Abstract
PCR has been widely used in different fields including molecular biology, pathogen detection, medical diagnosis, food detection and etc. However, the difficulty of promoting PCR in on-site point-of-care testing reflects on challenges relative to its speed, convenience, complexity, and even cost. With the emerging state-of-art of microfluidics, rapid PCR can be achieved with more flexible ways in micro-reactors. PCR plays a critical role in the detection of SARS-CoV-2. Under this special background of COVID-19 pandemic, this review focuses on the latest rapid microfluidic PCR. Rapid PCR is concluded in two main features, including the reactor (type, size, material) and the implementation of thermal cycling. Especially, the compromise between speed and sensitivity with microfluidic PCR is explored based on the system ratio of (thermal cycling time)/(reactor size). Representative applications about the detection of pathogens and SARS-CoV-2 viruses based on rapid PCR or other isothermal amplification are discussed as well.
Keywords: Rapid PCR, Thermal cycling, Pathogen detection, COVID-19, Microfluidics, Point-of-care testing (POCT)
1. Introduction
As a critical molecular biological technology which was invented by Mullis [1], polymerase chain reaction (PCR) is widely applied in different fields, for example, pathogen detection [2,3], food and water safety [4,5], forensics [6] and etc. PCR is performed with repeated thermal cycling which provides the required temperatures for different reaction stages (denaturation, annealing, and extension) in one amplification cycle. Under different conditions, PCR mix including primers, enzymes, targeted DNA templates and specific buffers should be properly prepared and even optimized to ensure efficient amplification [7].
Typically, conventional PCR is performed in a volume from 25 to 50 μL with 200 μL tubes in a normal PCR instrument [8], which cannot achieve ultra-fast ramping rate because of both the big reaction size and the large thermal resistance between the heating block and the reaction tube (with high thermal capacity). Therefore, even for the latest commercial PCR instruments with advanced configuration, the ramping rate is normally limited to 4–6°C/s even when thermoelectric modules with high quality are adopted [9].
Since the invention of PCR, many efforts have been made to reduce the required amplification time. For example, instead of using a typical 200 μL polypropylene tube, a thin capillary tube with low thermal capacity is heated by hot air to achieve remarkably high ramping rates [10]. On the other side, microfluidics provide a potential solution to improve PCR speed since small or elaborate PCR reactors can be properly fabricated. Different from tube-based PCR, when the size of microfluidic PCR reactors is down to a couple of μL, or even less than 1 μL, PCR can be performed with ultra-high ramping rates due to low system thermal capacity. Or alternatively, ultra-fast ramping rates can be achieved in elaborately designed PCR reactors based on state-of-art of thermal cycling implementation.
For successful commercialization and application, rapid PCR with high sensitivity is especially preferred in the detection of infectious diseases. In principle, sensitivity is directly affected by the size of a PCR reactor, or the available nucleic acid templates. Normally, to achieve high sensitivity, the required amplification time with a large reactor tends to be prolonged. As it is well known, there is always a compromise between speed and sensitivity with microfluidic PCR in the detection of infectious diseases. Considering this compromise, an evaluation parameter, e.g., the system ratio of (thermal cycling time: min)/(reactor size: μL), γ is defined for a microfluidic PCR system.
As a hot topic in the field of nucleic acid diagnostics, various PCR-related microfluidic systems have been systematically summarized and reviewed from different aspects, e.g., microPCR and isothermal systems based on microfluidics [11], diverse microfluidic PCR devices with or without sample preparation [12], a brief summarization of PCR technology from its past to present and to future [13], and a broad review of POC miniature PCR devices including field requirements, system characteristics, integration, applications, and practical issues [14]. In this review, a more specifically focused topic, rapid PCR powered by microfluidics mostly in the last 5 years has been summarized and discussed. A couple of critical factors relative to rapid PCR, including both reactor and implementation of thermal cycling, are discussed in details. The characteristics (commercialization, application) of different microfluidic PCR reactors can be more specifically illustrated with the above predefined system ratio, γ. Representative applications about the detection of pathogens based on rapid PCR are discussed as well. Especially, witnessing the global COVID-19 pandemic, this review also emphasizes the key contribution of rapid PCR or isothermal amplification to immediate detection of SARS-CoV-2.
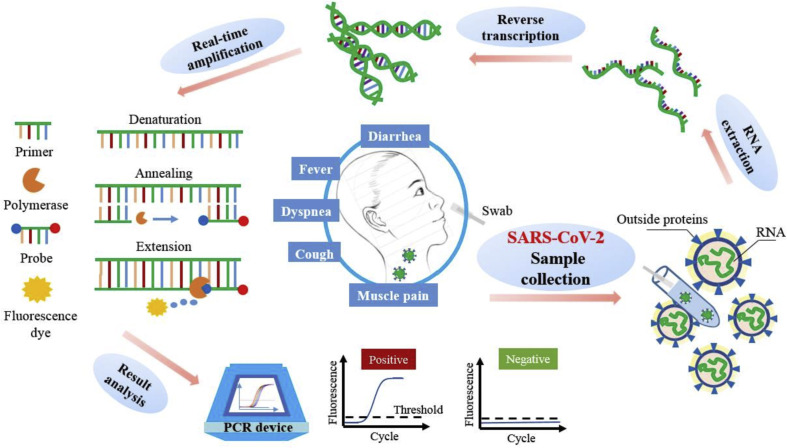
According to WHO's (World Health Organization) laboratory testing guidance, as a gold standard, real-time reverse-transcription PCR (RT-PCR) is the routine confirmation test for COVID-19, and the targeted viral genes include the ORF1ab/RdRp, E, N, and S genes [15]. As a representative example, Fig. 1 shows the procedure of the detection to SARS-CoV-2 or other pathogens with PCR.
Fig. 1.
Procedure of the detection to SARS-CoV-2 with PCR.
2. Reactor of rapid PCR
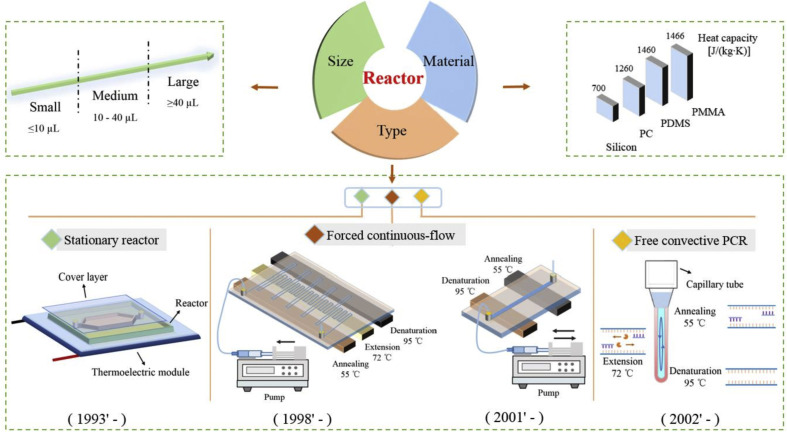
As shown in Fig. 2 , PCR reactor can be discussed from three aspects, including reactor type, size and material. To satisfy different applications, PCR reactor should be properly designed to accommodate critical system performances including detection time, sensitivity, specificity and even cost.
