Abstract
Background
Currently, there is no biologically based rationale for drug selection in migraine prophylactic treatment.
Methods
To investigate the genetic variation underlying treatment response to verapamil prophylaxis, we selected 225 patients from a longitudinally established, deeply phenotyped migraine database (N = 5983), and collected uninterrupted quantitated verapamil treatment response data and DNA for these 225 cases. We recorded the number of headache days in the four weeks preceding treatment with verapamil and for four weeks, following completion of a treatment period with verapamil lasting at least five weeks. Whole‐exome sequencing (WES) was applied to a discovery cohort consisting of 21 definitive responders and 14 definitive non‐responders, and the identified single nucleotide polymorphisms (SNPs) showing significant association were genotyped in a separate confirmation cohort (185 verapamil treated patients). Statistical analysis of the WES data from the discovery cohort identified 524 SNPs associated with verapamil responsiveness (p < 0.01); among them, 39 SNPs were validated in the confirmatory cohort (n = 185) which included the full range of response to verapamil from highly responsive to not responsive.
Results
Fourteen SNPs were confirmed by both percentage and arithmetic statistical approaches. Pathway and protein network analysis implicated myo‐inositol biosynthetic and phospholipase‐C second messenger pathways in verapamil responsiveness, emphasizing the earlier pathogenic understanding of migraine. No association was found between genetic variation in verapamil metabolic enzymes and treatment response.
Conclusion
Our findings demonstrate that genetic analysis in well‐characterized subpopulations can yield important pharmacogenetic information pertaining to the mechanism of anti‐migraine prophylactic medications.
Keywords: genomic, migraine, phospholipase‐C, prophylactic, verapamil
Whole exome sequencing in 21 definitive verapamil responders and 14 definitive non‐responders identified 524 SNPs showing significant (p < 0.01) association with verapamil responsiveness. Genotyping the 524 significant SNPs in 185 other verapamil treated patients identified 39 validated SNP's. Pathway analysis based on the 39 significant SNPs implicated the related myo‐inositol and phospholipase‐C second messenger pathways in verapamil responsiveness.
1. INTRODUCTION
Migraine has an estimated global prevalence of 14.7% (Vos et al., 2012). Its estimated cost to the United States economy is over 78 billion dollars annually (Gooch et al., 2017). Delayed therapeutic responsiveness from “trial and error” approaches among six pharmacologically distinct classes of prophylactic treatments increases both the economic burden and the cost in human suffering caused by migraine. Identification of genetic factors that correlate with drug treatment response in migraine would allow for developing models predictive of individual response to one class of treatment versus another and perhaps to the identification of the relevant therapeutic action of the drug in treatment responsiveness. Recent research has identified specific alterations in trigeminal sensory pathways (Noseda & Burstein, 2013) and other fundamental molecular processes important in migraine pathogenesis,(Kowalska et al., 2016; Takeshima, 2009). The identification of molecular mechanisms underlying migraine susceptibility might aid the choice of a particular drug in prophylactic treatment. To this end, we investigated the genetic influences on the responsiveness of a subset of migraine patients to verapamil, a papaverine derivative L‐type calcium channel blocker used for decades and shown in small controlled trials to be effective in migraine prophylactic treatment (Markley et al., 1984; Solomon et al., 1983). This study is the first of its kind in migraine.
2. METHODS
2.1. Ethical compliance
The study was approved by the Mayo Clinic Institutional Review Board and all patients that participated in this study have given written consent. DNA samples from consenting migraine patients were collected under Mayo Clinic IRB #06 002002 at the Mayo Biospecimen Processing Core (CLIA certified lab).
We first carried out whole‐exome sequencing (WES) in a discovery cohort of migraine patients who were either highly responsive or not responsive to prophylactic treatment with verapamil. Then we performed a series of genotyping analyses on identified single nucleotide polymorphisms (SNP’s) in a separate, deeply phenotyped validation cohort of patients treated with verapamil who showed the full range of possible responses to further assess genetic predisposition to therapeutic responsiveness, while also investigating the role of metabolism genetics as a factor in drug response. Finally, we conducted pathway and protein network analyses to assess how the identified SNP’s exert synergetic biological impact.
2.2. Study population
To begin a new genomic approach to migraine prophylaxis, we carefully selected a cohort of patients from whom we had comprehensive headache occurrence data and prospectively documented scalable drug response.
Since 2001, we have employed a digital management application to gather, store and analyze phenotypic and treatment response data in the Headache section of the Department of Neurology at Mayo Clinic in Rochester, MN. In this system, headache patients completed a detailed phenotyping questionnaire which yielded up to 55 separate data elements relating to their headaches including baseline headache frequency and severity in the 4 weeks prior to their period of treatment (see Figure 1). All of the data were stored in an elemental searchable database. At each subsequent visit, the number of days with headaches in the 4 weeks prior to their return visit, the subjective average pain intensity (0–10), functional severity (0–3), maximum and current doses of preventative medications and the appearance of side effects were recorded and stored. At the time of subject selection for this study, there were 5983 migraine patients whose elemental migraine phenotype was stored in the database. A subset of these patients had headaches of sufficient frequency and severity to warrant prophylactic treatment and who wished to be followed in the headache practice at Mayo Clinic. The patients were provided and instructed in the use of a headache diary which they were asked to complete over the course of their treatment. These patients took treatments such NSAIDS, acetaminophen, and triptans for breakthrough headaches but were not using any analgesic more than two days per week on average during the period of assessment of verapamil response. They were then treated with verapamil or another standard antimigraine prophylactic medication based largely on the presence or absence of factors that would preclude the use of a specific medication. The patients were then titrated using a standard protocol specific to the medication provided. They returned for follow‐up after at least 12–14 weeks. This system allowed treatment response to a given migraine prophylactic monotherapy to be identified prospectively by searching the database. Individuals selected in the cohorts were taking no other medication known to affect headache frequency and were not over‐using analgesic medications. Subjects unwilling to keep a diary or who were unable to remain on the prophylactic medication at the target dose for at least 5 weeks were excluded. Prospectively identified change in the number of headache days from baseline compared to the 4 weeks prior to return was used as the primary determinant for response to treatment (Figure 1).
