Abstract
Post-transcriptional gene regulation, including that by RNA binding proteins (RBPs), has recently been described as an important mechanism in cancer. We had previously identified a set of RBPs that were highly dysregulated in B-cell acute lymphoblastic leukemia (B-ALL) with MLL translocations, which carry a poor prognosis. Here, we sought to functionally characterize these dysregulated RBP genes by performing a focused CRISPR dropout screen in B-ALL cell lines, finding dependencies on several genes including EIF3E, EPRS and USO1. Validating our findings, CRISPR/Cas9-mediated disruption of USO1 in MLL-translocated B-ALL cells reduced cell growth, promoted cell death, and altered the cell cycle. Transcriptomic analysis of USO1-deficient cells revealed alterations in pathways related to mTOR signaling, RNA metabolism, and targets of MYC. In addition, USO1-regulated genes from these experimental samples were significantly and concordantly correlated with USO1 expression in primary samples collected from B-ALL patients. Lastly, we found that loss of Uso1 inhibited colony formation of MLL-transformed in primary bone marrow cells from Cas9-EGFP mice. Together, our findings demonstrate an approach to performing focused sub-genomic CRISPR screens and highlight a putative RBP vulnerability in MLL-translocated B-ALL, thus identifying potential therapeutic targets in this disease.
Subject terms: Leukaemia, Cancer, Cancer genetics
Introduction
B-ALL is the most common type of leukemia in the pediatric population, and is characterized by a number of recurrent chromosomal rearrangements1–4. Among these, the t (4;11) MLL-AF4 (KMT2A-AFF1) translocation gives rise to a highly aggressive form of B-ALL5,6. Patients with MLL-rearranged B-ALL have a dismal prognosis, with 5-year event-free survival rates hovering at 33.6% for infants7 and 50% for older children and adults8. Most of these patients are resistant to conventional treatment with chemotherapy and steroids9, with bone marrow transplantation being the only curative therapeutic alternative10. Although recent developments such as CAR-T therapy11 and anti-CD19 based therapy such as Blinatumomab12 have raised hope for such patients13, antigen escape and lineage infidelity in MLL-translocated leukemia have proved problematic14. Therefore, there is an urgent need to better characterize potential therapeutic targets with high specificity.
The MLL-AF4 translocation engenders a unique transcriptional profile, as the fusion protein juxtaposes a histone methyltransferase (MLL, also known as KMT2A) with a protein that is involved in transcriptional regulation (AF4, or AFF1). Recently our lab carried out a study examining the expression of RBPs in B-ALL15, including both known and predicted RBPs16. In our analysis, we identified 36 RBPs that are highly upregulated in MLL-AF4 translocated B-ALL15. To study the importance of these genes in B-ALL, we implemented the powerful gene editing technique, CRISRP/Cas9, to perform a rapid and medium-throughput assessment of gene function. Genome wide CRISPR/Cas9 screens on AML cell lines have identified multiple gene targets critical for cell proliferation and survival17, but similar studies have not been performed in MLL-AF4 translocated B-ALL.
In the present study, we performed a sub-genomic CRISPR/Cas9 dropout screen using 36 highly upregulated RBPs in primary human B-ALL and identified several novel vulnerabilities that included three putative RBPs. Of these, USO1, a putative RBP and a known regulator of vesicular transport, was identified as a MLL-AF4 target gene. CRISPR/Cas9-mediated disruption of USO1 significantly altered cell growth and the cell cycle in B-ALL cell lines; as well as inhibited the colony forming potential of MLL-transformed primary murine bone marrow cells. USO1 depletion regulated the expression of genes related to mTOR signaling, metabolism of RNA, and MYC targets. Together, our studies provide a comprehensive rubric to functionally evaluate putative targets identified from expression profiling, and the identification of a novel potential target in MLL-rearranged leukemia.
Results
CRISPR/Cas9 screen identifies potential vulnerabilities in MLL-AF4 leukemic cell growth
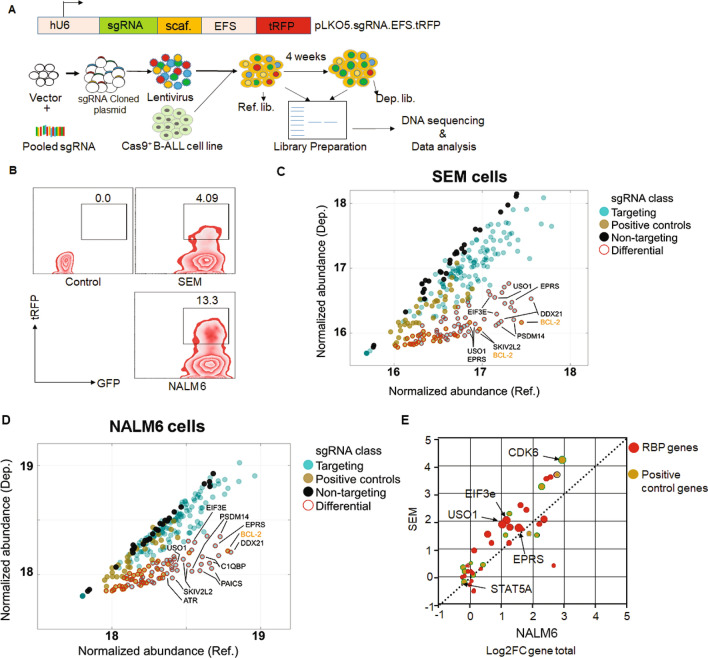
Previously, we identified 36 putative RBPs that were significantly dysregulated in primary human MLL-rearranged B-ALL15. To directly query the functional relevance of RBP dysregulation in MLL-rearranged B-ALL, we performed a sub-genomic CRISPR screen (Fig. 1A). The library consisted of sgRNAs targeting 36 RBP genes, 12 “positive control” genes, representing known vulnerabilities in MLL-translocated leukemia17, and 28 non-targeting (NT) sgRNAs. The screen was designed using a two-vector lentiviral system. First, B-ALL cells with and without MLL-AF4 translocation (SEM and NALM6, respectively) were stably transduced with a Cas9-P2A-EGFP transgene, followed by FACS sorting of the transduced cells based on GFP positivity. Next, Cas9-GFP+ SEM and NALM6 cells were transduced at a MOI of < 0.3 with the pooled sgRNA lentiviral library, comprised of 268 unique sgRNAs. Cells were subsequently FACS sorted 48 h following transduction for GFP and tRFP double positivity (Fig. 1B). 2 × 106 cells were sorted, of which 106 cells were used to isolate genomic DNA for the reference (REF) libraries and the remainder were cultured to maintain 3700 × coverage until collection of genomic DNA for the depletion (DEP) libraries following 28 days in culture. Experimental replicates of REF and DEP libraries showed a high degree of concordance in abundance of both individual sgRNAs and for total sgRNAs per gene (Supplementary Fig. 1D & 1E). In addition, the SEM and NALM6 REF libraries showed overall similar rates of individual gRNA incorporation, as measured by the abundance of each gRNA in each of the cell lines and biological replicates (Supplementary Fig. 1F). As expected, a majority of “positive control” genes, including BCL2, COA5, CDK6, and MYC, were significantly downregulated in the DEP libraries in both NALM6 and SEM cells. This is not surprising as many of these are known oncogenic genes, particularly in B-ALL. Non-targeting or “negative control” sgRNAs were consistently unchanged between the REF and DEP libraries (Fig. 1C, D). Comparing the results across cell lines, we found that sgRNAs targeting three genes, USO1, EIF3E and EPRS were significantly depleted in SEM cells (p < 0.001), when compared to NALM6 cells (Fig. 1E). Interestingly, sgRNA dropout in general was more readily observed in SEM cells than in NALM6 cells, potentially due to the fact that these genes were selected based on high expression in patient B-ALL samples with MLL-AF4 translocation. Of these three genes, USO1 had previously been detected in a genome-wide CRISPR screen as a vulnerability in MV-4–11 cells, which also harbor the MLL-AF4 fusion gene17.
Figure 1.
