Abstract
In metazoans, protein O-fucosylation of Ser/Thr residues was only found in secreted or cell surface proteins, and this post-translational modification is catalyzed by ER-localized protein O-fucosyltransferases (POFUTs) in the GT65 family. Recently, a novel nucleocytoplasmic POFUT, SPINDLY (SPY), was identified in the reference plant Arabidopsis thaliana to modify nuclear transcription regulators DELLAs, revealing a new regulatory mechanism for gene expression. The paralog of AtSPY, SECRET AGENT (SEC), is an O-link-N-acetylglucosamine (GlcNAc) transferase (OGT), which O-GlcNAcylates Ser/Thr residues of target proteins. Both AtSPY and AtSEC are tetratricopeptide repeat-domain-containing glycosyltransferases in the GT41 family. The discovery that AtSPY is a POFUT clarified decades of miss-classification of AtSPY as an OGT. SPY and SEC play pleiotropic roles in plant development, and the interactions between SPY and SEC are complex. SPY-like genes are conserved in diverse organisms, except in fungi and metazoans, suggesting that O-fucosylation is a common mechanism in modulating intracellular protein functions.
Keywords: protein O-fucosylation, protein O-GlcNAcylation, nucleocytoplasmic protein O-fucosylation, SPINDLY, POFUT
Graphical Abstract
Discovery of nucleocytoplasmic protein O-fucosyltransferase SPINDLY in plants
The discovery of the nucleocytoplasmic protein O-fucosyltransferase (POFUT) came from the studies of SPINDLY (SPY) in Arabidopsis. AtSPY was initially identified as a negative regulator of plant hormone gibberellin (GA) signaling because the hypomorphic mutations in AtSPY partially rescue the seed germination defect and dwarf phenotypes caused by chemical-induced GA deficiency or genetic mutations in GA biosynthesis [1,2]. In addition, SPY represses other aspects of GA-regulated processes, including floral induction, anther development and pollen tube growth [1-3].
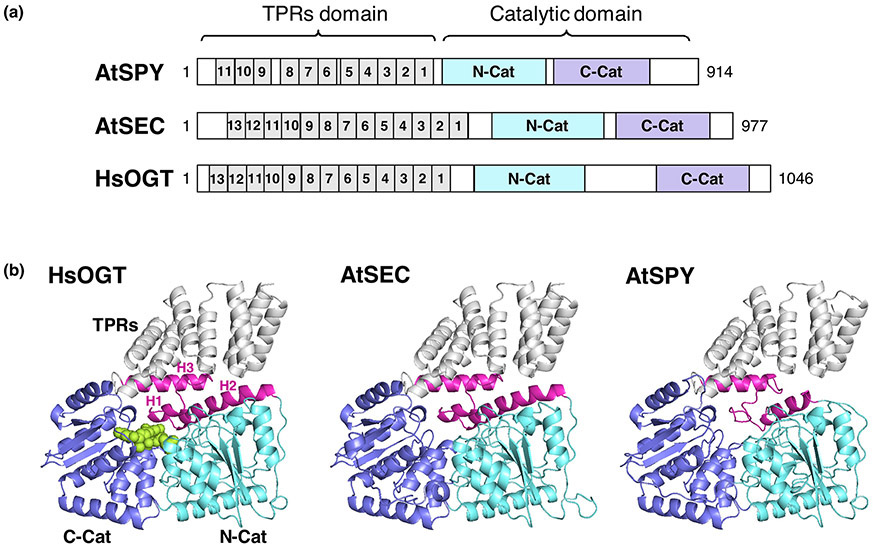
Based on sequence comparison, both AtSPY and its paralog AtSEC (SECRET AGENT) were predicted to be OGTs, with a tetratricopeptide-repeat (TPR) domain and a putative OGT catalytic domain (Fig. 1A) [4-7]. The TPR domain of SPY and SEC functions as a protein-protein interaction domain for recruiting target proteins, and overexpression of the TPR domain of AtSPY has a dominant negative effect that confers a spy-like phenotype [8,9]. Recombinant AtSEC expressed in E. coli was shown to exhibit OGT activity [5], but the enzymatic activity of AtSPY was not detected conclusively in a similar in vitro assay.
Figure 1. Structure comparison among human OGT, Arabidopsis SEC and SPY.
(a) Diagrams of HsOGT, AtSEC and AtSPY. TPRs are in grey. N-terminal catalytic domains, N-Cat, are in cyan. C-terminal catalytic domains, C-Cat, are in blue. (b) 3D structures of HsOGT (PDB ID: 4N3C, containing 4.5-TPRs)[71], and predicted 3D structures of Arabidopsis SEC and SPY using SWISS MODEL[72,73]. The HsOGT crystal structure (PDB ID: 4N3C)[71] was used as scaffold to predict AtSEC and AtSPY structures. The color schemes for HsOGT, AtSEC and AtSPY are as in (a). In (b), UDP-GlcNAc in HsOGT is shown as spheres (in lime-green). In the HsOGT structure in (b), the transitional helix (H3) between TPRs and N-Cat, and the first 2 α-helices (H1 and H2) of N-Cat are highlighted in magenta. The long intervening domain between N-Cat and C-Cat of HsOGT is omitted from the structure because this domain is uniquely present in the animal OGTs. This figure was modified from Zentella et al. [14].
