Abstract
Testicular development and function rely on interactions between somatic cells and the germline, but similar to other organs, regenerative capacity declines in aging and disease. Whether the adult testis maintains a reserve progenitor population remains uncertain. Here, we characterize a recently identified mouse testis interstitial population expressing the transcription factor Tcf21. We found that TCF21lin cells are bipotential somatic progenitors present in fetal testis and ovary, maintain adult testis homeostasis during aging, and act as potential reserve somatic progenitors following injury. In vitro, TCF21lin cells are multipotent mesenchymal progenitors which form multiple somatic lineages including Leydig and myoid cells. Additionally, TCF21+ cells resemble resident fibroblast populations reported in other organs having roles in tissue homeostasis, fibrosis, and regeneration. Our findings reveal that the testis, like other organs, maintains multipotent mesenchymal progenitors that can be potentially leveraged in development of future therapies for hypoandrogenism and/or infertility.
Subject terms: Germline development, RNA sequencing, Transcriptomics, Reproductive biology, Mesenchymal stem cells
Whether the adult testis harbours a somatic progenitor population is unknown. Here, the authors provide evidence that the testis interstitial cells expressing the transcription factor Tcf21 maintain adult testis homeostasis during aging, and act as potential reserve somatic progenitors following injury.
Introduction
Sexual reproduction relies on the generation of distinct sexes to increase biological diversity. The core of this strategy rests with a bipotential gonadal primordium that supports development of sex-specific reproductive organs with functionally distinct gonadal cell types. The gonadal primordium is comprised of primordial germ cells and mesenchymal cells that originate from two sources: the coelomic epithelium1–5 and the mesonephros6. In early development, the coelomic epithelial cells give rise to multiple cell lineages, including interstitial cells and Sertoli cells1, but later become restricted to the interstitial compartment of the testis. In contrast, mesonephric-derived cells migrating into the gonad contribute only to fetal/adult Leydig and interstitial cells, but not Sertoli cells7,8. Hence, the gonadal mesenchyme is heterogeneous on the molecular and cellular scales, and certain somatic lineages (e.g. Leydig and myoid) have multiple cells of origin9, pointing to a complex developmental programming of reproductive organs (reviewed in10–12).
Establishment of a functional somatic microenvironment is essential for continuous sperm production, germ cell homeostasis, and regeneration13–15. Disruptions in somatic cell populations can dramatically alter germ cell development and testis function. For instance, genetic ablation of macrophages in the adult testis leads to disruption of spermatogonial proliferation and differentiation16. Others have shown that testicular endothelial cells14 as well as lymphatic endothelial cells15 support human and mouse spermatogonial stem cell survival and expansion, and can also modulate spermatogonial cell homeostasis and regeneration15. Therefore, these studies underscore the importance of defining the interstitial cell composition and function.
The testis interstitial cells are believed to be postmitotic (not actively proliferating). However, studies in rats have demonstrated that adult Leydig cells can regenerate after ethane dimethane sulfonate (EDS)-induced cell death (reviewed in17). Multiple interstitial cell (CD90/PDGFRA/COUPTFII/NESTIN) populations were shown to re-enter the cell cycle upon EDS-induced Leydig cell death, suggesting that the interstitial compartment contains a reserve Leydig or a general somatic cell progenitor population that can be activated in response to damage18–26. Without single-cell RNA-seq analysis it remains difficult to tease apart whether these markers are observed in a single homogenous population of Leydig stem cells or if these cells are a heterogenous pool of progenitors. Furthermore, it is also unclear if a common somatic progenitor could give rise to additional cell types other than Leydig, and what the role of such stem/progenitors would be in the normal adult testis.
Using single-cell RNA sequencing (scRNA-seq) we previously identified a mesenchymal cell population in the adult mouse testis that expresses the transcription factor Tcf21 and appears in vivo as rare spindle-shaped cells surrounding the seminiferous tubule13. The Tcf21 gene has known roles in the development of multiple organs, including the testis27–29. Loss of Tcf21 Tcf21 promotes feminization of external genitalia in karyotypically male mice30, while overexpression of Tcf21 in primary embryonic ovary cells leads to in vitro sex-reversal via aberrant anti-Mullerian hormone expression31. These results provide evidence for a role of TCF21 in male sex determination and testis somatic cell differentiation. Recent reports of single-cell sequencing during sex determination and cell linage specification also identified Tcf21 expression among subsets of gonadal somatic cells in both male and female, although these experiments were limited to NR5A1-eGFPcells32,33. The similarity between the adult Tcf21+ population and fetal somatic progenitors32,33 suggests that the Tcf21+ population that persists in adulthood may retain fetal developmental or functional properties.
In this study, we examine the role of TCF21+ cells in the developing and adult mouse testis. To answer this question, we utilize genetic lineage tracing to mark and follow the potential and fate of the TCF21lin population both in vitro and in vivo. First, in directed in vitro differentiation paradigms, we find that flow-sorted TCF21lin cells possess mesenchymal stem cell (MSC)-like properties and can be directed to differentiate to either myoid or Leydig cell lineages, thus acting as true multipotent progenitors in vitro. In vivo, fetal TCF21lin cells are bipotential somatic progenitors, contributing to all known somatic cell populations in the fetal and adult testis and ovary. In the adult testis, the TCF21lin cells replenish somatic populations in response to injury as well as in normal aging. Furthermore, the adult testis Tcf21+ cells resemble resident fibroblast populations in multiple organs which have been implicated in tissue homeostasis, fibrosis, and regeneration. In summary, our work demonstrates the first evidence for a reserve somatic cell population in the adult testis, representing a potential targetable cell population for development of treatments for gonad-related defects and disease.
Results
The Tcf21+ population is a molecularly heterogenous mesenchymal cell population that is transcriptomically similar to myoid and Leydig cells
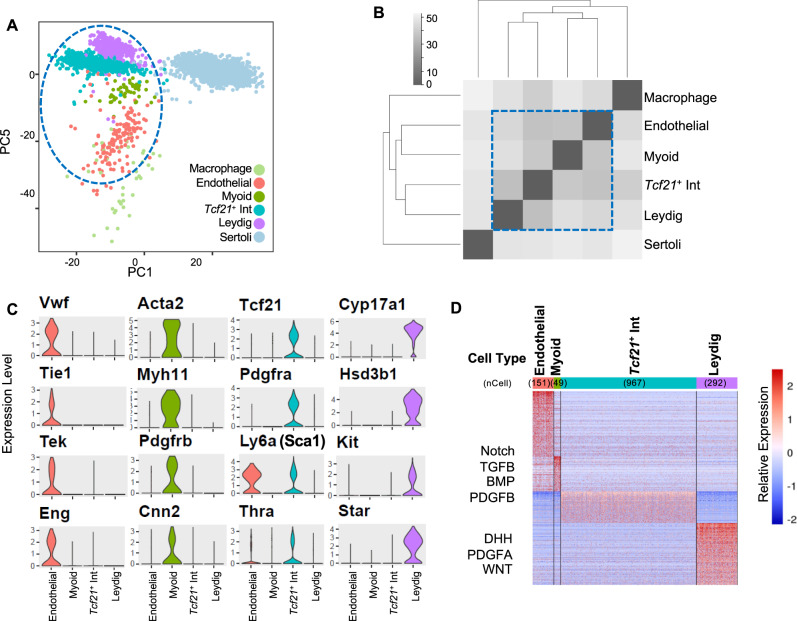
We recently employed single-cell RNA-seq (scRNA-seq) to generate a cell atlas of the mouse testis13,34. Our analysis of ~35,000 cells identified all known somatic cell types as well as an unexpected Tcf21+ population (Fig. 1A)13. To better understand its potential function, we first examined if the Tcf21+ population has molecular similarity to other known somatic cells in the testis. By using a pairwise dissimilarity matrix for somatic cell centroids, we find that the Tcf21+ population was distinct from macrophage and Sertoli lineages but transcriptomically similar to myoid, Leydig, and endothelial cells (Fig. 1B). In contrast with these cell types, Tcf21+ cells did not express any of their terminally differentiated markers (Fig. 1C). Rather, this population was uniquely demarcated by the expression of Tcf21, Pdgfra, CoupTFII, and mesenchymal progenitor cell (MP) markers including Sca1 (Fig. 1C, Supplementary Data 1), Arx, and Vim13. To define molecular properties of Tcf21+ cells more broadly we identified differentially expressed genes between the Tcf21+ population and its most similar cell types (using >2-fold change and FDR < 5%). Gene ontology analysis suggested that the Tcf21+ population is of mesenchymal origin, and likely involved in extracellular matrix (ECM) biology, tissue injury, and repair processes (Fig. 1D; Supplementary Data 1). Although the ECM was once considered a passive support scaffold, a wealth of data now suggest an active role for ECM in many aspects of biology, from tissue maintenance, regeneration, cell differentiation, to fibrosis and cancer35,36.
Fig. 1. Identification and characterization of a Tcf21-expressing interstitial somatic cell in the adult testis.
A Visualization of 6 somatic cell types in PC space. Data reprocessed from Green et al.13. Note somatic cell populations were enriched using a combination of cell surface markers or transgenic lines, therefore cell frequencies in PCA plot are not representative of in vivo. B Heatmap of dissimilarity matrix of somatic cell types illustrates high similarity for the Tcf21+ interstitial population with 3 somatic cell types—myoid, Leydig, and endothelial cells. Gray scale indicates Euclidean distance. C Violin plots of representative markers for endothelial, myoid, Tcf21+ interstitial population, and Leydig cells. D Heatmap of differentially expressed markers for each of the 4 closely related somatic cell types—endothelial, myoid, Tcf21+ interstitial population, and Leydig cells.
Given the identification of multiple mesenchymal progenitor markers in the Tcf21+ population, we next sought to validate expression of these markers in vivo. We took advantage of previously generated Tcf21mCrem mice, in which a tamoxifen inducible Cre recombinase inserted at the Tcf21 locus enables the long-term tracing of the descendant populations of the Tcf21-expressing cells, whether in the gonads, heart, kidney, and cranial muscle37. Specifically, testes were collected from Tcf21mCrem:R26RtdTom mice after 3 doses of tamoxifen (tdTom+ labeled cells referred to as TCF21lin), dissociated, stained for a comprehensive panel of mouse MSC markers (SCA1, CD73, CD29, CD34, and THY1) while excluding mature Leydig (cKIT+) or immune (CD45+) cells, and analyzed using flow cytometry (Supplementary Fig. 1A–F). Since our Tcf21+ cells were initially discovered by enriching SCA1+ cells in the testis, we examined the heterogeneity of the SCA1+ and TCF21lin cells. As expected, the SCA1+ population has a broader representation in the testis than TCF21lin (~3–5% vs. ~1–2%), with about 45% of SCA1+ cells being also positive for TCF21lin (Supplementary Fig. 1G)—a proportion consistent with our estimates from scRNA-seq data. Within the SCA1+ population, we identified two TCF21lin subpopulations (blue and purple in Supplementary Fig. 1B, C) based on co-expression of MSC markers: a larger TCF21lin population (13.8%, blue) that strongly expressed CD29 (ITGB1), CD73, and CD34, and a smaller discrete TCF21lin population (0.77%, purple) that expressed all MSC markers. In contrast, 80% of the TCF21lin population expresses SCA1 (Supplementary Fig. 1G), and within the TCF21lin populations we identify multiple subtypes that are molecularly heterogenous with respect to MSC marker expression (Supplementary Fig. 1D–F) (see below for the functional assessment of TCF21lin heterogeneity in vitro).
Given the heterogeneity observed within the TCF21lin population, we asked whether TCF21lin subtypes could be further defined by co-expression of previously described interstitial markers including PDGFRA, COUPTFII, CD34, and FGF5, respectively. To answer this question, we co-stained testes from Tcf21mCrem:R26RtdTom or Tcf21mCrem:R26RtdTom:PdgfraDGFRAGFP mice with COUPTFII, CD34, and FGF5 antibodies (Supplementary Fig. 1H), but we did not detect a preferred segregation of TCF21lin cells within the PDGRA, COUPTFII, CD34 or FGF5 subpopulation (Supplementary Fig. 1H), suggesting that these populations are heterogeneous on both the cellular and molecular level. Furthermore, for all markers analyzed we find co-stained cells both in the interstitium or surrounding tubules—suggesting that the cellular heterogeneity is not a result of spatial location.
The TCF21lin/SCA1+ population has mesenchymal progenitor properties in vitro and can be differentiated to Leydig and myoid cell fates in vitro
Mesenchymal progenitors are typically characterized by their capacity for forming adherent fibroblast-like colonies on plastic (measured as CFU-F: fibroblastic colony-forming units) and for differentiating into adipocytes, osteocytes, and chondrocytes in vitro (reviewed in38). Such populations have been isolated from multiple human and mouse organs, including juvenile or adult testes18,39–41. To examine whether the adult SCA1+ and TCF21lin cells have mesenchymal progenitor-like properties in vitro, we sorted four types of cells: SCA1−/cKIT+ interstitial cells (control), SCA1+/cKIT−, SCA1+/TCF21lin, or TCF21lin cells from Tcf21mCrem:R26RtdTom animals (Supplementary Fig. 2A) and plated these cells at single-cell density to measure clonogenic potential in vitro. Although SCA1+/cKIT−, SCA1+/TCF21lin, and TCF21lin populations all formed colonies in vitro, clonogenic potential differed across populations. The TCF21lin and SCA1+/TCF21lin double-positive cells formed the highest number of colonies—~100 colonies per 1000 plated cells, and this was followed by SCA1+/cKIT− cells (regardless of the TCF21lin status) and cKIT+ cells with ~40 and 10 colonies, respectively (Supplementary Fig. 2B). Altogether, these data demonstrate that SCA1+and SCA1+/TCF21lin populations have characteristics of mesenchymal progenitors in vitro, but selecting for the TCF21lin population significantly increases colony formation potential.
