Abstract
Introduction
The modification of lysine crotonylation (Kcr) is another biological function of histone in addition to modification of lysine acetylation (Kac), which may play a specific regulatory role in diseases.
Objectives
This study compared the expression levels of Kcr and proteome between patients with chronic obstructive pulmonary disease (COPD) combined with type II respiratory failure (RF) to study the relationship between Kcr, proteome, and COPD.
Methods
We tested the Kcr and proteome of COPD combined with type II RF and normal control (NC) using croton acylation enrichment technology and liquid chromatography tandem mass spectrometry (LC-MS/MS) with high resolution.
Results
We found that 32 sites of 23 proteins were upregulated and 914 sites of 295 proteins were downregulated. We performed Kyoto Encyclopedia of Genes and Genomes (KEGG), protein domain, and Gene Ontology (GO) analysis on crotonylated protein. In proteomics research, we found that 190 proteins were upregulated and 151 proteins were downregulated. Among them, 90 proteins were both modified by differentially expressed crotonylation sites and differentially expressed in COPD combined with type II RF and NC.
Conclusion
Differentially expressed crotonylation sites may be involved in the development of COPD combined with type II RF. 90 proteins modified by crotonylation and differentially expressed in COPD combined with type II RF can be used as markers for the study of the molecular pathogenesis of COPD combined with type II RF.
1. Introduction
Chronic obstructive pulmonary disease (COPD) is a common respiratory disease caused by incomplete reversible and progressive development of airflow limitation [1]. The main causes of COPD are inhalation of smoke, sensitization of respiratory tract, occupational exposure, and air pollution [2]. Respiratory failure (RF) is a condition in which the respiratory system fails in one or both of its gas exchange functions. The cause is hypoxemia caused by lung failure or hypoxemia caused by pump failure or alveolar hypoventilation and hypercapnia caused by pump failure [3]. RF is divided into type I and type II. RF is a serious complication of COPD, and the main cause of death from COPD is type II RF. Our national statistics of research found that patients with COPD combined with type II are older than type I. At present, the clinical treatment methods for COPD patients mainly include smoking cessation, drugs, exercise rehabilitation, mechanical ventilation, and lung volume reduction surgery. However, due to delayed patient visits, the rate of pulmonary function tests is low, and patients often do not receive timely and effective treatment. Therefore, early diagnosis and treatment of COPD combined with type II RF is of great significance to patients and families.
Lysine acetylation (Kac) is the earliest and most studied type of modification [4]. The lysine crotonylation (Kcr) is a newly identified biological function of posttranslational modification, which mainly occurs in the lysine residues of histones [5]. The Kcr is similar to the Kac modification in the structure, the regulatory enzyme system, and the recognition protein. However, the crotonylation modification is more potent for gene expression than the acetylation modification, and the balance of histone crotonylation modification and acetylation modification has an effect on gene expression [6]. At present, studies have confirmed that Kcr has the functions of regulating gene transcription, participating in sperm formation and stress protection of acute kidney injury [6–10]. Montellier et al. found that the Kcr modification had a high level in the round sperm chromosome, and the Kcr modification sites were abundantly present at the transcription initiation site, thereby promoting the expression levels of the gene [11]. Xu et al. found that crotonylated modified nonhistones in H1299 cells may be involved in multiple signaling pathways and cellular functions, such as involvement in transport, formation of ribosomes, and Parkinson's disease pathways [12]. Recently, a high degree of agreement between crotonylated proteins and crotonylation sites in zebrafish embryos and humans was reported, and Kcr regulates muscle contraction and protein synthesis [13]. Therefore, Kcr may have an effect on many other diseases and may be a novel class of biomarkers for diagnosis, evaluation, and treatment targets. This study hopes to identify some Kcr of new and reliable biomarkers, so as to provide a data basis for the clinical research of COPD combined with type II RF.
2. Materials and Methods
2.1. Samples Collection
Peripheral blood samples were collected from 8 patients with COPD combined with type II RF and 36 healthy individuals from the Shenzhen People's Hospital (Shenzhen, China). 36 healthy individuals belonged to the normal controls (NC) in this study. All 8 patients (5 males and 3 females; mean age, 75.75 ± 12.02) had long-term symptoms of cough, shortness of breath, and lung infection. The mean value of PaCO2 (mmHg) in blood gas analysis is 52.96 ± 22.63, and PaO2 (mmHg) in blood gas analysis is 80.70 ± 24.78. PaCO2 (mmHg) > 50 mmHg and PaO2 (mmHg) < 60 mmHg were considered type II RF. Among them, 5 males have mean pack-years of smoking 20.00 ± 7.07 years and quit smoking. They did not have severe liver and kidney disease and family genetic history and had not received hormone therapy recently. The clinical characteristics of patients are summarized in Table 1. All of the peripheral blood samples were obtained after receiving informed consent from the participating subject. And the peripheral blood was obtained when the patients were admitted to the hospital because of the deterioration of the condition. This study was performed according to the guidelines of the Shenzhen People's Hospital, which abided by the Helsinki Declaration on ethical principles for medical research involving human subjects. Then, peripheral blood mononuclear cells (PBMCs) were isolated by density gradient centrifugation using Ficoll-Hypaque (GE Healthcare Bio-Sciences AB, Uppsala, Sweden). PBMCs were lysed with TRIzol reagent (Invitrogen, Carlsbad, CA) and stored at −80°C.
Table 1.
