Abstract
Loss of cell differentiation is a hallmark for the progression of oral squamous cell carcinoma (OSCC). Archival Formalin-Fixed Paraffin-Embedded (FFPE) tissues constitute a valuable resource for studying the differentiation of OSCC and can offer valuable insights into the process of tumor progression. In the current study, we performed LC–MS/MS-based quantitative proteomics of FFPE specimens from pathologically-confirmed well-differentiated, moderately-differentiated, and poorly-differentiated OSCC cases. The data were analyzed in four technical replicates, resulting in the identification of 2376 proteins. Of these, 141 and 109 were differentially expressed in moderately-differentiated and poorly differentiated OSCC cases, respectively, compared to well-differentiated OSCC. The data revealed significant metabolic reprogramming with respect to lipid metabolism and glycolysis with proteins belonging to both these processes downregulated in moderately-differentiated OSCC when compared to well-differentiated OSCC. Signaling pathway analysis indicated the alteration of extracellular matrix organization, muscle contraction, and glucose metabolism pathways across tumor grades. The extracellular matrix organization pathway was upregulated in moderately-differentiated OSCC and downregulated in poorly differentiated OSCC, compared to well-differentiated OSCC. PADI4, an epigenetic enzyme transcriptional regulator, and its transcriptional target HIST1H1B were both found to be upregulated in moderately differentiated and poorly differentiated OSCC, indicating epigenetic events underlying tumor differentiation. In conclusion, the findings support the advantage of using high-resolution mass spectrometry-based FFPE archival blocks for clinical and translational research. The candidate signaling pathways identified in the study could be used to develop potential therapeutic targets for OSCC.
Supplementary Information
The online version of this article contains supplementary material available at 10.1007/s12079-021-00609-3
Keywords: Cancer pathology, Pressure cycling technology, Molecular medicine, Cancer grade, Quantitative proteomics, Tumor differentiation
Introduction
Cancers of the lip and oral cavity arise primarily from epithelial cells, and 90% of these are composed of oral squamous cell carcinoma (OSCC) by origin (Miranda-Filho and Bray 2020). OSCC arises in various anatomical locations within the oral cavity, including the tongue, buccal mucosa, gingiva, palate, lip, and floor of the mouth. Carcinoma of the oral cavity results from a multi-step process triggered by various stimuli such as tobacco, alcohol consumption, and infections with high-risk types of human papillomavirus (HPV) (Argiris et al. 2008). Despite the continuous advancement in the multimodality treatment, satisfactory survival rates have not been achieved, owing to diagnosis at advanced stages of the disease, resulting in poor outcomes (Forastiere et al. 2001).
Molecular and histopathological features play an essential role in prognostic evaluation of oral squamous cell carcinoma (OSCC), which is based on the TNM staging system (primary tumor, regional lymph node metastasis, and distant metastasis) developed by the American Joint Committee on Cancer (AJCC). OSCC is categorized into three histopathological types based on the degree of differentiation, such as well-differentiated OSCC (Well-OSCC), moderately-differentiated OSCC (Moderate-OSCC), and poorly-differentiated OSCC (Poor-OSCC). Such histological grading using Broder’s classification is subjective, and attempts have been made to add more rigor to the grading by taking into account other parameters such as mitotic index, DNA content, Ki-67 expression among others (Akhter et al. 2011). Well-OSCC, also referred to as low-grade OSCC, resembles normal squamous mucosa with a good prognosis. Moderate-OSCC displays intermediated forms of tumors and has less keratinization, whereas Poor-OSCC is more aggressive, resulting in a worse prognosis (Barnes et al. 2005). Cancer metastasis is a multi-step process that involves the invasion of neighboring tissues by malignant cells resulting in loss of cell-to-cell adhesion and apical-basal polarity. It is assumed that such morphological changes serve as an indicator of epithelial-mesenchymal transition (EMT) where Well-OSCC, Moderate-OSCC, and Poor-OSCC derive its architecture owing to such EMT transition (Guarino et al. 2007). Accurate identification and categorization could play a vital role in significantly improving the prognosis of OSCC.
In the post-genomic era, proteomics has become an effective approach to identify distinct protein expression signatures associated with the cancer phenotype and enhanced our understanding of tumor-specific changes. Frozen tumor tissue is a preferred biological material, as it is a reservoir of signaling components from diverse cell compartments (Poschmann et al. 2009). Besides, most tumor tissues are stored as formalin-fixed paraffin-embedded (FFPE) tissues as a part of routine pathological investigations. These are readily available as a resource for research and can be used to identify candidate biomarkers for diagnosis or treatment outcome. Because of formalin fixation-induced intermolecular and intramolecular cross-linking of proteins, FFPE samples are not generally preferred for proteomics investigations (Ahram et al. 2003). However, several efforts have been made to develop a clinically applicable high-throughput mass spectrometry approach, to mine the FFPE tissue proteome to identify candidate biomarkers (Hwang et al. 2007; Mertins et al. 2016; Negishi et al. 2009; Patel et al. 2008; Pozniak et al. 2016; Tyanova et al. 2016; Xiao et al. 2010; Zhang et al. 2014, 2016). In recent years, workflows for extracting proteins from FFPE specimens for mass spectrometry (MS)-based proteomic analysis have been developed (Hwang et al. 2007; Patel et al. 2008) and these, in turn, have enabled numerous other studies. In 2014, quantitative proteomics analysis of FFPE tissue samples from OSCC patients was carried out, which resulted in the identification of galectin-7 as a potential predictive marker of chemo- and/or radiotherapy resistance in OSCC (Matsukawa et al. 2014). Similarly, two-dimensional liquid chromatography-tandem mass spectrometry coupled with an isobaric tag for relative and absolute quantification (iTRAQ) labeling technique was used by Xiao et al. for the analysis of nasopharyngeal carcinoma specimens (Xiao et al. 2010). The use of pressure cycling technology (PCT) has been suggested to be efficient in improving protein recovery and overall protein identifications from FFPE blocks compared to heat and detergent-based denaturation methods (Fowler et al. 2010).
In the current study, we utilized PCT technology to extract proteins from FFPE tissue specimens of three histopathological types based on the degree of differentiation of OSCC (Well-OSCC, Moderate-OSCC, and Poor-OSCC). The samples were subjected to tandem mass tags (TMT)-labeling followed by tandem mass spectrometry analysis to identify and quantify the candidate proteins across these subtypes. We attempted to study the underlying molecular mechanisms of OSCC tumor differentiation and look for potential discriminant proteins suggesting histological type. This, in turn, could help identify clinically relevant biomarkers specific to these subtypes, which will further help improve therapeutic outcomes. Our analysis revealed differential expression of proteins associated with extracellular matrix and cell adhesion, with Moderate-OSCC showing higher expression. In comparison, Poor-OSCC revealed a lower expression of the same class of proteins.
Materials and methods
Study design
A total of 3 sample sets of FFPE tissue specimens, including 6 cases of each differentiation type of oral cancer, namely well-, moderate-, and poorly-differentiated, were retrieved from the archive of Burdwan Dental College and Hospital, Burdwan, West Bengal, India. The study was reviewed and approved by the Institutional Ethical Committee, Burdwan Dental College and Hospital, Burdwan, West Bengal, India (dated 02/01/2017), and informed consent was obtained from each patient for the study. The details of the tissues used are provided in Supplementary Table S1.
Protein extraction and estimation
For analysis, 10 μm tissue sections were considered. The deparaffinization of FFPE sections involved three cycles of xylene treatment for 2 min each. These sections were rehydrated in 90% and 70% ethanol, followed by water for 1 min. The rehydrated tissue sections were resuspended in lysis buffer (4% Sodium Dodecyl Sulfate, 100 mM DTT, and 50 mM TEABC (Sigma-Aldrich)) and transferred to PCT microtubes. Barocycler was used for protein extraction with a temperature of 95ºC and 60 cycles of alternating pressure (1cycle = 40,000 psi for 50 s and 5000 psi for 10 s). The lysates were cooled at room temperature and centrifuged at 12,000 rpm for 20 min. The supernatant was separated, and the protein concentration was determined using the bicinchoninic acid assay kit (Thermo Scientific Pierce).
In-solution digestion and labeling with TMT reagents
Briefly, 100 µg of protein from each condition was reduced using 10 mM dithiothreitol (DTT) (Sigma-Aldrich) at 60 °C for 30 min. Alkylation was done using 20 mM iodoacetamide in the dark for 10 min at room temperature. Buffer exchange was carried out using 30 kDa filters to remove SDS as it interferes with trypsin action. Digestion was carried using trypsin as a proteolytic enzyme using a barocycler. After digestion, the peptides were dried using speedVac and kept at -80 ºC until TMT labeling.
