Abstract
Background:
After heart transplantation, Endomyocardial biopsy (EMBx) is used to monitor for acute rejection (AR). Unfortunately, EMBx is invasive and its conventional histologic interpretation has limitations. This is a validation study to assesses the performance of a sensitive blood biomarker— percent donor-derived cell-free DNA (%ddcfDNA) — for detection of AR in cardiac transplant recipients.
Methods:
This multicenter, prospective cohort study recruited heart transplant subjects and collected plasma samples contemporaneously with EMBx for %ddcfDNA measurement by shotgun sequencing. Histopathology data was collected to define AR, its two phenotypes (acute cellular rejection, ACR, and antibody-mediated rejection, AMR) and controls without rejection. The primary analysis was to compare %ddcfDNA levels (median and interquartile range – IQR) for AR, AMR and ACR to controls and to determine %ddcfDNA test characteristics using receiver-operator characteristics analysis.
Results:
The study included 171 subjects with median post-transplant follow-up of 17.7 months (IQR: 12.1-23.6), with 1,392 EMBx, and 1,834 ddcfDNA measures available for analysis. Median %ddcfDNA levels decayed after surgery to 0.13% (0.03-0.21) by 28 days. %ddcfDNA increased again with AR compared to controls values (0.38, IQR=0.31-0.83, vs. 0.03, IQR=0.01-0.14 p<0.001). The rise was detected 0.5 and 3.2 months before histopathological diagnosis of ACR and AMR. The area-under-the- receiver-operator characteristics curve (AUROC) for AR was 0.92. A 0.25 %ddcfDNA threshold had a negative predictive value (NPV) for AR of 99% and would have safely eliminated 81% of EMBx. %ddcfDNA showed distinctive characteristics comparing AMR to ACR, included 5-fold higher levels (pAMR ≥2 1.68, IQR=0.49-2.79 vs. ACR grade ≥2R 0.34, IQR=0.28-0.72), higher AUROC (0.95 vs. 0.85), higher guanosine-cytosine content, and higher percentage of short ddcfDNA fragments.
Conclusion:
%ddcfDNA detected AR with a high AUROC and NPV. Monitoring with ddcfDNA, demonstrated excellent performance characteristics for both ACR and AMR and led to earlier detection than the EMBx-based monitoring. This study supports the use of %ddcfDNA to monitor for AR in heart transplant patients and paves the way for a clinical utility study.
Clinical Trial Registration:
http://clinicaltrials.gov/ct2/show/NCT02423070 (NCT#02423070)
Keywords: cardiac transplantation, biomarkers, cell-free DNA, allograft rejection, graft injury
Introduction
Early diagnosis and treatment of acute rejection (AR) is a major pillar in the management of heart transplant recipients. The goal is to preserve graft function, extend patient survival, and avoid cardiac allograft vasculopathy (CAV), a leading cause of long-term allograft failure and mortality.1, 2 Histological grading of AR on endomyocardial biopsy plus histopathology (EMBx) is the standard method for diagnosing AR, assessing its severity, and the response to therapy. However, AR has two phenotypes, acute cellular rejection (ACR) and antibody-mediated rejection (AMR), which challenges the histopathologic diagnosis. Both have internationally accepted guidelines for grading,3, 4 but their initial presentation is often clinically silent. Further the histopathologic interpretation of the EMBx has low sensitivity to detect early rejection, high interobserver variability,5 and the invasive nature of an EMBx pose further limitations to early diagnosis and treatment. Therefore, a reliable non-invasive marker to detect AR prior to the development of graft dysfunction would possibly result in better outcomes for those patients who develop allograft rejection.
We previously developed a novel non-invasive genomic blood test — donor-derived cell-free DNA (ddcfDNA) — as a potential biomarker for heart transplant rejection.6, 7 When allograft cells die, they release short DNA fragments (ddcfDNA) into the recipient circulation. Through sequencing and single nucleotide polymorphism assessment, ddcfDNA fragments are easily identified and quantitated. The percentage of ddcfDNA (%ddcfDNA) describes the amount of ddcfDNA compared with the total cfDNA found in the blood. Our single-center studies showed that the percentage of %ddcfDNA increased with allograft rejection and decreased after treatment. Few studies have reported on the utility of %ddcfDNA for diagnosis of AMR. In one multicenter study of renal transplant recipients, the median %ddcfDNA was significantly higher in AMR than in patients with ACR or no rejection.8 Among lung transplant recipients, we showed that allograft injury detected by ddcfDNA preceded clinical or histopathological signs of AMR by a median of 2.8 months.9 Taken together, these data suggest that ddcfDNA reflects the severity of allograft injury and, thus, might serve as a clinically useful biomarker of early AMR.
Validation of these initial findings is critical prior to a clinical utility study. In 2015, the National Heart, Lung, and Blood Institute (NHBLI) established a multicenter prospective cohort study to validate the characteristics %ddcfDNA to detect AR.10 The initial studies from the Genomic Research Alliance for Transplantation (GRAfT), confirmed reproducibility of the ddcfDNA assay across multiple platforms, different cohorts, and in heart and lung transplant recipients.11 In this study, our objective was to determine the suitability of ddcfDNA as a biomarker of cardiac allograft injury. We hypothesized that higher levels of ddcfDNA would be detected with AMR than ACR as seen in other solid organ transplant populations. In addition, we hypothesized that %ddcfDNA would be elevated prior to the clinical diagnosis of AR. To test these hypotheses, we designed a prospective cohort study to determine: 1) the performance characteristics of ddcfDNA for diagnosis of AR that meets standard criteria for treatment (i.e., ACR grade ≥ 2 or AMR grade ≥ 1; 2) the relationship between %ddcfDNA and rejection grade (i.e., severity of histological findings on biopsy); 3) the ability of ddcfDNA to distinguish AMR from ACR; and 4) assess the prevalence of biopsy-negative allograft injury.
Methods
Data Sharing
The data, analytical methods, and study materials will be made available to other researchers for the purposes of reproducing results or replicating procedures. Please contact Dr. Sean Agbor-Enoh with specific requests sean-agbor.enoh@nih.gov.
Study Design
This prospective cohort study was conducted by GRAfT (NCT#02423070). GRAfT includes five transplant centers (The Johns Hopkins Hospital, University of Maryland Medical Center, Inova Heart and Vascular Institute, Medstar Washington Hospital Center, Virginia Commonwealth University) within geographic proximity to the NHLBI. GRAfT was established to validate the test characteristics of %ddcfDNA for acute rejection and to determine the ability of the test to predict long term outcomes after heart transplantation, including CAV, graft failure, and mortality.10 Subjects who were 18 years of age or older were recruited from the heart transplant waitlist and monitored after transplantation. Surveillance monitoring at pre-specified post-transplant time points included EMBx for histopathology, donor specific antibodies (DSA) and cytomegalovirus (CMV) testing, as well as monitoring immunosuppression trough levels. Clinically indicated monitoring often includes EMBx, DSA, echocardiography, and the other tests performed when patients present with unexplained signs or symptoms of allograft dysfunction. The GRAfT study longitudinally tracked clinical data and collected plasma samples coincident to both surveillance and clinically indicated monitoring (post-transplant surveillance protocol, induction and maintenance immunosuppression regimen Tables I and II in the Supplement). Clinical data was used to define the primary endpoint, pre-specified by GRAfT Steering Committee. Plasma samples were used to quantitate %ddcfDNA by unbiased shotgun sequencing.11 The performance characteristics of %ddcfDNA to detect AR was determined by the area-under-the-receiver-operator-characteristics curve (AUROC). The institutional review boards of all five centers and the NHLBI approved the study.
Primary and secondary endpoints:
The GRAfT Heart Steering Committee pre-specified the study endpoint. The Committee include heart transplant cardiologists (n=6), immunogeneticists (n=2), pathologists (n=2), a statistician (n=1), and genomics experts (n=2). The primary endpoint was AR defined by international standards,3, 4 as a composite endpoint of ACR or AMR, defined based on individual center histologic readings to be consistent with usual care and included the histopathology grades treated at GRAfT centers — ACR grade ≥2 and AMR grade ≥1. AMR grade 1 was defined based on either histologic (e.g. AMR1h) or immunohistochemical findings (e.g. AMR1i). Hemodynamics, DSA test results and echocardiographic findings were not required to make the diagnosis of AMR. The secondary endpoints were the two AR phenotypes (ACR grade ≥2 and AMR grade ≥1). Endpoints were defined based on histopathology reads by individual center’s pathologists to be consistent with usual care. Allograft dysfunction was defined as a reduction of left ventricular ejection fraction (LVEF) by ≥5%, and further stratified by severity based on the magnitude of LVEF decline as no (<5%), mild (5 – <10%), moderate (≥10 – <15%) or severe (≥15%) allograft dysfunction.
