Abstract
Over the past four decades, research on the natural history of HIV infection has described how HIV wreaks havoc on human immunity and causes AIDS. HIV host genomic research, which aims to understand how human genetic variation affects our response to HIV infection, has progressed from early candidate gene studies to recent multi-omic efforts, benefiting from spectacular advances in sequencing technology and data science. In addition to invading cells and co-opting the host machinery for replication, HIV also stably integrates into our own genome. The study of the complex interactions between the human and retroviral genomes has improved our understanding of pathogenic mechanisms and suggested novel preventive and therapeutic approaches against HIV infection.
Subject terms: Infectious diseases, Genomics, Genome-wide association studies, Genetic variation
McLaren and Fellay review our current understanding of the effects of human genetic variation on HIV infection and disease progression and how this knowledge is contributing to preventative and therapeutic approaches.
Introduction
HIV-1 is the human retrovirus responsible for the HIV/AIDS pandemic, which has claimed more than 30 million lives over the past four decades. HIV infection continues to be a major global public health issue, with currently around 40 million people living with HIV (PLWH). Lifelong antiretroviral therapy (ART) has transformed the disease into a manageable chronic health condition. When available, ART enables PLWH to lead long and healthy lives but there is still no effective vaccine and no cure.
Early in the pandemic, it became clear that the risk of HIV acquisition is highly variable across humans. Socioeconomic and behavioural factors played a central role in this variability with some risk groups, such as intravenous drug users and men who have sex with men (MSM), being disproportionately affected1. Still, even among the most highly exposed individuals, a fraction remained HIV negative2,3. Similarly, important differences in the natural course of HIV infection (such as time from infection to AIDS diagnosis and the occurrence of opportunistic infections or malignancies) were only partially explained by known variables such as age and comorbidities4. Taken together, these clinical and epidemiological observations suggested a role for additional factors in the modulation of the individual response to HIV, including inherited variation in the genes and pathways involved in the retroviral life cycle and in innate or adaptative immunity against the infection.
HIV enters its main target cell, the CD4+ T lymphocyte, by binding to its receptor CD4 and to the co-receptor CC-chemokine receptor 5 (CCR5)5. This binding event triggers the fusion of the viral and human cell membranes, initiating a complex intracellular life cycle that will lead to the production of new viruses (Fig. 1). The natural immune response against HIV infection relies mostly on CD8+ T cells, also called cytotoxic T lymphocytes (CTLs). Upon primary infection, intense HIV replication results in a very high plasma viral load, measured as copies of the HIV RNA genome per millilitre of plasma, which is then partly controlled by the specific CD8+ T cell response. The very diverse human leucocyte antigen (HLA) class I molecules play a central role in this immune response by presenting small viral fragments, called epitopes, at the surface of infected cells. The recognition of these epitopes by CTL leads to the elimination of HIV-infected cells. A more efficient immune response is linked to a lower viral load during the chronic phase of an untreated infection and to slower disease progression, though it is unable to eliminate the virus6.
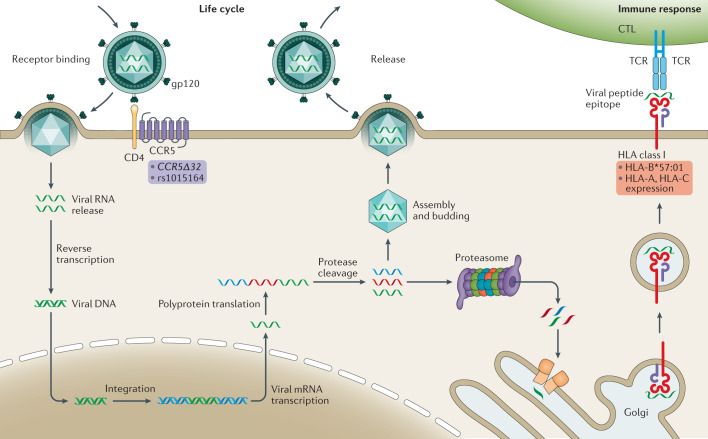
Fig. 1. Schematic representation of the HIV life cycle and the HLA-mediated host response.
The viral envelope glycoprotein gp120 binds CD4 and CC-chemokine receptor 5 (CCR5) on the surface of target cells triggering the fusion of the viral and host cell membranes; host genomic studies have implicated genetic variants in CCR5 (purple background) listed as modifiers of infectivity. Reverse transcription of the single-stranded RNA genome into double-stranded DNA (dsDNA) occurs using proteins carried by the infecting virion. Viral dsDNA is trafficked to the nucleus, where it is integrated into the human genome. The transcription of viral dsDNA results in viral gene expression and genome replication. Viral mRNA is translated into polyproteins, which are cleaved by the viral protease. Functional proteins assemble with copies of the viral genome at the cell membrane and mature virions bud from the surface. In parallel, as part of the immune response, viral proteins are digested by the host proteasome and processed through tapasin I and II (orange rectangles) into the Golgi, where the epitopes are loaded in the human leucocyte antigen (HLA) class I molecules. The peptide-loaded HLA protein is trafficked to the cell surface and presented to cytotoxic T lymphocytes (CTLs). Variability in epitope presentation by HLA-B alleles, such as the protective allele B*57:01, and in expression levels of HLA-C and HLA-A modify the response to infection and the set point viral load (red background). TCR, T cell receptor.
As a retrovirus, HIV can be described as a genomic pathogen. Indeed, it not only uses the molecular machinery of the infected cell for replication and dissemination but it also has the remarkable capacity to integrate a DNA copy of its RNA genome into a host cell chromosome. By becoming part of the human genome, HIV can persist in long-term cellular reservoirs for decades, making it extremely challenging to develop therapeutic strategies resulting in complete eradication7.
To better fight HIV infection, we must once again consider the old Delphic maxim: ‘know thyself’. Because HIV is an expert at hijacking human cells and immunity, we have no choice but to improve our understanding of our inner machinery, starting with the most fundamental layer of biological information — the human genome. The exploration of human diversity at the DNA level, long hampered by technological limitations, has been fuelled by the development of new and more powerful tools over the past decades8. Thanks to progress in our understanding of human genetic diversity, in genotyping and sequencing technology, as well as in bioinformatics and data science, it became possible to search for genetic factors that modulate the individual response to HIV, including the resistance and susceptibility to infection and the natural history of the disease in PLWH9.
In this Review, we first present an overview of the technological and conceptual developments that have fuelled HIV host genomic research. We then describe the major genetic factors modulating the natural history of HIV infection — in the HLA class I region and the CCR5 locus. Next, we highlight the recent convergence of human and HIV genomics, which allows longitudinal analyses of host–pathogen genetic interactions. Finally, we explain how genomic knowledge is poised to have a positive impact on PLWH, notably through pharmacogenomic interventions and stratification of care based on polygenic risk scores (PRS) before discussing the short-term and long-term perspectives for translational research and clinical applications of human genomics in the field of HIV.
Discovering host genetic variants
The search for human genetic differences that have an impact on HIV-related outcomes was first motivated by clinical observations, namely the striking variability in individual trajectories of patients in the absence of treatment. It was further propelled by a desire to uncover fundamental physiopathological mechanisms by the careful exploration of genomic variants and their impact on host and viral molecular processes.