Fig. 2.
A brief summarization of typical PCR reactors.
In principle, the reactor size and especially the system ratio γ (thermal cycling time: min)/(reactor size: μL) are critical parameters which can be used to evaluate its adoptability to clinical application for an existing PCR system. Obviously, it is difficult for a rapid but tiny PCR reactor (e.g., size ≤ 1 μL) to achieve desired detection sensitivity due to insufficient involved nucleic acid templates in the detection of infectious diseases. Therefore, in this review, the system ratio γ is defined to demonstrate two dimensional characteristics, e.g., the reactor size and the required thermal cycling time from two different aspects to properly evaluate the performance of a rapid PCR system.
2.1. Reactor type
In order to achieve rapid PCR, different types of microfluidic reactors have been developed. As shown in Fig. 2, based on different mechanisms of thermal cycling, three types of representative PCR reactors have been invented respectively at different time. Type #1 is the stationary reactor [16], where PCR amplification is achieved when the reagent inside the stationary chamber is heated to different temperatures for thermal cycling. Type #2 is the forced continuous-flow PCR [17], where PCR amplification is achieved when the reagent is pushed through a long (often serpentine) channel with distinct zones maintained at different temperatures needed for melting, annealing, and extension, or alternatively, pushed back and forth iteratively inside a short channel with different heating temperatures. Type #3 is the free convection PCR (CPCR) [18], where PCR amplification is achieved when a continuous, sustained circulation of reactants between hot and cool zones of the reaction is induced due to free thermal convection for spatially separate and stable melting, annealing, and extension. More details about working mechanisms of different reactors see the below part ‘Implementation of thermal cycling’.
2.2. Reactor size
Typically, with different fabrication technologies, the volume of the PCR reactor can be shifted from μL to nL [19,20]. Smaller reactor with less thermal capacity is able to achieve high ramping rates and therefore reduce detection time. In contrast, larger reactor allows more nucleic acid templates to be utilized and therefore improve detection sensitivity. In this review, as shown in Fig. 2, three types of reactors with different size ranges are defined as following: small-size (≤10 μL), medium-size (10–40 μL), and large-size (≥40 μL).
To achieve ultra-fast thermal cycling, in most of existing rapid PCR systems, the reactor size is normally not larger than 10 μL. With lithography, the size of a stationary reactor can even be scaled down to nL. For example, it just takes ~3.7 s for a tiny reactor (~20 nL) to complete a single thermal cycle [19]. For this reactor, its system ratio γ is 108. However, since a tiny reactor can't accommodate enough nucleic acid templates, its detection sensitivity especially to clinical samples, for example infectious diseases, can't be guaranteed. Comparing to small-size reactor, medium-size reactor is able to perform more sensitive detection with more involved nucleic acid templates, which is normally achieved with the sacrifice of longer detection time. A medium-size disc-type reactor (25 μL) is able to perform 33-cycle thermal cycling in relatively long time (76 min) [20]. For this reactor, its system ratio γ is 3.04. Large-size reactor can be fabricated for specific purposes, for example, to increase the sensitivity of the detection to infectious diseases, which should be reasonably designed based on highly efficient thermal cycling format, for example, free thermal convection to save the amplification time. Qiu et al. used a 19 mm-long capillary tube reactor (40 μL) for the detection to influenza A (H1N1) virus based on CPCR with a limit of detection of 1.0 within 30 min [21]. For this reactor, its system ratio γ is 0.75.
2.3. Reactor material
Different materials can be adopted by PCR reactors for different applications. Beside the heating efficiency of the thermal cycling system itself, the system ramping rate also heavily depends on the thermal capacity of the reactor material. As an example, thermal capacity of four typical materials has been shown in Fig. 2. When the PCR reactor is made from material with lower thermal capacity, e.g., silicon, higher ramping rate can be more easily achieved. For a specific PCR reactor, the relationship among ‘material-fabrication-size’ should be considered. Typically, when the PCR reactor is made from silicon substrate with lithography and covered by silicon oil or a glass plate, its size is normally not larger than 10 μL to achieve rapid thermal cycling [22]. To reduce the cost of the reactor, the PCR reactor can also be made from PDMS (polydimethylsiloxane) with lithography, which is able to perform PCR with a large size range (nL to tens of μL) [23]. However, gas permeability of PDMS will cause sample loss in PCR amplification due to reagent evaporation. A couple of methods have been developed to avoid liquid evaporation with PDMS chip, for example, to insert a polyethylene (PE) barrier layer [19], or to coat a Parylene-C film [24], which is helpful to ensure smooth PCR amplification. Furthermore, to simplify the fabrication procedure, the PCR reactor can be made from transparent plastic substrate (PC(polycarbonate), PMMA (poly (methyl methacrylate)), COC (cyclic olefin copolymer), and etc.) with CNC machining or laser cutting, which is more suitable to fabricate reactors with medium- or large-size [25]. For commercial application, plastic substrate is preferred because it is compatible with injection-molding-based mass-production.
3. Implementation of thermal cycling
From its invention, PCR can be implemented in two typical ways, including space- and time-domain based thermal cycling [26]. At the beginning of PCR application, space-domain based thermal cycling was normally performed by manually switching a reaction tube among three independent hot water sinks which were heated at three different temperatures respectively for melting, annealing, and extension. Time-domain based thermal cycling, which is normally perform in stationary reactors, has become popular after thermoelectric modules were invented.