Sample size calculation
The sample size was computed to estimate the area under the receiver operating characteristic curve (AUROC), where the predictor is %ddcfDNA value, and the outcome variable is AR episode (yes/no) concurrent with the %ddcfDNA measurement. The sample-size calculation is based on achieving an pre-specified AUROC with a narrow confidence interval.12 Prior observations showed high immediately post-transplant %ddcfDNA levels that decay logarithmically to a low stable baseline level until the onset of AR, when %ddcfDNA levels increase again.6 So, the sample size was computed based on equations (6.2) and (6.5) from Zhou, Obuchowski and McClish.13 To obtain a two-sided 95% confidence interval of width 2L, we need N subjects with an AR episode, N being determined by: N = (3.84 * V)/L, where V is the variance of the AUROC estimate. Considering reported %ddcfDNA test AUROC of 0.83 (95% CI = 0.78-0.91) to diagnose AR, then, 13 AR episodes will correspond to a 95% confidence interval of length 2L = 0.25 when the AUROC = 0.78 (assuming the lower limit of the AUROC). Since we estimate that approximately 20% of the post-heart transplant subjects will have an AR episode, we conclude that 65 post-heart transplant subjects will be a reasonable sample size.
Quantitation of %ddcfDNA
%ddcfDNA was measured by shotgun sequencing.11 Briefly, prior to transplant donors and recipients were genotyped using DNA extracted from whole blood. Genotype data was compared for each donor-recipient pair to identify donor and recipient single nucleotide polymorphisms (SNPs) for the pair. After transplantation, plasma cfDNA was extracted to prepare DNA libraries for paired end shotgun sequencing. Sequence reads were surveyed for donor and recipient SNPs. %ddcfDNA was computed as percentage of reads with donor SNPs to total reads for donor plus recipient SNPs
Determining %ddcfDNA fragment length in rejection subtypes
To determine the fragment lengths, we leveraged paired-end shotgun sequencing, which sequence cfDNA fragments from both ends. Sequence reads were aligned to the human genome reference sequence (Hg19). Ends of properly aligned paired end reads were identified. The number of base pairs (bps) between allied paired ends on the reference genome were computed as the cfDNA fragment length.6, 14, 15 The distribution of fragment lengths were plotted, and based on this, three fragment length thresholds (140 bp, 120 bp and 100 bp) were used to define short cfDNA fragments. For each sample time-point, the percentage of short cfDNA fragments was computed as the number of short cfDNA fragments to total aligned cfDNA. The percentage of short cfDNA fragments for AMR or ACR was compared to controls; controls being defined as within-subject time-points with no rejection. All three short cfDNA thresholds produced similar results; thus, the threshold of 100 bp is reported in the results section. We also compared the percentage of guanosine and cytosine bases at AMR, ACR and no rejection controls.
The composition of genomic elements in rejection subtypes
Next, in a subset of patients, we compared the genomic composition of cfDNA in AMR and ACR by analyzing the variability in genomic elements identified within the allograft cfDNA. Genomic elements of interest include genomic regions associated with gene expression (exons, introns, promoters, transcriptional start sites) and intergenic regions. To determine the genomic composition, cfDNA was aligned to the reference genome to identify and annotate properly aligned sequences. The ratio of the number of reads annotated to each genomic element compared to the total number of aligned reads was computed and compared between AMR, ACR, and within subject controls.
Statistical Analysis
The primary analysis was to compute the area under the receiver operator characteristics curve (AUROC) with %ddcfDNA as predictor and AR as outcome. Only %ddcfDNA with concurrent EMBx were included. Controls for this analysis was defined as time-points not meeting AR definition (ACR/AMR grade 0 or ACR grade 1). To compute the AUROC, we first determined the post-transplant decay kinetics of %ddcfDNA assuming a one-phase logarithmic decay pattern based on a previous report.6 Median %ddcfDNA values at different post-transplant intervals were also computed and plotted to define changes in baseline allograft injury over time. Given post-transplant %ddcfDNA decay, we first determined change of the AUROC over-time post-transplantation using time points pre-specified by the steering committee to reflect clinical benchmarks (days 7, 14, 28 and 45). As an example, for the day 28 analysis, EMBx and %ddcfDNA data before Day 28 were excluded. The EMBx and %ddcfDNA data set with the highest AUROC was selected for all subsequent analyses (day 28). This included analyses to determine the correlation between %ddcfDNA and grades of acute rejection or severity of allograft dysfunction. In addition to using non-parametric tests (Mann Whitney U test or Kruskal-Willis test) to compare median %ddcfDNA between groups, we used the generalized estimating equation approach16 to compare %ddcfDNA levels between different groups, while accounting for the correlation among repeated %ddcfDNA measurements in the same subject. %ddcfDNA was log-transformed as log2(x+0.01) to reduce the skewness. For AMR and ACR events, we also plotted %ddcfDNA at diagnosis (time 0) and at pre-specified intervals preceding diagnosis. Next, we defined allograft injury as a %ddcfDNA ≥ 0.25% based on prior reports,6 and our experience, and then assessed the test characteristics for EMBx to detect allograft injury.
Results
Patient population
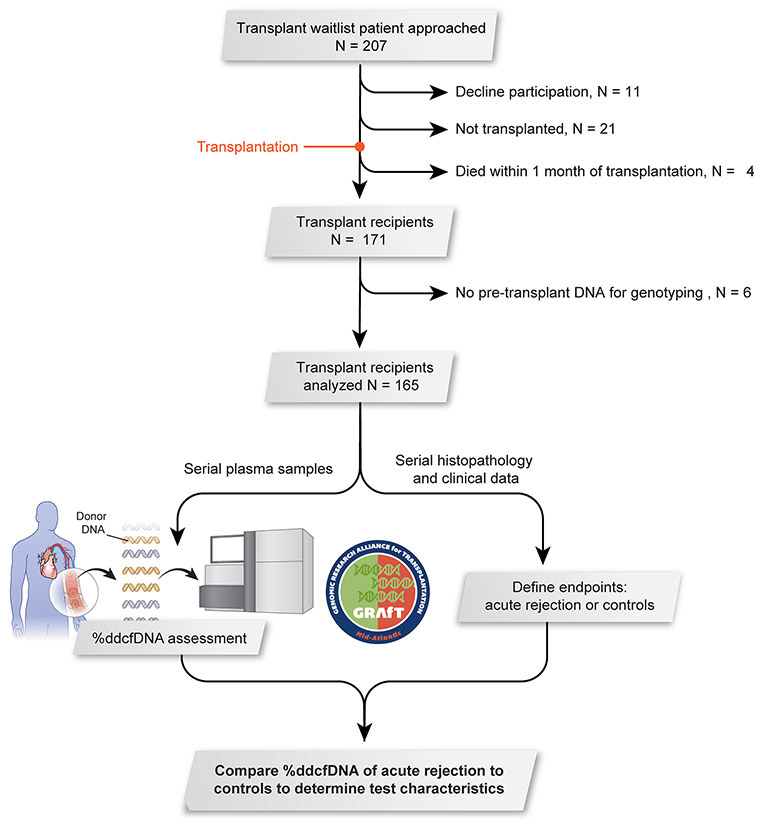
The cohort included 165 subjects with an average age of 53 years (range = 20–70 years) and similar proportion of patients with European or African ancestry (Figure 1). Females accounted for 28% of the study population. Non-ischemic cardiomyopathy (53%) was the most common reason of transplantation, and about two-thirds of patients had a ventricular assist device at the time of transplantation (Table 1). The median post-transplant follow-up was 17.7 months (IQR = 12.1 – 23.6).
Figure 1: Study design.
Patients were recruited from five regional transplant centers. Serial plasma samples were collected at the time of routine surveillance procedures after transplant (e.g. endomyocardial biopsy or echocardiogram) and when clinically indicated monitoring was performed (e.g. graft dysfunction or suspected rejection, Table I in the Supplement). A total of 165 patients were included in this analysis and 1,867 ddcfDNA measures.