In addition to expanding in culture, mesenchymal progenitors, under appropriate conditions, can be directed to differentiate into adipocytes, chondrocytes, and osteoblasts. To examine if TCF21lin cells maintain such properties in vitro, we sorted either SCA1+/cKIT− or cKIT+ cells from Tcf21mCrem:R26RtdTom animals and examined whether the TCF21lin within the SCA1+ population contribute to all three lineages (Supplementary Fig. 2A). Importantly, we found that SCA1+ cells robustly differentiated into adipocytes, chondrocytes, and osteocytes as shown by Oil red, Alizarin red, and Alcian blue staining, respectively (Supplementary Fig. 2C). Furthermore, co-immunofluorescence of Osterix (osteocytes), Perilipin (adipocytes), and SOX9 (chondrocytes) with tdTom+ (TCF21lin) cells demonstrates that the TCF21lin cells within the SCA1+ population can contribute to all three lineages in vitro (Supplementary Fig. 2D).
Given the ability of the SCA1+/TCF21lin to generate multiple mesenchymal cell types in vitro and the transcriptomic relationship of Tcf21+ cells with both Leydig and myoid cells (Fig. 1B), we next asked whether the TCF21lin/SCA1+ population can be directed to differentiate to both Leydig and myoid cells in vitro and if the TCF21lin/SCA1+ population serves as a multipotent progenitor for Leydig and myoid lineages. To this end, we sorted SCA1+/cKIT− (regardless of TCF21 status) or TCF21lin/SCA1+/cKIT− cells from Tcf21mCrem:R26RtdTom adult male testes and directed their differentiation to either myoid or Leydig cells using a set of growth factors based on the repertoire of receptors expressed in our scRNA-seq datasets and earlier in vivo genetic findings of Leydig or myoid cell specification (Supplementary Fig. 2E–K)42–44. After treating the bulk SCA1+ or SCA1+/TCF21lin cells with a myoid differentiation cocktail which includes Smoothened agonist (SAG, an activator of Desert Hedgehog), PDGFAA, PDGFBB, ACTIVINA, BMP2, BMP4, and Valproic Acid (outlined in Supplementary Fig. 2E), we observed a morphological conversion of spindle-shaped cells to flattened and striated cells resembling smooth muscle cells (Supplementary Fig. 2F). This conversion was confirmed by expression of smooth muscle cell markers such as smooth muscle actin (Supplementary Fig. 2G, H).
Previous in vivo and in vitro experiments uncovered that Desert Hedgehog (DHH), FGF, and PDGF signaling are stimulatory to Leydig cell differentiation, while Notch signaling and other factors are inhibitory42–44. By incorporating these findings from the literature with our scRNA-seq data, we developed a 14-day differentiation protocol that includes PDGFAA, PDGFBB, SAG, FGF2, LiCl2, and DAPT (Notch inhibitor) (detailed in Supplementary Fig. 2I). This protocol enables successful differentiation of SCA1+/cKIT− or SCA1+/TCF21lin cells to Leydig cells (Supplementary Fig. 2J–K). Importantly, the in vitro-derived Leydig cells expressed steroidogenic factor 1 (SF1) (Supplementary Fig. 2K, L) and secreted testosterone (Supplementary Fig. 2M, N). Notably, Leydig cell generation and testosterone secretion was achieved in vitro independent of LH control (Supplementary Fig. 2M, N). Consistent with absence of LH regulation, we detected only low levels of luteinizing hormone/choriogonadotropin receptor (Lhcgr) transcripts in day 14 Leydig cells (Supplementary Data 2).
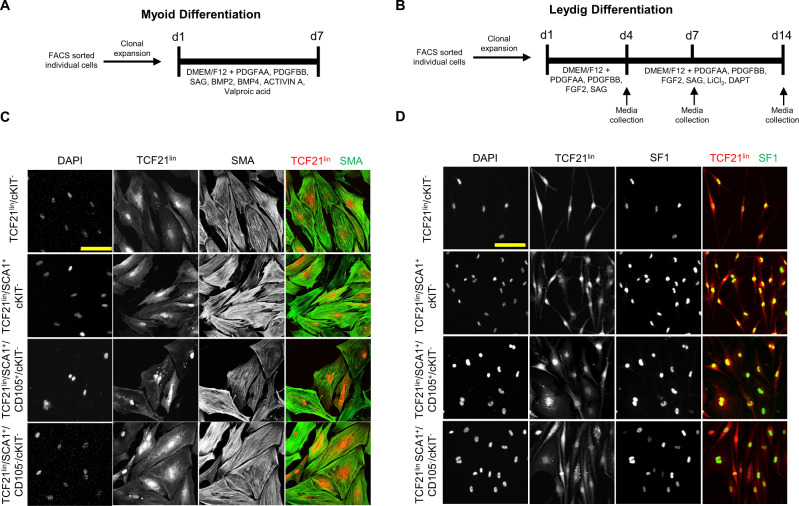
As the SCA1+/TCF21lin cells were labeled as a population, the results described above could not distinguish between two scenarios: (1) each TCF21lin/SCA1+ cell is a multipotent progenitor, capable of adopting any of multiple fates, or (2) these cells are heterogeneous, comprised of multiple subtypes of progenitors that each have been cryptically committed to differentiate into a different somatic lineage. To distinguish between these two scenarios, we sorted individual TCF21lin/SCA1+ cells into separate wells on 96-well plates and allowed each to expand into individual clones. Following clonal expansion, clones were randomly assigned to either the myoid or Leydig cell differentiation protocol (Fig. 2A, B). Therefore, if these cells have already been primed for one or the other lineage, we would expect that some clones, but not all, that undergo the myoid-inducing treatment will differentiate to the myoid lineage, and likewise some clones in the Leydig treatment group will follow the Leydig lineage. Co-immunostaining with either SMA for myoid cells or SF1 for Leydig cells reveals that all TCF21lin/SCA1+ clones in either group differentiate to adopt the induced fate 100% of the time (number of clones provided in Supplementary Table 1), suggesting that the TCF21lin/SCA1+ cells are individually multipotent in vitro (Fig. 2C, D; Supplementary Table 1). However, since the TCF21lin/SCA1+ population can be further stratified by the expression of CD105 in vivo, we examined whether TCF21lin/SCA1+ multipotency may be restricted to a specific subset of TCF21lin/SCA1+ cells. To address this, we sorted and differentiated TCF21lin/SCA1+/CD105+ or TCF21lin/SCA1+/CD105− cells to Leydig or myoid cells. Consistent with our earlier finding using TCF21lin/SCA1+ cells, both the TCF21lin/SCA1+/CD105+ and the TCF21lin/SCA1+/CD105− clonal cells differentiated to both lineages with 100% efficiency (Fig. 2C, D; Supplementary Table 1). These observations suggest that multipotency is not due to heterogeneous subpopulations of TCF21lin/SCA1+ cells identified by flow cytometry (Supplementary Fig. 1E, F). We caution, however, since these clonal cell experiments were performed in vitro, the multipotency of these cells in vivo remains to be confirmed in future studies.
Fig. 2. The adult TCF21lin cells are multipotent and can be directed to differentiate to Leydig and smooth muscle cells in vitro.
Schematic representation of experimental timelines for myoid (A) and Leydig cell (B) directed differentiation. Gating strategy used for FACS is presented in Supplementary Fig. 2G. C Immunofluorescence staining of smooth muscle actin (SMA) in clonal TCF21lin cells after 7 days of in vitro culture in the presence of differentiation media. Representative images of n = 13 technical replicates of TCF21lin/cKit−, n = 6 technical replicates of TCF21lin/SCA1+/cKit−, n = 6 technical replicates of TCF21lin/SCA1+/cKit−/CD105+, and n = 14 technical replicates of TCF21lin/SCA1+/cKit−/CD105. Scale bar: 100 μm for all images in the panel. D Immunofluorescence staining of steroidogenic factor 1 (SF1) in clonal TCF21lin cells after 14 days of in vitro culture in the presence of differentiation media. Representative images of n = 19 technical replicates of TCF21lin/cKit−, n = 17 technical replicates of TCF21lin/SCA1+/cKit−, n = 12 technical replicates of TCF21lin/SCA1+/cKit−/CD105+, and n = 22 technical replicates of TCF21lin/SCA1+/cKit−/CD105−. Scale bar: 100 μm for all images in the panel.
Single-cell time-course analysis reveals timing and diverging trajectories during Leydig cell differentiation
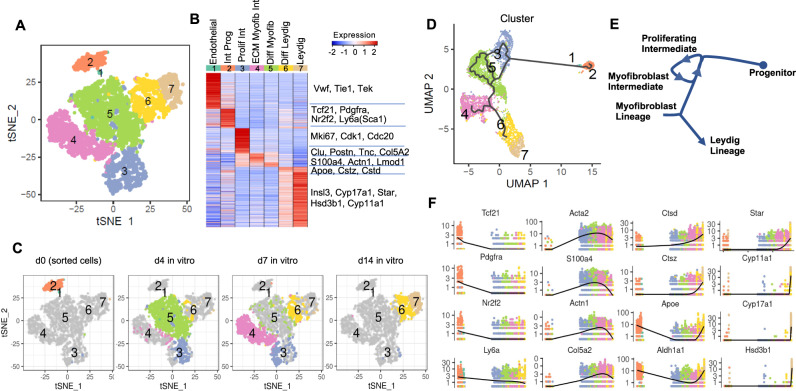
We next characterized the Leydig cell differentiation process in vitro using scRNA-seq. To this end, we collected and analyzed ~6500 cells across four time-points along the in vitro differentiation process (d0 sorted SCA1+/cKIT−, d4, d7, and d14 in culture). By clustering the merged dataset and using gene expression and marker gene analysis we identified seven distinct clusters (Fig. 3A, B). When overlaying time points on the different clusters, we find that freshly sorted SCA1+/cKIT− cells at d0 contribute to clusters 1 and 2 (Fig. 3C). A small number of cells in cluster 1 express von Willebrand factor (Vwf) and the receptor tyrosine kinase Tie-1, indicating that some endothelial cells also express Sca1 (Fig. 1C, 3B; Supplementary Data 2). However, cluster 2, which constitutes the majority of SCA1+/cKIT− cells, is the interstitial progenitor population that expresses Tcf21, Pdgfra, and CoupTFII (Fig. 3B; Supplementary Data 2).
Fig. 3. scRNA-seq differentiation trajectory of in vitro derived Leydig cells.
A Single-cell RNA-seq time course analysis of in vitro Leydig differentiation (days 0, 4, 7, and 14) identifies seven clusters, as visualized in t-SNE space. Note: n = 1 replicate per timepoint, but in vitro differentiation was successfully completed 8 times in prior experiments. B Heatmap of differentially expressed markers across the seven cluster centroids. C Visualization of the contribution of each individual time point at day 0, 4, 7, or 14 to the 7 clusters in t-SNE space. D Pseudotime ordering of cells from the 4 time points (days 0, 4, 7, and 14) by Monocle3 in UMAP space, colored by 7 clusters. E Schematic annotation of differentiation trajectory as defined by Monocle. F Expression profiles of selected markers across the differentiation pseudotime.
Four days after exposure to expansion media, cells dominate in clusters 3 and 5 with a smaller number of cells appearing in clusters 4 and 6 (Fig. 3C). Cells in cluster 3 are actively proliferating, as reflected by expression of proliferative marker Mki67, cyclins Cdk1 and Cdc20, and mitotic microtubule associated proteins (Fig. 3B; Supplementary Data 2). Cells in cluster 5 are postmitotic myofibroblasts expressing genes involved in actin cytoskeleton dynamics and remodeling (e.g. S100a4, Actn1, Lmod1, Nexn, Acta2) (Fig. 3B; Supplementary Data 2). Three days (d7) after transition to differentiation media, cells have become directed to an ECM-depositing myofibroblast cell state (expressing: Clu, Postn, Tnc, Col5A2; cluster 4) or a progenitor Leydig cell state (cluster 6) (Fig. 3B, C; Supplementary Data 2). Although cells in cluster 4 do not ultimately contribute to Leydig differentiation, they express fibroblast markers (Col5a2, Postn, Tnc) as well as extracellular-matrix related proteins involved in tissue remodeling (Fig. 3B, C; Supplementary Data 2). Interestingly, this population also expresses Pdgfa, raising the possibility that cluster 4 cells serve as an intermediate supportive cell population required to promote continued differentiation of Leydig cells.
By day 14 (10 days of exposure to differentiation media), cells in clusters 6 and 7 have become more differentiated (Fig. 3C). Cells in cluster 6 appear to prepare for steroidogenesis by increasing expression of lysosome/exosome genes, likely employing autophagy to degrade cellular components into steroid building blocks like cholesterol and fats (Fig. 3B; Supplementary Data 2). Previously, autophagy in Leydig cells was shown to be a rate-limiting step for testosterone synthesis45. Apolipoprotein E (ApoE) is also expressed at this time, indicating that LDL uptake is occurring which is critical for steroidogenesis, in line with genetic evidence in ApoE/Ldlr knockout mice46. Finally, in cluster 7, steroidogenic enzymes Cyp17A1, Hsd3B1, StAR, Cyp11A1 are expressed, as well as the mature Leydig cell factor Insl3, indicating a cellular state with functional steroidogenesis (Fig. 3B, F; Supplementary Data 2). The in vitro developmental progression based on time point sampling also confirmed Monocle3 pseudotime analysis (Fig. 3D–F), where sorted progenitors give rise to a cycle of proliferating and differentiating intermediates. Cells then branch into two differentiation trajectories, one aborting in cluster 4, which is an ECM producing myofibroblast, possibly a support intermediate cell, while the remaining cells proceed through clusters 6 and 7 which lead to differentiated Leydig cells (Fig. 3D–F).
Given our success with generating molecularly functional (testosterone secreting) Leydig cells in vitro, we next asked whether the in vitro derived Leydig cells bear resemblance to in vivo Leydig cells by comparing to previously published adult and fetal somatic cell states13,33. Notably, the in vitro intermediate states (clusters 2–5) in Leydig cell differentiation correlate with early interstitial progenitors in the fetal gonad, whereas clusters 6 and 7 have a higher correlation to fetal Leydig cells (Supplementary Fig. 3A). When comparing to the adult testis, the in vitro intermediate states (Clusters 2–5) correlate more closely with the Tcf21-expressing interstitial population, whereas clusters 6 and 7 have the highest correlation to adult Leydig cells (Supplementary Fig. 3B). Interestingly, overall the in vitro derived Leydig cells have higher correlation to adult Leydig cells than fetal Leydig cells (r = 0.84 vs. 0.58, respectively) (Supplementary Fig. 3A, B).
