Abstract
The currently largest genome-wide association study (GWAS) for age-related macular degeneration (AMD) defines disease association with genome-wide significance for 52 independent common and rare genetic variants across 34 chromosomal loci. Overall, these loci contain over 7200 variants and are enriched for genes with functions indicating several shared cellular processes. Still, the precise mechanisms leading to AMD pathology are largely unknown. Here, we exploit the phenomenon of epistatic interaction to identify seemingly independent AMD-associated variants that reveal joint effects on gene expression. We focus on genetic variants associated with lipid metabolism, organization of extracellular structures, and innate immunity, specifically the complement cascade. Multiple combinations of independent variants were used to generate genetic risk scores allowing gene expression in liver to be compared between low and high-risk AMD. We identified genetic variant combinations correlating significantly with expression of 26 genes, of which 19 have not been associated with AMD before. This study defines novel targets and allows prioritizing further functional work into AMD pathobiology.
Subject terms: Functional genomics, Gene expression, Gene regulation, Genetic association study, Genetic interaction
Introduction
A first successful genome-wide association study (GWAS) was reported in 2005 and identified with genome-wide significance genetic variants at the CFH locus associated with age-related macular degeneration (AMD), a complex disease which is a frequent cause of progressive vision loss in the elderly population1. Since then, the list of AMD-associated genetic variation has grown exponentially, presently bringing the total to 52 independent common and rare variants across 34 chromosomal loci2. In an initial effort to extract biological meaning from the latter association data, enrichment analyses were done to determine molecular pathways from the 368 genes located within the immediate AMD loci as defined by index variants and proxies at r2 ≥ 0.5 within ± 100 kb of the lead signal. This broadly emphasized the lipid metabolism, extracellular matrix organization and assembly as well as the complement pathway to play a critical role in AMD pathogenesis2.
A more refined dissection of the biological mechanisms triggering AMD disease is challenging, although it is a crucial step to translate genetic findings into a clinical context. Understanding the functional impact of synonymous or non-synonymous variants in exonic sequences appears rather straightforward as they likely exert a direct effect on protein translation or stability3. However, in AMD over 90% of associated genetic variation is estimated to be located in intronic or intergenic regions2, which hampers an intuitive access to understanding their pathological impact. A growing number of studies now suggests that variation in noncoding regions likely affect mRNA expression of nearby or distal genes through promoter, enhancer or silencer effects, or modulate transcription factor binding affinities, alternative splicing, and/or other epigenetic modifications4. To some extent, such an altered gene expression can be addressed by mapping expression quantitative trait loci (eQTL)5. In AMD, eQTL analyses were conducted in several tissues. For example, Ratnapriya and colleagues investigated eQTL in the retina, and detected altered gene expression for 20 potentially AMD-associated genes6, while in liver a total of 15 genes were identified7. Further, a transcriptome-wide association study included 27 different tissues and identified 106 genes with an expression profile linked to the genetic risk of AMD8. Although primary pathology of AMD manifests in the back of the eye, eQTL findings in various tissues reveal genes potentially influencing general tissue integrity and homeostasis9–11. This is strengthened by pleiotropy analyses, demonstrating a common genetic basis of AMD and several other phenotypes, such as cardiovascular diseases or metabolic traits12,13. Although the retinal pigment epithelium (RPE) may be the cell type of interest in AMD pathology14, it is likely that circulating molecules ultimately contribute to local manifestations15–17. Against this background, liver tissue is of particular interest as it is the major expression site for secreted proteins involved in at least two major pathways of AMD pathology, including the lipid metabolism and the complement system18–20. Finally, a gene expression dataset with nearly 600 individuals is available for liver tissue providing sufficient statistical power to perform advanced analyses.
So far, eQTL mapping in AMD was directed exclusively on studying effects of single genetic variants6,7. Additive models including the integration of combinations of genetic variation on gene expression have not been considered. Here, we focused on genetic variation in loci which were assigned to one of three likely biological pathways in AMD aetiology, including lipid metabolism, extracellular matrix organization, and the complement cascade2,9. We combined customized genetic risk scores (GRS) and eQTL mapping and evaluated an influence on gene expression in liver tissue for genetic variants individually or in combination with each other. Such an approach more closely resembles a true biological situation, where AMD patients likely harbor multiple risk variants with possible synergistic or antagonistic effects on a defined outcome. Taken together, exploiting possible epistatic interactions of AMD-associated variants on gene expression regulation has revealed at least 19 genes of interest which have not been found in the available AMD datasets so far. This could open new avenues to learn about biological pathways and mechanisms underlying AMD disease.
Results
Defining pathway-related clusters of independent AMD-associated variants
To search for epistatic effects of independent AMD-associated genetic variants on gene expression in liver tissue, we first selected tentative biological routes to disease mechanisms represented by three likely AMD-associated pathways including the lipid metabolism, extracellular matrix organization, and the complement cascade2. We used the g:Profiler database21 to assign gene ontology terms (GO-terms) to the 386 genes within the 34 genome-wide associated AMD loci as defined by Fritsche et al. (index variants and proxies at r2 ≥ 0.5 within ± 100 kb of lead variants)2. GO-terms used for locus categorization were “cholesterol transport” (GO:0030301), “regulation of plasma lipoprotein” (GO:0097006), “extracellular structure organization” (GO:0043062), and “immune system process” (GO:0002376). Loci were assigned to multiple pathways, if the corresponding genes mapped to several GO-terms. In total, seven of the 34 independent AMD loci were assigned to the lipid metabolism pathway, 16 to the extracellular matrix term, and 23 to the complement system (Fig. 1a). Six of the 34 AMD loci were not assigned to any of the selected GO-terms.
Figure 1.