Candidate gene studies
In the candidate gene approach, population-level associations are sought between HIV-related phenotypes and specific genetic variants in genes that have been selected based on previous biological knowledge or functional work. The selected variants are typically typed using targeted genotyping assays or Sanger sequencing of the region of interest. This framework was first applied to HIV host genetics in the early 1990s in analyses of allelic variants in genes known or suspected to play a role in HIV pathogenesis or in the antiretroviral immune response. Therefore, genetic associations were reported in two broad categories: genes coding for proteins involved in the HIV life cycle (such as PPIA10 and TSG101 (ref.11)) and immune-related genes encoding molecules implicated in innate and adaptive immune pathways (such as MBL2 (ref.12) and TLR9 (ref.13)) as well as in specific antiretroviral defence mechanisms (such as APOBEC3G14 and TRIM5 (ref.15)). Dozens of genes were tested in multiple cohorts. Unfortunately, as has been the case in the broader field of human genetics, most reported associations turned out to be false positives, notably owing to the small sizes of the studied cohorts, population stratification and the lack of correction for multiple testing. Replication attempts in larger cohorts, where these factors could be better controlled, showed no association for the vast majority of variants16–18. In fact, only two major discoveries remain from the candidate gene era: the protective effect of a homozygous 32 bp deletion in CCR5 (CCR5Δ32) against HIV acquisition19–21 and the modulating effect of HLA alleles on HIV progression for which early studies, largely in MSM of European ancestry in the United States, noted a strong impact of the HLA-B alleles B*57 and B*27 on delaying time to AIDS onset22.
Genome-wide association studies
Advances in genotyping and sequencing technologies progressively transformed human genetic analyses during the first decade of this century. In particular, the commercial availability of genome-wide genotyping arrays marked the beginning of the era of genome-wide association studies (GWAS). The principle of a GWAS is to simultaneously test very large numbers of genetic variants throughout the genome for potential associations with a phenotype of interest23. This truly agnostic approach finally allowed for a more comprehensive exploration of the human genome. To date, most GWAS have been based on the genotyping of single nucleotide polymorphisms (SNPs) followed by imputation, a process that leverages the linkage disequilibrium property of the human genome to statistically infer the genotypes that are not directly measured. This approach allows near-comprehensive testing of common variants (that is, variants with a minor allele frequency of >1%) in most human populations24.
The first GWAS of any infectious disease focused on the level of detectable viral genetic material in the blood of untreated, chronically infected individuals during the period of HIV latency25. This phenotype, known as set point viral load (spVL), was selected because of its relative ease of measurement and its known correlation to the rate of progression to AIDS26 and transmission potential27. The spectrum of interrogated variants was limited by early DNA genotyping arrays, yet genome-wide significant associations were identified in the HLA class I region, the most polymorphic locus in the human genome, known to have a crucial role in the modulation of T cell immunity (see ‘HLA variation in HIV control’, below). These findings were soon validated and expanded by other GWAS performed in independent cohorts, which demonstrated that the genetic architecture of HIV spVL is comparable between the general population of PLWH16,28–30 and a particular group of individuals able to maintain low viral loads for prolonged periods of time in the absence of ART, the so-called HIV controllers17,31. The absence of specific genetic factors explaining the HIV controller phenotype was a disappointment in terms of potential therapeutic development. However, it is consistent with what has been found for many complex human traits and diseases — that individuals at the extremes of the phenotypic distribution are more likely to carry multiple common variants with weak effects rather than rare, high-impact variants32. Beyond genotyping, a single exome sequencing study has been published so far in the HIV field33 also indicating that rare coding variants with large effect sizes are unlikely to make a major contribution to host control of HIV infection.
GWAS were less successful in the search for determinants of HIV resistance, with no definitive evidence found of human genetic polymorphisms conferring an altered susceptibility to HIV apart from CCR5 variation18,34. However, recent genome sequencing studies of extreme exposure phenotypes35 have shown promising associations in CD101, a gene encoding an immunoglobulin superfamily member implicated in regulatory T cell function36, and in UBE2V1, which encodes a ubiquitin-conjugating enzyme involved in pro-inflammatory cytokine expression37,38 that associates with the HIV restriction factor TRIM5α38. Although both CD101 and UBE2V1 are plausible candidates, further functional studies are required to validate their role in HIV susceptibility. Finally, analyses of GWAS data provide evidence for residual heritability owing to additive genetic effects beyond CCR5 (ref.18) and genetic overlap with behavioural and socioeconomic traits39. These results suggest that larger genomic studies of HIV acquisition may identify additional loci that impact susceptibility and warrant further investigation, potentially in large biobanks.
Several intrinsic limitations make it difficult to investigate the genetic mechanisms potentially involved in HIV resistance. For example, sample sizes are usually small (in the tens or hundreds) because studies need to be performed on highly exposed yet uninfected individuals such as patients with haemophilia exposed to HIV through contaminated blood products34, sex workers in hyper-endemic areas40 or serodiscordant couples (stable heterosexual couples where one partner has HIV infection and the other is seronegative for HIV at enrolment)29. Frailty (or survival) bias is a limitation in cross-sectional studies of HIV cohorts with long-term follow-up, as these cohorts are enriched for genetic factors protecting against HIV disease progression. Another limitation is misclassification bias in studies comparing the genomes of patients with HIV infection to unselected controls from the general population, in which most individuals are in fact susceptible to HIV infection18. The identification of additional genetic determinants of individual susceptibility to HIV infection will require increased sample sizes (ideally in the thousands) as well as the use of sequencing approaches to characterize the rare functional variants that are not interrogated in studies based on genotyping arrays.
HLA variation in HIV control
The HLA locus in infectious diseases
The human major histocompatibility complex (MHC) located on chromosome 6 is one of the most genetically diverse loci in the genome41. The extended MHC occupies ~7.6 Mb of the human genome42 and encodes more than 400 genes, many of which are key mediators of the innate and adaptive immune responses. Within this locus, alleles at the classical class I (HLA-A, HLA-B, HLA-C) and class II (HLA-DR, HLA-DQ, HLA-DP) genes have been associated with numerous autoimmune, inflammatory and infectious diseases (reviewed in refs43,44) with recent preprints demonstrating extensive disease associations in large biobanks from multiple populations45,46. In the context of infectious disease, class I HLA proteins present endogenous peptides on the surface of infected cells for recognition by CTLs, triggering the development of an adaptive response. As discussed below, the variability in epitope specificity of HLA proteins and expression levels of HLA class I alleles has a dramatic impact on the progression of HIV disease.
Effects of amino acid variability encoded by HLA alleles
An individual’s genotype at class I HLA genes has been consistently demonstrated to be the major host genetic determinant of HIV spVL and rate of disease progression across geographic contexts and ancestries17,22,47–50. This observation was put in the genome-wide context by the first GWAS of HIV spVL25 and HIV controllers17 that exclusively identified SNPs in strong linkage disequilibrium with classical HLA-B alleles. Although array-based techniques for the genotyping of DNA samples do not allow for the direct resolution of classical HLA alleles, computational methods leveraging linkage disequilibrium structure between SNPs and sequence-based HLA types in reference populations allow for accurate imputation of classical HLA types from GWAS data51. The application of this technique to a sample of >6,000 PLWH of European ancestry underscored the dramatic effect of HLA-B*57:01 on reducing viral load, which was, on average, ~0.8log10 RNA copies/ml lower in individuals carrying this allele52. This study also demonstrated strong associations at multiple other classical class I HLA alleles that had a range of effects, from decreasing spVL (B*57:01, B*27:05, B*13:02, B*14:02, C*06:02, C*08:02, C*12:02) to increasing spVL (B*07:02, B*08:01, C*07:01, C*07:02, C*04:01).