TCF21lin cells contribute to somatic lineages in the male gonad in vivo
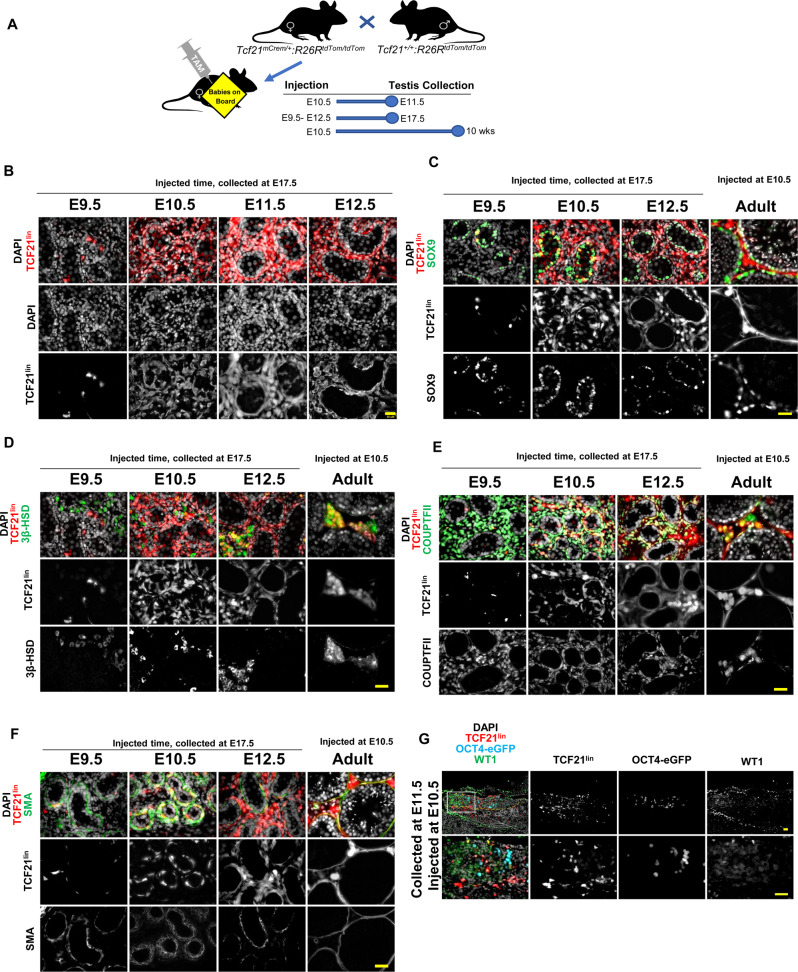
Given our ability to differentiate TCF21lin cells to Leydig or myoid cells in vitro, we next asked if the Tcf21 lineage can serve as a somatic progenitor in vivo. To this end, we performed lineage-tracing from early developmental time points in Tcf21mCrem:R26RtdTom mice and analyzed fully formed testes at E17.5 and adult testes at 10 weeks. Specifically, timed pregnant females were given a single dose of tamoxifen at either gestational days E9.5, E10.5, E11.5 or E12.5 (Fig. 4A). We verified that TCF21lin labeling was absent in embryonic gonads harvested from vehicle-treated timed pregnant females, confirming tight regulation of the tamoxifen inducible Cre (Supplementary Fig. 4A). Additionally, lineage traced cells did not overlap with Vasa, a germ cell marker, confirming specificity of the Tcf21mCrem line (Supplementary Fig. 4B).
Fig. 4. The TCF21lin population contributes to multiple somatic lineages in the fetal and adult testis.
A Experimental timeline used for Tcf21 lineage tracing analysis. Tcf21mCrem:R26RtdTom timed pregnant females were injected with a single dose of Tamoxifen at E9.5, 10.5, E11.5 or 12.5 and testes were analyzed at E17.5 or 10 weeks. B The TCF21lin cells at E10.5 (n = 17) and E11.5 (n = 11) contribute to all major somatic cell populations in the fetal gonad, whereas the E12.5 (n = 11) TCF21lin cells give rise only to testis interstitial cells. C–F Co-immunostaining of fetal or fostered adult TCF21lin testis cross-sections with Sertoli cell marker SOX9 (green; C, n = 5 for E9.5 and E12.5, n = 3 for E10.5), Leydig cell marker 3β-HSD (green; D, n = 5 for E9.5, n = 3 for E10.5 and E12.5), interstitial cell marker COUPTFII (NR2F2; green in E, n = 5 for E9.5, E10.5, and E12.5), and a myoid cell marker alpha smooth muscle actin (SMA, green in F, n = 5 for E9.5, n = 3 for E10.5, n = 4 for E12.5). G The TCF21lin in the fetal testis of Tcf21mCrem; R26RtdTom; Oct4-eGFP embryos are present in both the coelomic epithelium and mesonephros. Colocalization of WT1+ (Green) cells in the E11.5 gonad with TCF21lin cells (n = 2). In all panels the nuclear counterstain is DAPI (white B–G, n = 2). Scale bars for all B–G: 20 μm.
Immunofluorescence and histological analysis of E17.5 male gonads revealed marked differences in the extent of labeling across the different tamoxifen injection time points (Fig. 4B). We observed relatively fewer TCF21lin cells from E9.5 injected animals, yet those cells co-localized with multiple somatic lineages, as evidenced by colocalization with markers for Sertoli cells (SOX9), interstitial cells (COUPTFII) and, to a lower extent, fetal Leydig cells (3BHSD), and myoid (SMA) cells (Fig. 4C–F, Supplementary Fig. 4C–F). In contrast, a greater number of TCF21lin cells are observed in the E17.5 gonads collected from animals treated with tamoxifen at E10.5, E11.5, and E12.5, possibly due to broader expression of Tcf21 in multiple somatic progenitors at these later injection timepoints (Fig. 4C–F, Supplementary Fig. 4C–F). The TCF21lin cells at these different timepoints again contributed to Sertoli (SOX9), fetal Leydig (3BHSD), interstitial (COUPTFII, PDGFRA, and GLI), and myoid (SMA) cells (Fig. 4C–F, Supplementary Fig. 4C–H). Curiously, a significant fraction of E9.5 or E10.5 TCF21lin cells contribute to Sertoli cells, but these cells account for only a fraction of all SOX9+ cells in the fetal and postnatal testis (Supplementary Fig. 4C). This suggests either incomplete labeling of TCF21lin cells or Sertoli cells arising from multiple somatic progenitor populations.
To determine the origin of TCF21lin+ cells in the embryonic gonad and potential overlap with the WT1+ population—a somatic progenitor previously shown to give rise to Sertoli and interstitial populations including adult Leydig cells47—we injected timed pregnant Tcf21mCrem:R26RtdTom;Oct4-eGFP females with a single dose of tamoxifen at E10.5 and collected and stained whole mount gonads with WT1 at E11.5. In the E11.5 gonads, the TCF21lin cells localize both to the coelomic epithelium and the mesonephros, making it difficult to determine if TCF21lin cells truly originate from the coelomic epithelium or mesonephros, or are present in either location. However, we find that TCF21lin cells partially overlap with the WT1+ cells in the coelomic epithelium, but many cells are either TCF21lin or WT1+, suggesting these are possibly two distinct populations (Fig. 4G).
Unlike most somatic cell populations in the testis, the steroid-producing Leydig cells are unique in that they arise in two distinct waves. To ascertain whether the fetal-derived TCF21lin cells persist in the postnatal testis and give rise to adult Leydig cells, Tcf21mCrem:R26RtdTom time pregnant females received a single injection of tamoxifen at E10.5 and the pups were fostered and matured to adulthood (see Methods; Fig. 4A). In 10-week-old male testes, we found that a fraction of adult Sertoli, peritubular myoid, endothelial cells, and interstitial cells are tdTom+ (i.e., derived from TCF21lin) (Fig. 4C–F). Furthermore, we observe overlap between TCF21lin and 3BHSD in the adult testis, suggesting that the fetal TCF21lin population gives rise to adult Leydig cells (Fig. 4D). Taken together, we demonstrate that the fetal TCF21lin contributes to all adult somatic lineages of the testis. However, since the overlap is incomplete in all somatic populations this raises two possibilities: incomplete labeling or an alternative progenitor source population.
Although the fetal TCF21lin cells in vivo contribute to multiple somatic progenitor populations, the limitations of labeling a single somatic progenitor in the fetal testis has hampered our ability to conclusively determine whether the fetal TCF21 population is a heterogeneous population already committed to different fates, or if each cell is individually multipotent.
The TCF21lin gives rise to multiple fetal and adult ovarian somatic cell types
Prior to sex determination in mammals, the gonadal primordium is bipotential, meaning that the gonadal mesenchyme has the ability to give rise to either male or female support cell and steroidogenic cell lineages32. Once Sry expression is turned on during a critical window of fetal development, testis differentiation is initiated48. Despite an early commitment to either male or female somatic cell types, genetic studies in mouse have shown that terminally differentiated cell types must be actively maintained throughout life. For example, loss of either Dmrt1 in Sertoli cells or Foxl2 in granulosa cells can trigger reciprocal cell fate conversions (from Sertoli to granulosa cell fate or granulosa to Sertoli cell fate, respectively)49–51. Taking into account the known gonadal mesenchyme plasticity and the ability of TCF21lin cells to give rise to multiple somatic lineages in the testis, we next asked if TCF21lin cells are present in the fetal ovary and whether they might give rise to analogous cell types in females. For our female gonad experiments, we injected timed pregnant female Tcf21mCrem:R26RtdTom;Oct4-eGFP mice with a single dose of tamoxifen at E10.5 and collected female embryonic gonads at E11.5. Our analysis shows that TCF21lin cells are present in the coelomic epithelium and mesonephros, similarly to what we had observed in male embryonic gonads (Supplementary Fig. 4K). We then injected tamoxifen in timed-pregnant females at E10.5, E11.5, or E12.5 Tcf21mCrem:R26RtdTom mice and found broad somatic cell labeling in the female gonads at E17.5 (Supplementary Fig. 4L). Co-staining ovarian cross-sections with terminally differentiated markers shows that TCF21lin cells overlap with markers for granulosa cells (FOXL2+), interstitial cells (COUPTFII+), and smooth muscle cells (SMA+), and does not overlap with the germ cell markers VASA or OCT4 (Supplementary Fig. 4M–P).
In the postnatal mouse ovary, the fetal TCF21lin population contributes to multiple adult somatic lineages including granulosa cells (FOXL2+ and WT1+) of primordial and growing follicles, and endothelial cells (PECAM+), but not germ cells (Supplementary Fig. 4Q). Furthermore, we find that the TCF21lin cells surround the follicles and co-expresses the theca cell marker 3BHSD+52. Previous studies have shown that theca cells are derived postnatally from GLI1+ and/or WT1+ populations53,54. Therefore, we asked if the fetal TCF21lin cells co-express WT1. To this end, we examined E11.5 embryonic gonads from timed pregnant Tcf21mCrem:R26RtdTom;Oct4-eGFP mice and co-stained gonad sections for WT1 (Supplementary Fig. 4K). WT1 and TCF21lin appear to be largely in distinct populations with the exception of a few cells in the coelomic epithelium (Supplementary Fig. 4K). Therefore, the fetal TCF21lin population is largely distinct from the WT1 population, yet, it contributes to all somatic lineages in the adult ovary including: granulosa cells (FOXL2+ and WT1+) of primordial and growing follicles, endothelial cells (PECAM+), and theca cells (3BHSD+), suggesting that the female gonad, like the male gonad, may have multiple somatic progenitors.
Tcf21lin cells regenerate Leydig cells in the adult testis in response to chemical ablation
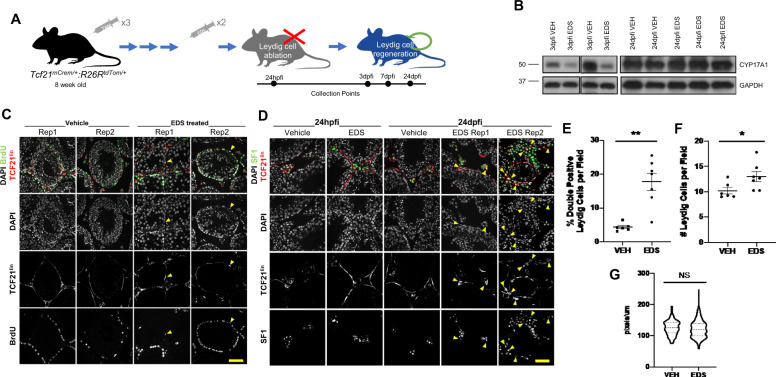
Although somatic cells of the testis are considered to be postmitotic and do not naturally turnover, our steady state scRNA-seq datasets identified rare Tcf21+ cells that express low levels of either steroidogenic acute regulatory protein (StAR) or Sma, suggesting that a rare subset of Tcf21+ cells transition to either Leydig or myoid cells, respectively. We then examined if the adult TCF21lin is capable of serving as a somatic progenitor at least for adult Leydig cells in the testis. Specifically, animals were treated with EDS to reduce Leydig cell numbers and the testes were collected and assessed to determine (1) if regeneration does occur and (2) if TCF21lin contributes to the regeneration.
EDS treatment was previously used in rats to selectively ablate Leydig cells55–58. Although there is strong species specificity and animal-to-animal variation in Leydig cell sensitivity to EDS59, two 300 mg/kg EDS injections in C57BL/6 mice spaced 48 h apart (Supplementary Fig. 5A) resulted in a reduction of mature Leydig cells in the adult testis as evidenced by cell death and CYP17A1 protein levels (Supplementary Fig. 5B, C). At 12 h post final injection (hpfi), we detected TUNEL positive Leydig cells, but given the spacing of 48 h between the first and second injection, the majority of apoptotic events likely preceded the time of analysis and could therefore not be quantified (Supplementary Fig. 5B). However, consistent with Leydig cell loss, we find that CYP17A1 protein expression decreased as early as 12 hpfi and had largely recovered by 14 dpfi (Supplementary Fig. 5C). We confirmed that the recovery of CYP17A1 protein expression was not simply due to Leydig cell hypertrophy, as the Leydig cell diameter is similar between the EDS- and vehicle-treated animals at 14 dpfi (Supplementary Fig. 5D).