Sub-genomic CRISPR screen identifies functionally important genes in MLL-AF4-translocated B-ALL. (A) Schematic of sub-genomic CRISPR screen. (B) FACS contour plots reflecting sorting strategy based on high expression of Cas9 (GFP) and pooled guide RNA library (tRFP) in B-ALL cell lines. (C, D) Variance-stabilized normalized abundance for individual sgRNAs in Reference (Ref; x-axis) and Depletion (Dep; y-axis) libraries in SEM (C) and NALM6 (D) cell lines. Dots are colored by sgRNA class (dark yellow: positive controls; black: non-targeting negative controls; teal: targeting sgRNAs). Dots highlighted with a red border were classified as differentially abundant (Log2 fold change [Log2FC] > 1, Wald adjusted p value < 0.001). Genes with three or more differential sgRNAs are highlighted in the inset and colored by class. (E) Differential expression of sgRNAs aggregated by gene (Log2FC gene total), in NALM6 (x-axis) vs SEM (y-axis) cell lines. Dots in the upper left represent genes with a higher fold-change in SEM cells, while those on the lower right represent genes with a higher fold-change in NALM6 cells. Dots are colored by sgRNA class and sized according to the number of individual sgRNAs that showed significant differential representation in the Dep libraries.
USO1 is directly regulated by MLL-AF4
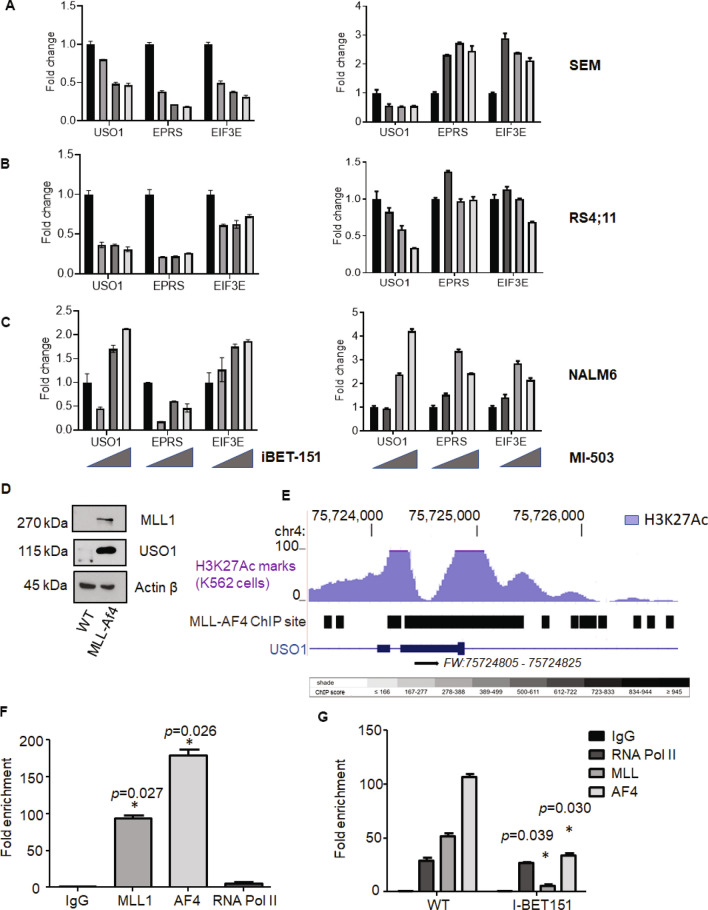
USO1, EPRS and EIF3E expression was assessed in, SEM, RS4;11 (another MLL-AF4 translocated cell line), and NALM6 (Supplementary Fig. 2A & 2B). To query their regulation by the MLL-AF4 fusion protein, we analyzed their expression following inhibition with I-BET15118, a BRD4 BET domain inhibitor, MI-503 menin-MLL1 inhibitor19 and EPZ5676, a DOT1L inhibitor20, all known to inhibit MLL-dependent gene expression regulation. With increasing doses of I-BET151, MI-503 and EPZ5676 there was a decrease in the USO1 mRNA expression level in SEM (Fig. 2A), and RS4;11 cells (Fig. 2B), but not in NALM6 cells (Fig. 2C as well as Supplementary Fig. 2C, 2D & 2E), suggesting that USO1 expression is MLL-AF4 dependent. A consistent reduction in EIF3E and EPRS was not observed in the MLL translocated cell lines. We further queried the MLL dependence of USO1 by over-expressing the MLL-Af4 transgene21 in murine bone marrow cells. Western Blot and RT-qPCR analysis showed upregulation of MLL1 and USO1 in MLL-Af4 transduced cells compared to control cells (Fig. 2D and Supplementary Fig. 2F). Using publicly available data21, we found that the USO1 and EIF3E genes demonstrated multiple MLL-AF4 binding sites within the 5’UTR, the first exon, and the first intron (Fig. 2E and Supplementary Fig. 2G). In contrast, the EPRS gene did not show strong MLL-AF4 binding sites (Supplementary Fig. 2H). EIF3e and EPRS were previously reported to be “common essential genes”, per the depmap portal22, whereas USO1 was not such a gene, suggesting its potential utility as a novel clinical target. To confirm our findings of USO1 dependence on MLL-AF4, we designed chromatin immunoprecipitation (ChIP) experiments, designing primers for the regulatory regions of USO1 including the 5’ UTR within the first exon. We found significant enrichment of the USO1 regulatory regions in both the MLL1 and AF4 pulldowns (Fig. 2F). Treatment of the cells with I-BET151 inhibited the association of both MLL1 and AF4 with the promoter region of USO1 (Fig. 2G). Together, these findings indicate that USO1 is a direct target of the MLL-AF4 transcriptional program.
Figure 2.
Dependence of RBP gene expression on MLL-AF4 translocation. (A–C) Effect of I-BET 151 and MI-503 treatment on mRNA expression levels of USO1,EPRS and EIF3E, measured by RT-qPCR, in SEM (A), RS4;11 (B), and NALM6 (C). The cells were treated with increasing concentrations of I-BET151 (DMSO only, 0.5, 1 and 2 μM) or with increasing concentrations of the menin inhibitor, MI-503 (DMSO only, 0.12, 0.25, and 0.5 μM). RT-qPCR was performed with an optimized set of primers, normalized to 18S, and then represented as fold-change from vehicle-treated control. D. Western blot analysis of murine bone marrow cells with and without transduction with MLL-Af4 (WT versus MLL-Af4), for MLL1 (top), USO1 (middle) and β-actin (lower). (E) UCSC genome browser shot of the USO1 locus showing the MLL-AF4 ChIP site(s), as identified from the ChIP-Seq data from Lin et al.21, in a gene expression regulatory region; Courtesy: UCSC Genome Browser. Shown are the H3K27Ac track in hematopoietic K562 cells (Blue), and MLL-AF4 binding sites represented as a grayscale score, with black indicating the highest score/highest number of reads from the dataset. (F) Chromatin immunoprecipitation with indicated antibodies (MLL1, AF4, and RNA Pol II), followed by qPCR (ChIP-qPCR) analysis for quantitation of bound USO1 promoter/regulatory region to MLL1 and AF4 pulldown samples. Shown is the fold-enrichment for qPCR of the USO1 regulatory site over background (t test; *P < 0.05) (G) SEM cells treated with 1 µM of I-BET151 for 48 h. and subjected to ChIP qPCR with MLL1 and AF4 antibodies as in (F) (t test; *P < 0.05).
