Summary
Rank signaling enhances stemness in mouse and human mammary epithelial cells (MECs) and mediates mammary tumor initiation. Mammary tumors initiated by oncogenes or carcinogen exposure display high levels of Rank and Rank pathway inhibitors have emerged as a new strategy for breast cancer prevention and treatment. Here, we show that ectopic Rank expression in the mammary epithelia unexpectedly delays tumor onset and reduces tumor incidence in the oncogene-driven Neu and PyMT models. Mechanistically, we have found that ectopic expression of Rank or exposure to Rankl induces senescence, even in the absence of other oncogenic mutations. Rank leads to DNA damage and senescence through p16/p19. Moreover, RANK-induced senescence is essential for Rank-driven stemness, and although initially translates into delayed tumor growth, eventually promotes tumor progression and metastasis. We uncover a dual role for Rank in the mammary epithelia: Rank induces senescence and stemness, delaying tumor initiation but increasing tumor aggressiveness.
Keywords: breast cancer, mammary gland, receptor activator of NFkB, senescence, senolytics, stemness, metastasis
Graphical abstract

Highlights
-
•
Rank expression delays mammary tumor onset in oncogene-driven models
-
•
Activation of Rank signaling induces senescence through p16/p19
-
•
Rank-induced senescence in luminal cells increases basal and luminal stemness
-
•
Rank-induced senescent cells promote mammary tumor growth
Rank pathway inhibitors have emerged as therapeutic options for breast cancer prevention and treatment. Benitez et al. show that activation of Rank signaling induces senescence, which paradoxically delays tumor onset and incidence but promotes tumor aggressiveness. Treatment with senolytics eliminates Rank-induced senescent cells, reducing stemness and breast cancer growth.
Introduction
Cellular senescence is a potent tumor-suppressive mechanism (Braig and Schmitt, 2006; Collins and Sedivy, 2003), induced by stress factors such as dysfunctional telomeres, DNA damage, and certain oncogenes (Campisi, 2003). Mutations in p53 or p16lnk4a/pRB pathways cause resistance to senescence and increase cancer risk (Collado and Serrano, 2005). In mice and humans, the senescence response prevents premalignant lesions from progressing to malignant cancers (Krtolica et al., 2001). But senescent cells can also act as tumor-promoting agents (Coppé et al., 2010; Lujambio, 2016; Sun et al., 2018), secreting factors that alter the tissue microenvironment; a feature called senescence-associated secretory phenotype (SASP) (Malaquin et al., 2019). The long-term exposure to senescent cells is potentially detrimental, and their elimination may be fundamental for the treatment of cancer and other diseases (Zhu et al., 2016). Several senolytic drugs, such as ABT263 (navitoclax), have been shown to preferentially kill senescent cells with a wide range of beneficial effects (Baker et al., 2011; Wang and Bernards, 2018; Zhu et al., 2016, 2015).
Rank and its ligand Rankl are key regulators of mammary gland (MG) development (Fata et al., 2000; Gonzalez-Suarez et al., 2007) and act as paracrine mediators of progesterone in mouse (Beleut et al., 2010; Fata et al., 2000) and human mammary epithelium (Tanos et al., 2013). Rank plays a dual role during MG development since both Rank deletion and Rank overexpression impair lactation (Fata et al., 2000; Gonzalez-Suarez et al., 2007). Early in gestation, Rank acts as a positive mediator of progesterone, essential for mammary epithelial cell (MEC) proliferation, survival, and alveologenesis. However, at mid-gestation, Rank interferes with prolactin signaling, preventing lactogenesis (Cordero et al., 2016; Fata et al., 2000; Gonzalez-Suarez et al., 2007).
Rank overexpression in the MG under the mouse mammary tumor virus (MMTV) promoter (MMTV-Rank or Rank+/tg) impairs MEC differentiation and leads to an expansion of mammary stem cells (MaSCs)—with increased ability to reconstitute the MG—and luminal progenitors—with increased colony-forming ability (Pellegrini et al., 2013). Multiparous Rank+/tg mice spontaneously develop mammary adenocarcinomas containing embryonic-like cells—dual positive for cytokeratins CK14/CK8 (Pellegrini et al., 2013)—and show a shorter tumor latency and increased tumor incidence compared with wild-type (WT) after carcinogenic protocols (Gonzalez-Suarez et al., 2010). Conversely, genetic or pharmacological inhibition of Rank signaling prevents or attenuates mammary tumor initiation driven by carcinogens (Gonzalez-Suarez et al., 2010; Schramek et al., 2010).
MMTV-Neu (Neu+/−) mice—which express Neu, the rat ortholog of human HER2 (ErbB2)—and MMTV-PyMT (PyMT+/−) mice—which express the middle T protein of the polyomavirus—develop aggressive multifocal adenocarcinomas with high incidence of lung metastasis (Muller et al., 1988; Guy et al., 1992; Herschkowitz et al., 2007; Maglione et al., 2001). Rank protein is expressed focally in Neu and PyMT non-transformed MGs, and its levels increase in mammary preneoplastic lesions and invasive adenocarcinomas (Gonzalez-Suarez et al., 2010; Yoldi et al., 2016). In contrast, Rankl is expressed in progesterone-receptor-positive (PR+) cells in the non-transformed MGs (Fernandez-Valdivia et al., 2009), but in Neu and PyMT preneoplastic lesions and adenocarcinomas Rankl levels are low or undetectable, which is in accordance with the loss of hormone receptor expression during tumor progression (Lin et al., 2003). We previously showed that pharmacologic or genetic loss of Rank signaling in Neu+/− and PyMT+/− models delays tumor onset and decreases tumor and metastasis incidence (Gonzalez-Suarez et al., 2010; Yoldi et al., 2016).
Here, our studies reveal that in non-transformed mammary epithelia, Rank ectopic expression induces senescence. Rank-driven senescence recapitulates the main characteristics of “oncogene-induced senescence” (Braig and Schmitt, 2006) not only in MECs, but also in mouse embryonic fibroblasts (MEFs): reduced proliferation, increased senescence-associated β-galactosidase (SA-β-gal) activity (Dimri et al., 1995), dependency on p16/p19 tumor suppressors, DNA damage, SASP induction, and sensitivity to senolytics. Rank-driven senescence initially delays tumor onset in oncogene-driven models; however, in later stages of tumor progression it promotes stemness tumor growth and metastasis.
Results
Ectopic Rank expression in the mammary epithelia induces senescence and delays mammary tumor onset
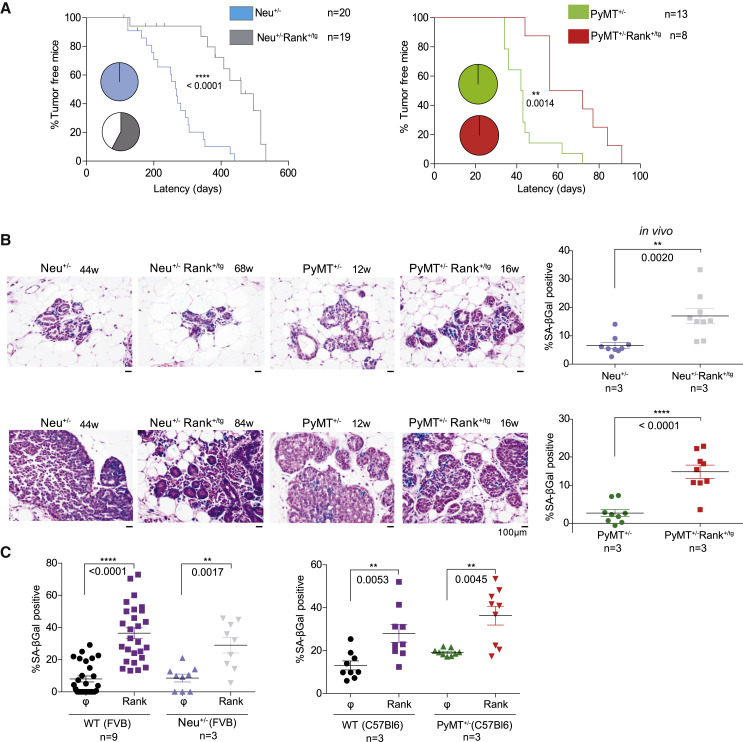
Mice with ectopic MMTV-driven Rank expression, Rank+/tg mice, develop spontaneous breast tumors, but only after multiple pregnancies and with long latency (Pellegrini et al., 2013), suggesting that additional oncogenic events may be required for full transformation. To test this hypothesis, Rank+/tg mice were crossed with the oncogene-driven Neu+/− or PyMT+/− mice (Neu+/−Rank+/tg or PyMT+/−Rank+/tg). Unexpectedly, Rank ectopic expression delayed tumor onset driven by Neu and PyMT oncogenes: Neu+/−Rank+/tg females develop tumors later (414 ± 115 days) than Neu+/− mice (261 ± 87 days). In PyMT+/−Rank+/tg, tumor onset is also delayed (67 ± 16 days) compared with PyMT+/− females (41 ± 7 days) (Figure 1A). Moreover, 42% of Neu+/−Rank+/tg females died without tumors, whereas all Neu+/− mice developed invasive carcinomas (Figure 1A).
Figure 1.
Rank overexpression in oncogenic Neu+/− and PyMT+/− backgrounds delays tumor onset and induces senescence
(A) Kinetics of palpable tumor onset with age in the indicated genotypes. Statistical difference between groups was evaluated by log-Rank test. Pie charts represent the total frequency of mice with tumors at sacrifice. Note that some Neu+/−Rank+/tg mice died or had to be sacrificed due to humane endpoints unrelated to tumor development.
(B) Representative images of H&E with SA-β-gal staining and quantifications (frequency of SA-β-gal-positive cells relative to total MECs) in ducts or preneoplastic lesions of indicated age and genotype. Three representative pictures of the indicated number of MGs/mice were quantified.
(C) Frequency of SA-β-gal-positive cells in non-transformed MECs isolated from Neu+/− and PyMT+/− mice 6 days after the infection with Rank-overexpressing (Rank) or control (ϕ) vectors. Total number of independent experiments/MG is shown. Quantifications were performed in triplicates and each dot indicates a replica.
(B and C) Mean, SEM and t test p values are shown. See also Figure S1.
Senescence can be an efficient anti-tumor mechanism (Collado and Serrano, 2010); we thus hypothesize that high levels of Rank may trigger senescence, delaying tumor initiation. Indeed, a clear increase in the frequency of SA-β-gal-positive cells was found in the hyperplasic lesions and early mammary intraepithelial neoplasia (MIN) of Neu+/−Rank+/tg and PyMT+/−Rank+/tg MGs compared with the corresponding single Neu or PyMT MGs (Figure 1B). Similar to the MGs of single Rank+/tg mice (Pellegrini et al., 2013), multiple hyperplasias were found in the MGs of double transgenic Neu+/−Rank+/tg and PyMT+/−Rank+/tg mice (Figure S1A), supporting that senescence was preceded by highly proliferative mammary epithelia. Despite the high levels of proliferation found in the hyperplastic lesions, no overlap between senescence and the proliferation marker Ki67 was observed, further supporting the senescent nature of these lesions (Figure S1B).
To confirm that Rank expression could induce senescence in mammary epithelia, non-transformed Neu+/−and PyMT+/− MECs and their corresponding WT controls were infected with Rank-overexpressing lentivirus (Figure S1C). MECs from PyMT+/− C57Bl6 were used, since tumor onset is significantly delayed in this background compared with FVB. The frequency of senescent cells in control-infected MECs was similar in all genotypes (10%–15%). However, a significant increase in the frequency of senescent cells after Rank overexpression was observed in Neu, PyMT, and WT MECs (Figures 1C and S1D). These results support that Rank overexpression induces senescence in the mammary epithelia, even in the absence of additional oncogenes. Accordingly, the non-transformed and hyperplastic epithelium of Rank+/tg MGs and their derived cultured MECs also contained multiple senescent cells (Figures 2A, 2B, S1E, and S1F). Senescent cells were mainly located in the luminal compartment, in hormone-receptor-negative cells, as no co-staining of SA-β-gal with PR expression was observed (Figure S1G). In contrast, overexpression of PyMT or Neu oncogenes neither triggered senescence in WT MECs nor increased senescence levels in Rank+/tg MECs (Figures S1F and S1H). These results demonstrate that Rank signaling is a potent inducer of senescence in the mammary epithelia.
Figure 2.
Rank signaling induces senescence in MECs in vivo and in vitro
(A) Representative images of H&E with SA-β-gal staining and quantifications in vivo (frequency of SA-β-gal-positive cells relative to total MECs) in MGs of the indicated age. Three representative pictures of each MGs were quantified.
(B) Representative images of SA-β-gal staining in WT and Rank+/tg MECs cultured for 8 days and corresponding quantification ex vivo. Quantifications were performed in triplicates and each dot indicates a replica.
(C) Representative images of H&E with SA-β-gal staining and quantification of in MGs from WT females after 14 days of Rankl (n = 3) or mock in vivo treatment (n = 3). Quantifications were done in triplicates and each dot indicates a replica.
(D) Quantification of SA-β-gal staining of MECs isolated from Rankl-treated (as in C) WT females (n = 3) cultured for 8 days (ex vivo).
