Summary
In ribosomal DNA (rDNA) repeats, sequences encoding small subunit (SSU) rRNA precede those encoding large subunit (LSU) rRNAs. Processing the composite transcript and subunit assembly requires >100 subunit-specific nucleolar assembly factors (AFs). To investigate the functional organization of the nucleolus, we localized AFs in S. cerevisiae in which the rDNA axis was “linearized” to reduce its dimensionality, thereby revealing its coaxial organization. In this situation rRNA synthesis and processing continue. The axis is embedded in an inner layer/phase of SSU AFs that is surrounded by an outer layer/phase of LSU AFs. When subunit production is inhibited, subsets of AFs differentially relocate between the inner and outer layers, as expected if there is a cycle of repeated relocation whereby “latent” AFs become “operative” when recruited to nascent subunits. Recognition of AF cycling and localization of segments of rRNA make it possible to infer the existence of assembly intermediates that span between the inner and outer layers, and to chart the cotranscriptional assembly of each subunit. AF cycling also can explain how having more than one protein phase in the nucleolus makes possible “vectorial 2-phase partitioning” as a driving force for relocation of nascent rRNPs. Since nucleoplasmic AFs are also present in the outer layer, we propose that critical surface remodeling occurs at this site, thereby partitioning subunit precursors into the nucleoplasm for post-transcriptional maturation. Comparison to observations on higher eukaryotes shows that the coaxial paradigm is likely to be applicable for the many other organisms that have rDNA repeats.
Keywords: Nucleolus, yeast, rDNA, ribosomal subunit biogenesis, assembly factors, nucleolar subcompartments, nucleolar domain separation, 2-phase partitioning
Graphical Abstract
eTOC Blurb
Tartakoff et al. document the coaxial organization of the yeast nucleolus in which the rDNA axis is surrounded by two dynamic layers/phases of subunit assembly factors that alternate between latent and operative states during each transcription cycle.
Introduction
Transcription of the ~ 150 rDNA repeats on chromosome XII in S. cerevisiae produces the composite RNA precursor of ribosomal subunits (SSUs, LSUs). The primary transcript undergoes folding, modification, processing and compaction, along with assembly of ribosomal proteins, within the specialized environment of the nucleolus1–4. The nascent transcript associates with many factors (AFs) that promote its assembly and - as we suggest - govern the phase compatibility and vectorial transport of maturing particles. The large majority of AFs are implicated in assembly of only one type of subunit (SSU, LSU). We therefore refer to SSU AFs and to LSU AFs.
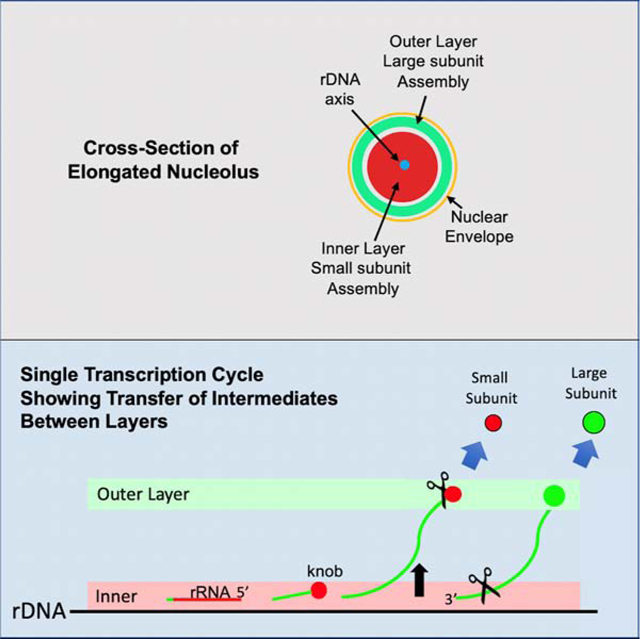
The 5’ segment of each rDNA repeat gives rise to SSU rRNA, followed by an internal transcribed spacer (ITS1) and the segment encoding LSU rRNAs (Figure 1A,B). Terminal “knobs” form at the 5’ ends of these nascent transcripts, as viewed in EM spreads5. These knobs are precursors of the “processome” that includes selected SSU AFs and ribosomal proteins, as well as snoRNAs6–8. Cleavage in ITS1 and release of the knobs are largely co-transcriptional in rapidly-growing yeast. Continued elongation, assembly into LSU intermediates, and cleavage within the 3’ external transcribed spacer at site Bo then releases LSU precursor particles (Figure 1B). Assembly intermediates have been purified and characterized at near-atomic resolution2, 4, 9.
Figure 1. In Arrested Cells the rDNA Axis is Colinear with Hmo1 and Processing of pre-rRNA Continues.
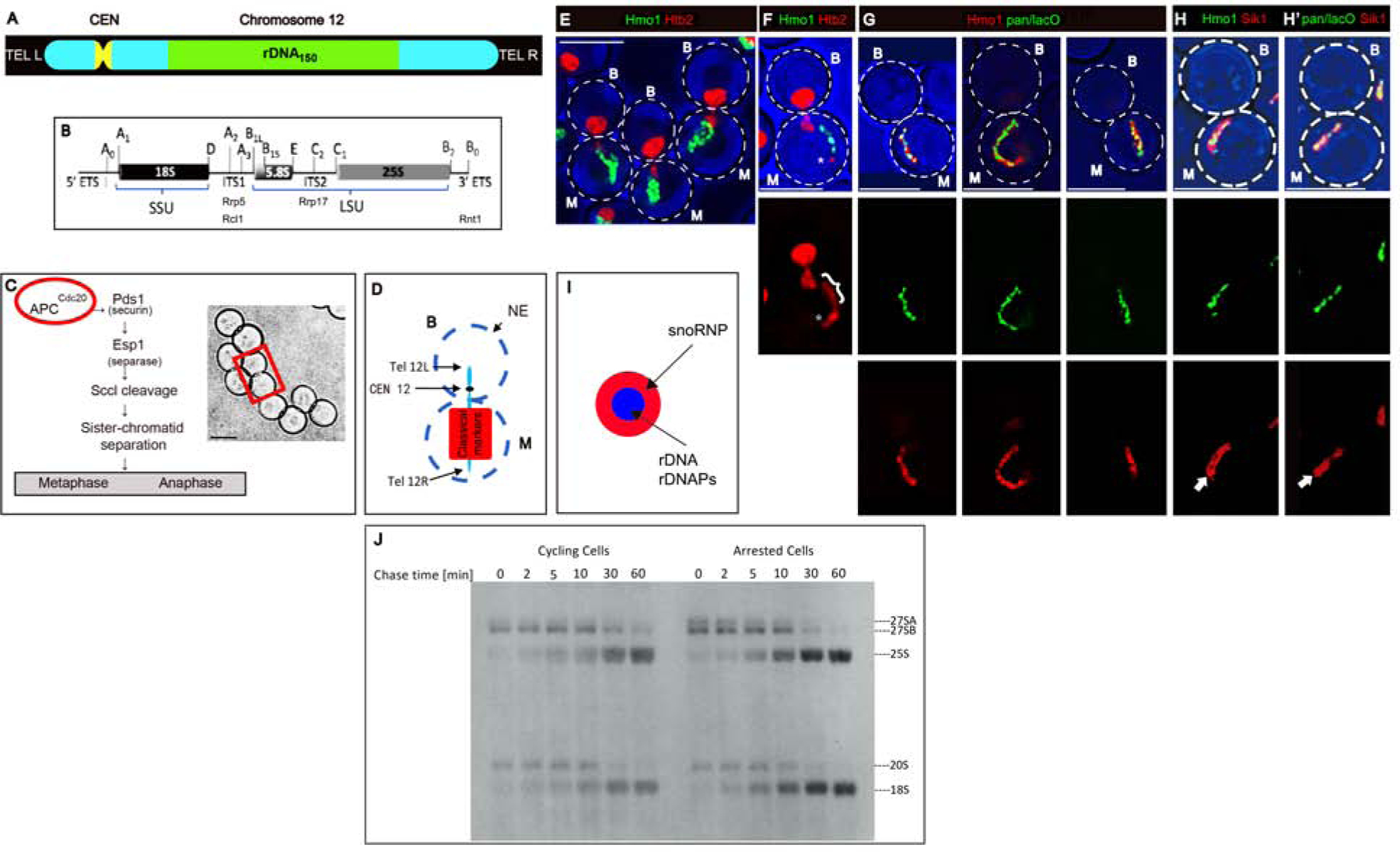
A) Organization of chromosome XII.
B) A single rDNA repeat, indicating segments that code for SSU and LSU rRNAs and the sites of binding (Rrp5) or potential cleavage (Rcl1, Rrp17, Rnt1).
C) Left: Overview of the consequences of activation of the anaphase promoting complex. Right: Arrested cells. The rectangle encloses a typical mother/bud pair.
D) Diagram of the position of chromosome XII and classical markers in arrested cells24. For all panels, M: mother; B: bud. The scale bar in all figures is 5 microns.
E) Projected image of arrested cells that express the histone, Htb2-mRFP, and Hmo1-GFP. Strain: ATY10569.
F) Single image plane of an arrested cell showing that the elongated rDNA segment has a Htb2-mRFP at the distal extremity (*). The bracket designates the Hmo1-GFP-positive segment. Strain: ATY10569.
G) Single image planes of arrested cells that express pan-lacO-tagged rDNA (green) and Hmo1-Apple (red). Strain: ATY10682.
H) Comparison of the filamentous Hmo1-Apple (H) and pan-lacO (H’) with the broader Sik1/Nop56-mRFP signal. In each case, the Sik1-mRFP domain includes a non-fluorescent central element (arrows) that approximately coincides with rDNA. Strains: ATY10342, ATY10747.
I) Scheme indicating that rDNA and rDNAPs define an axis internal to snoRNP proteins.
J) To compare the synthesis and turnover of pre-rRNAs in cycling cells and arrested cells, metaphaseCdc20 cells carrying the URA3 plasmid, pRS316, were pre-grown in medium lacking methionine and uracil. Half of the culture was transferred to synthetic media including methionine for 3 hours. Both samples were pulse-labeled for 10 min with [5.6-3 H]-uridine and chased with an excess of unlabeled uridine for the indicated times. Pre-rRNAs and mature rRNA species are labeled. Due to long pulse, the earliest pre-rRNA processing intermediates (35S, 32S) had already been processed to intermediates. Strain: ATY10402 [pRS316].
Related to Figure S1.
The intermixed subcompartments of nucleoli are thought to constitute distinct protein phases10, 11. They have traditionally been referred to as the FC (fibrillar center), the surrounding DFC (dense fibrillar component), and the more peripheral GC (granular component). rDNA transcription occurs along the FC/DFC interface, and the DFC and GC are considered responsible for “RNA processing” and “subunit assembly” or “late processing,” respectively. Prior to the localization of multiple AFs, these insightful conjectures have lacked an extensive biochemical counterpart12–17.
In nucleoli that are minimally active in transcribing rDNA - e.g. extrachromosomal nucleoli of Xenopus oocytes - selected “marker” AFs, while remaining contiguous, become segregated from each other18. Segregation can also be detected upon inhibition of rRNA synthesis or treatment with chemotherapeutic agents. It is not known whether segregated domains have the same composition as subcompartments in cells that are making subunits. In yeast, some subregions of the nucleolus have also been detected, e.g.19.
We have used yeast AFs as potential markers of subcompartments, to ascribe functions to subcompartments, and to chart the itineraries of subunit precursors. We conclude that the axis is surrounded by an inner layer/phase and an outer layer/phase that are enriched in SSU AFs and LSU AFs, respectively. This coaxial model is based on observations that 1) Many AFs, processing nucleases, and successive segments of rRNA localize to distinct layers, 2) The distribution of many AFs between layers depends on whether new subunits are being assembled, and 3) Stratification of AFs is orthogonal to the polarity of rDNA.
Our approach takes into consideration much of the in vivo complexity of subunit production. We also integrate biochemical and structural studies of subunit maturation with the concept of “vectorial phase partitioning,” according to which particulate intermediates can be stable only if their surface matches the surrounding milieu. Correspondingly, cotranscriptional events endow particles with surfaces that are densely-covered with AFs. As post-transcriptional steps in subunit genesis entail further remodeling, we hypothesize that the make-up of their surfaces is critical for entry into the nucleoplasm.
