Abstract
Uterine tumors resembling ovarian sex cord tumor (UTROSCT) are rare mesenchymal neoplasms, of uncertain biologic potential, that were recently reported to exhibit recurrent gene fusions involving NCOA2–3. The purpose of this study was to, using a larger sample size, better characterize the histopathologic and molecular diversity of UTROSCT. Twenty-six cases of UTROSCT from five institutions were selected for further study. Fluorescence in situ hybridization for NCOA1, NCOA2, NCOA3, ESR1 and GREB1, and targeted RNA sequencing was performed on seventeen and eight UTROSCTs, respectively. Eight cases underwent massively parallel sequencing to detect single nucleotide variants (SNV), copy number variations (CNV), and structural variants using a targeted hybrid-capture based assay. NCOA1–3 rearrangement was identified in 81.8% (18/22) of cases. The most common fusion was ESR1-NCOA3, occurring in 40.9% (9/22). GREB1-NCOA1 (n=4), ESR1-NCOA2 (n=3), and GREB1-NCOA2 (n=1) rearrangements were also identified. No recurrent SNVs were identified and no tumor had SNVs in FOXL2, DICER1, STK11, or AKT1, which can be seen in ovarian sex cord-stromal tumors. CNVs were infrequent. Clinical follow-up was available for eleven cases with a mean follow-up interval of 94.4 (range 1 – 319) months. Only one case had a recurrence 66 months after the initial diagnosis and this was the single case with a with a GREB1-NCOA2 fusion. This study reports the morphologic spectrum of UTROSCT and confirms the recently reported recurrent NCOA2/3 gene fusions, in addition to identifying novel rearrangements involving NCOA1 in these tumors.
INTRODUCTION
Uterine tumor resembling ovarian sex cord tumor (UTROSCT) is a rare mesenchymal neoplasm showing predominantly sex cord-like differentiation with a variable histologic appearance and polyphenotypic immunoprofile.1,2 Although the name had not yet been coined, descriptions of UTROSCT date back to 1945 when Morehead and Bowman described a uterine tumor resembling a granulosa cell tumor.3 This was further elucidated in 1976 by Clement and Scully, when they reported 14 cases of endometrial stromal tumors with epithelial-like differentiation resembling ovarian sex-cord tumors. They divided these tumors into two groups, Type 1 and Type 2.4 Type 1 tumors were typical endometrial stromal neoplasms with focal areas of sex-cord like differentiation and Type 2 tumors were predominantly or exclusively epithelial in appearance resembling ovarian sex cord tumors and contained minimal to no stromal component. These Type 2 tumors were designated UTROSCT.4 The current World Health Organization (WHO) classification includes these tumors under the category of “Endometrial Stromal and Related Tumors.”1 However, they are polyphenotypic with ultrastructural epithelial and sex cord-like differentiation,5 immunohistochemical features of sex-cord, epithelial and smooth muscle differentiation,2,6–8 and their relationship to stromal neoplasms has not been confirmed.
UTROSCT usually occur in middle-aged women with an average age of 50 years who present with abnormal bleeding, or pelvic pain.9 Occasionally, the tumors are discovered incidentally.9 This neoplasm is considered to be of low malignant potential and typically exhibits benign biological behavior.1,4,9,10 However in one study, up to 23.5% had extra-uterine recurrence, which may in part reflect some referral bias.11 Hysterectomy with or without bilateral salpingo-oophorectomy is considered standard treatment, although studies have reported uterine-sparing treatment for those who wish to preserve fertility.12–14
Few studies have examined the molecular alterations in UTROSCT.15–22 Despite their morphologic resemblance to ovarian sex cord-stromal tumors, mutations in genes such as FOXL2 or DICER1, that are known to occur in comparable ovarian tumors, have not been found in UTROSCT.18,20 In addition, although originally postulated as potentially representing stromal tumors with pure sex-cord differentiation, molecular abnormalities characteristic of stromal neoplasia (e.g., JAZF1-SUZ12 gene fusion) have not been identified.16,17,22 Until recently, no recurrent molecular alterations in UTROSCT had been identified. A recent study by two coauthors showed recurrent NCOA2/3 gene fusions in four UTROSCTs.21 This was followed by another study showing a GREB1-CTNNB1 fusion in a single case.19 In light of these findings, the intent of this study was to assess NCOA1–3, GREB1, and ESR1 rearrangements in UTROSCT to better characterize the molecular alterations present in these tumors.
MATERIALS AND METHODS
Case Collection and Review
Twenty-nine cases of UTROSCT were retrieved from the archives of five institutions. None of the cases were previously reported. Histologic and immunohistochemical slides of all cases of UTROSCT were reviewed and the diagnosis was confirmed by three gynecologic pathologists. Morphologic features including architectural pattern, cell shape, nuclear size and shape, cytoplasmic quality, and mitotic index per 10 high power fields (0.55 mm field diameter) were identified in each tumor and recorded along with immunohistochemical data. Two cases were excluded from the study as the diagnosis could not be confirmed, bringing the total number of cases to twenty-seven. Institutional Research Ethics Board approval was obtained for this study.
Immunohistochemistry
Various immunohistochemical panels were performed on the UTROSCT cases at the time of diagnosis. For the immunohistochemistry performed at Brigham and Women’s Hospital, 4 μm thick tissue sections from formalin-fixed paraffin-embedded blocks were stained for calretinin (1:500 dilution; clone DC8; Zymed, San Francisco, CA), inhibin (1:20 dilution; clone R1; Bio-Rad Antibodies, Hercules, CA), desmin (1:5000 dilution; clone DE-U-10; Sigma, St. Louis, MO), AE1/AE3 (1:200 dilution, AE1 and AE3 clones; Dako, Carpinteria, CA), WT-1 (1:75 dilution; clone 6F-H2; Dako, Carpinteria, CA), CD56 (1:100 dilution; clone 123C3.D5; Cell Marque, Rocklin, CA), SF-1 (1:100 dilution; clone N1665; R&D systems, Tokyo, Japan), estrogen receptor (ER; 1:50 dilution; clone SP1; Fisher Scientific, Hampton, NH), progesterone receptor (PR; 1:50 dilution; clone PgR 636; Dako, Carpinteria, CA), SMA (dilution 1:20 000; clone 1A4; Sigma, St. Louis, MO), caldesmon (1:300 dilution; h-CD clone; Dako, Carpinteria, CA), and CD10 (1:20 dilution; 56C6 clone; Cell Marque, Rocklin, CA) using standard techniques. Slides were stained using Dako Autostainer Plus and the Envision Plus detection system (Dako, Carpinteria, CA).
Molecular testing
Next Generation Sequencing (NGS)
Eight UTROSCTs underwent massively parallel sequencing to detect single nucleotide variants (SNV), copy number variations (CNV), and structural variants using a targeted hybrid-capture based assay that assesses alterations in exons of 447 genes and 191 regions across 60 genes for rearrangement detection (see supplemental digital content 1 for the complete list of genes in the assay). The procedure has been previously described in detail.23,24 In brief, DNA was isolated from unstained formalin-fixed paraffin-embedded tissue slides. Indexed sequencing libraries were enriched for genes of interest with solution-based hybrid capture (Agilent SureSelect; Agilent Technologies). Massively parallel sequencing was performed using Illumina HiSeq 2500 instruments (Illumina, Inc). Data were analyzed by a custom bioinformatics pipeline and interpreted by a pathologist (FD).
Fluorescence In Situ Hybridization (FISH)
Seventeen cases underwent FISH studies to identify molecular rearrangements. FISH for ESR1, GREB1, NCOA1, NCOA2, NCOA3, and NCOA4 using custom break-apart bacterial artificial chromosome (BAC) probes was performed as previously described.21 Nick translation was used to fluorochrome label the DNA BAC probes (see Table S1, supplemental digital content 2, for the list of DNA BAC probes). Standard staining techniques were used and included deparaffizing, pretreating, and hybridizing denatured probes to the unstained slides. Following an overnight incubation period, the slides were rinsed and stained with 4’,6-diamidino-2-phenylindole (DAPI), mounted, and examined under a Zeiss fluorescence microscope (Zeiss Axioplan, Oberkochen, Germany).
