Abstract
In the 21st century, we have witnessed three coronavirus outbreaks: SARS in 2003, MERS in 2012, and the ongoing pandemic coronavirus disease 2019 (COVID-19). The search for efficient vaccines and development and repurposing of therapeutic drugs are the major approaches in the COVID-19 pandemic research area. There are concerns about the evolution of mutant strains (e.g., VUI – 202012/01, a mutant coronavirus in the United Kingdom), which can potentially reduce the impact of the current vaccine and therapeutic drug development trials. One promising approach to counter the mutant strains is the “development of effective broad-spectrum antiviral drugs” against coronaviruses. This study scientifically investigates potent food bioactive broad-spectrum antiviral compounds by targeting main protease (Mpro) and papain-like protease (PLpro) proteases of coronaviruses (CoVs) using in silico and in vitro approaches. The results reveal that phycocyanobilin (PCB) shows potential inhibitor activity against both proteases. PCB had the best binding affinity to Mpro and PLpro with IC50 values of 71 and 62 μm, respectively. Also, in silico studies with Mpro and PLpro enzymes of other human and animal CoVs indicate broad-spectrum inhibitor activity of the PCB. As with PCB, other phycobilins, such as phycourobilin (PUB), phycoerythrobilin (PEB), and phycoviolobilin (PVB) show similar binding affinity to SARS-CoV-2 Mpro and PLpro.
Keywords: food bioactive constituents, broad-spectrum inhibitors, coronaviruses, SARS-CoV-2, COVID-19, main protease, papain-like protease
Introduction
Coronaviruses (CoVs) belongs to the subfamily of Orthocoronavirinae, family Coronavidae, order Nidovirales. They are large (average diameter of 120 nm), enveloped, positive-sense single-stranded RNA viruses with a genome size of ∼26 to 32 kb (Woo et al., 2010). Based on antigen cross-reactivity and genetic makeup, four subgroups (alpha, beta, gamma, and delta) are subdivided into 26 different species of CoVs (Cleri et al., 2010). CoVs cause diseases in mammals and birds; alpha and beta group CoVs are pathogenic to humans (Paules et al., 2020). The seven CoVs that can cause infectious diseases in humans are HCoV-229E, HCoV-NL63, HCoV-OC43, HCoV-HKU1, severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory virus coronavirus (MERS-CoV), and 2019-nCoV (2019-novel coronavirus) or SARS-CoV-2 (Hamre and Procknow, 1966; McIntosh et al., 1967; Drosten et al., 2003; van der Hoek et al., 2004; Woo et al., 2005; Bermingham et al., 2012; Wu F. et al., 2020). The first four common CoVs persistently circulate in humans and are responsible for 10–30% of common colds (Paules et al., 2020). The other three deadly viruses are etiological agents of fatal respiratory syndromes SARS, MERS, and coronavirus disease 2019 (COVID-19), respectively. The SARS epidemic in 2003 ended with 8098 reported cases, 774 deaths (fatality rate 9.7%), whereas the MERS outbreak in 2012 caused 2494 reported cases, 858 deaths (fatality rate 34%) (World Health Organization (WHO), 2003; Alfaraj et al., 2019). COVID-19, the current pandemic outbreak, first identified in 2019, report >37.1 million confirmed cases with >1.07 million deaths (fatality rate 2.9%) as of October 12, 2020 (World Health Organization (WHO), 2020). Avian infectious bronchitis virus (IBV), feline infectious peritonitis virus (FIPV), canine CoV, and porcine transmissible gastroenteritis virus (TGEV) cause respiratory and enteric diseases in farm and domestic pet animals (Pratelli, 2006; Cavanagh, 2007; Pedersen, 2009; Odend’hal, 2012).
Till now, there are no approved vaccines and therapeutic drugs for COVID-19 or other human coronavirus infections and a lack of enough clinical trial data to make treatment decisions. Although vaccines have been developed against animal viruses IBV, canine CoV, and TGEV to help prevent serious diseases (Park et al., 1998; Carmichael, 1999; Liu and Kong, 2004), there are some potential problems, such as recombination events between field and vaccine strains, the emergence of novel serotypes, and antibody-dependent enhancement remain. The rapid development of vaccines and repurposing of approved antivirals drugs (e.g., remdesivir) are major clinical approaches of a pandemic preparedness plan. The development of broad-spectrum antiviral agents that are effective against a wide range of CoVs and other classes of viruses, including emerging ones, could be a promising strategy (Bekerman and Einav, 2015; Fauci and Morens, 2016; Cho and Glenn, 2020).
Broad-spectrum antiviral targeting strategies can be classified into two categories: (i) entry inhibitors that interact with existing virus particles outside of cells and prevent infection (Hangartner et al., 2006) and (ii) replication inhibitors aimed at stopping viral genome replication to curtail production of new virus particles (De Clercq, 2004). The S glycoprotein of coronaviruses, the main determinant of host cell attachment and viral entry, is not well conserved between HCoVs (Totura and Bavari, 2019). On the other hand, CoV non-structural proteins (nsps) are highly conserved components of the coronavirus life cycle that mediate viral replication (Totura and Bavari, 2019). Literature studies report the following SARS-CoV-2 nsp targets; main protease (Mpro), papain-like protease (PLpro), Nsp3, RdRp, Helicase, Nsp14, Nsp15, Nsp16, N protein to inhibit virus replication (Wu C. et al., 2020). Proteolytic processing of viral polyproteins into functional nsps by two viral proteases, the Mpro and PLpro, is an important event of the CoV life cycle. The Mpro acts on minimum 11 cleavage sites of replicase 1ab, ∼790 kDa; at recognition sequence Leu-Gln↓ (Ser, Ala, Gly) (↓ indicates cleavage site), most cleavage sites block viral replication (Zhang et al., 2020). PLPro enzyme hydrolyses the peptide bond at the carboxyl side of glycine (P1 position) and releases nsp1, nsp2, and nsp3 functional proteins, which play a key role in viral replication (Rut et al., 2020). Therefore, these proteases would be potential targets for the development of broad-spectrum antiviral drugs. CoVs Mpro and PLpro crystal structures are available for public access in the protein data bank (PDB).
Natural food bioactive compounds are gaining importance as supplementary antiviral therapeutic compounds in the modern healthcare sector because of their lower toxicity and fewer side effects, additional health benefits (antioxidant, anti-inflammatory, and immunomodulation activities), and potential use in conjunction with preexisting therapies. Several literature studies report antiviral properties of food bioactive compounds against CoVs and other viruses (Table 1; Ghildiyal et al., 2020; Mani et al., 2020). In view of the issues posed above, identifying natural food bioactive broad-spectrum antiviral agents against the CoVs is a more reasonable and attractive prospect and could provide an effective first line of defense against future emerging CoVs related diseases. Herein, we report the phycobilins as potent food bioactive broad-spectrum inhibitor compounds against Mpro and PLpro of SARS-CoV-2 and other CoVs via in silico and in vitro approaches.
TABLE 1.
