Abstract
Epithelial ovarian cancer (EOC), a gynecological tumor, is associated with high mortality. MicroRNAs (miRs) serve a crucial role in EOC; however, the mechanisms underlying the effect of miRNA-193a-5p in EOC are not completely understood. Therefore, the present study aimed to investigate the expression levels of miR-193a-5p in serum samples of patients with EOC and to determine the role of miR-193a-5p in EOC. Reverse transcription-quantitative PCR was used to analyze the expression levels of miR-193a-5p in serum samples of patients with EOC and EOC cell lines. The effects of miR-193a-5p and RB binding protein 6, ubiquitin ligase (RBBP6) on the biological functions of EOC were determined by conducting a series of in vitro cell function experiments. The results indicated that the expression levels of miR-193a-5p were significantly decreased in serum samples obtained from patients with EOC and EOC cell lines compared with healthy individuals and normal cells, respectively. Further investigations indicated that RBBP6 was a target gene of miR-193a-5p. The expression levels of RBBP6 were significantly increased in patients with EOC compared with healthy individuals. In addition, in vitro analysis suggested that miR-193a-5p mimic significantly decreased SKOV3 cell proliferation, migration and invasion, and promoted SKOV3 cell apoptosis compared with the control and mimic-negative control groups. In addition, RBBP6 overexpression reversed miR-193a-5p mimic-mediated effects. In conclusion, the results of the present study suggested that downregulated expression levels of miR-193a-5p may serve an inhibitory role in EOC by inhibiting cell proliferation and metastasis, and promoting apoptosis.
Keywords: epithelial ovarian cancer, microRNA-193a-5p, RB binding protein 6, ubiquitin ligase
Introduction
Ovarian cancer is the most common cause of death among patients with female genital tumors, and consists of a variety of pathological types, of which the most common subtype is epithelial ovarian cancer (EOC) (1,2). Due to the ovary being located deep in the pelvic cavity, the early diagnosis of EOC remains difficult, and currently, there is a lack of effective screening methods; therefore, the majority of patients are diagnosed with EOC upon reaching the advanced stages (3–5). In addition, ovarian cancer is prone to extensive pelvic, abdominal and lymph node metastasis, resulting in EOC displaying the highest mortality rate among tumors of the reproductive system (6–8). Tumor initiation and development are affected by numerous factors, including cell movement, the aberrant expression of tumor suppressor genes or oncogenes, and the abnormal regulation of migration (9,10). The current treatment strategy for ovarian cancer is surgery supplemented with platinum-based chemotherapy; however, despite the majority of patients initially responding to treatment, a significant proportion of patients eventually die from recurrence and resistance (11,12). Therefore, investigation of the molecular mechanisms underlying EOC to identify novel treatment targets and improve the survival rate of patients with EOC is required.
MicroRNAs (miRNAs/miRs) are non-coding, endogenous, small single-stranded RNAs that are 20–24 nucleotides in length, which regulate numerous basic biological processes in vivo (13,14). Although miRNAs are non-coding RNAs, by binding with the 3′-untranslated region (3′-UTR) of target mRNAs, miRNAs promote mRNA cleavage or block mRNA translation, thereby controlling the expression of certain target proteins in cells (14,15). In addition, miRNAs also serve important roles during the occurrence, development and metastasis of tumors (16,17). Numerous studies have reported the tumor-suppressive role of miR-193a-5p. For example, miR-193a-5p inhibits HT-29 colon cancer cell metastasis (18). Moreover, miR-193a-5p targeted SPARC (osteonectin), cwcv and kazal like domains proteoglycan 1 to inhibit liver cancer cell migration and proliferation, while promoting apoptosis (18). miR-193a-5p overexpression in vitro and in vivo inhibited the formation of pulmonary metastases in non-small cell lung cancer, as well as inhibiting cell migration, invasion and epithelial-mesenchymal transition (19). Additionally, phosphoinositide-3-kinase regulatory subunit 3 (PIK3R3) and mTOR were identified as direct target genes of miR-193a-5p, thereby indicating that miR-193a-5p inhibited the AKT/mTOR signaling pathway (19,20). However, to the best of our knowledge, the role of miR-193a-5p in EOC has not been previously reported. Therefore, the aim of the present study was to investigate the expression levels of miR-193a-5p in serum samples from patients with EOC and to determine the role of miR-193a-5p in EOC.
Materials and methods
Clinical sample collection
A total of 60 blood samples were collected from 30 female patients with EOC (age, 41.33±3.01 years) and 30 female healthy individuals (age, 43.21±2.47 years) who attended Beijing Ditan Hospital Capital Medical University (Beijing, China) between January 2018 and July 2019. The present study was approved by the Ethics Committee of Beijing Ditan Hospital Capital Medical University. Written informed consent was obtained from all participants. The blood samples were left standing at 4°C for 1 h, then centrifuged at 3,000 × g at 4°C for 5 min to obtain the serum samples.
Cell culture and transfection
EOC cell lines (SKOV3, A2780, HEY, OVCAR3 and Es2) and the normal cell line IOSE386 were purchased from the American Type Culture Collection. Cells were cultured in RPMI-1640 (Invitrogen; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (HyClone; Cytiva) and 100 U/ml penicillin and 100 µg/ml streptomycin (Hyclone; Cytiva), and maintained in a humidified atmosphere of 5% CO2 at 37°C.
After 6 h starvation, SKOV3 cells were cultured in 6-well plates to 70–80% confluence and transfected with miR-negative control (NC) mimic (100 nM; 5′-UUCUCCGAACGUGUCACGUTT-3′; Guangzhou RiboBio Co., Ltd.) or miR-193a-5p mimic (50 nM; 5′- UGGGUCUUUGCGGGCGAGAUGA−3′; Guangzhou RiboBio Co., Ltd.). The pcDNA3.1 RBBP6 overexpression vector (oe-RBBP6; 100 nM; Guangzhou RiboBio Co., Ltd.) or an empty pcDNA3.1 vector (oe-NC; 100 nM; Guangzhou RiboBio Co., Ltd.) using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). Following transfection for 48 h in a humidified atmosphere of 5% CO2 at 37°C, cells were harvested for use in subsequent experiments.