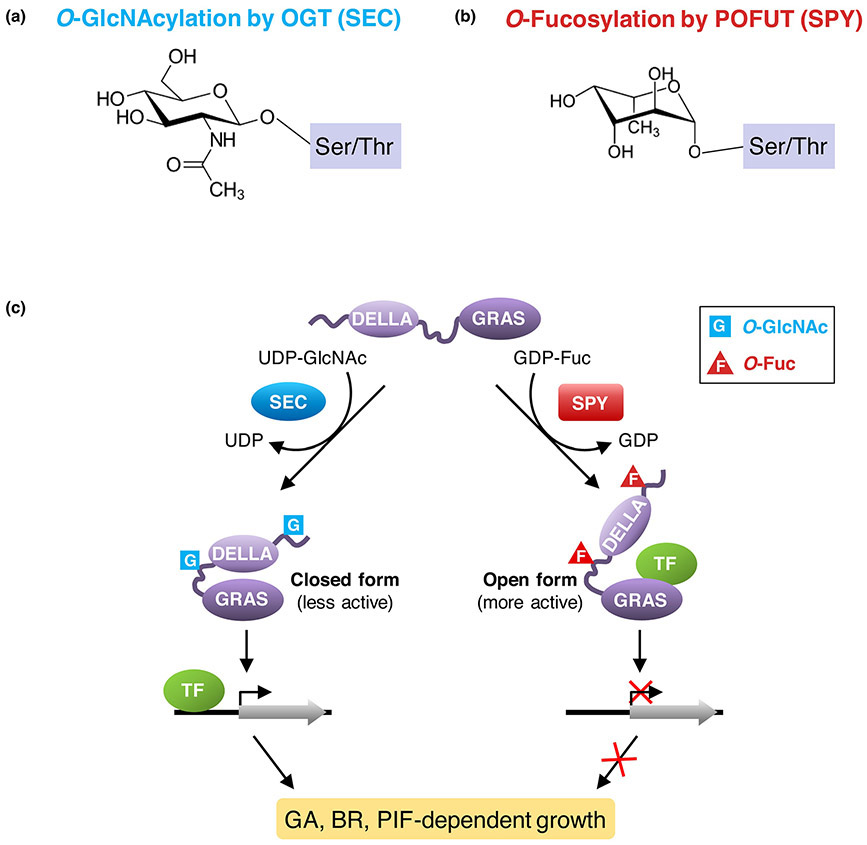
Because spy displays elevated GA signaling, and the presence of putative O-GlcNAc sites in the nuclear DELLA repressors (also known as GA-signaling repressors), AtSPY was long proposed to activate AtDELLAs by O-GlcNAcylation [10-12]. Through a combination of electron transfer dissociation (ETD)-MS/MS analysis, in vitro enzyme assays and genetic studies, AtSEC was shown to be an OGT that O-GlcNAcylates DELLAs using UDP-GlcNAc as its donor substrate [13]. Surprisingly, AtSPY was found to be a novel POFUT, which is highly selective to GDP-fucose as its donor substrate and catalyzes the transfer of O-Fucose monosaccharide to the hydroxyl oxygen on Ser and Thr residues of DELLA proteins (acceptor substrates) [14] (Fig. 2).
Figure 2. Model for the opposing roles of O-fucosylation and O-GlcNAcylation of DELLA in regulating plant growth.
(a) O-GlcNAcylation by OGT (SEC). (b) O-Fucosylation by POFUT (SPY). (c) The nuclear growth repressor DELLA proteins are activated by O-Fucosylation, and repressed by O-GlcNAcylation. Each DELLA protein contains an N-terminal DELLA domain and a C-terminal GRAS domain. O-Fucosylation (labeled as F) by SPY may induce the DELLA protein to an open conformation that is a more active growth repressor; this open form promotes binding of the GRAS domain to interacting transcription factors (e.g., BZR1 and PIFs), which leads to down-regulated expression of target genes of BZR1 and PIFs to restrict plant growth. In contrast, O-GlcNAcylation (labeled as G) by SEC may cause the DELLA protein to fold into a closed conformation that is less active because this form reduces its binding affinity to BZR1 and PIFs so that growth-related target genes can be activated. TF, DELLA-interacting transcription factor. The figure (c) was modified from Zentella et al. [14].
Predicted 3D structure of AtSPY is unrelated to ER-localized POFUTs, but is similar to OGTs
Sequence alignment and three dimension (3D) protein structure modeling indicate that AtSPY is distinct from the ER-localized POFUTs, which belong to GlycosylTransferase Family 65 (GT65 [15], http://www.cazy.org) and modify secreted cell surface proteins in animals [16-18], Instead, AtSPY’s 3D model is highly similar to the TPR domain-containing OGTs, members of the GT41 family [14,19] (Fig. 1B). Moreover, like OGTs, AtSPY is localized to both cytoplasm and nucleus [20]. AtSPY is the first nucleocytoplasmic-localized POFUT found in any organism.
Multiple O-Fuc and O-GlcNAc sites identified in AtDELLA are clustered within two structurally disordered polyS/T sequences flanking the conserved DELLA domain [13,14], suggesting that AtSPY and AtSEC may modify target proteins via a similar mechanism as in HsOGT, which modifies flexible sequences of its target proteins by binding to the substrate amide backbone [19,21]. The critical residue(s) that contribute to the distinct substrate selectivity of AtSPY have not been identified experimentally, although some differences between AtSPY and OGTs have been noted through sequence alignment and 3D model comparison. The H3 transition helix and the H1 and H2 helices of N-Cat are more divergent in AtSPY (Fig 1B). Moreover, two key His residues (H498-H499 in HsOGT and F540-H541 in AtSEC) that are crucial for OGT activity, are absent in AtSPY [13,14,19,22,23].
Opposing roles of AtSPY and AtSEC in regulating DELLA function and activities of multiple signaling pathways
Intriguingly, genetic and biochemical studies further showed that O-GlcNAc and O-Fuc modifications by the two paralogs AtSEC and AtSPY display opposite effects on DELLA function and GA signaling activity [13,14]. DELLAs are master growth repressors, which integrate multiple signaling activities by protein-protein interactions with key transcription factors to coordinate plant growth with internal and external cues [12,24]. For example, BRASSINAZOLE-RESISTANT1 (BZR1) and PHYTOCHROME-INTERACTING-FACTORs (e.g., PIF3 and PIF4) are key transcription factors that promote hypocotyl elongation in response to the phytohormone brassinosteroid (BR) and external light conditions, whereas DELLAs inhibit hypocotyl growth by antagonistic interactions with BZR1 and PIFs to repress expression of BZR1- and PIFs-target genes [25-27]. The null sec mutant shows reduced GA responses with a shorter hypocotyl and internode length than the wild-type Arabidopsis plant [13]. These results indicate that AtSEC is an activator of GA signaling, which is in contrast to the repressive role of AtSPY in GA signaling. By deduction, AtSEC may reduce DELLA activity and AtSPY may increase DELLA activity to achieve their effects on GA signaling activity. Indeed, in vitro pulldown assays showed that O-fucosylation by AtSPY enhances DELLA interactions with BZR1 and PIFs [14]. In contrast, O-GlcNAcylation by AtSEC reduces DELLA interactions with these key transcription factors in BR and light signaling pathways [13]. Furthermore, spy mutations confer increased responses to BR and elevated transcript levels of target genes of BZR1 and PIFs, whereas the sec null allele shows an opposite effect. Therefore, these two distinct O-glycosyl modifications of DELLAs by AtSPY and AtSEC differentially modulate GA, BR and light signaling pathways to regulate plant growth and development. The identified O-GlcNAc and O-Fuc sites in DELLA are partially overlapping or nearby. A model was proposed in which highly O-GlcNAcylated DELLA may lock into a “closed form” that interferes with binding of target proteins. Increasing O-fucosylation may convert DELLA conformation to an “open form” that enhances interaction with target proteins (Fig. 2C).