To determine if regenerating Leydig cells were derived from the adult testis TCF21lin we injected 8-week-old Tcf21mCrem:R26RtdTom mice with three 2 mg tamoxifen injections, and then treated the mice with two 300 mg/kg EDS injections spaced 48 h apart (Fig. 5A). Similar to EDS-treated C57BL/6 animals, CYP17A1 protein levels in Tcf21mCrem:R26RtdTom EDS-treated animals decreased at 3 dpfi and recovered by 24 dpfi (Fig. 5B). To ensure that the reduction in CYP17A1 is due to Leydig cell loss, we quantified the number of SF1+ cells in EDS- and vehicle-treated animals at 3 dpfi. In 3 dpfi animals, ~20% of all cells in testis cross-sections are SF1+ in the vehicle-treated animal, whereas the number of SF1+ cells is reduced to 5% in the EDS-treated animals, suggesting many Leydig cells were lost (n = 3 vehicle and n = 3 EDS animals). To test whether the TCF21lin population contributes to Leydig cell regeneration, we examined if TCF21lin cells proliferate in response to injury and contribute to the regenerated Leydig cells. At 3 dpfi, TCF21lin cells surrounding the tubules re-entered the cell cycle as detected by colocalization of BrdU and TCF21lin (Fig. 5C). Importantly, by 24 dpfi in EDS-treated animals, ~18% of SF1+ Leydig cells are Tcf21lin positive as compared to ~5% in the vehicle-treated animals (Fig. 5D, E). Furthermore, we find that the EDS-treated animals have a higher number of SF1+ cells as compared to controls (Fig. 5F), which is consistent with an overcompensation in CYP17A1 protein levels observed in EDS-treated animals (Fig. 5B) and the absence of Leydig cell hypertrophy (Fig. 5G).
Fig. 5. The TCF21lin population regenerates Leydig cells in vivo after injury.
A Schematic representation of the experimental method used to ablate Leydig cells in the Tcf21mCrem:R26RtdTom background. B Representative CYP17A1 protein immunoblots from EDS- and vehicle-treated animals at 3 dpfi and 24 dpfi. A total of n = 8 independent experiments were performed. C Representative images of BrdU+ cells in the testis of vehicle- and EDS-treated animals (representative from n = 2 vehicle, n = 3 EDS). D Colocalization of TCF21lin (tdTom) and Leydig cell marker SF1 in EDS- or vehicle-treated Tcf21mCrem:R26RtdTom animals at 24 h post final injection (24 hpfi) or 24 days post final injection (24 dpfi). Representative images from n = 6 vehicle, n = 7 EDS animals. E Quantification of the percentage of Leydig cells expressing both SF1 and TCF21lin (tdTom) per field at 24 dpfi (Note—each circle represents the percentage per animal. A total of n = 6 vehicle, n = 7 EDS were analyzed). Data are presented as mean ± SEM. P = 0.0018. F Quantification of Leydig cell number per field of view at 24 dpfi (n = 6 vehicle, n = 7 EDS). Data are presented as mean ± SEM. P = 0.042. G Average Leydig cell diameter measurements in Tcf21mCrem:R26RtdTom EDS or vehicle-injected animals, after Leydig cell recovery at 24 dpfi (n = 3 per condition). Lines indicate mean and quartiles. P = 0.092. All statistical tests were performed using Welch’s unpaired, two-sided t-tests. Scale bars: 20 μm for all images in C, D.
To independently validate the role of TCF21lin cells in testis somatic cell regeneration, we performed allogenic transplants. Specifically, we sorted adult SCA1+ cells from Tcf21mCrem:R26RtdTom animals and transplanted them into the testis interstitium of EDS-treated C57BL/6 animals where no Leydig cells in the host animal will be TdTom+ (Supplementary Fig. 5E). We reasoned that if the transplanted TCF21lin population contributed to Leydig cell regeneration, then we should detect TCF21lin/SF1 double-positive cells in the C57BL/6 EDS-treated animal. By 24 hpfi, we found TCF21lin cells homed to the basement membrane, and by 7 dpfi, we began detecting TCF21lin/SF1+ cells (Supplementary Fig. 5F), suggesting that the Tcf21lin cells engrafted in the ablated C57BL/6 mouse testis and gave rise to SF1+ cells.
Therefore, by using the Tcf21mCrem:R26RtdTom EDS model and the TCF21lin transplant approach in EDS-treated C57BL/6 mice, we demonstrate that the adult Tcf21lin population in the testis can at least serve as a reserve Leydig progenitor in response to Leydig cell loss.
Peritubular myoid cells of the testis can regenerate after injury in adult testis
While we demonstrated that Leydig cells can be regenerated in response to tissue injury, it is unclear whether additional somatic cells in the testis can do the same. Previous studies have shown that Sertoli cells can be replaced by transplantation but cannot be regenerated following targeted diphtheria toxin (DTX) treatment60–62, but the regenerative ability of peritubular myoid cells has not been assessed. To examine whether (1) peritubular myoid cells regenerate and (2) if TCF21lin cells contribute to the regeneration, we treated 6–13-week-old Myh11cre-eGFP; Rosa26iDTR/+ mice (referred to as MYH11-cre:iDTR hereafter) with multiple low doses of DTX to balance animal survival and myoid cell ablation and collected testes at 12 hpfi and 4 dpfi (Supplementary Fig. 6A). By 12 hpfi, TUNEL positive cells were present on the tubule basement membrane in Myh11cre-eGFP; Rosa26iDTR/+ mice but absent in control mice but were no longer detectable by 4 dpfi (Supplementary Fig. 6B). However, at 4 dpfi the testis cross-sections of Myh11cre-eGFP; Rosa26iDTR/+ animals continue to display vacuoles and disordered tubules, whereas, these histological features were absent from controls (Supplementary Fig. 6C). By 4 dpfi, we detect BrdU positive smooth muscle cells surrounding the basement membrane (Supplementary Fig. 6D, yellow arrow) as well as BrdU positive cells on the tubule surface which lacked SMA expression (Supplementary Fig. 6D, white arrow), indicating both neighboring peritubular smooth muscle cells and nearby interstitial cell progenitors re-enter the cell cycle to regenerate the basement membrane in the DTX-treated Myh11cre-eGFP; Rosa26iDTR/+mice. However, since our TCF21 antibody did not yield clear immunofluorescence staining, it could not be determined whether proliferating cells are TCF21+.
In an attempt to overcome these limitations, we sorted SCA1+/TCF21lin cells from Tcf21mCrem:R26RtdTom animals and transplanted these cells into the interstitial space of Myh11cre-eGFP; Rosa26iDTR/+ DTX-treated animals. Within 24 h after transplant, we were able to detect TCF21lin cells surrounding the damaged tubules (Supplementary Fig. 6E, F), but failed to observe any TCF21lin become SMA+ at this time point (Supplementary Fig. 6G). Therefore, unlike with Leydig cells, for which we relied on EDS to induce cell-specific ablation, our effort to ablate myoid cells relied on the use of Myh11cre-egfp; Rosa26iDTR/+ which is expressed in multiple organs, leading to lower animal viability and preventing the analysis of the fate of TCF21lin transplanted cells at later time-points.
In summary, we have demonstrated that peritubular myoid cells can be regenerated. Unlike in the case of Leydig cells described above, whether myoid cells can be derived from TCF21lin cells could not be ascertained at this time and will require additional tools.
Adult TCF21lin cells contribute to somatic turnover in the testis during natural aging
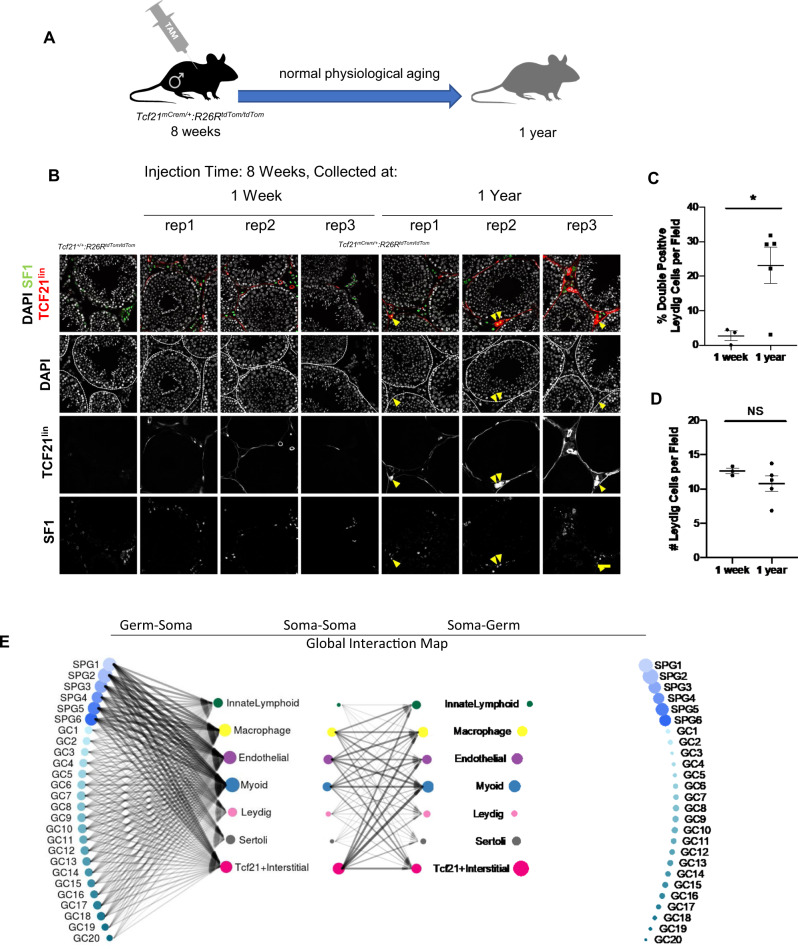
Once established, the somatic cells of the testis are maintained throughout a male’s reproductive age. However, a study in rats using [3H]thymidine labeling to detect proliferation suggested that Leydig and possibly peritubular myoid cells may undergo rare events of cellular turnover during an animal’s natural lifespan63. We then asked to what extent adult mouse testis somatic cells turnover during natural aging, and secondly, whether new cells would be derived from TCF21lin progenitors. To answer this question, we performed a long-term lineage tracing experiment where 8-week-old (adult) Tcf21mCrem:R26RtdTom animals were injected with a single dose of 2 mg tamoxifen. Animals were euthanized either 1-week past final injection or 1-year past final injection (aged mice) (Fig. 6A). In animals with 1-week labeling we detected rare TCF21lin cells surrounding the basement membrane, and these cells do not significantly overlap with Leydig cell markers such as SF1 (Fig. 6B). In aged mice, we detected a significant increase inTCF21lin and SF1 double-positive cells (Fig. 6C) as well as more labeling around peritubular cells suggesting that peritubular cells may also be replenished (Fig. 6B). This extent of labeling varied by individual animal which could be due to tamoxifen injection efficiency or true natural biological variability in inbred mice. Interestingly, while there is no significant difference in Leydig cell number between short-term labeled and aged individuals, there is more variability among aged individuals (Fig. 6D). Nevertheless, these data indicate that somatic cells of the testis naturally turnover, possibly at low rates, and the TCF21lin population contributes to the replenishment and maintenance of the adult Leydig cell population, and likely peritubular myoid cells as well.
Fig. 6. The TCF21lin population maintains testis tissue homeostasis during aging.
A Schematic representation of TCF21 lineage tracing in the aging testis. B Colocalization of TCF21lin (tdTom) and Leydig cell marker SF1 (green) at 1 week or 1 year following a single injection of tamoxifen in 8-week-old adults. Scale bar: 20 μm all panels. DAPI is used as the nuclear counterstain (white). (Oil only control n = 2, demonstrating the tight control of TCF21-Cre, n = 3 for 1 week, n = 5 for 1 year). C Quantification of the percent of Leydig cells expressing both SF1 and TCF21lin (tdTom) per field (Note—each circle/square represents the percentage per animal; n = 3 for 1 week, n = 5 for 1 year). P = 0.016. Data are presented as mean ± SEM. All statistical tests were performed using Welch’s unpaired, two-sided t-tests. D Quantification of Leydig cell number per field (Note—each circle/square represents the percentage per animal; n = 3 for 1 week, n = 5 for 1 year). P = 0.19. Data are presented as mean ± SEM. All statistical tests were performed using Welch’s unpaired, two-sided t-tests. E Summary of putative ligand–receptor interactions in the mouse testis between the germline and soma (left), within soma (center), and soma and germline (right). Arrows summarize top 5% of all interactions. ScRNA-seq data from42, processed as described in34. Symbol size indicates the number of receptor–ligand interactions contributed by a cell type and line width shows the number of interactions between the two cell types.
Additional intercellular interactions suggested by scRNA-seq data
To gain a sense of cellular crosstalk between Tcf21+ cells and germ/somatic cells, we focused on previously documented ligand–receptor (L–R) pairs that are highly variable among major cell types in our scRNA-seq data, and calculated Interaction Scores between germ cells and somatic cells, or among somatic cells of the testis (Fig. 6E, Supplementary Data 3). We previously showed that the Tcf21+ population has more potential interactions with spermatogonial populations than other germ cells34 (Fig. 6E, right panel), Similarly, we find that spermatogonia can potentially signal back to the Tcf21+ population (Fig. 6E, left panel) via PDGFA, various ADAMs, calmodulins, FGFs, and guanine nucleotide binding proteins (Supplementary Data 3).
Within the somatic compartment, Tcf21+ cells have the greatest potential interactions with endothelial, myoid, and macrophages (Fig. 6E, middle panel) involving receptor–ligand signaling systems such as Lrp1 (Cd91), a multifunctional, endocytic receptor capable of binding a vast array of ligands64 and known to regulate the levels of signaling molecules by endocytosis, as well as directly participate in signaling for cell migration, proliferation, and vascular permeability (reviewed in65). Like other tissue mesenchymal progenitors, Tcf21+ cells may also modulate local inflammatory responses, as they express Thrombomodulin (Tbhd) which interacts with and can proteolytically cleave the pro-inflammatory molecule Hmgb1 (reviewed in66). Additionally, there are several more cell-specific interactions with myoid cells (Tgfb2-Tgfbr3;Gpc3-Cd81), endothelial cells (Cxcl12-Itgb1; Pdgfa-Pdgfra; and Vegfa-Itgb1), and macrophages (Igf1-Igf1r; F13a1-Itgb1) (Supplementary Data 3). Several of these putative interactions are involved in growth, wound healing, phagocytosis, and matrix remodeling in various mesenchymal cell types67. These putative interactions will need to be validated by spatial analysis and/or functional perturbations of individual signaling pathways.