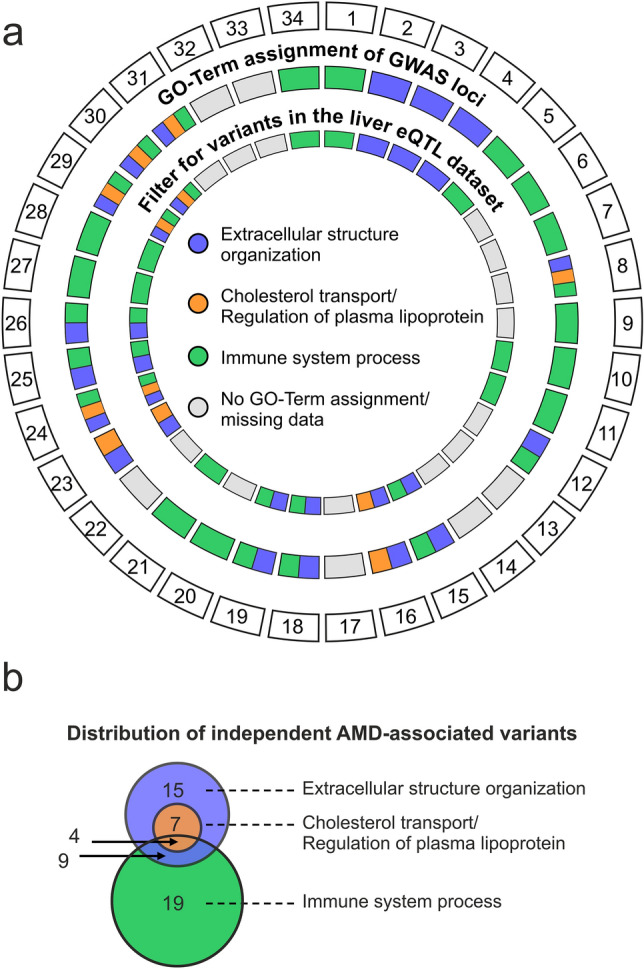
Categorizing independent AMD-associated variants. (a) AMD-associated loci as defined in ref. 2 were assigned to three biological pathways discussed to be relevant in AMD pathobiology, particularly the “Extracellular structure organization”, “Cholesterol transport” and “Regulation of plasma lipoprotein”, and “Immune system process”. A locus was assigned to a pathway if a gene located in this locus (locus definition: index variant(s) and proxies (r2 > 0.5; +/- 100 kb)2 is functionally associated by GO-Terms. Subsequently, the independent signals within these loci were correlated with the liver eQTL dataset reported by Strunz et al.7. (b) Distribution of independent AMD-associated variants over the three biological pathways identified. Overlapping circles indicate independent variants that were assigned to more than one category. Details are given in Supplementary Table S1.
We then filtered the liver eQTL dataset reported by Strunz et al.7 for independent lead variants within the 28 pathway-associated AMD loci. This resulted in 21 loci for further analysis, corresponding to 31 independent genetic variants. Specifically, seven variants at 5 loci were assigned to the lipid metabolism pathway, 15 variants at 13 loci to the extracellular matrix pathway, and 19 variants at 16 loci to the complement system pathway (Fig. 1b and Supplementary Table S1). Interestingly, the five loci assigned to the lipid metabolism pathway are also associated with the extracellular matrix pathway.
Defining test parameters in liver tissue
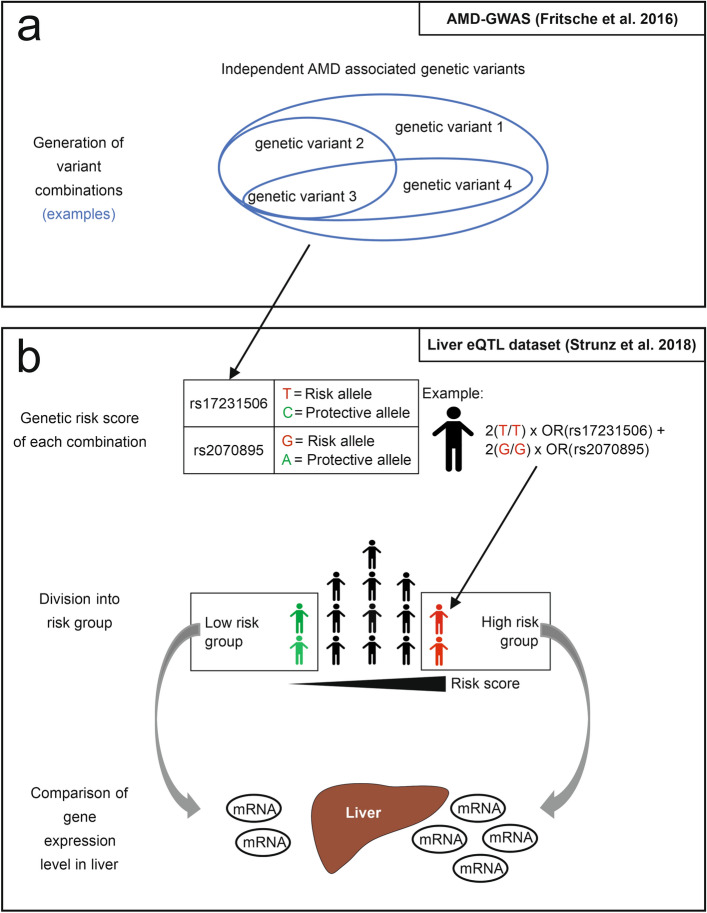
The category “lipid metabolism” was used to initially define the test parameters for the identification of significant variant combinations (eCombinations) with a joint effect on gene expression regulation in liver tissue. In total, 5 loci containing 7 independent variants were included in this category (Fig. 2a). The 7 variants were organized in 127 unique combinations.
Figure 2.
Schematic project workflow. (a) AMD-associated genetic variants were used to generate unique variant combinations. (b) Based on each combination, a genetic risk score (GRS) was calculated for 588 individuals represented in the liver eQTL dataset7. According to the individual genetic risk profile, each person was graded into risk groups, with a low-risk group containing the 30%-quantile of the lowest scores and a high-risk group containing the 30%-quantile of the highest scores. Consequently, expression of 24,123 genes in liver tissue were compared between the risk groups.
Effects on the regulation of gene expression were determined based on the GRS approach, which categorizes individuals into low and high-risk groups for the respective variant combination. Subsequently, gene expression was compared between low and high-risk individuals (Fig. 2b). Group size was defined based on the ability to detect differently expressed genes, which were previously reported to be influences by AMD-associated variants (eGenes)7. For this purpose, a GRS including all seven variants assigned to the “lipid metabolism” category was calculated. Afterwards, different risk group sizes, ranging between 5 and 35% of the corresponding lowest or highest risk quantiles, were tested for the ability to detect four known eGenes, including LIPC, CETP, ALDH1A2 and ADAM107, after correction for multiple testing by a false discovery rate (FDR, Q-value) smaller than 5%. Consequently, low and high-risk groups were defined at the 30% highest and lowest quantiles, respectively (Supplementary Figure S1). These two groups each contain 176 liver tissue samples.
To define the significance threshold, we considered the expression of 24,123 genes for 127 combinations requiring over 3 million tests in the category “lipid metabolism” (Table 1). Quantile–quantile plots with all P-values of tests performed in the respective categories allow adjustment for multiple testing (Supplementary Figure S2a). We defined the FDR to be under 5%, as this threshold produces comparable results to the thresholds that can be derived from the quantile–quantile plots.
Table 1.