TMT labeling was carried out using reagents from a TMT 10-plex kit (Thermo Fisher Scientific) as per the manufacturer’s protocol (TMT channel details: Well-OSCC: 126; Moderate-OSCC: 128C; Poor-OSCC: 131). The labeling reactions were quenched using 5% hydroxylamine. Labeled peptides were then pooled and treated with C18 clean-up before fractionation using HPLC. Peptides were fractionated into 6 fractions, each using the reverse-phase liquid chromatography (bRPLC) method to reduce the sample complexity (Verma et al. 2017).
Tandem mass spectrometry analysis (MS/MS)
Analysis of TMT-labelled samples was carried out on an Orbitrap Fusion Tribrid mass spectrometer (Thermo Scientific, Bremen, Germany) interfaced with Easy-nLC-1200 (Thermo Scientific, Bremen, Germany). The vacuum-dried peptides obtained after C18 cleaning were resuspended in 0.1% formic acid (Solvent A) and loaded onto the trap column (75 µm × 2 cm, nanoViper, 3 µm, 100 A°) filled with C18 at a flow rate of 4 µl/min with Solvent A. The peptides were further resolved onto the analytical column (15 cm × 50 µm, nanoViper, 2 µm). Each fraction was injected four times into the mass spectrometer to represent technical replicates. Data were acquired using data-dependent acquisition mode with the following parameters- MS/MS analysis at a scan range of 110–2000, with top ten intense precursor ions selected for each duty cycle, higher collision energy dissociation with 33% normalized collision energy. The fragmented ions were detected using the Orbitrap mass analyzer at a resolution of 30,000 with a maximum injection time of 200 ms. Synchronous precursor selection MS3 scanning for TMT labels was performed at a scan range of 400–1600 in positive mode with a maximum injection time 200 ms and an automatic gain control (AGC) target value of 5.0 × e5 with quadrupole isolation enabled. The fragment ions for MS3 scans were based on the precursor selection range of 250–1600 m/z, and isobaric tag loss exclusion for TMT. The top ten intense precursors were selected for MS3 scans with a maximum fill time of 150 ms, 60,000 resolution at m/z 200, with MS3 scans acquired in profile mode.
Data processing of LC–MS/MS analysis
Mass spectrometry derived data were searched against the Human RefSeq 81 protein database (consisting of 110,386 protein entries along with 116 common contaminants) in Proteome Discoverer 2.1 (Thermo Scientific, Bremen, Germany) using SequestHT and Mascot (version 2.5.1, Matrix Science, London, UK) as search algorithms. The search parameters included trypsin as a proteolytic enzyme with a maximum of two missed cleavages. Cysteine carbamidomethylation, TMT at lysine, and peptide N-termini were specified as fixed modifications. Acetylation of protein N-terminus and methionine oxidation were set as variable modifications with a minimum length of 7 amino acids as peptide length. Precursor ion mass tolerance and fragment ion mass tolerance were allowed with 10 ppm and 0.05 Da, respectively. Quantitation was carried out using the reporter ion quantifier node in Proteome Discoverer 2.1 from MS3 scans using an integration tolerance of 20 ppm with the most confident centroid setting. The data were searched against the decoy database with a 1% FDR cut-off at PSM and peptide levels.
Bioinformatics analysis
The datasets from Proteome Discoverer were exported, and identified proteins with missing values and contaminants were removed before bioinformatics analysis. Only the proteins identified across all technical replicates were considered for further analysis. Protein intensities were subjected to TMT channel-wise median- normalization. Median values were calculated from replicates for each condition, followed by computation of fold-change ratios and p-values between differentiation states. Proteins with fold-change ratios of ≥ 1.5 and p-value ≤ 0.05 were considered to be significantly upregulated, while those with fold-change ratios of ≤ 0.66 and p-value ≤ 0.05 were considered to be significantly downregulated. The list of proteins identified were compared with gene lists for protein classes and functions such as protein kinases, phosphatases, tumor suppressor genes, oncogenes, epigenetic regulators, hypoxia, oxidative stress, and metabolism obtained from the Molecular Signatures Database (MSigDB, v7.0, https://www.gsea-msigdb.org/gsea/msigdb) or compiled from existing literature as previously described (Subbannayya et al. 2020, 2019). Gene Ontology (GO) enrichment into biological process, sub-cellular localization, and molecular function categories and Reactome-based pathway enrichment analysis was conducted for differentially expressed proteins using Enrichr (https://maayanlab.cloud/Enrichr) (Chen et al. 2013).
Data availability
The mass spectrometry data have been made publicly available by depositing it to ProteomeXchange Consortium Proteomics IDEntifications (PRIDE) partner repository with dataset identifier PXD019456 (Vizcaino et al. 2014).
Results
We analyzed the protein expression in OSCC tissue sections from three histopathological types based on the degree of differentiation (well, moderate, and poorly differentiated), as illustrated in Supplementary Fig. 1a. Six FFPE tissue sections, each of Well-OSCC, Moderate-OSCC, and Poor-OSCC, were analyzed on Orbitrap Fusion Tribrid mass spectrometer after pooling. We identified 4367 proteins, of which 2376 proteins (Supplementary Table S2) were quantified across all four technical replicates in Well-OSCC, Moderate-OSCC, and Poor-OSCC. Among these, 199 proteins were identified with a single peptide with single PSM support and were excluded from further analysis.
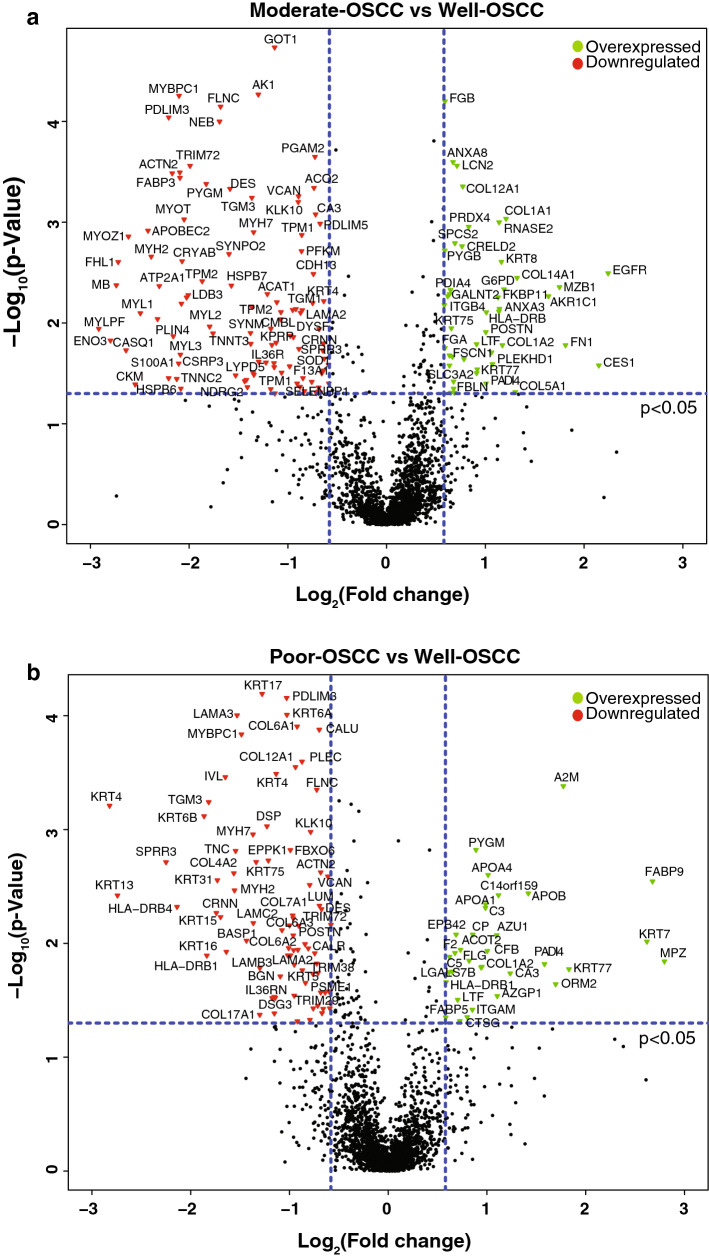
Fig. 1.
