Abstract
Background
COVID‐19 can present with lymphopenia and extraordinary complex multiorgan pathologies that can trigger long‐term sequela.
Aims
Given that inflammasome products, like caspase‐1, play a role in the pathophysiology of a number of co‐morbid conditions, we investigated caspases across the spectrum of COVID‐19 disease.
Materials & Methods
We assessed transcriptional states of multiple caspases and using flow cytometry, the expression of active caspase‐1 in blood cells from COVID‐19 patients in acute and convalescent stages of disease. Non‐COVID‐19 subject presenting with various comorbid conditions served as controls.
Results
Single‐cell RNA‐seq data of immune cells from COVID‐19 patients showed a distinct caspase expression pattern in T cells, neutrophils, dendritic cells, and eosinophils compared with controls. Caspase‐1 was upregulated in CD4+ T‐cells from hospitalized COVID‐19 patients compared with unexposed controls. Post‐COVID‐19 patients with lingering symptoms (long‐haulers) also showed upregulated caspase‐1activity in CD4+ T‐cells that ex vivo was attenuated with a select pan‐caspase inhibitor. We observed elevated caspase‐3/7levels in red blood cells from COVID‐19 patients compared with controls that was reduced following caspase inhibition.
Discussion
Our preliminary results suggest an exuberant caspase response in COVID‐19 that may facilitate immune‐related pathological processes leading to severe outcomes. Further clinical correlations of caspase expression in different stages of COVID‐19 will be needed.
Conclusion
Pan‐caspase inhibition could emerge as a therapeutic strategy to ameliorate or prevent severe COVID‐19.
Keywords: caspase, COVID‐19, emricasan, red blood cell, T‐helper cell
This study assesses transcriptional states of multiple caspases and the expression of active caspase‐1 in blood cells from COVID‐19 patients in acute and convalescent stages of disease. Elevated caspase‐3/7 levels in red blood cells is observed in COVID‐19 patients compared to controls. Post‐COVID‐19 patients with lingering symptoms show up‐regulated caspase‐1 activity in CD4+ T‐cells that is attenuated ex vivo with a select pan‐caspase inhibitor. An exuberant caspase response in COVID‐19 that may facilitate immune‐related pathological processes leading to severe outcomes.
Abbreviations: APC, antigen‐presenting cell; COVID‐19, coronavirus disease 2019; ICU, intensive care unit; PBMC, peripheral blood mononuclear cell; RBC, red blood cell; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2

1. INTRODUCTION
Coronavirus Disease 2019 (COVID‐19) is the latest global health threat and, as in two preceding instances of the emergence of coronavirus respiratory disease, poses critical challenges for the public health, research, and medical communities. 1 , 2 Although the pathology of COVID‐19 is now well described, the mechanisms underlying disease progression remain unclear. While a robust vaccination campaign and the further development of vaccines against SARS‐CoV‐2, the causal agent of COVID‐19, are underway, a variety of investigational therapeutic approaches are also being explored. 3 Dexamethasone, plitidepsin, and monoclonal antibody therapies, such astoclilizumab and eculizumab, have shown promise in lowering soluble inflammatory markers part of the cytokine storm and reducing severe outcomes inCOVID‐19. 4 , 5 , 6 , 7 Further elucidating effector molecules responsible for disease progression to determine effective interventions earlier in the course of the disease is needed to help design effective therapies to ameliorate disease manifestations and its complications. 8 , 9 , 10
The scope and severity of COVID‐19 vary among those infected. Some patients exhibit no or minor flu‐like symptoms and quick recovery, some have sustained fever and have persistent fatigue with a postviral syndrome, while others experience serious lung involvement that requires hospitalization that may lead to death. 11 Although the respiratory and the gastrointestinal system are initial targets for SARS‐CoV‐2, there clearly is a systemic nature to this disease in severe cases that may be driven by micro‐emboli and inflammatory processes. 12 , 13 While follow‐up in natural history studies will likely uncover additional postinfection sequelae, the notable impairment in type‐I interferon responses, neutrophils, chemokines and the rapid lymphopenia clearly plays a role in disease severity 14 , 15 , 16 highlighting the need for novel therapeutics that take into consideration the mechanism(s) of infection, viral replication, and effector pathways that lead to COVID‐19‐associated pathologies. 17 , 18
Inflammasome activation in peripheral immune cells and tissues was recently observed in COVID‐19 patients and the level of inflammasome‐derived products, including active caspase‐1, associated with disease severity and poor outcomes. 19 We recently reported that caspase‐1 expression in lymphocytes and serum IL‐18 levels are increased in liver transplant patients acutely ill with SARS‐CoV‐2 infection, suggesting that pyroptosis mechanisms may play a role in severe COVID‐19. 20 A recent study showed that SARS‐CoV‐2 infection of rhesus macaques led to an upregulation of caspase‐1molecular signature in peripheral blood cells as early as day 2 post‐inoculation. 21 Pyroptosis, also known as caspase‐1‐dependent cell death, is inherently inflammatory, triggered by various pathological stimuli (i.e., stroke, heart attack, and cancer), crucial for controlling microbial infections, 22 , 23 , 24 and characterized by rapid plasma‐membrane rupture and the release of proinflammatory intracellular contents, 25 , 26 a marked contrast to the regulated death process of apoptosis. 27 Insights into the complex activation and regulation of the inflammasome complex and the way in which COVID‐19 intersects with this pathway are an area of significant investigation 28 Thus, strategies targeting the inflammasome/pyroptosis pathway upstream of the production of the effector cytokines may be a novel approach to reverse COVID‐19‐induced immune perturbations 29 Building on our previous findings, we sought to expand our analysis to investigate caspase‐1 activity in SARS‐CoV‐2 infection, as well as the role of other caspases, including in red blood cells (RBCs) given the significance of COVID‐19‐associated coagulopathies 30 , 31
2. METHODS
2.1. Subjects
COVID‐19 patient blood samples used for immunophenotyping were obtained during patients’ visit or hospitalizations at SUNY Downstate Medical Center in New York from May through to July 2020. Patients were defined as (1) non‐hospitalized, with and without presentation of COVID‐19 symptoms and (2) hospitalized with presentation of COVID‐19 symptoms. Total of 10 ml peripheral blood from venipuncture was drawn into EDTA and Heparin‐coated vacutainer tubes for immunophenotyping and processed within 48 h of blood draw. Control blood samples from healthy volunteers without SARS‐CoV‐2 infection or comorbid conditions were collected after obtaining written informed consent. Demographics of the patients are shown in Tables 1 and 2. None of the patients or controls were HIV positive. There were no significant differences in demographics and comorbidities of the patients.
TABLE 1.
Demographics and comorbidities of COVID‐19 patients and healthy controls
| Hospitalized critical/ICU | SARS‐CoV‐2 PCR + non‐critical/non‐ICU | Healthy SARS‐CoV‐2 Ab Neg | |
|---|---|---|---|
| n | 18 | 11 | 28 |
| Age (median) | 66 | 71.5 | 52 |
| 80+ | 5 | 1 | 5 |
| 71–80 | 4 | 3 | 6 |
| 61–70 | 2 | 2 | 5 |
| 51–60 | 3 | 3 | 4 |
| 41–50 | 2 | 1 | 5 |
| 0–40 | 2 | 1 | 3 |
| Ethnicity | |||
| African/American | 11 | 4 | 10 |
| Other | 7 | 7 | 18 |
| Gender | |||
| Male | 9 | 5 | 13 |
| Female | 9 | 6 | 14 |
| Mean BMI | 31 | 29 | 26 |
| Comorbid conditions | |||
| Asthma | 3 | 3 | 0 |
| Autoimmune disease | 3 | 2 | 0 |
| Cancer | 1 | 3 | 0 |
| COPD | 8 | 3 | 0 |
| Coronary artery disease | 7 | 6 | 0 |
| Congestive heart failure | 6 | 5 | 0 |
| CKD without dialysis | 2 | 4 | 0 |
| CKD/ESRD with dialysis | 7 | 2 | 0 |
| Diabetes mellitus | 6 | 4 | 0 |
| Hyperlipidemia | 4 | 5 | 0 |
| Hypertension | 11 | 5 | 0 |
| Immune suppression | 3 | 2 | 0 |
TABLE 2.