Statistical overview of variant interactions investigated.
| Lipid metabolism pathway | Extracellular matrix pathway | Immune pathway | |
|---|---|---|---|
| Independent signalsa | 7 | 15 | 19 |
| Combinations | 127 | 32,767 | 524,287 |
| Tests (24,123 genes) | 3 M | 790 M | 12.6 B |
| Significance threshold | Q-value < 0.05 | Q-value < 0.05 | P-value < 10–5 |
| Significant results | 2 | 0 | 641,016 |
| eUnique eCombinations | 2 | 0 | 326,144 |
| Unique eGenes | 1 | 0 | 1725 |
aIndependent signals = independent AMD-associated genetic variants as defined by Fritsche et al.2
Interaction of assigned variants in the lipid metabolism pathway
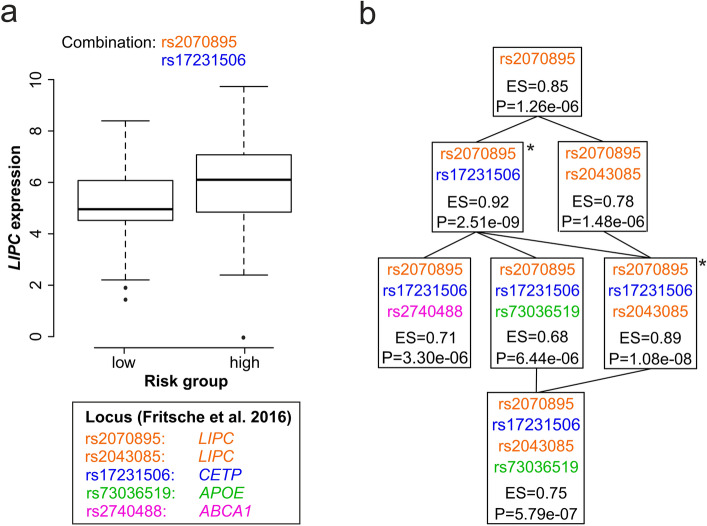
We identified two eCombinations (rs2070895/rs17231506; rs2043085/rs2070895/rs17231506) in the lipid metabolism pathway significantly influencing gene expression in liver. Both eCombinations correlate with the expression of the LIPC gene (Supplementary Table S2). The eCombination with the highest significance include the variant rs2070895 (LIPC locus, independent signal 23.2 in ref.2) and rs17231506 (CETP locus) (Fig. 3a,b). Based on the two variants individuals with a high GRS showed a significantly increased expression of LIPC, in comparison to individuals with a low GRS (effect size (ES) = 0.92, standard error (SE) = 0.15, Q-value = 7.7 × 10–3). The second eCombination with a significant impact on LIPC expression includes the same variants as the first eCombination, and additionally rs2043085 (LIPC locus, independent signal 23.1 in ref.2). Again, individuals with a high GRS have a higher LIPC expression (ES = 0.89, SE = 0.15, Q-value = 1.65 × 10–2).
Figure 3.
eCombinations altering expression of LIPC. (a) The boxplot compares LIPC expression between the AMD low and high-risk groups based on the genetic profile of the two genetic variants rs2070895 (orange, located in the LIPC locus) and rs17231506 (blue, located in the CETP locus). The y-axis corresponds to the relative level of gene expression. (b) The tree chart shows related variant combinations influencing the expression of LIPC. The chart starts with a single variant combination at the top and adds one variant per step downwards. Every box represents a set of variants used to determine risk groups. The P-value (P) and the effect size (ES) display the relationship between calculated risk groups and gene expression. Combinations highlighted with an asterisk remain significant after adjustment for multiple testing (Q-value < 0.05). Color code of variants refer to their locus affiliation.
Next, we investigated additional eCombinations and their effect on gene expression differences between risk groups. We modified the combinations step-wise by adding or removing genetic variants. This identified less strong, and less significant effects (Fig. 3b). Of note, in our dataset the single variant rs2070895, which contributes to a known liver eQTL with LIPC7, revealed no significant effect after correcting for multiple testing.
Interaction of assigned variants in the extracellular matrix pathway
Fifteen independent genetic variants were assigned to the extracellular matrix pathway (Fig. 1b), resulting in 32,767 unique combinations. We then analyzed the effect of each combination on the expression of 24,123 genes, which required over 790 million tests (Table 1). Applying the same threshold as described above, no significant correlation was observed (Q-value < 0.05) (Supplementary Table S3). This was confirmed by a quantile–quantile-plot, which showed no P-value differing from those expected by chance (Supplementary Figure S2b). Interestingly, the two eCombinations significant in the lipid metabolism pathway were included in this analysis, but failed to remain significant due to the higher burden of multiple testing.
Interaction of assigned variants in the complement pathway
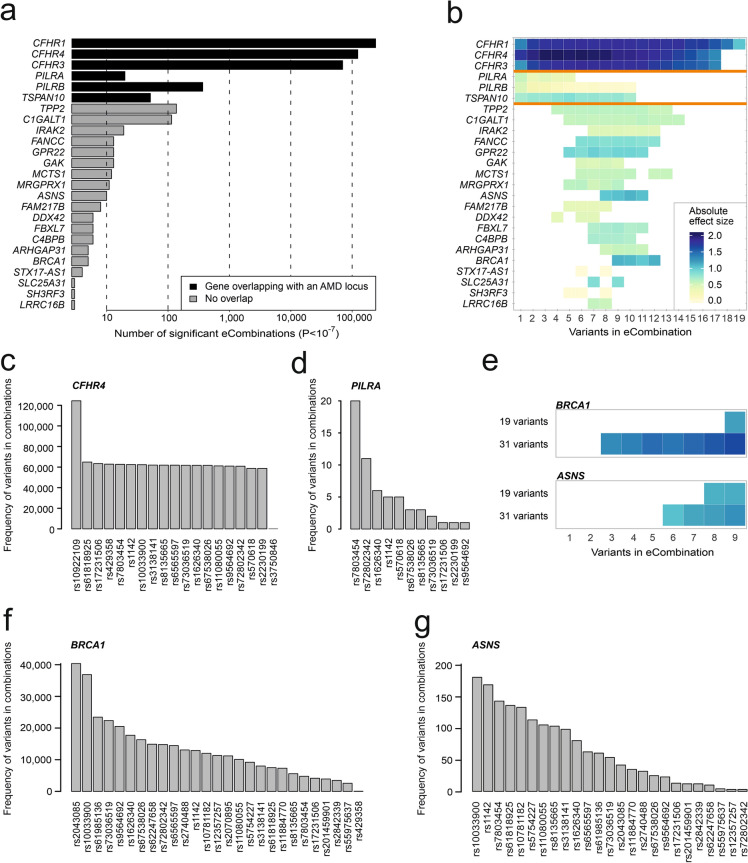
The 19 variants assigned to the complement pathway category result in 524,287 unique combinations which would require over 12 billion tests. A quantile–quantile-plot demonstrated that P-values smaller than 10–5 did not occur by chance (Supplementary Figure S2c). Altogether 641,016 significant results were observed, including 326,144 unique eCombinations and 1725 eGenes, whereby eCombinations could influence the expression of more than one eGene (Table 1, Supplementary Table S4). To focus on the most significant effects, the significance threshold was lowered to a P-value < 10–7 and only eGenes detected by at least three different eCombinations were analyzed (Supplementary Table S5). The filtering reduced the significant results to 438,219, including 243,302 unique eCombinations and 25 eGenes (Fig. 4a). Interestingly, over 99% of all significant results correspond to expression alterations of the complement factor H related genes 1, 3 and 4 (CFHR1: 242,652 eCombinations, CFHR4: 124,372 eCombinations, and CFHR3: 70,362 eCombinations). To further evaluate significant eCombinations, we initially focused on the number of variants included in the various combinations. This defined three categories of genes (Fig. 4b):
Figure 4.