3.1. Space-domain based thermal cycling
3.1.1. Continuous-flow PCR
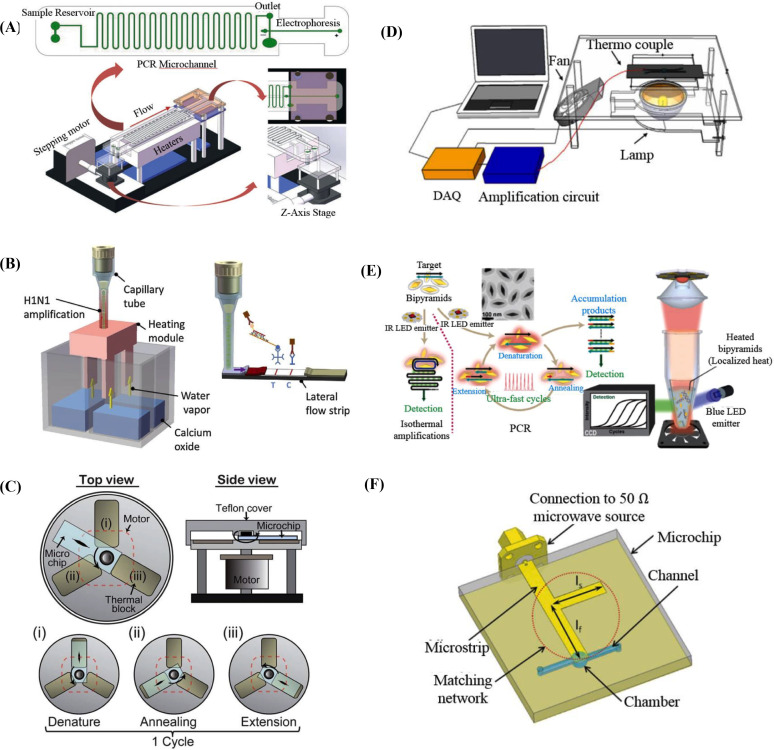
Based on a long serpentine channel or a short straight channel, forced continuous-flow PCR aims to achieve rapid PCR by actively pushing reagent with an outside pump to different pre-heated zones to avoid slow transition stage when different reaction temperatures are switched [27]. To optimize the PCR performance, the cross-section, the width-to-depth ratio, and the length ratio of three temperature zones of the micro-channels and their effect to thermal and flow distribution of fluid in reactor were systematically investigated with finite element analysis [28]. As shown in Fig. 3 (A), in a streamlined microfluidic ship, Li et al. combines continuous-flow PCR (size: 25 μL) with electrophoresis to amplify and detect treponema denticola totally in 6 min 14 s for 35 cycles [29]. For this reactor, its system ratio γ is 0.09. Alternatively, in a bidirectional-flow micro-channel, Kopparthy et al. developed real-time continuous-flow PCR (size: 10 μL) with melting curve analysis to successfully detect specific targets with 25 cycles in 30 min [30]. For this reactor, its system ratio γ is 3. To achieve consistent continuous-flow PCR, reagent has to be elaborately driven with a stable flow rate. Because of the extraordinarily high surface to volume ratio of long micro-channels, PCR amplification efficiency can be deteriorated due to surface inhibition. Because of both the inconvenience to operation and the complicated procedure of reactor pre-treatment, just few of continuous-flow PCR systems have been recently applied in commercial applications.
Fig. 3.
(A) Continuous-flow chip including PCR and electrophoresis [29]. (B) Instrument-free CPCR [36]. (C) Rotary PCR system [39]. (D) Non-contact photonic heating to the PCR reagent itself [47]. (E) Non-contact photonic heating to AuBPs mixed with PCR reagent [48]. (F) Microwave heating to PCR reagent [49].
3.1.2. Convection PCR
In the form of a capillary tube, a capillary loop, or even a thin disk, CPCR is able to perform rapid nucleic acid amplification with naturally-induced free thermal convection without any outside pump. When the capillary tube is heated with one or two fixed temperatures, its axial stable temperature field causes the difference in fluid density and induces spontaneous thermal convection. Comparing to the low temperature area, the reagent density in the high temperature area is relatively low, therefore the resulting buoyancy forces the reagent to move from the high temperature area to the low temperature area [31].
It has been found that the aspect ratio (h/d, h and d are height and diameter of a cylindrical reactor, respectively) of a CPCR reactor affects the temperature and velocity fields, and therefore decides doubling time and amplification efficiency. Muddu et al. found that ultra-fast CPCR amplification (~10 min) can be achieved when chaotic advection is introduced in the reactor [32]. Smartphone-based CPCR reactors have been developed to read the fluorescence signal from the capillary tube through the smartphone camera and meanwhile analyze the signal with custom Java applications [[33], [34], [35]]. As shown in Fig. 3(B), an instrument-free CPCR system has been developed by heating the reactor (size: 40 μL) with a disposable chemical heater for 30 min and detecting the amplicons with one-step nucleic acid dipstick assay [36]. For this reactor, its system ratio γ is 0.75. Beside vertical CPCR, a horizontal real-time CPCR system is developed by the same group to heat the capillary tube reactor at one end to achieve efficient thermal cycling based on horizontal thermal convection [37]. With the disk-based CPCR reactor (size: 25 μL), Khodakov et al. present the donut PCR platform to perform high multiplexing, precise quantitative nucleic acid detection with single nucleotide discrimination in 20 min [38]. For this reactor, its system ratio γ is 0.8.
Comparing to continuous-flow PCR, ultra-fast thermal cycling can be achieved in CPCR reactors based on free thermal convection without any outside pump. Comparing to stationary PCR, sensitive diagnosis can be achieved in large-size CPCR reactors with pseudo-isothermal manner without sacrifice of detection time. Therefore, CPCR can be regarded as a competitive, easy-to-operate nucleic acid diagnosis platform especially in point-of-care testing.
3.1.3. Spatially-switched PCR
As shown in Fig. 3(C), Jung et al. proposed a rotary PCR genetic analyzer to achieve multiple RT-PCR amplification by combining the characteristics of the flow-through and stationary PCR [39]. Similar to the flow-through PCR, according to a predefined cycle, the sample is spatially switched between the adjacent blocks which are maintained at different temperatures needed for melting, annealing, and extension. For each heating block, high ramping rates are achieved due to the low thermal mass like the stationary PCR system. Ultra-fast PCR amplification in a tiny reactor (1 μL) can be achieved for totally 34 cycles within 25.5 min. For this reactor, its system ratio γ is 25.5. It has to be pointed out that, with such a small reactor (size down to 1 μL), it is difficult to achieve highly sensitive pathogen detection especially for clinical samples of infectious diseases. Or alternatively, more like benchtop, ultra-fast PCR with a large sample volume (50 μL) can be achieved by continually transporting a normal PCR tube between the heating and cooling blocks with a complicated two dimensional moving stage, which is able to complete 40 cycles in 13.8 min [40]. For this reactor, its system ratio γ is 0.276.
3.2. Time-domain based thermal cycling
In principle, a stationary microfluidic reactor for time-domain based thermal cycling can be regarded as an updated version of conventional tube-based PCR, and the former one normally has a reduced reaction size to achieve significantly high ramping rates. With contact heating, when the size of the micro-reactor is quite small, ultra-fast thermal cycling can be achieved especially when its substrate has good thermal conductivity, and on the other side, with non-contact heating, ultra-fast thermal cycling can be achieved when more elaborate heating methods are adopted to achieve highly efficient energy conversion [41].
3.2.1. Contact heating
In contact heating, there have two different major strategies for thermal cycling. One relies on popular thermoelectric modules, and the other use the thin-film heater and the fan forced air respectively for heating and cooling. Qiu et al. developed a large volume (from 10 μL to 100 μL), portable, real-time PCR reactor by utilizing a double-sided heater that features a master, thermoelectric element and a thermal waveguide connected to a second thermoelectric element for efficiently and uniformly heating [42]. Neuzil et al. developed an ultra-fast PCR reactor (size: 100 nL) with a thin-film heater, which is able to complete 40 cycles within 6 min with remarkably high heating (175°C/s) and cooling rates (125°C/s) [43]. For this reactor, its system ratio γ is 60. Jeong et al. applied dual platinum thin-film heaters which respectively located on the bottom and top of a PCR reactor (size: 8.5 μL) in a sandwiched structure to complete 30 cycles within 50 min [44]. For this reactor, its system ratio γ is 5.88. To heat the PCR reaction chamber (size: 6 μL) of a TOPAS@ chip, in a concise version, Bu et al. developed a silicon heater with integrated platinum thin film resistive heater and temperature sensor patterned on the same bottom side, which is able to complete 30 cycles within 20 min [45]. For this reactor, its system ratio γ is 3.33. For contact heating, its ramping rate is easily to be limited because of the low efficiency of energy conversion and heat conduction unless tiny PCR reactors with much-low thermal capacity are adopted.