Antiviral properties of selected food bioactive constituents.
| Bioactive compound | Antiviral activity | References |
| Phycocyanobilin | Spirulina extract exhibited anti-flu efficacy against wide range of influenza viruses with EC50 values from 0.58 to 1.17 mg/mL. | Chen et al., 2016 |
| Quercetin | Inhibited hepatitis C virus production almost completely (>95%) at concentration of 10 μM. | Bachmetov et al., 2012 |
| Riboflavin | In combination with poly r(A-U) showed 7 to 12-fold antiviral activity against human foreskin fibroblast-vesicular stomatitis virus | Jamison et al., 1990 |
| Cyanidin | Cyanidin-3-sambubiocide was found to be a potent inhibitor for H1N1 neuraminidase (NA) activity with IC50 value 72 μM. | Kannan and Kolandaivel, 2018 |
| Daidzein | Exhibited anti-dengue activity with IC50 = 142.6 μg mL–1 against DENV-2. | Zandi et al., 2011 |
| Genistein | Reduced hepatitis B virus production with an IC50 value of 33 and 46 μM for human and macaque fibroblasts, respectively. | LeCher et al., 2019 |
| Catechin | Catechins (−)-epigallocatechin gallate (EGCG), (−)-epicatechin gallate (ECG) were identified as potent inhibitors of influenza virus replication with EC50 of 22–28 and 22–40 μM, respectively. | Song et al., 2005 |
| Resveratrol | MERS-CoV titer reduced 4 logs by resveratrol treatment at 250 μM concentration after 48 h of infection. | Lin et al., 2017 |
| Curcumin | Curcumin and its derivatives showed antiviral effects on HSV-1 in cell culture with IC50 values in range of 13.9–33.0 μg/mL. | Zandi et al., 2010 |
| Astaxanthin | Pre-treatment of Vero cells with 75 μg mL–1 of Haematococcus pluvialis ethanol extract with carotenoids inhibited Herpes simplex virus type 1 (HSV-1) infection by approximately 85%. | Santoyo et al., 2012 |
| β-carotene | Pre-treatment of Vero cells with 75 μg mL-1 of H. pluvialis ethanol extract with carotenoids inhibited Herpes simplex virus type 1 (HSV-1) infection by approximately 85%. | Santoyo et al., 2012 |
| Capsaicin | Methanolic extract of Capsicum annuum exhibited a considerable anti-HSV-1 and anti-HSV-2 activities at the concentration of 25 μg/mL. | Hafiz et al., 2017 |
| Gingerol | Fresh Zingiber officinale inhibited human respiratory syncytial virus (HRSV) with IC50 of 144.9 μg/mL in HEp-2 cells and 73.3 μg/ml in A549 cells. | San Chang et al., 2013 |
| Vanillin | MY21 (a vanillin derivative) had the IC50 of 50 μM against H1N1 neuraminidase (NA). | Hariono et al., 2016 |
| Eugenol | IC50 values for the anti-HSV effects of eugenol were 25.6 and 16.2 μg/mL for HSV-1 and HSV-2, respectively. | Benencia and Courreges, 2000 |
| Thymol | Exhibited significant antiviral activity with an IC50 of 7 μM against herpes simplex virus type I. | Lai et al., 2012 |
Materials and Methods
In silico Screening of Inhibitor Compounds
Preparation of Protein and Ligand for Docking
The crystal structures of Mpro (PDB ID – 6LU7) and PLpro of SARS-CoV-2 (PDB ID – 6WUU) and other CoVs used in this study were obtained from the RCSB PDB. Ligand structures were obtained from Pubchem and Chemical Entities of Biological Interest (ChEBI) as SDF format, Open Babel was used for format transformation or 3-D coordinate generation for the uploaded files (O’Boyle et al., 2011). The MGLTools were used to delete other chains, and heteroatoms (included water), adding missing atoms, hydrogens, and charges. Further, the pdbqt files were prepared for proteins and ligands binding.
Molecular Docking and Molecular Simulation Studies
Autodock Vina was used as a docking engine. It is critical to define the docking grid box appropriately due to the small molecule docking procedure (Trott and Olson, 2010). The docking box is defined as the center of native ligand coordinates with 40 Å × 40 Å × 40 Å in length to include the residues of the entire cavity, and the exhaustiveness level was set on 12 with number of modes 10. For visualization, the docking results PDBQT files were exported, and docked protein-ligand complex structures were visualized using Pymol. Active site residues within 3 or 3.5 Å of ligand and polar contacts were determined with this same tool. The ligand docking procedure was validated by redocking of the native ligand with the same protocol and the grid parameters as used for food bioactive compounds. The redocked ligand was then superimposed onto the reference co-crystallized ligand complex using Pymol, and the root mean square deviation (RMSD) was analyzed.
Molecular dynamic simulations were performed using NAMD (Phillips et al., 2005). The parameters, structure, and topology files for the ligand were generated using the CHARMM-GUI Web server (Jo et al., 2008). Visual molecular dynamics (VMD) was used to generate protein structure (PSF) files (Humphrey et al., 1996). Each protein-ligand docked complex was solvated and ionized with 0.15 M ions (Na+ and Cl−) to neutralize the charge and electrostatic screening. The systems were subjected to 2000 steps of steepest descent energy minimization before a production run at the NPT of 0.5 ns (250,000 steps). The temperature (310 K) and pressure (1 atm) were controlled by the Langevin and Langevin piston methods (Feller et al., 1995). VMD software was used to visualize simulations and to analyze average ligand-RMSD and protein-RMSD and hydrogen bonds.
In vitro Enzymatic Assays
For enzyme inhibition studies, selected phytochemicals, PCB, Quercetin, Riboflavin, Cyanidin, Daidzein, and Genistein, were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, United States). Enzyme assay kits, 3CL Protease, MBP-tagged (SARS-CoV-2) assay (Catalog #79955), and papain-like protease (SARS-CoV-2) assay kit: protease activity (Catalog #79995), were purchased from BPS Bioscience (San Diego, CA, United States).
Mpro Assay
Fluorescence resonance energy transfer (FRET)-based cleavage assay (Zhu et al., 2011) was used for in vitro enzyme inhibition study. SARS-CoV-2 Mpro or 3CL Protease, GenBank Accession No. YP_009725301, amino acids 1-306 (full length), with an N-terminal MBP-tag, expressed in an Escherichia coli and its fluorescent substrate with cleavage site (indicated by ↓) DABCYL-KTSAVLQ↓SGFRKME-EDANS, inhibitor control (GC376), and the assay buffer composed of 20 mM Tris, 100 mM NaCl, 1 mM EDTA, 1 mM DTT, pH 7.3 were used. Initially, 15 μL of the SARS-CoV-2 Mpro in reaction buffer at the final concentration of 10 ng/μL and 5 μL of inhibitor control (GC376, final concentration 50 μM)/test inhibitor (10–600 μM)/inhibitor solvent (positive control) was pipetted into a 384-well plate. Stock solutions of the compounds were prepared with 100% DMSO. Afterward, the plate was preincubated for 30 min at room temperature with slow shaking. The enzymatic reaction was then initiated by adding of 5 μL of the substrate dissolved in the reaction buffer to 25 μL final volume (final concentration 50 μM) and incubated at room temperature for 4 h. The fluorescence signal of the Edans generated due to the cleavage of the substrate by the Mpro was monitored at excitation at 360 nm with an emission wavelength of 460 nm, using a spectrophotometric microplate reader (Synergy H1 Hybrid Multi-Mode Reader; BioTek Instruments, Inc., Winooski, VT, United States).