Demographics of post‐COVID‐19 healthcare workers (HCWs)
| HCW | Comorbidities* | |
|---|---|---|
| n | 36 | |
| Age (median) | 64 | |
| 71–80 | 3 (8) | 2 (67) |
| 61–70 | 4 (11) | 2 (50) |
| 51–60 | 16 (44) | 8 (50) |
| 41–50 | 4 (11) | 1 (25) |
| 18–40 | 9 (25) | 2 (22) |
| Ethnicity | ||
| African/American | 12 (33) | 7 (58) |
| Other | 24 (67) | 8 (33) |
| Gender | ||
| Male | 15 (42) | 9 (60) |
| Female | 21 (58) | 6 (28) |
(*) Comorbidities are the same as listed in Table 1. Age group, ethnicity, and gender percentages of the total HCW numbers are shown in parenthesis in the second column. Number of comorbidities for each age group, ethnicity, and gender are shown in the third column, along with the respective percentages in parenthesis.
2.2. Flow cytometry
Whole blood was stained as per the clinical standard immunophenotyping protocols (Amerimmune LLC, Fairfax, VA). The samples were stained with the multiple antibody combinations for 30 min at 4°C. RBCs were lyzed using BD FACS lysis solution (BD Bioscience, San Jose, CA) as per manufacturer's directions. In brief, freshly obtained peripheral blood mononuclear cells (PBMC) were separated from 2 ml of whole blood within 24 h of collection and diluted 1:1 with phosphate‐buffered saline pH 7.2 (PBS) (Thermo Fisher Scientific, Carlsbad, CA) using Lymphoprep (Stem cell Technologies, Cambridge, MA) and Accuspin tubes (Sigma‐Aldrich, St. Louis, MO) as per manufacturer's directions. PBMCs were washed in PBS and resuspended in 0.5 ml PBS; 100 µl of the PBMCs was immunostained with a mixture of antibodies at 4℃ for 1 h. Cells were washed and resuspended in PBS prior to acquisition. Antibodies used for the immune phenotyping of patient samples are detailed in the Appendix S1 section of the manuscript (Appendix S1).
Apoptosis (caspase 3/7) and pyroptosis (caspase‐1) were measured by flow cytometry using fluorescent‐labeled inhibitors of caspase probe assay (FLICA; Immunochemistry Technologies, Minneapolis, MN). As a control, PBMCs were stimulated with nigericin for 2 h. FAM‐FLICA probes specific for caspase‐1 or caspase 3/7 were added to 50 µl PBMC or whole blood and incubated for 1 h at 37℃. Cells were subsequently washed and stained with a cocktail of fluorescently conjugated antibodies against CD45 PE‐CY7 [HI30], CD3 AF700 [UCHT1], CD4 PE [RPA‐T4], CD45RO PerCP‐EF710 [UCHL1], and Viability Dye 780 (Thermo Fisher Scientific, Carlsbad, CA). Red blood cells were identified as CD235‐positive cells in the experiments where caspase3/7 activity was measured. Samples were acquired on a 3 laser BD FACS Canto 10. CS&T beads (BD Bioscience, San Jose, CA) were acquired daily to ensure consistent performance of the BD FACS Canto 10 Canto10. The BD FACS Canto 10 utilized for this study has been validated for T, B, NK and dendritic cell immunophenotyping clinical diagnostic testing. Denovo FCS Express v6 clinical edition (De Novo Software, Pasadena, CA) was used for flow cytometric analyses.
2.3. Plasma experiments
Plasma was separated from whole blood following centrifugation at 960 RCF. Cells (RBC and WBC) were either incubated at 37°C alone or in the presence of trypsin for 1 h, then washed with 10 packed cell volumes of RPMI 1640 incomplete medium. Plasma was either held at room temperature (18–25°C) or heat inactivated at 56°C for 1 h. Plasma was added back to the RBC/WBC in a 1:1 ratio and incubated overnight, rocking at room temperature. RBC caspase 3/7 activity was measured as described under flow cytometry.
2.4. Public SARS‐CoV‐2 and COVID‐19 transcriptome analyses
Single‐cell RNA‐Seq data from three COVID‐19 participants that were ventilated and diagnosed with acute respiratory distress syndrome at 2–16 days after symptom onset and from 6 healthy controls were accessed from GEO 32 RNA‐Seq data from cell lines infected in vitro with SARS‐CoV‐2 was accessed from GEO: GSE147507 33 Expression values for caspase genes were normalized by DESeq2.
2.5. Ex vivo stimulation studies
Active caspase‐1 in COVID‐19 infected patient samples. Whole blood from a COVID‐19‐positive patient was either (A) untreated or (B) treated with the pan‐caspase inhibitor emricasan, EMR (Sigma Aldrich, MO, SML2227‐5MG) or selective caspase‐1 inhibitor VX765 overnight at 37 degrees in a water bath at 1 and 5 μM concentrations. Subsequently, PBMCs were purified (Accuspin System – Histopaque 1077; Sigma Aldrich, MO, A6929) and incubated with nigericin (Immunochemistry Technologies, MN) for 2 h. A Fam‐FLICA probe specific for active caspase‐1 was added to 50 µl PBMC and incubated for 1 h at 37℃. PMBCs were washed with cell wash buffer (Immunochemistry Technologies, MN) and stained with a cocktail of fluorescently conjugated antibodies against CD45 PE‐CY7 [HI30], CD3 AF700 [UCHT1], CD4 PE [RPA‐T4], CD45RO PerCP‐EF710 [UCHL1], and Viability Dye 780 (Thermo Fisher Scientific, Carlsbad, CA). Lymphocytes were identified using a standard gating schematic, which incorporated gating of lymphocytes on an FSC/SSC plot and singlets on a FSC‐A/FSC‐H plot. Lymphocytes were further identified as CD45+ on a CD45/SSC plot and subsequent CD3‐, CD3+ and CD3+CD4+ cells were identified on a CD45+ CD3/CD4 plot.
2.6. Statistical analysis
Demographic and HIV‐related characteristics were described using the median, first quartile (Q1) and third quartile (Q3) for continuous variables and frequency for categorical variables. Differences among continuous variables were evaluated by either the Mann–Whitney, student t test or Kruskal–Wallis test with Dunn's multiple comparisons. Relationships among parameters were examined by Pearson correlation for continuous variables. All statistical tests were performed with GraphPad Prism version 8.0 (Graphpad Software Inc., CA, USA). Statistical significance is indicated as *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. p‐values ≤0.100, but not significant, are noted as statistical trends.
2.7. Study approval
All clinical investigations were conducted according to Declaration of Helsinki principles. All human studies were approved by institutional review boards (IRB 269846‐10 and 1285028 protocols from State University of New York Downstate Medical Center and Amerimmune respectively). Written informed consent was received from participants prior to inclusion in the study.
3. RESULTS
3.1. Changes in intracellular active Caspase‐1 levels in immune cells in hospitalized patients with COVID‐19 disease
Transcriptional support of our previous findings showing increased gene expression of caspase‐1 in CD4+ T cells was observed using a published single‐cell RNA‐Seq immune profiling dataset of patients with moderate–severe COVID‐19 (Figure S1). 32 , 33 CD4+ T cells also showed an upregulation of caspase‐7 and ‐9 upon IFN stimulation. Interestingly, there were altered caspase gene expression in other cellular subsets, including neutrophils (all inflammatory and apoptotic caspases), plasmacytoid dendritic cells (caspases 7 and 9), and eosinophils (caspase 6). Examination of caspase gene expression levels in public transcriptome profiling datasets of in vitro SARS‐CoV‐2 infection models (Figure S2) shows further evidence of caspase gene expression upregulation upon SARS‐CoV‐2 infection in target cells.