USO1 depletion alters B-ALL cell proliferation, survival and cell cycle
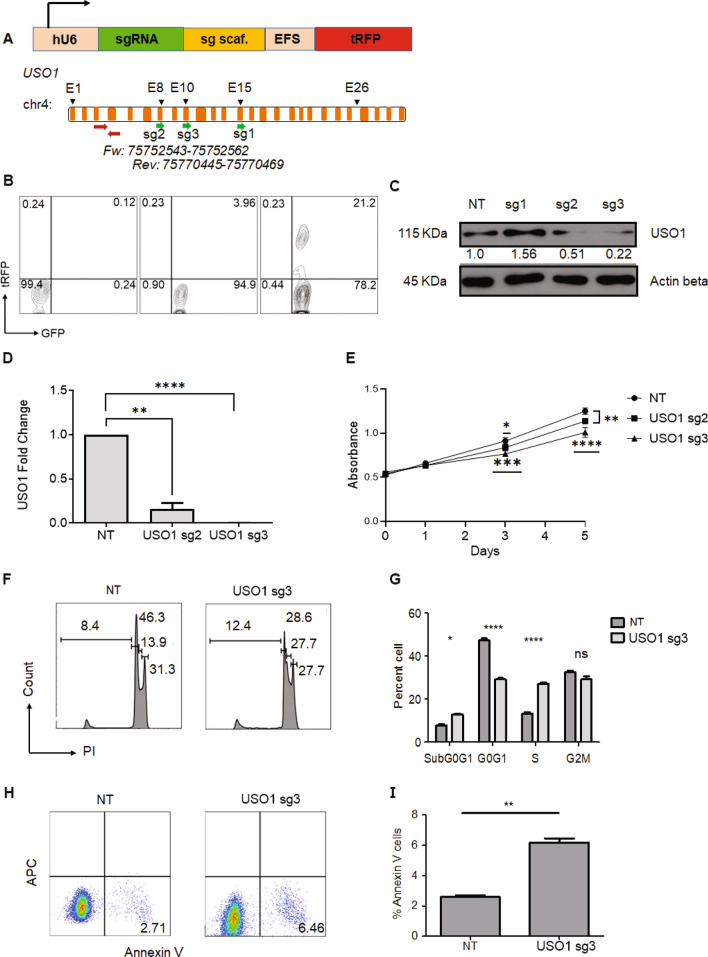
USO1 expression is upregulated in several types of cancer including B-ALL with MLL-AF4 translocations23–25. To characterize the functional role of USO1 in B-ALL suggested by our CRISPR screen, we utilized the previously mentioned two-vector lentivirus system to transduce SEM cells with three different lentiviral constructs containing sgRNAs that target different regions of the USO1 gene (Fig. 3A, B). We found that two sgRNAs (sg2 and sg3) caused a significant downregulation of USO1 protein and mRNA expression by approximately 80% in SEM cells (Fig. 3C, D). Cas9-mediated frameshift mutation in USO1 was confirmed using TIDE assay26 (Supplementary Fig. 3A). The lack of complete ablation was a reproducible finding in all cell lines we tested, as multiple single-cell cloning experiments failed to produce cells with complete knockout and multiple bulk cultures experiments showed retained partial expression of < 25%. Nonetheless, we observed that bulk USO1-depleted SEM cells from sg2 and sg3 showed reduced proliferation by MTS assay (Fig. 3E). Since sg3 showed near-total ablation of USO1 protein expression, we later used sg3 to target USO1 in RS4;11 cells (Supplementary Fig. 3B & 3C), finding downregulation, but not complete depletion, of USO1 protein. Propidium Iodide (PI) based cell cycle analysis on USO1-depleted cells showed an increased percentage of cells in the G0/G1 stage, suggesting cell cycle arrest, and more cells in Sub-G0/G1, suggesting increased apoptosis (Fig. 3F, G). Increased cell death was also observed in the USO1-depleted cells by Annexin V staining (Fig. 3H, I). Interestingly, USO1-depleted cells treated with I-BET151 also showed increased cell cycle arrest and apoptosis (Supplementary Fig. 3D–3G), suggesting an additive effect with this inhibitor of BRD4. To confirm our findings in an orthogonal system, we introduced siRNAs targeting USO1 using nucleofection. In these short-term assays, we found that there was partial reduction of USO1 mRNA, as expected with siRNA-mediated knockdown (Supplementary Fig. 4A) and increased Annexin V staining (Supplementary Fig. 4B, C). There were also increased cells in Sub-G0/G1 and a modest reduction in cell proliferation as measured by the MTS assay (Supplementary Fig. 4E, F). Together, these observations confirm the importance of USO1 in regulation of cell cycle and survival of B-ALL cells.
Figure 3.
Depletion of USO1 leads to decreased cell growth, cell cycle arrest and increased apoptosis. (A) Schematic representation of the pLKO5.sgRNA.EFS.tRFP lentiviral vector. Abbreviations, hU6, human U6 promoter; sgRNA, short guide RNA; sg scaf, sgRNA scaffold; tRFP, turbo red fluorescent protein. (B) Sample FACS plots of SEM cells transduced sequentially with Cas9 vector and sgRNA containing vector. Left, non-transduced SEM cells; middle, transduced with pLentiCas9-GFP; right, cells transduced with both pLenti-Cas9-GFP and pLKO5 vector containing USO1-targeting sgRNA. (C) Western blot for USO1 in SEM cells following CRISPR/Cas9-mediated disruption of the USO1 gene using three different sgRNAs (sg1-3) and NT, non-targeting sgRNAs. (D) RT-qPCR measurement of USO1 in control (NT) and USO1 (sg2 & sg3) SEM cells (t test; **P < 0.01; ****P < 0.0001) (E) MTS assay to study the cell growth of USO1-depleted cells (sg2 & sg3), measured as Absorbance at 490 nM (t test; **P < 0.01; ***P < 0.001). (F, G) Cell cycle analysis using propidium iodide (PI) staining of control cells and USO1-depleted cells (F) and quantitation of cells from cell cycle analysis (Two-way Annova with Bonferroni correction; *P < 0.05; ****P < 0.0001). (H, I) FACS plots of Annexin V positivity in control versus USO1-depleted cells (H), Quantitation of cells with Annexin V positivity (t test; **P < 0.01) (I).
USO1 impacts pathways related to cellular proliferation and RNA homeostasis
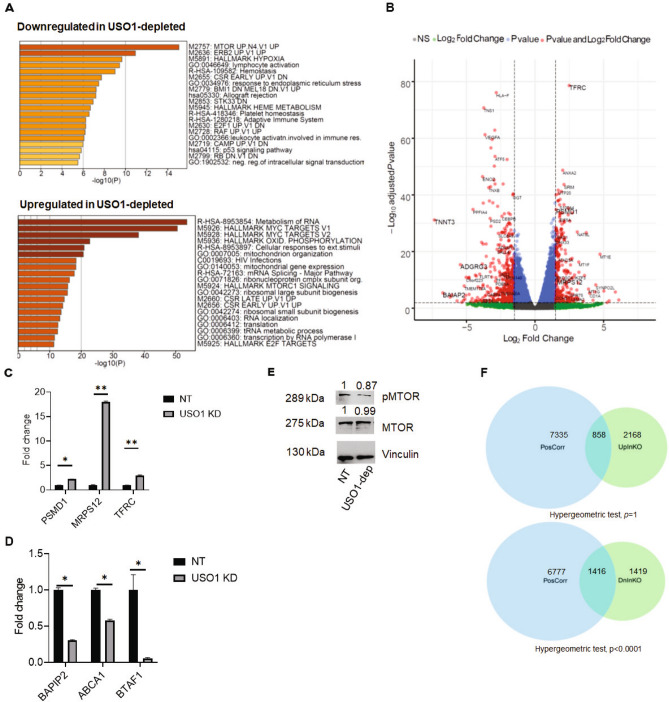
To evaluate the effect of USO1 depletion on the overall gene expression pattern in B-ALL cells, we performed RNA-Seq on SEM cells that were depleted of USO1 by CRISPR/Cas9 (three biological replicates per condition). Following differential expression analysis of USO1-depleted and control cells, we utilized the Metascape algorithm and Gene set enrichment analysis (GSEA) to assess pathway enrichment27,28. We found that significantly downregulated pathways included MTOR, ERB2, and Hallmark Hypoxia, while upregulated terms included Metabolism of RNA and Hallmark MYC targets (Fig. 4A and Supplementary Fig. 5A). Analysis of Gene Ontology -Molecular Function gene sets revealed positive enrichment of several gene sets related to RNA homeostasis in the USO1-depleted cells (Supplementary Fig. 5B). Selected examples of up- and down regulated genes (log2FC > 1.5 and Padj. value < 0.01), from the enriched pathway identified by GSEA are highlighted in the Volcano plot for RNA-seq data (Fig. 4B). RT-qPCR was used to confirm these same significantly upregulated (TFRC, MRPS12, and PSMD1) (Fig. 4C) and downregulated genes (BAPIP2, ABCA1 and BTAF1) (Fig. 4D) in USO1-depleted SEM cells. This led us to hypothesize that there were alterations in expression and activation of MTOR in USO1-depleted cells using western blotting. We observed there was a mild downregulation of p-MTOR (Ser2481) in the USO1-depleted cells (Fig. 4E). Hence, it appears that USO1 regulates several pathways that are known to play a role in cell survival and cell death29.
Figure 4.
