Abstract
Background The lack of machine-interpretable representations of consent permissions precludes development of tools that act upon permissions across information ecosystems, at scale.
Objectives To report the process, results, and lessons learned while annotating permissions in clinical consent forms.
Methods We conducted a retrospective analysis of clinical consent forms. We developed an annotation scheme following the MAMA (Model-Annotate-Model-Annotate) cycle and evaluated interannotator agreement (IAA) using observed agreement ( A o ), weighted kappa ( κ w ), and Krippendorff's α .
Results The final dataset included 6,399 sentences from 134 clinical consent forms. Complete agreement was achieved for 5,871 sentences, including 211 positively identified and 5,660 negatively identified as permission-sentences across all three annotators ( A o = 0.944, Krippendorff's α = 0.599). These values reflect moderate to substantial IAA. Although permission-sentences contain a set of common words and structure, disagreements between annotators are largely explained by lexical variability and ambiguity in sentence meaning.
Conclusion Our findings point to the complexity of identifying permission-sentences within the clinical consent forms. We present our results in light of lessons learned, which may serve as a launching point for developing tools for automated permission extraction.
Keywords: informed consent, consent forms, natural language processing
Background and Significance
The informed consent process is woven into the fabric of health care ethics, and documentation of informed consent must be included in patients' records as evidence of express permissions for treatment or clinical procedures. 1 2 Although there are benefits to eConsent, 3 4 5 the reality is that consent forms remain largely paper-based in health care settings. 6 Permissions are typically interpreted through manual review on a case-by-case basis; this presents significant issues in terms of scalability and consistency of interpretation. Consent forms are also largely scanned, limiting the usability and/or transferability of the forms.
Machine-interpretable representations of consent permissions are needed to support development of tools that act upon permissions across information ecosystems at scale. While several machine-interpretable representations of consent have been developed, 3 7 8 these efforts are centered on consent for research rather than consent in clinical contexts. Moreover, tools for processing real-world consent forms—a necessary precursor to linking consent form content to machine-interpretable representations—remain underdeveloped.
An annotation scheme serves as a human-readable blueprint to guide manual discovery of a given phenomenon and is a fundamental step toward automation. An iterative approach is often used for development of annotation schemes, starting with an initial guideline and updating it after multiple rounds of small sample annotations. 9
Objective
This case report presents the process, results, and lessons learned while developing and testing an annotation scheme to identify permission-sentences in clinical consent forms.
Methods
Design
This was a retrospective analysis of clinical consent forms. The principal investigator (PI), a nurse scientist, a trained research assistant (RA), who contributed a health care consumer perspective, and a practicing registered nurse (RN) were involved throughout the entirety of the analysis and annotation process. A data scientist with experience in text processing supported technical aspects of the study. Institutional review board review was not required because human subjects were not involved. Only blank consent forms were collected and analyzed.
Recruitment and Sampling
Consent forms were collected through (1) direct contribution by health care facilities and (2) systematic web searching. The Michigan Health Information Management Association (MHIMA) sent an email to 29 directors of health information management departments requesting direct contribution of clinical consent forms. The systematic web search identified publicly available consent forms using search terms for 200 randomly selected hospitals registered with the Centers for Medicare and Medicaid Services (CMS), 10 50 randomly selected ambulatory surgical centers participating in the Ambulatory Surgical Center Quality Reporting Program, 11 and 63 health care facilities affiliated with all Clinical and Translational Science Award (CTSA) hubs funded during 2014 to 2018 (1:1 match for CTSA hub to health care facility). 12 All facilities in the sample of clinical consent forms are described in Supplementary Appendix A (available in the online version).
Consent Form Management
Directly contributed consent forms were emailed to the PI or MHIMA contact; one facility allowed the PI to download consent forms from their internal Web site. Web searches for consent forms were retrieved from facility Web sites and Google searches. Forms were included if their primary purpose was consent for a clinical care process or procedure. We excluded duplicate forms, forms used for hospital operations or nonclinical purposes, and forms that were written in languages other than English or were not human-readable after conversion to .txt formats. The RA created records for facilities and forms in Excel spreadsheets and mapped metadata assigned by CMS (e.g., unique identifiers, name, location, facility type) to each form. Fig. 1 summarizes the data collection and screening procedures.
Fig. 1.