It is unclear how SPY and SEC activities are regulated, although they appear to be unaffected by the GA status in the plant [13,14]. In animals, OGT functions as a nutrient sensor because its activity is tightly correlated with the levels of its donor substrate UDP-GlcNAc, which is derived from several key metabolites in the cell via the hexosamine biosynthesis pathway [7,28,29]. It was proposed that dynamic O-GlcNAc vs O-Fuc modifications of DELLAs (and additional regulatory proteins) may help to coordinate the metabolic status of the plant with its growth and development in response to internal and external cues, although specific glucosidases have not been identified [14].
Protein O-fucosylation and O-GlcNAcylation play diverse roles in plant development
The interplay between AtSPY and AtSEC during Arabidopsis development is complex. Although SPY and SEC play opposite roles in regulating DELLA-mediated signaling activities as described above, these two protein glycosyltransferases may interact differently in DELLA-independent cellular processes in plants. Both AtSPY and AtSEC regulate embryogenesis and flowering time [1,5,30], whereas each enzyme displays unique roles in a subset of developmental processes. For example, SPY is a positive regulator of phytohormone cytokinin signaling [31,32], and regulates the circadian clock [33,34]. In contrast, the sec mutations do not alter cytokinin responses or circadian rhythms. On the other hand, SEC but not SPY promotes Plum Pox Virus (PPV) infection by O-GlcNAcylating the coat protein of PPV [35,36]. In vitro assays suggest that O-GlcNAcylation regulates protein trafficking through plasmodesmata by altering their interactions with the Nicotiana tabacum NON-CELL-AUTONOMOUS PATHWAY PROTEIN1 [37]. Mechanisms of cellular processes regulated by both SPY and SEC, and those that are uniquely regulated by SPY are described below.
Embryo development
While AtSPY and AtSEC play opposite roles in regulating DELLA to modulate multiple signaling activities [13,14], the spy sec double mutant is embryo lethal [5,38]. This synthetic lethal phenotype of spy sec indicates that AtSPY and AtSEC regulate unidentified essential process(es) during embryogenesis. The knockout OGT mutants in mouse and in Drosophila are embryo lethal [39-41]. The OGTs in animals are known to regulate intracellular functions including altering gene expression at the epigenetic and transcription levels as well as modulating protein synthesis, stability, activity or subcellular localization [42,43]. In contrast, the functions of OGT (SEC) and POFUT (SPY) in plants are much less understood. Recently, proteomic studies have identified a large number of O-GlcNAcylated proteins in Arabidopsis (262) and in winter wheat Triticum aestivum (168), many of which function in epigenetic and transcriptional regulation, RNA processing, translation and metabolic processes [44,45], suggesting that OGT in plants also play diverse roles as the animal OGTs do. So far, the known protein substrates of AtSPY only include DELLAs and PSEUDO RESPONSE REGULATOR 5 (PRR5, a circadian clock component) (see below), although several other interacting proteins have been identified by Y2H or co-IP assays (including MYB, NAC-like, TCP and ZIM domain transcription factors, a circadian clock regulator GIGANTIA, and SWI3C, a subunit of the chromatin remodeling complexes) [33,46-49]. Considering that O-Fuc and O-GlcNAc sites in DELLA largely overlap [13,14], SPY and SEC may share additional common targets in plants. However, the interaction between O-GlcNAcylation and O-fucosylation may be different depending on the target proteins because the embryo-lethal phenotype of the spy sec mutant suggests an additive interaction, which is in contrast to their antagonistic interaction in modulating DELLA activity. In addition, AtSPY plays unique roles in a subset of cellular processes, which will be discussed below.
Flowering time
The hypomorphic spy mutants in Arabidopsis flower earlier than WT in both long-day and short-day conditions, indicating that AtSPY negatively regulates floral induction [1,10]. One way for AtSPY to delay flowering is by enhancing DELLA activity to repress GA-induced flowering. Additionally, AtSPY interacts with a core circadian clock protein GIGANTIA (GI) that promotes flowering in long day [33]. The gi mutant is late flowering, whereas spy gi double mutant is early flowering, indicating that spy is epistatic to gi, although the role of SPY-GI interaction in flowering time control is unclear. Interestingly, O-GlcNAcylation catalyzed by AtSEC also delays flowering in Arabidopsis. AtSEC upregulates the expression of the major flowering repressor FLOWERING LOCUS C (FLC) [30]. Further analysis of the chromatin around the FLC locus indicates that H3 lysine 4 trimethylation (H3K4me3, an active chromatin mark) is reduced significantly in the sec mutant. Importantly, AtSEC O-GlcNAcylates the histone methyltransferase ATX1 in planta, and this modification enhances ATX1’s activity to methylate H3 in vitro. Thus, AtSEC induces expression of FLC to delay flowering, at least in part by O-GlcNAcylation and activation of the histone methyltransferase ATX1 to increase the H3K4me3 active chromatin mark at the FLC locus. In contrast, O-GlcNAcylation in Triticum aestivum (winter wheat) mediates vernalization-induced flowering [50]. Vernalization (prolonged cold period) promotes flowering in winter wheat by enhancing expression of a flowering activator TaVRN1 (a MADS-box transcription factor). Without vernalization, TaVRN1 mRNA processing is inhibited by an RNA binding protein TaGRP2 that binds to the first intron of TaVRN1 pre-mRNA. Vernalization increases TaVRN1 mRNA levels by inducing O-GlcNAcylation of TaGRP2, which in turn promotes sequestration of TaGRP2 by a vernalization-induced lectin VERN2.
Cytokinin responses
In addition to an elevated GA-response phenotype, the spy single mutants in Arabidopsis display other pleiotropic phenotypes, including abnormal cotyledon numbers, altered phyllotaxy, reduced leaf serration, and decreased trichomes on sepals [10,32,51]. The reduced leaf serration and sepal trichome formation in spy mutants are caused by reduced responses to another phytohormone cytokinin, indicating that AtSPY is a positive regulator of cytokinin signaling [31,32]. Screening and characterization of AtSPY-interacting proteins identified two bHLH transcription factors TCP14 and TCP15 that are involved in AtSPY-regulated cytokinin responses [49]. The tcp14 tcp15 double mutant shows reduced cytokinin responses, whereas overexpression of TCP14 displays enhanced cytokinin responses. The GFP-TCP14 protein accumulates to a lower level in the spy mutant than that in WT, but this reduced protein stability of TCP14 can be reversed by treatment with the 26S proteasome inhibitor MG132 or in the mutant cul1 background (CULLIN1 encodes a component of the SCF E3 ligase complex) [52]. It is likely that AtSPY stabilizes TCP14 by O-fucosylation, although this has not been demonstrated directly.
