Abstract
Hepcidin is a peptide hormone that targets the iron exporter ferroportin, thereby limiting iron entry into the bloodstream. It is generated in hepatocytes mainly in response to increased body iron stores or inflammatory cues. Iron stimulates expression of bone morphogenetic protein 6 (BMP6) from liver sinusoidal endothelial cells, which in turn binds to BMP receptors on hepatocytes and induces the SMAD signaling cascade for transcriptional activation of the hepcidin-encoding HAMP mRNA. SMAD signaling is also essential for inflammatory HAMP mRNA induction by the IL-6/STAT3 pathway. Herein, we utilized human Huh7 hepatoma cells and primary murine hepatocytes to assess the effects of iron perturbations on signaling to hepcidin. Iron chelation appeared to slightly impair signaling to hepcidin. Subsequent iron supplementation not only failed to reverse these effects, but drastically reduced basal HAMP mRNA and inhibited HAMP mRNA induction by BMP6 and/or IL-6. Thus, treatment of cells with excess iron inhibited basal and BMP6-mediated SMAD5 phosphorylation and induction of HAMP, ID1 and SMAD7 mRNAs in a dose-dependent manner. Iron also inhibited IL-6-mediated STAT3 phosphorylation and induction of HAMP and SOCS3 mRNAs. These responses were accompanied by induction of GCLC and HMOX1 mRNAs, known markers of oxidative stress. We conclude that hepatocellular iron overload suppresses hepcidin by inhibiting the SMAD and STAT3 signaling pathways downstream of their respective ligands.
Introduction
Hepcidin is a hormonal regulator of systemic iron traffic [1, 2]. It is highly expressed in hepatocytes as pre-pro-hepcidin, which undergoes proteolytic processing to yield a mature peptide of 25 amino acids. Hepcidin operates by binding to the iron exporter ferroportin in target cells, such as tissue macrophages, duodenal enterocytes and hepatocytes. The binding of hepcidin directly inhibits iron efflux from ferroportin into plasma, and also targets ferroportin for lysosomal degradation. Hepcidin-mediated inhibition of iron entry into plasma serves as a homeostatic response to increased body iron stores, or as an innate immune response to infection [3, 4].
Expression of hepcidin-encoding HAMP mRNA is primarily induced in response to iron or inflammatory signals. The underlying mechanisms are fairly well characterized; however, critical aspects of iron sensing remain elusive [2, 5]. Hepatic iron loading triggers expression and release of bone morphogenetic protein 6 (BMP6) from liver sinusoidal endothelial cells [6]; these cells also constitutively express BMP2 [7]. BMP6 binds as homodimer to ALK2 and as heterodimer with BMP2 to ALK3 [8], which are type I BMP receptors on hepatocytes. Binding of the ligand activates SMAD signaling via a mechanism that is enhanced by hemojuvelin (HJV), a BMP co-receptor and other auxiliary factors. The signaling cascade involves phosphorylation of SMAD1/5/8, recruitment of SMAD4 and formation of a complex that translocates to the nucleus for binding to the HAMP promoter and activation of HAMP mRNA transcription.
The inflammatory cytokine IL-6 is another potent inducer of hepcidin. The binding of IL-6 to its receptor on hepatocytes triggers phosphorylation of STAT3 by JAK1/2 kinases, which in turn translocates to the nucleus to transcriptionally induce hepcidin [9, 10]. It is well established that this pathway requires active BMP/SMAD signaling [11, 12].
Treatment of hepatoma cells or primary murine hepatocytes with BMP6, BMP2 or IL-6 recapitulates hepcidin induction [8, 12–14]. However, while dietary, genetic or pharmacological iron loading readily induce hepcidin expression in vivo [15–17], exposure of cells to iron-loaded transferrin or iron salts was shown to rather suppress hepcidin [14, 18, 19]. Herein, we explore the mechanisms by which iron affects hepcidin expression in cultured cells. We demonstrate that iron loading suppresses HAMP mRNA levels by inhibiting basal SMAD signaling. Moreover, excess iron inhibits ligand-induced SMAD and STAT3 signaling and antagonizes HAMP mRNA induction by BMP6 or IL-6.
