Abstract
Introduction:
The PD-L1 immune checkpoint inhibitors atezolizumab and durvalumab have received regulatory approval for the first-line treatment of patients with extensive-stage small cell lung cancer. However, when used in combination with platinum-based chemotherapy, these PD-L1 inhibitors only improve overall survival by 2–3 months. This may be due to the observation that <20% of SCLC tumors express PD-L1 at >1%. Evaluating the composition and abundance of checkpoint molecules in SCLC may identify molecules beyond PD-L1 that are amenable to therapeutic targeting.
Methods:
We analyzed RNA-Seq data from SCLC cell lines (n=108) and primary tumor specimens (n=81) for expression of 39 functionally validated, inhibitory checkpoint ligands. Further, we generated tissue microarrays containing SCLC cell lines and SCLC patient specimens to confirm expression of these molecules by immunohistochemistry. We annotated patient outcomes data, including treatment response and overall survival.
Results:
The checkpoint protein B7-H6 (NCR3LG1) exhibited increased protein expression relative to PD-L1 in cell lines and tumors (P < 0.05). Higher B7-H6 protein expression correlated with longer progression-free survival (P = 0.0368) and increased total immune infiltrates (CD45+) in patients. Furthermore, increased B7-H6 gene expression in SCLC tumors correlated with a decreased activated NK cell gene signature, suggesting a complex interplay between B7-H6 expression and immune signature in SCLC.
Conclusions:
We investigated 39 inhibitory checkpoint molecules in SCLC and found that B7-H6 is highly expressed and associated with progression-free survival. In addition, 26/39 immune checkpoint proteins in SCLC tumors were more abundantly expressed than PD-L1, indicating an urgent need to investigate additional checkpoint targets for therapy in addition to PD-L1.
Keywords: small cell lung cancer (SCLC), immunotherapy, checkpoint molecules, immune checkpoint inhibitor, B7-H6
Introduction
Small cell lung cancer (SCLC) is a high-grade neuroendocrine tumor that affects ~15% of lung cancer patients and causes ~200,000 deaths annually worldwide.1 Approximately 70% of SCLC patients present with distant metastases at initial diagnosis and median overall survival is ~1 year for this disease.1 SCLC treatment has consisted of a combination of platinum-based chemotherapy plus etoposide for the last 30 years with no significant advances in therapeutic paradigms.2 However, in 2019, the United States Food and Drug Administration (FDA) approved the addition of the PD-L1 inhibitor, atezolizumab, in combination with chemotherapy for first-line treatment of patients with extensive-stage SCLC (ES-SCLC).3 Nearly one year later, another PD-L1 inhibitor, durvalumab, received an analogous approval.4 Chemotherapy (platinum/etoposide) plus immune checkpoint inhibitors (ICIs) is now the accepted standard of care for ES-SCLC. However, addition of PD-L1 inhibitors in this setting only improves overall survival by 2–3 months.3, 5 This may be due to the fact that despite high tumor mutation burden,6 SCLC tumors express minimal PD-L1,7-10 although additional factors may also explain the poor response. In addition, large clinical trials utilizing alternative checkpoint inhibitors such as anti-PD-111-14 and anti-CTLA-415 have either been negative or achieved minimal response rates (10–30%).10, 12, 16 Notably, these trials did not stratify patients based on PD-L1 expression, and as a result, it is unclear who benefits from current FDA-approved ICIs in SCLC. These data reveal a significant knowledge gap in the SCLC clinical realm; immune biomarkers which are standardly accepted in other tumor types, including non-small cell lung cancer (NSCLC), are either 1) inappropriate targets or 2) not predictive of response to immune checkpoint blockade in SCLC.17
There is an urgent need to define the immune landscape in SCLC in order to identify the best biomarker(s) of response and resistance to immune checkpoint blockade, maximize the utility of ICIs, and personalize medicine for patients with SCLC. Through these studies, we sought to characterize inhibitory checkpoint molecules in SCLC in an effort to identify targets amenable to ICIs.
Materials and Methods
RNA-Seq Datasets
Details of the RNA-seq datasets and analyses are provided in the Supplementary Methods.
SCLC cell line tissue microarray
A tissue microarray (TMA) was constructed using formalin-fixed and paraffin-embedded (FFPE) cell pellets from 19 unique SCLC cell lines (DMS53, DMS114, COR-L51, H69, H82, H128, H187, H196, H209, H211, H524, H526, H740, H841, H889, H1048, H1607, H2195, H2679) gifted from John Minna, M.D. (University of Texas Southwestern), Pierre Massion, M.D. (Vanderbilt University Medical Center), and Vito Quaranta, M.D. (Vanderbilt University). The TMA consisted of 1 mm diameter tissue cores from 19 SCLC cell lines, each in at least 3 replicates (n=66), human lung adenocarcinoma (n=3), and control tissues from normal human lung (n=3) and brain cortex (n=3). 5μm thick sections were cut from the array and mounted onto charged slides for immunohistochemistry (IHC) analyses. A sequential section was stained with H&E to confirm presence of the histological feature of interest (normal or tumor). The actual number of samples reported for each marker was lower than the total samples analyzed due to unavoidable tissue loss.
SCLC patients and tissue microarray
FFPE tumor samples from 51 SCLC patients treated at Vanderbilt University Medical Center (VUMC) were used to construct a TMA. Patient demographics can be found in Supplementary Table 1. All tissue was used in accordance with the VUMC institutional review board (IRB) under protocols #030763 and #160769, which approved the patient consent forms or waiver of consent. Archival tissue blocks were stained with H&E and reviewed by a pathologist. The TMA consisted of 0.6 mm diameter tissue cores from 51 unique patients, each in at least 2 replicates (n=164), and 7 control tissues including normal human tonsil (n=9), lung (n=2), brain cortex (n=2), pancreas (n=2), adrenal (n=2), thyroid (n=2), and parathyroid (n=2). Clinicopathological characteristics of the patients were compiled from medical records. 5μm thick sections were cut from the array and mounted onto charged slides for IHC analyses. A sequential section was stained with H&E to confirm presence of the histological feature of interest (normal or tumor). The actual number of samples reported for each marker was lower than the total samples analyzed due to unavoidable tissue loss and absence of or limited tumor cells.
B7-H3, B7-H6, PD-L1, and immune cell quantification in SCLC cell lines and tumors.
Quantification of immunohistochemistry staining is described in detail in Supplementary Methods.
Statistical Analyses
Statistics were performed as indicated using Python, R, or GraphPad Prism. P < 0.05 was considered statistically significant for all studies. For paired analyses, a 2-tailed paired t-test was utilized. The Chi-square (χ2) test was used to analyze the correlation of B7-H6 expression and clinical parameters. The Kaplan-Meier method and Cox regression were used to estimate survival and hazard ratios, and survival between groups was analyzed for significance using log-rank (Mantel-Cox) test. Where error bars are presented, all data are mean ± SD.