Dual-luciferase reporter assay
To determine the targets of miR-193a-5p, TargetScan (http://www.targetscan.org/mamm_31) was used to predict the target gene of RBBP6. Subsequently, a dual-luciferase reporter assay was performed to verify the findings. Briefly, SKOV3 cells (4×104) were seeded into 24-well plates and cultured for 24 h in a humidified atmosphere of 5% CO2 at 37°C. A total of 4×104 cells/well were plated into 60-mm cell culture dishes and cultured for 24 h until reaching 60–80% confluence. The putative miR-193a-5p binding site in the 3′-UTR of RBBP6 [wild-type (WT) or mutant (MUT)] was cloned into psi-CHECK (Promega Corporation) downstream of the firefly luciferase 3′-UTR. The psi-CHECK vector also provided Renilla luciferase as the normalization signal. Subsequently, 100 ng psi-CHECK-RBBP6-WT or psi-CHECK-RBBP6-MUT luciferase plasmids were co-transfected into SKOV3 cells alongside 100 nM miR-193a-5p mimic or mimic-NC using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). Following incubation for 24 h at 37°C, luciferase activities were detected using a Dual-Luciferase Reporter Assay system (Promega Corporation). Firefly luciferase activities were normalized to Renilla luciferase activities.
Cell Counting Kit-8 (CCK-8) assay
SKOV3 cells were plated (3×103 cells/ml/well) into 96-well plates and separated into two groups: i) Mimic-NC group and ii) miR-193a-5p mimic group. To assess cell proliferation, 10 µl CCK-8 solution was added to each well and incubated for 2 h at 37°C. The absorbance of each well was measured at a wavelength of 450 nm using a microplate reader.
Colony formation assay
SKOV3 cells were plated (3×103 cells/well) into 6-well plates and incubated at 37°C with 5% CO2 for 8–16 days. Following incubation, cells were fixed with 75% methanol for 30 min and stained with 0.5% crystal violet for 15 min, both at 37°C. Stained cells were visualized using an IX51 optical microscope (magnification, ×10; Olympus Corporation) to calculate the number of cell colonies (>50 cells) formed.
Wound healing assay and Transwell assay for determining of cell migration and invasion, respectively
Cell migration and invasion were measured by performing a wound healing and Transwell assay, respectively, as previously described (21).
For the wound healing assay, 5×105 SKOV3 cells/well were plated into six-well plates and cultured until 100% confluence in DMEM supplemented with 10% FBS at 37°C. Subsequently, the monolayer of cells was scratched with a 200-µl sterile pipette tip and the cells were incubated in serum-free DMEM for 24 h at 37°C. The migratory distance of the cells was observed under a light microscope (magnification, ×200; Olympus Corporation) and analyzed using ImageJ version 1.49 software (National Institutes of Health).
For the cell invasion assay, the upper chambers of Transwell plates (BD Biosciences) were precoated with Matrigel (BD Biosciences) at 37°C for 2 h, then cells (5×106 cells/ml) were seeded into the upper chambers in serum-free DMEM (Invitrogen; Thermo Fisher Scientific, Inc.) supplemented with 2.5% FBS was plated into the lower chambers. Following culture for 24 h at 37°C, cells on the upper surface of the Transwell membrane were removed with a cotton swab. Cells on the lower surface of the Transwell membrane were washed with PBS and fixed with 4% paraformaldehyde at room temperature for 30 min. Subsequently, the paraformaldehyde was discarded, and cells were stained with 0.1% crystal violet at room temperature for 15 min, prior to being observed in five fields of view using a microscope (Olympus Corporation; magnification, ×200). The total number of cells in each field of view was recorded and the mean number of cells was calculated.
TUNEL assay
SKOV3 cells (3×103 cells/well) were fixed in 4% formaldehyde at room temperature for 10 min and washed with PBS buffer solution twice. SKOV3 cell apoptosis was investigated using a TUNEL assay kit (cat. no. ab206386; Abcam), according to the manufacturer's protocol. TUNEL-positive cells presented with yellow nuclei following visualization using a fluorescence microscope (Nikon Eclipse 80i; Nikon Corporation; magnification, ×200) in five randomly selected fields of view.
Western blotting
Total protein was extracted from SKOV3 cells using RIPA lysis buffer (Beyotime Institute of Biotechnology) at 4°C for 1 h. Total protein was quantified using a Pierce™ BCA Protein assay kit (Thermo Fisher Scientific, Inc.) and protein (30 µg/lane) was separated via 10% SDS-PAGE and transferred onto PVDF membranes. The membranes were blocked with 5% skim milk in TBS-0.05% Tween-20 (TBST) for 1 h at room temperature. Subsequently, the membranes were incubated overnight with the following primary antibodies in blocking buffer at 4°C: Bcl2 (1:1,000; cat. no. ab182858; Abcam), Bax (1:1,000; cat. no. ab32503; Abcam), cleaved-caspase 3 (1:1,000; cat. no. ab49822; Abcam), cleaved-caspase 7 (1:1,000; cat. no. ab32042; Abcam), cleaved-caspase 9 (1:1,000; cat. no. ab2324; Abcam), RBBP6 (1:1,000; cat. no. ab237514; Abcam), Ki67 (1:1,000; cat. no. ab15580; Abcam), proliferating cell nuclear antigen (PCNA; 1:1,000; cat. no. 13110; Cell Signaling Technology, Inc.), matrix metallopeptidase (MMP)-2 (1:1,000; cat. no. 40994; Cell Signaling Technology, Inc.), MMP-9 (1:1,000; cat. no. 13667; Cell Signaling Technology, Inc.) and GAPDH (1:2,000; cat. no. ab181602; Abcam). Following primary incubation, the membranes were washed with TBST and incubated with a horseradish peroxidase-conjugated secondary antibody (1:10,000; cat. no. ab205718; Abcam) for 2 h at room temperature. Proteins bands were visualized using an ECL reagent (Invitrogen; Thermo Fisher Scientific, Inc.). Densitometric analysis was performed using ImageJ software (version 1.49v; National Institutes of Health).