Table 1:
Donor and recipient demographic factors (N = 165)
| Recipient Factors | |
|---|---|
| Mean Age (Range) | 53 (20-70) |
| Sex (%) | |
| Male | 72 |
| Female | 28 |
| Race (%) | |
| Black or African American | 44 |
| White | 49 |
| American Indian or Alaskan Native | 0 |
| Asian | 4 |
| Other | 3 |
| History of Smoking (%) | |
| Never | 46 |
| Past | 54 |
| BMI (%) | |
| BMI<30 | 63 |
| BMI >=30 | 37 |
| Primary Diagnosis (%) | |
| NICM | 53 |
| ICM | 22 |
| Hypertrophic CM | 1 |
| Restrictive CM | 1 |
| Myocarditis | 1 |
| Other | 22 |
| Ventricular assist device use (%) | 64 |
| UNOS Status (%) | |
| 1A | 47 |
| 1B | 53 |
| Donor Factors | |
| Mean Age (Range) | 32 (12-57) |
| Sex (%) | |
| Male | 69 |
| Female | 31 |
| Race (%) | |
| Black or African Americans | 29 |
| White or European Americans | 60 |
| American Indian or Alaskan Native | 3 |
| Asian | 4 |
| Other | 5 |
| History of Smoke (%) | |
| Yes | 11 |
| No | 89 |
| History of Chest Trauma (%) | |
| Yes | 12 |
| No | 88 |
| Donor Cause of Death (%) | |
| Anoxia | 43 |
| CVA | 9 |
| Head Trauma | 43 |
| Other | 6 |
| BMI (%) | |
| BMI<30 | 70 |
| BMI >=30 | 30 |
BMI: body-mass index, CVA: cerebrovascular accident, ICM: ischemic cardiomyopathy, NICM: non-ischemic cardiomyopathy, UNOS: United Network for Organ Sharing,
Post-transplant %ddcfDNA decay
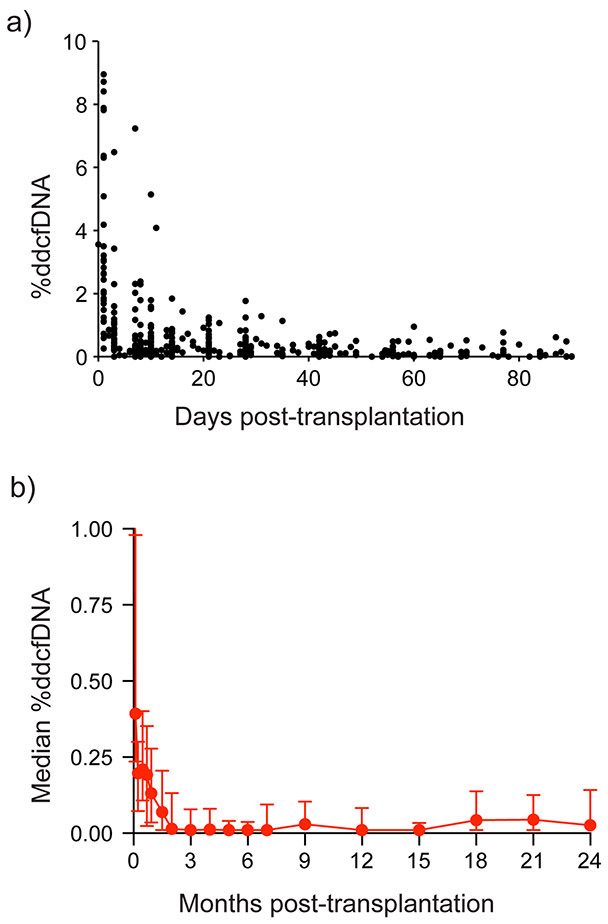
Thirty-seven of the 1904 samples were excluded because the sequencing depth below 10 million reads. The remaining 1,867 plasma samples were sequenced to a median sequencing depth was ~30 million reads. After removing poorly aligned or low-quality reads, 66% of reads were used for downstream quantitation of ddcfDNA (Table IIIa in the Supplement). cfDNA fragments were predominantly mononucleosomal and had a DNA periodicity of 10. Median %ddcfDNA was high after heart transplant surgery (median = 2.83%, IQR: 1.68 – 4.03) (Figure 2a). Levels then followed a one-phase logarithmic decay (Table IIIb in the Supplement) to 0.21% (IQR: 0.03 – 0.21) on Day 28 of transplant and to lowest baseline levels of 0.01 – 0.02% (IQR: 0.01 – 0.13) by Day 60. Thereafter, levels remain stable until Month 18 when a small increase to ~0.05% was observed (IQR: 0.01 – 0.14, Figure 2b, Table IIIc in the Supplement).
Figure 2: %ddcfDNA exponential decay early after transplant and longitudinal %ddcfDNA measures after transplant.
A) Exponential decay of %ddcfDNA after transplant. Black dots represent individual %ddcfDNA measures. Decay parameters are in Table IIIb in the Supplement.
B) Median %ddcfDNA overtime. Box-and-whisker plot is the median value (red dot), the lower quartile and the upper quartile of a given set of data. Median values are in Table IIIc in the Supplement.
%ddcfDNA correlate with severity of acute rejection grades and allograft dysfunction
Over the median 17.7 months follow-up, ~8 EMBx were performed per patient (total = 1,392 EMBx, IQR: 4 - 13), and each EMBx containing an average of 4.3 endomyocardial tissue fragments (range: 1–10). Only EMBx with a concurrent ddcfDNA measurement were included in the rejection analysis. On histology, 95% of EMBx showed no rejection (ACR0, pAMR0, 55%, n=749) or mild rejection (ACR1, pAMR = 0, 40%, n = 570), with only 5% (n = 73) showing the primary endpoint of AR (ACR grade ≥2, n=38, pAMR ≥2, n= 15, pAMR1, n= 17, or mixed AMR/ACR = 3). AR incidence showed a trimodal distribution with peak incidence between 0 and 3 months of transplantation, and two smaller peak incidences at 9 to 12 months and 15 to 21 months. Beyond 21 months, AR was rare; only 1 of the 37 biopsies beyond 21 months showed acute rejection. 82% of EMBx had concurrent %ddcfDNA data (n = 1141, ~7 per patient).
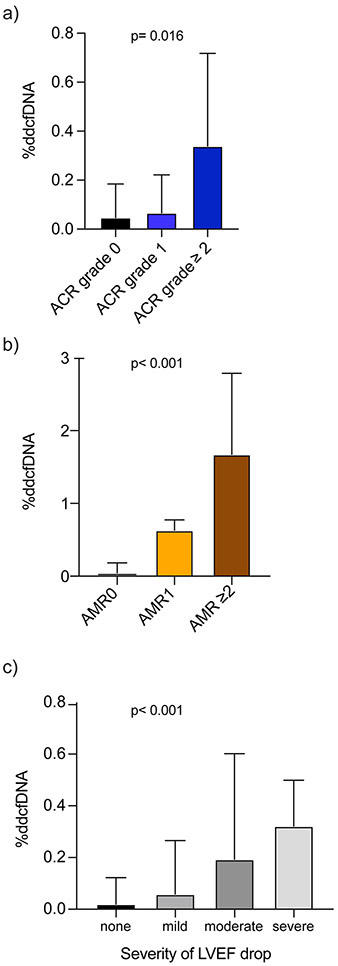
Based on our pre-specified time points post-transplant and due to the logarithmic decay of ddcfDNA after transplant (Figure 2a, Table IIb in the Supplement), we performed our analyses from post-transplant day 28 onwards. AR showed 13-fold higher %ddcfDNA than for mild-to-no rejection (0.38% vs. 0.03%, p <0.001, Table 2). Median %ddcfDNA levels were ~2X higher for ACR grade 1 than for grade 0 rejection (0.04 vs. 0.02%, p = 0.023), and ~9 times higher for ACR ≥ grade 2 than for ACR grade 1 (0.34% vs. 0.04%, p < 0.006, Figure 3a, Table 2). %ddcfDNA also correlated with severity of AMR grade on histopathology; levels were ~30 times higher for pAMR1 than for grade 0 rejection (0.63% vs. 0.02%, p <0.001) and ~5 times higher for pAMR ≥ 2 than pAMR1 (1.68% vs. 0.63%, p = 0.039, Figure 3b, Table 2).
