Summary
SARS-CoV-2 is responsible for the global COVID-19 pandemic. Angiotensin converting enzyme 2 (ACE2) is the membrane-delimited receptor for SARS-CoV-2. Lung, intestine, and kidney, major sites of viral infection, express ACE2 that harbors an intracellular, carboxy-terminal PDZ-recognition motif. These organs prominently express the PDZ protein Na+/H+ exchanger regulatory factor-1 (NHERF1). Here, we report NHERF1 tethers ACE2 and augments SARS-CoV-2 cell entry. ACE2 directly binds both NHERF1 PDZ domains. Disruption of either NHERF1 PDZ core-binding motif or the ACE2 PDZ recognition sequence eliminates interaction. Proximity ligation assays establish that ACE2 and NHERF1 interact at constitutive expression levels in human lung and intestine cells. Ablating ACE2 interaction with NHERF1 accelerated SARS-CoV-2 cell entry. Conversely, elimination of the ACE2 C-terminal PDZ-binding motif decreased ACE2 membrane residence and reduced pseudotyped virus entry. We conclude that the PDZ interaction of ACE2 with NHERF1 facilitates SARS-CoV-2 internalization. β-Arrestin is likely indispensable, as with G protein-coupled receptors.
Subject areas: molecular biology, virology
Graphical abstract
Highlights
-
•
The SARS-CoV-2 receptor ACE2 C-terminal PDZ-recognition motif 802QTSF805 binds to NHERF1
-
•
NHERF1 and ACE2 interact directly in SARS-CoV-2-susceptible lung and intestine cells
-
•
NHERF1 expression correlates with SARS-CoV-2 entry by regulating ACE2 membrane abundance
-
•
β-Arrestins may cooperate with NHERF1 to promote ACE2-mediated SARS-CoV-2 cell entry
Molecular biology; Virology
Introduction
SARS-CoV and the novel SARS-CoV-2 infections initiate viral attachment, proteolytic processing of SARS-CoV spike protein, membrane fusion, and RNA genome release into the host cells. Most attention has focused on the extracellular multi-step coordinated actions during virus entry involving the viral spike protein and host cell surface proteins angiotensin converting enzyme 2 (ACE2), TMPRSS2, and ADAM17 (Heurich et al., 2014; Hoffmann et al., 2020). However, little information is available regarding intracellular factors affecting SARS-CoV-2 cell entry.
An early report showed that deletion of the ACE2 C-terminal tail causes significant change in SARS-CoV incorporation (Inoue et al., 2007), implying an important role for the carboxy-terminus region and its potential interaction with unidentified intracellular proteins mediating viral entry. While the present study was in progress, a report postulated that the interacting partners associated with the cytoplasmic tail of ACE2 are likely to be PDZ-containing (Singh et al., 2009) (PDZ is an acronym for the first three identified proteins sharing a highly similar structural domain: Postsynaptic density 95/Disc large/Zonula occludens-1) proteins SNX27, NHERF3, or SHANK based on bioinformatics analysis and fluorescence polarization assay using FITC-labeled C-terminal peptides of ACE2 and individual truncated PDZ domains from the three mentioned proteins (Kliche et al., 2021). Functional evidence for that proposition was not provided.
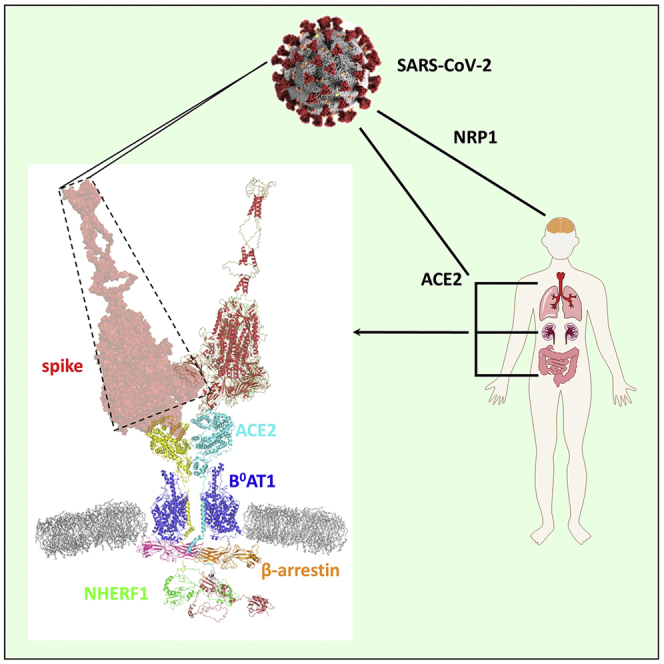
ACE2 is distributed ubiquitously in diverse cells particularly those such as pulmonary epithelial cells and intestinal enterocytes (Hamming et al., 2004) that are exposed to the external environments and serves as the SARS-CoV-2 receptor for binding to viral spike protein (Hoffmann et al., 2020; Li et al., 2003; Shang et al., 2020). ACE2 is a carboxypeptidase, primarily metabolizing angiotensin II (Ang II) to a heptapeptide Ang-(1-7) that contributes to the cardiorenal control of blood pressure (Ferrario et al., 2010). ACE2 is not a classical membrane-spanning receptor such as the G protein-coupled receptors or receptor tyrosine kinases that transduce binding of an extracellular ligand to intracellular signals that trigger a cascade of biologically appropriate responses (Drake et al., 2006; Wacker et al., 2017). ACE2 is a hybrid, single-pass membrane-delimited convertase with a short intracellular tail with no known signaling capacity. The ACE2 intracellular carboxy terminal sequence, -Thr-Ser-Phe, forms a canonical recognition motif facilitating binding to PDZ proteins such as Na+/H+ exchange regulatory factor-1 (NHERF1, SLC9A3R1), which is highly expressed in lung, intestine, and other organs where ACE2 is found. NHERF1 belongs to the larger NHERF family consisting of four PDZ-containing isoforms: NHERF1, NHERF2, NHERF3, and NHERF4 (Ardura and Friedman, 2011; Donowitz et al., 2005). Two forms, NHERF1 and NHERF2, possess two PDZ domains, whereas NHERF3 and NHERF4 contain four. Each PDZ domain includes about 90 amino acid residues and a strictly conserved GYGF or GLGF core-binding motif, where the hydrophobic pocket is formed with the surrounding residues, facilitating its interaction with the last hydrophobic residue (P0 position) of the canonical C terminus or within internal PDZ-binding motifs from the interacting partners. In the case of NHERF1, through its two tandem PDZ domains and Ezrin-binding domain, it serves as an adapter assembling short-lived functional units (Romero et al., 2011) frequently with membrane receptors and transporters (Figure 1A). Compared with the other forms, NHERF1 exhibits a more extensive tissue distribution than NHERF2 (kidney, intestine, lung, and brain), NHERF3 (kidney, intestine, and liver), and NHERF4 (kidney and intestine) (Ardura and Friedman, 2011). In addition, NHERF1 knockouts seem to have a far more impactful phenotype than other NHERF scaffold protein as reported in the regulation of the cystic fibrosis transmembrane conductance regulator (CFTR) by NHERF1 (Ardura and Friedman, 2011; Hynes, 2011; Jeong et al., 2019; Seidler et al., 2009; Singh et al., 2009; Wheeler et al., 2011).
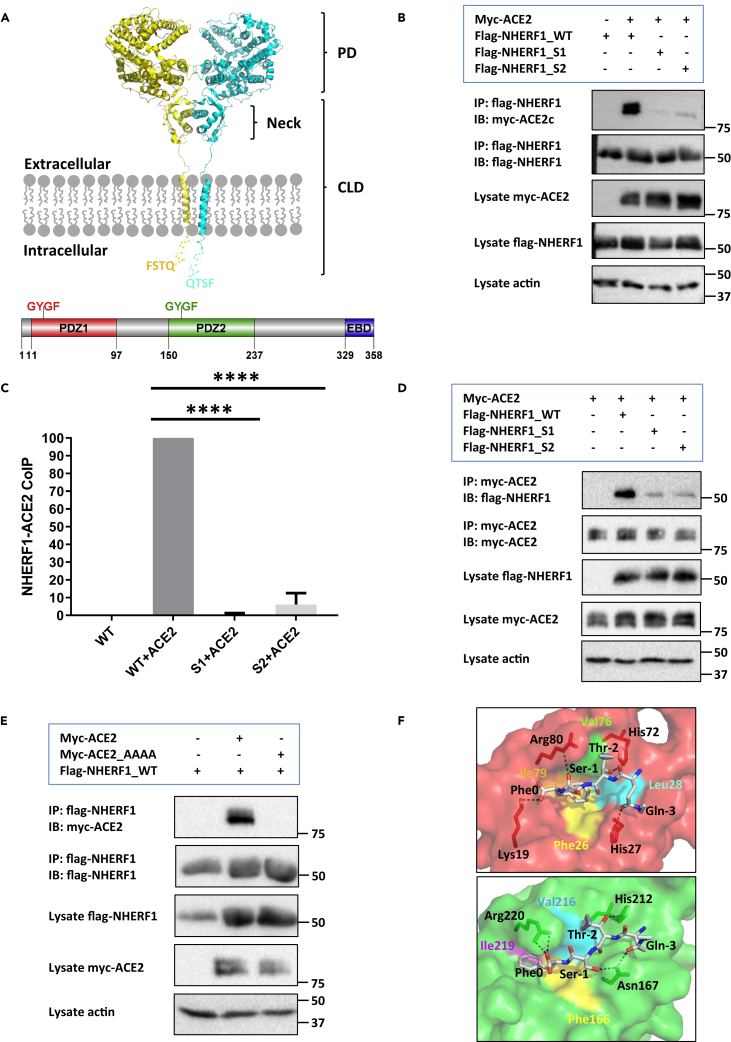
Figure 1.