Interaction of genetic variants assigned to the immune pathway. (a) Number of eCombinations (P-value < 10–7) influencing the expression of the respective eGene given on the Y-axis. eCombinations were generated from genetic variants assigned to the immune pathway by Fritsche et al.2 and only eGenes which were detected at least three times are shown. Applying these thresholds, 438,219 significant results were obtained and related to expression changes of 25 eGenes. Black bars highlight eGenes that overlap with a known AMD locus2. (b) Color-coded effect sizes (ES) reflecting the correlation of eCombinations and gene expression. Colored fields show the absolute ES of an eCombination. White fields indicate a non-significant combination. The X-axis displays the number of genetic variants included in the respective eCombination. The orange line separates three subtypes of results, specifically one group including the CFH-related genes with one genetic variant being sufficient for eQTL detection and high absolute ES (highest absolute ES > 1.8), one group with a single genetic variant being sufficient for eQTL detection but lower ES (highest absolute ES < 1), and one group where an effect on gene expression can be observed by combining several genetic variants. (c,d) Contribution of different genetic variants to combinatory effects on gene expression of CFHR4 (c, 124,372 eCombinations) and PILRA (d, 20 eCombinations). (e) Comparison of testing combinatory effects of all 31 AMD-associated variants and testing combinatory effects of 19 variants assigned to the category immune system process. The heatmap functionally refers to (b). (f,g) Contribution of different variants to combinatory effects on gene expression of BRCA1 (f, 40,903 eCombinations) and ASNS (g, 192 eCombinations) with all 31 variants included in the analysis. Results refer to a P-value threshold < 10–7.
eGenes influenced by single variants and combinatory effects with almost any variant
This group refers to CFH-related genes CFHR1, CFHR3, and CFHR4, which also show the strongest differences in expression between low and high-risk individuals (highest absolute ES > 1.8) (Fig. 4b). To identify effect-driving variants, we examined how often each variant is represented among all eCombinations for the respective eGene (Supplementary Table S6). For example, CFHR4 expression is significantly altered by 124,372 eCombinations, and each one of these combinations includes the AMD-associated variant rs10922109 (CFH locus) (Fig. 4c). In contrast, variant rs3750846 (locus ARMS2/HTRA) contributes to 103 of the 124,372 eCombinations while all other variants in this category contribute to approximately 50% of eCombinations, including variants rs570618 and rs61818925 located at the CFH locus (Supplementary Table S6).
eGenes influenced by single variants and combinations including a maximum of 10 variants
The second group of eGenes includes the genes PILRA, PILRB, and TSPAN10. Overall, the observed ESs are smaller than in the first group of eGenes (highest absolute ES < 1). A single variant (rs7803454, PILRB/PILRA locus) drives the strongest effect on expression of PILRA (ES = 0.66, SE = 0.09, Q-value = 9.21 × 10–11) and PILRB (ES = 0.59, SE = 0.04, Q-value = 7.97 × 10–29) (Supplementary Table S5). Also, this variant contributes to every eCombination identifying PILRA (Fig. 4d), although the ES decreases when additional variants join the eCombination (Fig. 4b). In contrast, an eCombination of 5 variants shows the strongest effect on TSPAN10 expression (ES = − 0.93, SE = 0.14, Q-value = 2.77 × 10–9) (Fig. 4b).
eGenes exclusively influenced by combinatorial effects
Nineteen eGenes within the third group reveal changes in gene expression due to eCombinations of at least 4 and a maximum of 14 variants (Fig. 4b). BRCA1 and ASNS show the highest ESs in this group with downregulation of BRCA1 (ES = − 1.18, SE = 0.21, Q-value = 1.55 × 10–7; 12 variants), and upregulation of ASNS (ES = 1.17, SE = 0.20, Q-value = 9.36 × 10–8; 10 variants) (Supplementary Table S5).
Validation of regulatory effects of variants assigned to the complement pathway
To validate the methodological approach, we established a two-step protocol based on data of the genotype-tissue expression (GTEx) project. In a first step, we tested whether our findings are liver-specific. Consequently, we selected the five eCombinations with the strongest absolute ES for the 25 eGenes identified (Fig. 4a) and analyzed their regulatory effect on the respective gene in three tissues with comparable sample sizes to the liver data, namely skeletal muscle (584 samples), whole blood (556 samples) and subcutaneous adipose tissue (477 samples) (Supplementary Table S7). Out of the 25 genes, four (CFRH4, SLC25A31, GPR22 and MRGPRX1) were not expressed in the selected tissues. Of the remaining 21 genes, the regulatory effects on genes known for their local eQTL (CFHR1, CFHR3, PILRA, PILRB and TSPAN10) were regularly detectable with the exception of PILRA in whole blood for any of the tested eCombinations including the single eVariant rs7803454 (representing a local eQTL). Regulatory effects on the remaining genes, which are not known for their local eQTL and mostly exhibited smaller ESs in the initial analysis in liver tissue, were observed only sporadically in the three additional tissues examined, and not always with the same effect direction. This indicates that the observed regulatory interaction effects of AMD-associated variants on gene expression represent mainly tissue-specific effects.