PLpro Assay
Severe acute respiratory syndrome coronavirus-2 PLpro (papain-like protease), GenBank Accession No. QHD43415, amino acids 1564–1882, with N-terminal His-tag, expressed in an E. coli and its fluorescent substrate Z-Arg-Leu-Arg-Gly-Gly-AMC, inhibitor control (GRL0617) and the assay buffer (40 mM Tris pH 8, 110 mM NaCl, 1 mM DTT) was used for the inhibition assay. Briefly, 30 μL of the SARS-CoV-2 PLpro in reaction buffer at the final concentration of 0.44 ng/μL and 10 μL of inhibitor control (GRL0617, final concentration 100 μM)/test inhibitor (10–600 μM)/inhibitor solvent (positive control) was pipetted into a 96-well plate. Afterward, the plate was preincubated for 30 min at room temperature with slow shaking. The enzymatic reaction was then initiated by the addition of 10 μL of the substrate dissolved in the reaction buffer to 50 μL final volume (final concentration 25 μM), incubated at room temperature for 40–60 min. The fluorescence signal of the substrate after the enzymatic reaction was monitored at an excitation at 360 nm with an emission wavelength of 460 nm, using a spectrophotometric microplate reader (Synergy H1 Hybrid Multi-Mode Reader; BioTek Instruments, Inc., Winooski, VT, United States). Triplicate experiments (N = 3) were performed for both Mpro and PLpro assays, and the mean value was presented with ± standard deviation (SD).
Results
Selection of Phytochemicals for the Study
A total of 16 phytochemicals from different chemical classes were selected based on the previous reports of their potent antiviral effects (Table 1): linear tetrapyrrole – phycocyanobilin (PCB), flavonols – quercetin, catechin, flavin – riboflavin, anthocyanin – cyanidin, isoflavones – daidzein, genistein, stilbenoid phenol – resveratrol, linear diarylheptanoid – curcumin, Xanthophyll – astaxanthin, carotenes – β-carotene, phenolic alkaloid – capsaicin, phenolic ketone – gingerol, phenolic aldehyde – vanillin, allylbenzene – eugenol, monoterpenoid phenol – thymol.
In silico Binding Interaction Studies of Selected Phytochemical Compounds With SARS-CoV-2 Mpro and PLpro
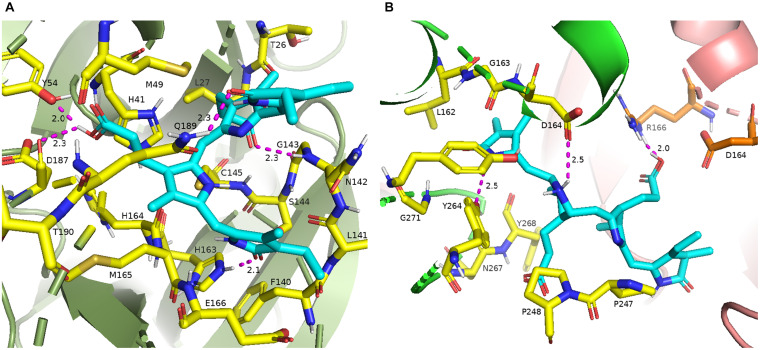
The 16 selected phytochemicals were docked into the active site pocket of SARS-CoV-2 Mpro and PLpro. Table 2 depicts the source, docking score, and polar contacts of selected phytochemical bioactive compounds with binding site amino acid residues of SARS-CoV-2 proteases. For Mpro, the results show PCB docked with the best score or binding energy of −8.6 Kcal/mol followed by Riboflavin (−7.9 Kcal/mol), Cyanidin (−7.9 Kcal/mol), Daidzein (−7.8 Kcal/mol), and Genistein (−7.6 Kcal/mol). Twelve key active-site amino acid residues (Tyr 54, Gly 143, His 163, Asp 187, Gln 189, Glu 166, Cys 145, Leu 141, Ser 144, Thr 26, Gln 192, and Thr 190) of SARS-CoV-2 Mpro involved in polar interactions at a distance of ≤3 Å with ligand phytochemical compounds. Specific polar contacts of each phytochemical compound are shown in Table 2. In the case of PLpro, as the reported peptide inhibitor VIR250 is bound to the dimer interface in the crystal structure of 6WUU (Rut et al., 2020), the docking studies were performed with dimer form. Similarly, PCB docked with the best score or binding energy of −9.8 Kcal/mol followed by Astaxanthin, (−9.3 Kcal/mol), β-carotene (−9.2 Kcal/mol), Daidzein (−8.9 Kcal/mol), Riboflavin (−8.5 Kcal/mol), and Genistein (−8.3 Kcal/mol). Eleven key active site amino acid residues (Asp 164, Tyr 264, Gln 269, Arg 166, Tyr 273, Glu 161, Tyr 268, Lys 157, Leu 162, Gly 266, and Ser 170) in chain A and 13 amino acid residues (Arg 166, Gln 174, Met 208, Glu 161, Glu 167, Cys 155, Lys 232, Met 206, Arg 183, Glu 203, Tyr 268, Tyr 273, and Thr 301) in chain B of SARS-CoV-2 PLpro are involved in polar interactions at a distance of ≤3 Å with ligand phytochemical compounds. Table 2 illustrates the specific polar contacts between phytochemical compounds and proteases. Figure 1 shows a 3-D representation of the binding pocket of Mpro and PLpro with top score model pose of PCB. The co-crystalized structure of native 6LU7-N3 and 6WUU-VIR250 complexes and polar contacts are represented in Supplementary Figures 1a, 2a. The docking validation studies reveal that both N3 peptide inhibitor and VIR250 bound exactly to the active site of 6LU7 and 6WUU, respectively. Superimposed redocked N3 on to the native co-crystallized N3 show a low RMSD of 1.82 Å was observed, whereas redocked VIR250 had a RMSD of 2.096 Å (Supplementary Figures 1b, 2b). These results show less variation in comparison with the native binding pose of ligands in co-crystallized form.
TABLE 2.
Molecular docking results of food bioactive compounds with COVID-19 main protease (Mpro), papain-like protease (PLpro).