Structural determinants for ACE2 binding to the PDZ-scaffold protein NHERF1
(A) Domain organization of ACE2 and NHERF1. ACE2 is shown as a dimer (PDB code 6M18) prepared by PyMol (Schrödinger). Monomeric ACE2 colored in yellow or cyan is a single-pass transmembrane protein containing an N-terminal peptidase domain (PD) and a C-terminal collectrin-like domain (CLD). The missing intracellular C-terminal tail containing a typical PDZ-binding motif 802QTSF805 from the solved structure is represented as yellow or cyan dotted lines assuming a random coil conformation. NHERF1 possesses a carboxy-terminal Ezrin/Radixin/Moesin/Merlin-binding domain (EBD) and two N-terminal PDZ domains, each of which has a same conserved GYGF core-binding motif. The schematic diagram of NHERF1 was created by Domain Graph (DOG) (Ren et al., 2009).
(B) Co-immunoprecipitation of ACE2 with wild-type (WT), S1 where GYGF was mutated to GAGA in PDZ1, or S2 with GAGA substitution for GYGF. Proteins from HEK293 GnTI− cell lysates were immunoprecipitated with anti-FLAG M2 agarose beads followed by immunoblot analysis using FLAG, myc, or actin antibodies. The molecular markers (kDa) are shown on the right.
(C) Quantification analysis of Figure 1B by ImageJ (Schneider et al., 2012). Intensity of ACE2 binding to wild-type NHERF1 was set to 100%. Relative binding of various NHERF1 to ACE2 was calculated and plotted using GraphPad Prism 9. Data are means ± S.D. (error bars) of n = 3 independent experiments (∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001; ∗∗∗∗, p < 0.0001).
(D) Reverse co-immunoprecipitation analysis to that shown in Figure 1B. Samples were immunoprecipitated with anti-c-Myc agarose beads followed by immunoblot analysis using FLAG, myc, or actin antibodies. The illustrated immunoblots are representative of three replicates. Quantification analysis is shown in Figure S1.
(E) Co-immunoprecipitation of NHERF1 with wild-type ACE2 or a mutant with quadruple Ala replacement for the PDZ -QTSF recognition motif. Purified proteins from HEK293 GnTI− cell lysates were immunoprecipitated with anti-FLAG M2 agarose beads followed by immunoblot analysis using FLAG, myc, or actin antibodies. Results represent three independent experiments.
(F) Computational modeling of NHERF1 PDZ1 (red) or PDZ2 (green) structure complexed with the C-terminal eight-residue tail of ACE2 by molecular dynamics simulation as detailed previously (Mamonova et al., 2015). For the sake of simplicity, only the last four resides 802QTSF805 were shown in the structures. The hydrophobic pocket is formed by Phe26 (yellow), Leu28 (cyan), Val76 (green), and Ile79 (orange) in PDZ1 and Phe166 (yellow), Val216 (cyan), and Ile219 (purple) in PDZ2. The hydrogen bonds are shown as dashed lines.
In addition to NHERF1, the intracellular adaptor protein β-arrestin also promotes 5-HT2R-mediated (G protein-coupled serotonin 5-hydroxytryptamine subfamily 2 receptors) JCPyV (JC polyomavirus) cell entry (Mayberry et al., 2019). In the case of the G protein-coupled purinergic receptor (P2Y12R), both β-arrestin and NHERF1 support efficient receptor internalization, where the presence of an intact PDZ-binding motif is required to form the β-arrestin-NHERF1-receptor complex (Nisar et al., 2012). In this work, we set out to determine ACE2 interaction with NHERF1 and their critical interacting structural determinants and further evaluate the functional definition of NHERF1 in ACE2-mediated SARS-CoV-2 entry. Our results confirm that NHERF1 binds to the C-terminal tail of ACE2 through both PDZ1 and PDZ2. The interaction was validated at endogenous expression levels in SARS-CoV-2 susceptible cells including lung (Calu-3) and intestine (Caco-2). The PDZ interaction was evaluated in mediating SARS-CoV-2 cell entry by introducing engineered NHERF1 constructs harboring discrete variations into NHERF1-lacking and ACE2-expressing HK2 cells, or ACE2 mutants in NHERF1-expressing and ACE2-deficient HEK293T cells. β-Arrestin was indeed identified in binding to ACE2 with high affinity with no preference for isoform 1 or 2. The described results will help to fully understand the molecular biology of SARS coronavirus by bridging the gap between virus attachment and genome release and may lead to the development of a novel druggable target against SARS-CoV-2 human infections.
Results
ACE2 binding to NHERF1 through direct PDZ interaction
Preliminary bioinformatic analysis indicates that ACE2 harbors a typical NHERF1-preferred type I PDZ-binding motif 802QTSF805 (Figure 1A), implying a possible role of NHERF1 in ACE2-mediated SARS-CoV-2 cell entry. To verify ACE2 binding to NHERF1 and identify the required structural determinants for each, myc-ACE2 was coexpressed in the high-protein yield-generating HEK293 GnTI− cells (Chaudhary et al., 2012) with wild-type FLAG-NHERF1 or its various mutants wherein PDZ1 (S1) or PDZ2 (S2) was scrambled (GYGF → GAGA) (Figure 1B). As predicted, ACE2 is captured by wild-type NHERF1 using anti-FLAG M2 agarose beads. Unexpectedly, disrupting either PDZ1 or PDZ2 core-binding determinants significantly diminished ACE2 coupling to NHERF1 (Figure 1C). The findings were corroborated by reversing the coimmunoprecipitation experiments using anti-c-myc agarose beads, where myc-ACE2 is immunoprecipitated and FLAG-NHERF1 immunodetected (Figures 1D and S1). In addition, to confirm the requirement for the ACE2-QTSF PDZ-recognition motif for binding to NHERF1, wild-type ACE2 or a mutant with quadruple Ala replacement for QTSF was coexpressed with FLAG-NHERF1 in HEK293 GnTI− cells. As shown in Figure 1E, NHERF1 immunoprecipitates with wild-type ACE2 while binding to the mutant ACE2 is abolished, suggesting that ACE2 binds NHERF1 through a classic PDZ interaction. The unbiased PDZ binding preference observed for ACE2 may facilitate better NHERF1 scaffolding of the SARS coronavirus receptor ACE2 (Li et al., 2003). We generated a computational structural model by docking the carboxy-terminal eight-residue peptide of ACE2 into NHERF1 PDZ1 or PDZ2 domains. The map provides key atomic binding details as illustrated in Figure 1F. Phe0 (position 0, -1, -2, and -3 referring to the C-terminal residues of PDZ ligands) inserts into a deep hydrophobic cavity formed by Phe26, Leu28, Val76, and Ile79 in PDZ1 and corresponding Phe166, Val216, and Ile219 in PDZ2. Hydrogen bonds are formed between Lys19 and Phe0, Arg80 and Ser-1, His72 and Thr-2, His27 and Gln-3 in PDZ1; Arg220 and Phe0, Asn167 and Ser-1, His212 and Thr-2, Asn167 and Gln-3 in PDZ2. The equivalent hydrogen bond interactions observed in PDZ1 and PDZ2 explain why ACE2 binds comparably with either PDZ domain of NHERF1, notably different from the PDZ1-preferred binding exhibited by the type 2 sodium phosphate cotransporter (NPT2A), where despite the above-mentioned identical hydrogen bonds, the PDZ-binding motif 636ATRL639 forms an extra-strong Arg-1/Glu43 salt bridge (Mamonova et al., 2015) that is absent in the second PDZ domain.
Similar PDZ interactions manifested by ACE2-NHERF1 have also been observed between ACE2 and NHERF3, a putative ACE2-interacting partner (Kliche et al., 2021). As shown in Figure S2, full-length myc-ACE2 overexpressed in HEK293 GnTI− cells coimmunoprecipitates with NHERF3-FLAG as observed for NHERF1. However, NHERF3 expression exhibits limited tissue distribution (Figure S3) and is seemingly limited to kidney HK2 and intestine Caco-2 cells, in agreement with previous descriptions (Ardura and Friedman, 2011). In contrast, Calu-3 lung cells, a primary target of SARS-CoV-2, displayed no detectable NHERF3 rendering it unsuitable for supporting ACE2 membrane residence.