The second validation step applied an undirected approach. Essentially, we repeated the interaction analysis to identify eGenes influenced by interaction of complement pathway assigned variants in the GTEx tissue skeletal muscle. Applying a threshold of P < 10–5, we observed 160,466 significant results, including 102,293 unique eCombinations and 1425 unique eGenes (Supplementary Table S8). A direct comparison with the results obtained at the same threshold in liver tissue (P-value < 10–5, Supplementary Table S4) revealed 80 eGenes identified in both tissues (out of 1725 unique eGenes in liver and 1425 unique eGenes in muscle). Moreover, screening for the eGenes with the most robust effects in liver (25 eGenes, P-value < 10–7 and detected by at least three eCombinations, Supplementary Table S5), three eGenes were also detected in muscle (Supplementary Table S8), namely PILRA, PILRB and IRAK2. While PILRA and PILRB both represent eGenes with known strong local eQTL effects, IRAK2 was solely detected in liver when a risk profile including 7 to 12 AMD-associated variants was examined (Supplementary Table S5, Fig. 4b). In muscle, a regulatory effect on IRAK2 was detected by 4 eCombinations, including 4 to 6 variants (Supplementary Table S8).
Applying the same critical thresholds for muscle tissue as was done for liver to address the strongest effects (P-value < 10–7 and only eGenes detected by at least three eCombinations), 41,206 significant results were obtained, including 17,836 unique eCombinations and 17 unique eGenes (Supplementary Table S9). Comparing these results with liver data for the same thresholds (Supplementary Table S5), identified two eGenes detected in both tissues, namely PILRA and PILRB.
Together, the methodological validation demonstrates that the combination of GRS and eQTL analyses represents a valid approach to investigate regulatory effects of a defined genetic risk profile on gene expression in independent datasets and multiple tissues.
Resolution of regulatory effects on BRCA1 and ASNS expression in liver
The combination of GRS and eQTL analyses has the power to identify novel potentially disease-relevant eGenes based on epistatic effects of available eCombination data. In liver tissue strongest effects for novel AMD eGenes were observed for BRCA1 and ASNS. These genes become evident when testing variant combinations for a joint effect on gene expression within the complement pathway. There is currently no known involvement of the BRCA1 or ASNS gene in this pathway, or more specifically in AMD pathology. To gain a deeper insight by refining the risk profile required for a change of expression, we assessed combinations of all 31 AMD-associated genetic variants included in our dataset (Fig. 4e) and restricted testing for combinations consisting of a maximum of 9 variants due to the large computational and multiple testing burden. A P-value threshold of < 1 × 10–7 resulted in 40,903 eCombinations with impact on BRCA1 expression (Supplementary Table S10), and 192 eCombinations with impact on ASNS expression (Supplementary Table S11). Interestingly, the analysis including the 31 variants demonstrates even stronger ESs when compared with the results exclusively obtained with the complement-related variants (Supplementary Table S5). For example, BRCA1 expression reveals an ES of − 1.61 (SE = 0.22, Q-value = 5.04 × 10–6) in contrast to the initially strongest ES of − 1.18 (SE = 0.21, Q-value = 1.55 × 10–7). ASNS also shows a small increase of ESs from the initially strongest ES of 1.17 (SE = 0.20, Q-value = 9.36 × 10–8) to an ES of 1.25 (SE = 0.20, Q-value = 8.99 × 10–6) (Fig. 4e).
Regarding BRCA1 expression, our analysis identifies two effect-driving variants with rs2043085 (LIPC locus) in 98.7% (40,366) and rs10033900 (CFI locus) in 90.2% (36,873) of the 40,903 eCombinations (Fig. 4f). 25 variants appear less often (0.10–57.31%) and 4 variants do not contribute to any significant eCombination, namely rs10922109 (CFH locus), rs570618 (CFH locus), rs3750846 (ARMS2/HTRA1 locus) and rs2230199 (C3 locus). Interestingly, the odds ratios (ORs) in the AMD GWAS of contributing variants vary between 1.09 and 1.49 while the ORs of non-contributing variants vary between 1.47 and 2.932.
ASNS expression is mostly driven by eCombinations containing rs10033900 (CFI locus) (182 eCombinations, 94.8%), followed by rs1142 (KMT2E/SRPK2 locus) (170 eCombinations, 88.54%) (Fig. 4g). The frequency of the other 23 variants in the eCombinations vary between 75% (144 eCombinations) and 1.56% (3 eCombinations). Six variants exert no influence on ASNS expression, including rs2070895 (LIPC locus) and rs429358 (APOE locus) as well as the same 4 variants which also do not contribute to BRCA1 expression.
Discussion
Within the past 15 years, GWAS have deepened our knowledge on the genetic factors conferring risk to develop complex phenotypes. Still, enormous challenges remain to better understand the complexities and interactions of the genetic contributions to disease. Specifically, functional consequences arising from multiple genetic variants, individually often contributing only small effects, need to be addressed. The latter issue is key insofar as it reflects a situation where a person almost always harbors various combinations of multiple risk variants. With this in mind, we aimed for an approach which tests epistatic effects of independent GWAS variants associated with AMD. We used variant combinations to generate genetic risk models in a dataset of 588 individuals with unknown AMD status. The genetic information was then correlated with gene expression in a model tissue, namely liver. This approach revealed 26 genes with a significantly altered expression in high-risk versus low-risk variant combinations. Of these, seven eGenes, namely LIPC, CFHR1, CFHR4, CFHR3, PILRA, PILRB, and TSPAN10 were previously reported to be regulated by single AMD-associated variants7 while 19 eGenes represent exciting new candidates becoming evident only due to additive effects of AMD-associated genetic variant combinations.
Choosing three biologically relevant pathways in AMD2,6,14,20, we interrogated previous GWAS data to determine 21 functionally associated loci corresponding to 31 independent genetic variants. Testing all possible combinations would have generated a multiple testing burden of over 52 trillion tests, likely obscuring true positive signals22. Even reducing the number of genetic variants by pathway allocation only remotely reduces the impact of multiple testing. For example, the analysis of seven variants within the lipid metabolism pathway-related loci required around 3 million tests which, after correction for multiple testing, resulted in two eCombinations correlated with LIPC expression. Already the combination of 15 variants located in extracellular matrix pathway-related loci resulted in 790 million tests and revealed no significant result, even though all combinations of the lipid metabolism pathway approach were included in this analysis. This demonstrates the need to initially define a precise hypothesis to keep the number of tests within a reasonable magnitude consequently avoiding false-negative results due to excessive test corrections.