The Tcf21+ population in the testis resembles resident fibroblast populations in other tissues
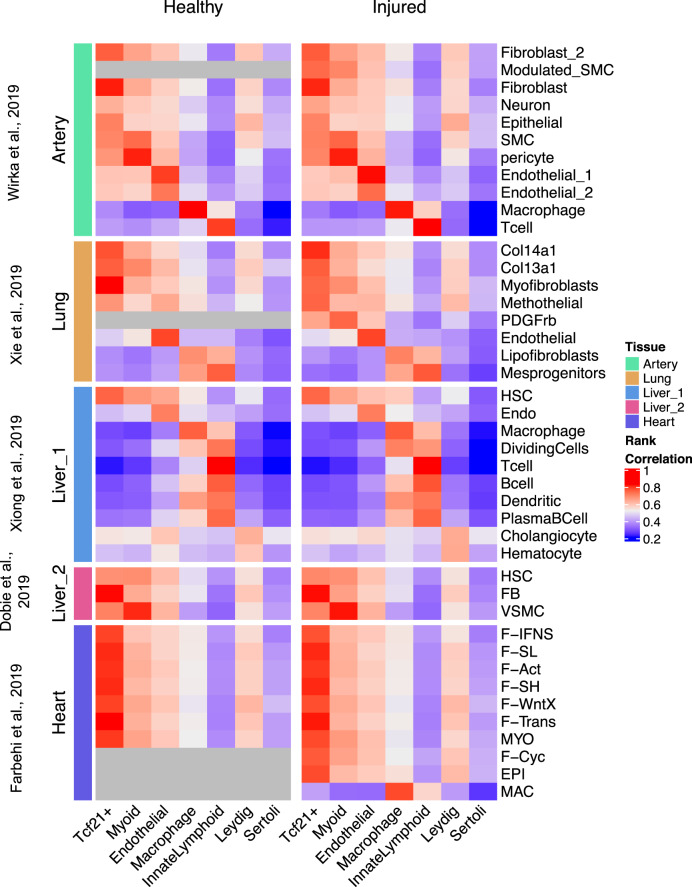
Finally, given the essential role of Tcf21+ cells in mesenchymal development of many tissues, including the heart, lung, and kidney27,29,37,68, we sought to understand whether the adult testis Tcf21+ population resembles other Tcf21+ mesenchymal populations found in single-cell analyses of other tissues. Our comparison of testis somatic cells with the publicly available scRNA-seq datasets from coronary artery, heart, lung, and liver69–73 find that the testis Tcf21+ population most closely resembled the resident fibroblast or myofibroblast populations (Fig. 7, left), as well as the fibroblast/myofibroblast populations that appear transiently after tissue injury (Fig. 7, right). These diverse fibroblast cell types have been documented across tissues and implicated in fibrotic damage and tissue regeneration, even when they may differ with respect to cellular markers or nomenclature (reviewed in74). Our results indicate that they are transcriptomically similar to the testicular Tcf21+ population characterized here, suggesting that they collectively represent an emerging class of resident adult progenitor cells playing similar roles in tissue maintenance and repair across multiple organ systems.
Fig. 7. The adult Tcf21+ interstitial population resembles fibroblasts in other tissues.
A Rank correlation of scRNA-seq cluster centroids of somatic cells from wild-type adult mouse testes with other tissues in healthy adult mice (left panels) or after injury (right panels). Gray blocks indicate cell types not present in healthy datasets.
Discussion
Tcf21 is a basic helix-loop-helix transcription factor, known to have roles in the development of numerous organs, including the testis27–29. During testis development, Tcf21 is expressed in the bipotential gonadal ridge at E10.5 similar to other key transcription factors, including WT1 and GATA475, and loss of Tcf21 results in gonadal dysgenesis29,30. Here, our lineage tracing data show that the fetal TCF21lin population is a bipotential gonadal progenitor giving rise to most somatic cell types including steroid-producing cells (fetal and adult), stromal/interstitial cells, and supporting cells, as well as vasculature, consistent with a common bipotential progenitor model recently described32,33. Gonadal organogenesis is a complex process with multiple somatic progenitors have been described in both males and females including: WT1, COUPTFII, NESTIN, and PDGFRA18,21,23,47,76. To a certain extent, our data reconcile these findings by showing that TCF21lin overlaps with many of these markers, but our data supports a multi-progenitor model for both interstitial and Sertoli cell populations that are molecularly and cellularly heterogeneous.
Furthermore, we demonstrate that the TCF21lin cells share characteristics with adult mesenchymal progenitors (MPs) for example: TCF21lin/SCA1+cells can self-renew in vitro and possess numerous traits like homing ability, secretion of molecules with anti-inflammatory and immunoregulatory effects, and have multi-lineage potential77,78. Furthermore, we show that the SCA1+/TCF21lin cells can be directed to differentiate to myoid and Leydig cells in vitro. Importantly, the in vitro derived Leydig cells secrete testosterone and highly resemble in vivo-derived Leydig cells. The availability of such a robust differentiation protocol makes it possible to use in vitro-derived cells to study how environmental toxicants effect steroidogenesis or Leydig cell function/biology.
Testosterone is essential for the development and maintenance of male characteristics and fertility. Reduced serum testosterone affects millions of men and is associated with numerous pathologies including infertility, cardiovascular diseases, metabolic syndrome, and decreased sexual function. Although exogenous replacement therapies are largely successful in ameliorating these symptoms, they carry increased risks of cardiovascular and prostate disease or infertility79–81. Therefore, identifying a progenitor population and/or natural mechanism to restore testosterone levels in vivo and combat hypogonadism or age-related decline in testosterone levels is critical82,83. Here, we show that resident TCF21lin cells or TCF21lin allogenic transplants can be activated to support Leydig cell regeneration and replenish Leydig cells upon injury or aging. To date there have been several putative stem Leydig cell populations described in the fetal and early postnatal testis of multiple species. These likely analogous populations are demarcated by diverse cell surface markers that are often species specific, like CD90 for rat and CD51 for mouse (reviewed in82,84–86) and exhibit species specific properties. For example, PDGFRA+ cells isolated from rat can be differentiated to Leydig cells, whereas, although the PDGFRA+ isolated cells from the human testes exhibit aspects of MSC characteristics in vitro, they are unable to fully differentiate into Leydig cells, nor can they produce testosterone41.
Finally, given the critical role of Tcf21 in the development of other tissues, as well as in aging and disease models87,88, we examined commonalities between testis Tcf21+ cells and similar populations in other organ systems and disease states69–73. The Tcf21+ population in the adult testis molecularly resembles Tcf21+ fibroblast or fibroblast-like populations that have functional roles in normal tissue maintenance, injury, or disease in other organs such as the heart, lung, and liver. While the response to tissue injury is often context dependent, resulting in fibrosis vs. regeneration, it remains unknown if a single resident mesenchymal population is activated to promote either response depending on the levels of damage or signaling pathways activated or if multiple populations respond and leading to divergent outcomes. Here in response to EDS, the TCF21lin restores Leydig cells but in many cases, fibrosis is often observed in men with impaired spermatogenesis89–93. Therefore, whether dysregulation of the TCF21+ population may be involved in the pathogenesis of testis fibrosis in certain contexts of infertility remains to be examined. A greater understanding of this population and its regulation may contribute to more informed strategies to restore testis function, as well as the tissue regeneration and tissue repair therapies for other organs94.
Methods
Lead contact and materials availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Saher Sue Hammoud (hammou@med.umich.edu).
Mice
All animal experiments were carried out with prior approval of the University of Michigan Institutional Committee on Use and Care of Animals (Animal Protocols: PRO00006047, PRO00008135), in accordance with the guidelines established by the National Research Council Guide for the Care and Use of Laboratory Animals. Mice were housed in the University of Michigan animal facility, in an environment controlled for light (12 h on/off) and temperature (21–23 °C) with ad libitum access to water and food (Lab Diet #5008 for breeding pairs, #5LOD for non-breeding animals).
Colony founders for Rosa26iDTR (Stock #007900), Oct4-eGFP (Stock #004654), PdgfraDGFRAEGFP (Stock007669), R26RtdTom mice (Stock #007909), and Myh11cre-egfp (Stock #007742, maintained on a B6 background) were obtained from Jackson Labs. The Tcf21mCrem and the Gli1EGFP were generously provided by Michelle Tallquist and Deb Gumuccio, respectively. EDS injection studies were performed on age-matched C57BL/6 mice obtained from Jackson Labs (Stock #000664) or Tcf21mCrem:R26RtdTom animals. For detailed mouse strain information, see below. All primers used for genotyping are provided in Supplementary Data 4.
Interstitial populations single-cell data analysis
Single-cell RNA-sequencing analysis comparing somatic cell types in the adult mouse testis
Somatic cells (N = 3622) and their cell type classifications were defined by Green et al.13. To compare somatic cell populations of the testis we obtained the Euclidean distances for the somatic cell centroids then ordered the cell types using the optimal leaf ordering (OLO) algorithm in R Package Seriation. Based on this analysis, we discovered that the Tcf21+ population is highly correlated to endothelial, myoid and Leydig cells, but distinct from immune cells and Sertoli cells. To get a better understanding of the functional role of the Tcf21+ population, we called differentially expressed genes in each somatic cell type using a nonparametric binomial test. The differentially expressed genes have: (1) At least 20% difference in detection rate; (2) a minimum of 2-fold change in average expression levels, and (3) p value < 0.01 in the binomial test. Pathway Enrichment analysis for the differentially expressed genes was performed with PANTHER tool v.15 (http://www.pantherdb.org)95. Significance of the over- and under-representation of GO Complete Biological Process categories was calculated using Fisher’s exact test and multiple testing correction with the false discovery rate.
Ligand–receptor analysis
We used a previously published curated list of ligand–receptor (LR) pairs34. We limited the analysis to LR pairs that have either highly variable ligand genes among the 7 somatic cell centroids, or highly variable receptor gene among the 26 germ cell centroids (6 SPG and 20 non-SPG clusters). Thresholds were set for both genes mean and variance across the clusters according to the density. For each ligand–receptor pair we calculated its apparent signaling strength as an “Interaction Score”, defined as the product of the mean expression level of the ligand in one cell type and that of the receptor in another cell type. In all, we calculated such an Interaction Score matrix of cell type pairs for germ (ligand)-soma (receptor) interaction, soma (ligand)-soma (receptor) interaction and soma (ligand)-germ (receptor) interaction (reproduced with permission from34, respectively. To extract the general signaling pattern for each interaction matrix, we defined “strong interactions” for each matrix by keeping the highest 5% Interaction Scores for each matrix. We then calculated the number of such strong L–R interactions for each pair of cell types as their overall interaction strength and displayed them as the line width of arrows in the pairwise interaction plots.
Cell type correlations across tissues
We downloaded the single-cell counts data from GEO for artery, lung, heart and two liver datasets69–73. For the datasets providing cluster information including our testis datasets, we generated expression centroid for each cell type. We then calculated the spearman rank correlation for all cell type pairs between testis and other tissues. For the artery and the liver datasets, the cell type clusters were not provided, so for these datasets we re-analyzed the raw data using Seurat—following the analysis descriptions from the original papers. For parameters that are not specified, we either used default values or set accordingly. To regenerate the clusters for these raw datasets, we used Louvain clustering in Seurat and assigned cell types according to markers listed in the two papers. We then followed the same procedure to calculate cell type expression centroid and spearman rank correlations with cell types from testis. Summary of all correlations are illustrated in the heatmap.
In vitro differentiation assays
Flow cytometry
Testes were collected from adult C57BL/6 (JAX®mice, stock #000664) mice and enzymatically and mechanically dissociated into a single-cell suspension. Briefly, testes from adult mice were excised and the tunica albuginea was removed. Seminiferous tubules were transferred to 10 ml of digestion buffer 1 (comprised of Advanced DMEM:F12 media (ThermoFisher Scientific), 200 mg/ml Collagenase IA (Sigma), and 400 units/ml DNase I (Worthington Biochemical Corp)). Tubules were dispersed by gently shaking by hand and a 5-min dissociation at 35 °C/215 rpm. To enrich for interstitial cells, tubules were allowed to settle, and the supernatant was collected, quenched with the addition of fetal bovine serum (FBS) (ThermoFisher Scientific), filtered through a 100 μm strainer and pelleted at 600 g for 5 min. The remaining tubules were then transferred to digestion buffer 2 (200 mg/ml trypsin (ThermoFisher Scientific) and 400 units/ml DNase I (Worthington Biochemical Corp) dissolved in Advanced DMEM:F12 media) and dissociated at 35 °C/215 rpm for 5 min each and quenched with the addition of FBS (ThermoFisher Scientific). The cell pellets from multiple digests were combined and filtered through a 100 μm strainer, washed in phosphate-buffered saline (PBS), pelleted at 600 g for 3 min, and re-suspended in MACS buffer containing 0.5% BSA (Miltenyi Biotec).
The single-cell suspensions were stained with a single antibody or combination of antibodies depending on the experiment. The antibodies used include anti-Ly6a-AlexaFluor 488 (1:100; Biolegend, Cat#108115), Biotinylated anti-Ly6a (1:200; Biolegend, Cat#108103), streptavidin conjugated AlexaFluor 488 (1:1000; Life Technologies Cat# S11223; RRID: AB_2336881), anti-CD73-APC (1:300; Biolegend, Cat#127209), anti-CD90.1-Brilliant Violet 650 (1:300; Biolegend, Cat#202533), anti-CD29-PE/Dazzle 594 (1:300; Biolegend, Cat#102231), anti-CD105-PerCP/Cy5.5 (1:300; Biolegend, Cat#120415), anti-CD45-Brilliant Violet 510 (1:300; Biolegend, Cat#), anti-CD117-PE/Cy7 (c-KIT) (1:300; Biolegend, Cat#105813), and anti-CD34-PE (1:300; Biolegend, Cat#128609).