To better understand how functional variation in HLA class I proteins can impact HIV spVL, recent studies have tested variable amino acid positions within these proteins to fine-map the classical allele associations. In a GWAS performed by the International HIV Controllers study, this technique was applied to demonstrate that previously identified associations between HIV control and classical HLA alleles such as B*57:01 could be explained by variability across a small number of amino acid positions within the HLA-B protein17. The strongest effect was observed at position 97 of the protein, which accommodates six alternative amino acids, including valine, which is unique to B*57 haplotypes. A recent preprint describing the comprehensive analysis of the impact of HLA amino acid polymorphisms on spVL in a multi-ethnic sample of >12,000 PLWH identified three amino acid positions in HLA-B (positions 67, 97 and 156) and one in HLA-A (position 77) as independently associating with spVL53. The positions within HLA-B map to classical HLA alleles known to impact spVL, whereas the HLA-A position suggests that HLA-A functions independently of HLA-B. Interestingly, all four positions are located in the peptide-binding groove of the respective HLA protein, supporting the hypothesis that epitope presentation is key for the natural suppression of HIV replication. Furthermore, there was no substantial evidence that the effects of these polymorphic positions differed across ancestry groups, suggesting biological relevance across global contexts.
Several mechanisms of action have been proposed to explain why different alleles of the same HLA gene have differential effects on HIV progression. Studies of epitope specificity have shown that certain protective alleles, including B*57:01 (which uniquely carries valine at position 97) and B*27:05 (which carries the protective cysteine and asparagine residues at positions 67 and 97, respectively), drive compensatory mutations in the HIV genome leading to reduced viral fitness54–56. In addition to differential epitope specificity, the CTL effector function induced by epitope presentation has been implicated in HIV control, with CTLs in carriers of some protective HLA alleles exhibiting an enhanced proliferative capacity and more polyfunctional responses57–59.
Within-host diversity and epitope presentation
In addition to the impact of specific class I HLA alleles on HIV progression at the population level, the within-host diversity of HLA alleles may be important at the individual level. An early study looking at the impact of allele combinations revealed that maximum heterozygosity at HLA class I genes (that is, individuals carrying two different alleles at all three class I genes) was associated with a reduced time to AIDS47. This observation was supported by a GWAS that showed that individuals carrying different HLA alleles at each class I gene had a significantly lower viral load than homozygous individuals, even after accounting for the additive effect at each allele60. This heterozygote advantage likely comes from the ability to present multiple HIV epitopes, supporting the hypothesis that the breadth of presentation is beneficial in preventing HIV progression. To further test this hypothesis, a recent in silico study used novel algorithms to predict the binding affinity of all possible 9-mer peptides in the HIV proteome to HLA proteins encoded by the different class I alleles61. Coupling these predicted affinities to clinical and genetic data demonstrated that spVL was negatively correlated with the breadth of the peptide repertoire bound by an individual’s HLA protein isoforms. Moreover, HLA-B isoforms had the largest predicted breadth of epitope recognition and conferred the strongest reduction of viral load (Fig. 2a). However, the quantity of epitopes alone is unlikely to fully explain the protective capacity of an individual’s HLA alleles, as subsets of epitopes that are uniquely presented by protective HLA isoforms explained more of the observed variance in spVL than the entire predicted set. This observation is further supported by an in silico and functional study that demonstrated that HIV epitopes that encode structurally important residues are preferentially targeted by protective HLA isoforms and associate with elite control of replication62. Thus, the quantity and quality of HIV epitopes presented by combinations of HLA isoforms are the key drivers of spVL.
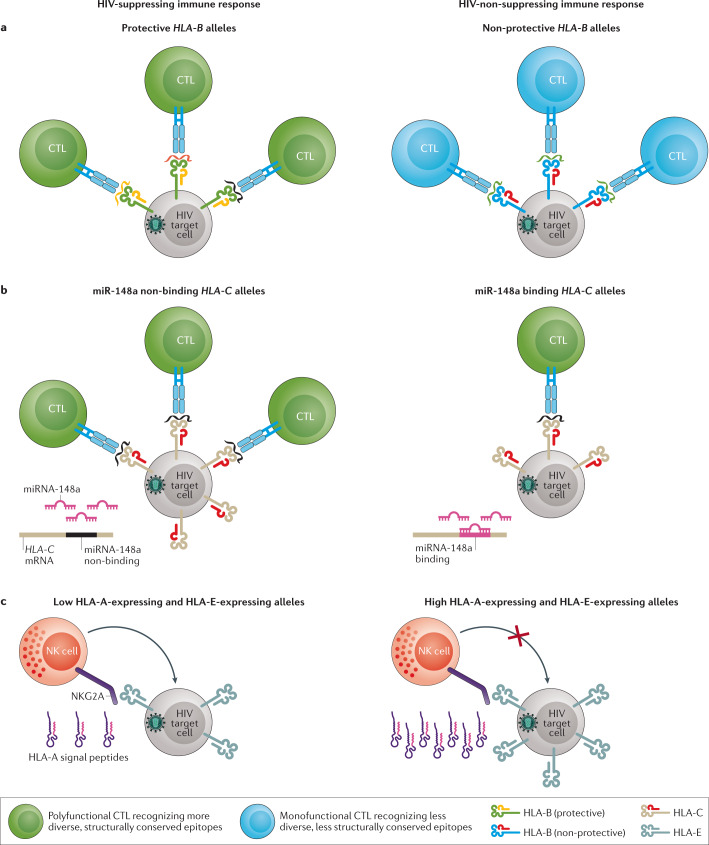
Fig. 2. Classical and non-classical effects of HLA class I on HIV suppression.
a | HIV-infected cells expressing protective HLA-B alleles tend to present a more diverse and more structurally conserved set of HIV epitopes compared to non-protective alleles. Interactions with protective alleles tend to produce a more polyfunctional cytotoxic T lymphocyte (CTL) response. b | HLA-C protein isoforms vary broadly in their level of expression on the surface of infected cells. HLA-C alleles that do not have a binding site for microRNA-148a (miRNA-148a) in the 3′ untranslated region of their mRNAs escape suppression and present more peptide on the cell surface than alleles with an miRNA-148a binding site, resulting in the initiation of stronger CTL responses. c | Different HLA-A alleles express different amounts of HLA-A signal peptide, which positively correlates with HLA-E peptide expression. HLA-E interacts with the NKG2A receptor on the surface of natural killer (NK) cells and, when highly expressed, inhibits the killing of infected cells.
Non-classical effects of HLA variation
In addition to the classical effects of HLA genes on peptide presentation, several studies have suggested that non-classical effects may play a part in limiting HIV replication in vivo. In particular, the variable expression levels of classical HLA-C alleles have been linked to HIV control, with those expressed at high levels conferring protection against disease progression63. This effect has been observed across ancestries and has been linked to the absence of a variable microRNA-148a (miR-148a) binding site in the 3′ untranslated region of HLA-C64. The proposed model suggests that mRNA from alleles lacking the miR-148a binding site escape suppression by miR-148a; as a consequence, proteins encoded by these alleles and loaded with HIV epitopes are expressed at higher levels on infected cells, allowing for greater rates of detection by CTLs64 (Fig. 2b). Similarly, proteins encoded by HLA-A alleles are also expressed at variable levels on the cell surface65. However, in contrast to HLA-C, HLA-A alleles expressed at high levels associate with poorer control of viral replication and with faster disease progression66. A combination of genetic and functional studies indicated that increased HLA-A expression levels correlated with higher viraemia in a combined cohort of more than 9,000 PLWH from sub-Saharan Africa and the United States. It was proposed that this effect may be the result of enhanced production of the HLA class I signal peptide that regulates HLA-E expression, a hypothesis that was supported by a correlation between HLA-A expression and HLA-E expression among 58 healthy donors tested66. HLA-E is a ligand for natural killer group protein 2A (NKG2A) and their interaction results in strong inhibition of natural killer (NK) cell degranulation (Fig. 2c). Thus, the enhanced production of the HLA class I signal peptide in individuals carrying highly expressing HLA-A alleles may lead to enhanced inhibition of immune responses in infected individuals, resulting in poorer clinical outcomes.