RNA sequencing (RNA-Seq)
RNA-Seq was performed as previously described in detail.21 Briefly, RNA was extracted from formalin-fixed paraffin-embedded tissue using the ExpressArt FFPE Clear RNA Ready kit (Amsbio, Cambridge, MA, USA). Total RNA was quantified, and RNA-Seq libraries were prepared following the manufacturer’s instructions using an input of 20 to 100 ng RNA and the TruSight RNA Fusion Panel (Illumina, San Diego, CA, USA). Cases 7, 20, 22, 23, 24, and 25 were analyzed using both the STAR aligner and Manta fusion caller, and the JAFFA fusion caller utilizing BOWTIE2 aligner; cases 26 and 27 were analyzed using the STAR aligner and STAR fusion caller. A complete list of genes covered by the TruSight RNA fusion panel is in supplemental digital content 3.
RESULTS
Clinical Features
Twenty-seven cases diagnosed as UTROSCT based on their histomorphology and immunohistochemical profile were examined in this study and the clinical, histological, and molecular characteristics are summarized in Table 1. Case 5 has been excluded from statistics in the results due to the finding of a JAZF1-SUZ12 translocation by NGS that would suggest a stromal neoplasm. The mean age of patients with UTROSCT in our series was 49.6 (range 20 to 74) years. The specimens reviewed were hysterectomies (65.4%, 17/26), myomectomies (11.5%, 3/26), and endometrial biopsies or curettings (23.1%, 6/26). Follow-up information was available for eleven patients (42.3%, 11/26), with a mean follow-up interval of 94.4 (range 1 – 319) months. Of these, one tumor recurred in the pelvis 66 months after initial diagnosis of UTROSCT (case 16). The other ten patients are alive without evidence of disease.
Table 1:
Clinical, histological, and molecular characteristics of UTROSCT.
| Case | Patient age (years) | Specimen type | Tumor size (cm) | Microscopic tumor margins | Tumor architecture | Cytomorphology | Mitoses (per 10 HPF) | NCOA1–3 fusion | NCOA1–3 fusion detection method | JAZF1/SUZ12/PHF1 rearrangement | Follow-Up (duration in months) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 51 | hysterectomy | 6 | Infiltrative | Sertoliform | Spindle and epithelioid | 4 | -- | -- | Negative | -- |
| 2 | 45 | EMC | at least 1.3 cm | Infiltrative | Corded, trabecular | Spindle and epithelioid | <1 | NCOA2 | FISH | Negative | -- |
| 3 | 48 | hysterectomy | 0.7 | Infiltrative | Corded, sertoliform | Spindle and epithelioid | <1 | ESR1-NCOA3 | FISH | Negative | NED (78.4) |
| 4 | 52 | hysterectomy | 0.5 | Infiltrative | Corded | Spindle and epithelioid | <1 | -- | -- | Negative | -- |
| 6 | 65 | hysterectomy | -- | Circumscribed | Nested, corded, trabecular | Epithelioid | 2 | -- | -- | Negative | -- |
| 7 | 71 | hysterectomy | 6.2 | Infiltrative | Nested, corded, whorled | Spindle and epithelioid | 1 | GREB1-NCOA1 | FISH and RNA-Seq |
Negative | NED (54.3) |
| 8 | 56 | hysterectomy | 0.9 | Infiltrative | Nested, corded, trabecular, sertoliform, retiform | Spindle and epithelioid | 1 | -- | -- | Negative | -- |
| 9 | 49 | hysterectomy | 2.7 | Infiltrative | Corded | Spindle and epithelioid (patchy signet ring cell morphology) | 16 | Negative | FISH | -- | -- |
| 10 | 31 | EMC | -- | Infiltrative | Nested, corded | Spindle and epithelioid | 3 | ESR1-NCOA3 | FISH | -- | -- |
| 11 | 49 | hysterectomy | 10 | Infiltrative | Corded, whorled, cystic | Spindle and epithelioid | 2 | Negative | FISH | -- | -- |
| 12 | 29 | myomectomy | -- | Infiltrative | Nested, sertoliform | Spindle and epithelioid (focally rhabdoid) | 2 | ESR1-NCOA3 | FISH | -- | -- |
| 13 | 20 | myomectomy | 7.7 | Infiltrative | Nested, corded | Epithelioid (rhabdoid) | 6 | ESR1-NCOA2 | FISH | -- | -- |
| 14 | 39 | hysterectomy | 3.2 | Infiltrative | Corded, trabecular, sertoliform | Spindle and epithelioid | 4 | ESR1-NCOA2 | FISH | -- | -- |
| 15 | 71 | EMC | -- | Infiltrative | Sertoliform, retiform | Spindle and epithelioid | <1 | GREB1-NCOA1 | FISH | -- | -- |
| 16 | 74 | hysterectomy | 13 | Infiltrative | Corded, trabecular, sertoliform, retiform | Spindle and epithelioid | 1 | GREB1-NCOA2 | FISH | -- | Recurrence to pelvis (66) |
| 17 | 57 | EMB | -- | Infiltrative | Nested | Epithelioid | <1 | ESR1-NCOA3 | FISH | -- | -- |
| 18 | 38 | EMC | 2 | Infiltrative | Retiform | Spindle and epithelioid | <1 | ESR1-NCOA3 | FISH | -- | NED (50) |
| 19 | 34 | hysterectomy | 3 | Infiltrative | Corded, trabecular | Spindle and epithelioid | <1 | ESR1-NCOA3 | FISH | -- | NED (319) |
| 20 | 55 | hysterectomy | 6 | Circumscribed | Corded, trabecular | Spindle and epithelioid | <1 | Negative | FISH and RNA-Seq |
Negative | NED (228) |
| 21 | 44 | hysterectomy | 2.5 | Infiltrative | Sertoliform, retiform | Spindle and epithelioid | <1 | ESR1-NCOA3 | FISH | -- | NED (23) |
| 22 | 45 | hysterectomy | 15 | Infiltrative | Sertoliform, retiform | Spindle and epithelioid | <1 | ESR1-NCOA3 | RNA-Seq | Negative | NED (207.2) |
| 23 | 54 | hysterectomy | 0.7 | Infiltrative | Corded, sertoliform, retiform | Spindle and epithelioid | <1 | ESR1-NCOA3 | RNA-Seq | Negative | NED (2) |
| 24 | 43 | hysterectomy | 0.4 | Infiltrative | Sertoliform, retiform | Spindle and epithelioid | <1 | Negative | RNA-Seq | Negative | NED (1) |
| 25 | 74 | EMB | at least 2.5 cm | Infiltrative | Sertoliform | Spindle and epithelioid | 3 | GREB1-NCOA1 | RNA-Seq | Negative | -- |
| 26 | 61 | hysterectomy | 8 | Infiltrative | Nested, corded, whorled | Spindle and epithelioid | 2 | GREB1-NCOA1 | RNA-Seq | Negative | NED (10) |
| 27 | 35 | myomectomy | -- | Infiltrative | Whorled | Spindle and epithelioid | <1 | ESR1-NCOA2 | RNA-Seq | Negative | -- |
| ///////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////// | |||||||||||
| *5 | 61 | hysterectomy | 11.5 | Circumscribed | Sertoliform, retiform | Spindle and epithelioid | <1 | Negative | FISH | JAZF1-SUZ12 | -- |
EMC (endometrial curetting); EMB (endometrial biopsy); HPF (high power fields, 400x); NED (no evidence of disease)
Excluded from statistics due to molecular translocation suggesting an endometrial stromal neoplasm
-- Not available/performed
Pathologic Features
Gross Features
The average tumor size was 5.1 (range 0.5 – 15) cm. Macroscopically, most (10/15, 67%) tumors were well-circumscribed, tan-white-yellow, intramural masses (Figure 1), however a minority were submucosal with polypoid protrusion into the endometrial cavity (4/15, 27%), and one was an ill-defined, partially solid and cystic, intramural mass (1/15, 6%) (case 11).
Figure 1:
Gross appearance of UTROSCT showing a well-circumscribed, tan-yellow, intramural mass (case 20).
Histologic Features
Despite the majority appearing grossly circumscribed, microscopic infiltration into the myometrium was common and seen in 91.7% (22/24). UTROSCT has a variable histologic appearance. On low magnification, most exhibited diffuse growth (73.1%, 19/26), with the remaining having a nodular appearance. At higher magnification, the architectural appearance varied from nested, trabecular, corded, whorled, sertoliform and retiform, with the tumors often exhibiting multiple patterns (Figure 2).