Results
PD-L2 and B7-H6 emerge as potential candidates of immune checkpoint blockade in SCLC
To evaluate inhibitory checkpoint molecules in SCLC, we first performed an extensive literature review and assembled a list of checkpoint molecules that were 1) expressed by tumors, 2) T cell suppressive, and 3) potentially amenable to ICI or other targeted therapies (Supplementary Tables 2 and 3). We evaluated the presence of these inhibitory checkpoint molecules (n=39) in SCLC cell lines (n=108) using bulk RNA-Seq compiled from three publicly available datasets: the Cancer Cell Line Encyclopedia (CCLE),18 Genomics of Drug Sensitivity in Cancer (GDSC),19 and SCLC cBioPortal for Cancer Genomics20, 21 (Fig. 1A). Previous studies report that most SCLC tumors exhibit low PD-L1 expression with <1% PD-L1 positive tumor and immune cells.7-10 Quantified expression of each inhibitory checkpoint molecule revealed that 18/38 (47%) had significantly increased gene expression (P < 0.01) relative to PD-L1 (Fig. 1B).
Figure 1: Expression of immune checkpoint molecules in SCLC cell lines.
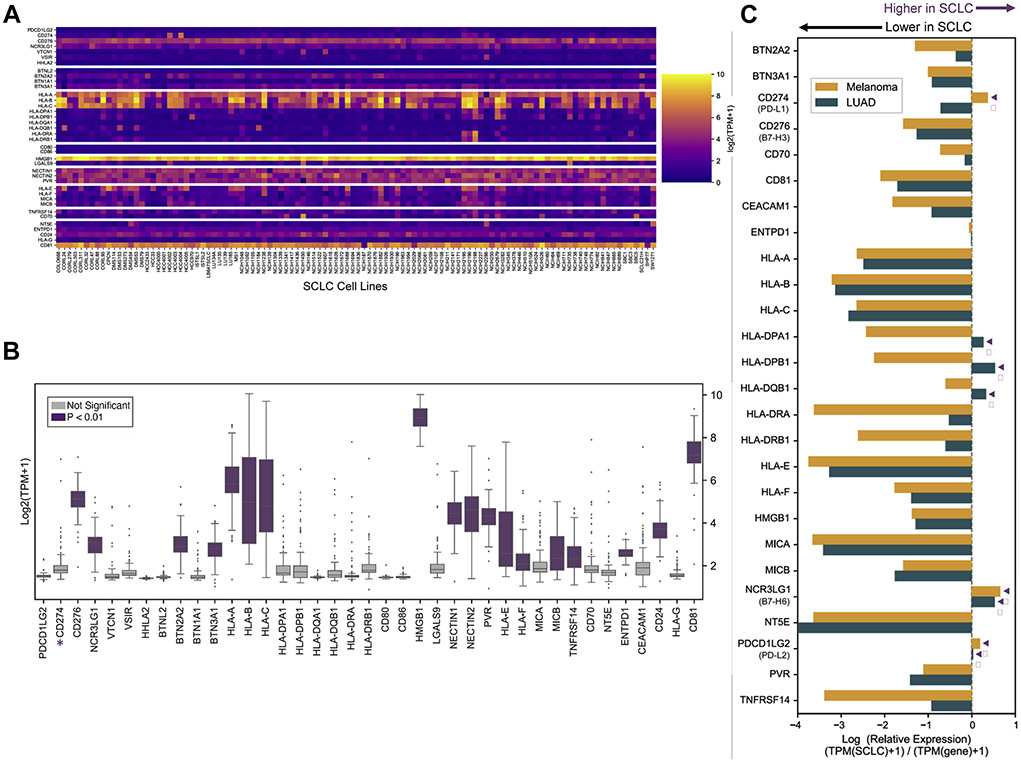
(A) Heatmap depicting log-transformed absolute gene expression of checkpoint molecules (n=39) described in Supplementary Table 2 in SCLC cell lines (n=108). Color scale bar represents absolute expression on a log scale. (B) Boxplots of log-transformed absolute expression of checkpoint molecules quantified in all SCLC cell lines. Genes that are significantly increased (P < 0.01) compared to PD-L1 (CD274, starred) are highlighted in purple. (C) Analysis of SCLC checkpoint molecule expression relative to melanoma (n=49) and LUAD (n=188) CCLE cell lines. Relative expression of log-transformed median gene expression of inhibitory checkpoint molecules with SCLC as baseline log(SCLC+1/[melanoma or LUAD]+1). Only genes present in all three datasets are shown. SCLC, small cell lung cancer; LUAD, lung adenocarcinoma. Arrowheads, increased relative expression in SCLC.
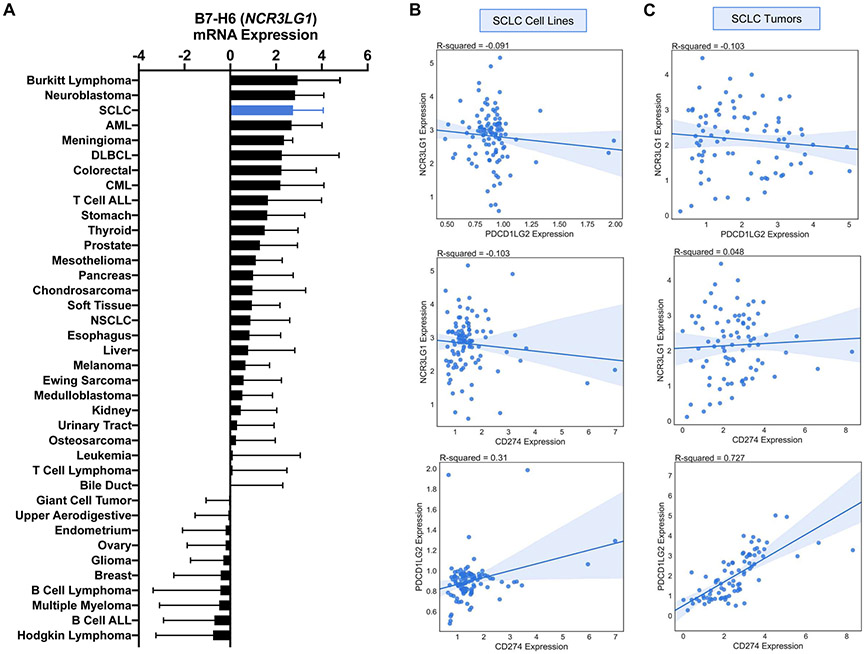
Lung adenocarcinoma (LUAD) and melanoma have each been reported to have high response rates to immune checkpoint blockade therapy.22, 23 We reasoned that genes that show similar or increased expression levels in SCLC compared to these tumor types would be more likely to have clinical relevance. Using the CCLE dataset, we compared gene expression of inhibitory molecules in SCLC cell lines (n=50) to LUAD (n=188) and melanoma (n=49) cell lines. Many checkpoint ligands (n=16/39, 41%) exhibited reduced gene expression in SCLC cell lines (Fig. 1C). CD274 expression was higher in SCLC relative to melanoma but not LUAD. However, PDCD1LG2 (PD-L2) and NCR3LG1 (B7-H6) exhibited increased expression relative to both LUAD and melanoma cell lines (Fig. 1c). Additional comparison of B7-H6 expression of cell lines in the CCLE revealed an increased expression in SCLC relative to most tumor types (Fig. 2A). PD-L2 showed decreased expression relative to other tumor types (data not shown). Additionally, we examined the potential co-expression of PDCD1LG2 (PD-L2) and NCR3LG1 (B7-H6) with CD274 (PD-L1) by assessing their correlation. NCR3LG1 (B7-H6) demonstrated minimal correlation with CD274 (PD-L1) and PDCD1LG2 (PD-L2) in SCLC cell lines (Fig. 2B).