Detail clinical information for all COPD combined with type II RF.
| No. | Gender | Age | PaCO2 (mmHg) | PaO2 (mmHg) | Smoke (pack-years) |
|---|---|---|---|---|---|
| P1 | Male | 79 | 64.3 | 57.1 | 20 |
| P2 | Male | 66 | 89.9 | 127.0 | 30 |
| P3 | Male | 50 | 30.0 | 74.8 | 10 |
| P4 | Male | 82 | 49.0 | 74.7 | 20 |
| P5 | Male | 87 | 33.5 | 82.0 | 20 |
| P6 | Female | 79 | 28.9 | 108.0 | — |
| P7 | Female | 82 | 77.2 | 62.0 | — |
| P8 | Female | 81 | 50.9 | 60.0 | — |
2.2. Quantitative Analysis of Kcr and Proteomics
2.2.1. Protein Extraction
Samples were removed from −80°C and added to 4 volumes of lysis buffer (8 M urea, 1% protease inhibitor cocktail, 3 μM TSA, 50 mM NAM, and 2 mM EDTA) for sonication. After centrifugation at 12,000 g for 10 min at 4°C, the cell debris was removed, the supernatant was transferred to a new centrifuge tube, and the protein concentration was determined using a BCA kit.
2.2.2. Trypsin Digestion
Dithiothreitol was added to the protein solution to a final concentration of 5 mM and reduced at 56°C for 30 min. Iodoacetamide was then added to a final concentration of 11 mM and incubated for 15 min at room temperature in the dark. Finally, the urea concentration of the sample is diluted to less than 2 M. Trypsin was added at a mass ratio of 1 : 50 (pancreatin: protein), digested overnight at 37 °C, added at a mass ratio of 1 : 100 (pancreatin: protein), and continued to digest for 4 h.
2.2.3. Relative and Absolute Quantitation (TMT/iTRAQ) Labeling
The trypsin-digested peptide was desalted with Strata X C18 (Phenomenex) and vacuum-dried. The peptide was solubilized with 0.5 M TEAB and the peptide was labeled according to the TMT kit instructions.
2.2.4. High-Performance Liquid Chromatography (HPLC) Fractionation
For proteome experiment, the tryptic peptides were fractionated into fractions by high pH reverse-phase HPLC using Agilent 300Extend C18 column (5 μm particles, 4.6 mm ID, and 250 mm length).
2.2.5. The Kcr Modification Enrichment
The peptides were dissolved in IP buffer solution (100 mM NaCl, 1 mM EDTA, 50 mM Tris-HCl, 0.5% NP-40, and pH 8.0), and the supernatant was transferred to the prewashed crotonylated resin (PTM503, from Hangzhou Jingjie Biotechnology Co., Ltd., PTM Bio) and placed on a rotary shaker at 4 °C. The resin was washed 4 times with IP buffer solution and twice with deionized water after incubation. At that time, the 0.1% trifluoroacetic acid eluent eluted the resin-bound peptide three times. Finally, the eluate was collected and dried in vacuo. After draining, the salt of peptides was removed according to C18 ZipTips instructions for liquid chromatography tandem mass spectrometry (LC-MS/MS) analysis.
2.2.6. LC-MS/MS Analysis
The peptide was dissolved in liquid phase A phase (0.1% aqueous formic acid) and separated using an EASY-nLC 1000 ultrahigh-performance liquid chromatography (UPLC) system. The gradient was comprised of solvent B. Liquid phase gradient setting was 0∼38 min, 8%–22% solvent B; 38∼52 min, 22%–35% solvent B; 52∼56 min, 35%–80% solvent B; 56∼60 min, 80% solvent B.
The peptides were dissociated by a UPLC system and injected into an NSI ion source for ionization and analyzed by Q Exactive™ Plus (Thermo).
2.2.7. Database Search
Secondary mass spectral data was retrieved using MaxQuant (v1.5.2.8).
For searching Kcr database, the effect was set to trypsin/P (the number of missed sites is set to 4). The minimum length of the peptide is set to 7 amino acid residues, and the maximum number of modifications of the peptide is set to 5. The mass tolerance for precursor ions of first search and main search was set to 20 ppm and 5 ppm, respectively, and the mass error tolerance of the secondary fragment ions was 0.02 Da. Carbamidomethyl on Cys was designated as fixed modification. Acetylation modification, Kcr modification, and oxidation on Met were designated as variable modifications. FDR was adjusted to <1% and the minimum score for modified peptides was set to >40.
For searching proteome database, the effect was set to trypsin/P (the number of missed sites is set to 2). The minimum length of the peptide is set to 7 amino acid residues, and the maximum number of modifications of the peptide is set to 5. The mass tolerance for precursor ions of first search and main search was set to 20 ppm and 5 ppm, respectively, and the mass error tolerance of the secondary fragment ions was 0.02 Da. Carbamidomethyl on Cys was designated as fixed modification and oxidation on Met was designated as variable modifications. FDR was adjusted to <1% and the minimum score for peptides was set to >40.
2.2.8. Gene Ontology (GO) Annotation
GO annotation proteome was derived from the UniProt-GOA database (www. http://www.ebi.ac.uk/GOA/). Firstly, convert identified protein ID to UniProt ID and then map it to GO IDs by protein ID. Then proteins were classified by GO annotation based on three categories: biological process, cellular component, and molecular function. GO with a corrected value of P < 0.05 was considered significant.
2.2.9. Domain Annotation
The protein domain annotations were performed on the identified proteins using the protein sequence algorithm-based software InterProScan and the corresponding InterPro domain database. The InterPro domain database (http://www.ebi.ac.uk/interpro/) is a free web database that integrates information including protein family classification, protein domain classification, and protein functional site classification. Protein domains with a corrected value of P < 0.05 were considered significant.
2.2.10. Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Annotation
We used the KEGG pathway database to annotate the protein pathway. First, the submitted protein was annotated using the KEGG online service tool KAAS (http://www.genome.jp/tools/kaas/), and the annotated protein was then matched into the corresponding pathway in the database by the KEGG mapper. The pathway with a corrected value of P < 0.05 was considered significant. These pathways were classified into hierarchical categories according to KEGG website.