Finally, it has also been observed that the combination of HLA genotype and the expression of particular killer cell immunoglobulin-like receptor (KIR) proteins variably modulated HIV disease course67. The KIR proteins are a highly variable set of cell-surface receptors expressed on NK cells (and some T cells) that, when engaged by their cognate receptors, either activate or inhibit NK cell-mediated killing (recently reviewed in ref.68). In particular, the combination of the activating KIR3DS1 allele with a set of HLA-B alleles that carry isoleucine in the Bw4 epitope (Bw4-I80) is highly associated with HIV control69. Taken together, these results demonstrate the complex interplay between epitope presentation, HLA protein expression and NK inhibition.
CCR5 variation in HIV infection
CCR5Δ32 and resistance against HIV infection
Perhaps the most highly touted example of human genetic variability restricting infectious diseases is the observation that individuals carrying two copies of a loss-of-function variant in the gene encoding the cell receptor CCR5 are highly resistant to infection by HIV. CCR5 is a chemokine receptor expressed on the surface of multiple subsets of monocytes and lymphocytes, including CD4+ T cells, the major HIV target cells. At the earliest stages of infection, the HIV envelope protein gp120 binds CD4 and CCR5 on the cell surface, resulting in fusion of the viral and host cell membranes and in the release of the viral genome into the target cell. The discovery that individuals who carry homozygous loss-of-function alleles at CCR5 are resistant to infection was first made in a group of MSM that were multiply exposed to the virus but remained uninfected19. It was determined that these men all shared a 32-bp deletion in the CCR5 gene (the CCR5Δ32 allele) that leads to the production of a non-functional protein and the absence of functional CCR5 on the cell surface prevents HIV from entering target cells (Fig. 3a). The CCR5Δ32 allele is observed at ~10% frequency in individuals of European ancestry (homozygosity occurs at a frequency of 1%), at a reduced frequency in southern Europeans compared to those in the north70 and is not observed at an appreciable frequency in other continental populations. Compound heterozygotes (that is, individuals carrying one copy of CCR5Δ32 and a second loss-of-function CCR5 variant) are also resistant to infection, although these individuals are exceedingly rare71.
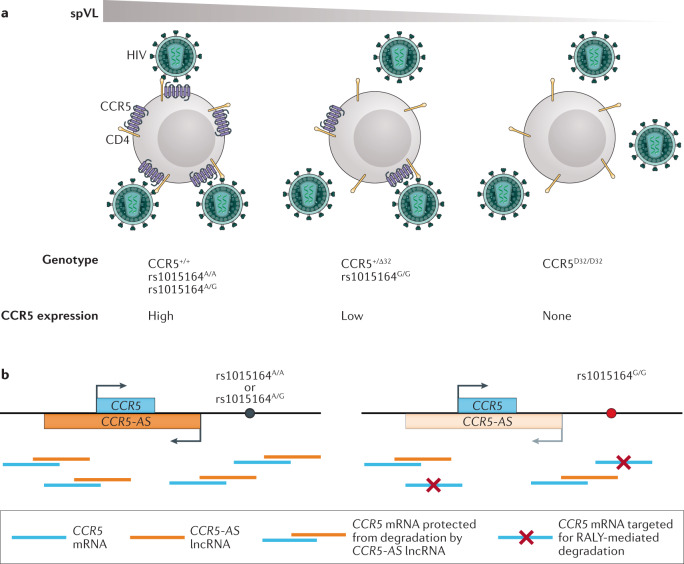
Fig. 3. CCR5 expression modifies HIV progression.
a | A 32-bp deletion in CCR5 (CCR5Δ32) results in the reduced expression of CC-chemokine receptor 5 (CCR5) on the surface of target cells. Heterozygous individuals exhibit reduced CCR5 expression, lower set point viral loads (spVLs) and slower disease progression. Individuals carrying two defective copies of the CCR5 gene show no surface expression and are highly resistant to HIV infection. Additionally, rs1015164, a single nucleotide polymorphism downstream of CCR5, affects cell surface levels of CCR5. In homozygous reference (A/A) and heterozygous (A/G) individuals, the surface expression of CCR5 is normal, whereas G/G homozygous individuals have a lower CCR5 surface expression and lower spVLs. b | rs1015164 regulates the expression of an antisense RNA termed CCR5-AS. When CCR5-AS (orange) is expressed at high or intermediate levels in homozygous reference (A/A) and heterozygous (A/G) individuals, respectively, CCR5 mRNA (blue) is protected from RALY-mediated degradation and results in normal levels of surface expression. In G/G homozygous individuals, CCR5-AS expression is diminished and CCR5 mRNA is degraded, resulting in lower surface expression at the cellular level and a lower spVL overall. lncRNA, long non-coding RNA.
The observation that individuals lacking CCR5 expression are resistant to HIV infection directly led to the development of the antiviral drug Maraviroc, a CCR5 antagonist72, as well as to the world’s first ethically fraught attempt at human embryo engineering73. Perhaps most interestingly, bone marrow transplants between CCR5Δ32 homozygous donors and recipients with HIV infection have resulted in the only two confirmed cases of long-term HIV cure74,75. Although promising, this effect has been difficult to replicate in engineered autologous stem cell models76 and is unlikely to be scalable to the level necessary to stem the pandemic. Additionally, the protection is not absolute, as several confirmed cases of infection in CCR5Δ32 homozygotes have been reported (reviewed in ref.77), presumably by viruses that utilize the minor co-receptor CXCR4 or by dual-tropic viruses.
Associations between CCR5 variation and spontaneous HIV control
In addition to the impact of homozygosity on preventing infection, individuals with a single CCR5Δ32 copy exhibit lower spVL and delayed disease progression compared to those with two functional copies19,20,78, likely because the reduced levels of CCR5 protein on the cell surface lower the efficiency of HIV entry into target cells (Fig. 3a). The CCR5 locus was also identified in GWAS, first in a study of ~2,500 PLWH in Europe16 and then in an expanded set of 6,300 individuals from across the globe52. However, the CCR5Δ32 allele was not directly assayed on the genotyping platforms used in these studies, thus only proxy SNPs were identified. In a combined analysis of GWAS data and direct CCR5Δ32 genotyping, it was observed that the CCR5Δ32 allele was not the most strongly associated variant in the region, suggesting that multiple independent genetic effects occur at this locus. Conditional analysis accounting for the effect of CCR5Δ32 showed that an additional marker, rs1015164, was also strongly associated with spVL. Functional analysis of this variant showed that it regulates the expression of an antisense long non-coding RNA called CCR5-AS, which overlaps the CCR5 gene79. This study further showed that the increased expression of CCR5-AS resulted in increased CCR5 expression because CCR5-AS interfered with the RALY-mediated degradation of CCR5 mRNA. Moreover, the knockdown of CCR5-AS reduced the susceptibility of CD4+ T cells to HIV-1 infection ex vivo (Fig. 3b). These results demonstrate that the clinical course of untreated HIV infection is directly influenced by the innate level of CCR5 expression within the infected individual. Whether additional functional polymorphisms in CCR5 have similar effects remains an open question.
Host and pathogen genetic variation
Pathogen sequence variation as an indicator of host genetic pressure
Most studies performed so far in the field of host genetics focused on clinically defined outcomes such as the susceptibility to infection or disease progression. However, intermediate phenotypes have been shown to be very valuable in identifying subtle genetic association signals that are not always detectable using more complex clinical outcomes. A particularly promising intermediate phenotype, unique by its nature to infectious diseases, is variation in the pathogen genome (Fig. 4). HIV is a highly variable virus that establishes a lifelong infection. Therefore, it represents an ideal model to search for the potential effects of intra-host selective pressure on a human pathogen. While some of the variants observed in the HIV genomic sequence are present in the transmitted/founder virus, another fraction is acquired during the course of the disease resulting, at least partially, from the selective pressure exerted by the host response to infection. Signs of host-driven selection are clearly visible in the HIV genome. In particular, specific variants have been described in key viral epitopes presented by HLA class I molecules and targeted by CTL responses80. Mutations have also been reported in regions targeted by KIR, suggesting the escape from immune pressure by NK cells81,82. A non-negligible fraction of the HIV-1 genome (~12%) is under positive selection but only about half of the positively selected sites map to canonical CD8+ T cell epitopes83, indicating that additional host factors could be driving evolution in non-epitope sites.