Figure 2: Co-expression of PD-L1, PD-L2, and B7-H6 in SCLC cell lines and tumor.
(A) Quantification of mean log-transformed B7-H6 expression in CCLE cell lines. Error bars are standard deviation. Co-expression of NCR3LG1 (B7-H6), CD274 (PD-L1), and PDCD1LG2 (PD-L2) in (B) SCLC cell lines and (C) SCLC tumors with associated R-squared value (Pearson correlation) and 95% confidence interval for fit using linear regression. SCLC, small cell lung cancer; AML, acute myeloid leukemia; DLBCL, diffuse large B cell lymphoma; CML, chronic myelogenous leukemia; ALL, acute lymphocytic leukemia; NSCLC, non-small cell lung cancer.
To validate the findings from SCLC cell lines, a publicly available SCLC tumor RNA-Seq dataset (EGAS0000100092524; n=81) was analyzed for expression of inhibitory checkpoint molecules (Supplementary Table 2) in SCLC tumors (Supplementary Data 1A). Quantified expression of inhibitory checkpoint molecules revealed that 26/38 (68%) had significantly increased expression (P < 0.01) relative to PD-L1 (Supplementary Data 1B). The TCGA dataset was used to compare gene expression of inhibitory molecules in SCLC tumors (n=81) to LUAD (n=576) and melanoma (n=473) tumors. Analyses did not validate increased PDCD1LG2 (PD-L2) and NCR3LG1 (B7-H6) gene expression in SCLC tumors relative to LUAD and melanoma tumors (Supplementary Data 1C) as observed in SCLC cell lines (Fig 1C). In fact, most checkpoint ligands (n=32/33, 97%) exhibited reduced gene expression in SCLC tumors. PDCD1LG2 (PD-L2) and CD274 (PD-L1) showed strong correlation (R2=0.727) in SCLC tumors, indicating a pattern of co-expression (Fig. 2C). A weaker correlation was observed in SCLC cell lines (R2 = 0.310, Fig. 2B). As observed in SCLC cell lines, NCR3LG1 (B7-H6) demonstrated little correlation with CD274 (PD-L1) and PDCD1LG2 (PD-L2) in SCLC tumors (Fig. 2C).
Immunohistochemistry validates B7-H6 as a potential therapeutic target for immune checkpoint blockade
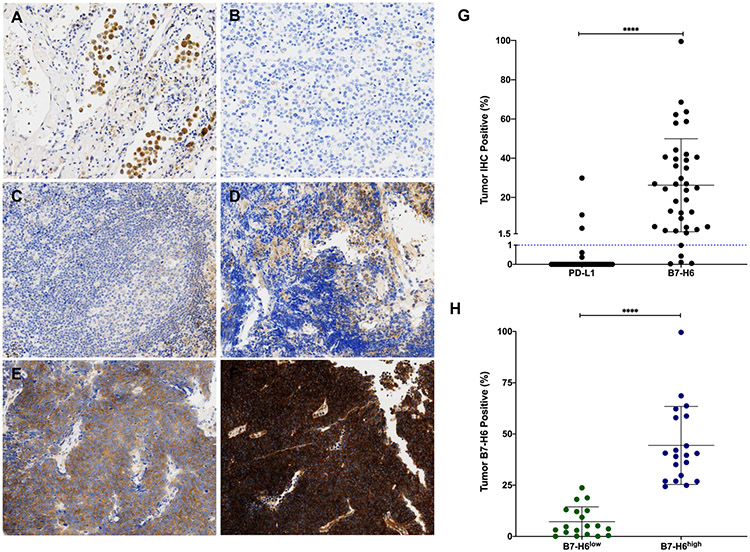
Although NCR3LG1 (B7-H6) did not demonstrate increased gene expression relative to CD274 (PD-L1) in SCLC tumors (Supplementary Data 1A, B) and relative to LUAD and melanoma tumors (Supplementary Data 1C), it displayed increased expression in SCLC cell lines (Fig. 1B) and increased gene expression relative to both LUAD and melanoma cell lines (Fig. 1C). Therefore, we chose to validate B7-H6 protein expression in SCLC. We examined B7-H6 (NCR3LG1) via immunohistochemistry (IHC) in SCLC cell lines (n=21) and clinical specimens (n=120). For comparison, we also analyzed SCLC cell lines for the presence of PD-L1 and B7-H3 protein; B7-H3 (CD276) is an inhibitory checkpoint ligand previously identified to exhibit high expression in SCLC7 that showed increased gene expression relative to PD-L1 (Fig. 1b and Supplementary Data 1B). Correlating with the RNA-Seq data (Fig. 1B and Supplementary Data 1B) and previous studies, total PD-L1 (CD274) positivity was low (< 20% tumor cells) (Supplementary Data 2A, B). Over half (59%, n=10) of cell lines had >1% total cells positive for PD-L1 immunoreactivity (Supplementary Data 2B). Although B7-H6 average protein expression was similar to PD-L1, >1% total cells positive were detected in all (n=17) cell lines (Supplementary Data 2B). In addition, median B7-H6 cell positivity was 10-fold higher than PD-L1 (data not shown). Consistent with gene expression data (Fig. 1B and Supplementary Data 1B), B7-H3 was highly expressed at the protein level and showed >1% positive staining in all cell lines (Supplementary Data 2B).
Primary normal human lung tissue was assessed for B7-H6 staining and we observed no expression (Fig. 3A). Cells showing immunoreactivity are hemosiderin-laden macrophages. Negative staining (Fig. 3B), weak staining (Fig. 3C), moderate staining (Fig. 3D), and strong staining (Fig. 3E-F) were observed in SCLC tumors. Similar to SCLC cell lines, most (87%, n=34/39) SCLC tumors showed >1% positive staining. B7-H6 expression was also more abundant in tumors (P < 0.0001) when compared to PD-L1 (Fig. 3G).
Figure 3: B7-H6 immunohistochemistry staining in normal lung and SCLC tissues.
Representative images, 40X magnification; (A) No staining in normal lung (cells showing immunoreactivity are hemosiderin-laden macrophages). (B) Negative staining in SCLC. (C) Weak staining in SCLC. (D) Moderate staining in SCLC. (E, F) Strong staining in SCLC. (G) Comparison of PD-L1 and B7-H6 positive staining in SCLC tumors. Line represents 1% tumor positive IHC staining. (H) Stratification of SCLC tumors into B7-H6low (n=19) and B7-H6high (n=20) based on median expression (24.3%). ****, P < 0.0001; SCLC, small cell lung cancer.