2.2.11. Subcellular Localization
We used WoLF PSORT, a subcellular localization prediction software, to predict subcellular localization.
3. Result
3.1. Quantitative Analysis of Kcr Modification in COPD and NC
A total of 946 crotonylation sites containing quantitative information of differential expression were identified by LC-MS/MS in 318 proteins between COPD combined with type II RF and NC The fold change was more than 1.2 times as a significant upregulation and less than 1/1.2 as a criterion for significant downregulation. Among them, 32 sites of 23 proteins were upregulated and 914 sites of 295 proteins were downregulated. The quality control test standard for mass spectrometry (MS) data is that the mass error is concentrated below 0 and 10 ppm, indicating that the mass accuracy of the MS data meets the requirements. Most peptides ranged in length from 8 to 20 amino acids, which were consistent with the rule of trypsin-digesting peptides, indicating that sample preparation is up to standard. The quality control of the peptides of all samples in this study reached the standard.
3.2. GO Functional Annotation and Subcellular Localization of Crotonylation Modification Sites Corresponding to Proteins
We performed a GO functional classification of biological processes, cellular components, and molecular functions of quantitative proteins. In upregulated crotonylated proteins, most crotonylated proteins were involved in response to stimulus, single-organism process, organelle, cell, and binding, accounting for 11%, 11%, 18%, 18%, and 48% of all crotonylated proteins, respectively (Figures 1(a)–1(c)). In downregulated crotonylated proteins, most crotonylated proteins were involved in cellular process, single-organism process, cell, organelle, extracellular region, binding, and catalytic activity, accounting for 13%, 12%, 20%, 20%, 16%, 52%, and 23% of all crotonylated proteins, respectively (Figures 1(d)–1(f)). The upregulated crotonylated proteins and downregulated crotonylated proteins most were involved in organelle, cell, and binding.
Figure 1.
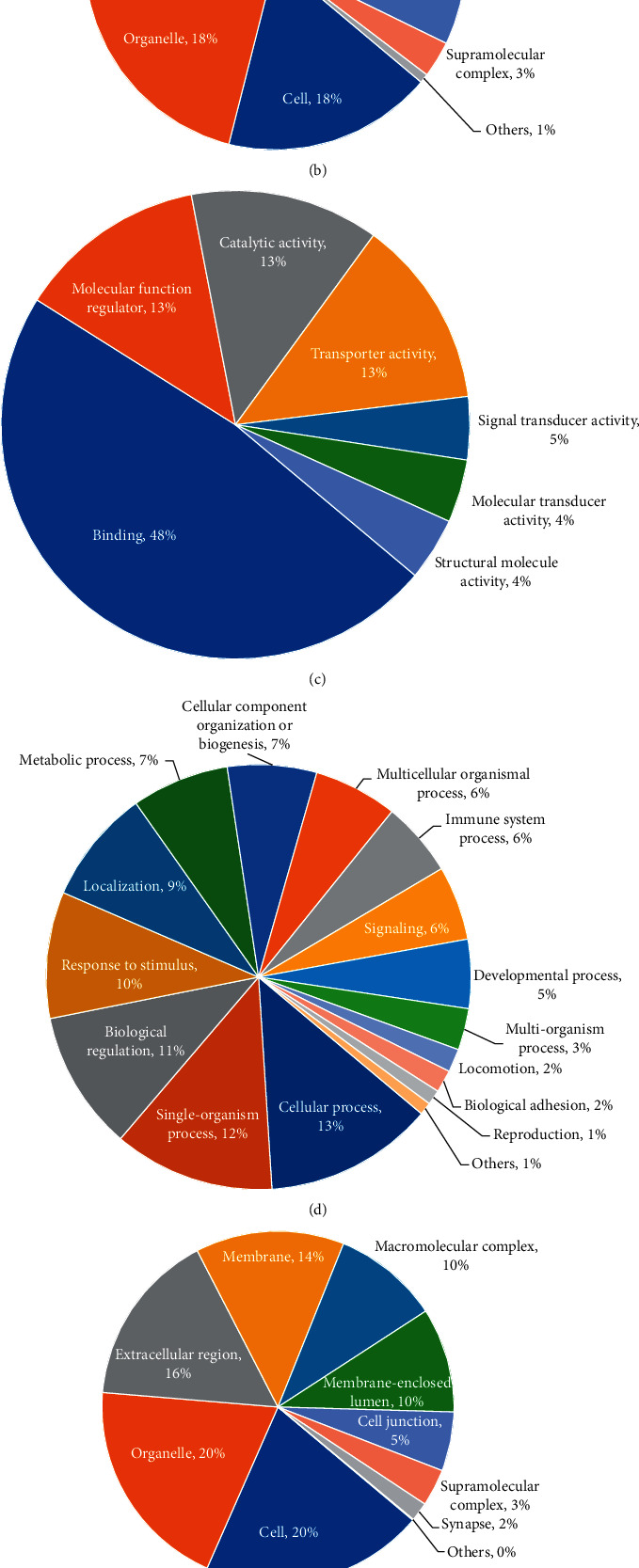
GO functional annotation of crotonylated proteins. GO analysis of upregulated crotonylated proteins shows biological processes (a), cellular component (b), and molecular function (c). GO analysis of downregulated crotonylated proteins shows biological processes (d), cellular component (e), and molecular function (f).
We used WoLF PSORT to predict the subcellular localization of crotonylated proteins. Most upregulated crotonylated proteins were distributed in the cytoplasm (48%) and extracellular (22%). Most downregulated proteins were distributed in the cytoplasm (54%), nucleus (12%), and mitochondria (10%) (Figures 2(a) and 2(b)). The upregulated crotonylated proteins and downregulated crotonylated proteins most were distributed in the cytoplasm.
Figure 2.
