Abstract
Purpose:
Myofiber strain, Eff, is a mechanistically relevant metric of cardiac cell shortening and is expected to be spatially uniform in healthy populations, making it a prime candidate for the evaluation of local cardiomyocyte contractility. In this study, a new, efficient pipeline was proposed to combine microstructural cDTI and functional DENSE data in order to estimate Eff in vivo.
Methods:
Thirty healthy volunteers were scanned with 3 long-axis (LA) and 3 short-axis (SA) DENSE slices using 2D displacement encoding and one SA slice of cDTI. The total acquisition time was 11 min ± 3 min across volunteers. The pipeline first generates 3D SA displacements from all DENSE slices which are then combined with cDTI data to generate a cine of myofiber orientations and compute Eff. The precision of the post-processing pipeline was assessed using a computational phantom study. Transmural myofiber strain was compared to circumferential strain, Ecc, in healthy volunteers using a Wilcoxon sign rank test.
Results:
In vivo, computed Eff was found uniform transmurally compared to Ecc (−0.14[−0.15,−0.12] vs −0.18 [−0.20,−0.16], p<0.001, −0.14 [−0.16, −0.12] vs −0.16 [−0.17,−0.13], p<0.001 and −0.14 [−0.16,−0.12] vs Ecc_C=−0.14 [−0.15,−0.11], p=0.002, Eff_C vs Ecc_C in the endo, mid, and epi layers respectively).
Conclusion:
We demonstrate that it is possible to measure in vivo myofiber strain in a healthy human population in 10 min per subject. Myofiber strain was observed to be spatially uniform in healthy volunteers making it a potential biomarker for the evaluation of local cardiomyocyte contractility in assessing cardiovascular dysfunction.
Keywords: Myofiber strain, DENSE, cDTI
Introduction
Cardiac function arises from the complex deformation of billions of cardiac muscle cells (cardiomyocytes) during the cardiac cycle (1). Aggregated cardiomyocytes form so-called “myofibers” that change orientation from epicardium to endocardium in a helical fashion (2). This unique microstructural architecture transforms uniaxial myofiber shortening into the circumferential, longitudinal, and radial deformation of the left ventricle (LV). Despite the mechanistic importance of myofiber strain (Eff) to heart function, it has remained very difficult to measure in vivo owing to the challenges of acquiring and integrating both functional motion and microstructural cardiac data.
Cardiac deformation can be characterized using motion tracking methods that enable the measurement of the tissue displacement field. In particular, the Displacement ENcoding with Stimulated Echoes (DENSE) (3,4) approach provides spatiotemporally resolved maps of myocardial tissue displacement. DENSE encodes each component of tissue displacement from a reference state to a deformed state into the phase of the complex-valued MRI signal. The tissue displacement data is then used to estimate cardiac strain. However, since myofiber orientation data is typically not available, cardiac strains are usually computed assuming a geometry dependent cylindrical coordinate system along the longitudinal (Ell), circumferential (Ecc), and radial (Err) directions of the LV. These descriptive strains are useful, but they are not mechanistically linked to cardiac contraction – except in the LV midwall where it is assumed that Ecc accord with myofiber strain (Eff) owing to the presumed circumferential orientation of midwall myofibers. A direct measure of myofiber orientations across the myocardium is needed to eliminate this widely used assumption and enable Eff calculation throughout the LV.
Currently, cardiac Diffusion Tensor imaging (cDTI) is the only approach that is able to interrogate cardiac microstructure in vivo. cDTI probes the mobility of water molecules within the myocardium, where they are hindered by the surrounding microstructure and undergo higher diffusivity in the direction of the cardiomyocyte long-axis. Therefore, cDTI provides a direct measure of the myofiber orientation (5). Recently, the development of first and second-order motion-compensated gradient waveform designs have enabled microstructural imaging in vivo despite the inherent respiratory and cardiac bulk motion (6–8). This approach has been used to characterize the in vivo microstructural remodeling in hypertrophic cardiomyopathy (9,10), dilated cardiomyopathy (9), and amyloidosis (11).
Measuring Eff in vivo requires both microstructural and functional information, which is technically challenging and has only been reported in a few studies. Tseng et al. (12) produced the first measures of in vivo Eff in healthy humans using a STEAM-based sequence for measuring cardiac microstructure and to measure cardiac velocity information. Wang et al. (13) used a finite element model to estimate Eff by fusing in vivo 2D tagging and an ex vivo atlas of myofiber orientation. More recently, Perotti et al. (14) and Verzhbinsky et al. (15) proposed a post-processing approach to measure Eff that combines in vivo cDTI and 3D displacement encoded DENSE data. However, the clinical translation of the approaches described above remains limited as they rely either on velocity rather than displacement data, integration of ex vivo cDTI data, or long 3D displacement encoded DENSE acquisitions.
In this work, we propose a new method that combines cDTI and a volume of short- and long-axis DENSE slices with 2D displacement encoding to enable the rapid measurement of in vivo myofiber strain (Eff) in humans. The accuracy of the approach for measuring Eff was evaluated using a computational phantom for a range of myofiber orientations. Finally, in vivo Eff values were measured and reported for thirty (N=30) healthy volunteers.
Theory
Cardiac deformation can be assessed from displacement encoding methods such as DENSE. Like most motion encoding approaches, the displacement encoding in DENSE relies on a pair of gradients to encode and decode the cardiac motion into the phase of the complex-valued MR signal. However, the DENSE approach stores the magnetization along the longitudinal axis (Mz) after motion encoding and then restores it at each cardiac phase using a stimulated echo before motion decoding. Consequently, only the relative displacement between encoding and decoding is reflected in the MR phase. In practice, this encoded displacement corresponds to an Eulerian description of the displacement field. In order to measure tissue strains, the Eulerian description of the displacement field is first converted to a Lagrangian description using spatial and temporal fitting approaches (16). These tissue displacements in the Lagrangian description are spatially discretized to a set of nodes (one per pixel), and can be expressed as follows:
| [1] |
where is the position vector for the -th node in the reference configuration corresponding to the first cardiac phase measured using DENSE, represents the deformed position vector at time , and is the Lagrangian displacement field at time .
To obtain a continuously defined is subsequently interpolated spatially using the scattered interpolant function (17) (Matlab, The MathWorks, Natick, MA, USA) with a ‘natural’ interpolation approach. Once is continuously defined, the deformation gradient tensor at each location and time t can be expressed as
| [2] |
In practice, is computed numerically using a central finite difference scheme. The cardiac strain components along a chosen direction are then evaluated as
| [3] |
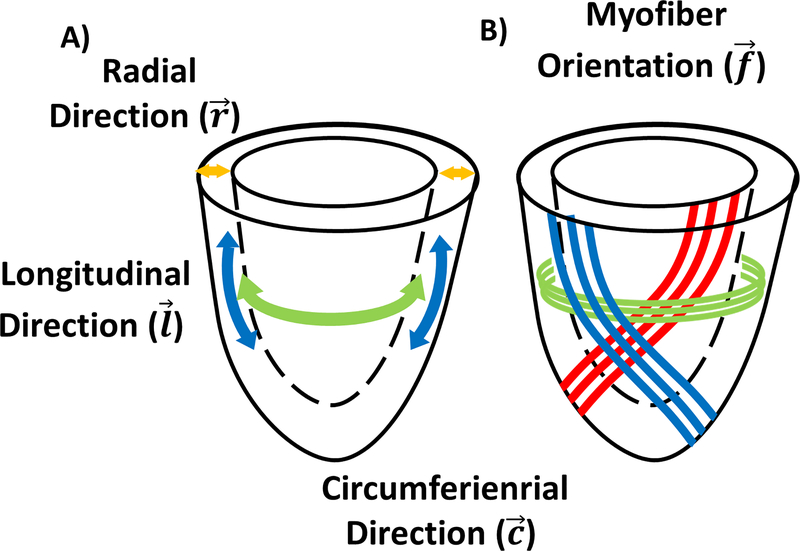
where is the Green-Lagrange strain tensor , is the right Cauchy-Green deformation tensor and I is the identity matrix. As shown in Figure 1, cardiac strains are traditionally measured along the longitudinal (), circumferential (), and radial () directions of the LV, thereby producing the longitudinal (Ell), circumferential (Ecc), and radial (Err) strain components. In this work, the myofiber orientation measured using cDTI, represented by the vector , is used to compute the myofiber strain (Eff). To do so, we have developed an acquisition and analysis pipeline to compute Eff across the LV in humans combining cDTI and DENSE methods.
Figure 1:
(A) Arbitrary cylindrical coordinate system along the longitudinal (l), circumferential (c), and radial (r) directions of the left ventricle used to project the strain. (B) Representation of the helical orientation of the myofiber (f) from endocardium to epicardium. Please note that myofibers are not continuous throughout the heart.
Methods
Imaging Protocol Overview
Thirty healthy volunteers (N=30) were recruited for an MRI exam after obtaining signed statements of informed consent under an IRB approved protocol. Images were acquired on a single 3T MRI scanner (Prisma, Siemens Healthineers, Erlangen), using an 18-channel body phased-array coil and a 32-channel spine array coil.