Differential expression of proteins across OSCC histopathological types based on the degree of differentiation. Volcano plots depicting differential expression of proteins in a Moderate-OSCC versus Well-OSCC and b Poor-OSCC versus Well-OSCC. Red and green dots represent significantly (p-value ≤ 0.05) downregulated and overexpressed proteins, respectively
Overview of protein expression across different grades of oral cancer
We compared the differentially expressed proteins from all three grades of differentiation. Proteins with fold-change ratios of ≥ 1.5 and p-value ≤ 0.05 were considered to be significantly upregulated, while those with fold-change ratios of ≤ 0.66 and p-value ≤ 0.05 were considered to be significantly downregulated. Employing this cut-off, we identified 141 proteins (44 overexpressed and 97 downregulated) differentially regulated in Moderate-OSCC; and 109 proteins (34 overexpressed and 75 downregulated) significantly altered in Poor-OSCC in comparison to Well-OSCC (Supplementary Fig. 1b). Comparison of protein expression profiles across differentiation grades revealed an overlap of 41 (19.6%) proteins while 100 proteins (47.8%) were specific to Moderate-OSCC vs. Well-OSCC, and 68 proteins (32.5%) were specific to Poor-OSCC vs. Well-OSCC (Supplementary Fig. 1b and d). The dysregulated proteins are graphically represented as volcano plots for moderate-OSCC cases (Fig. 1a) and poor-OSCC cases (Fig. 1b) compared to well-OSCC. Principal component analysis PCA analysis was performed using the dysregulated proteins, which revealed that these three grades show distinct protein expression patterns (Supplementary Fig. 1c). A partial list of overexpressed and downregulated proteins in Moderate-OSCC and Poor-OSCC as compared to Well-OSCC are provided in Tables 1 and 2, respectively.
Table 1.
A partial list of differentially expressed proteins in Moderate-OSCC as compared to Well-OSCC (p ≤ 0.05)
| Gene Symbol | Gene ID | Protein Description | Fold-change ratio (Moderate-OSCC vs. Well-OSCC) |
|---|---|---|---|
| Upregulated proteins | |||
| EGFR | 1956 | Epidermal growth factor receptor | 4.73 |
| CES1 | 1066 | Liver carboxylesterase 1 | 4.43 |
| KRT3 | 3850 | Keratin, type II cytoskeletal 3 | 3.67 |
| FN1 | 2335 | Fibronectin | 3.51 |
| THBS1 | 7057 | Thrombospondin-1 | 2.67 |
| Downregulated proteins | |||
| MURC | 347,273 | Muscle-related coiled-coil protein | 0.48 |
| GOT1 | 2805 | Aspartate aminotransferase, cytoplasmic | 0.46 |
| KRT13 | 3860 | Keratin, type I cytoskeletal 13 | 0.48 |
| CRABP2 | 1382 | Cellular retinoic acid-binding protein 2 | 0.46 |
| KLHL40 | 131,377 | Kelch-like protein 40 | 0.15 |
Table 2.
A partial list of differentially expressed proteins in Poor-OSCC as compared to Well-OSCC (p ≤ 0.05)
| Gene symbol | Gene ID | Protein description | Fold-change ratio (Poor-OSCC vs. Well-OSCC) |
|---|---|---|---|
| Upregulated proteins | |||
| AGR2 | 10,551 | Anterior gradient protein 2 | 18.96 |
| BPIFB2 | 80,341 | BPI fold-containing family B member 2 | 11.92 |
| MUC5B | 727,897 | Mucin-5B | 10.48 |
| MPZ | 4359 | Myelin protein P0 isoform L-MPZ | 6.95 |
| FABP9 | 646,480 | Fatty acid-binding protein 9 | 6.39 |
| Downregulated proteins | |||
| HLA-DQA1 | 3117 | HLA class II histocompatibility antigen | 0.47 |
| CD9 | 928 | CD9 antigen | 0.52 |
| KRT14 | 3861 | Keratin, type I cytoskeletal 14 | 0.46 |
| GBP5 | 115,362 | Guanylate-binding protein 5 | 0.50 |
| CMA1 | 1215 | Chymase | 0.41 |
A comparison of differentially expressed proteins against gene lists of selected processes and functions was carried out. Among the known protein kinases, we found EGFR, a receptor tyrosine kinase as well as a widely known oncogene to be highly overexpressed in Moderate-OSCC (4.73-fold) whereas CSK (1.81-fold) was overexpressed in Poor-OSCC compared to Well OSCC. OBSCN, a protein kinase involved in organization of myofibrils (0.60-fold), was found to be downregulated in Moderate-OSCC. Additionally, we observed alterations in several well-established oncogenes and tumor suppressor genes between Moderate-OSCC and Poor-OSCC samples, which are enlisted in Supplementary Table S3.
We wanted to understand if the tumor differentiation is guided by epigenetic events and sought to identify epigenetic regulators in our data. Comparison with a list of epigenetic regulators revealed upregulation of PADI4 (2.01-fold), HIST1H1B (1.64-fold), and HMGN1 (1.57-fold) in Moderate-OSCC and PADI4 (3-fold) in Poor-OSCC as compared to Well-OSCC. Comparing with a list of cytokines and chemokines to look at cytokine activity in the process of differentiation identified the downregulation of ILRN and IL36RN receptor antagonists in both Moderate-OSCC and Poor-OSCC while there were no changes in the levels of identified interleukins- IL16 and IL18. These findings suggest the presence of epigenetic events and absence or low detectable limits of cytokine activity.
We also compared our data with a list of metabolic pathways to identify evidence of underlying metabolic reprogramming. While several enzymes of glycolysis, including ADH1B, ENO3, PDHA1, PFKM, PGM1, PGAM2 were downregulated in Moderate-OSCC; ADH1B was upregulated in Poor-OSCC (Highlighted in green in Supplementary Table S2). Further, there were stark differences between moderate and poorly differentiated-OSCC samples with respect to lipid metabolism. Lipid metabolism genes including ACAA2, ACO2, ACOT2, CA2, ENO3, GPD1, HADH, and PDHA1 were downregulated in Moderate-OSCC while ACOT2 was upregulated in Poor-OSCC (Highlighted in blue in Supplementary Table S2). These suggest a significant difference between the extent of metabolic reprogramming across OSCC tumor grades. Metabolic pathways, including Amino acid metabolism, TCA cycle, and oxidative phosphorylation, showed minor changes across cancer grades. No significant trends were observed between differentiation states with respect to proteins belonging to processes such as cell cycle, hypoxia, oxidative stress, and oxidative phosphorylation.
Functional analysis of differentially expressed proteins
Reactome pathway and Gene Ontology analyses of differentially expressed proteins were carried out using Enrichr to identify significantly altered signaling pathways and biological processes in Moderate-OSCC and Poor-OSCC compared to Well-OSCC (Supplementary Figs. 2, 3).
Consistent with differentiation grade, extracellular matrix (ECM) organization, collagen formation, muscle contraction, and glucose metabolism pathways were mainly dysregulated in Moderate-OSCC as compared to Well-OSCC. Altered metabolism is often observed in tumors where cancer cells rely on glycolysis, also known as the Warburg effect (Warburg 1956). In the current study, we observed increased levels of glycolytic enzymes in Well-OSCC, but they were comparatively decreased in Moderate-OSCC. Recent findings highlighted such metabolic changes tend to modulate HNSCC and have potential in the development of adjuvant anti-cancer therapy (Hsieh et al. 2019; Sur et al. 2019). Similarly, activation of complement components, lipid digestion, ECM-receptor interaction pathways were observed to be disrupted in the Poor-OSCC phenotype when compared to Well-OSCC.
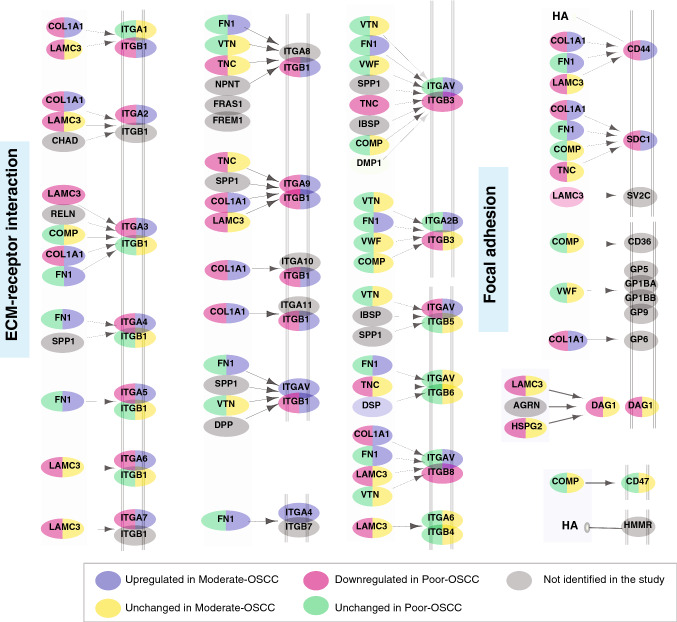
The most striking functional difference between Moderate-OSCC and Poor-OSCC was the significant enrichment of ECM organization having opposite trend across two differentiation grades (Fig. 2, Supplementary Fig. 2). This finding was also supported by GO analysis (Supplementary Fig. 3) In the oral cavity, ECM is composed of collagens, laminins, fibronectins, glycoproteins, and proteoglycans. It provides structural integrity and regulates cellular activities such as cellular adhesion, proliferation, and migration and has been shown to exhibit aberrant behavior in cancer, facilitating tumor progression. We identified altered expression of several members of the collagen family (Table 3). These findings are in line with previous results where altered expression of collagens have been reported to be associated with increased cellular migration, differentiation, and progression of OSCC (Fang et al. 2014). In addition, the laminin family of proteins, which constitute an integral part of ECM, were represented by LAMA2 (2.55 -fold in Moderate-OSCC and 0.61 -fold in Poor-OSCC), LAMA3 (0.35-fold in Poor-OSCC), LAMB3(0.41-fold in Poor-OSCC), LAMC2 (0.39-fold in Poor-OSCC).