Materials and methods
Cell culture
Human Huh7 hepatoma cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% heat-inactivated fetal bovine serum (Wisent), non-essential amino acids, 100 U/ml penicillin and 100 μg/ml streptomycin. Primary hepatocytes were isolated from livers of C57BL/6 wild type mice (22–25 g) by using a two-step collagenase perfusion technique as previously described [12]. The experimental procedure was performed at the animal facility of the Lady Davis Institute for Medical Research and was approved by the Animal Care Committee of McGill University (protocol 4966). The cells were seeded onto tissue culture dishes (26,000 cells/cm2) in Williams’ medium E (Gibco) supplemented with 10% heat-inactivated fetal bovine serum, 2 mM L-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, and allowed 90 min to attach before washing and overnight culture. The serum-containing medium was removed the next morning, and the cells were subjected to different culture conditions in serum-free Williams’ medium E. In control groups, cells were incubated with medium alone for the indicated time of the experiment.
Materials
Deferoxamine (DFO), FeSO4 and ferric ammonium citrate (FAC) were purchased from Sigma-Aldrich. Following recombinant proteins were used: murine IL-6 (#5216, Cell Signaling), human IL-6 (#I1395, Sigma-Aldrich), human BMP6 (#507-BP, R&D Systems), and human holo-transferrin (#T4132, Sigma). Salicylaldehyde isonicotinoyl hydrazone (SIH) was a gift of the late Dr. Prem Ponka. Fe-SIH was prepared by mixing SIH with ferric citrate in 2:1 ratio [20].
Quantitative real-time PCR (qPCR)
The cells were washed twice with phosphate-buffered-saline and RNA was extracted by using the RNeasy kit (Qiagen). cDNA was synthesized from 1 μg RNA by using the OneScript cDNA Synthesis Kit (ABM). Gene-specific primers pairs (S1 Table) were validated by dissociation curve analysis and demonstrated amplification efficiency between 90–110%. SYBR Green (Bioline) and primers were used to amplify products under following cycling conditions: initial denaturation 95°C 10 min, 40 cycles of 95°C 5 s, 58°C 30 s, 72°C 10 s, and final cycle melt analysis between 58°-95°C. Relative mRNA expression was calculated by the comparative Ct method. Data were normalized to human ribosomal protein RPS18 for Huh7 cells or murine Rpl19 for primary murine hepatocytes. Data are reported as fold changes compared to untreated control cells. Original Ct values can be found in S1 Dataset.
Western blotting
Following 2x wash with phosphate-buffered-saline, the cells were lysed as previously described [21]. Cell lysates containing 30 μg of proteins were analyzed by SDS-PAGE on 10% gels and proteins were transferred onto nitrocellulose membranes (BioRad). The blots were blocked in 10% bovine serum albumin or 10% fat-free skim milk in tris-buffered saline (TBS) containing 0.1% (v/v) Tween-20 (TBS-T), and probed overnight with 1:1000-diluted (unless otherwise indicated) primary antibodies against pSMAD5 (phospho-S463/465, #92698 rabbit monoclonal; Abcam), SMAD1 (#9743 rabbit polyclonal; Cell Signaling), pSTAT3 (phospho-Y705, #9138, mouse monoclonal; Cell Signaling), STAT3 (#9139, mouse monoclonal; Cell Signaling), ferritin heavy chain (1:500-diluted, #NB600-920, Novus), or β-actin (Sigma). Following a 3x wash with TBS-T, the membranes were incubated with 1:5000-diluted anti-mouse or 1:20000-diluted anti-rabbit peroxidase-coupled secondary antibodies for 2 h. Immunoreactive bands were detected by enhanced chemiluminescence with the Western Lightning ECL Kit (Perkin Elmer). Original Western blot images can be found in S1 Raw images.
Statistical analysis
Statistical analysis was performed by using the Prism GraphPad software (version 7.0e). Multiple groups were subjected to analysis of variance (ANOVA) with Tukey’s post-test comparison (one-way ANOVA) or Sidak’s multiple comparison (two-way ANOVA). A probability value p<0.05 was considered statistically significant.