| Source | Compounds | Mpro |
PLpro |
||
| Dock score | Polar contacts | Dock score | Polar contacts | ||
| Cyanobacteria | Phycocyanobilin | −8.6 | Y54, G143, H163, D187, Q189 | −9.8 | D164 (A), R166 (B), Y264 (A) |
| Fruits, vegetables, seeds, and grains | Quercetin | −7.8 | Y54, Q189 | −8 | R166 (B), Q269 (A) |
| Eggs, meat, fruits, and vegetables | Riboflavin | −7.9 | E166, C145, H163, L141, S144 | −8.5 | R166 (A), Y264 (A), Y273 (A) |
| Grapes and berries | Cyanidin | −7.8 | S144, H163 | −7.9 | E161 (A), Y268 (A) |
| Legumes | Daidzein | −7.8 | T26, E166, Q192, T190 | −8.9 | K157 (A), D164 (A), R166 (B), Q174 (B) |
| Legumes | Genistein | −7.6 | E166 | −8.3 | K157 (A), L162 (A), Q174 (B), M208 (B) |
| Green tea | Catechin | −7.3 | L141, H163 | −7.1 | E161 (B), R166(A) |
| Grapes and berries skin | Resveratrol | −7 | L141, H163, D187 | −7.2 | R166 (A), E167 (B), C155 (B) |
| Turmeric | Curcumin | −7 | G143, S144, C145 | −8 | K157 (A), K232 (B), Y264 (A) |
| Microalgae | Astaxanthin | −7 | None | −9.3 | G266 (A), M206 (B) |
| Fruits and vegetables | β-carotene | −6.5 | None | −9.2 | None |
| Chili pepper | Capsaicin | −6.3 | E166, T190, Q192 | −6.5 | K157 (A), M208 (B) |
| Ginger | Gingerol | −6.1 | G143, S144, C145, H163, E166 | −6.4 | R183 (B), E203 (B), R183 (B) |
| Vanilla | Vanillin | −5 | G143, S144, C145, H163, E166 | −5.4 | Y268 (B), Y273 (B), T301 (B) |
| Cloves | Eugenol | −4.9 | L141, G143, S144, C145, H163 | −5.6 | S170 (A), C155 (B) |
| Thyme | Thymol | −4.8 | None | −5.4 | E203 (B) |
FIGURE 1.
(A) 3-D binding pocket of SARS-CoV-2 Mpro with top model PCB (cyan color), surrounding active site amino acid residues (yellow color) within 3 Å; remaining residues are represented as a cartoon; (B) 3-D binding pocket of SARS-CoV-2 PLpro with top model PCB (cyan color), surrounding active site amino acid residues (chain A, yellow color; chain B, orange color) within 3 Å; remaining residues are represented as a cartoon (chain A, green color; chain B, light pink color). Polar interactions are represented as magenta color.
To evaluate the reliability of the molecular docking and stability of docked complexes, we conducted molecular dynamics simulations with a traditional force field (CHARMM36m), starting with the docking-generated pose of Mpro-ligand complexes with binding energy cutoff values of −7.6 Kcal/mol, PLpro-ligand complexes with binding energy cut-off values of −8.0 Kcal/mol accompanying with a higher binding affinity toward Mpro. Root mean square deviation (RMSD) was measured to measure the structural conformation differences between the initial and final positions of proteins and ligands. Table 3 depicts average ligand and protein RMSD and occupancy of major hydrogen bonds for food bioactive compounds in traditional MD simulations. The data show ligand RMSD values are in the range of 0.285 ± 0.090 to 1.743 ± 0.219, protein RMSD values are in the range of 1.370 ± 0.164 to 2.298 ± 0.353 in both Mpro and PLpro complexes. These lower RMSD values indicate greater stability of protein-ligand docked complexes.
TABLE 3.
Average ligand and protein RMSD and occupancy of major hydrogen bonds for food bioactive compounds in traditional MD simulations.
| Compound | Ligand-RMSD (Å) | Protein-RMSD (Å) | Major hydrogen bonds and its occupancy (%) |
| Mpro | |||
| PCB | 1.743 ± 0.219 | 1.720 ± 0.201 | G143 (38.5), N119 (38.7), S46 (25.6), Y54 (12) |
| Quercetin | 0.342 ± 0.104 | 1.372 ± 0.143 | Y54 (18.7), E166 (10.9), S144 (4.5) |
| Cyanidin | 0.832 ± 0.252 | 1.462 ± 0.207 | H163 (34.6), H164 (13.5), G143 (6.7), N (5.9) |
| Daidzein | 0.513 ± 0.145 | 1.407 ± 0.148 | E166 (51.5), R188 (23.9), T190 (25.7), T26 (24.2), G143 (17.4) |
| Genistein | 0.391 ± 0.088 | 1.370 ± 0.164 | D187 (32), E166 (26.8), G143 (9.2), Q189 (4.2) |
| Riboflavin | 1.131 ± 0.193 | 1.482 ± 0.297 | H163 (33.9), N142 (24.7), E166 (15.9), R188 (10.2) |
| PLpro | |||
| PCB | 1.452 ± 0.125 | 2.226 ± 0.125 | D164(C) (82.1), R166(C) (57.4), D164(A) (51.9), G271(A) (21.6) |
| Quercetin | 0.875 ± 0.118 | 2.298 ± 0.353 | E203(C) (54.5), E167(A) (27.6), S170(C) (22.7), M208(C) (21.4), Y171(A) (8.1), K157(A) (5.5), |
| Cyanidin | 0.285 ± 0.090 | 1.988 ± 0.316 | E203(C) (73.7), Y264(A) (15.1) |
| Daidzein | 0.794 ± 0.088 | 2.02 ± 0.486 | D164(A) (46.6), C155(A) (30.2), K157(A) (19.2), R166(A) (18.1) |
| Genistein | 1.072 ± 0.196 | 1.864 ± 0.254 | Q269(A) (51.9), M208(C) (21), Y171(A) (16.8), K157(A) (11.3) |
Values in parentheses denotes hydrogen bonds occupancy (%).
In vitro Enzymatic Assay Studies to Screen Potent Phytochemical Inhibitor Compounds Against SARS-CoV-2 Mpro and PLpro
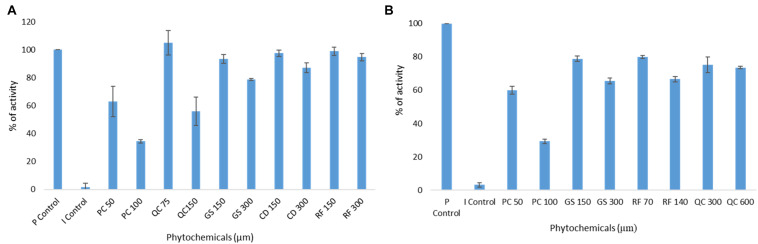
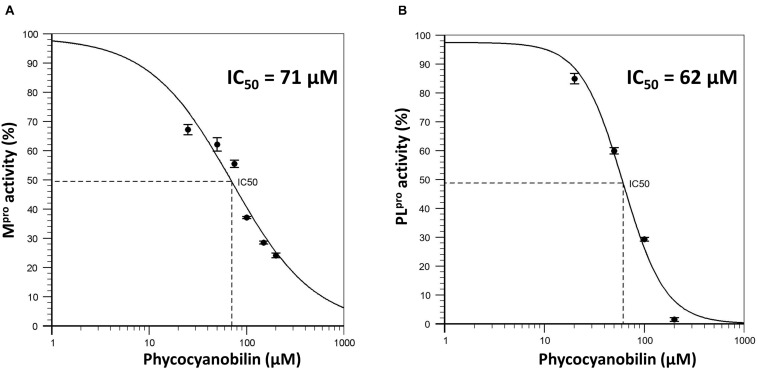
To further validate the molecular docking and molecular dynamics studies, in vitro enzymatic studies were conducted. A positive control without the inhibitor compound in the reaction mixture, an inhibitor control that contains authentic inhibitors GC376 (for Mpro), GRL0617 (for PLpro) were used in this study. The enzyme’s relative activity in the presence of inhibitors was estimated by considering positive control activity as 100%. Based on in silico studies, we selected the top six phytochemicals (PCB, quercetin, riboflavin, cyanidin, daidzein, and genistein) with a binding energy cutoff value of −7.6 Kcal/mol for Mpro enzymatic assay studies. Initial screening results revealed that PCB had higher inhibitor activity followed by quercetin, genistein, cyanidin, and riboflavin (p < 0.05) (Figure 2). Further, the IC50 value of top two compounds, PCB and quercetin, was determined, and the results show an effective IC50 value of 71 μM for PCB (Figure 3) than quercetin (145 μM) (Supplementary Figure 3). For PLpro, four compounds (Phycocyanobilin, Riboflavin, Genistein, and Quercetin) with a binding energy cutoff value of −8.0 Kcal/mol, accompanying higher inhibitor activity toward Mpro, were selected for the in vitro inhibitor activity assay (Figure 2). It was envisaged that PCB showed potent inhibitor activity compared to other compounds (Figure 2), with an IC50 value of 62 μM (Figure 3). Overall, in silico docking and in vitro enzyme inhibitor activity data show PCB as a potent inhibitor against SARS-CoV-2 Mpro and PLpro.