The correlation of LIPC expression and AMD pathology was established before7,23. One study showed an increased LIPC expression in AMD by applying a machine learning approach known as PrediXcan, which predicts gene expression based on genetic variation23. This approach, however, does not allow a resolution of the association down to a distinct variant. In a second study applying eQTL analysis in liver tissue, LIPC expression was directly correlated with local risk variant rs20708957. Interestingly, the current analysis revealed a significant correlation between an increased LIPC expression and a risk genotype containing rs2070895 (LIPC locus, chromosome 15q21.3) and rs17231506 (CETP locus, chromosome 16q13). The combination of these two variants displayed an even higher ES than rs2070895 alone. The mechanism of the combined effect of these two variants on LIPC expression needs to be clarified, specifically since LIPC and CETP are attributed to distinct processes within the lipid metabolism pathway7,24–26.
Gene expression regulation by genetic variants at immune-related loci initially revealed a remarkably high number of over 1700 eGenes. We therefore focused on the most significant results with P-value < 1 × 10–7 and filtered for eGenes which are correlated with at least three eCombinations leaving 25 eGenes of interest. Remarkably, three of these 25 eGenes were also detected by applying our two-step approach in an independent dataset of skeletal muscle tissue from the GTEx project. This suggests that combining GRS and eQTL analyses yield valid and robust results. Six of the eGenes identified in liver have been linked to AMD before and are well-known eQTL genes, namely CFHR1, CFHR3 and CFHR4, as well as PILRA, PILRB, and TSPAN106,7,23,27. The major effect driving variant for the three CFH-related genes is rs10922109 which is located within the CFH locus. The addition of further variants to the genetic risk model reveals only little impact on ESs. In fact, including rs3750846, a variant localized within the ARMS2/HTRA1 locus, eliminates the effect on the expression of CFH-related genes. This could suggest that CFH-related genes and relevant genes in the ARMS2/HTRA1 interval are involved in independent mechanisms leading to AMD pathology.
Of the 26 eGenes with differential expression in high-risk versus low-risk variant combinations, 19 eGenes have not been associated with AMD pathogenesis before. We speculate that the novel findings are the result of a summation of small unidirectional effects of single genetic variants. Of note, the corresponding variants are often located in distant (trans) loci, such as eCombination rs2043085 (chromosome 15) and rs10033900 (chromosome 4), which in combination with further variants influence the expression of BRCA1 (chromosome 17). Some of such distant variant combinations have ESs on gene expression that are even stronger than previously reported cis-eQTL variants7. Of note, a validation of our approach in different tissues available in the GTEx dataset highlight that most of the 19 eGenes associated with AMD for the first time, likely represent liver-specific findings.
Following our eCombination risk approach, the unexpected findings for BRCA1 and ASNS expression were substantiated by generating combinations from the full set of 31 independent AMD-associated variants included in our dataset. Considering the burden of multiple testing, we explored a maximum of nine variants per combination. This resulted in even stronger effects than in our initial findings which were limited to results from loci pre-assigned to the complement pathway. Loss of function mutations in BRCA1 are foremost known to be correlated with an increased risk to develop breast and ovarian cancer28. Functionally, BRCA1 is involved in repairing double strand breaks but has also been reported to regulate telomere length and stability29. Shortened telomere length is related to cell senescence, and double strand breaks are considered to be highly relevant in aging30,31. As age is a major risk factor for the development of AMD, this is a potential confounder which was adjusted for in the current study. Our data now point to downregulation of BRCA1 expression as part of the AMD pathomechanism which is further supported by a previous study that implicated DNA damage and a reduced DNA repair potential in AMD etiology32. Another remarkable finding in our analysis relates to the eGene ASNS, whose protein product catalyzes the conversion of aspartate to asparagine in the presence of glutamine33. A recent study examined the role of asparagine and glutamine in angiogenesis, and demonstrated that silencing of ASNS inhibited endothelial cell sprouting34. Our data now demonstrate that an increased AMD risk is significantly correlated with an upregulation of ASNS expression. This could make ASNS a pro-angiogenic factor possibly involved in neovascular complications of late-stage AMD.
It is important to note that the present study is not suited to identify novel genetic variants associated with AMD as conventional GWAS usually do. In fact, our approach facilitates the detection of effects on so far undetected gene expression that only becomes apparent if an individual carries a distinct combination of known AMD-associated risk variants. This represents a unique approach to find disease-associated genes that are potentially hidden in existing GWAS data. It allows the identification of individuals at risk who might especially benefit from a certain prevention strategy or therapy. Although most of our findings show small to moderate ES, the altered regulation of gene expression likely exerts a lifetime effect and it is conceivable that the effects on gene expression even increase in combination with other factors, like ageing or smoking.
Taken together, our study presents a novel approach to investigate joint effects of genetic variants on gene expression by combining GRS and eQTL mapping. We replicate a number of previous eQTL findings in AMD GWAS data, and report 19 novel genes correlating with the genetic risk to develop AMD. All genes were identified by jointly analyzing several seemingly independent AMD-associated signals, which is perfectly in-line with the idea that the signals underlying GWAS associations contribute to shared biological mechanisms.
Methods
AMD risk variants and dataset
52 independent genetic variants associated with AMD and their respective ORs were extracted from the most recent GWAS dataset2. Genotype and gene expression data of liver tissue from 588 individuals of European ancestry were provided by Strunz and colleagues7. The latter study combined data from four different studies35–38 and contains expression information on 24,123 genes. From genotype data, variants with (1) unavailable information, (2) a minor allele frequency less than 5% and (3) multi-allelic variants, were excluded. Thus, genotypes of 31 of the 52 AMD-associated variants were available for our study (Supplementary Table S1).
Category definition
The independent AMD variants were assigned to three superordinate biological processes according to biological pathways relevant in AMD pathology. To this end, 368 genes located in AMD loci as defined by Fritsche and colleagues (index variant and proxies (r2 > 0.5 and ± 100 kb) were included2. Functional profiling21 of these genes revealed 48 significantly enriched biological processes (g:SCS algorithm corrected for multiple testing, user threshold: 0.05). The superordinate biological processes were detected by connecting all significant processes in ancestor charts (https://www.ebi.ac.uk/QuickGO/, accessed November 2019). Two processes, “Cholesterol transport” and “Regulation of plasma lipoprotein”, were located in independent ancestor trees and were merged to one category (“lipid metabolism pathways”) due to their high similarities. An AMD locus was assigned to a category when at least one gene overlapping with the locus was assigned to the respective biological process.
Variant combination and GRS calculation
To generate the variant combinations, we used the R39 function combn39. Every combination consists of a sequence of variants that is unique in its composition. The number of variants in a combination varies between 1 and the total number of variants used for calculation. For every combination of variants a unique GRS was calculated as reported elsewhere40. In short, the GRS is defined as the sum of risk alleles of one individual weighted by the respective ES of each variant, which was obtained from the latest AMD GWAS2. As a consequence, the OR was transformed by the natural logarithm. Instead of centering the GRS, we normalized it by dividing it through the average ES of all included variants as reported elsewhere13.