USO1 depletion significantly affects gene expression and pathways related to cell survival and proliferation. (A) Enrichment plots generated by the Metascape gene list enrichment analysis webtool on for the RNA-seq data from USO1-depleted versus NT control SEM cells. The top and bottom panels show pathways that are downregulated and upregulated in USO1-depleted SEM cells, respectively. (B) Volcano plot representing differentially expressed genes (Padj < 0.01 and Log2FC > 1.5) with several examples highlighted from the pathways enriched in (A). (C, D) RT-qPCR validation of differentially expressed genes identified in (B) (t test; *P < 0.05; **P < 0.01) (E) Western blot showing mildly reduced expression of p-MTOR (Ser2481) expression in USO1-depleted cells, compared to NT control, while MTOR remains unchanged. (F) Venn diagrams showing the number of shared genes between USO1-positively correlated genes in Target-Phase II ALL dataset with the genes that are significantly upregulated (top) or downregulated (bottom) in USO1-depleted SEM cells. A hypergeometric test was utilized to compare the overlaps between the datasets using a genome size of 24,278 genes. Total and shared number of genes are indicated.
To assess whether the gene expression changes caused by USO1-depletion had any clinical relevance, we turned to the Target Phase II Acute lymphoblastic leukemia dataset, accessed via the cBioPortal interface30,31. We calculated correlation coefficients between USO1 and 24,278 genes detected by RNA-seq in 203 samples from 154 patients. Using a cut-off q value of 0.001, we then overlapped genes up- or down-regulated in USO1-depleted with genes that had either a positive or negative correlation co-efficient. We found a highly significant overlap between genes that were positively correlated with USO1 across the B-ALL samples and downregulated in USO1-depleted cells (Hypergeometric test; p < 0.0001), but not in genes that were upregulated in USO1-depleted cells (non-significant p value) (Fig. 4F, Supplementary 5C & 5D). This suggests that USO1 regulates gene expression in patient samples of B-ALL. As concrete examples, differentially expressed genes confirmed by RT-qPCR (Fig. 4C, D) showed a direct correlation with USO1 expression in the ALL dataset from cBioPortal (Supplementary Fig. 6A–6D). These findings strongly suggest that USO1 plays a role in gene regulation in human B-ALL.
USO1 inhibits the MLL-AF4-driven leukemogenesis in primary murine bone marrow cells
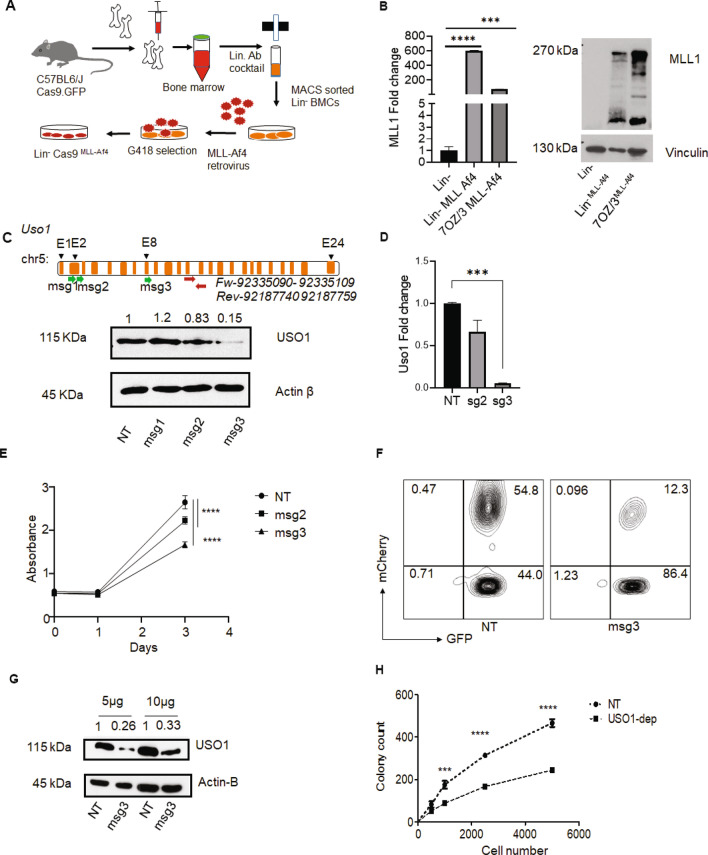
Having demonstrated that USO1 is required for the survival and growth of MLL-AF4+ cells in culture, we wanted to determine if USO1 was required in an experimentally induced primary cell model of MLL-AF4-driven leukemia. Briefly, Lin- cells from the bone marrow of Cas9-EGFP mice32 were transduced with MLL-Af4 retrovirus33 (Fig. 5A) and selected with G418. We confirmed expression of the MLL-Af4 transgene, finding overexpression by RT-qPCR and western blot analysis (Fig. 5B). As expected, we observed rapid proliferation and expansion of the Lin- Cas9MLL-Af4 cells. In order to deplete Uso1 from these cells, we designed and cloned three Uso1 murine specific sgRNAs (msg2 and msg3) into our internally designed MSCV.EFs.mCherry retroviral vector. To determine their effectiveness, we sorted GFP+ mCherry+ 70Z/3 cells transduced with the MSCV retroviruses, briefly expanded them in culture, and then queried USO1 expression. Western blot and RT-qPCR demonstrated that msg2 and msg3 both resulted in significant reduction of USO1 protein and mRNA expression (Fig. 5C, D). Uso1-depleted 7OZ/3 cells also had reduced cell growth by MTS assay which correlated with the different levels of USO1 depletion for msg2 and msg3 (Fig. 5E). From these data, the msg3 retrovirus was selected for transduction of Lin- Cas9MLL-Af4 cells. After sorting GFP+ mCherry+ transduced Lin- Cas9MLL-Af4 cells (Fig. 5F), USO1-depletion was confirmed by western blot, in which protein expression was reduced to 26% (Fig. 5G), and the cells were subsequently used in a colony formation assay. Functionally, USO1-depletion resulted in significantly fewer colonies in Lin-Cas9MLL-Af4 cells compared to NT control cells at 12 days. This change was maintained at several different starting numbers of Lin-Cas9MLL-Af4 cells (Fig. 5H). Hence, the function of USO1 is preserved in not only in human B-ALL cell lines but also in primary murine MLL-Af4 transformed cells.
Figure 5.
USO1 depletion in transformed bone marrow cells shows reduced proliferation and colony forming potential. (A) Schematic of an in vitro model system to transform Lin- bone marrow cells from Cas9-egfp mice using overexpression of MLL-Af4 transgene in. (B) Analysis of overexpression of MLL-Af4 by RT-qPCR and western blot in retrovirally transduced Lin-Cas9MLL-Af4 cells, Lin-Cas9 cells were used as negative control and 70Z/3 cells transduced with MLL-Af4 were used as positive control. RT qPCR was performed with an optimized set of primers, normalized to L32, and represented as fold-change from an internal control for each experiment. Western Blotting was performed with an antibody to MLL1. Vinculin was used as a high molecular weight loading control. (C) Upper panel, schematic of murine Uso1 depletion experiments, showing location of sgRNAs relative to the gene, and RT-qPCR primer location. Bottom panel, western blot analysis of 70Z/3 cells transduced with three different sgRNAs targeting Uso1 (msg1-3) cloned in the MSCV.sgRNA.mCherry.v1 vector. (D) RT-qPCR analysis of Uso1 depletion in 70Z/3 cells. (E) MTS assay (Absorbance at 490 nm) to measure proliferation in Uso1-depleted 70Z/3 cells compared to NT control cells. (F) FACS plot showing gating schema to sort the Lin-Cas9MLL-Af4 GFP+ mCherry+ population following transduction with the Uso1 msg3 vector. (G) Western blot was used to confirm the reduction in USO1 expression in sorted Lin-Cas9MLL-Af4 cells. (H). Colony formation assay using Lin-Cas9MLL-Af4 cells in methylcellulose assay as described in methods with titration of input cell number (t test, ***P < 0.001; ****P < 0.0001).