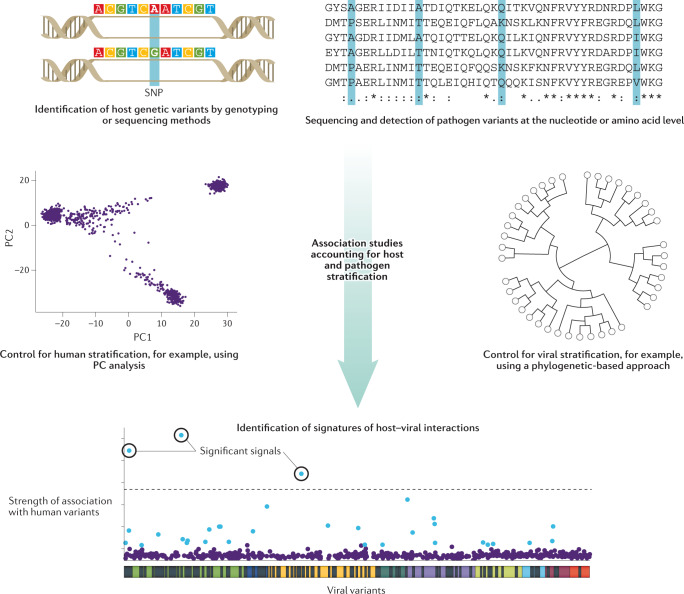
Fig. 4. Detecting genomic signatures of host–pathogen interactions in matched host and virus samples.
First, genetic variants in the host (human) and pathogen (viral) genomes are identified from genome-wide genotyping or sequencing data and catalogued. A genome-wide search for associations between human polymorphisms and viral variants is then performed, which needs to consider the risk of systematic signal inflation owing to population stratification. On the human side, this can be addressed by methods that infer genetic ancestry, such as principal component (PC) analysis followed by the inclusion of the top PCs as covariates, or by the use of mixed models that incorporate the full covariance structure of the study population130. On the pathogen side, various phylogenetic-based and model-based approaches have been proposed85. Significant associations, after correction for multiple testing, reveal the loci involved in host–pathogen genomic conflicts. The plot showing the signatures of viral–host interactions is adapted from ref.84, CC BY 3.0 (https://creativecommons.org/licenses/by/3.0/).
Genome-to-genome studies
Computational approaches developed over the past decade have allowed more comprehensive analyses of the reciprocal genetic signals resulting from the host–pathogen interaction84,85. Joint analyses of human and HIV sequence variation start with the generation of large-scale genomic data from paired samples. The retroviral genome can be isolated and sequenced either as native RNA during replicative infection or as proviral DNA, integrated into the host genome, during latent infection. Human genomic information can be obtained using genotyping or sequencing technology. The principle of genome-to-genome (G2G) studies is then to perform a systematic search for associations between human genetic polymorphisms and viral sequence variants, at the nucleotide or amino acid levels. Because of the very large number of models run in parallel — one GWAS for each viral variant — this approach requires stringent correction for multiple testing. By mapping all interacting loci, G2G studies have the potential to uncover the most important genes and pathways involved in specific responses to infectious agents, thereby revealing novel diagnostic or therapeutic targets. In addition to identifying the sites of genetic interplay between virus and host, this study design makes it possible to estimate the biological consequences of such interactions and to estimate the relative impact of human and viral genetic variation on phenotypic outcomes by assessing associations between human-driven escape at viral sites and a quantitative clinical phenotype. In spite of these promises, it must be acknowledged that G2G studies have not led, as of today, to the identification of novel HIV restriction factors in the human genome86. Future studies will require larger sample sizes to increase power but also more diversity with a strong focus on the inclusion of PLWH of non-European ancestries.
Nevertheless, studies based on the combined analysis of host and pathogen genomic variation have already demonstrated their potential in other infections. In particular, the use of a similar study design in chronic hepatitis C virus infection highlighted the evolutionary pressure exerted by both innate (interferon-λ) and acquired (HLA class II) immune defence mechanisms87,88. The intra-host evolution of DNA viruses can also be investigated using a G2G approach, as shown in a recent study that revealed several associations between human and Epstein–Barr virus sequence variation in immunosuppressed PLWH89.
HIV precision medicine
Expanding antiretroviral therapy to eradicate HIV
With the now accepted knowledge that PLWH who do not have detectable plasma viral loads cannot transmit the virus to others90, the Joint United Nations Programme on HIV/AIDS (UNAIDS) set an ambitious 90–90–90 target91, where 90% of infected people know their status, 90% of those are on antiviral therapy and 90% of those are suppressing the virus below the level of detection. This aspirational treatment target would practically mean, given currently available technologies, that more than 34 million people would be on lifelong chemotherapy. Although this treatment as prevention approach would undoubtedly result in decreases in transmission and dramatic increases in life expectancy for the population with HIV infection, it also requires a deeper understanding of how human genetic variation relates to variability in drug toxicity and response to long-term therapy.
HIV pharmacogenetics
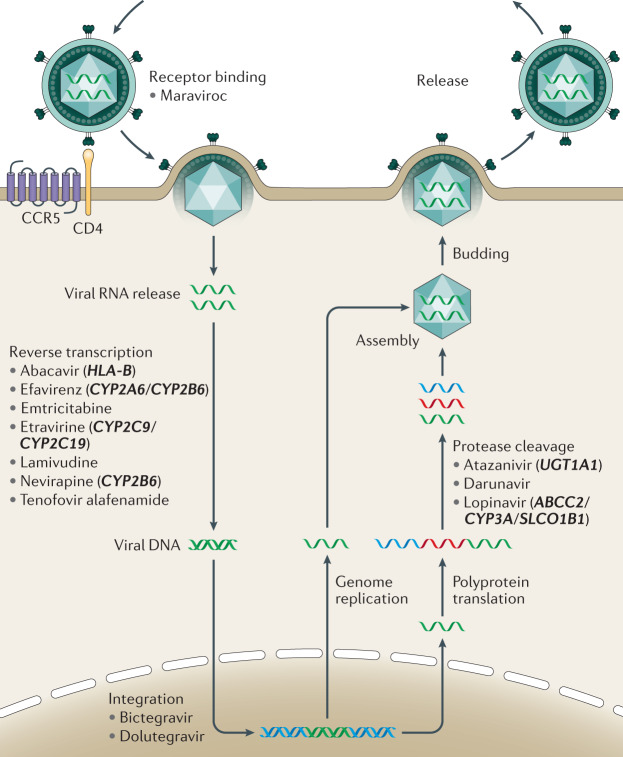
In addition to affecting HIV disease progression in untreated individuals, human genetic variability has also been implicated in modifying the response to treatment. A major achievement in the fight against HIV has been the development of multiple, effective therapeutics that target several stages of the viral life cycle. These include entry inhibitors, which prevent the binding of the viral spike protein gp120 to host cell receptors and fusion of the virus with host cell membranes; nucleoside and non-nucleoside reverse transcriptase inhibitors, which prevent the reverse transcription of the viral RNA genome into DNA; integrase inhibitors, which prevent the integration of the viral DNA product into the host genome; and protease inhibitors, which prevent the cleavage of viral polyproteins into their functional subunits (Fig. 5). For several classes of anti-HIV therapy, human genetic variability is known to influence response to the drug, which in some cases leads to severe adverse events and treatment discontinuation92. Paradoxically, the HLA-B allele B*57:01, most notably associated with the control of infection, also predisposes carriers to a severe hypersensitivity reaction to the nucleoside reverse transcriptase inhibitor abacavir93,94; the high specificity binding of abacavir alters the binding pocket of HLA-B*57:01 and triggers reactivity to self-peptides95. Similarly, variants in genes encoding the drug-metabolizing enzymes CYP2B6 (refs96–98), CYP2A6 (ref.99), CYP2C9 (ref.100), CYP2C19 (ref.100), CYP3A101 and ABCC2 (ref.101) have all been associated with slow metabolization kinetics of their cognate drugs (Table 1), in some cases leading to drug accumulation in the brain, psychiatric complications and treatment stoppage99. The frequency of many of these polymorphisms varies depending on ancestral background, leading to reduced drug tolerance and therefore reduced efficacy in some populations. For example, the allele CYP2B6*6 (rs3745274), which results in the slow metabolism of efavirenz and nevirapine, two non-nucleoside reverse transcriptase inhibitors recommended for first-line use by WHO until recently, has an approximately twofold higher frequency in some African populations compared to Europeans102. This increased frequency and the resulting adverse events led to thousands of cases of treatment discontinuation in Zimbabwe when the nation adopted a single-pill efavirenz-containing regimen103. This example highlights the need to not only tailor the therapy to the individual but, in some cases, to the population as well. Newer generations of HIV therapies, such as integrase inhibitors and advanced nucleoside reverse transcriptase inhibitors, have more favourable pharmacokinetic and safety profiles104. However, the effects of long-term treatment with these drugs and any potential interactions with human genetic variability remain to be understood.