To follow up on our previous findings of increased T‐cell caspase‐1 expression in COVID‐19, we analyzed intracellular active caspase‐1 in CD4+ T cells of non‐ICU and ICU patients with COVID‐19 and healthy individuals for comparison (Table 1) using our laboratory‐developed test (LDT) that has been analytically validated in a CLIA‐certified and CAP‐accredited flow cytometry laboratory. Frequency of caspase‐1+ CD4+ T cells was significantly elevated at baseline in hospitalized (both ICU and non‐ICU) COVID‐19 patients compared with healthy participants (Figure 1A–D; all p‐value <0.0001). Nigericin was used as a positive control, as it is crucial for oligomerization of the NLRP3 inflammasome and activation of caspase‐1, and we found that with nigericin stimulation, hospitalized COVID‐19 patients still had a higher frequency of active caspase‐1 in CD4+ T cells compared with controls (all p‐value <0.0001; Figure 1A–D). Similar findings were obtained for CD8T cells (Figure S3). However, we also found that the levels of pannexin‐1, an intermediatory protein involved in nigericin signaling‐induced caspase‐1 activation and IL‐1ß processing and release, were also elevated in COVID‐19 patients (Figure S4). Variation in the expression levels of pannexin‐1 between individual healthy and COVID‐19 subjects may explain the differences in the nigericin response to upregulate T‐cell caspase‐1.
FIGURE 1.
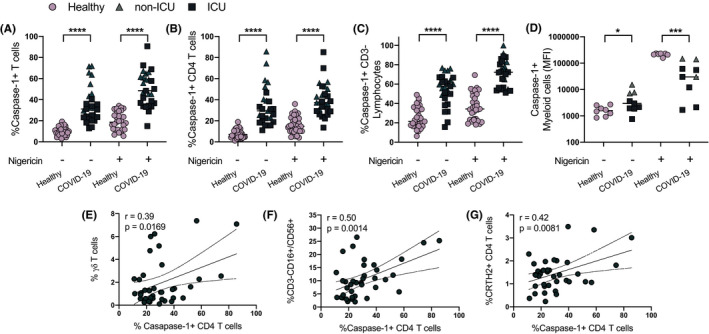
Caspase‐1 expression in immune cells. Caspase‐1 expression is shown for total (CD3+) T cells, CD4+ T cells, non‐CD3+ T‐cell lymphocytes, and myeloid cells (A‐D). Caspase‐1 expression in CD4+ T cells is correlated with γ/δ T cells, NK cells (CD3‐CD56/16+), and CRTH2+ CD4+ T cells (E‐G). Individual patient data are shown and the frequency of caspase‐1+ CD4+ T cells is significantly elevated at baseline in COVID‐19 patients (n = 29) compared with healthy (n = 28) participants with and without nigericin stimulation. All p‐values are by unpaired and 2‐tailed Student's t test or linear regression analysis. *p < 0.05, ***p < 0.001, ****p < 0.0001
We next correlated active caspase‐1 with cellular subsets and cytokines associated with its activation. We observed that active caspase‐1 expression is predominant in the CD45RO+ memory population and showed a weak but statistically significant correlation with older age, a finding that might potentially explain advanced age as one of the biggest risk factors for poor outcomes in COVID‐19 (Table S1). Furthermore, our preliminary data show that CD4+ T‐cell active caspase‐1 levels in patients with COVID‐19 were correlated with CRTH2+ T‐cells, γ/δT‐cells, CD3‐CD16+/CD56+ lymphocytes, and plasmacytoid dendritic cells (Figure 1E–G and Table S1). CD4+ T‐cell active caspase‐1 expression directly correlated with elevated serum levels of IL‐18 in hospitalized COVID‐19 individuals (Figure S5).
Next, we wanted to determine if elevated active caspase‐1 activity in CD4+ T cells is unique to COVID‐19 patients. We assessed caspase‐1 activity in pediatric or adult non‐COVID‐19 patients (n = 104), including those who presented chronic sinusitis, asthma, common variable immune deficiency, or chronic idiopathic urticaria based on ICD‐10 diagnosis codes indicated in the patient's chart, in which T‐cell caspase‐1 measurement was performed as a part of patient care during immunological work‐up (Figure 2). Among adults, there was a statistically significant elevation of baseline CD4 T‐cell caspase‐1 in only asthmatics (p < 0.001), and nigericin stimulated CD4 T‐cell caspase in asthmatics (p < 0.0001) and chronic rhinosinusitis (p < 0.05). There was no elevation of baseline CD4 T‐cell caspase‐1 in any of the disease categories in the pediatric population; however, nigericin stimulated CD4 T‐cell caspase was elevated in pediatric asthma and common variable immune deficiency (p < 0.001), further providing preliminary evidence on the role of active caspase‐1 in this high‐risk population.
FIGURE 2.
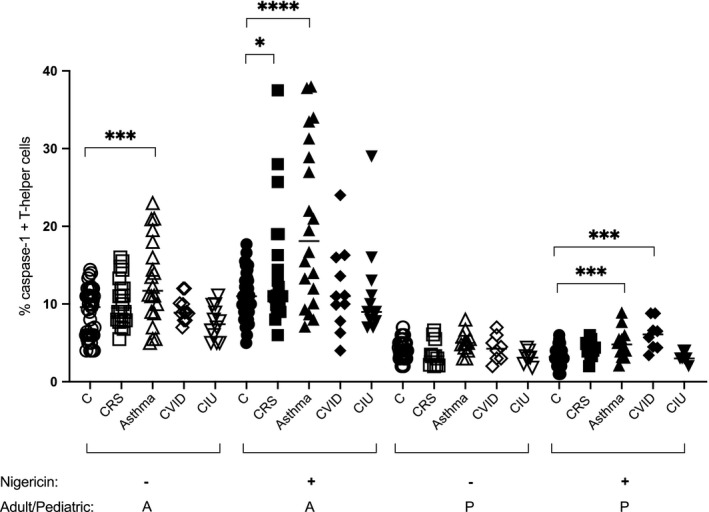
Caspase‐1 expression in CD4+ T cells of non‐COVID‐19 patients (unexposed and uninfected adult and pediatric patients with allergic/immunological disorders). Open symbols are resting nonstimulated CD4+ T cells. Closed symbols represent caspase‐1 expression in nigericin‐stimulated CD4+ T cells. Different symbols represent different disease states. Adults (>18 years) are represented with an A (n = 65) and pediatric subjects (<18 years) represented with a P (n = 39) in the bottom of the graph. CRS; chronic rhinosinusitis, CVID; common variable immune deficiency, and CIU; chronic idiopathic urticaria. The diagnosis and T‐cell caspase‐1 data are retrospective data from medical records of patients presenting to an allergy immunology clinic for an immunological evaluation. Control patient data were generated during clinical assay validation of T‐helper cell caspase‐1 assay (n = 45 for adults and n = 39 for pediatrics). All p‐values are by unpaired and 2‐tailed Student's t test. *p < 0.05, ***p < 0.001, ****p < 0.0001
3.2. Caspase‐1 upregulation is not limited to the acute stage of COVID‐19 disease
Up to 87% of inpatients and 35% of outpatients who recover from COVID‐19 report persistence of at least 1 symptom, particularly fatigue and dyspnea 34 , 35 Although preliminary reports describe this new feature as “post‐COVID‐19 syndrome” [also known as long COVID or Post‐Acute Sequelae of SARS‐CoV‐2 infection (PASC)], its mechanisms and natural history remain unknown. We assayed caspase‐1 activity inCD4+ T cells of healthcare workers (HCWs) with persistent respiratory and/or neurological (fatigue) symptoms at least 90 days post‐SARS‐CoV‐2 infection (Table 2). There was a statistically significant upregulation of baseline as well as nigericin‐stimulated CD4 T‐cell caspase‐1 levels only in symptomatic “post‐COVID‐19” HCWs, also known as long haulers (Figure 3). An asymptomatic group of patients with history of PCR+virus infection and no flu‐like illness preceding the PCR test and absent seroconversion showed elevated CD4 T‐cell caspase‐1 expression. Interestingly, PCR‐negative symptomatic HCWs with history of flu‐like illness in early 2020 as well as those with positive IgG to SARS‐CoV‐2 also had increase caspase‐1 expression. The level of expression of nigericin‐stimulated caspase‐1 was comparable to those with active infection as seen in Figure 2, although the baseline caspase‐1 levels are lower in long haulers. Non‐exposed control subjects showed no T‐cell caspase‐1 overexpression. The non‐exposed subjects were identified from a different geographical area (Fairfax, Virginia, USA) at a time when the pandemic was at its lowest numbers (August–September 2020). These subjects were also exercising strict self‐isolation.