Results
The rDNA Axis is Colinear with Hmo1
To study the structure of the nucleolus, we arrested the cell cycle at metaphase using cells that carry a methionine-repressible MET3-CDC20 chromosomal integrant. Such cells are grown in methionine-free medium prior to reincubation in methionine-containing medium to deplete Cdc20 (Figure 1C). After 3 hr, DNA has replicated, the nuclear envelope (NE) extends across the bud neck, bulk chromatin transits between the mother and bud, and the bud is nearly as large as the mother20. To detect rDNA, we used strains in which each of the > 100 rDNA repeats is adjacent to lacO repeats (“pan-lacO” rDNA). We also visualized fluorescent fusions of proteins implicated in rDNA organization, including the HMG-like protein, Hmo1, that associates with rDNA21–23.
Nucleolar markers remain in the mother domain in arrested cells (Figure 1D)24. Moreover, especially when chromatin (Htb2-mRFP) is in the bud, rDNA and Hmo1 form an elongated filament in the mother domain (Figure 1E), whose length appears comparable to the length of rDNA in cycling cells25. Histones (Htb2-mRFP) can be detected along the gnarled Hmo1-GFP-positive filament, and occasionally highlights the end of the filament, presumably near the right telomere of chromosome XII (Figure 1A, F). The pan-lacO signal and Hmo1-Apple are nearly coincident (Figure 1G).
In arrested cells, Hmo1-Apple also aligns with other rDNAPs (rDNA-associated proteins): topoisomerases (Top1, Top2), proteins that contribute to rDNA recombination (Csm1, Lrs4), condensins (Smc4, Ycs4), a cohesin (Scc3), Fob1 and the rDNA transcription factors, Rrn6, Rrn7 and Rrn9 (Figure S1A and not shown).
In cycling cells that express the mRFP-tagged snoRNP protein, Sik1/Nop56, the pan-lacO signal forms a few dots throughout the nucleolar crescent (Figure 2A, panels 1–2). The dots are likely cross-sections of the folded rDNA filament11, 26.
Figure 2. Nucleolar Assembly Factors Localize to Coaxial Layers.
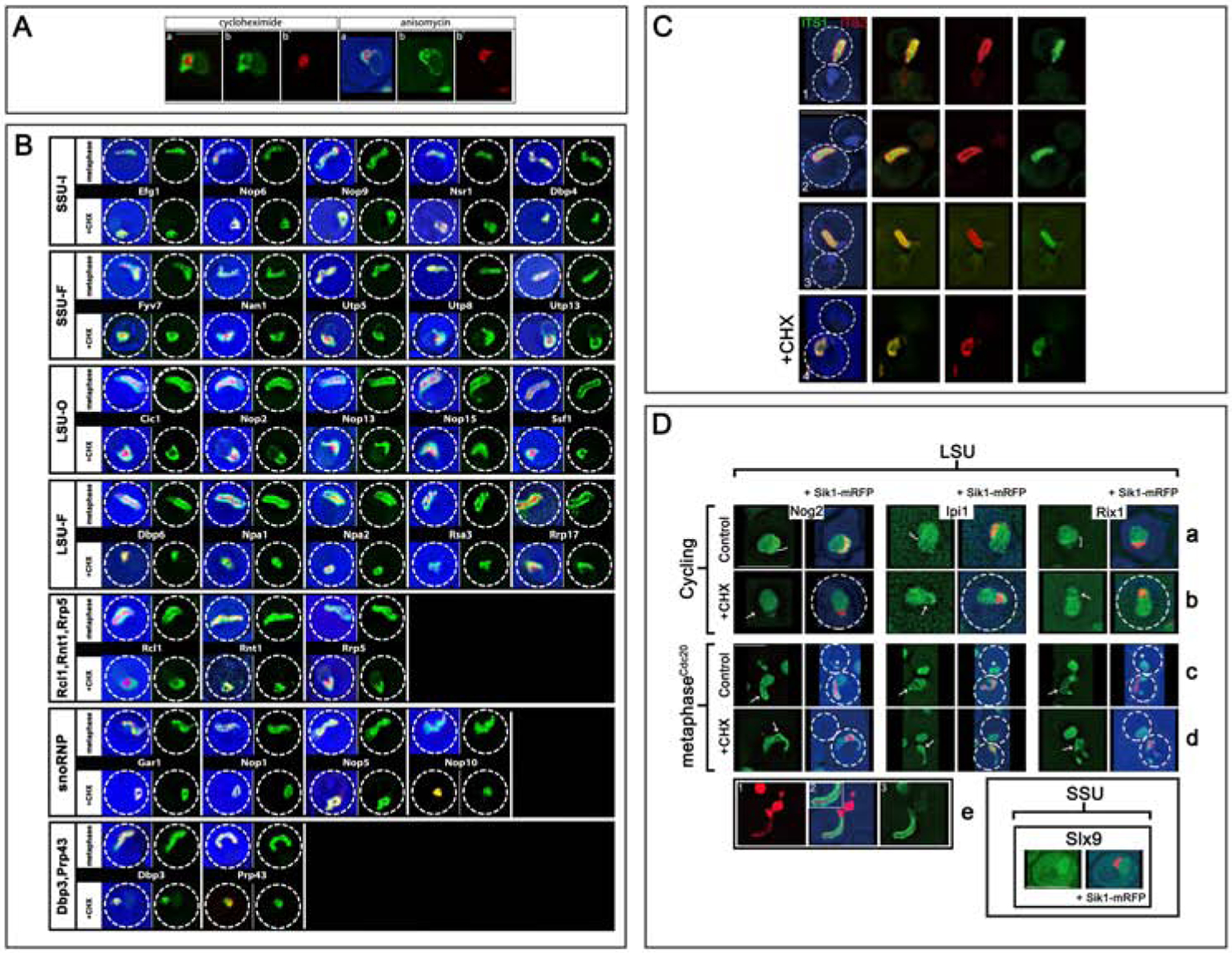
A) Distributions of AFs in Cycling Cells. In panels (1–5), Sik1-mRFP is included to define the crescent. Panels (1–2): pan-lacO dots are throughout the crescent. Panels (3–5) illustrate AFs that localize either to the crescent with a weak signal along the NE and a weak signal throughout the nucleoplasm (Mak11), to the nucleoplasm (Rea1), or exclusively to the crescent (Rrp9). In (3), the small boxes illustrate the single colors. Panel (6) illustrates Nmd3, that localizes largely to the cytoplasm, co-expressed with a marker of the NE/ER: mRFP-HDEL. Strains: ATY10659, ATY8300, ATY7833, ATY7838, ATY8119.
B) In arrested cells, the snoRNP, GFP-Nop1, colocalizes with Sik1-mRFP. The panels show the two colors separately or combined. Strain: ATY10412.
C) In arrested cells, the SSU AF, Efg1-GFP, colocalizes with Sik1-mRFP. Strain: ATY10383.
D) In arrested cells, the LSU AF, Mak11-GFP, surrounds Sik1-mRFP. Strain: ATY8297.
E) Comparison of the LSU AF, Ytm1-Apple, to the SSU AF, Utp30-GFP. (E’) comparison of Mak11-GFP to Utp30-CFP, all in arrested cells. Strains: ATY10589, ATY10400.
F) Comparison of LSU AFs, Ytm1-Apple and Mak11-GFP, to markers of the NE/ER: GFP-HDEL and mRFP-HDEL, all in arrested cells. Strains: ATY10717, ATY10722.
G) Scheme of the coaxial organization of the nucleolar cable.
H/I) Schematic of Distributions of AFs in Cells that are Making Subunits.
Left) This two-phase diagram (T-diagram) depicts the inner and outer layers/phases as two separated horizontal bands (pink, green). Inner layer AFs are snoRNP proteins, Rpa subunits, Dbp3, Prp43, SSU AFs, Rrp5, and Rnt1. Among outer layer AFs are Rcl1 and many LSU AFs, including Rrp17. SSU AFs are red and LSU AFs are green. Proteins that contribute to both subunits have both colors.
Right) This related diagram includes the nucleoplasmic compartment in blue and the axis in yellow. As is described later in this text, multiple AFs that are conspicuous in the nucleoplasm (dark blue) are also detected in the outer layer for arrested cells. Htb2 is found both along the axis and throughout the nucleoplasm. By contrast, Hmo1 (green) is associated with the axis but is not detected in the nucleoplasm.
Related to Figure S2.
rRNA Synthesis and Processing Continue in Arrested Cells
To learn whether rRNA transcription continues in arrested cells, we performed 3H-uridine pulse-chase experiments. As shown in Figure 1J, synthesis and cleavage of rRNA do continue, as compared to controls. There is no indication of alternative cleavage at site A3, as occurs upon stress or other conditions leading to slow growth27.
Nucleolar Assembly Factors and rRNA Segments Localize to Coaxial Layers
In the following sections we localize a physically and functionally diverse group of AFs, both when they are operative (when associated with nascent subunits) and also when they are latent (when they are not associated with nascent subunits) (Figures 2–3, Figure S2). Latency has seldom been investigated28; however, we envisage it as a recurrent intermediate during each cycle of rDNA transcription. We have approximated this state by studying cells treated with cycloheximide. These binary distributions (Table 1) provide a basis for reasoning with regard to sequential AF engagement during rRNA transcription. We began by focusing on “nucleolar” AFs, i.e. GFP-tagged proteins that are concentrated in the nucleolar crescent and are much less evident in the nucleoplasm and cytoplasm.
Figure 3. Localizations of Nucleolar Assembly Factors and rRNA.
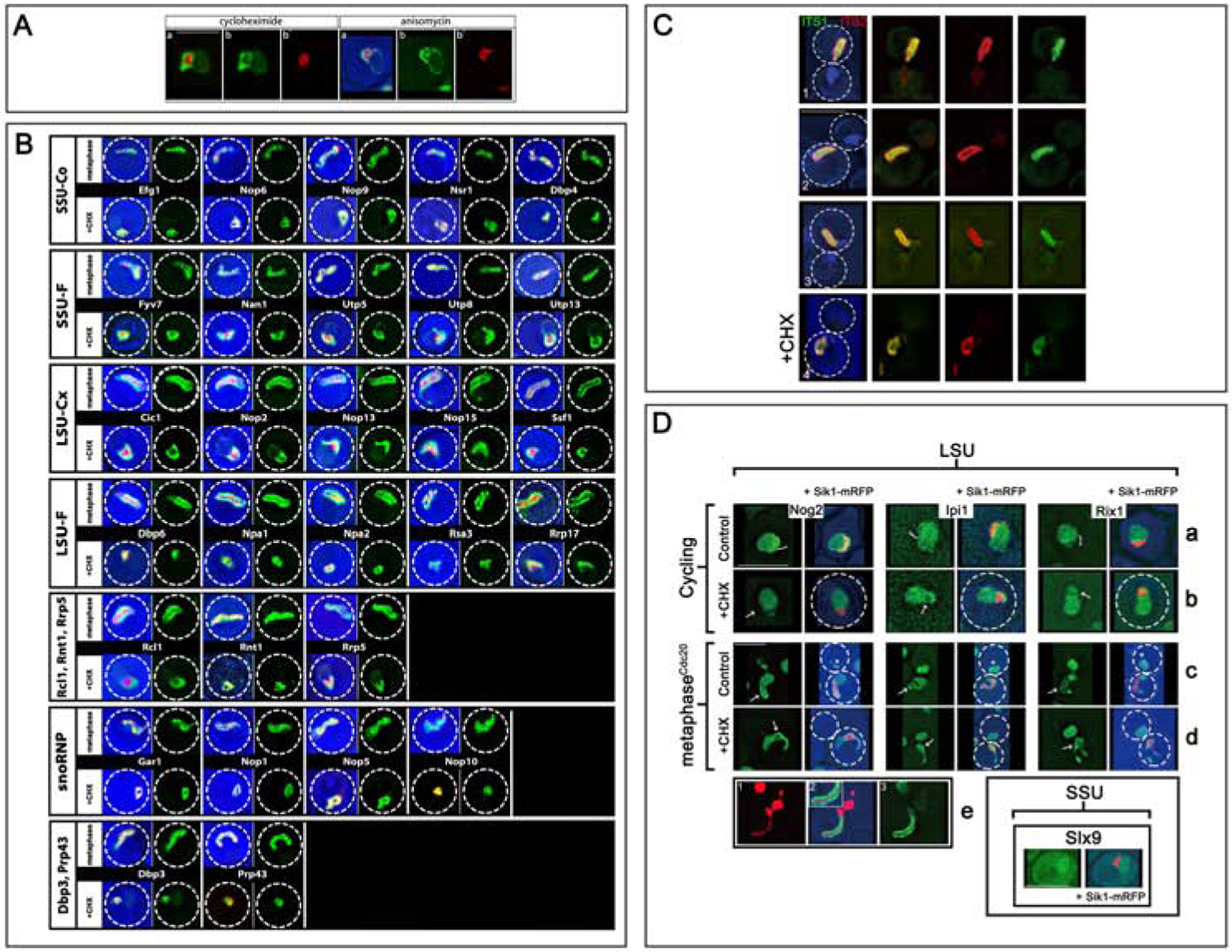
A) Cycling cells expressing Mak11-GFP and Sik1-mRFP were treated with cycloheximide or anisomycin for 30 min. Note the conspicuous domain separation of the two colors, with GFP surrounding the red signal. Before treatment, the two colors were extensively intermixed. Note that a faint GFP signal extends around the nucleus. Strain: ATY8112.