Table 2:
%ddcfDNA for primary and secondary endpoints from Day 28 onwards
| Clinical endpoint | Number of events |
Subjects with events |
Median %ddcfDNA |
%ddcfDNA interquartile range (%) |
p-value | |
|---|---|---|---|---|---|---|
| Controls (ACR0, ACR1, pAMR0) | 1072 | 165 | 0.03 | 0.01 – 0.14 | - | |
| Acute rejection (AR) | 49 | 31 | 0.38 | 0.31 – 0.83 | <0.001* | |
| ACR | ||||||
| Grade 0 | 618 | 165 | 0.02 | 0.01 – 0.13 | ||
| Grade 1 | 454 | 165 | 0.04 | 0.01 – 0.17 | 0.023† | |
| Grade ≥2 | 28 | 21 | 0.34 | 0.28 – 0.72 | <0.001† | |
| AMR | ||||||
| pAMR0 | 1072 | 165 | 0.03 | 0.01 – 0.14 | ||
| pAMR1 | 14 | 9 | 0.63 | 0.34 – 0.77 | <0.001‡ | |
| pAMR2 | 11 | 9 | 1.68 | 0.49 – 2.79 | <0.001‡ | |
| Allograft dysfunction | ||||||
| None | 866 | 165 | 0.02 | 0.01 – 0.12 | ||
| mild | 168 | 83 | 0.06 | 0.01 – 0.27 | 0.068‖ | |
| moderate | 62 | 49 | 0.19 | 0.01 – 0.60 | 0.018‖ | |
| severe | 38 | 28 | 0.32 | 0.05 – 0.47 | <0.001‖ | |
p-values obtained using generalized estimating equation approach to compare groups accounting for repeated measures at individual subject level.
p-value comparing AR to control
p-value comparing ACR grades 1 and 2 to ACR grade 0
p-values comparing pAMR grades 1 and 2 to grade 0
p-values comparing severities of allograft dysfunction to no allograft dysfunction. Subjects may have more than 1 category of endpoints.
Figure 3: Correlation %ddcfDNA measures with biopsy graded rejection and allograft dysfunction by echocardiography.
a) %ddcfDNA in relation to severity of acute cellular rejection (ACR) by histopathologic interpretation of endomyocardial biopsy. Grade 0 rejection includes both ACR grade 0 and pAMR 0. P-value obtained by generalized estimating equation approach comparing all categories.
b) %ddcfDNA in relation to severity of antibody-mediated rejection (AMR) by histopathologic interpretation of the endomyocardial biopsy. Grade 0 rejection includes both ACR grades 0 or 1 and pAMR 0. P-value obtained by generalized estimating equation approach comparing all categories.
c) %ddcfDNA in relation to severity of allograft dysfunction measured by echocardiography. Allograft dysfunction was defined as a reduction of left ventricular ejection fraction (LVEF) by ≥5%, and further stratified by severity based on the magnitude of LVEF decline as no (<5%), mild (5 – <10%), moderate (≥10 – <15%) or severe (≥15%) allograft dysfunction. P-value obtained by generalized estimating equation approach comparing all categories.
About 9 echocardiograms recordings were captured per patient (IQR: 6 - 18). 81% of these echocardiograms had concurrent %ddcfDNA data. One-fifth of echocardiograms showed allograft dysfunction, defined as a ≥ 5% reduction in LVEF from the prior echocardiogram (n = 188), which were categorized as mild (n=119, 63.3%), moderate (n=44, 23.4%) or severe (n=25, 13.3%). %ddcfDNA levels correlated with severity of allograft dysfunction; median %ddcfDNA levels for no, mild, moderate, and severe allograft dysfunction were 0.02%, 0.06%, 0.19%, and 0.32%, respectively (Figure 3c, Table 2).
Performance Characteristics of %ddcfDNA to detect AR
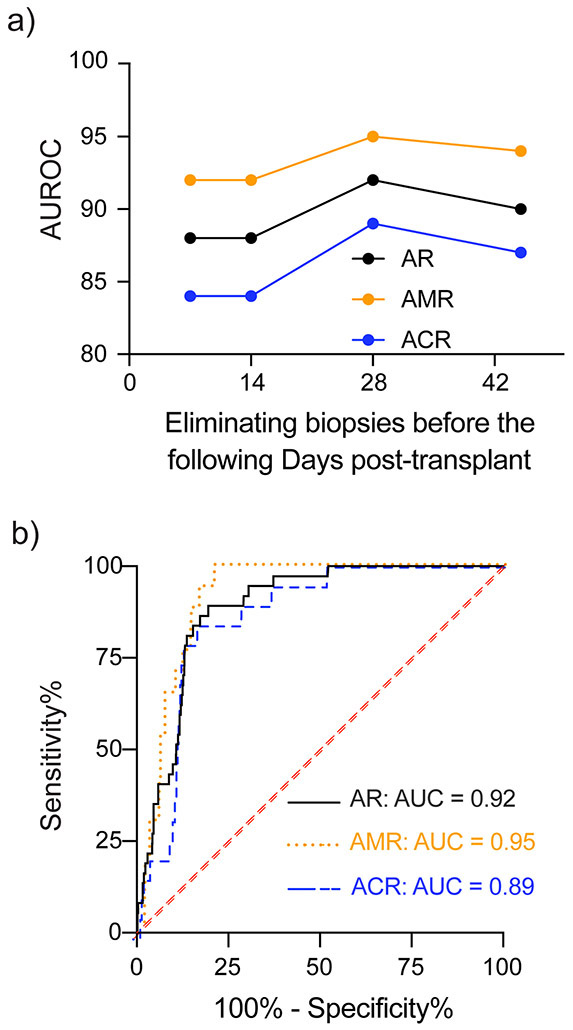
The AUROC calculations included %ddcfDNA measures concurrent to EMBx. Elimination of ddcfDNA measures and EMBx performed before the pre-determined early post-transplant Days (7, 14, 28, 45) show that the AUROC increased from Day 7 to Day 28 (0.88 to 0.92), and reduced thereafter (Figure 4a, Table 3, Table III in the Supplement). The maximal AUROC was identified for day 28 and %ddcfDNA ≥ 0.25% detected AR with an AUROC of 0.92 (Figure 4b), a sensitivity of 81%, a specificity of 85%, a positive predictive value (PPV) of 19.6%, and a negative predictive value (NPV) of 99.2%. Test characteristics for other %ddcfDNA thresholds are presented in Table 3.
Figure 4: Receiver operator characteristics curve for %ddcfDNA to detect endomyocardial biopsy-diagnosed acute rejection.
a) The area under the receiver operator characteristics curve (AUROC) for %ddcfDNA overtime for acute rejection (AR), AMR and ACR. Maximal test performance characteristics were noted from day 28. ROC characteristics for each pre-specified time are presented in Table 3 and Table IV in the Supplement.
b) AUROC of %ddcfDNA to detect acute rejection (AR), antibody mediated rejection (AMR, pAMR ≥1) or acute cellular rejection (ACR, grade ≥2R). Primary analysis excluded endomyocardial biopsies before day 28. 923 endomyocardial biopsies and concurrent ddcfDNA measures were included in this analysis. ROC characteristics are presented in Table 3.
Table 3:
%ddcfDNA test characteristics to detect biopsy-positive acute rejection: Eliminated biopsies before Day 28 after transplantation
| %ddcfDNA threshold | Sensitivity (%) |
Specificity (%) |
AUROC (95% CI) |
|||||
|---|---|---|---|---|---|---|---|---|
| 0.1 | 0.25 | 0.5 | 0.1 | 0.25 | 0.5 | |||
| AR diagnosis | ||||||||
| AR | 95 | 81 | 41 | 68 | 85 | 92 | 0.92 (0.88 – 0.95) | |
| AMR | 100 | 88 | 65 | 68 | 85 | 92 | 0.95 (0.90 – 0.99) | |
| ACR | 89 | 79 | 21 | 68 | 85 | 92 | 0.89 (0.83 – 0.95) | |
Since maximal test performance occurred from day 28 on and assuming a %ddcfDNA ≥ 0.25% triggers an EMBx to evaluate the source of allograft injury, monitoring patients with this strategy avoids 8-out-of-every 10 EMBx (751/923, 81%). The NPV of 99.2% was attributed to the 0.25 %ddcfDNA threshold, with only 6 episodes of rejection out of the 751 EMBx with %ddcfDNA below 0.25 (i.e. false negative test); 1/6 was AMR1 and 5/6 were ACR grade 2, with only one of the six rejection episodes being associated with a drop in LVEF. The PPV of 19.6% was attributed to 135 elevations in ddcfDNA above the 0.25% threshold. These elevations in ddcfDNA showed no histopathologic rejection but were of clinical relevance as they often preceded overt histopathologic rejection or were associated with allograft dysfunction in the absence of rejection. These “false positives” are reviewed in detail below.