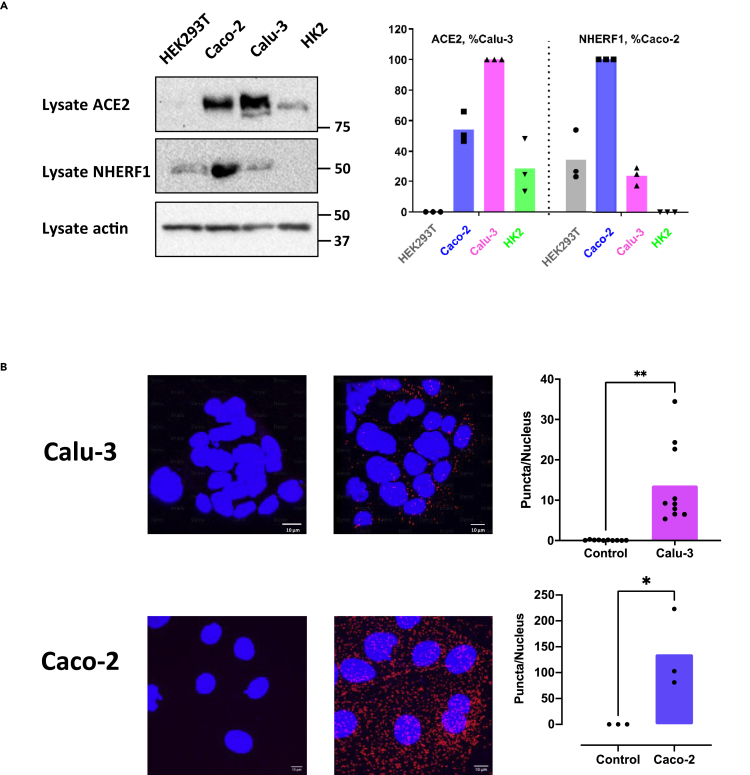
The observations thus far rely upon heterologous overexpression of ACE2 and NHERF1 in HEK293 GnTI− cells. To authenticate that this interaction occurs at endogenous levels in SARS-CoV-2-favored tissues lung and intestine (Gupta et al., 2020), proximity ligation assays (PLAs) were applied to characterize ACE2 interactions with NHERF1 in human Calu-3 pulmonary epithelial cells and Caco-2 intestinal enterocytes. Both cells display high endogenous ACE2 expression with scattered amounts of NHERF1 as shown in Figure 2A, providing supportive evidence for the following PLA. As illustrated in Figure 2B, ACE2 and NHERF1 exhibited significant puncta per nucleus, confirming direct PDZ interactions at endogenous ACE2 and NHERF1 expression levels both in Calu-3 and Caco-2 cells. More puncta observed in Caco-2 than in Calu-3 correlates with ACE2 and NHERF1 expression levels as shown in Figure 2A, where Caco-2 cells exhibit not only high levels of ACE2 but also of NHERF1 that is significantly lower in Calu-3. In contrast to both ACE2- and NHERF1-expressing Caco-2 and Calu-3 cells, no puncta are observed (Figure S4) in either NHERF1-lacking HK2 or ACE2-deficient HEK293T cells as demonstrated in Figure 2A.
Figure 2.
Direct interaction between endogenous ACE2 and NHERF1 in lung and intestine cells susceptible to SARS-CoV-2 infection
(A) Constitutive expression of ACE2 and NHERF1 in HEK293T, Caco-2, Calu-3, and HK2 cells detected by western blotting (left panel). Molecular markers are indicated in kDa on the right. Relative protein expression of ACE2 and NHERF1 from the whole-cell lysate was normalized to actin and plotted in the right panel, where the highest level of ACE2 in Calu-3 (red) and NHERF1 in Caco-2 (blue) was set to 1, respectively. Shown are representative immunoblots of three experiments with similar results. Data are means ± S.D. (error bars).
(B) Visualization of ACE2/NHERF1 interactions in Calu-3 and Caco-2 cells labeled by PLA probes (red) and DAPI (blue). Representative images of confocal microscopy were shown on the left panel. Scale bar (white), 10 μm. Quantification of PLA signals per cell was graphed in the right panel (error bars are SD).
NHERF1 regulates ACE2-mediated SARS-CoV-2 cell entry
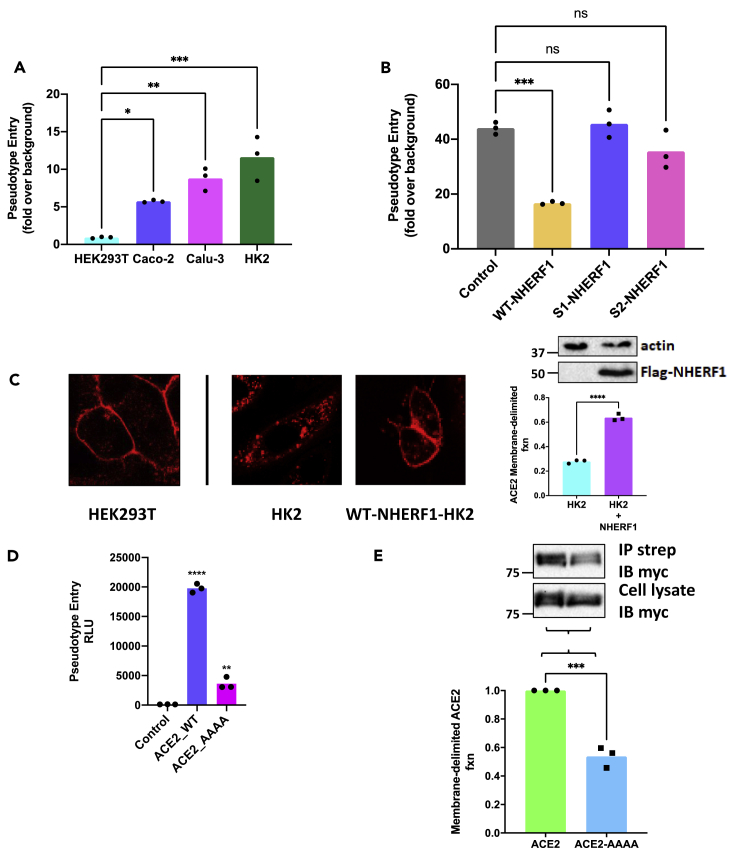
We next sought to define the functional role of NHERF1 in ACE2-mediated SARS-CoV-2 cell entry. These studies used lentivirus-based SARS-CoV-2 spike pseudotyped virions containing a firefly luciferase gene as a reporter to mimic SARS-CoV-2 spike-driven cell entry (Schmidt et al., 2020). As shown in Figure 3A, Caco-2, Calu-3, and HK2 cells constitutively expressing ACE2 are sensitive to SARS-CoV-2-spike-facilitated pseudovirus infection as indicated by luciferase reporter activity (Figure 3A), whereas HEK293T cells lacking ACE2 expression are essentially unsusceptible. Intriguingly, HK2 cells lacking NHERF1 displayed highly efficient virus particle incorporation. We therefore selected HK2 cells to characterize the structural elements within NHERF1 that contribute to ACE2-mediated SARS-CoV-2 entry using wild-type or discrete NHERF1 mutants. As shown in Figure 3B, overexpression of wild-type NHERF1 in HK2 cells significantly decreased SARS-CoV-2 spike pseudotyped virion incorporation, consistent with the described observations in Figure 3A, where abundant NHERF1 expression in Caco-2 cells impeded virus entry. HK2 cells expressing (S1) PDZ1- or (S2) PDZ2-scrambled NHERF1, where PDZ interaction was eradicated, displayed substantial particle incorporation as illustrated in Figures 1B–1D. We speculate that under conditions of comparable basal level of endogenous ACE2, overexpressed wild-type NHERF1 binds to ACE2 by PDZ interactions, thereby constraining receptor mobility and retarding pseudotyped virion internalization. NHERF1 impacts sequestration of various receptors, particularly GPCRs (Weinman et al., 2006). As shown in Figure 3C, overexpressed wild-type NHERF1 in HK2 cells strongly scaffolds RFP-ACE2 at the plasma membrane demonstrated by the red fluorescence ring as similarly observed in HEK293T cells with endogenous NHERF1 expression (Figure 2A), but distinct from HK2 cells lacking NHERF1, where less RFP-ACE2 accumulated at the plasma membrane as quantified in Figure 3C (right panel). Conversely, in the case of equivalent endogenous NHERF1 expression in HEK29T cells, destruction of the ACE2 PDZ-binding motif -QTSF to quadruple Ala remarkably decreased SARS-CoV-2 spike pseudotyped virion incorporation compared with wild-type ACE2-expressing cells (Figure 3D), likely due to diminished membrane residence of ACE2-AAAA in the absence of PDZ interaction. Membrane abundance of either wild-type or quadruple-Ala mutant ACE2 detected by biotinylation confirms that the cell surface fraction of total ACE2-AAAA is much lower than its wild-type form (Figure 3E).
Figure 3.
Functional assessment of PDZ-mediated ACE2-NHERF1 interaction in SARS-CoV-2 pseudovirus infection
(A) SARS-CoV-2 spike pseudovirus infectivity assays in different cell lines. Pseudotyped entry representing viral infectivity was evaluated by measuring luciferase activity in cell lysates. Signals from the bald pseudovirion without envelope spike glycoprotein were used as a negative control (background). Error bars indicate SD.
(B) Functional role of NHERF1 in SARS-CoV-2 spike pseudovirus infectivity assayed in HK2 cells without or with various overexpressed NHERF1. Error bars are SD.