B) Cells expressing the indicated AF-GFP fusions, as well as Sik1-mRFP, were either arrested (metaphase) or, when cycling, were treated with cycloheximide (+ CHX). In each case, we include both a 3-color image and an image of the GFP-tagged protein by itself. For the arrested cells, only the mother domain is included. The dotted circle indicates the cell perimeter in all figures. 16 further examples of SSU-F and LSU-Ou AFs are in Figure S2.
C) To localize different portions of rRNA in arrested cells, we used fluorescent in situ hybridization. As illustrated for three representative cells (1–3), sequences upstream of cleavage site A2 in ITS1 emphasize the inner layer (green signal), while sequences within ITS2 have more external localization (red signal). The blue signal is DAPI. The final row of images (4) illustrates the impact of cycloheximide (30 min) before fixation. Note that the two signals have become coincident. Strain: ATY10402.
D) Nucleoplasmic AFs fill the outer layer but chromatin is absent. Rows (a-d): The distributions of three LSU AFnpls (Nog2, Ipi1, Rix1) are illustrated +/− Sik1-mRFP. Each protein is abundant in the nucleoplasm and can also be detected (weakly) in the nucleolar crescent (bracket) where the interface with the nucleoplasm is often highlighted (row (a)). The SSU AF, Slx9, that is also present in the cytoplasm, is included in the separate panel in the lower right. Upon arrest (row c), Nog2, Ipi1 and Rix1 continue to be visible throughout the nucleoplasm (asterisks) and along the outer layer (arrows). They do not coincide with Sik1-mRFP. When cycling cells are treated with cycloheximide (row (b) vs row (a)) these LSU AFnpls still fill the nucleoplasm. It is unclear whether they become depleted from the region occupied by Sik1-mRFP (arrows).
In arrested cells, the nucleoplasmic signal persists, as does highlighting of the outer layer (arrows). It is not obvious that the intensity of this highlighting is accentuated by comparison to controls (row d vs row c). There is little or no overlap with the condensed Sik1-mRFP. Strains: ATY8105, ATY8126, ATY8280, ATY10770, ATY10774, ATY10785. Row (e). Cells expressing Mak11-GFP and Htb2-mRFP were arrested and imaged. Note the elongated domain (in the mother) in which the Htb2-mRFP-labeled axis is flanked by the GFP signal. Strain: ATY10157.
Related to Figures S2 and S5.
Table 1.
Distributions of Assembly Factors
| Specificity Identity | Property subset | Site of Accumulation | Names of Factorsx | Suggested Function | |
|---|---|---|---|---|---|
| Operative* | Latent** | ||||
| Small Subunit AF | Constitutive SSU-In | Inner layer | Inner layer | Dbp4, Efg1, Nop6, Nop9, Nsr1 | Retention in the inner layer |
| Facultative SSU-F | Inner layer | Outer layer | Dbp8, Dhr2, Fyv7, Rrp7, Utp5A, Utp6B, Utp8A, Utp9A, Utp13B, Utp15A, Utp17A/Nan1, Utp18B, Utp19/Noc4, Utp21dB, Utp25, Utp30 | SSU knob formation | |
| Large Subunit AF | Constitutive LSU-Ou | Outer layer | Outer layer | Brx1, Cic1/Nsa3, Fpr3, Mak5, Mak11, Mrt4, Noc1, Noc2, Noc3, Nog1, Nop2, Nop4, Nop7, Nop13, Nop15, Nop16, Puf6, Rix7, Rrp1, Rrp17+, Ssf1, Tif6, Ytm1 | LSU knob formation |
| Facultative LSU-F | Outer layer | Inner layer | Dbp6, Npa1/Urb1, Npa2/Urb2, Rsa3 | Retention in the inner layer | |
| Endo-nucleases | Constitutive | Outer layer | Outer layer | Rcl1 | Release of SSU knobs ? |
| Inner layer | Inner layer | Rnt1 | Removal of full-length transcripts | ||
| Rrp5 | Facultative | Inner layer | Outer layer | Rrp5 | Binds ITS1 |
| snoRNP | Constitutive | Inner layer | Inner layer | Gar1, Nop1, Nop5/Nop58, Nop10, Nop56/Sik1 | Modify rRNA |
| Various | Constitutive | Inner layer | Inner layer | Dbp3, Prp43 | Shared steps |
This table summarizes the distributions of assembly factors, both in cells that are making subunits and in cells that have been treated with cycloheximide.
The suffixes have the following meaning: -In (constitutively at the inner layer), -Ou (constitutively along the outer layer), -F (facultative, i.e. redistributes upon addition of cycloheximide).
MetaphaseCdc20 cells. Cells were transferred into medium with methionine for 3 hr. By examining cells in which Sik1-mRFP was elongated, images were classified as to whether the GFP signal coincided with Sik1-mRFP.
Cycling cells after treatment with cycloheximide for 30 min. Images were classified according to whether the GFP signal defined a cortical arc (outer layer) or overlapped extensively with the condensed Sik1-mRFP (inner layer).
The superscripts designate SSU AFs that are part of the UTP-A or UTP-B subcomplexes.
Rrp17 is an exonuclease that resects the 3’ end of ITS2.
In cycling cells that express both the LSU AF, Mak11-GFP, and Sik1-mRFP, the distribution of these AFs generally overlaps throughout the nucleolar crescent (Figure 2A panel 3), although inhomogeneities can be seen, perhaps due to lack of transcription of some rDNA repeats29, 30. Panels (3–6) of Figure 2A illustrate the distributions of additional AFs that localize to the crescent, the nucleoplasm, or to the cytoplasm.
To follow the distribution of GFP-tagged AFs more accurately we used arrested cells in which Sik1-mRFP in the mother domain is elongated (Figure 1H). As explained below, it is surrounded by an outer layer. A layered distribution of AFs can also be seen as cycling cells progress through metaphase (not shown).
Individual AFs that function in assembly of both 40S and 60S subunits localize to the inner layer (Figure 2B, Figure 3B). These AFs include all snoRNP proteins that we have examined (5/5), the two DExD/H-box proteins, Dbp3 and Prp43, and Rrp5, that binds ITS131.
Surprisingly, subunit-specific AFs have either of two distinct localizations (inner vs outer layers) and these localizations correspond to the two subunits:
SSU AFs that bind the 5’-ETS or to SSU sequences (Table S1) overlap extensively with the inner layer that is highlighted by Sik1-mRFP (21/21 examined) (Figure 2C, Figure 3B, Figure S2, Tables 1A/1B).
LSU AFs that associate with each domain of 25S rRNA2, 32, 33 are along the elongated outer layer that flanks the inner layer (27/27 examined) (Figure 2D–E’, Figure 3B, Figure S2). The characteristic gap in their fluorescent signal can be tortuous but is visible in > 75% of cells for which Sik1-mRFP (or Utp30) is elongated. In further affirmation of the importance of the outer layer for LSU assembly, the Rrb1 chaperone of a LSU ribosomal protein is also concentrated in the outer layer34 (Figure S2). The outer layer aligns closely with the NE (Figure 2F).
The distributions of key nucleases are characteristically different from each other: Rcl1 (that can cut at site A2 in vitro35) and Rrp17 (that resects sequences within ITS236) are along the outer layer. Rnt1 (that cleaves site Bo as well as precursors of U3 and other snoRNAs) localizes to the inner layer (Figure 3B)37. Utp24 cuts at A1 and may cut at A238, 39; however, we have not localized it (see STAR Methods).
There thus are four coaxial layers (Figure 2G/H).
In parallel in situ hybridization studies of arrested cells, we find that rRNA sequences upstream of A2 in ITS1 localize primarily to the inner layer, while sequences from ITS2 localize to the outer layer - Figure 3C (1–3).
Organization of the Nucleolus When Subunits are not Produced
To investigate the importance of ongoing subunit assembly for rDNA organization and stratification of AFs, we eliminated synthesis of ribosomal proteins for 30 minutes using cycloheximide, an inhibitor of translation elongation whose impact is rapid and reversible. It seems likely that addition of cycloheximide to growing cells will perturb most stages of maturation of nascent rRNPs since ribosomal proteins are added during many steps of assembly2, 4, 40–42. Correspondingly, 35S precursor rRNA is known to be present upon treatment of cells with cycloheximide but rRNA processing intermediates appear to be absent40, 41, 43, 44.
In arrested cells, cycloheximide causes the pan-lacO signal and multiple rDNAPs to condense within 30 min, generally forming a cluster (Figure S1B, C). Moreover, the condensed pan-lacO (and Hmo1) signal retains contact with the Sik1, which appears reduced in size (Figure S1). Deletion of topoisomerase 1 also condenses the pan-lacO signal (Figure S1D).
To identify the latent distributions of nucleolar AFs, we treated cycling cells that expressed the LSU AF, Mak11-GFP, and Sik1-mRFP with cycloheximide for 30 min. By contrast to their intermixing in cycling cells (Figure 2A panel 3), after treatment Mak11-GFP forms a massive arc outside the compacted inner mass of Sik1-mRFP (Figure 3A). Within 30 min ~ 80% of cells are affected and this distribution continues for at least 3 hr. Comparable outer vs inner domain separation is seen when cells are exposed to the translation elongation inhibitor, anisomycin45 (Figure 3A) and in arrested cells exposed to cycloheximide (Figure S3).
Since AFs turn over slowly, we expect that cycloheximide has only minimal impact on their abundance46. It therefore seems plausible to attribute the effects of cycloheximide primarily to the sudden absence of newly-synthesized ribosomal proteins. Inhibition of both translation and rRNA synthesis (by removal of sugar47) also causes segregation of Mak11-GFP vs Sik1-mRFP, but without having the green signal enclose the red. Inhibition of synthesis of ribosomal proteins and rRNA synthesis (with rapamycin40) or inhibition of RNA polymerase 1 (with BMH-2148), causes lesser and less uniform color separation over 30–60 min. Similar observations were made using thiolutin, that inhibits RNA polymerases 1, 2 and 349 (not shown).
The following paragraphs describe the generality of the impact of cycloheximide on AFs, as summarized in Table 1. Two subsets of AFs are designated either with the suffix, - Ou (if they remain outside/outer layer +/− cycloheximide) or the suffix, - In (if they remain internal/inner layer +/− cycloheximide). The suffix -F (facultative) designates AFs that relocate from the inner to the outer layer or vice versa when cycloheximide is added. Images of the distributions of 43 AFs are in Figure 3B and Figure S2. In no case does treatment with cycloheximide cause AFs to relocate to the chromatin-filled nucleoplasm or cytoplasm.
Each of the snoRNP proteins, as well as Dbp3 and Prp43 - that also are required to produce both SSU and LSU - remains associated with the inner layer. For the snoRNPs, this could ensure their ability to modify nascent segments of SSU and LSU rRNA50.
Two responses are seen for SSU AFs: Many of these factors relocate from the inner layer to the outer layer, while a limited subset (Dbp4, Efg1, Nop6, Nop9, Nsr1) remains along the inner layer.
An inverse pair of responses is seen for LSU AFs - a limited subset relocates to the inner layer (Dbp6, Npa1/Urb1, Npa2/Urb2, Rsa3), while most remain along the outer layer. The chaperone, Rrb1, also remains along the outer layer (Figure S2).
Rcl1 and Rrp17 remain along the outer layer, while Rnt1 remains in the inner layer. By contrast, Rrp5 is comparable to SSU-F AFs, relocating to the outer layer.
Upon treatment of arrested cells, the in situ hybridization signals for ITS1 and ITS2 become nearly coincident (Figure 3C (row 4, + CHX)) and are condensed, as for rDNAPs (Figure S3B). Such colocalization is expected since precursor 35S rRNA is not processed in this situation.