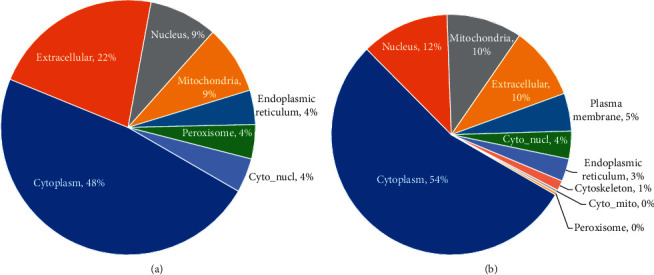
Subcellular localization of upregulated (a) and downregulated (b) crotonylated proteins.
3.3. Functional Enrichment Analysis of Crotonylation Modification Sites Corresponding to Proteins
In order to detect whether the differential expression has a significant enrichment trend in some functional types, we performed enrichment analysis of functional annotation types such as GO, KEGG, and protein domains for the corresponding proteins of crotonylation modification sites. The P-value obtained by the enrichment test (Fisher's exact test) is converted by negative logarithm (−log10). The larger the converted value is, the more significant the richness of this functional type is.
The enrichment analysis of the cellular components revealed that the upregulated crotonylated proteins were highly enriched in the extracellular space, cytoplasmic vesicle part, vesicle lumen, and cytoplasmic vesicle lumen. The enrichment results of the molecular function category showed that the upregulated crotonylated proteins were highly enriched in the iron ion binding, glycoprotein binding, and enzyme binding. The enrichment results of the biological processes showed that the upregulated crotonylated proteins were highly enriched in the regulation of protein stability, cell activation, regulation of nitric oxide biosynthetic process, positive regulation of nitric oxide metabolic process, and positive regulation of nitric oxide biosynthetic process (Figure 3(a)). The KEGG pathway enrichment analysis showed that the upregulated crotonylated proteins were mainly enriched in the hsa04657 IL-17 signaling pathway and hsa04612 antigen processing and presentation (Figure 3(b)). The upregulated crotonylated protein domains were significantly enriched in the serpin domain, histidine kinase-like ATPase, C-terminal domain, heat shock protein Hsp90, N-terminal, and ribosomal protein S5 domain 2-type fold (Figure 3(c)). It is suggested that the Kcr plays an important role in these processes. However, there is no downregulation of crotonylation protein enrichment in GO analysis, KEGG pathway, and protein domains.
Figure 3.
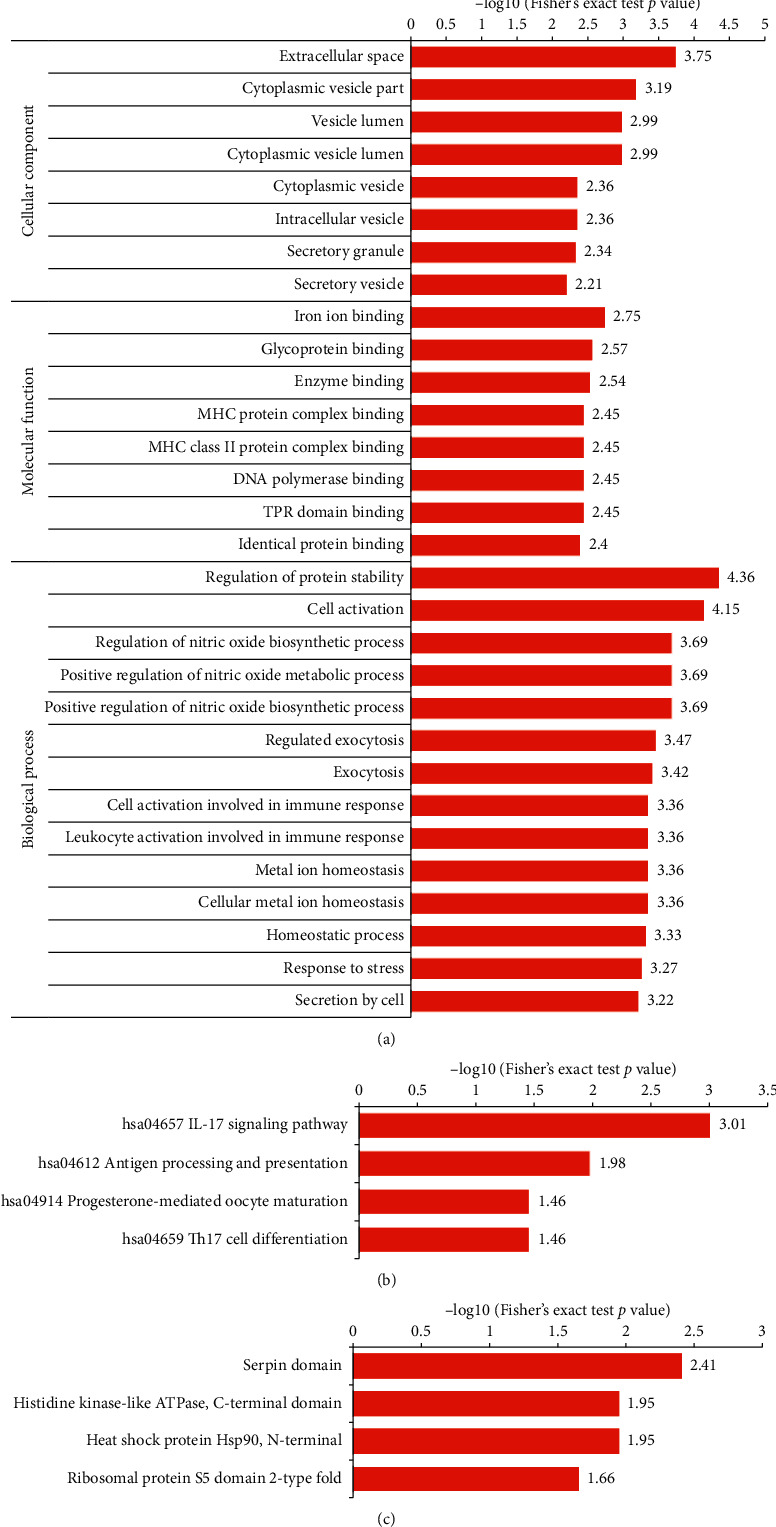
Functional enrichment analysis of crotonylation modification sites corresponding to proteins. (a) Enrichment of GO analysis of upregulated crotonylated protein. (b) Enrichment of KEGG pathway analysis of upregulated crotonylated protein. (c) Enrichment of protein domain analysis of upregulated crotonylated protein.