DENSE acquisition
Six 2D spiral cine DENSE slices were acquired in three short-axis (SA) views and three long-axis (LA) views. SA slices corresponded to the base, mid, and apex locations of the LV. LA slices were acquired in 2, 4, and 3 chambers views. The sequence parameters were TE=1.08ms, TR=15ms, Flip Angle 15°, 2.2×2.2×8mm, Matrix 90×90, FOV 200×200mm with a reduced zonal excitation of 120×120mm. The multi-shot spiral readout was composed of four interleaves; two spiral interleaves were acquired per cardiac phase; 2784 samples per leave. 2D displacements were acquired using balanced 3-point displacement encoding with ke=0.1 cycle/mm. The acquisition was ECG triggered and acquired during a breath-hold that required 14 RR-intervals per slice. At least 30 cardiac phases were acquired per volunteer using the beginning of systole as the reference configuration and with a temporal resolution of 15ms. The total acquisition time for the six slices was ~3 minutes.
The DENSE displacements were reconstructed off-line using the DENSE analysis tool (version 0.5.1, repository: https://github.com/denseanalysis/denseanalysis) in Matlab (MathWorks, Natick, MA, USA). Within the tool, DENSE images were segmented using motion guided segmentation with manual seeding (18). The voxel-wise Lagrangian displacement field was calculated using a 10th order polynomial temporal model and a linear spatial model with smoothness equal to 0.9. Using the DENSE analysis tool, the measured circumferential (Ecc_M) and radial (Err_M) strains were reported for the SA slices as well as the longitudinal (Ell_M) strain for the LA slices.
cDTI acquisition
A single mid-ventricular short-axis slice was imaged during free-breathing with a mid-systolic trigger delay (TD~250 ms) using a custom motion-compensated single-shot spin-echo EPI cDTI sequence. The sequence parameters were TE=61 ms, inner-volume excitation, FOV 200×160 mm, acquisition matrix 128×104 leading to a resolution 1.6×1.6×8 mm, which was interpolated in-plane to 0.8×0.8×8 mm. Parallel imaging with 2x-GRAPPA, partial Fourier 6/8, and a readout bandwidth of 1860 Hz/px were used to reduce the EPI echo-train length in order to limit the image distortion. Two b-values (0 and 350 s/mm2) were acquired using six diffusion-encoding directions and ten repetitions each. The diffusion encoding gradient waveform was composed of four gradient lobes designed symmetrically around the 180° refocusing pulse to null both the first and second-order gradient moments (6). Gradient durations were 6.33/10.47/10.47/6.33 ms with a ramp time of 1.52 ms and a maximum amplitude of 74.37 mT/m. The single-shot acquisition was ECG and end-respiratory triggered using a navigator located on the right liver dome resulting in a TR of one breathing cycle (~4000 ms). A total of 70 cDTI images were obtained at mid-systole corresponding to an acquisition time of ~6 minutes.
cDTI reconstruction was performed off-line using a custom Matlab framework (https://github.com/KMoulin/DiffusionRecon) that included a rigid registration, an average of the ten repetitions, and a Fourier transform zero-filling interpolation. The non-diffusion weighted image and the six diffusion weighted directions were then fit to the second-order tensor using a non-linear least-squares approach (19). cDTI metrics including mean diffusivity (MD), fractional anisotropy (FA), and eigenvectors (), were directly extracted from the tensor model. After manual segmentation of the LV, the first eigenvector () representing the myofiber orientation was projected to the epicardial LV surface (10,20) and used to measure the helix angle (HA).
The uncertainty of the in vivo cDTI experiment was estimated using a bootstrapping approach (21). Among the ten repetitions acquired, a subset of five repetitions was randomly selected and averaged to reconstruct the diffusion tensor. This bootstrapping operation was repeated 1000 times to produce a distribution of reconstructed diffusion metrics. Finally, the uncertainty of the cDTI metrics (dMD, dFA, dHA) were estimated pixel-wise taking the 95% confidence (95CI) interval of each distribution.
Combination of cDTI and DENSE
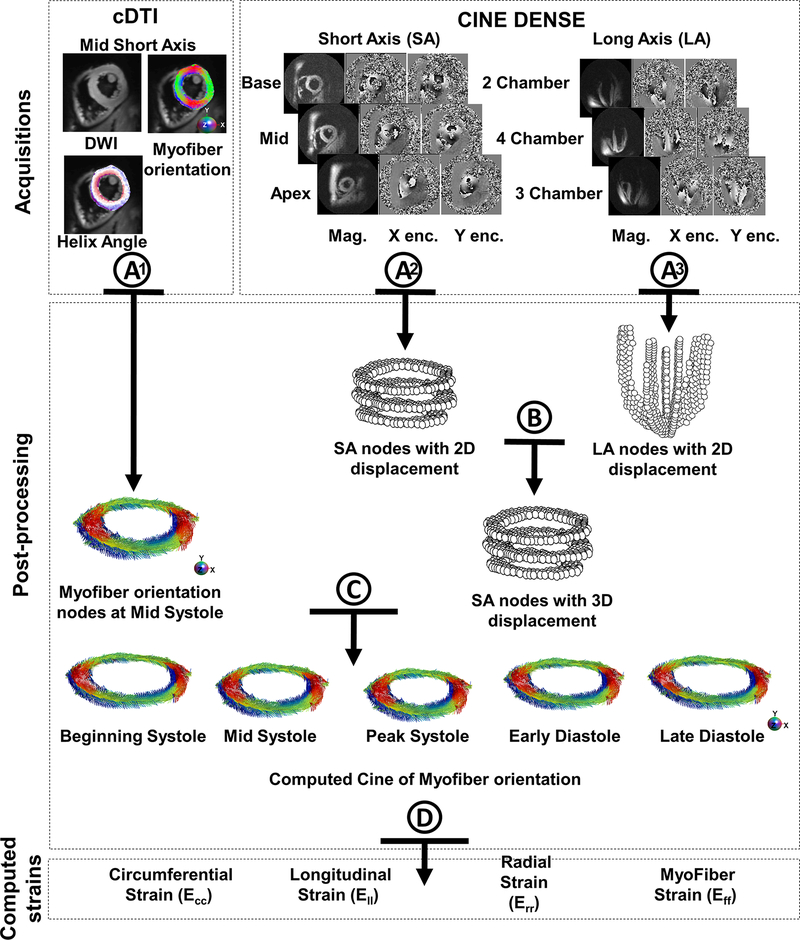
The analysis pipeline used to compute Eff in humans with combined in vivo cDTI and DENSE is shown in Figure 2 and illustrated in Supporting Video S1. It is composed of four steps:
-
A)
cDTI voxels within the segmented LV were discretized as a set of nodes (one per voxel). About 2200 voxels per slice were identified. The Lagrangian DENSE displacements were also defined using a set of nodes. About 300 voxels per DENSE slice were identified. All nodes were expressed in a global coordinate system using their respective image quaternions. At the matching cardiac phase, the cDTI nodes were registered in-plane to the SA mid-ventricular DENSE nodes using a center of mass approach to account for the potential misalignment between the two acquisitions.
-
B)
The LA displacements are generally smooth. Therefore, the 2D displacement fields from LA and SA DENSE slices can be combined to obtain a SA 3D displacement field. 2D displacements from the LA were projected along the direction orthogonal to the SA slice and applied to the SA nodes using linear interpolation.
-
C)
The 3D SA displacement field was interpolated and differentiated to calculate the deformation gradient tensor F at each cDTI node (Equation 2). F was then used to reorient the myofiber orientation field from its initial configuration (mid-systole) to the DENSE reference configuration (beginning of systole). The same operation was repeated to define for each cardiac phase.
-
D)
Cardiac strains were then computed along the circumferential (Ecc_C), radial (Err_C), longitudinal (Ell_C), and myofiber (Eff_C) directions (Equation 3). Computed strain rates (strain temporal derivatives) were also reported in the circumferential (SRcc_C) and myofiber (SRff_C) directions.
Figure 2:
Post-processing steps used to combine DENSE and cDTI data and calculate myofiber strain Eff. After cDTI reconstruction (A1), myofiber orientations were represented as nodes using one node per voxel. Cardiac displacement fields were also represented using nodes after the DENSE reconstruction (A2 & A3). (B) 2D Displacement fields from long-axis (LA) and short-axis (SA) are combined to obtain a SA 3D displacement field. (C) The SA 3D displacement field is interpolated and differentiated to generate a deformed cine of myofiber orientation during the cardiac cycle. (D) Finally, cardiac strains are calculated from the SA 3D displacement field and the myofiber orientations and using the beginning of systole as the reference configuration.
All analyses were performed in Matlab and the code for the post-processing pipeline to combine cDTI and DENSE is available at https://github.com/KMoulin/Eff.