The challenge - which we address in the Discussion - is to fit these observations on the distributions of both AFs and rRNA into a model of the contranscriptional itinerary followed by subunit precursors.
Nucleoplasmic Assembly Factors are in the Outer Layer
In cycling cells, a subset of AFs (AFnpls) is conspicuous throughout the nucleoplasm and is less evident in the nucleolar crescent. This group comprises AFs that associate with preribosomes in the nucleolus and travel with them to the nucleoplasm. For the LSU this group includes the GTPases, Nog2/Nug2 and Nug1, and the dynein-like ATPase, Rea1, as well as Alb1, Arx1, Bud20, Ipi1 and Rix1 (Figure 3D, Figure S5). Judging from the literature, Ipi3, Rsa4 and Sda1 are also present2. The only SSU AF that we find to be conspicuous throughout the nucleoplasm is Slx9, that is also visible in the cytoplasm (Figure 3D - lower right).
The distributions of Ipi1, Nog2 and Rix1 in cycling cells are illustrated in Figure 3D (row a). In arrested cells (row c), these proteins are again visible throughout the nucleoplasm and, interestingly, along the outer layer. Indeed, all additional LSU AFnpls that we have followed (Arx1, Bud20, Nug1, Rea1) are also present both in the nucleoplasm and along the outer layer in arrested cells (Figure 2I and not shown).
Cycloheximide treatment has little impact on the distribution of Ipi1, Nog2 and Rix1 between the nucleolar crescent and nucleoplasm, both in cycling cells and after arrest (Figure 3D row (b) vs A and row (d) vs row (c), Figure S5). Arx1, Bud20 and Rea1 also are not obviously affected (not shown).
Interestingly - although AFnpls are both in the nucleoplasm and along the outer layer - the histone, Htb2, is not found along the outer layer (Figure 3D - panels (e), Figure 2I). Moreover, many AF’s that localize primarily to the outer layer (e.g. LSU-Ou AFs) are not visible in the nucleoplasm (Figure 2 and 3B, Figure S2).
Cytoplasmic Assembly Factors Do Not Relocate Upon Inhibition of Subunit Assembly
In a survey of cytoplasmic GFP-tagged AFs [Drg1L, Efl1L, Jjj1L, Lsg1L, Ltv1S, Ngl2L, Nmd3L, Nop8L, Rei L, Rio2S, Sdo1L and Yvh L] we see no indication of relocation into the nucleus upon addition of cycloheximide (Figure S5). [Superscripts indicate subunit specificity.]
Nucleolar Assembly Factors and rDNAPs Leave the Nucleolus
Since many AFs redistribute when subunit assembly is inhibited and since they may normally relocate during each cycle of transcription (see below), we asked whether AFs can normally access the entire nucleus. For this purpose, we developed an intranuclear shuttling assay based on the observation that when the nuclei of mating partners fuse, their nucleoli remain separate until the first anaphase (Figure S4)51. All AFs that we have tested (58 SSU and LSU AFs, 9 rDNAPs and 5 snoRNP proteins) do equilibrate between the nucleoli over the 2 hrs required for the assay (Table S2). This group includes several AFs that do not relocate between the inner and outer layers when cells are treated with cycloheximide. This mobility is critical for understanding the itinerary of AFs during each transcription cycle.
Discussion
Our goal has been to learn whether the repertoire of AFs can be used to identify subcompartments of the nucleolus, to attribute function to them, and to chart the itinerary of each subunit during its assembly. These observations show that AFs are distributed concentrically around the rDNA axis and that that the localization of distinct subsets of AFs depends on whether they are engaged in making subunits. It has therefore been possible to formulate a model according to which subunit assembly intermediates transfer between concentric layers and to rationalize the presence of distinct protein phases within the nucleolus.
Broad Features of Topography
The nucleolus and chromatin do not obviously intermix and may constitute distinct phases52, 53. Indeed, AFs do not intermix with chromatin even in the absence of subunit assembly (Figure 3A/B, Figure S2). Immiscibility may also account for the traces of AFs along the entirety of the NE (Figure 2A panel 3, Figure 3, Figure S2). Moreover, selected AFs do bind NE proteins and nucleoporins54, 55.
Dynamic Stratification of Assembly Factors
rDNA and rDNAPs form an axis that is surrounded by an inner layer that contributes to the construction of SSUs and an outer layer that includes AFs that are dedicated to assembling LSUs. Table 1 summarizes the distributions of AFs in cells that are making subunits and in cells that are not. Two assumptions underly our interpretations.
Since SSU-F and LSU-F AFs relocate when subunit assembly is interrupted, we assume that they relocate as part of their recycling during each transcription cycle, being primarily in the operative state in the layers where they normally are seen, and being in their latent state, transiently, when not associated with rRNPs (Figure 4). It seems reasonable to think that SSU-In and LSU-Ou AFs - although they do not visibly relocate - also cycle between operative and latent states.
Figure 4. Differential Relocation of Subunit-Specific Assembly Factors.
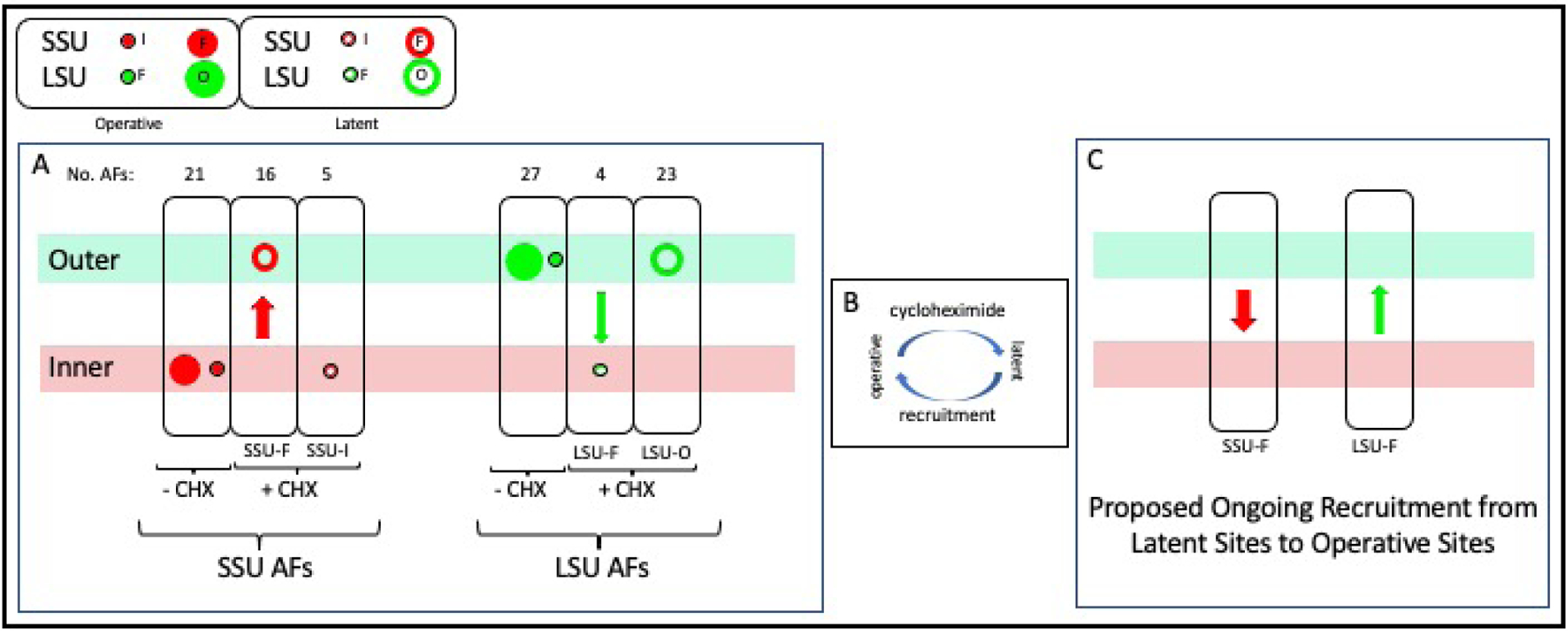
A) Three ovals at the left: In cells that are making subunits, both SSU-In and SSU-F AFs localize to the inner layer (first oval). When assembly is inhibited, SSU-F AFs relocate (red arrow) to the outer layer while SSU-In AFs remain in the inner layer. Second group of three ovals: LSU-Ou and LSU-F AFs concentrate along the outer layer when subunits are being produced. The LSU-F subset relocates (green arrow) to the inner layer when assembly is inhibited, while the LSU-Ou subset remains along the outer layer. The size of the symbols reflects the relative number of AFs in each group.
B) We hypothesize that each AF repeatedly cycles between an operative state and a latent state and that cycloheximide stops recruitment to the operative state.
C) We propose that relocation of SSU-F AFs from the inner layer to the outer layer upon addition of cycloheximide (and inverse relocation of LSU-F AFs) signifies that these AFs normally transit in the opposite directions during each cycle of transcription.
Related to Figure S3 and Figure S2B.
The realization that AFs cycle calls attention to the significance of the colocalization of latent SSU-F and latent (as well as operative) LSU-Ou AFs in the outer layer. This suggests that both groups of AFs are progressively recruited from this common reservoir when binding sites emerge along nascent rRNA.
Transcription begins within the inner layer, so one might expect that the SSU knobs would be removed within that compartment. If fact, it is only when transcription has reached well into sequences that encode the LSU RNAs that they are removed - (site (b) in Figure 5A)5. This could make sense if the nascent rRNP had relocated from the inner layer to the outer layer at this point, considering that the nuclease Rrp17, as well as many LSU AFs, localize to the outer layer (Figure 3B). (The nuclease, Rcl1, is also in the outer layer; however, there is dispute as to whether it normally cuts within ITS1.) These AFs could be added to nascent subunits either just before arrival or upon relocation. We correspondingly assume that a segment of the nascent transcript transfers (“lifts-off”) from the inner to the outer layer prior to being cleaved and releasing the processome itself. Corresponding displacement is not seen in EM spreads, presumably because of the hypotonic alkaline conditions used for their preparation. In support of the proposed transfer event, in situ hybridization data document the shift from inner layer localization to outer layer localization using probes complementary to ITS1 vs ITS2 (Figure 3C).
Figure 5. Transcript Maturation, the Polarity of rDNA and Energy Relations.