(E) Representative images and quantification (right panel) of SA-β-gal staining in WT MECs cultured in vitro in the presence of Rankl. Quantifications were performed in triplicates. Each dot indicates a replica of 6 independent experiments. In all panels mean, SEM, and t test p values are shown. See also Figure S1.
We next asked whether stimulation of Rank signaling with Rankl in the WT mammary epithelia with physiological levels of Rank also induced senescence. Multiple senescent cells and hyperplasic structures were found in the MGs of WT mice treated with Rankl for 14 days, recapitulating the phenotype observed in the Rank+/tg MGs (Figure 2C). Quantification of SA-β-gal staining in cultured MECs isolated from Rankl-treated WT mice (ex vivo) revealed senescence in 20% of the cells compared with 3% in non-treated controls (Figure 2D). Accordingly, WT MECs cultured with Rankl show senescence levels similar to Rank+/tg MECs (Figures 2E and 2A). These results highlight the physiological relevance of Rank-induced senescence and confirm that activation of Rank signaling, either by ectopic expression of the receptor or Rankl stimulation, promotes senescence in vivo and in vitro.
Rank overexpression leads to senescence in MECs and MEFs through p16/p19
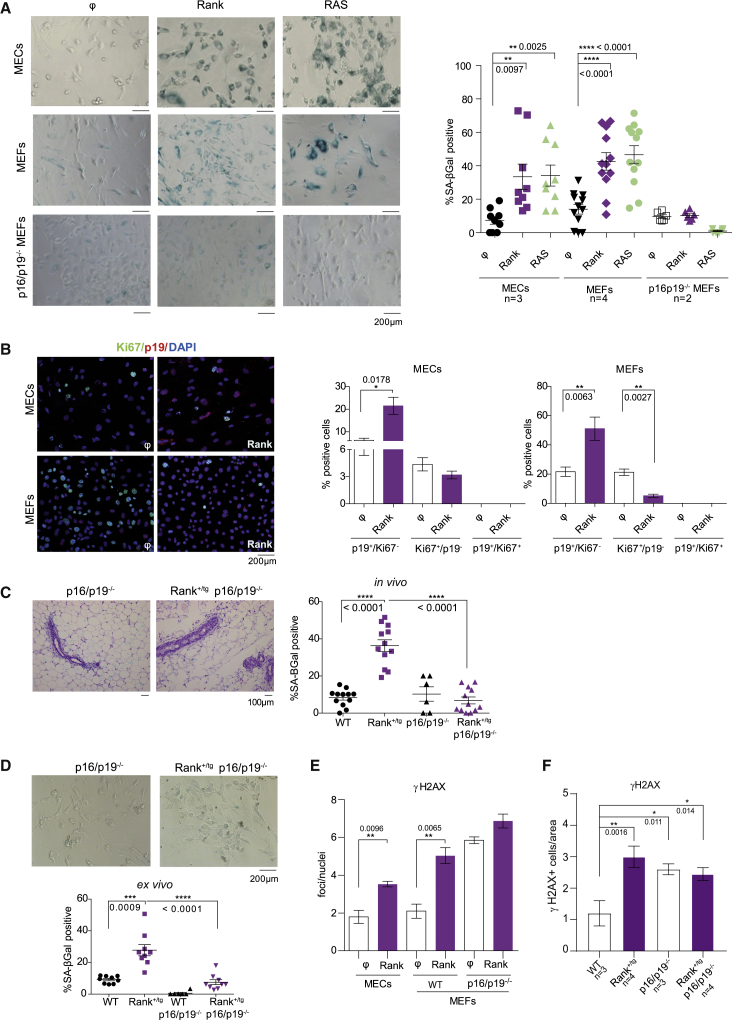
Our data suggest that Rank overexpression leads to senescence in MECs. Overexpression of the activated oncogene RAS (RASV12) in MEFs constitutes the best characterized in vitro model of OIS (Bringold and Serrano, 2000; Campisi, 2001; Kodama et al., 2001; Serrano et al., 1997). Rank and RASV12 were overexpressed by viral infection in MECs and MEFs isolated from WT mice and analyzed for SA-β-gal staining (Figures 3A and S2A). Rank overexpression in MECs and MEFs led to the activation of downstream pathways in the absence of Rankl (Figure S2B) and induced senescence to a similar extent than RASV12, 35% approximately in MECs and 40% in MEFs (Figure 3A). Even low levels of Rank expression (achieved using alternative constitutive or inducible vectors) induced senescence in MECs and MEFs (Figures S2C and S2D).
Figure 3.
Rank overexpression induces senescence through p16/p19 (Ink4a/Arf) in MECs and MEFs
(A) Representative images and quantification of SA-β-gal staining in MECs isolated from WT MGs and WT and p16/p19−/− MEFs 6 days after infection with Rank or RASV12 overexpressing and control (ϕ) vectors. Number of independent experiments is shown. Quantifications were performed in triplicates and each dot represents a replica.
(B) Representative images and quantifications of Ki67+ (green) and p19+ (red) cells within total nuclei (DAPI, blue) in WT MECs and MEFs 6 days after infection with Rank-overexpressing and control (ϕ) lentivirus.
(C) Representative images and quantification of the H&E with SA-β-gal staining in MGs of the indicated genotypes. Three representative pictures of the indicated number of MGs were quantified.
(D) Representative images and quantification of SA-β-gal staining in Rank+/tg and Rank+/tg p16/p19−/− MECs (ex vivo) cultured for 8 days. Quantifications were performed in triplicates and each dot represents a replica of three independent MGs/mice.
(E) Quantification of γH2AX+ foci per nuclei in the indicated cells 6 days after infection with control (ϕ) and Rank-overexpressing virus.
(F) Quantification of γH2AX+ cells relative to the epithelial area in the MGs of WT, Rank+/tg, p16/p19−/−, and Rank+/tg p16/p19−/− females. The number of mice analyzed is indicated for each genotype. Representative images are shown in Figure S3A. In all panels mean, SEM and t test p values are shown. See also Figures S2 and S3.
Rank- and RAS-overexpressing MECs and MEFs showed higher levels of p16(Ink4a) and p19(Arf) transcripts, encoded in the Cdkn2a locus and involved in senescence (Collado and Serrano, 2005), compared with control cells (Figure S2E). Moreover, an increased expression of p19 protein was detected in Rank+/tg compared with WT-derived MECs (Figure S2F). Co-staining of Ki67 and p19 in Rank-transduced MECs and MEFs revealed no overlap between p19+ and Ki67+ and an increase in p19+ cells compared with corresponding controls (Figure 3B). Proliferation determined by Ki67 staining was very low in MECs; in MEFS, the percentage of Ki67+ cells decreased from 20% to 3% after Rank overexpression, consistent with a senescent growth-arrested phenotype (Figure 3B). Besides, neither Rank nor RASV12 were able to induce senescence in MEFs in the absence of p16 and p19 (Figures 3A and S2A).
To corroborate that Rank-induced senescence in MECs was also mediated by p16/p19, we generated Rank+/tg p16/p19−/− mice. The frequency of SA-β-gal-positive cells in the MGs from Rank+/tg p16/p19−/− was comparable with that of WT mice and lower than in Rank+/tg MGs (Figure 3C). Quantification of SA-β-gal staining in cultured MECs isolated from the different genotypes corroborated these findings (Figure 3D).
The senescence induced by RASV12 is associated with the activation of the DNA-damage response (Di Micco et al., 2011; Tu et al., 2012), and DNA damage might cause OIS (d'Adda di Fagagna, 2008). A significant increase in γH2AX was found upon Rank overexpression in MGs, MECs and MEFs compared with the corresponding WT controls, but not in p16/p19−/− MEFs, which show higher basal γH2AX levels (Figures 3E, 3F, S3A, and S3B). Accordingly, γH2AX+ cells were found at a similarly higher frequency in the MGs of p16/p19−/−, Rank+/tg p16/p19−/−, and Rank+/tg compared with WT mice (Figure 3F). Thus, Rank overexpression induces DNA damage and senescence through p16/p19 in MECs and MEFs.
Finally, we wondered whether, similar to activated RAS, Rank was able to transform MEFs in the absence of p16/p19−/−. As expected, in p16/p19−/− MEFs, RASV12 overexpression rapidly led to the growth of multiple colonies in soft agar, whereas only a few and much smaller colonies grew upon Rank overexpression (Figure S3C). These results suggest that Rank overexpression may lead to mammary tumorigenesis in p16/p19−/− mice. The MGs of aged Rank+/tg p16/p19−/− showed periductal fibrotic thickening to a greater extent than the MGs of the single mutants Rank+/tg and p16/p19−/− (Figure S3D), but no epithelial hyperplasias and/or neoplasias were found in any of the mice analyzed. We conclude that, although Rank and RASV12 induce similar levels of senescence, Rank oncogenic potential is much lower than that of activated RAS.
Rank induces senescence in luminal cells and stemness in basal and luminal cells
We previously found that Rank enhances stemness properties in mouse (Pellegrini et al., 2013) and human MECs (Palafox et al., 2012), as well as in breast cancer cells (Yoldi et al., 2016). Rank expression leads to the counterintuitive concurrence of stem cell properties and senescence features.
RANK expression under the MMTV promoter is predominantly located in luminal cells (Pellegrini et al., 2013) and, accordingly, Rank-induced senescence is restricted to the luminal compartment in particular HR-negative cells (Figures 1B, 1C, and S1G). We previously demonstrated that both Rank+/tg luminal and basal MECs show an increased colony-forming ability compared with the corresponding WT MECs. Moreover, Rank+/tg basal MECs are able to regenerate MGs in limiting dilution assays more frequently than WT basal MECs (Pellegrini et al., 2013). Thus, while Rank-induced senescence in Rank+/tg localizes within the MG luminal compartment, both basal and luminal MECs show stemness features, indicating that Rank-driven paracrine senescence mechanisms such as SASP might favor stemness (Coppé et al., 2008; Malaquin et al., 2016). Indeed, interrogation of the cytokine profiles of mammary epithelial lysates revealed higher levels of proteins associated with senescence such as Mmp2, Timp2, and Cxcl15 (a murine IL8 homolog), as well as Mmp3, bFgf, Il7, and Ccl20 (Özcan et al., 2016) in Rank+/tg MECs compared with control cells (Figure S4A), indicating that Rank overexpression induces the SASP.
To gain mechanistic insights into the ability of Rank to simultaneously induce stemness and senescence, RNA-seq data were obtained from luminal and basal WT and Rank+/tg MECs. A 14-fold increase in Rank mRNA expression was found in Rank+/tg luminal cells, and 1,124 genes were differentially expressed between Rank+/tg and WT luminal MECs (897 up and 227 down in Ranktg/+) (Table S1). Luminal Rank+/tg MECs have increased expression of Cdkn2a transcripts (encoding p16/p19 proteins) than WTs, corroborating their senescence phenotype (Table S1). Gene set enrichment analysis (GSEA) revealed a decrease in the luminal mature signature in Rank+/tg compared with WT luminal MECs (Figure 4A), supportive of the stemness properties of Rank+/tg MECs (Pellegrini et al., 2013). In accordance, increased expression of the basal Krt14-, Pdpn-, the Wnt-related genes Rspo1 and Axin2, and reduction in the luminal differentiation markers Pr, PrlR, and Csn was found in Rank+/tg luminal MECs compared with WTs (Table S1). According to GSEA, the signatures increased in luminal Rank+/tg were related with cell cycle, proliferation, p53 senescence/apoptotic pathway, Rb pathway, and DNA repair (Table S1; Figure 4A). Some of the most prominent pathways found in luminal Rank+/tg MECs include TLX-negative targets (the orphan nuclear receptor TLX inhibits senescence and is a key regulator of stemness; O'Loghlen et al., 2015; Wu et al., 2015; Zhang et al., 2008) and EZH2 targets, suggestive of epigenetic reprograming mechanisms (Onder et al., 2012) (Table S1). These results corroborate that Rank drives senescence and stemness in luminal MECs.