Computational Phantom Validation
An in silico experiment was designed to validate the accuracy of the proposed cDTI and DENSE pipeline to estimate Eff. A computational LV phantom was used to produce a cardiac-like displacement field. Given an initial LV geometry and a rule-based myofiber orientation field (), the phantom numerically solves an optimization problem to define a displacement field of a chosen form that approximates as best as possible target Ell, Ecc, Err, and Eff strains. All simulations were performed in Matlab and the code to generate and analyze the computational phantom is available at: https://github.com/luigiemp/CardiacKinematicsPhantom.
The original mid-ventricular phantom was modified to include an apex-to-base LV geometry. The reference phantom configuration was a 3D LV shape, 85mm high from apex to base, with epicardial (epi) and endocardial (endo) radii of 35 mm and 25 mm at the base that are then linearly reduced toward the apex. The target endocardial and epicardial strains were [−0.15, −0.15] for Ell; [−0.20, −0.16] for Ecc; [0.45, 0.30] for Err; and [−0.15, −0.15] for Eff. A quadratic transmural distribution of myofiber orientation is adopted with corresponding HA ranging from 37° at endocardium, −9° in the mid-wall, and −45° at epicardium (22). Once the displacement field was simulated, the numerical “ground-truth” circumferential (Ecc_GT), radial (Err_GT), and longitudinal (Ell_GT) strains were computed.
From the simulated displacement field, six cine DENSE simulated images were then generated,as described in (15), in the short-axis (SA) and long-axis (LA) views with sequence parameters matched to the in vivo balanced 3-point encoding cine DENSE protocol. These synthetic DENSE images underwent the same reconstruction steps as described for the in vivo processing. Similarly, the measured circumferential (Ecc_M) and radial (Err_M) strains were reported for the SA slices as well as the longitudinal (Ell_M) strain for the LA slices. From the defined myofiber orientation field, synthetic cDTI orientation maps were generated with identical resolution as the in vivo cDTI protocol. Both simulated cDTI and cine DENSE images were then processed using the proposed pipeline from which the computed circumferential (Ecc_C), radial (Err_C), longitudinal (Ell_C), and myofiber (Eff_C) strains were extracted.
Finally, the robustness of the proposed pipeline to estimate in vivo Eff was assessed with regard to the uncertainty of the myofiber orientation field. Variability was added to the myofiber orientation maps before combining them with DENSE to include variance of the transmural HA distribution.
Quantitative Analysis
All quantitative values (MD, FA, HA, dMD, dFA, dHA, Ecc, Ell, Err, Eff, SRcc, and SRff) were reported as median [Quartile 1, Quartile 3] across all volunteers. HA as well as mid systolic SRcc_C, SRff_C, and peak systolic Ecc_C and Eff_C were also reported as a function of the transmural wall depth by dividing the wall into nine equally spaced bins. Peak-systole was determined as the cardiac phase corresponding to the lowest median Ecc value across all volunteers. Similarly, strain rates were reported at mid-systole, determined using the lowest median SRcc across volunteers. Finally, the time-resolved median of Ecc_C, Eff_C, SRcc_C, and SRff_C, were reported throughout the cardiac cycle at three equally spaced transmural depths (endo, mid, epi).
All statistical analyses were carried out in SPSS (Version 26, IBM Corp.) using a non-parametric pair-wise Wilcoxon sign rank test. p<0.05 was considered statistically significant.
Results
Computational Phantom Validations
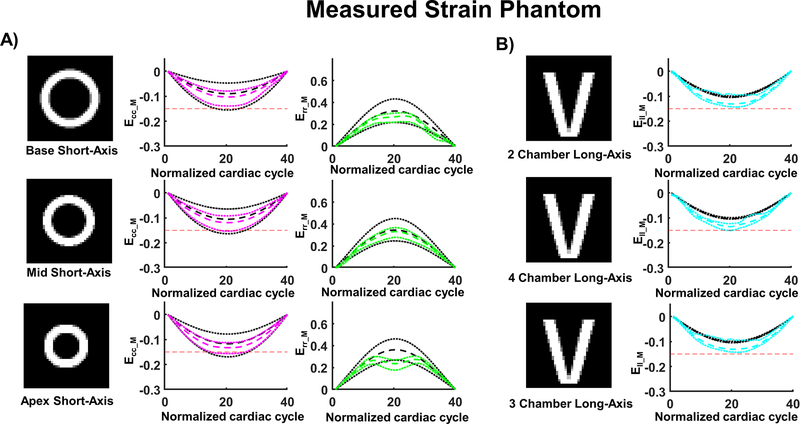
A comparison between ground truth strains and the strains extracted from the synthetic DENSE acquisitions for all slices are shown in Figure 3. For the SA slices at peak systole, the magnitude of measured circumferential strain was slightly higher than the ground truth strain (Ecc_GT= −0.09 [−0.16, −0.05], −0.13 [−0.18, −0.10], −0.12 [−0.17, −0.08] vs. Ecc_M= −0.10 [−0.14, −0.08], −0.12 [−0.15, −0.09], −0.13 [−0.16, −0.11] at the base, mid and apex, respectively). In contrast, the magnitude of measured radial strain was lower than the ground truth strain (Err_GT= 0.32 [0.22, 0.43], 0.38 [0.30, 0.47], 0.36 [0.27, 0.46] vs Err_M= 0.27 [0.22, 0.30], 0.33 [0.28, 0.37], 0.24 [0.18, 0.27] at the base, mid and apex, respectively). For all the LA slices at peak systole, the magnitude of measured longitudinal strain was higher than ground truth strain (Ell_GT= −0.10 [−0.11, −0.10] vs Ell_M= −0.13 [−0.14, −0.09]).
Figure 3:
Cardiac strains measured in the computational phantom using synthetic DENSE images. (A) Measured circumferential (Ecc_M) and radial strain (Err_M) are calculated from DENSE using 2D displacement encoding in a short-axis view at base, mid, and apical levels. (B) Measured longitudinal strain (Ell_M) was calculated from DENSE using 2D displacement encoding in long axis 2-, 4-, and 3- chamber views. The red dashed line provides a −0.15 strain reference; the colored dashed and dotted lines represent the median strain and the first and third quartiles across the slice. The black dashed and dotted lines represent the median ground truth strain and the first and third quartile across the slice.
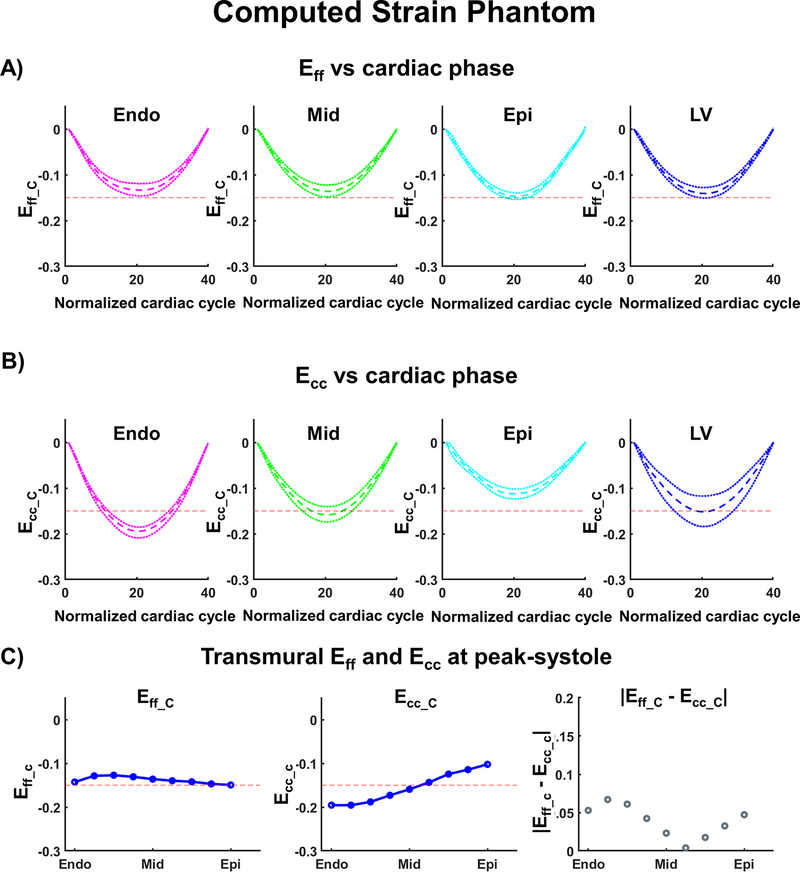
To evaluate the accuracy of computed Eff, synthetic cDTI orientation maps were generated from a quadratic transmural distribution of myofiber orientations and combined with synthetic DENSE data using the proposed pipeline. The resulting Eff_C are shown in Figure 4 and compared to Ecc_C. (Eff_C= −0.13 [−0.15, −0.12], −0.14 [−0.15, −0.12], −0.15 [−0.15, −0.14] vs Ecc_C= −0.19 [−0.21, −0.19], −0.16 [−0.17, −0.14], −0.11 [−0.12, −0.10] in the endo, mid, and epi layers, respectively). At peak systole for the whole LV, the computed Eff was slightly higher than Ecc (Eff_C= −0.14 [−0.15, −0.13] vs Ecc_C= −0.15 [−0.18, −0.12]). At peak systole, the computed longitudinal strain was Ell_C= −0.16 [−0.16, −0.16] and the computed radial strain Err_C= 0.27 [0.22, 0.29].