3.4. Interaction between Differential Expression of Kcr and Proteins
In this project experiment, 341 proteins' quantitative information was obtained by LC-MS/MS between COPD combined with type II RF and NC. The fold change was more than 1.2 times as a significant upregulation and less than 1/1.2 as a criterion for significant downregulation. Among them, 190 proteins were upregulated and 151 proteins were downregulated.
We analyzed the expression profiles of differentially expressed crotonylation site corresponding protein and proteome and conducted a correlation analysis between Kcr and proteome. As a result, there were 90 identical proteins in the crotonylated proteins and proteome with quantitative information. This result showed that these 90 proteins were both modified by differentially expressed crotonylation sites and differentially expressed in COPD combined with type II RF and NC (Figure 4 and Table 2).
Figure 4.
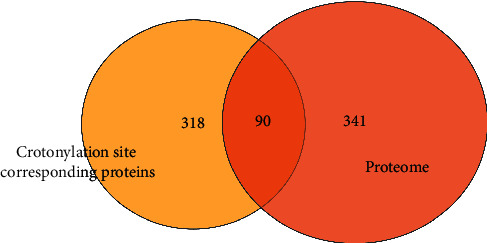
Common proteins of differentially expressed crotonylation site corresponding proteins and proteome between COPD and NC.
Table 2.
90 protein expression fold change value.
| Protein description | Protein fold change | Protein-regulated type |
|---|---|---|
| FYN-binding protein OS = Homo sapiens GN = FYB | 0.788 | Down |
| Flotillin-1 OS = Homo sapiens GN = FLOT1 | 1.452 | Up |
| Haptoglobin OS = Homo sapiens GN = HP | 0.462 | Down |
| Carbonic anhydrase 1 OS = Homo sapiens GN = CA1 | 2.024 | Up |
| Carbonic anhydrase 2 OS = Homo sapiens GN = CA2 | 1.39 | Up |
| Alpha-1-antitrypsin OS = Homo sapiens GN = SERPINA1 | 0.475 | Down |
| Ig kappa chain C region OS = Homo sapiens GN = IGKC | 0.411 | Down |
| Ig gamma-1 chain C region OS = Homo sapiens GN = IGHG1 | 0.386 | Down |
| Ig gamma-2 chain C region OS = Homo sapiens GN = IGHG2 | 0.468 | Down |
| “Spectrin alpha chain, erythrocytic 1 OS = Homo sapiens GN = SPTA1” | 1.522 | Up |
| Apolipoprotein A-I OS = Homo sapiens GN = APOA1 | 0.299 | Down |
| Apolipoprotein A-II OS = Homo sapiens GN = APOA2 | 0.197 | Down |
| Fibrinogen alpha chain OS = Homo sapiens GN = FGA | 0.803 | Down |
| Fibrinogen beta chain OS = Homo sapiens GN = FGB | 0.819 | Down |
| Fibrinogen gamma chain OS = Homo sapiens GN = FGG | 0.791 | Down |
| Platelet basic protein OS = Homo sapiens GN = PPBP | 0.715 | Down |
| Serotransferrin OS = Homo sapiens GN = TF | 0.271 | Down |
| Lactotransferrin OS = Homo sapiens GN = LTF | 1.463 | Up |
| Catalase OS = Homo sapiens GN = CAT | 1.416 | Up |
| Apolipoprotein B-100 OS = Homo sapiens GN = APOB | 0.668 | Down |
| Cytochrome b-245 heavy chain OS = Homo sapiens GN = CYBB | 1.361 | Up |
| Protein S100-A8 OS = Homo sapiens GN = S100A8 | 1.42 | Up |
| Myeloperoxidase OS = Homo sapiens GN = MPO | 1.348 | Up |
| Glucose-6-phosphate isomerase OS = Homo sapiens GN = GPI | 1.307 | Up |
| Protein disulfide-isomerase OS = Homo sapiens GN = P4HB | 1.531 | Up |
| Annexin A2 OS = Homo sapiens GN = ANXA2 | 0.792 | Down |
| Annexin A6 OS = Homo sapiens GN = ANXA6 | 0.782 | Down |
| Heat shock protein HSP 90-beta OS = Homo sapiens GN = HSP90AB1 | 1.316 | Up |
| Leukotriene A-4 hydrolase OS = Homo sapiens GN = LTA4H | 1.264 | Up |
| Heat shock 70 kDa protein 1B OS = Homo sapiens GN = HSPA1B | 1.67 | Up |
| 78 kDa glucose-regulated protein OS = Homo sapiens GN = HSPA5 | 1.266 | Up |
| “Solute carrier family 2, facilitated glucose transporter member 1 OS = Homo sapiens GN = SLC2A1” | 1.919 | Up |
| “C-1-Tetrahydrofolate synthase, cytoplasmic OS = Homo sapiens GN = MTHFD1” | 1.365 | Up |
| Alcohol dehydrogenase class-3 OS = Homo sapiens GN = ADH5 | 0.825 | Down |
| Annexin A3 OS = Homo sapiens GN = ANXA3 | 2.256 | Up |
| X-ray repair cross-complementing protein 5 OS = Homo sapiens GN = XRCC5 | 0.777 | Down |
| “HLA class I histocompatibility antigen, A-11 alpha chain OS = Homo sapiens GN = HLA-A” | 1.296 | Up |
| Endoplasmin OS = Homo sapiens GN = HSP90B1 | 1.214 | Up |
| Bactericidal permeability-increasing protein OS = Homo sapiens GN = BPI | 1.773 | Up |
