Abstract
In recent years, many studies have been conducted to investigate the influence of terahertz (THz) radiation on the gene expression in various cell types, but the underling molecular mechanism has not yet been fully revealed. In this study, we explored the effects of 0.1 THz radiation on the gene expression in primary neuron cells through RNA-seq analysis. 111 up-regulated and 54 down-regulated genes were identified. Several biomolecule binding related categories such as “long-chain fatty acid binding”, “tropomyosin binding”, “BMP receptor binding”, as well as “GTPase binding” and “phospholipid binding” were enriched by GO analysis. Moreover, the GSEA analysis indicated that genes encoding protein biosynthetic machinery ribosome were up-regulated by 0.1 THz irradiation. In addition, we demonstrated that the binding efficiency of a transcription factor (TF) AP-1 with its transcription factor binding site (TFBS) in DNA was reduced by THz irradiation, which suggested that THz irradiation might affect the interaction between TFs with DNA and consequently regulate the gene expression. Our results provide new insights into the biological effects of terahertz irradiation.
1. Introduction
The electromagnetic radiation of the Terahertz (THz) band that corresponds to the wavelength interval of 30 µm – 3 mm, has attracted considerable interest in both fundamental and applied research field in the last decades. Taking the advantage of its relative high penetrating capacity and low photon energy, THz has been increasingly introduced into biomedical applications in various fields such as medicine, security and tumor imaging [1–5]. In the meantime, although THz radiation does not cause damage to biomacromolecules by breaking covalent bonds as the way ultraviolet light or ionizing radiation does [4], it does elicit various cellular responses such as changing the gene expression patterns, altering the structure and integrity of the genome and affecting the assembling process of cytoskeleton [6–8]. Boian et al. reported that THz radiation affected the expression of certain genes in mouse stem cells with unaffected heat shock proteins expression [6]. The similar phenomenon was also found in human embryonic stem cells, artificial human skin and human keratinocytes while exposed to THz irradiation [9–11]. As for the underlying mechanism, the DeoxyriboNucleic Acid (DNA) breathing model was first applied to explain and predict the effect of THz radiation on gene expression. THz radiation may create new open states in the DNA helix through nonlinear resonance and therefore influence gene expression [12–14]. The computational simulation results suggested the existence of large transient bubbles at the transcription start sites in the core promoter regions of the peroxisome proliferators-activated receptors γ (PPARγ) and solute carrier organic anion transporter family member 4a1 (Slco4a1) genes under the given THz irradiation exposure condition, which facilitated the transcription initiation [12,13]. In addition to the DNA double strain dynamic, gene expression is also regulated by many other factors such as the transcription factors (TFs) and their interaction with their specific transcription factor binding sites (TFBSs) [15,16]. It was reported that the frequency of THz waves matches the conformational oscillation of biomolecules, which may excite the energy levels of vibration and rotation of biological macromolecules, resulting in changes of molecular conformation and affecting the interactions among biological macromolecules [4]. It’s reasonable to speculate that the activity of various TFs or their recognition and binding to specific TFBSs might be influenced by THz radiation [17]. Recently, Tachizaki et al. reported that the gene networks in human induced pluripotent stem cells could be altered by THz irradiation [17]. They found that many of those altered genes were regulated by zinc-dependent TFs and therefore hypothesized that THz irradiation might impact the gene transcription by affecting those zinc-dependent TFs [17]. However, there is still no direct evidence to illustrate whether the binding of TFs on their corresponding TFBSs could be affected by THz radiation, which is obviously one of the most important mechanisms affecting gene expression.
In this study, we investigated the effect of 0.1 THz (33 mW/cm2 power density) radiation on the gene expression in primary hippocampal neurons. RNA-sequencing (RNA-seq) was performed to identify the expression of genes in response to THz irradiation. These differentially expressed genes (DEGs) between the irradiation group and the control group were used to find the related pathway involved in the THz irradiation response. Considering the gene expression was regulated by TFs, the electrophoretic mobility shift assay (EMSA) was further used to analyse the binding between nuclear proteins with the DNA probe for the TF activator protein-1 (AP-1). The results showed that THz radiation did alter the binding activity of AP-1 with its DNA probe. Our findings provided a potential mechanism to explain the THz induced changing of gene expression and highlighted several biological functions that might be affected by THz irradiation.
2. Materials and methods
2.1. Isolation and culture of hippocampal neurons
Neonates of Sprague-Dawley (SD) rats (at postnatal day 1) were purchased from Experimental Animal Center of Xi'an Jiaotong University Health Science Center and sacrificed for its brain. The primary neurons were isolated from the hippocampi of neonatal SD rats as described previously [18]. The hippocampi were dissected and immersed into D-Hank’s solution. Subsequently, the tissue was cut into pieces, digested with 0.25% trypsin at 37 °C for 15 min and then terminated by fetal bovine serum (FBS, PAA, USA). The digested cells were centrifuged at 1000 rpm for 5 min. Then the supernatant was discarded and the pellet was re-suspended in Dulbecco’s Modified Eagle’s Medium (DMEM)/F12 medium supplemented with 10% FBS (HyClone, USA). Cells were then plated on the poly-L-lysine (Sigma-Aldrich, USA) pre-coated cell culture dishes. The cells were incubated for 4-6 h in complete DMEM/F12 media and then replaced with Neurobasal medium supplemented with 2% B27 and 1% Gln. The purity of neurons was identified by MAP2-immunfluorescence.
2.2. THz sources and irradiation
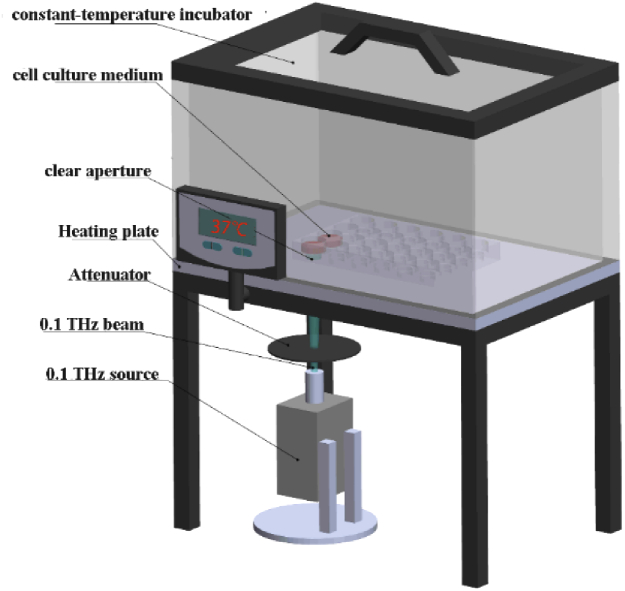
The THz source (0.1 THz, Terasense Group Inc.) was placed under the thermostatic cell incubator and the schematic representation of the exposure set-up was shown in Fig. 1. The cells planted in the culture plate and put inside the incubator were exposed to the THz irradiation from the bottom of the culture dish and the culture medium was 500 µL. The temperature changes of cell culture medium between irradiated group and control group were measured by hand-held infrared thermometer (TN40ALC, ZyTemp). For the irradiation experiment, 6-day cultured primary hippocampal neurons were exposed to 0.1 THz (average power density 33 mW/cm2) irradiation once for 20 min, and then cultured for another 2 h in CO2 incubator for the next experiment.