Results and discussion
Iron overload abrogates BMP/SMAD and IL-6/STAT signaling to hepcidin in Huh7 cells
We have reported that iron-depleted mice fail to mount an appropriate inflammatory induction of hepcidin and this response can be restored by iron supplementation [12]. This prompted us to evaluate the potential role of hepatocellular iron status on hepcidin regulation. Human Huh7 hepatoma cells were treated with IL-6 and/or BMP6 for 4 h following pre-treatment (or not) with the iron chelator deferoxamine (DFO) for 18 h. The conditions for IL-6 and BMP6 treatments in terms of dose and duration were optimized in previous experiments [12]. Likewise, conditions for iron chelation were based on previous literature [22, 23]. DFO did not affect IL-6-mediated induction of HAMP mRNA or STAT3 phosphorylation but modestly mitigated BMP6-mediated HAMP mRNA induction and SMAD5 phosphorylation (Fig 1A and 1B). Moreover, it negatively affected levels of ID1 mRNA (Fig 1C), a known target of BMP6/SMAD signaling [24]. Surprisingly, administration of ferric ammonium citrate (FAC) together with IL-6 and/or BMP6 (following DFO wash) did not restore but rather strongly suppressed HAMP and ID1 mRNA induction, as well as SMAD5 phosphorylation by BMP6. In addition, FAC abolished HAMP mRNA induction by IL-6. FAC is widely used as an iron donor in cell culture experiments [25, 26]. The underlying iron transport pathway is not fully understood but experiments in primary rat hepatocytes suggested a facilitated diffusion mechanism [27, 28].
Fig 1. Iron overload inhibits SMAD and STAT signaling downstream of BMP6 and IL-6 in Huh7 hepatoma cells.
Huh 7 cells were treated with 100 μM DFO for 18 hours before washing and supplementation with 50 μM FAC. After 2 hours, cells were treated with 20 ng/ml human IL-6, 5 ng/ml BMP6, or both over 4 hours. (A) qPCR analysis of HAMP mRNA. (B) Western blot analysis of pSTAT3, STAT3, pSMAD5, SMAD5, ferritin and β-actin. (C-F) qPCR analysis of ID1, TFRC, GCLC and HMOX1 mRNAs. All data in graphs are presented as the mean ± SEM from three independent experiments for (A) and two independent experiments for (B-F). Statistical analysis was performed by one-way ANOVA. Statistically significant differences (p<0.05) across iron treatments (compared to respective values from iron-unperturbed cells shown in black bars) are indicated by *.
As expected, the DFO treatment resulted in increased expression of TFRC mRNA (Fig 1D), which encodes transferrin receptor 1 (TfR1), and serves as marker of iron deficiency [29]. Furthermore, FAC administration for 6 h prevented TFRC mRNA induction by DFO, suggesting that it terminated cellular iron deficiency. Notably, under these experimental conditions, ferritin expression remained suppressed (Fig 1B), in spite of excess iron. Increased ferritin steady-state levels are only evident after longer (48 h) treatments with FAC (S1 Fig). Considering that ferritin operates by storing and detoxifying excess iron [30], the lack of ferritin induction after a relatively short (6 h) treatment with FAC (Fig 1B and S1 Fig), and the reported rapid kinetics (25x106 atoms/cell/min) of FAC uptake by hepatocytes [27] are consistent with accumulation of redox-active “free” iron in the cells that can trigger oxidative stress [31]. In support to this interpretation, the FAC treatment increased expression of GCLC mRNA (Fig 1E), an oxidative stress marker that encodes glutamate-cysteine ligase catalytic subunit [32, 33]. Moreover, FAC also induced expression of the heme oxygenase 1-encoding HMOX1 mRNA (Fig 1F), another established marker of oxidative stress [33, 34]. The above data suggest that hepatocellular iron overload acts as a potent inhibitor of HAMP mRNA induction by the BMP6/SMAD and also IL-6/STAT signaling pathways.
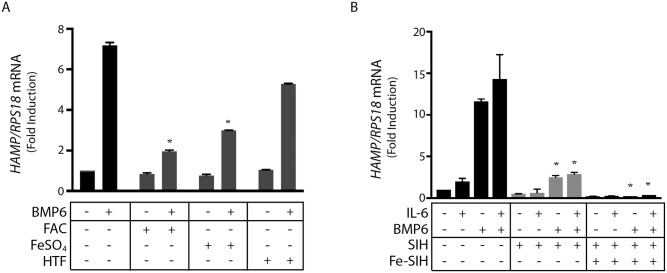
The inhibitory effects of FAC could be reproduced with FeSO4, an additional source of inorganic iron (Fig 2A). Holo-transferrin was less efficient in inhibiting BMP6-mediated HAMP mRNA induction (Fig 2A), possibly because it elicits modest iron loading compared to inorganic iron salts. Similar results with FAC and FeSO4 were obtained by using the iron donor Fe-SIH (Fig 2B), which is generated by mixing ferric citrate with the iron chelator SIH [20]. Notably, SIH inhibited induction of HAMP by either BMP6 or IL-6 (Fig 2B), even though DFO did not significantly antagonize the effects of IL-6 (Fig 1A). SIH acts fast and is a cell permeable iron chelator [35]; by contrast, DFO is a slow-acting chelator that enters cells via fluid-phase endocytosis [36].