Determination of risk group sizes
To determine the risk group sizes, all seven variants assigned to the superordinate biological process “Cholesterol transport/Regulation of plasma lipoprotein” were used to generate a GRS. According to their GRS, individuals were classified into risk groups. To determine the size of the risk groups ensuring sufficient power for subsequent tests, we performed eQTL analysis in low and high-risk groups with different group sizes (5%, 10%, 15%, 20%, 25%, 30% and 35%) of the entire dataset. eQTL analysis were performed with Matrix eQTL41. The group size was regarded to be sufficient in cases in which a replication could be shown for four previously detected eGenes, specifically LIPC, CETP, ALDH1A2 and ADAM10, in the same tissue7 with a FDR42 smaller than 5%. A proportion of 30% (176 samples) of the total cohort size (588 samples) was determined to be the minimal group size (Supplementary Figure S1).
Regression analysis, adjustment for multiple testing and trans-eQTL analysis
A multivariate linear regression was performed using Matrix eQTL41 to examine the relationship between AMD risk group and gene expression. The covariates age, gender, the original study representing the data source, and the first five genotype based principle components were included as provided by Strunz and colleagues7. The variants included in each initial category were analyzed in independent test series. We adjusted for multiple testing in every test series separately by controlling the FDR to be smaller than 5%. The FDR was calculated using the R function p.adjust (method “fdr”)39.
Validation of approach in GTEx
To validate our combined approach of GRS and eQTL, we used the genotype and gene expression data of the GTEx project (version 8). Whole genome sequencing data were retrieved from dbGaP (accession ID: phs000424.v8.p2) in VCF format43. Detailed information about the genotype processing and quality control (QC) protocols are provided elsewhere44. To determine ethnicity of samples, a principal component analysis (PCA) was carried out in R (version 3.3.1)45 using the snpgdsPCA46 function based on 100,000 random genetic variants of each sample and the corresponding genotype information of the 1000 Genomes Project reference panel (Phase 3, release 20130502)47. The first two principal components were plotted to determine the ethnicity. Only samples clustering next to the European reference individuals were included to consider the known variation of haplotype structures between populations. Gene expression and covariate data of three tissues were downloaded from the GTEx Portal48 and filtered for European individuals based on the genotype PCA. Altogether, genotype, gene expression, and covariate data from 584 muscle skeletal, 556 whole blood, and 477 adipose subcutaneous samples were included in the validation step. The AMD-associated variant rs61818925 (AMD signal 1.6)2 was not covered in the GTEx dataset and was replaced by the proxy rs61818924, which shows a R2 to rs61818925 of 0.8 in Europeans.
Supplementary Information
Acknowledgements
We thank Patricia Berber (Institute of Human Genetics, University of Regensburg) for critically reading the manuscript. The study was supported in part by institutional funds (TG77) of the Institute of Human Genetics Regensburg, Germany, and by a grant from the Helmut Ecker Foundation (Ingolstadt, Germany) to B.H.F.W. (No. 05/17). The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. The data used for the analyses described in this manuscript were obtained from the GTEx Portal under dbGaP accession number phs000424.v8.p2.
Author contributions
C.K. and T.S. contributed to study design, supervised the analysis and the writing of the manuscript. C.A.N. carried out the computational analysis and contributed to writing of a first draft of the manuscript. S.S. contributed to study and computer code design. T.S. performed the validation analysis based on the GTEx data. B.H.F.W. was responsible for study design, coordination of the study, and finalizing the manuscript. All authors reviewed the manuscript before submission.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Data availability
Genotype and gene expression data of the liver eQTL dataset were provided by Strunz and colleagues and are available in public databases as reported7. GWAS information for genetic variants, locus names and ORs were taken from Fritsche et al.2. Genotype, gene expression, and covariate data of the GTEx project are available in dbGaP (accession ID: phs000424.v8.p2) or the GTEx portal (http://www.gtexportal.org).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Christina Kiel, Christoph A. Nebauer and Tobias Strunz.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-92351-4.
References
- 1.Klein RJ, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fritsche LG, et al. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat. Genet. 2016;48:134–143. doi: 10.1038/ng.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rathje M, et al. Genetic variants in the bipolar disorder risk locus SYNE1 that affect CPG2 expression and protein function. Mol. Psychiatry. 2019 doi: 10.1038/s41380-018-0314-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Albert FW, Kruglyak L. The role of regulatory variation in complex traits and disease. Nat. Rev. Genet. 2015 doi: 10.1038/nrg3891. [DOI] [PubMed] [Google Scholar]
- 5.Battle A, Montgomery SB. Determining causality and consequence of expression quantitative trait loci. Hum. Genet. 2014;133:727–735. doi: 10.1007/s00439-014-1446-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ratnapriya R, et al. Retinal transcriptome and eQTL analyses identify genes associated with age-related macular degeneration. Nat. Genet. 2019;51:606–610. doi: 10.1038/s41588-019-0351-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strunz T, et al. A mega-analysis of expression quantitative trait loci (eQTL) provides insight into the regulatory architecture of gene expression variation in liver. Sci. Rep. 2018;8:5865. doi: 10.1038/s41598-018-24219-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strunz T, et al. A transcriptome-wide association study based on 27 tissues identifies 106 genes potentially relevant for disease pathology in age-related macular degeneration. Sci. Rep. 2020;10:1–16. doi: 10.1038/s41598-020-58510-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kiel C, Lastrucci C, Luthert PJ, Serrano L. Simple and complex retinal dystrophies are associated with profoundly different disease networks. Sci. Rep. 2017;7:41835. doi: 10.1038/srep41835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Camelo S. Potential sources and roles of adaptive immunity in age-related macular degeneration: Shall we rename AMD into autoimmune macular disease? Autoimmune Dis. 2014;2014:1–11. doi: 10.1155/2014/532487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morohoshi K, Goodwin AM, Ohbayashi M, Ono SJ. Autoimmunity in retinal degeneration: Autoimmune retinopathy and age-related macular degeneration. J. Autoimmun. 2009;33:247–254. doi: 10.1016/j.jaut.