FIGURE 2.
Initial screening of phytochemicals (selected based on docking score and our availability) by in vitro enzymatic assays; (A) Mpro assay; (B) PLpro assay; P control, positive control; I control, inhibitor control; PC, phycocyanobilin; QC, quercetin; GS, genistein; CD, cyaniding; RF, riboflavin.
FIGURE 3.
(A) Dose response curve of Phycocyanobilin versus Mpro activity; (B) Dose response curve of Phycocyanobilin versus PLpro activity.
In silico Insights of Broad-Spectrum Inhibitor Activity of PCB Against Mpro and PLpro
The broad-spectrum efficacy of PCB against CoVs was evaluated by molecular docking studies with available crystal PDB structure of various human and animal CoVs. Table 4 shows the PDB identification code and top docking scores of PCB with Mpro and PLpro enzymes of human and animal CoVs. Due to the limitation on the availability of crystal PDB structures of PLpro, both dimer and monomeric forms were used in docking studies. For Mpro, docking scores are in the range of −8.3 to −9.3 Kcal/mol. PCB showed higher binding affinity with docking score (−9.3 Kcal/mol) for MERS Mpro followed by HCoV NL63 (−9.0 Kcal/mol) and IBV (−8.9 Kcal/mol). For PLpro, docking scores were in the range of −8.9 to −7.6 Kcal/mol. The results reveal that PCB had a higher binding affinity to the dimer form of PLpro enzymes than monomeric forms. When compared monomers only, PCB had best docking score for MERS-CoV (−8.5 Kcal/mol) followed by TGEV (−8.1 Kcal/mol) and SARS-CoV-2 (−8.0 Kcal/mol). Supplementary Figures 4, 5 show polar contacts of PCB with binding pocket key amino acid residues of Mpro and PLpro enzymes of human and animal CoVs. Surprisingly, the docking results suggest PCB as a promising broad-spectrum food bioactive inhibitor compound against CoVs proteases.
TABLE 4.
Molecular docking results of phycocyanobilin with proteases of other pathogenic human and animal CoVs.
| CoVs | PDB ID | Dock score | Polar contacts |
| Mpro | |||
| SARS-CoV-1 | 1WOF | −8.5 | Y54, N142, G143, S144, T190 |
| MERS-CoV | 5C3N | −9.3 | H41 (2), Q167, K191 (2), Q195 (2) |
| MHV | 6JIJ | −8.4 | F138, H161, E164, Q187, Q190 |
| TGEV | 2AMP | −8.3 | V26, H41, H162 |
| FIPV | 5EU8 | −8.5 | H41, T47, H162, H163, G167, Q191 |
| IBV | 2Q6F | −8.9 | F46, G141, A142, C143, E187, Q190 |
| HCoV 229 E | 3DBP | −8.3 | I140, H162, E165, G167 |
| HCoV NL63 | 5DWY | −9 | Y53, G142 (2), A143, H163, Q164 |
| HCoV HKU1 | 3D23 | −8.4 | E166 (2), S168 |
| PLpro | |||
| SARS-CoV-1 (dimer) | 2FE8 | −8.9 | K158 (A), D165 (A), E168 (A), H172 (B) |
| SARS-CoV-1 (monomer) | 2FE8 | −7.6 | L163, G164, Y269, T302 |
| SARS-CoV-2 (monomer) | 6LU7 | −8.0 | R166, G266 |
| MERS-CoV (monomer) | 4RNA | −8.5 | D164, D165, G248, G277, Y279 |
| TGEV (monomer) | 3MP2 | −8.1 | D80, H153, Q180, G182, Y184 |
| IBV (monomer) | 4 × 2Z | −7.8 | D150, F151 (2), S152, D153 |
In silico Insights Into Inhibitor Activities of Other Phycobilins
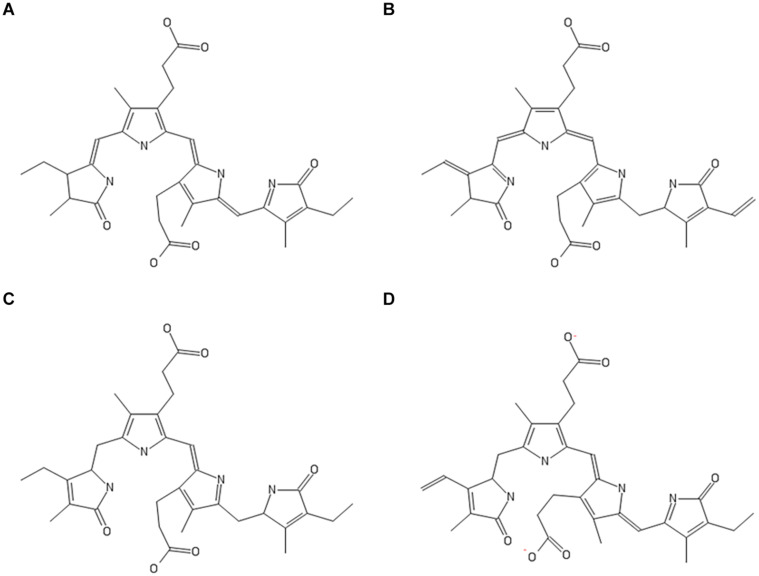
Phycobilins are linear tetrapyrrole chromophore compounds found in certain photosynthetic organisms (cyanobacteria, red algae, glaucophytes, and some cryptomonads) and covalently linked to phycobiliproteins (Beale, 1993). Four types of phycobilins are identified: (i) phycoerythrobilin (PEB), (ii) phycourobilin (PUB), (iii) phycoviolobilin (PVB), and (iv) PCB. Figure 4 represents the molecular structures of phycobilins. Based on the PCB results, the other phycobilin inhibitor activity against SARS-CoV-2 proteases via molecular docking approach was demonstrated and docking scores, polar contacts are given in Table 5. All phycobilins show strong binding affinity to key amino acids of Mpro and PLpro binding pockets. The docking scores were in the order of PUB (−8.7 Kcal/mol) >PCB (−8.6 Kcal/mol) >PEB (−8.2 Kcal/mol) >PVB (−8.2 Kcal/mol) for Mpro, whereas in the case of PLpro, the order was PCB (−9.8 Kcal/mol) = PEB (−9.8 Kcal/mol) >PUB (−9.6 Kcal/mol) >PVB (−9.5 Kcal/mol). Nine key binding pocket amino acids (Y54, L141, G143, S144, C145, H163, E166, D187, and Q189) of Mpro participated in polar contacts with phycobilins, and specific polar contacts of each phycobilin are shown in Supplementary Figure 6. Ten key binding pocket amino acids [D164 (A), Y264 (A), R166 (A), G266 (A), E161 (A), L162 (A), G271 (A), K232 (A), R166 (B), and T301 (B)] of PLpro participated in polar contacts with phycobilins; specific polar contacts are shown in Supplementary Figure 6.