Fig 2. Iron salts but not transferrin-bound iron inhibit hepcidin induction by BMP6 and/or IL-6 in Huh7 hepatoma cells.
(A) Huh 7 cells were pretreated with either 50 μM FAC, 50 μM FeSO4, or 30 μM holo-transferrin (HTF) for 2 hours and then treated with BMP6 for 4 hours; HAMP mRNA was measured by qPCR. (B) Huh 7 cells were treated with 100 μM SIH for 18 hours before washing and supplementation with 50 Fe-SIH. After 2 hours, cells were treated with 20 ng/ml IL-6, 5ng/ml BMP6, or both over 4 hours. HAMP mRNA was measured by qPCR. All data in graphs are presented as the mean ± SEM from three independent experiments. Statistical analysis was performed by one-way ANOVA. Statistically significant differences (p<0.05) across iron treatments (compared to respective values from iron-unperturbed cells shown in black bars) are indicated by *.
Iron inhibits basal and BMP6-mediated SMAD signaling to hepcidin in Huh7 cells in a dose-dependent manner
The SMAD pathway is a central node for signaling to hepcidin in response to iron, inflammatory stimuli and other cues. Having established that iron overload suppresses BMP6-mediated hepcidin induction in Huh7 cells, we performed dose-curve experiments to determine the minimal inhibitory iron concentrations. The data in Fig 3A show that FAC inhibits BMP6-dependent HAMP mRNA expression even at low (2–5 μM) doses. Similar results were obtained with ID1 mRNA (Fig 3B), while inhibition of BMP6-dependent SMAD5 phosphorylation was evident at 50 μM FAC (Fig 3C).
Fig 3. Dose-dependent inhibitory effects of iron on basal and BMP6-mediated SMAD signaling and hepcidin expression in Huh7 hepatoma cells.
Huh7 cells were treated with increasing doses of FAC over 2 hours. When indicated, the experiment was either terminated or the cells were washed and further incubated with 25 ng/ml BMP6 (A and C) or 5 ng/ml BMP6 (B, D-F) for 4 hours. (A and B) qPCR analysis of HAMP and ID1 mRNAs. (C) Western blot analysis of pSMAD5, SMAD5 and β-actin. (D-F) qPCR analysis of HAMP, ID1 and GCLC mRNAs. All data in graphs are presented as the mean ± SEM from three independent experiments. Statistical analysis was performed by one-way ANOVA (A, B, D and E) two-way ANOVA (F). Statistically significant differences (p<0.05) across iron treatments (compared to respective values from iron-unperturbed cells shown in black bars) are directly shown or indicated by *.
Interestingly, FAC also inhibited basal HAMP (Fig 3D) and ID1 (Fig 3E) mRNAs in a dose-dependent manner, which is indicative of inhibition in basal SMAD signaling. The data in Fig 3F show that GCLC mRNA was very sensitive to FAC treatment and was maximally induced under these experimental conditions independently of the iron dose. This further supports the notion that exposure of Huh7 cells to FAC triggers oxidative stress.
Iron-dependent inhibition of BMP/SMAD and IL-6/STAT signaling to hepcidin in primary murine hepatocytes
We used primary murine hepatocytes to address whether the sensitivity of hepcidin signaling pathways to iron perturbations is preserved in another cell model from a different species. Iron chelation by DFO did not significantly affect basal or stimulated expression of hepcidin or other markers of Bmp/Smad or IL-6/Stat signaling in this setting (Fig 4). The data in Fig 4A recapitulate the potent inhibitory effects of FAC on Hamp mRNA induction via BMP6 and/or IL-6. Moreover, FAC inhibited basal, as well as BMP6- and IL-6-dependent phosphorylation of Smad5 and Stat3, respectively (Fig 4B). In line with these findings, FAC drastically suppressed BMP6-mediated induction of Id1 (Fig 4C) and Smad7 (Fig 4D) mRNAs, both targets of Bmp/Smad signaling [24]. Additionally, FAC antagonized IL-6-mediated induction of Socs3 mRNA (Fig 4E), a target of IL-6/Stat signaling [37].
Fig 4. Iron overload inhibits Smad and Stat signaling downstream of BMP6 and IL-6 in primary murine hepatocytes.