Fig. 1.
Schematic diagram of the irradiation device. The exposure of primary hippocampus neuron to terahertz radiation was carried out using a terahertz source (Terasense Group Inc.). The frequency was 0.1 THz, the spot diameter was 1.1 cm and the average power was 33 mW/cm2. The cells were cultured in the 37 °C constant temperature incubator, and irradiated by terahertz radiation from the bottom of the culture dish. The cell culture medium was 500 µL.
2.3. RNA-seq and data analysis
Total RNA was individually extracted from the 3 irradiated cell samples and their respective controls using Trizol reagent (Takara, Japan) according to the manufacturer’s instructions, quantified and qualified using an Agilent 2100 Bioanalyzer (Agilent Technologies, USA).1 µg total RNA with OD260/280 within 1.8∼2.2 and RIN value > 7 was used to purified by oligo-dT magnetic beads and then the DNA libraries were constructed and loaded on an Illumina HiSeq instrument (Illumina, USA) according to the manufacturer’s instructions. Sequencing was carried out using a 2 × 125 paired-end configuration to get FastQ data and FastQC was conducted to evaluate the quality of sequencing data. Raw data of adaptors and low quality of the sequences were wiped by TrimGalore. Clean data were aligned with Rnor6 as the reference genome using HISAT2 software [19]. Then the number of fragments per kilobase of transcript per million fragments mapped reads (FPKM) was calculated by Stringtie. DEGs, that is, those with p values < 0.05 and |log2(fold change) | >1, were identified using DESeq2 software [20]. Subsequently, Gene Ontology (GO) analysis was performed and Kyoto Encyclopedia of Genes and Genomes (KEGG) was examined to describe the function of DEGs by using R package clusterProfiler. Gene Set Enrichment Analysis (GSEA) with the C5 (Ontology gene sets) was used to analyze the GO gene sets [21].
2.4. qRT-PCR
The remaining RNA after sequencing was used for quantification of genes which were changed in the RNAseq data. These RNA were reverse transcribed into cDNA by Transcriptor First STRAND cDNA synthesis kit (Vazyme). Real-time PCR was performed using AceQ qPCR SYBR Master Mix (Vazyme). The sequence of primers used in this experiment are listed in Table 1. GAPDH was served as the internal control. The 2-△△Ct method was used to analyze the relative gene expression.
Table 1. Sequences of primers for Real-time PCR.
| Target | Forward primer | Reverse primer |
|---|---|---|
| AQP5 | 5′- taacctggccgtcaatgc-3′ | 5′- gggctagctggaaagtcaag -3′ |
| MMP3 | 5′- ttgtccttcgatgcagtcag-3′ | 5′- ggggtcctgagagattttcg -3′ |
| ITPR1 | 5′- tgacgaaaggcgaagagaat- 3′ | 5′- agatctcatcacgttgctgct -3′ |
| ERF | 5′- ctgatctccagcacgctaca -3′ | 5′- acggctctcaggaatctcg-3′ |
| APOF | 5′- ttacttctcaatggctggactatatg -3′ | 5′- gaattgatgtcgaattgagtctga-3′ |
| ATP2b4 | 5′- tccacctcttctgccgttac-3′ | 5′- aatgacgtctcggaaactatgaa-3′ |
| GAPDH | 5′-aagtatgatgacatcaaaaggtggt-3′ | 5′-agcccaggatgccctttagt -3′ |
2.5. Electrophoretic mobility shift assay (EMSA)
The electrophoretic mobility shift assay (EMSA) is a rapid and sensitive method to detect protein-nucleic acid interactions [22]. Nuclear extracts of HT22 cells were prepared according to the protocol of Nuclear and Cytoplasmic Protein Extraction Kit (Beyotime Biotechnology, China). The sequence of biotin-labeled AP-1 consensus oligonucleotide is as follows: 5′- CGC TTG ATG ACT CAG CCG GAA -3′. For the irradiation experiment, 5 µg nuclear extracts and 1 µL biotin-labeled AP-1 probe with EMSA/Gel-Shift binding buffer (total volume of 15 µL) were exposed to 0.1 THz source for 20 min, and the following step was performed according to the protocol of EMSA/Gel-Shift kit (Beyotime Biotechnology, China) requires. The analysis of shift band was processed using the Quantity One software.
2.6. Data analysis
The statistical significance of differences was analyzed using Prism 5 (GraphPad Software, USA) with Student’s t test. The data are shown as mean ± S.E.M. The value of p < 0.05 was considered to be statistically significant.
3. Results
3.1. Temperature variation during THz irradiation
During the experiment the bottom of the culture dish is right above the THz source, and the temperature of the medium in the sham group (only placed at the through hole without irradiation for 20 minutes) and 0.1 THz irradiation group (average power density 33.0 mW/cm2 for 20 minutes) was measured by hand-held thermometer. The detected temperatures of each group are shown in Fig. 2, and the results showed that there was no significant difference in the temperature between the culture medium of the sham group (n=8) and the radiation group (n=9). These results showed that there was no obvious temperature variation due to irradiation (Fig. 2).
Fig. 2.

Temperature variation of the sham group and the irradiated group. The temperature of culture medium of the sham group (n = 8) and the irradiated group (n = 9) were measured by a hand-held infrared thermometer after sham exposure and 20 minutes THz irradiation, respectively.
3.2. Gene Expression Analysis
According to the p values < 0.05 and |log2(fold change) | >1 for cut-off to select the DEGs, totally 165 genes were considered being differentially expressed between THz-irradiated and non-irradiated groups, among which 111 genes were upregulated and 54 genes were downregulated by THz irradiation (Table 2). 48157 transcripts were obtained from the RNA-seq and the differential expressed transcripts (DETs) analysis showed about 5% of the gene expression were changed by THz irradiation. Among them, 1199 were up-regulated and 1170 were down-regulated by THz irradiation (Table 2).