Circadian clock
The animal OGTs have been shown to regulate the circadian clock by rhythmic O-GlcNAcylation of key components of the clock. In Drosophila and mammals, the transcription repressor PERIOD binds to and inhibits transcription activators CLOCK and BMAL, whereas CLOCK/BMAL induces transcription of PERIOD. O-GlcNAcylation of PERIOD by OGT inhibits the activity of PERIOD by promoting its degradation and preventing its translocation to the nucleus [53,54]. In addition, O-GlcNAcylation of BMAL and CLOCK stabilizes these transcription activators [55]. Reducing OGT expression results in a longer circadian period [53]. OGT in Arabidopsis, however, does not play a significant role in regulating the circadian clock as the sec mutants do not show abnormal circadian phenotypes [34]. Instead, AtSPY was found to regulate circadian clock. The spy mutants display a longer circadian period in comparison to that of WT [33,34]. The circadian period phenotype of spy is rescued more effectively by Pspy:GFP-SPY-NLS (nuclear localization signal) than by Pspy:GFP-SPY-NES (nuclear export signal), suggesting that AtSPY mainly functions in the nucleus to modulate circadian clock speed. Intriguingly, expression of the cytoplasmic SPY fusion protein (GFP-SPY-NES) is required to inhibit GA responses (e.g., in seed germination, leaf expansion, floral induction) and to promote cytokinin signaling (leaf serration) [31,34]. These results suggest that cytoplasmic-and nuclear-localized AtSPY regulates distinct cellular responses, although it remains to be determined whether DELLAs and TCP14 are O-fucosylated in the cytoplasm or nucleus.
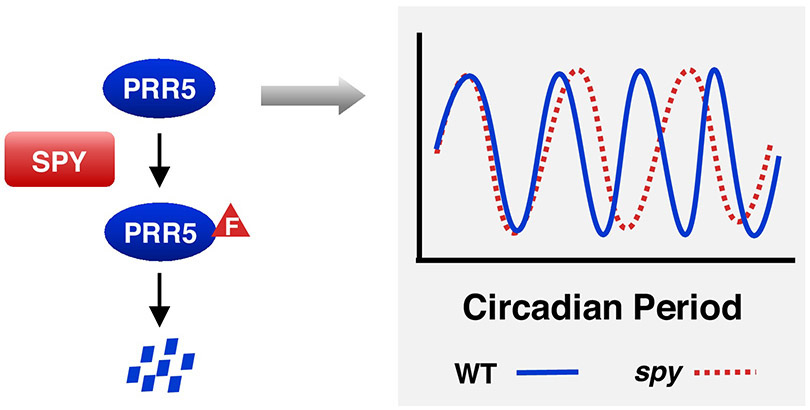
How does AtSPY regulate circadian clock? PRR5, a transcription repressor that is a key circadian clock component, was identified recently to be an interactor of AtSPY by MS analysis of proteins that were co-immunoprecipitated with SPY using Pspy:GFP-SPY and Pspy:GFP-SPY-NLS transgenic lines [34]. Theprr5 mutant shows a reduced circadian period, andprr5 partially rescues the longer circadian period phenotype of spy, whereas overexpression of PRR5 confers a longer circadian period. Transient co-expression of PRR5 and AtSPY in N. benthamiana showed that PRR5 is O-fucosylated by AtSPY. In addition, PRR5 protein levels are elevated in the spy mutant. Taken together, these results indicate that the nuclear-localized AtSPY modulates circadian clock speed by promoting PRR5 degradation via O-fucosylation (Fig. 3).
Figure 3. SPY regulates circadian period by inducing PRR5 degradation.
O-Fucosylation (labeled as F) of the transcription repressor PRR5 by SPY promotes PRR5 degradation. The spy mutant has a longer circadian period than WT.
Plant architecture
Alteration of SPY function in Arabidopsis and petunia has been shown to affect plant height and leaf shape through changes in GA and cytokinin signaling activities [9,10,32]. In addition, RNAi silencing of SPINDLY in Oryza sativa (OsSPY in rice) results in an increase in the leaf angle (due to increased bending of the lamina joint), which resembles an elevated BR response [56]. The OsSPY knockdown plants accumulate slightly elevated BR levels, suggesting that OsSPY may inhibit BR biosynthesis. If OsSPY and AtSPY are functionally conserved, it is also possible that OsSPY may repress BR signaling by enhancing DELLA-BZR1 interaction in rice.
A recent genome-wide association study (GWAS) identified OsSPY as a key factor in regulating rice architecture, including stem (culm) height, and size and numbers of panicles (branched flower clusters) [57]. In comparison to haplotype I, two polymorphisms in haplotype II, which result in S9T and R833L substitutions in OsSPY, correlate with taller stem and increased panicle size, but lower numbers of panicles. Importantly, the R833L substitution in the conserved POFUT catalytic domain was shown to reduce OsSPY activity by an in vitro enzyme assay. Further studies revealed that the effect of altered OsSPY activity on rice architecture is mainly through its regulation of GA signaling. This GWAS analysis also indicates that the enhanced OsSPY allele with R833 (in haplotype I) has a selective advantage through recent breeding programs because it confers a semidwarf and larger panicle-number phenotype in response to chemical fertilizer.
Root development
The spy mutants show root development defects, including formation of premature middle cortex (an extra layer of cortex) [58], and ectopic root hairs [59]. SPY may inhibit extra cortex formation by modulating redox homeostasis in the root meristem and elongation zone because H2O2 induces middle cortex formation in WT seedlings and the spy mutants accumulate higher amounts of H2O2 in their root tips than WT [60]. The precise mechanism of SPY-regulated root hair cell patterning is unclear, although SPY functions upstream of WEREWOLF and GLABRA2, which are two transcription factors that promote non-hair cell fate in the developing epidermal cells of the root [59].
Abiotic and biotic stresses
In addition to regulation of plant development, SPY also functions in plant’s responses to abiotic and biotic stresses. The hypomorphic spy mutants in Arabidopsis are more tolerant to high salt and drought conditions than WT, whereas SPY overexpression confers reduced drought tolerance [61]. These results suggest that SPY negatively regulates plant’s responses to these abiotic stresses. On the other hand, the spy mutants display enhanced susceptibility to a bacterial pathogen, P. syringae [62]. The quadruple della mutant, however, was previously shown to be more resistant to this pathogen infection [63], suggesting that SPY promotes plant defense responses by regulating pathways that are independent of GA and DELLAs.