Figure 2:
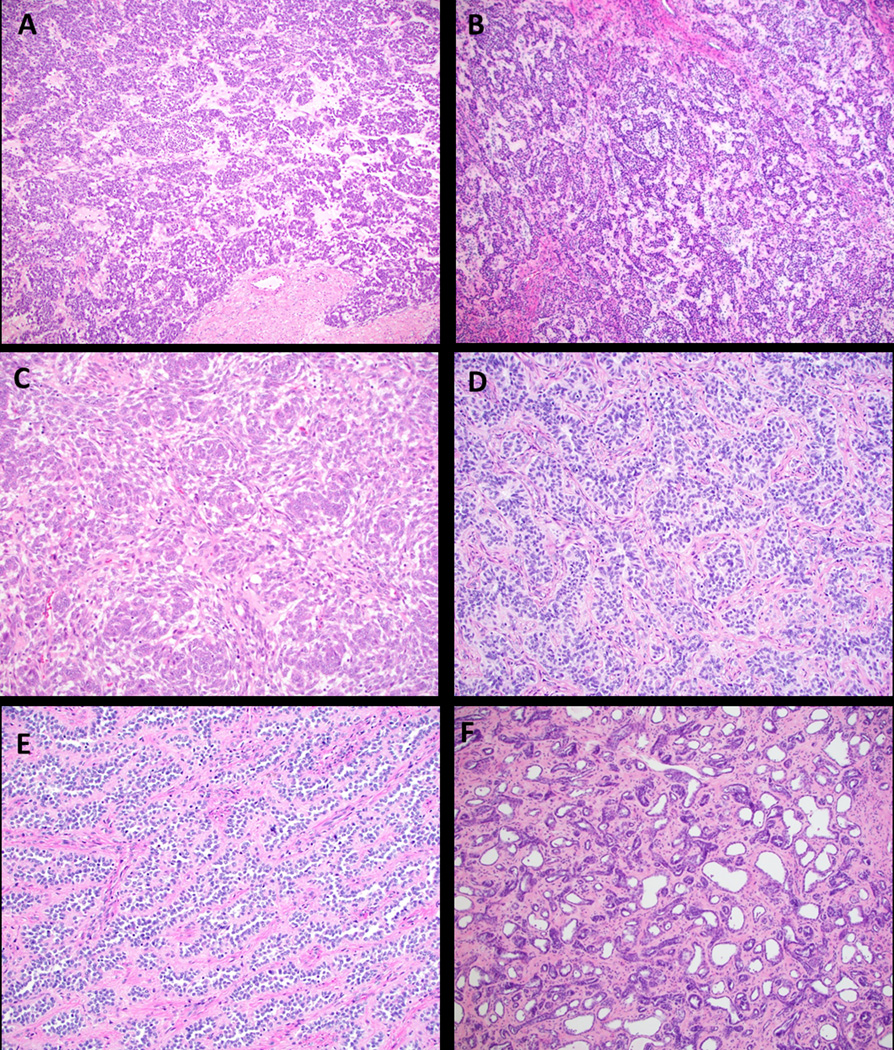
Various architectural patterns in UTROSCT. A. Nested pattern (case 7, H&E, 40x); B. Corded and trabecular pattern (case 20, H&E, 40x); C. Whorled architecture (case 1, H&E, 40x); D, E, F. Sertoliform and retiform patterns (D. case 14, H&E, 200x; E. case 21, H&E, 100x; F. case 15, H&E, 40x).
The neoplasms were mostly composed of a combination of spindle and epithelioid cells with three (11.5%, 3/26) having epithelioid morphology only. The nuclei varied in size, ranging from 1 to 4 times the size of a lymphocyte, but were most commonly two times the size (57.7%, 15/26). Nuclear contours were generally a combination of round/oval to irregular (57.7%, 15/26) with the remaining exhibiting either round, oval or irregular nuclei. The tumors cells exhibited both inconspicuous (61.5%, 16/26) and prominent (38.5%, 10/26) nucleoli with 53.8% (14/26) also having nuclear grooves or clefts, at least focally (Figure 3).
Figure 3:
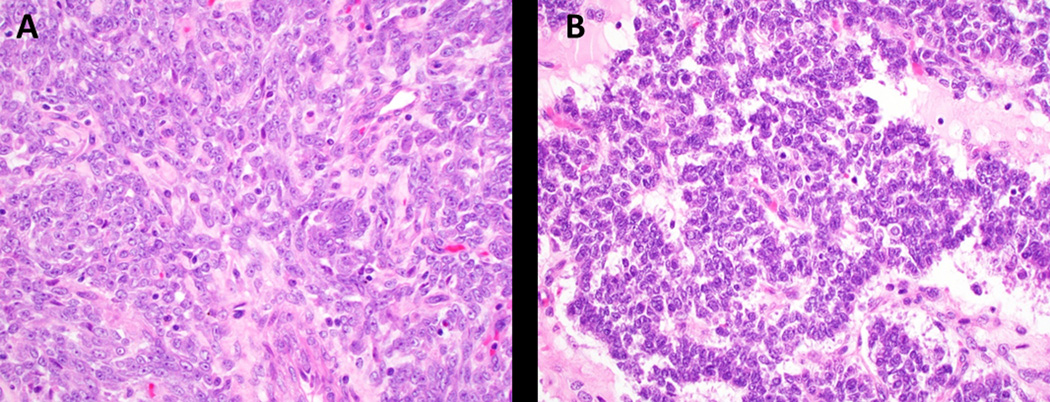
Nuclear cytomorphology in UTROSCT. A. Prominent nucleoli (case 1, H&E, 400x); B. Nuclear grooves (case 7, H&E, 400x).
The neoplastic cells usually contained a small to moderate amount of pale, eosinophilic cytoplasm (Figure 4A) with two (cases 12 and 13, 7.7%) showing rhabdoid morphology, one of which was prominent (Figure 4B) and the other focal. Seven (26.9%) contained clear or vacuolated/lipidized cytoplasm at least focally, and one (case 9, 3.8%) showed microcystic and signet ring cell-like change (Figure 4C–E). Mitotic activity was generally low to absent (< 5 mitotic figures per 10 high power fields (HPF)), except case 13, which had 6 mitotic figures per 10 HPF and case 9, which had 16 mitotic figures per 10 HPF. Atypical mitotic figures were not seen in any of the cases. Six (23.1%) showed collagen/hyalinized stromal change in the form of thin-to-thick septa (Figure 5A). Myxoid change or stromal edema was seen in 19.2% (5/26) and a lymphocytic infiltrate was prominent in 19.2% (5/26) of cases (Figure 5B). The majority had a delicate vascular network of small capillaries; however, two cases exhibited hemangiopericytoma-like vessels. Lymphovascular invasion was present in two cases (cases 1 and 6).