FIGURE 3.
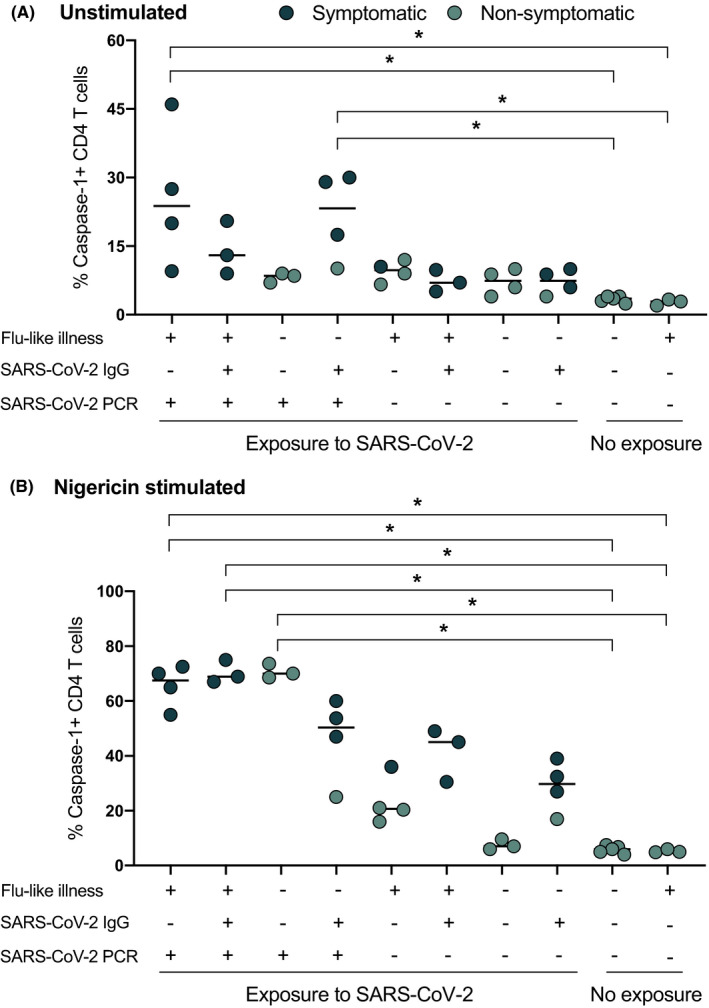
Caspase‐1 expression on CD4+T cells in post‐COVID‐19 Health Care Workers. Blood samples were analyzed at least 90 days after SARS‐CoV‐2 exposure in healthcare workers. Patients with no exposure history and negative PCR to SARS‐CoV‐2 were used as controls. Solid black circles represent symptomatic patients and green circles represent nonsymptomatic patients. Exposure indicates being in close proximity to SARS‐CoV‐2‐infected patients in the absence of personal protection equipment. Persistent post‐COVID19 symptoms correlated with elevated caspase‐1 expression in T‐helper cells (p < 0.05)
3.3. Pan‐caspase inhibitor suppresses elevated caspase‐1 activity in CD4+T cells derived from moderate–severe COVID‐19 patients
To assess whether CD4+Tcell caspase‐1 activity can be suppressed by small‐molecule caspase inhibitors, we incubated whole‐blood samples with either the oral pan‐caspase inhibitor emricasan (EMR) 36 or the selective orally active ICE/caspase‐1 inhibitor VX765, 37 followed 24 h later with or without nigericin stimulation. We found that EMR suppressed CD4+ T‐cell caspase‐1 activity in COVID‐19 samples or prevented its upregulation in healthy subjects (Figure 4), while VX765 showed only minimal suppressive effect.
FIGURE 4.
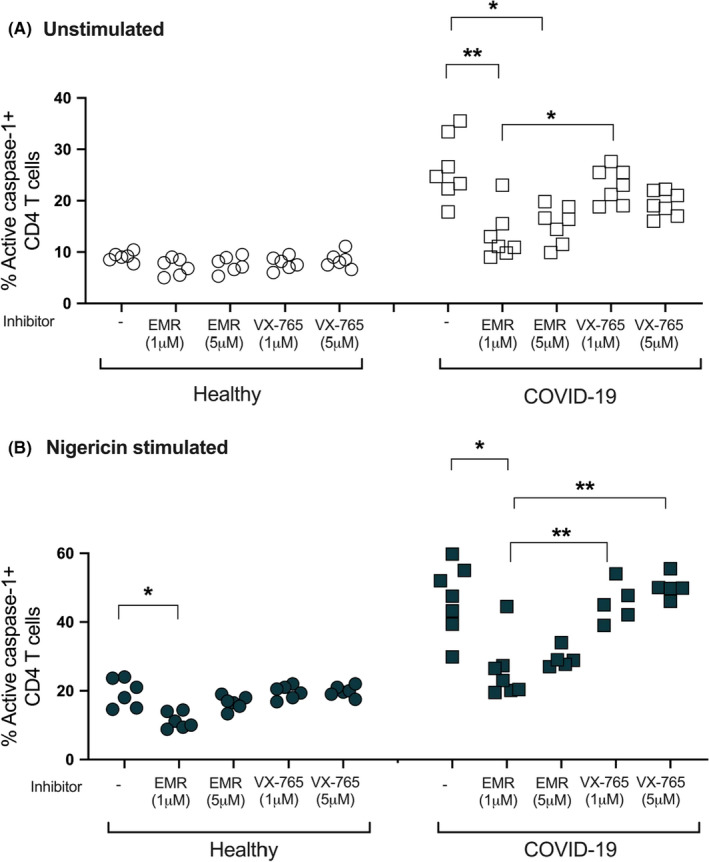
Effect of caspase inhibition on CD4+ T cells in COVID‐19 patients. Samples from healthy and COVID‐19 subjects incubated with caspase inhibitors: EMR or VX765. Activated caspase‐1 was measured by flow cytometry using a Fam‐FLICA probe. Emricasan (EMR) at 1 μM concentration induces the strongest suppression of CD4+ T‐cell caspase‐1 in unstimulated cells (p < 0.01), whereas the selective caspase‐1 inhibitor VX‐765 does not induce a similar effect. Kruskal–Wallis ANOVA test with Tukey multiple comparisons for >2 group comparisons was used. p values are as follows: *p < 0.05, **p < 0.01. Experiments represent n = 3
3.4. Red Blood cells show increased caspase‐3/7 activity in COVID‐19 disease which is suppressed by a pan‐caspase inhibitor
Recent reports suggest abnormalities in the RBCs in patients with COVID‐19. 38 , 39 , 40 In the process of Ficoll separation, we observed a layer of RBCs contaminating the PBMC layer that was universally present in all samples from COVID‐19 individuals (Figure 5A). Although we did not quantify the degree of RBC contamination, this finding was also present in up to 80% of COVID‐19 convalescent subjects. Plasma from acutely infected COVID‐19 subjects induced a similar finding when incubated overnight with plasma‐depleted whole blood of healthy patients. Treatment of the plasma samples with trypsin, DNAse, or heat inactivation did not abolish this effect (data not shown), suggesting a cell intrinsic process rather than cell surface changes. Cellular caspases are not limited to immune cells. RBCs do not express caspase‐1, but have been shown to have detectable caspase‐3 that increases with various disorders. 38 We found that RBCs from acute COVID‐19 subjects showed upregulation of caspase‐3/7 activity compared with healthy controls (Figure 5B). Plasma from these patients also upregulated caspase‐3 in healthy subjects’ RBCs. When healthy subjects’ RBCs were incubated with plasma from influenza‐infected patients, this effect was not observed, although a similar RBC contamination was observed in these samples after Ficoll separation. Furthermore, EMR suppressed the caspase‐3 upregulation in samples incubated with COVID‐19 patient‐derived plasma, but did not change the baseline expression levels in influenza‐plasma incubated samples.