Discussion
The molecular mechanism of MLL-AF4 driven leukemogenesis remains incompletely understood, and this subtype of B-ALL is highly aggressive5,7. Although there are several fusion partners for MLL in acute leukemia, downstream transcriptional dysregulation is a common feature15,34,35. In this study, we sought to understand whether overexpression of putative RBPs, which we identified previously, contributes to the pathogenesis of MLL-AF4+ B-ALL15. We performed a focused sub-genomic CRISPR/Cas9 dropout screen to specifically address whether these putative RBPs had a functional role in leukemia cell growth. Indeed, we identified three genes (EIF3E, EPRS, and USO1 ) that appeared to be required in the MLL-AF4+ cell line, SEM. These genes showed slightly higher rates of dropout in SEM cells than in NALM6 cells. Of these, USO1 expression showed a dependence on MLL-AF4, whereas EIF3E and EPRS did not show the same dependence and were previously reported to be “common essential genes”, per the depmap portal22. Follow-up studies confirmed a role for USO1, both in cell lines and in a model of MLL-AF4 driven leukemia in primary murine bone marrow cells. RNA sequencing revealed that USO1 regulates numerous pathways, including mTOR, MYC targets, as well as elements of RNA homeostasis.
One of the challenges of genomic-scale CRISPR screens is that genes with a small average effect size on the phenotype of interest can be quite difficult to identify and can frequently be “drowned out” by genes with a larger effect size36. As we were particularly interested in the role that RBPs play in B-ALL, and their effect size in cell lines is unknown, we chose to perform a sub-genomic essentiality screen targeting a pre-defined set of RBPs known to be highly expressed in B-ALL. Therefore, under the hypothesis that a significant proportion of the sgRNAs would have a negative effect on cell viability, our design included a substantial number of both positive and negative control sgRNAs in order to properly model the null effect distribution and compare the effect size of functional RBPs to that of known essential genes. Secondly, the use of a non-MLL-AF4 control cell line allowed us to identify those proteins whose expression might be most important in MLL-AF4 leukemia. Supporting the idea that such specific effects may not be seen in genome scale studies, USO1 dropout was detected in one prior genome-scale analysis, whereas it was not observed in two other genome scale studies that queried vulnerabilities in MLL-translocated leukemia17,37,38. Thus, our approach to perform this type of CRISPR/Cas9 screen may help inform the design of future forward genetic screens.
Our group is interested in understanding RNA binding proteins in B-ALL, and we recently described the functional role of IGF2BP3 in pathologic expansions of cells within the hematopoietic system and its requirement for survival and growth in B-ALL cell lines15. Here, we focused on USO1, based on its identification in this screen as well as a prior study in MV-4–11 AML cells that had identified USO1 as a factor required for AML survival17. A recent study reported a KMT2A(MLL)-USO1 fusion gene in a secondary AML, hinting at a further connection between MLL-driven leukemia and USO139. More generally, USO1 has been shown to be of functional importance in cancer23–25. USO1 was recently reported to have RNA-binding function in studies utilizing high-throughput biochemical techniques16,40,41, despite a canonical role in the regulation of vesicular transport42,43. Adding to these prior descriptions of USO1 function, we validated USO1 as a MLL-AF4-induced gene, and found that it was functionally required in cell lines and in primary bone marrow for MLL-AF4 dependent phenotypes. This work firmly establishes the significance of this protein in acute leukemia, which was not previously appreciated.
Recent studies in multiple myeloma have shown that USO1-deficient cells showed have reduced cell proliferation and increased apoptosis via regulation of Erk pathway activity24. Transcriptome analysis of USO1-depleted SEM cells in our study demonstrated a mixed picture, with the downregulation of certain cancer and cell growth-related pathways, including mTOR and ERB2, but concurrent downregulation of RNA metabolism and MYC targets. Curiously, there was also upregulation of the mTORC1 hallmark pathway in USO1-depleted cells (as opposed to downregulation of mTOR generally), perhaps indicating a specific effect on mTORC2. Nonetheless, we observed decreased phospho-mTOR, which is consistent with the effect seen on cell growth and cell cycle. Additionally, our data suggests that USO1 is associated with other molecular functions of gene regulation, such as RNA homeostasis. Interestingly, the pathways noted to be deregulated show similarity to those deregulated upon inhibition of the RNA demethylase FTO44. It is important to note, however, that we have not characterized its function as a RBP. It is tempting to speculate that USO1 is a bifunctional protein with roles in vesicular transport and RNA binding, perhaps in a manner similar to YBX145. YBX1 appears to bind to and sort microRNAs, specifically, miR-223, into exosomes. By regulating this process, YBX1 can impact cellular homeostasis. Hence, further work to assess the molecular role of USO1 as a putative RBP in MLL-AF4 translocated leukemia is warranted.
Overall, our study successfully queried the functional relevance of a set of genes identified from primary patient samples using expression profiling. Here, we provide a rubric for how to functionally analyze a prioritized list of genes in leukemogenesis, or in other pathogenetic processes. In addition, we establish a role for the putative RBP, USO1, in leukemogenesis. Given the broader range of cancer types that show USO1 dysregulation, our work may have implications beyond those in B-ALL. Furthermore, understanding how non-canonical RBP may participate in leukemogenesis may open up new avenues in developing novel strategies for the diagnosis, prognosis, and treatment of B-ALL.
Methods
Cell lines and cell culture
All the cell lines involved in the study were maintained at 37 °C in a humidified incubator at 5% CO2. RS4;11 (ATCC CRL-1873), NALM6 (ATCC CRL-3273) were cultured in RPMI 1640 supplemented with 10% FBS. 70Z/3 (ATCC TIB 158) cells were cultured in RPMI 1640 supplemented with 10% FBS and 0.05 mM 2-mercaptoethanol. SEM cells (DMZ-ACC 546), MV-4–11 (ATCC CRL-9591) were cultured in Iscove’s Modified Dulbecco’s Medium (IMDM) supplemented with 10% FBS. Mouse bone marrow derived Lineage (Lin-) cells were cultured in IMDM media supplemented with 15% FBS, 20 ng/mL mTPO, 20 ng/FLT3 ligand and 50 ng/mL mSCF.
Sub-genomic CRISPR screen
A sub-genomic CRISPR/Cas9 screen was designed to target 36 RBP genes and 12 “positive control genes”, and included 28 negative control (or non-targeting) single guide RNAs (sgRNAs). The positive control genes, representing known vulnerabilities in MLL-translocated acute leukemia, were selected from top 100 genes dysregulated in Genome wide CRISPR screen in MV-4–11 cells17. These genes are expected to “drop out” in a CRISPR screen of MLL-translocated leukemia, but it is not known whether they are specific for MLL-translocated leukemia. This design was adapted to provide enough non-targeting controls in the context of a sub-genomic screen, where a significant proportion of targeting sgRNAs may be expected to change. Five sgRNAs were designed for each RBP or positive control genes, using sgRNA design tools from Broad Institute46. pLKO5.sgRNA.EFS.tRFP is a lentiviral vector, which contains EF-1 alpha binding sequence (EFS) upstream of tRFP, was obtained from Addgene (#57,823)47. The 268 pooled sgRNA were then cloned into pLKO5.sgRNA.EFS.tRFP lentiviral vector using standard protocols17. Prior to CRISPR/Cas9 screening, B-ALL cell lines with MLL-AF4 translocation (SEM)33 and without MLL-AF4 translocation (NALM6)48 were stably transduced with pLentiCas9-GFP49 lentivirus and sorted on GFP positivity, with subsequent confirmation of Cas9 expression (Supplementary Fig. 1A). pLKO5.sgRNA.EFS.tRFP lentiviral pool titers were calculated from SEM and NALM6 cell transduction. For experiments, bulk GFP+ SEM and NALM6 cells were infected at < 0.3 MOI and 2 × 106 cells GFP+ tRFP+ were sorted by FACS after 48 h of infection (Supplementary Fig. 1B). Genomic DNA (gDNA) isolated from 106 cells was used for construction of the Reference (REF) library sample, and the other 106 cells were cultured and expanded. Cells were split every five days and 106 cells were reseeded for culture50 to maintain a sgRNA representation of 3700X. Following 28 days of culture, cells were harvested and gDNA was isolated for the Depletion (DEP) library sample preparation.
Library preparation, DNA sequencing, and analysis
Sequencing libraries were prepared from both the Reference (REF) and Depleted (DEP) genomic DNA (gDNA) samples obtained at days 0 and 28 of CRISPR screen experiment, respectively17,50. Libraries were prepared from 200 ng of input DNA, by using Q5 high-fidelity DNA polymerase (#M0492S, NEB) and Illumina adapted primers to amplify the sgRNA target region from the gDNA, as previously described17 (Supplementary Fig. 1C). The purified PCR product was quantified using Qubit and quality control was done using Bioanalyzer and sequenced on HiSeq 3000 at the Technology Center for Genomic and Bioinformatics at UCLA. Adapter sequences were removed using in-house scripts. Candidate reads (those containing a valid primer sequence and with a minimum length of 20 bp after trimming) were aligned to the sgRNA library using bowtie v0.12.851 with a maximum tolerance of one mismatch. Counts tables for both individual sgRNAs and gene-level summaries were compiled from non-ambiguous hits for both the Reference and Depletion libraries in each experiment and for each cell line. Count tables were processed with DESeq248 to obtain variance-stabilized normalized abundance and rank sgRNAs and genes based on differential abundance (moderated fold change and adjusted Wald test p value).