Primary murine hepatocytes were maintained in serum-free William’s Medium E over the course of experiments. The cells were pretreated with 100 μM DFO for 18 hours and then washed and supplemented with 50 μM FAC over 2 hours. The cells were then treated with 20 ng/ml murine IL-6, 25 ng/ml BMP6, or both over 4 hours. (A) qPCR analysis of Hamp mRNA. (B) Western blotting of pSTAT3, STAT3, pSMAD5, SMAD1, ferritin, and β-actin. (C-F) qPCR analysis of Id1, Smad7, Socs3 and Tfrc mRNAs. All data in graphs are presented as the mean ± SEM from two independent experiments. Statistical analysis was performed by one-way ANOVA. Statistically significant differences (p<0.05) across iron treatments (compared to respective values from iron-unperturbed cells shown in black bars) are indicated by *.
We noted that the induction of Hamp mRNA in primary hepatocytes by BMP6 (Fig 4A) was more potent compared to the respective HAMP mRNA induction in Huh 7 cells (Fig 1A); this may be related to the different doses used, species-specific variations, and/or differences between primary hepatocytes and hepatoma cell lines. We did not examine the effects of iron perturbations on signaling to Hamp2, a second hepcidin-encoding gene that is expressed in rodents and fish, because Hamp2 does not appear to significantly affect systemic iron homeostasis [38, 39]. Furthermore, Hamp2 expression does not respond to inflammatory stimuli [40].
As expected, exposure of primary murine hepatocytes to the iron chelator DFO elicited strong induction of Tfrc mRNA, while subsequent administration of FAC neutralized this response (Fig 4F). By contrast, the FAC treatment failed to induce ferritin, or even neutralize its suppression by DFO (Fig 2B). Thus, under these experimental conditions, excess iron was apparently not scavenged and remained redox-active.
Conclusions
We show herein that pharmacological iron chelation slightly impaired hepcidin induction by BMP6 and/or IL-6 primarily in human Huh7 hepatoma cells. Nevertheless, iron supplementation completely abolished BMP6- and/or IL-6-mediated induction of hepcidin in Huh7 cells and primary murine hepatocytes. None of these treatments affected cell viability. The data in Figs 1 and 4 reveal that cellular iron inhibits both BMP/SMAD and JAK/STAT signaling pathways. Considering the relatively short time frame of the experiments that did not allow accumulation of the iron storage protein ferritin, and consequently iron detoxification, we speculate that the inhibitory effects are caused by iron-induced oxidative stress. Experimental support is provided by the induction of GCLC and HMOX1 mRNAs, markers of oxidative stress [32–34], in cells exposed to FAC (Figs 1 and 2). Our findings are consistent with the known sensitivity of both BMP/SMAD and JAK/STAT signaling pathways to oxidants [41, 42], and the previously described suppression of hepcidin in iron-loaded [14, 18, 19] and iron-loaded/IL-6-treated [43] cultured hepatic cells. Moreover, they provide evidence that the iron-mediated suppression of hepcidin in cultured cells is caused by inhibition of basal SMAD signaling. The in vivo relevance of these findings remains to be explored.
Supporting information
Huh7 cells were treated with either 100 μM DFO for 18 hours or 50 μM FAC for the indicated time intervals. Ferritin and β-actin expression in cell lysates were assessed by Western blotting. Western data are representative of two independent experiments.
(TIF)
(DOCX)
(XLSX)
(PDF)
Data Availability
All relevant data are within the manuscript and its Supporting information files.
Funding Statement
This work was supported by a grant from the Canadian Institutes of Health Research (CIHR; PJT-159730). EC was funded by a fellowship from the Natural Sciences and Engineering Research Council of Canada (NSERC) and is currently a recipient of a fellowship from the Fonds de recherche du Québec – Santé (FRQS).