3.2.2. Non-contact heating
Comparing to contact heating, non-contact heating methods are developed by reasonably combining energy conversion and heat conduction together for highly efficient heating for PCR thermal cycling. There have three typical strategies for non-contact heating including induction heating [46], photonic heating [47,48], microwave heating [49].
When a coil powered with an AC power supply is used to generate the varying magnetic field, the eddy currents produced by electromagnetic induction will cause heat generation inside the metal plate, and accordingly, a microfluidic PCR reactor above this metal plate can be heated for thermal cycling [41]. Bio Molecular Systems developed a fast qPCR system to heat sample based on a patented magnetic induction technology and fan forced air for cooling, which is able to perform tube-based PCR (size: 5–30 μL) with 35 cycles in under 25 min [46]. For this reactor, its system ratio γ is 0.83–5. In photonic heating, the energy of light can be converted into heat for thermal cycling. For direct photonic heating, tungsten or halogen lamps with broad wavelengths are used to heat PCR reagent itself. Accompanied cooling is achieved with fan forced air. As shown in Fig. 3(D), the Landers group utilized a tungsten or halogen lamp combined with a spatial filter made of aluminum foil to heat PCR reagent directly, which is able to perform ultra-fast PCR (size: 10 μL) for 30 cycles within 10 min [47]. For this reactor, its system ratio γ is 1. For indirect photonic heating, lasers or light-emitting diodes (LEDs) with a specific wavelength are adopted to heat nanoparticles in PCR reagent. As shown in Fig. 3(E), Lee et al. developed a low-energy gold bipyramid nanoparticle (AuBP)-based PPT (plasmonic photothermal) real-time PCR device, which is able to perform ultra-fast PCR (size: 10 μL) for 40 cycles within 7.5 min [48]. For this reactor, its system ratio γ is 0.75. Gold bipyramid nanoparticles (AuBPs) mixed with the PCR reagent were used to produce heat energy when activated with an IR LED (wavelength: 846 nm). Since the spatially-distributed AuBPs are able to absorb the photonic energy and heat the surrounded PCR reagent immediately, highly efficient energy conversion and heat transfer can be achieved with volumetric heating. Based on microwave signal, microwave heating is able to directly heat the PCR solution with a high transmission efficiency (up to 95%) and a uniform heating profile. As depicted in Fig. 3(F), Marchiarullo et al. demonstrated an ultra-fast microfluidic PCR reactor (size: 1.3 μL) based on microwave heating, which is able to perform 30 cycles within 26.5 min [49]. For this reactor, its system ratio γ is 20.38. To achieve non-contact heating, elaborate physical mechanisms, which are able to efficiently perform energy conversion and heat transfer, have to be properly incorporated into PCR systems.
4. Applications and discussions
4.1. Pathogen detection
For immediate pathogen detection, elaborate, state-of-art of PCR reactors have to be developed and adopted to facilitate fast thermal cycling without sacrifice of sensitivity in point-of-care testing [50]. Detection to foodborne pathogens based on nucleic acid amplification is a critical tool in food safety monitoring. Song et al. used a CPCR reactor (size: 20 μL) to detect 1% adulteration in raw and thermally processed meats in 24 min [5]. For this reactor, its system ratio γ is 1.2. Rapid PCR is also widely used for pathogen detection in diagnostics of infectious diseases. Liu et al. developed an ultra-fast PCR reactor (size: 20 μL) based on photonic heating, which is able to amplify HPV viruses with 30 cycles within 50 min, and the nucleic acid amplicons are detected with a lateral flow strip [2]. For this reactor, its system ratio γ is 2.5. Qiu et al. used a CPCR reactor (size: 40 μL) to detect influenza A viruses with a limit of detection of 1.0 TCID50/ml within 30 min. Two different detection methods including real-time fluorescence monitoring and dipstick assay are analyzed and compared between each other [51]. For this reactor, its system ratio γ is 0.75.
4.2. SARS-CoV-2 detection
As an ultra-dangerous infectious disease, COVID-19 has already caused a serious public health problem. Globally, there had 115, 289, 961 confirmed cases and 2,564,560 deaths due to COVID-19 by March 5, 2021 (WHO) [52]. To efficiently control the COVID-19 pandemic, PCR or isothermal amplification has been widely used for immediate detection to SARS-CoV-2 viruses. Especially, with rapid PCR or isothermal amplification, the turnaround time after sample loading can be significantly shortened to improve the diagnosis and confirmation efficiency of COVID-19 [53].
A couple of commercialized devices for rapid detection to SARS-CoV-2 viruses have been successfully developed based on PCR or isothermal amplification, and each of them is able to perform rapid nucleic acid amplification in a reactor with a size of tens of μL. As shown in Fig. 4 , Cepheid developed a ‘sample-to-answer’ PCR system, Xpress SARS-CoV-2 based on their existing technologies. With an integrated cartridge for liquid handling, the GeneXpert system is able to perform SARS-CoV-2 detection in 45 min (30 min for positive test) with sensitivity of 98% and specificity of 100% [54]. Similarly, BioFire developed another ‘sample-to-answer’ system, BioFire COVID-19 based on their existing technologies with multiplexed nested PCR. With a disposable cassette integrated with assay-specific ‘pouches’, the FilmArray system is able to perform SARS-CoV-2 detection in 45 min with satisfied sensitivity and specificity [55]. Abbott developed a ‘sample-to-answer’ system, ID NOW COVID-19 based on their existing technologies with LAMP (Loop-Mediated Isothermal Amplification). With three disposable cartridge modules, the ID NOW system is able to perform SARS-CoV-2 detection in 13 min (5 min for high positive test) with sensitivity of 94.7% and specificity of 98.6% [56,57].
Fig. 4.
Representative commercialized devices for rapid detection to SARS-CoV-2 viruses.
Especially, there have two representative devices which allow for home-based SARS-CoV-2 test since they can be easily and conveniently operated without any assisting devices or tools. As shown in Fig. 4, Lucira developed an ‘all-in-one’ LAMP device, Lucira COVID-19 based on their existing technologies. The Lucira COVID-19 is able to perform SARS-CoV-2 detection in 30 min (11 min for positive test) [58]. Visby Medical developed an ‘all-in-one’ detection device, Visby Medical COVID-19 based on continuous-flow PCR [59]. The Visby Medical COVID-19 is able to perform SARS-CoV-2 detection in 30 min [60]. It is believed that home-based POC SARS-CoV-2 test is able to implement more immediate diagnosis and confirmation comparing to other typical detection methods.