Cell line treatment with transcription inhibitors
B-ALL cell lines were plated at 0.5 × 106 cells/mL density 24 h before treatment, and harvested 48 h after initiating treatment with the chemical inhibitor. I-BET151, a BRD4 inhibitor was reconstituted in DMSO (10 mM), was diluted in complete media and added to the cells at a concentration of 0.5 µM, 1.0 µM and 2.0 µM. MI-503, a menin-MLL inhibitor, was used to treat the cells at 0.12 µM, 0.25 µM and 0.5 µM. EPZ5676, a DOT1L inhibitor, was used to treat cells at a concentration of 0.5 µM of EPZ5676.
Cell proliferation, cell cycle and apoptosis assays
Cell proliferation assay was performed using standard MTS assay protocol. 5000 cells were plated in 100 µL volume of media in a single well of 96-well tissue culture plates. Cells were harvested at different time points of day 0, day1, day 3 and day 5. MTS reagent mix was prepared by adding 100 µL of PMS solution (0.21 mg/mL) to 1 ml of MTS reagent (0.33 mg/mL) and 20 µL reagent mix was added to each well. The plate was incubated at 37 °C for 2 h and absorbance was taken at 490 nm in a microplate reader.
Cell cycle analysis was performed using propidium iodide (PI). Cells were harvested and washed in PBS and fixed in 70% ethanol overnight at − 20 °C. Fixed cells were washed with PBS and centrifuged at 2500 rpm. PI solution (2 mg/10 mL) was diluted in PBS and added with 0.2 mg/mL of DNase free RNase A. Nearly, 300 µL of the PI solution was added to each tube and incubated at RT for 2 h. The stained samples were analyzed by flow cytometer.
Annexin V staining was performed to study the apoptosis in the cells using standard protocol. Briefly, cells treated with inhibitors/siRNAs were harvested and washed in PBS before resuspending in binding buffer (106/mL). 100 µL of cell suspension was stained with 0.5 µL of Annexin V antibody conjugated to Pacific blue and incubated at RT for 30 min. After incubation, 300 µL of binding buffer was added to the sample and analyzed by flow cytometer.
siRNA knockdown of cell lines
siRNA transfection was performed using standard Nucleofection program provided by the manufacturer. SEM cells were Nucleofected using the 4D Nucleofector System (Lonza, Cologne, Germany). Cells were washed with phosphate-buffered saline and then resuspended in nucleofection solution (SF Cell Line 4D-Nucleofector X Kit, Lonza, Cologne, Germany), at a final concentration of 2 × 106 cells/100 µL reaction. Cells were nucleofected with 30 pmol of control siRNA, USO1 siRNA1, or USO1 siRNA 4, in 100 µL cuvettes using program CV-104. Immediately after nucleofection, 500 µL of pre-warmed, antibiotic-free media was added to the cuvette and incubated for 10 min at RT. After incubation cells were transferred to a 12 well plate containing 1.5 mL of media. Nucleofected cells were maintained at 37 °C and 5% CO2 prior to harvesting for analysis.
RT-qPCR assays
Previous protocols were adapted for RT-qPCR, based on our prior work15. A full list of RT-qPCR primers is presented in Table S2. For normalization, we utilized RT-qPCR primers for 18S (human) and L32 (mouse).
Western Blotting
Western Blotting was performed as previously described15. The blots were developed and imaged on ECL film or on a Bio-Rad Chemidoc digital imager using Super signal West Pico PLUS chemiluminescent reagent. EPRS (#A303-957A), EIF3E (#A302-984A), USO1 (#A304-513A) antibodies were purchased from Bethyl laboratories. USO1 (13,509–1-AP) antibody to detect mouse USO1 was purchased from Proteintech. MTOR (#2972 s) and Phospho-MTOR (Ser2481) (#2971) antibodies were procured from Cell Signaling Technology. Vinculin (Santa Cruz Biotechnology, # sc-73614) and Anti-β-Actin (Sigma Aldrich, #A1978) were used for loading controls.
Chromatin immunoprecipitation (ChIP)
SEM cells were cultured with and without I-BET151 (Sigma Aldrich,# SML0666) and DMSO for 48 h at 37 °C52. ChIP was performed using EZ-Magna ChIP kit (Millipore, # 17–408) with MLL1 antibody (Bethyl laboratories, #A300-374A) and AF4 antibody (Abcam, #ab31812). The purified DNA was used as input for qPCR and binding was quantitated as previously described52.
Immortalization of Lin- bone marrow cells
All mice used in this study were obtained from Jackson Labs and were genotyped according to JAX protocols and maintained in the UCLA Division of Laboratory Animal Medicine. Bone marrow cells from C57BL6/J Cas9-EGFP mice53 (Jackson Laboratories, # 026,179) were isolated by flushing the bones from the mice and creating a single cell suspension. Cells were incubated with a lineage antibody cocktail and depleted for Lineage+ cells using MACS technology (Miltenyi Biotech). Lin- cells were spin-infected and transduced with MLL-Af4 retroviral preparation33. The MSCV-MLL-flag-Af4 plasmid was the kind gift via MTA by Dr. Michael Thirman (University of Chicago, Department of Medicine). After four rounds of transduction, cells were selected in 400 µg/mL G418 supplemented media for 7 days.
MSCV sgRNA vectors
We generated a novel MSCV vector that can overexpress an individual sgRNA in addition to an mCherry reporter. In brief, the MSCV.mU6.sgRNA-EFs.mCherry.v1 retroviral vector was constructed by replacing a 2.1 kb EGFP-PGK.Puro fragment from the pMGP vector with a 2.7 kb sequence containing mU6.BbsI-stuffer-BbsI-scaffold-spacer-EFs.mCherry via BglII/ClaI digest. The sgRNA scaffold and EF-1α short (EFs) promoter elements were derived from the pLentiCRISPRv2 vector. The mU6 promoter was designed from the GenBank sequence NC_000076.6 (nt 79,908,880–79,909,195). A silent mutation was incorporated into the mCherry reporter element to remove an internal BbsI restriction site. The 1.2 kb stuffer sequence was derived from portions of the 1.8 kb firefly luciferase gene. The sgRNA sequences targeting mouse were designed as above and directionally cloned between the mU6 promoter and sgRNA scaffold sequence via BbsI. Detailed methods and vector maps are available upon request.
Colony forming unit assay
A colony forming unit assay was performed to study the effect of USO1-depletion on the colony forming potential of Lin-Cas9MLL-Af4 cells. The assay was performed using the Methocult colony forming media (STEMCELL Technologies, #M3434)54. Briefly, approximately 5,000 Lin-Cas9MLL-Af4 USO1 depleted cells were mixed in 3.2 ml of overnight thawed Methocult media and plated in two 35 mm dishes along with the NT controls and cultured for 12 days. After 12 days of culture, individual 35 mm dishes were counted for both total number and morphologic subtypes of colonies formed by USO1 depletion and NT control cells.
RNA-Seq library preparation and analysis
Libraries for RNA-Seq were prepared with Nugen Universal plus mRNA-Seq Kit to generate strand-specific RNA-seq libraries. Sequencing was performed on Illumina HiSeq 3000 SR 1 × 50 bp run. Data quality check was done on Illumina SAV. Demultiplexing was performed with Illumina Bcl2fastq2 v 2.19.1.403 program. The STAR ultrafast universal RNA-seq aligner v2.7.0d55 was used to align the reads to a genome index that included both the genome sequence (GRC38 human primary assembly) and the exon/intron structure of known human gene models (Gencode v29 genome annotation). Alignment files were used to generate strand-specific, gene-level count summaries with STAR's built-in gene counter. Independent filtering was applied as before56,57: genes with less than 6 total counts across all samples, count outliers, or low mappability (< 50 bp) were filtered out for downstream analyses. Expression estimates were computed in units of fragments per kilobase of mappable length and million counts (FPKMs). Differential expression analyses between USO1 depletion and non-targeted controls was performed with DESeq227 and genes were ranked based on moderated fold change and adjusted Wald test p value. Functional enrichment for selected genes was performed with Metascape36.