B7-H6 correlation with clinicopathological features in SCLC
Previous reports in other tumor types have indicated that B7-H6 protein expression correlates with prognoses and other clinicopathological parameters (tumor progression, stage, metastasis, treatment response, and tumor differentiation).25-30 Therefore, we investigated the association of B7-H6 expression with clinicopathological features of SCLC (Table 1). Since current clinical practices utilize PD-L1 positivity and not H-score to determine ICI administration, we stratified patients based on B7-H6 tumor positivity. In addition, since no clinical guidelines exist for B7-H6 positivity in tumors, we based stratification on the median B7-H6 tumor positive percentage of 24.3% in our cohort. Patients with B7-H6 tumor positive < median (n=19, B7-H6low) or ≥ median (n=20, B7-H6high) were analyzed for SCLC clinical parameters (Fig. 3H). Strong correlation was seen between B7-H6 and gender, with males exhibiting higher B7-H6 expression (P < 0.0154). All other clinical parameters analyzed were insignificant as noted in Table 1.
Table 1.
Correlation Between B7-H6 Expression and Clinicopathologic Parameters of Patients With SCLC
| Tumor B7-H6 Positive % |
|||||
|---|---|---|---|---|---|
| Clinical Parameters | Cases | < Median (B7-H6low) | ≥ Median (B7-H6high) | Chi-Square | p Value |
| Median survival (range) | 39 | 7.65 mo (1.44-85.53) | 15.86 mo (0.10-51.20) | – | 0.4576 |
| Gender | |||||
| Male | 18 | 5 | 13 | 5.867 | 0.0154a |
| Female | 21 | 14 | 7 | ||
| Age at diagnosis | |||||
| < 65 y | 25 | 11 | 14 | 0.6205 | 0.4309 |
| ≥ 65 y | 14 | 8 | 6 | ||
| Stage at diagnosis | |||||
| Limited-stage SCLC | 18 | 7 | 11 | 1.293 | 0.2556 |
| Extensive-stage SCLC | 21 | 12 | 9 | ||
| Stage at biopsy | |||||
| Limited-stage SCLC | 14 | 5 | 9 | 1.216 | 0.2241 |
| Extensive-stage SCLC | 25 | 14 | 11 | ||
| Site of biopsy | |||||
| Lung | 16 | 5 | 11 | 3.327 | 0.1895 |
| Lymph node | 8 | 5 | 3 | ||
| Otherb | 15 | 9 | 6 | ||
| Distant metastases | |||||
| M0 | 18 | 7 | 11 | 1.293 | 0.2556 |
| M1 | 21 | 12 | 9 | ||
| Survival time | |||||
| <1 y | 24 | 14 | 10 | 1.520 | 0.1286 |
| ≥ 1 y | 15 | 5 | 10 | ||
| Response to first-line therapy | |||||
| CR | 9 | 3 | 6 | 2.272 | 0.5180 |
| PR | 13 | 8 | 5 | ||
| Stable disease | 3 | 2 | 1 | ||
| POD | 8 | 5 | 3 | ||
| Did not receive therapy | 5 | 1 | 4 | ||
| Data not available | 1 | 0 | 1 | ||
Signifies p < 0.05.
Liver, bone, brain, neck, bowel, and adrenal.
CR, complete response; PR, partial response; POD, progression of disease.
B7-H6 is associated with longer progression-free survival and increased immune infiltrates in SCLC
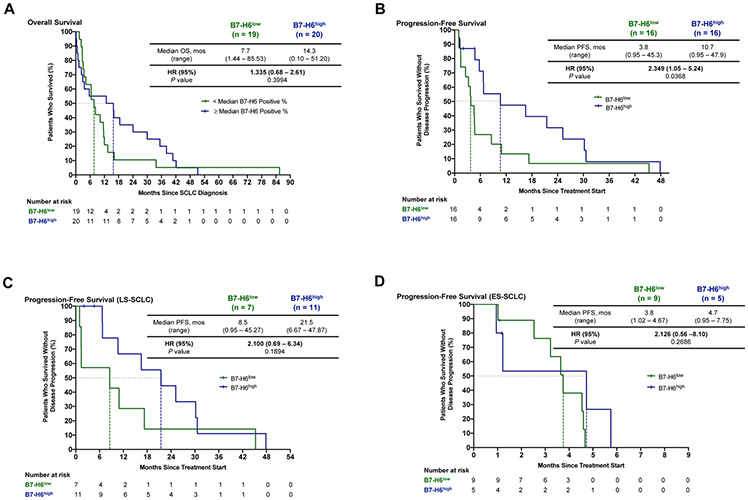
In order to further investigate the prognostic value of B7-H6 in SCLC, we conducted survival analyses based on B7-H6 tumor expression. B7-H6 expression had no significant correlation (P = 0.3994) with overall survival (OS) (Fig. 4A). Interestingly, median OS was nearly 2-fold higher (14.3 versus 7.7 months – CI) in B7-H6high patients (Fig. 4A). When evaluating progression-free survival (PFS) from start of treatment, B7-H6high patients experienced a significantly longer disease control (P = 0.0368) and a more than 3-fold improvement in median PFS (10.7 versus 3.8 months) compared to B7-H6low patients (Fig. 4B). Considering that stage is a major factor of OS and PFS in SCLC, we further assessed PFS in stage-matched patients. B7-H6high patients with limited-stage SCLC (LS-SCLC) had a nearly 3-fold higher median PFS than B7-H6low low patients (21.5 versus 8.5 months), although this difference was not statistically significant (Fig. 4C). Median PFS was similar when comparing patients with B7-H6high and B7-H6low ES-SCLC (4.7 versus 3.8 months, respectively; Fig. 4D). These data suggest B7-H6 expression is a contributing factor in length of PFS, although stage cannot be completely excluded.
Figure 4: Overall survival (OS) and progression-free survival (PFS) in B7-H6high and B7-H6low patients with SCLC.
Kaplan-Meier estimates of (A) OS and (B) PFS in patients with SCLC according to low or high B7-H6 expression. Kaplan-Meier estimates of PFS limited to only patients who received systemic therapy with (C) limited-stage or (D) extensive-stage SCLC. SCLC, small cell lung cancer; OS, overall survival; PFS, progression-free survival; HR, hazard ratio; LS, limited-stage; ES, extensive-stage.