Table 2. DEGs and DETs.
| All DEGs | Up DEGs | Down DEGs | All DETs | Up DETs | Down DETs |
|---|---|---|---|---|---|
| 165 | 111 | 54 | 2369 | 1199 | 1170 |
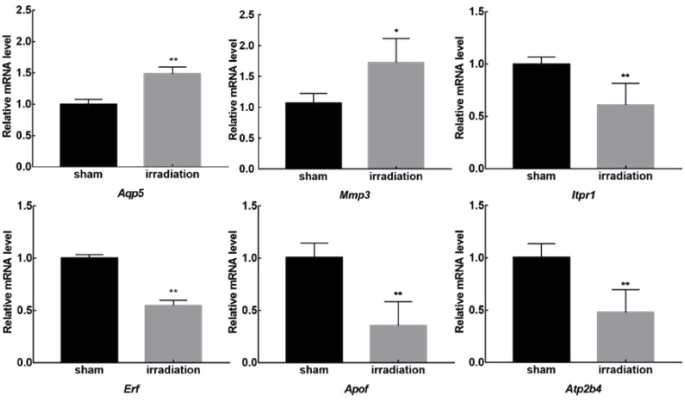
According to the significance and fold change value of DEGs/DETs from RNA-seq data, six genes were selected and verified by the qRT-PCR method. Among them, the expression of Aqp5 which encodes for the aquaporin 5 (AQP5), and Mmp3 which encodes for the matrix metallopeptidase 3, were increased more than 1.5 times. The remaining four genes Erf (encodes for a transcriptional repressor that binds to the Ets2 promoter), Apof (encodes for a kind of apolipoprotein), Atp2b4 (encodes the plasma membrane calcium ATPase isoform 4) and Itpr1 (encodes an intracellular receptor for inositol 1,4,5-trisphosphate, IP3R) were suppressed by THz irradiation. The variation trends of these verified genes were all consistent with the sequencing results, which confirmed the reliability of RNA-seq data (Fig. 3).
Fig. 3.
Verification of Selected DEGs in Response to THz irradiation. The primary neurons were radiated with or without 0.1 THz for 20 min followed another culture for 2 h. The Aqp5, Mmp3, Itpr1, Erf, Apof and Atp2b4 mRNA levels were detected by qRT-PCR in primary neurons. Data are shown as mean ± SEM, ∗p < 0.05, ∗∗p < 0.01.
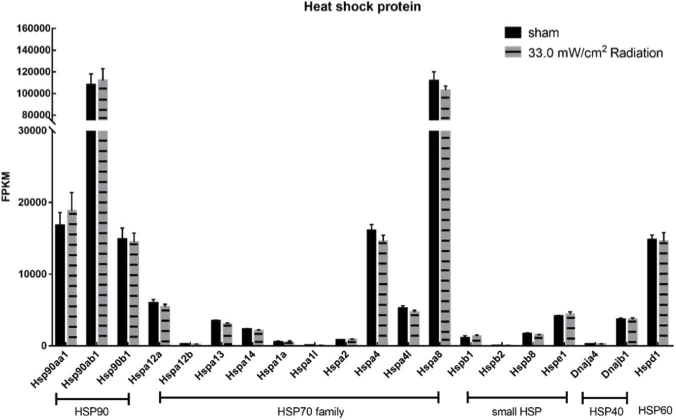
Moreover, to further confirm the altered gene expression pattern wasn’t due to the thermal effects of THz irradiation, the expression levels of several heat shock proteins (HSPs) genes were specifically compared according to the original RNA-seq data. The result indicated that no heat-shock related genes in the HSP90 family, HSP 70 family, HSP40 and small HSP family was significantly up-regulated by THz irradiation (Fig. 4). It suggested that the cells which were exposed to 0.1 THz irradiation for 20 min didn’t undergo thermal stress. It was also consistent with the aforementioned temperature detection results. Therefore, it’s more reasonable to consider that the DEGs revealed by RNA-seq were resulted from the non-thermal effects of 0.1 THz radiation.
Fig. 4.
No heat-shock response genes were up-regulated after THz irradiation. The FPKMs were used to calculate the expression of HSP90, HSP70, small HSP, HSP40 and HSP60 gene of THz irradiated group and control group.
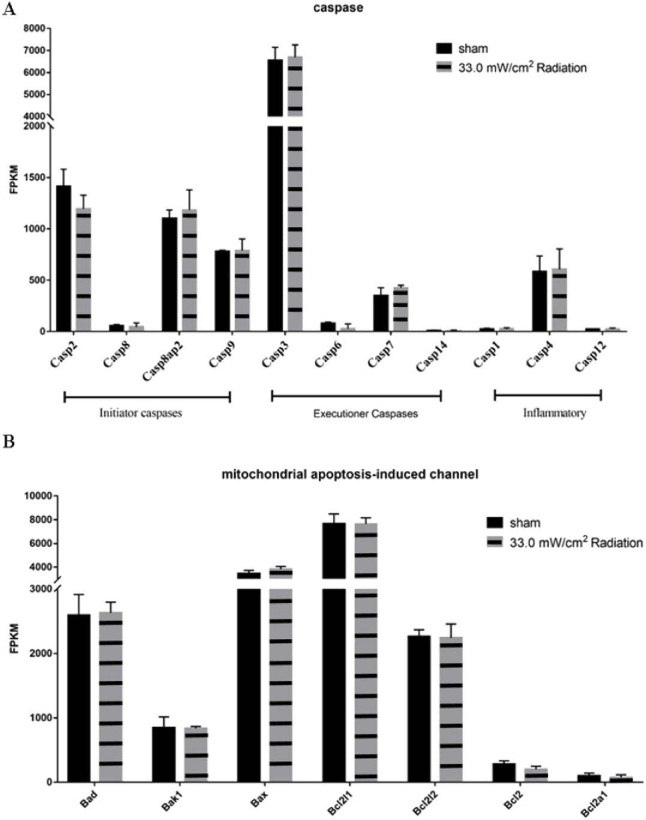
In addition to the HSP genes, the apoptosis-related genes were also specifically concerned. The activation of caspases is considered as the marker for eukaryotic cell apoptosis. In the RNA-seq data, there was no significant difference among the expression levels of initial caspases (Caspase2, Caspase8, Caspase9), executive caspases (Caspase3, Caspase6, Caspase7, Caspase14), and inflammatory activated caspases (Caspase1, Caspase4, Caspase12), which showed that the apoptosis pathway was not triggered by 33 mW/cm2 0.1 THz irradiation exposure (Fig. 5). Moreover, the expression levels of Bcl-2 family proteins which was involved in the mitochondrial apoptotic pathway were also compared based on the original RNA-seq data. The Bcl-2 family proteins, such as the anti-apoptotic Bcl-2 and the pro-apoptotic members Bad, Bak and Bax, control apoptosis by regulating outer mitochondrial membrane permeabilization. In the RNA-seq data, there was no significant difference between the irradiated group and the non-irradiated group in the Bcl-2 family-related genes (Fig. 5).