Performance of the endomyocardial biopsy to detect allograft injury measured by %ddcfDNA
To clarify the PPV of ddcfDNA and to understand the performance characteristics of the EMBx to detect allograft injury as measured by ddcfDNA, we performed analyses with EMBx as the test and ddcfDNA as the reference injury marker. From day 28 on, 1-in-5 %ddcfDNA measures had allograft injury as defined by a %ddcfDNA ≥0.25% (168/923). The sensitivity of EMBx to detect these episodes was low at 19.6%, specificity was 99.2% and the NPV was 85% (Table 4). So, over three quarters of allograft injury episodes as defined by ddcfDNA were associated with a negative EMBx (n=135, or a false negative test). The majority of these allograft injury+/EMBx- were of clinical relevance; 20.7% were associated with concurrent allograft dysfunction (n=28), and about half represent a %ddcfDNA rise preceding AR (n = 46, 27.4%) or allograft dysfunction (n=28, 16.7%). Certain patients had a slow post-transplant %ddcfDNA decay that extended beyond 28 days without evidence of allograft rejection (n=19, 14.1%). Finally, 14 patients beyond 15 months post-transplant had a rise in %ddcfDNA without allograft dysfunction on echocardiography (n=8, 5.9%), or had an isolated and transient %ddcfDNA elevation (n=6).
Table 4:
Performance Characteristics of the Endomyocardial Biopsy to detect allograft injury measured via %ddcfDNA
| Biopsy + Histopathology |
%ddcfDNA | |
|---|---|---|
| ≥0.25% | <0.25% | |
| % (n) | % (n) | |
| Positive | 19.6 (33) | 0.8 (6) |
| Negative | 80.4 (135) | 99.2 (745) |
| Total | 100 (168) | 100 (751) |
Distinguishing ddcfDNA features in AMR and ACR.
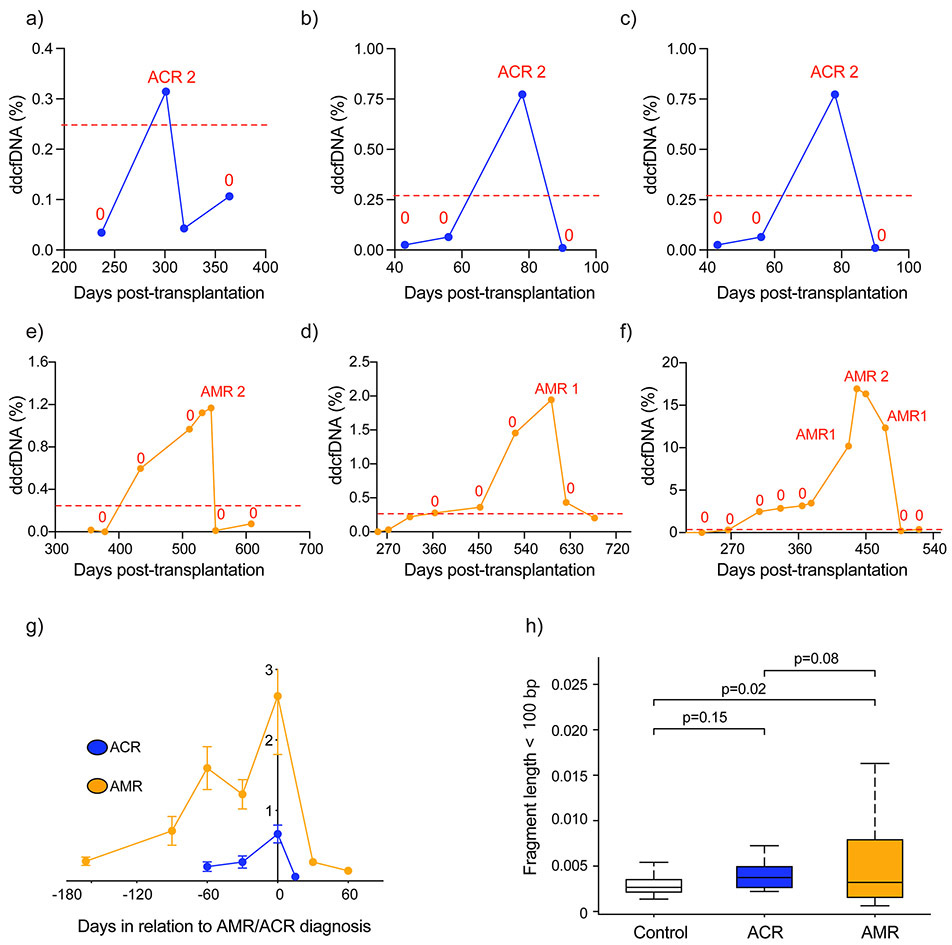
AMR showed three distinct %ddcfDNA patterns compared to ACR. First, for similar histopathology grades, AMR showed quantitatively higher %ddcfDNA levels compared to ACR. For example, pAMR1 showed ~11X higher %ddcfDNA than ACR grade 1, and pAMR ≥2 showed 5X higher %ddcfDNA than ACR grade ≥2 (Figure 3, Table 2). Consequently, %ddcfDNA had a higher AUROC for AMR compared to ACR (Figure 4b, Table 3, Table III in the Supplement). Second, the %ddcfDNA trends before diagnosis were different (Figures 5 a – g); a %ddcfDNA rise preceding diagnosis was not commonly observed with ACR (Figures 5 a – c); only 2/17 ACR episodes evaluated showed %ddcfDNA rise before ACR diagnosis. On the other hand, a %ddcfDNA rise was commonly detected before AMR diagnosis; 12/15 AMR episodes with sufficient data to assess (Figure 5 d – f) showed a rise in %ddcfDNA that was detected at a median 3.2 months before AMR diagnosis (Figure 5g). Third, the fragment length of sequenced cfDNA was distinct in AMR and ACR. In the setting of AMR, the cfDNA fragments were shorter than ACR or controls with no-rejection. The percentage of short cfDNA fragments (read length below < 120 bp) was highest in AMR, and similar in ACR and controls (Figure 5h). Similarly, the percentage of guanosine-cytosine bases were highest in AMR compared to ACR or no rejection controls. Finally, ACR, AMR and controls show similar percentage of reads mapping as exons. However, other genomic elements showed different patterns. For example, both AMR and ACR showed higher percentage of reads mapping as promoters compared to controls, and lower percentages of reads mapping as introns and intergenic region compare to control (Figure I in the Supplement).
Figure 5: Distinct %ddcfDNA patterns in AMR and ACR.
a – c %ddcfDNA against time plots for prototype subjects with acute cellular rejection, “0” = ACR grade 0 or grade 1 and pAMR0
d – f %ddcfDNA against time plots for prototype subjects with antibody-mediated rejection. “0” = ACR grade 0 or grade 1 and pAMR0
g. Trends of %ddcfDNA measures in all patients preceding a histopathologic diagnosis of AMR or ACR; mean and standard deviations are shown. For AMR, there is a longer lead-time and larger quantitative ddcfDNA release of ddcfDNA prior to diagnosis. In addition, 12/15 cases of AMR had elevations of ddcfDNA preceding the clinical diagnosis. In ACR the ddcfDNA elevations preceding clinical diagnosis are rarer (2/17), lower quantitatively and are detected for a shorter time preceding overt diagnosis.
h. Different %ddcfDNA characteristics based on cfDNA length in ACR and AMR. Short DNA fragments were defined based on total read length < 100bp. In AMR, the % short cfDNA fragments is much higher than in ACR or controls without rejection.
Discussion
This study documents five key findings that are potentially relevant for the use of ddcfDNA as a biomarker of graft injury from acute rejection. First, %ddcfDNA levels were significantly higher in AMR than ACR, overall and at all grades of rejection. Second, although increases in ddcfDNA preceded both forms of AR, elevations occurred earlier in AMR than in ACR and both the fragment length and genomic composition of ddcfDNA in AMR varied from ACR. Third, despite both forms of AR being predicted by ROC of %ddcfDNA, the AUROC was higher for AMR. This finding is particularly important in clinical practice since ACR and AMR have different treatment approaches and long-term implications. Fourth, %ddcfDNA was higher with increasing severity of allograft dysfunction as measured by echocardiography. Finally, the EMBx had limited ability to detect early stages of allograft injury that preceded overt rejection. Taken together, these findings support our hypothesis that higher levels of ddcfDNA would be detected with AMR than ACR. Moreover, these results may provide insights into the mechanism underlying allograft dysfunction in the setting of AMR. Below we discuss each of these findings from the perspective ddcfDNA as non-invasive tool for monitoring rejection and as a biomarker of graft injury to study mechanisms underlying cardiac allograft failure.
