CONFLICT OF INTEREST
M. F. Bachmann is a board member of Saiba AG, involved in the development of RBD‐CuMV a vaccine against COVID‐19. All other authors declare no conflict of interest.
To the Editor,
The receptor binding domain (RBD) of SARS‐CoV‐2 spike (S) glycoprotein, which is involved in virus attachment and cell entry, is the primary target for neutralizing antibodies. 1 Immunization against full‐length S or parts of it may result in more potent and longer‐lasting antibody responses than natural viral infection. 2 Therefore, global vaccination programs based on induction of neutralizing antibodies are the most promising strategy for controlling the COVID‐19 pandemic.
A worrying factor, however, has been the emerging variants capable to escape immunity produced by vaccination or infection. Three variants have attracted attentions due to their abnormally high rates of propagation: B.1.1.7 (N501Y, D614G); B.1.351 (K417 N, E484 K, N501Y, D614G); and P.1 (K417 N/T, E484 K, N501Y). Limited knowledge of the presence of cross‐neutralizing antibodies induced by natural infection or vaccination is a key gap in current understanding of the spread of SARS‐CoV‐2. It is imperative to determine the impact of these mutations on the responses induced by currently marketed vaccines.
A study has shown that E484 K mutation is associated with reduced neutralization, by SARS‐CoV‐2 infection or BNT162b2‐elicited sera. 3 Whether reduced neutralization was due to impaired binding was, however, not analyzed. Here, we assessed the presence of such cross‐reactive antibodies in convalescent sera and sera from individuals immunized with mRNA‐based BNT162b2 vaccine.
To this end, we generated four mutant RBDs:K417 N (RBD417), E484 K (RBD484), N501Y (RBD501), and one triple mutated version with all three mutations (RBDtrip) (Figure 1A). Two of these mutations, E484 K and N501Y, are localized within the receptor binding motif, 4 directly interacting with ACE2. Our ELISA results show that binding of convalescent sera was strongly reduced for RBD417 and RBD501 and, however, essentially abolished for RBD484 and RBDtrip (Figure 1B, left). In contrast, BNT162b2‐elicited antibodies exhibited only weakly reduced binding to RBD417 and RBD501 (2.5–3‐fold), but 10‐fold reduced binding to RBD484 and RBDtrip (Figure 1B, right). Figure 1C summarizes the antibody titers and Figure 1D quantifies the reduced binding of vaccine‐induced sera to the mutant RBDs compared to RBDwt (wild‐type RBD). Interestingly, the E484 K mutation was equally potent at reducing antibody binding as in RBDtrip, indicating that the mutation E484 K is particularly problematic, perhaps because it involves a change from positive to negative charge. This is consistent with recent data showing less neutralization titer against a variant containing E484 K mutation, however, only with 1.41‐fold reduction. 5
FIGURE 1.
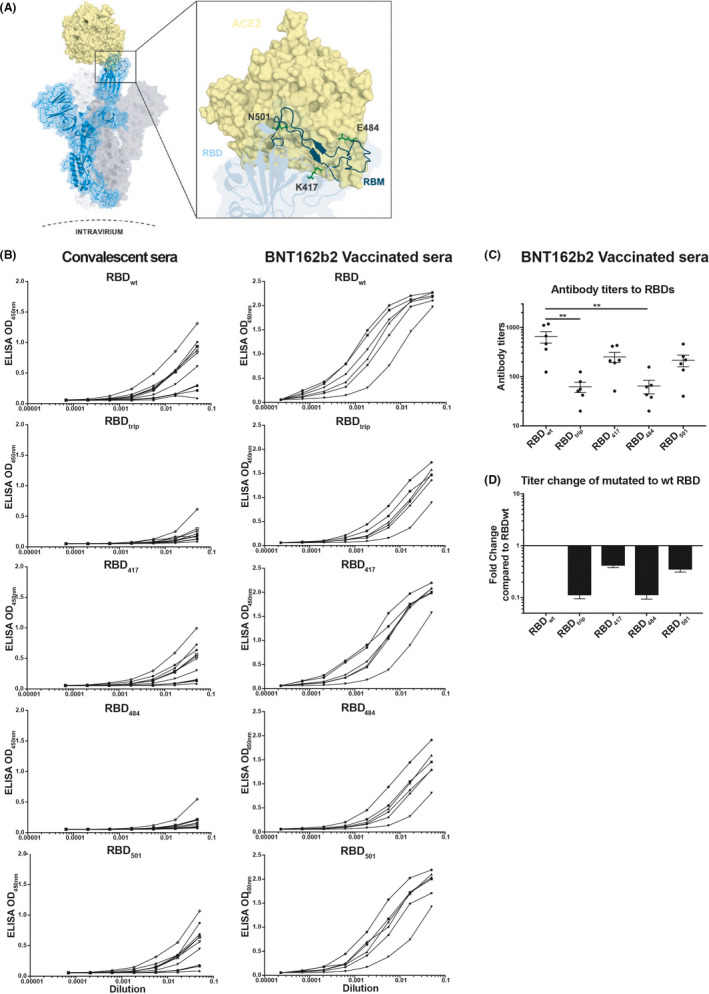
Strongly reduced recognition of mutant RBDs by convalescent and BNT162b2 vaccinated human sera. (A) Structure of RBD and location of the individual mutations used in this study (E484 K, K417 N, N501Y, or all 3 mutations combined). (B) Titration on RBDs of sera from convalescent patients (left panel) or from BNT162b2 vaccinated individuals (right panel). (C) Antibody titers (OD50) of sera from 6 BNT162b2 vaccinated individuals on RBDs. (D) Fold reduction of mutant RBDs compared to wild‐type RBD‐recognition by sera of BNT162b2 vaccinated individuals
We performed assays to estimate the antibodies’ avidity for RBD and mutants. Interestingly, virus‐induced antibodies were of limited avidity for RBDwt, and binding to mutant RBDs was essentially abolished with 7 M urea wash, indicating that the antibodies binding to mutant RBDs were all of low avidity (Figure 2A shows results for RBDwt). In contrast, BNT162b2‐induced antibodies were of significantly higher avidity (Figure 2B). In addition, there was some residual binding to mutant RBDs, indicating, however, overall low avidity as well (Figure 2C). The avidity index allows to quantify the loss in binding caused by the 7 M urea wash and therefore reflects the “quality” of the antibodies. Indeed, the avidity index of vaccine‐induced antibodies is much higher for RBDwt and mutants than those induced by infection. This reduced affinity of antibodies induced by infection is consistent with the notion that individual RBDs are spaced by 25 nm on SARS‐CoV‐2, too large for inducing optimal antibodies. 6
FIGURE 2.