| Phosphoglycerate mutase 1 OS = Homo sapiens GN = PGAM1 | 1.257 | Up |
| Nucleolin OS = Homo sapiens GN = NCL | 0.748 | Down |
| Neutrophil cytosol factor 2 OS = Homo sapiens GN = NCF2 | 1.415 | Up |
| “V-type proton ATPase subunit B, brain isoform OS = Homo sapiens GN = ATP6V1B2” | 1.262 | Up |
| Nonspecific lipid-transfer protein OS = Homo sapiens GN = SCP2 | 1.359 | Up |
| Nucleoside diphosphate kinase B OS = Homo sapiens GN = NME2 | 1.328 | Up |
| cAMP-dependent protein kinase catalytic subunit beta OS = Homo sapiens GN = PRKACB | 0.649 | Down |
| Cofilin-1 OS = Homo sapiens GN = CFL1 | 0.77 | Down |
| Cathepsin S OS = Homo sapiens GN = CTSS | 1.219 | Up |
| Protein S100-P OS = Homo sapiens GN = S100P | 3.417 | Up |
| Erythrocyte band 7 integral membrane protein OS = Homo sapiens GN = STOM | 1.392 | Up |
| Calreticulin OS = Homo sapiens GN = CALR | 1.243 | Up |
| Transketolase OS = Homo sapiens GN = TKT | 1.392 | Up |
| Flavin reductase (NADPH) OS = Homo sapiens GN = BLVRB | 1.609 | Up |
| “Peptidyl-prolyl cis-trans isomerase F, mitochondrial OS = Homo sapiens GN = PPIF” | 0.812 | Down |
| Coronin-1A OS = Homo sapiens GN = CORO1A | 0.822 | Down |
| Protein S100-A11 OS = Homo sapiens GN = S100A11 | 1.698 | Up |
| Peroxiredoxin-2 OS = Homo sapiens GN = PRDX2 | 1.967 | Up |
| Transaldolase OS=Homo sapiens GN = TALDO1 | 1.25 | Up |
| Macrophage-capping protein OS = Homo sapiens GN = CAPG | 1.339 | Up |
| “Malate dehydrogenase, cytoplasmic OS = Homo sapiens GN = MDH1” | 0.702 | Down |
| Myeloid cell nuclear differentiation antigen OS = Homo sapiens GN = MNDA | 1.259 | Up |
| LIM and senescent cell antigen-like-containing domain protein 1 OS = Homo sapiens GN = LIMS1 | 0.799 | Down |
| “6-Phosphogluconate dehydrogenase, decarboxylating OS = Homo sapiens GN = PGD” | 1.609 | Up |
| Histone H4 OS = Homo sapiens GN = HIST1H4A | 0.309 | Down |
| Tubulin alpha-4A chain OS = Homo sapiens GN = TUBA4A | 0.721 | Down |
| Hemoglobin subunit beta OS = Homo sapiens GN = HBB | 1.475 | Up |
| Neutrophil gelatinase-associated lipocalin OS = Homo sapiens GN = LCN2 | 1.714 | Up |
| “Phosphate carrier protein, mitochondrial OS = Homo sapiens GN = SLC25A3” | 0.719 | Down |
| 14-3-3 protein eta OS = Homo sapiens GN = YWHAH | 1.271 | Up |
| Selenium-binding protein 1 OS = Homo sapiens GN = SELENBP1 | 1.342 | Up |
| Integrin-linked protein kinase OS = Homo sapiens GN = ILK | 0.819 | Down |
| Stromal interaction molecule 1 OS = Homo sapiens GN = STIM1 | 0.764 | Down |
| Four and a half LIM domains protein 1 OS = Homo sapiens GN = FHL1 | 0.653 | Down |
| Protein disulfide-isomerase A6 OS = Homo sapiens GN = PDIA6 | 1.223 | Up |
| Septin-7 OS = Homo sapiens GN = SEPT7 | 0.822 | Down |
| Putative elongation factor 1-alpha-like 3 OS = Homo sapiens GN = EEF1A1P5 | 1.222 | Up |
| Phospholipase B-like 1 OS = Homo sapiens GN = PLBD1 | 1.748 | Up |
| Olfactomedin-4 OS = Homo sapiens GN = OLFM4 | 3.945 | Up |
| Fermitin family homolog 3 OS = Homo sapiens GN = FERMT3 | 0.772 | Down |
| Histone H2A type 1-C OS = Homo sapiens GN = HIST1H2AC | 0.592 | Down |
| Sarcoplasmic/endoplasmic reticulum calcium ATPase 3 OS = Homo sapiens GN = ATP2A3 | 0.833 | Down |
| WW domain-binding protein 2 OS = Homo sapiens GN = WBP2 | 1.24 | Up |
| EF-hand domain-containing protein D2 OS = Homo sapiens GN = EFHD2 | 0.78 | Down |
| Histone H2A type 1-J OS = Homo sapiens GN = HIST1H2AJ | 0.732 | Down |
| Beta-parvin OS = Homo sapiens GN = PARVB | 0.815 | Down |
| Alpha-hemoglobin-stabilizing protein OS = Homo sapiens GN = AHSP | 1.824 | Up |
| Bridging integrator 2 OS = Homo sapiens GN = BIN2 | 0.824 | Down |
| Coronin-1C OS = Homo sapiens GN = CORO1C | 0.808 | Down |
| Protein kinase C and casein kinase substrate in neurons protein 2 OS = Homo sapiens GN = PACSIN2 | 1.23 | Up |
| Voltage-dependent anion-selective channel protein 3 OS = Homo sapiens GN = VDAC3 | 0.694 | Down |
The fold change was more than 1.2 times as a significant upregulation and less than 1/1.2 as a criterion for significant downregulation.