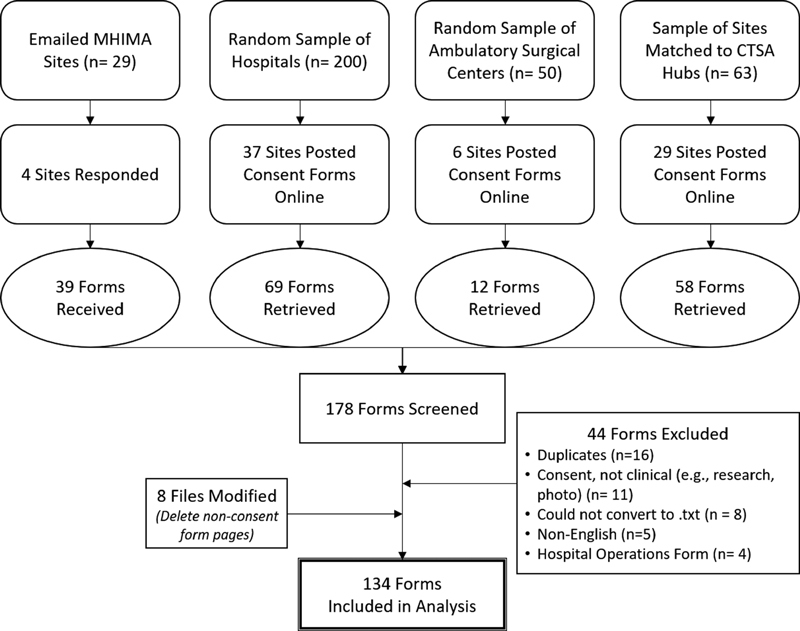
Data collection and management procedures.
Annotation Scheme Development
We followed Pustejovsky and colleagues' MAMA (Model-Annotate-Model-Annotate) cycle for annotation scheme development. 13 We iteratively annotated unique sets of five randomly selected consent forms at a time. The study team met after each round. We manually compared output after each iteration, qualitatively examined themes for differences between annotators, and adapted the annotation scheme and guideline accordingly to clarify its specifications. The annotation scheme was stable after five iterations.
The final annotation scheme and guideline is provided in Supplementary Appendix B (available in the online version). A permission-sentence was formally defined as a “statement(s) that, upon signature of the consent form, authorizes any new action or activity that may, must, or must not be done.” This definition enabled discrimination of permission-sentences from those which did not allow or forbid some new action or activity (e.g., descriptions of care, agreements for payment, statements of patients' rights). The tag Positive was used to markup sentences as permission-sentences (i.e., This is a permission-sentence ). The tag Indeterminate (i.e., This might be a permission-sentence ) indicated uncertainty by annotators due to ambiguous or inconsistent language in the consent form; this tag allowed annotators to group those uncertain sentences into this category rather than forcing a binary decision. All remaining sentences were tagged with Negative (i.e ., This is not a permission-sentence ).
Annotation and Data Preprocessing
Consent forms were converted from their original formats (.pdf, .doc) to text files (.txt) using document format conversion tools built into Adobe Acrobat DC and MS Word. Permission-sentences were identified and tagged in the text files by three annotators using an open-source annotation platform. 14 Considerable preprocessing was required prior to analysis. We used an open-source software library for natural language processing 15 without any case-specific optimization to parse and generate a list of all sentences for each informed consent form. We enforced standard character encoding (ASCII), and all non-ASCII characters were removed. We excluded “sentences” (i.e., text strings) that lacked English alphabet characters, were less than nine characters long, or were less than three words long.
Analysis of Interannotator Agreement
We calculated observed or raw agreement ( A o ) by summing the count of agreed-upon annotations for all tags and dividing by the count of all sentences. We also calculated weighted kappa ( κ w ) and Krippendorff's α to account for the degree of difference between tags (i.e., there is greater distance between Positive and Negative than between either tag and Indeterminate ) and demonstrate interannotator agreement (IAA) beyond what was attributable to chance. 16 All analyses were performed using Python 3.7 or R for Statistical Computing. 17
Results
The final dataset included 134 clinical consent forms from 62 health care facilities. The consent form files have been made publicly available. 18 These consent forms include 6,399 total sentences. Complete agreement was achieved for 5,871 sentences, including 211 positively identified and 5,660 negatively identified as permission-sentences across all three annotators ( A o = 0.944, Krippendorff's α = 0.599). Pairwise agreement was highest between PI and RN ( κ w = 0.655).