FIGURE 1.
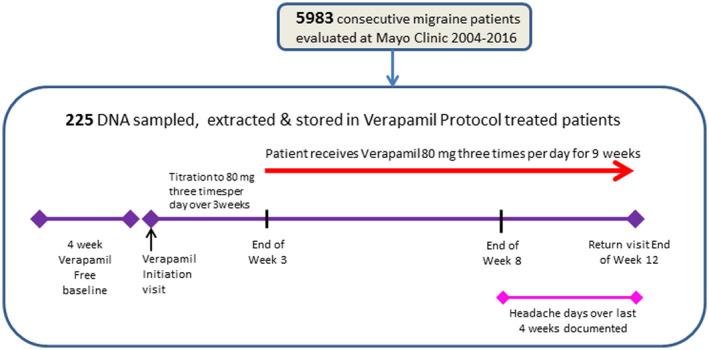
Of 5983 migraine patients evaluated at Mayo clinic from 2004 to 2016, pre‐ and post‐verapamil treatment data were obtained using the protocol shown in 380 patients, 225 of whom provided DNA samples
2.3. Sample selection
DNA was requested and obtained from willing patients either from a blood or saliva sample. At the time of subject selection, 3323 subjects had provided DNA.
2.4. Whole‐exome sequencing and bioinformatics analysis
We used the Agilent SureSelect Whole Exome enrichment system to capture exons and promoter regions for high fold coverage sequencing, and analyzed the data set with bioinformatics procedures utilizing available human genome databases. Libraries enriched for exon and exon‐flanking (i.e., promoters, UTRs and splice sites) regions were prepared from genomic DNA using the TruSeq Exome Enrichment Kit from Illumina. Sequencing was performed on an Illumina HiSeq 2000 sequencer in the Medical Genomics Facility Sequencing Core at Mayo Clinic to generate sufficient average coverage depth (150‐200X) for reliable variant calling. Secondary analyses included: alignment, variant calling (single nucleotide variation) and variant annotation. Novoalign was used to align raw reads to the human reference genome (hg19/GRCh37). Realignment and recalibration were performed using GATK. Germline variant calling and annotation were carried out using Haplotyper. We performed variant calling using the Variant Quality Score Recalibration (VQSR) package.
2.5. Sequenom MassARRAY and OpenArray genotyping
PCR primers and extension probes of Sequenom panels were designed using the Sequenom MassARRAY Design 3.0 software. PCR amplification and single‐base extension were performed. Products were then dispensed into a Spectro‐CHIP bioarray and analyzed using a MALDI‐TOF spectrometer. Genotypes were interpreted using the MassARRAY workstation software (version 3.3). OpenArray genotyping was carried out using a custom OpenArray format on a QuantStudio 12K Flex Real‐Time PCR System (ThermoFisher Scientific). Variant analysis of QuantStudio results was carried out using TaqMan Genotyper Software v1.3.
2.6. Statistical analysis
The Armitage trend test was used to assess variants in the initial WES cohort of extreme responders and non‐responders. In the subsequent larger cohort where there was the full range of potential responses, linear regression analysis in an additive genetic model was used to assess the change in the number of headache days. The SNPs were coded as the number of copies of the minor allele based on the assumption that one copy of the allele has an intermediate effect that is between the effect of no copies and two copies. Two different endpoints were used: the arithmetic change in the number of headache days pre‐ to post‐treatment (Pre‐treatment − Post‐treatment) adjusting for the pre‐treatment value in the model and the percentage change (Pre‐treatment − Post‐treatment)/(Pre‐treatment values).
2.7. Pathway and protein network analysis
We employed Ingenuity Pathway Analysis (IPA, Qiagen), a manually curated content of the Ingenuity Knowledge Base. IPA facilitated the interpretation of the genotyping data in the context of biological processes, pathways, and networks. The protein interactions were largely derived from previously published work and computational modeling. The canonical pathways included both metabolic pathways and signaling pathways. This pathway analysis also generated networks based on SNP data where the differentially regulated genes can be linked based on previously known associations and functions of the molecules involved.
3. RESULTS
3.1. WES identified 524 SNPs that significantly associated with Verapamil treatment response
We studied 5983 migraine patients (ICHD‐2 criteria) (Headache Classification Subcommittee of the International Headache Society, 2004) who were seen at Mayo Clinic and had treatment follow‐up data from 2004 to 2016. For 380 of them, we had recorded the number of headache days during the four weeks preceding treatment with verapamil, and for the four weeks following completion of a treatment period lasting at least 5 weeks with verapamil at a dose of 240 mg per day (80 mg three times per day). Among these 380 patients with verapamil‐treatment response data, we were able to obtain DNA samples from 225. A greater than 50% decrease in the number of headache days from the pre‐treatment baseline compared to the number of headache days in the month prior to follow‐up was considered to be a positive response. A decrease of ≤20% or an increase in headache days was considered to be a non‐response. Finally, a reduction of 21% to 49% was considered an intermediate response. We set out to identify genetic variants that have a significant impact on the treatment response on the genome‐wide scale. We identified a smaller cohort of extreme responders and non‐responders: 21 patients who experienced a definite, significant positive response (74% mean reduction from baseline headache frequency) after verapamil treatment, and 14 patients in whom there was little, if any, reduction (4% mean reduction) after treatment with verapamil under the same titration protocol (Figure 2). WES was then carried out in these patients and 226,585 SNPs were identified with high confidence. We subsequently applied the Armitage trend test to identify genotype differentials between the 21 definite responders and 14 definite non‐responders, and 524 SNPs were found to have a significant association with treatment response with p‐value <0.01 (Figure 3). There was no significant difference in the exposure time to the medication either in the WES subjects (responders 100.4 days SD 13.3; non‐responders 99.9 days SD 18.3) or in the genotyped cohort (responders 96.6 days SD 16.3; non‐responders 97.7 days SD 14.7).