FIGURE 5.
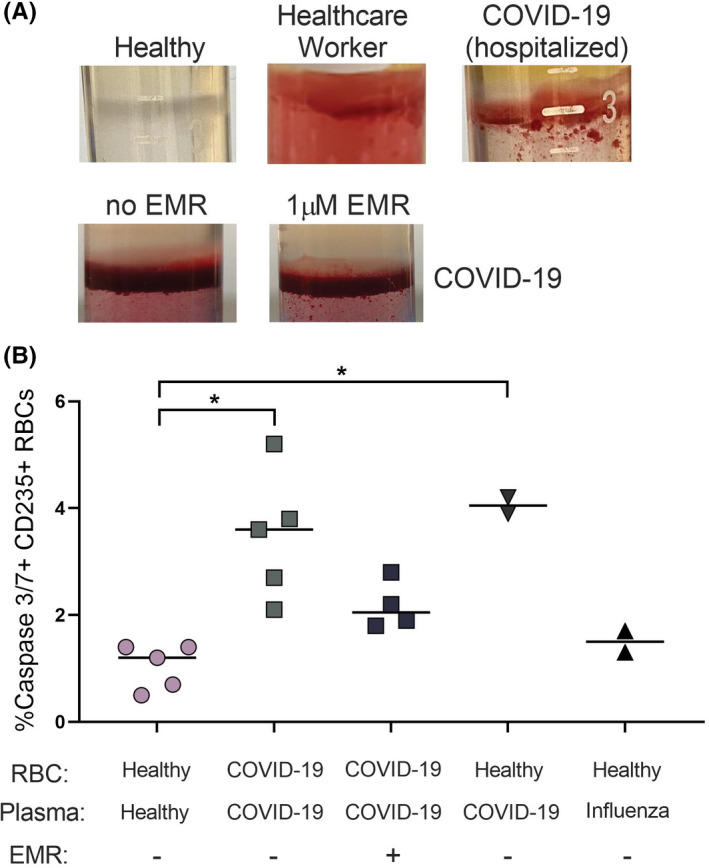
Caspase 3/7 activity in red blood cells (RBC) derived from COVID‐19 patients. Blood samples were analyzed from hospitalized patients with SARS‐CoV‐2 infection. (A) RBC contamination of the PBMC layer after Ficoll separation. (B) Analysis of caspase 3/7 activity in COVID‐19 patients and healthy controls. Some experiments were performed using plasma from COVID‐19 or subjects with influenza incubated with RBCs from healthy uninfected donors as indicated. COVID‐19 patients’ RBCs show elevated caspase 3/7 (p < 0.01) and EMR has a significant suppressive activity on this expression (p < 0.05). Plasma from hospitalized COVID‐19 patients induces caspase 3/7 in healthy RBCs on overnight incubation (p < 0.01). Kruskal–Wallis ANOVA test with Tukey multiple comparisons for >2 group comparisons was used. p‐values are as follows: *p < 0.05, **p < 0.01. Experiments represent n = 3. The photos are representative and have been reproduced in the relevant patient categories
4. DISCUSSION
While COVID‐19 disease presents primarily with respiratory symptoms, for many patients including children, it is a systemic disease with a wide range of effects on many organs. 41 , 42 , 43 In this report, we show preliminary evidence for association of caspase molecules that play a role in cell death and immunity, not only in the acute phase but also in late stages of COVID19. 44 The changes are seen in multiple caspase molecules, and a number of different circulating blood cells, a finding that will further lead to exploration of the systemic nature of this disease.
Caspase‐1 has been proposed to play a role in the pathophysiology of COVID‐19. 45 In addition to leading to a lytic form of cell death called pyroptosis, caspase‐1 induces the formation of biologically active IL‐18 and IL‐1β. 28 , 46 IL‐18 induces an IFN‐γ response, while IL‐1β induces neutrophil influx and activation, T‐ and B‐cell activation, cytokine and antibody production, and promotes Th17 differentiation. 47 , 48 , 49 , 50 Elevated levels of IL‐18, IL‐1β, and other proinflammatory cytokines were observed from the lungs and sera of COVID‐19 patients. 51 Although activation of the inflammasome enhances immunity against pathogens, the accompanying danger and inflammatory signals originating from pyroptosing immune system cells (e.g., T‐cell and macrophage/dendritic cells) can be damaging to the host in several ways. First, it will result in immune cell lymphopenia, such as that observed with T cells, a pathognomonic feature for SARS‐CoV‐2, creating an adaptive immune defect. Second, the host will have difficulty controlling the inflammation created in the setting of this immune deficiency as the “danger signals” would also be originating from dying immune system cells. 52 , 53 , 54 The end result is likely a self‐damaging shut down of the immune system, resulting in acute virus‐induced immune deficiency (AVID). Preventing the pyroptotic lymphocyte death by using caspase inhibitors may lead to better success rather than inhibition of the inflammatory response from the cell death itself. The failure of cytokine‐targeted therapies could be as a result of that adaptive immune dysfunction caused by AVID weighing more heavily than an inflammatory response in disease progression. 20
In our activeCD4 T‐cell caspase‐1 assay, we analyzed the sensitivity to nigericin stimulation. This provided further information on the cell surface pannexin‐1 expression, which is upregulated by cellular caspases and can play a role in disease pathogenesis. Also, cell surface expression level of pannexin‐1 can vary between healthy controls, which can explain the differences in response to nigericin. We also found that EMR is effective in reducing active caspase‐1 CD4 T cells from COVID‐19 patients, while VX765 failed to significantly do so. VX765 is a prodrug that needs hydrolyzation to form into its active form and is a reversible inhibitor of caspase‐1, as opposed to EMR which is an irreversible inhibitor. Furthermore, EMR is transported into cells via active transport with cell membrane channels, whereas VX765 is internalized by passive diffusion. All these factors may explain the differences we see between these two caspase inhibitors. Further studies are needed to explain the differences in inhibition between the two molecules, particularly in the context of SARS‐CoV‐2 infection.