Figure 4:
Cardiac strain values for the computational phantom calculated after combining cDTI and DENSE. (A) Computed Myofiber strain (Eff_C) and (B) circumferential strain (Ecc_C) at endo, mid, epi layers, and across the left ventricular (LV) wall. The red dashed line provides a −0.15 strain reference; the colored dashed and dotted lines represent the median strain and the first and third quartiles across the corresponding layer. (C) Transmural distribution of Eff_C, Ecc_C, and the absolute difference between Eff_C and Ecc_C at peak systole. Blue solid lines represent the transmural median.
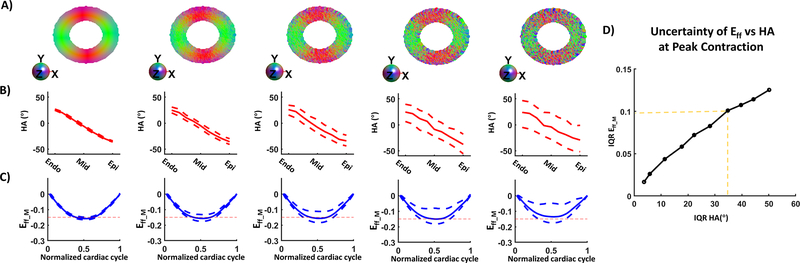
The robustness of the proposed pipeline to estimate Eff was assessed with regard to the uncertainty in the myofiber orientation field. Variability was added to the myofiber orientation maps which resulted in an increase of IQR for the transmural HA distribution (Figure 5). Small variations in the HA (HA= 25.7 [24.1, 19.6] and −35.6 [−36.3, −34.2] ° at endo and epi) corresponded to tight range of Eff_C at peak systole (Eff_C= −0.16 [−0.17, −0.15]). As the spread of HA increased (HA= 19.6 [−3.4, 38.2], −29.2 [−48.6 −8.15] ° at endo and epi), so did the spread of Eff (Eff_C= −0.14 [−0.18, −0.06]).
Figure 5:
A) Myofiber orientation colored maps with no variance at the extreme left and with an added variance to the right. (B) Corresponding transmural HA distribution. C) Computed Eff after combining DENSE with the myofiber orientation maps shown in A). D) Inter-quartile range (Quartile 1 – Quartile 3) of Eff vs the Inter-quartile range of HA.
In vivo acquisition
A total of thirty healthy volunteers were scanned (8 males and 22 females, 54 ± 18 years old, 72 ± 13 beats per minute). The average acquisition time for one DENSE slice was 12 ± 2 s and 7 ± 3.5 min for the free-breathing cDTI SA slice. The total acquisition time was 11 min ± 3 min across volunteers. After manual segmentation of the LV, 311 [278, 355], 295 [274, 340] and 254 [210 291] voxels were identified for the DENSE acquisition in the Basal, Mid, and Apical slices, respectively. 2171 [1881 2520] voxels were identified for the Mid LV cDTI acquisition.
DENSE Strains
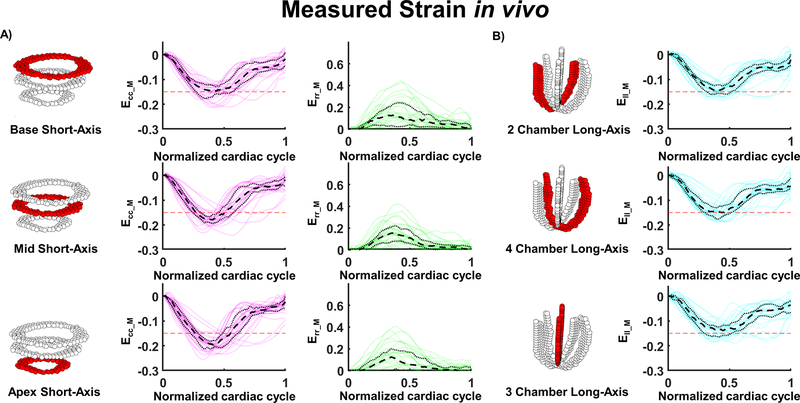
Before combining with cDTI data, cardiac strains were measured by using solely the DENSE acquisitions. Measured strains for each volunteer are displayed throughout the cardiac cycle in Figure 6. For the SA slices and at peak systole, the magnitude of measured circumferential strain (Ecc_M) was found to increase from base to apex (Ecc_M= −0.15 [−0.17, −0.12], −0.18 [−0.19, −0.16], −0.19 [−0.21, −0.18]) in the base, mid, and apex, respectively. Radial strain was similar for the three slices (Err_M= 0.11 [0.02, 0.24], 0.14 [0.08, 0.22], 0.10 [0.01, 0.19]) at base, mid, and apex, respectively). Longitudinal strain was also similar across slices (Ell_M= −0.15 [−0.16, −0.12], −0.15 [−0.18, −0.13], −0.14 [−0.16, −0.12] in the 2-, 4-, and 3-chamber view respectively).
Figure 6:
Measured cardiac strains in healthy adults (N=30). (A) Circumferential (Ecc) and radial strain (Err) are calculated from DENSE using 2D displacement encoding in a short-axis view at base, mid, and apex levels. (B) Longitudinal strain (Ell) was calculated from DENSE using 2D displacement encoding in long axis 2-, 4-, and 3-chambers views. The red dashed line provides a −0.15 strain reference.
cDTI Metrics
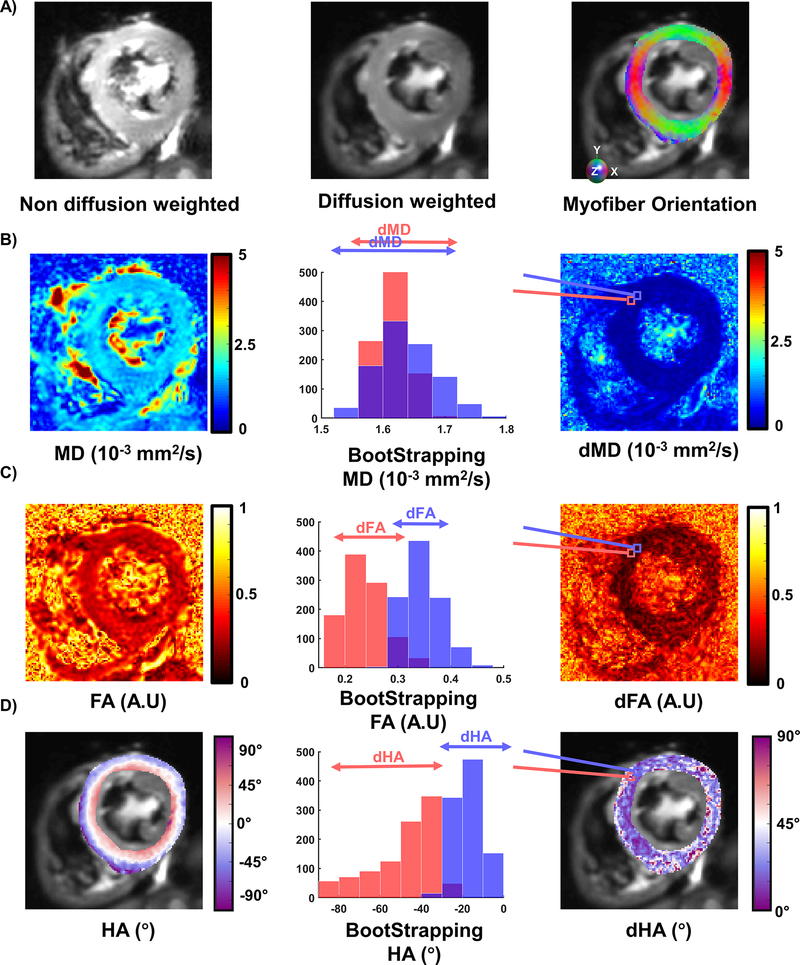
An example of cDTI acquisition and uncertainty maps are shown in Figure 7. cDTI metrics and corresponding uncertainty estimations per volunteer are given in Supporting Information Figure S1. In the SA mid-wall, MD was 1.52 [1.45, 1.59] x 10−3 mm2/s across volunteers (N=30) with corresponding dMD 0.22 [0.20, 0.31] x 10−3 mm2/s which was estimated using a bootstrapping approach. For all volunteers, FA was 0.36 [0.34, 0.38] A.U. and the uncertainty dFA was 0.23 [0.20 0.26] A.U.
Figure 7:
Example of DTI metrics and uncertainty estimation. A) Non-diffusion weighted, diffusion weighted and myofiber orientation colored maps. B) Mean diffusivity (MD) maps, two bootstrapped distributions displayed for two adjacent pixels exhibiting extreme differences. The 95% confidence interval of these distributions correspond to the uncertainty MD (dMD) maps displayed on the right. C) Fraction of anisotropy maps, bootstrapped distribution for two voxels, and corresponding dFA maps. D) Helix Angle (HA) maps, bootstrapped example and dHA maps.