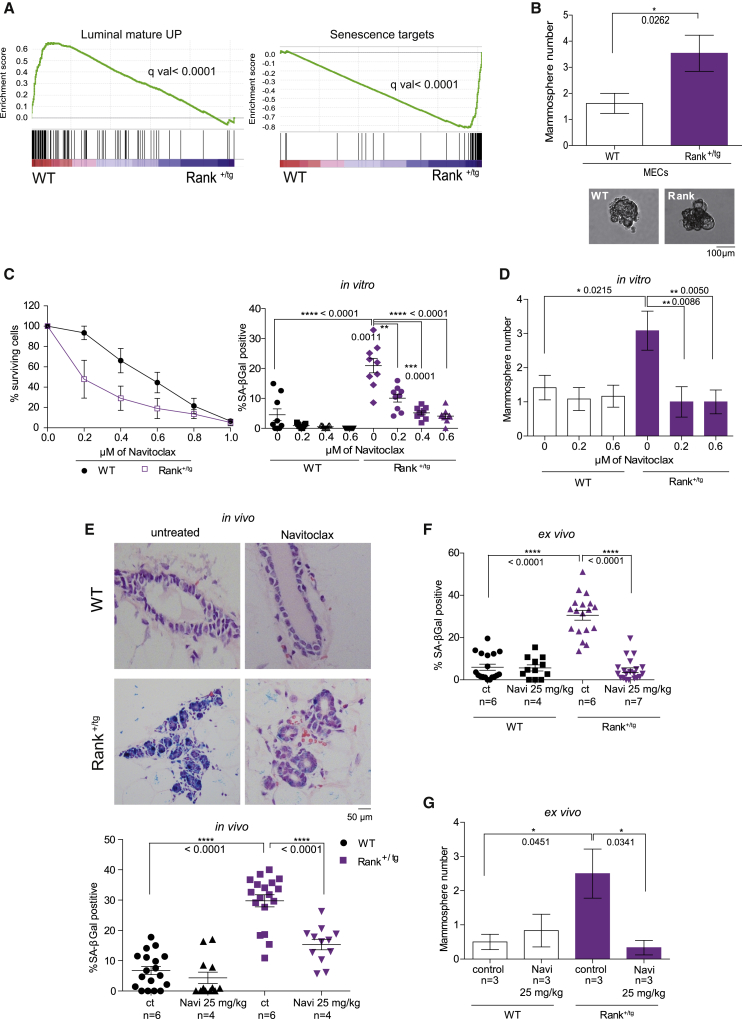
Figure 4.
Rank-induced senescent MECs are sensitive to navitoclax and required for Rank-driven stemness
(A) GSEA graphical outputs for the association analysis between WT and Rank+/tg in luminal MECs. Each signature was analyzed using the gene subsets corresponding to over or underexpression (Table S1). The GSEA enrichment score and nominal p values are shown.
(B) Number of secondary mammospheres and representative images derived from WT and Rank+/tg EpCAM+ MECs. Mean and t test p values for 5 independent experiments are shown. For each experiment, pools of MECs from 2–3 mice were used (13 mice per genotype).
(C) Percentage of surviving cells (left panel) and SA-β-gal-positive cells (right) of WT and Rank+/tg MECs treated 48 h in vitro with the indicated doses of navitoclax. Quantifications were performed in triplicates (each dot represents a replica) and mean and SEM for 3 independent experiments are shown.
(D) Number of secondary mammospheres formed by WT and Rank+/tg EpCAM+ MECs after in vitro treatment with the indicated concentrations of navitoclax. Mean, SEM, and t test p values for 4 independent experiments are shown.
(E) Representative images and quantifications (in vivo) of H&E with SA-β-gal staining in WT and Rank+/tg MGs after 14 days of navitoclax treatment. Three representative pictures of the indicated number of MGs were quantified and mean and t test p values are shown.
(F) Quantification (ex vivo) of SA-β-gal staining of WT and Rank+/tg MECs isolated from navitoclax-treated mice and cultured for 8 days. Quantifications were performed in triplicates and each dot represents a replica. Mean, SEM, and t test p values are shown.
(G) Number of secondary mammospheres formed by WT and Rank+/tg EpCAM+ MECs isolated from females treated with navitoclax in vivo for 14 days. MECs from 3 mice per genotype and treatment were pooled for each experiment. Quantifications for each experiment were done in triplicates. Mean, SEM, and t test p values for 2 independent experiments are shown. See also Figure S4 and Table S1.
In the basal compartment, even though no significant differences in the expression of Rank were found between Rank+/tg and WT MECs, 476 genes were differentially expressed, highlighting the relevance of the paracrine communication between basal and luminal cells. Cdkn2a transcripts were not expressed in Rank+/tg basal cells and, compared to WT cells, E2F and EZH2 targets (cell cycle) were upregulated, whereas p53 and ROS pathways, and p53 senescence targets, including Cdkn1a (p21), were downregulated (Table S1). Moreover, in Rank+/tg basal cells the TNF signaling pathway, recently found to restrict basal bipotency (Centonze et al., 2020), was downregulated (Table S1). In line with a stemness phenotype, Rank+/tg MECs gave rise to more secondary mammospheres than WT MECs did, confirming the increased self-renewal potential of Rank+/tg basal cells, but no differences in mammosphere size, indicative of proliferation, were found (Figure 4B).
Together these results support that in Rank+/tg MECs senescence is restricted to the luminal compartment, whereas increased stemness features are found in both basal and luminal populations.
Rank-induced senescence is required for Rank-driven stemness
Next, we tested whether Rank-induced senescence is causally involved in the activation/acquisition of stemness properties. To this end, we used the senolytic drug navitoclax (ABT263) (Chang et al., 2016). In accordance with the presence of senescent cells, Rank+/tg MECs are more sensitive than WTs to navitoclax treatment in vitro and a reduction in the frequency of SA-β-gal positivity was observed in Rank+/tg MECs with increasing doses of navitoclax (Figure 4C). Besides, in the presence of navitoclax, the increased frequency of secondary mammosphere formation in Rank+/tg compared with WT MECs was no longer observed (Figure 4D).
Accordingly, senescence was not detected in the MGs of Rank+/tg mice treated in vivo during 14 days with 25 mg/kg of navitoclax (Figure 4E), and the total number of viable MECs isolated from navitoclax-treated mice was significantly reduced in Rank+/tg mice compared with WT (Figure S4B). SA-β-gal analysis of cultured WT and Rank+/tg MECs isolated from navitoclax-treated mice demonstrated comparable levels of senescent cells between genotypes (Figures 4F and S4C). Rank+/tg MGs showed a reduction in the relative frequency of basal (CK5+) cells in the epithelia and PR+ positive cells within the luminal compartment, but navitoclax treatment did not change the luminal/basal and HR+/HR− ratios neither in WT nor in Rank+/tg MGs (Figures S4E and S4F). Again, after in vivo treatment with navitoclax, a significant reduction in mammosphere formation ability was observed in MECs derived from Rank+/tg but not from WT mice. In fact, WT and Rank+/tg MECs isolated from navitoclax-treated mice gave rise to similar numbers of mammospheres, in contrast with those derived from mock-treated mice (Figure 4G). These results demonstrate that Rank-induced senescence in MECs is essential for stemness.
Rank ectopic expression increases breast cancer stemness
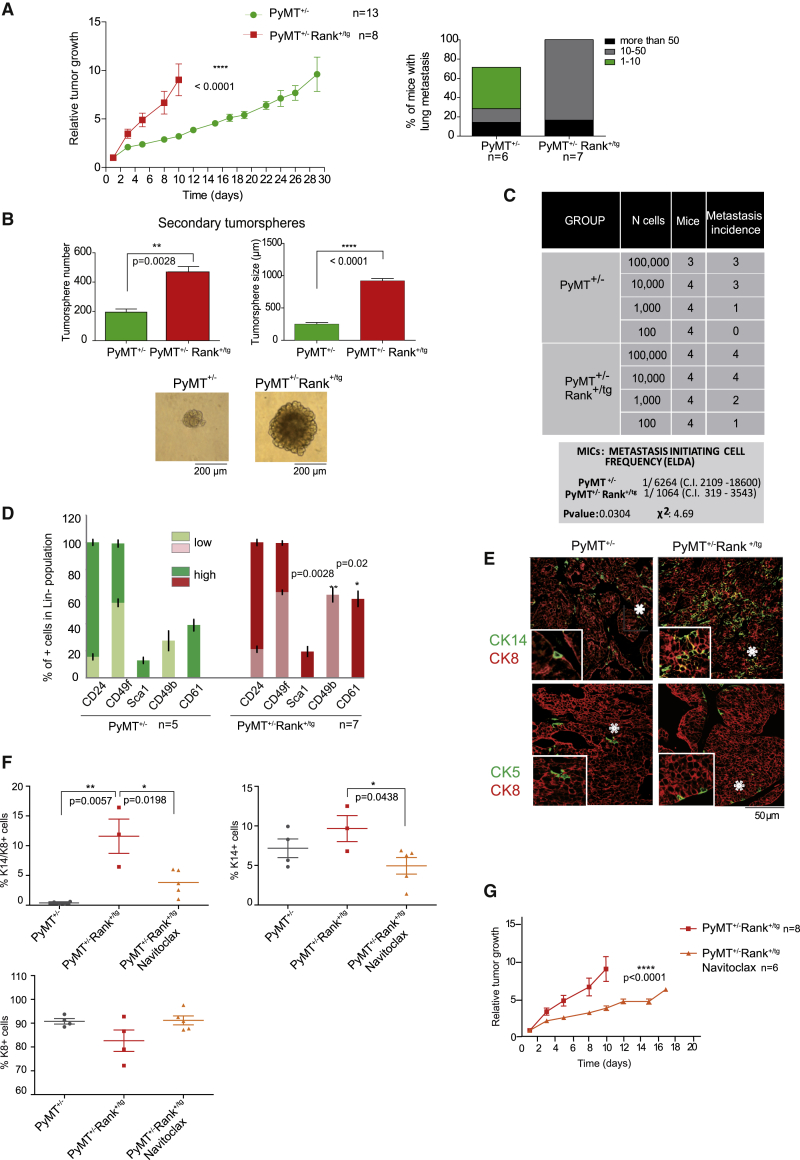
Based on the results obtained in non-transformed epithelia and previous data supporting the oncogenic role of Rank (Gonzalez-Suarez et al., 2010; Palafox et al., 2012; Pellegrini et al., 2013), we hypothesized that high levels of Rank could enhance stemness in the context of breast cancer. Indeed, despite their delayed tumor onset, tumors in PyMT+/−Rank+/tg mice grew faster and developed more lung metastasis than single PyMT mice developed (Figure 5A). Rank mRNA expression increases in PyMT+/− adenocarcinomas compared with non-tumorigenic mammary epithelia (Yoldi et al., 2016) and PyMT+/−Rank+/tg tumors showed even higher levels of Rank (Figure S5A). More and larger secondary tumorspheres (Figure 5B), as well as a significantly higher frequency of metastasis initiating cells (MICs) (1 in 1,064 cells) was observed in PyMT+/−Rank+/tg compared with PyMT+/− MGs (1 in 6,264 cells) (Figure 5C), confirming that high levels of Rank increases breast cancer stemness. FACS analysis revealed a significant increase in the luminal progenitor populations CD61+ and CD49b+ cells (Oakes et al., 2008; Shehata et al., 2012) in PyMT+/−Rank+/tg compared with PyMT+/− tumors (Figure 5D).
Figure 5.
Rank ectopic expression increases breast cancer aggressiveness
(A) Relative tumor growth and the percentage of lung metastasis in the indicated genotypes. Mean tumor volume ± SEM at each time point relative to their volume on the first day of detection and statistical t test values are shown. The percentage of females with the indicated number of lung metastasis and total number of mice is indicated.
(B) Number, size and representative pictures of secondary tumorspheres derived from PyMT+/− and PyMT+/−Rank+/tg tumor cells. Each bar represents a pool of 3 independent tumors/experiments. Mean, SEM, and t test p values are shown.
(C) Metastasis initiating cell frequencies (with confidence intervals) calculated by ELDA, p- and chi-square values of PyMT+/− and PyMT+/−Rank+/tg tumor cells are shown. Cells from 2 independent tumors per genotype were pooled for injections and metastasis were scored 8 weeks later.
(D) Frequency of indicated cells in the Lin− (CD45− CD31−) population found in PyMT+/− and PyMT+/−Rank+/tg spontaneous tumors (n = 5–7) analyzed by FACS. Positive/negative and high/low populations were set according to cell populations in the normal MG. Mean, SEM, and t test p values are shown.
(E) Representative CK14 or CK5 (green) and CK8 (red) immunostaining in PyMT+/− and PyMT+/−Rank+/tg spontaneous adenocarcinomas. Asterisks indicate magnifications (2×) to highlight CK14+/CK8+ cells. No CK5+/CK8+ cells were found.
(F) Quantification of CK8+, CK14+ and CK14+/CK8+ cells in spontaneous adenocarcinomas of PyMT+/− and PyMT+/−Rank+/tg mice pretreated or not with navitoclax.
(G) Relative tumor growth curves in PyMT+/−Rank+/tg mice treated or not with navitoclax for 14 days. Mean tumor volume ± SEM at each time point relative to their volume on the first day of detection and statistical t test p values are shown. The number of mice is indicated.
(F and G) Navitoclax treatment started (at 4 weeks) before palpable lesions were detected. See also Figure S5.
Mice from both genotypes developed luminal-like tumors expressing CK8 (mRNA and protein) (Figures 5E and S5A). Higher expression of Krt14 mRNA was found in PyMT+/−Rank+/tg tumors, and an increase in the number of CK14+ and double CK14+/CK8+ cells compared with PyMT+/− tumors was found, whereas CK5 cells were scarce in both genotypes (Figures 5E, 5F, and S5A). These results indicate that Rank overexpression leads to an accumulation of the embryonic-like dual positive CK14/CK8 cells in the PyMT tumor-prone model (Figures 5E and 5F), as previously observed in spontaneous Rank+/tg tumors (Pellegrini et al., 2013). Thus, Rank overexpression in the PyMT+/− mouse model promotes the accumulation of bipotent CK14+/CK8+ MECs, an increase in CD49b+ and CD61+ progenitor-like cells and an expansion of the CSC pool, resulting in more aggressive tumors with faster growth and metastatic-initiation potential, compared with PyMT+/− mice.