(C) Overexpressed NHERF1 retains RFP-ACE2 at the cell surface as analyzed by confocal microscopy (left panel). HEK293T cells with endogenous NHERF1 were transduced with RFP-ACE2 BacMam and were used as a positive control. HK2 cells transfected with or without FLAG-NHERF1 (as indicated in top immunoblots of the right panel. Numbers at the left side of the blots indicate the position of molecular mass markers) were transduced with RFP-ACE2 BacMam as detailed in methods. Fluorescence intensity was quantified by ImageJ (Schneider et al., 2012). Plasma membrane fluorescence intensity was obtained by subtracting the cytosolic signal from the total fluorescence measured in a single cell (Boutte et al., 2013; Platre et al., 2019). Membrane fraction of RFP-ACE2 was defined as membrane fluorescence intensity divided by total fluorescence intensity for each cell (right panel). Error bars indicate SD.
(D) Intact C-tail PDZ-binding motif is required for high SARS-CoV-2 infectivity. HEK293T cells with endogenous NHERF1 expression were transfected with wild-type or mutant ACE2 where the last four residues 802QTSF805 were mutated to AAAA. The resulting relative luciferase activity from the bald pseudovirion without envelope spike glycoprotein was used as a negative control. Error bars are SD.
(E) Surface biotinylation of wild-type or mutant ACE2 receptor expressed in HEK293T cells. Receptor biotinylation was performed using cleavable EZ-Link sulfo-NHS-SS-biotin as described before (Zhang et al., 2018). After unreacted biotin is quenched by ice-cold PBS containing 100 mM glycine, cells were extensively washed with PBS and lysed. Biotinylated surface ACE2 receptors were precipitated by streptavidin beads and detected by immunoblotting. The molecular markers are indicated in kDa on the left. The relative amount of mutant ACE2_AAAA receptor to its wild-type form was calibrated by their individual expression from the whole-cell lysate. Data are means ± SD (error bars).
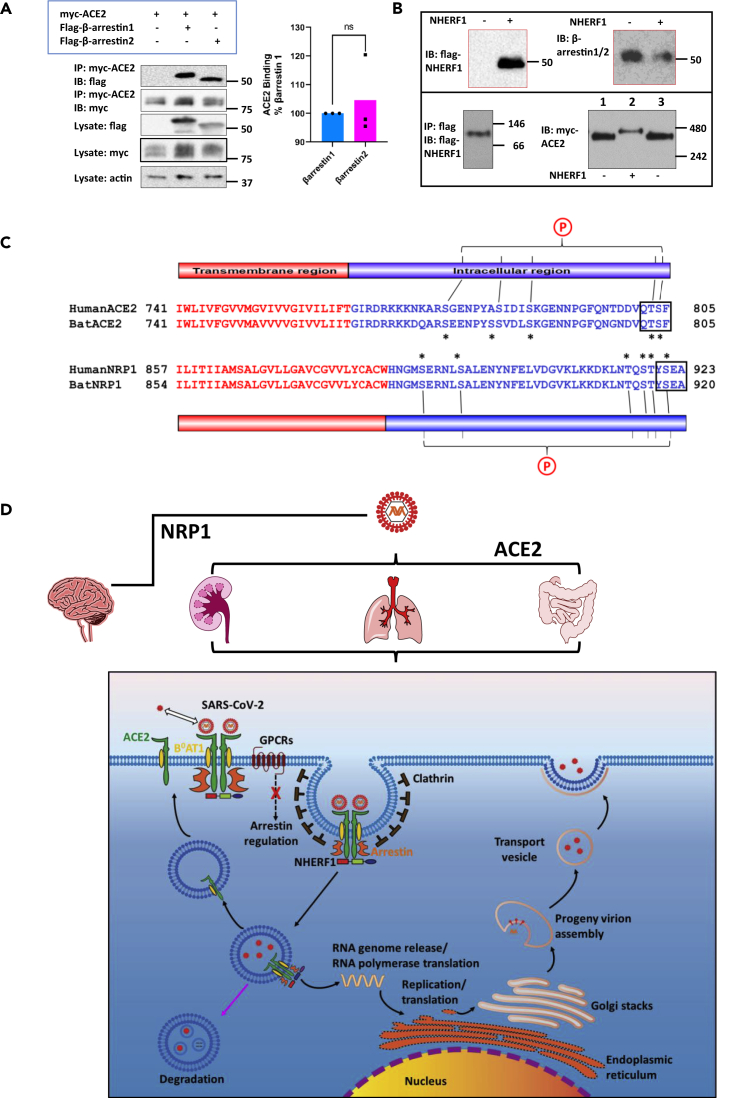
NHERF1 does not promote ACE2 dimerization but supports β-arrestin binding to ACE2
The pseudovirus assays described above demonstrate that NHERF1 plays an important role in ACE2-mediated SARS-CoV-2 cell entry despite the absence of evidence of authentic SARS-CoV-2 infection. NHERF1 is also an important workbench for assembling macromolecular complexes that perform various cellular functions, including receptor sequestration. NHERF1 facilitates internalization of the G protein-coupled P2Y12 and parathyroid hormone receptors (Nisar et al., 2012; Wang et al., 2009) by promoting association with β-arrestin adapter proteins. β-Arrestins, which are commonly required for GPCR internalization (Ferguson et al., 1996), interact with ACE2 with virtually equal affinities for β-arrestin 1 and β-arrestin 2 (Figure 4A). Considering that β-arrestins are expressed in SARS-CoV-2 targeted pulmonary Calu-3 and intestinal Caco-2 cells (Figure S3), this finding suggests that β-arrestin participates in ACE2 internalization and thereby SARS-CoV-2 invasion. GPCR internalization associated with β-arrestins is mostly clathrin dependent (Gurevich and Gurevich, 2013). Indeed, clathrin mediates SARS-CoV-2 endocytosis (Armin Bayati and Vincent Francis, 2021). β-Arrestins control a variety of cellular activities ranging from cytoskeletal rearrangement to transcriptional regulation, cell growth and survival, and GPCR endocytosis, trafficking and GPCR signaling (Ahn et al., 2020). The unbiased occupancy of β-arrestin by SARS-CoV-2 may further terminate β-arrestin-dependent signal transduction to desensitize the infected cells in response to external stimuli. This would explain why COVID-19 patients lose their sense of taste or smell mediated by specific taste or olfactory GPCRs, in addition to the reported hyperinflammatory response (Merad and Martin, 2020), where replication of SARS-CoV-2 induces cellular stress challenges.
Figure 4.
Proposed SARS-CoV-2 cell entry mechanism regulated by PDZ-mediated interaction
(A) β-Arrestin 1 or β-arrestin 2 binds to ACE2 with nearly equivalent affinity. FLAG-β-arrestin 1 or 2 was transiently coexpressed in HEK293 GnTI cells with Myc-ACE2. The cleared cell lysates were incubated with anti-c-Myc agarose beads. Proteins are eluted by 2x Laemmli sample buffer (Bio-Rad) supplemented with β-mercaptoethanol and detected by western blotting using the indicated specific antibodies (left panel). β-Actin was used as a loading control. The molecular markers are indicated in kDa on the right. Given the equivalent expression of myc-ACE2 observed in the cell lysate and immunoprecipitates, pull-down of β-arrestin 1 or β-arrestin 2 by myc-ACE2 was quantified by normalizing the signal intensities to their individual lysate expression, respectively (right panel). Error bars indicate SD.
(B) NHERF1 is not required to promote ACE2 dimerization. FLAG-NHERF1 and Myc-ACE2 were transiently transfected into HK2 cells. Forty-eight hours post transfection, the cell lysates were cleared by centrifugation and incubated with anti-c-Myc agarose beads (only the sample in the lower left panel was prepared using anti-FLAG agarose beads and therefore eluted by 3x FLAG peptide). Purified proteins by c-Myc peptide elution were detected by the indicated antibodies. The boxed blots in red are from SDS-PAGE (top panel) and the blots in the lower panel are from blue native PAGE performed as described before (Dewson, 2015). Molecular weights are indicated at the right side of the blots. In the lower right panel, the same samples were loaded in the first and third lane to better show the slight difference of protein size from different samples.
(C) Sequence alignment of ACE2 and NRP1 from SARS-CoV-2 hosts human (Protein ID: NP_001358344.1 for ACE2 and ADN93470.1 for NRP1) and bat (Protein ID: NP_001231902.2 for ACE2 and KAF6372190.1 for NRP1). Human and bat ACE2/Ace2 and NRP1/Nrp1 are used because they are primary hosts of SARS-CoV-2 (Burki, 2020). Both identified SARS-CoV-2 receptors are single-pass transmembrane proteins and have a typical type I PDZ-binding motif (QTSF and YSEA) implying a possible common mechanism by which NHERF1 regulates SARS-CoV-2 receptor internalization. The single-pass transmembrane regions are colored in red, and the intracellular C-terminal tail is colored in blue. The conserved serine and threonine residues are indicated by asterisks and are likely to be potential phosphorylation sites denoted by an encircled P. These sites may mediate SARS-CoV-2 receptor binding to β-arrestin as in the case of β2AR, where the phosphorylated receptor C terminus is required for binding to β-arrestin and receptor internalization (Nobles et al., 2011).