Tri-lineage differentiation assay
Testes were collected from adult C57BL/6 or Tcf21mCrem:R26RtdTom mice and dissociated into a single-cell suspension and sorted for SCA1+/cKITit− or SCA1+/TCF21lin/cKITit− cells, respectively. For adipogenic differentiation, 3 × 104 cells were plated in a monolayer, cultured for 10 days (Stem ProAdipogenic differentiation kit) and stained with either Oil Red O or Perilipin (1:250, Sigma, Cat#P1873). For chondrogenic differentiation, 5 × 104 cells were plated in micromass, cultured for 21 days (Stem ProChondrogenic differentiation kit) and stained with either Alcian blue or SOX9 (1:250, EMD Millipore, Cat#ABE571). For osteogenic differentiation, 1 × 104 cells were plated in micromass, cultured for 14 days (StemProOsteogenicdifferentiationkit), and stained with either Alizarin red or Osterix (1:250, Abcam, Cat#ab22552)96.
CFU-assay
Colony-forming unit assays were performed as previously described97. Briefly, testes were collected from C57BL/6 or adult Tcf21mCrem:R26RtdTom males following 3 injections of tamoxifen (2 mg) every other day. Following single-cell dissociations, SCA1+/cKITit−, SCA1+/TCF21lin/cKITit−, TCF21lin/cKITit−, or cKIT+/SCA1− cells were plated into Corning Primaria 6 well plates at a density of 1000 cells/well. Cells were cultured for 14 days in Mesen Cult MSC medium (StemCell Technologies) and colonies were stained using Giemsa. Colonies were defined as clumps having either >20 or >50 cells.
In vitro directed differentiation to Leydig and myoid cells
Testes were collected from adult C57BL/6 or Tcf21mCrem:R26RtdTom males, dissociated into a single-cell suspension, and sorted for SCA1+/cKit− cells. Cells were plated into a 24 well, Matrigel-coated plate at a density of 100,000 cells/well in DMEM/F12 supplemented with 10% FBS and 1X Normocin. For myoid cell differentiation, after 18 h media was replaced with differentiation media- DMEM/F12 supplemented with 1X Penicillin/streptomycin, 10 ng/ml PDGFAA, 10 ng/ml PDGFBB, 0.5 µM SAG, 10 ng/ml BMP2, 10 ng/ml BMP4, 10 ng/ml ActivinA, and 1 mM Valproic acid. After the 7-day differentiation protocol, cells were stained for SMA (1:200, Sigma, Cat#A5228). For Leydig cell differentiation, the FACs sorted cells were initially recovered for 18 h in DMEM+10%FBS and then the cells were expanded for 3 days in DMEM/F12 supplemented with 1X Normocin, 10 ng/ml PDGFAA, 10 ng/ml PDGFBB, 0.5 µM SAG, and 10 ng/ml FGF2. After 3 days, the expansion media was replaced with differentiation media- DMEM/F12 supplemented with 1X Penicillin/streptomycin, 10 ng/ml PDGFAA, 10 ng/ml PDGFBB, 0.5 µM SAG, 10 ng/ml FGF2, 5 mM LiCl2, and 10 µM DAPT. After 10 days in differentiation media, cells were stained for SF1 (1:100, CosmoBio, Cat#KAL-KO610). Media was collected every other day for testosterone measurements.
Clonal expansion and directed differentiation of TCF21lin cells
To assess multipotency of TCF21lin cells, testes were collected from adult Tcf21mCrem:R26RtdTom males and dissociated into a single-cell suspension. Individual TCF21lin/cKit−, SCA1+/TCF21lin/cKit−, SCA1+/TCF21lin/cKit−/Cd105+ or SCA1+/TCF21lin/cKit−/Cd105− cells were sorted into Corning Primaria 96-well plates and cultured in Mesen Cult mouse MSC media for ~3 weeks to allow for colony formation. Individual colonies were then directed to differentiate to either a myoid or Leydig cell fate following the directed differentiation protocols described above. Cells were then stained for either SMA (for myoid cells, 1:200, Sigma, Cat#A5228) or SF1 (Leydig cells, 1:100, CosmoBio, Cat#KAL-KO610).
Drop-seq analysis of the Leydig cell differentiation time-course analysis
Drop-seq was performed on cells collected from various points of differentiation, where a single sample per timepoint was diluted to 280 cells/µl and processed as described previously98. Briefly, cells, barcoded microparticle beads (MACOSKO-2011-10, Lots 113015B and 090316, ChemGenes Corporation), and lysis buffer were co-flown into a microfluidic device and captured in nanoliter-sized droplets. After droplet collection and breakage, the beads were washed, and cDNA synthesis occurred on the bead using Maxima H-minus RT (ThermoFisher Scientific) and the Template Switch Oligo. Excess oligos were removed by exonuclease I digestion. cDNA amplification was done for 15 cycles from pools of 2000 beads using Hot Start Ready Mix (Kapa Biosystems) and the SMART PCR primer. Individual PCRs were purified and pooled for library generation. A total of 600 pg of amplified cDNA was used for a NexteraXT library preparation (Illumina) with the New-P5-SMARTPCR hybrid oligo, and a modified P7 Nexteraoligo with 10 bp barcodes. Sequencing was performed on a NovaSeq (Illumina) for read 2 length of 94 nt with the Read1 Custom Seq primer. Oligosequences are the same as previously described13,98.
Single-cell RNA-seq analysis across time points
The paired-end Drop-seq data from days 4, 7, and 14 of in vitro Leydig cell differentiation were sequenced in the same batch and were processed using Drop-seq tools v1.13 from the McCarroll laboratory as previously described98,99. Specifically, the reads were aligned to the mouse reference genome (GRCm38, version 38) using STAR v2.7.1a100. The pipeline generated digital gene expression matrices with genes as rows and cells as columns that served as the starting point for downstream analyses.
The cells from each time point were first filtered by cell size and integrity—cells with <500 detected genes or with >10% of transcripts corresponding to mitochondria-encoded genes were removed, resulting in 973–2980 pass-QC cells for the three time points, for a total of 6124 cells. Among the retained cells, the average number of detected genes per cell was ~1888, and the average number of UMIs was ~4838. For each cell, we normalized transcript counts by (1) dividing by the total number of UMIs per cell and (2) multiplying by 10,000 to obtain a transcripts-per-10K measure, and then log-transformed it by E = ln(transcripts-per-10K+1).
For each time point, we standardized the expression level of each gene across cells by using (E-mean(E))/sd(E) and performed PCA using highly variable genes (HVG). We obtained 4 clusters using Louvain-Jaccard clustering with top PCs by R package Seurat (v2.3.4). We calculated cluster centroids and ordered the clusters by minimizing pairwise Euclidean distance of cluster centroids in R package Seriation. We evaluated batch effect by comparing the top PC placements and rank correlation of ordered cluster centroids across the 3 time points. Differentially expressed markers for each cluster were obtained by comparing it against all other clusters using a nonparametric binomial test, requiring at least 20% higher detection rate, a minimum of 1.5-fold higher average expression level, and p value < 0.01. Clusters were defined based on known markers.
We extracted the somatic cells from the INT4 dataset from13. This dataset was enriched for the Tcf21+ interstitial population and was used as the starting time point: day 0. We merged the 3 datasets of in vitro Leydig differentiation with the somatic cells of INT4, for 6619 good-quality cells and 24,698 detected genes. These cells on average have 1837 detected genes and 4623 UMIs per cell. We selected 2344 HVG genes in the merged dataset and did PCA using HVG. We performed t-SNE, UMAP and Louvain-Jaccard clustering using top PCs. We obtained 10 clusters initially, and ordered the clusters as described above. Based on differentially expressed markers for each cluster and rank correlation across the cluster centroids, we decided to merge 3 clusters (clusters 4–6) and identified them as ECM myofibroblast. We merged 2 other clusters (clusters 7–8) as differentiating myofibroblast. This led to the identification of 7 cell types for in vitro Leydig differentiation—(1) Endothelial, (2) Interstitial progenitor, (3) Proliferating progenitor, (4) ECM myofibroblasts, (5) Differentiating myofibroblasts, (6) Differentiating Leydig, and (7) Leydig. We did pseudotemporal ordering of cells from the 4 time points (days 0, 4, 7, and 14) by Monocle3 and visualized the single-cell trajectory in UMAP space.
We compared our in vitro Leydig differentiation data with those of fetal mouse gonads and adult mouse testis somatic cells. Specifically, we calculated the rank correlation between our 7 cell type centroids of Leydig differentiation data with the 6 cell type centroids from NR5A1-eGFP+ progenitor cells from E10.5 to E16.5 fetal male mouse gonads33 using markers present in both data (N = 2692). We also calculated the rank correlation between our 7 cell type centroids with the 7 cell type centroids from adult mouse testis (Green et al.13) using the union of markers present in both datasets (N = 1769).
TCF21 lineage tracing
TCF21 lineage tracing analysis and colocalization with immunofluorescence in fetal and adult testis and ovary
Tcf21mCrem:R26RtdTom or Tcf21mCrem:R26RtdTom; Oct4-eGFP timed-pregnant females were administrated with a single dose of 1 mg tamoxifen via gavage at E9.5, E10.5, E11.5, or E12.5. Embryos were obtained at E11.5, E17.5, or E19.5 via C-section. Tail clippings from the embryos were used to identify sex and genotype. Embryonic gonads were fixed in 4% paraformaldehyde (PFA) at 4 °C for 1 h, transferred to 30% sucrose in 1xPBS at 4 °C overnight and embedded in OCT (Surgipath cryo-gel, Leica #39475237). To analyze the TCF21lin contribution in adult testis and ovaries, the E19.5 pups obtained by C-section were fostered to CD1 females. The foster mice were euthanized at 10 weeks. Adult testes and ovaries were fixed in 4% PFA at 4 °C overnight, transferred to 30% sucrose in 1xPBS at 4 °C for overnight and embedded in OCT.
For immunofluorescence, 7–10 micron thick OCT sections were cut using a Leica CM3050S cryostat and sections were refixed with 4% PFA for 10 min and permeabilized by incubation in 0.1% Triton in PBS for 15 min. Sections were blocked in 1xPBS supplemented with 3% BSA and 500 mM glycine for 1 h at room temperature and co-stained with FGF5, SOX9, 3β-HSD, CD34, SMA, VASA, COUPTFII, CD31, SF1, WT1, FOXL2, and DsRed antibodies. The primary antibodies and concentrations used are summarized in Supplementary Data 5. All secondary antibodies (Alexa-488-, Alexa-568-, and Alexa-647-conjugated secondary antibodies; Life Technologies/MolecularProbes) were all used at a 1:1000 dilution. DAPI was used as a nuclear counterstain at 1:1000. Representative images were taken with a ZeissAX10 epifluorescence microscope, a Leica SP8 confocal microscope, or a Nikon A1R-HD25 confocal microscope and processed with ImageJ.
Quantification of immunofluorescence colocalization
Tissue sections were stained for immunofluorescence as described above and >20 images per testis were imaged with a ×40 1.2NA objective on a ZeissAX10 epifluorescence microscope, all at a single z-section. The percent overlap between the TCF21lin population and several marker proteins was done using accustom written ImageJ macro (available upon request). Briefly, nuclear regions of interest (ROIs) were created from DAPI staining by blurring with a Gaussian filter, making the image binary, separating overlapping nuclei with a watershed function, then saving the outline of each binary nucleus to ImageJ’s ROI manager. The signal from TCF21lin and the immunostaining were made binary using ImageJ’s automatic thresholding function and the overlap of the binary stain and the nuclear ROI was measured using Image J’s Measurement function. Cells positive for each stain and double-positive cells were sorted and identified in Microsoft Excel. The quantification from each testis was the sum of all quantified images taken from a single testis. The macro was optimized by contrasting its results to manual quantification from at least five images per immunostaining.
Lineage-tracing analysis of the Tcf21+ population in the aged testis
Five or eight-week-old Tcf21mCrem:R26RtdTom males were injected with a single dose of 2 mg tamoxifen intraperitoneally. The testes were collected 1 week (as control) or 1-year past injection. The testes were fixed in 4% PFA at 4 °C overnight, transferred to 30% sucrose in 1xPBS at 4 °C overnight and embedded in OCT. The sections were co-stained for SF1 (1:100, CosmoBio, Cat#KAL-KO610). Overlap of SF1 and tdTomato were counted manually per field; ~25–100 fields per biological replicate per condition were collected.
In vivo ablation of Leydig cells
Ethane dimethane sulfonate (EDS) injections
EDS (AABlocks, Cat. No: 4672-49-5) was dissolved in DMSO at a concentration of 150 mg/mL. Working solutions of EDS were further diluted in PBS to a final concentration of 50 mg/mL. A total of 300 mg/kg of EDS was injected intraperitoneally every other day for 2 days into C57BL/6 age-matched or Tcf21mCrem:R26RtdTom mice. Testes and sera were collected 12 h, 24 h, 4 days, 7 days, 14 days, or 21 days past final injections. Negative controls were given vehicle treatment of DMSO: PBS without EDS.
Diphtheria toxin Injections
DTX (Sigma) was diluted in PBS at a concentration of 2 mg/ml. A final concentration of 150 ng DTX was injected intraperitoneally every day for 3 or 4 days into Myh11cre-egfp;Rosa26DTR/+ male mice. Testes were collected 12 h, and 4 days past final injections. Littermates that are Myh11-Cre-recombinase negative were given DTX and served as controls.
TUNEL staining
Testes were collected, fixed in 4% PFA for ~16 h at 4 °C, dehydrated in ethanol wash series, and embedded in paraffin. Five-micron FFPE tissue sections were deparaffinized, rehydrated, and permeabilized in 20 μg/ml Proteinase K solution for 15 min at room temperature. Samples were further processed following the Promega Dead End Colorimetric TUNEL kit according to the manufacturer instructions. All images collected used a Leica Leitz DMRD microscope.
Hormone measurements
Testosterone measurements were performed by the University of Virginia Center for Research in Reproduction Ligand Assay and Analysis Core.