Fig. 5.
No apoptosis-related genes were changed by THz irradiation. The FPKMs were used to calculate the expression of caspase related gene (A) and mitochondrial apoptosis related gene (B) of THz irradiated group and control group.
3.3. Functional enrichment of DEGs & DETs by GO and KEGG pathway analysis
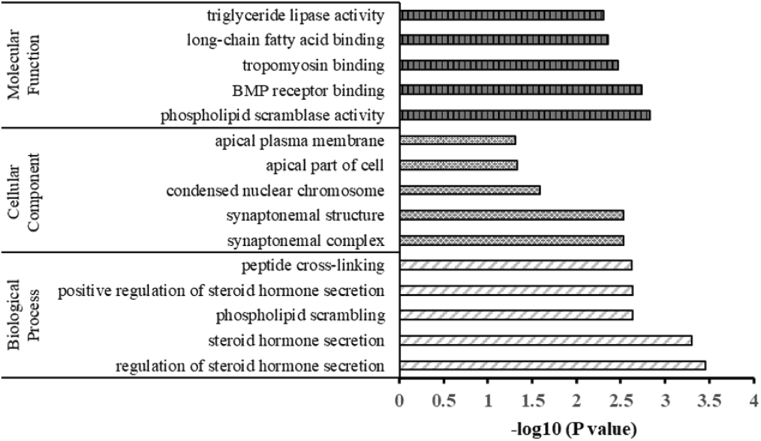
The DEGs were further applied to GO enrichment analysis to identify the significant pathways affected by THz irradiation in primary hippocampal neurons. The top 5 clusters of enriched genes in three main GO categories were summarized in Fig. 6. It could be noticed that the “synapsis” and “synaptonemal complex” were significantly enriched in the category of biological process and the cellular component, respectively. The “phospholipid scrambling” and “phospholipid scramblase activity” were significantly enriched in the biological process category and the molecular function category, respectively. Besides, the “long-chain fatty acid binding”, “tropomyosin binding”, “BMP receptor binding”, “regulation of steroid hormone secretion”, as well as the “apical plasma membrane” which contain the previously verified Aqp5 gene, were also significantly enriched. It suggested that THz irradiation had the possibility in affecting the cell mitosis, phospholipid distribution and the biomacromolecule interaction processes, which was worthy for the further investigation.
Fig. 6.
The GO analysis of DEGs (the top 5 for each category). The three main GO categories: Molecular Function, Cellular Component and Biological Process were shown in the figure.
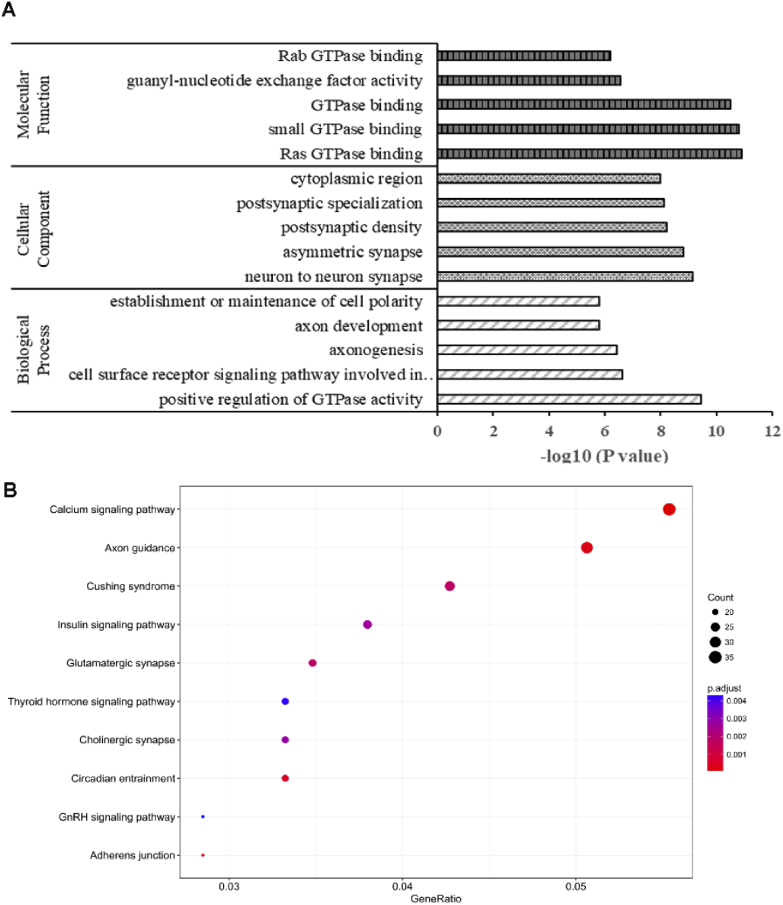
Since the number of DEGs was relatively limited, we also performed the GO and KEGG enrichment analysis for the DETs. The molecular function (MF) of the DETs mainly enriched in the categories of GTPase binding, phospholipid binding, voltage-gated ion channel activity and RAS guanylate exchange factor. These DETs were closely involved in the biological processes of axon development, axonogenesis, regulation of vesicle−mediated transport and especially the small GTPase mediated signal transduction. Additionally, the localization of the differential transcripts products was mainly situated in the synapses (postsynapse, asymmetric synapse and neuron to neuron synapse), cell leading edge and postsynaptic density zone (Fig. 7(A)). KEGG signaling pathway analysis also indicated that the DETs mainly participated in the calcium signaling pathway, axon guidance, glutamatergic synapse and cholinergic synapse (Fig. 7(B)). It was also consistent with the GO analysis result that THz irradiation influenced the transcripts that regulate the synapse function. These DETs enrichment analysis results strongly implied that 0.1 THz irradiation has great potential to affect the synapse development and neural signaling transduction, which were also closely related to the molecular interactions especially the small GTPase binding.