FIGURE 2.
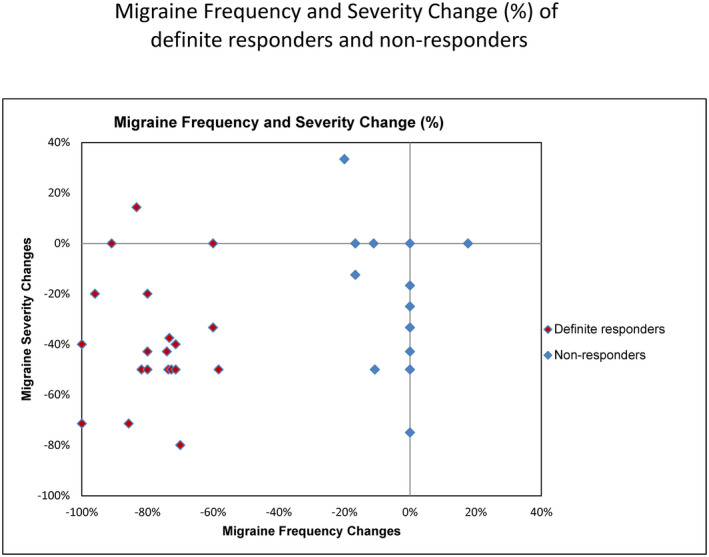
Distribution of the extreme responders and non‐responders to verapamil treatment studied in the WES phase shows a clear separation of the two groups. Subjects are plotted using the % change in the number of headache days pre‐treatment to the number of headache days post‐treatment versus % change in headache severity (0–10 visual analog scale) pre‐treatment to post‐treatment
FIGURE 3.
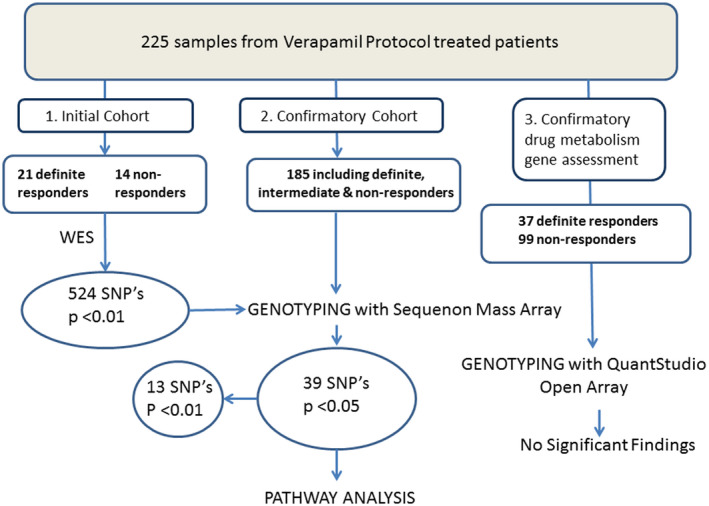
Schematic summary of the study procedures: Whole‐exome sequencing was carried out in 21 definite responders and 14 definite non‐responders to verapamil and 524 SNPs were found to have a significant association with treatment response with p‐value < 0.01. The 524 identified SNP’s were then genotyped using Sequenom custom panel approach in much larger, separate cohort of 185 patients. After genotyping 39 SNP’s attained significance of p < 0.05 and of those 13 reached of p‐value <0.01. Ingenuity pathway analysis was then performed using SNP’s with p < 0.05 and Ingenuity Protein network analysis was carried out using the SNP’s with p < 0.01. To assess whether the observed treatment response to verapamil might be related to variation in the genes involved in verapamil metabolism or cellular transport, all the definite responders and non‐responders from the combined WES and larger Sequenom MassARRAY genotyped cohorts (37 definite responders and 99 definite non‐responders) were genotyped for variants in CYP2C8, CYP3A4 and CYP3A5 as well as in the transporter gene ABCB1 (c.3435T>C, count C; rs2032582 c.2677G>T/A, count A; c.2677G>T/A, count T) with QuantStudio OpenArray
3.2. Further selection of associated variants using a separate confirmation cohort
To further evaluate and filter the impact of 524 SNPs from the discovery phase, we selected a new cohort that included 185 different patients for whom we had pre‐verapamil and post‐verapamil monotherapy data (Figure 4), and genotyped the 524 SNPs employing a Sequenom custom panel approach. There was no difference in the age at the time of treatment (39.6 years SD 12.5 responders; 38.6 SD 12.8) or the gender. (Male 17.8% responders; 14.1% non‐responders p = 0.51 Fisher's exact test) of the responder and non‐responder groups. Nor was there any difference as to the presence of migraine aura 42.9% in responders; 42.8% in non‐responders), the presence of trigeminal autonomic symptoms (33.3% in responders vs. 28.9% in non‐responders) or the pretreatment diagnosis of episodic (<15 headache days per month) versus chronic migraine (≥15 headache days per month). (Among responders: 20.7% episodic versus among non‐responders: 22.6% episodic migraine). When this much larger and new cohort was studied; only a small percentage from the initially identified 524 SNPs passed the statistical significance cutoff. Using the adjusted arithmetic results, 25 SNPs had a p‐value <0.05, among which 6 SNPs had a p‐value <0.01. In the percentage change results, 28 SNPs had a p‐value <0.05, and 10 SNPs had a p‐value <0.01. Fourteen SNPs were overlapping and present in both arithmetic and percentile analyses. Together, 39 SNPs yielded p < 0.05 and 13 SNPs yielded p < 0.01. The 13 SNPs which were highly correlated (p < 0.01) with the changes in the number of migraine headache days (post‐verapamil versus pre‐verapamil treatment) are shown in Tables 1 and 2. See Table S1 for the number of variant alleles in each sample in the confirmation cohort ranked by percent reduction post‐treatment in the subject providing the sample.