In healthy individuals, the majority of cfDNA is released from hematopoietic cells, with less than 1% release from the heart. The increased amount of cfDNA observed with AR suggests higher levels of allograft injury. The finding that %ddcfDNA levels were significantly higher at all grades of rejection validates our previous single-center experience and supports %ddcfDNA as a reliable, quantitative biomarker to monitor cardiac allograft health.6 Increases in ddcfDNA preceded both forms of AR, but elevations occurred earlier in AMR than in ACR, suggesting that %ddcfDNA is a sensitive marker to detect allograft injury weeks-to-months before current clinical tools. The median ddcfDNA levels were notably higher in patients AMR than ACR, reflecting that differences in the pathogenesis of AMR and ACR likely contribute to the quantitative differences in %ddcfDNA levels observed. AMR is associated with DSA and results in diffuse macro- and microvascular injury and progresses to pan-carditis and cellular DNA release, as opposed to ACR, which is associated with more localized lymphocytic infiltration and injury often contained within the interstitium and perivascular tissue spaces. Moreover, since the earliest histological features of AMR is vascular inflammation,17 we anticipate that early indicators of AMR would include cardiac dysfunction with a concomitant rise in ddcfDNA. This would be consistent with our observations in a xenograft model of AMR where the injury is predominantly targeted to the microvasculature.18 Our findings demonstrating higher levels of ddcfDNA with AMR (1.68%) compared to ACR (0.34%) are consistent with prior studies showing that ddcfDNA with concomitant graft dysfunction occurs more frequently in AMR (37% of patients), compared to only 5% in ACR patients.19, 20 Accurate discrimination of AMR and ACR is critical as the initial therapeutic strategy varies significantly and the long-term implications of rejection are quite different, with more significant graft dysfunction, higher incidence of recurrent rejection, greater risk for CAV and death with AMR.21
Using a threshold value of 0.25% %ddcfDNA we showed that the test has an AUROC of 89-95% and a NPV of 99%; these findings support the use of %ddcfDNA as a non-invasive biomarker of AR. We propose that after day 28 of surgery, heart transplant recipients could be monitored for AR using %ddcfDNA levels; specifically, if the levels remain below 0.25% surveillance biopsies would no longer be required. Employing a ddcfDNA threshold of 0.25% from day 28 onwards would have safely avoided 81% of all routine EMBx (751/923), with less than 1% (n=6) of these being “missed” cases of rejection, - the majority of which were ACR and not associated with graft dysfunction. The majority of ddcfDNA elevations (80%) were associated with a negative EMBx leading to a high “false positive” rate when the EMBx is used as the “gold standard”. This suggests that ddcfDNA detected episodes of allograft injury that were histologically silent on the EMBx. These episodes were felt to be of clinical significance as they were associated with allograft dysfunction or preceded histopathologic rejection. Our results suggest that patients with elevated %ddcfDNA in the absence of rejection may benefit from closer monitoring. In addition, the longer lead-time before the clinical diagnosis of AMR supports the concept of subclinical AMR,21, 22 where there is antibody deposition and complement fixation that lead to vascular injury without overt graft dysfunction. It is important to note that ddcfDNA levels are distinct and complementary to DSA testing. Despite the development of de novo DSA in ~ 1/3 of heart transplant patients,23, 24 only 54% of patients with DSA develop pAMR; particularly in the presence of class II antibodies.19 We suspect that the immune-mediated allograft injury associated with deleterious DSA begins at a molecular level in the microvasculature, which is detected by %ddcfDNA well before AMR becomes apparent on histopathology or clinically.
Comparing performance characteristics of ddcfDNA across studies is difficult due to inherent differences in the study populations and protocols; for example, our study included a higher proportion of African Americans who have heightened immune system activity and are at higher risk for AMR.23, 25-27 In addition, the prior studies used a case-control format and enriched for cases of allograft rejection, while our cohort include all adult heart transplant recipients at the GRAfT centers who provided informed consent and included serial ddcfDNA measures from time of transplant irrespective of rejection status. Nevertheless, there are some meaningful comparisons worth discussing across the studies. The median ddcfDNA levels for controls in our study was 0.03% was lower than prior publications in adult and pediatric heart transplantation, that reported basal levels of 0.07% and 0.10%.26, 28 This may again reflect enrichment for patients at higher risk for rejection in the prior studies. Nevertheless, the ddcfDNA threshold to diagnose AR identified in these previous analyses is similar to ours ranging from 0.2% to 0.23%, and all studies reported higher allograft injury levels with AMR than ACR. In addition, the NPV of ddcfDNA ranged from 93% in the pediatric study to 97% in both adult studies suggesting non-invasive monitoring with ddcfDNA appears to be a safe surveillance strategy in heart transplant recipients.
In our study 20% of patients had graft dysfunction based on standard echocardiographic analysis. Of these patients only 14% had concurrent ACR or AMR, and the remainder had biopsy-negative rejection (BNR) with mild to severe graft dysfunction. These patients may have had early rejection or subclinical AMR, and a better understanding of the cause of graft dysfunction in these patients is needed. A recent study by Loupy et al. suggests that subclinical AMR and the associated immune-mediated injury are significant factors leading to long-term allograft failure.17 We hypothesize that these patients with echocardiographic graft dysfunction and elevated %ddcfDNA levels may have subclinical AMR or AMR that was missed by the EMBx. Subclinical AMR may result in ongoing endothelial cell injury and inflammation that leads to endothelial dysfunction and perturbations in microvasculature resistance that have been shown to precede the overt development of CAV.29, 30 The role of subclinical AMR in CAV and graft dysfunction will need to be explored in future studies.
The concept of BNR has been reported in cardiac transplantation for many decades.31 Early cases of BNR in the 1980’s and early 1990’s likely represented humoral rejection or AMR, before it was a clinically accepted entity within the heart transplant community.32 With the advent of immunostaining for complement fixation, the reported incidence of BNR has diminished significantly as the histopathologic diagnosis of AMR has improved. Further, the EMBx only identified 20% of allograft injury episodes as defined by ddcfDNA ≥0.25. These allograft injury episodes were clinically significant, and the majority were associated with either allograft dysfunction or preceded overt ACR or AMR.
In addition to using ddcfDNA as a quantitative biomarker of allograft injury, our data suggests that there are qualitative differences in ddcfDNA between AMR and ACR. In this study, AMR also showed shorter cfDNA fragments and different composition of some genomic elements compared to ACR. These differences were similarly observed comparing AMR to within-subject no rejection controls. These, and similar observations in hepatocellular carcinoma,33 other cancers,34 and potentially other conditions suggest that cfDNA characteristics are not random, but rather dependent on the pathogenesis leading to their production. The diagnostic value of these distinguishing cfDNA features observed in AMR and ACR warrants further investigation.
STUDY LIMITATIONS
Several limitations of our study are noteworthy. First, variabilities in the management of post-operative immunosuppression among the five GRAfT centers could have affected the incidence of allograft rejection, its detection, and the levels of ddcfDNA. In addition, the prospective cohort study design used is beneficial because it provides a real-world analysis of the performance of ddcfDNA, however this might have contributed to variabilities in patient care. Second, although our results indicate significantly higher levels of ddcfDNA levels in patients with systolic dysfunction on echocardiography and rejection on biopsy, concomitant measurements of allograft function and histology were only available for one third of the study subjects.
A third and important limitation is the use of a suboptimal “gold standard,” the EMBx, to assess a new paradigm-shifting modality, ddcfDNA. The explosion of next generation sequencing technologies coupled with computational pipelines for data processing has tremendous impact in basic science research. These advancements are being translated to highly effective diagnostic tools for application to clinical care as evidence by prenatal diagnosis.35 However, the advent of a paradigm-shifting scientific advancements poses challenges to conventional statistical methods that employ sensitivity and specificity measures to assess the effectiveness of a new diagnostic against an established standard, in this case the EMBx. The concept of a paradigm shift in science was first described by American physicist and philosopher Thomas Kuhn, as a “fundamental change in the basic concepts and experimental practices of a scientific discipline.”36 The ddcfDNA offers a fundamental change in the basic concept of allograft rejection by shifting away from histologic evidence of inflammatory infiltrate and myocyte damage, to a direct quantitative measure of donor-derived DNA in the recipient’s plasma. Our results indicating that ddcfDNA is elevated several weeks before histologic features of rejection are apparent, creates an incorrect assignment as “false positive” and an inaccurate assessment of the test characteristics which leads to a low positive predictive value. Therefore, clinical researchers should question the validity of the biopsy as the “gold standard” for rejection in the current era of highly precise genomic-based assays and consider alternative approaches to assessing the ability of a new biomarker such as ddcfDNA to identify patients at the earliest stages of acute rejection.