Fig. 5. Antiretroviral drugs target multiple stages of the HIV life cycle.
Commonly used antiretroviral drugs target receptor binding, reverse transcription, integration and protease cleavage. Genetic variation in several human genes (in bold) have been shown to modify drug metabolism and contribute to adverse drug reactions (Table 1). CCR5, CC-chemokine receptor 5.
Table 1.
Genetic variants known to affect the pharmacokinetics of anti-HIV drugs
| Drugs | Class | Genes | Variants | Effect on patients |
|---|---|---|---|---|
| Abacavir | NRTI | HLA-B | rs2395029 (*57:01) | Hypersensitivity reaction |
| Atazanavir | PI | UGT1A1 | rs8175347 (*28,*37) | Predisposition to hyperbilirubinemia |
| Efavirenz | NNRTI | CYP2A6 | rs1801272 (*2), rs5031016 (*7,*10,*19), rs28399433 (*9) | Predisposition to high drug plasma levels and treatment discontinuation |
| Efavirenz and Nevirapine | NNRTI | CYP2B6 | rs3745274 (*6), rs12721655 (*8,*13), rs35303484 (*11), rs36060847 (*12), rs35773040 (*14), rs35979566 (*15), rs28399499 (*16,*18) | Predisposition to high drug plasma levels |
| Etravirine | NNRTI | CYP2C9, CYP2C19 | rs1057910 (*3), rs4424285 (*2) | Predisposition to high drug plasma levels |
| Lopinavir | PI | ABCC2, CYP3A | rs717620 (T), rs6945984 (C) | Predisposition to high drug plasma levels |
| Lopinavir | PI | SLCO1B1 | rs4149056 (*5), rs17329885 (*4) | Predisposition to low drug plasma levels |
NNRTI, non-nucleoside reverse transcriptase inhibitor; NRTI, nucleoside reverse transcriptase inhibitor; PI, protease inhibitor.
Complex trait genomics in HIV medicine
In addition to the direct interactions between host genotype and drug metabolism, patients on long-term HIV therapy also experience early onset of several chronic diseases, including cardiovascular disease105–107, metabolic syndrome108, kidney disease109,110 and liver fibrosis111. These conditions are all known to have high heritability in the HIV uninfected population and genetic risk factors for type 2 diabetes mellitus112 and cardiovascular disease113 have been shown to be enhanced in PLWH on therapy. Recently, there has been a push to develop PRS in the general population. These scores, built by summing the additive effects of dozens to thousands of genetic variants within an individual, have been shown to have a strong predictive ability for multiple metabolic, inflammatory, tumoural and cardiovascular conditions32. Investigations of PRS in the specific context of HIV infected individuals receiving long-term antiretroviral therapy have just begun, with the recent demonstrations that the prediction of chronic kidney disease can be improved through the addition of a PRS to the known clinical and pharmacological risk factors114,115 and that a PRS can be useful to stratify PLWH at a high risk of cardiometabolic diseases who may benefit from preventive therapies116. An important caveat is that PRS are not necessarily transferable across ancestral groups and, as in all areas of genomics, attention should be paid to enhancing diversity and ensuring equity in precision medicine approaches.
Conclusion and future perspectives
Host genomic studies have advanced our understanding of HIV biology in several important ways. Firstly, the demonstration of the dominant impact of HLA variation on HIV progression in the context of the whole genome reinforced the need to focus on T cell responses in vaccine design. Moreover, the ability to accurately infer HLA allele types and protein-level variability from genotyping array data, an approach first piloted in HIV genomic studies, has greatly increased our understanding of how amino acid variability in HLA molecules contributes to multiple medically important traits. Secondly, dense genotyping and large sample sizes enabled the discovery of multiple, independent signals in the CCR5 locus, which provided a deeper understanding of how the expression of CCR5 is regulated and how it modulates HIV infection beyond the known impact of the CCR5Δ32 allele. Finally, amassing genome-wide data for large cohorts of PLWH has enabled the validity of previous candidate gene associations to be assessed, providing a new standard for identifying novel loci of HIV restriction.
In recent years, there have been several barriers to further advancing our understanding of how host genomics affects HIV susceptibility and progression. Firstly, current studies have predominantly included individuals of European ancestry, mirroring the lack of diversity in genomics in general117, which is particularly problematic because the vast majority of PLWH are non-White. The example of the population-specific CCR5Δ32 allele further highlights the need to stretch beyond European cohorts to determine if other population-specific effects may exist. Attaining the large sample sizes required for genomic discovery in non-European populations will require a substantial investment of resources and building of capacity in low-income and middle-income countries. Furthermore, understanding the potential function of genetic variants identified in diverse samples will require a shift towards inclusivity across genomics databases118. Secondly, with improvements in HIV care and broad adoption of test and treat strategies, the focus of host genomics studies has necessarily shifted away from the natural history of infection phenotypes to intermediate phenotypes, pharmacogenomics of long-term therapy, comorbidities or vaccine response. Thirdly, understanding other classes of genetic variation that are not well captured by genotyping arrays, for example, diversity of KIR alleles and T cell receptor usage, the other partner in the HLA interaction, should be investigated to better understand how genetic variation in key innate and adaptive immune genes impact disease outcomes. However, capturing these types of variation requires in-depth sequencing to resolve genetic diversity and, in the case of T cell receptor variation, targeted immune assays to capture the relevant cells. Progress on computational methods for inferring variation at some complex loci from genotyping array data51,119 or next-generation sequencing data120–122 will greatly aid these efforts.
The full translational potential of host genomics discovery in HIV has yet to be realised. Although the association between HLA allele type, epitope binding and HIV control have been well established, this knowledge has yet to be translated into an effective preventative or therapeutic vaccine. As mentioned above, treatment of PLWH with CCR5-deficient cells has shown potential as an HIV cure but several technological improvements in autologous cell editing will be required before it becomes a scalable strategy. In addition to targeting host genes for editing, in vitro studies have also shown that it is feasible to directly target and excise the integrated proviral genome123,124. Although an extremely promising strategy, the delivery of the necessary machinery to latently infected cells remains a challenge.