SPY orthologs are present in diverse organisms
Phylogenetic analysis indicates that SPY-like genes are evolutionarily conserved, and are found in diverse organisms, including prokaryotes, protists, algae and all plants [4]. Both SPY and SEC genes are present in genomes of all plants, and in red algae. Different lineages of bacteria and protists contain either a SPY-like or a SEC-like gene [4]. Intriguingly, animal and fungi kingdoms only contain SEC-like (OGT) genes, but not the SPY-like genes. In addition, protein O-GlcNAcylation by OGT in animals is a dynamic modification that is reversible by O-GlcNAcase (OGA), whereas no OGA orthologs have been identified in plant genomes.
Although SPY orthologs have long been assumed to be OGTs based on sequence similarity, the finding that AtSPY is a POFUT raised the question whether SPY-like proteins in non-plant organisms are also POFUTs. Consistent with this hypothesis, recent studies in Toxoplasma gondii (a parasitic protist) indicate that TgSPY also encodes a POFUT [64]. Phylogenetic analysis suggests that TgSPY is a SPY-like gene [4]. Importantly, Bendini et al. (2016) identified O-fucosylated nucleocytoplasmic proteins in T. gondii by affinity purification using a terminal fucose-specific Aleuria aurantia lectin (AAL) and MS/MS analysis [65]. Predicted functions of these O-fucosylated proteins in T. gondii include nucleoporins, transcription regulators, and components involved in mRNA processing and signaling, suggesting that O-fucosylation may regulate nuclear targeting and gene expression in T. gondii [65]. The knockout TgSPY mutant generated by CRISPR-Cas9 approach failed to exhibit any intracellular signals by AAL staining [66]. Furthermore, the POFUT activity of TgSPY was demonstrated recently by in vitro enzyme assays [67]. Mutant analysis further showed that TgSPY plays a role in promoting the accumulation of its target proteins, and T. gondii proliferation in vitro and in mice.
Besides AtSPY and TgSPY, two additional SPY-like proteins from Cryptosporidium parvum (a parasitic protist) and Synechococcus elongatus (a cyanobacterium) have been reported in earlier studies to hydrolyze UDP-GlcNAc in vitro, although the specific glycosyltransferase activity was not demonstrated directly [68,69]. Another study reported the crystal structures of TtOGT in Thermobaculum terrenum (a thermophilic bacterium) and the TtOGT-UDP complex [70]. However, TtOGT did not exhibit any OGT activity in vitro, and MS analysis of the T. terrenum proteome failed to identify any O-GlcNAcylated proteins. Sequence alignment suggests that TtOGT is more similar to SPYs than to OGTs [14]. It remains to be determined whether these SPY-like proteins are POFUTs. Alternatively, they may display both OGT and POFUT activities, or novel glycosyltransferase activity with distinct donor substrate selectivity.
Future perspectives
The discovery of AtSPY-catalyzed protein O-fucosylation reveals a novel mechanism for regulating nucleocytoplasmic protein functions in plants. Our understanding of SPY- and SEC-regulated plant growth and development is only the tip of the iceberg. Future studies using multifaceted approaches including proteomics, chemical biology, genomics and metabolomics will help to elucidate the global functions of SPY and SEC, and the interplay between protein O-fucosylation and O-GlcNAcylation in regulating plant development. In addition, it is important to determine whether and how O-GlcNAc and O-Fuc modifications serve as sensors of metabolic status in plants and how these PTMs are modulated to integrate external conditions with internal programs. Recent characterization of TgSPY, the AtSPY ortholog in the human parasite T. gondii, supports the notion that intracellular protein O-fucosylation by SPY orthologs may regulate a wide range of biological processes in diverse organisms. The knowledge gain from studying how SPY functions in plants has broader implication in illuminating the molecular mechanisms by which nucleocytoplasmic protein O-fucosylation regulates gene expression and other cellular processes.
Acknowledgements
I thank Yan Wang for her help with Figure 3. This work was supported by the National Institutes of Health (2R01 GM100051-05A1), the National Science Foundation (MCB-1818161), US Department of Agriculture (2018-67013-27395), and Department of Energy (DE-SC0019393).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Intellectual Property
We confirm that we have given due consideration to the protection of intellectual property associated with this work and that there are no impediments to publication, including the timing of publication, with respect to intellectual property. In so doing we confirm that we have followed the regulations of our institutions concerning intellectual property.
Research Ethics
We further confirm that any aspect of the work covered in this manuscript that has involved human patients has been conducted with the ethical approval of all relevant bodies and that such approvals are acknowledged within the manuscript.
IRB approval was obtained (required for studies and series of 3 or more cases)
Written consent to publish potentially identifying information, such as details or the case and photographs, was obtained from the patient(s) or their legal guardian(s).