References
- 1.Ganz T. Systemic iron homeostasis. Physiol Rev. 2013;93(4):1721–41. Epub 2013/10/19. doi: 10.1152/physrev.00008.2013 . [DOI] [PubMed] [Google Scholar]
- 2.Wang CY, Babitt JL. Liver iron sensing and body iron homeostasis. Blood. 2019;133(1):18–29. Epub 2018/11/08. doi: 10.1182/blood-2018-06-815894 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coffey R, Ganz T. Iron homeostasis: An anthropocentric perspective. J Biol Chem. 2017;292(31):12727–34. Epub 2017/06/16. doi: 10.1074/jbc.R117.781823 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ganz T, Nemeth E. Iron homeostasis in host defence and inflammation. Nat Rev Immunol. 2015;15(8):500–10. doi: 10.1038/nri3863 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Katsarou A, Pantopoulos K. Basics and principles of cellular and systemic iron homeostasis. Mol Aspects Med. 2020;75:100866. Epub 2020/06/23. doi: 10.1016/j.mam.2020.100866 . [DOI] [PubMed] [Google Scholar]
- 6.Canali S, Zumbrennen-Bullough KB, Core AB, Wang CY, Nairz M, Bouley R, et al. Endothelial cells produce bone morphogenetic protein 6 required for iron homeostasis in mice. Blood. 2017;129(4):405–14. doi: 10.1182/blood-2016-06-721571 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koch PS, Olsavszky V, Ulbrich F, Sticht C, Demory A, Leibing T, et al. Angiocrine Bmp2 signaling in murine liver controls normal iron homeostasis. Blood. 2017;129(4):415–9. Epub 2016/12/03. doi: 10.1182/blood-2016-07-729822 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang CY, Xu Y, Traeger L, Dogan DY, Xiao X, Steinbicker AU, et al. Erythroferrone lowers hepcidin by sequestering BMP2/6 heterodimer from binding to the BMP type I receptor ALK3. Blood. 2020;135(6):453–6. Epub 2019/12/05. doi: 10.1182/blood.2019002620 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Verga Falzacappa MV, Vujic Spasic M, Kessler R, Stolte J, Hentze MW, Muckenthaler MU. STAT3 mediates hepatic hepcidin expression and its inflammatory stimulation. Blood. 2007;109:353–8. doi: 10.1182/blood-2006-07-033969 . [DOI] [PubMed] [Google Scholar]
- 10.Pietrangelo A, Dierssen U, Valli L, Garuti C, Rump A, Corradini E, et al. STAT3 is required for IL-6-gp130-dependent activation of hepcidin in vivo. Gastroenterology. 2007;132:294–300. doi: 10.1053/j.gastro.2006.10.018 . [DOI] [PubMed] [Google Scholar]
- 11.Mayeur C, Lohmeyer LK, Leyton P, Kao SM, Pappas AE, Kolodziej SA, et al. The type I BMP receptor Alk3 is required for the induction of hepatic hepcidin gene expression by interleukin-6. Blood. 2014;123(14):2261–8. doi: 10.1182/blood-2013-02-480095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fillebeen C, Wilkinson N, Charlebois E, Katsarou A, Wagner J, Pantopoulos K. Hepcidin-mediated hypoferremic response to acute inflammation requires a threshold of Bmp6/Hjv/Smad signaling. Blood. 2018;132(17):1829–41. Epub 2018/09/15. doi: 10.1182/blood-2018-03-841197 . [DOI] [PubMed] [Google Scholar]
- 13.Andriopoulos B Jr, Corradini E, Xia Y, Faasse SA, Chen S, Grgurevic L, et al. BMP6 is a key endogenous regulator of hepcidin expression and iron metabolism. Nat Genet. 2009;41(4):482–7. Epub 2009/03/03. doi: 10.1038/ng.335 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nemeth E, Valore EV, Territo M, Schiller G, Lichtenstein A, Ganz T. Hepcidin, a putative mediator of anemia of inflammation, is a type II acute-phase protein. Blood. 2003;101:2461–3. doi: 10.1182/blood-2002-10-3235 [DOI] [PubMed] [Google Scholar]