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from serum samples and SKOV3 cells with TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). Total RNA was reverse transcribed using RevertAid reverse transcriptase (Invitrogen; Thermo Fisher Scientific, Inc.) at 42°C for 1 h, according to the manufacturer's protocol. qPCR was performed using the LightCycler 480 SYBR Green I Master kit (Roche Diagnostics) on a LightCycler 480 II (Roche Diagnostics). The following thermocycling conditions were used for qPCR: Initial denaturation at 95°C for 5 min; followed by 45 cycles of amplification, including denaturation at 94°C for 10 sec, annealing at 60°C for 20 sec and a final extension at 72°C for 30 sec. The following primers were used for qPCR: miR-193a-5p forward, 5′-ACACTCCAGCTGGGTGGGTCTTTGCGGGCG-3′ and reverse, 5′-TGGTGTCGTGGAGTCG-3′; RBBP6 forward, 5′-CTCCCCATACACTTCCTCTCC-3′ and reverse, 5′-TTCTTTTAGTCGTCGCTGCTC-3′; GAPDH forward, 5′-GAGCCATGAGGGAGGCTG-3′ and reverse, 5′-CAGTTGAATCAGATGGATCC-3′; and U6 forward, 5′-ATCGTCCGTGATCAGCGT-3′ and reverse, 5′-GCAGGTGGTCGGTCGA-3′. miRNA and mRNA expression levels were quantified using the 2−∆∆Cq method and normalized to the internal reference genes U6 and GAPDH, respectively (22).
Statistical analysis
Statistical analyses were performed using SPSS software (version 18.0; SPSS, Inc.). Data are presented as the mean ± SD. Statistical differences among groups were determined using a one-way ANOVA followed by a Tukey's or Dunnett's post hoc test. P<0.05 was considered to indicate a statistically significant difference. All experiments were repeated at least three times.
Results
miR-193a-5p is abnormally expressed in EOC
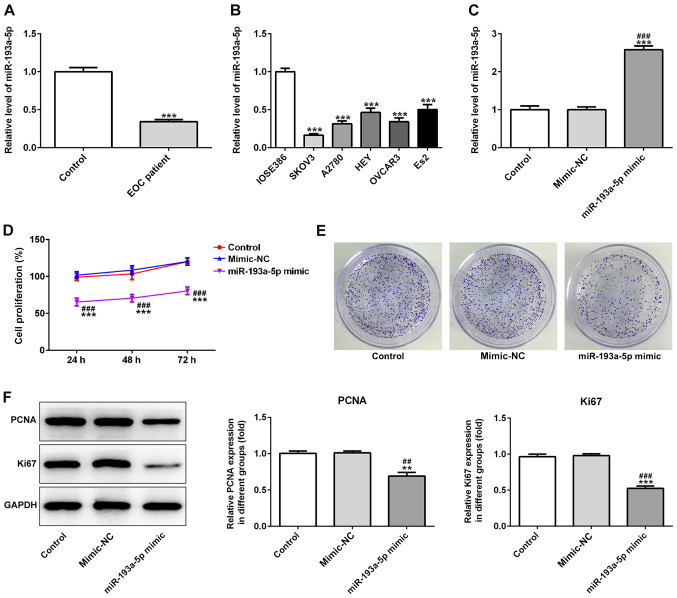
The expression levels of miR-193a-5p in 30 serum samples obtained from patients with EOC were significantly decreased compared with serum samples obtained from healthy individuals (Fig. 1A). To investigate the effects of miR-193a-5p, the expression levels of miR-193a-5p were analyzed in EOC cells. The results indicated that miR-193a-5p expression levels were decreased in EOC cells compared with normal cells (Fig. 1B).
Figure 1.
Expression of miR-193a-5p in EOC. miR-193a-5p expression levels in (A) serum samples obtained from patients with EOC and (B) EOC cell lines. (C) Transfection efficiency of miR-193a-5p mimic. (D) Cell Counting Kit-8 and (E) colony formation assays were performed to investigate the effect of miR-193a-5p overexpression on cell proliferation. Magnification, ×10. (F) Protein expression levels of proliferation-related proteins (PCNA and Ki67). **P<0.01 and ***P<0.001 vs. control or IOSE386. ##P<0.01 and ###P<0.001 vs. mimic-NC. miR, microRNA; EOC, epithelial ovarian cancer; PCNA, proliferating cell nuclear antigen; NC, negative control.
miR-193a-5p regulates EOC cell progression
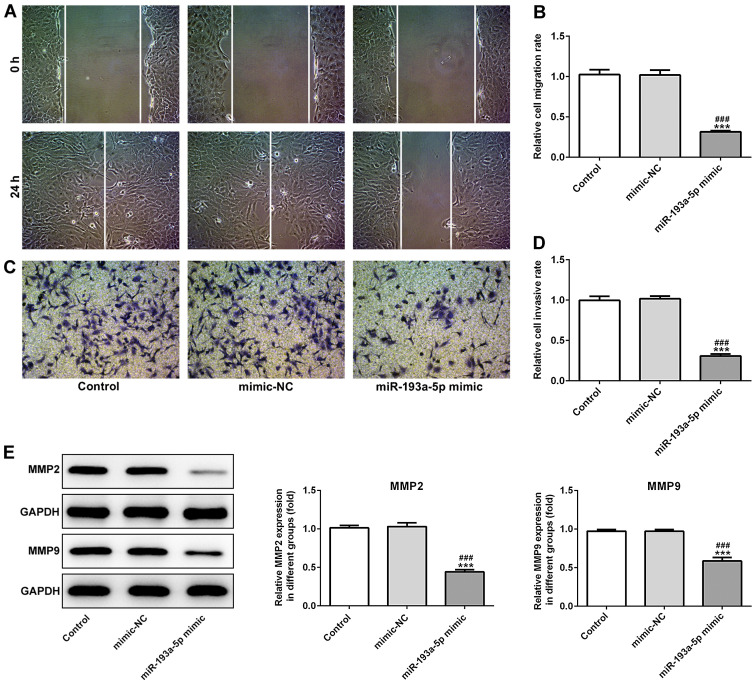
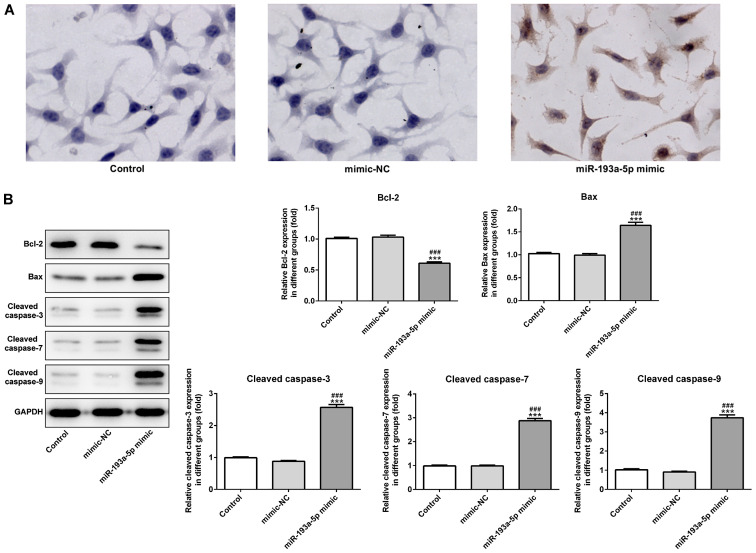