Fig. 2.
Role of extracellular matrix–receptor (ECM-receptor) interaction pathway in OSCC. The ECM-receptor interaction pathway was enriched based on proteins dysregulated in Moderate-OSCC and Poor-OSCC compared to Well-OSCC. Proteins highlighted in grey were not identified in our study. Pathway enrichment analysis was performed using Reactome pathway analysis carried out using Enrichr
Table 3.
Expression of collagen family members in Moderate-OSCC and Poor-OSCC as compared to Well-OSCC
| Gene symbol | Moderate-OSCC/Well-OSCC (Fold-change) | Poor-OSCC/Well-OSCC (Fold-change) |
|---|---|---|
| COL1A1 | 2.31 (Up) | 1.32 (No change) |
| COL1A2 | 2.25 (Up) | 1.93 (Up) |
| COL3A1 | 2.14 (Up) | 1.02 (No change) |
| COL4A2 | 1.33 (No change) | 0.34 (Down) |
| COL5A1 | 2.47(Up) | 0.76 (Down) |
| COL5A2 | 2.18(Up) | 0.69 (Down) |
| COL6A1 | 1.04 (No change) | 0.53 (Down) |
| COL6A2 | 1.01 (No change) | 0.47 (Down) |
| COL6A3 | 1.57 (Up) | 0.58 (Down) |
| COL7A1 | 1.19 (No change) | 0.51 (Down) |
| COL12A1 | 1.71 (Up) | 0.52 (Down) |
| COL14A1 | 2.5 (Up) | 0.84 (Down) |
| COL17A1 | 0.89 (No change) | 0.44 (Down) |
The current analysis also demonstrated significant downregulation of muscle contraction-related proteins in the Moderate-OSCC compared to Well-OSCC. In particular, muscle contraction proteins such as troponins TNNI1 (0.39-fold), TNNI2 (0.24-fold), TNNT1(0.35-fold), TNNT3 (0.30-fold), TNNC2 (0.23-fold)), myosins (MYL1 (0.18-fold), MYL2 (0.29-fold), MYL3 (0.24-fold), MYLPF (0.13-fold), MYBPC1 (0.23-fold)) and tropomyosins (TPM1 (0.55-fold), TPM2 (0.27-fold)) were found to be downregulated. This finding was also supported by GO analysis, where proteins participating in muscle contraction-related processes were downregulated in both Moderate-OSCC and Poor-OSCC compared to Well-OSCC (Supplementary Fig. 3). Troponin and tropomyosin are Ca2+-binding proteins and known to regulate striated muscle contraction. There are reports implicating the role of troponins and its isoforms in various human tumors (Casas-Tinto et al. 2016). Taken together, our results indicate diverse expression profiles of multiple cellular pathways between Moderate and Poor-OSCC.
Discussion
Formalin-fixed, paraffin-embedded tumor tissue blocks are a valuable resource for clinical research as they are routinely prepared in clinical settings for pathological investigations. FFPE specimens were initially thought to be problematic for proteomic analysis because of formalin-induced protein cross-links and modifications hindering separation, visualization, and characterization of individual proteins (Palmer-Toy et al. 2005). However, advances in proteomics methods have made FFPE tissue specimens a useful resource for molecular discovery, and several studies have highlighted the feasibility of biomarker discovery from FFPE tissue proteins by shotgun proteomics approach (Xiao et al. 2010), (Azimzadeh et al. 2010; Palmer-Toy et al. 2005; Sprung et al. 2009). The current study used a TMT-labeling based quantitative proteomics approach to study OSCC FFPE tissue specimens across three differentiation grades.
Malignant transformation involves activation of underlying connective tissue and generation of phenotypically altered and specific tumor stroma, with a possibility to influence tumor cells (Kellermann et al. 2008). Epithelial tumor growth and the tumorigenesis process are regulated by tumor stroma comprising of inflammatory cells, endothelial cells, fibroblasts, and myofibroblasts (Baglole et al. 2006). Myofibroblasts-based activity modulated by cancer cells promotes the secretion of extracellular matrix proteins and is involved in the growth, adhesion, migration, and differentiation of tumor cells (Kellermann et al. 2008; Mukaratirwa et al. 2005; Pourreyron et al. 2003). ECM is responsible for providing structural integrity and regulating cellular activities such as cellular adhesion, proliferation, and migration and has been shown to exhibit aberrant behavior in cancer, facilitating tumor progression (Bissell and Hines 2011; Lu et al. 2012; Nissen et al. 2019; Oh et al. 2012; Walker et al. 2018). In the current study, we identified proteins belonging to the ECM organization pathway to be upregulated in Moderate-OSCC compared to Well-OSCC. On the contrary, proteins belonging to the same pathway were found to be downregulated in Poor-OSCC compared to Well-OSCC. This was also reflected in the results of the GO analysis. This suggests that ECM pathway remodeling status is probably dependent on the differentiation state of the tumor cells.
Our results indicate that the main components of the basement membrane, including collagen family members, fibrinogens (FGA, FGB), fibronectin (FN1), and fibulin 1 (FBLN1), were upregulated in Moderate-OSCC. These proteins have been implicated in stromal impairment facilitating tumorigenesis (Chu et al. 2019; Sun et al. 2020; Xiao et al. 2014). FN1 is an extracellular matrix glycoprotein and plays an essential role in cell adhesion, wound healing, proliferation, metastasis, and epithelial-mesenchymal transition (EMT) (Pankov and Yamada 2002). Its aberrant expression is reported in multiple cancer types, including OSCC, colorectal cancer, esophageal SCC, nasopharyngeal carcinoma, thyroid cancer, and gastric cancer (Cai et al. 2018; Nakagawa et al. 2014; Sponziello et al. 2016; Sun et al. 2020; Wang et al. 2017; Xiao et al. 2018). FBLN1 is also a glycoprotein and its aberrant expression has been associated with tumor progression (Harikrishnan et al. 2020; Xiao et al. 2014).
Notably, our results indicated ECM-related proteins, including collagen family members and laminins, to be significantly downregulated in Poor-OSCC compared to Well-OSCC, which probably allows cancerous cells to detach from the site of their origin, resulting in invasion and metastasis. The role of laminins in ECM-remodeling in cancer is well known. Further, the expression for laminins has been found to correlate with cell differentiation in oral cancer (Kulkarni et al. 2019). We also identified tenascin-C (TNC), whose expression is mainly observed in the microenvironment of tumors (Orend and Chiquet-Ehrismann 2006). Similarly, LUM (Lumican) expression has been linked to tumor growth in various malignancies; however, its precise mechanism remains elusive to date (Nikitovic et al. 2008). Alterations in its levels have been reported in multiple cancers where its diverse role in homeostasis and pathological processes makes it a potential candidate for targeted therapy in cancer (Brezillon et al. 2013). Taken together, most of the ECM-related proteins were dysregulated and showed opposite expression across different histological grades of OSCC. Such alterations contribute to tumor invasiveness and metastasis and play a crucial role in cancer progression. This strongly implicates that targeting the effectors of this pathway will prove useful for therapeutic intervention.