Figure 4:
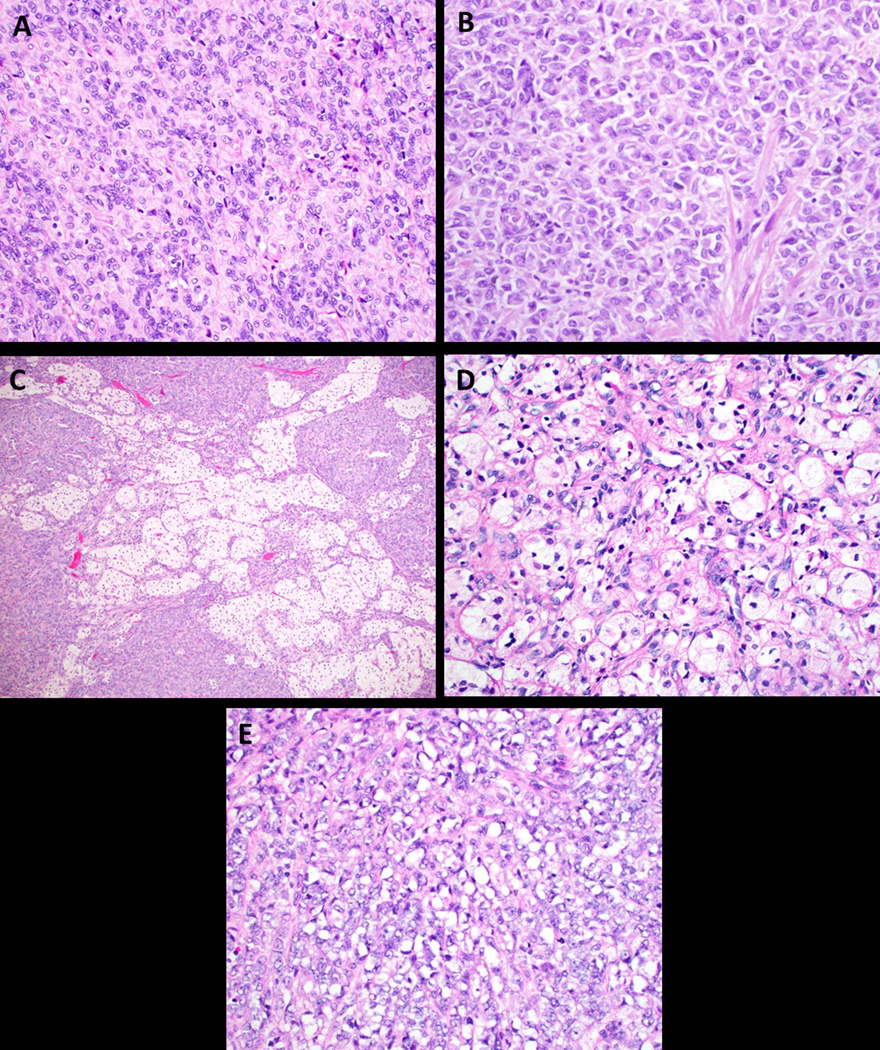
Cytoplasmic features in UTROSCT. A. Most have a small to moderate amount of pale, eosinophilic cytoplasm as seen in case 14 (H&E, 400x); B. Rhabdoid morphology in case 13 (H&E, 400x); C and D. Vacuolated/lipidized cytoplasm (C. case 2, H&E, 100x; D. case 12, H&E, 400x); E. Signet ring morphology in a single case (case 9, H&E, 400x).
Figure 5:
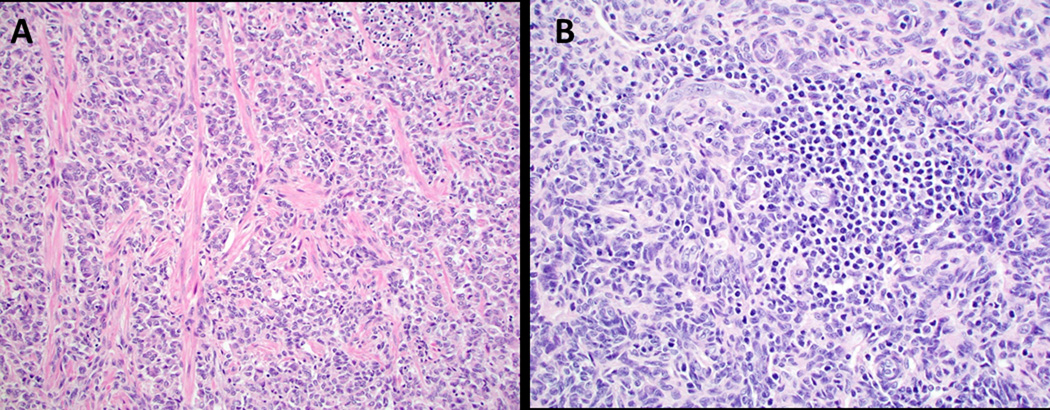
Collagen bands and lymphocytic infiltrate were present in a minority of cases. A. Thick hyaline septa (case 13, H&E, 200x); B. Lymphocytic infiltrate was brisk in a minority of tumors (case 4, H&E, 400x).
Immunohistochemical Features
UTROSCT are polyphenotypic neoplasms that express epithelial, smooth muscle, and sex-cord markers, as well as hormone receptors (Table 2). A variety of immunohistochemical panels had been previously performed on the cases in our series as part of the initial work-up. 16/23 (69.6%) were inhibin positive. Of the seven cases that were inhibin negative, five were positive for at least one other sex-cord marker (calretinin, WT-1, and/or CD56), one was negative for calretinin, and one did not have additional sex-cord markers performed. Of those that were inhibin positive, all were also positive for AE1/AE3 (8/8) and desmin (12/12). Hormone receptors were typically positive with 81.8% (9/11) expressing ER and 100% (9/9) expressing PR. Of note, SF-1 was performed on only three cases, all of which were negative.
Table 2:
Immunohistochemical features of cases.
| Case Number | Inhibin | Calretinin | WT1 | CD56 | SF-1 | AE1/3 | Desmin | SMA | Caldesmon | CD10 | ER | PR |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | + | + | + | + | n/a | + | + | - | - | - | + | n/a |
| 2 | + | + | + | n/a | n/a | + | + | + | - | - | n/a | n/a |
| 3 | + | n/a | n/a | n/a | n/a | n/a | + | + | + | + | n/a | n/a |
| 4 | + | n/a | n/a | n/a | n/a | n/a | + | + | n/a | - | n/a | n/a |
| 6 | - | - | + | n/a | n/a | n/a | + | n/a | + | n/a | n/a | |
| 7 | - | + | n/a | + | n/a | + | + | + | - | + | n/a | n/a |
| 8 | + | + | n/a | n/a | n/a | + | + | + | n/a | - | - | + |
| 9 | + | + | n/a | n/a | - | n/a | n/a | n/a | + | n/a | n/a | |
| 10 | + | + | + | + | n/a | n/a | + | + | - | + | n/a | |
| 11 | + | - | + | n/a | n/a | n/a | + | - | - | + | n/a | n/a |
| 12 | + | + | n/a | n/a | n/a | +* | + | + | - | + | n/a | n/a |
| 13 | + | + | + | n/a | n/a | + | + | + | - | + | n/a | n/a |
| 14 | + | + | n/a | n/a | n/a | +* | + | + | n/a | - | n/a | n/a |
| 15 | + | + | + | n/a | - | + | n/a | n/a | + | + | + | |
| 16 | - | + | + | + | - | - | + | - | - | + | + | + |
| 17 | + | + | + | n/a | n/a | + | + | + | n/a | + | + | + |
| 18 | + | + | n/a | - | n/a | + | n/a | - | n/a | - | + | + |
| 19 | n/a | n/a | n/a | n/a | n/a | + | n/a | + | n/a | n/a | n/a | n/a |
| 20 | n/a | + | n/a | n/a | n/a | + | n/a | + | n/a | n/a | n/a | n/a |
| 21 | + | + | n/a | n/a | n/a | + | n/a | + | n/a | n/a | n/a | n/a |
| 22 | - | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| 23 | + | + | + | n/a | n/a | +* | + | n/a | - | - | + | + |
| 24 | - | - | n/a | n/a | n/a | –* | n/a | n/a | - | n/a | n/a | n/a |
| 25 | n/a | + | - | n/a | n/a | +* | - | - | n/a | n/a | + | + |
| 26 | - | + | + | - | n/a | + | - | - | - | - | - | + |
| 27 | - | + | n/a | n/a | n/a | + | - | + | - | - | + | + |
| ///////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////////// | ||||||||||||
| ^5 | - | + | n/a | + | n/a | + | + | + | + | + | n/a | n/a |
Pankeratin; AE1/3 not done
Excluded from statistics due to molecular translocation suggesting an endometrial stromal neoplasm (JAZF1-SUZ12)
n/a (not available)
Molecular Features
Twenty-two cases had either FISH (14/22) or RNA-Seq (6/22) performed to detect NCOA1, NCOA2, NCOA3, ESR1 and GREB1 rearrangement, and two cases (2/22) had both FISH and RNA-Seq performed. Recurrent NCOA1, NCOA2, or NCOA3 gene fusions were identified in 81.8% (18/22) of cases (Table 1). The most common fusion was ESR1-NCOA3, occurring in 40.9% (9/22) (Figure 6). Four (18.2%), three (13.6%), and one (4.5%) had GREB1-NCOA1, ESR1-NCOA2, and GREB1-NCOA2 fusions (Figure 7A–B), respectively. There was one additional case in which a NCOA2 rearrangement was detected by FISH, however the fusion partner was not identified. For the cases that had fusions detected by RNA-Seq, the tumors with an NCOA2 or NCOA3 rearrangement involved exon 14, which fused with ESR1 exon 3 (cases 22, 23, and 27) and the tumors that had a NCOA1 rearrangement involved exon 13, which fused with GREB1 exon 6 (cases 7, 25 and 26).