Approvals and Compliance
This study was carried out in compliance with the ARRIVE guidelines. All animal experiments were carried out in accordance with relevant guidelines governing the use of animals in research. In addition, all experiments involving animals were approved by the University of California, Los Angeles, Chancellor’s Animal Research Committee (ARC), which was established for compliance with Public Health Service (PHS) guidelines on animal research.
Supplementary information
Acknowledgements
We thank Drs. Jayanth Palanichamy and Jennifer King for helpful discussions and technical assistance with experiments performed herein. This work was supported by the NIH/NCI R01CA166450 (D.S.R), NIH/NCI R03CA251845 (D.S.R), NIH/NIAID R21AI132869 (D.S.R), Tumor Cell Biology Training Grant T32 CA009056 (T.M.T.), Tumor Immunology Training Grant T32CA009120 (T.L.L.), , and a grant from the Margaret E. Early Trust (D.S.R.). Flow cytometry was performed in the Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research UCLA Flow Cytometry Core Resource and the UCLA JCCC/CFAR Flow Cytometry Core Facility that is supported by NIH AI-28697, P30CA016042, the JCCC, the UCLA AIDS Institute, and the David Geffen School of Medicine at UCLA. The results published here are in part based upon data generated by the Therapeutically Applicable Research to Generate Effective Treatments (https://ocg.cancer.gov/programs/target) initiative, phs000218. The data used for this analysis are available at https://portal.gdc.cancer.gov/projects.
Author contributions
A.K.J.: Experimental Design, Experimentation, Data Analysis and Interpretation, Manuscript Preparation. H.T., T.L.L: Experimentation, Data Analysis and Interpretation. T.M.T.: Experimentation, Data Analysis and Interpretation, Manuscript Preparation. D.C.: Data Analysis and Interpretation. M.O.A.: Experimentation. D.S.R.: Experimental Design, Data Analysis and Interpretation, Manuscript Preparation, Funding.
Data availability
All sequencing data have been deposited in the Sequence Read Archive (PRJNA658354). All research materials will be made available in accordance with UCLA policy.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-92448-w.
References
- 1.Mullighan CG. Molecular genetics of B-precursor acute lymphoblastic leukemia. J. Clin. Invest. 2012;122(10):3407–3415. doi: 10.1172/JCI61203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takeuchi S, Seriu T, Bartram CR, Golub TR, Reiter A, Miyoshi I, et al. TEL is one of the targets for deletion on 12p in many cases of childhood B-lineage acute lymphoblastic leukemia. Leukemia. 1997;11(8):1220–1223. doi: 10.1038/sj.leu.2400743. [DOI] [PubMed] [Google Scholar]
- 3.Luong-Gardiol N, Siddiqui I, Pizzitola I, Jeevan-Raj B, Charmoy M, Huang Y, et al. Gamma-catenin-dependent signals maintain BCR-ABL1(+) B cell acute lymphoblastic leukemia. Cancer Cell. 2019;35(4):649–663. doi: 10.1016/j.ccell.2019.03.005. [DOI] [PubMed] [Google Scholar]
- 4.Stong RC, Korsmeyer SJ, Parkin JL, Arthur DC, Kersey JH. Human acute leukemia cell line with the t(4;11) chromosomal rearrangement exhibits B lineage and monocytic characteristics. Blood. 1985;65(1):21–31. doi: 10.1182/blood.V65.1.21.21. [DOI] [PubMed] [Google Scholar]
- 5.Dreyer ZE, Dinndorf PA, Camitta B, Sather H, La MK, Devidas M, et al. Analysis of the role of hematopoietic stem-cell transplantation in infants with acute lymphoblastic leukemia in first remission and MLL gene rearrangements: A report from the Children's Oncology Group. J. Clin. Oncol. 2011;29(2):214–222. doi: 10.1200/JCO.2009.26.8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Behm FG, Raimondi SC, Frestedt JL, Liu Q, Crist WM, Downing JR, et al. Rearrangement of the MLL gene confers a poor prognosis in childhood acute lymphoblastic leukemia, regardless of presenting age. Blood. 1996;87(7):2870–2877. doi: 10.1182/blood.V87.7.2870.bloodjournal8772870. [DOI] [PubMed] [Google Scholar]
- 7.Hilden JM, Dinndorf PA, Meerbaum SO, Sather H, Villaluna D, Heerema NA, et al. Analysis of prognostic factors of acute lymphoblastic leukemia in infants: Report on CCG 1953 from the Children's Oncology Group. Blood. 2006;108(2):441–451. doi: 10.1182/blood-2005-07-3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moorman AV, Ensor HM, Richards SM, Chilton L, Schwab C, Kinsey SE, et al. Prognostic effect of chromosomal abnormalities in childhood B-cell precursor acute lymphoblastic leukaemia: Results from the UK Medical Research Council ALL97/99 randomised trial. Lancet Oncol. 2010;11(5):429–438. doi: 10.1016/S1470-2045(10)70066-8. [DOI] [PubMed] [Google Scholar]
- 9.Witkowski MT, Hu Y, Roberts KG, Boer JM, McKenzie MD, Liu GJ, et al. Conserved IKAROS-regulated genes associated with B-progenitor acute lymphoblastic leukemia outcome. J. Exp. Med. 2017;214(3):773–791. doi: 10.1084/jem.20160048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miglino M, Berisso G, Grasso R, Canepa L, Clavio M, Pierri I, et al. Allogeneic bone marrow transplantation (BMT) for adults with acute lymphoblastic leukemia (ALL): Predictive role of minimal residual disease monitoring on relapse. Bone Marrow Transplant. 2002;30(9):579–585. doi: 10.1038/sj.bmt.1703659. [DOI] [PubMed] [Google Scholar]
- 11.Prasad V. Immunotherapy: Tisagenlecleucel—The first approved CAR-T-cell therapy: implications for payers and policy makers. Nat. Rev. Clin. Oncol. 2018;15(1):11–12. doi: 10.1038/nrclinonc.2017.156. [DOI] [PubMed] [Google Scholar]
- 12.Gokbuget N, Dombret H, Bonifacio M, Reichle A, Graux C, Faul C, et al. Blinatumomab for minimal residual disease in adults with B-cell precursor acute lymphoblastic leukemia. Blood. 2018;131(14):1522–1531. doi: 10.1182/blood-2017-08-798322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turtle CJ, Hanafi LA, Berger C, Gooley TA, Cherian S, Hudecek M, et al. CD19 CAR-T cells of defined CD4+:CD8+ composition in adult B cell ALL patients. J. Clin. Invest. 2016;126(6):2123–2138. doi: 10.1172/JCI85309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Majzner RG, Mackall CL. Tumor antigen escape from CAR T-cell therapy. Cancer Discov. 2018;8(10):1219–1226. doi: 10.1158/2159-8290.CD-18-0442. [DOI] [PubMed] [Google Scholar]
- 15.Palanichamy JK, Tran TM, Howard JM, Contreras JR, Fernando TR, Sterne-Weiler T, et al. RNA-binding protein IGF2BP3 targeting of oncogenic transcripts promotes hematopoietic progenitor proliferation. J. Clin. Invest. 2016;126(4):1495–1511. doi: 10.1172/JCI80046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Castello A, Fischer B, Frese CK, Horos R, Alleaume AM, Foehr S, et al. Comprehensive identification of RNA-binding domains in human cells. Mol. Cell. 2016;63(4):696–710. doi: 10.1016/j.molcel.2016.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wallace J, Hu R, Mosbruger TL, Dahlem TJ, Stephens WZ, Rao DS, et al. Genome-wide CRISPR-Cas9 screen identifies microRNAs that regulate myeloid leukemia cell growth. PLoS ONE. 2016;11(4):e0153689. doi: 10.1371/journal.pone.0153689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478(7370):529–533. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Borkin D, He S, Miao H, Kempinska K, Pollock J, Chase J, et al. Pharmacologic inhibition of the Menin-MLL interaction blocks progression of MLL leukemia in vivo. Cancer Cell. 2015;27(4):589–602. doi: 10.1016/j.ccell.2015.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Daigle SR, Olhava EJ, Therkelsen CA, Basavapathruni A, Jin L, Boriack-Sjodin PA, et al. Potent inhibition of DOT1L as treatment of MLL-fusion leukemia. Blood. 2013;122(6):1017–1025. doi: 10.1182/blood-2013-04-497644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin S, Luo RT, Ptasinska A, Kerry J, Assi SA, Wunderlich M, et al. Instructive role of MLL-fusion proteins revealed by a model of t(4;11) Pro-B acute lymphoblastic leukemia. Cancer Cell. 2016;30(5):737–749. doi: 10.1016/j.ccell.2016.10.008. [DOI] [PubMed] [Google Scholar]