Apart from the ECM family of proteins, our data revealed significant enrichment of muscle-contraction related proteins, which were downregulated in Moderate-OSCC compared to Well-OSCC. Muscle wasting is observed among cancer patients resulting in disruptions of mitochondrial homeostasis and reduced oxidative capacity, which has been associated with systemic inflammation (Hardee et al. 2020). Loss of muscle mass has been linked with reduced response to anti-cancer treatments. Loss of adipose tissue and skeletal muscle majorly accounts for muscle wasting and represents an early characteristic in cancer patients (Penna et al. 2014). Cancer-induced muscle wasting (sarcopenia) represents one of the hallmarks of cachexia. A recent study also demonstrated the presence of cachexia, marked by the loss of adipose tissues and skeletal mass in OSCC patients (Yoshimura et al. 2020). Preferential losses of myosin and actin proteins are known to play an essential role in cancer-induced muscle wasting resulting due to altered translational regulation or protein degradation (Banduseela et al. 2007). There are studies demonstrating the role of troponins and their isoforms as striated muscle-specific proteins and their role in tumor growth (Johnston et al. 2018; Sheng and Jin 2016). The existing literature states loss of skeletal muscle is accompanied by chronic respiratory disease, cardiac failure, chronic kidney disease, and sepsis, further resulting in poor overall survival in cancer patients (Bowen et al. 2015). Hence, further studies exploring the role of signaling pathways resulting in muscle contraction and loss are warranted for the identification of candidate drug targets.
In addition to ECM and muscle contraction proteins, we observed altered metabolic reprogramming and epigenetic modulation across OSCC tumor grades. Increased glucose uptake, aerobic glycolysis, and fermentation of glucose are characteristic features of cancer cells. The switching of tumor cells from oxidative phosphorylation to glycolysis for their energy needs despite the presence of functioning mitochondria is known as the Warburg effect (Warburg 1956), and this phenomenon has been suggested to promote tumorigenesis (Liberti and Locasale 2016). The findings from our study were consistent with the Warburg effect. Similarly, increased uptake and metabolism of lipid metabolism are a feature of tumor cells (Koundouros and Poulogiannis 2020; Munir et al. 2019). However, the extent of the change in expression of glycolytic enzymes varied across the tumor grade. Both glycolysis and lipid-metabolism associated proteins were found to be downregulated in Moderate-OSCC compared to Well-OSCC.
Peptidyl arginine deiminases (PADI).catalyze the process of citrullination, a post-translational modification that is known to modulate epigenetic events and regulate transcription (Beato and Sharma 2020). PADI2 and PADI4 have been previously shown to citrullinate histones bringing about changes in transcription activities (Christophorou et al. 2014; Guertin et al. 2014; Zhang et al. 2012). PADI4, in particular, was found to citrullinate H1 histone resulting in its dissociation from chromatin and causing chromatin decondensation in embryonic stem cells (Christophorou et al. 2014). In addition, PADI4 has been shown to modulate arginine methylation through methylation in HL-60 granulocytes and regulate transcription of estrogen-responsive genes such as pS2 (or trefoil factor 1) in MCF-7 breast cancer cells (Wang et al. 2004). In the current study, we identified PADI4 as well as its transcriptional target- HIST1H1B to be overexpressed in both Moderate-OSCC and Poor-OSCC as compared to Well-OSCC. This suggests a possible association of PADI4-induced epigenetic events across tumor differentiation states. In addition, citrullination events could also be modulating the ECM remodeling events observed in the current study. This hypothesis is supported by a previous study where increased ECM localization of PADI4 was observed in liver metastasis of colorectal cancer (Yuzhalin et al. 2018). In addition, ECM citrullination altered adhesion, motility and epithelial to mesenchymal transition (EMT) of colorectal cancer cells, and pharmacological inhibition of PADIs resulted in decreased metastasis. This suggests that PADI4 could be a potential therapeutic target for OSCC, and more investigations are required to confirm this.
Conclusions
In conclusion, proteomic characterization of various grades of OSCC provided novel insights into the underlying molecular mechanisms associated with differentiation. The data obtained from the study indicate significant differences across various grades with respect to ECM organization proteins as well as metabolic pathways and epigenetic events. These differences could be utilized to develop prognostic signature panels or identify potential therapeutic targets for OSCC. Altered proteins identified in the current study need to be further validated in a larger cohort of patient samples. The lack of validation of candidate proteins and the limited number of patient samples used constitute the limitations of the study. Finally, the study shows the power of utilizing archival FFPE tissues for high-throughput clinical omics investigations to unravel molecular mechanisms of cancer.
Supplementary Information
Supplementary Figure 1: (a) Schematic representation of the proteomic analysis of oral FFPE tissues. FFPE blocks were collected and proteins were extracted using Pressure Cycling Technology (PCT)-based Baorcycler approach. The proteins were subjected to in-solution tryptic digestion. Digested peptides were labeled with TMT and were analyzed using a high-resolution Orbitrap Fusion Tribrid mass spectrometer in four technical replicates. The data were analyzed in Proteome Discoverer 2.1 version, against NCBI Human RefSeq 81 using Mascot and SequestHT. (b) Results of the proteomic analysis of FFPE tissues from Poor-, Moderate- and Well-differentiated OSCC cases. (c) Principle component analysis (PCA) of proteins expression demonstrating variations between three grades of OSCC tumors. The replicates were clustered using ClustVis (https://biit.cs.ut.ee/clustvis/). All four replicates from Well-OSCC, Moderate-OSCC, and Poor-OSCC are separated from each other, with the first two components accounting for about 88% of the total variation. (d) Venn diagram showing a significant overlap of 41 proteins where 100 proteins were exclusive to Moderate-OSCC vs. Well-OSCC, and 68 proteins were exclusive to Poor-OSCC vs. Well-OSCC. Dysregulated proteins (overexpressed and downregulated proteins, p≤0.05) these cohorts were considered for this analysis. (EPS 3866 kb)
Supplementary Figure 2: Pathway analysis of differentially expressed proteins in (a) Moderately-differentiated OSCC as compared to Well-differentiated OSCC and (b) Poorly-differentiated OSCC as compared to Well-differentiated OSCC. (EPS 2443 kb)
Supplementary Figure 3: Gene Ontology analysis of differentially expressed proteins in (a) Moderately-differentiated OSCC as compared to Well-differentiated OSCC and (b) Poorly-differentiated OSCC as compared to Well-differentiated OSCC. (EPS 2869 kb)
Acknowledgments
We thank Karnataka Biotechnology and Information Technology Services (KBITS), Government of Karnataka, for the support to the Center for Systems Biology and Molecular Medicine at Yenepoya (Deemed to be University) under the Biotechnology Skill Enhancement Programme in Multiomics Technology (BiSEP GO ITD 02 MDA 2017). We thank Yenepoya (Deemed to be University), Mangalore, India, and Institute of Bioinformatics, Bangalore, India, for access to instrumentation. Varshasnata Mohanty is a recipient of the Women Scientist-A (SR/WOS-A/LS-32/2018) award from the Department of Science and Technology (DST), Government of India.
Abbreviations
- DTT
Dithiothreitol
- FDR
False Discovery Rate
- FFPE
Formalin-Fixed Paraffin Embedded
- GO
Gene Ontology
- IAA
Iodoacetamide
- MS/MS
Tandem Mass Spectrometry
- OSCC
Oral Squamous Cell Carcinoma
- PCT
Pressure Cycling Technology
- PSM
Peptide Spectrum Match
- SDS
Sodium Dodecyl Sulfate
- TEABC
Triethyl Ammonium Bicarbonate
- TMT
Tandem Mass Tags
Author contributions
AC, TSKP, HG, and DS conceptualized the study, designed the experiments, and reviewed the manuscript. RA, SB, SR, NM, and JGR provided pathological evaluation of FFPE for the study. VNP, VM, MAN, and KD carried out sample preparation, fractionation, and mass spectrometry. VM, YS, and AC analyzed the mass spectrometry data. VM, YS, and AC prepared figures, tables and wrote the manuscript. SP, SMP, DS, HG critically reviewed the manuscript. All the authors read and revised the manuscript for important intellectual content and approved the final manuscript.
Funding
This work was supported by the Department of Science and Technology (DST), Government of India (SR/WOS-A/LS-32/2018).
Data availability
The data have been made publically available by depositing it to ProteomeXchange Consortium Proteomics IDEntifications (PRIDE) partner repository with dataset identifier PXD019456.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
The study was performed in accordance with the ethical standards as laid down in the 1964 Declaration of Helsinki. The study was reviewed an approved by the Institutional Ethical Committee, Burdwan Dental College and Hospital, Burdwan, West Bengal, India (dated 02/01/2017) and informed consent was obtained from each patient for the study.
Footnotes
Varshasnata Mohanty and Yashwanth Subbannayya contributed equally to the manuscript.
Contributor Information
Varshasnata Mohanty, Email: varsham@yenepoya.edu.in.
Yashwanth Subbannayya, Email: yashwanth.subbannayya@ntnu.no.