Figure 6:
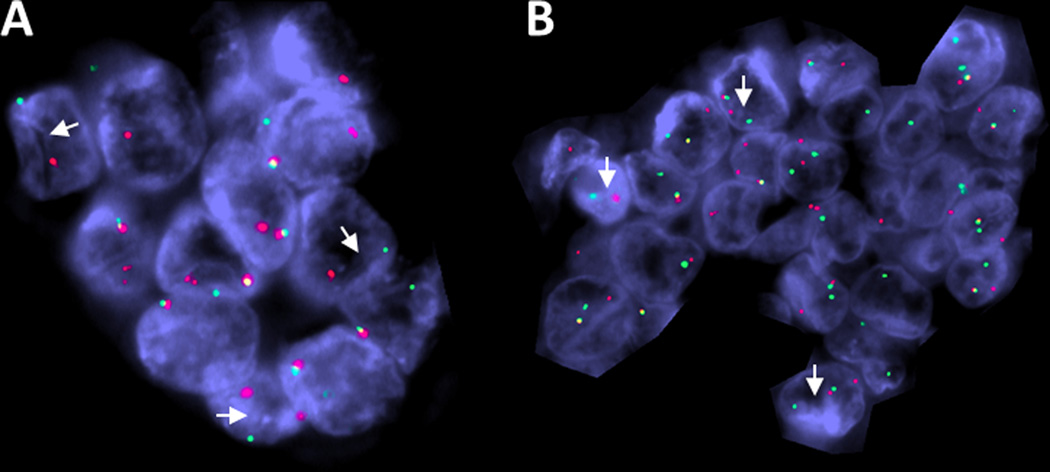
ESR1-NCOA3 fusion was the most common fusion in UTROSCT in this series. FISH showing break-apart signals for: A. ESR1 and B. NCOA3 (case 21) (white arrows: red, centromeric; green, telomeric).
Figure 7:
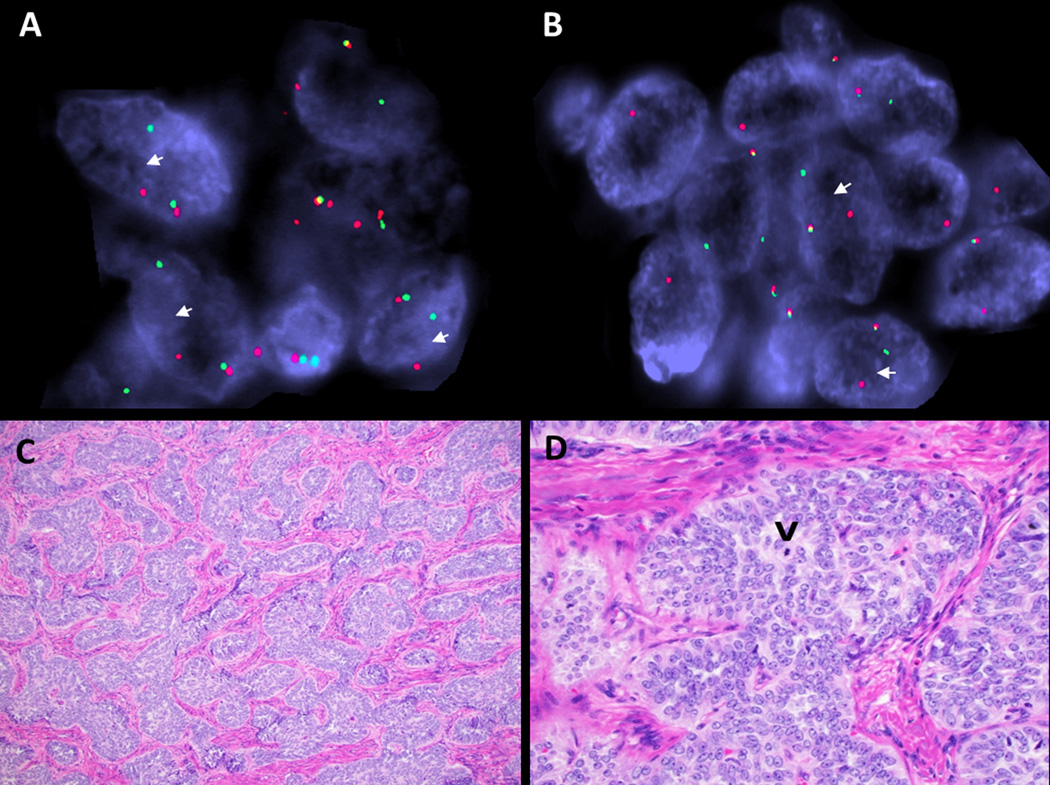
Recurrent UTROSCT (case 16) was the only tumor in series with GREB1-NCOA2 rearrangement. FISH showing break-apart signals for: A. GREB1, and B. NCOA2 (white arrows: red, centromeric; green, telomeric). The tumor showed: C. Sertoliform architecture (H&E,100x) and D. prominent nucleoli and low mitotic index (single mitosis indicated by arrowhead) (H&E, 400x).
18.1% (4/22) did not show a NCOA1–3 fusion, but had morphologic and immunophenotypic features of UTROSCT (Figures 2B and 8 and Table 2). As noted above, an additional tumor (case 5) was subjected to FISH and massively parallel sequencing and found to have a JAZF1-SUZ12 translocation and was negative for NCOA1–3 fusions. Morphologically this tumor showed pure sex-cord differentiation (sertoliform and retiform architecture) with no areas typical of endometrial stromal differentiation (Figure 9). Interestingly, case 16 was the only case with a GREB1-NCOA2 rearrangement in our series, which recurred in the pelvis 66 months after initial diagnosis. The primary tumor involved the serosa and showed nodular growth, sertoliform architecture, prominent nucleoli and low mitotic index (Figure 6C–D). No distinct morphologic features were identified to distinguish NCOA1–3 fusion-positive tumors from fusion-negative tumors.
Figure 8:
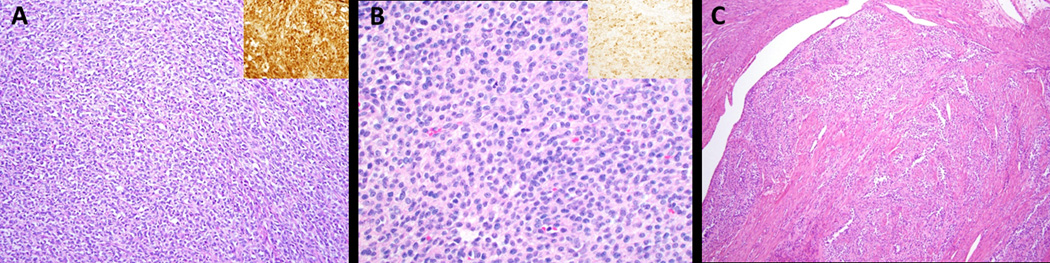
UTROSCT lacking NCOA1–3 fusions have similar morphology to fusion-positive UTROSCT. A. Case 9 showing corded architecture (H&E, 200x) and calretinin positivity (inset: calretinin IHC, 400x); B. Case 11 showing monotonous cells with round to oval nuclei and a moderate amount of pale eosinophilic cytoplasm (H&E, 400x) and inhibin positivity (inset: inhibin IHC, 400x); C. Case 24 showing sertoliform and retiform architecture (H&E, 100x).
Figure 9:
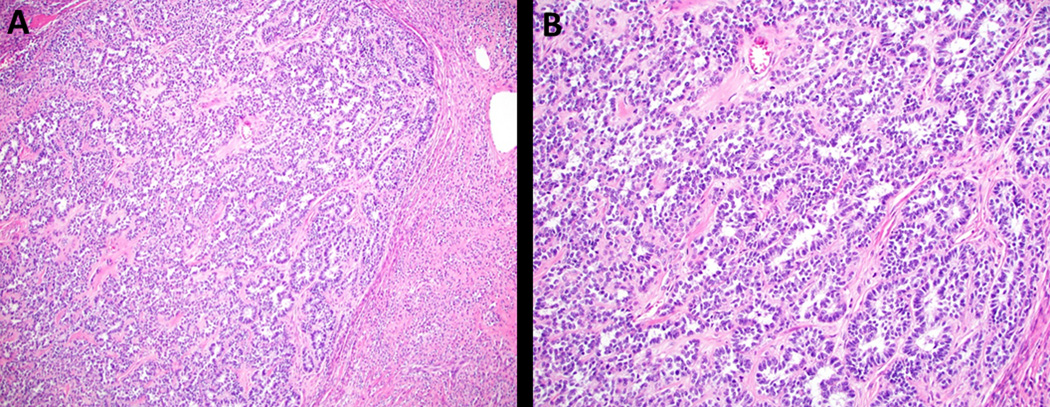
Case 5 is a tumor morphologically consistent with UTROSCT, but contains a JAZF1-SUZ12 translocation typically found in endometrial stromal neoplasia. This tumor had pure sex-cord differentiation (sertoliform and retiform architecture) with no areas typical of low-grade endometrial stromal sarcoma (H&E, A. 40x and B. 100x).