CONCLUSIONS
In summary, our data supports the use of %ddcfDNA as a “liquid biopsy,” to monitor allograft health in heart transplant patients. %ddcfDNA is reliable and reproducible, varies both quantitatively and qualitatively in AMR and ACR, has excellent biomarker performance characteristics, and unmasks pathology earlier than existing tools. This work paves the way for clinical utility and mechanistic studies in heart transplantation.
Supplementary Material
Clinical Perspective.
What Is New?
In a multicenter, prospective study of cardiac transplant patients, performance characteristics of the non-invasive biomarker, donor-derived cell-free DNA (ddcfDNA) were excellent. The area under the curve for ddcfDNA to detect allograft rejection day 28 onwards (both acute cellular rejection (ACR) and antibody-mediated rejection (AMR)) was 0.92.
Allograft injury as measured by ddcfDNA was quantitatively higher with higher grades of rejection, in AMR vs. ACR and was often elevated months before clinical detection.
When ddcfDNA was used as the new gold standard to detect allograft injury, traditional histopathologic interpretation of endomyocardial biopsy samples had limited sensitivity to detect allograft injury (19%).
What Are the Clinical Implications?
ddcfDNA measures vary between AMR and ACR in lead time, quantitative levels and cfDNA fragment type/length. This provides a framework to use ddcfDNA to differentiate AMR and ACR.
Starting at 28 days after heart transplant, the use of a liquid biopsy with a ddcfDNA threshold of ≥ 0.25% had excellent sensitivity and specificity for diagnosis of rejection, safely avoiding 81% of all surveillance endomyocardial biopsies.
Acknowledgements
Clinical coordinators at participating centers for recruiting and monitoring study subjects, organ procurement organizations (Living Legacy Foundation, Washington Regional Transplant Consortium, and LifeHealth Net) for providing donor blood samples for genotyping, National Heart, Lung and Blood Institute Sequencing Core for shotgun sequencing, National Cancer Institute sequencing core for genotyping, Kelly Byrne for editorial support, and Erina He for graphing support.
Sources of Funding
This study is supported by National Heart, Lung and Blood Institute Division of Intramural Research (HHSN268201300001C). Palak Shah is supported by an American Heart Association / Enduring Hearts Foundation Scientist Development Grant (17SDG33660431).
Non-Standard Abbreviations and Acronyms:
- ACR
acute cellular rejection
- AMR
antibody mediated rejection
- AR
acute rejection
- CAV
cardiac allograft vasculopathy
- CMV
cytomegalovirus
- ddcfDNA
donor-derived cell-free DNA
- DSA
donor specific antibodies
- GRAfT
Genomic Research Alliance for Transplantation
- LVEF
left ventricular ejection fraction
- NPV
negative predictive value
Footnotes
Disclosures:
The authors have no relevant disclosures.
References
- 1.Mehra MR, Crespo-Leiro MG, Dipchand A, Ensminger SM, Hiemann NE, Kobashigawa JA, Madsen J, Parameshwar J, Starling RC and Uber PA. International Society for Heart and Lung Transplantation working formulation of a standardized nomenclature for cardiac allograft vasculopathy-2010. J Heart Lung Transplant. 2010;29:717–27. [DOI] [PubMed] [Google Scholar]
- 2.Costanzo-Nordin MR. Cardiac allograft vasculopathy: relationship with acute cellular rejection and histocompatibility. J Heart Lung Transplant. 1992;11:S90–103. [PubMed] [Google Scholar]
- 3.Stewart S, Winters GL, Fishbein MC, Tazelaar HD, Kobashigawa J, Abrams J, Andersen CB, Angelini A, Berry GJ, Burke MM, Demetris AJ, Hammond E, Itescu S, Marboe CC, McManus B, Reed EF, Reinsmoen NL, Rodriguez ER, Rose AG, Rose M, Suciu-Focia N, Zeevi A and Billingham ME. Revision of the 1990 working formulation for the standardization of nomenclature in the diagnosis of heart rejection. J Heart Lung Transplant. 2005;24:1710–20. [DOI] [PubMed] [Google Scholar]
- 4.Berry GJ, Burke MM, Andersen C, Bruneval P, Fedrigo M, Fishbein MC, Goddard M, Hammond EH, Leone O, Marboe C, Miller D, Neil D, Rassl D, Revelo MP, Rice A, Rene Rodriguez E, Stewart S, Tan CD, Winters GL, West L, Mehra MR and Angelini A. The 2013 International Society for Heart and Lung Transplantation Working Formulation for the standardization of nomenclature in the pathologic diagnosis of antibody-mediated rejection in heart transplantation. J Heart Lung Transplant. 2013;32:1147–62. [DOI] [PubMed] [Google Scholar]
- 5.Marboe CC, Billingham M, Eisen H, Deng MC, Baron H, Mehra M, Hunt S, Wohlgemuth J, Mahmood I, Prentice J and Berry G. Nodular endocardial infiltrates (Quilty lesions) cause significant variability in diagnosis of ISHLT Grade 2 and 3A rejection in cardiac allograft recipients. J Heart Lung Transplant. 2005;24:S219–26. [DOI] [PubMed] [Google Scholar]
- 6.De Vlaminck I, Valantine HA, Snyder TM, Strehl C, Cohen G, Luikart H, Neff NF, Okamoto J, Bernstein D, Weisshaar D, Quake SR and Khush KK. Circulating cell-free DNA enables noninvasive diagnosis of heart transplant rejection. Sci Transl Med. 2014;6:241ra77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Snyder TM, Khush KK, Valantine HA and Quake SR. Universal noninvasive detection of solid organ transplant rejection. Proc Natl Acad Sci U S A. 2011;108:6229–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bloom RD, Bromberg JS, Poggio ED, Bunnapradist S, Langone AJ, Sood P, Matas AJ, Mehta S, Mannon RB, Sharfuddin A, Fischbach B, Narayanan M, Jordan SC, Cohen D, Weir MR, Hiller D, Prasad P, Woodward RN, Grskovic M, Sninsky JJ, Yee JP, Brennan DC and Circulating Donor-Derived Cell-Free DNAiBfDARiKTRSI. Cell-Free DNA and Active Rejection in Kidney Allografts. J Am Soc Nephrol. 2017;28:2221–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Agbor-Enoh S, Jackson AM, Tunc I, Berry GJ, Cochrane A, Grimm D, Davis A, Shah P, Brown AW, Wang Y, Timofte I, Shah P, Gorham S, Wylie J, Goodwin N, Jang MK, Marishta A, Bhatti K, Fideli U, Yang Y, Luikart H, Cao Z, Pirooznia M, Zhu J, Marboe C, Iacono A, Nathan SD, Orens J, Valantine HA and Khush K. Late manifestation of alloantibody-associated injury and clinical pulmonary antibody-mediated rejection: Evidence from cell-free DNA analysis. J Heart Lung Transplant. 2018;37:925–932. [DOI] [PubMed] [Google Scholar]
- 10.Agbor-Enoh S, Fideli UD J., Zhu J, Tunc I, Shah P, Russell S, Feller E, Shah K, Rodrigo M, Shah P, Pham S, Iacono A, Nathan S, Orens J and Valantine HA. Genomic Research Alliance for Transplantation (GRAfT): A Model for Long Term Transplant Studies in Thoracic Organ Transplantation. J Heart Lung Transplant. 2016;35:S161. [Google Scholar]
- 11.Agbor-Enoh S, Tunc I, De Vlaminck I, Fideli U, Davis A, Cuttin K, Bhatti K, Marishta A, Solomon MA, Jackson A, Graninger G, Harper B, Luikart H, Wylie J, Wang X, Berry G, Marboe C, Khush K, Zhu J and Valantine H. Applying rigor and reproducibility standards to assay donor-derived cell-free DNA as a non-invasive method for detection of acute rejection and graft injury after heart transplantation. J Heart Lung Transplant. 2017;36:1004–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanley JA and McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982;143:29–36. [DOI] [PubMed] [Google Scholar]
- 13.Zhou X-h, McClish DK and Obuchowski NA. Statistical methods in diagnostic medicine. 2nd ed. Hoboken, N.J.: Wiley; 2011. [Google Scholar]
- 14.De Vlaminck I, Martin L, Kertesz M, Patel K, Kowarsky M, Strehl C, Cohen G, Luikart H, Neff NF, Okamoto J, Nicolls MR, Cornfield D, Weill D, Valantine H, Khush KK and Quake SR. Noninvasive monitoring of infection and rejection after lung transplantation. Proc Natl Acad Sci U S A. 2015;112:13336–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Agbor-Enoh S, Tunc I, De Vlaminck I, Davis A, Gorham S, Jang M, Cuttin K, Fideli U, Marshita A, Wylie J, Luikart H, Khush K and Valantine H. Early Graft Injury Measured by Donor-Derived Cell-Free DNA Predicts Early Mortality in Lung Transplant Recipients. J Heart Lung Transplant. 2017;36:S65. [Google Scholar]