Unlike the increased frequency of senescent cells observed in preneoplastic lesions from PyMT+/−Rank+/tg compared with PyMT+/− MGs, the levels of senescence in tumor cells were comparable between genotypes (Figure S5B). Thus, we evaluated whether the selective elimination of Rank-induced senescent cells would affect tumor aggressiveness. To avoid misinterpreting the effects of navitoclax on tumor cell survival, treatments were performed before palpable lesions were detected (4–6 weeks of age), and the mice were monitored for tumor growth. As we are not avoiding senescence, no differences in tumor onset were observed (Figure S5C); however, tumor growth rates were attenuated in PyMT+/−Rank+/tg mice pretreated with the senolytic drug navitoclax compared with untreated mice, indicating that the senescent cells present in the preneoplastic lesions are indeed favoring tumor growth (Figure 5G). Moreover, the frequency of CK14/CK8-dual-positive embryonic-like cells and CK14+ cells was reduced in PyMT+/−Rank+/tg mice pretreated with navitoclax (Figure 5F). In parallel we observed that when PyMT tumor cells were orthotopically implanted in the MGs from Rank+/tg and WT female littermates, 17 out of 20 tumors (85%) grew in the senescent environment of the Rank+/tg MGs, compared with 7 out of 18 tumors in WT glands. PyMT-derived tumor cells also grew slightly faster in the Rank+/tg compared with WT MGs (Figures S5D and S5E).
Together, these results demonstrate that ectopic Rank expression enhances mammary tumor aggressiveness and suggest that non-tumorigenic senescent cells support tumor growth and stemness features.
Rank-induced senescence increases stemness in human breast cancer cells through paracrine mediators
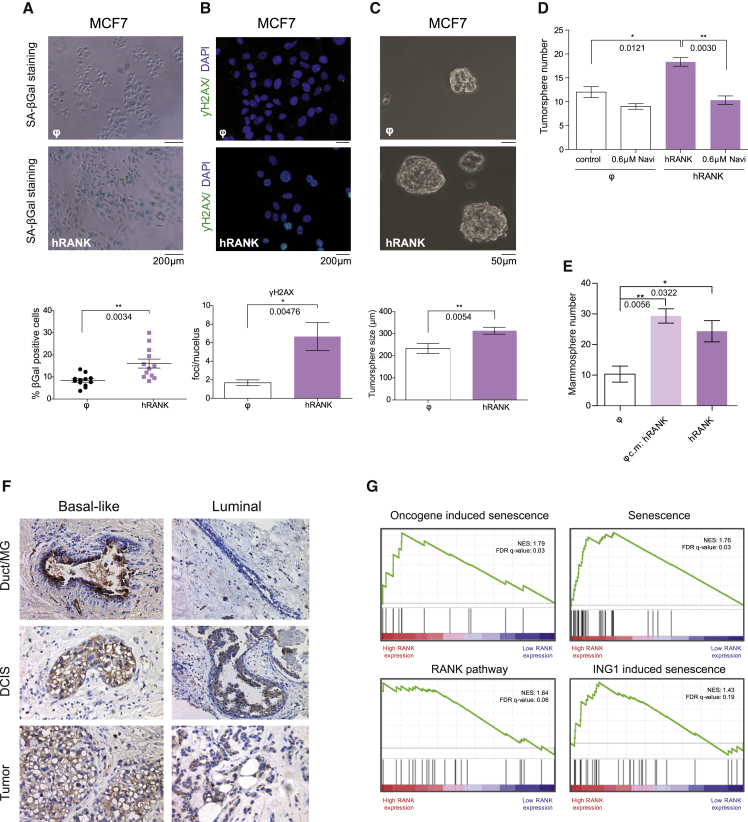
We next interrogated the functional consequences of overexpressing RANK in the human luminal breast cancer cell line MCF7. Regardless of the levels achieved, RANK-induced senescence (Figures 6A, S6A, S6B, and S6C) and increased the number of γH2AX foci per cell (Figure 6B) and the frequency of secondary tumorspheres (Figures 6C and S6D) in MCF7 cells. A significant reduction in MCF7 tumorsphere formation ability was found in RANK-expressing MCF7 cells after treatment with navitoclax (Figure 6D). Strikingly, control MCF7 cells showed an increased frequency of secondary tumorsphere formation when cultured in the conditioned media of RANK-induced senescent MCF7 cells (Figures 6E and S6D). Together these results confirm the functional link between senescence and stemness in human breast cancer and denote that RANK-induced senescent breast cancer cells enhance cancer stemness through paracrine mediators.
Figure 6.
RANK-driven senescence in human breast cancer cells promotes stemness by paracrine mechanisms
(A–C) Representative images and quantifications of SA-β-gal+ cells (A), γH2AX (green) foci per nuclei (DAPI, blue) (B), and size of secondary tumorspheres (C) of MCF7 human breast cancer cells 2 weeks after infection with RANK-overexpressing (hRANK) and control (ϕ) vectors. Quantifications for each experiment were done in triplicates. Each dot indicates a replica. Mean, SEM, and t test p values are shown.
(D) Number of secondary tumorspheres in RANK-overexpressing (hRANK) and control (ϕ) MCF7 cells after in vitro treatment with navitoclax.
(E) Quantification of secondary tumorspheres of control (ϕ) and RANK-overexpressing MCF7 cells cultured in conditioned media (cm) of senescent RANK-overexpressing MCF7 cells. All quantifications were performed in triplicates and mean, SEM, and t test p values for 3 independent experiments are shown.
(F) Representative images of RANK detection by immunohistochemistry in a subset of non-transformed breast (duct/MG), preneoplastic human lesions (ductal carcinoma in situ, DCIS), and tumors from basal-like and luminal breast cancer.
(G) Breast cancer TCGA samples classified as luminal A or luminal B were selected and ranked by RANK expression, top and bottom 15% quantile samples were selected as high and low rank expression, respectively. GSEA was used to perform GSEA of the described gene signatures. Genes were ranked based on Signal2Noise metric and called significant under an FDR q value of 25%. See also Figure S6 and Table S2.
Finally, to evaluate the clinical relevance of our findings, we analyzed RANK protein expression during breast cancer progression in patients' samples. RANK is expressed in a subset of basal and luminal invasive breast adenocarcinomas, as previously reported (Palafox et al., 2012; Pfitzner et al., 2014) (Figure 6F). Importantly, high levels of RANK protein are also detected in preneoplastic human lesions (ductal carcinoma in situ, DCIS) from both luminal and basal-like breast cancer (Figure 6F), recapitulating the increase in RANK expression levels found in preneoplastic MINs from Neu and PyMT-oncogene-driven mouse models (Figure S6E).
We previously described that high levels of RANK expression in luminal tumors from the TCGA dataset were positively correlated with gene sets that characterize mammary stem cells and luminal progenitors (Pellegrini et al., 2013). To determine whether RANK may also induce senescence in human breast cancer patients, we further interrogated the TCGA dataset and found an association between high RANK expression in human breast tumors from the luminal subtype and several gene sets associated with senescence (Figure 6G; Table S2). Thus, high levels of RANK expression are associated to senescence and stemness in clinical breast cancer.
Discussion
Despite multiple evidences supporting a pro-tumorigenic role for RANK pathway activation, here, we reveal that ectopic expression of Rank in Neu or PyMT-oncogene-driven mouse models promotes a delay in tumor onset and reduces tumor incidence, as observed in loss-of-function approaches (Gonzalez-Suarez et al., 2010; Yoldi et al., 2016). Hence, Rank loss and Rank overexpression both delay and reduce tumor initiation through different mechanisms. Decreased proliferation, survival, and stemness are observed in Rank−/− MECs, preventing tumorigenesis (Fata et al., 2000; Gonzalez-Suarez et al., 2010; Yoldi et al., 2016). Now we show that the activation of Rank signaling leads to a potent senescence response that acts as a barrier for tumorigenesis (Vergel and Carnero, 2014; Prieur and Peeper, 2008). Rank or RASV12 overexpression leads to senescence in MECs and MEFs to a similar extent and share multiple characteristics including induction of p16/p19, DNA damage, and SASP (Serrano et al., 1997; Tu et al., 2012). In contrast, neither Neu nor PyMT overexpression, despite being strong oncogenes, induced senescence in MECs. However, activated RAS shows a greater transformation ability than Rank in MEFs and MGs (Sarkisian et al., 2007; Serrano et al., 1997). The fibrotic lesions observed in the virgin MGs of Rank+/tg p16/p19−/− females reinforce the anti-fibrotic role of p16/p19 (Krizhanovsky et al., 2008; Meyer et al., 2016). We previously determined that Rank expression in the mammary epithelia increases during aging (Pellegrini et al., 2013) and in preneoplastic lesions (Gonzalez-Suarez et al., 2010; Yoldi et al., 2016), and now we show that RANK is highly expressed in human DCIS. We also demonstrate that the activation of the Rank signaling pathway, either by expression of the receptor or treatment with the ligand, is enough to induce senescence. Together, these results support a role for Rank-driven senescence during MG development and to prevent breast cancer in mice and humans.
High levels of Rank in mouse and human MECs, and breast cancer cells, interfere with differentiation resulting in an accumulation of stem and progenitor cells, (Cordero et al., 2016; Gonzalez-Suarez et al., 2007; Palafox et al., 2012; Pellegrini et al., 2013). Moreover, RANK overexpression enhances mammosphere- and tumorsphere-forming ability and leads to the accumulation of stem/progenitor populations. Rank loss in PyMT invasive carcinomas significantly reduces tumor and metastasis initiating ability (Yoldi et al., 2016), demonstrating that Rank signaling contributes to the maintenance of the cancer stem cell population. Accordingly, double transgenic PyMT+/−Rank+/tg mice show a higher incidence of lung metastasis. Inhibition of Rank signaling or senolytic drug treatment to eliminate the senescent cells could help to attenuate tumor progression or metastasis in this context. Indeed, RANK induces senescence and stemness in MCF7 cells and high levels of RANK expression associate with stemness (Pellegrini et al., 2013) and senescence in human luminal breast cancer, highlighting the relevance of our findings in the clinical setting.
The fact that Rank concomitantly increases senescence and stemness seems contradictory; however, recent studies confirm a link between both states, with senescent cells promoting the occurrence of stem cells in their vicinity by paracrine mechanisms (Ritschka et al., 2017). Senescent cells can enhance MEF reprogramming through SASP (Mosteiro et al., 2018), while senescence enhances plasticity in muscle stem cells (Chiche et al., 2017; Ocampo et al., 2016). The reversibility of senescence has been a subject of debate (Martínez-Zamudio et al., 2020). Several studies have identified genes that allow bypassing senescence, whereas less evidence in support of a true escape of senescence is available (Wang et al., 2017). There is a cellular state of light senescence, characterized by low levels of p16 that allows cells to resume proliferation following p53 inactivation (Beauséjour et al., 2003), different from a deep senescence state which is irreversible. Cells in a state of senescence express a latent adult stem cell signature and present stem cell properties (Milanovic et al., 2018). Here, we show that Rank-induced senescence is essential for Rank-driven stemness: upon elimination of senescent cells located in the luminal compartment, the enhanced stemness phenotype is lost. TLX and EZH2 targets are enriched in luminal Rank+/tg MECs, suggesting that the stemness/senescence phenotypes may be controlled by epigenetic mechanisms (Franzen et al., 2016; Lee and Schmitt, 2019; Orioli and Dellambra, 2018; Yang and Sen, 2018). Of note, multiple genes were differentially expressed in Rank+/tg basal cells, including downregulation of the p53 target p21, indicative of a luminal/basal crosstalk. Functional assays from WT- and Rank+/tg-derived MECs prove enhanced progenitor/stemness activity on both basal (increased mammary reconstitution ability) and luminal compartments (increased colony-forming ability) (Pellegrini et al., 2013), pointing to paracrine mechanisms regulating the link between senescence and stemness upon Rank expression. Despite the enhanced senescence observed in early stages, tumors in PyMT+/−Rank+/tg are more aggressive, displaying faster tumor growth and more metastasis than PyMT+/− tumors, while tumor cells derived from PyMT+/−Rank+/tg mice show increased metastasis initiating ability than PyMT+/−-derived tumor cells. Indeed, Rank+/tg mice are more susceptible to mammary tumorigenesis driven by carcinogens and multiparous females spontaneously develop mammary tumors (Gonzalez-Suarez et al., 2010; Pellegrini et al., 2013). These results are in agreement with the findings describing that senescent cells that escape senescence upon inactivation of Suv39h1 or p53 exhibit higher tumor initiation potential (Milanovic et al., 2018). We observe no differences in tumor onset upon elimination of senescent cells by navitoclax, as we are not avoiding senescence, but tumor growth is attenuated. Moreover, increased tumor growth is observed when PyMT tumor cells are transplanted in the senescent environment of Rank+/tg MGs, and the senescence conditioned media favors human breast cancer stemness. These results suggest that senescent MECs in the vicinity of PyMT+/−Rank+/tg tumors contribute to tumor growth probably through paracrine mediators as reported in other systems (Coppé et al., 2010; Schosserer et al., 2017). However, we cannot discard that tumor-cell-intrinsic mechanisms also contribute to the faster tumor growth, metastasis, and stemness observed in double transgenic PyMT+/−Rank+/tg mice. Lineage tracing studies will help to determine whether there is a direct conversion of senescent cells into mammary stem or cancer stem cells.