In principle, PCR normally has high sensitivity and specificity. However, false-negative results will somehow occur due to variability or mismatches between the primers and probes and the target sequences. To further improve the sensitivity and specificity of PCR, multiple target gene amplification can be utilized to simultaneously amplify multiple genomes from the same target template with multiple pairs of primers and probes. For example, in the detection of SARS-CoV-2, two different genomes, N and human RNAse P genes are simultaneously amplified in the same reaction to achieve sensitive detection (limit of detection<5 copies/reaction) [61]. Here, human RNAse P gene is amplified as an internal control to confirm successful and smooth PCR amplification. Alternatively, even three different genomes, N, E, and human ABL1 (internal control) genes can be simultaneously amplified in the detection of SARS-CoV-2 to ensure the detection specificity [62].
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) represents a family of nucleic acid sequences found in prokaryotic organisms, such as bacteria. These sequences can be recognized and cut by a set of bacterial enzymes, called CRISPR-associated enzymes, exemplified by Cas9, Cas12, and Cas13. Certain enzymes in the Cas12 and Cas13 families can be programmed to target and cut viral RNA sequences [56,63]. The detection sensitivity and specificity can be further improved by combining PCR with CRISPR. For example, Huang et al. utilized a custom CRISPR Cas12a/gRNA complex and a fluorescent probe to detect target amplicons produced by standard RT-PCR or isothermal recombinase polymerase amplification (RPA), to allow highly sensitive and specific detection of SARS-CoV-2 samples (Limit of detection: 2 copies/sample) [64].
It should be noted that POC nucleic acid diagnosis has been remarkably promoted because of the emerging technologies and products for the detection to SARS-CoV-2. No matter for rapid PCR, or isothermal amplification, an integrated ‘sample-to-answer’ system with the function of sample processing will be one of important directions with the development of nucleic acid diagnosis technology.
4.3. Discussions
To better show the relationship among reactor size, thermal cycling time and detection sensitivity, the system ratio and the corresponding limit of detection of the mentioned devices in this review have been summarized in Table 1 .
Table 1.
The system ratio γ and the corresponding limit of detection of different devices.
| Category | Characteristics | Thermal cycling time (min) | Reactor size (μL) | System ratio γ (min/μL) | Limit of detection | Reference |
|---|---|---|---|---|---|---|
| γ ≥ 5.88 | Low compatibility with clinical diagnosis of infectious diseases | ~ 2.16 | ~ 0.02 | 108 | 75 copies/single well | [19] |
| 6 | 0.1 | 60 | NA | [43] | ||
| 26.5 | 1.3 | 20.38 | 0.1 ng/μL | [49] | ||
| 25.5 | 1 | 25.5 | 12 ag (~2 copy number) | [39] | ||
| 50 | 8.5 | 5.88 | 80 ng/μL | [44] | ||
| γ < 5.88 | High compatibility with clinical diagnosis of infectious diseases | 24 | 20 | 1.2 | 1 pg | [5] |
| 10 | 10 | 1 | 1 ng/μL | [47] | ||
| 30 | 40 | 0.75 | 1.0 TCID50/mL | [36] | ||
| 20 | 25 | 0.8 | 10 copies | [38] | ||
| 7.5 | 10 | 0.75 | 1 pg/μL | [48] | ||
| 30 | 40 | 0.75 | 1.0 TCID50/mL | [51] | ||
| 30 | 40 | 0.75 | 1.0 TCID50/mL | [21] | ||
| 6.14 | 25 | 0.09 | 125 cfu/μL | [29] | ||
| 13.8 | 50 | 0.276 | 10−3 ng/μL | [40] | ||
| 20 | 6 | 3.33 | NA | [45] | ||
| 30 | 10 | 3 | 108 copies/μL | [30] | ||
| 76 | 25 | 3.04 | NA | [20] | ||
| 50 | 20 | 2.5 | NA | [2] | ||
| Commercialization | 25 | 5–30 | 0.83–5 | 200 copies/μL | [46] | |
| 30 | 25–50 | 0.75 | 1112 copies/mL | [60] | ||
| ≤11 (positive) ≤30 (negative) |
25–50 | 0.22–1.2 | 900 copies/mL | [58] | ||
| ~ 30 (positive) ≤45 (negative) |
25–50 | 0.6–1.8 | 0.0200 PFU/mL | [54] | ||
| ≤5 (positive) ≤13 (negative) |
25–50 | 0.1–0.52 | 125 genome equivalents/mL | [57] | ||
| ~ 45 | 25–50 | 0.9–1.8 | Heat-inactivated virus ATCC VR-1986HK 500 copies/mL Infectious virus 160 copies/mL |
[53,55] |
As shown in Table 1, it can be concluded that, when the system ratio γ is too large (for example, ≥ 5.88 (min/μL)), e.g., rapid PCR with small reactors (reactor size: not larger than 1.3 μL), the PCR system is not suitable for detection of clinical samples with infectious diseases due to limited nucleic acid templates. On the other side, when the system ratio γ is reasonably low (for example, ≤ 1.2 (min/μL)), e.g., rapid PCR with large reactors (reactor size: not less than 20 μL), rapid and sensitive PCR can be applied in clinical diagnosis of infectious diseases because enough nucleic acid templates can be accommodated by the large reactor. Therefore, for the rapid, large PCR reactor with a smaller system ratio γ, it has higher compatibility with clinical diagnosis of infectious diseases.
As one of hot divisions of PCR techniques, digital PCR is a platform to perform PCR in an ultra-small volume, e.g., in a droplet with a single nucleic acid template to facilitate absolute quantitative analysis comparing to conventional PCR. For digital PCR, instead of the efficiency of thermal cycling, uniformity and homogeneity of multiple micro-PCR reactor units, e.g., droplets or micro-wells are intensively studied topics. Since this review focuses on rapid PCR based microfluidics, digital PCR is not included accordingly.
5. Conclusions and outlook
Since PCR was invented by Mullis, remarkable achievements have been made to keep pushing PCR technology to a higher level. Combined with BioMEMS or microfluidics, miniaturized microfluidic PCR has become one of important directions of PCR technology. By utilizing the advantages of microfluidics, rapid PCR can be performed in such a portable device with high convenience, which is highly desired in POC diagnostics. Especially, many efforts have been made to push the PCR technology toward the limit of speed, which is helpful to save the total detection time in nucleic acid analysis.
As described before, for different rapid PCR systems, the system ratio γ can be regarded as an important parameter to evaluate their value to commercialization and applications. For a quite small PCR reactor even with rapid thermal cycling (e.g., with a large γ), its detection sensitivity cannot be ensured due to limited nucleic acid templates in the detection of infectious diseases. Considering its compatibility with mostly used methods for nucleic acid extraction and simple heating strategy, CPCR (γ: 0.75–1.2, reactor size: 20–40 μL) can be regarded as one of competitive rapid PCR platforms in point-of-care testing due to its ultra-high efficiency of thermal cycling with a reasonably large reactor.