- 15.Corradini E, Meynard D, Wu Q, Chen S, Ventura P, Pietrangelo A, et al. Serum and liver iron differently regulate the bone morphogenetic protein 6 (BMP6)-SMAD signaling pathway in mice. Hepatology. 2011;54(1):273–84. Epub 2011/04/14. doi: 10.1002/hep.24359 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramos E, Kautz L, Rodriguez R, Hansen M, Gabayan V, Ginzburg Y, et al. Evidence for distinct pathways of hepcidin regulation by acute and chronic iron loading in mice. Hepatology. 2011;53(4):1333–41. Epub 2011/04/12. doi: 10.1002/hep.24178 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daba A, Gkouvatsos K, Sebastiani G, Pantopoulos K. Differences in activation of mouse hepcidin by dietary iron and parenterally administered iron dextran: compartmentalization is critical for iron sensing. J Mol Med (Berl). 2013;91:95–102. Epub 2012/08/01. doi: 10.1007/s00109-012-0937-5 . [DOI] [PubMed] [Google Scholar]
- 18.Gehrke SG, Kulaksiz H, Herrmann T, Riedel HD, Bents K, Veltkamp C, et al. Expression of hepcidin in hereditary hemochromatosis: evidence for a regulation in response to serum transferrin saturation and non-transferrin-bound iron. Blood. 2003;102:371–6. doi: 10.1182/blood-2002-11-3610 [DOI] [PubMed] [Google Scholar]
- 19.Guttmann S, Dewald ET, Wohlfarth C, Muller JC, Karst U, Schmidt HH, et al. Compound-specific adaptation of hepatoma cell lines to toxic iron. Metallomics. 2019;11(11):1836–46. Epub 2019/09/26. doi: 10.1039/c9mt00202b . [DOI] [PubMed] [Google Scholar]
- 20.Fillebeen C, Pantopoulos K. Iron inhibits replication of infectious hepatitis C virus in permissive Huh7.5.1 cells. J Hepatol. 2010;53(6):995–9. Epub 2010/09/04. doi: 10.1016/j.jhep.2010.04.044 . [DOI] [PubMed] [Google Scholar]
- 21.Fillebeen C, Muckenthaler M, Andriopoulos B, Bisaillon M, Mounir Z, Hentze MW, et al. Expression of the subgenomic hepatitis C virus replicon alters iron homeostasis in Huh7 cells. J Hepatol. 2007;47:12–22. doi: 10.1016/j.jhep.2007.01.035 . [DOI] [PubMed] [Google Scholar]
- 22.Wang J, Chen G, Muckenthaler M, Galy B, Hentze MW, Pantopoulos K. Iron-mediated degradation of IRP2: an unexpected pathway involving a 2-oxoglutarate-dependent oxygenase activity. Mol Cell Biol. 2004;24:954–65. doi: 10.1128/MCB.24.3.954-965.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fillebeen C, Chahine D, Caltagirone A, Segal P, Pantopoulos K. A phosphomimetic mutation at Ser-138 renders iron regulatory protein 1 sensitive to iron-dependent degradation. Mol Cell Biol. 2003;23:6973–81. doi: 10.1128/MCB.23.19.6973-6981.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kautz L, Meynard D, Monnier A, Darnaud V, Bouvet R, Wang RH, et al. Iron regulates phosphorylation of Smad1/5/8 and gene expression of Bmp6, Smad7, Id1, and Atoh8 in the mouse liver. Blood. 2008;112(4):1503–9. Epub 2008/06/10. doi: 10.1182/blood-2008-03-143354 . [DOI] [PubMed] [Google Scholar]
- 25.Newman SA, Pan Y, Short JL, Nicolazzo JA. Increasing Intracellular Levels of Iron with Ferric Ammonium Citrate Leads to Reduced P-glycoprotein Expression in Human Immortalised Brain Microvascular Endothelial Cells. Pharm Res. 2021;38(1):97–111. Epub 2021/02/04. doi: 10.1007/s11095-021-03006-y . [DOI] [PubMed] [Google Scholar]
- 26.Che J, Lv H, Yang J, Zhao B, Zhou S, Yu T, et al. Iron overload induces apoptosis of osteoblast cells via eliciting ER stress-mediated mitochondrial dysfunction and p-eIF2alpha/ATF4/CHOP pathway in vitro. Cell Signal. 2021;84:110024. Epub 2021/04/27. doi: 10.1016/j.cellsig.2021.110024 . [DOI] [PubMed] [Google Scholar]
- 27.Baker E, Baker SM, Morgan EH. Characterisation of non-transferrin-bound iron (ferric citrate) uptake by rat hepatocytes in culture. Biochim Biophys Acta. 1998;1380(1):21–30. Epub 1998/04/18. doi: 10.1016/s0304-4165(97)00120-7 . [DOI] [PubMed] [Google Scholar]
- 28.Graham RM, Morgan EH, Baker E. Characterisation of citrate and iron citrate uptake by cultured rat hepatocytes. J Hepatol. 1998;29(4):603–13. Epub 1998/11/21. doi: 10.1016/s0168-8278(98)80156-6 . [DOI] [PubMed] [Google Scholar]