Most sentences in the consent forms did not create new contracts of what could or could not be done. Of the sentences that at least one annotator believed may serve a contractual purpose ( n = 739), 28.6% had full agreement and 47.5% were identified by at least two annotators ( n = 351). Table 1 presents IAA measures for all combinations of annotators. Table 2 depicts the count and proportion of permission-sentences as the threshold for IAA was relaxed.
Table 1. Interannotator agreement measures among subgroups of and all annotators.
| A o | κ w | 95% CI | |
| PI–RA | 0.944 | 0.604 | 0.415–0.793 |
| RA–RN | 0.937 | 0.580 | 0.372–0.787 |
| RN–PI | 0.951 | 0.655 | 0.459–0.851 |
|
Krippendorff's α | 95% CI | |
| PI–RA–RN | 0.944 | 0.599 | 0.566–0.631 |
Abbreviations: CI, confidence interval. PI, principal investigator; RA, research assistant; RN, registered nurse.
Note: IAA is reported using all labels (Positive, Indeterminate, and Negative).
A
o
is observed agreement, and
κ
w
is a weighted Kappa coefficient, both of which are used to measure IAA between pairs of annotators.
 and Krippendorff's
α
are used to measure IAA across all three annotators.
and Krippendorff's
α
are used to measure IAA across all three annotators.
Table 2. Identification of permission-sentences based on number of annotators.
| Agreement by 3 annotators | Agreement by ≥2 annotators | Annotation by ≥1 annotator | |
|---|---|---|---|
| Positive: this is a permission-sentence | 211 | 351 | 635 |
| 211/739 (28.6%) | 351/739 (47.5%) | 635/739 (85.9%) | |
| 211/6,399 (3.3%) | 351/6,399 (5.5%) | 635/6,399 (9.9%) | |
| Indeterminate: this might be a permission-sentence | 0 | 5 | 139 |
| 0/739 (0%) | 5/739 (0.7%) | 139/739 (18.8%) | |
| 0/6,399 (0%) | 5/6,399 (0.0%) | 139/6,399 (2.1%) | |
| Negative: this is not a permission-sentence | 5,660 | 6,028 | 6,168 |
| – | – | – | |
| 5,660/6,399 (88.5%) | 6,028/6,399 (94.8%) | 6,168/6,399 (96.4%) | |
| Total sentences 6,399 |
Note: Percentages indicate the proportion of identified sentences to possible permission-sentences ( n = 739) and the entire corpus ( n = 6,399).
We found some consistency in the language used in permission-sentences. Table 3 lists the top 10 verbs, including their frequency and an example, across the 211 completely agreed-upon permission-sentences. These verbs largely reflect either the act of giving permission (authorize, consent, may, request, agree, give) or otherwise refer to the actor consent is being given to or the action begin consented for (perform, named, use, receive). This provided our identifying and modeling processes a common structure of permission-sentences, which is reported elsewhere. 19 However, these common words and structure alone were not enough to discriminate permission-sentences with complete consistency, eliminating the possibility of simple rule-based extraction. Beyond instances of missingness (i.e., one or more annotators did not annotate a permission-sentence), disagreements emerged for several reasons. Example sentences for sources of disagreements are provided in Table 4 .
Table 3. Most common verbs in the 211 agreed upon permission-sentences, their frequency, and example of use.
| Verb | n (%) | Example permission-sentence |
|---|---|---|
| Authorize | 73 (34.6%) | I authorize medical evaluation and treatment, and release of information for insurance/medical purposes concerning my illness and treatment. |
| Consent | 65 (30.8%) | I also consent to diagnostic studies, tests, anesthesia, X-ray examinations, and any other treatment or courses of treatment relating to the diagnosis or procedure described herein. |
| May | 39 (18.5%) | If any unforeseen circumstances should arise which … require deviation from the original anesthetic plan, I further authorize that whatever other anesthetics or emergency procedures deemed advisable by them may be administered or performed. |
| Request | 25 (11.8%) | I, request and consent to the start or induction of my labor by my provider: [sic] and other assistants as may be selected by him/her. |
| Agree | 23 (10.9%) | I agree that any excess tissue, fluids, or specimens removed from my body during my outpatient visit or hospital stay … may be used for such educational purposes and research, including research on the genetic materials (DNA). |
| Perform | 21 (10.0%) | I, (Wife) authorize the Strong Fertility and Reproductive Science Center to perform one or more artificial inseminations on me with sperm obtained from an anonymous donor for the purpose of making me pregnant. |
| Give | 18 (8.5%) | I give permission to my responsible practitioner to do whatever may be necessary if there is a complication or unforeseen condition duringmy [sic] procedure. |
| Named | 13 (6.2%) | I give permission to the hospital and the above-named practitioner to photograph and/or visually record or display the procedure(s) for medical, scientific, or educational purposes. |
| Use | 12 (5.7%) | I request and consent to use of anonymous donor sperm in hopes of achieving a pregnancy. |
| Receive | 12 (5.7%) | … I voluntarily consent to receive medical and health care services that may include diagnostic procedures, examination, and treatment. |
Table 4. Explanations and examples for common sources of disagreement.