FIGURE 4.
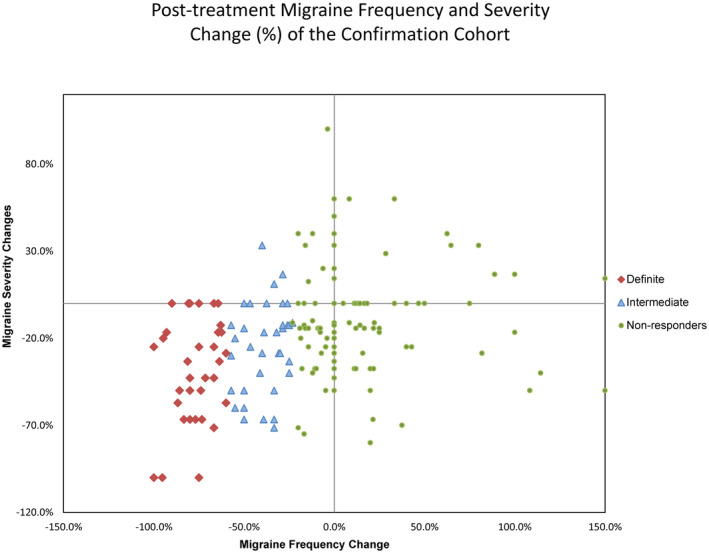
Distribution of change in pre‐ to post‐verapamil treatment data in a separate cohort of 185 different subjects in whom we genotyped the 524 SNP’s identified using WES in the discovery phase. Subjects are plotted using the % change in the number of headache days pre‐treatment to the number of headache days post‐treatment versus % change in headache severity (0–10 visual analog scale) pre‐treatment to post‐treatment
TABLE 1.
Adjusted arithmetic reduction pre‐ to post‐treatment
| SNP | Ref | Alt | Gene (GenBank Reference sequence#) | CHR | BP | MAF | Mean WT HA days reduced |
Mean_ Carrier HA days reduced |
p value | Resp. |
|---|---|---|---|---|---|---|---|---|---|---|
| rs144645569 | C | A | SRP72 (NC_000004.12) | 4 | 57,371,694 | 0.04 | 2.70 | 8.31 | 0.00809 | + |
| rs17844444 | G | A | PCDHB6 (NC_000005.10) | 5 | 140,532,165 | 0.18 | 3.98 | 1.13 | 0.00774 | − |
| rs3733694 | A | G |
PCDHB7 |
5 | 140,558,528 | 0.18 | 4.14 | 1.16 | 0.00769 | − |
| rs676947 | T | C | STRIP2 (NC_000007.14) | 7 | 129,125,825 | 0.06 | 2.40 | 8.05 | 0.00337 | + |
| rs6591182 | T | G | EHBP1L1 (NC_000011.10) | 11 | 65,349,756 | 0.49 | 1.84 | 3.53 | 0.00813 | + |
| rs2230433 | G | C | ITGAL (NC_000016.10) | 16 | 30,518,041 | 0.28 | 1.96 | 4.25 | 0.00448 | + |
The table shows for each Single Nucleotide Polymorphism, the reference and base alteration, the Gene in which the alteration resides, the chromosome, base pair, the minor allele frequency, the mean decrease in headache days from pre‐treatment to post‐treatment in individuals who had no copy of the alternate allele (reference DNA base) and the mean decrease in headache days in individuals who had at least one copy of the minor allele (DNA base alteration). These values show the magnitude and direction of change in headache days from pre‐treatment to post‐treatment in the two groups. In the arithmetic model (Table 1) the value represents the number of days reduced and in the percentage model (Table 2), the percent reduction. In treatment response column, “plus” indicates verapamil treatment results in decreased headache days.
TABLE 2.
Percentage reduction pre‐ to post‐treatment change
| SNP | Ref | Alt | Gene (GenBank Reference sequence#) | CHR | BP | MAF | Mean_WT %HA days reduced |
Mean_Carrier %HA days reduced |
P value | Resp. |
|---|---|---|---|---|---|---|---|---|---|---|
| rs10779261 | C | T | USH2A (NC_000001.11) | 1 | 216,595,306 | 0.27 | 0.20 | −0.01 | 0.00408 | − |
| rs6443266 | A | G | TTLL3 (NC_000003.12) | 3 | 9,841,989 | 0.28 | 0.22 | −0.01 | 0.00434 | − |
| rs17844444 | G | A | PCDHB6 (NC_000005.10) | 5 | 140,532,165 | 0.18 | 0.17 | −0.04 | 0.00081 | − |
| rs3733694 | A | G | PCDHB7 (NC_000005.10) | 5 | 140,558,528 | 0.18 | 0.17 | −0.03 | 0.00111 | − |
| rs17096961 | G | A | PCDHB7 (NC_000005.10) | 5 | 140,559,849 | 0.19 | 0.17 | −0.02 | 0.00340 | − |
| rs1982151 | A | G | RMI1 (NC_000009.12) | 9 | 86,617,265 | 0.27 | 0.21 | −0.03 | 0.00439 | − |
| rs10882386 | G | A | PLCE1 (NC_000010.11) | 10 | 95,790,669 | 0.24 | 0.00 | 0.23 | 0.00529 | + |
| rs1531394 | T | A | ANO3 (NC_000011.10) | 11 | 26,353,643 | 0.40 | 0.25 | 0.03 | 0.00818 | − |
| rs116903927 | A | G | KDM2A (NC_000011.10) | 11 | 67,022,766 | 0.02 | 0.12 | −0.43 | 0.00912 | − |
| rs2230433 | G | C | ITGAL (NC_000016.10) | 16 | 30,518,041 | 0.28 | 0.01 | 0.20 | 0.00266 | + |
The table shows for each Single Nucleotide Polymorphism, the reference and base alteration, the Gene in which the alteration resides, chromosome, the base pair, the minor allele frequency, the mean decrease in headache days from pre‐treatment to post‐treatment in individuals who had no copy of the alternate allele (reference DNA base) and the mean decrease in headache days in individuals who had at least one copy of the minor allele (DNA base alteration). These values show the magnitude and direction of change in headache days from pre‐treatment to post‐treatment in the two groups. In the arithmetic model (Table 1) the value represents the number of days reduced and in the percentage model (Table 2), the percent reduction. In treatment response column, “plus” indicates verapamil treatment results in decreased headache days.
