & Shen YQ (2018). Dysbiosis of gut microbiota and microbial metabolites in Parkinson’s Disease. Ageing Res Rev 45, 53–61. [DOI] [PubMed] [Google Scholar]
- Takeuchi O & Akira S (2001). Toll-like receptors; their physiological role and signal transduction system. Int Immunopharmacol 1, 625–635. [DOI] [PubMed] [Google Scholar]
- Tan MS, Tan L, Jiang T, Zhu XC, Wang HF, Jia CD & Yu JT (2014). Amyloid-β induces NLRP1-dependent neuronal pyroptosis in models of Alzheimer’s disease. Cell Death Dis 5, e1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist PH, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J & Ponten F (2015). Tissue-based map of the human proteome. Science 347, 1260419. [DOI] [PubMed] [Google Scholar]
- Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, Ley R, Wakeland EK & Hooper LV (2011). The antibacterial lectin RegIIIγ promotes the spatial segregation of microbiota and host in the intestine. Science 334, 255–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villani AC, Lemire M, Fortin G, Louis E, Silverberg MS, Collette C, Baba N, Libioulle C, Belaiche J, Bitton A, Gaudet D, Cohen A, Langelier D, Fortin PR, Wither JE, Sarfati M, Rutgeerts P, Rioux JD, Vermeire S, Hudson TJ & Franchimont D (2009). Common variants in the NLRP3 region contribute to Crohn’s disease susceptibility. Nat Genet 41, 71–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh D, McCarthy J, O’Driscoll C & Melgar S (2013). Pattern recognition receptors—molecular orchestrators of inflammation in inflammatory Bowel disease. Cytokine Growth Factor Rev 24, 91–104. [DOI] [PubMed] [Google Scholar]
- Walter S, Letiembre M, Liu Y, Heine H, Penke B, Hao W, Bode B, Manietta N, Walter J, Schulz-Schuffer W & Fassbender K (2007). Role of the toll-like receptor 4 in neuroinflammation in Alzheimer’s disease. Cell Physiol Biochem 20, 947–956. [DOI] [PubMed] [Google Scholar]
- Wasko NJ, Kulak MH, Paul D, Nicaise AM, Yeung ST, Nichols FC, Khanna KM, Crocker S, Pachter JS & Clark RB (2019). Systemic TLR2 tolerance enhances central nervous system remyelination. J Neuroinflammation 16, 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasko NJ, Nichols F & Clark RB (2020). Multiple sclerosis, the microbiome, TLR2, and the hygiene hypothesis. Autoimmun Rev 19, 102430. [DOI] [PubMed] [Google Scholar]
- Weber AN, Morse MA & Gay NJ (2004). Four N-linked glycosylation sites in human toll-like receptor 2 cooperate to direct efficient biosynthesis and secretion. J Biol Chem 279, 34589–34594. [DOI] [PubMed] [Google Scholar]
- Zhan X, Stamova B, Jin LW, DeCarli C, Phinney B & Sharp FR (2016). Gram-negative bacterial molecules associate with Alzheimer disease pathology. Neurology 87, 2324–2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HX, Wang ZT, Lu XX, Wang YG, Zhong J & Liu J (2014). NLRP3 gene is associated with ulcerative colitis (UC), but not Crohn’s disease (CD), in Chinese Han population. Inflamm Res 63, 979–985. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Lu M, Du RH, Qiao C, Jiang CY, Zhang KZ, Ding JH & Hu G (2016). MicroRNA-7 targets Nod-like receptor protein 3 inflammasome to modulate neuroinflammation in the pathogenesis of Parkinson’s disease. Mol Neurodegener 11, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang H, Cheng L, Wang Y, Zhang YK, Zhao MF, Liang GD, Zhang MC, Li YG, Zhao JB, Gao YN, Zhou YJ & Liu SL (2019a). Dysbiosis of the gut microbiome in lung cancer. Front Cell Infect Microbiol 9, 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang L, Chen H, Zhang S, Zhuang J, Li Q & Feng Z (2019b). Intestinal microbiota in early life and its implications on childhood health. Genomics Proteomics Bioinformatics 17, 13–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler G, Freyer D, Harhausen D, Khojasteh U, Nietfeld W & Trendelenburg G (2011). Blocking TLR2 in vivo protects against accumulation of inflammatory cells and neuronal injury in experimental stroke. J Cereb Blood Flow Metab 31, 757–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilbauer M, Dorrell N, Elmi A, Lindley KJ, Schuller S, Jones HE, Klein NJ, Nunez G, Wren BW & Bajaj-Elliott M (2007). A major role for intestinal epithelial nucleotide oligomerization domain 1 (NOD1) in eliciting host bactericidal immune responses to Campylobacter jejuni. Cell Microbiol 9, 2404–2416. [DOI] [PubMed] [Google Scholar]