In addition, eight cases were subjected to massively parallel sequencing including two that had NCOA1–3 fusions (cases 3 and 7). No recurrent SNVs were identified and no tumor had SNVs in FOXL2, DICER1, STK11, or AKT1, which are genes that can be mutated in ovarian sex cord-stromal tumors. CNVs were infrequent, but 25% (2/8) had loss of one copy of chromosome 11 and 25% (2/8) had gains in chromosome 1q. One tumor (case 4) had focal gain in FOXL2 and another (case 1) had a single copy loss of DICER1.
DISCUSSION
This study describes the morphologic, immunophenotypic and molecular characteristics of 26 UTROSCTs. It confirms the recently reported findings of recurrent NCOA2–3 gene fusions in UTROSCT21 and identifies novel NCOA1 translocations. In this study, the identified fusion partners in the NCOA1–3 translocations were Estrogen Receptor 1 (ESR1) and Growth Regulation by Estrogen in Breast Cancer (GREB1).
NCOA genes encode a family of nuclear hormone receptor coactivators, that when bound to nuclear receptors, stimulate transcription in a hormone-dependent manner. Their critical role in hormone-dependent uterine function and dysfunction have been described in murine models.25 ESR1 is a ligand dependent transcription factor and GREB1, which is expressed in all ESR1-expressing tissues, is a mediator of estrogen activity.26 Of note, GREB1 is important for tumor growth and is overexpressed in ovarian, breast and prostate carcinomas.27 The proposed tumorigenic mechanism of the recurrent fusion in UTROSCT is based on the principle that ESR1 and GREB1 show a high level expression in uterine tissue. ESR1-NCOA2/3 fusion oncoproteins retain the estrogen receptor zinc finger domains of ESR1 and the nuclear receptor coactivator domain of NCOA2/3. The active promoter region of the ESR1 gene is “hijacked” to result in dysregulated expression of the nuclear receptor coactivator domain of NCOA2/3.21
NCOA1–3 fusions have been identified in a variety of tumors including rhabdomyosarcomas,28–30 mesenchymal chondrosarcomas,31,32 leukemias,33 soft tissue angiofibromas34,35 and more recently, lung adenocarcinoma36 and biphenotypic sinonasal sarcoma.37 In the gynecological tract, NCOA1–3 fusions have been reported in Mullerian adenosarcoma (ESR1-NCOA2/3)38 and undifferentiated uterine sarcoma (UUS) (GREB1-NCOA2 and GREB1-NCOA1).39,40 In the two Mullerian adenosarcomas identified to have either an NCOA2 or NCOA3 fusion, only the tumor with an ESR1-NCOA2 had sex-cord like differentiation.38
The recurrent UTROSCT in our series was the only one to have a GREB1-NCOA2 rearrangement; this tumor showed nodular growth with sertoliform architecture (Figure 7). A GREB1-NCOA2 fusion has been previously reported in one UTROSCT,21 but in contrast to our case, it had morphological overlap with the UUS reported to have the same fusion, showing diffuse sheet-like fascicular and herringbone pattern of spindle to polygonal cells with only some tubular pattern.21 A recent study reports four GREB1-rearranged uterine sarcomas, which had variable and mostly inconspicuous sex-cord differentiation;40 this is in contrast to the GREB1-rearranged UTROSCTs in our current series, which had prominent sex-cord differentiation (Figures 2F and 7). These cases emphasize the translocation-independent morphologic heterogeneity of UTROSCT. Furthermore, fusion-negative tumors were morphologically and immunohistochemically indistinguishable from the fusion positive cases. Other gene fusion(s), which were not detected with the assays used in this study, may be present in UTROSCT.
In one recent study, Croce et al. reported a recurrent UTROSCT with a GREB1-CTNNB1 fusion, in which the authors postulated that the fusion may have contributed to the aggressive behavior of the tumor on the basis of overexpression of a truncated β-catenin protein, and not due to alteration to GREB1.19 CTNNB1 rearrangements were not identified in eleven other UTROSCT cases studied by Croce et al. Interestingly, the recurrent case (case 16) in our study had a GREB1-NCOA2 fusion. This suggests that although CTNNB1 fusion may have contributed to the aggressive behavior in the previously reported UTROSCT, it is not the sole determinant and further elucidation of molecular alterations that confer higher risk for recurrence or metastases remains to be determined. We report the third case of a GREB1-rearranged uterine tumor that has recurred.19,39 Similar to the recent study of GREB1-rearranged sarcomas, we found that GREB1-rearranged UTROSCT occurred in older women (mean age 70 (range 61–74) years) when compared to ESR1-rearranged UTROSCT (mean age 39.5 (range 20–57) years, p < 0.0001); however in contrast, statistically significant differences in tumor size and mitotic activity were not observed (p = 0.16 and p = 0.88, respectively).40 Further studies are required to determine if the GREB1 rearrangement has prognostic significance.
Case 5 in this series which showed a JAZF1-SUZ12 rearrangement and was negative for NCOA1–3 fusions raises an interesting discussion about the relationship of UTROSCT and endometrial stromal neoplasia. The WHO categorizes UTROSCT under “Endometrial Stromal and Related Tumors”1; however, the histogenesis of UTROSCT remains controversial with three potential scenarios: 1) these tumors result from progressive overgrowth of the sex-cord stromal elements in endometrial stromal tumors, 2) UTROSCT are a subset of endometrial stromal tumors that are composed predominantly of sex-cord elements from initiation of the tumor, such that endometrial stromal elements are not identified, and 3) they are a distinct entity that is separate from endometrial stromal tumors.2,16 The majority of evidence supports the third theory. Staats et al. showed that a series of 24 UTROSCT lack the JAZF1-SUZ12 gene fusion16 that is found in approximately 50% of endometrial stromal neoplasms41 and more recently, the discovery of recurrent NCOA1–3 fusions in UTROSCT21 and our study support the notion that it is a distinct entity. Case 5 was an 11.5 cm, adequately sampled mass, that showed pure sex-cord differentiation without typical areas of stromal differentiation (Figure 9) and was polyphenotypic (positive for calretinin, WT-1, CD56, AE1/3, desmin, SMA, and caldesmon). This suggests that in rare instances, UTROSCT may be impossible to morphologically distinguish from endometrial stromal neoplasia showing pure sex cord differentiation and emphasizes the potential use of the newly found molecular distinction in the diagnosis of these two tumors. Despite this difference in underlying molecular abnormalities, preliminary RNA expression data suggests that a subset of low-grade endometrial stromal sarcomas and UTROSCTs cluster together, raising the possibility of a potential relationship between these two neoplasms.21
Several histologically similar entities are in the differential diagnosis of UTROSCT, especially when the surgical specimen is an endometrial biopsy, curetting, or myomectomy. These include endometrial carcinoma with sex cord-like growth, endometrial stromal neoplasm with sex cord elements, adenosarcoma with extensive sex cord-like differentiation and epithelioid smooth muscle neoplasm. Carcinoma with sex cord-like growth typically has areas of low grade adenocarcinoma merging with the sex cord-like areas and are often sex-cord and smooth muscle marker negative.42 Similarly, adenosarcoma is usually biphasic and contains a benign epithelial glandular component in addition to a low grade stromal component that is typically strongly CD10 positive.43 Epithelioid smooth muscle neoplasms usually lack prominent sex-cord morphology and marker expression.44 Endometrial stromal neoplasms typically have conventional stromal areas that are negative for sex-cord markers and often harbor a JAZF1-SUZ12 fusion or PHF1 rearrangement.45,46 In our study, 53.8% (14/26) of UTROSCTs were analyzed for alterations characteristic of low grade stromal neoplasms (JAZF1-SUZ12 translocations and PHF1 rearrangements) by either NGS or RNA-Seq, all of which were negative. Of these, 80% (8/10) contained a NCOA1–3 fusion. Similarly, case 5, containing the JAZF1-SUZ12 translocation did not harbor a NCOA1–3 fusion. This further supports that these rearrangements are mutually exclusive and that NCOA1–3 fusion detection may be a useful ancillary test for assisting in distinguishing UTROSCT from stromal neoplasms, however larger comparative studies are required. Considering the four NCOA1–3 fusion negative UTROSCTs, two (cases 20 and 24) were not found to harbor a JAZF1 or PHF rearrangement by RNA-Seq, in one RNA-Seq was attempted, but failed (case 11), and there was insufficient material available to test the last one (case 9).