The host genomics approach established in HIV research has since been applied to several other infectious diseases, including those posing substantial threats to human health such as hepatitis C virus125,126, tuberculosis127, malaria128 and even SARS-CoV-2 (ref.129), among others. These studies have time and again uncovered novel therapeutic targets and mechanisms to identify the individuals who are most vulnerable to specific infections. As the world struggles with a novel pandemic-causing RNA virus, the lessons we can learn from how the human genome contributes to variability in outcome have never been more important.
Glossary
- HIV/AIDS
HIV is the causative agent of AIDS, which is a state of severe immune deficiency defined as an HIV infection with either a CD4+ T cell count <200 cells/µl or the occurrence of a specific AIDS-defining illness.
- CC-chemokine receptor 5
(CCR5). A β-chemokine receptor that is involved in lymphocyte trafficking. In combination with CD4, CCR5 is the major host cell receptor for HIV and interacts with gp120 in the viral envelope to promote cell entry and infection.
- Cytotoxic T lymphocytes
(CTLs). Cytotoxic T cells (also called CD8+ T cells) are effector T lymphocytes that specifically kill target cells that express an appropriate peptide–MHC class I complex recognised by its T cell receptor.
- Human leucocyte antigen
(HLA). A protein, encoded by one of a group of HLA genes, that presents antigens that train the adaptive immune response. HLA genes are highly variable and allelic variants encode proteins that are differentially able to present antigens based on the amino acid sequences in the peptide-binding grooves.
- Epitopes
Parts of an antigen that make contact with a particular antibody or T cell receptor and are thus capable of stimulating an immune response.
- Polygenic risk scores
(PRS). Statistics that are calculated by enumerating the number of risk alleles associated with a particular phenotype (often weighted by their population-level effect sizes) that are present in a single individual and comparing the individual’s score to the distribution of risk scores in the population.
- Population stratification
Presence of systematic differences in allele frequencies between population subgroups owing to systematic differences in ancestry.
- HIV progression
The natural disease course of HIV infection in untreated individuals, characterized by an acute phase, a chronic phase and the development of AIDS. The rate of HIV progression varies dramatically in the infected population.
- HIV latency
The long-term persistence of HIV in an integrated but transcriptionally inactive form in the host genome. Because latent HIV resides in memory T cells, it persists indefinitely even in patients on suppressive antiretroviral therapy. This latent reservoir is a major barrier to curing HIV infection.
- Set point viral load
(spVL). Mean log viral load (HIV RNA copies per millilitre of plasma) measured in an individual with HIV infection during the chronic phase of infection. The spVL varies substantially within a population and correlates with disease progression.
- Genetic architecture
Underlying genetic basis of a given trait, in terms of variant number, effect size, allele frequency and interactions.
- HIV controllers
A group of people living with HIV whose plasma HIV RNA load is spontaneously maintained at very low levels for several years (usually at least 3–5 years) in the absence of antiretroviral therapy.
- Restriction factor
A host cellular protein that participates in antiviral defence by interfering with specific steps of the viral replication cycle.
- Killer immunoglobulin-like receptor
(KIR). A family of highly polymorphic activating and inhibitory receptors that serve as key regulators of human natural killer cell function.
- HIV target cells
The cells primarily infected by HIV, namely CD4+ T cells and macrophages, both of which are key components of a healthy immune system.
- RALY-mediated degradation
A mechanism in which the RALY protein binds to the 3′ untranslated region of an mRNA to promote its degradation.
- Transmitted/founder virus
The single viral variant that is responsible for a new infection after being transferred from an infected individual to an uninfected individual. Sexually transmitted HIV infections are typically established from a single transmitted/founder virus.
- Genome-to-genome (G2G) studies
Methods that test the hypothesis that host genetic variability causes pathogen genetic variability and can indicate novel host restriction factors.
- Treatment as prevention
Strategy to prevent the sexual transmission of HIV through the prescription of HIV medication. It is a highly effective option for preventing HIV transmission. People living with HIV who take antiretroviral drugs and maintain an undetectable viral load have effectively no risk of sexually transmitting the virus to their HIV-negative partners.
Author contributions
The authors contributed equally to all aspects of the article.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Friedland GH, Klein RS. Transmission of the human immunodeficiency virus. N. Engl. J. Med. 1987;317:1125–1135. doi: 10.1056/NEJM198710293171806. [DOI] [PubMed] [Google Scholar]
- 2.Kulkarni PS, Butera ST, Duerr AC. Resistance to HIV-1 infection: lessons learned from studies of highly exposed persistently seronegative (HEPS) individuals. AIDS Rev. 2003;5:87–103. [PubMed] [Google Scholar]
- 3.Horton RE, McLaren PJ, Fowke K, Kimani J, Ball TB. Cohorts for the study of HIV-1-exposed but uninfected individuals: benefits and limitations. J. Infect. Dis. 2010;202(Suppl. 3):S377–S381. doi: 10.1086/655971. [DOI] [PubMed] [Google Scholar]
- 4.Sabin CA, Lundgren JD. The natural history of HIV infection. Curr. Opin. Hiv. AIDS. 2013;8:311–317. doi: 10.1097/COH.0b013e328361fa66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen B. Molecular mechanism of HIV-1 entry. Trends Microbiol. 2019;27:878–891. doi: 10.1016/j.tim.2019.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Letvin NL, Walker BD. Immunopathogenesis and immunotherapy in AIDS virus infections. Nat. Med. 2003;9:861–866. doi: 10.1038/nm0703-861. [DOI] [PubMed] [Google Scholar]
- 7.Dufour C, Gantner P, Fromentin R, Chomont N. The multifaceted nature of HIV latency. J. Clin. Invest. 2020;130:3381–3390. doi: 10.1172/JCI136227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Claussnitzer M, et al. A brief history of human disease genetics. Nature. 2020;577:179–189. doi: 10.1038/s41586-019-1879-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Naranbhai V, Carrington M. Host genetic variation and HIV disease: from mapping to mechanism. Immunogenetics. 2017;69:489–498. doi: 10.1007/s00251-017-1000-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.An P, et al. Regulatory polymorphisms in the cyclophilin A gene, PPIA, accelerate progression to AIDS. PLoS Pathog. 2007;3:e88. doi: 10.1371/journal.ppat.0030088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bashirova AA, et al. Consistent effects of TSG101 genetic variability on multiple outcomes of exposure to human immunodeficiency virus type 1. J. Virol. 2006;80:6757–6763. doi: 10.1128/JVI.00094-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boniotto M, et al. Polymorphisms in the MBL2 promoter correlated with risk of HIV-1 vertical transmission and AIDS progression. Genes Immun. 2000;1:346–348. doi: 10.1038/sj.gene.6363685. [DOI] [PubMed] [Google Scholar]
- 13.Bochud P-Y, et al. Polymorphisms in Toll-like receptor 9 influence the clinical course of HIV-1 infection. AIDS. 2007;21:441–446. doi: 10.1097/QAD.0b013e328012b8ac. [DOI] [PubMed] [Google Scholar]
- 14.An P, et al. APOBEC3G genetic variants and their influence on the progression to AIDS. J. Virol. 2004;78:11070–11076. doi: 10.1128/JVI.78.20.11070-11076.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Javanbakht H, et al. Effects of human TRIM5alpha polymorphisms on antiretroviral function and susceptibility to human immunodeficiency virus infection. Virology. 2006;354:15–27. doi: 10.1016/j.virol.2006.06.031. [DOI] [PubMed] [Google Scholar]