FIGURE 4.
Molecular structures of phycobilins; (A) PCB; (B) PEB; (C) PUB; (D) PVB.
TABLE 5.
Molecular docking results of other phycobilins with proteases of SARS-CoV-2.
| Other phycobilins | Dock score | Polar contacts |
| Mpro | ||
| Phycoerythrobilin | −8.2 | L141, H163, E166, Q189 (2) |
| Phycourobilin | −8.7 | G143, S144, C145, H163 |
| Phycoviolobilin | −7.7 | L141, G143, S144, C145, E166 |
| PLpro | ||
| Phycoerythrobilin | −9.8 | R166 (A), Y264 (A), T301 (B) |
| Phycourobilin | −9.6 | R166 (A) (2), G266 (A) |
| Phycoviolobilin | −9.5 | E161 (A), L162 (A), G271 (A), R166 (B) (2), K232 (A) |
Discussion
Several SARS-CoV-2 mutants are spreading globally; most notably, mutants emerged in the United Kingdom (B.1.1.7), South Africa (B.1.351), and Brazil (P.1) (CDC, 2021). Hence, the efficacy of currently developed vaccines against these variants is questionable. The development of broad-spectrum antiviral drugs against existing viruses is an attractive approach that could provide first-line defense against emerging viral variants. The selection of highly conserved drug targets is an important step to develop broad-spectrum antiviral drugs. Sequence alignment analysis of SARS-CoV-2 and other CoVs (SARS-CoV and MERS) reveals that Mpro and PLpro are highly conserved, especially in the functional regions, which makes them potential targets for COVID-19 drug targets (Wu C. et al., 2020).
Several target-based in silico screening approaches were performed to find promising protease inhibitors among repurposed drugs, natural phytochemicals, and herbal medicinal compounds against COVID-19 (Amin et al., 2020; Wu C. et al., 2020; Xian et al., 2020). In this study, we focused on screening natural bioactive compounds for potential inhibitor activity against SARS-CoV-2 proteases. Amin et al. (2020) report the list of key amino acids involved in catalysis and substrate binding for Mpro (H41, C145, M49, G143, S144, H163, H164, M165, E166, L167, D187, R188, Q189, T190, A191, and Q192) and PLpro (C111, H272, D286, Y268, M208, P247, P248, T301, P248, Y264, N267, Q269, L162, C270, G271, and Y273). Our molecular docking results with natural compounds also show polar interactions in these specific substrate-binding regions for both proteases. Further in vitro validation studies found a good correlation between docking and enzymatic assay results. Both in silico and in vitro approaches show that PCB has strong inhibitor activity against both SARS-CoV-2 proteases. Shih et al. (2003) report direct antiviral activity of allophycocyanin against enterovirus 71 in human rhabdomyosarcoma cells and African green monkey kidney cells. In another study, El-Morsi et al. (2016) demonstrate the reduction of ΦX174 and MS2 by extracted phycobiliproteins from cyanobacterium Synechococcus cedrorum Sauvageau.
Though numerous research studies report potential inhibitors with major emphasis on SARS-CoV-2, limited literature is available on broad-spectrum inhibitors against a wide range of CoVs, including human and animal CoVs. For instance, Sheahan et al. (2020) report broad-spectrum antiviral activity of ribonucleoside analog β-D-N4-hydroxycytidine (NHC; EIDD-1931) against SARS-CoV-2, SARS-CoV, MERS-CoV, and bat-CoVs. This study reports the broad-spectrum activity of natural phytochemical compound PCB against 11 CoVs (seven human CoVs and four animal CoVs). The computed physical properties of phycocyanobilin show a rotatable bond count of 10, hydrogen bond donor count of five, and hydrogen bond acceptor count of seven (National Center for Biotechnology Information (NCBI), 2020), which makes multiple hydrogen bond interactions between the compound and specific amino acid residues located at the active site of the pocket of the wide range of protease enzymes. Molecular docking studies indicated that propionic carboxyl and lactam ring carbonyl oxygens of PCB are involved in polar interactions with proteases’ amino acid residues.
To investigate other structurally similar phycobilin (PEB, PUB, and PVB) inhibitor activity, we attempted molecular docking with SARS-CoV-2 proteases. The results reveal that, like PCB, all phycobilins show similar binding affinity toward Mpro and PLpro of SARS-CoV-2. Besides this, potent therapeutic properties, such as peroxy radical scavenging, inhibition of cancer cell proliferation, and platelet aggregation are reported for phycobilins (Watanabe et al., 2014). Phycobilin compounds can be directly administered orally as phycobiliproteins (a complex of phycobilins and protein). For instance, when phycocyanin is administered orally to humans, it can be digested and free phycocyanobilin released in the gastrointestinal tract (Watanabe et al., 2014). Thus, noticed therapeutic properties of phycobiliproteins might reflect the effects of their phycobilins (chromophores).