miR-193a-5p mimic significantly increased the expression levels of miR-193a-5p in SKOV3 cells compared with the control and mimic-NC groups (Fig. 1C). In addition, the CCK-8 assay indicated that miR-193a-5p overexpression significantly decreased SKOV3 cell proliferation compared with the control and mimic-NC groups (Fig. 1D). Moreover, the colony formation assay indicated that miR-193a-5p overexpression notably decreased SKOV3 colony formation compared with the control and mimic-NC groups (Fig. 1E). Furthermore, miR-193a-5p overexpression significantly decreased the expression levels of proliferation-related proteins, PCNA and Ki67, compared with the control and mimic-NC groups, which also suggested that miR-193a-5p regulated EOC cell proliferation (Fig. 1F). Similarly, miR-193a-5p overexpression significantly inhibited SKOV3 cell migration compared with the control and mimic-NC groups, as indicated by the Transwell and wound healing assays (Fig. 2A-D). Moreover, miR-193a-5p overexpression significantly decreased the expression levels of MMP2 and MMP9 compared with the control and mimic-NC groups (Fig. 2E), suggesting that miR-193a-5p may potentially modulate cell migration. Furthermore, the effects of miR-193a-5p on SKOV3 cell apoptosis were analyzed by performing a TUNEL assay and western blotting. The TUNEL assay indicated that miR-193a-5p overexpression notably increased SKOV3 cell apoptosis compared with the control and mimic-NC groups (Fig. 3A). Furthermore, the expression levels of proapoptotic proteins, Bax, cleaved caspase-3, −7 and −9, were significantly increased in the miR-193a-5p mimic group compared with the control and mimic-NC group. By contrast, miR-193a-5p overexpression significantly decreased the expression levels of the antiapoptotic protein Bcl2 compared with the control and mimic-NC groups (Fig. 3B).
Figure 2.
miR-193a-5p regulates SKOV3 cell migration and invasion. Cell migration was (A) determined by performing the wound healing assay (magnification, ×200) and (B) quantified. Cell migration was also (C) assessed by performing the Transwell assay (magnification, ×200) and (D) quantified. (E) Western blotting was performed to assess the expression levels of MMP2 and MMP9. ***P<0.001 vs. control; ###P<0.001 vs. mimic-NC. miR, microRNA; MMP, matrix metallopeptidase; NC, negative control.
Figure 3.
miR-193a-5p regulates SKOV3 cell apoptosis. (A) The TUNEL assay was performed to assess the effect of miR-193a-5p on SKOV3 cell apoptosis. Magnification, ×400. (B) Western blotting was performed to investigate the expression of apoptosis-related proteins. ***P<0.001 vs. control; ###P<0.001 vs. mimic-NC. miR, microRNA; NC, negative control.
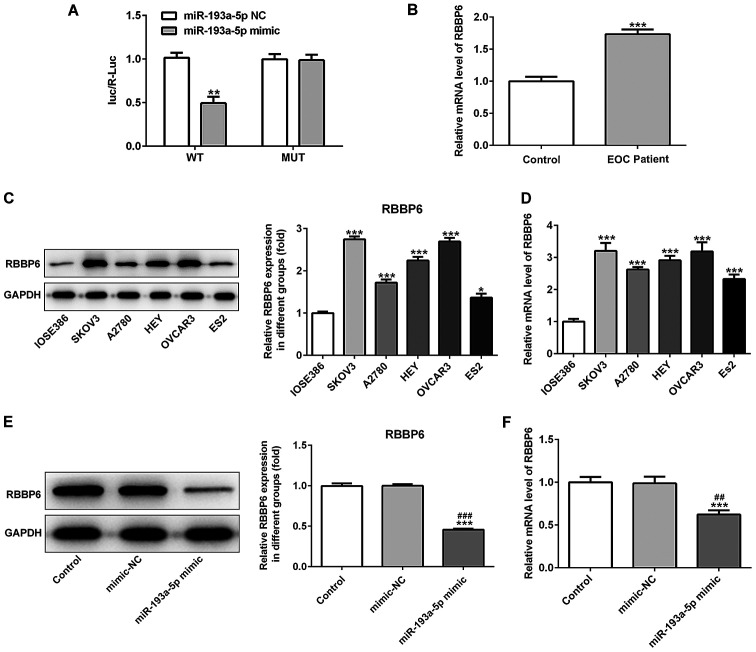
RBBP6 is a target gene of miR-193a-5p
RBBP6 was predicted as the target gene of miR-193a-5p using TargetScan. Subsequently, a dual-luciferase reporter assay was performed to verify RBBP6 as a target gene of miR-193a-5p. The reporter vector containing RBBP6-WT exhibited markedly decreased luciferase activity in SKOV3 cells co-transfected with the miR-193a-5p mimic compared with cells co-transfected with the miR-193a-5p NC (Fig. 4A). However, the statistical difference between cells co-transfected with the miR-193a-5p mimic or miR-193a-5p NC and MUT reporters was not significant. Taken together, these results showed that RBBP6 was the direct target gene of miR-193a-5p. The mRNA expression levels of RBBP6 in serum samples obtained from patients with EOC were significantly increased compared with blood samples obtained from healthy individuals (Fig. 4B). Furthermore, the protein and mRNA expression levels of RBBP6 in EOC cell lines were significantly increased compared with IOSE386 cells (Fig. 4C and D). In addition, RBBP6 expression levels were significantly decreased in the miR-193a-5p mimic group compared with the control and mimic-NC groups (Fig. 4E and F).
Figure 4.