There are 3 highly significant SNPs (p‐value < 0.008) in both the arithmetic and percentage change models: rs2230433 within the Integrin Subunit Alpha L gene (ITGAL) [OMIM#153370], rs17844444 in Protocadherin Beta 6 gene (PCDHB6) [OMIM#606332] and rs3733694 in Protocadherin Beta 7 gene (PCDHB7) [OMIM#606333]. The minor allele of rs2230433 was associated with a mean reduction in headache days of about 20%. For the two other SNPs, rs17844444 (PCDHB6) and rs3733694 (PCDHB7), the minor alleles predicted non‐response to verapamil.
Other SNPs whose minor alleles are associated with a reduction in mean headache days included: rs144645569 located in Signal Recognition Particle 72 gene (SRP72) [OMIM#602122], rs676947 in Striatin Interacting Protein 2 Member B gene (STRIP2) [OMIM#617919], rs6591185 in the EH Domain Binding Protein 1 Like 1 gene (EHBP1L1) [OMIM#609922] and rs10882386 in the Phospholipase C Epsilon 1 gene (PLCE1) [OMIM#608414]. For all other highly significant SNPs the minor allele corresponded to verapamil non‐response.
3.3. Protein network and pathway analysis
To identify the canonical pathways containing proteins impacted by significant SNPs, we carried out pathway analysis using Ingenuity Pathway Analysis (IPA) based on 39 SNPs from the larger, genotyped cohort with p < 0.05. The analysis ranked the associated canonical pathways by p‐value significance and ratio value (number of molecules in a given pathway that has been implicated, divided by the total number of molecules in the pathway). Pathway analysis showed the Myo‐inositol biosynthetic and Phospholipase C cascades as top pathways associated with the response to verapamil in migraine prophylaxis (Figure S1).
To better understand the molecular mechanism of the proteins implicated in verapamil treatment response, we applied IPA protein network analysis to assess the connections and networks of these proteins. Specific protein−protein interactions play important roles in the understanding of how a multiprotein network links to a canonical pathway. Thus, mapping these interactions is a key to a systems‐level understanding of molecular mechanisms. The resultant protein interaction network indicated that the genes containing identified SNPs (Tables 1 and 2) are closely linked, and ITGAL gene appears to be a highly connected player in this network (Figures S2 and S3).
3.4. Assessing genes involved in verapamil metabolism
A number of genes in the Cytochrome P450 Family (CYP2C8 [OMIM#601129], CYP3A4 [OMIM#124010], CYP3A5 [OMIM#605325]) and the transporter gene ABCB1 [OMIM#171050] are known to be involved in drug metabolism or intracellular transport of verapamil (Tracy et al., 1999; Zhao et al., 2009). To assess the impact of these genes in verapamil metabolism, we carried out a genotyping study to specifically interrogate SNPs present in these 4 genes. We identified a third cohort that included 37 definite responders and 99 non‐responders from the combined WES and Sequenom MassARRAY genotyped cohorts (14 definite responders from the WES cohort and 23 definite responders from the Sequenom MassARRAY genotyped cohort; 21 definite non‐responders from the WES cohort and 78 definite non‐responders from genotyped cohort) and then genotyped them for variants in CYP2C8, CYP3A4, CYP3A5 and the transporter gene ABCB1 (Figure 3). Specifically, 2 SNPs in ABCB1 (rs1045642 and rs2032582), 6 SNPs in CYP2C8 (rs11572080, rs72558196, rs72558195, rs1058930, rs11572103, and rs10509681), 9 SNPs in CYP3A4 (rs72552799, rs35599367, rs12721627, rs4987161, rs138105638, rs28371759, rs67784355, rs12721629, and rs4986909), and 6 SNPs in CYP3A5 (rs55817950, rs776746, rs55965422, rs10264272, rs28383479, and rs41303343) were genotyped.
Using the SNP genotyping results, haplotypes were assigned for CYP2C8, CYP3A4, and CYP3A5 based on allele structure information available through the Pharmacogene Variation Consortium (www.PharmVar.org). ABCB1 haplotypes are not well‐established, so data for each SNP was treated independently. Finally, based on the diplotype a predicted phenotype was assigned for each patient for CYP2C8, CYP3A4, and CYP3A5. The predicted phenotypes were assigned using standard clinical processes and ranged from poor metabolizer to normal metabolizer. Interestingly, our results showed there is no correlation between response to verapamil prophylaxis and genotypes of SNP’s in the genes coding for verapamil metabolic enzymes (CYP2C8 p > 0.064; CYP3A4 p > 0.916; CYP3A5 p > 0.508, or transporters (ABCB1 c.3435T>C, count C p > 0.263; ABCB1 c.2677G>T/A, count A p > 0.401; ABCB1 c.2677G>T/A, count T p.0.450). The results confirm our WES analysis results of 35 samples, which did not identify any significant associations between variation in these genes and verapamil response.