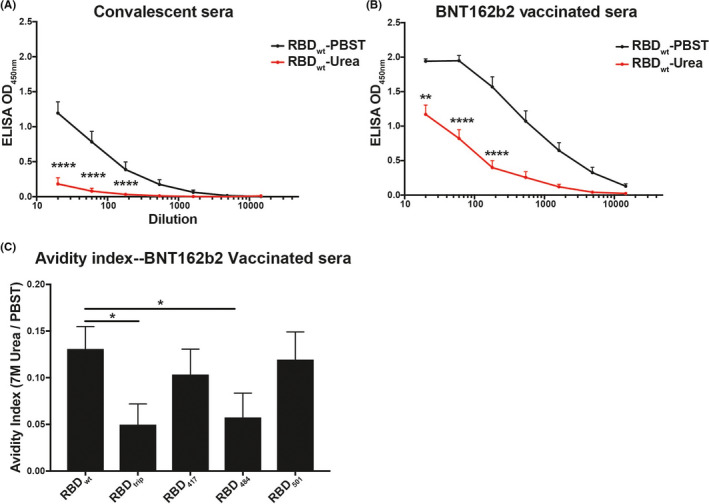
Increased avidity of BNT162b2‐induced antibodies compared to antibodies induced by infection. (A, B) Average recognition of RBDwt by convalescent (A) or vaccine‐induced sera (B) by avidity ELISA. Samples washed with PBS‐Tween (black) and 7 M urea (red). (C) Avidity indexes are shown for recognition of mutant RBDs by vaccine‐induced sera. These indexes are calculated by dividing the area under the curve (AUC) of urea washed samples by AUC of PBS‐Tween washed samples. Note that recognition of mutant RBDs by convalescent sera was too low to calculate a meaningful avidity index
In conclusion, BNT162b2‐induced antibodies recognize mutant RBDs better than those by natural infection. Recognition may, however, be 10‐fold reduced for the variants B.1.351/P.1, suggesting that development of a new vaccine may be warranted. E484 K mutation is shown here to be a key hurdle for immune recognition. Hence, monoclonal antibody therapy and serological assays based on wildtype sequence may therefore be seriously impaired.
ACKNOWLEDGEMENTS
We thank Marianne Zwicker for production of mutant RBDs.
Mohsen and Bachmann equal contribution.
Funding information
The work was supported by Saiba AG, the Swiss National Science Foundation (SNF grants 31003A 149925 and 310030‐179459) and the Inselspital, Bern, Switzerland.
Contributor Information
Mona O. Mohsen, Email: mona.mohsen@dbmr.unibe.ch.
Martin F. Bachmann, Email: martin.bachmann@dbmr.unibe.ch.
REFERENCES
- 1. Tai W, He L, Zhang X, et al. Characterization of the receptor‐binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell Mol Immunol. 2020;17:613‐620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tay MZ, Poh CM, Renia L, MacAry PA, Ng LFP. The trinity of COVID‐19: immunity, inflammation and intervention. Nat Rev Immunol. 2020;20:363‐374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dejnirattisai W, Zhou D, Supasa P, Ren J, Stuart DI, Screaton GR. Article antibody evasion by the P.1 strain of SARS‐CoV‐2 antibody evasion by the P.1 strain of SARS‐CoV‐2. Cell. 2021;184:1‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Greaney AJ, Starr TN, Gilchuk P, et al. Complete mapping of mutations to the SARS‐CoV‐2 spike receptor‐binding domain that escape antibody recognition. Cell Host Microbe. 2021;29:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Xie X, Liu Y, Liu J, et al. Neutralization of SARS‐CoV‐2 spike 69/70 deletion, E484K and N501Y variants by BNT162b2 vaccine‐elicited sera. Nat Med. 2021;27:620‐621. [DOI] [PubMed] [Google Scholar]
- 6. Bachmann MF, Mohsen MO, Zha L, Vogel M, Speiser DE. SARS‐CoV‐2 structural features may explain limited neutralizing‐antibody responses. NPJ Vaccines. 2021;6(1):2. [DOI] [PMC free article] [PubMed] [Google Scholar]