Collectively, these results demonstrate that rank induces senescence and stemness in non-transformed cells and in breast cancer. Rank-induced senescence acts as a tumor suppressor mechanism in early stages of tumorigenesis but later increases tumor aggressiveness. Our findings point to a physiological role for Rank-induced senescence and stemness during MG development, tumor initiation, and metastasis, which may have implications for breast cancer prevention and treatment.
Limitations of the study
One of the limitations of our current Rank+/tg mouse models is that Rank expression is under the MMTV promoter, which is mainly but not exclusively expressed in luminal cells and it is regulated by multiple factors, including hormones and even Rankl. Moreover, since it is not feasible to isolate senescent cells for further functional assays, we cannot determine whether Rank-induced senescent luminal cells may hold stemness properties. Tracing strategies and superior genetic models that allow to direct and control Rank expression specifically to basal or luminal MECs will help to discern the particular contribution of Rank-driven senescence within each cell compartment to stemness, and in a cancer context, to tumor outcome.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Keratin 5 rabbit polyclonal AF-138 | Covance Biolegend | Cat#905501; RRID: AB_2565050 |
| Keratin 5 chicken | Biolegend | Cat#905901; RRID: AB_2565054 |
| Mouse Keratin 8 | Biologend | Cat#904801; RRID: AB_2565043 |
| Rat monoclonal anti-CK8 | Deposited to the Developmental Studies Hybridoma Bank (NIH), Univ. of Iowa (USA) by Brulet, P. / Kemler, R | Cat#TROMA-I |
| Anti-CK14 AF-64 | Covance Biolegend | Cat#PRB-155P; RRID: AB_292096 |
| Anti phospho-H2AX (Ser139) | Millipore | Cat#05-636-I |
| Ki67 | Cell Signaling Technologies | Cat#12202 |
| Ki67 | Thermo Fisher Scientific | RM9106S1 Cat#12603707 |
| Progesterone Receptor clone SP2 | Thermo Fisher Scientific | RM-9102-R7 Cat#12683667 |
| Mouse Rank | R&D Systems | Cat#AF692 |
| Human Rank | Amgen | Cat#N-1H8 |
| Biotinylated goat anti-mouse | Vector Laboratories | Cat#BA-9200 |
| CDKN2A/P19 ARF clone 5-C3-1 | Santa Cruz | Cat#sc-32748 |
| Goa anti-mouse IgG Alexa 546 conjugated | Molecular Probes | Cat#A-11010; RRID: AB_2534077 |
| Donkey anti-rabbit IgG Alexa 647 conjugated | Molecular Probes | Cat A-31573; RRID: AB_2536183 |
| Phospho-NF-κB p65 (Ser536) (clone 93H1) | Cell Signaling Technologies | Cat#3033 |
| Tubulin | Abcam | Cat#Ab21058 |
| Biological samples | ||
| Human breast cancer | University Hospital Bellvitge (Spain) | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Navitoclax ABT-263 | Selleck Chemicals | Cat#S1001 |
| Rankl-lz | Amgen | N/A |
| Diva Deckloader Solution | Biocare Medical | Cat#DV2004G1 |
| Protease XXIV | Sigma-Aldrich | Cat#P8038-100MG |
| streptavidin-horseradish peroxidase | Vector Laboratories | Cat#SA-5004-1 |
| DAB substrate | DAKO | Cat#K3468 |
| TSA Blocking Reagent | Perkin Elmer | Cat# FP1012 |
| Prolong Gold Antifade reagent | Thermo Fisher Scientific | Cat#P36930 |
| 5-bromo-4-chloro-3-indolyl beta-D-galactoside (X-Gal) | Apollo Scientific | Cat#BIMB1001 |
| Mycoplasma test detection kit | Biotools | Cat#4542 |
| Epidermal Growth Factor (EGF) | Sigma | Cat#E9644 |
| Insulin | Sigma | Cat#I0516 |
| Cholera toxin | Sigma | Cat#C8052-.5MG |
| PEG400 | MERK | Cat#81172-1L |
| Matrigel matrix | BD Biosciences | Cat# 354234 |
| Collagenase A | Sigma | Cat#11088793001 |
| Dispase I | Life Technologies | Cat#17105041 |
| Hypotonic lysis buffer | Lonza Ibérica | Cat#10-548E |
| DNase I | Roche | Cat#11284932001 |
| Blasticidin | Thermo Fisher Scientific | Cat#A1113903 |
| Puromycin | SC-108071 | Cat#SC-108071 |
| EpCAM microbeads | Miltenyi Biotec | Cat#130-105-958 |
| LS columns | Miltenyi Biotec | Cat#130042401 |
| B27 supplement | Gibco | Cat#17504044 |
| N2 supplement | Gibco | Cat#17502048 |
| Accutase | Life Technologies | Cat#A1110501 |
| TriPure Isolation Reagent | Roche | Cat#11667165001 |
| SuperScript® II Reverse Transcriptase | Thermo Fisher Scientific | Cat#18064022 |
| PhosSTOP™20 TABLETS | Roche | Cat#4906837001 |
| cOmplete™, Mini Protease Inhibitor Cocktail | Merck | Cat#11836153001 |
| Critical commercial assays | ||
| LightCycler® 480 SYBR green MasterMix | Roche | Cat#04707516001 |
| RNeasy Mini kit | Qiagen | Cat#50974134 |
| Vectastain Elite ABC system | Vector | Cat#PK-6105 Cat#PK-4002 |
| DC Protein Assay Reagent | Bio-Rad | Cat#5000112 |
| Mouse Cytokine Array C100 Kit | RayBiotech | Cat# AAM-CYT-1000-2 |
| Deposited data | ||
| RNA-seq o f luminal and basal MECs | GEO ID: GSE139675 | N/A |
| Experimental models: cell lines | ||
| MCF7 | ATCC | HTB-22 |
| Mouse MEFs | This paper | N/A |
| Mouse MECs | This paper | N/A |
| HEK-293T | ATCC | CRL-3216 |
| Experimental models: organisms/strains | ||
| Mouse: FVB/N-Tg(MMTVneu)202Mul/J | The Jackson Laboratory | Cat#002376 |
| Mouse: FVB/N-Tg(MMTV-PyVT)634Mul/J | The Jackson Laboratory | Cat#002374 |
| Mouse: MMTV-Rank (FVB) | Amgen | N/A |
| Mouse: INK4a null | IRB (Manuel Serrano) | N/A |
| Mouse: Athymic nude, Foxn1nu | Harlan Envigo | Cat#069 |
| Oligonucleotides | ||
| See Table S3 for genotyping and qPCR analyses (SYBR green) | ||
| Recombinant DNA | ||
| pDONR 201 | Invitrogen | Cat#12536017 |
| pENTR223.1 | This paper | N/A |
| pwpi-GW | modified from the original Addgene plasmid # 12254 by Howard Kendrick and M Smalley | N/A |
| psD69 | M Bentires-Alj and S Duss | N/A |
| pLenti6/V5-DEST | Invitrogen | Cat#V49610 |
| pCW57.1 | Addgene | Cat#41393 |
| pEIZ-PyMT | modified from the Addgene plasmid #18121 HIV-ZSGreen by B Welm, Huntsman Cancer Institute, USA | N/A |
| pwpi-NeuN | M Smalley, Cardiff, UK | N/A |
| HRAS (G12V)-pcw107 | modified from the original Addgene plasmid #64603, by M Collado, IDIS, Spain | N/A |
| Software and algorithms | ||
| ImageJ | (Rueden et al., 2017) | https://imagej.nih.gov/ij/ |
| LAS AF Lite | Leica | https://leica-las-af-lite.software.informer.com/4.0/ |
| LightCycler 480 software | Roche | Cat#04994884001 |
| STAR/ 2.6.1b | University of Chicago, USA | https://rcc.uchicago.edu/docs/software/modules/STAR/midway2/2.6.1b.html |
| Package subread version 1.5 | (Liao et al., 2014). | https://bioconductor.org/packages/release/bioc/html/Rsubread.html |
| Limma package ver 3.40.6 | (Smyth, 2005) | https://www.bioconductor.org/packages/release/bioc/html/limma.html |
| R version 3.6.0 | R Core Team (2020). R Foundation for Statistical Computing, Vienna, Austria | https://www.R-project.org/ |
| edgeR package | (Robinson et al., 2010) | https://bioconductor.org/packages/release/bioc/html/edgeR.html |
| GSEA software | UC San Diego and Broad Institute | https://www.gsea-msigdb.org/gsea/index.jsp |
| UCSC Xernabrowser | (Goldman et al., 2020) | https://xenabrowser.net/ |
| BioRender.com | N/A | |
| Smart (Servier Medical Art) | LES LABORATOIRES SERVIER | https://smart.servier.com/ |
| Other | ||
| 8-well chamber slides | LabTek | Cat#154534 |
| McIlwain tissue chopper | Campden Instrument Ltd. | Cat# TC752 |
| Leica TCS SP5 confocal microscope | Leica | N/A |
| PreCellys® tissue homogenizer | Berting Technologies | Cat# P000669-PR240-A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Eva González-Suárez (egonzalez@cnio.es; egsuarez@idibell.cat).
Materials availability
All plasmids, cell lines and mouse lines generated in this work can be requested from the lead contact's laboratory.
Data and code availability
Original gene expression data by RNAseq from luminal and basal MECs from Rank+/tg and WT mice have been submitted to GEO ID: GSE139675. Table S1 includes differentially expressed genes between WT and Rank+/tg MECs and GSEA analyses. Table S2 includes the associations found in the TCGA dataset between RANK expression levels and senescence gene sets.
Experimental model and subject details
In vivo animal studies
All research involving animals was performed at the IDIBELL and CNIO animal facilities in compliance with protocols approved by the IDIBELL and CNIO Committees on Animal Care and following national and European Union regulations. MMTV-Neu mice (N202 Mul; FVB background) and MMTV-PyMT (FVB/N-Tg(MMTV-PyMT)634Mul) were obtained from Jackson laboratory and have been described previously) (Guy et al., 1992). MMTV-Rank mice (FVB background) were obtained through an MTA from Amgen (Gonzalez-Suarez et al., 2007). MMTV-Neu (Neu+/-) and MMTV-PyMT (PyMT-/+) were crossed with MMTV-Rank+/tg (Rank+/tg) to obtain the double mutant PyMT+/-Rank+/tg and Neu+/-Rank+/tg mice. p16/p19-/- mice were obtained through a collaboration with M Serrano (IRB Barcelona) and bred with Rank+/tg mice. Given the fast tumor onset in PyMT+/- (FVB) females, PyMT+/- males are bred with Rank+/tg female mice and after birth the pups were fed by a foster mother, as Rank+/tg females cannot lactate (Cordero et al., 2016; Gonzalez-Suarez et al., 2007). Mice were monitored for tumor formation 3 times per week and tumors bigger than 1 cm diameter were considered as endpoint criteria for sacrifice. PyMT+/- (FVB) were backcrossed to C57Bl6 for at least 15 generations. MEFs were obtained from embryos at day E13. For obtaining MECs and tumor cells, adult females (≥ 8 weeks-old) were used. MECs from PyMT+/- (C57Bl6) were used for lentiviral infections as tumor onset is delayed in this background.
Cell lines and primary cultures
MEFs were prepared essentially as described (Serrano et al., 1997). Briefly, each embryo was dispersed and trypsinized in a 10 cm diameter plate and incubated for 2 days. At this point, most of the attached cells had the appearance of fibroblasts. These cells were frozen in aliquots and considered passage 0 (P0). Human breast cancer MCF7 cell line was purchased from the American Type Culture Collection (Rockville, MD). ATCC provides molecular authentication is support of their collection through their genomics and proteomic cores by using DNA barcoding and species identification, quantitative gene expression and transcriptomic analyses. Lines were expanded and frozen within 2 weeks of purchase and used for a maximum of 4 months after being unfrozen. Cell lines were routinely tested for mycoplasma (Biotools, B & M Labs) and before each experiment they were confirmed to be free of contamination. MECs were derived from adult female mice (≥ 8 weeks-old) as described below and were cultured in DMEM/F12, 5% FBS (Gibco), epidermal growth factor (EGF) 100 μg/ml (Sigma), insulin 10 mg/ml (Sigma), cholera toxin 1 mg/ml and 1X Penicillin/Streptomycin (ThermoFisher Scientific). MEFs and human breast carcinoma cells were cultured in DMEM high glucose containing 10% FBS and 1X Penicillin/Streptomycin. All cells were incubated at 37°C in 5% CO2 incubators. Primary p16/p19-/- MEFs were provided by M Collado. For Rankl treatments in vitro, 1 μg/ml RANKL-LZ (Amgen) was used.
Human tumor samples
Samples for immunohistochemical analysis from consented breast cancer patients were collected from the University Hospital of Bellvitge (detailed in (Palafox et al., 2012)), using protocols approved by the IDIBELL Ethics Committee and according to Declaration of Helsinki.