Shankargouda Patil, Email: dr.ravipatil@gmail.com.
Vinuth N. Puttamallesh, Email: vinuthnp@gmail.com
Mohd. Altaf Najar, Email: altaf@yenepoya.edu.in.
Keshava K. Datta, Email: keshava.k.datta@gmail.com
Sneha M. Pinto, Email: sneha.pinto@ntnu.no
Sameera Begum, Email: zabiyasheik@gmail.com.
Neeta Mohanty, Email: dr.neetamohanty@gmail.com.
Samapika Routray, Email: drroutray.samapika@gmail.com.
Riaz Abdulla, Email: rizdent@yenepoya.edu.in.
Jay Gopal Ray, Email: jaygopalray60@gmail.com.
David Sidransky, Email: dsidrans@jhmi.edu.
Harsha Gowda, Email: harshahc@gmail.com.
T. S. Keshava Prasad, Email: keshav@yenepoya.edu.in.
Aditi Chatterjee, Email: aditixchatterjee@gmail.com.
References
- Ahram M, Flaig MJ, Gillespie JW, Duray PH, Linehan WM, Ornstein DK, Niu S, Zhao Y, Petricoin EF, Emmert-Buck MR. Evaluation of ethanol-fixed, paraffin-embedded tissues for proteomic applications. Proteomics. 2003;3:413–421. doi: 10.1002/pmic.200390056. [DOI] [PubMed] [Google Scholar]
- Akhter M, Hossain S, Rahman QB, Molla MR. A study on histological grading of oral squamous cell carcinoma and its co-relationship with regional metastasis. J Oral Maxillofac Pathol. 2011;15:168–176. doi: 10.4103/0973-029X.84485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argiris A, Karamouzis MV, Raben D, Ferris RL. Head and neck cancer. Lancet. 2008;371:1695–1709. doi: 10.1016/S0140-6736(08)60728-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azimzadeh O, Barjaktarovic Z, Aubele M, Calzada-Wack J, Sarioglu H, Atkinson MJ, Tapio S. Formalin-fixed paraffin-embedded (FFPE) proteome analysis using gel-free and gel-based proteomics. J Proteome Res. 2010;9:4710–4720. doi: 10.1021/pr1004168. [DOI] [PubMed] [Google Scholar]
- Baglole CJ, Ray DM, Bernstein SH, Feldon SE, Smith TJ, Sime PJ, Phipps RP. More than structural cells, fibroblasts create and orchestrate the tumor microenvironment. Immunol Invest. 2006;35:297–325. doi: 10.1080/08820130600754960. [DOI] [PubMed] [Google Scholar]
- Banduseela V, Ochala J, Lamberg K, Kalimo H, Larsson L. Muscle paralysis and myosin loss in a patient with cancer cachexia. Acta Myol. 2007;26:136–144. [PMC free article] [PubMed] [Google Scholar]
- Barnes L, Eveson JW, Reichart P, Sidransky D (eds) (2005) Pathology and genetics of head and neck tumours. In: Kleihues P, Sobin LH, series eds. World Health Organization Classification of Tumours. IARC Press, Lyon, France (Paperback, ISBN:9283224l 75, Sl ID, www.iarc.fr)
- Beato M, Sharma P. Peptidyl arginine deiminase 2 (PADI2)-mediated arginine citrullination modulates transcription in cancer. Int J Mol Sci. 2020 doi: 10.3390/ijms21041351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissell MJ, Hines WC. Why don't we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nature Medicine. 2011;17:320–329. doi: 10.1038/nm.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen TS, Schuler G, Adams V. Skeletal muscle wasting in cachexia and sarcopenia: molecular pathophysiology and impact of exercise training. J Cachexia Sarcopenia Muscle. 2015;6:197–207. doi: 10.1002/jcsm.12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brezillon S, Pietraszek K, Maquart FX, Wegrowski Y. Lumican effects in the control of tumour progression and their links with metalloproteinases and integrins. FEBS J. 2013;280:2369–2381. doi: 10.1111/febs.12210. [DOI] [PubMed] [Google Scholar]
- Cai X, Liu C, Zhang TN, Zhu YW, Dong X, Xue P. Down-regulation of FN1 inhibits colorectal carcinogenesis by suppressing proliferation, migration, and invasion. J Cell Biochem. 2018;119:4717–4728. doi: 10.1002/jcb.26651. [DOI] [PubMed] [Google Scholar]
- Casas-Tinto S, Maraver A, Serrano M, Ferrus A. Troponin-I enhances and is required for oncogenic overgrowth. Oncotarget. 2016;7:52631–52642. doi: 10.18632/oncotarget.10616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma'ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophorou MA, Castelo-Branco G, Halley-Stott RP, Oliveira CS, Loos R, Radzisheuskaya A, Mowen KA, Bertone P, Silva JC, Zernicka-Goetz M, Nielsen ML, Gurdon JB, Kouzarides T. Citrullination regulates pluripotency and histone H1 binding to chromatin. Nature. 2014;507:104–108. doi: 10.1038/nature12942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu HW, Chang KP, Hsu CW, Chang IY, Liu HP, Chen YT, Wu CC. Identification of salivary biomarkers for oral cancer detection with untargeted and targeted quantitative proteomics approaches. Mol Cell Proteomics. 2019;18:1796–1806. doi: 10.1074/mcp.RA119.001530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang M, Yuan J, Peng C, Li Y. Collagen as a double-edged sword in tumor progression. Tumour Biol. 2014;35:2871–2882. doi: 10.1007/s13277-013-1511-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forastiere A, Koch W, Trotti A, Sidransky D. Head and neck cancer. N Engl J Med. 2001;345:1890–1900. doi: 10.1056/NEJMra001375. [DOI] [PubMed] [Google Scholar]
- Fowler CB, Chesnick IE, Moore CD, O'Leary TJ, Mason JT. Elevated pressure improves the extraction and identification of proteins recovered from formalin-fixed, paraffin-embedded tissue surrogates. PloS one. 2010;5:e14253. doi: 10.1371/journal.pone.0014253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarino M, Rubino B, Ballabio G. The role of epithelial-mesenchymal transition in cancer pathology. Pathology. 2007;39:305–318. doi: 10.1080/00313020701329914. [DOI] [PubMed] [Google Scholar]
- Guertin MJ, Zhang X, Anguish L, Kim S, Varticovski L, Lis JT, Hager GL, Coonrod SA. Targeted H3R26 deimination specifically facilitates estrogen receptor binding by modifying nucleosome structure. PLoS Genet. 2014;10:e1004613. doi: 10.1371/journal.pgen.1004613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardee JP, Fix DK, Koh HJ, Wang X, Goldsmith EC. Carson JA (2020) Repeated eccentric contractions positively regulate muscle oxidative metabolism and protein synthesis during cancer cachexia in mice. J Appl Physiol. 1985;128:1666–1676. doi: 10.1152/japplphysiol.00908.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harikrishnan K, Joshi O, Madangirikar S, Balasubramanian N. Cell derived matrix fibulin-1 associates with epidermal growth factor receptor to inhibit its activation, localization and function in lung cancer calu-1 cells. Front Cell Dev Biol. 2020;8:522. doi: 10.3389/fcell.2020.00522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh YT, Chen YF, Lin SC, Chang KW, Li WC. Targeting cellular metabolism modulates head and neck oncogenesis. Int J Mol Sci. 2019 doi: 10.3390/ijms20163960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang SI, Thumar J, Lundgren DH, Rezaul K, Mayya V, Wu L, Eng J, Wright ME, Han DK. Direct cancer tissue proteomics: a method to identify candidate cancer biomarkers from formalin-fixed paraffin-embedded archival tissues. Oncogene. 2007;26:65–76. doi: 10.1038/sj.onc.1209755. [DOI] [PubMed] [Google Scholar]
- Johnston JR, Chase PB, Pinto JR. Troponin through the looking-glass: emerging roles beyond regulation of striated muscle contraction. Oncotarget. 2018;9:1461–1482. doi: 10.18632/oncotarget.22879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellermann MG, Sobral LM, da Silva SD, Zecchin KG, Graner E, Lopes MA, Kowalski LP, Coletta RD. Mutual paracrine effects of oral squamous cell carcinoma cells and normal oral fibroblasts: induction of fibroblast to myofibroblast transdifferentiation and modulation of tumor cell proliferation. Oral Oncol. 2008;44:509–517. doi: 10.1016/j.oraloncology.2007.07.001. [DOI] [PubMed] [Google Scholar]