Fig. 7.
The GO and KEGG analysis for DETs. (A) The three main GO categories: Molecular Function, Cellular Component and Biological Process were shown in the figure (the top 5 for each category). (B) Selected top 10 significantly enriched KEGG pathways
3.4. Gene Set Enrichment Analysis
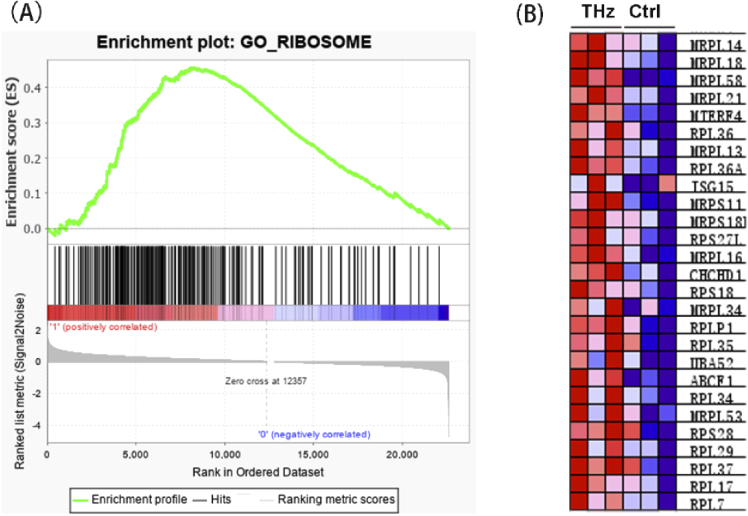
A major drawback of GO enrichment analysis is that it only considers the DEGs screened by a certain p-value/FDR threshold, which may result in the loss of some meaningful information due to the threshold setting. Therefore, we further used Gene Set Enrichment Analysis (GSEA) method to calculate whether a predefined gene set showed significant consistent up-regulation or down-regulation trend in the THz-irradiated group compared with the non-irradiated group. The result showed that the gene sets “ribosome” were significantly enriched with the up-regulation trend after THz irradiation (Fig. 8) and the gene sets mainly include the genes coding small subunit (Rps18 and Rps28) and large subunit (Rpl36, Rpl36A, and Rpl35) of ribosomal protein and mitoribosomal Mrps (Mrps18c, Mrps15 and Mrps11) and Mrpl (Mrpl14, Mrpl18, Mrpl58. And Mrpl21). These results intensively revealed that THz radiation might affects the protein biosynthetic process through increasing ribosome genes expression.
Fig. 8.
Ribosome related gene were up-regulated by 0.1 THz radiation. (A) GSEA analysis revealed that ribosome is significantly enriched. NES = 2.02, FDR < 0.25. (B) The heatmap of genes in the ribosome gene set.
3.5. Analysis of the interaction of AP-1 with DNA
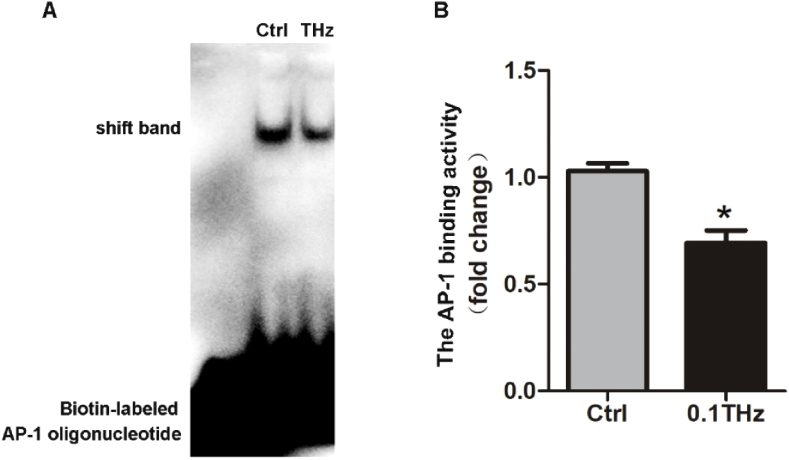
As we know that gene expression is under the control of various TFs. Binding between the activated TFs and their corresponding DNA TFBSs is a pivotal step to initiate an effective gene transcription. Among the six genes verified in our study, Itpr1 and Atp2b4 were reported to be regulated by AP-1 [23,24] and the expression of these two genes were both reduced by THz irradiation. AP-1 is a heterodimeric transcription factor that regulates gene expression response to a variety of stimuli [25]. Thus, the effect of THz radiation on the interaction between AP-1 and its TFBS in the promoter region of these genes should be studied. Due to the limited irradiation area, it is difficult to obtain sufficient cell counts for ChIP-qPCR experiments to quantify the intracellular binding between AP-1 and its TFBS. Therefore, EMSA experiment was performed to detect the binding ability of AP-1 protein with biotin-labeled AP-1 consensus oligonucleotide probe which represented the binding site of AP-1. As shown in Fig. 9(A and (B)), the AP-1 binding efficiency was reduced after THz irradiation. This encouraging result indicated that AP-1 may participate in the gene expression change induced by THz irradiation.
Fig. 9.
The interaction of AP-1 with DNA was down-regulated by THz irradiation. (A) Nuclear extracts from HT22 cells were prepared and incubated with or without biotin-labeled AP-1 probe for THz radiation and the binding efficiency was analyzed by gel. (B) Quantitative analysis of immoblots, which is shown in (A). n=3, Data are shown as mean ± SEM, Student’s t test, ∗p < 0.05.
4. Discussion
In this study, we investigated the effects of 0.1 THz irradiation with 33 mW/cm2 power density on genes expression in primary hippocampal neuron cells. In the process of irradiation, the temperature of cell culture media was not changed significantly. The basic screen for the expression of both HSP genes and the apoptosis related genes indicated that the cells did not undergo obvious heat stress and serious apoptosis response, which is promising in suggesting the relative safety of the 0.1 THz exposure to neuron cells.