RBBP6 is a target gene of miR-193a-5p. (A) The dual-luciferase reporter assay was performed to verify that RBBP6 was a target gene of miR-193a-5p. (B) mRNA expression levels of RBBP6 in serum samples obtained from patients with EOC. (C) Protein and (D) mRNA expression levels of RBBP6 in epithelial ovarian cancer cell lines. Effect of miR-193a-5p overexpression on (E) protein and (F) mRNA expression levels of RBBP6. *P<0.05, **P<0.01 and ***P<0.001 vs. control or IOSE386; ##P<0.01 and ###P<0.001 vs. mimic-NC. RBBP6, RB binding protein 6, ubiquitin ligase; miR, microRNA; NC, negative control; WT, wild-type; MUT, mutant; luc, luciferase; R, Renilla.
miR-193a-5p exerts antitumor effects following RBBP6 overexpression
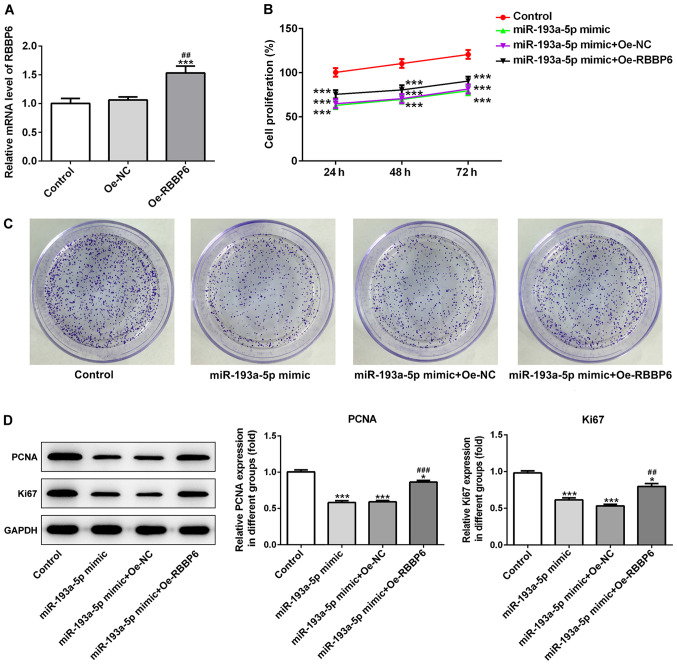
Subsequently, the RBBP6 overexpression vector was transfected into SKOV3 cells to determine whether RBBP6 served a role in the effects of miR-193a-5p on cell proliferation, migration and apoptosis. The transfection efficacy of RBBP6 overexpression in SKOV3 cells was determined via RT-qPCR (Fig. 5A). RBBP6 overexpression partially reversed miR-193a-5p mimic-mediated effects on cell proliferation (Fig. 5B-D), migration (Fig. 6A-C) and apoptosis (Fig. 7A and B). Collectively, the results suggested that miR-193a-5p may inhibit cell proliferation, migration and apoptosis by regulating RBBP6.
Figure 5.
RBBP6 overexpression reverses miR-193a-5p mimic-mediated effects in SKOV3 cells. (A) Transfection efficiency of RBBP6 overexpression. (B) Cell Counting Kit-8 and (C) colony formation assays were performed to investigate cell proliferation. Magnification, ×10. (D) Western blotting was performed to investigate the expression of proliferation-related proteins (PCNA and Ki67). *P<0.05 and ***P<0.001 vs. control; ##P<0.01 and ###P<0.001 vs. miR-193a-5p mimic + Oe-NC. RBBP6, RB binding protein 6, ubiquitin ligase; miR, microRNA; PCNA, proliferating cell nuclear antigen; Oe, overexpression; NC, negative control.
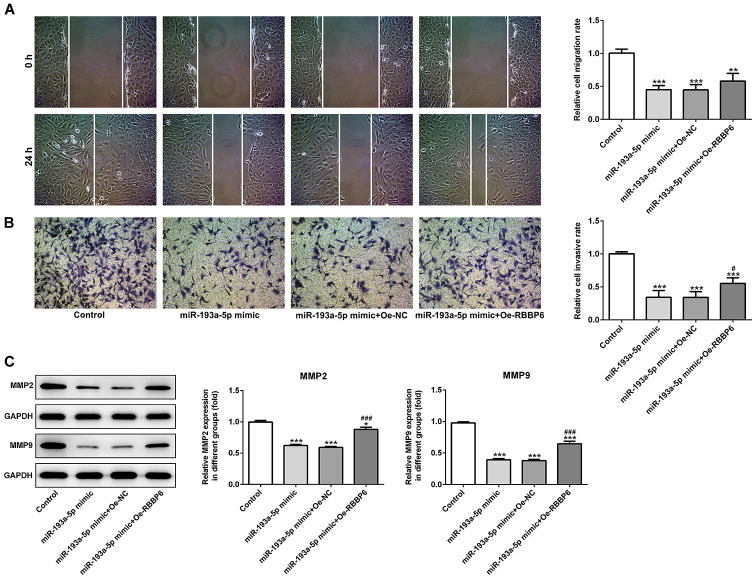
Figure 6.
miR-193a-5p-mediated regulation of SKOV3 cell migration and invasion is mediated by RBBP6 overexpression. (A) Wound healing and (B) Transwell assays were conducted to assess cell migration and invasion, respectively. Magnification, ×200. (C) Western blotting was performed to assess the expression levels of MMP2 and MMP9. *P<0.05, **P<0.01 and ***P<0.001 vs. control; #P<0.05 and ###P<0.001 vs. miR-193a-5p mimic + Oe-NC. miR, microRNA; RBBP6, RB binding protein 6, ubiquitin ligase; MMP, matrix metallopeptidase; Oe, overexpression; NC, negative control. Magnification, ×200.
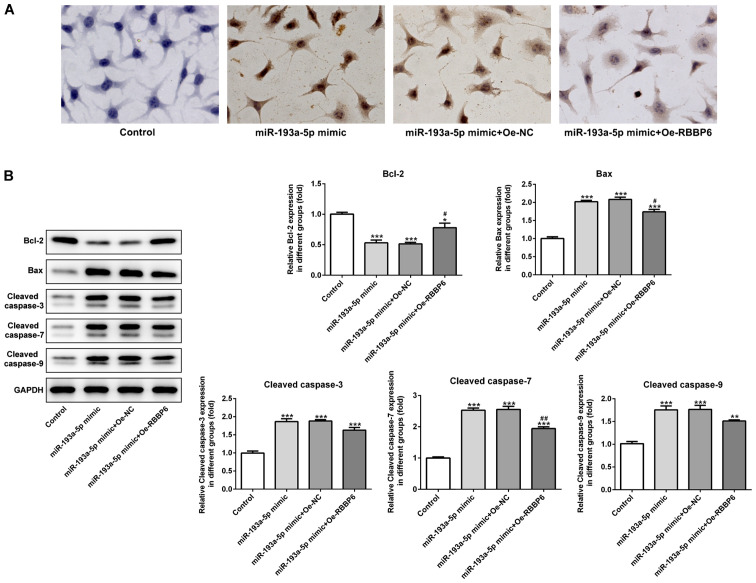
Figure 7.
miR-193a-5p-mediated regulation of SKOV3 cell apoptosis is mediated by RBBP6 overexpression. (A) The TUNEL assay was performed to assess the effect of miR-193a-5p and RBBP6 on SKOV3 cell apoptosis. Magnification, ×400. (B) Western blotting was performed to investigate the expression of apoptosis-related proteins. *P<0.05, **P<0.01 and ***P<0.001 vs. control; #P<0.05 and ##P<0.01 vs. miR-193a-5p mimic + Oe-NC. miR, microRNA; RBBP6, RB binding protein 6, ubiquitin ligase; Oe, overexpression; NC, negative control.