In conclusion, by using in silico (molecular docking and MD simulations), in vitro enzymatic assay screenings, we discovered PCB as potent phytochemical inhibitors to Mpro and PLpro proteases of SARS-CoV-2. Phycocyanobilin had IC50 values of 71 and 62 μM for SARS-CoV-2 Mpro and PLpro, respectively. Further PCB docking studies with other CoVs Mpro and PLpro proteases revealed its broad-spectrum inhibitor activity. A similar binding affinity of other phycobilins (PEB, PUB, and PVB) to these proteases were observed. However, in vitro enzymatic studies with Mpro and PLpro of other CoVs and in vivo studies on the inhibition of CoVs infectivity using human cells and animal models are needed. Further structure-guided development of phycobilin lead compounds could rapidly lead to discovering a single agent with clinical potential against existing and possible future emerging CoV-associated diseases.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
BP: conceptualization, methodology, investigation, visualization, data curation, and writing original draft preparation. AP: conceptualization, supervision, writing, reviewing, and editing. CD: reviewing and editing. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.645713/full#supplementary-material
References
- Alfaraj S. H., Al-Tawfiq J. A., Assiri A. Y., Alzahrani N. A., Alanazi A. A., Memish Z. A. (2019). Clinical predictors of mortality of Middle East Respiratory Syndrome Coronavirus (MERS-CoV) infection: a cohort study. Travel Med. Infect. Dis. 29 48–50. 10.1016/j.tmaid.2019.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin S. A., Banerjee S., Ghosh K., Gayen S., Jha T. (2020). Protease targeted COVID-19 drug discovery and its challenges: insight into viral main protease (Mpro) and papain-like protease (PLpro) inhibitors. Bioorg. Med. Chem. 29:115860. 10.1016/j.bmc.2020.115860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmetov L., Gal-Tanamy M., Shapira A., Vorobeychik M., Giterman-Galam T., Sathiyamoorthy P., et al. (2012). Suppression of hepatitis C virus by the flavonoid quercetin is mediated by inhibition of NS3 protease activity. J. Viral Hepat. 19 e81–e88. [DOI] [PubMed] [Google Scholar]
- Beale S. I. (1993). Biosynthesis of phycobilins. Chem. Rev. 93 785–802. 10.1021/cr00018a008 [DOI] [Google Scholar]
- Bekerman E., Einav S. (2015). Combating emerging viral threats. Science 348 282–283. 10.1126/science.aaa3778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benencia F., Courreges M. C. (2000). In vitro and in vivo activity of eugenol on human herpesvirus. Phytother. Res. 14 495–500. [DOI] [PubMed] [Google Scholar]
- Bermingham A., Chand M. A., Brown C. S., Aarons E., Tong C., Langrish C., et al. (2012). Severe respiratory illness caused by a novel coronavirus, in a patient transferred to the United Kingdom from the Middle East, September 2012. Eurosurveillance 17:20290. [PubMed] [Google Scholar]
- Carmichael L. E. (1999). Canine viral vaccines at a turning point—a personal perspective. Adv. Vet. Med. 41 289–307. 10.1016/s0065-3519(99)80022-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D. (2007). Coronavirus avian infectious bronchitis virus. Vet. Res. 38 281–297. 10.1051/vetres:2006055 [DOI] [PubMed] [Google Scholar]
- CDC (2021). Emerging SARS-CoV-2 Variants. Available online at: https://www.cdc.gov/coronavirus/2019-ncov/more/science-and-research/scientific-brief-emerging-variants.html (accessed January 28, 2021). [Google Scholar]
- Chen Y. H., Chang G. K., Kuo S. M., Huang S. Y., Hu I. C., Lo Y. L., et al. (2016). Well-tolerated Spirulina extract inhibits influenza virus replication and reduces virus-induced mortality. Sci. Rep. 6:24253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho N. J., Glenn J. S. (2020). Materials science approaches in the development of broad-spectrum antiviral therapies. Nat. Mater. 19 813–816. 10.1038/s41563-020-0698-4 [DOI] [PubMed] [Google Scholar]
- Cleri D. J., Ricketti A. J., Vernaleo J. R. (2010). Severe acute respiratory syndrome (SARS). Infect. Dis. Clin. 24 175–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clercq E. (2004). Antivirals and antiviral strategies. Nat. Rev. Microbiol. 2 704–720. 10.1038/nrmicro975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C., Günther S., Preiser W., van der Werf S., Brodt H. R., Becker S., et al. (2003). Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348 1967–1976. [DOI] [PubMed] [Google Scholar]
- El-Morsi A. A., Abd El-Hamid M. I., Abd El-Fatah W. M. (2016). Anti-hepatitis viruses of phycobiliprotiens aqueous extract of the cyanobacterium Synechococcu scedrorum Sauvageau using bacteriophages MS-2/ΦX-174 as model systems. J. Environ. Sci. 45 119–130. [Google Scholar]
- Fauci A. S., Morens D. M. (2016). Zika virus in the Americas—yet another arbovirus threat. N. Engl. J. Med. 374 601–604. 10.1056/nejmp1600297 [DOI] [PubMed] [Google Scholar]
- Feller S. E., Zhang Y., Pastor R. W., Brooks B. R. (1995). Constant pressure molecular dynamics simulation: the Langevin piston method. J. Chem. Phys. 103 4613–4621. 10.1063/1.470648 [DOI] [Google Scholar]
- Ghildiyal R., Prakash V., Chaudhary V. K., Gupta V., Gabrani R. (2020). “Phytochemicals as antiviral agents: recent updates,” in Plant-Derived Bioactives, ed. Swamy M. (Singapore: Springer; ), 279–295. 10.1007/978-981-15-1761-7_12 [DOI] [Google Scholar]
- Hafiz T. A., Mubaraki M., Dkhil M., Al-Quraishy S. (2017). Antiviral activities of Capsicum annuum methanolic extract against herpes simplex virus 1 and 2. Pak. J. Zool. 49 251–255. 10.17582/journal.pjz/2017.49.1.251.255 [DOI] [Google Scholar]
- Hamre D., Procknow J. J. (1966). A new virus isolated from the human respiratory tract. Proc. Soc. Exp. Biol. Med. 121 190–193. 10.3181/00379727-121-30734 [DOI] [PubMed] [Google Scholar]
- Hangartner L., Zinkernagel R. M., Hengartner H. (2006). Antiviral antibody responses: the two extremes of a wide spectrum. Nat. Rev. Immunol. 6 231–243. 10.1038/nri1783 [DOI] [PubMed] [Google Scholar]
- Hariono M., Abdullah N., Damodaran K. V., Kamarulzaman E. E., Mohamed N., Hassan S. S., et al. (2016). Potential new H1N1 neuraminidase inhibitors from ferulic acid and vanillin: molecular modelling, synthesis and in vitro assay. Sci. Rep. 6:38692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W., Dalke A., Schulten K. (1996). VMD: visual molecular dynamics. J. Mol. Graph. 14 33–38. 10.1016/0263-7855(96)00018-5 [DOI] [PubMed] [Google Scholar]
- Jamison J. M., Krabill K., Hatwalkar A., Jamison E., Tsai C. C. (1990). Potentiation of the antiviral activity of poly r (AU) by xanthene dyes. Cell Biol. Int. Rep. 14 1075–1084. 10.1016/0309-1651(90)90015-q [DOI] [PubMed] [Google Scholar]
- Jo S., Kim T., Iyer V. G., Im W. (2008). CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 29 1859–1865. 10.1002/jcc.20945 [DOI] [PubMed] [Google Scholar]
- Kannan S., Kolandaivel P. (2018). The inhibitory performance of flavonoid cyanidin-3-sambubiocide against H274Y mutation in H1N1 influenza virus. J. Biomol. Struct. Dyn. 36 4255–4269. 10.1080/07391102.2017.1413422 [DOI] [PubMed] [Google Scholar]
- Lai W. L., Chuang H. S., Lee M. H., Wei C. L., Lin C. F., Tsai Y. C. (2012). Inhibition of herpes simplex virus type 1 by thymol-related monoterpenoids. Planta Med. 78 1636–1638. 10.1055/s-0032-1315208 [DOI] [PubMed] [Google Scholar]