- 22.Tsherniak A, Vazquez F, Montgomery PG, Weir BA, Kryukov G, Cowley GS, et al. Defining a cancer dependency map. Cell. 2017;170(3):564–76.e16. doi: 10.1016/j.cell.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sui J, Li X, Xing J, Cao F, Wang H, Gong H, et al. Lentivirus-mediated silencing of USO1 inhibits cell proliferation and migration of human colon cancer cells. Med. Oncol. 2015;32(8):218. doi: 10.1007/s12032-015-0658-z. [DOI] [PubMed] [Google Scholar]
- 24.Jin Y, Dai Z. USO1 promotes tumor progression via activating Erk pathway in multiple myeloma cells. Biomed. Pharmacother. 2016;78:264–271. doi: 10.1016/j.biopha.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 25.Howley BV, Link LA, Grelet S, El-Sabban M, Howe PH. A CREB3-regulated ER-Golgi trafficking signature promotes metastatic progression in breast cancer. Oncogene. 2018;37(10):1308–1325. doi: 10.1038/s41388-017-0023-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sentmanat MF, Peters ST, Florian CP, Connelly JP, Pruett-Miller SM. A survey of validation strategies for CRISPR-Cas9 editing. Sci. Rep. 2018;8(1):888. doi: 10.1038/s41598-018-19441-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019;10(1):1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Riggelen J, Yetil A, Felsher DW. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat. Rev. Cancer. 2010;10(4):301–309. doi: 10.1038/nrc2819. [DOI] [PubMed] [Google Scholar]
- 30.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6(269):11. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159(2):440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin S, Luo RT, Shrestha M, Thirman MJ, Mulloy JC. The full transforming capacity of MLL-Af4 is interlinked with lymphoid lineage commitment. Blood. 2017;130(7):903–907. doi: 10.1182/blood-2017-04-777185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xia ZB, Popovic R, Chen J, Theisler C, Stuart T, Santillan DA, et al. The MLL fusion gene, MLL-AF4, regulates cyclin-dependent kinase inhibitor CDKN1B (p27kip1) expression. Proc. Natl. Acad. Sci. USA. 2005;102(39):14028–14033. doi: 10.1073/pnas.0506464102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andersson AK, Ma J, Wang J, Chen X, Gedman AL, Dang J, et al. The landscape of somatic mutations in infant MLL-rearranged acute lymphoblastic leukemias. Nat. Genet. 2015;47(4):330–337. doi: 10.1038/ng.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bodapati S, Daley TP, Lin X, Zou J, Qi LS. A benchmark of algorithms for the analysis of pooled CRISPR screens. Genome Biol. 2020;21(1):62. doi: 10.1186/s13059-020-01972-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tzelepis K, Koike-Yusa H, De Braekeleer E, Li Y, Metzakopian E, Dovey Oliver M, et al. A CRISPR dropout screen identifies genetic vulnerabilities and therapeutic targets in acute myeloid leukemia. Cell Rep. 2016;17(4):1193–1205. doi: 10.1016/j.celrep.2016.09.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Erb MA, Scott TG, Li BE, Xie H, Paulk J, Seo HS, et al. Transcription control by the ENL YEATS domain in acute leukaemia. Nature. 2017;543(7644):270–274. doi: 10.1038/nature21688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jin W, Chen L, Liu Y, Chen Q, Zhao M, Tan Y, et al. A novel KMT2A–USO1 fusion gene-induced de novo secondary acute myeloid leukaemia in a patient initially diagnosed with acute promyelocytic leukaemia. Br. J. Haematol. 2021;192:e32–e36. doi: 10.1111/bjh.17183. [DOI] [PubMed] [Google Scholar]
- 40.Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149(6):1393–1406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 41.Beckmann BM, Horos R, Fischer B, Castello A, Eichelbaum K, Alleaume AM, et al. The RNA-binding proteomes from yeast to man harbour conserved enigmRBPs. Nat. Commun. 2015;6:10127. doi: 10.1038/ncomms10127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakamura N, Lowe M, Levine TP, Rabouille C, Warren G. The vesicle docking protein p115 binds GM130, a cis-Golgi matrix protein, in a mitotically regulated manner. Cell. 1997;89(3):445–455. doi: 10.1016/S0092-8674(00)80225-1. [DOI] [PubMed] [Google Scholar]
- 43.Allan BB, Moyer BD, Balch WE. Rab1 recruitment of p115 into a cis-SNARE complex: Programming budding COPII vesicles for fusion. Science. 2000;289(5478):444–448. doi: 10.1126/science.289.5478.444. [DOI] [PubMed] [Google Scholar]
- 44.Huang Y, Su R, Sheng Y, Dong L, Dong Z, Xu H, et al. Small-molecule targeting of oncogenic FTO demethylase in acute myeloid leukemia. Cancer Cell. 2019;35(4):677–691. doi: 10.1016/j.ccell.2019.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shurtleff MJ, Temoche-Diaz MM, Karfilis KV, Ri S, and Schekman R. Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. Elife.5, e19276 (2016). [DOI] [PMC free article] [PubMed]
- 46.Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 2016;34(2):184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heckl D, Kowalczyk MS, Yudovich D, Belizaire R, Puram RV, McConkey ME, et al. Generation of mouse models of myeloid malignancy with combinatorial genetic lesions using CRISPR-Cas9 genome editing. Nat. Biotechnol. 2014;32(9):941–946. doi: 10.1038/nbt.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Benito JM, Godfrey L, Kojima K, Hogdal L, Wunderlich M, Geng H, et al. MLL-Rearranged acute lymphoblastic leukemias activate BCL-2 through H3K79 methylation and are sensitive to the BCL-2-specific antagonist ABT-199. Cell Rep. 2015;13(12):2715–2727. doi: 10.1016/j.celrep.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang S, Chim B, Su Y, Khil P, Wong M, Wang X, et al. Enhancement of LIN28B-induced hematopoietic reprogramming by IGF2BP3. Genes Dev. 2019;33(15–16):1048–1068. doi: 10.1101/gad.325100.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Doench JG. Am I ready for CRISPR? A user's guide to genetic screens. Nat. Rev. Genet. 2018;19(2):67–80. doi: 10.1038/nrg.2017.97. [DOI] [PubMed] [Google Scholar]
- 51.Langmead B. Aligning short sequencing reads with Bowtie. Curr. Protoc. Bioinform. 2010; Chapter 11:Unit 11.7. [DOI] [PMC free article] [PubMed]
- 52.Ptasinska A, Assi SA, Martinez-Soria N, Imperato MR, Piper J, Cauchy P, et al. Identification of a dynamic core transcriptional network in t(8;21) AML that regulates differentiation block and self-renewal. Cell Rep. 2014;8(6):1974–1988. doi: 10.1016/j.celrep.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.LaFleur MW, Nguyen TH, Coxe MA, Yates KB, Trombley JD, Weiss SA, et al. A CRISPR-Cas9 delivery system for in vivo screening of genes in the immune system. Nat. Commun. 2019;10(1):1668. doi: 10.1038/s41467-019-09656-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McKenzie MD, Ghisi M, Oxley EP, Ngo S, Cimmino L, Esnault C, et al. Interconversion between tumorigenic and differentiated states in acute myeloid leukemia. Cell Stem Cell. 2019;25(2):258–272. doi: 10.1016/j.stem.2019.07.001. [DOI] [PubMed] [Google Scholar]
- 55.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Casero D, Sandoval S, Seet CS, Scholes J, Zhu Y, Ha VL, et al. Long non-coding RNA profiling of human lymphoid progenitor cells reveals transcriptional divergence of B cell and T cell lineages. Nat. Immunol. 2015;16(12):1282–1291. doi: 10.1038/ni.3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data have been deposited in the Sequence Read Archive (PRJNA658354). All research materials will be made available in accordance with UCLA policy.