Conflict of Interest
No conflict of interest exists
References and recommended reading
• of special interest
•• of outstanding interest
- 1.Jacobsen SE, Olszewski NE: Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. Plant Cell 1993, 5:887–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Silverstone AL, Mak PYA, Casamitjana Martínez E, Sun T-p: The new RGA locus encodes a negative regulator of gibberellin response in Arabidopsis thaliana. Genetics 1997, 146:1087–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singh DP, Jermakow AM, Swain SM: Gibberellins are required for seed development and pollen tube growth in Arabidopsis. Plant Cell 2002, 14:3133–3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olszewski NE, West CM, Sassi SO, Hartweck LM: O-GlcNAc protein modification in plants: Evolution and function. Biochim Biophys Acta 2010, 1800:49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hartweck LM, Scott CL, Olszewski NE: Two O-Linked N-acetylglucosamine transferase genes of Arabidopsis thaliana L. Heynh. have overlapping functions necessary for gamete and seed development. Genetics 2002, 161:1279–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jacobsen SE, Binkowski KA, Olszewski NE: SPINDLY, a tetratricopeptide repeat protein involved in gibberellin signal transduction in Arabidopsis. Proc Natl Acad Sci U S A 1996, 93:9292–9296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hart GW, Slawson C, Ramirez-Correa G, Lagerlof O: Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu Rev Biochem 2011, 80:825–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tseng T-s Swain SM, Olszewski NE: Ectopic expression of the tetratricopeptide repeat domain of SPINDLY causes defects in gibberellin response. Plant Physiol 2001,126:1250–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Izhaki A, Swain SM, Tseng T-s, Borochov A, Olszewski NE, Weiss D: The role of SPY and its TPR domain in the regulation of gibberellin action throughout the life cycle of Petunia hybrida plants. Plant J 2001, 28:181–190. [DOI] [PubMed] [Google Scholar]
- 10.Silverstone AL, Tseng T-S, Swain S, Dill A, Jeong SY, Olszewski NE, Sun T-p: Functional analysis of SPINDLY in gibberellin signaling in Arabidopsis. Plant Physiol 2007, 143:987–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daviere JM, Achard P: Gibberellin signaling in plants. Development 2013, 140:1147–1151. [DOI] [PubMed] [Google Scholar]
- 12.Sun TP: The molecular mechanism and evolution of the GA-GID1-DELLA signaling module in plants. Curr Biol 2011, R338–345. [DOI] [PubMed] [Google Scholar]
- 13.Zentella R, Hu J, Hsieh WP, Matsumoto PA, Dawdy A, Barnhill B, Oldenhof H, Hartweck LM, Maitra S, Thomas SG, et al. : O-GlcNAcylation of master growth repressor DELLA by SECRET AGENT modulates multiple signaling pathways in Arabidopsis. Gems Dev 2016, 30:164–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zentella R, Sui N, Barnhill B, Hsieh WP, Hu J, Shabanowitz J, Boyce M, Olszewski NE, Zhou P, Hunt DF, et al. : The Arabidopsis O-fucosyltransferase SPINDLY activates nuclear growth repressor DELLA. Nat Chem Biol 2017, 13:479–485.•• In contrast to AtSEC (an OGT), AtSPY was shown in this study to be a novel nucleocytoplasmic POFUT. AtSPY and AtSEC play opposite roles in modulating multiple signaling pathways by affecting the activity of nuclear growth repressors DELLAs, revealing a new regulatory mechanism of gene expression.
- 15.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B: The carbohydrateactive enzymes database (CAZy) in 2013. Nucleic Acids Res 2014, 42:D490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holdener BC, Haltiwanger RS: Protein O-fucosylation: structure and function. Curr Opin Struct Biol 2019, 56:78–86.• An excellent review on regulatory mechanisms of ER-localized POFUTs in animals.
- 17.Li Z, Han K, Pak JE, Satkunarajah M, Zhou D, Rini JM: Recognition of EGF-like domains by the Notch-modifying O-fucosyltransferase POFUT1. Nat Chem Biol 2017, 13:757–763. [DOI] [PubMed] [Google Scholar]
- 18.Valero-Gonzalez J, Leonhard-Melief C, Lira-Navarrete E, Jimenez-Oses G, Hernandez-Ruiz C, Pallares MC, Yruela I, Vasudevan D, Lostao A, Corzana F, et al. : A proactive role of water molecules in acceptor recognition by protein O-fucosyltransferase 2. Nat Chem Biol 2016, 12:240–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lazarus MB, Nam Y, Jiang J, Sliz P, Walker S: Structure of human O-GlcNAc transferase and its complex with a peptide substrate. Nature 2011, 469:564–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swain SM, Tseng T-s, Thornton TM, Gopalraj M, Olszewski N, : SPINDLY is a nuclear-localized repressor of gibberellin signal transduction expressed throughout the plant. Plant Physiol 2002, 129:605–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pathak S, Alonso J, Schimpl M, Rafie K, Blair DE, Borodkin VS, Albarbarawi O, van Aalten DMF: The active site of O-GlcNAc transferase imposes constraints on substrate sequence. Nat Struct Mol Biol 2015, 22:744–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schimpl M, Zheng X, Borodkin VS, Blair DE, Ferenbach AT, Schuttelkopf AW, Navratilova I, Aristotelous T, Albarbarawi O, Robinson DA, et al. : O-GlcNAc transferase invokes nucleotide sugar pyrophosphate participation in catalysis. Nat Chem Biol 2012, 8:969–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martinez-Fleites C, Macauley MS, He Y, Shen DL, Vocadlo DJ, Davies GJ: Structure of an O-GlcNAc transferase homolog provides insight into intracellular glycosylation. Nat Struct Mol Biol 2008, 15:764–765. [DOI] [PubMed] [Google Scholar]
- 24.Daviere JM, Achard P: A pivotal role of DELLAs in regulating multiple hormone signals. Mol Plant 2016, 9:10–20. [DOI] [PubMed] [Google Scholar]
- 25.de Lucas M, Daviere JM, Rodriguez-Falcon M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blazquez MA, Titarenko E, Prat S: A molecular framework for light and gibberellin control of cell elongation. Nature 2008, 451:480–484. [DOI] [PubMed] [Google Scholar]
- 26.Feng S, Martinez C, Gusmaroli G, Wang Y, Zhou J, Wang F, Chen L, Yu L, Iglesias-Pedraz JM, Kircher S, et al. : Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 2008, 451:475–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bai MY, Shang JX, Oh E, Fan M, Bai Y, Zentella R, Sun T-p, Wang Z-Y: Brassinosteroid, gibberellin, and phytochrome signalling pathways impinge on a common transcription module in Arabidopsis. Nature Cell Biol 2012, 14:810–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanover JA, Krause MW, Love DC: The hexosamine signaling pathway: O-GlcNAc cycling in feast or famine. Biochim Biophys Acta 2010, 1800:80–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang X, Qian K: Protein O-GlcNAcylation: emerging mechanisms and functions. Nat Rev Mol Cell Biol 2017, 18:452–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xing L, Liu Y, Xu S, Xiao J, Wang B, Deng H, Lu Z, Xu Y, Chong K: Arabidopsis O-GlcNAc transferase SEC activates histone methyltransferase ATX1 to regulate flowering. EMBO J 2018, 37: e98115.•• Previously, O-GlcNAcylation was shown to mediate vernalization-induced flowering in winter wheat (Xiao et al., 2014). In contrast, this study shows that OGT in Arabidopsis (AtSEC) induces expression of flowering repressor FLC to delay flowering, at least in part by O-GlcNAcylation and activation of the histone methyltransferase ATX1 to increase the H3K4me3 active chromatin mark at the FLC locus.