Discussion
The poor prognosis of ovarian cancer is associated with the fact that the majority of patients with EOC are diagnosed at an advanced stage; therefore, the early diagnosis of ovarian cancer may aid with controlling disease progression and reducing mortality rates (23). miRNAs affect tumor cell proliferation and apoptosis by regulating cytokines (24,25). For example, miR-193a-5p expression levels are downregulated in numerous types of cancer, including colon cancer, non-small cell lung cancer, human endometrioid endometrial adenocarcinoma and prostate cancer, which has been reported to be associated with tumor progression (18–20,26,27). A previous study also reported an improvement to membranous nephropathy following miR-193a inhibition, which affected podocytosis by targeting WT1 transcription factor (WT1) (28). Jin et al (29) demonstrated that miR-193a-5p exerted a tumor-suppressive role in glioblastoma via modulating NOVA alternative splicing regulator 1. In addition, Shirafkan et al (18) identified that miR-193a-5p inhibited human HT-29 colon cancer cell migration by suppressing the metastatic pathway. miR-193a-5p also suppressed human non-small-cell lung cancer metastasis by downregulating the erb-b2 receptor tyrosine kinase 4/PIK3R3/mTOR/ribosomal protein S6 kinase B2 signaling pathway (20). In human endometrioid endometrial carcinoma, a novel miR-193a-5p-YY1 transcription factor- APC regulator of WNT signaling pathway regulatory axis was identified (26). By contrast, miR-193a-5p knockdown increased the chemosensitivity of prostate cancer cells to docetaxel (30). Furthermore, miR-193a downregulation contributes to non-small cell lung cancer metastasis by targeting the WT1/E-cadherin axis (31).
miRNAs serve a role in cells and other parts of the body via circulating in peripheral blood and other body fluids (32). Previous studies have demonstrated that serum and plasma miRNAs may serve as non-invasive biomarkers due to their availability and long-term stability (33–35); therefore, miRNAs may be used for the early diagnosis of various types of cancer (36). miR-193a-3p is associated with cancer. For example, miR-193a-3p overexpression in esophageal squamous cell carcinoma regulates cancer cell proliferation, migration and apoptosis. Lin et al (37) demonstrated that the expression level of miR-193a-5p was negatively correlated with metastasis and poor prognosis in patients with esophageal squamous cell carcinoma. The serum expression level of miR-193a-5p can reflect the expression level of miR-193a-5p in tissues and cells; therefore, serum miR-193a-5p expression levels can be used as a noninvasive diagnostic indicator for the prognosis of esophageal cancer (37). The present study indicated that miR-193a-5p expression levels were significantly decreased in serum samples obtained from patients with EOC and EOC cell lines compared with serum samples obtained from healthy individuals and normal cells, respectively. The results also suggested a potential diagnostic value and molecular mechanism underlying miR-193a-5p in EOC. Further investigations predicted that RBBP6 was a target gene of miR-193a-5p. RBBP6 has been reported to be associated with various types of cancer. For example, Yoshitake et al (38) reported that RBBP6 expression levels were significantly upregulated in invasive esophageal cancer, suggesting that RBBP6 may promote tumor cell proliferation and may serve as a promising target for immunotherapy. In addition, RBBP6 knockdown in breast cancer cells significantly inhibited cell proliferation (39). Similarly, the increased expression levels of RBBP6 reported in lung cancer tissues suggested that RBBP6 may be involved in promoting malignancy in lung tissues, and inhibition of RBBP6 target genes may render cancer cells sensitive to apoptosis (40). RBBP6 has also been identified as a potential marker of apoptosis and cell cycle arrest in cervical cancer (41). The present study demonstrated that the expression levels of RBBP6 were significantly increased in patients with EOC compared with healthy individuals. Moreover, in vitro analysis indicated that miR-193a-5p mimic significantly decreased SKOV3 cell viability, migration and invasion, but promoted SKOV3 cell apoptosis compared with the control and mimic-NC groups. However, RBBP6 overexpression reversed miR-193a-5p overexpression-mediated effects.