As it is expected, important achievements with rapid PCR are beneficial to not only the fully integrated, ‘sample-to-answer’ nucleic acid diagnostic system, but also the multiplexed nucleic acid analysis system. For example, with rapid PCR, the time to result in the detection to SARS-CoV-2 viruses can be significantly reduced, which is quite helpful to efficiently control the COVID-19 pandemic. In principle, for a commercialized POCT system, its reliance on a large reactor could be relieved by more efficient and sensitive nucleic acid amplification methods based on PCR or isothermal amplification, which will in return further improve the speed of detection due to a size-decreased reactor.
Declaration of competing interest
The authors declare that they have no conflict of interest.
Acknowledgements
The work was supported by the National Natural Science Foundation of China (No. 81871505, 61971026), the Fundamental Research Funds for the Central Universities (XK1802-4), the National Science and Technology Major Project (2018ZX10732101-001-009), and the research fund to the top scientific and technological innovation team from Beijing University of Chemical Technology (No. buctylkjcx06).
References
- 1.Mullis K.B., Faloona F.A., Scharf S.J., Saiki R.K., Erlich H.A. Specific enzymatic amplification of DNA in vitro: the polymerase chain reaction. Cold Spring Harbor Symp. Quant. Biol. 1986;51(Pt 1):263–273. doi: 10.1101/SQB.1986.051.01.032. [DOI] [PubMed] [Google Scholar]
- 2.Liu W., Zhang M., Liu X., Sharma A., Ding X. A Point-of-Need infrared mediated PCR platform with compatible lateral flow strip for HPV detection. Biosens. Bioelectron. 2017;96:213–219. doi: 10.1016/j.bios.2017.04.047. [DOI] [PubMed] [Google Scholar]
- 3.Qiu G., Gai Z., Tao Y., Schmitt J., Kullak-Ublick G.A., Wang J. Dual-functional plasmonic photothermal biosensors for highly accurate severe acute respiratory syndrome coronavirus 2 detection. ACS Nano. 2020;14:5268–5277. doi: 10.1021/acsnano.0c02439. [DOI] [PubMed] [Google Scholar]
- 4.Kim T.-H., Hwang H.J., Kim J.H. Ultra-fast on-site molecular detection of foodborne pathogens using a combination of convection polymerase chain reaction and nucleic acid lateral flow immunoassay. Foodb. Pathog. Dis. 2019;16:144–151. doi: 10.1089/fpd.2018.2500. [DOI] [PubMed] [Google Scholar]
- 5.Song K.-Y., Hwang H.J., Kim J.H. Ultra-fast DNA-based multiplex convection PCR method for meat species identification with possible on-site applications. Food Chem. 2017;229:341–346. doi: 10.1016/j.foodchem.2017.02.085. [DOI] [PubMed] [Google Scholar]
- 6.Gu Y., Zhuang B., Han J., Li Y., Song X., Zhou X., Wang L., Liu P. Modular-based integrated microsystem with multiple sample preparation modules for automated forensic DNA typing from reference to challenging samples. Anal. Chem. 2019;91:7435–7443. doi: 10.1021/acs.analchem.9b01560. [DOI] [PubMed] [Google Scholar]
- 7.Wu H., Zhang S., Chen Y., Qian C., Liu Y., Shen H., Wang Z., Ping J., Wu J., Zhang Y., Chen H. Progress in molecular detection with high-speed nucleic acids thermocyclers. J. Pharmaceut. Biomed. Anal. 2020;190:113489. doi: 10.1016/j.jpba.2020.113489. [DOI] [PubMed] [Google Scholar]
- 8.Cho B., Lee S.H., Song J., Bhattacharjee S., Feng J., Hong S., Song M., Kim W., Lee J., Bang D., Wang B., Riley L.W., Lee L.P. Nanophotonic cell lysis and polymerase chain reaction with gravity-driven cell enrichment for rapid detection of pathogens. ACS Nano. 2019;13:13866–13874. doi: 10.1021/acsnano.9b04685. [DOI] [PubMed] [Google Scholar]
- 9.Chung J.H., Jeong S. Experimental performance analysis of a small thermoelectric system applicable to real-time PCR devices. Symmetry. 2020;12 doi: 10.3390/sym12121963. [DOI] [Google Scholar]
- 10.LightCycler 2.0 Instrument. https://lifescience.roche.com/en_cn/products/lightcycler14301-20-instrument.html#overview (Accessed on 05 March 2021).
- 11.Ahmad F., Hashsham S.A. Miniaturized nucleic acid amplification systems for rapid and point-of-care diagnostics: a review. Anal. Chim. Acta. 2012;733:1–15. doi: 10.1016/j.aca.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 12.Ahrberg C.D., Manz A., Chung B.G. Polymerase chain reaction in microfluidic devices. Lab Chip. 2016;16:3866–3884. doi: 10.1039/C6LC00984K. [DOI] [PubMed] [Google Scholar]
- 13.Zhu H., Zhang H., Xu Y., Laššáková S., Korabečná M., Neužil P. PCR past, present and future. Biotechniques. 2020;69:317–325. doi: 10.2144/btn-2020-0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu H., Zhang H., Ni S., Korabečná M., Yobas L., Neuzil P. The vision of point-of-care PCR tests for the COVID-19 pandemic and beyond. TrAC Trends Anal. Chem. (Reference Ed.) 2020;130:115984. doi: 10.1016/j.trac.2020.115984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Kasteren P.B., van der Veer B., van den Brink S., Wijsman L., de Jonge J., van den Brandt A., Molenkamp R., Reusken C.B.E.M., Meijer A. Comparison of seven commercial RT-PCR diagnostic kits for COVID-19. J. Clin. Virol. 2020;128:104412. doi: 10.1016/j.jcv.2020.104412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mendoza-Gallegos R.A., Rios A., Garcia-Cordero J.L. An affordable and portable thermocycler for real-time PCR made of 3D-printed parts and off-the-shelf electronics. Anal. Chem. 2018;90:5563–5568. doi: 10.1021/acs.analchem.7b04843. [DOI] [PubMed] [Google Scholar]
- 17.Jafek A.R., Harbertson S., Brady H., Samuel R., Gale B.K. Instrumentation for xPCR incorporating qPCR and HRMA. Anal. Chem. 2018;90:7190–7196. doi: 10.1021/acs.analchem.7b05176. [DOI] [PubMed] [Google Scholar]