- 31.Maymon I, Greenboim-Wainberg Y, Sagiv S, Kieber JJ, Moshelion M, Olszewski N, Weiss D: Cytosolic activity of SPINDLY implies the existence of a DELLA-independent gibberellin-response pathway. Plant J 2009, 58:979–988. [DOI] [PubMed] [Google Scholar]
- 32.Greenboim-Wainberg Y, Maymon I, Borochov R, Alvarez J, Olszewski N, Ori N, Eshed Y, Weiss D: Cross talk between gibberellin and cytokinin: the Arabidopsis GA response inhibitor SPINDLY plays a positive role in cytokinin signaling. Plant Cell 2005, 17:92–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tseng TS, Salome PA, McClung CR, Olszewski NE: SPINDLY and GIGANTEA interact and act in Arabidopsis thaliana pathways involved in light responses, flowering, and rhythms in cotyledon movements. Plant Cell 2004, 16:1550–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y, He Y, Su C, Zentella R, Sun TP, Wang L: Nuclear localized O-fucosyltransferase SPY facilitates PRR5 proteolysis to fine-tune the pace of Arabidopsis circadian clock. Mol Plant 2020, 13:446–458.•• AtSPY directly O-fucosylates PRR5 protein, which induces PRR5 degradation and modulates circadian clock speed. The cytoplasmic- and nuclear-localized AtSPY appears to regulate distinct cellular responses, and only the nuclear-localized AtSPY regulates circadian period.
- 35.Chen D, Juarez S, Hartweck L, Alamillo JM, Simon-Mateo C, Perez JJ, Fernandez-Fernandez MR, Olszewski NE, Garcia JA: Identification of SECRET AGENT as the O-GlcNAc transferase that participates in Plum pox virus infection. J Virol 2005, 79:9381–9387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Jesus Perez J, Udeshi ND, Shabanowitz J, Ciordia S, Juarez S, Scott CL, Olszewski NE, Hunt DF, Garcia JA: O-GlcNAc modification of the coat protein of the potyvirus Plum pox virus enhances viral infection. Virology 2013, 442:122–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taoka K, Ham BK, Xoconostle-Cazares B, Rojas MR, Lucas WJ: Reciprocal phosphorylation and glycosylation recognition motifs control NCAPP1 interaction with pumpkin phloem proteins and their cell-to-cell movement. Plant Cell 2007, 19:1866–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hartweck LM, Genger RK, Grey WM, Olszewski NE: SECRET AGENT and SPINDLY have overlapping roles in the development of Arabidopsis thaliana L. Heyn. J Exp Bot 2006, 57:865–875. [DOI] [PubMed] [Google Scholar]
- 39.Shafi R, Iyer SP, Ellies LG, O'Donnell N, Marek KW, Chui D, Hart GW, Marth JD: The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc Natl Acad Sci U S A 2000, 97:5735–5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gambetta MC, Oktaba K, Muller J: Essential role of the glycosyltransferase sxc/Ogt in polycomb repression. Science 2009, 325:93–96. [DOI] [PubMed] [Google Scholar]
- 41.Sinclair DA, Syrzycka M, Macauley MS, Rastgardani T, Komljenovic I, Vocadlo DJ, Brock HW, Honda BM: Drosophila O-GlcNAc transferase (OGT) is encoded by the Polycomb group (PcG) gene, super sex combs (sxc). Proc Natl Acad Sci U S A 2009, 106:13427–13432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hart GW: Nutrient regulation of signaling and transcription. J Biol Chem 2019, 294:2211–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bond MR, Hanover JA: A little sugar goes a long way: the cell biology of O-GlcNAc. J Cell Biol 2015, 208:869–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu SL, Chalkley RJ, Maynard JC, Wang W, Ni W, Jiang X, Shin K, Cheng L, Savage D, Huhmer AF, et al. : Proteomic analysis reveals O-GlcNAc modification on proteins with key regulatory functions in Arabidopsis. Proc Natl Acad Sci U S A 2017, 114:E1536–E1543.•• This is the first report of O-GlcNAcylated proteome in plants. 262 O-GlcNAcylated proteins were identified by affinity purification and MS analysis using Arabidopsis flower clusters. Some of these proteins are also phosphorylated. The data generated in this study are an important resource for investigating the roles of protein O-GlcNAcylation in plants.
- 45.Xu S, Xiao J, Yin F, Guo X, Xing L, Xu Y, Chong K: The Protein Modifications of O-GlcNAcylation and Phosphorylation Mediate Vernalization Response for Flowering in Winter Wheat. Plant Physiol 2019, 180:1436–1449.• By affinity purification and MS analysis, 168 O-GlcNAcylated proteins were identified in winter wheat (including TaGRP2, an RNA binding protein that was shown previously by Xiao et al. 2014) to induce flowering upon O-GlcNAcylation in response to vernalization. Some of these proteins are also phosphorylated. The data generated in this study are an important resource for investigating the roles of protein O-GlcNAcylation in plants.
- 46.Robertson M: Two transcription factors are negative regulators of gibberellin response in the HvSPY-signaling pathway in barley aleurone. Plant Physiol 2004, 136:2747–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sarnowska EA, Rolicka AT, Bucior E, Cwiek P, Tohge T, Femie AR, Jikumaru Y, Kamiya Y, Franzen R, Schmelzer E, et al. : DELLA-interacting SWI3C core subunit of switch/sucrose nonfermenting chromatin remodeling complex modulates gibberellin responses and hormonal cross talk in Arabidopsis. Plant Physiol 2013, 163:305–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cuellar Perez A, Nagels Durand A, Vanden Bossche R, De Clercq R, Persiau G, Van Wees SC, Pieterse CM, Gevaert K, De Jaeger G, Goossens A, et al. : The non-JAZ TIFY protein TIFY8 from Arabidopsis thaliana is a transcriptional repressor. PLoS One 2014, 9:e84891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Steiner E, Efroni I, Gopalraj M, Saathoff K, Tseng TS, Kieffer M, Eshed Y, Olszewski N, Weiss D: The Arabidopsis O-inked N-acetylglucosamine transferase SPINDLY interacts with Class I TCPs to facilitate cytokinin responses in leaves and flowers. Plant Cell 2012, 24:96–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiao J, Xu S, Li C, Xu Y, Xing L, Niu Y, Huan Q, Tang Y, Zhao C, Wagner D, et al. : O-GlcNAc-mediated interaction between VER2 and TaGRP2 elicits TaVRN1 mRNA accumulation during vernalization in winter wheat. Nat Commun 2014, 5:4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Swain SM, Tseng T-s, Olszewski NE: Altered expression of SPINDLY affects gibberellin response and plant development. Plant Physiol 2001, 126:1174–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Steiner E, Livne S, Kobinson-Katz T, Tal L, Pri-Tal O, Mosquna A, Tarkowska D, Mueller B, Tarkowski P, Weiss D: The putative O-inked N-acetylglucosamine transferase SPINDLY inhibits Class I TCP proteolysis to promote sensitivity to cytokinin. Plant Physiology 2016, 171:1485–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim EY, Jeong EH, Park S, Jeong HJ, Edery I, Cho JW: A role for O-GlcNAcylation in setting circadian clock speed. Genes Dev 2012, 26:490–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li YH, Liu X, Vanselow JT, Zheng H, Schlosser A, Chiu JC: O-GlcNAcylation of PERIOD regulates its interaction with CLOCK and timing of circadian transcriptional repression. PLoS Genet 2019, 15:e1007953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li MD, Ruan HB, Hughes ME, Lee JS, Singh JP, Jones SP, Nitabach MN, Yang X: O-GlcNAc signaling entrains the circadian clock by inhibiting BMAL1/CLOCK ubiquitination. Cell Metab 2013, 17:303–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shimada A, Ueguchi-Tanaka M, Sakamoto T, Fujioka S, Takatsuto S, Yoshida S, Sazuka T, Ashikari M, Matsuoka M: The rice SPINDLY gene functions as a negative regulator of gibberellin signaling by controlling the suppressive function of the DELLA protein, SLR1, and modulating brassinosteroid synthesis. Plant J 2006, 48:390–402. [DOI] [PubMed] [Google Scholar]
- 57.Yano K, Morinaka Y, Wang F, Huang P, Takehara S, Hirai T, Ito A, Koketsu E, Kawamura M, Kotake K, et al. : GWAS with principal component analysis identifies a gene comprehensively controlling rice architecture. Proc Natl Acad Sci U S A 2019, 116:21262–21267.••OsSPY was identified by GWAS to modulate stem height, as well as the size and numbers of panicles (branched flower clusters), all of which affect grain yield in rice.