In conclusion, the results of the present study suggested that upregulated expression levels of miR-193a-5p may serve an inhibitory role in EOC via inhibiting cell proliferation and migration, and promoting apoptosis. Moreover, the effects of miR-193a-5p may be partly mediated by RBBP6. Collectively, the results indicated that miR-193a-5p may serve as a potential therapeutic target for EOC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
JL and JH collected and analyzed the data. SZ and NY performed the experiments, conceived and designed the study, confirm the authenticity of all the raw data and drafted and revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Beijing Ditan Hospital Capital Medical University. Written informed consent was obtained from all participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.De A, De A, Sharma R, Suo W, Sharma M. Sensitization of Carboplatinum- and Taxol-Resistant High-Grade Serous Ovarian Cancer Cells Carrying p53, BRCA1/2 Mutations by Emblica officinalis (Amla) via Multiple Targets. J Cancer. 2020;11:1927–1939. doi: 10.7150/jca.36919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chua KJC, Patel RD, Trivedi R, Greenberg P, Beiter K, Magliaro T, Patel U, Varughese J. Accuracy in Referrals to Gynecologic Oncologists Based on Clinical Presentation for Ovarian Mass. Diagnostics (Basel) 2020;10:106. doi: 10.3390/diagnostics10020106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang D, Gaul DA, Nan H, Kim J, Fernández FM. Deep Metabolomics of a High-Grade Serous Ovarian Cancer Triple-Knockout Mouse Model. J Proteome Res. 2019;18:3184–3194. doi: 10.1021/acs.jproteome.9b00263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giampaolino P, Della Corte L, Foreste V, Vitale SG, Chiofalo B, Cianci S, Zullo F, Bifulco G. Unraveling a difficult diagnosis: The tricks for early recognition of ovarian cancer. Minerva Med. 2019;110:279–291. doi: 10.23736/S0026-4806.19.06086-5. [DOI] [PubMed] [Google Scholar]
- 5.Leskela S, Romero I, Cristobal E, Pérez-Mies B, Rosa-Rosa JM, Gutierrez-Pecharroman A, Santón A, Gonzalez BO, López-Reig R, Hardisson D, et al. The Frequency and Prognostic Significance of the Histologic Type in Early-stage Ovarian Carcinoma: A Reclassification Study by the Spanish Group for Ovarian Cancer Research (GEICO) Am J Surg Pathol. 2020;44:149–161. doi: 10.1097/PAS.0000000000001365. [DOI] [PubMed] [Google Scholar]
- 6.Della Corte L, Giampaolino P, Fabozzi A, Cieri M, Zizolfi B, Morra I, Bifulco G. Breast metastasis two years after pelvic surgery and adjuvant chemotherapy for serous ovarian cancer. Gynecol Endocrinol. 2019;35:211–213. doi: 10.1080/09513590.2018.1521795. [DOI] [PubMed] [Google Scholar]
- 7.Buensuceso A, Ramos-Valdes Y, DiMattia GE, Shepherd TG. AMPK-Independent LKB1 Activity Is Required for Efficient Epithelial Ovarian Cancer Metastasis. Mol Cancer Res. 2020;18:488–500. doi: 10.1158/1541-7786.MCR-19-0530. [DOI] [PubMed] [Google Scholar]
- 8.Tato-Varela S, Kuhn W. Impact of retroperitoneal lymph node dissection in ovarian cancer - time for paradigm shift? Horm Mol Biol Clin Investig. 2019;41:20190020. doi: 10.1515/hmbci-2019-0020. [DOI] [PubMed] [Google Scholar]
- 9.Kontomanolis EN, Fasoulakis Z, Papamanolis V, Koliantzaki S, Dimopoulos G, Kambas NJ. The Impact of microRNAs in Breast Cancer Angiogenesis and Progression. MicroRNA. 2019;8:101–109. doi: 10.2174/2211536607666181017122921. [DOI] [PubMed] [Google Scholar]
- 10.Mucha O, Podkalicka P, Mikulski M, Barwacz S, Andrysiak K, Biela A, Mieczkowski M, Kachamakova-Trojanowska N, Ryszawy D, Białas A, et al. Development and characterization of a new inhibitor of heme oxygenase activity for cancer treatment. Arch Biochem Biophys. 2019;671:130–142. doi: 10.1016/j.abb.2019.07.002. [DOI] [PubMed] [Google Scholar]
- 11.Grayson K, Gregory E, Khan G, Guinn BA. Urine Biomarkers for the Early Detection of Ovarian Cancer - Are We There Yet? Biomark Cancer. 2019;11:1179299X19830977. doi: 10.1177/1179299X19830977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stewart D, Cristea M. Antibody-drug conjugates for ovarian cancer: Current clinical development. Curr Opin Obstet Gynecol. 2019;31:18–23. doi: 10.1097/GCO.0000000000000515. [DOI] [PubMed] [Google Scholar]
- 13.Zou X, Zhu D, Zhang H, Zhang S, Zhou X, He X, Zhu J, Zhu W. MicroRNA expression profiling analysis in serum for nasopharyngeal carcinoma diagnosis. Gene. 2020;727:144243. doi: 10.1016/j.gene.2019.144243. [DOI] [PubMed] [Google Scholar]
- 14.Jain N, Das B, Mallick B. Restoration of microRNA-197 expression suppresses oncogenicity in fibrosarcoma through negative regulation of RAN. IUBMB Life. 2020;72:1034–1044. doi: 10.1002/iub.2240. [DOI] [PubMed] [Google Scholar]
- 15.Tani S, Kusakabe R, Naruse K, Sakamoto H, Inoue K. Genomic organization and embryonic expression of miR-430 in medaka (Oryzias latipes): Insights into the post-transcriptional gene regulation in early development. Gene. 2010;449:41–49. doi: 10.1016/j.gene.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 16.Liu Z, Wang Y, Dou C, Sun L, Li Q, Wang L, Xu Q, Yang W, Liu Q, Tu K. MicroRNA-1468 promotes tumor progression by activating PPAR-γ-mediated AKT signaling in human hepatocellular carcinoma. J Exp Clin Cancer Res. 2018;37:49. doi: 10.1186/s13046-018-0717-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Dong D, Mu Z, Wei N, Sun M, Wang W, Xin N, Shao Y, Zhao C. Long non-coding RNA ZFAS1 promotes proliferation and metastasis of clear cell renal cell carcinoma via targeting miR-10a/SKA1 pathway. Biomed Pharmacother. 2019;111:917–925. doi: 10.1016/j.biopha.2018.12.143. [DOI] [PubMed] [Google Scholar]
- 18.Shirafkan N, Shomali N, Kazemi T, Shanehbandi D, Ghasabi M, Baghbani E, Ganji M, Khaze V, Mansoori B, Baradaran B. MicroRNA-193a-5p inhibits migration of human HT-29 colon cancer cells via suppression of metastasis pathway. J Cell Biochem. 2018 Dec 2; doi: 10.1002/jcb.28164. (Epub ahead of print). doi: 10.1002/jcb.28164. [DOI] [PubMed] [Google Scholar]