| Explanation of disagreement | Example sentence |
|---|---|
| Use of “may” which could indicate either possibility or allowability of some action | A. I/my child/my fetus (circle one) will be tested for genetic indicators that may be linked to the following genetic disease or condition (insert general description of disease/condition). B. This may include performing exams under anesthesia that are relevant to my procedures. |
| Broad statements of agreement to all form content | I have read and agree to the contents of this form. |
| Statements of understanding or necessity rather than new allowability | I understand that blood and urine specimens will need to be collected to determine my care. |
| Statements of allowable actions in the second person as opposed to first person | You are allowing the clinic to use this material for quality control purposes before being discarded in accordance with normal laboratory procedures and applicable laws. |
| Statements of unwanted or irrelevant actions, which may or may not be forbidden | A. I do not wish medical care of any kind except emergency care to be provided. B. Transfusion is not applicable to my operation |
| Tiered consent, when annotators may markup text differently (e.g., stem only, stem + options, options only, etc.) |
The following text includes four consecutive parsed sentences for tiered consent: sentence stem, first option, description of first option, second option.
1. In the event the patient dies prior to use of all the embryos, we agree that the embryos should be disposed of in the following manner (check only one box): 2. Award to patient s [sic] spouse or partner, which gives complete control for any purpose, including implantation, donation for research, or destruction. 3. This may entail maintaining the embryos in storage, and the fees and other payments due the clinic for these cryopreservation services. 4. Award for research purposes, including but not limited to embryonic stem cell research, which may result in the destruction of the embryos but will not result in the birth of a child. |
Discussion
We report on development and testing of an annotation scheme to identify permission-sentences in clinical consent forms. This is a first step toward developing tools to automate permission extraction and machine interpretation of consent form content. One among the challenges in developing the annotation scheme was the need to generate a definition of a permission-sentence that was stable for use across multiple documents and under review by three annotators. With the definition, we achieved a level of agreement that is encouraging as a foundation for future work by us or others. While it is known that reader comprehension of clinical consent forms is an ongoing challenge, 20 our findings may also point to such complexity and obfuscation within the forms that even two clinicians (PI and RN) had difficulty identifying permission-sentences within the sample of forms.
Table 5 outlines lessons learned during this study to improve future annotation and machine-interpretability of permission-sentences. It is important to acknowledge that establishing content standards for defining and interpreting the details of permissions within clinical consent forms requires the involvement of those who author the form, those who sign forms, those who review forms, and those who approve and regulate consent forms at federal, state, and organizational levels. The emergence of single institutional review board reviews and efforts to improve procedural inefficiencies of data use agreements may cast a bright light on the need for some standard content about permissions in consent documents. The primary goal of consent forms, however, is to serve as a tool for communication between providers and patients, and secondarily to provide enduring documentation of the agreements between patients and providers regarding the allowability of actions that, absent a consent form, would not be allowable. A balance must be struck between standardly written content and standards for expressing content.