The changes in caspase expression are not only limited to CD4 T cells, as we show changes in caspase‐3 in RBCs and caspase‐5 in neutrophils. Caspase activation has been shown to induce changes in the RBC morphology, 55 , 56 , 57 which can explain the contamination of the PBMC layer during cell separation as a result of a reduction in their density. Furthermore, their overexpression of caspase 3/7 can subsequently contribute to the formation or advancement of inflammatory microvascular thrombi, which is prominently found in the lung, kidney, and heart of patients with COVID‐19. 58 , 59 Although viral illnesses typically will impact the function or the life cycle of lymphocytes, presenting with either lymphocytosis, such as in CMV, influenza, varicella, or more rarely, lymphopenia, as in H5N1, H1N1, HIV, the finding of neutrophilia in the setting of moderate‐to‐severe COVID‐19 has been a common, but intriguing finding. 60 In the absence of significant overexpression of apoptotic caspases, the increase in the inflammatory caspase‐5 in neutrophils may play a part in the neutrophilia observed with COVID‐19. Furthermore, the production of IL‐10 by neutrophils with increase caspase activity, can further suppress the proliferation of T‐lymphocytes, hence contributing to the adaptive immune deficiency. The differential expression of caspases and their interplay in different blood and/or tissue cells need further identification, as it may impact the acute and chronic complications from this disease.
Caspase molecules have been studied extensively in many forms of inflammatory conditions, such as obesity, diabetes and nonalcoholic statohepatitis (NASH). 61 , 62 , 63 Caspase‐1‐dependent inflammasome activation has been shown to have a crucial function in the establishment of diabetic nephropathy. 64 In an animal model of hypertension, apoptosis of myocardial cells was demonstrated, and the apoptosis becomes more serious with the constantly elevated level and prolonged duration of hypertension. The activity of caspase‐3 was shown to have a close correlation with cardiomyocyte apoptosis. 65 Our data showing increased expression of active caspase‐1 in T‐helper cells of patients with asthma and immune deficiencies correlate with their high‐risk classification for severe COVID‐19 as provided by the centers for disease control (CDC). Perhaps, the changes in cellular caspases seen in COVID‐19 may not only explain the multisystem involvement in this disease, but may allow for identification for those at risk for complications, including long haulers, based on caspase expression in blood cells.
Our findings suggest a novel alternate therapeutic approach against COVID‐19 through the use of caspase inhibition early on in the course of infection to alleviate or prevent disease progression. As an oral formulation, EMR has been shown to reduce serum markers of apoptosis (caspase‐3/7), liver enzymes, function (e.g., reducing alanine aminotransferase, model for end‐stage liver disease & Child‐Pugh scores, international normalized ratio, and total bilirubin), and inflammatory biomarkers (CK‐18) in patients w/ hepatitis C virus and NASH. 66 Although there was no improvement in liver histology, it is possible that the pathology of this disease has mechanisms that are caspase‐independent or with the timing of therapy. 67 , 68 Although SARS‐CoV‐2 does not seem to infect immune system cells (with the possible exception of macrophage or dendritic cells), the outcome of T‐cell depletion in severe forms of the disease seems to be through caspase‐1 activation, a mechanism also proposed in HIV. 69 A better understanding of the impact of different comorbid conditions on T‐cell caspase expression at baseline, before exposure to SARS‐CoV‐2, may identify those who are at highest risk for developing severe disease. There is a large body of evidence pointing out an activated inflammasome in a wide variety of disorders that overlap with high‐risk conditions for severe COVID‐19. 15 , 53 , 54 , 70 Ultimately in vivo clinical data are necessary to test the hypothesis of whether pan‐caspase inhibition can prevent inflammasome activation in early onset SARS‐CoV‐2 patients and subsequent lymphopenia and sequelae development. Furthermore, the pan‐caspase inhibitor, EMR, has been shown in a bioinformatics computational screen to bind to the COVID‐19 receptor ACE2, suggesting a potential block to cell entry. 71 In a separate unrelated study, a screening of ~6,070 drugs with a known 28 previous history of use in humans was conducted to identify compounds that inhibit the activity of SARS‐CoV‐2 main protease Mpro in vitro. 72 EMR was shown to be among 50 compounds with activity against Mpro with an overall hit rate <0.75%. Preliminary evidence on this multimodal therapeutic effect of EMR raises a relevant key question that will need to be answered through a randomized clinical trial in the setting of COVID‐19 (Graphical Abstract).
Caspase plays important role in the initiation of immune responses. The interplay of caspases during the acute stage of a viral infection, however, involves a complex set of events that can have a positive or a negative impact on the immunity against the infection as well as the damage created on the host. 73 There are a number of mouse model studies suggesting that CD8+ T cells early on involve a number of cell expansion cycles that are under control of caspase 8. 74 At the same time, caspase‐1 is a key component in inflammasome, strongly promoting severe inflammation, while the proapoptotic caspases, such as caspase 3/6/7/9, lead to cell death, including that of WBCs. The latter mechanisms may be responsible for the leukopenia seen on severe COVID‐19. While the ultimate answer lies in a carefully monitored clinical study of caspase inhibitors, such as emricasan, in mild COVID‐19 patients, there are a number of clinical studies with emricasan in liver disease, including hepatitis patients that suggest otherwise. These studies hint that even in severe liver and kidney function compromised patients, emricasan treatment was well tolerated and even after prolonged treatment and at high doses did not result in an elevated infection risk. 75
An important aspect of our study is the demonstration of caspase‐1 expression well past the acute stage of COVID‐19, suggesting a role in the convalescent phase or disease sequelae. Such persistent changes cannot only be limited to immune system cells but also can be seen in tissues such as endothelial cells, which could be a causal impact on multiple organ systems. 21 , 41 , 76 Assessing the sequelae, such as fatigue, dyspnea, cough, joint pain, anosmia, among others, 34 , 77 in correlation with the changes in caspase molecules in natural history studies, is warranted. Sequelae targeting populations where caspase elevations are more common, such as the elderly, and those with other comorbid conditions, such as heart disease, diabetes, hypertension, provide further evidence for the association of caspases with poor outcomes from COVID‐19. 1 Dampening the inflammatory response with a caspase inhibitor early in the disease process may be a strategy to prevent sequelae, such as in rheumatic fever, where treating streptococcus early on in the disease through the co‐treatment of penicillin and antiinflammatories can prevent severe disease sequelae. Currently, as of April 2021, a phase 1 clinical trial, the PACE study, on the safety and tolerability of the pan‐caspase inhibitor emricasan, is enrolling patients with mild COVID‐19 (NCT04803227).
AUTHOR CONTRIBUTIONS
MP, TV, KL and PC conducted experiments and acquired data. MP, OA, TAP, and ZB analyzed the data. MP, OA, MJC, KL, PC, TV, SM, TAP, APSP, ZB, STY, THE, GN, ML, PM, AS, NR, DL, LCN, and RG wrote the manuscript. OA, MP, LCN and RG contributed to the study design and concept. All authors reviewed the manuscript.
Supporting information
App S1
ACKNOWLEDGEMENTS
The authors wish to acknowledge all the study participants and multiple parties who have advanced the science around caspase biology. This study is dedicated in memory of those who did not survive COVID‐19.