- 29.Wang J, Pantopoulos K. Regulation of cellular iron metabolism. Biochem J. 2011;434(3):365–81. Epub 2011/02/26. doi: 10.1042/BJ20101825 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arosio P, Levi S. Cytosolic and mitochondrial ferritins in the regulation of cellular iron homeostasis and oxidative damage. Biochim Biophys Acta. 2010;1800(8):783–92. Epub 2010/02/24. doi: 10.1016/j.bbagen.2010.02.005 . [DOI] [PubMed] [Google Scholar]
- 31.Galaris D, Barbouti A, Pantopoulos K. Iron homeostasis and oxidative stress: An intimate relationship. Biochim Biophys Acta Mol Cell Res. 2019;1866(12):118535. Epub 2019/08/26. doi: 10.1016/j.bbamcr.2019.118535 . [DOI] [PubMed] [Google Scholar]
- 32.Krejsa CM, Franklin CC, White CC, Ledbetter JA, Schieven GL, Kavanagh TJ. Rapid activation of glutamate cysteine ligase following oxidative stress. J Biol Chem. 2010;285(21):16116–24. Epub 2010/03/25. doi: 10.1074/jbc.M110.116210 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loboda A, Damulewicz M, Pyza E, Jozkowicz A, Dulak J. Role of Nrf2/HO-1 system in development, oxidative stress response and diseases: an evolutionarily conserved mechanism. Cell Mol Life Sci. 2016;73(17):3221–47. Epub 2016/04/22. doi: 10.1007/s00018-016-2223-0 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bauer M, Bauer I. Heme oxygenase-1: redox regulation and role in the hepatic response to oxidative stress. Antioxid Redox Signal. 2002;4(5):749–58. Epub 2002/12/10. doi: 10.1089/152308602760598891 . [DOI] [PubMed] [Google Scholar]
- 35.Glickstein H, El RB, Shvartsman M, Cabantchik ZI. Intracellular labile iron pools as direct targets of iron chelators: a fluorescence study of chelator action in living cells. Blood. 2005;106(9):3242–50. Epub 2005/07/16. doi: 10.1182/blood-2005-02-0460 . [DOI] [PubMed] [Google Scholar]
- 36.Doulias PT, Christoforidis S, Brunk UT, Galaris D. Endosomal and lysosomal effects of desferrioxamine: protection of HeLa cells from hydrogen peroxide-induced DNA damage and induction of cell-cycle arrest. Free Radic Biol Med. 2003;35:719–28. doi: 10.1016/s0891-5849(03)00396-4 . [DOI] [PubMed] [Google Scholar]
- 37.Babon JJ, Nicola NA. The biology and mechanism of action of suppressor of cytokine signaling 3. Growth Factors. 2012;30(4):207–19. Epub 2012/05/12. doi: 10.3109/08977194.2012.687375 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu S, Seravalli J, Harrison-Findik D. Inductively coupled mass spectrometry analysis of biometals in conditional Hamp1 and Hamp1 and Hamp2 transgenic mouse models. Transgenic Res. 2015;24(4):765–73. Epub 2015/04/24. doi: 10.1007/s11248-015-9879-3 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neves JV, Ramos MF, Moreira AC, Silva T, Gomes MS, Rodrigues PNS. Hamp1 but not Hamp2 regulates ferroportin in fish with two functionally distinct hepcidin types. Sci Rep. 2017;7(1):14793. Epub 2017/11/03. doi: 10.1038/s41598-017-14933-5 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krijt J, Cmejla R, Sykora V, Vokurka M, Vyoral D, Necas E. Different expression pattern of hepcidin genes in the liver and pancreas of C57BL/6N and DBA/2N mice. J Hepatol. 2004;40(6):891–6. Epub 2004/05/26. doi: 10.1016/j.jhep.2004.02.029 . [DOI] [PubMed] [Google Scholar]
- 41.Chen L, Ma B, Liu X, Hao Y, Yang X, Liu M. H2O2 induces oxidative stress damage through the BMP-6/SMAD/hepcidin axis. Dev Growth Differ. 2020;62(2):139–46. Epub 2020/02/06. doi: 10.1111/dgd.12650 . [DOI] [PubMed] [Google Scholar]
- 42.Ahmed ST, Ivashkiv LB. Inhibition of IL-6 and IL-10 signaling and Stat activation by inflammatory and stress pathways. J Immunol. 2000;165(9):5227–37. Epub 2000/10/25. doi: 10.4049/jimmunol.165.9.5227 . [DOI] [PubMed] [Google Scholar]
- 43.Villarroel P, Le Blanc S, Arredondo M. Interleukin-6 and lipopolysaccharide modulate hepcidin mRNA expression by HepG2 cells. Biol Trace Elem Res. 2012;150(1–3):496–501. Epub 2012/10/16. doi: 10.1007/s12011-012-9522-6 . [DOI] [PubMed] [Google Scholar]