Table 5. Lessons learned to improve on future permission annotation tasks.
| Lessons learned |
|---|
| • Assess multiple tools (e.g., file conversion tool, annotation software) for a given task and select according to task-specific performance. |
| • Aim for a diverse sample on which to develop the annotation guideline and schema. |
| • Text file data should be cleaned and parsed prior to annotation to prevent disagreements based on differences in annotation boundaries. |
| • Collect rationale for each annotator's decisions, particularly during the schema development phase and when an annotator assigns the indeterminate tag. Document resolution. |
| • A single annotator may not be sufficient to identify instances of a given phenomenon amidst lexical variability, ambiguity of meaning, and complexity of the task. Using more than one annotator reduces errors of missingness and enables discussions around boundaries and rules for annotation. |
There is a clear need for informatics-based standards in this domain, related to clearly defined structures, language use, and encoding the meaning of these patient permissions in documents that are constructed in machine-interpretable formats. A working group within Health Level Seven (HL7) is currently developing a Fast Healthcare Interoperability Resource intended to provide interoperability standards that will address three types of consents: privacy consent directives, medical treatment consent directives, and research consent directives. 21 HL7 provides a tremendous opportunity for collaboration among those with “real-world” expertise in consent forms and their use and those developing standards for the exchange, integration, sharing, and retrieval of information within the consent forms. In the contemporary environment of digital documents and electronic health records, standards-based representations of consent, at the appropriate level of granularity, are essential to transparency into the permissions authorized by patients when signing consent forms. Future collaborative work should include development and refinement of a gold standard list of permission-sentences and annotations that can be used for automated- or semiautomated annotation that requires extensive programing or training of the tagging system. 15 While our IAA reflected moderate to substantial agreement, 22 these results should be interpreted cautiously. The scheme we propose may not yet be sufficient for information retrieval tasks. We believe that our agreement metrics would be higher with increased clarity and consistency in language used in consent forms.
This study has several limitations. First, it is not known whether the sample is broadly representative of clinical consent forms, nor whether web-retrieved consent forms are current. Second, additional levels of error variance were not accounted for as we did not nest permission-sentences by form or facility. It is possible that certain facilities used language that was highly agreed or disagreed upon. Readability of the consent forms was also not assessed. Lastly, use of the PI as an annotator, who was both lead annotation scheme developer and trainer of the RA, may have introduced bias. The annotation scheme should be further refined in future studies or reused by others for similar annotation tasks.
Conclusion
We developed, tested, and shared an annotation scheme for classifying permission-sentences within clinical consent forms that performed with moderate to substantial reliability among three annotators. Our findings point to the complexity of identifying permission-sentences within the clinical consent forms. We present our results in light of lessons learned, which may serve as a launching point for developing tools for automated permission extraction. Future research should examine the understandability of consent permissions across stakeholders, and potentially standardization of clinical consent form structures and content, with emphasis on increasing their understandability by both human and system users.
Clinical Relevance Statement
Informed consent is foundational to respecting patients' autonomy. However, consent information presently relies on human interpretation which is increasingly problematic as the health information ecosystem grows more interconnected. This study and our lessons learned serve as a launching point for future permission extraction, which are the precursors for tools to interpret and act upon permissions at scale.
Multiple Choice Questions
-
Which measure of agreement does not take agreement due to chance or random error into account?
Observed agreement ( A o )
Weighted kappa κ w
Krippendorff's α
Fleiss κ
Correct Answer: The correct answer is a. Observed agreement is a simple ratio of all items agreed upon by annotators to all possible items. It does not take random error into account.
-
Who should clinical consent forms be understandable by?
Clinicians
Patients
Compliance officers and lawyers
All of the above
Correct Answer: The correct answer is option d. Consent forms must be consistently understood by those obtaining consent (clinicians), those granting consent (patients), and those who develop and oversee the use of consent forms (compliance officers and lawyers).
Funding Statement
Funding E. Umberfield was supported in part by the Robert Wood Johnson Foundation Future of Nursing Scholar's Program predoctoral training program. She is presently funded as a Postdoctoral Research Fellow in Public and Population Health Informatics at Fairbanks School of Public Health and Regenstrief Institute, supported by the National Library of Medicine of the National Institutes of Health under award number 5T15LM012502-04. The study was further supported by the National Human Genome Research Institute of the National Institutes of Health under award number 5U01HG009454-03, the Rackham Graduate Student Research Grant, and the University of Michigan Institute for Data Science. The content of this publication is solely the responsibility of the authors and does not necessarily represent the official views of the funding entities.
Conflict of Interest None declared.
Protection of Human and Animal Subjects
Institutional Review Board review was not required because human subjects were not involved. Only blank consent forms were collected and analyzed.