Plassmeyer M, Alpan O, Corley MJ, et al. Caspases and therapeutic potential of caspase inhibitors in moderate–severe SARS‐CoV‐2 infection and long COVID. Allergy. 2022;77:118–129. 10.1111/all.14907
Funding information
None for the study
REFERENCES
- 1. Fauci AS, Lane HC, Redfield RR. Covid–19 ‐ Navigating the uncharted. N Engl J Med. 2020;382(13):1268‐1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Paules CI, Marston HD, Fauci AS. Coronavirus infections‐more than just the common cold. JAMA. 2020;323(8):707‐708. [DOI] [PubMed] [Google Scholar]
- 3. Barlow A, Landolf KM, Barlow B, et al. Review of emerging pharmacotherapy for the treatment of coronavirus disease 2019. Pharmacotherapy. 2020;40(5):416‐437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Group RC , Horby P, Lim WS, Emberson JR, et al. Dexamethasone in hospitalized patients with Covid‐19 ‐ preliminary report. N Engl J Med. 2020;384(8):693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Xu X, Han M, Li T, et al. Effective treatment of severe COVID‐19 patients with tocilizumab. Proc Natl Acad Sci USA. 2020;117(20):10970‐10975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Spinner CD, Gottlieb RL, Criner GJ, et al. Effect of Remdesivir vs standard care on clinical status at 11 days in patients with moderate COVID‐19: a randomized clinical trial. JAMA. 2020;324(11):1048‐1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. White KM, Rosales R, Yildiz S, et al. Plitidepsin has potent preclinical efficacy against SARS‐CoV‐2 by targeting the host protein eEF1A. Science. 2021;371(6532):926‐931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Alijotas‐Reig J, Esteve‐Valverde E, Belizna C, et al. Immunomodulatory therapy for the management of severe COVID‐19. Beyond the anti‐viral therapy: a comprehensive review. Autoimmun Rev. 2020;19(7):102569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Baum A, Fulton BO, Wloga E, et al. Antibody cocktail to SARS‐CoV‐2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020;369(6506):1014‐1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Soy M, Keser G, Atagunduz P, Tabak F, Atagunduz I, Kayhan S. Cytokine storm in COVID‐19: pathogenesis and overview of anti‐inflammatory agents used in treatment. Clin Rheumatol. 2020;39(7):2085‐2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Becker RC. COVID‐19 update: Covid‐19‐associated coagulopathy. J Thromb Thrombolysis. 2020;50(1):54‐67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hasoksuz M, Kilic S, Sarac F. Turk J Med Sci. 2020;50(SI‐1):549‐556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pouletty M, Borocco C, Ouldali N, et al. Paediatric multisystem inflammatory syndrome temporally associated with SARS‐CoV‐2 mimicking Kawasaki disease (Kawa‐COVID‐19): a multicentre cohort. Ann Rheum Dis. 2020;79(8):999‐1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wu JT, Leung K, Bushman M, et al. Estimating clinical severity of COVID‐19 from the transmission dynamics in Wuhan, China. Nat Med. 2020;26(4):506‐510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497‐506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mehta P, McAuley DF, Brown M, et al. COVID‐19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395(10229):1033‐1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Guéant JL, Guéant‐Rodriguez RM, Fromonot J, et al. Elastase and exacerbation of neutrophil innate immunity are involved in multi‐visceral manifestations of COVID‐19. Allergy. 2021;76(6):1846‐1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pairo‐Castineira E, Clohisey S, Klaric L, et al. Genetic mechanisms of critical illness in COVID‐19. Nature. 2021;591(7848):92‐98. [DOI] [PubMed] [Google Scholar]
- 19. Rodrigues TS, de Sa KSG, Ishimoto AY, et al. Inflammasomes are activated in response to SARS‐CoV‐2 infection and are associated with COVID‐19 severity in patients. J Exp Med. 2021;218(3):e20201707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kroemer A, Khan K, Plassmeyer M, et al. Inflammasome activation and pyroptosis in lymphopenic liver patients with COVID‐19. J Hepatol. 2020;73(5):1258‐1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Aid M, Busman‐Sahay K, Vidal SJ, et al. Vascular disease and thrombosis in SARS‐CoV‐2‐infected rhesus macaques. Cell. 2020;183(5):1354‐1366.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gao YL, Zhai JH, Chai YF. Recent advances in the molecular mechanisms underlying pyroptosis in sepsis. Mediators Inflamm. 2018;2018:5823823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jia C, Chen H, Zhang J, et al. Role of pyroptosis in cardiovascular diseases. Int Immunopharmacol. 2019;67:311‐318. [DOI] [PubMed] [Google Scholar]
- 24. Man SM, Karki R, Kanneganti TD. Molecular mechanisms and functions of pyroptosis, inflammatory caspases and inflammasomes in infectious diseases. Immunol Rev. 2017;277(1):61‐75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kovacs SB, Miao EA. Gasdermins: effectors of pyroptosis. Trends Cell Biol. 2017;27(9):673‐684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang Y, Chen X, Gueydan C, Han J. Plasma membrane changes during programmed cell deaths. Cell Res. 2018;28(1):9‐21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fleisher TA. Apoptosis. Ann Allergy Asthma Immunol. 1997;78(3):245‐250; quiz 9‐50. [DOI] [PubMed] [Google Scholar]
- 28. Moretti J, Blander JM. Increasing complexity of NLRP3 inflammasome regulation. J Leukoc Biol. 2020;109(3):561‐571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jamilloux Y, Henry T, Belot A, et al. Should we stimulate or suppress immune responses in COVID‐19? Cytokine and anti‐cytokine interventions. Autoimmun Rev. 2020;19(7):102567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Al‐Samkari H, Karp Leaf RS, Dzik WH, et al. COVID‐19 and coagulation: bleeding and thrombotic manifestations of SARS‐CoV‐2 infection. Blood. 2020;136(4):489‐500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Grobler C, Maphumulo SC, Grobbelaar LM, et al. Covid‐19: the rollercoaster of fibrin(Ogen), D‐Dimer, von willebrand factor, P‐selectin and their interactions with endothelial cells, platelets and erythrocytes. Int J Mol Sci. 2020;21(14):5168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wilk AJ, Rustagi A, Zhao NQ, et al. A single‐cell atlas of the peripheral immune response in patients with severe COVID‐19. Nat Med. 2020;26(7):1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Blanco‐Melo D, Nilsson‐Payant BE, Liu WC, et al. Imbalanced host response to SARS‐CoV‐2 drives development of COVID‐19. Cell. 2020;181(5):1036‐1045 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Carfi A, Bernabei R, Landi F, Gemelli Against C‐P‐ACSG. Persistent symptoms in patients after acute COVID‐19. JAMA. 2020;324(6):603‐605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Garg P, Arora U, Kumar A, Wig N. The, "post‐COVID" syndrome: how deep is the damage? J Med Virol. 2020;93(2):673‐674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Linton SD, Aja T, Armstrong RA, et al. First‐in‐class pan caspase inhibitor developed for the treatment of liver disease. J Med Chem. 2005;48(22):6779‐6782. [DOI] [PubMed] [Google Scholar]
- 37. Stack JH, Beaumont K, Larsen PD, et al. IL‐converting enzyme/caspase‐1 inhibitor VX‐765 blocks the hypersensitive response to an inflammatory stimulus in monocytes from familial cold autoinflammatory syndrome patients. J Immunol. 2005;175(4):2630‐2634. [DOI] [PubMed] [Google Scholar]
- 38. Foy BH, Carlson JCT, Reinertsen E, et al. Association of red blood cell distribution width with mortality risk in hospitalized adults with SARS‐CoV‐2 infection. JAMA Netw Open. 2020;3(9):e2022058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Maellaro E, Leoncini S, Moretti D, et al. Erythrocyte caspase‐3 activation and oxidative imbalance in erythrocytes and in plasma of type 2 diabetic patients. Acta Diabetol. 2013;50(4):489‐495. [DOI] [PubMed] [Google Scholar]