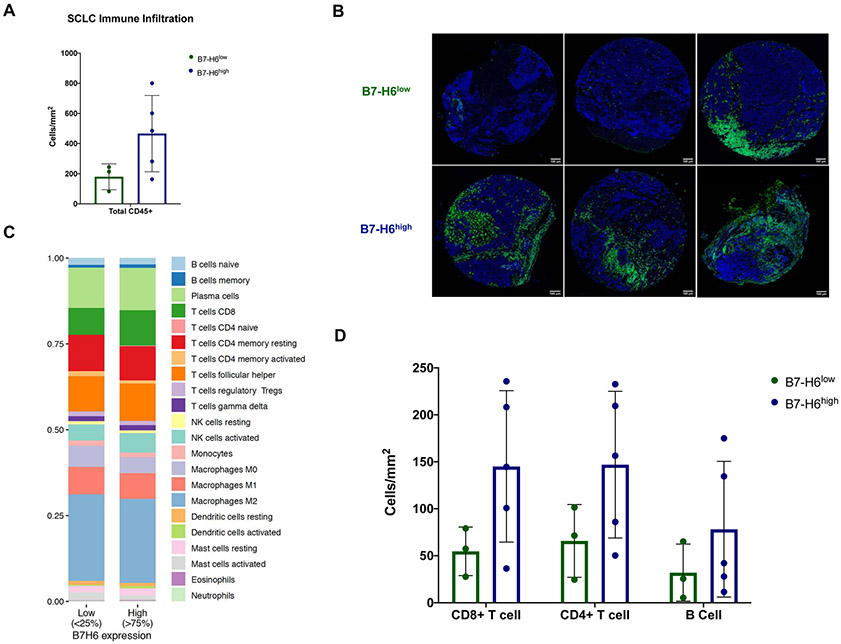
Based on the observation of increased PFS in B7-H6high patients, we next sought to investigate the influence of B7-H6 expression on immune complexity. Further investigation of B7-H6high/low cohorts via IHC revealed increased leukocyte infiltration (CD45+) in primary SCLC lung tumors of B7-H6high patients (n=7), a feature which has been reported to be predictive of improved patient outcomes and response to immunotherapy31-33 (Fig. 5A, B). To establish the relationship between B7-H6 expression and immune infiltration in SCLC tumors, we estimated the inferred relative proportions of immune cell populations using CIBERSORTx34 and bulk tumor RNA-Seq data (EGAS0000100092524; n=81). High B7-H6 expression was associated with increased CD8+ T cells, plasma cells, and follicular helper T cells, which have been reported to be associated with better patient prognosis31, 35, 36 (Fig. 5C and Supplementary Data 3A, B). We confirmed the increased presence of these immune cell types in B7-H6high SCLC primary lung tumors via IHC (Fig. 5D). Although NK cell relative proportions did not differ between the B7-H6high/low cohorts, we further examined the relationship between B7-H6 and its only known cognate receptor, NKp30,37 on inferred NK cell populations identified using the CIBESORTx34 methodology and the LM22 gene signature. We also examined the relationship between B7-H6 expression and abundance of NK cells. The analyses revealed a significant negative correlation between resting NK cells and NKp30 (NCR3) expression (R = −0.29, P = 0.0085; Supplementary Data 4A), but a significant positive correlation between activated NK cells and NKp30 expression (R = 0.64, P = 9.9E-11; Supplementary Data 4B). B7-H6 (NCR3LG1) expression was also inversely correlated with abundance of activated NK cells (R = −0.23, P = 0.043; Supplementary Data 4C), indicating a potential NK-inhibitory role for B7-H6 in SCLC.
Figure 5: Correlation of B7-H6 and immune signature in SCLC tumors.
(A) Quantification of tumor-infiltrating immune cells (CD45+) in B7-H6low (n=3) and B7-H6high (n=5) primary lung SCLC tumors and (B) representative images showing nuclei (blue) and CD45+ infiltration (green); scale bar, 100 μm. (C) Relative abundance of immune cell populations determined by CIBERSORTx34 methodology in B7-H6low (n=20) and B7-H6high SCLC tumors (n=20). (D) Quantification of CD8+ and CD4+ T cells, and B cells in B7-H6low (n=3) and B7-H6high (n=5) primary lung SCLC tumors. SCLC, small cell lung cancer.
SCLC cell lines and tumors display subtype-specific expression of inhibitory checkpoint molecules
Developing in parallel to these practice-changing clinical trials involving use of ICIs in SCLC are large-scale - omics efforts to bring more precision to the diagnosis and treatment of SCLC at the molecular level. At present, SCLC is approached clinically as a single disease with no standard biomarker assessments (such as tumor DNA sequencing) to personalize care. Attempts to better define specific patient populations and understand SCLC heterogeneity at the molecular level have led to the identification of SCLC subtypes within the previously described, broader neuroendocrine (NE) and non-neuroendocrine (non-NE) classifications.38-40 These subtypes are defined by differential gene expression of four main transcription regulators—Achaete-Scute Complex Homolog-Like 1 (ASCL1), Neurogenic Differentiation Factor 1 (NEUROD1), POU Class 2 Homeobox 3 (POU2F3), and Yes-Associated Protein-1 (YAP1)—and exhibit potentially unique therapeutic susceptibilities.41, 42 At present, little is known about the relationship between the immune microenvironment and SCLC subtypes.
SCLC cell lines exhibited distinct subtypes—ASCL1high (SCLC-A), NEUROD1high (SCLC-N), POU2F3high (SCLC-P), YAP1high (SCLC-Y)—based on relative RNA-Seq expression of four well-defined transcription regulators42 (Supplementary Data 5A and Supplementary Table 4). Since therapeutic implications amongst individual subtypes have not been fully clarified, the more well-elucidated classifications of neuroendocrine (NE; SCLC-A and SCLC-N) and non-neuroendocrine (non-NE; SCLC-P and SCLC-Y) were used for downstream analyses. Insulinoma-associated 1 (INSM1) gene expression, which is a marker of neuroendocrine tumors,43 was also quantified as a measure of subtyping validation (Supplementary Data 5A). Subtyping analyses classified most SCLC cell lines as NE (n=92) and the rest as non-NE (n=16). Quantified expression of inhibitory checkpoint molecules in NE and non-NE subtypes revealed differential expression of several genes (Supplementary Data 5B). Notably, PD-L1 and B7-H6 were not differentially expressed amongst SCLC subtypes. Checkpoint genes that were validated in downstream patient analyses are displayed in Supplementary Data 5C. Notably, CD70 (TNFSF7), CD81 (CD81), HLA-B (MHCI), HLA-E (HLA-E), MICA (MICA), and VTCN1 (B7-H4) all showed increased expression (P < 0.05) in the non-NE subtype (Supplementary Data 5C). Similar to SCLC cell lines, tumors were first classified into NE (n=67) and non-NE (n=14) subtypes, with NE being the predominant subtype (Supplementary Data 6A). NE and non-NE tumors were then analyzed for expression of checkpoint molecules (Supplementary Data 6b). Differentially expressed markers in SCLC cell lines (Supplementary Data 5C) were validated (P < 0.05) in tumor samples (Supplementary Data 6C). The trend of increased gene expression in non-NE subtypes observed in cell lines was also evident in tumor samples.