Methods details
Rankl and Navitoclax treatments
Treatments with the Bcl-2/Bcl-xL/Bcl-w-inhibitor Navitoclax, also known as ABT-263 (Selleckchem), were performed by daily oral gavage during 14 days at 25 mg/kg (dissolved in 15% DMSO/PEG400). Treatments with Rankl (Amgen) were performed 3 times per week by intraperitoneal injection, during 2 weeks at 0.75 mg/kg.
MECs and mammary tumor cell isolation and orthotopic transplants
Single cells were isolated from MGs or tumors as previously described (Smalley, 2010). Briefly, fresh tissues were mechanically dissected with McIlwain tissue chopper and enzymatically digested with appropriate medium (DMEM F-12, 0.3% collagenase A, 2.5 U/ml dispase, 20 mM HEPES, and Penicillin/Streptomycin) 45 min at 37°C. Samples were washed with Leibowitz L15 medium 10% fetal bovine serum (FBS) between each step. Erythrocytes were eliminated by treating samples with hypotonic lysis buffer (Lonza Iberica). For MEC isolation, fibroblasts were excluded by incubation with DMEM high glucose containing 10% FBS for 1 hour at 37°C (the majority of fibroblasts attach to the tissue culture plate while most of the epithelial organoids do not). Single MECs were then isolated by treating with trypsin for 2 min at 37°C followed by an incubation with 2.5 U/ml dispase I (GIBCO), 20 U/ml DNaseI (Roche) for 5 min at 37°C. Cell aggregates were removed by filtering the cell suspensions with 40 μm filters before counting.
For tumor cell isolation, samples were treated with trypsin for 2 min at 37°C. Cell aggregates were removed by filtering the cell suspension with 70 μm filters and 300.000 cells counted. For orthotopic transplants, tumor cells isolated from PyMT mice were mixed 1:1 with Matrigel matrix (BD Biosciences) and orthotopically implanted in the inguinal mammary gland of adult virgin syngeneic FVB Rank+/tg females and WT controls. Mammary tumor growth was monitored by palpation and caliper measurement 3 times per week.
Metastasis limiting dilution assays
For limiting dilution assays, mammary tumor cells isolated from 3 primary tumors of 2 PyMT+/- and 2 PyMT-/+Rank+/tg tumor bearing mice were pooled, resuspended in 200 μl of cold PBS and injected intravenously (tail vein) in limiting dilutions (1.000.000, 10.000, 1.000, 100 and 10 cells) in 5 weeks old Foxn1nu mice (Harlan) (4 mice per dilution). Mice were sacrificed 8 weeks after cell injection and lungs were collected for histological analyses.
Tissue histology and immunostaining
For histological analyses, 3 μm sections from formalin fixed, paraffin embedded mammary glands were cut and stained with hematoxylin and eosin (H&E). Entire lungs were step-sectioned every 75/100 μm and individual metastases identified histologically based on nuclear morphology and similarity with primary tumors.
For immunohistochemistry or immunofluorescence staining of CK5 (AF-138, Covance; 905901 Biolegend), CK8 (904801, Biolegend; TROMAI, obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at The University of Iowa, Department of Biology, Iowa City, IA, USA), CK14 (AF-64, Covance), γH2AX (05-636-I, Millipore), Ki67 (12202, Cell Signaling), and PR (SP2, 12683667, Fisher), 3 μm sections from paraffin or SA-βGal embedded mammary glands were sectioned, slides were deparaffinized and rehydrated. Antigen heat retrieval was performed with citrate buffer (pH 6) in a pressure cooker (20 min) prior to overnight antibody incubation at 4°C. Anti-mouse Rank (R&D Systems AF692) immunostaining was performed pretreating sections with Protease XXIV 5 U/ml (Sigma-Aldrich) for 5 min at room temperature. The antigen-antibody complex was detected with streptavidin-horseradish peroxidase (Vector Laboratories) and revealed with DAB substrate (DAKO). Anti-human RANK immunohistochemistry was carried out as detailed in (Gonzalez-Suarez et al., 2010). Briefly, antigen heat retrieval was performed with Diva Deckloader solution (Biocare Medical) and samples were blocked with avidin and biotin blocking (DAKO) in consecutive incubations for 15 min at room temperature. Upon overnight incubation with hRANK (N-1H8, Amgen) diluted in TNB blocking solution (Perkin Elmer), slides were incubated with biotinylated goat anti-mouse antibody (Vector Laboratories) for 30 min. For detection, Vectastin Elite ABC system (Vector) and DAB substrate (Invitrogen) were used and counterstained samples were mounted with DPX.
For immunofluorescence of MG tissue sections, suitable fluorochrome-conjugated secondary antibodies and DAPI were added after overnight primary antibody incubations at 4°C and slides were mounted with Prolong Gold Antifade reagent (ThermoFisher Scientific).
Senescence-associated β-galactosidase (SA-βGal) staining
For whole-mount SA-βGal staining, MGs and tumors were washed in PBS, fixed for 30 min (room temperature) in 2% formaldehyde/0.2% glutaraldehyde, washed, and incubated overnight at 37°C with fresh SA-βGal staining solution: 1 mg of 5-bromo-4-chloro-3-indolyl beta-D-galactoside (X-Gal) per ml (Apollo Scientific), 40 mM citric acid/sodium phosphate pH 5.5 (in case of human samples pH 6), 5 mM K3Fe[CN]6, 5 mM K4Fe[CN]6, 150 mM NaCl, and 2 mM MgCl2 (Dimri et al., 1995). Stained MGs/tumors were dehydrated in ethanol and embedded in paraffin. For SA-βGal staining of cells in culture, cells were washed in PBS, fixed for 15 min (room temperature) in 2% formaldehyde/0.2% glutaraldehyde, washed and incubated overnight at 37°C with fresh SA-βGal staining solution: 1 mg of X-Gal per ml (Fisher Scientific), 40 mM citric acid/sodium phosphate pH 5.5, 5 mM K3Fe[CN]6, 5 mM K4Fe[CN]6, 150 mM NaCl, and 2 mM MgCl2. For MCF7 cells the same protocol was used except for the 40 mM citric acid/sodium phosphate whose pH was 6.0. SA-βGal stained mammary glands and cells were photographed on a bright field microscope and the number of positive cells was quantified using ImageJ software.
Immunofluorescence analyses
MECs, MEFs and human breast cancer MCF7 cells were seeded in eight-wells chamber slides (LabTek) with growth medium (50.000 cells/well for MECs and 25.000 cells/well for MEFs and human cell lines). After 48 hours, the medium was removed, and cells were fixed in 2% paraformaldehyde and permeabilized using 0.5% Triton X-100 before blocking. Cells were incubated overnight at room temperature with the primary antibodies; Ki67 (ThermoFisher Scientific), p19 ARF (5-C3-1, Santa Cruz), Anti-phospho-Histone γH2A.X (Ser139) (Millipore) and then with Alexa-564-647 conjugated secondary antibodies (1:750; Molecular Probes) for 45 min at room temperature and DAPI for nuclear staining. Slides were mounted with Prolong Gold Antifade Reagent (ThermoFisher Scientific). Confocal analysis was carried out using a Leica confocal microscope. Images were captured using LasaF software (Leica). The number of positive foci was quantified using ImageJ software.
Plasmid generation
Rank overexpressing plasmids were generated using Gateway® cloning system strategy (Katzen, 2007) following manufacture instructions. The BP Reaction allows the recombination of an attB-plasmid (Rank/RANK) with an attP donor vector (pDONR 201, Invitrogen). Briefly, the ORFeome collaboration sequence of human or mouse RANK inserted in a pENTR223.1 vector were transferred to the following attR destination vectors by a LR reaction: pwpi-GW (modified from the original Addgene plasmid # 12254 to insert the gateway cassette by H Kendrick and M Smalley), psd69 (provided by M Bentires-Alj and S Duss), pLenti6/V5-DEST (Invitrogen #V49610) or pCW57.1 (Addgene plasmid # 41393). The expression plasmids were verified by restriction enzymes and qRT-PCR analyses upon cell transduction. Lentiviral vectors with promoters of different strength were used in order to drive different levels for Rank/RANK overexpression: pwpi (EF1a, contains GFP), plenti6/V5-DEST (CMV, blasticidin selection), psd69 (PGK, puromycin selection) and inducible vector pCW57.1 (puromycin selection).
In addition, the following plasmids were generously provided: (i) pEIZ-PyMT (Ma et al., 2012) (modified from the Addgene plasmid #18121 HIV-ZSGreen) by B Welm (Huntsman Cancer Institute, UT, USA); (ii) pwpi-NeuN (pSV2) by M Smalley (Stem Cells Institute, Cardiff, UK); and HRAS (G12V)-pcw107, modified from the original Addgene plasmid #64603, by M Collado (IDIS, Santiago de Compostela, Spain).
Lentivirus production and cell infection
Lentiviral infection was done following the manufacturer’s indications (Invitrogen). HEK 293T cells were transfected with lentiviral empty, Rank or RAS overexpressing plasmids and packaging (gag-pol, vsvg, rev) plasmids (Addgene) by calcium phosphate method. 25 mM HEPES was added 16 hours later. The virus was harvested 72 hours post-transfection, centrifugation at 1500 rpm for 5 min and filtered with 0.22 μm filters. Non-transformed MECs, MEFs, tumor cells derived from Neu+/- or PyMT+/- tumors and human breast cancer cell lines were transduced in a ratio 1:4 of the virus with fresh growth medium and with 8 μg/ml of polybrene. Plates were centrifuged 1 hour at 600 g at 32°C to improve the infection, infected cells were selected with 5 μg/ml of blasticidin (pLenti6/V5-DEST) or 1.5 μg/ml of puromycin (psd69, HRas (G12V)-pcw107 and the inducible vector). After selection infected cells were maintained in 0.75 μg/ml of puromycin or 2.5 μg/ml of blasticidin.
Mammosphere and tumorsphere assays
MECs or tumor cells were isolated as described and filtered to obtain single cells. As Rank+/tg MGs contain more epithelia than WT MGs, CD326+ (EPCAM+) cells were isolated using the EpCAM Microbeads and LS Columns (Miltenyi Biotec), to make sure that the same number of epithelial cells was plated. 25.000 cells/ml were cultured in 2 ml of serum-free mammosphere medium DMEM/F12, 20 ng/ml of EFG, 2% B27, 1% N2, 1% P/S and 1% glutamine onto ultra-low attachment multiwell culture plates (Sigma). Medium was replenished 3 times a week with or without Navitoclax (0.2 or 0.6 μM) or conditioned media. Conditioned media was collected from RANK overexpressing MCF7 cells or controls cultured in 2D for 8 days. After 7 days, mammospheres or tumorspheres were isolated by 5 min treatment with accutase (Life Technologies) at 37°C and 25.0000 cells plated for secondary mammospheres or tumorsphere formation in triplicates. Individual spheres from each replicate well were counted under a bright field microscope and pictures were taken for size analyses.
Soft agar colony formation assays
Infected MEFs were selected with the appropriate antibiotics and 5.000-40.000 cells were plated in a 0.6% agar upper layer over a bottom layer of 1.2% agar in triplicates following the protocol detailed in (Borowicz et al., 2014).
RNA isolation, RT-PCR and gene expression analysis
Total RNA of frozen tumor pieces was prepared with Tripure Isolation Reagent (Roche) following the manufacturer’s instructions. Frozen tumors tissues were fractionated using glass beads (Sigma-Aldrich) and PreCellys® tissue homogenizer (Berting Technologies). cDNA was produced by reverse transcription using 1 μg of RNA following kit instructions (Applied Biosystems). Quantitative PCR was performed using LightCycler® 480 SYBR green MasterMix (Roche).
Total RNA from MECs, MEFs, mouse and human breast cancer cells was extracted using the RNeasy Mini Kit (Qiagen), sorted cells with RNAeasy Micro kit (Qiagen) and reverse-transcribed with SuperScript® II Reverse Transcriptase (ThermoFisher Scientific). Quantitative RT-PCR (qRT-PCR) was performed using the LightCycler® 480 SYBR green. Primer sequences are indicated below. Ct analysis was performed using LightCycler 480 software (Roche).
Western blot analyses
Protein extracts were prepared with modified RIPA buffer (50 mM Tris pH 7.4, 150 mM NaCl, 1% Triton NP-40, 0.25% sodium deoxycholate) containing 1x PhosSTOP and Complete protease inhibitor cocktail (Roche), and protein concentrations determined with DC protein assay reagents (BIO-RAD). 15 μg protein were resolved by SDS-PAGE and blotted into Immobilon-P 0.45 μm membranes (Millipore). Antibodies against the following proteins were used for probing: Rank (R&D Systems AF692), p-p65 (#3033) and tubulin (Abcam ab21058).