Supplementary Material
References
- 1.American Medical Association Informed Consent American Medical Association. Published 2020. Accessed January 4, 2020 at:https://www.ama-assn.org/delivering-care/ethics/informed-consent
- 2.Hertzum M. Electronic health records in Danish home care and nursing homes: inadequate documentation of care, medication, and consent. Appl Clin Inform. 2021;12(01):27–33. doi: 10.1055/s-0040-1721013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chalil Madathil K, Koikkara R, Obeid J. An investigation of the efficacy of electronic consenting interfaces of research permissions management system in a hospital setting. Int J Med Inform. 2013;82(09):854–863. doi: 10.1016/j.ijmedinf.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reeves J J, Mekeel K L, Waterman R S. Association of electronic surgical consent forms with entry error rates. JAMA Surg. 2020;155(08):777–778. doi: 10.1001/jamasurg.2020.1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen C, Lee P-I, Pain K J, Delgado D, Cole C L, Campion T R., Jr Replacing paper informed consent with electronic informed consent for research in academic medical centers: a scoping review. AMIA Jt Summits Transl Sci Proc. 2020;2020:80–88. [PMC free article] [PubMed] [Google Scholar]
- 6.Litwin J.Engagement shift: informed consent in the digital era Appl Clin Trials 201625(6/7):26, 28, 30, 32 [Google Scholar]
- 7.Obeid J, Gabriel D, Sanderson I. A biomedical research permissions ontology: cognitive and knowledge representation considerations. Proc Gov Technol Inf Policies. 2010;2010:9–13. doi: 10.1145/1920320.1920322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin Y, Harris M R, Manion F J.Development of a BFO-based Informed Consent Ontology (ICO)In: 5th ICBO Conference Proceedings; Houston, Texas; October 6–10, 2014
- 9.Chapman W W, Dowling J N. Inductive creation of an annotation schema for manually indexing clinical conditions from emergency department reports. J Biomed Inform. 2006;39(02):196–208. doi: 10.1016/j.jbi.2005.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Centers for Medicare & Medicaid Services Hospital general informationData.gov. Published February 23, 2019. Accessed November 7, 2019 at:https://data.cms.gov/provider-data/dataset/xubh-q36u
- 11.Centers for Medicare & Medicaid Services Ambulatory surgical quality measures – facilityData.gov. Published October 31, 2019. Accessed November 7, 2019 at:https://data.cms.gov/provider-data/dataset/wue8-3vwe
- 12.CTSA Program Hubs National center for advancing translational sciencesPublished March 13, 2015. Accessed November 26, 2019 at:https://ncats.nih.gov/ctsa/about/hubs
- 13.Pustejovsky J, Bunt H, Zaenen A. Dordrecht: Springer; 2017. Designing annotation schemes: from theory to model; pp. 21–72. [Google Scholar]
- 14.Dataturks Trilldata Technologies Pvt Ltd; 2018. Accessed May 11, 2021 at:https://github.com/DataTurks [Google Scholar]
- 15.Honnibal M, Montani I.spaCy 2: Natural language understanding with Bloom embeddings, convolutional neural networks and incremental parsingPublished online 2017
- 16.Artstein R. Dordrecht: Springer; 2017. Inter-annotator Agreement; pp. 297–313. [Google Scholar]
- 17.R Core Team R: A Language and Environment for Statistical Computing Vienna, Austria: R Foundation for Statistical Computing; 2013. Accessed April 23, 2021 at:http://www.R-project.org/
- 18.Umberfield E, Ford K, Stansbury C, Harris M R.Dataset of Clinical Consent Forms [Data set]University of Michigan - Deep Blue. Accessed May 11, 2021 at:https://doi.org/10.7302/j17s-qj74
- 19.Umberfield E, Stansbury C, Ford K.Evaluating and Extending the Informed Consent Ontology for Representing Permissions from the Clinical Domain(Under Review) [DOI] [PMC free article] [PubMed]
- 20.Eltorai A EM, Naqvi S S, Ghanian S. Readability of invasive procedure consent forms. Clin Transl Sci. 2015;8(06):830–833. doi: 10.1111/cts.12364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.HL7 6.2 Resource Consent - Content. HL7 FHIR Release 5, Preview 3Published 2021. Accessed March 11, 2021 at:http://build.fhir.org/consent.html
- 22.Landis J R, Koch G G. The measurement of observer agreement for categorical data. Biometrics. 1977;33(01):159–174. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