4. DISCUSSION
One of the biggest hurdles faced in the management of patients with frequent migraine attacks is the selection of the optimal prophylactic drug therapy from several unrelated classes of medications. As a consequence of current empiric treatment, patients often must endure a series of trials with medications that frequently have unpleasant side effects, are costly and often result in limited if any improvement in their migraine. We hypothesize that response to a migraine prophylactic medication is due to its compensatory or stabilizing effect on the underlying molecular driver(s) of migraine susceptibility. Therefore, the pattern of therapeutic response is tied to the underlying cause of an individual's migraine vulnerability and is a part of their unique migraine phenotype. The use of prophylactic treatments in patients is often driven by comorbidities that might also improve with treatment with the medication. For example, the use of verapamil in migraine patients with chronic diarrhea or Raynaud's disease might be advantageous. However, whether there is any correlation to response in migraine and the presence of such comorbidities is unknown and should be the focus of future study.
Verapamil might seem an unlikely choice for the first drug to study with a pharmacogenomic approach rather than one of the treatments with higher ratings in the clinical guidelines. While important in clinical practice, guidelines and published meta‐analyses are necessarily based on available trial evidence which automatically skews the recommendations in favor of drugs introduced after the mid 1990’s when a financial interest in obtaining an FDA indication for migraine treatment necessitated having evidence from larger well‐powered clinical trials. Lack of the evidence from a large trial never conducted because of a lack of funding for such trials in generic medications is not evidence of lack of beneficial effect in an individual patient. Verapamil has been used quite successfully in a subset of patients for migraine prophylaxis since the 1980s. In our practice, verapamil is frequently offered to patients who have not responded to or been intolerant to several of the treatments recommended by the guidelines. In the verapamil responsive patients, we used in the WES phase of the study, the change after treatment was often dramatic and persisted for months and in some patients for years. In fact in the WES cohort, responders had been given slightly more unsuccessful treatments prior to receiving verapamil than the non‐responder group (See Table S2) making a large placebo effect which might be seen in treatment naïve patients unlikely. This line of research rests on the hypothesis that, like other common complex genetic disorders, migraine is based on heterogeneous genetics which is reflected in the wide array of chemically varied medications that can have a beneficial therapeutic effect. Here we investigate the reason why in some patients with migraine verapamil is clearly beneficial. We are gathering samples from patients who received treatment with representatives of each of the major classes of standard migraine prophylaxis. Verapamil was chosen for study prior to current migraine prophylactic guidelines and we hope to expand our discovery approach to additional medications considered as first‐line, for example, topiramate and beta‐blockers.
The SNP most highly correlated with the reduction in headache days post‐treatment in both the arithmetic and percentage models was rs2230433 in the ITGAL gene which encodes the integrin alpha L chain of a heterodimeric integral membrane receptor protein that functions in costimulatory signaling and intercellular adhesion. The binding of a ligand to integrins triggers an “outside‐in” (Schwarz & Ginsberg, 2002) activation of a signal transduction pathway that mediates regulation of the cell cycle and movement of new receptors to the cell membrane (Giancotti & Ruoslahti, 1999). Integrins allow rapid and flexible responses to events at the cell surface. RNA expression studies have shown ITGAL is highly expressed in multiple tissues including the immune system and cerebral cortex (Sterner et al., 2012).
Also significantly associated with headache reduction was rs10882386 in the PLCE1 gene. PLCE1 encodes a phospholipase C enzyme that catalyzes the hydrolysis of 1‐phosphatidyl‐1D‐myo‐inositol 4,5‐bisphosphate (PIP2) to generate two second messengers: inositol 1,4,5‐triphosphate (IP3) and diacylglycerol (DAG) (Lopez et al., 2001; Smrcka & Brown, 2012). These second messengers, in part through modulation of intracellular calcium, regulate various processes affecting gene expression, cell growth, and differentiation. PLCE1 mRNA is also highly expressed in cerebellum and frontal cerebral cortex (Zhou & Hildebrandt, 2009).
Pathway analysis identified two molecular cascades which included the greatest number of proteins coded by genes containing SNPs highly correlated to change in headache frequency after verapamil treatment: the myo‐inositol synthetic pathway when SNPs with a correlation of p < 0.05 were included, and the phospholipase C (PLC) signaling cascade when the SNPs were further restricted to those with a correlation of p < 0.01. Interestingly, the myo‐inositol pathway produces PIP2, the substrate for the phospholipase C epsilon pathway supporting a functional connection of the two pathways in the therapeutic action of verapamil in migraine. PLC activity has been observed to increase in the cerebrospinal fluid of migraineurs during acute attacks when compared both to the interictal state and to non‐migraine control subjects (Fonteh et al., 2011). In addition, U73122, a PLC inhibitor, strongly decreases induced mast cell degranulation (Baun et al., 2012), a known inflammatory component of trigeminal activation that has been implicated in migraine pathophysiology (Dimitriadou et al., 1991). The fact that the extended myo‐inositol/phospholipase C functional molecular pathway ultimately modulates oscillations in cytosolic calcium concentration, through the release of endoplasmic reticulum calcium stores, is even more interesting when one remembers that verapamil, through its actions at L type calcium channels in the cell membrane, also modulates intracellular calcium concentration. The oscillation in cytosolic calcium, in turn, plays an important role in the activation of the protein kinase C (PKC) second messenger system. PKC pathway activation has been linked to meningeal nociception in animal models of migraine (Galeotti & Ghelardini, 2013). In addition to its known activity as an L‐type calcium channel blocker, verapamil also directly inhibits PKC activity (DePetrillo et al., 1994; Hoffman et al., 1998; Tanaka et al., 1991), suggesting two ways by which verapamil might dampen or compensate for instability caused by alterations in the myo‐inositol or PLC pathways.
Our data suggest that genetically determined instability in the myo‐inositol/phospholipase C functional molecular pathways and the subsequent instability in PKC activity underlie migraine susceptibility in the subset of migraineurs responsive to verapamil. The speculative nature of these suggestions will require basic laboratory functional assay confirmation. Although at present there is not a simple functional test for altered myo‐inositol/phospholipase C pathway function in migraine patients, the measurement of myoinositol levels in the anterior cingulate cortex has been proposed as a potential biomarker for depression in schizophrenia (Chiappelli et al., 2015). The development of a comparable functional biomarker would move us a step closer to precision medicine in migraine.