Discussion
ICIs have significantly altered survival outcomes for some cancer patients with demonstrated success in many tumor types.44 The FDA has approved PD-L1 inhibitors for use in more than 10 cancer types,45 including small cell lung cancer, in which it is approved for first-line treatment of advanced disease. However, the number of patients who respond is few, and the benefit gained by responders is minimal. Even blockade of alternative inhibitory checkpoints molecules—CTLA-4 and PD-1—does not substantially prolong overall survival in SCLC patients.11-15 One explanation could be low or absent expression of these molecules in SCLC,7-10 precluding their use as viable targets for immune checkpoint blockade. Alternatively, expression of these checkpoint molecules might not serve as predictive biomarkers for ICI administration. Nonetheless, existing ICIs could prove successful if administered within the proper tumor context. To date, there are no guidelines surrounding the administration of ICIs in SCLC and no predictive biomarkers have been established.
In these studies, we aimed to provide context for ICI administration by considering inhibitory checkpoint molecules beyond PD-L1 for immune checkpoint blockade. Additionally, using large datasets, we have highlighted the necessity of considering subtypes in SCLC immune checkpoint blockade by identifying subtype-specific, differential gene expression of inhibitory checkpoint molecules—CD70 (CD70), CD81 (CD81), HLA-B (MHCI), HLA-E (HLA-E), MICA (MICA), VTCN1 (B7-H4)—which are putative targets for immune-based therapies (Supplementary Table 2). Further, we identified two potential candidates for immune checkpoint inhibition in SCLC—NCR3LG1 (B7-H6) and PDCD1LG2 (PD-L2)—and validated B7-H6 as a potential target. PD-L2 has been investigated in SCLC but showed no association with clinicopathological factors or prognosis in initial studies.46 However, the strong correlation of PD-L2 (PDCD1LG2) and PD-L1 (CD274) gene expression might indicate that a combined PD-L1 / PD-L2 ICI regimen would be more efficacious. B7-H6 is a novel ligand belonging to the B7 family. Several B7 family members, including PD-L1, have been revealed to be overexpressed in tumors and implicated in tumor progression and overall worse prognosis. Distinct from other family members, B7-H6 is abnormally expressed in tumor tissues29, 47 with little to no expression in normal human tissues, making it an attractive target for existing immunotherapies such as ICI, chimeric antigen receptor (CAR) T cells,48-50 or bispecific T cell engagers (BiTEs).51 Aberrant B7-H6 overexpression has been reported in a breadth of tumor types including glioma, hepatocellular carcinoma, triple-negative breast cancer, and non-small cell lung cancer.27, 30, 47, 52 However, unlike PD-L1, B7-H6 can also be co-stimulatory, by binding to NKp30 and enhancing antitumor NK cell cytotoxicity and cytokine secretion.37 Due to its dual immune regulatory nature, the clinical implications of abnormal B7-H6 expression in human cancer remain elusive. While some studies report strong correlation of B7-H6 expression with tumor progression26, 52-55 and chemoresistance55, 56 in multiple tumor types, others do not.30, 57
Although our study herein revealed a positive correlation of B7-H6 expression with progression-free survival and increased immune infiltration, we observed a negative correlation with B7-H6 and activated NK cells. These findings support previous reports of a context-specific, immune regulatory role for B7-H6 in SCLC. Of note, we did not assess for the presence of soluble B7-H6, which has been demonstrated to be inhibitory to NK cell function.54, 58 Higher expression of B7-H6 on tumor cells could be indicative of lower serum amounts of soluble B7-H6, which could explain the longer PFS in B7-H6high SCLC patients. We also do not know how the expression of B7-H6 changes dynamically over time and in response to chemotherapy. Furthermore, the comparison of inhibitory checkpoint molecules in SCLC to PD-L1 at the gene expression level might not correlate directly with therapeutic relevance due to various factors that affect protein expression, including protein stability and membrane trafficking. Our studies also assume that higher protein expression correlates with ICI effectiveness, which might not be the case for all checkpoint molecules.
In conclusion, our studies revealed that several evaluated inhibitory checkpoint molecules were more highly expressed than PD-L1, indicating that globally PD-L1 may not be the most optimal target for ICIs in SCLC. We also report that SCLC cell lines could serve as valid models for checkpoint ligand studies in lieu of the relative paucity of SCLC tissues available for research. Future studies will seek to validate B7-H6 as a target amenable to immune checkpoint inhibition and investigate checkpoint molecules beyond PD-L1 as candidates for immune checkpoint blockade in SCLC.
Supplementary Material
Acknowledgments
The authors thank all of the patients who made this study possible and all of the health care providers who cared for these patients; Dr. John Minna, Dr. Pierre Massion, and Dr. Vito Quaranta for gifting SCLC cell lines; the team in the Translational Pathology Shared Resource facility; Nell Kirchberger and Giovanney Gonzalez for technical support related to IHC staining and quantification; Dr. Ann Richmond, Dr. Jonathan Irish, Dr. Andries Zijlstra, Dr. Pierre Massion, Dr. Maria Lima, Dr. Fernando Villalta, and the Vanderbilt U54 SCLC Consortia for all the helpful feedback; Jerome Arceneaux, Stephen Williams, James Mungin, Jr., Samantha Beik, Dr. Zhenfang Zhang, Dr. Huan Qiao, Ron Lovly, and Dr. Henry Henderson for reviewing the manuscript. This study was supported in part by a Vanderbilt-Ingram Cancer Center Young Ambassadors Award and a Lung Cancer Foundation of America/International Association for the Study of Lung Cancer Lori Monroe Scholarship. Dr. Lovly was also supported by National Institutes of Health [grant numbers U54CA217450-01, U01CA224276-01, P30-CA086485, UG1CA233259]. Portia Thomas was supported by grant S21MD000104 and the Ann Melly Summer Scholarship in Oncology. Dr. Iams was supported by the National Institutes of Health (NIH) and National Cancer Institute (NCI) Vanderbilt Clinical Oncology Research Career Development Award (VCORCDP) 2K12CA090625-17 and an American Society of Clinical Oncology/Conquer Cancer Foundation Young Investigator Award. Dr. Tyson was supported by the NIH grants: R50CA243783 and U54CA217450. Dr. Coussens was supported by the National Institutes of Health (1U01 CA224012, U2C CA233280, R01 CA223150, R01 CA226909, R21 HD099367), the Knight Cancer Institute, and the Brenden-Colson Center for Pancreatic Care at OHSU. All data storage for this research project used Vanderbilt’s Redcap data storage infrastructure, funded by the NCATS/NIH grant, UL1TR000445. The results published here are in whole or part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga.