(D) NHERF1 functions in ACE2-mediated SARS-CoV-2 cell entry in lung, kidney, and intestine and possibly NRP1-mediated virus entry in brain. NHERF1 enhances plasma membrane localization of B0AT1-stabilized ACE2 receptor and regulates its internalization and hence, ACE2-mediated SARS-CoV-2 cell entry. Except NHERF1, the virion-receptor complex also requires β-arrestin to facilitate its internalization process in which clathrin is involved. Once internalized, SAS-CoV-2 is either degraded in lysosomes (pink arrow) or released into the cytosol where the viral genomic RNA is released and immediately translated into viral RNA polymerase. The ACE2 receptor may be recycled back to the plasma membrane. Viral RNA genome and its structural nucleocapsid protein is replicated and synthesized in the cytoplasm. Other viral structural proteins are translated in the endoplasmic reticulum and further glycosylated in the Golgi. Mature progeny virions are then formed and bud out from plasma by exocytosis.
ACE2 functions as a strong heterodimer with a sodium-dependent neutral amino acid transporter B0AT1 (SLC6A19, Solute Carrier Family 6 Member 19) in mediating SARS-CoV-2 cell entry (Yan et al., 2020). B0AT1 is responsible for the uptake of neutral amino acids. ACE2 serves as an interacting partner to support B0AT1 membrane trafficking and coordinates peptide digestion and neutral amino acid uptake in the intestine and kidney (Kowalczuk et al., 2008). The solved structure of ACE2/B0AT1 complex indicates that B0AT1 interacts with the extracellular neck and transmembrane region of ACE2 (Figure 1A) (Yan et al., 2020), which stabilizes ACE2 membrane localization. This engagement is critical for ACE2 particularly given that it is a single-pass transmembrane protein. We speculated that NHERF1 promotes heterodimerization to enhance ACE2 cell membrane localization. In NHERF1-lacking HK2 cells, co-immunoprecipitation of myc-ACE2 by anti-c-myc agarose beads captures NHERF1 (Figure 4B upper left panel), consistent with the described observation in Figure 1B. Unexpectedly, endogenous β-arrestin was also found in the resulting immunoprecipitate in the absence or presence of NHERF1, suggesting very robust binding between ACE2 and β-arrestin (Figure 4B, upper right panel). In immunoblots obtained from blue native PAGE of FLAG eluates from HK2 cells expressing FLAG-NHERF1, NHERF1 migrated at 100 kDa, implicating previously identified dimeric NHERF1 (Lau and Hall, 2001) in the cellular context (Figure 4B, lower left panel). In addition to NHERF1 and β-arrestin, B0AT1 with ACE2 as mentioned above is also found in kidney HK2 cells used in Figure 4B and co-immunoprecipitates with myc-ACE2 at endogenous expression levels (Figure S5) as reported (Kowalczuk et al., 2008). In the absence of NHERF1, the ACE2 (92 kDa) complex with its strong binding partners B0AT1 (71 kDa) and β-arrestin (50 kDa) would be expected to generate a dimeric 426-kDa ACE2 band in blue native PAGE, in agreement with the observation shown in Figure 4B (Lane 1 and 3 in the lower right panel). In the presence of dimeric NHERF1 overexpressed in HK2 cells, ACE2 would be predicted to display an ∼526-kDa band, close to the 480-kDa reference indicator (Lane 2 in the lower right panel). No monomeric ACE2 band with a predicted molecular weight of 213 kDa (92-kDa ACE2, 71-kDa B0AT1, and 50-kDa β-arrestin) was observed in native blue PAGE, suggesting that NHERF1 does not promote ACE2 dimerization but serves as a scaffold to regulate its cell membrane abundance. Of interest, NHERF1 and ACE2 (Figure 2A), β-arrestin, and B0AT1 (Figure S3) are all variably expressed in SARS-CoV-2-targeted lung Calu-3 and intestine Caco-2 cells, supporting the likelihood that their resulting super-complex promotes SARS-CoV-2 cell entry.
Discussion
In this work, we demonstrate that NHERF1 enhances ACE2 membrane residence by direct interaction between NHERF1 PDZ domains and the ACE2 C-terminal PDZ-binding motif, with consequential increases of ACE-mediated SARS-CoV2 cell entry. Of interest, a secondary SARS-CoV-2 receptor neuropillin-1 (NRP1) recently found in the nervous system (Cantuti-Castelvetri et al., 2020; Daly et al., 2020) also contains a classic type I PDZ binding-motif 920YSEA923 at its short intracellular C terminus and like ACE2 is a single-pass transmembrane protein (Figure 4C). Direct interaction of NHERF1 and NRP1 is predictable but cannot presently be confirmed owing to unavailability of its human construct. If confirmed for NRP1, the observation that two identified SARS-CoV-2 receptors in both humans and bats contain a prototypical PDZ-binding motif likely underwrites a common structural mechanism involved in establishing SARS-CoV-2 cell entry by common PDZ interactions.
As shown in Figure 4D, in tissues highly susceptible to SARS-CoV-2 infection such as intestine, lung, and kidney, NHERF1 enhances ACE2 membrane localization and forms a half-megadalton super-complex with ACE2, B0AT1, and β-arrestin in the high-affinity state as reported elsewhere (Yan et al., 2020) and observed here (Figure 4B). PDZ interactions formed by NHERF1 and ACE2 may play a central role in tethering this super-complex to facilitate ACE2 receptor internalization as observed for the ternary complex β-arrestin-NHERF1-P2Y12R (Nisar et al., 2012). We propose that the super-complex carries the SARS-CoV-2 virion by ACE2-mediated recognition of the viral surface spike protein, which facilitates viral attachment and ensuing clathrin-assisted internalization into endosomes. The viral entry process hijacks the host cell's endocytic machinery and specifically impairs β-arrestin-dependent GPCR internalization as implicated in the β2-adrenergic receptor (β2AR) (Ferguson et al., 1996) and calcium-sensing receptor (CaSR) (Mos et al., 2019) and, as a consequence, diverse GPCR signaling. For instance, impaired GPCR signaling due to lack of β-arrestin may desensitize the SARS-CoV-2 infected cells in response to various stimuli as mentioned above. In this case, the infected cells appear normal, which in turn helps them survive without being targeted by the host immune system. In addition, utilization of β-arrestin in SARS-CoV-2 entry may depress β-arrestin-mediated cell signaling and affect other β-arrestin-regulated biological processes (Ahn et al., 2020; Latorraca et al., 2020). In summary, introduction of NHERF1 enhances ACE2 plasma membrane localization and slows SARS-CoV-2 penetration in NHERF1-lacking HK2 cells (Figure 3B). Experimentally eliminating PDZ interaction between NHERF1 and ACE2 by mutating the PDZ-binding motif dramatically reduces SARS-CoV-2 incorporation into NHERF1-expressing HEK293T cells (Figure 3D). Thus, NHERF1 regulates ACE2-mediated SARS-CoV-2 cell entry, possibly through a mechanism controlling the kinetics of ACE2 internalization as with the P2Y12 receptor (Nisar et al., 2012). Once internalized, SARS-CoV-2 may be transported to lysosomes for degradation (Yang and Shen, 2020) or in most cases escape endo-lysosomes to release its RNA genome to the cytosol. The unloaded ACE2 may recycle to plasma membrane. Viral structural proteins are synthesized in the endoplasmic reticulum and glycosylated in Golgi. Mature progeny virions are then formed and bud from plasma membranes by exocytosis (Harrison et al., 2020). Of note, the described scenario may also proceed in brain with NRP1-mediated SARS-CoV-2 infection by a common PDZ interaction shared with ACE2-NHERF1, although for the purpose of simplicity NRP1 is omitted in the proposed mechanism as illustrated in the lower panel of Figure 4D. The identified PDZ interaction between ACE2 and NHERF1 shown here provides a mechanistic explanation of novel intracellular factors, NHERF1 and β-arrestin, involved in mediating SARS-CoV-2 entry. These insights bridge gaps in understanding SARS-CoV-2 internalization through but not limited to ACE2.
Future work will focus on functionally characterizing the structural determinants of β-arrestin binding to ACE2, investigating SARS-CoV-2-stressed GPCR trafficking and signaling, and extending the present observations to NRP1 and DPP4 (Raj et al., 2013), the other two described SARS/MERS receptors that also express carboxy-terminal PDZ-binding sequences. Given the role of DPP4 in glucose and amino acid homeostasis and the prevalence of type II diabetes associated with Covid-19 (Valencia et al., 2020), DPP4 (CD26) is especially intriguing considering that its carboxy tail resides in the extracellular space (Yan et al., 2007). In addition to host cell surface receptors including ACE2, NRP1, and DPP4, viral proteins E, 3A, and N from SARS-CoV-2 itself also possess PDZ-binding motifs (Caillet-Saguy et al., 2021), suggesting PDZ interaction possibly defines SARS-CoV-2 invasion from either intra- or extracellular sides of infected host cells. The related outcomes from these characterizations may delineate a broad role for PDZ proteins in SARS/MERS infections and help us better understand the pathogenic mechanisms of SARS coronavirus and, consequently, serve as a foundation for developing new therapeutic strategies to restrict SARS-CoV-2 infection.