- 19.Chen J, Gao S, Wang C, Wang Z, Zhang H, Huang K, Zhou B, Li H, Yu Z, Wu J, et al. Pathologically decreased expression of miR-193a contributes to metastasis by targeting WT1-E-cadherin axis in non-small cell lung cancers. J Exp Clin Cancer Res. 2016;35:173. doi: 10.1186/s13046-016-0450-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu T, Li J, Yan M, Liu L, Lin H, Zhao F, Sun L, Zhang Y, Cui Y, Zhang F, et al. MicroRNA-193a-3p and −5p suppress the metastasis of human non-small-cell lung cancer by downregulating the ERBB4/PIK3R3/mTOR/S6K2 signaling pathway. Oncogene. 2015;34:413–423. doi: 10.1038/onc.2013.574. [DOI] [PubMed] [Google Scholar]
- 21.Ding N, Sun X, Wang T, Huang L, Wen J, Zhou Y. miR-378a-3p exerts tumor suppressive function on the tumorigenesis of esophageal squamous cell carcinoma by targeting Rab10. Int J Mol Med. 2018;42:381–391. doi: 10.3892/ijmm.2018.3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen Td. Analysis of relative gene expression data using real-time quantitative Pcr and the 2(−delta deltac(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Hou M, Cheng Z, Shen H, He S, Li Y, Pan Y, Feng C, Chen X, Zhang Y, Lin M, et al. Correction: High expression of CTHRC1 promotes EMT of epithelial ovarian cancer (EOC) and is associated with poor prognosis. Oncotarget. 2020;11:825–826. doi: 10.18632/oncotarget.27370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie Y, Zhao J, Liang Y, Chen M, Luo Y, Cui X, Jiang B, Peng L, Wang X. MicroRNA-10b controls the metastasis and proliferation of colorectal cancer cells by regulating Krüppel-like factor 4. Artif Cells Nanomed Biotechnol. 2019;47:1722–1729. doi: 10.1080/21691401.2019.1606006. [DOI] [PubMed] [Google Scholar]
- 25.Hu S, Zang R, Wang Y, Liang Y, Mu J, Zhang Y, Ma J. Highly expressed microRNA-124 inhibits migration and promotes apoptosis of esophageal cancer cells by degrading PDCD6. J BUON. 2019;24:805–812. [PubMed] [Google Scholar]
- 26.Yang Y, Zhou L, Lu L, Wang L, Li X, Jiang P, Chan LK, Zhang T, Yu J, Kwong J, et al. A novel miR-193a-5p-YY1-APC regulatory axis in human endometrioid endometrial adenocarcinoma. Oncogene. 2013;32:3432–3442. doi: 10.1038/onc.2012.360. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y, Jiang F, He H, Ye J, Mao X, Guo Q, Wu SL, Zhong W, Wu CL, Lin N. Identification of a novel microRNA-mRNA regulatory biomodule in human prostate cancer. Cell Death Dis. 2018;9:301. doi: 10.1038/s41419-018-0293-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J, Chen Y, Shen L, Deng Y. Improvement of membranous nephropathy by inhibition of miR-193a to affect podocytosis via targeting WT1. J Cell Biochem. 2019;120:3438–3446. doi: 10.1002/jcb.27616. [DOI] [PubMed] [Google Scholar]
- 29.Jin L, Li H, Wang J, Lin D, Yin K, Lin L, Lin Z, Lin G, Wang H, Ying X, et al. MicroRNA-193a-5p exerts a tumor suppressor role in glioblastoma via modulating NOVA1. J Cell Biochem. 2019;120:6188–6197. doi: 10.1002/jcb.27906. [DOI] [PubMed] [Google Scholar]
- 30.Yang Z, Chen JS, Wen JK, Gao HT, Zheng B, Qu CB, Liu KL, Zhang ML, Gu JF, Li JD, et al. Silencing of miR-193a-5p increases the chemosensitivity of prostate cancer cells to docetaxel. J Exp Clin Cancer Res. 2017;36:178. doi: 10.1186/s13046-017-0649-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Finetti F, Moglia A, Schiavo I, Donnini S, Berta GN, Di Scipio F, Perrelli A, Fornelli C, Trabalzini L, Retta SF. Yeast-Derived Recombinant Avenanthramides Inhibit Proliferation, Migration and Epithelial Mesenchymal Transition of Colon Cancer Cells. Nutrients. 2018;10:1159. doi: 10.3390/nu10091159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stepien A, Knop K, Dolata J, Taube M, Bajczyk M, Barciszewska-Pacak M, Pacak A, Jarmolowski A, Szweykowska-Kulinska Z. Posttranscriptional coordination of splicing and miRNA biogenesis in plants. Wiley Interdiscip Rev RNA. 2017 May 8; doi: 10.1002/wrna.1403. (Epub ahead of print). doi: 10.1002/wrna.1403. [DOI] [PubMed] [Google Scholar]
- 33.Movafagh S, Crook S, Vo K. Regulation of hypoxia-inducible factor-1a by reactive oxygen species: New developments in an old debate. J Cell Biochem. 2015;116:696–703. doi: 10.1002/jcb.25074. [DOI] [PubMed] [Google Scholar]
- 34.Sideris M, Papagrigoriadis S. Molecular biomarkers and classification models in the evaluation of the prognosis of colorectal cancer. Anticancer Res. 2014;34:2061–2068. [PubMed] [Google Scholar]
- 35.Maroof H, Salajegheh A, Smith RA, Lam AK. Role of microRNA-34 family in cancer with particular reference to cancer angiogenesis. Exp Mol Pathol. 2014;97:298–304. doi: 10.1016/j.yexmp.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 36.Wang MJ, Xu YY, Huang RY, Chen XM, Chen HM, Han L, Yan YH, Lu CJ. Role of an imbalanced miRNAs axis in pathogenesis of psoriasis: Novel perspectives based on review of the literature. Oncotarget. 2017;8:5498–5507. doi: 10.18632/oncotarget.12534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin CH, Tsai CH, Yeh CT, Liang JL, Hung WC, Lin FC, Chang WL, Li HY, Yao YC, Hsu TI, et al. MiR-193a-5p/ERBB2 act as concurrent chemoradiation therapy response indicator of esophageal squamous cell carcinoma. Oncotarget. 2016;7:39680–39693. doi: 10.18632/oncotarget.9444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoshitake Y, Nakatsura T, Monji M, Senju S, Matsuyoshi H, Tsukamoto H, Hosaka S, Komori H, Fukuma D, Ikuta Y, et al. Proliferation potential-related protein, an ideal esophageal cancer antigen for immunotherapy, identified using complementary DNA microarray analysis. Clin Cancer Res. 2004;10:6437–6448. doi: 10.1158/1078-0432.CCR-04-0841. [DOI] [PubMed] [Google Scholar]
- 39.Moela P, Choene MM, Motadi LR. Silencing RBBP6 (Retinoblastoma Binding Protein 6) sensitises breast cancer cells MCF7 to staurosporine and camptothecin-induced cell death. Immunobiology. 2014;219:593–601. doi: 10.1016/j.imbio.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 40.Motadi LR, Bhoola KD, Dlamini Z. Expression and function of retinoblastoma binding protein 6 (RBBP6) in human lung cancer. Immunobiology. 2011;216:1065–1073. doi: 10.1016/j.imbio.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 41.Moela P, Motadi LR. RBBP6: A potential biomarker of apoptosis induction in human cervical cancer cell lines. OncoTargets Ther. 2016;9:4721–4735. doi: 10.2147/OTT.S100964. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.