To better understand how the 0.1 THz irradiation influences the neuron cells, we performed a global analysis of the transcriptome (RNA-sequencing) to identify the DEGs in response to THz irradiation. The results reported that 111 up-regulated genes and 54 down-regulated genes were found under the irradiation of THz. The DEGs, DETs analysis and the GO/KEGG enrichment results revealed significant enrichment in the categories of biomacromolecule interaction processes such GTPase binding, phospholipid binding, tropomyosin binding, BMP receptor binding and long-chain fatty acid binding. These enrichment results promisingly implied that THz is able to broadly impact the biomolecules interactions [26]. Moreover, the expression of transcripts which was related to neuron function especially synapse development, axonogenesis, synaptic membrane and postsynaptic density were also found to be enriched. This finding supported the theoretical conjecture that THz irradiation regulates the neuron function by inducing interference of those biomacromolecule interaction processes [27–30]. Furthermore, GSEA analysis showed an up-regulation of the gene set that encodes ribosome, the protein translation machine, under the effect of THz irradiation. Since the deficits of ribosomal biogenesis may disturb neurodevelopment by reducing neuronal connectivity [30], a robust ribosomal apparatus is required to carry out protein synthesis to sustain dendritic growth and maintenance. Interestingly, there was a similar case reported by Bogomazova et. al, in whose work showed that the ribosome genes were up-regulated by 2.3 THz irradiation in human embryonic stem cell [9]. There was no clear inspiration could be drawn from previous reports to make a convincing speculation for the detailed mechanism. It’s still an attractive topic to understand the THz triggering conditions for the enhancement of ribosome biosynthesis in various cell types.
Besides, quantitative analysis of six genes, Aqp5, Mmp3, Itpr1, Erf, Apof and Atp2b4 carried out by qRT-PCR showed the consistent changing trends with that of the RNA-seq results. Although these genes were chosen for validating the RNA-seq results, their changes in the expression level still insinuated the functional outcome induced by 0.1 THz irradiation. The products of these gene were involved in various aspect of cellular process in neurons. Itpr1 and Atp2b4 code for the key proteins of calcium signaling pathway. The physiological function of PMCA, a protein encoded by Atp2b4, is to pump out intracellular Ca2+ against the concentration gradient [31]. When PMCA expression is reduced, it is conducive to intracellular calcium accumulation. IP3R, the product of Itpr1 gene, is a receptor of second messenger IP3, contributing to the release of endoplasmic reticulum (ER) calcium [32]. The decreased expression of Itpr1 implies the possible reducement of ER calcium outflow. It has been reported that THz irradiation increased the intracellular calcium concentration in sperm and neuroblastoma cells, while this effect could not be completely eliminated upon the inhibition of calcium channels [33]. Our findings provide a potential explanation for THz induced intracellular calcium elevation. Furthermore, the qPCR result indicated that the expression of Mmp3 was increased by THz irradiation. The coding product of Mmp3, matrix metallopeptidase 3 (MMP-3), is a zinc-dependent peptidase degrading various extracellular molecules. In addition to be involved in many inflammatory and immune responses, recent study has revealed that MMP-3 also contributed to the regulation of L-type calcium channel-dependent LTP in the CA3-CA1 hippocampal projection, promoting the hippocampal neurons plasticity and activity via the regulating NMDAR function and calcium flux [34]. Thus, by combining with the GO analysis results which enriched “neuron to neuron synapse” in the “Cellular Components” category, further study is also quite necessary to discover the impact of THz on the intracellular calcium homostasis and the consequent synapse function in neurons. The functions of the protein products of the other verified genes, Apof, Erf and Aqp5, were involved in the cellular metabolism, proliferation and water transport, respectively [35–37]. The coding product of Apof gene is one of the minor apolipoproteins. This protein forms complexes with lipoproteins and is involved in transport and/or esterification of cholesterol [35]. Erf gene codes for a transcriptional repressor that binds to the Ets2 promoter, which regulates the genes involved in cellular proliferation [38]. Erf is required to inhibit proliferation of neural progenitors to allow differentiation, whereas overexpression of Erf leads to an increase in the number of primary hippocampal neurons [39]. AQP5 forms a water-specific channel and plays an important role in regulating the movement of water through the plasma membrane of secretory and absorptive cells in response to osmotic gradients [40]. Although the function of AQP5 in hippocampal neurons has not been intensively studied, considering the GO enrichment in the categories of “lipid metabolic process”, “lipid transport”, “synaptonemal structure” and “apical plasma membrane”, it would be interesting to evaluate the effects of THz irradiation on cellular lipid metabolism, neuronal differentiation and water homeostasis with further detailed and specialized research.
Additionally, as we know that the THz irradiation may influence the molecular conformation and consequently affects the interaction among biological macromolecules [41]. As for the effect of THz on gene expression, numerous biomolecules interactions might be affected by THz radiation and the DNA breathing model just provide one of the many possible underlying mechanism. Especially, the interactions between transcription factors (TFs) and their TFBSs which play a vital role in activating the transcription initiation might also be the targets of THz radiation. Tachizaki et al. reported that THz pulses could affect the expression of many genes regulated by the zinc dependent transcription [17]. Although they didn’t verify their speculation with any experiment, they highlighted the possibility of THz on influencing gene expressions via regulating the function of transcription factors. In our study, we provided a direct in vitro evidence to illustrate a reduced binding between AP-1, a typical transcription factor, with its corresponding biotin-labeled DNA probe under the effect of 0.1 THz irradiation via the classical EMSA experiment. Although the actual in vivo regulation process of gene transcription is much more complicated, our finding indeed proved that THz could influence the binding between TF and the TFBS, which was very likely to be involved in the gene expression changes induced by THz irradiation. At least, the expressions of Itpr1 and Atp2b4, which are regulated by AP-1, were decreased by THz irradiation. This result consists with the deduction obtained from EMSA experiment. According to the observation in our study, further direct in vivo evidence are still needed to demonstrate the mechanism. CHIP-qPCR analysis is an efficient way to reveal the in vivo binding situation of AP-1 on a specific promoter. However, the number of cells required for the CHIP-qPCR experiment is far more than what we could obtain in our present study due to the relative limited irradiating spot area. Therefore, novel THz source that fits to a wider range irradiation is necessary to pursue the answer.
Overall, in this study we used a large scale RNA-seq analysis to reveal the differentially expressed genes, transcripts and pathways in primary hippocampal neurons exposed to 0.1 THz. Several biomolecule binding related categories such as “long-chain fatty acid binding”, “tropomyosin binding”, “BMP receptor binding”, as well as “GTPase binding” and “phospholipid binding” were significantly found enriched by GO analysis. The GSEA analysis indicated that the genes that encode protein biosynthetic machinery ribosome were up-regulated by THz irradiation. These results suggested that THz irradiation affected various biomolecules interaction. The EMSA experiment result provided an encouraging evidence that the binding efficiency between the transcription factor AP-1 with its biotin-labeled DNA probe could be diminished by THz irradiation. These results enriched the mechanism of THz in regulating gene expression and also highlighted several interesting aspects of THz induced functional bioeffects in neurons.