Limitations of the study
The SARS-CoV-2 receptor NRP1 is currently unavailable. Additional work needs to be performed to extend the present findings to NRP1-mediated SARS-CoV-2 invasion.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Myc-tag (71D10) rabbit mAb | Cell Signaling Technology | Cat#2278S |
| β-arrestin 1/2 (D24H9) rabbit mAb | Cell Signaling Technology | Cat#4674S |
| Anti-flag rabbit antibody | Sigma-Aldrich | Cat#F7425 |
| Mouse monoclonal anti-β-actin antibody | Sigma-Aldrich | Cat#A1978 |
| Rabbit polyclonal anti-ACE2 antibody | Abcam | Cat#ab15348 |
| Mouse monoclonal anti-EBP50/NHERF1 antibody | Abcam | Cat#9526 |
| Recombinant Anti-PDZK1 antibody (NHERF3) | Abcam | Cat#ab92491 |
| Recombinant Anti-SLC6A19 antibody (B0AT1) | Abcam | Cat#ab180516 |
| Rabbit polyclonal anti-NHERF1/EBP50 antibody | Alomone Labs | Cat#APZ-006 |
| Pierce anti-c-myc agarose | ThermoFisher Scientific. | Cat#20168 |
| Anti-flag M2 affinity gel | Sigma-Aldrich | Cat#A2220 |
| Streptavidin sepharose high performance medium | GE Healthcare Life Sciences | Cat#GE17-5113-01 |
| Bacterial and virus strains | ||
| pCEP4-myc-ACE2 | Chan et al. (2020) | Addgene#141185 |
| pcDNA3.1(+)-PDZK1-Flag (NM_002614.4, NHERF3) | GenScript | Clone#Ohu21391 |
| Critical commercial assays | ||
| Angiotensin converting enzyme 2 (ACE2)-Red | Montana Molecular | Cat#C1100R |
| PierceTM BCA Protein Assay Kit | ThermoFisher Scientific | Cat#23225 |
| Duolink in situ proximity ligation assay (PLA) kit | Sigma-Aldrich | Cat#DUO92001, DUO92005 |
| Spike (SARS-CoV-2) Pseudotyped Lentivirus (Luciferase Reporter) | BPS Bioscience | Cat#79942-1 |
| Bald Lentiviral Pseudovirion (Luciferase Reporter) | BPS Bioscience | Cat#79943 |
| One-StepTM Luciferase Assay System | BPS Bioscience | Cat#60690-1 |
| Experimental models: Cell lines | ||
| Calu-3 (ATCC® HTB-55TM) | ATCC | HTB-55 |
| Caco-2 (ATCC® HTB-37TM) | ATCC | HTB-37 |
| Software and algorithms | ||
| PyMOL | Schrödinger | https://pymol.org/2/ |
| Domain Graph (DOG) | Ren et al. (2009) | http://dog.biocuckoo.org/ |
| ImageJ | Schneider et al. (2012) | https://imagej.nih.gov/ij/ |
| GraphPad Prism 9 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| Imaris software | Oxford Instruments | https://imaris.oxinst.com/ |
Resource avilability
Lead contact
Further information and requests for resources and data should be directed to the lead contact, Qiangmin Zhang (zhqiangm@outlook.com).
Materials availability
All materials generated in this work will be available from the lead contact upon request.
Data and code availability
The published article includes all data generated or analysed in this work. This paper does not report original code.
Experimental model and subject details
Cell lines
All cell lines including experimental models Calu-3 and Caco-2 are detailed under the section “Cell Culture and Transfection”.
Method details
Plasmids and antibodies
The plasmid pCEP4-myc-ACE2 (Chan et al., 2020) expressing human ACE2 with an N-terminal c-myc tag was purchased from Addgene (Plasmid# 141185); Quadruple Ala replacement for 802QTSF805 was introduced into the pCEP4-myc-ACE2 by PCR to prepare pCEP4-myc-ACE2_AAAA using the QuikChange Site-Directed mutagenesis kit (Agilent). N-terminal Flag-tagged NHERF1 in the form of Flag-NHERF1_WT, Flag-NHERF1_S1 (GYGF was mutated to GAGA in PDZ1) and Flag-NHERF1_S2 (GYGF was mutated to GAGA in PDZ2) was constructed into pcDNA3.1+ (ThermoFisher Scientific). N-terminal Flag-tagged β-arrestin 1 or β-arrestin 2 was also constructed into pcDNA3.1+. The BacMam vector expressing RFP-ACE2 was purchased from Montana Molecular (#C1100R).
Myc-tag (71D10) rabbit mAb (#2278) and β-arrestin 1/2 (D24H9) rabbit mAb (#4674) were purchased from Cell Signaling Technology; Anti-flag rabbit antibody (#F7425) and mouse monoclonal anti-β-actin antibody (#A1978) were purchased from Sigma-Aldrich; Rabbit polyclonal anti-ACE2 antibody (#ab15348), mouse monoclonal anti-EBP50/NHERF1 antibody (#ab9526), recombinant anti-PDZK1 antibody (NHERF3, #ab92491) and recombinant anti-SLC6A19 antibody (B0AT1, #ab180516) were purchased from Abcam; Rabbit polyclonal anti-NHERF1/EBP50 antibody (#APZ-006) was purchased from Alomone Labs. Pierce anti-c-myc agarose (#20168) was purchased from ThermoFisher Scientific. Anti-flag M2 affinity gel (#A2220) was purchased from Sigma-Aldrich. Streptavidin sepharose high performance medium (#GE17-5113-01) was purchased from GE Healthcare Life Sciences. All antibodies were used at 1:1000 dilution unless otherwise stated.
Cell culture and transfection
HEK293 GnTI- and HEK293T cells were cultured in high-glucose Dulbecco’s modified Eagle’s medium (DMEM; Corning Cellgro, 10-013-CV) supplemented with 5% heat-inactivated FBS and 1% penicillin and streptomycin. Calu-3 human lung epithelial cells (ATCC® HTB-55TM) and Caco-2 human intestinal epithelial cells (ATCC® HTB-37TM) from ATCC (American Type Culture Collection) were cultured in Eagle’s Minimum Essential Medium (EMEM; ATCC® 30-2003™) supplemented with 5% FBS and 1% penicillin-streptomycin according to ATCC instructions. HK2 human proximal convoluted tubule cells were grown in DMEM/F12 medium (Corning Cellgro, 10-090-CV) supplemented with 5% FBS and 1% penicillin-streptomycin. All cell lines were grown at 37°C in a humidified atmosphere containing 5% CO2. Cells were transfected with the indicated plasmids using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s directions.
Immunoprecipitation and Western Blotting
Cells were plated in 6-cm dishes and transfected next day with 2 μg of each indicated plasmid. 48 h post-transfection, cells were washed once with PBS, collected and lysed in a lysis buffer containing 50 mM Tris-HCl, pH8.0, 150 mM NaCl, 1% NP40 and 2X protease inhibitors (MedChem Express) for 30 min on ice. The cell lysates were cleared by high-speed centrifugation at 13,200 rpm for 30 min at 4°C, and the resulting supernatant were incubated with the corresponding agarose beads while rotating at 4°C overnight. The resin was washed 4 times with a buffer containing 50 mM Tris-HCl, pH8.0, 150 mM NaCl and 0.5% NP40 and mixed with 2X Laemmli sample buffer containing 5% β-mercaptoethanol. The eluted protein samples were heated at 37°C for 30 min, separated on 12% SDS-polyacrylamide gel and transferred onto Immobilon-P polyvinylidene difluoride membrane (EMD Millipore) using Bio-Rad Trans-Blot SD Semi-Dry Transfer Cell. For endogenous protein detection, the concentration of total protein from the indicated cell lysate was determined by Pierce BCA protein assay and equal amount of total protein for each sample was used for western blotting. The resulting membranes were blocked with TBS blocking buffer containing 50 mM Tris-HCl (pH7.5), 150 mM NaCl, 0.1 % Tween20, 1% BSA and 5% non-fat dry milk at room temperature for 1 h. Membranes were then incubated with primary antibodies in the blocking buffer at 4°C overnight. After washing 4 times for 10 min each, membranes were incubated in the blocking buffer containing horseradish peroxidase (HRP)-conjugated secondary antibodies with agitation for 1 h at room temperature. Membranes were subsequently developed with ProSignal Pico ECL reagent (Genesee Scientific) and chemiluminescent signals were acquired using Bio-Rad ChemiDoc imager.
Proximity ligation assay (PLA)
50,000 cells were seeded on 22 mm, collagen-coated glass coverslips (Corning, #354089) for 24 h. Cells were then rinsed with PBS and fixed with 4% paraformaldehyde/PBS for 15 min at room temperature. Cells were washed again 3 times with PBS and 100 mM Glycine was added for 10 min to quench PFA. The cells were permeabilized with 0.1% TritionX-100 in PBS for 10 min, and washed 3 times with PBS. The cells were then blocked in Duolink blocking solution and incubated with the following antibodies: mouse anti-NHERF1 (Abcam, #ab9526) and rabbit anti-ACE-2 (Abcam, #ab15348) diluted in Duolink blocking solution. PLA probes (Sigma-Aldrich, #Duo92001 and #Duo92005) were then added and the assay was performed as per the manufacturer’s instructions. Coverslips were then mounted in Duolink mounting medium with DAPI. Images were acquired using an Olympus FluoView FV1000 confocal microscope and PLA signals were analysed using Imaris Software. Results are expressed as the absolute number of PLA puncta per nucleus (cell).