The transmural HA distributions as a function of LV are shown in Figure 7–C. Transmural HA distribution was 40.4 [21.7, 49.8] °, 2.0 [−0.1, 5.8] °, and −26.5 [−33.3, 4.6] ° at endo, mid, and epi respectively. Across volunteers, the estimated uncertainty dHA was 34.5 [29.7, 48.3] ° overall and 36.6 [28.8, 45.7] °, 33.8 [24.6, 42.1] °, and 50.6 [46.3, 62.1] ° at endo, mid, and epi respectively.
In vivo Myofiber Strain and Strain Rate
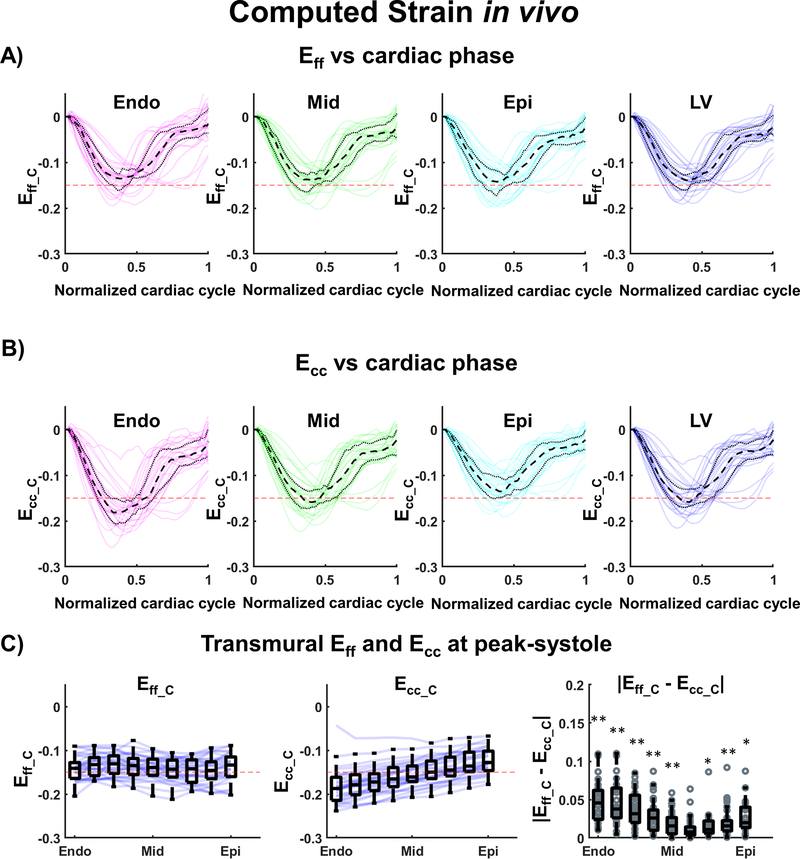
After combining cDTI and DENSE, strains were computed at the mid-ventricular SA slice level as shown in Supporting Information Figure S2. Figure 8A and 8B show a comparison of computed myofiber and circumferential strains during the cardiac cycle within endo, mid, and epi layers and over the entire LV. At peak systole, Eff_C was smaller in magnitude than Ecc_C in the endo layer (Eff_C= −0.14 [−0.15, −0.12] vs Ecc_C= −0.18 [−0.20, −0.16], p<0.001) and in the mid layer (Eff_C= −0.14 [−0.16, −0.12] vs Ecc_C= −0.16 [−0.17, −0.13], p<0.001) and slightly larger in the epi layer (Eff_C= −0.14 [−0.16, −0.12] vs Ecc_C= −0.14 [−0.15, −0.11], p=0.002). Across the whole SA LV at peak systole, statistical differences were observed between Eff_C and Ecc_C (Eff_C= −0.14 [−0.16, −0.13] vs Ecc_C= −0.16 [−0.17, −0.13], p<0.001). The distributions of Eff_C and Ecc_C are shown in Figure 8C. Eff_C is shown to be transmurally uniform, which indicates uniform myofiber shortening across the heart, while a transmural gradient is evident for Ecc_C. The transmural values for Eff_C and Ecc_C as well as the statistical differences are reported in Table 1.
Figure 8:
Cardiac strain values across volunteers (N=30) calculated after combining cDTI and DENSE. (A) Computed Myofiber strain (Eff_C) and (B) circumferential strain (Ecc_C) at endo, mid, epi layers, and across the left ventricular (LV) wall. The black dashed and dotted lines represent the median and the first and third quartiles across volunteers; the red dashed line provides a −0.15 strain reference; the colored solid lines are individual medians per volunteers. (C) Transmural distribution of Eff_C, Ecc_C, and of the absolute difference of Eff_C and Ecc_C at peak-systole. Blue solid lines represent the transmural median per volunteer and the box plots represent the median and IQR across volunteers. Statistical differences between Eff_C and Ecc_C have been assessed using a pair-wise Wilcoxon sign rank test: p-values<0.01 are represented by (**) and p-values<0.05 by (*).
Table 1:
Transmural values and mid left ventricle (LV) values for computed myofiber strain (Eff_C), circumferential strain (Ecc_C), myofiber strain rate (SRff_C), and circumferential strain rate (SRcc_C).
| Wall Segment | 1 Endo | 2 | 3 | 4 | 5 Mid | 6 | 7 | 8 | 9 Epi | LV |
|---|---|---|---|---|---|---|---|---|---|---|
| Eff_C | −0.14 [−0.16, −0.13] | −0.13 [−0.15, −0.12] | −0.13 [−0.15, −0.11] | −0.14 [−0.15, −0.12] | −0.14 [−0.16, −0.12] | −0.15 [−0.16, −0.12] | −0.14 [−0.17, −0.13] | −0.15 [−0.17, −0.13] | −0.14 [−0.16, −0.12] | −0.14 [−0.17, −0.13] |
| Ecc_C | −0.18 [−0.21, −0.17] | −0.18 [−0.20, −0.16] | −0.17 [−0.19, −0.15] | −0.17 [−0.18, −0.14] | −0.16 [−0.17, −0.14] | −0.15 [−0.16, −0.12] | −0.14 [−0.16, −0.12] | −0.14 [−0.15, −0.11] | −0.13 [−0.15, −0.10] | −0.16 [−0.17, −0.13] |
| p-value | <0.001 (**) | <0.001 (**) | <0.001 (**) | <0.001 (**) | <0.001 (**) | 0.797 | 0.019 (*) | 0.004 (**) | 0.010 (*) | <0.001 (**) |
| SRff_C (s−1) | −0.69 [−0.88, −0.57] | −0.67 [−0.88, −0.56] | −0.72 [−0.87, −0.55] | −0.72 [−0.89, −0.58] | −0.75 [−0.84, −0.65] | −0.76 [−0.83, −0.58] | −0.73 [−0.80, −0.64] | −0.71 [−0.79, −0.65] | −0.64 [−0.81, −0.60] | −0.70 [−0.81, −0.62] |
| SRcc_C (s−1) | −0.95 [−1.08, −0.83] | −0.89 [−1.03, −0.83] | −0.89 [−1.00, −0.76] | −0.83 [−0.97, −0.72] | −0.81 [−0.89, −0.68] | −0.77 [−0.86, −0.61] | −0.70 [−0.78, −0.55] | −0.68 [−0.76, −0.52] | −0.60 [−0.67, −0.47] | −0.80 [−0.87, −0.63] |
| p-value | <0.001 (**) | <0.001 (**) | <0.001 (**) | <0.001 (**) | 0.01 (*) | 0.171 | 0.530 | 0.185 | 0.185 | 0.012 (*) |
Strains are reported at peak systole, and strain rate at mid systole. p-values are reported after pair-wise Wilcoxon sign rank test.
shows statistical differences for p<0.05 and
for p<0.01.