One limitation of this study is that nine cases (34.6%) were from non-hysterectomy specimens. UTROSCT is often difficult to diagnose with certainty in endometrial biopsies, curettings or myomectomies; however histologic features, such as sex-cord morphology in the absence of typical low-grade endometrial stromal morphology, along with a polyphenotypic immunoprofile, allow for a diagnosis of UTROSCT to be favored. The non-hysterectomy cases in our study were originally diagnosed using descriptive terms such as “spindle and epithelioid neoplasm with sex-cord differentiation, favor UTROSCT”. Of note, all of the non-hysterectomy specimens in this study harbored a NCOA1–3 fusion. In instances where the diagnosis is equivocal, FISH or RNA-Seq for these fusions may be helpful.
In conclusion we confirmed, in the largest series to date, that the majority (81.8% in our series) of UTROSCTs are characterized by recurrent NCOA1–3 fusions and emphasize the translocation independent morphologic heterogeneity and immunohistochemical profile of these tumors. This provides further support that UTROSCT is a distinct entity in which identification of these fusions can be used to help distinguish this tumor from other entities that routinely fall into the differential diagnosis, such as endometrial stromal neoplasia with sex-cord like differentiation.
Supplementary Material
REFERENCES
- 1.Kurman RJ, Carcangiu ML, Herrington CS, Young RH. WHO Classification of Tumours of Female Reproducitve Organs. Lyon: International Agency for Research on Cancer; 2014. [Google Scholar]
- 2.Hurrell DP, McCluggage WG. Uterine tumour resembling ovarian sex cord tumour is an immunohistochemically polyphenotypic neoplasm which exhibits coexpression of epithelial, myoid and sex cord markers. J Clin Pathol. 2007;60(10):1148–1154. doi: 10.1136/jcp.2006.044842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morehead R, Bowman M. Heterologous Mesodermal Tumors of the Uterus. Report of a Neoplasm Resembling a Granulosa Cell Tumor. Am J Pathol. 1945;21(1):53–61. http://www.ncbi.nlm.nih.gov/pubmed/19970803. Accessed April 20, 2019. [PMC free article] [PubMed] [Google Scholar]
- 4.Clement PB, Scully RE. Uterine tumors resembling ovarian sex-cord tumors. A clinicopathologic analysis of fourteen cases. Am J Clin Pathol. 1976;66(3):512–525. [DOI] [PubMed] [Google Scholar]
- 5.Gupta M, de Leval L, Selig M, Oliva E, Nielsen GP. Uterine tumors resembling ovarian sex cord tumors: an ultrastructural analysis of 13 cases. Ultrastruct Pathol. 2010;34(1):16–24. doi: 10.3109/01913120903506603 [DOI] [PubMed] [Google Scholar]
- 6.de Leval L, Lim GSD, Waltregny D, Oliva E. Diverse Phenotypic Profile of Uterine Tumors Resembling Ovarian Sex Cord Tumors. Am J Surg Pathol. 2010;34(12):1749–1761. doi: 10.1097/PAS.0b013e3181f8120c [DOI] [PubMed] [Google Scholar]
- 7.Irving JA, Carinelli S, Prat J. Uterine tumors resembling ovarian sex cord tumors are polyphenotypic neoplasms with true sex cord differentiation. Mod Pathol. 2006;19(1):17–24. doi: 10.1038/modpathol.3800475 [DOI] [PubMed] [Google Scholar]
- 8.Stewart CJR, Crook M, Tan A. SF1 immunohistochemistry is useful in differentiating uterine tumours resembling sex cord-stromal tumours from potential histological mimics. Pathology. 2016;48(5):434–440. doi: 10.1016/j.pathol.2016.03.013 [DOI] [PubMed] [Google Scholar]
- 9.Czernobilsky B. Uterine Tumors Resembling Ovarian Sex Cord Tumors. Int J Gynecol Pathol. 2008;27(2):229–235. doi: 10.1097/PGP.0b013e3181569a21 [DOI] [PubMed] [Google Scholar]
- 10.Clement PB, Scully RE. Uterine tumors with mixed epithelial and mesenchymal elements. Semin Diagn Pathol. 1988;5(2):199–222. http://www.ncbi.nlm.nih.gov/pubmed/2840723. Accessed April 20, 2019. [PubMed] [Google Scholar]
- 11.Moore M, McCluggage WG. Uterine tumour resembling ovarian sex cord tumour: first report of a large series with follow-up. Histopathology. 2017;71(5):751–759. doi: 10.1111/his.13296 [DOI] [PubMed] [Google Scholar]
- 12.Schraag SM, Caduff R, Dedes KJ, Fink D, Schmidt A- M. Uterine Tumors Resembling Ovarian Sex Cord Tumors - Treatment, recurrence, pregnancy and brief review. Gynecol Oncol reports. 2017;19:53–56. doi: 10.1016/j.gore.2017.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blake EA, Sheridan TB, Wang KL, et al. Clinical characteristics and outcomes of uterine tumors resembling ovarian sex-cord tumors (UTROSCT): a systematic review of literature. Eur J Obstet Gynecol Reprod Biol. 2014;181:163–170. doi: 10.1016/j.ejogrb.2014.07.050 [DOI] [PubMed] [Google Scholar]
- 14.Jeong KH, Lee HN, Kim MK, Kim M- L, Seong SJ, Shin E. Successful delivery after conservative resectoscopic surgery in a patient with a uterine tumor resembling ovarian sex cord tumor with myometrial invasion. Obstet Gynecol Sci. 2015;58(5):418–422. doi: 10.5468/ogs.2015.58.5.418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J, Blakey GL, Zhang L, Bane B, Torbenson M, Li S. Uterine tumor resembling ovarian sex cord tumor: report of a case with t(X;6)(p22.3;q23.1) and t(4;18)(q21.1;q21.3). Diagn Mol Pathol. 2003;12(3):174–180. http://www.ncbi.nlm.nih.gov/pubmed/12960700. Accessed March 28, 2019. [DOI] [PubMed] [Google Scholar]
- 16.Staats PN, Garcia JJ, Dias-Santagata DC, et al. Uterine tumors resembling ovarian sex cord tumors (UTROSCT) lack the JAZF1-JJAZ1 translocation frequently seen in endometrial stromal tumors. Am J Surg Pathol. 2009;33(8):1206–1212. doi: 10.1097/PAS.0b013e3181a7b9cf [DOI] [PubMed] [Google Scholar]
- 17.Umeda S, Tateno M, Miyagi E, et al. Uterine tumors resembling ovarian sex cord tumors (UTROSCT) with metastasis: clinicopathological study of two cases. Int J Clin Exp Pathol. 2014;7(3):1051–1059. http://www.ncbi.nlm.nih.gov/pubmed/24696722. Accessed July 31, 2018. [PMC free article] [PubMed] [Google Scholar]
- 18.Chiang S, Staats PN, Senz J, et al. FOXL2 Mutation is Absent in Uterine Tumors Resembling Ovarian Sex Cord Tumors. Am J Surg Pathol. 2015;39(5):618–623. doi: 10.1097/PAS.0000000000000367 [DOI] [PubMed] [Google Scholar]
- 19.Croce S, Lesluyes T, Delespaul L, et al. GREB1-CTNNB1 fusion transcript detected by RNA-sequencing in a uterine tumor resembling ovarian sex cord tumor (UTROSCT): a novel CTNNB1 rearrangement. Genes, Chromosom Cancer. October 2018. doi: 10.1002/gcc.22694 [DOI] [PubMed] [Google Scholar]
- 20.Croce S, de Kock L, Boshari T, et al. Uterine Tumor Resembling Ovarian Sex Cord Tumor (UTROSCT) Commonly Exhibits Positivity With Sex Cord Markers FOXL2 and SF-1 but Lacks FOXL2 and DICER1 Mutations. Int J Gynecol Pathol. 2016;35(4):301–308. doi: 10.1097/PGP.0000000000000240 [DOI] [PubMed] [Google Scholar]