Abbreviations:
- ASCL1
Achaete-Scute Complex Homolog-Like 1
- CTLA-4
Cytotoxic T-lymphocyte-associated protein 4
- FPKM
Fragments Per Kilobase of transcript per Million
- GDSC
Genomics of Drug Sensitivity in Cancer
- ICI
Immune checkpoint inhibitor
- IHC
Immunohistochemistry
- INSM1
Insulinoma-associated 1
- LUAD
Lung adenocarcinoma
- NE
Neuroendocrine
- NEUROD1
Neurogenic Differentiation Factor 1
- NSCLC
Non-small cell lung cancer
- OS
Overall survival
- PD-1
Programmed cell death protein 1
- PD-L1
Programmed death-ligand 1
- PFS
Progression-free survival
- RNA-Seq
RNA sequencing
- SCLC
Small cell lung cancer
- TMA
Tissue microarray
- TPM
Transcripts per million
- YAP1
Yes-Associated Protein-1
Footnotes
Conflict of interest: CML is a consultant/advisory board member for Pfizer, Novartis, AstraZeneca, Genoptix, Sequenom, Ariad, Takeda, Blueprints Medicine, Cepheid, Foundation Medicine, Roche, Achilles Therapeutics, Genentech, Syros, Amgen, EMD-Serono, and Eli Lilly and reports receiving commercial research grants from Xcovery, AstraZeneca, and Novartis. WTI reports consulting for Genentech, Outcomes Insights, Jazz Pharmaceuticals, and Defined Health and clinical trial funding from EMD Serono. VQ is an Academic co-Founder of Parthenon Therapeutics, Inc. LMC is a paid consultant for Cell Signaling Technologies, Shasqi, Inc., and AbbVie, Inc., received reagent and/or research support from Plexxikon, Inc., Pharmacyclics, Inc., Acerta Pharma, LLC, Deciphera Pharmaceuticals, LLC, Genentech, Inc., Roche Glycart AG, Syndax Pharmaceuticals, Inc., Innate Pharma, and NanoString Technologies, and serves on Advisory Boards of Syndax Pharmaceuticals, Carisma Therapeutics, Zymeworks, Inc., Verseau Therapeutics, Cytomix Therapeutics, Inc., Kineta, Inc, and Hibercell Inc. The remaining authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bernhardt EB, Jalal SI. Small Cell Lung Cancer. Cancer Treat Res 2016;170:301–322. [DOI] [PubMed] [Google Scholar]
- 2.Sabari JK, Lok BH, Laird JH, et al. Unravelling the biology of SCLC: implications for therapy. Nat Rev Clin Oncol 2017;14:549–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Horn L, Mansfield AS, Szczesna A, et al. First-Line Atezolizumab plus Chemotherapy in Extensive-Stage Small-Cell Lung Cancer. N Engl J Med 2018;379:2220–2229. [DOI] [PubMed] [Google Scholar]
- 4.Paz-Ares L, Dvorkin M, Chen Y, et al. Durvalumab plus platinum-etoposide versus platinum-etoposide in first-line treatment of extensive-stage small-cell lung cancer (CASPIAN): a randomised, controlled, open-label, phase 3 trial. Lancet 2019;394:1929–1939. [DOI] [PubMed] [Google Scholar]
- 5.Paz-Ares LG, Dvorkin M, Chen Y, et al. Durvalumab ± tremelimumab + platinum-etoposide in first-line extensive-stage SCLC (ES-SCLC): Updated results from the phase III CASPIAN study. Journal of Clinical Oncology 2020;38:9002–9002. [Google Scholar]
- 6.Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature 2013;500:415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carvajal-Hausdorf D, Altan M, Velcheti V, et al. Expression and clinical significance of PD-L1, B7-H3, B7-H4 and TILs in human small cell lung Cancer (SCLC). J Immunother Cancer 2019;7:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonanno L, Pavan A, Dieci MV, et al. The role of immune microenvironment in small-cell lung cancer: Distribution of PD-L1 expression and prognostic role of FOXP3-positive tumour infiltrating lymphocytes. Eur J Cancer 2018;101:191–200. [DOI] [PubMed] [Google Scholar]
- 9.Zhao X, Kallakury B, Chahine JJ, et al. Surgical Resection of SCLC: Prognostic Factors and the Tumor Microenvironment. J Thorac Oncol 2019;14:914–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Antonia SJ, Lopez-Martin JA, Bendell J, et al. Nivolumab alone and nivolumab plus ipilimumab in recurrent small-cell lung cancer (CheckMate 032): a multicentre, open-label, phase 1/2 trial. Lancet Oncol 2016;17:883–895. [DOI] [PubMed] [Google Scholar]
- 11.Gadgeel SM, Pennell NA, Fidler MJ, et al. Phase II Study of Maintenance Pembrolizumab in Patients with Extensive-Stage Small Cell Lung Cancer (SCLC). J Thorac Oncol 2018;13:1393–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ott PA, Elez E, Hiret S, et al. Pembrolizumab in Patients With Extensive-Stage Small-Cell Lung Cancer: Results From the Phase Ib KEYNOTE-028 Study. J Clin Oncol 2017;35:3823–3829. [DOI] [PubMed] [Google Scholar]
- 13.Rudin CM, Awad MM, Navarro A, et al. KEYNOTE-604: Pembrolizumab (pembro) or placebo plus etoposide and platinum (EP) as first-line therapy for extensive-stage (ES) small-cell lung cancer (SCLC). Journal of Clinical Oncology 2020;38:9001–9001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leal T, Wang Y, Dowlati A, et al. Randomized phase II clinical trial of cisplatin/carboplatin and etoposide (CE) alone or in combination with nivolumab as frontline therapy for extensive-stage small cell lung cancer (ES-SCLC): ECOG-ACRIN EA5161. Journal of Clinical Oncology 2020;38:9000–9000. [Google Scholar]
- 15.Reck M, Luft A, Szczesna A, et al. Phase III Randomized Trial of Ipilimumab Plus Etoposide and Platinum Versus Placebo Plus Etoposide and Platinum in Extensive-Stage Small-Cell Lung Cancer. J Clin Oncol 2016;34:3740–3748. [DOI] [PubMed] [Google Scholar]
- 16.Ready N, Farago AF, de Braud F, et al. Third-Line Nivolumab Monotherapy in Recurrent SCLC: CheckMate 032. J Thorac Oncol 2019;14:237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iams WT, Porter J, Horn L. Immunotherapeutic approaches for small-cell lung cancer. Nat Rev Clin Oncol 2020;17:300–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barretina J, Caponigro G, Stransky N, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012;483:603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang W, Soares J, Greninger P, et al. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res 2013;41:D955–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ackermann CJ, Reck M, Paz-Ares L, et al. First-line immune checkpoint blockade for advanced non-small-cell lung cancer: Travelling at the speed of light. Lung Cancer 2019;134:245–253. [DOI] [PubMed] [Google Scholar]
- 23.Mahoney KM, Freeman GJ, McDermott DF. The Next Immune-Checkpoint Inhibitors: PD-1/PD-L1 Blockade in Melanoma. Clin Ther 2015;37:764–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.George J, Lim JS, Jang SJ, et al. Comprehensive genomic profiles of small cell lung cancer. Nature 2015;524:47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou H, Dong J, Guo L, et al. The prognostic value of B7-H6 in esophageal squamous cell carcinoma. Sci Rep 2019;9:18122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou Y, Xu Y, Chen L, et al. B7-H6 expression correlates with cancer progression and patient's survival in human ovarian cancer. Int J Clin Exp Pathol 2015;8:9428–9433. [PMC free article] [PubMed] [Google Scholar]