- 18.Espulgar W.V., Saito M., Takahashi K., Ushiro S., Yamamoto N., Akeda Y., Hamaguchi S., Tomono K., Tamiya E. Deskilled and rapid drug-resistant gene detection by centrifugal force-assisted thermal convection PCR device. Sensors. 2021;21 doi: 10.3390/s21041225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee S.H., Song J., Cho B., Hong S., Hoxha O., Kang T., Kim D., Lee L.P. Bubble-free rapid microfluidic PCR. Biosens. Bioelectron. 2019;126:725–733. doi: 10.1016/j.bios.2018.10.005. [DOI] [PubMed] [Google Scholar]
- 20.Pishbin E., Eghbal M., Navidbakhsh M., Zandi M. Localized air-mediated heating method for isothermal and rapid thermal processing on lab-on-a-disk platforms. Sensor. Actuator. B Chem. 2019;294:270–282. doi: 10.1016/j.snb.2019.05.039. [DOI] [Google Scholar]
- 21.Qiu X., Ge S., Gao P., Li K., Yang Y., Zhang S., Ye X., Xia N., Qian S. A low-cost and fast real-time PCR system based on capillary convection. SLAS Technol. Transl. Life Sci. Innovat. 2017;22:13–17. doi: 10.1177/2211068216652847. [DOI] [PubMed] [Google Scholar]
- 22.Jie J., Hu S., Liu W., Wei Q., Huang Y., Yuan X., Ren L., Tan M., Yu Y. Portable and battery-powered PCR device for DNA amplification and fluorescence detection. Sensors. 2020;20 doi: 10.3390/s20092627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Houssin T., Cramer J., Grojsman R., Bellahsene L., Colas G., Moulet H., Minnella W., Pannetier C., Leberre M., Plecis A., Chen Y. Ultrafast, sensitive and large-volume on-chip real-time PCR for the molecular diagnosis of bacterial and viral infections. Lab Chip. 2016;16:1401–1411. doi: 10.1039/C5LC01459J. [DOI] [PubMed] [Google Scholar]
- 24.Shin Y.S., Cho K., Lim S.H., Chung S., Park S.-J., Chung C., Han D.-C., Chang J.K. PDMS-based micro PCR chip with parylene coating. J. Micromech. Microeng. 2003;13:768. doi: 10.1088/0960-1317/13/5/332. [DOI] [Google Scholar]
- 25.Amadeh A., Ghazimirsaeed E., Shamloo A., Dizani M. Improving the performance of a photonic PCR system using TiO2 nanoparticles. J. Ind. Eng. Chem. 2020;94:195–204. doi: 10.1016/j.jiec.2020.10.036. [DOI] [Google Scholar]
- 26.Zhu H., Fohlerová Z., Pekárek J., Basova E., Neužil P. Recent advances in lab-on-a-chip technologies for viral diagnosis. Biosens. Bioelectron. 2020;153:112041. doi: 10.1016/j.bios.2020.112041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kulkarni M., Goel S. Advances in continuous-flow based microfluidic PCR devices -A review. Eng. Res. Express. 2020;2:1–22. doi: 10.1088/2631-8695/abd287. [DOI] [Google Scholar]
- 28.Thomas S., Orozco R.L., Ameel T. Microscale thermal gradient continuous-flow PCR: a guide to operation. Sensor. Actuator. B Chem. 2017;247:889–895. doi: 10.1016/j.snb.2017.03.005. [DOI] [Google Scholar]
- 29.Li Z., Ju R., Sekine S., Zhang D., Zhuang S., Yamaguchi Y. All-in-one microfluidic device for on-site diagnosis of pathogens based on an integrated continuous flow PCR and electrophoresis biochip. Lab Chip. 2019;19:2663–2668. doi: 10.1039/C9LC00305C. [DOI] [PubMed] [Google Scholar]
- 30.Kopparthy V.L., Crews N.D. A versatile oscillating-flow microfluidic PCR system utilizing a thermal gradient for nucleic acid analysis. Biotechnol. Bioeng. 2020;117:1525–1532. doi: 10.1002/bit.27278. [DOI] [PubMed] [Google Scholar]
- 31.Miao G., Zhang L., Zhang J., Ge S., Xia N., Qian S., Yu D., Qiu X. Free convective PCR: from principle study to commercial applications—a critical review. Anal. Chim. Acta. 2020;1108:177–197. doi: 10.1016/j.aca.2020.01.069. [DOI] [PubMed] [Google Scholar]
- 32.Muddu R., Hassan Y.A., Ugaz V.M. Chaotically accelerated biochemistry in microscale convective flows. Miniaturized Syst. Chem. Life Sci. 2010:2126–2128. doi: 10.1002/anie.201004217. [DOI] [Google Scholar]
- 33.Rajendran V.K., Bakthavathsalam P., Bergquist P.L., Sunna A. A portable nucleic acid detection system using natural convection combined with a smartphone. Biosens. Bioelectron. 2019;134:68–75. doi: 10.1016/j.bios.2019.03.050. [DOI] [PubMed] [Google Scholar]
- 34.Rajendran V.K., Bakthavathsalam P., Bergquist P.L., Sunna A. Smartphone detection of antibiotic resistance using convective PCR and a lateral flow assay. Sensor. Actuator. B Chem. 2019;298:126849. doi: 10.1016/j.snb.2019.126849. [DOI] [Google Scholar]
- 35.Qiu X., Ge S., Gao P., Li K., Yang S., Zhang S., Ye X., Xia N., Qian S. A smartphone-based point-of-care diagnosis of H1N1 with microfluidic convection PCR. Microsyst. Technol. 2017;23:2951–2956. doi: 10.1007/s00542-016-2979-z. [DOI] [Google Scholar]
- 36.Qiu X., Zhang S., Xiang F., Wu D., Guo M., Ge S., Li K., Ye X., Xia N., Qian S. Instrument-free point-of-care molecular diagnosis of H1N1 based on microfluidic convective PCR. Sensor. Actuator. B Chem. 2017;243:738–744. doi: 10.1016/j.snb.2016.12.058. [DOI] [Google Scholar]
- 37.Qiu X., Shu J.I., Baysal O., Wu J., Qian S., Ge S., Li K., Ye X., Xia N., Yu D. Real-time capillary convective PCR based on horizontal thermal convection. Microfluid. Nanofluidics. 2019;23:39. doi: 10.1007/s10404-019-2207-0. [DOI] [Google Scholar]
- 38.Khodakov D., Li J., Zhang J.X., Zhang D.Y. Donut PCR: a rapid, portable, multiplexed, and quantitative DNA detection platform with single-nucleotide specificity. BioRxiv. 2020 doi: 10.1101/2020.04.24.058453. [DOI] [Google Scholar]
- 39.Jung J.H., Choi S.J., Park B.H., Choi Y.K., Seo T.S. Ultrafast Rotary PCR system for multiple influenza viral RNA detection. Lab Chip. 2012;12:1598–1600. doi: 10.1039/C2LC21269B. [DOI] [PubMed] [Google Scholar]
- 40.Lee D., Kim D., Han J., Yun J., Lee K.-H., Kim G.M., Kwon O., Lee J. Integrated, automated, fast PCR system for point-of-care molecular diagnosis of bacterial infection. Sensors. 2021;21:377. doi: 10.3390/s21020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sreejith K.R., Ooi C.H., Jin J., Dao D.V., Nguyen N.-T. Digital polymerase chain reaction technology – recent advances and future perspectives. Lab Chip. 2018;18:3717–3732. doi: 10.1039/C8LC00990B. [DOI] [PubMed] [Google Scholar]