Although the details of the relationship between the pathway instabilities and migraine await further study, our findings demonstrate that genetic analysis in well‐characterized subpopulations can yield important pharmacogenetic information pertaining to the mechanism of anti‐migraine prophylactic medications.
Our findings are consistent with several examples from other diseases of an association between genetic variation and therapeutic effect (Mega JL et al., 2010; Backman et al., 2017; Lu et al., 2014; Mega et al., 2009; Muir et al., 2014; Scott et al., 2013; Wheeler et al., 2013).
CONFLICT OF INTEREST
F. Michael Cutrer reports royalties for UptoDate as author, honorarium for Advisory Board for Biohaven, no conflicts for this study. Ann M. Moyer reports no disclosures. Elizabeth Atkinson reports no disclosures. Liguo Wang reports no disclosures. Shulan Tian reports no disclosures. Yanhong Wu reports no disclosures. Ivan Garza reports royalties from UpToDate as an author no conflicts for this study. Carrie E. Robertson reports honoraria from UpToDate as an author. Advisory board for Alder, Amgen, and Eli‐Lilly no conflicts for this study. Carey Huebert reports no disclosures. Brenda E. Moore reports no disclosures. Christopher J. Klein has received educational honorarium from Akcea therapeutics no conflicts for this study.
Supporting information
Fig S1
Fig S2
Fig S3
Table S1
Table S2
ACKNOWLEDGEMENTS
Center for Individualized Medicine, Mayo Clinic. Mary Ella Jerome. Edward and Anne Armfield. Migraine Research Foundation.
DATA AVAILABILITY STATEMENT
Anonymized data not appearing in the publication will be shared by request from any qualified investigator.
REFERENCES
- .Mega, J. L. , Simon, T. , Collet, J.‐P. , Anderson, J. L. , Antman, E. M. , Bliden, K. , Cannon, C. P. , Danchin, N. , Giusti, B. , Gurbel, P. , Horne, B. D. , Hulot, J.‐S. , Kastrati, A. , Montalescot, G. , Neumann, F.‐J. , Shen, L. , Sibbing, D. , Steg, P. G. , … Sabatine, M. S. (2010). Reduced‐function CYP2C19 genotype and risk of adverse clinical outcomes among patients treated with clopidogrel predominantly for PCI: A meta‐analysis. JAMA, 304, 1821–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backman, J. D. , O’Connell, J. R. , Tanner, K. , Peer, C. J. , Figg, W. D. , Spencer, S. D. , Mitchell, B. D. , Shuldiner, A. R. , Yerges‐Armstrong, L. M. , Horenstein, R. B. , & Lewis, J. P. (2017). Genome‐wide analysis of clopidogrel active metabolite levels identifies novel variants that influence antiplatelet response. Pharmacogenetics and Genomics, 27(4), 159–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baun, M. , Pedersen, M. H. , Olesen, J. , & Jansen‐Olesen, I. (2012). Dural mast cell degranulation is a putative mechanism for headache induced by PACAP‐38. Cephalalgia, 32(4), 337–345. [DOI] [PubMed] [Google Scholar]
- Chiappelli, J. , Rowland, L. M. , Wijtenburg, S. A. , Muellerklein, F. , Tagamets, M. , McMahon, R. P. , Gaston, F. , Kochunov, P. , & Hong, L. E. (2015). Evaluation of myo‐inosital as a potential biomarker for depression in schizophrenia. Neuropsychopharmacology, 40, 2157–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePetrillo, P. B. , Abernethy, D. R. , Wainer, I. W. , & Andrawis, N. S. (1994). Verapamil decreases lymphocyte protein kinase C activity in humans. Clinical Pharmacology and Therapeutics, 55(1), 44–49. [DOI] [PubMed] [Google Scholar]
- Dimitriadou, V. , Buzzi, M. G. , & Moskowitz, M. A. (1991). Theoharides TC Trigeminal sensory fiber stimulation induces morphological changes reflecting secretion in rat dura mater mast cells. Neuroscience, 44(1), 97–112. [DOI] [PubMed] [Google Scholar]
- Fonteh, A. N. , Chung, R. , Sharma, T. L. , Fisher, R. D. , Pogoda, J. M. , Cowan, R. , & Harrington, M. G. (2011). Cerebrospinal fluid phospholipase C activity increases in migraine. Cephalalgia, 4, 456–462. [DOI] [PubMed] [Google Scholar]
- Galeotti, N. , & Ghelardini, C. (2013). Inhibition of the PKCγ‐ε pathway relieves from meningeal nociception in an animal model: an innovative perspective for migraine therapy? Neurotherapeutics: the Journal of the American Society for Experimental NeuroTherapeutics, 10(2), 329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giancotti, F. G. , & Ruoslahti, E. (1999). Integrin signaling. Science, 285, 1028–1032. [DOI] [PubMed] [Google Scholar]
- Gooch, C. L. , Pracht, E. , & Borenstein, A. R. (2017). The burden of neurological disease in the United States: A summary report and call to action. Annals of Neurology, 81(4), 479–484. 10.1002/ana.24897 [DOI] [PubMed] [Google Scholar]
- Headache Classification Subcommittee of the International Headache Society . The International classification of headache disorders, 2nd edition. Cephalalgia, 2004, 24(Supplement 1), 1–160. [DOI] [PubMed] [Google Scholar]
- Hoffman, S. , Gopalakrishna, R. , Gundimeda, U. , Murata, T. , Spee, C. , Ryan, S. J. , & Hinton, D. R. (1998). Verapamil inhibits proliferation, migration and protein kinase C activity in human retinal pigment epithelial cells. Experimental Eye Research, 67(1), 45–52. [DOI] [PubMed] [Google Scholar]