- LeCher J. C., Diep N., Krug P. W., Hilliard J. K. (2019). Genistein has antiviral activity against herpes b virus and acts synergistically with antiviral treatments to reduce effective dose. Viruses 11:499. 10.3390/v11060499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S. C., Ho C. T., Chuo W. H., Li S., Wang T. T., Lin C. C. (2017). Effective inhibition of MERS-CoV infection by resveratrol. BMC Infect. Dis. 17:144. 10.1186/s12879-017-2253-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Kong X. (2004). A new genotype of nephropathogenic infectious bronchitis virus circulating in vaccinated and non-vaccinated flocks in China. Avian Pathol. 33 321–327. 10.1080/0307945042000220697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani J. S., Johnson J. B., Steel J. C., Broszczak D. A., Neilsen P. M., Walsh K. B., et al. (2020). Natural product-derived phytochemicals as potential agents against coronaviruses: a review. Virus Res. 284:197989. 10.1016/j.virusres.2020.197989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh K., Becker W. B., Chanock R. M. (1967). Growth in suckling-mouse brain of “IBV-like” viruses from patients with upper respiratory tract disease. Proc. Natl. Acad. Sci. U.S.A. 58 2268–2273. 10.1073/pnas.58.6.2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Center for Biotechnology Information (NCBI) (2020). PubChem Compound Summary for CID 365902, Phycocyanobilin. Available online at: https://pubchem.ncbi.nlm.nih.gov/compound/Phycocyanobilin (accessed November 9, 2020) [Google Scholar]
- O’Boyle N. M., Banck M., James C. A., Morley C., Vandermeersch T., Hutchison G. R. (2011). Open Babel: an open chemical toolbox. J. Cheminform. 3:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odend’hal S. (2012). Porcine transmissible gastroenteritis virus. Geogr. Distrib. Anim. Viral Dis. 1983 329–331. 10.1016/b978-0-12-524180-9.50089-3 [DOI] [Google Scholar]
- Park S., Sestak K., Hodgins D. C., Shoup D. I., Ward L. A., Jackwood D. J., et al. (1998). Immune response of sows vaccinated with attenuated transmissible gastroenteritis virus (TGEV) and recombinant TGEV spike protein vaccines and protection of their suckling pigs against virulent TGEV challenge exposure. Am. J. Vet. Res. 59 1002–1008. [PubMed] [Google Scholar]
- Paules C. I., Marston H. D., Fauci A. S. (2020). Coronavirus infections—more than just the common cold. JAMA 323 707–708. 10.1001/jama.2020.0757 [DOI] [PubMed] [Google Scholar]
- Pedersen N. C. (2009). A review of feline infectious peritonitis virus infection: 1963–2008. J. Feline Med. Surg. 11 225–258. 10.1016/j.jfms.2008.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips J. C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., et al. (2005). Scalable molecular dynamics with NAMD. J. Comput. Chem. 26 1781–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli A. (2006). Genetic evolution of canine coronavirus and recent advances in prophylaxis. Vet. Res. 37 191–200. 10.1051/vetres:2005053 [DOI] [PubMed] [Google Scholar]
- Rut W., Lv Z., Zmudzinski M., Patchett S., Nayak D., Snipas S. J., et al. (2020). Activity profiling and crystal structures of inhibitor-bound SARS-CoV-2 papain-like protease: a framework for anti–COVID-19 drug design. Sci. Adv. 6:eabd4596. 10.1126/sciadv.abd4596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Chang J., Wang K. C., Yeh C. F., Shieh D. E., Chiang L. C. (2013). Fresh ginger (Zingiber officinale) has anti-viral activity against human respiratory syncytial virus in human respiratory tract cell lines. J. Ethnopharmacol. 145 146–151. 10.1016/j.jep.2012.10.043 [DOI] [PubMed] [Google Scholar]
- Santoyo S., Jaime L., Plaza M., Herrero M., Rodriguez-Meizoso I., Ibañez E., et al. (2012). Antiviral compounds obtained from microalgae commonly used as carotenoid sources. J. Appl. Phycol. 24 731–741. 10.1007/s10811-011-9692-1 [DOI] [Google Scholar]
- Sheahan T. P., Sims A. C., Zhou S., Graham R. L., Pruijssers A. J., Agostini M. L., et al. (2020). An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 12:eabb5883. 10.1126/scitranslmed.abb5883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih S. R., Tsai K. N., Li Y. S., Chueh C. C., Chan E. C. (2003). Inhibition of enterovirus 71-induced apoptosis by allophycocyanin isolated from a blue-green alga Spirulina platensis. J. Med. Virol. 70 119–125. [DOI] [PubMed] [Google Scholar]
- Song J. M., Lee K. H., Seong B. L. (2005). Antiviral effect of catechins in green tea on influenza virus. Antiviral Res. 68 66–74. 10.1016/j.antiviral.2005.06.010 [DOI] [PubMed] [Google Scholar]
- Totura A. L., Bavari S. (2019). Broad-spectrum coronavirus antiviral drug discovery. Expert Opin. Drug Discov. 14 397–412. 10.1080/17460441.2019.1581171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O., Olson A. J. (2010). AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoek L., Pyrc K., Jebbink M. F., Vermeulen-Oost W., Berkhout R. J., Wolthers K. C., et al. (2004). Identification of a new human coronavirus. Nat. Med. 10 368–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe F., Yabuta Y., Bito T. (2014). “Tetrapyrrole compounds of cyanobacteria,” in Studies in Natural Products Chemistry, Vol. 42 ed. Atta-ur-Rahman (Amsterdam: Elsevier; ), 341–351. 10.1016/b978-0-444-63281-4.00011-2 [DOI] [Google Scholar]
- Woo P. C., Huang Y., Lau S. K., Yuen K. Y. (2010). Coronavirus genomics and bioinformatics analysis. Viruses 2 1804–1820. 10.3390/v2081803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P. C., Lau S. K., Chu C. M., Chan K. H., Tsoi H. W., Huang Y., et al. (2005). Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 79 884–895. 10.1128/jvi.79.2.884-895.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (WHO) (2003). Consensus Document on the Epidemiology of Severe Acute Respiratory Syndrome (SARS) (No. WHO/CDS/CSR/GAR/2003.11). Geneva: World Health Organization. [Google Scholar]
- World Health Organization (WHO) (2020). Coronavirus Disease 2019 (COVID-19). Available online at: https://www.who.int/docs/default-source/coronaviruse/situation-reports/20201012-weekly-epi-update-9.pdf (accessed October 12, 2020). [Google Scholar]
- Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y., et al. (2020). Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B 10 766–788. 10.1016/j.apsb.2020.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y. M., Wang W., Song Z. G., et al. (2020). A new coronavirus associated with human respiratory disease in China. Nature 579 265–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xian Y., Zhang J., Bian Z., Zhou H., Zhang Z., Lin Z., et al. (2020). Bioactive natural compounds against human coronaviruses: a review and perspective. Acta Pharma. Sin. B 10 1163–1174. 10.1016/j.apsb.2020.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zandi K., Ramedani E., Mohammadi K., Tajbakhsh S., Deilami I., Rastian Z., et al. (2010). Evaluation of antiviral activities of curcumin derivatives against HSV-1 in Vero cell line. Nat. Prod. Commun. 5 1935–1938. [PubMed] [Google Scholar]
- Zandi K., Teoh B. T., Sam S. S., Wong P. F., Mustafa M. R., AbuBakar S. (2011). Antiviral activity of four types of bioflavonoid against dengue virus type-2. Virol. J. 8:560. 10.1186/1743-422x-8-560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., et al. (2020). Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science 368 409–412. 10.1126/science.abb3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., George S., Schmidt M. F., Al-Gharabli S. I., Rademann J., Hilgenfeld R. (2011). Peptide aldehyde inhibitors challenge the substrate specificity of the SARS-coronavirus main protease. Antiviral Res. 92 204–212. 10.1016/j.antiviral.2011.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.