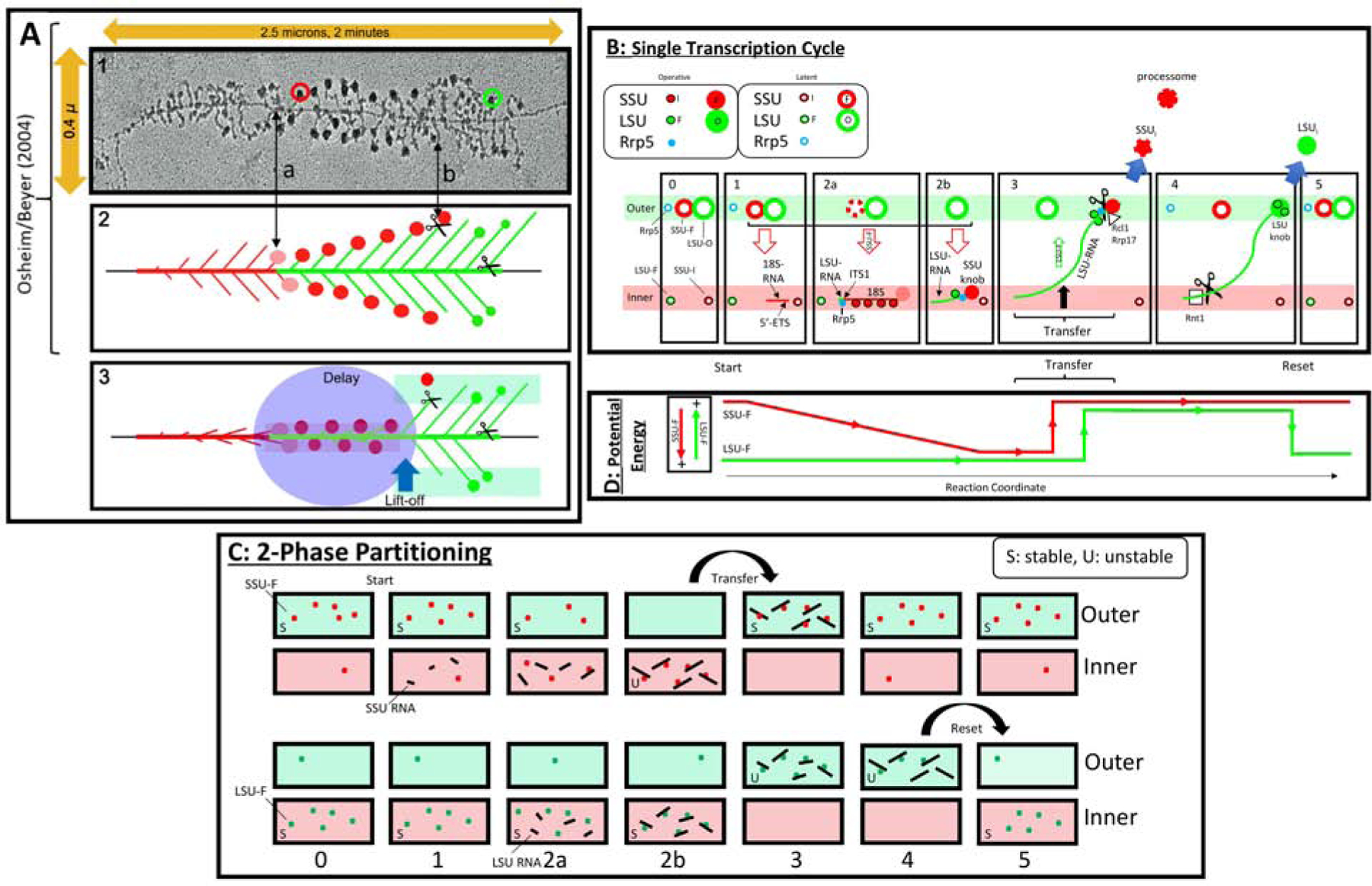
A) Transcript elongation. Panels (1–2) are modified from5. The rDNA and rRNA segments are color-coded: red for 5’ETS and SSU rRNA/DNA, green for LSU rRNA/DNA.
Panel 1: EM of a spread of yeast rDNA during transcription. Note the horizontal rDNA axis and the lateral emergence of transcripts. A SSU terminal knob is circled in red, and a putative LSU knob is circled in green. ITS1 is indicated as (a). The removal of SSU knobs occurs at (b).
Panel 2: Once transcription reaches ITS1, SSU pre-knobs (pink) and knobs (red) appear. They persist until transcription has reached into LSU sequences.
Panel 3: Since cleavage at ITS1 is delayed well beyond the point at which the 3’ extremity of the SSU rRNA coding sequences has been reached, we propose that removal requires transfer to the outer layer. This could allow them to undergo further maturation and, likely, to be cleaved by Rcl1 or Utp24. Further elongation, processing, and formation of particulate intermediates would also occur along the outer layer. Final cleavage occurs at site Bo near the 3’ extremity.
B) Suggested sequential processing of rRNA. See the text for a detailed description of these T-Diagrams. In frames (2a) and (3), the interruption of the perimeter of the large red circles designates progressive release of AFs. AFs that are required for both subunits are not included. The solid circular symbols imply that the indicated AFs are associated with maturing subunits. When not associated, the symbols have a white center.
C) Vectorial 2-Phase Partitioning. Schematic of the relocation of SSU-F AFs and LSU-F AFs. The upper two rows pertain to SSU maturation and the lower two rows pertain to LSU maturation. We consider the localization of these AFs after cycloheximide treatment to be an indication of the phase in which they are most stable (S). By contrast, once they become associated with nascent transcripts, they localize to phases in which they are relatively unstable (U).
D) Energy Relations. During a single cycle, the present observations suggest that 5’-ETS AFs and SSU-F AFs begin in the outer layer and relocate to the inner layer as they load onto nascent rRNA. Phase transfer then allows them to return to the outer layer. This is energetically downhill. Reciprocally, the LSU-F AFs (green line) begin in the inner layer, but shift to the outer layer, perhaps in conjunction with transfer of SSU-Fs. At the end of the cycle, their downhill return to the inner layer resets the system for repeated use. The vertical arrows at the left designate the energy relations. Coupling of inner-to-outer flux of both SSU precursors and LSU-F AFs (states 2b/3) could make their reciprocal flux energetically neutral.
Related to Figure S6
Latent AFs along the inner layer must have characteristic(s) that ensure their proximity to the axis. A minimal hypothesis is that proximity reflects the relative off-rate of their association with the axis and that mutual affinities among these AFs become massively cooperative due to the juxtaposition of rDNA repeats53. There are two such groups of AFs: SSU-In (Dbp4, Efg1, Nop6, Nop9, Nsr1) and LSU-F (Dbp6, Npa1, Npa2, Rsa3 - the “Npa1 complex”). Latent AFs along the outer layer accordingly lack affinity for the axis, yet cannot intermix with chromatin.
Given the invariant localization of SSU-In AFs to the inner layer, they must be along the inner layer prior to transcription of rRNA. They therefore could help retain the relatively 5’ segments of transcripts with which they associate (Table S1). Indeed, Dbp4 and Efg1 bind nascent rRNA before formation of the processome and both they and the other members of this group are released from transcripts as elongation continues into the 3’-minor domain of 18S rRNA56–59. LSU-F AFs are also found in the inner layer upon cycloheximide treatment and may help retain nascent LSU transcripts within the inner layer. Consistent with such a role, they form a complex that can be recovered in an early precursor of LSUs60, 61.
Steps of Cotranscriptional Assembly
The EM spread of nascent rRNA transcripts in yeast (Figure 5A, panel 1)5 and the author’s interpretive diagram (panel 2) sketch the growth of the nascent rRNA. Terminal SSU knobs appear only when transcription has reached the end of SSU rRNA (arrow (a) in Figure 5A), consistent with recent structural studies62.
Based on the binary information summarized in Table 1 and published observations2, 4, 58, 63, we propose step-wise coordination between elongation of transcripts, their association with distinct AFs, and their transfer between the inner and outer layers. In this cyclic process, we propose that AFs are recruited to nascent transcripts, thereby becoming operative, and ultimately are released to latent pools.
In the single cycle diagrammed in Figure 5B, all AFs begin in their latent states (as after cycloheximide) - frame (0). We have not included snoRNP proteins since they always localize to the inner layer. The key steps are as follows:
Transcription of the 5’-ETS (red horizonal line in frame (1)) leads to association with SSU-In AFs along the inner layer (small red circles in frame (2a)) as well as recruitment of UtpA and UtpB AFs (large red arrow in frames (1–2b)) from the outer layer.
These events and synthesis of ITS1 (frame (2a) are followed by synthesis of initial LSU sequences (frames (2a-b)). The presence of ITS1 allows recruitment of Rrp5 from the outer layer (frame (2a)). Rrp5 binds SSU precursors and is required for knob formation, thereby explaining why knobs do not appear during earlier steps of 18S transcription.
Further recruitment of SSU-F AFs from the outer layer, formation of the characteristic terminal knob (pink, then red in frames (2a-b)).
Binding of the Npa1 complex of latent LSU-F AFs (small green circle in frame (2b)) to nascent rRNA/rRNPs (green horizontal line in frame (2b)) generates early LSU intermediates.
After further elongation, sequences including ITS1 transfer to the outer layer (frame (3) and Figure 5A2/3) along with SSU-F AFs, whereupon ITS1 is cleaved, and the processome (including the 20S SSU rRNA) is severed from the 5’-ETS (frame (3))7. LSU-F AFs (small green circles) also may transfer to the outer layer at this time as indicated. AFs that dissociate from the 5’-ETS and from SSU precursors (SSUI) in the outer layer can then be recruited to the inner layer. The driving force for transfer is discussed below.
The nascent LSU segment remains along the outer layer during further elongation, cleavage (by enzymes including Rrp17), and particle maturation through a sequence of states (frame (4))2, 3, 32. In this process, the intermediates recruit latent LSU-Ou AFs from the outer layer. When transcripts extend far enough to include site Bo they are cut by Rnt1, in the inner layer (frame (4)). They then undergo surface remodeling along the outer layer (see below).
When no longer associated with nascent rRNPs, LSU-F AFs return to the inner layer and LSU-Ou AFs remain along the outer layer, thereby resetting the coaxial distribution of AFs as in frame (0).
An alternative schematic summarizing AF flux and assembly is in Figure S6.
Energy Considerations for Transfer
When transcription begins, the nascent rRNA that elongates in the inner layer is expected to recruit 5’-ETS AFs and SSU-F AFs whose latent forms are intrinsically stable along the outer layer. The potential energy characteristic of their (mis)localization to the inner layer (Figure 5C/D) therefore could subsequently promote return of the maturing SSU precursor particle to the outer layer. This consideration may rationalize the presence of the massive size of the 5’-ETS once it becomes laden with AFs. Correspondingly, if LSU-F AFs relocate from the inner layer to the outer layer, this mislocalization could drive their ultimate downhill return to the inner layer. Such a “2-phase vectorial partitioning model” is the only model that rationalizes the importance of there being more than a single protein phase in the nucleolus.
Post-Transcriptional Steps of Assembly
Following cleavage at sites A0, A1, and A2, the 90S processome precursor is converted to a pre-40S particle that arrives in the nucleoplasm7, 64. For the LSU, upon termination of transcription and cleavage at Bo, precursors also are remodeled prior to exit from the nucleolus9, 33, 65, 66. A key step is the removal of Ytm1 and other AFs by the AAA ATPase, Rea133. Subsequently, Alb1, Arx1, Nog2 and Rsa4 assemble on this intermediate, forming the Nog2 particle9. Based on the steady-state localization of these proteins (Figure 3D, Figure S5), these Nog2 particles appear to spend only a short time in the nucleolus and then exit to the nucleoplasm.
Remodeling by Rea1 alters the surface distribution of both AFs and rRNA domains of pre-60S particles. We therefore propose that these events allow LSU precursors to become compatible with the nucleoplasmic phase, into which they can be diluted. This remodeling seems likely to occur in the outer layer, where Ytm1 and multiple other AFnpls are found. In this view, the importance of remodeling is analogous to the entry of exportins into nuclear pores67. The overall transit of subunit precursors thus can be represented by four consecutive cycles (Figure 6).
Figure 6. Proposed Steps of Translocation of Immature Subunits and Assembly Factors.
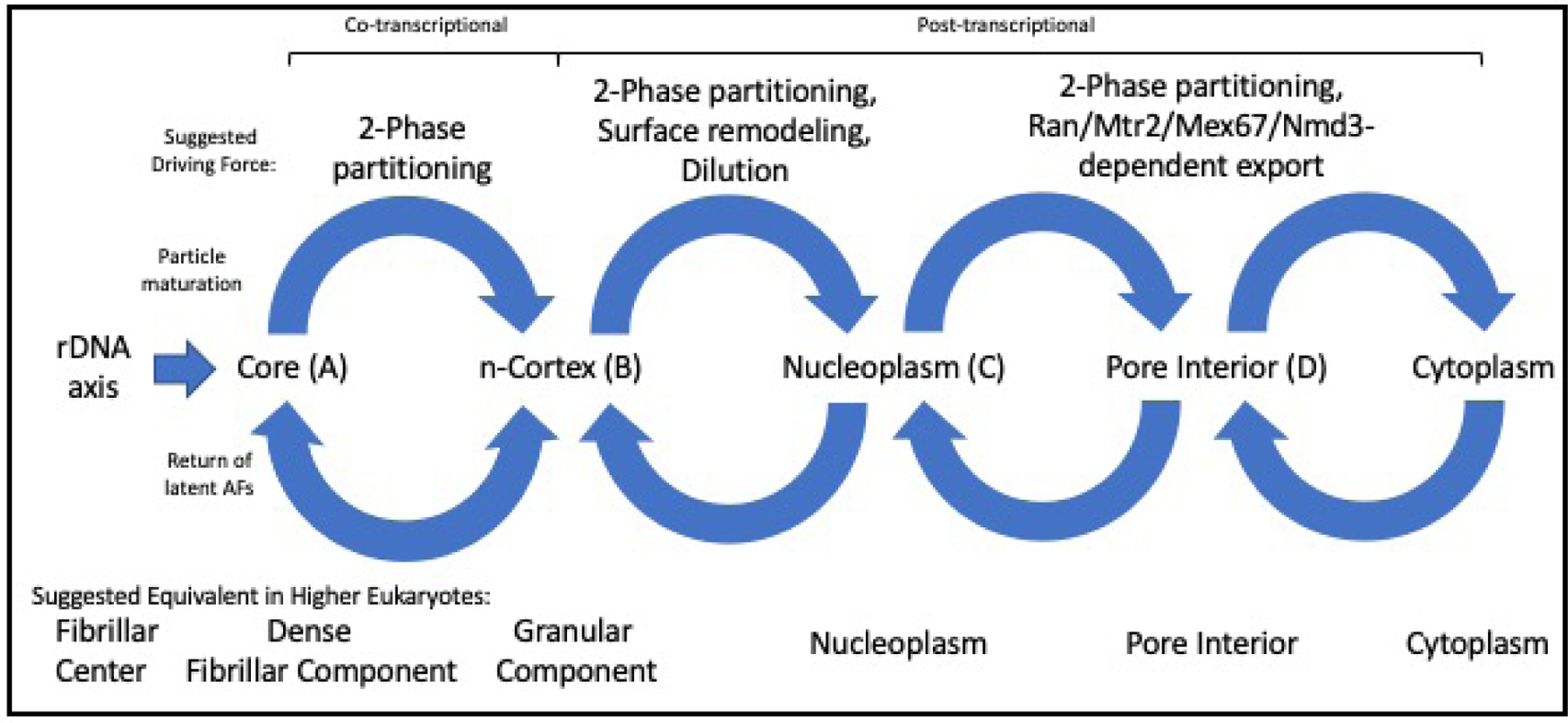
Cycle I can be attributed to vectorial 2-phase partitioning. It is bidirectional since latent SSU-F and latent LSU-F AFs relocate in opposite directions between inner and outer layers. Only during this first cycle do the immature subunits include nascent rRNAs. Cycle II relies on surface AFs that make it possible for subunit precursors to intermix with chromatin. Cycle III involves binding to export factors (exportins, Mex67/Mtr2, Nmd3) that confer compatibility with the interior of the nuclear pore. Cycle IV corresponds to release into the cytoplasm and return of export factors.