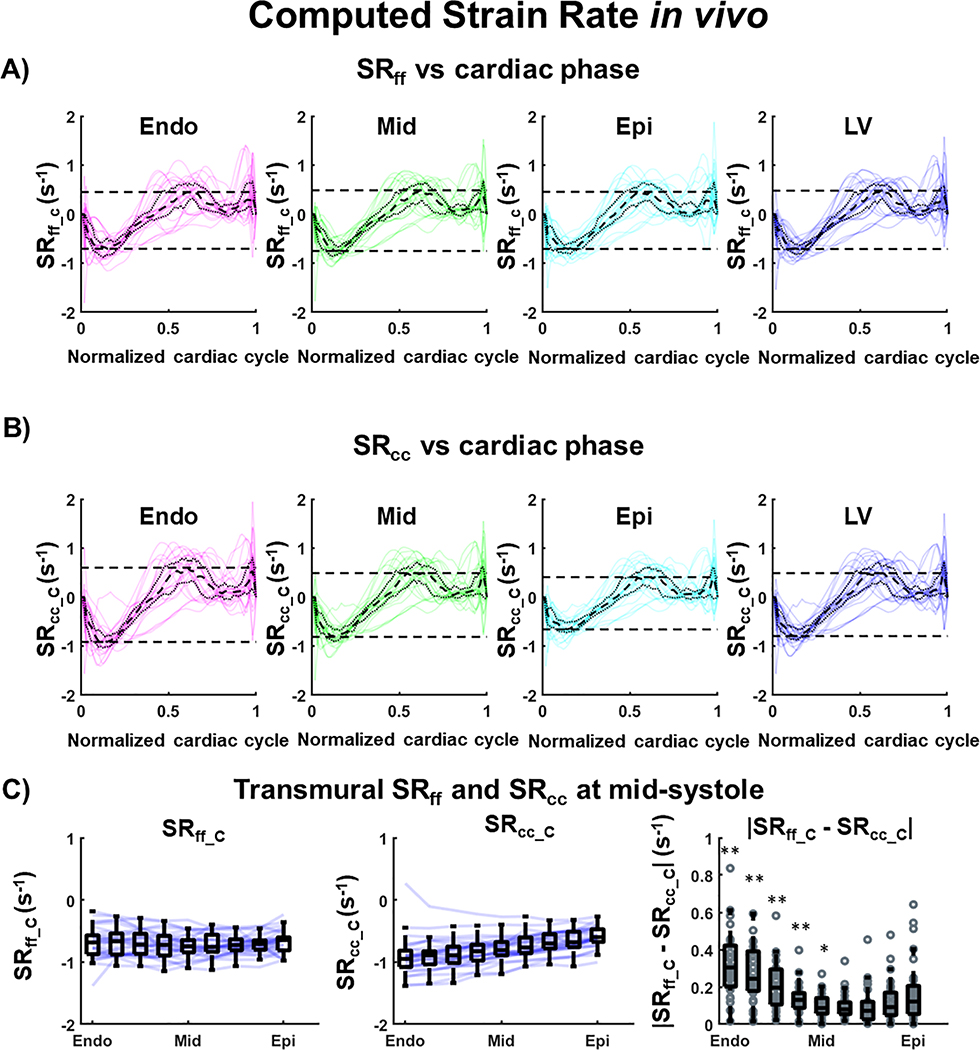
Strain rate comparisons between SRff_C and SRcc_C during the cardiac cycle are shown in Figure 9A and 9B. Across volunteers at mid-systole, SRff_C was lower in magnitude than SRcc_C in the endo layer (SRff_C= −0.68 [−0.88, −0.56] s−1 vs SRcc_C= −0.90 [−1.04, −0.80] s−1, p<0.001) and in the mid layer (SRff_C= −0.75 [−0.84, −0.63] s−1 vs SRcc_C= −0.80 [−0.90, −0.67], p=0.008) and larger in the epi layer (SRff_C= −0.70 [−0.79, −0.64] s−1 vs SRcc_C= −0.64 [−0.72, −0.52], p=0.116). In mid-systole across the whole SA LV, statistical differences were observed between SRff_C and SRcc_C (SRff_C= −0.70 [−0.81, −0.62] s−1 vs SRcc_C= −0.80 [−0.87, −0.63], p=0.012). As shown in Figure 9C, SRff_C was transmurally uniform compared to SRcc_C, which exhibited a transmural gradient. Transmural distribution and statistical differences between SRff_C and SRcc_C are reported in Table 1.
Figure 9:
Cardiac strain values across volunteers (N=30) calculated after combining cDTI and DENSE. (A) Computed Myofiber strain rate (SRff_C) and (B) circumferential strain rate (SRcc_C) at endo, mid, epi layers, and across the left ventricular (LV) wall. The black dashed and dotted lines represent the median and the first and third quartile across volunteers; the red dashed line provides a −0.15 strain reference; the colored solid lines are individual medians per volunteers. (C) Transmural distribution of SRff_C, SRcc_C, and of the absolute difference of SRff_C and SRcc_C at mid-systole. Blue solid lines represent the transmural median per volunteer and the box plots represent the median and IQR across volunteers. Statistical differences between SRff_C and SRcc_C have been assessed using a pair-wise Wilcoxon sign rank test: p-values<0.01 are represented by (**) and p-values<0.05 by (*).
Discussion
This work builds on previous efforts to combine cDTI and DENSE data and represents one of the first approaches to measure Eff in vivo in humans. In this study, a new, efficient pipeline was proposed to combine microstructural and functional data in order to estimate myofiber strains. The result is the largest report (N=30) to date on in vivo Eff in humans. At peak systole in the LV, Eff was found to be transmurally uniform, while Ecc displayed a transmural gradient. The precision of the proposed post-processing pipeline was also assessed using a computational phantom, which demonstrated that the uncertainty in myofiber strain may be impacted by an uncertainty in myofiber orientation.
DENSE acquisition
Cardiac strains measured using only DENSE imaging were comparable to those in previous reports. In our work, the magnitude of measured circumferential strain was shown to increase from the base through the mid LV and to the apex (Ecc_M= −0.15 [−0.17, −0.12], −0.18 [−0.19, −0.16], −0.19 [−0.21, −0.18] respectively), which was also observed by Del-Cantos et al. (23) using MR Tagging (Ecc= −17.2±3.1, −20.1±3.1, −20.3±3.0 in base, mid, and apex, respectively). The magnitude of longitudinal strain across slices (Ell_M= −0.15 [−0.16, −0.12], −0.15 [−0.18, −0.13], −0.14 [−0.16, −0.12]) corresponded to previous report of Ell using DENSE (4) (Ell= −0.16±0.02), but were slightly lower than global longitudinal strain (GLS) measured with feature tracking (24) (GLS= −0.20, CI: −0.21, −0.19) or Ultrasound imaging (GLS=−0.17±0.05/−0.16±0.05 in women/men) (25). Radial strain, Err (Err_M = 0.11 [0.02, 0.24], 0.14 [0.08, 0.22], 0.10 [0.01, 0.19] in apex, mid, and base, respectively) was found significantly lower than other reports using DENSE (Err=0.42±0.11) in Moore et al. (26) and (Err=0.33±0.10) in Zhong et al. (4). Cao et al. (27) showed a poor agreement in Err between DENSE (Err=0.4 ± 0.28), tagging (Err=0.32 ± 0.24) and three feature tracking methods (Err=0.47 0.26, Err=0.64 ± 0.33, Err=0.23 ± 0.9). The DENSE processing pipeline has been shown in previous in silico work (15,28) to induce significant error in computing Err, which may account for the poor reproducibility of Err (29–32), and the wide range of radial strain values reported in the literature.
cDTI acquisition
The cDTI acquisitions were acquired with a high-resolution protocol (1.6×1.6×8mm) compared to previous studies. Tensor invariants, such as MD and FA, and myofiber HA were reported. Across the thirty volunteers MD (1.59 [1.47, 1.67] x 10−3 mm2/s) and FA (0.38 [0.34 0.41] A.U.) were both in the range of previous reports using a similar motion compensated spin-echo approach (7,8,21,33,34).
The HA transmural distribution measured here (HA= 40.4 [21.7, 49.8] °, 2.0 [−0.1, 5.8] ° and −26.5 [−33.3, 4.6] ° at endo, mid, and epi, respectively) was also comparable to ex vivo (5) (HA= 66 ± 15, - 41 ± 26 ° at endo and epi, respectively) and in vivo reports (35) (HA= 37.2 [IQR: 3.9] −47.5 [IQR: 15.1] ° at endo, and epi, respectively).
Combination of cDTI and DENSE acquisitions
The proposed post-processing approach to combine cDTI and DENSE is in continuity of the study done by Verzhbinsky et al. (15). In this previous study, two SA slices of 3D displacement encoding DENSE were acquired immediately above and below one SA cDTI to estimate Eff. In this work instead, a stack of 2D SA and LA DENSE slices were combined with the cDTI data. Unlike previous work, this pipeline employs a DENSE protocol that can be acquired in a single breath-hold per slice, which results in shorter total acquisition times. Additionally, because this approach relies on 2D displacements in the SA and LA planes, it is compatible with other 2D displacement encoding methods, such as tagging and feature tracking. However, an additional post-processing step is required to transform the 2D displacement fields into a registered 3D displacement field. For each of the post-processing steps described in Figure 2, noise may be introduced in the measures and may affect the accuracy of the proposed pipeline to estimate Eff.