- 21.Dickson BC, Childs TJ, Colgan TJ, et al. Uterine Tumor Resembling Ovarian Sex Cord Tumor. A Distinct Entity Characterized by Recurrent NCOA2/3 Gene Fusions. Am J Surg Pathol. 2019;43(2):178–186. doi: 10.1097/PAS.0000000000001153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nucci M, Schoolmeester J, Sukov W et al. Uterine tumors resembling ovarian sex cord tumor (UTROSCT) lack rearrangement of PHF1 by FISH. Mod Pathol. 2014;27:298A. [Google Scholar]
- 23.Sholl LM, Do K, Shivdasani P, et al. Institutional implementation of clinical tumor profiling on an unselected cancer population. JCI insight. 2016;1(19):e87062. doi: 10.1172/jci.insight.87062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia EP, Minkovsky A, Jia Y, et al. Validation of OncoPanel: A Targeted Next-Generation Sequencing Assay for the Detection of Somatic Variants in Cancer. Arch Pathol Lab Med. 2017;141(6):751–758. doi: 10.5858/arpa.2016-0527-OA [DOI] [PubMed] [Google Scholar]
- 25.Szwarc MM, Kommagani R, Lessey BA, Lydon JP. The p160/steroid receptor coactivator family: potent arbiters of uterine physiology and dysfunction. Biol Reprod. 2014;91(5):122. doi: 10.1095/biolreprod.114.125021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hodgkinson K, Forrest LA, Vuong N, Garson K, Djordjevic B, Vanderhyden BC. GREB1 is an estrogen receptor-regulated tumour promoter that is frequently expressed in ovarian cancer. Oncogene. 2018;37(44):5873–5886. doi: 10.1038/s41388-018-0377-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hodgkinson KM, Vanderhyden BC. Consideration of GREB1 as a potential therapeutic target for hormone-responsive or endocrine-resistant cancers. Expert Opin Ther Targets. 2014;18(9):1065–1076. doi: 10.1517/14728222.2014.936382 [DOI] [PubMed] [Google Scholar]
- 28.Alaggio R, Zhang L, Sung Y- S, et al. A Molecular Study of Pediatric Spindle and Sclerosing Rhabdomyosarcoma: Identification of Novel and Recurrent VGLL2-related Fusions in Infantile Cases. Am J Surg Pathol. 2016;40(2):224–235. doi: 10.1097/PAS.0000000000000538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mosquera JM, Sboner A, Zhang L, et al. Recurrent NCOA2 gene rearrangements in congenital/infantile spindle cell rhabdomyosarcoma. Genes, Chromosom Cancer. 2013;52(6):538–550. doi: 10.1002/gcc.22050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arnold MA, Barr FG. Molecular diagnostics in the management of rhabdomyosarcoma. Expert Rev Mol Diagn. 2017;17(2):189–194. doi: 10.1080/14737159.2017.1275965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Panagopoulos I, Gorunova L, Bjerkehagen B, Boye K, Heim S. Chromosome aberrations and HEY1-NCOA2 fusion gene in a mesenchymal chondrosarcoma. Oncol Rep. 2014;32(1):40–44. doi: 10.3892/or.2014.3180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang L, Motoi T, Khanin R, et al. Identification of a novel, recurrent HEY1-NCOA2 fusion in mesenchymal chondrosarcoma based on a genome-wide screen of exon-level expression data. Genes Chromosomes Cancer. 2012;51(2):127–139. doi: 10.1002/gcc.20937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carapeti M, Aguiar RC, Goldman JM, Cross NC. A novel fusion between MOZ and the nuclear receptor coactivator TIF2 in acute myeloid leukemia. Blood. 1998;91(9):3127–3133. http://www.ncbi.nlm.nih.gov/pubmed/9558366. Accessed April 21, 2019. [PubMed] [Google Scholar]
- 34.Jin Y, Möller E, Nord KH, et al. Fusion of the AHRR and NCOA2 genes through a recurrent translocation t(5;8)(p15;q13) in soft tissue angiofibroma results in upregulation of aryl hydrocarbon receptor target genes. Genes Chromosomes Cancer. 2012;51(5):510–520. doi: 10.1002/gcc.21939 [DOI] [PubMed] [Google Scholar]
- 35.Arbajian E, Magnusson L, Mertens F, Domanski HA, Vult von Steyern F, Nord KH. A novel GTF2I/NCOA2 fusion gene emphasizes the role of NCOA2 in soft tissue angiofibroma development. Genes Chromosomes Cancer. 2013;52(3):330–331. doi: 10.1002/gcc.22033 [DOI] [PubMed] [Google Scholar]
- 36.Cao Q, Liu Z, Huang Y, Qi C, Yin X. NCOA1–ALK: a novel ALK rearrangement in one lung adenocarcinoma patient responding to crizotinib treatment. Onco Targets Ther. 2019;Volume 12:1071–1074. doi: 10.2147/OTT.S192367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andreasen S, Bishop JA, Hellquist H, et al. Biphenotypic sinonasal sarcoma: demographics, clinicopathological characteristics, molecular features, and prognosis of a recently described entity. Virchows Arch. 2018;473(5):615–626. doi: 10.1007/s00428-018-2426-x [DOI] [PubMed] [Google Scholar]
- 38.Piscuoglio S, Burke KA, Ng CK, et al. Uterine adenosarcomas are mesenchymal neoplasms. J Pathol. 2016;238(3):381–388. doi: 10.1002/path.4675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brunetti M, Panagopoulos I, Gorunova L, Davidson B, Heim S, Micci F. RNA-sequencing identifies novel GREB1-NCOA2 fusion gene in a uterine sarcoma with the chromosomal translocation t(2;8)(p25;q13). Genes Chromosomes Cancer. 2018;57(4):176–181. doi: 10.1002/gcc.22518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee C, Kao Y, Lee W, et al. Clinicopathologic Characterization of GREB1-rearranged Uterine Sarcomas With Variable Sex-Cord Differentiation. Am J Surg Pathol. 2019;00(00):1–15. doi: 10.1097/PAS.0000000000001265 [DOI] [PubMed] [Google Scholar]
- 41.Hodge JC, Bedroske PP, Pearce KE, Sukov WR. Molecular Cytogenetic Analysis of JAZF1, PHF1, and YWHAE in Endometrial Stromal Tumors Discovery of Genetic Complexity by Fluorescence in Situ Hybridization. J Mol Diagnostics. 2016;18(4). doi: 10.1016/j.jmoldx.2016.02.001 [DOI] [PubMed] [Google Scholar]
- 42.Murray SK, Clement PB, Young RH. Endometrioid carcinomas of the uterine corpus with sex cord-like formations, hyalinization, and other unusual morphologic features: A report of 31 cases of a neoplasm that may be confused with carcinosarcoma and other uterine neoplasms. Am J Surg Pathol. 2005. doi: 10.1097/01.pas.0000149704.89463.05 [DOI] [PubMed] [Google Scholar]
- 43.McCluggage WG. Mullerian adenosarcoma of the female genital tract. Adv Anat Pathol. 2010. doi: 10.1097/PAP.0b013e3181cfe732 [DOI] [PubMed] [Google Scholar]
- 44.Toledo G, Oliva E. Smooth muscle tumors of the uterus: a practical approach. Arch Pathol Lab Med. 2008;132(4):595–605. doi: 10.1043/1543-2165(2008)132[595:SMTOTU]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- 45.Lee C- H, Nucci MR. Endometrial stromal sarcoma - the new genetic paradigm. Histopathology. 2015;67(1):1–19. doi: 10.1111/his.12594 [DOI] [PubMed] [Google Scholar]
- 46.Nucci MR. Practical issues related to uterine pathology: Endometrial stromal tumors. Mod Pathol. 2016;29(S1):S92–S103. doi: 10.1038/modpathol.2015.140 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