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 12.Wu J, Uchino M, Sastry SM, Schaumberg DA. Age-related macular degeneration and the incidence of cardiovascular disease: A systematic review and meta-analysis. PLoS ONE. 2014;9:e89600. doi: 10.1371/journal.pone.0089600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grassmann F, et al. Genetic pleiotropy between age-related macular degeneration and 16 complex diseases and traits. Genome Med. 2017;9:29. doi: 10.1186/s13073-017-0418-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ambati J, Fowler BJ. Mechanisms of age-related macular degeneration. Neuron. 2012 doi: 10.1016/j.neuron.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clark SJ, McHarg S, Tilakaratna V, Brace N, Bishop PN. Bruch’s membrane compartmentalizes complement regulation in the eye with implications for therapeutic design in age-related macular degeneration. Front. Immunol. 2017;8:1778. doi: 10.3389/fimmu.2017.01778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Betzler BK, Rim TH, Sabanayagam C, Cheung CMG, Cheng C-Y. High-density lipoprotein cholesterol in age-related ocular diseases. Biomolecules. 2020;10:645. doi: 10.3390/biom10040645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kersten E, et al. Systemic and ocular fluid compounds as potential biomarkers in age-related macular degeneration. Surv. Ophthalmol. 2018;63:9–39. doi: 10.1016/j.survophthal.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 18.Alper CA, Myron Johnson A, Birtch AG, Moore FD. Human C3: Evidence for the liver as the primary site of synthesis. Science. 1969;163:286–288. doi: 10.1126/science.163.3864.286. [DOI] [PubMed] [Google Scholar]
- 19.Morgan BP, Gasque P. Extrahepatic complement biosynthesis: Where, when and why? Clin. Exp. Immunol. 1997;107:1–7. doi: 10.1046/j.1365-2249.1997.d01-890.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lewis GF, Rader DJ. New insights into the regulation of HDL metabolism and reverse cholesterol transport. Circ. Res. 2005;96:1221–1232. doi: 10.1161/01.RES.0000170946.56981.5c. [DOI] [PubMed] [Google Scholar]
- 21.Reimand J, et al. g:Profiler-a web server for functional interpretation of gene lists (2016 update) Nucleic Acids Res. 2016;44:W83–W89. doi: 10.1093/nar/gkw199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bender R, Lange S. Adjusting for multiple testing—When and how? J. Clin. Epidemiol. 2001;54:343–349. doi: 10.1016/S0895-4356(00)00314-0. [DOI] [PubMed] [Google Scholar]
- 23.Strunz T, et al. A transcriptome-wide association study based on 27 tissues identifies 106 genes potentially relevant for disease pathology in age-related macular degeneration. Sci. Rep. 2020;10:1584. doi: 10.1038/s41598-020-58510-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okamoto H, et al. A cholesteryl ester transfer protein inhibitor attenuates atherosclerosis in rabbits. Nature. 2000;406:203–207. doi: 10.1038/35018119. [DOI] [PubMed] [Google Scholar]
- 25.Neale BM, et al. Genome-wide association study of advanced age-related macular degeneration identifies a role of the hepatic lipase gene (LIPC) Proc. Natl. Acad. Sci. 2010;107:7395–7400. doi: 10.1073/pnas.0912019107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burgess S, Davey Smith G. Mendelian randomization implicates high-density lipoprotein cholesterol-associated mechanisms in etiology of age-related macular degeneration. Ophthalmology. 2017;124:1165–1174. doi: 10.1016/j.ophtha.2017.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Orozco LD, et al. Integration of eQTL and a single-cell atlas in the human eye identifies causal genes for age-related macular degeneration. Cell Rep. 2020;30:1246–1259.e6. doi: 10.1016/j.celrep.2019.12.082. [DOI] [PubMed] [Google Scholar]
- 28.Boulton SJ. Cellular functions of the BRCA tumour-suppressor proteins. Biochem. Soc. Trans. 2006;34:633–645. doi: 10.1042/BST0340633. [DOI] [PubMed] [Google Scholar]
- 29.Rosen EM. BRCA1 in the DNA damage response and at telomeres. Front. Genet. 2013;4:1–14. doi: 10.3389/fgene.2013.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.White RR, Vijg J. Do DNA double-strand breaks drive aging? Mol. Cell. 2016;63:729–738. doi: 10.1016/j.molcel.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shay JW. Role of telomeres and telomerase in aging and cancer. Cancer Discov. 2016;6:584–593. doi: 10.1158/2159-8290.CD-16-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chu XK, et al. In-depth analyses unveil the association and possible functional involvement of novel RAD51B polymorphisms in age-related macular degeneration. Age. 2014;36:1453–1462. doi: 10.1007/s11357-014-9627-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patterson MK, Orr GR. Asparagine biosynthesis by the Novikoff Hepatoma isolation, purification, property, and mechanism studies of the enzyme system. J. Biol. Chem. 1968;243:376–380. doi: 10.1016/S0021-9258(18)99303-8. [DOI] [PubMed] [Google Scholar]
- 34.Huang H, et al. Role of glutamine and interlinked asparagine metabolism in vessel formation. EMBO J. 2017;36:2334–2352. doi: 10.15252/embj.201695518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aguet, F. et al. Genetic effects on gene expression across human tissues. Nature550, 204–213 (2017). [DOI] [PMC free article] [PubMed]
- 36.Schadt EE, et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Innocenti F, et al. Identification, replication, and functional fine-mapping of expression quantitative trait loci in primary human liver tissue. PLoS Genet. 2011;7:e1002078. doi: 10.1371/journal.pgen.1002078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schröder A, et al. Genomics of ADME gene expression: Mapping expression quantitative trait loci relevant for absorption, distribution, metabolism and excretion of drugs in human liver. Pharmacogenomics J. 2013;13:12–20. doi: 10.1038/tpj.2011.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.R Development Core Team, R. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, vol. 1, (2011).
- 40.Grassmann F, Fritsche LG, Keilhauer CN, Heid IM, Weber BHF. Modelling the genetic risk in age-related macular degeneration. PLoS ONE. 2012;7:e37979. doi: 10.1371/journal.pone.0037979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shabalin AA. Matrix eQTL: Ultra fast eQTL analysis via large matrix operations. Bioinformatics. 2012;28:1353–1358. doi: 10.1093/bioinformatics/bts163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B. 1995;57:289–300. [Google Scholar]
- 43.Danecek P, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aguet F, et al. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369:1318–1330. doi: 10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.R Team Core. A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2017).
- 46.Zheng X, et al. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics. 2012;28:3326–3328. doi: 10.1093/bioinformatics/bts606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Altshuler DM, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.GTEx Portal datasets. https://www.gtexportal.org/home/datasets (Accessed 7 July 2020)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genotype and gene expression data of the liver eQTL dataset were provided by Strunz and colleagues and are available in public databases as reported7. GWAS information for genetic variants, locus names and ORs were taken from Fritsche et al.2. Genotype, gene expression, and covariate data of the GTEx project are available in dbGaP (accession ID: phs000424.v8.p2) or the GTEx portal (http://www.gtexportal.org).