- 58.Cui H, Benfey PN: Interplay between SCARECROW, GA and LIKE HETEROCHROMATIN PROTEIN 1 in ground tissue patterning in the Arabidopsis root. Plant J 2009, 58:1016–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mutanwad KV, Zangl I, Lucyshyn D: The Arabidopsis O-fucosyltransferase SPINDLY regulates root hair patterning independently of gibberellin signaling. Development 2020, 147: 10.1242/dev.192039.••This study shows that SPY plays a role in regulating root hair cell fate, and it functions upstream of the key transcription factors, WEREWOLF and GLABRA2.
- 60.Cui H, Kong D, Wei P, Hao Y, Torii KU, Lee JS, Li J: SPINDLY, ERECTA, and its ligand STOMAGEN have a role in redox-mediated cortex proliferation in the Arabidopsis root. Mol Plant 2014, 7:1727–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Qin F, Kodaira KS, Maruyama K, Mizoi J, Tran LS, Fujita Y, Morimoto K, Shinozaki K, Yamaguchi-Shinozaki K: SPINDLY, a negative regulator of gibberellic acid signaling, is involved in the plant abiotic stress response. Plant Physiol 2011, 157:1900–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang L, Paasch BC, Chen J, Day B, He SY: An important role of l-fucose biosynthesis and protein fucosylation genes in Arabidopsis immunity. New Phytol 2019, 222:981–994.••The results of this study suggest that SPY promotes plant defense responses by regulating GA/DELLA-independent pathways.
- 63.Navarro L, Bari R, Achard P, Lison P, Nemri A, Harberd NP, Jones JD: DELLAs control plant immune responses by modulating the balance of jasmonic acid and salicylic acid signaling. Curr Biol 2008, 18:650–655. [DOI] [PubMed] [Google Scholar]
- 64.Bandini G, Albuquerque-Wendt A, Hegermann J, Samuelson J, Routier FH: Protein O- and C-glycosylation pathways in Toxoplasma gondii and Plasmodium falciparum. Parasitology 2019, 146:1755–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bandini G, Haserick JR, Motari E, Ouologuem DT, Lourido S, Roos DS, Costello CE, Robbins PW, Samuelson J: O-fucosylated glycoproteins form assemblies in close proximity to the nuclear pore complexes of Toxoplasma gondii. Proc Natl Acad Sci U S A 2016, 113:11567–11572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gas-Pascual E, Ichikawa HT, Sheikh MO, Serji MI, Deng B, Mandalasi M, Bandini G, Samuelson J, Wells L, West CM: CRISPR/Cas9 and glycomics tools for Toxoplasma glycobiology. J Biol Chem 2019, 294:1104–1125.••This study used CRISPR-Cas9 and glycomics approaches to examine the functions of 33 putative glycosyltransferases in T gondii. The TgSPY mutant was found to be viable, although its proliferation is impaired in cell culture. This mutant failed to show any intracelluar AAL staining, suggesting that TgSPY is a POFUT.
- 67.Bandini G, Agop-Nersesian C, van der Wei H, Mandalasi M, Kim HW, West CM, Samuelson J: The nucleocytosolic O-fucosyltransferase Spindly affects protein expression and virulence in Toxoplasma gondii. J Biol Chem 2020, 10.1074/jbc.RA120.015883.•• This study presents direct evidence for the POFUT activity of TgSPY in vitro. The TgSPY knockout mutation affects T. gondii proliferation in cell culture and in mice. The levels of selected reporter proteins of TgSPY were much lower in the TgSPY knockout mutant.
- 68.Sokol KA, Olszewski NE: The putative eukaryote-like O-GlcNAc transferase of the cyanobacterium Synechococcus elongatus PCC 7942 hydrolyzes UDP-GlcNAc and is involved in multiple cellular processes. J Bacteriol 2015, 197:354–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Banerjee S, Robbins PW, Samuelson J: Molecular characterization of nucleocytosolic O-GlcNAc transferases of Giardia lamblia and Cryptosporidiumparvum. Glycobiology 2009, 19:331–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ostrowski A, Gundogdu M, Ferenbach AT, Lebedev AA, van Aalten DM: Evidence for a functional O-Linked N-acetylglucosamine (O-GlcNAc) system in the thermophilic Bacterium Thermobaculum terrenum. J Biol Chem 2015, 290:30291–30305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lazarus MB, Jiang J, Kapuria V, Bhuiyan T, Janetzko J, Zandberg WF, Vocadlo DJ, Herr W, Walker S: HCF-1 is cleaved in the active site of O-GlcNAc transferase. Science 2013, 342:1235–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Arnold K, Bordoli L, Kopp J, Schwede T: The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 2006, 22:195–201. [DOI] [PubMed] [Google Scholar]
- 73.Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Gallo Cassarino T, Bertoni M, Bordoli L, et al. : SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 2014, 42:W252–258. [DOI] [PMC free article] [PubMed] [Google Scholar]