- 16.Liang K-Y and Zeger SL. Longitudinal data analysis using generalized linear models. Biometrika. 1986;73:13–22. [Google Scholar]
- 17.Loupy A, Toquet C, Rouvier P, Beuscart T, Bories MC, Varnous S, Guillemain R, Pattier S, Suberbielle C, Leprince P, Lefaucheur C, Jouven X, Bruneval P and Duong Van Huyen JP. Late Failing Heart Allografts: Pathology of Cardiac Allograft Vasculopathy and Association With Antibody-Mediated Rejection. Am J Transplant. 2016;16:111–20. [DOI] [PubMed] [Google Scholar]
- 18.Agbor-Enoh S, Chan JL, Singh A, Tunc I, Gorham S, Zhu J, Pirooznia M, Corcoran PC, Thomas ML, Lewis BGT, Jang MK, Ayares DL, Horvath KA, Mohiuddin MM and Valantine H. Circulating cell-free DNA as a biomarker of tissue injury: Assessment in a cardiac xenotransplantation model. J Heart Lung Transplant. 2018;37:967–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clerkin KJ, Farr MA, Restaino SW, Zorn E, Latif F, Vasilescu ER, Marboe CC, Colombo PC and Mancini DM. Donor-specific anti-HLA antibodies with antibody-mediated rejection and long-term outcomes following heart transplantation. J Heart Lung Transplant. 2017;36:540–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mills RM, Naftel DC, Kirklin JK, Van Bakel AB, Jaski BE, Massin EK, Eisen HJ, Lee FA, Fishbein DP and Bourge RC. Heart transplant rejection with hemodynamic compromise: a multiinstitutional study of the role of endomyocardial cellular infiltrate. Cardiac Transplant Research Database. J Heart Lung Transplant. 1997;16:813–21. [PubMed] [Google Scholar]
- 21.Colvin MM, Cook JL, Chang P, Francis G, Hsu DT, Kiernan MS, Kobashigawa JA, Lindenfeld J, Masri SC, Miller D, O'Connell J, Rodriguez ER, Rosengard B, Self S, White-Williams C, Zeevi A, American Heart Association Heart F, Transplantation Committee of the Council on Clinical C, American Heart Association Heart F, Transplantation Committee of the Council on Cardiopulmonary Critical Care P, Resuscitation, American Heart Association Heart F, Transplantation Committee of the Council on Cardiovascular Disease in the Y, American Heart Association Heart F, Transplantation Committee of the Council on Clinical Cardiology CoC, Stroke N, American Heart Association Heart F, Transplantation Committee of the Council on Cardiovascular R, Intervention, American Heart Association Heart F, Transplantation Committee of the Council on Cardiovascular S and Anesthesia. Antibody-mediated rejection in cardiac transplantation: emerging knowledge in diagnosis and management: a scientific statement from the American Heart Association. Circulation. 2015;131:1608–39. [DOI] [PubMed] [Google Scholar]
- 22.Kfoury AG, Renlund DG, Snow GL, Stehlik J, Folsom JW, Fisher PW, Reid BB, Clayson SE, Gilbert EM, Everitt MD, Bader FM, Singhal AK and Hammond ME. A clinical correlation study of severity of antibody-mediated rejection and cardiovascular mortality in heart transplantation. The Journal of heart and lung transplantation : the official publication of the International Society for Heart Transplantation. 2009;28:51–7. [DOI] [PubMed] [Google Scholar]
- 23.Cole RT, Gandhi J, Bray RA, Gebel HM, Yin M, Shekiladze N, Young A, Grant A, Mahoney I, Laskar SR, Gupta D, Bhatt K, Book W, Smith A, Nguyen D, Vega JD and Morris AA. Racial differences in the development of de-novo donor-specific antibodies and treated antibody-mediated rejection after heart transplantation. J Heart Lung Transplant. 2018;37:503–512. [DOI] [PubMed] [Google Scholar]
- 24.Smith JD, Banner NR, Hamour IM, Ozawa M, Goh A, Robinson D, Terasaki PI and Rose ML. De novo donor HLA-specific antibodies after heart transplantation are an independent predictor of poor patient survival. Am J Transplant. 2011;11:312–9. [DOI] [PubMed] [Google Scholar]
- 25.Khush KK, Pham MX, Teuteberg JJ, Kfoury AG, Deng MC, Kao A, Anderson AS, Cotts WG, Ewald GA, Baran DA, Hiller D, Yee J and Valantine HA. Gene expression profiling to study racial differences after heart transplantation. J Heart Lung Transplant. 2015;34:970–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khush KK, Patel J, Pinney S, Kao A, Alharethi R, DePasquale E, Ewald G, Berman P, Kanwar M, Hiller D, Yee JP, Woodward RN, Hall S and Kobashigawa J. Noninvasive detection of graft injury after heart transplant using donor-derived cell-free DNA: A prospective multicenter study. Am J Transplant. 2019;19:2889–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.North PE, Ziegler E, Mahnke DK, Stamm KD, Thomm A, Daft P, Goetsch M, Liang HL, Baker MA, Vepraskas A, Rosenau C, Dasgupta M, Simpson P, Mitchell ME and Tomita-Mitchell A. Cell-free DNA donor fraction analysis in pediatric and adult heart transplant patients by multiplexed allele-specific quantitative PCR: Validation of a rapid and highly sensitive clinical test for stratification of rejection probability. PloS one. 2020;15:e0227385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richmond ME, Zangwill SD, Kindel SJ, Deshpande SR, Schroder JN, Bichell DP, Knecht KR, Mahle WT, Wigger MA, Gaglianello NA, Pahl E, Simpson PM, Dasgupta M, North PE, Hidestrand M, Tomita-Mitchell A and Mitchell ME. Donor fraction cell-free DNA and rejection in adult and pediatric heart transplantation. J Heart Lung Transplant. 2020;39:454–463. [DOI] [PubMed] [Google Scholar]
- 29.Lee JH, Okada K, Khush K, Kobayashi Y, Sinha S, Luikart H, Valantine H, Yeung AC, Honda Y and Fearon WF. Coronary Endothelial Dysfunction and the Index of Microcirculatory Resistance as a Marker of Subsequent Development of Cardiac Allograft Vasculopathy. Circulation. 2017;135:1093–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang HM, Khush K, Luikart H, Okada K, Lim HS, Kobayashi Y, Honda Y, Yeung AC, Valantine H and Fearon WF. Invasive Assessment of Coronary Physiology Predicts Late Mortality After Heart Transplantation. Circulation. 2016;133:1945–50. [DOI] [PubMed] [Google Scholar]
- 31.Fishbein MC and Kobashigawa J. Biopsy-negative cardiac transplant rejection: etiology, diagnosis, and therapy. Curr Opin Cardiol. 2004;19:166–9. [DOI] [PubMed] [Google Scholar]
- 32.Hammond EH, Yowell RL, Nunoda S, Menlove RL, Renlund DG, Bristow MR, Gay WA Jr., Jones KW and O'Connell JB. Vascular (humoral) rejection in heart transplantation: pathologic observations and clinical implications. J Heart Transplant. 1989;8:430–43. [PubMed] [Google Scholar]
- 33.Jiang P, Chan CW, Chan KC, Cheng SH, Wong J, Wong VW, Wong GL, Chan SL, Mok TS, Chan HL, Lai PB, Chiu RW and Lo YM. Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc Natl Acad Sci U S A. 2015;112:E1317–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen SY, Singhania R, Fehringer G, Chakravarthy A, Roehrl MHA, Chadwick D, Zuzarte PC, Borgida A, Wang TT, Li T, Kis O, Zhao Z, Spreafico A, Medina TDS, Wang Y, Roulois D, Ettayebi I, Chen Z, Chow S, Murphy T, Arruda A, O'Kane GM, Liu J, Mansour M, McPherson JD, O'Brien C, Leighl N, Bedard PL, Fleshner N, Liu G, Minden MD, Gallinger S, Goldenberg A, Pugh TJ, Hoffman MM, Bratman SV, Hung RJ and De Carvalho DD. Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nature. 2018;563:579–583. [DOI] [PubMed] [Google Scholar]
- 35.Bianchi DW and Chiu RWK. Sequencing of Circulating Cell-free DNA during Pregnancy. N Engl J Med. 2018;379:464–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuhn TS. The structure of scientific revolutions. Chicago: University of Chicago Press; 1962. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.