Relevance to Other Organisms
In prokaryotes lacking rDNA repeats and in mitochondria, there are few AFs and a nucleolus is not evident68–70 It therefore seems plausible that the appearance of rDNA repeats in evolution cooperatively promoted mutual associations among AFs, that in turn constituted the nucleolus68.
For yeast, we conclude that there are three coaxial nucleolar subcompartments. In higher eukaryotes only few proteins have been assigned to different subcompartments11, 14, 71, 72. On the basis of their relative distance from rDNA and a few protein homologies, we propose that the classical FC, DFC and GC correspond roughly to the yeast axis, inner and outer layers (Tartakoff et al. in preparation).
Earlier studies have not addressed the possible subunit specificity of subcompartments. The present observations indicate that the functional significance of subcompartments pertains to the coordinated assembly of both nascent subunits, that each subunit is largely assembled in a distinct layer/phase, that assembly intermediates transfer between the layers/phases, and that latent AFs cycle between them.
In higher eukaryotes, extrusion of the 5’ segment of nascent rRNA from the FC/DFC interface into the DFC requires the disordered domain of the Nop1 homolog, fibrillarin, in the DFC11. The present analysis emphasizes vectorial 2-phase partitioning for transfer between layers, and a variant of this process for transfer into the nucleoplasm. This provides a framework for understanding the logic that governs genesis of ribosomal subunits in the context of the organization of the nucleolus.
In most higher eukaryotic cells, rDNA repeats are found on several chromosomes, cleavage of precursor rRNA is post-transcriptional, and nucleoli have little contact with the NE. There seems no reason to expect these considerations to interfere with coaxial organization.
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Alan Tartakoff (amt10@case.edu)
Materials Availability
To request strains, protocols, further data or any images generated in this study, please contact the lead contact.
Data and Code Availability
To request related data or any images generated in this study, please contact the lead author.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cells and Cell Culture
Yeast cells were grown to A600 < 1 in complete synthetic medium (CSM) - or variants of CSM - with 2% glucose at 23°C with constant shaking unless indicated otherwise. They were studied with the OD600 as < 1.0. Strain constructions were by standard methods. All strains were from a S288C or W303 background.
To localize nucleolar proteins, we used the Invitrogen/Life Technologies S288C-based strain collection in which each protein carries a C-terminal GFP(S65T)-tag. In nearly all cases the tagged proteins are unique chromosomal integrants, implying that - for the large majority that are essential - the tagged copies are functional. Some proteins could not be studied since corresponding GFP fusion strains are not in this collection or because the corresponding signals were too weak. Since this strain collection does not include a strain with tagged Nop1, we localized GFP-Nop1 by expressing it from a corresponding URA3/CEN plasmid. The Apple-tagged version of Hmo1 was generated by integrative transformation. The Invitrogen/Life Technologies strain collection does not include tagged Utp24 and we have not been able to generate such a strain. Many constructions required crosses of strains from the GFP collection with a partner that expressed Sik1-mRFP (ATY1513) to produce the diploid strains that were used for experiments with cycloheximide.
To study metaphaseCdc20 cells, GFP-tagged strains were crossed with a strain that expressed either Hmo1-Apple or Sik1-mRFP and carried the MET3-CDC20 cassette (ATY10567, ATY10343). The resulting diploids were sporulated. Spores that arrested upon transfer to methionine-containing medium were grown in methionine-free medium and used for imaging after 3 hr in medium containing methionine (−/+ a further incubation for 30 min with addition of cycloheximide). Replating assays showed that survival was excellent upon subsequent return to medium without methionine.
Strains in which AFs were tagged at their C-terminae were from Invitrogen. They are not listed individually. Corresponding diploid strains were constructed by crossing with ATY1513 (MATα Sik1-mRFP), with ATY10567 (MATα Hmo1-Apple), ATY3847 (MATα Htb2-mRFP) or with ATY3196 (MATα mRFP-HDEL), or with equivalent strains in which pMET3-CDC20 had been integrated by transformation with pAT1520. To generate metaphaseCdc20 strains, these diploids were sporulated.
To evaluate the reversibility of the change of AF distributions caused by cycloheximide, strain ATY8300 (Mak11-GFP, Sik1-mRFP) was exposed to cycloheximide for 30 min in growth medium (CSM-glucose) and then sedimented, washed twice in growth medium and reincubated with shaking in growth medium. Samples were removed over 3 hr and photographed. At the end of cycloheximide treatment > 90 % of cells (n = 200) showed the characteristic outer/inner layer distribution of Mak11-GFP and Sik1-mRFP. In representative experiments, the corresponding figures during the chase were 81 +/− 15% (1 hr), 31 +/− 16% (2 hr) and 15 +/− 13% (3 hr) (n = 3). In parallel experiments, the distribution of Utp30-GFP normalized over a comparable period of time.
To evaluate the impact of transferring cells to a medium without glucose, rapidly growing cells were sedimented, washed twice in growth medium lacking sugar and then examined on pads lacking sugar.
METHOD DETAILS
Imaging
0.5–1 μl samples of pellets of living cells were applied to 1.5% agarose pads on microscope slides including medium identical to that in which they had been incubated (e.g. with inclusion of 100 μg/ml cycloheximide). Through-focal series were examined in all cases. Superresolution imaging did not obviously improve the resolution of the inner layer vs outer layer distribution of AFs.
For imaging after cycloheximide treatment, at least 50 cells were examined in each of 3–5 replicate experiments. Typically, three independent fields of cells were scored by comparing the distribution of the GFP-tagged AF to the distribution of condensed inner layer marker, Sik1-mRFP. Independent blinded observers assigned images of the GFP-tagged proteins to categories: a) broadly overlapping with the condensed Sik1-mRFP, or b) showing a more cortical distribution (external to Sik1-mRFP). Each tagged AF had a characteristic localization (inner layer or outer layer) that was evident in >80% of cells examined (detailed tabulations can be provided upon request). The fluorescent signals that overlapped with the inner layer marker often had a complex internal structure, likely dictated by the compacted filament of rDNA.
To learn whether additional strategies could be used to stop addition of ribosomal proteins, we also examined the consequences of transferring yeast to 37°C, abruptly changing carbon source, deliberately overexpressing single AFs, or growing cells to near-saturation. Each of these conditions perturbs the distribution of Mak11 and Sik1 by comparison to their distribution under normal growth conditions (OD600 < 1.0, 23°C, 2% glucose), but the end point is much more variable than upon addition of cycloheximide. In no condition do these AFs relocate to the nucleoplasm.
After cell cycle arrest, we examined cells for which the elongated Sik1-mRFP signal in the mother had an axial ratio of at least 3 and independent blinded observers assigned images of the GFP-tagged proteins to categories: a) broadly overlapping with Sik1-mRFP, or b) showing a cortical distribution, i.e. two parallel faces separated by an obvious gap whose transverse dimension was comparable to the width of the Sik1-mRFP-positive domain. The contour of the elongated segment of the nucleolus was often tortuous and did not lie uniformly in a single plane of section.
We typically examined through-focal 0.2 μ stacks of images from three equivalent fields, each with >25 cells to make these assignments. 3–5 replicate experiments were performed and >75% of cells fell into the predominant category (detailed tabulations can be provided upon request). For GFP-tagged proteins that overlapped with Sik1-mRFP, the green/red match often showed internal inhomogeneity, but there was no indication that the green signal included two parallel faces separated by a gap.
Samples of living cells were examined in a Deltavision RT epifluorescence microscope with an automated stage (Applied Precision, Inc) and a 100x oil immersion objective (Olympus UPlanApo 100x/1.40; ∞/0.17/FN26.5). z-stacks were captured at 0.2–0.5 μ intervals using a CCD digital camera (Photometrics CoolSnap HQ). Out-of-focus light was removed using the Softworks deconvolution software (20 cycles) (Applied Precision, Inc).
Pulse-Chase Experiments
Synthesis and turnover of pre-RNA and rRNA were measured by pulse-chase analysis, with the following modifications. MET-CDC20 strain was transformed with pRS316 to allow for growth in media lacking uracil. Cells were grown at room temperature in synthetic media lacking methionine and uracil to OD600=0.98, or grown in the same media to OD600=0.75, and shifted for 3 hours to synthetic media containing methionine and lacking uracil, to arrest cells in metaphase, Final OD600 after 3 hours of shift was 1.08, comparable to cycling cells. Cells were pulse-labeled with [5,6-3H]-uridine to a final concentration of 20 uCi/ml (Perkin Elmer, cat # NET367) for 20 minutes (cycling cells) or 10 minutes (arrested cells). Pulse times were previously determined for each culture. Chase was performed with excess of unlabeled uridine (Acros Organics) to a final concentration of 2 mM, at time points 2, 5, 10, 30, and 60 minutes. Cells for each time point were harvested and frozen in dry ice.
RNA was extracted by breaking cells with glass beads in the presence of RNA buffer (500 mM NaCl, 200 mM Tris-HCl pH7.5, 10 mM EDTA) and phenol:chloroform:isoamyl alcohol (50:49:1), precipitated with 100% ethanol, washed with 70% ethanol, resuspended in 15 ul of RNAse-free water, and mixed with two volumes of sample buffer (8% formaldehyde, 1.3× MOPS buffer, 65% formamide, 0.02% xylene cyanol, and 0.1% bromophenol blue). Samples were subjected to electrophoresis on a 23-cm 1.2% agarose gel containing 6% formaldehyde and 1X MOPS buffer [1 mM sodium EDTA, 20 mM 3–(N-morpholino propane sulfonic acid pH 7.5, 8 mM sodium acetate)] for 24 h at 55 V or 4.4–6 h at 150 V with continuous recirculation of 1X MOPS buffer. RNAs were visualized using ethidium bromide (0.5 μg/mL). Following electrophoresis, gels were washed in 1X TBE for 10 min, and RNA was transferred from the gel to a Zeta-Probe blotting membrane (Bio-rad) by capillary action. The RNA was cross-linked to the membrane using UV Stratalinker 2400 (Stratagene). The membrane was exposed to the Biomax MS film (Carestream) using BioMax Transcreen LE Intensifying screen (Kodak), at −80°C for 30 days.
Shuttling Assays
Equal volumes of actively growing cultures were mixed, sedimented, resuspended in CSM-glucose and 50 μl samples were applied to the surface of Petri dishes containing solid 2% agar-CSM-glucose. After 2 hr at room temperature, cells were washed off using growth medium, sedimented and observed on agarose pads. In these protocols, cell encounters, fusion and karyogamy are asynchronous.