FFPE immunofluorescence
Whole testes were fixed in 4% PFA overnight at 4 °C and processed for formalin fixed paraffin embedding as described in (Fisher et al. 2008). Five-micron FFPE tissue sections were deparaffinized by incubation in Histoclear 3× for 5 min, followed by incubation in 100% EtOH 2× for 5 min, 95% EtOH 2× for 5 min, 80% EtOH 1× for 5 min, 70% EtOH 1× for 5 min, 50% EtOH 1× for 5 min, 30% EtOH 1× for 5 min, and deionized water 2× for 3 min each. Tissue sections were permeabilized by incubation in 0.1% Triton in PBS for 15 min. For all antibodies, antigen retrieval was performed by boiling in 10 mM sodium citrate, pH 6.0 for 30 min. Sections were blocked in 1xPBS supplemented with 3% BSA and 500 mM glycine for 3 h at room temperature. Endogenous peroxidases and alkaline phosphatases were blocked by a 10-min incubation in BloxAll solution (VectorLabs, Cat. No: SP-6000). The primary antibodies and concentrations used are listed below. Alexa-488-, Alexa- 555-, and Alexa-647-conjugated secondary antibodies (Life Technologies/MolecularProbes) were all used at 1:1000. DAPI was used as a nuclear counterstain. For quantification, the overlap of SF1 and tdTomato were counted manually per field; ~25–50 fields per biological replicate per condition were collected.
Cell diameter measurements
For all cell diameter measurements, FFPE slides were stained as described above and co-stained with SF1 to mark Leydig cells (1:100, CosmoBio, Cat#KAL-KO610) and 488-wheat germ agglutinin (WGA, Biotium Cat. No: 29022-1) to mark cell perimeter. A tubule was centered in the field of view at ×40 magnification and an image was taken. At least 70 well-defined Leydig cells, marked by both SF1 and clear visible cell perimeter by WGA, were counted per condition. Cell diameter was measured manually using ImageJ’s Line and Measure functions.
FFPE immunohistochemistry
Whole testes were fixed in 4% PFA overnight at 4 °C, processed and deparaffinized as described above. The primary antibodies and concentrations used are listed in Supplementary Data 5. Horseradish peroxidase and alkaline phosphatase conjugated secondary antibodies (Abcam) were all used at a 1:100 concentration and left to incubate for 1-h at room temperature. Slides were rinsed of developing solution under running DI water for 1 min and mounted in per mount.
Transplantation of TCF21lin cells into the testis of WT EDS treated or Myh11cre-egfp;Rosa26DTR/+ animals
6–18 weeks Tcf21mCrem:R26RtdTom male mice were injected with 1 mg tamoxifen intraperitoneally every other day for a total of three injections. Testes were dissociated into a single-cell suspension and the SCA1+/cKITit− stained cells were collected by FACs, as described above. For each animal ~65,000–150,000 cells were diluted in 10 μl of MEM media plus Trypan Blue and were injected into the interstitium via the rete testes of either EDS-treated C57BL/6 animals 24 hpfi of EDS or into Myh11cre-egfp;Rosa26DTR/+ mice 24 hpfi of DTX. As a control, 10 μL of MEM media plus Trypan Blue was injected into the contralateral testis of each experimental animal. Testes were collected at 24 h post transplant (hpt), 4 days post transplant (dpt) and 7 dpt for FPPE processing as described above.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
We thank members of the Hammoud, Li and Yamashita Labs for scientific discussions and manuscript comments. We thank Drs. Barry Zirkin and Haolin Chen for EDS compound. This research was supported by National Institute of Health (NIH) grants 1R21HD090371-01A1 (S.S.H., J.Z.L.), 1DP2HD091949-01 (S.S.H.), R01 HD092084 (K.E.O., S.S.H), F30HD097961 (A.N.S), F31HD100124 (G.L.M.), training grants 5T32HD079342 (A.N.S.), 5T32GM007863 (A.N.S.), NSF 1256260 DGE (L.M.), Rackham Predoctoral Fellowship (L.M.), T32GM007315 (L.M), T32HD007505 (G.L.M.), T32GM007315 (G.L.M.), and Michigan Institute for Data Science (MIDAS) grant for Health Sciences Challenge Award (J.Z.L., S.S.H.), Open Philanthropy Grant 2019-199327 (5384) (S.S.H.).
Source data
Author contributions
S.S.H., H.L., and A.N.S. provided overall project design. Y.S., H.L., A.N.S., L.M., G.L.M., M.S., M.C., and C.S. performed experiments. Q.M. J.Z.L, and X.Z. analyzed data. S.J.G. performed flow cytometry experiments and analysis. A.N.S. and S.S.H. wrote the manuscript with input from H.C., J.R.S., K.E.O, M.T. and J.Z.L. Q.M. and G.L.M. contributed equally. Comments from all authors were provided.
Data availability
Single-cell RNA-seq data files are available in “GSE151337”. All other relevant data supporting the key findings of this study are available within the article and its Supplementary Information files or from the corresponding author upon reasonable request. A reporting summary for this article is available as a Supplementary Information file. Source data are provided with this paper.
Code availability
Codes used in this analysis were deposited onto GitHub: 10.5281/zenodo.4743036.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information Nature Communications thanks Miles Wilkinson and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yu-chi Shen, Adrienne Niederriter Shami, Lindsay Moritz, Hailey Larose.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-021-24130-8.
References
- 1.Karl J, Capel B. Sertoli cells of the mouse testis originate from the coelomic epithelium. Dev. Biol. 1998;203:323–333. doi: 10.1006/dbio.1998.9068. [DOI] [PubMed] [Google Scholar]
- 2.Hatano O, Takakusu A, Nomura M, Morohashi K. Identical origin of adrenal cortex and gonad revealed by expression profiles of Ad4BP/SF-1. Genes Cells. 1996;1:663–671. doi: 10.1046/j.1365-2443.1996.00254.x. [DOI] [PubMed] [Google Scholar]
- 3.Morohashi K. The ontogenesis of the steroidogenic tissues. Genes Cells. 1997;2:95–106. doi: 10.1046/j.1365-2443.1997.1060304.x. [DOI] [PubMed] [Google Scholar]
- 4.Ikeda Y, Shen WH, Ingraham HA, Parker KL. Developmental expression of mouse steroidogenic factor-1, an essential regulator of the steroid hydroxylases. Mol. Endocrinol. 1994;8:654–662. doi: 10.1210/mend.8.5.8058073. [DOI] [PubMed] [Google Scholar]
- 5.Lin YT, Barske L, DeFalco T, Capel B. Numb regulates somatic cell lineage commitment during early gonadogenesis in mice. Development. 2017;144:1607–1618. doi: 10.1242/dev.144113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martineau J, Nordqvist K, Tilmann C, Lovell-Badge R, Capel B. Male-specific cell migration into the developing gonad. Curr. Biol. 1997;7:958–968. doi: 10.1016/S0960-9822(06)00415-5. [DOI] [PubMed] [Google Scholar]
- 7.Buehr M, McLaren A, Bartley A, Darling S. Proliferation and migration of primordial germ cells in We/We mouse embryos. Dev. Dyn. 1993;198:182–189. doi: 10.1002/aja.1001980304. [DOI] [PubMed] [Google Scholar]
- 8.Tilmann C, Capel B. Mesonephric cell migration induces testis cord formation and Sertoli cell differentiation in the mammalian gonad. Development. 1999;126:2883–2890. doi: 10.1242/dev.126.13.2883. [DOI] [PubMed] [Google Scholar]
- 9.DeFalco T, Takahashi S, Capel B. Two distinct origins for Leydig cell progenitors in the fetal testis. Dev. Biol. 2011;352:14–26. doi: 10.1016/j.ydbio.2011.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brennan J, Capel B. One tissue, two fates: molecular genetic events that underlie testis versus ovary development. Nat. Rev. Genet. 2004;5:509–521. doi: 10.1038/nrg1381. [DOI] [PubMed] [Google Scholar]
- 11.Rotgers E, Jorgensen A, Yao HH. At the crossroads of fate-somatic cell lineage specification in the fetal gonad. Endocr. Rev. 2018;39:739–759. doi: 10.1210/er.2018-00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cederroth CR, Pitetti JL, Papaioannou MD, Nef S. Genetic programs that regulate testicular and ovarian development. Mol. Cell Endocrinol. 2007;265-266:3–9. doi: 10.1016/j.mce.2006.12.029. [DOI] [PubMed] [Google Scholar]
- 13.Green CD, et al. A comprehensive roadmap of murine spermatogenesis defined by single-cell RNA-seq. Dev. Cell. 2018;46:651–667 e610. doi: 10.1016/j.devcel.2018.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhang DH, et al. Testicular endothelial cells are a critical population in the germline stem cell niche. Nat. Commun. 2018;9:4379. doi: 10.1038/s41467-018-06881-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kitadate Y, et al. Competition for mitogens regulates spermatogenic stem cell homeostasis in an open niche. Cell Stem Cell. 2019;24:79–92 e76. doi: 10.1016/j.stem.2018.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeFalco T, et al. Macrophages contribute to the spermatogonial niche in the adult testis. Cell Rep. 2015;12:1107–1119. doi: 10.1016/j.celrep.2015.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith LB, O’Shaughnessy PJ, Rebourcet D. Cell-specific ablation in the testis: what have we learned? Andrology. 2015;3:1035–1049. doi: 10.1111/andr.12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang MH, et al. Characterization of Nestin-positive stem Leydig cells as a potential source for the treatment of testicular Leydig cell dysfunction. Cell Res. 2014;24:1466–1485. doi: 10.1038/cr.2014.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davidoff MS, et al. Progenitor cells of the testosterone-producing Leydig cells revealed. J. Cell Biol. 2004;167:935–944. doi: 10.1083/jcb.200409107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kilcoyne KR, et al. Fetal programming of adult Leydig cell function by androgenic effects on stem/progenitor cells. Proc. Natl Acad. Sci. USA. 2014;111:E1924–E1932. doi: 10.1073/pnas.1320735111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qin J, Tsai MJ, Tsai SY. Essential roles of COUP-TFII in Leydig cell differentiation and male fertility. PLoS One. 2008;3:e3285. doi: 10.1371/journal.pone.0003285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ge RS, et al. In search of rat stem Leydig cells: identification, isolation, and lineage-specific development. Proc. Natl Acad. Sci. USA. 2006;103:2719–2724. doi: 10.1073/pnas.0507692103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Landreh L, Stukenborg JB, Soder O, Svechnikov K. Phenotype and steroidogenic potential of PDGFRalpha-positive rat neonatal peritubular cells. Mol. Cell Endocrinol. 2013;372:96–104. doi: 10.1016/j.mce.2013.03.019. [DOI] [PubMed] [Google Scholar]
- 24.Chen H, Huhtaniemi I, Zirkin BR. Depletion and repopulation of Leydig cells in the testes of aging brown Norway rats. Endocrinology. 1996;137:3447–3452. doi: 10.1210/endo.137.8.8754773. [DOI] [PubMed] [Google Scholar]
- 25.Chen H, Stanley E, Jin S, Zirkin BR. Stem Leydig cells: from fetal to aged animals. Birth Defects Res C. Embryo Today. 2010;90:272–283. doi: 10.1002/bdrc.20192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar DL, DeFalco T. A perivascular niche for multipotent progenitors in the fetal testis. Nat. Commun. 2018;9:4519. doi: 10.1038/s41467-018-06996-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu J, et al. The basic helix-loop-helix transcription factor capsulin controls spleen organogenesis. Proc. Natl Acad. Sci. USA. 2000;97:9525–9530. doi: 10.1073/pnas.97.17.9525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu JR, et al. Control of facial muscle development by MyoR and capsulin. Science. 2002;298:2378–2381. doi: 10.1126/science.1078273. [DOI] [PubMed] [Google Scholar]
- 29.Quaggin SE, et al. The basic-helix-loop-helix protein pod1 is critically important for kidney and lung organogenesis. Development. 1999;126:5771–5783. doi: 10.1242/dev.126.24.5771. [DOI] [PubMed] [Google Scholar]
- 30.Cui S, et al. Disrupted gonadogenesis and male-to-female sex reversal in Pod1 knockout mice. Development. 2004;131:4095–4105. doi: 10.1242/dev.01266. [DOI] [PubMed] [Google Scholar]
- 31.Bhandari RK, Sadler-Riggleman I, Clement TM, Skinner MK. Basic helix-loop-helix transcription factor TCF21 is a downstream target of the male sex determining gene SRY. PLoS One. 2011;6:e19935. doi: 10.1371/journal.pone.0019935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stevant I, et al. Dissecting cell lineage specification and sex fate determination in gonadal somatic cells using single-cell transcriptomics. Cell Rep. 2019;26:3272–3283 e3273. doi: 10.1016/j.celrep.2019.02.069. [DOI] [PubMed] [Google Scholar]
- 33.Stevant I, et al. Deciphering cell lineage specification during male sex determination with single-cell RNA sequencing. Cell Rep. 2018;22:1589–1599. doi: 10.1016/j.celrep.2018.01.043. [DOI] [PubMed] [Google Scholar]
- 34.Shami AN, et al. Single-cell RNA sequencing of human, macaque, and mouse testes uncovers conserved and divergent features of mammalian spermatogenesis. Dev. Cell. 2020;54:529–547. doi: 10.1016/j.devcel.2020.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hynes RO. The extracellular matrix: not just pretty fibrils. Science. 2009;326:1216–1219. doi: 10.1126/science.1176009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bonnans C, Chou J, Werb Z. Remodelling the extracellular matrix in development and disease. Nat. Rev. Mol. Cell Biol. 2014;15:786–801. doi: 10.1038/nrm3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Acharya A, Baek ST, Banfi S, Eskiocak B, Tallquist MD. Efficient inducible Cre-mediated recombination in Tcf21 cell lineages in the heart and kidney. Genesis. 2011;49:870–877. doi: 10.1002/dvg.20750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uccelli A, Moretta L, Pistoia V. Mesenchymal stem cells in health and disease. Nat. Rev. Immunol. 2008;8:726–736. doi: 10.1038/nri2395. [DOI] [PubMed] [Google Scholar]
- 39.Gonzalez R, et al. A putative mesenchymal stem cells population isolated from adult human testes. Biochem Biophys. Res Commun. 2009;385:570–575. doi: 10.1016/j.bbrc.2009.05.103. [DOI] [PubMed] [Google Scholar]
- 40.Ahmed M, Ghabriel M, Amleh A. Enrichment, propagation, and characterization of mouse testis-derived mesenchymal stromal cells. Cell Reprogram. 2017;19:35–43. doi: 10.1089/cell.2016.0022. [DOI] [PubMed] [Google Scholar]