- 40. Thomas T, Stefanoni D, Dzieciatkowska M, et al. Evidence for structural protein damage and membrane lipid remodeling in red blood cells from COVID‐19 patients. J Proteome Res. 2020. 10.1101/2020.06.29.20142703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cheung EW, Zachariah P, Gorelik M, et al. Multisystem inflammatory syndrome related to COVID‐19 in previously healthy children and adolescents in New York City. JAMA. 2020;324(3):294‐296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jiang L, Tang K, Levin M, et al. COVID‐19 and multisystem inflammatory syndrome in children and adolescents. Lancet Infect Dis. 2020;20(11):e276‐e288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Singh‐Grewal D, Lucas R, McCarthy K, et al. Update on the COVID‐19‐associated inflammatory syndrome in children and adolescents; paediatric inflammatory multisystem syndrome‐temporally associated with SARS‐CoV‐2. J Paediatr Child Health. 2020;56(8):1173‐1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Richardson S, Hirsch JS, Narasimhan M, et al. Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID‐19 in the New York city area. JAMA. 2020;323(20):2052‐2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yap JKY, Moriyama M, Iwasaki A. Inflammasomes and pyroptosis as therapeutic targets for COVID‐19. J Immunol. 2020;205(2):307‐312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sutterwala FS, Haasken S, Cassel SL. Mechanism of NLRP3 inflammasome activation. Ann N Y Acad Sci. 2014;1319:82‐95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Akdis M, Aab A, Altunbulakli C, et al. Interleukins (from IL‐1 to IL‐38), interferons, transforming growth factor beta, and TNF‐alpha: Receptors, functions, and roles in diseases. J Allergy Clin Immunol. 2016;138(4):984‐1010. [DOI] [PubMed] [Google Scholar]
- 48. Arend WP, Palmer G, Gabay C. IL‐1, IL‐18, and IL‐33 families of cytokines. Immunol Rev. 2008;223:20‐38. [DOI] [PubMed] [Google Scholar]
- 49. He Y, Hara H, Nunez G. Mechanism and regulation of NLRP3 inflammasome activation. Trends Biochem Sci. 2016;41(12):1012‐1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Latz E, Xiao TS, Stutz A. Activation and regulation of the inflammasomes. Nat Rev Immunol. 2013;13(6):397‐411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhou Z, Ren L, Zhang L, et al. Heightened innate immune responses in the respiratory tract of COVID‐19 patients. Cell Host Microbe. 2020;27(6):883‐390 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Labzin LI, Lauterbach MA, Latz E. Interferons and inflammasomes: cooperation and counterregulation in disease. J Allergy Clin Immunol. 2016;138(1):37‐46. [DOI] [PubMed] [Google Scholar]
- 53. Matzinger P. Tolerance, danger, and the extended family. Annu Rev Immunol. 1994;12:991‐1045. [DOI] [PubMed] [Google Scholar]
- 54. Matzinger P. The danger model: a renewed sense of self. Science. 2002;296(5566):301‐305. [DOI] [PubMed] [Google Scholar]
- 55. Carelli‐Alinovi C, Pirolli D, Giardina B, Misiti F. Protein kinase C mediates caspase 3 activation: a role for erythrocyte morphology changes. Clin Hemorheol Microcirc. 2015;59(4):345‐354. [DOI] [PubMed] [Google Scholar]
- 56. Firat U, Kaya S, Cim A, et al. Increased caspase‐3 immunoreactivity of erythrocytes in STZ diabetic rats. Exp Diabetes Res. 2012;2012:316384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rinalducci S, Ferru E, Blasi B, Turrini F, Zolla L. Oxidative stress and caspase‐mediated fragmentation of cytoplasmic domain of erythrocyte band 3 during blood storage. Blood Transfus. 2012;10(Suppl 2):s55‐s62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zini G, Bellesi S, Ramundo F, d'Onofrio G. Morphological anomalies of circulating blood cells in COVID‐19. Am J Hematol. 2020;95(7):870‐872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Liu Y, Zhang X, Qiao J, et al. A controllable inflammatory response and temporary abnormal coagulation in moderate disease of COVID‐19 in Wuhan, China. J Clin Med Res. 2020;12(9):590‐597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tomar B, Anders HJ, Desai J, Mulay SR. Neutrophils and neutrophil extracellular traps drive necroinflammation in COVID‐19. Cells. 2020;9(6):1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wiewiora M, Piecuch J, Sedek L, Mazur B, Sosada K. The effects of obesity on CD47 expression in erythrocytes. Cytometry B Clin Cytom. 2017;92(6):485‐491. [DOI] [PubMed] [Google Scholar]
- 62. Marini JJ, Gattinoni L. Management of COVID‐19 respiratory distress. JAMA. 2020;323(22):2329‐2330. [DOI] [PubMed] [Google Scholar]
- 63. Neupane K, Ahmed Z, Pervez H, Ashraf R, Majeed A. Potential treatment options for COVID‐19: a comprehensive review of global pharmacological development efforts. Cureus. 2020;12(6):e8845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Shahzad K, Bock F, Al‐Dabet MM, et al. Caspase‐1, but not Caspase‐3, promotes diabetic nephropathy. J Am Soc Nephrol. 2016;27(8):2270‐2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Communal C, Sumandea M, de Tombe P, Narula J, Solaro RJ, Hajjar RJ. Functional consequences of caspase activation in cardiac myocytes. Proc Natl Acad Sci USA. 2002;99(9):6252‐6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Frenette CT, Morelli G, Shiffman ML, et al. Emricasan improves liver function in patients with cirrhosis and high model for end‐stage liver disease scores compared with placebo. Clin Gastroenterol Hepatol. 2019;17(4):774‐783 e4. [DOI] [PubMed] [Google Scholar]
- 67. Barreyro FJ, Holod S, Finocchietto PV, et al. The pan‐caspase inhibitor Emricasan (IDN‐6556) decreases liver injury and fibrosis in a murine model of non‐alcoholic steatohepatitis. Liver Int. 2015;35(3):953‐966. [DOI] [PubMed] [Google Scholar]
- 68. Harrison SA, Goodman Z, Jabbar A, et al. A randomized, placebo‐controlled trial of emricasan in patients with NASH and F1–F3 fibrosis. J Hepatol. 2020;72(5):816‐827. [DOI] [PubMed] [Google Scholar]
- 69. Doitsh G, Galloway NL, Geng X, et al. Cell death by pyroptosis drives CD4 T‐cell depletion in HIV‐1 infection. Nature. 2014;505(7484):509‐514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Schurink B, Roos E, Radonic T, et al. Viral presence and immunopathology in patients with lethal COVID‐19: a prospective autopsy cohort study. Lancet Microbe. 2020;1(7):e290‐e299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kim J, Zhang J, Cha Y, et al. Advanced bioinformatics rapidly identifies existing therapeutics for patients with coronavirus disease‐2019 (COVID‐19). J Transl Med. 2020;18(1):257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Baker JD, Uhrich RL, Kraemer GC, Love JE, Kraemer BC. A drug repurposing screen identifies hepatitis C antivirals as inhibitors of the SARS‐CoV‐2 main 1 protease; 2020. [DOI] [PMC free article] [PubMed]
- 73. Rongvaux A, Jackson R, Harman CC, et al. Apoptotic caspases prevent the induction of type I interferons by mitochondrial DNA. Cell. 2014;159(7):1563‐1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Feng Y, Daley‐Bauer LP, Roback L, et al. Caspase‐8 restricts antiviral CD8 T cell hyperaccumulation. Proc Natl Acad Sci USA. 2019;116(30):15170‐15177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Garcia‐Tsao G, Bosch J, Kayali Z, et al. Randomized placebo‐controlled trial of emricasan for non‐alcoholic steatohepatitis‐related cirrhosis with severe portal hypertension. J Hepatol. 2020;72(5):885‐895. [DOI] [PubMed] [Google Scholar]
- 76. Yaqinuddin A, Kashir J. Novel therapeutic targets for SARS‐CoV‐2‐induced acute lung injury: targeting a potential IL‐1beta/neutrophil extracellular traps feedback loop. Med Hypotheses. 2020;143:109906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Mitrani RD, Dabas N, Goldberger JJ. COVID‐19 cardiac injury: Implications for long‐term surveillance and outcomes in survivors. Heart Rhythm. 2020;17(11):1984‐1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
App S1