Uncertainty in cDTI
Measuring microstructural organization in the beating heart remains a major challenge. Both respiratory and cardiac bulk motion are sources of error that can lead to uncertainty in the cDTI metrics, which may be reflected in the computed Eff. By adding variance to the myofiber orientation field, myofibers no longer lie exactly in the circumferential-longitudinal plane. As a result, strains along the perturbed myofibers include a component of radial thickening, leading to a positive shift in the Eff estimation. This can be seen in the in silico experiment where the overall IQR of an idealistic transmural HA distribution increases (Figure 5). In vivo, the uncertainty in myofiber orientation cannot be directly measured through the HA IQR and instead has to be estimated using a bootstrapping approach. Here, MD and FA uncertainty (dMD= 0.31 [0.21, 0.41] x 10−3 mm2/s, dFA= 0.23 [0.20, 0.26] A.U.) were found comparable to the work of Aliotta et al. (21) (dMD= 0.38 ± 0.02 × 10−3mm2/s, dFA=0.20 ± 0.01 A.U.). However, Aliotta et al. also reported a lower uncertainty of the first eigenvector (dE1= 15.5 ± 1.2 °) compared to the one measured on HA here (dHA= 34.5 [29.7, 48.3] °). This difference may be due to the fact that only 6 diffusion encoding directions were acquired in this work, which has been shown to increase uncertainty in myofiber orientation (36). According to our in silico experiment, this uncertainty in the myofiber orientation may result in a more positive shift of Eff and thus the Eff magnitude computed in this work may be slightly underestimated.
Myofiber Strain in vivo
Using the proposed pipeline to combine cDTI and DENSE, myofiber strain Eff was computed in thirty volunteers in vivo. At peak systole and across volunteers Eff was estimated to Eff_C= −0.14 [−0.16, −0.13]. Early work by Tseng et al. (12) found a slightly lower magnitude of myofiber strain (Eff= −0.12 ± 0.01) using cDTI and velocity encoding. McGowan et al. (37) found a similar strain (Eff= −0.14±0.01) while Wang et al. (13) found a higher magnitude (Eff= −0.18 ± 0.03), using both MRI tagging imaging and ex vivo myofiber orientations. Very similar results were found in our previous work (15) in healthy swine (Eff= −0.14 [−0.16, −0,14]) using in vivo cDTI and 3D displacement encoded DENSE. Slightly lower Eff magnitudes were obtained in healthy human in vivo (Eff= −0.13 ± 0.02) using a single DENSE slice with an imposed incompressibility constraint (14).
Previous computational modeling work (38) has used an “adaptation hypothesis” to demonstrate that the observed transmural myofiber orientation gradient is optimal for minimizing the spatial variance of myofiber shortening. This work confirms that myofiber shortening has, in fact, a low spatial variance while in contrast, circumferential strain displays a transmural gradient. This work has also demonstrated that the circumferential strain can only be assumed equivalent to the myofiber strain in a fine layer between the mid-wall and epicardium, where the HA is approximately 0° and the myofiber and circumferential directions are aligned.
Limitations
One limitation of this study was the limited heart coverage of the cDTI acquisition, which only permitted analysis of myofiber strains at a single mid-ventricular LV position. Thus, it was not possible to estimate the myofiber strain distribution in the full LV from base to apex. However, several approaches have been proposed to increase the spatial coverage of cDTI (20,39,40) and should be considered in future work to develop Eff maps that span the entire LV.
Another limitation is that myofiber strains were estimated through a post-processing pipeline that combines information acquired with two different sequences. These two datasets were measured with different readouts (Spiral vs EPI), spatial resolutions (2.5×2.5×8mm vs 1.6×1.6×8mm), and contrasts (TE/TR=1.08/15ms vs TE/TR=55/4000ms). Even though care was taken to co-register the two datasets retrospectively, the acquisitions may present some geometrical and spatial differences and may not overlap perfectly. This problem would be substantially alleviated by sequences that encode inherently registered displacement and diffusion information into the same sequence (12).
In this work, we use the commonly referenced term “myofiber”, which may incorrectly imply continuous fiber bundles that run throughout the heart. The heart is composed of a continuously branching syncytium of cardiomyocytes. The cDTI method doesn’t measure directly individual cardiomyocyte cell orientations, but rather the orientation of an aggregate of cardiomyocytes within a voxel. In general, in vivo and ex vivo cDTI studies have reported lower HA range compared to histology reports. Streeter et al. (2) showed that transmural HA distributions are mostly linear in most of the mid-wall and become steeper at the extreme endocardium and epicardium regions. Even though high-resolution cDTI was acquired here, HA distributions obtained were mostly linear. Additional work is needed to better understand the differences between some previous histologic reports and the data from in vivo cDTI.
Conclusion
We proposed an efficient acquisition and analysis method to combine microstructural cDTI and functional cine DENSE to estimate in vivo myofiber strain (Eff). We demonstrated that it is possible to measure in vivo myofiber strain in a healthy human population in 10 minutes per subject. Myofiber strain is a mechanistically relevant metric that is spatially uniform in healthy populations, making it an intriguing candidate for the evaluation of local cardiomyocyte contractility in assessing cardiovascular dysfunction.
Supplementary Material
Supporting Information Video S1: Post-processing steps used to combine DENSE and cDTI data and calculate myofiber strain Eff. After cDTI reconstruction (A1), myofiber orientations were represented at nodes using one node per voxel. Cardiac displacement fields were also represented using nodes after the DENSE reconstruction (A2 & A3). (B) 2D Displacement fields from long axis (LA) and short axis (SA) are combined to obtain a SA 3D displacement field. (C) The SA 3D displacement field is interpolated and differentiated to generate a deformed cine of myofiber orientations during the cardiac cycle. (D) Finally, cardiac strains are calculated from the SA 3D displacement field and the myofiber orientations and using the beginning of systole as the reference configuration.
Acknowledgments
This work was supported by NIH/NHLBI R01-HL131975, R01-HL131823, K25-HL135408 grants and AHA post-doctoral fellowship AHA-20POST35210644. This work was supported by the HCL Actions Incitatives (69HCL15_744), and performed within the framework of the RHU MARVELOUS (ANR-16-RHUS-0009) of l’Université Claude Bernard Lyon 1 (UCBL), within the program “Investissements d’Avenir“ operated by the French National Research Agency (ANR).
The authors thank SIEMENS healthineers for their technical support and Dr. Epstein (University of Virginia) for providing the DENSE sequence used in this study.
References
- 1.Axel L, Wedeen VJ, Ennis DB. Probing dynamic myocardial microstructure with cardiac magnetic resonance diffusion tensor imaging. J. Cardiovasc. Magn. Reson 2014;16 doi: 10.1186/s12968-014-0089-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Streeter DD, Spotnitz HM, Patel DP, Ross J, Sonnenblick EH. Fiber Orientation in the Canine Left Ventricle during Diastole and Systole. Circ. Res 1969;24:339–347 doi: 10.1161/01.RES.24.3.339. [DOI] [PubMed] [Google Scholar]
- 3.Aletras AH, Ding S, Balaban RS, Wen H. DENSE: Displacement Encoding with Stimulated Echoes in Cardiac Functional MRI. J. Magn. Reson 1999;137:247–252 doi: 10.1006/jmre.1998.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhong X, Spottiswoode BS, Meyer CH, Kramer CM, Epstein FH. Imaging three-dimensional myocardial mechanics using navigator-gated volumetric spiral cine DENSE MRI. Magn. Reson. Med 2010;64:1089–1097 doi: 10.1002/mrm.22503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lombaert H, Peyrat J, Croisille P, et al. Human Atlas of the Cardiac Fiber Architecture: Study on a Healthy Population. IEEE Trans. Med. Imaging 2012;31:1436–1447 doi: 10.1109/TMI.2012.2192743. [DOI] [PubMed] [Google Scholar]
- 6.Welsh CL, DiBella EVR, Hsu EW. Higher-Order Motion-Compensation for In Vivo Cardiac Diffusion Tensor Imaging in Rats. IEEE Trans. Med. Imaging 2015;34:1843–1853 doi: 10.1109/TMI.2015.2411571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stoeck CT. Second order motion compensated spin-echo diffusion tensor imaging of the human heart. 2015:3. [DOI] [PubMed]
- 8.Aliotta E, Wu HH, Ennis DB. Convex optimized diffusion encoding (CODE) gradient waveforms for minimum echo time and bulk motion–compensated diffusion-weighted MRI. Magn. Reson. Med 2017;77:717–729 doi: 10.1002/mrm.26166. [DOI] [PubMed] [Google Scholar]