- Koundouros N, Poulogiannis G. Reprogramming of fatty acid metabolism in cancer. Br J Cancer. 2020;122:4–22. doi: 10.1038/s41416-019-0650-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni S, Abdulla R, Jose M, Adyanthaya S, Rex DB, Patil AH, Pinto SM, Subbannayya Y. Omics data-driven analysis identifies laminin-integrin-mediated signaling pathway as a determinant for cell differentiation in oral squamous cell carcinoma. Indian J Pathol Microbiol. 2019;62:529–536. doi: 10.4103/IJPM.IJPM_1_19. [DOI] [PubMed] [Google Scholar]
- Liberti MV, Locasale JW. The Warburg effect: how does it benefit cancer cells? Trends Biochem Sci. 2016;41:211–218. doi: 10.1016/j.tibs.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P, Weaver VM, Werb Z. The extracellular matrix: a dynamic niche in cancer progression. J Cell Biol. 2012;196:395–406. doi: 10.1083/jcb.201102147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsukawa S, Morita K, Negishi A, Harada H, Nakajima Y, Shimamoto H, Tomioka H, Tanaka K, Ono M, Yamada T, Omura K. Galectin-7 as a potential predictive marker of chemo-and/or radio-therapy resistance in oral squamous cell carcinoma. Cancer Med. 2014;3:349–361. doi: 10.1002/cam4.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertins P, Mani DR, Ruggles KV, Gillette MA, Clauser KR, Wang P, Wang X, Qiao JW, Cao S, Petralia F, Kawaler E, Mundt F, Krug K, Tu Z, Lei JT, Gatza ML, Wilkerson M, Perou CM, Yellapantula V, Huang KL, Lin C, McLellan MD, Yan P, Davies SR, Townsend RR, Skates SJ, Wang J, Zhang B, Kinsinger CR, Mesri M, Rodriguez H, Ding L, Paulovich AG, Fenyo D, Ellis MJ, Carr SA. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature. 2016;534:55–62. doi: 10.1038/nature18003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda-Filho A, Bray F. Global patterns and trends in cancers of the lip, tongue and mouth. Oral Oncol. 2020;102:104551. doi: 10.1016/j.oraloncology.2019.104551. [DOI] [PubMed] [Google Scholar]
- Mukaratirwa S, Koninkx JF, Gruys E, Nederbragt H. Mutual paracrine effects of colorectal tumour cells and stromal cells: modulation of tumour and stromal cell differentiation and extracellular matrix component production in culture. Int J Exp Pathol. 2005;86:219–229. doi: 10.1111/j.0959-9673.2005.00425.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munir R, Lisec J, Swinnen JV, Zaidi N. Lipid metabolism in cancer cells under metabolic stress. Br J Cancer. 2019;120:1090–1098. doi: 10.1038/s41416-019-0451-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa Y, Nakayama H, Nagata M, Yoshida R, Kawahara K, Hirosue A, Tanaka T, Yuno A, Matsuoka Y, Kojima T, Yoshitake Y, Hiraki A, Shinohara M. Overexpression of fibronectin confers cell adhesion-mediated drug resistance (CAM-DR) against 5-FU in oral squamous cell carcinoma cells. Int J Oncol. 2014;44:1376–1384. doi: 10.3892/ijo.2014.2265. [DOI] [PubMed] [Google Scholar]
- Negishi A, Masuda M, Ono M, Honda K, Shitashige M, Satow R, Sakuma T, Kuwabara H, Nakanishi Y, Kanai Y, Omura K, Hirohashi S, Yamada T. Quantitative proteomics using formalin-fixed paraffin-embedded tissues of oral squamous cell carcinoma. Cancer Sci. 2009;100:1605–1611. doi: 10.1111/j.1349-7006.2009.01227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikitovic D, Katonis P, Tsatsakis A, Karamanos NK, Tzanakakis GN. Lumican, a small leucine-rich proteoglycan. IUBMB Life. 2008;60:818–823. doi: 10.1002/iub.131. [DOI] [PubMed] [Google Scholar]
- Nissen NI, Karsdal M, Willumsen N. Collagens and Cancer associated fibroblasts in the reactive stroma and its relation to Cancer biology. J Exp Clin Cancer Res CR. 2019;38:115. doi: 10.1186/s13046-019-1110-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh ES, Seiki M, Gotte M, Chung J. Cell adhesion in cancer. Int J Cell Biol. 2012;2012:965618. doi: 10.1155/2012/965618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orend G, Chiquet-Ehrismann R. Tenascin-C induced signaling in cancer. Cancer Lett. 2006;244:143–163. doi: 10.1016/j.canlet.2006.02.017. [DOI] [PubMed] [Google Scholar]
- Palmer-Toy DE, Krastins B, Sarracino DA, Nadol JB, Jr, Merchant SN. Efficient method for the proteomic analysis of fixed and embedded tissues. J Proteome Res. 2005;4:2404–2411. doi: 10.1021/pr050208p. [DOI] [PubMed] [Google Scholar]
- Pankov R, Yamada KM. Fibronectin at a glance. J Cell Sci. 2002;115:3861–3863. doi: 10.1242/jcs.00059. [DOI] [PubMed] [Google Scholar]
- Patel V, Hood BL, Molinolo AA, Lee NH, Conrads TP, Braisted JC, Krizman DB, Veenstra TD, Gutkind JS. Proteomic analysis of laser-captured paraffin-embedded tissues: a molecular portrait of head and neck cancer progression. Clin Cancer Res. 2008;14:1002–1014. doi: 10.1158/1078-0432.CCR-07-1497. [DOI] [PubMed] [Google Scholar]
- Penna F, Baccino FM, Costelli P. Coming back: autophagy in cachexia. Curr Opin Clin Nutr Metab Care. 2014;17:241–246. doi: 10.1097/MCO.0000000000000048. [DOI] [PubMed] [Google Scholar]
- Poschmann G, Sitek B, Sipos B, Hamacher M, Vonend O, Meyer HE, Stuhler K. Cell-based proteome analysis: the first stage in the pipeline for biomarker discovery. Biochim Biophys Acta. 2009;1794:1309–1316. doi: 10.1016/j.bbapap.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Pourreyron C, Dumortier J, Ratineau C, Nejjari M, Beatrix O, Jacquier MF, Remy L, Chayvialle JA, Scoazec JY. Age-dependent variations of human and rat colon myofibroblasts in culture: Influence on their functional interactions with colon cancer cells. Int J Cancer. 2003;104:28–35. doi: 10.1002/ijc.10898. [DOI] [PubMed] [Google Scholar]
- Pozniak Y, Balint-Lahat N, Rudolph JD, Lindskog C, Katzir R, Avivi C, Ponten F, Ruppin E, Barshack I, Geiger T. System-wide clinical proteomics of breast cancer reveals global remodeling of tissue homeostasis. Cell Syst. 2016;2:172–184. doi: 10.1016/j.cels.2016.02.001. [DOI] [PubMed] [Google Scholar]
- Sheng JJ, Jin JP. TNNI1, TNNI2 and TNNI3: Evolution, regulation, and protein structure-function relationships. Gene. 2016;576:385–394. doi: 10.1016/j.gene.2015.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sponziello M, Rosignolo F, Celano M, Maggisano V, Pecce V, De Rose RF, Lombardo GE, Durante C, Filetti S, Damante G, Russo D, Bulotta S. Fibronectin-1 expression is increased in aggressive thyroid cancer and favors the migration and invasion of cancer cells. Mol Cell Endocrinol. 2016;431:123–132. doi: 10.1016/j.mce.2016.05.007. [DOI] [PubMed] [Google Scholar]
- Sprung RW, Jr, Brock JW, Tanksley JP, Li M, Washington MK, Slebos RJ, Liebler DC. Equivalence of protein inventories obtained from formalin-fixed paraffin-embedded and frozen tissue in multidimensional liquid chromatography-tandem mass spectrometry shotgun proteomic analysis. Mol Cell Proteomics. 2009;8:1988–1998. doi: 10.1074/mcp.M800518-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbannayya Y, Pinto SM, Bosl K, Prasad TSK, Kandasamy RK. Dynamics of dual specificity phosphatases and their interplay with protein kinases in immune signaling. Int J Mol Sci. 2019 doi: 10.3390/ijms20092086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbannayya Y, Haug M, Pinto SM, Mohanty V, Meas HZ, Flo TH, Prasad TSK, Kandasamy RK. The proteomic landscape of resting and activated CD4+ T cells reveal insights into cell differentiation and function. Int J Mol Sci. 2020 doi: 10.3390/ijms22010275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Zhao C, Ye Y, Wang Z, He Y, Li Y, Mao H. High expression of fibronectin 1 indicates poor prognosis in gastric cancer. Oncol Lett. 2020;19:93–102. doi: 10.3892/ol.2019.11088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sur S, Nakanishi H, Flaveny C, Ippolito JE, McHowat J, Ford DA, Ray RB. Correction to: Inhibition of the key metabolic pathways, glycolysis and lipogenesis, of oral cancer by bitter melon extract. Cell Commun Signal CCS. 2019;17:151. doi: 10.1186/s12964-019-0475-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyanova S, Albrechtsen R, Kronqvist P, Cox J, Mann M, Geiger T. Proteomic maps of breast cancer subtypes. Nat Commun. 2016;7:10259. doi: 10.1038/ncomms10259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Pinto SM, Patil AH, Advani J, Subba P, Kumar M, Sharma J, Dey G, Ravikumar R, Buggi S, Satishchandra P, Sharma K, Suar M, Tripathy SP, Chauhan DS, Gowda H, Pandey A, Gandotra S, Prasad TS. Quantitative Proteomic and Phosphoproteomic Analysis of H37Ra and H37Rv Strains of Mycobacterium tuberculosis. J Proteome Res. 2017;16:1632–1645. doi: 10.1021/acs.jproteome.6b00983. [DOI] [PubMed] [Google Scholar]