- 42.Qiu X., Mauk M.G., Chen D., Liu C., Bau H.H. A large volume, portable, real-time PCR reactor. Lab Chip. 2010;10:3170–3177. doi: 10.1039/C0LC00038H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neuzil P. Ultrafast miniaturized real-time PCR: 40 cycles in less than six minutes. Nucleic Acids Res. 2006;34:e77. doi: 10.1093/nar/gkl416. e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jeong S., Lim J., Kim M.-Y., Yeom J., Cho H., Lee H., Shin Y.-B., Lee J.-H. Portable low-power thermal cycler with dual thin-film Pt heaters for a polymeric PCR chip. Biomed. Microdevices. 2018;20:14. doi: 10.1007/s10544-018-0257-9. [DOI] [PubMed] [Google Scholar]
- 45.Bu M., Perch-Nielsen I.R., Sørensen K.S., Skov J., Sun Y., Duong Bang D., Pedersen M.E., Hansen M.F., Wolff A. A temperature control method for shortening thermal cycling time to achieve rapid polymerase chain reaction (PCR) in a disposable polymer microfluidic device. J. Micromech. Microeng. 2013;23 doi: 10.1088/0960-1317/23/7/074002. [DOI] [Google Scholar]
- 46.Bio Molecular Systems Mic qPCR Cycler https://biomolecularsystems.com/mic-qpcr/
- 47.Ouyang Y., Duarte G.R.M., Poe B.L., Riehl P.S., dos Santos F.M., Martin-Didonet C.C.G., Carrilho E., Landers J.P. A disposable laser print-cut-laminate polyester microchip for multiplexed PCR via infra-red-mediated thermal control. Anal. Chim. Acta. 2015;901:59–67. doi: 10.1016/j.aca.2015.09.042. [DOI] [PubMed] [Google Scholar]
- 48.Lee J.-H., Cheglakov Z., Yi J., Cronin T.M., Gibson K.J., Tian B., Weizmann Y. Plasmonic photothermal gold bipyramid nanoreactors for ultrafast real-time bioassays. J. Am. Chem. Soc. 2017;139:8054–8057. doi: 10.1021/jacs.7b01779. [DOI] [PubMed] [Google Scholar]
- 49.Marchiarullo D.J., Sklavounos A.H., Oh K., Poe B.L., Barker N.S., Landers J.P. Low-power microwave-mediated heating for microchip-based PCR. Lab Chip. 2013;13:3417–3425. doi: 10.1039/c3lc50461a. [DOI] [PubMed] [Google Scholar]
- 50.Paul R., Ostermann E., Wei Q. Advances in point-of-care nucleic acid extraction technologies for rapid diagnosis of human and plant diseases. Biosens. Bioelectron. 2020;169:112592. doi: 10.1016/j.bios.2020.112592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qiu X., Zhang S., Mei L., Wu D., Guo Q., Li K., Ge S., Ye X., Xia N., Mauk M.G. Characterization and analysis of real-time capillary convective PCR toward commercialization. Biomicrofluidics. 2017;11 doi: 10.1063/1.4977841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.W.H.O. 5 March 2021. Coronavirus Disease (COVID-19) Pandemic Numbers at A Glance.https://www.who.int/emergencies/diseases/novel-coronavirus-2019 [Google Scholar]
- 53.Song Q., Sun X., Dai Z., Gao Y., Gong X., Zhou B., Wu J., Wen W. Point-of-care testing detection methods for COVID-19. Lab Chip. 2021;21:1634–1660. doi: 10.1039/D0LC01156H. [DOI] [PubMed] [Google Scholar]
- 54.GenxXpert, Xpert® Xpress SARS-CoV-2 Instructions for Use. https://www.cepheid.com/Package%20Insert%20Files/Xpert%20Xpress%20SARS-CoV-2%20Assay%20ENGLISH%20Package%20Insert%20302-3787%20Rev.%20B.pdf (Accessed on 05 March 2021).
- 55.BioFire ® COVID-19 Test Instructions for Use. https://www.fda.gov/media/136353/download. (Accessed on 05 March 2021).
- 56.Carter L.J., Garner L.V., Smoot J.W., Li Y., Zhou Q., Saveson C.J., Sasso J.M., Gregg A.C., Soares D.J., Beskid T.R., Jervey S.R., Liu C. Assay techniques and test development for COVID-19 diagnosis. ACS Cent. Sci. 2020;6:591–605. doi: 10.1021/acscentsci.0c00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abbott ID NOW COVID-19. https://www.globalpointofcare.abbott/en/product-details/id-now-covid-19-ous.html. (Accessed on 05 March 2021).
- 58.LuciraTM COVID-19 All-In-One Test Kit Instruction for Use. https://www.lucirahealth.com/wp-content/uploads/2020/11/Lucira-HCP-Instructions-For-Use-IFU.pdf (Accessed on 05 March 2021).
- 59.Click Diagnostics, Inc., Devices and Methods for Molecular Diagnostic Testing. 2018. https://pdfpiw.uspto.gov/.piw?PageNum=0&docid=10052629&IDKey=9B96E763FA9D%0D%0A&HomeUrl=http%3A%2F%2Fpatft.uspto.gov%2Fnetahtml%2FPTO%2Fpatimg.htm US. [Google Scholar]
- 60.Visby Medical COVID-19. https://www.visbymedical.com/static/Visby-Medical-COVID-19-Point-of-Care-Package-Insert-Instructions-for-Use-7a90c6413463be36f720e26c7c07b991.pdf (Accessed on 05 March 2021).
- 61.Petrillo S., Carrà G., Bottino P., Zanotto E., De Santis M.C., Margaria J.P., Giorgio A., Mandili G., Martini M., Cavallo R., Barberio D., Altruda F. A novel multiplex qRT-PCR assay to detect SARS-CoV-2 infection: high sensitivity and increased testing capacity. Microorganisms. 2020;8 doi: 10.3390/microorganisms8071064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ishige T., Murata S., Taniguchi T., Miyabe A., Kitamura K., Kawasaki K., Nishimura M., Igari H., Matsushita K. Highly sensitive detection of SARS-CoV-2 RNA by multiplex rRT-PCR for molecular diagnosis of COVID-19 by clinical laboratories. Clin. Chim. Acta Int. J. Clin. Chem. 2020;507:139–142. doi: 10.1016/j.cca.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rahimi H., Salehiabar M., Barsbay M., Ghaffarlou M., Kavetskyy T., Sharafi A., Davaran S., Chauhan S.C., Danafar H., Kaboli S., Nosrati H., Yallapu M.M., Conde J. CRISPR systems for COVID-19 diagnosis. ACS Sens. 2021;6:1430–1445. doi: 10.1021/acssensors.0c02312. [DOI] [PubMed] [Google Scholar]
- 64.Huang Z., Tian D., Liu Y., Lin Z., Lyon C., Lai W., Fusco D., Drouin A., Yin X., Hu T., Ning B. Ultra-sensitive and high-throughput CRISPR-Powered COVID-19 diagnosis. Biosens. Bioelectron. 2020:112316. doi: 10.1016/j.bios.2020.112316. [DOI] [PMC free article] [PubMed] [Google Scholar]