4. Discussion
Since COPD has been defined as a heterogeneous disease [14], more and more researchers have conducted molecular biology research, including genomics [15–19] and metabolomics [20, 21]. For proteomics, Braido et al. [22] found that there is a possible link between the innate immune system and the worsening of COPD infectivity by discovering that the immune-associated Clara cell 16 (CC-16) is differentially expressed in COPD. Leuzzi et al. [23] found that high baseline C-reactive protein (CRP) levels are significantly associated with late mortality in COPD patients. Recently, Xiang et al. [24] found that YKL-40 proteins were differentially expressed in COPD, suggesting that YKL-40 may be involved in bronchial inflammation and remodeling of COPD and may be a useful biomarker for the diagnosis and monitoring of COPD. Although the relationship between proteomics and COPD has been the focus of research, the relationship between Kcr and COPD combined with type II RF is largely unknown.
In the GO enrichment, the upregulated crotonylated proteins were enriched in biological processes, cell structural components, and cell-binding functional types, indicating that Kcr participated in pathologically related cell and molecular life activities in COPD combined with type II RF. The functional enrichment of Kcr in KEGG analysis revealed IL-17 signaling pathway, antigen processing, and presentation, all of which have an association with immunology [25, 26]. Mario and Maria [27] have also shown that IL-17 plays an important proinflammatory effect in COPD. The lung infection of COPD is mainly in the large and small airway position, with bronchial mucosal edema, inflammation, and hypersecretion of gland as the main clinical manifestations [28]. It is suggested that the differentially expressed Kcr sites enriched in IL-17 signaling pathway and antigen processing and presentation may regulate the expression of related proteins in the immune response of COPD lung infection.
We used TMT labeling, crotonylation enrichment technology, and the quantitative proteomics research strategy of high-resolution LC-MS/MS to quantitatively study Kcr and proteomics in COPD combined with type II RF and NC. We found 32 and 914 sites, corresponding to 23 and 295 proteins, which were significantly upregulated and downregulated, respectively. We also conducted differential expression studies on proteomics. We found that 90 proteins were both modified by differentially expressed crotonylation sites and differentially expressed in COPD and NC. These 90 proteins were differentially expressed between COPD combined with type II RF and NC and may be biomarkers for the diagnosis of COPD combined with type II RF.
The present study has some limitations. Firstly, the small number of samples may cause certain errors in the research results. Secondly, this research provides a data basis for the correlation between Kcr and COPD combined with type II RF, which is only the initial stage. Therefore, we will consider collecting more samples for subsequent research. In a later study, we will consider performing site-directed mutagenesis in vitro and introduce site mutation for proteins and sites that modify levels of difference. Immunofluorescence (IF), fluorescent resonance energy transfer (FRET), and other fluorescent labeling methods will be used to analyze the subcellular localization and real-time dynamic changes of the target molecule, which will provide a basis for functional mechanism research and protein interaction.
5. Conclusion
We found that differentially expressed Kcr may be involved in the modification of COPD-related proteins and is involved in the pathogenesis of COPD combined with type II RF. Our research provides an idea for studying the association of Kcr, proteins, and COPD to identify biomarkers of COPD combined with type II RF.
Acknowledgments
The authors gratefully acknowledge the financial support by the Fund of Guangxi Key Laboratory of Metabolic Diseases Research (No. 20-065-76), Shenzhen Science and Technology Project (Nos. JCYJ20170307095606266, JCYJ20170413093032806, and JCYJ20150403101146305), and Science and Technology Planning Project of Guangdong Province, China (No. 2017A020214016). The authors thank the patients and healthy volunteers who participated in this study.
Contributor Information
Yingyun Fu, Email: yingyunf2013@163.com.
Yong Dai, Email: daiyong22@aliyun.com.
Data Availability
The raw datasets used to support the findings of this study are available from the corresponding author upon request.
Disclosure
Authors' institution 1 and institution 2 contributed equally to this work.
Conflicts of Interest
The authors declare no conflicts of interest.
Authors' Contributions
Qing Gan and Donge Tang contributed equally to this work.