RNA sequencing
MECs were isolated as previously described; luminal (CD24hi CD49flo) and basal (CD24lo CD49fhi) cells within the Lin- population were FACs-sorted. Raw sequencing reads in the fastq files were mapped with STAR version 2.6.1b (Dobin et al., 2013) to the Gencode release 17 based on the GRCm38.p6 reference genome and the corresponding GTF file. The table of counts was obtained with FeatureCounts function in the package subread, version 1.5.(Liao et al., 2014). The differential expression gene analysis (DEG) was assessed with voom+limma in the limma package version 3.40.6 (Smyth, 2005) and R version 3.6.0. Genes having less than 10 counts in at least 3 samples were excluded from the analysis. Raw library size differences between samples were treated with the weighted “trimmed mean method” TMM (Robinson and Oshlack, 2010) implemented in the edgeR package (Robinson et al., 2010). The normalized counts were used in order to make unsupervised analysis, PCA and clusters. For the differential expression (DE) analysis, read counts were converted to log2-counts-per-million (logCPM) and the mean-variance relationship was modelled with precision weights using voom approach in limma package. RNAseq data has been submitted to GEO ID: GSE139675.
GSEA
Pre-Ranked Gene Set Enrichment Analysis (GSEA) (GSEA 2017) was used in order to retrieve functional pathways. The ranked list of genes was generated using the -log(p.val)∗signFC for each gene from the statistics obtained in the DE analysis with limma (Smyth, 2005). Functional annotation is obtained based on the enrichment of gene sets belonging to gene set collections (MSigDB 2017) c2.all. Gene sets collected from various sources such as online pathway databases, publications in PubMed, and knowledge of domain experts. Breast carcinomas TCGA raw counts were downloaded from UCSC Xenabrowser, lowly expressed genes across samples and count normalization was performed using edgeR. Breast carcinomas samples classified as Luminal A or Luminal B were selected and ranked by RANK expression, top and bottom 15% quantile samples were selected as High and Low RANK expression respectively. GSEA was used to perform gene-set enrichment of the described gene signatures, genes were ranked based in Signal2Noise metric and called significant under an FDR q-value of 25%.
Mouse cytokine array
The cytokine array was performed using the Mouse Cytokine Array C100 Kit (RayBiotech) to measure the relative levels of proteins in mammary epithelial cells lysates. The assay was performed according to the manufacturer’s instructions. Cell lysates were pooled and an equal amount of total protein (500 μg) was loaded onto the array kit. Blot images were analyzed with ImageJ software.
Statistical analysis
Statistical analyses were performed using GraphPad Prism software. Analysis of the differences between mouse cohorts or conditions was performed with a two-tailed Student’s t-test or one-way ANOVA. Two-way ANOVA was used to analyze tumor growth curves. ∗∗∗∗ p < 0.0001; ∗∗∗ p < 0.001; ∗∗ p < 0.01; ∗ p < 0.05; n.s. not significant. Estimation of tumor-initiating cells in limiting dilutions was calculated using the extreme limiting dilution assay (ELDA) (Hu and Smyth, 2009).
Acknowledgments
This work was supported by grants to E. González-Suárez by the Agencia Estatal de Investigación (AEI) (SAF2014-55997-R, SAF2017-86117-R) co-funded by FEDER funds/European Regional Development Fund (ERDF) (a way to build Europe), by the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement no. 682935), and by the Fundació La Marató de TV3. We thank CERCA Programme/Generalitat de Catalunya for institutional support. S. Benítez was a recipient of an FPI, A. Collado-Solé holds an FPU from the MICINN and J Redondo-Pedraza is a recipient of an FPI Severo Ochoa Fellowship. We are grateful to Amgen for providing Rankl and Rankt+/tg mice. Plasmids were generously provided by B. Welm (Huntsman Cancer Institute, Utah, USA), Howard Kendrich and M. Smalley (Stem Cells Institute, Cardiff, UK), and S. Duss and M. Bentires-Alj (University of Basel, Basel, Switzerland). We thank the IDIBELL and CNIO Animal Facilities for their assistance with mouse colonies.
Author contributions
Collection and/or assembly of data, S.B., A.C., P.G.S., J.R.-P., A.S.R., M.J., A.S.-M., A.C.-S., G.Y., J.C.S., I.D.B., S.D.S.-Á., N.A., K.T., G.G.L., and E.G.-S.; data analyses and interpretation, all authors; manuscript writing, S.B., A.C., P.G.S., and E.G.-S.; conception and design, financial support, E.G.-S.; final approval of the manuscript, all authors.
Declaration of interests
E.G.-S. has served on advisory boards for Amgen and has received research funding from Amgen. M.S. is co-founder and advisor of Senolytic Therapeutics, S.L. (Spain) and Senolytic Therapeutics (USA) aimed at developing senolytic therapies.
Published: May 17, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.devcel.2021.04.022.
Supplemental information
References
- Baker D.J., Wijshake T., Tchkonia T., LeBrasseur N.K., Childs B.G., van de Sluis B., Kirkland J.L., van Deursen J.M. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature. 2011;479:232–236. doi: 10.1038/nature10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauséjour C.M., Krtolica A., Galimi F., Narita M., Lowe S.W., Yaswen P., Campisi J. Reversal of human cellular senescence: roles of the p53 and p16 pathways. EMBO J. 2003;22:4212–4222. doi: 10.1093/emboj/cdg417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beleut M., Rajaram R.D., Caikovski M., Ayyanan A., Germano D., Choi Y., Schneider P., Brisken C. Two distinct mechanisms underlie progesterone-induced proliferation in the mammary gland. Proc. Natl. Acad. Sci. USA. 2010;107:2989–2994. doi: 10.1073/pnas.0915148107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowicz S., Van Scoyk M., Avasarala S., Karuppusamy Rathinam M.K., Tauler J., Bikkavilli R.K., Winn R.A. The soft agar colony formation assay. J. Vis. Exp. 2014;(92):e51998. doi: 10.3791/51998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braig M., Schmitt C.A. Oncogene-induced senescence: putting the brakes on tumor development. Cancer Res. 2006;66:2881–2884. doi: 10.1158/0008-5472.CAN-05-4006. [DOI] [PubMed] [Google Scholar]
- Bringold F., Serrano M. Tumor suppressors and oncogenes in cellular senescence. Exp. Gerontol. 2000;35:317–329. doi: 10.1016/s0531-5565(00)00083-8. [DOI] [PubMed] [Google Scholar]
- Campisi J. From cells to organisms: can we learn about aging from cells in culture? Exp. Gerontol. 2001;36:607–618. doi: 10.1016/s0531-5565(00)00230-8. [DOI] [PubMed] [Google Scholar]
- Campisi J. Cancer and ageing: rival demons? Nat. Rev. Cancer. 2003;3:339–349. doi: 10.1038/nrc1073. [DOI] [PubMed] [Google Scholar]
- Centonze A., Lin S., Tika E., Sifrim A., Fioramonti M., Malfait M., Song Y., Wuidart A., Van Herck J., Dannau A. Heterotypic cell-cell communication regulates glandular stem cell multipotency. Nature. 2020;584:608–613. doi: 10.1038/s41586-020-2632-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J., Wang Y., Shao L., Laberge R.M., Demaria M., Campisi J., Janakiraman K., Sharpless N.E., Ding S., Feng W. Clearance of senescent cells by ABT263 rejuvenates aged hematopoietic stem cells in mice. Nat. Med. 2016;22:78–83. doi: 10.1038/nm.4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiche A., Le Roux I., von Joest M., Sakai H., Aguín S.B., Cazin C., Salam R., Fiette L., Alegria O., Flamant P. Injury-induced senescence enables in vivo reprogramming in skeletal muscle. Cell Stem Cell. 2017;20:407–414.e4. doi: 10.1016/j.stem.2016.11.020. [DOI] [PubMed] [Google Scholar]
- Collado M., Serrano M. The senescent side of tumor suppression. Cell Cycle. 2005;4:1722–1724. doi: 10.4161/cc.4.12.2260. [DOI] [PubMed] [Google Scholar]
- Collado M., Serrano M. Senescence in tumours: evidence from mice and humans. Nat. Rev. Cancer. 2010;10:51–57. doi: 10.1038/nrc2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins C.J., Sedivy J.M. Involvement of the INK4a/Arf gene locus in senescence. Aging Cell. 2003;2:145–150. doi: 10.1046/j.1474-9728.2003.00048.x. [DOI] [PubMed] [Google Scholar]
- Coppé J.P., Desprez P.Y., Krtolica A., Campisi J. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu. Rev. Pathol. 2010;5:99–118. doi: 10.1146/annurev-pathol-121808-102144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppé J.P., Patil C.K., Rodier F., Sun Y., Muñoz D.P., Goldstein J., Nelson P.S., Desprez P.Y., Campisi J. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008;6:2853–2868. doi: 10.1371/journal.pbio.0060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero A., Pellegrini P., Sanz-Moreno A., Trinidad E.M., Serra-Musach J., Deshpande C., Dougall W.C., Pujana M.A., González-Suárez E. Rankl impairs lactogenic differentiation through inhibition of the prolactin/Stat5 pathway at midgestation. Stem Cells. 2016;34:1027–1039. doi: 10.1002/stem.2271. [DOI] [PubMed] [Google Scholar]
- d'Adda di Fagagna F. Living on a break: cellular senescence as a DNA-damage response. Nat. Rev. Cancer. 2008;8:512–522. doi: 10.1038/nrc2440. [DOI] [PubMed] [Google Scholar]
- Di Micco R., Sulli G., Dobreva M., Liontos M., Botrugno O.A., Gargiulo G., dal Zuffo R., Matti V., d'Ario G., Montani E. Interplay between oncogene-induced DNA damage response and heterochromatin in senescence and cancer. Nat. Cell Biol. 2011;13:292–302. doi: 10.1038/ncb2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimri G.P., Lee X., Basile G., Acosta M., Scott G., Roskelley C., Medrano E.E., Linskens M., Rubelj I., Pereira-Smith O. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fata J.E., Kong Y.Y., Li J., Sasaki T., Irie-Sasaki J., Moorehead R.A., Elliott R., Scully S., Voura E.B., Lacey D.L. The osteoclast differentiation factor osteoprotegerin-ligand is essential for mammary gland development. Cell. 2000;103:41–50. doi: 10.1016/s0092-8674(00)00103-3. [DOI] [PubMed] [Google Scholar]
- Fernandez-Valdivia R., Mukherjee A., Ying Y., Li J., Paquet M., DeMayo F.J., Lydon J.P. The RANKL signaling axis is sufficient to elicit ductal side-branching and alveologenesis in the mammary gland of the virgin mouse. Dev. Biol. 2009;328:127–139. doi: 10.1016/j.ydbio.2009.01.019. [DOI] [PubMed] [Google Scholar]
- Franzen J., Wagner W., Fernandez-Rebollo E. Epigenetic modifications upon senescence of mesenchymal stem cells. Curr. Stem Cell Rep. 2016;2:248–254. doi: 10.1007/s40778-016-0051-7. [DOI] [Google Scholar]
- Goldman M.J., Craft B., Hastie M., Repečka K., McDade F., Kamath A., Banerjee A., Luo Y., Rogers D., Brooks A.N. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol. 2020 doi: 10.1038/s41587-020-0546-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Suarez E., Branstetter D., Armstrong A., Dinh H., Blumberg H., Dougall W.C. RANK overexpression in transgenic mice with mouse mammary tumor virus promoter-controlled RANK increases proliferation and impairs alveolar differentiation in the mammary epithelia and disrupts lumen formation in cultured epithelial acini. Mol. Cell. Biol. 2007;27:1442–1454. doi: 10.1128/MCB.01298-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Suarez E., Jacob A.P., Jones J., Miller R., Roudier-Meyer M.P., Erwert R., Pinkas J., Branstetter D., Dougall W.C. RANK ligand mediates progestin-induced mammary epithelial proliferation and carcinogenesis. Nature. 2010;468:103–107. doi: 10.1038/nature09495. [DOI] [PubMed] [Google Scholar]
- Guy C.T., Cardiff R.D., Muller W.J. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol. Cell. Biol. 1992;12:954–961. doi: 10.1128/mcb.12.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschkowitz J.I., Simin K., Weigman V.J., Mikaelian I., Usary J., Hu Z., Rasmussen K.E., Jones L.P., Assefnia S., Chandrasekharan S. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8:R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Smyth G.K. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods. 2009;347:70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- Katzen F. Gateway® recombinational cloning: a biological operating system. Expert Opin Drug Discov. 2007;2:571–589. doi: 10.1517/17460441.2.4.571. [DOI] [PubMed] [Google Scholar]