Disclosures
The authors declare no competing financial interests.
We declare that we do not have any commercial or associative interest that represents a conflict of interest in connection with the work submitted.
Data availability
Data underlying the results presented in this paper are not publicly available at this time but may be obtained from the authors upon reasonable request.
References
- 1.Liu H. B., Zhong H., Karpowicz N., Chen Y., Zhang X. C., “Terahertz spectroscopy and imaging for defense and security applications,” Proc. IEEE 95(8), 1514–1527 (2007). 10.1109/JPROC.2007.898903 [DOI] [Google Scholar]
- 2.Wolbarst A. B., Hendee W. R., “Evolving and experimental technologies in medical imaging,” Radiology 238(1), 16–39 (2006). 10.1148/radiol.2381041602 [DOI] [PubMed] [Google Scholar]
- 3.Yamaguchi S., Fukushi Y., Kubota O., Itsuji T., Yamamoto M. D. S., Ouchi T., “Terahertz spectroscopy and detection of brain tumor in rat fresh-tissue samples,” Proc. SPIE 9321, 93210O (2015). 10.1117/12.2078564 [DOI] [Google Scholar]
- 4.Wang M., Yang G., Li W., Wu Q., “An overview of cancer treatment by terahertz radiation,” in 2013 IEEE MTT-S International Microwave Workshop Series on RF and Wireless Technologies for Biomedical and Healthcare Applications (IMWS-BIO), 2014). [Google Scholar]
- 5.Woodward R. M., Cole B. E., Wallace V. P., Pye R. J., Arnone D. D., Linfield E. H., Pepper M., “Terahertz pulse imaging in reflection geometry of human skin cancer and skin tissue,” Phys. Med. Biol. 47(21), 3853–3863 (2002). 10.1088/0031-9155/47/21/325 [DOI] [PubMed] [Google Scholar]
- 6.Alexandrov B. S., Rasmussen KØ, Bishop A. R., Usheva A., Rodriguez G., “Non-thermal effects of terahertz radiation on gene expression in mouse stem cells,” Biomed. Opt. Express 2(9), 2679–2689 (2011). 10.1364/BOE.2.002679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamazaki S., Harata M., Ueno Y., Tsubouchi M., Konagaya K., Ogawa Y., Isoyama G., Otani C., Hoshina H., “Propagation of THz irradiation energy through aqueous layers: demolition of actin filaments in living cells,” Sci. Rep. 10(1), 9008 (2020). 10.1038/s41598-020-65955-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamazaki S., Harata M., Idehara T., Konagaya K., Yokoyama G., Hoshina H., Ogawa Y., “Actin polymerization is activated by terahertz irradiation,” Sci. Rep. 8(1), 9990 (2018). 10.1038/s41598-018-28245-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bogomazova A. N., Vassina E. M., Goryachkovskaya T. N., Popik V. M., Sokolov A. S., Kolchanov N. A., Lagarkova M. A., Kiselev S. L., Peltek S. E., “No DNA damage response and negligible genome-wide transcriptional changes in human embryonic stem cells exposed to terahertz radiation,” Sci. Rep. 5(1), 7749 (2015). 10.1038/srep07749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Titova L. V., Ayesheshim A. K., Golubov A., Rodriguez-Juarez R., Woycicki R., Hegmann F. A., Kovalchuk O., “Intense THz pulses down-regulate genes associated with skin cancer and psoriasis: a new therapeutic avenue?” Sci. Rep. 3(1), 2363 (2013). 10.1038/srep02363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bourne N., Clothier R. H., D’Arienzo M., Harrison P., “The effects of terahertz radiation on human keratinocyte primary cultures and neural cell cultures,” ATLA, Altern. Lab. Anim. 36(6), 667–684 (2008). 10.1177/026119290803600610 [DOI] [PubMed] [Google Scholar]
- 12.Bock J., Fukuyo Y., Kang S., Phipps M. L., Alexandrov L. B., Rasmussen K., Bishop A. R., Rosen E. D., Martinez J. S., Chen H. T., Rodriguez G., Alexandrov B. S., Usheva A., “Mammalian stem cells reprogramming in response to terahertz radiation,” PLoS One 5(12), e15806 (2010). 10.1371/journal.pone.0015806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alexandrov B. S., Phipps M. L., Alexandrov L. B., Booshehri L. G., Erat A., Zabolotny J., Mielke C. H., Chen H. T., Rodriguez G., Rasmussen K., Martinez J. S., Bishop A. R., Usheva A., “Specificity and heterogeneity of terahertz radiation effect on gene expression in mouse mesenchymal stem cells,” Sci. Rep. 3(1), 1184 (2013). 10.1038/srep01184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alexandrov B. S., Gelev V., Bishop A. R., Usheva A., Rasmussen K. O., “DNA breathing dynamics in the presence of a terahertz field,” Phys. Lett. A 374(10), 1214–1217 (2010). 10.1016/j.physleta.2009.12.077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alexandrov B. S., Gelev V., Yoo S. W., Alexandrov L. B., Fukuyo Y., Bishop A. R., Rasmussen K., Usheva A., “DNA dynamics play a role as a basal transcription factor in the positioning and regulation of gene transcription initiation,” Nucleic Acids Res. 38(6), 1790–1795 (2010). 10.1093/nar/gkp1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Z., Tjian R., “Visualizing transcription factor dynamics in living cells,” J. Cell Biol. 217(4), 1181–1191 (2018). 10.1083/jcb.201710038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tachizaki T., Sakaguchi R., Terada S., Kamei K. I., Hirori H., “Terahertz pulse-altered gene networks in human induced pluripotent stem cells,” Opt. Lett. 45(21), 6078–6081 (2020). 10.1364/OL.402815 [DOI] [PubMed] [Google Scholar]
- 18.Hu E., Du H., Zhu X., Wang L., Shang S., Wu X., Lu H., Lu X., “Beta-hydroxybutyrate promotes the expression of BDNF in hippocampal neurons under adequate glucose supply,” Neuroscience 386, 315–325 (2018). 10.1016/j.neuroscience.2018.06.036 [DOI] [PubMed] [Google Scholar]