Intranuclear shuttling was judged by determining whether – when one parent initially expressed a tagged AF – this signal had become equally visible at both extremities of the elongated zygotic nucleus. In order to restrict attention to relatively early events, we examined zygotes which showed no evidence of budding. 2–3 experiments were performed for each GFP-tagged protein. The possible contribution of nucleocytoplasmic shuttling was judged by following GFP-tagged AFs in crosses with a kar1Δ15 mutant that delays nuclear fusion. This partner also expressed mRFP-HDEL, that served as an indicator of fusion of the parental cells and the lack of fusion of their nuclei. In all crosses, cells were examined only if the HDEL signal had equilibrated. Judging from our earlier studies, nuclei fuse in [kar1 × wt] zygotes with a t½ of ≈ 5 hr51.
Drug Stocks
Drug stocks were 100–1000x concentrated. They were prepared in water (cycloheximide) or in DMSO (all other drugs).
in situ Hybridization
For in situ hybridization, ≈ 6 × 107 cells at A600 < 0.3 were fixed by addition of 1/10 volume of 37% formaldehyde for 15 min with shaking, sedimented, and further fixed by addition of 4% paraformaldehyde in PBS for 1.5 hr at room temperature with shaking. They were then washed 2x in 0.1 M potassium phosphate, pH7.5 with 1.2M sorbitol (buffer A). For spheroplasting, they were resuspended in 1% β-mercaptoethanol and 80 μg/ml freshly dissolved zymolyase 20T in 250 μl buffer A for 1 hr at 37°C. After addition of 1/500 volume of vanadyl complex (protect RNA RNase inhibitor, Sigma R7397) two washes with buffer A, and resuspension in 20–200 μl of buffer A, they were spotted for 30 min onto ethanol-cleaned slides (Carlson Scientific, Inc. Printed Microscope Slides) pretreated with 0.1% poly-L-lysine. After two washes in buffer A, they received 70% ethanol for >30 min. After air drying, they received 50 μl of 2 × SSC/10% formamide (solution B) for 5 min at room temperature before being transferred to the hybridization solution (2 × SSC/10% formamide/10% dextran sulfate, overnight at 37°C with probe(s)) in a water-tight chamber. After two washes with hybridization solution lacking probe and a 15 min incubation in this solution at 37°C, they were transferred to solution B, stained with DAPI in solution B for 5 min, washed with B, mounted and examined.
QUANTIFICATION AND STATISTICAL ANALYSIS
Experimental verification was obtained by repeating experiments at least three times using independent clones of cells. As needed, standard deviations were calculated. Although all GFP-tagged strains originated from unique samples in the Invitrogen strain collection, multiple strains were derived from them.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals | ||
| Anisomycin | Sigma Chemical | A-9789 |
| BMH-21 | MedChemExpress | HY-12484/CS-3557 |
| Cycloheximide | Sigma-Aldrich | C7698 |
| Rapamycin | Millipore | 553211 |
| Thiolutin | N. Belcher, Pfizer | lot 1325-56-1 |
| Zymolyase 20T | MP Biochemicals | 320921 |
| [5,6-3H]-uridine | Perkin-Elmer | cat # NET367 |
| dimethylsulfoxide (DMSO) | Fisher | D128 |
| glucose | Sigma | G8270 |
| NaCl | Sigma | S7653 |
| EDTA | Sigma | E6511 |
| MOPS | Sigma | M1254 |
| formamide | Sigma-Aldrich | 221198 |
| Agarose | Invitrogen | 15510–027 |
| paraformaldehyde | Sigma-Aldrich | 158127 |
| formaldehyde (37%) | Sigma-Aldrich | 252549 |
| vanadate complex (protect RNA RNase inhibitor) | Sigma | R7397 |
| poly-L-lysine | Sigma-Aldrich | P4707 |
| dextran sulfate | Sigma | S8906 |
| xylene cyanol | Sigma-Aldrich | X4126 |
| bromphenol blue | Sigma-Aldrich | B0126 |
| Software and Algorithms | ||
| SoftWorX deconvolution software | Applied Precision, Inc. | http://www.sussex.ac.uk/gdsc/intranet/pdfs/softWoRx%20user%20manual.pdf |
| Excel (for calculating standard deviations) | Microsoft | N/A |
| Experimental Models: Strains of S. cerevisiae | ||
| S. cerevisiae panel of strains expressing single GFP-tagged proteins MATa His+ his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 | Invitrogen/Thermo Fisher | Yeast Deletion and Yeast GFP Collections |
| MATα Sik1-mRFP G418R | W-K. Huh | ATY1513 |
| MATα ss-mRFP-HDEL | this study | ATY3196 |
| MATα Htb2-mRFP | this study | ATY3847 |
| kar1Δ15 MAT α CAN1 RNR1 lys2Δ his3–11,15 ura3–1 trp1–1 leu2–3,112 RAD5 W303 | R. Rothstein W2108–14C | ATY4263 |
| MATα kar1Δ15 mRFP-HDEL | this study | ATY6618 |
| MET3-CDC20 (SAC) | this study | ATY10402 |
| MATα Hmo1-Apple | this study | ATY10567 |
| pan-lacO/rDNA MAT a leu2–3,112 trp1–1 can1–100 ura3–1 ade2–1 his3–11,15 ade2:: pAFS144-wtGFP (ADE2) rDNA::pTM-lacO50 (URA3) W303 | T. Kobayashi, TMY3 | ATY10658 |
| pan-lacO/rDNA Sik1-mCherry | As for ATY10658 Sik1-mCherry (KANr) | ATY10659 |
| MATα SAC Hmo1-Apple pan-lacO | this study | ATY10682 |
| MATa SAC Hmo1-Apple pan-lacO | this study | ATY10688 |
| SAC Hmo1-Apple Utp15-CFP Mak11-GFP | this study | ATY10694 |
| SAC Ytm1-Apple mRFP-[pGFP-HDEL] | this study | ATY10717 |
| SAC Mak11-GFP mRFP-HDEL | this study | ATY10722 |
| MATα top1::HIS top4–2 can1–100 his3–11, 15 leu2–3, 112 trp1–1 ura3–1 W303 | R. Rothstein, W1477–5B | ATY10730 |
| MATa SAC pan-lacO Sik1-mRFP | this study | ATY10747 |
| top1Δ top2–4 pan-lacO Sik1-mCherry | this study | ATY10795 |
| Oligonucleotides | ||
| 5’-CTTGCCCAGTAAAAGCTCTCATGCTCTTGC/3Cy3 | IDT | ITS1 probe |
| 5’-CATTATACCTCAAGCACGCAGAGAAACCTC/3Cy5 | IDT | ITS2 probe |
| 5’-AAAGAAGAAGAAGAAGAAGA AGAAGGATAAGA AGAAGGACAAATCCAACTCTTCTATTGGTGACGGT GCTGGTTTA-3’ | Forward primer | for C-terminal tagging Hmo1 with Apple |
| 5’-TTATTATTATATTTATTTTAGAAAGACAGTAGAGT AATAGTAACGAGTTTGTCCGTCCATCGATGAATTCG AGCTCG-3’ | Reverse primer | |
| Plasmids | ||
| pKar2Frag-mRFP-HDEL (URA3/Yip) | N. Dean | pTi-kmRFP |
| pSS-GFP-HDEL (URA3/CEN) | W. Prinz | PWP1055 |
| pMET3-CDC20 (LEU2/Yip22) | R. Uhlmann | p640 |
| pGFP-Nop1 (YCPLAC33 URA3/CEN) | C. Horigome | pNop1 |
| yoApple HIS3 | K.Thorn/M.Garabedian | Apple tag |
Highlights.
The yeast nucleolus is organized as a coaxial cable around an axis of rDNA.
The axis is surrounded by two layers/phases of assembly factors.
Nascent subunit precursors pass from the inner to outer layer.
Subunit assembly factors cycle between latent and operative states.
Acknowledgements
Thanks to the NIH for R01GM089872 (A.T.), R01GM028301 (J.W.), RO1HL126626 and HL141423 (G.M.) and P30CA43703-12 and thanks to the Visconsi family for support. This publication was made possible by the Clinical and Translational Science Collaborative of Cleveland, UL1TR002548 from the National Center for Advancing Translational Sciences (NCATS) component of the National Institutes of Health and NIH Roadmap for Medical Research. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH. Thanks for technical help to David Dulce, Alexis Kerr, Dalya Khalife, Jelena Micic, Kyle Parker and Julianne Walker. Thanks for materials and advice to V.Boerner, N.Dean, M.Garabedian, C.Horigome, W-K.Huh, F.Kizito, T.Kobayashi, B.Luttge, D.MacDonald, W.Prinz, M.Rose, and F.Uhlmann.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests: The authors declare no competing interests.
References
- 1.Nerurkar P, Altvater M, Gerhardy S, Schutz S, Fischer U, Weirich C, et al. Eukaryotic Ribosome Assembly and Nuclear Export. Int Rev Cell Mol Biol. 2015;319:107–40. [DOI] [PubMed] [Google Scholar]
- 2.Klinge S, Woolford JL Jr. Ribosome assembly coming into focus. Nature reviews Molecular cell biology. 2019;20(2):116–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pena C, Hurt E, Panse VG. Eukaryotic ribosome assembly, transport and quality control. Nature structural & molecular biology. 2017;24(9):689–99. [DOI] [PubMed] [Google Scholar]
- 4.Bassler J, Hurt E. Eukaryotic Ribosome Assembly. Annu Rev Biochem. 2019;88:281–306. [DOI] [PubMed] [Google Scholar]
- 5.Osheim YN, French SL, Keck KM, Champion EA, Spasov K, Dragon F, et al. Pre-18S ribosomal RNA is structurally compacted into the SSU processome prior to being cleaved from nascent transcripts in Saccharomyces cerevisiae. Molecular cell. 2004;16(6):943–54. [DOI] [PubMed] [Google Scholar]
- 6.Vincent NG, Charette JM, Baserga SJ. The SSU processome interactome in Saccharomyces cerevisiae reveals novel protein subcomplexes. RNA. 2018;24(1):77–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng J, Lau B, La Venuta G, Ameismeier M, Berninghausen O, Hurt E, et al. 90S pre-ribosome transformation into the primordial 40S subunit. Science. 2020;369(6510):1470–6. [DOI] [PubMed] [Google Scholar]
- 8.Barandun J, Hunziker M, Klinge S. Assembly and structure of the SSU processome-a nucleolar precursor of the small ribosomal subunit. Current opinion in structural biology. 2018;49:85–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu S, Tutuncuoglu B, Yan K, Brown H, Zhang Y, Tan D, et al. Diverse roles of assembly factors revealed by structures of late nuclear pre-60S ribosomes. Nature. 2016;534(7605):133–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lafontaine DLJ, Riback JA, Bascetin R, Brangwynne CP. The nucleolus as a multiphase liquid condensate. Nature reviews Molecular cell biology. 2020. [DOI] [PubMed] [Google Scholar]
- 11.Yao RW, Xu G, Wang Y, Shan L, Luan PF, Wang Y, et al. Nascent Pre-rRNA Sorting via Phase Separation Drives the Assembly of Dense Fibrillar Components in the Human Nucleolus. Molecular cell. 2019;76(5):767–83 e11. [DOI] [PubMed] [Google Scholar]
- 12.Puvion-Dutilleul F, Bachellerie JP, Puvion E. Nucleolar organization of HeLa cells as studied by in situ hybridization. Chromosoma. 1991;100(6):395–409. [DOI] [PubMed] [Google Scholar]
- 13.Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI. The multifunctional nucleolus. Nature reviews Molecular cell biology. 2007;8(7):574–85. [DOI] [PubMed] [Google Scholar]
- 14.Hernandez-Verdun D, Roussel P, Thiry M, Sirri V, Lafontaine DL. The nucleolus: structure/function relationship in RNA metabolism. Wiley interdisciplinary reviews RNA. 2010;1(3):415–31. [DOI] [PubMed] [Google Scholar]
- 15.Lazdins IB, Delannoy M, Sollner-Webb B. Analysis of nucleolar transcription and processing domains and pre-rRNA movements by in situ hybridization. Chromosoma. 1997;105(7–8):481–95. [DOI] [PubMed] [Google Scholar]
- 16.Raska I, Shaw PJ, Cmarko D. Structure and function of the nucleolus in the spotlight. Current opinion in cell biology. 2006;18(3):325–34. [DOI] [PubMed] [Google Scholar]
- 17.Goessens G. Nucleolar structure. International review of cytology. 1984;87:107–58. [DOI] [PubMed] [Google Scholar]
- 18.Shah SB, Terry CD, Wells DA, DiMario PJ. Structural changes in oocyte nucleoli of Xenopus laevis during oogenesis and meiotic maturation. Chromosoma. 1996;105(2):111–21. [DOI] [PubMed] [Google Scholar]
- 19.Sloan KE, Gleizes PE, Bohnsack MT. Nucleocytoplasmic Transport of RNAs and RNA-Protein Complexes. Journal of molecular biology. 2016;428(10 Pt A):2040–59. [DOI] [PubMed] [Google Scholar]
- 20.Palmer RE, Koval M, Koshland D. The dynamics of chromosome movement in the budding yeast Saccharomyces cerevisiae. The Journal of cell biology. 1989;109(6 Pt 2):3355–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyazaki T, Kobayashi T. Visualization of the dynamic behavior of ribosomal RNA gene repeats in living yeast cells. Genes to cells : devoted to molecular & cellular mechanisms. 2011;16(5):491–502. [DOI] [PubMed] [Google Scholar]
- 22.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425(6959):686–91. [DOI] [PubMed] [Google Scholar]