Pseudovirus assays
For HK2 cells, 25,000 cells were seeded into the black-walled, clear bottom 96-well microplate (Greiner Bio-One, #655090) pretreated with 5 μg/ml rat tail collagen coating solution (Sigma-Aldrich) and allowed to attach for at least 30 min. Cells were transfected with 0.1 μg of pcDNA3.1-Flag-NHERF1 in the form of either wild-type, PDZ1-scrambled S1 or PDZ2-scrambled S2. For HEK293T cells, a similar experimental procedure was applied as mentioned above. Briefly, 10,000 cells (less number of cells than others were used because of their shorter time to reach confluence) were seeded into the described identical 96-well microplate pretreated with 50 μg/ml poly-D lysine. Upon cell attachment, cells were transfected with 0.05 μg of pCEP4-myc-ACE2 or its mutant form pCEP4-myc-ACE2_AAAA where the C-terminal four residues were replaced with quadruple Ala. 18-24 h post-transfection, the medium was replaced with 50 μl of fresh medium. For Caco-2 and Calu-3 cells, 25,000 cells were seeded into the 96-well collagen-coated plate 24 h prior to infection and medium was changed to 50 μl of fresh medium. All the cells as mentioned above were individually infected with 5 μl of SARS-CoV2 spike pseudotyped lentivirus (BPS Bioscience #79942-1) or bald lentiviral pseudovirion (BPS Bioscience #79943) that lacks the spike glycoprotein in the presence of 5 μg/ml polybrene as per manufacturer’s instructions. Cells were incubated at 37°C with 5% CO2 overnight. The medium containing the lentivirus were removed from the wells and replaced with 100 μl of fresh medium. Approximately 48-60 hours upon pseudovirus transduction, 50 μl of medium was removed and 50 μl of ONE-Step luciferase assay regent (BPS Bioscience #60690-1) was added to each well. Bioluminescence signal (relative luminescence units or RLU) was measured using the Spark 20M multimode plate reader (TECAN).
Fluorescence image analysis and quantification
HEK293T and HK2 cells were seeded in 6 cm dishes. HK2 cells were then transfected with 2 μg of Flag-NHERF1 and non-transfected HK2 cells were used as a control. 18 hours later, cells were detached by enzyme-free PBS-based cell dissociation buffer (Gibco, #13151014). HEK293T cells were seeded at 25,000 cells /well onto the poly-D-Lysine coated coverslip placed in the 6-well plate using a same coating protocol as mentioned above. HK2 cells were seeded at 25,000 cells/well onto the collagen-coated coverslip (Corning, #354089) placed in the 6-well plate. The remaining HK2 cells were lysed for immunoblot detection of NHERF1 expression. Once attached, cells were transduced with 30 μl of BacMam vector expressing RFP-ACE2 in the presence of 2 mM of sodium butyrate.16-20 hours later, coverslips were mounted in an Attofluor cell chamber (Life Technologies). Live cell images were acquired using an inverted Nikon Ti-E microscope equipped with A1 confocal unit and a 60 X/1.45 numeric aperture oil immersion objective. The regions of interest (ROI) representing the whole cell and its cytosolic part were manually outlined by 1-pixel freehand lines along the outer side or inner side of the plasma membrane, respectively. The integrated intensities for these two circled regions were employed as readouts (Platre et al., 2019) to quantify RFP immunofluorescence and measured using ImageJ (Schneider et al., 2012) as described previously (Boutte et al., 2013). Specifically, plasma membrane fluorescence intensity for each cell was obtained by subtracting the cytosolic intensity from the total fluorescence measurement. Membrane fraction of RFP-ACE2 was calculated by dividing membrane intensity by total fluorescence intensity. For each experiment at least 5 cells were measured for the purpose of quantification.
Quantification and statistical analysis
Experiments were performed in triplicate and repeated at least 3 times. Results were analysed using Prism version 9 (GraphPad, La Jolla, CA). Results represent the mean ± S.D. and were compared by analysis variance with post hoc testing using the Bonferonni procedure. Standard p-value threshold of < 0.05 was considered statistically significant.
Acknowledgments
We thank Karina Pena for assistance and discussion about confocal microscopy. This work was supported by National Institutes of Health Grant RO1 DK105811.
Author contributions
Q.Z. and P.A.F. designed and coordinated the experiments. J.G. performed PLA assays. W.B.S. and Q.Z. performed live-cell imaging analysis. T.M. performed the molecular dynamics simulations. Q.Z. performed experiments and data analysis. Q.Z. and P.A.F wrote the manuscript and prepared figures with input from the other authors.
Declaration of interests
The authors declare no competing interests.
Published: July 23, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.102770.
Contributor Information
Qiangmin Zhang, Email: zhqiangm@outlook.com.
Peter A. Friedman, Email: paf10@pitt.edu.
Supplemental information
References
- Ahn S., Shenoy S.K., Luttrell L.M., Lefkowitz R.J. SnapShot: beta-arrestin functions. Cell. 2020;182:1362–1362 e1361. doi: 10.1016/j.cell.2020.07.034. [DOI] [PubMed] [Google Scholar]
- Ardura J.A., Friedman P.A. Regulation of G protein-coupled receptor function by Na+/H+ exchange regulatory factors. Pharmacol. Rev. 2011;63:882–900. doi: 10.1124/pr.110.004176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armin Bayati R.K., Vincent Francis P.S.M. SARS-CoV-2 Uses Clathrin-Mediated Endocytosis to Gain Access into Cells. J. Biol. Chem. 2021;296:100306. doi: 10.1016/j.jbc.2021.100306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutte Y., Jonsson K., McFarlane H.E., Johnson E., Gendre D., Swarup R., Friml J., Samuels L., Robert S., Bhalerao R.P. ECHIDNA-mediated post-Golgi trafficking of auxin carriers for differential cell elongation. Proc. Natl. Acad. Sci. U S A. 2013;110:16259–16264. doi: 10.1073/pnas.1309057110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burki T. The origin of SARS-CoV-2. Lancet Infect. Dis. 2020;20:1018–1019. doi: 10.1016/S1473-3099(20)30641-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caillet-Saguy C., Durbesson F., Rezelj V.V., Gogl G., Tran Q.D., Twizere J.C., Vignuzzi M., Vincentelli R., Wolff N. Host PDZ-containing proteins targeted by SARS-CoV-2. FEBS J. 2021 doi: 10.1111/febs.15881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantuti-Castelvetri L., Ojha R., Pedro L.D., Djannatian M., Franz J., Kuivanen S., van der Meer F., Kallio K., Kaya T., Anastasina M. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020;370:856–860. doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K.K., Dorosky D., Sharma P., Abbasi S.A., Dye J.M., Kranz D.M., Herbert A.S., Procko E. Engineering human ACE2 to optimize binding to the spike protein of SARS coronavirus 2. Science. 2020;369:1261–1265. doi: 10.1126/science.abc0870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary S., Pak J.E., Gruswitz F., Sharma V., Stroud R.M. Overexpressing human membrane proteins in stably transfected and clonal human embryonic kidney 293S cells. Nat. Protoc. 2012;7:453–466. doi: 10.1038/nprot.2011.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly J.L., Simonetti B., Klein K., Chen K.E., Williamson M.K., Anton-Plagaro C., Shoemark D.K., Simon-Gracia L., Bauer M., Hollandi R. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science. 2020;370:861–865. doi: 10.1126/science.abd3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewson G. Blue native PAGE and antibody gel shift to assess bak and bax conformation change and oligomerization. Cold Spring Harb. Protoc. 2015;2015:485–489. doi: 10.1101/pdb.prot086488. [DOI] [PubMed] [Google Scholar]
- Donowitz M., Cha B., Zachos N.C., Brett C.L., Sharma A., Tse C.M., Li X. NHERF family and NHE3 regulation. J. Physiol. 2005;567:3–11. doi: 10.1113/jphysiol.2005.090399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake M.T., Shenoy S.K., Lefkowitz R.J. Trafficking of G protein-coupled receptors. Circ. Res. 2006;99:570–582. doi: 10.1161/01.RES.0000242563.47507.ce. [DOI] [PubMed] [Google Scholar]
- Ferguson S.S., Downey W.E., 3rd, Colapietro A.M., Barak L.S., Menard L., Caron M.G. Role of beta-arrestin in mediating agonist-promoted G protein-coupled receptor internalization. Science. 1996;271:363–366. doi: 10.1126/science.271.5247.363. [DOI] [PubMed] [Google Scholar]