- Vizcaino JA, Deutsch EW, Wang R, Csordas A, Reisinger F, Rios D, Dianes JA, Sun Z, Farrah T, Bandeira N, Binz PA, Xenarios I, Eisenacher M, Mayer G, Gatto L, Campos A, Chalkley RJ, Kraus HJ, Albar JP, Martinez-Bartolome S, Apweiler R, Omenn GS, Martens L, Jones AR, Hermjakob H. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat Biotechnol. 2014;32:223–226. doi: 10.1038/nbt.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker C, Mojares E, Del Rio Hernandez A. Role of extracellular matrix in development and cancer progression. Int J Mol Sci. 2018 doi: 10.3390/ijms19103028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wysocka J, Sayegh J, Lee YH, Perlin JR, Leonelli L, Sonbuchner LS, McDonald CH, Cook RG, Dou Y, Roeder RG, Clarke S, Stallcup MR, Allis CD, Coonrod SA. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science. 2004;306:279–283. doi: 10.1126/science.1101400. [DOI] [PubMed] [Google Scholar]
- Wang J, Deng L, Huang J, Cai R, Zhu X, Liu F, Wang Q, Zhang J, Zheng Y. High expression of Fibronectin 1 suppresses apoptosis through the NF-kappaB pathway and is associated with migration in nasopharyngeal carcinoma. Am J Transl Res. 2017;9:4502–4511. [PMC free article] [PubMed] [Google Scholar]
- Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Xiao Z, Li G, Chen Y, Li M, Peng F, Li C, Li F, Yu Y, Ouyang Y, Chen Z. Quantitative proteomic analysis of formalin-fixed and paraffin-embedded nasopharyngeal carcinoma using iTRAQ labeling, two-dimensional liquid chromatography, and tandem mass spectrometry. J Histochem Cytochem. 2010;58:517–527. doi: 10.1369/jhc.2010.955526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W, Wang J, Li H, Xia D, Yu G, Yao W, Yang Y, Xiao H, Lang B, Ma X, Guo X, Guan W, Xu H, Liu J, Zhang X, Ye Z. Fibulin-1 is epigenetically down-regulated and related with bladder cancer recurrence. BMC Cancer. 2014;14:677. doi: 10.1186/1471-2407-14-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Yang W, Xu B, Zhu H, Zou J, Su C, Rong J, Wang T, Chen Z. Expression of fibronectin in esophageal squamous cell carcinoma and its role in migration. BMC Cancer. 2018;18:976. doi: 10.1186/s12885-018-4850-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura T, Suzuki H, Takayama H, Higashi S, Hirano Y, Tezuka M, Ishida T, Ishihata K, Nishi Y, Nakamura Y, Imamura Y, Nozoe E, Nakamura N (2020) Impact of preoperative low prognostic nutritional index and high intramuscular adipose tissue content on outcomes of patients with oral squamous cell carcinoma. Cancers (Basel) 12(11):3167. 10.3390/cancers12113167 [DOI] [PMC free article] [PubMed]
- Yuzhalin AE, Gordon-Weeks AN, Tognoli ML, Jones K, Markelc B, Konietzny R, Fischer R, Muth A, O'Neill E, Thompson PR, Venables PJ, Kessler BM, Lim SY, Muschel RJ. Colorectal cancer liver metastatic growth depends on PAD4-driven citrullination of the extracellular matrix. Nat Commun. 2018;9:4783. doi: 10.1038/s41467-018-07306-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Bolt M, Guertin MJ, Chen W, Zhang S, Cherrington BD, Slade DJ, Dreyton CJ, Subramanian V, Bicker KL, Thompson PR, Mancini MA, Lis JT, Coonrod SA. Peptidylarginine deiminase 2-catalyzed histone H3 arginine 26 citrullination facilitates estrogen receptor alpha target gene activation. Proc Natl Acad Sci U S A. 2012;109:13331–13336. doi: 10.1073/pnas.1203280109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Wang J, Wang X, Zhu J, Liu Q, Shi Z, Chambers MC, Zimmerman LJ, Shaddox KF, Kim S, Davies SR, Wang S, Wang P, Kinsinger CR, Rivers RC, Rodriguez H, Townsend RR, Ellis MJ, Carr SA, Tabb DL, Coffey RJ, Slebos RJ, Liebler DC. Proteogenomic characterization of human colon and rectal cancer. Nature. 2014;513:382–387. doi: 10.1038/nature13438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Liu T, Zhang Z, Payne SH, Zhang B, McDermott JE, Zhou JY, Petyuk VA, Chen L, Ray D, Sun S, Yang F, Wang J, Shah P, Cha SW, Aiyetan P, Woo S, Tian Y, Gritsenko MA, Clauss TR, Choi C, Monroe ME, Thomas S, Nie S, Wu C, Moore RJ, Yu KH, Tabb DL, Fenyo D, Bafna V, Wang Y, Rodriguez H, Boja ES, Hiltke T, Rivers RC, Sokoll L, Zhu H, Shih Ie M, Cope L, Pandey A, Snyder MP, Levine DA, Smith RD, Chan DW, Rodland KD. Integrated proteogenomic characterization of human high-grade serous ovarian. Cancer Cell. 2016;166:755–765. doi: 10.1016/j.cell.2016.05.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: (a) Schematic representation of the proteomic analysis of oral FFPE tissues. FFPE blocks were collected and proteins were extracted using Pressure Cycling Technology (PCT)-based Baorcycler approach. The proteins were subjected to in-solution tryptic digestion. Digested peptides were labeled with TMT and were analyzed using a high-resolution Orbitrap Fusion Tribrid mass spectrometer in four technical replicates. The data were analyzed in Proteome Discoverer 2.1 version, against NCBI Human RefSeq 81 using Mascot and SequestHT. (b) Results of the proteomic analysis of FFPE tissues from Poor-, Moderate- and Well-differentiated OSCC cases. (c) Principle component analysis (PCA) of proteins expression demonstrating variations between three grades of OSCC tumors. The replicates were clustered using ClustVis (https://biit.cs.ut.ee/clustvis/). All four replicates from Well-OSCC, Moderate-OSCC, and Poor-OSCC are separated from each other, with the first two components accounting for about 88% of the total variation. (d) Venn diagram showing a significant overlap of 41 proteins where 100 proteins were exclusive to Moderate-OSCC vs. Well-OSCC, and 68 proteins were exclusive to Poor-OSCC vs. Well-OSCC. Dysregulated proteins (overexpressed and downregulated proteins, p≤0.05) these cohorts were considered for this analysis. (EPS 3866 kb)
Supplementary Figure 2: Pathway analysis of differentially expressed proteins in (a) Moderately-differentiated OSCC as compared to Well-differentiated OSCC and (b) Poorly-differentiated OSCC as compared to Well-differentiated OSCC. (EPS 2443 kb)
Supplementary Figure 3: Gene Ontology analysis of differentially expressed proteins in (a) Moderately-differentiated OSCC as compared to Well-differentiated OSCC and (b) Poorly-differentiated OSCC as compared to Well-differentiated OSCC. (EPS 2869 kb)
Data Availability Statement
The mass spectrometry data have been made publicly available by depositing it to ProteomeXchange Consortium Proteomics IDEntifications (PRIDE) partner repository with dataset identifier PXD019456 (Vizcaino et al. 2014).
The data have been made publically available by depositing it to ProteomeXchange Consortium Proteomics IDEntifications (PRIDE) partner repository with dataset identifier PXD019456.