- 23.Wittner M, Hamperl S, Stockl U, Seufert W, Tschochner H, Milkereit P, et al. Establishment and maintenance of alternative chromatin states at a multicopy gene locus. Cell. 2011;145(4):543–54. [DOI] [PubMed] [Google Scholar]
- 24.Rai U, Najm F, Tartakoff AM. Nucleolar asymmetry and the importance of septin integrity upon cell cycle arrest. PloS one. 2017;12(3):e0174306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lavoie BD, Hogan E, Koshland D. In vivo requirements for rDNA chromosome condensation reveal two cell-cycle-regulated pathways for mitotic chromosome folding. Genes & development. 2004;18(1):76–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deltour R, Motte P. The nucleolonema of plant and animal cells: a comparison. Biology of the cell / under the auspices of the European Cell Biology Organization. 1990;68(1):5–11. [DOI] [PubMed] [Google Scholar]
- 27.Kos-Braun IC, Kos M. Post-transcriptional regulation of ribosome biogenesis in yeast. Microb Cell. 2017;4(5):179–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Merl J, Jakob S, Ridinger K, Hierlmeier T, Deutzmann R, Milkereit P, et al. Analysis of ribosome biogenesis factor-modules in yeast cells depleted from pre-ribosomes. Nucleic acids research. 2010;38(9):3068–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Toussaint M, Levasseur G, Tremblay M, Paquette M, Conconi A. Psoralen photocrosslinking, a tool to study the chromatin structure of RNA polymerase I--transcribed ribosomal genes. Biochemistry and cell biology = Biochimie et biologie cellulaire. 2005;83(4):449–59. [DOI] [PubMed] [Google Scholar]
- 30.French SL, Osheim YN, Cioci F, Nomura M, Beyer AL. In exponentially growing Saccharomyces cerevisiae cells, rRNA synthesis is determined by the summed RNA polymerase I loading rate rather than by the number of active genes. Molecular and cellular biology. 2003;23(5):1558–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lebaron S, Segerstolpe A, French SL, Dudnakova T, de Lima Alves F, Granneman S, et al. Rrp5 binding at multiple sites coordinates pre-rRNA processing and assembly. Molecular cell. 2013;52(5):707–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen W, Xie Z, Yang F, Ye K. Stepwise assembly of the earliest precursors of large ribosomal subunits in yeast. Nucleic acids research. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kater L, Thoms M, Barrio-Garcia C, Cheng J, Ismail S, Ahmed YL, et al. Visualizing the Assembly Pathway of Nucleolar Pre-60S Ribosomes. Cell. 2017;171(7):1599–610 e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pausch P, Singh U, Ahmed YL, Pillet B, Murat G, Altegoer F, et al. Co-translational capturing of nascent ribosomal proteins by their dedicated chaperones. Nature communications. 2015;6:7494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horn DM, Mason SL, Karbstein K. Rcl1 protein, a novel nuclease for 18 S ribosomal RNA production. The Journal of biological chemistry. 2011;286(39):34082–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sahasranaman A, Dembowski J, Strahler J, Andrews P, Maddock J, Woolford JL, Jr. Assembly of Saccharomyces cerevisiae 60S ribosomal subunits: role of factors required for 27S pre-rRNA processing. The EMBO journal. 2011;30(19):4020–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Massenet S, Bertrand E, Verheggen C. Assembly and trafficking of box C/D and H/ACA snoRNPs. RNA biology. 2017;14(6):680–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wells GR, Weichmann F, Colvin D, Sloan KE, Kudla G, Tollervey D, et al. The PIN domain endonuclease Utp24 cleaves pre-ribosomal RNA at two coupled sites in yeast and humans. Nucleic acids research. 2016;44(11):5399–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tomecki R, Sikorski PJ, Zakrzewska-Placzek M. Comparison of preribosomal RNA processing pathways in yeast, plant and human cells - focus on coordinated action of endo- and exoribonucleases. FEBS letters. 2017;591(13):1801–50. [DOI] [PubMed] [Google Scholar]
- 40.Reiter A, Steinbauer R, Philippi A, Gerber J, Tschochner H, Milkereit P, et al. Reduction in ribosomal protein synthesis is sufficient to explain major effects on ribosome production after short-term TOR inactivation in Saccharomyces cerevisiae. Molecular and cellular biology. 2011;31(4):803–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Warner JR. Synthesis of ribosomes in Saccharomyces cerevisiae. Microbiological reviews. 1989;53(2):256–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gamalinda M, Ohmayer U, Jakovljevic J, Kumcuoglu B, Woolford J, Mbom B, et al. A hierarchical model for assembly of eukaryotic 60S ribosomal subunit domains. Genes & development. 2014;28(2):198–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tye BW, Commins N, Ryazanova LV, Wuhr M, Springer M, Pincus D, et al. Proteotoxicity from aberrant ribosome biogenesis compromises cell fitness. eLife. 2019;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Albert B, Kos-Braun IC, Henras AK, Dez C, Rueda MP, Zhang X, et al. A ribosome assembly stress response regulates transcription to maintain proteome homeostasis. eLife. 2019;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chan J, Khan SN, Harvey I, Merrick W, Pelletier J. Eukaryotic protein synthesis inhibitors identified by comparison of cytotoxicity profiles. RNA. 2004;10(3):528–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Christiano R, Nagaraj N, Frohlich F, Walther TC. Global proteome turnover analyses of the Yeasts S. cerevisiae and S. pombe. Cell reports. 2014;9(5):1959–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Torreira E, Louro JA, Pazos I, Gonzalez-Polo N, Gil-Carton D, Duran AG, et al. The dynamic assembly of distinct RNA polymerase I complexes modulates rDNA transcription. eLife. 2017;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wei T, Najmi SM, Liu H, Peltonen K, Kucerova A, Schneider DA, et al. Small-Molecule Targeting of RNA Polymerase I Activates a Conserved Transcription Elongation Checkpoint. Cell reports. 2018;23(2):404–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tipper DJ. Inhibition of yeast ribonucleic acid polymerases by thiolutin. J Bacteriol. 1973;116(1):245–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kos M, Tollervey D. Yeast pre-rRNA processing and modification occur cotranscriptionally. Molecular cell. 2010;37(6):809–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tartakoff AM, Jaiswal P. Nuclear fusion and genome encounter during yeast zygote formation. Molecular biology of the cell. 2009;20(12):2932–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Caragine CM, Haley SC, Zidovska A. Nucleolar dynamics and interactions with nucleoplasm in living cells. eLife. 2019;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McSwiggen DT, Mir M, Darzacq X, Tjian R. Evaluating phase separation in live cells: diagnosis, caveats, and functional consequences. Genes & development. 2019;33(23–24):1619–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oeffinger M, Zenklusen D, Ferguson A, Wei KE, El Hage A, Tollervey D, et al. Rrp17p is a eukaryotic exonuclease required for 5’ end processing of Pre-60S ribosomal RNA. Molecular cell. 2009;36(5):768–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Horigome C, Okada T, Shimazu K, Gasser SM, Mizuta K. Ribosome biogenesis factors bind a nuclear envelope SUN domain protein to cluster yeast telomeres. The EMBO journal. 2011;30(18):3799–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Soltanieh S, Osheim YN, Spasov K, Trahan C, Beyer AL, Dragon F. DEAD-box RNA helicase Dbp4 is required for small-subunit processome formation and function. Molecular and cellular biology. 2015;35(5):816–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shu S, Ye K. Structural and functional analysis of ribosome assembly factor Efg1. Nucleic acids research. 2018;46(4):2096–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang L, Wu C, Cai G, Chen S, Ye K. Stepwise and dynamic assembly of the earliest precursors of small ribosomal subunits in yeast. Genes & development. 2016;30(6):718–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hunziker M, Barandun J, Buzovetsky O, Steckler C, Molina H, Klinge S. Conformational switches control early maturation of the eukaryotic small ribosomal subunit. eLife. 2019;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Joret C, Capeyrou R, Belhabich-Baumas K, Plisson-Chastang C, Ghandour R, Humbert O, et al. The Npa1p complex chaperones the assembly of the earliest eukaryotic large ribosomal subunit precursor. PLoS genetics. 2018;14(8):e1007597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rosado IV, Dez C, Lebaron S, Caizergues-Ferrer M, Henry Y, de la Cruz J. Characterization of Saccharomyces cerevisiae Npa2p (Urb2p) reveals a low-molecular-mass complex containing Dbp6p, Npa1p (Urb1p), Nop8p, and Rsa3p involved in early steps of 60S ribosomal subunit biogenesis. Molecular and cellular biology. 2007;27(4):1207–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cheng J, Bassler J, Fischer P, Lau B, Kellner N, Kunze R, et al. Thermophile 90S Pre-ribosome Structures Reveal the Reverse Order of Co-transcriptional 18S rRNA Subdomain Integration. Molecular cell. 2019;75(6):1256–69 e7. [DOI] [PubMed] [Google Scholar]
- 63.Chaker-Margot M, Hunziker M, Barandun J, Dill BD, Klinge S. Stage-specific assembly events of the 6-MDa small-subunit processome initiate eukaryotic ribosome biogenesis. Nature structural & molecular biology. 2015;22(11):920–3. [DOI] [PubMed] [Google Scholar]
- 64.Du Y, An W, Zhu X, Sun Q, Qi J, Ye K. Cryo-EM structure of 90S small ribosomal subunit precursors in transition states. Science. 2020;369(6510):1477–81. [DOI] [PubMed] [Google Scholar]
- 65.Sanghai ZA, Miller L, Molloy KR, Barandun J, Hunziker M, Chaker-Margot M, et al. Modular assembly of the nucleolar pre-60S ribosomal subunit. Nature. 2018;556(7699):126–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou D, Zhu X, Zheng S, Tan D, Dong MQ, Ye K. Cryo-EM structure of an early precursor of large ribosomal subunit reveals a half-assembled intermediate. Protein Cell. 2019;10(2):120–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Frey S, Rees R, Schunemann J, Ng SC, Funfgeld K, Huyton T, et al. Surface Properties Determining Passage Rates of Proteins through Nuclear Pores. Cell. 2018;174(1):202–17 e9. [DOI] [PubMed] [Google Scholar]
- 68.Ebersberger I, Simm S, Leisegang MS, Schmitzberger P, Mirus O, von Haeseler A, et al. The evolution of the ribosome biogenesis pathway from a yeast perspective. Nucleic acids research. 2014;42(3):1509–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Davis JH, Williamson JR. Structure and dynamics of bacterial ribosome biogenesis. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 2017;372(1716). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Saurer M, Ramrath DJF, Niemann M, Calderaro S, Prange C, Mattei S, et al. Mitoribosomal small subunit biogenesis in trypanosomes involves an extensive assembly machinery. Science. 2019;365(6458):1144–9. [DOI] [PubMed] [Google Scholar]
- 71.Hozak P. Catching RNA polymerase I in Flagranti: ribosomal genes are transcribed in the dense fibrillar component of the nucleolus. Experimental cell research. 1995;216(2):285–9. [DOI] [PubMed] [Google Scholar]
- 72.Scheer U, Hock R. Structure and function of the nucleolus. Current opinion in cell biology. 1999;11(3):385–90. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
To request related data or any images generated in this study, please contact the lead author.