References
- 1.Minas M., Dimitropoulos K., Pastaka C., Papadopoulos D., Markoulis N., Gourgoulianis K. Global initiative for chronic obstructive lung disease for chronic obstructive pulmonary disease: GOLD opportunity for lung disorders. Preventive Medicine. 2005;40(3):274–277. doi: 10.1016/j.ypmed.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 2.Mannino D. M., Buist A. S. Global burden of COPD: risk factors, prevalence, and future trends. Lancet (London, England) 2007;370(9589):765–773. doi: 10.1016/S0140-6736(07)61380-4. North American Edition. [DOI] [PubMed] [Google Scholar]
- 3.Lamba T. S., Sharara R. S., Singh A. C., Balaan M. Pathophysiology and classification of respiratory failure. Critical Care Nursing Quarterly. 2016;39(2):85–93. doi: 10.1097/cnq.0000000000000102. [DOI] [PubMed] [Google Scholar]
- 4.Phillips D. The presence of acetyl groups in histones. Biochemical Journal. 1963;87(2):258–263. doi: 10.1042/bj0870258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tan M., Luo H., Lee S., et al. Identification of 67 histone marks and histone 1ysine crotonylation as a new type of histone modifcation. Cell. 2011;146(6):1016–1028. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sabari B. R., Tang Z., Huang H., et al. Intracellular crotonyl-CoA stimulates transcription through p300-catalyzed histone crotonylation. Molecular Cell. 2015;58(2):203–215. doi: 10.1016/j.molcel.2015.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sabari B. R., Zhang D., Allis C. D., et al. Metabolic regulation of gene expression through histone acylations. Nature Reviews Molecular Cell Biology. 2017;18(2):p. 90. doi: 10.1038/nrm.2016.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olga R. A., Dolores S. N. M., Pablo C. O., et al. Histone lysine crotonylation during acute kidney injury in mice. Disease Models & Mechanisms. 2016;9(6):633–645. doi: 10.1242/dmm.024455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu X., Wei W., Liu Y., et al. MOF as an evolutionarily conserved histone crotonyltransferase and transcriptional activation by histone acetyltransferasedefi cient and crotonyltransferase-competent CBP/p300. Cell Discovery. 2017;23(3):p. 17016. doi: 10.1038/celldisc.2017.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei W., Liu X., Chen J., et al. Class I histone deacetylases are major histone decrotonylases: evidence for critical and broad function of histone crotonylation in transcription. Cell Discovery. 2017;27(7):898–915. doi: 10.1038/cr.2017.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Montellier E., Rousseaux S., Zhao Y., Khochbin S. Histone crotonylation specifically marks the haploid male germ cell gene expression program: post-meiotic male-specific gene expression. Bioessays News & Reviews in Molecular Cellular & Developmental Biology. 2012;34(3):l87–93. doi: 10.1002/bies.201100141. [DOI] [PubMed] [Google Scholar]
- 12.Xu W., Wall J., Zhan J., et al. Global profiling of crotonylation on non-histone proteins. Cell Research. 2017;27(7):946–949. doi: 10.1038/cr.2017.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwon O. K., Kim S. J., Lee S. First profiling of lysine crotonylation of myofilament proteins and ribosomal proteins in zebrafish embryos. Scientific Reports. 2018;8(1):p. 3652. doi: 10.1038/s41598-018-22069-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agusti A., Sin D. D. Biomarkers in COPD. Clinics in Chest Medicine. 2014;35(1):131–141. doi: 10.1016/j.ccm.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 15.Bentley A. R., Emrani P., Cassano P. A. Genetic variation and gene expression in antioxidant related enzymes and risk of COPD: a systematic review. Thorax. 2008;63(11):956–961. doi: 10.1136/thx.2007.086199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Castaldi P. J., Cho M. H., Cohn M., et al. The COPD genetic association compendium: a comprehensive online database of COPD genetic associations. Human Molecular Genetics. 2010;19(3):526–534. doi: 10.1093/hmg/ddp519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nielsen A. O., Jensen C. S., Arredouani M. S., et al. Variants of the ADRB2 gene in COPD: systematic review and meta-analyses of disease risk and treatment response. COPD Journal of Chronic Obstructive Pulmonary Disease. 2017;14(4):1–10. doi: 10.1080/15412555.2017.1320370. [DOI] [PubMed] [Google Scholar]
- 18.Durme Y. V., Eijgelsheim M., Joos G. F., et al. Hedgehog-interacting protein is a COPD susceptibility gene: the Rotterdam study. European Respiratory Journal. 2010;36(1):p. 89. doi: 10.1183/09031936.00129509. [DOI] [PubMed] [Google Scholar]
- 19.Chen Z. H., Kim H. P., Ryter S. W., et al. Identifying targets for COPD treatment through gene expression analyses. International Journal of Chronic Obstructive Pulmonary Disease. 2008;3(3):359–370. doi: 10.2147/copd.s1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Q., Deeb R. S., Ma Y., et al. Serum metabolite biomarkers discriminate healthy smokers from COPD smokers. PLoS One. 2015;10(12) doi: 10.1371/journal.pone.0143937.e0143937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watson E. D., Sweeney C. R., Steensma K. A. Arachidonate metabolites in bronchoalveolar lavage fluid from horses with and without COPD. Equine Veterinary Journal. 1992;24(5):379–381. doi: 10.1111/j.2042-3306.1992.tb02859.x. [DOI] [PubMed] [Google Scholar]
- 22.Braido F., Riccio A. M., Guerra L., et al. Clara cell 16 protein in COPD sputum: a marker of small airways damage? Respiratory Medicine. 2007;101(10):2119–2124. doi: 10.1016/j.rmed.2007.05.023. [DOI] [PubMed] [Google Scholar]
- 23.Leuzzi G., Galeone C., Taverna F., Suatoni P., Morelli D., Pastorino U. C-reactive protein level predicts mortality in COPD: a systematic review and meta-analysis. European Respiratory Review. 2017;26(143):p. 160070. doi: 10.1183/16000617.0070-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiang T., Dongguang W., Sitong L., et al. The YKL-40 protein is a potential biomarker for COPD: a meta-analysis and systematic review. International Journal of Chronic Obstructive Pulmonary Disease. 2018;13:409–418. doi: 10.2147/COPD.S152655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korn T., Bettelli E., Oukka M., Kuchroo V. K. IL-17 and Th17 cells. Annual Review of Immunology. 2009;27(1):485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- 26.Germain R. N., Margulies D. H. The biochemistry and cell biology of antigen processing and presentation. Annual Review of Immunology. 1993;11(1):403–450. doi: 10.1146/annurev.iy.11.040193.002155. [DOI] [PubMed] [Google Scholar]
- 27.Mario C., Maria G. M. IL-17 in chronic obstructive pulmonary disease. Expert Review of Respiratory Medicine. 2012;6(2):135–138. doi: 10.1586/ers.12.7. [DOI] [PubMed] [Google Scholar]
- 28.Gotfried M. H. Macrolides for the treatment of chronic sinusitis, asthma, and COPD. Chest. 2004;125(2):52S–61S. doi: 10.1378/chest.125.2_suppl.52s. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw datasets used to support the findings of this study are available from the corresponding author upon request.