- 9.Nielles-Vallespin S, Khalique Z, Ferreira PF, et al. Assessment of Myocardial Microstructural Dynamics by In Vivo Diffusion Tensor Cardiac Magnetic Resonance. J. Am. Coll. Cardiol 2017;69:661–676 doi: 10.1016/j.jacc.2016.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferreira PF, Kilner PJ, McGill L-A, et al. In vivo cardiovascular magnetic resonance diffusion tensor imaging shows evidence of abnormal myocardial laminar orientations and mobility in hypertrophic cardiomyopathy. J. Cardiovasc. Magn. Reson 2014;16 doi: 10.1186/s12968-014-0087-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gotschy A, von Deuster C, van Gorkum RJH, et al. Characterizing cardiac involvement in amyloidosis using cardiovascular magnetic resonance diffusion tensor imaging. J. Cardiovasc. Magn. Reson 2019;21:56 doi: 10.1186/s12968-019-0563-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tseng W-YI, Reese TG, Weisskoff RM, Brady TJ, Wedeen VJ. Myocardial Fiber Shortening in Humans: Initial Results of MR Imaging1. Radiology 2000. doi: 10.1148/radiology.216.1.r00jn39128. [DOI] [PubMed] [Google Scholar]
- 13.Wang VY, Casta C, Zhu Y, et al. Image-Based Investigation of Human in Vivo Myofibre Strain. IEEE Trans. Med. Imaging 2016;35:2486–2496 doi: 10.1109/TMI.2016.2580573. [DOI] [PubMed] [Google Scholar]
- 14.Perotti LE, Magrath P, Verzhbinsky IA, Aliotta E, Moulin K, Ennis DB. Microstructurally Anchored Cardiac Kinematics by Combining In Vivo DENSE MRI and cDTI. In: Pop M, Wright GA, editors. Functional Imaging and Modelling of the Heart. Springer International Publishing; 2017. pp. 381–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Verzhbinsky IA, Perroti LE, Moulin K, Cork TE, Loecher M, Ennis DB. Estimating Aggregate Cardiomyocyte Strain Using In Vivo Diffusion and Displacement Encoded MRI. IEEE Trans. Med. Imaging 2019. doi: In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spottiswoode BS, Zhong X, Hess AT, et al. Tracking Myocardial Motion From Cine DENSE Images Using Spatiotemporal Phase Unwrapping and Temporal Fitting. IEEE Trans. Med. Imaging 2007;26:15–30 doi: 10.1109/TMI.2006.884215. [DOI] [PubMed] [Google Scholar]
- 17.Amidror I Scattered data interpolation methods for electronic imaging systems: a survey. J. Electron. Imaging 2002;11:157 doi: 10.1117/1.1455013. [DOI] [Google Scholar]
- 18.Spottiswoode BS, Zhong X, Lorenz CH, Mayosi BM, Meintjes EM, Epstein FH. Motion-guided segmentation for cine DENSE MRI. Med. Image Anal 2009;13:105–115 doi: 10.1016/j.media.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kingsley PB. Introduction to diffusion tensor imaging mathematics: Part III. Tensor calculation, noise, simulations, and optimization. Concepts Magn. Reson. Part A 2006;28A:155–179 doi: 10.1002/cmr.a.20050. [DOI] [Google Scholar]
- 20.Stoeck CT, Kalinowska A, von Deuster C, et al. Dual-Phase Cardiac Diffusion Tensor Imaging with Strain Correction Leemans A, editor. PLoS ONE 2014;9:e107159 doi: 10.1371/journal.pone.0107159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aliotta E, Moulin K, Magrath P, Ennis DB. Quantifying precision in cardiac diffusion tensor imaging with second-order motion-compensated convex optimized diffusion encoding. Magn. Reson. Med 2018;80:1074–1087 doi: 10.1002/mrm.27107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ennis DB, Nguyen TC, Riboh JC, et al. Myofiber angle distributions in the ovine left ventricle do not conform to computationally optimized predictions. J. Biomech 2008;41:3219–3224 doi: 10.1016/j.jbiomech.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Del‐Canto I, López‐Lereu MP, Monmeneu JV, et al. Characterization of normal regional myocardial function by MRI cardiac tagging. J. Magn. Reson. Imaging 2015;41:83–92 doi: 10.1002/jmri.24539. [DOI] [PubMed] [Google Scholar]
- 24.Vo HQ, Marwick TH, Negishi K. MRI-Derived Myocardial Strain Measures in Normal Subjects. JACC Cardiovasc. Imaging 2018;11:196–205 doi: 10.1016/j.jcmg.2016.12.025. [DOI] [PubMed] [Google Scholar]
- 25.Dalen H, Thorstensen A, Aase SA, et al. Segmental and global longitudinal strain and strain rate based on echocardiography of 1266 healthy individuals: the HUNT study in Norway. Eur. Heart J. - Cardiovasc. Imaging 2010;11:176–183 doi: 10.1093/ejechocard/jep194. [DOI] [PubMed] [Google Scholar]
- 26.Moore CC, Lugo-Olivieri CH, McVeigh ER, Zerhouni EA. Three-dimensional Systolic Strain Patterns in the Normal Human Left Ventricle: Characterization with Tagged MR Imaging. Radiology 2000;214:453–466 doi: 10.1148/radiology.214.2.r00fe17453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao JJ, Ngai N, Duncanson L, Cheng J, Gliganic K, Chen Q. A comparison of both DENSE and feature tracking techniques with tagging for the cardiovascular magnetic resonance assessment of myocardial strain. J. Cardiovasc. Magn. Reson 2018;20:26 doi: 10.1186/s12968-018-0448-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wehner GJ, Suever JD, Fielden SW, et al. Typical readout durations in spiral cine DENSE yield blurred images and underestimate cardiac strains at both 3.0 T and 1.5 T. Magn. Reson. Imaging 2018;54:90–100 doi: 10.1016/j.mri.2018.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haggerty CM, Kramer SP, Binkley CM, et al. Reproducibility of cine displacement encoding with stimulated echoes (DENSE) cardiovascular magnetic resonance for measuring left ventricular strains, torsion, and synchrony in mice. J. Cardiovasc. Magn. Reson 2013;15:71 doi: 10.1186/1532-429X-15-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin K, Meng L, Collins JD, Chowdhary V, Markl M, Carr JC. Reproducibility of cine displacement encoding with stimulated echoes (DENSE) in human subjects. Magn. Reson. Imaging 2017;35:148–153 doi: 10.1016/j.mri.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suever JD, Wehner GJ, Jing L, et al. Right Ventricular Strain, Torsion, and Dyssynchrony in Healthy Subjects Using 3D Spiral Cine DENSE Magnetic Resonance Imaging. IEEE Trans. Med. Imaging 2017;36:1076–1085 doi: 10.1109/TMI.2016.2646321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suever JD, Wehner GJ, Haggerty CM, et al. Simplified post processing of cine DENSE cardiovascular magnetic resonance for quantification of cardiac mechanics. J. Cardiovasc. Magn. Reson 2014;16:94 doi: 10.1186/s12968-014-0094-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scott AD, Nielles-Vallespin S, Ferreira PF, et al. An in-vivo comparison of stimulated-echo and motion compensated spin-echo sequences for 3 T diffusion tensor cardiovascular magnetic resonance at multiple cardiac phases. J. Cardiovasc. Magn. Reson 2018;20:1 doi: 10.1186/s12968-017-0425-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.von Deuster C, Stoeck CT, Genet M, Atkinson D, Kozerke S. Spin echo versus stimulated echo diffusion tensor imaging of the in vivo human heart. Magn. Reson. Med 2016;76:862–872 doi: 10.1002/mrm.25998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ferreira PF, Nielles‐Vallespin S, Scott AD, et al. Evaluation of the impact of strain correction on the orientation of cardiac diffusion tensors with in vivo and ex vivo porcine hearts. Magn. Reson. Med 2018;79:2205–2215 doi: 10.1002/mrm.26850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones DK. The effect of gradient sampling schemes on measures derived from diffusion tensor MRI: A Monte Carlo study†. Magn. Reson. Med 2004;51:807–815 doi: 10.1002/mrm.20033. [DOI] [PubMed] [Google Scholar]
- 37.MacGowan Guy A, Shapiro Edward P, Azhari Haim, et al. Noninvasive Measurement of Shortening in the Fiber and Cross-Fiber Directions in the Normal Human Left Ventricle and in Idiopathic Dilated Cardiomyopathy. Circulation 1997;96:535–541 doi: 10.1161/01.CIR.96.2.535. [DOI] [PubMed] [Google Scholar]
- 38.Rijcken J, Bovendeerd PHM, Schoofs AJG, van Campen DH, Arts T. Optimization of Cardiac Fiber Orientation for Homogeneous Fiber Strain During Ejection. Ann. Biomed. Eng 1999;27:289–297 doi: 10.1114/1.147. [DOI] [PubMed] [Google Scholar]
- 39.Moulin K, Croisille P, Feiweier T, et al. In vivo free-breathing DTI and IVIM of the whole human heart using a real-time slice-followed SE-EPI navigator-based sequence: A reproducibility study in healthy volunteers. Magn. Reson. Med 2016;76:70–82 doi: 10.1002/mrm.25852. [DOI] [PubMed] [Google Scholar]
- 40.Mekkaoui C, Reese TG, Jackowski MP, et al. Diffusion Tractography of the Entire Left Ventricle by Using Free-breathing Accelerated Simultaneous Multisection Imaging. Radiology 2017;282:850–856 doi: 10.1148/radiol.2016152613. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Video S1: Post-processing steps used to combine DENSE and cDTI data and calculate myofiber strain Eff. After cDTI reconstruction (A1), myofiber orientations were represented at nodes using one node per voxel. Cardiac displacement fields were also represented using nodes after the DENSE reconstruction (A2 & A3). (B) 2D Displacement fields from long axis (LA) and short axis (SA) are combined to obtain a SA 3D displacement field. (C) The SA 3D displacement field is interpolated and differentiated to generate a deformed cine of myofiber orientations during the cardiac cycle. (D) Finally, cardiac strains are calculated from the SA 3D displacement field and the myofiber orientations and using the beginning of systole as the reference configuration.