- Kodama S., Mori I., Roy K., Yang Z., Suzuki K., Watanabe M. Culture condition-dependent senescence-like growth arrest and immortalization in rodent embryo cells. Radiat. Res. 2001;155:254–262. doi: 10.1667/0033-7587(2001)155[0254:ccdslg]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Krizhanovsky V., Yon M., Dickins R.A., Hearn S., Simon J., Miething C., Yee H., Zender L., Lowe S.W. Senescence of activated stellate cells limits liver fibrosis. Cell. 2008;134:657–667. doi: 10.1016/j.cell.2008.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krtolica A., Parrinello S., Lockett S., Desprez P.Y., Campisi J. Senescent fibroblasts promote epithelial cell growth and tumorigenesis: a link between cancer and aging. Proc. Natl. Acad. Sci. USA. 2001;98:12072–12077. doi: 10.1073/pnas.211053698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Schmitt C.A. The dynamic nature of senescence in cancer. Nat. Cell Biol. 2019;21:94–101. doi: 10.1038/s41556-018-0249-2. [DOI] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Lin E.Y., Jones J.G., Li P., Zhu L., Whitney K.D., Muller W.J., Pollard J.W. Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am. J. Pathol. 2003;163:2113–2126. doi: 10.1016/S0002-9440(10)63568-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujambio A. To clear, or not to clear (senescent cells)? That is the question. BioEssays. 2016;38(suppl 1):S56–S64. doi: 10.1002/bies.201670910. [DOI] [PubMed] [Google Scholar]
- Ma J., Lanza D.G., Guest I., Uk-Lim C., Glinskii A., Glinsky G., Sell S. Characterization of mammary cancer stem cells in the MMTV-PyMT mouse model. Tumor Biol. 2012;33:1983–1996. doi: 10.1007/s13277-012-0458-4. [DOI] [PubMed] [Google Scholar]
- Maglione J.E., Moghanaki D., Young L.J., Manner C.K., Ellies L.G., Joseph S.O., Nicholson B., Cardiff R.D., MacLeod C.L. Transgenic polyoma middle-T mice model premalignant mammary disease. Cancer Res. 2001;61:8298–8305. [PubMed] [Google Scholar]
- Malaquin N., Martinez A., Rodier F. Keeping the senescence secretome under control: molecular reins on the senescence-associated secretory phenotype. Exp. Gerontol. 2016;82:39–49. doi: 10.1016/j.exger.2016.05.010. [DOI] [PubMed] [Google Scholar]
- Malaquin N., Tu V., Rodier F. Assessing functional roles of the senescence-Associated Secretory phenotype (SASP) Methods Mol. Biol. 2019;1896:45–55. doi: 10.1007/978-1-4939-8931-7_6. [DOI] [PubMed] [Google Scholar]
- Martínez-Zamudio R.I., Roux P.F., de Freitas J.A.N.L.F., Robinson L., Doré G., Sun B., Belenki D., Milanovic M., Herbig U., Schmitt C.A. AP-1 imprints a reversible transcriptional programme of senescent cells. Nat. Cell Biol. 2020;22:842–855. doi: 10.1038/s41556-020-0529-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K., Hodwin B., Ramanujam D., Engelhardt S., Sarikas A. Essential role for premature senescence of myofibroblasts in myocardial fibrosis. J. Am. Coll. Cardiol. 2016;67:2018–2028. doi: 10.1016/j.jacc.2016.02.047. [DOI] [PubMed] [Google Scholar]
- Milanovic M., Fan D.N.Y., Belenki D., Däbritz J.H.M., Zhao Z., Yu Y., Dörr J.R., Dimitrova L., Lenze D., Monteiro Barbosa I.A. Senescence-associated reprogramming promotes cancer stemness. Nature. 2018;553:96–100. doi: 10.1038/nature25167. [DOI] [PubMed] [Google Scholar]
- Mosteiro L., Pantoja C., de Martino A., Serrano M. Senescence promotes in vivo reprogramming through p16INK4a and IL-6. Aging Cell. 2018;17 doi: 10.1111/acel.12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller W.J., Sinn E., Pattengale P.K., Wallace R., Leder P. Single-step induction of mammary adenocarcinoma in transgenic mice bearing the activated c-neu oncogene. Cell. 1988;54:105–115. doi: 10.1016/0092-8674(88)90184-5. [DOI] [PubMed] [Google Scholar]
- Oakes S.R., Naylor M.J., Asselin-Labat M.L., Blazek K.D., Gardiner-Garden M., Hilton H.N., Kazlauskas M., Pritchard M.A., Chodosh L.A., Pfeffer P.L. The Ets transcription factor Elf5 specifies mammary alveolar cell fate. Genes Dev. 2008;22:581–586. doi: 10.1101/gad.1614608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocampo A., Reddy P., Martinez-Redondo P., Platero-Luengo A., Hatanaka F., Hishida T., Li M., Lam D., Kurita M., Beyret E. In vivo amelioration of age-associated hallmarks by partial reprogramming. Cell. 2016;167:1719–1733.e12. doi: 10.1016/j.cell.2016.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Loghlen A., Martin N., Krusche B., Pemberton H., Alonso M.M., Chandler H., Brookes S., Parrinello S., Peters G., Gil J. The nuclear receptor NR2E1/TLX controls senescence. Oncogene. 2015;34:4069–4077. doi: 10.1038/onc.2014.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onder T.T., Kara N., Cherry A., Sinha A.U., Zhu N., Bernt K.M., Cahan P., Marcarci B.O., Unternaehrer J., Gupta P.B. Chromatin-modifying enzymes as modulators of reprogramming. Nature. 2012;483:598–602. doi: 10.1038/nature10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orioli D., Dellambra E. Epigenetic regulation of skin cells in natural aging and premature aging diseases. Cells. 2018;7 doi: 10.3390/cells7120268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özcan S., Alessio N., Acar M.B., Mert E., Omerli F., Peluso G., Galderisi U. Unbiased analysis of senescence associated secretory phenotype (SASP) to identify common components following different genotoxic stresses. Aging. 2016;8:1316–1329. doi: 10.18632/aging.100971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palafox M., Ferrer I., Pellegrini P., Vila S., Hernandez-Ortega S., Urruticoechea A., Climent F., Soler M.T., Muñoz P., Viñals F. RANK induces epithelial-mesenchymal transition and stemness in human mammary epithelial cells and promotes tumorigenesis and metastasis. Cancer Res. 2012;72:2879–2888. doi: 10.1158/0008-5472.CAN-12-0044. [DOI] [PubMed] [Google Scholar]
- Pellegrini P., Cordero A., Gallego M.I., Dougall W.C., Muñoz P., Pujana M.A., Gonzalez-Suarez E. Constitutive activation of RANK disrupts mammary cell fate leading to tumorigenesis. Stem Cells. 2013;31:1954–1965. doi: 10.1002/stem.1454. [DOI] [PubMed] [Google Scholar]
- Pfitzner B.M., Branstetter D., Loibl S., Denkert C., Lederer B., Schmitt W.D., Dombrowski F., Werner M., Rüdiger T., Dougall W.C., von Minckwitz G. RANK expression as a prognostic and predictive marker in breast cancer. Breast Cancer Res. Treat. 2014;145:307–315. doi: 10.1007/s10549-014-2955-1. [DOI] [PubMed] [Google Scholar]
- Prieur A., Peeper D.S. Cellular senescence in vivo: a barrier to tumorigenesis. Curr. Opin. Cell Biol. 2008;20:150–155. doi: 10.1016/j.ceb.2008.01.007. [DOI] [PubMed] [Google Scholar]
- R Core Team . R Foundation for Statistical Computing, Vienna, Austria; 2020. R: A language and environment for statistical computing.http://www.R-project.org/ [Google Scholar]
- Ritschka B., Storer M., Mas A., Heinzmann F., Ortells M.C., Morton J.P., Sansom O.J., Zender L., Keyes W.M. The senescence-associated secretory phenotype induces cellular plasticity and tissue regeneration. Genes Dev. 2017;31:172–183. doi: 10.1101/gad.290635.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueden C.T., Schindelin J., Hiner M.C., DeZonia B.E., Walter A.E., Arena E.T., Eliceiri K.W. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics. 2017;18:529. doi: 10.1186/s12859-017-1934-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkisian C.J., Keister B.A., Stairs D.B., Boxer R.B., Moody S.E., Chodosh L.A. Dose-dependent oncogene-induced senescence in vivo and its evasion during mammary tumorigenesis. Nat. Cell Biol. 2007;9:493–505. doi: 10.1038/ncb1567. [DOI] [PubMed] [Google Scholar]
- Schosserer M., Grillari J., Breitenbach M. The dual role of cellular senescence in developing tumors and their response to cancer therapy. Front. Oncol. 2017;7:278. doi: 10.3389/fonc.2017.00278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramek D., Leibbrandt A., Sigl V., Kenner L., Pospisilik J.A., Lee H.J., Hanada R., Joshi P.A., Aliprantis A., Glimcher L. Osteoclast differentiation factor RANKL controls development of progestin-driven mammary cancer. Nature. 2010;468:98–102. doi: 10.1038/nature09387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano M., Lin A.W., McCurrach M.E., Beach D., Lowe S.W. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- Shehata M., Teschendorff A., Sharp G., Novcic N., Russell I.A., Avril S., Prater M., Eirew P., Caldas C., Watson C.J., Stingl J. Phenotypic and functional characterisation of the luminal cell hierarchy of the mammary gland. Breast Cancer Res. 2012;14:R134. doi: 10.1186/bcr3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalley M.J. Mouse Cell Culture. Humana Press; 2010. Isolation, culture and analysis of mouse mammary epithelial cells; pp. 139–170. [DOI] [PubMed] [Google Scholar]
- Smyth G.K. limma: linear models for microarray data. In: Gentleman R., Carey V.J., Huber W., Irizarry R.A., Dudoit S., editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Statistics for Biology and Health. Springer; New York, NY: 2005. [DOI] [Google Scholar]
- Sun Y., Coppé J.P., Lam E.W. Cellular senescence: the sought or the unwanted? Trends Mol. Med. 2018;24:871–885. doi: 10.1016/j.molmed.2018.08.002. [DOI] [PubMed] [Google Scholar]
- Tanos T., Sflomos G., Echeverria P.C., Ayyanan A., Gutierrez M., Delaloye J.F., Raffoul W., Fiche M., Dougall W., Schneider P. Progesterone/RANKL is a major regulatory axis in the human breast. Sci. Transl. Med. 2013;5:182ra55. doi: 10.1126/scitranslmed.3005654. [DOI] [PubMed] [Google Scholar]
- Tu Z., Aird K.M., Zhang R. RAS, cellular senescence and transformation: the BRCA1 DNA repair pathway at the crossroads. Small GTPases. 2012;3:163–167. doi: 10.4161/sgtp.19884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergel M., Carnero A. Cellular senescence as a barrier to environmental carcinogenesis. J. Carcinog. Mutagen. 2014:S3. doi: 10.4172/2157-2518.S3-004. [DOI] [Google Scholar]
- Wang L., Bernards R. Taking advantage of drug resistance, a new approach in the war on cancer. Front. Med. 2018;12:490–495. doi: 10.1007/s11684-018-0647-7. [DOI] [PubMed] [Google Scholar]
- Wang L., Leite de Oliveira R., Wang C., Fernandes Neto J.M., Mainardi S., Evers B., Lieftink C., Morris B., Jochems F., Willemsen L. High-throughput functional genetic and compound screens identify targets for senescence induction in cancer. Cell Rep. 2017;21:773–783. doi: 10.1016/j.celrep.2017.09.085. [DOI] [PubMed] [Google Scholar]
- Wu D., Yu S., Jia L., Zou C., Xu Z., Xiao L., Wong K.B., Ng C.F., Chan F.L. Orphan nuclear receptor TLX functions as a potent suppressor of oncogene-induced senescence in prostate cancer via its transcriptional co-regulation of the CDKN1A (p21(WAF1) (/) (CIP1) ) and SIRT1 genes. J. Pathol. 2015;236:103–115. doi: 10.1002/path.4505. [DOI] [PubMed] [Google Scholar]
- Yang N., Sen P. The senescent cell epigenome. Aging. 2018;10:3590–3609. doi: 10.18632/aging.101617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoldi G., Pellegrini P., Trinidad E.M., Cordero A., Gomez-Miragaya J., Serra-Musach J., Dougall W.C., Muñoz P., Pujana M.A., Planelles L., González-Suárez E. RANK signaling blockade reduces breast cancer recurrence by inducing tumor cell differentiation. Cancer Res. 2016;76:5857–5869. doi: 10.1158/0008-5472.CAN-15-2745. [DOI] [PubMed] [Google Scholar]
- Zhang C.L., Zou Y., He W., Gage F.H., Evans R.M. A role for adult TLX-positive neural stem cells in learning and behaviour. Nature. 2008;451:1004–1007. doi: 10.1038/nature06562. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Tchkonia T., Fuhrmann-Stroissnigg H., Dai H.M., Ling Y.Y., Stout M.B., Pirtskhalava T., Giorgadze N., Johnson K.O., Giles C.B. Identification of a novel senolytic agent, navitoclax, targeting the Bcl-2 family of anti-apoptotic factors. Aging Cell. 2016;15:428–435. doi: 10.1111/acel.12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Tchkonia T., Pirtskhalava T., Gower A.C., Ding H., Giorgadze N., Palmer A.K., Ikeno Y., Hubbard G.B., Lenburg M. The Achilles' heel of senescent cells: from transcriptome to senolytic drugs. Aging Cell. 2015;14:644–658. doi: 10.1111/acel.12344. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Original gene expression data by RNAseq from luminal and basal MECs from Rank+/tg and WT mice have been submitted to GEO ID: GSE139675. Table S1 includes differentially expressed genes between WT and Rank+/tg MECs and GSEA analyses. Table S2 includes the associations found in the TCGA dataset between RANK expression levels and senescence gene sets.