- 27.Chen L, Feng J, Xu B, et al. B7-H6 expression in human hepatocellular carcinoma and its clinical significance [corrected]. Cancer Cell Int 2018;18:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cao Y, Huo L, Zhou L, et al. Expression of B7-H6 in chronic myeloid leukemia and its clinical significance. Int J Clin Exp Pathol 2019;12:568–575. [PMC free article] [PubMed] [Google Scholar]
- 29.Sun J, Tao H, Li X, et al. Clinical significance of novel costimulatory molecule B7-H6 in human breast cancer. Oncol Lett 2017;14:2405–2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang X, Zhang G, Qin Y, et al. B7-H6 expression in non-small cell lung cancers. Int J Clin Exp Pathol 2014;7:6936–6942. [PMC free article] [PubMed] [Google Scholar]
- 31.Fridman WH, Zitvogel L, Sautes-Fridman C, et al. The immune contexture in cancer prognosis and treatment. Nat Rev Clin Oncol 2017;14:717–734. [DOI] [PubMed] [Google Scholar]
- 32.Kim HS, Lee JH, Nam SJ, et al. Association of PD-L1 Expression with Tumor-Infiltrating Immune Cells and Mutation Burden in High-Grade Neuroendocrine Carcinoma of the Lung. J Thorac Oncol 2018;13:636–648. [DOI] [PubMed] [Google Scholar]
- 33.Muppa P, Parrilha Terra SBS, Sharma A, et al. Immune Cell Infiltration May Be a Key Determinant of Long-Term Survival in Small Cell Lung Cancer. J Thorac Oncol 2019;14:1286–1295. [DOI] [PubMed] [Google Scholar]
- 34.Newman AM, Steen CB, Liu CL, et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol 2019;37:773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wouters MCA, Nelson BH. Prognostic Significance of Tumor-Infiltrating B Cells and Plasma Cells in Human Cancer. Clin Cancer Res 2018;24:6125–6135. [DOI] [PubMed] [Google Scholar]
- 36.Wculek SK, Cueto FJ, Mujal AM, et al. Dendritic cells in cancer immunology and immunotherapy. Nat Rev Immunol 2020;20:7–24. [DOI] [PubMed] [Google Scholar]
- 37.Brandt CS, Baratin M, Yi EC, et al. The B7 family member B7-H6 is a tumor cell ligand for the activating natural killer cell receptor NKp30 in humans. J Exp Med 2009;206:1495–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carney DN, Gazdar AF, Bepler G, et al. Establishment and identification of small cell lung cancer cell lines having classic and variant features. Cancer Res 1985;45:2913–2923. [PubMed] [Google Scholar]
- 39.Gazdar AF, Carney DN, Nau MM, et al. Characterization of variant subclasses of cell lines derived from small cell lung cancer having distinctive biochemical, morphological, and growth properties. Cancer Res 1985;45:2924–2930. [PubMed] [Google Scholar]
- 40.Gazdar AF, Bunn PA, Minna JD. Small-cell lung cancer: what we know, what we need to know and the path forward. Nat Rev Cancer 2017;17:765. [DOI] [PubMed] [Google Scholar]
- 41.Wooten DJ, Groves SM, Tyson DR, et al. Systems-level network modeling of Small Cell Lung Cancer subtypes identifies master regulators and destabilizers. PLoS Comput Biol 2019;15:e1007343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rudin CM, Poirier JT, Byers LA, et al. Molecular subtypes of small cell lung cancer: a synthesis of human and mouse model data. Nat Rev Cancer 2019;19:289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pedersen N, Pedersen MW, Lan MS, et al. The insulinoma-associated 1: a novel promoter for targeted cancer gene therapy for small-cell lung cancer. Cancer Gene Ther 2006;13:375–384. [DOI] [PubMed] [Google Scholar]
- 44.Granier C, Karaki S, Roussel H, et al. [Cancer immunotherapy: Rational and recent breakthroughs]. Rev Med Interne 2016;37:694–700. [DOI] [PubMed] [Google Scholar]
- 45.Sanmamed MF, Chen L. A Paradigm Shift in Cancer Immunotherapy: From Enhancement to Normalization. Cell 2018;175:313–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takamori S, Takada K, Azuma K, et al. Prognostic Impact of PD-L2 Expression and Association with PD-L1 in Patients with Small-cell Lung Cancer. Anticancer Res 2018;38:5903–5907. [DOI] [PubMed] [Google Scholar]
- 47.Jiang T, Wu W, Zhang H, et al. High expression of B7-H6 in human glioma tissues promotes tumor progression. Oncotarget 2017;8:37435–37447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu MR, Zhang T, DeMars LR, et al. B7H6-specific chimeric antigen receptors lead to tumor elimination and host antitumor immunity. Gene Ther 2015;22:675–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hua CK, Gacerez AT, Sentman CL, et al. Development of unique cytotoxic chimeric antigen receptors based on human scFv targeting B7H6. Protein Eng Des Sel 2017;30:713–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gacerez AT, Hua CK, Ackerman ME, et al. Chimeric antigen receptors with human scFvs preferentially induce T cell anti-tumor activity against tumors with high B7H6 expression. Cancer Immunol Immunother 2018;67:749–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu MR, Zhang T, Gacerez AT, et al. B7H6-Specific Bispecific T Cell Engagers Lead to Tumor Elimination and Host Antitumor Immunity. J Immunol 2015;194:5305–5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang B, Sun J, Yao X, et al. Knockdown of B7H6 inhibits tumor progression in triple-negative breast cancer. Oncol Lett 2018;16:91–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pesce S, Tabellini G, Cantoni C, et al. B7-H6-mediated downregulation of NKp30 in NK cells contributes to ovarian carcinoma immune escape. Oncoimmunology 2015;4:e1001224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Semeraro M, Rusakiewicz S, Minard-Colin V, et al. Clinical impact of the NKp30/B7-H6 axis in high-risk neuroblastoma patients. Sci Transl Med 2015;7:283ra255. [DOI] [PubMed] [Google Scholar]
- 55.Wu F, Wang J, Ke X. Knockdown of B7-H6 inhibits tumor progression and enhances chemosensitivity in B-cell non-Hodgkin lymphoma. Int J Oncol 2016;48:1561–1570. [DOI] [PubMed] [Google Scholar]
- 56.Rusakiewicz S, Perier A, Semeraro M, et al. NKp30 isoforms and NKp30 ligands are predictive biomarkers of response to imatinib mesylate in metastatic GIST patients. Oncoimmunology 2017;6:e1137418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen XJ, Shen J, Zhang GB, et al. B7-H6 protein expression has no prognostic significance in human gastric carcinoma. Pathol Oncol Res 2014;20:203–207. [DOI] [PubMed] [Google Scholar]
- 58.Gutierrez-Franco J, Hernandez-Gutierrez R, Bueno-Topete MR, et al. Characterization of B7H6, an endogenous ligand for the NK cell activating receptor NKp30, reveals the identity of two different soluble isoforms during normal human pregnancy. Immunobiology 2018;223:57–63. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.