- 41.Eliveld J, et al. Primary human testicular PDGFRalpha+ cells are multipotent and can be differentiated into cells with Leydig cell characteristics in vitro. Hum. Reprod. 2019;34:1621–1631. doi: 10.1093/humrep/dez131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li X, et al. Regulation of seminiferous tubule-associated stem Leydig cells in adult rat testes. Proc. Natl Acad. Sci. USA. 2016;113:2666–2671. doi: 10.1073/pnas.1519395113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clark AM, Garland KK, Russell LD. Desert hedgehog (Dhh) gene is required in the mouse testis for formation of adult-type Leydig cells and normal development of peritubular cells and seminiferous tubules. Biol. Reprod. 2000;63:1825–1838. doi: 10.1095/biolreprod63.6.1825. [DOI] [PubMed] [Google Scholar]
- 44.Tang H, et al. Notch signaling maintains Leydig progenitor cells in the mouse testis. Development. 2008;135:3745–3753. doi: 10.1242/dev.024786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao F, et al. Autophagy regulates testosterone synthesis by facilitating cholesterol uptake in Leydig cells. J. Cell Biol. 2018;217:2103–2119. doi: 10.1083/jcb.201710078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steinfeld K, et al. Low testosterone in ApoE/LDL receptor double-knockout mice is associated with rarefied testicular capillaries together with fewer and smaller Leydig cells. Sci. Rep. 2018;8:5424. doi: 10.1038/s41598-018-23631-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu C, Rodriguez K, Yao HH. Mapping lineage progression of somatic progenitor cells in the mouse fetal testis. Development. 2016;143:3700–3710. doi: 10.1242/dev.121731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koopman P, Munsterberg A, Capel B, Vivian N, Lovell-Badge R. Expression of a candidate sex-determining gene during mouse testis differentiation. Nature. 1990;348:450–452. doi: 10.1038/348450a0. [DOI] [PubMed] [Google Scholar]
- 49.Matson CK, et al. DMRT1 prevents female reprogramming in the postnatal mammalian testis. Nature. 2011;476:101–104. doi: 10.1038/nature10239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Uhlenhaut NH, et al. Somatic sex reprogramming of adult ovaries to testes by FOXL2 ablation. Cell. 2009;139:1130–1142. doi: 10.1016/j.cell.2009.11.021. [DOI] [PubMed] [Google Scholar]
- 51.Matson CK, Zarkower D. Sex and the singular DM domain: insights into sexual regulation, evolution and plasticity. Nat. Rev. Genet. 2012;13:163–174. doi: 10.1038/nrg3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Edson MA, Nagaraja AK, Matzuk MM. The mammalian ovary from genesis to revelation. Endocr. Rev. 2009;30:624–712. doi: 10.1210/er.2009-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Honda A, et al. Isolation, characterization, and in vitro and in vivo differentiation of putative thecal stem cells. Proc. Natl Acad. Sci. USA. 2007;104:12389–12394. doi: 10.1073/pnas.0703787104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu C, Peng J, Matzuk MM, Yao HH. Lineage specification of ovarian theca cells requires multicellular interactions via oocyte and granulosa cells. Nat. Commun. 2015;6:6934. doi: 10.1038/ncomms7934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kerr JB, Donachie K, Rommerts FF. Selective destruction and regeneration of rat Leydig cells in vivo. A new method for the study of seminiferous tubular-interstitial tissue interaction. Cell Tissue Res. 1985;242:145–156. doi: 10.1007/BF00225571. [DOI] [PubMed] [Google Scholar]
- 56.Molenaar R, de Rooij DG, Rommerts FF, van der Molen HJ. Repopulation of Leydig cells in mature rats after selective destruction of the existent Leydig cells with ethylene dimethane sulfonate is dependent on luteinizing hormone and not follicle-stimulating hormone. Endocrinology. 1986;118:2546–2554. doi: 10.1210/endo-118-6-2546. [DOI] [PubMed] [Google Scholar]
- 57.Morris ID, Phillips DM, Bardin CW. Ethylene dimethanesulfonate destroys Leydig cells in the rat testis. Endocrinology. 1986;118:709–719. doi: 10.1210/endo-118-2-709. [DOI] [PubMed] [Google Scholar]
- 58.Jackson H. Antispermatogenic agents. Br. Med Bull. 1970;26:79–86. doi: 10.1093/oxfordjournals.bmb.a070747. [DOI] [PubMed] [Google Scholar]
- 59.Ewing, L. L. & Keeney, D. S. Leydig Cells. Cell Mol. Biol. Testis, 137–165 (1993).
- 60.Yokonishi T, McKey J, Ide S, Capel B. Sertoli cell ablation and replacement of the spermatogonial niche in mouse. Nat. Commun. 2020;11:40. doi: 10.1038/s41467-019-13879-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rebourcet D, et al. Sertoli cells maintain Leydig cell number and peritubular myoid cell activity in the adult mouse testis. PLoS One. 2014;9:e105687. doi: 10.1371/journal.pone.0105687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rebourcet D, et al. Sertoli cells control peritubular myoid cell fate and support adult Leydig cell development in the prepubertal testis. Development. 2014;141:2139–2149. doi: 10.1242/dev.107029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Teerds KJ, De Rooij DG, Rommerts FF, van der Tweel I, Wensing CJ. Turnover time of Leydig cells and other interstitial cells in testes of adult rats. Arch. Androl. 1989;23:105–111. doi: 10.3109/01485018908986831. [DOI] [PubMed] [Google Scholar]
- 64.May P, Herz J. LDL receptor-related proteins in neurodevelopment. Traffic. 2003;4:291–301. doi: 10.1034/j.1600-0854.2003.00086_4_5.x. [DOI] [PubMed] [Google Scholar]
- 65.Lillis AP, Mikhailenko I, Strickland DK. Beyond endocytosis: LRP function in cell migration, proliferation and vascular permeability. J. Thromb. Haemost. 2005;3:1884–1893. doi: 10.1111/j.1538-7836.2005.01371.x. [DOI] [PubMed] [Google Scholar]
- 66.Li YH, Kuo CH, Shi GY, Wu HL. The role of thrombomodulin lectin-like domain in inflammation. J. Biomed. Sci. 2012;19:34. doi: 10.1186/1423-0127-19-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mitchell JL, Mutch NJ. Let’s cross-link: diverse functions of the promiscuous cellular transglutaminase factor XIII-A. J. Thromb. Haemost. 2019;17:19–30. doi: 10.1111/jth.14348. [DOI] [PubMed] [Google Scholar]
- 68.Robb, L. et al. epicardin: A novel basic helix-loop-helix transcription factor gene expressed in epicardium, branchial arch myoblasts, and mesenchyme of developing lung, gut, kidney, and gonads. Dev. Dyn.213, 105–113 (1998). [DOI] [PubMed]
- 69.Xie T, et al. Single-cell deconvolution of fibroblast heterogeneity in mouse pulmonary fibrosis. Cell Rep. 2018;22:3625–3640. doi: 10.1016/j.celrep.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wirka RC, et al. Atheroprotective roles of smooth muscle cell phenotypic modulation and the TCF21 disease gene as revealed by single-cell analysis. Nat. Med. 2019;25:1280–1289. doi: 10.1038/s41591-019-0512-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Farbehi, N. et al. Single-cell expression profiling reveals dynamic flux of cardiac stromal, vascular and immune cells in health and injury. Elife8, 10.7554/eLife.43882 (2019). [DOI] [PMC free article] [PubMed]
- 72.Dobie R, et al. Single-cell transcriptomics uncovers zonation of function in the mesenchyme during liver fibrosis. Cell Rep. 2019;29:1832–1847. doi: 10.1016/j.celrep.2019.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xiong X, et al. Landscape of intercellular crosstalk in healthy and NASH liver revealed by single-cell secretome gene analysis. Mol. Cell. 2019;75:644–660. doi: 10.1016/j.molcel.2019.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Swonger JM, Liu JS, Ivey MJ, Tallquist MD. Genetic tools for identifying and manipulating fibroblasts in the mouse. Differentiation. 2016;92:66–83. doi: 10.1016/j.diff.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Barsoum IB, Bingham NC, Parker KL, Jorgensen JS, Yao HH. Activation of the Hedgehog pathway in the mouse fetal ovary leads to ectopic appearance of fetal Leydig cells and female pseudohermaphroditism. Dev. Biol. 2009;329:96–103. doi: 10.1016/j.ydbio.2009.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brennan J, Tilmann C, Capel B. Pdgfr-alpha mediates testis cord organization and fetal Leydig cell development in the XY gonad. Genes Dev. 2003;17:800–810. doi: 10.1101/gad.1052503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dominici M, et al. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315–317. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- 78.Nauta AJ, Fibbe WE. Immunomodulatory properties of mesenchymal stromal cells. Blood. 2007;110:3499–3506. doi: 10.1182/blood-2007-02-069716. [DOI] [PubMed] [Google Scholar]
- 79.Yeap BB, Page ST, Grossmann M. Testosterone treatment in older men: clinical implications and unresolved questions from the Testosterone Trials. Lancet Diabetes Endocrinol. 2018;6:659–672. doi: 10.1016/S2213-8587(17)30416-3. [DOI] [PubMed] [Google Scholar]
- 80.Baburski AZ, Andric SA, Kostic TS. Luteinizing hormone signaling is involved in synchronization of Leydig cell’s clock and is crucial for rhythm robustness of testosterone productiondagger. Biol. Reprod. 2019;100:1406–1415. doi: 10.1093/biolre/ioz020. [DOI] [PubMed] [Google Scholar]
- 81.Patel AS, Leong JY, Ramos L, Ramasamy R. Testosterone is a contraceptive and should not be used in men who desire fertility. World J. Mens. Health. 2019;37:45–54. doi: 10.5534/wjmh.180036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stanley E, et al. Identification, proliferation, and differentiation of adult Leydig stem cells. Endocrinology. 2012;153:5002–5010. doi: 10.1210/en.2012-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ge RS, et al. Gene expression in rat leydig cells during development from the progenitor to adult stage: a cluster analysis. Biol. Reprod. 2005;72:1405–1415. doi: 10.1095/biolreprod.104.037499. [DOI] [PubMed] [Google Scholar]
- 84.Chen, P., Zirkin, B. R. & Chen, H. Stem Leydig cells in the adult testis: characterization, regulation and potential applications. Endocr. Rev.41, 10.1210/endrev/bnz013 (2020). [DOI] [PMC free article] [PubMed]
- 85.Ye L, Li X, Li L, Chen H, Ge RS. Insights into the development of the adult Leydig cell lineage from stem Leydig cells. Front. Physiol. 2017;8:430. doi: 10.3389/fphys.2017.00430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chen H, Wang Y, Ge R, Zirkin BR. Leydig cell stem cells: Identification, proliferation and differentiation. Mol. Cell Endocrinol. 2017;445:65–73. doi: 10.1016/j.mce.2016.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Maezawa Y, et al. Loss of the podocyte-expressed transcription factor Tcf21/Pod1 results in podocyte differentiation defects and FSGS. J. Am. Soc. Nephrol. 2014;25:2459–2470. doi: 10.1681/ASN.2013121307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kikuchi K, et al. tcf21+ epicardial cells adopt non-myocardial fates during zebrafish heart development and regeneration. Development. 2011;138:2895–2902. doi: 10.1242/dev.067041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Adashi, E. Y., Rock, J. A. & Rosenwaks, Z. Reproductive endocrinology, surgery, and technology. (Lippincott-Raven, 1996).
- 90.Meineke V, Frungieri MB, Jessberger B, Vogt H, Mayerhofer A. Human testicular mast cells contain tryptase: increased mast cell number and altered distribution in the testes of infertile men. Fertil. Steril. 2000;74:239–244. doi: 10.1016/S0015-0282(00)00626-9. [DOI] [PubMed] [Google Scholar]
- 91.Frungieri MB, et al. Number, distribution pattern, and identification of macrophages in the testes of infertile men. Fertil. Steril. 2002;78:298–306. doi: 10.1016/S0015-0282(02)03206-5. [DOI] [PubMed] [Google Scholar]
- 92.Frungieri MB, Weidinger S, Meineke V, Kohn FM, Mayerhofer A. Proliferative action of mast-cell tryptase is mediated by PAR2, COX2, prostaglandins, and PPARgamma: Possible relevance to human fibrotic disorders. Proc. Natl Acad. Sci. USA. 2002;99:15072–15077. doi: 10.1073/pnas.232422999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mayer C, et al. Sterile inflammation as a factor in human male infertility: Involvement of Toll like receptor 2, biglycan and peritubular cells. Sci. Rep. 2016;6:37128. doi: 10.1038/srep37128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li L, et al. Directing differentiation of human induced pluripotent stem cells toward androgen-producing Leydig cells rather than adrenal cells. Proc. Natl Acad. Sci. USA. 2019;116:23274–23283. doi: 10.1073/pnas.1908207116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mi H, Muruganujan A, Ebert D, Huang X, Thomas PD. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019;47:D419–D426. doi: 10.1093/nar/gky1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rux DR, et al. Regionally restricted Hox function in adult bone marrow multipotent mesenchymal stem/stromal cells. Dev. Cell. 2016;39:653–666. doi: 10.1016/j.devcel.2016.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Park D, et al. Endogenous bone marrow MSCs are dynamic, fate-restricted participants in bone maintenance and regeneration. Cell Stem Cell. 2012;10:259–272. doi: 10.1016/j.stem.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Macosko EZ, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shekhar K, et al. Comprehensive classification of retinal bipolar neurons by single-cell transcriptomics. Cell. 2016;166:1308–1323. doi: 10.1016/j.cell.2016.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
Single-cell RNA-seq data files are available in “GSE151337”. All other relevant data supporting the key findings of this study are available within the article and its Supplementary Information files or from the corresponding author upon reasonable request. A reporting summary for this article is available as a Supplementary Information file. Source data are provided with this paper.
Codes used in this analysis were deposited onto GitHub: 10.5281/zenodo.4743036.