- Kowalska, M. , Prendecki, M. , Kozubskin, W. , Lianeri, M. , & Dorszewska, J. (2016). Molecular factors in migraine. Oncotarget, 7(31), 50708–50718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez, I. , Mak, E. C. , Ding, J. , Hamm, H. E. , & Lomasney, J. W. (2001). A novel bifunctional phospholipase c that is regulated by Galpha 12 and stimulates the Ras/mitogen‐activated protein kinase pathway. Journal of Biological Chemistry, 276(4), 2758–2765. [DOI] [PubMed] [Google Scholar]
- Lu, D. Y. , Lu, T. R. , & Wu, H. Y. (2014). Personalized cancer therapy: A perspective. International Journal of Pharmacy Practice and Drug Research, 4(2), 108–118. [Google Scholar]
- Markley, H. , Cheronis, J. , & Piepho, R. (1984). Verapamil prophylactic therapy of migraine. Neurology, 34, 973–976. [DOI] [PubMed] [Google Scholar]
- Mega, J. L. , Close, S. L. , Wiviott, S. D. , Shen, L. , Hockett, R. D. , Brandt, J. T. , Walker, J. R. , Antman, E. M. , Macias, W. , Braunwald, E. , & Sabatine, M. S. (2009). Cyto‐chrome p‐450 polymorphisms and response to clopidogrel. New England Journal of Medicine, 360, 354–362. [DOI] [PubMed] [Google Scholar]
- Muir, A. J. , Gong, L. , Johnson, S. G. , Lee, M. M. , Williams, M. S. , Klein, T. E. , Caudle, K. E. , & Nelson, D. R. (2014). Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for IFNL3 (IL28B) genotype and PEG interferon‐α–based regimens. Clinical Pharmacology and Therapeutics, 95(2), 141–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noseda, R. , & Burstein, R. (2013). Migraine pathophysiology: anatomy of the trigeminovascular pathway and associated neurological symptoms, CSD, sensitization and modulation of pain. Pain, 154(Suppl 1), S44–S53: 10.1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz, M. A. , & Ginsberg, M. H. (2002). Networks and crosstalk: integrin signaling spreads. Nature Cell Biology, 4, E65–E68. [DOI] [PubMed] [Google Scholar]
- Scott, S. A. , Sangkuhl, K. , Stein, C. M. , Hulot, J.‐S. , Mega, J. L. , Roden, D. M. , Klein, T. E. , Sabatine, M. S. , Johnson, J. A. , & Shuldiner, A. R. (2013). Clinical Pharmacogenetics Implementation Consortium guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 update. Clinical Pharmacology and Therapeutics, 94(3), 317–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smrcka, A. V. , Brown, J. H. , & Holz, G. G. (2012). Role of phospholipase Ce in physiologicalphosphoinositide signaling networks. Cellular Signalling, 24(6), 1333–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon, G. D. , Steel, J. G. , & Spaccavento, L. J. (1983). Verapamil prophylaxis of migraine: A double‐blind placebo‐controlled study. JAMA, 250, 2500–2502. [PubMed] [Google Scholar]
- Sterner, K. N. , Weckle, A. , Chugani, H. T. , Tarca, A. L. , Sherwood, C. C. , Hof, P. R. , Kuzawa, C. W. , Boddy, A. M. , Abbas, A. , Raaum, R. L. , Grégoire, L. , Lipovich, L. , Grossman, L. I. , Uddin, M. , Goodman, M. , & Wildman, D. E. (2012). Dynamic gene expression in the human cerebral cortex distinguishes children from adults. PLoS One, 7(5), e37714. 10.1371/journal.pone.0037714. Published online 2012 May 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshima, T. (2009). Metabolic syndrome and prevention of migraine headache. Brain Nerve, 61(10), 1143–1153. [PubMed] [Google Scholar]
- Tracy, T. S. , Korzekwa, K. R. , Gonzalez, F. J. , & Wainer, I. W. (1999). Cytochrome P450 isoforms involved in metabolism of the enantiomers of verapamil and norverapamil. British Journal of Clinical Pharmacology, 47, 545–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos, T. , Flaxman, A. D. , Naghavi, M. , Lozano, R. , Michaud, C. , Ezzati, M. , Shibuya, K. , Salomon, J. A. , Abdalla, S. , Aboyans, V. , & Abraham, J. (2012). Years lived with disability (YLDs) for 1160 sequela of 289 diseases and injuries 1990–2010: A systematic analysis of the Global Burden of Disease Study 2010. The Lancet, 380, 2163–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler, H. E. , Maitland, M. L. , Dolan, M. E. , Cox, N. J. , & Ratain, M. J. (2013). Cancer pharmacogenomics: strategies and challenges. Nature Reviews Genetics, 14(1), 23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasushi, T. , Atsunori, K. , Tsutomu, O. , Nanami, A. , Takayuki, A. , Motoyoshi, I. , Yoshihumi, T. , & Yukio, S. (1991). Effect of verapamil on cardiac protein kinase C activity in diabetic rats. European Journal of Pharmacology, 200(2–3), 353–356. [DOI] [PubMed] [Google Scholar]
- Zhao, L. M. , He, X. J. , Qiu, F. , & SunYX, L.‐L. (2009). Influence of ABCB1 gene polymorphisms on the pharmacokinetics of verapamil among healthy Chinese Han ethnic subjects. British Journal of Clinical Pharmacology, 68(3), 395–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, W. , & Hildebrandt, F. (2009). Molecular cloning and expression of phospholipase C epsilon 1 in zebrafish. Gene Expression Patterns, 9(5), 282–288. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Fig S2
Fig S3
Table S1
Table S2
Data Availability Statement
Anonymized data not appearing in the publication will be shared by request from any qualified investigator.