- Ferrario C.M., Ahmad S., Joyner J., Varagic J. Advances in the renin angiotensin system focus on angiotensin-converting enzyme 2 and angiotensin-(1-7) Adv. Pharmacol. 2010;59:197–233. doi: 10.1016/S1054-3589(10)59007-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A., Madhavan M.V., Sehgal K., Nair N., Mahajan S., Sehrawat T.S., Bikdeli B., Ahluwalia N., Ausiello J.C., Wan E.Y. Extrapulmonary manifestations of COVID-19. Nat. Med. 2020;26:1017–1032. doi: 10.1038/s41591-020-0968-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich V.V., Gurevich E.V. Structural determinants of arrestin functions. Prog. Mol. Biol. Transl. Sci. 2013;118:57–92. doi: 10.1016/B978-0-12-394440-5.00003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamming I., Timens W., Bulthuis M.L., Lely A.T., Navis G., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison A.G., Lin T., Wang P. Mechanisms of SARS-CoV-2 transmission and pathogenesis. Trends Immunol. 2020;41:1100–1115. doi: 10.1016/j.it.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heurich A., Hofmann-Winkler H., Gierer S., Liepold T., Jahn O., Pohlmann S. TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein. J. Virol. 2014;88:1293–1307. doi: 10.1128/JVI.02202-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280 e278. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes R.O. Metastatic cells will take any help they can get. Cancer Cell. 2011;20:689–690. doi: 10.1016/j.ccr.2011.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue Y., Tanaka N., Tanaka Y., Inoue S., Morita K., Zhuang M., Hattori T., Sugamura K. Clathrin-dependent entry of severe acute respiratory syndrome coronavirus into target cells expressing ACE2 with the cytoplasmic tail deleted. J. Virol. 2007;81:8722–8729. doi: 10.1128/JVI.00253-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J., Kim W., Hens J., Dann P., Friedman P.A., Wysolmerski J. NHERF1 is required for proper localization of PMCA2 and suppression of early involution in the lactating mammary gland. Endocrinology. 2019;160:1797–1810. doi: 10.1210/en.2019-00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliche J., Kuss H., Ali M., Ivarsson Y. Cytoplasmic short linear motifs in ACE2 and integrin beta3 link SARS-CoV-2 host cell receptors to mediators of endocytosis and autophagy. Sci. Signal. 2021;14:eabf1117. doi: 10.1126/scisignal.abf1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczuk S., Broer A., Tietze N., Vanslambrouck J.M., Rasko J.E., Broer S. A protein complex in the brush-border membrane explains a Hartnup disorder allele. FASEB J. 2008;22:2880–2887. doi: 10.1096/fj.08-107300. [DOI] [PubMed] [Google Scholar]
- Latorraca N.R., Masureel M., Hollingsworth S.A., Heydenreich F.M., Suomivuori C.M., Brinton C., Townshend R.J.L., Bouvier M., Kobilka B.K., Dror R.O. How GPCR phosphorylation patterns orchestrate arrestin-mediated signaling. Cell. 2020;183:1813–1825 e1818. doi: 10.1016/j.cell.2020.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau A.G., Hall R.A. Oligomerization of NHERF-1 and NHERF-2 PDZ domains: differential regulation by association with receptor carboxyl-termini and by phosphorylation. Biochemistry. 2001;40:8572–8580. doi: 10.1021/bi0103516. [DOI] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamonova T., Zhang Q., Khajeh J.A., Bu Z., Bisello A., Friedman P.A. Canonical and noncanonical sites determine NPT2A binding selectivity to NHERF1 PDZ1. PLoS One. 2015;10:e0129554. doi: 10.1371/journal.pone.0129554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayberry C.L., Soucy A.N., Lajoie C.R., DuShane J.K., Maginnis M.S. JC polyomavirus entry by clathrin-mediated endocytosis is driven by beta-arrestin. J. Virol. 2019;93 doi: 10.1128/JVI.01948-18. e01948–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merad M., Martin J.C. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat. Rev. Immunol. 2020;20:355–362. doi: 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mos I., Jacobsen S.E., Foster S.R., Brauner-Osborne H. Calcium-sensing receptor internalization is β-arrestin-dependent and modulated by allosteric ligands. Mol. Pharmacol. 2019;96:463–474. doi: 10.1124/mol.119.116772. [DOI] [PubMed] [Google Scholar]
- Nisar S.P., Cunningham M., Saxena K., Pope R.J., Kelly E., Mundell S.J. Arrestin scaffolds NHERF1 to the P2Y12 receptor to regulate receptor internalization. J. Biol. Chem. 2012;287:24505–24515. doi: 10.1074/jbc.M112.347104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobles K.N., Xiao K., Ahn S., Shukla A.K., Lam C.M., Rajagopal S., Strachan R.T., Huang T.Y., Bressler E.A., Hara M.R. Distinct phosphorylation sites on the beta(2)-adrenergic receptor establish a barcode that encodes differential functions of beta-arrestin. Sci. Signal. 2011;4:ra51. doi: 10.1126/scisignal.2001707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platre M.P., Bayle V., Armengot L., Bareille J., Marques-Bueno M.D.M., Creff A., Maneta-Peyret L., Fiche J.B., Nollmann M., Miege C. Developmental control of plant Rho GTPase nano-organization by the lipid phosphatidylserine. Science. 2019;364:57–62. doi: 10.1126/science.aav9959. [DOI] [PubMed] [Google Scholar]
- Raj V.S., Mou H., Smits S.L., Dekkers D.H., Muller M.A., Dijkman R., Muth D., Demmers J.A., Zaki A., Fouchier R.A. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J., Wen L., Gao X., Jin C., Xue Y., Yao X. Dog 1.0: illustrator of protein domain structures. Cell Res. 2009;19:271–273. doi: 10.1038/cr.2009.6. [DOI] [PubMed] [Google Scholar]
- Romero G., von Zastrow M., Friedman P.A. Role of PDZ proteins in regulating trafficking, signaling, and function of GPCRs: means, motif, and opportunity. Adv. Pharmacol. 2011;62:279–314. doi: 10.1016/B978-0-12-385952-5.00003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt F., Weisblum Y., Muecksch F., Hoffmann H.H., Michailidis E., Lorenzi J.C.C., Mendoza P., Rutkowska M., Bednarski E., Gaebler C. Measuring SARS-CoV-2 neutralizing antibody activity using pseudotyped and chimeric viruses. J. Exp. Med. 2020;217:e20201181. doi: 10.1084/jem.20201181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidler U., Singh A., Chen M., Cinar A., Bachmann O., Zheng W., Wang J., Yeruva S., Riederer B. Knockout mouse models for intestinal electrolyte transporters and regulatory PDZ adaptors: new insights into cystic fibrosis, secretory diarrhoea and fructose-induced hypertension. Exp. Physiol. 2009;94:175–179. doi: 10.1113/expphysiol.2008.043018. [DOI] [PubMed] [Google Scholar]
- Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. U S A. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A.K., Riederer B., Krabbenhoft A., Rausch B., Bonhagen J., Lehmann U., de Jonge H.R., Donowitz M., Yun C., Weinman E.J. Differential roles of NHERF1, NHERF2, and PDZK1 in regulating CFTR-mediated intestinal anion secretion in mice. J. Clin. Invest. 2009;119:540–550. doi: 10.1172/JCI35541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia I., Peiro C., Lorenzo O., Sanchez-Ferrer C.F., Eckel J., Romacho T. DPP4 and ACE2 in diabetes and COVID-19: therapeutic targets for cardiovascular complications? Front. Pharmacol. 2020;11:1161. doi: 10.3389/fphar.2020.01161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacker D., Stevens R.C., Roth B.L. How ligands illuminate GPCR molecular pharmacology. Cell. 2017;170:414–427. doi: 10.1016/j.cell.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Yang Y., Abou-Samra A.B., Friedman P.A. NHERF1 regulates parathyroid hormone receptor desensitization: interference with beta-arrestin binding. Mol. Pharmacol. 2009;75:1189–1197. doi: 10.1124/mol.108.054486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinman E.J., Hall R.A., Friedman P.A., Liu-Chen L.Y., Shenolikar S. The association of NHERF adaptor proteins with g protein-coupled receptors and receptor tyrosine kinases. Annu. Rev. Physiol. 2006;68:491–505. doi: 10.1146/annurev.physiol.68.040104.131050. [DOI] [PubMed] [Google Scholar]
- Wheeler D.S., Barrick S.R., Grubisha M., Brufsky A.M., Friedman P.A., Romero G. Direct interaction between NHERF1 and Frizzled regulates β-catenin signaling. Oncogene. 2011;30:32–42. doi: 10.1038/onc.2010.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Vasudevan S., Nguyen H., Bork U., Sitaraman S., Merlin D. Extracellular interaction between hCD98 and the PDZ class II domain of hCASK in intestinal epithelia. J. Membr. Biol. 2007;215:15–26. doi: 10.1007/s00232-007-9001-8. [DOI] [PubMed] [Google Scholar]
- Yang N., Shen H.M. Targeting the endocytic pathway and autophagy process as a novel therapeutic strategy in COVID-19. Int. J. Biol. Sci. 2020;16:1724–1731. doi: 10.7150/ijbs.45498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Xiao K., Liu H., Song L., McGarvey J.C., Sneddon W.B., Bisello A., Friedman P.A. Site-specific polyubiquitination differentially regulates parathyroid hormone receptor-initiated MAPK signaling and cell proliferation. J. Biol. Chem. 2018;293:5556–5571. doi: 10.1074/jbc.RA118.001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published article includes all data generated or analysed in this work. This paper does not report original code.