- 19.Kim D., Langmead B., Salzberg S. L., “HISAT: a fast spliced aligner with low memory requirements,” Nat. Methods 12(4), 357–360 (2015). 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D. R., Pimentel H., Salzberg S. L., Rinn J. L., Pachter L., “Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks,” Nat. Protoc. 7(3), 562–578 (2012). 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P., “Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles,” Proc. Natl. Acad. Sci. U. S. A. 102(43), 15545–15550 (2005). 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hellman L. M., Fried M. G., “Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions,” Nat. Protoc. 2(8), 1849–1861 (2007). 10.1038/nprot.2007.249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mizuno K., Kurokawa K., Ohkuma S., “Regulation of type 1 IP3 receptor expression by dopamine D2-like receptors via AP-1 and NFATc4 activation,” Neuropharmacology 71, 264–272 (2013). 10.1016/j.neuropharm.2013.03.036 [DOI] [PubMed] [Google Scholar]
- 24.Du Y., Carlock L., Kuo T. H., “The mouse plasma membrane Ca2+ pump isoform 1 promoter: cloning and characterization,” Arch. Biochem. Biophys. 316(1), 302–310 (1995). 10.1006/abbi.1995.1041 [DOI] [PubMed] [Google Scholar]
- 25.Lin H., Han L., Blank M., Head M., Goodman R., “Magnetic field activation of protein-DNA binding,” J. Cell. Biochem. 70(3), 297–303 (1998). [DOI] [PubMed] [Google Scholar]
- 26.Romanenko S., Begley R., Harvey A. R., Hool L., Wallace V. P., “The interaction between electromagnetic fields at megahertz, gigahertz and terahertz frequencies with cells, tissues and organisms: risks and potential,” J. R. Soc., Interface 14(137), 20170585 (2017). 10.1098/rsif.2017.0585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fang Y., Durgerian S., Basarsky T. A., Haydon P. G., “GTP-binding proteins: necessary components of the presynaptic terminal for synaptic transmission and its modulation,” Adv. Second Messenger Phosphoprotein Res. 29, 121–132 (1994). 10.1016/S1040-7952(06)80011-X [DOI] [PubMed] [Google Scholar]
- 28.Lanuza M. A., Just-Borràs L., Hurtado E., Cilleros-Mañé V., Tomàs M., Garcia N., Tomàs J., “The impact of kinases in amyotrophic lateral sclerosis at the neuromuscular synapse: insights into BDNF/TrkB and PKC signaling,” Cells 8(12), 1578 (2019). 10.3390/cells8121578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao L., Michalski N., Kronander E., Gjoni E., Genoud C., Knott G., Schneggenburger R., “BMP signaling specifies the development of a large and fast CNS synapse,” Nat. Neurosci. 16(7), 856–864 (2013). 10.1038/nn.3414 [DOI] [PubMed] [Google Scholar]
- 30.Lee C. C., Huang C. C., Hsu K. S., “The phospholipid-binding protein SESTD1 negatively regulates dendritic spine density by interfering with Rac1-Trio8 signaling pathway,” Sci. Rep. 5(1), 13250 (2015). 10.1038/srep13250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bruce J. I. E., “Metabolic regulation of the PMCA: role in cell death and survival,” Cell Calcium 69, 28–36 (2018). 10.1016/j.ceca.2017.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiao W. C., Zhang J., Chen S. L., Shi Y. J., Xiao F., An W., “Alleviation of palmitic acid-induced endoplasmic reticulum stress by augmenter of liver regeneration through IP3R-controlled Ca(2+) release,” J. Cell. Physiol. 233(8), 6148–6157 (2018). 10.1002/jcp.26463 [DOI] [PubMed] [Google Scholar]
- 33.Bo W., Guo L., Yang Y., Ma J., Gong Y., “Numerical study of voltage-gated Ca2+ transport irradiated by terahertz electromagnetic wave,” IEEE Access 8, 10305–10315 (2020). 10.1109/ACCESS.2020.2964780 [DOI] [Google Scholar]
- 34.Wiera G., Nowak D., van Hove I., Dziegiel P., Moons L., Mozrzymas J. W., “Mechanisms of NMDA receptor- and voltage-gated L-Type calcium channel-dependent hippocampal LTP critically rely on proteolysis that is mediated by distinct metalloproteinases,” J. Neurosci. 37(5), 1240–1256 (2017). 10.1523/JNEUROSCI.2170-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen X. B., Huang L., Zhang S. H., Wang D. P., Wu Y. L., Chen W. N., Xu S. H., Lin X., “Transcriptional regulation of the apolipoprotein F (ApoF) gene by ETS and C/EBPα in hepatoma cells,” Biochimie 112, 1–9 (2015). 10.1016/j.biochi.2015.02.013 [DOI] [PubMed] [Google Scholar]
- 36.de Castro C. M., Rabe S. M., Langdon S. D., Fleenor D. E., Slentz-Kesler K., Ahmed M. N., Qumsiyeh M. B., Kaufman R. E., “Genomic structure and chromosomal localization of the novel ETS factor, PE-2 (ERF),” Genomics 42(2), 227–235 (1997). 10.1006/geno.1997.4730 [DOI] [PubMed] [Google Scholar]
- 37.Törnroth-Horsefield S., Hedfalk K., Fischer G., Lindkvist-Petersson K., Neutze R., “Structural insights into eukaryotic aquaporin regulation,” FEBS Lett. 584(12), 2580–2588 (2010). 10.1016/j.febslet.2010.04.037 [DOI] [PubMed] [Google Scholar]
- 38.Verykokakis M., Papadaki C., Vorgia E., Le Gallic L., Mavrothalassitis G., “The RAS-dependent ERF control of cell proliferation and differentiation is mediated by c-Myc repression,” J. Biol. Chem. 282(41), 30285–30294 (2007). 10.1074/jbc.M704428200 [DOI] [PubMed] [Google Scholar]
- 39.Janesick A., Abbey R., Chung C., Liu S., Taketani M., Blumberg B., “ERF and ETV3L are retinoic acid-inducible repressors required for primary neurogenesis,” Development 140(15), 3095–3106 (2013). 10.1242/dev.093716 [DOI] [PubMed] [Google Scholar]
- 40.Chai R. C., Jiang J. H., Wong A. Y., Jiang F., Gao K., Vatcher G., Hoi Yu A. C., “AQP5 is differentially regulated in astrocytes during metabolic and traumatic injuries,” Glia 61(10), 1748–1765 (2013). 10.1002/glia.22555 [DOI] [PubMed] [Google Scholar]
- 41.Homenko A., Kapilevich B., Kornstein R., Firer M. A., “Effects of 100 GHz radiation on alkaline phosphatase activity and antigen-antibody interaction,” Bioelectromagnetics 30(3), 167–175 (2009). 10.1002/bem.20466 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data underlying the results presented in this paper are not publicly available at this time but may be obtained from the authors upon reasonable request.