- 16.Fellay J, et al. Common genetic variation and the control of HIV-1 in humans. PLoS Genet. 2009;5:e1000791. doi: 10.1371/journal.pgen.1000791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.International HIV Controllers Study et al. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science. 2010;330:1551–1557. doi: 10.1126/science.1195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McLaren PJ, et al. Association study of common genetic variants and HIV-1 acquisition in 6,300 infected cases and 7,200 controls. PLoS Pathog. 2013;9:e1003515. doi: 10.1371/journal.ppat.1003515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dean M, et al. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE. Science. 1996;273:1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 20.Huang Y, et al. The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat. Med. 1996;2:1240–1243. doi: 10.1038/nm1196-1240. [DOI] [PubMed] [Google Scholar]
- 21.Liu R, et al. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–377. doi: 10.1016/S0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 22.Kaslow RA, et al. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat. Med. 1996;2:405–411. doi: 10.1038/nm0496-405. [DOI] [PubMed] [Google Scholar]
- 23.Tam V, et al. Benefits and limitations of genome-wide association studies. Nat. Rev. Genet. 2019;20:467–484. doi: 10.1038/s41576-019-0127-1. [DOI] [PubMed] [Google Scholar]
- 24.Das S, Abecasis GR, Browning BL. Genotype imputation from large reference panels. Annu. Rev. Genomics Hum. Genet. 2018;19:73–96. doi: 10.1146/annurev-genom-083117-021602. [DOI] [PubMed] [Google Scholar]
- 25.Fellay J, et al. A whole-genome association study of major determinants for host control of HIV-1. Science. 2007;317:944–947. doi: 10.1126/science.1143767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mellors JW, et al. Quantitation of HIV-1 RNA in plasma predicts outcome after seroconversion. Ann. Intern. Med. 1995;122:573–579. doi: 10.7326/0003-4819-122-8-199504150-00003. [DOI] [PubMed] [Google Scholar]
- 27.Quinn TC, et al. Viral load and heterosexual transmission of human immunodeficiency virus type 1. Rakai Project Study Group. N. Engl. J. Med. 2000;342:921–929. doi: 10.1056/NEJM200003303421303. [DOI] [PubMed] [Google Scholar]
- 28.Pelak K, et al. Host determinants of HIV-1 control in African Americans. J. Infect. Dis. 2010;201:1141–1149. doi: 10.1086/651382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lingappa JR, et al. Genomewide association study for determinants of HIV-1 acquisition and viral set point in HIV-1 serodiscordant couples with quantified virus exposure. PLoS ONE. 2011;6:e28632. doi: 10.1371/journal.pone.0028632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo M, et al. A genetic polymorphism of FREM1 is associated with resistance against HIV infection in the Pumwani Sex Worker Cohort. J. Virol. 2012;86:11899–11905. doi: 10.1128/JVI.01499-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mclaren PJ, et al. Fine-mapping classical HLA variation associated with durable host control of HIV-1 infection in African Americans. Hum. Mol. Genet. 2012;21:4334–4347. doi: 10.1093/hmg/dds226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khera AV, et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet. 2018;50:1219–1224. doi: 10.1038/s41588-018-0183-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McLaren PJ, et al. Evaluating the impact of functional genetic variation on HIV-1 control. J. Infect. Dis. 2017;216:1063–1069. doi: 10.1093/infdis/jix470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lane J, et al. A genome-wide association study of resistance to HIV infection in highly exposed uninfected individuals with hemophilia A. Hum. Mol. Genet. 2013;22:1903–1910. doi: 10.1093/hmg/ddt033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mackelprang RD, et al. Whole genome sequencing of extreme phenotypes identifies variants in CD101 and UBE2V1 associated with increased risk of sexually acquired HIV-1. PLoS Pathog. 2017;13:e1006703. doi: 10.1371/journal.ppat.1006703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bouloc A, Bagot M, Delaire S, Bensussan A, Boumsell L. Triggering CD101 molecule on human cutaneous dendritic cells inhibits T cell proliferation via IL-10 production. Eur. J. Immunol. 2000;30:3132–3139. doi: 10.1002/1521-4141(200011)30:11<3132::AID-IMMU3132>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 37.Xia Z-P, et al. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature. 2009;461:114–119. doi: 10.1038/nature08247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pertel T, et al. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature. 2011;472:361–365. doi: 10.1038/nature09976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Powell TR, et al. The behavioral, cellular and immune mediators of HIV-1 acquisition: new insights from population genetics. Sci. Rep. 2020;10:3304. doi: 10.1038/s41598-020-59256-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petrovski S, et al. Common human genetic variants and HIV-1 susceptibility: a genome-wide survey in a homogeneous African population. AIDS. 2011;25:513–518. doi: 10.1097/QAD.0b013e328343817b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shiina T, Hosomichi K, Inoko H, Kulski JK. The HLA genomic loci map: expression, interaction, diversity and disease. J. Hum. Genet. 2009;54:15–39. doi: 10.1038/jhg.2008.5. [DOI] [PubMed] [Google Scholar]
- 42.Horton R, et al. Gene map of the extended human MHC. Nat. Rev. Genet. 2004;5:889–899. doi: 10.1038/nrg1489. [DOI] [PubMed] [Google Scholar]
- 43.de Bakker PIW, Raychaudhuri S. Interrogating the major histocompatibility complex with high-throughput genomics. Hum. Mol. Genet. 2012;21:1–8. doi: 10.1093/hmg/dds384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kennedy AE, Ozbek U, Dorak MT. What has GWAS done for HLA and disease associations? Int. J. Immunogenet. 2017;44:195–211. doi: 10.1111/iji.12332. [DOI] [PubMed] [Google Scholar]
- 45.Venkataraman GR, et al. Pervasive additive and non-additive effects within the HLA region contribute to disease risk in the UK Biobank. Preprint at. bioRxiv. 2020 doi: 10.1101/2020.05.28.119669. [DOI] [Google Scholar]
- 46.Sakaue S, et al. A global atlas of genetic associations of 220 deep phenotypes. Preprint at. medRxiv. 2020 doi: 10.1101/2020.10.23.20213652. [DOI] [Google Scholar]
- 47.Carrington M, et al. HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science. 1999;283:1748–1752. doi: 10.1126/science.283.5408.1748. [DOI] [PubMed] [Google Scholar]
- 48.Migueles SA, et al. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc. Natl Acad. Sci. USA. 2000;97:2709–2714. doi: 10.1073/pnas.050567397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gao X, et al. Effect of a single amino acid change in MHC class I molecules on the rate of progression to AIDS. N. Engl. J. Med. 2001;344:1668–1675. doi: 10.1056/NEJM200105313442203. [DOI] [PubMed] [Google Scholar]
- 50.Kiepiela P, et al. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature. 2004;432:769–775. doi: 10.1038/nature03113. [DOI] [PubMed] [Google Scholar]
- 51.Jia X, et al. Imputing amino acid polymorphisms in human leukocyte antigens. PLoS ONE. 2013;8:e64683. doi: 10.1371/journal.pone.0064683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McLaren PJ, et al. Polymorphisms of large effect explain the majority of the host genetic contribution to variation of HIV-1 virus load. Proc. Natl Acad. Sci. USA. 2015;112:14658–14663. doi: 10.1073/pnas.1514867112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Luo Y, et al. A high-resolution HLA reference panel capturing global population diversity enables multi-ethnic fine-mapping in HIV host response. Preprint at. medRxiv. 2020 doi: 10.1101/2020.07.16.20155606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kelleher AD, et al. Clustered mutations in HIV-1 gag are consistently required for escape from Hla-B27–restricted cytotoxic T lymphocyte responses. J. Exp. Med. 2001;193:375–386. doi: 10.1084/jem.193.3.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martinez-Picado J, et al. Fitness cost of escape mutations in p24 Gag in association with control of human immunodeficiency virus type 1. J. Virol. 2006;80:3617–3623. doi: 10.1128/JVI.80.7.3617-3623.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schneidewind A, et al. Escape from the dominant HLA-B27-restricted cytotoxic T-lymphocyte response in Gag is associated with a dramatic reduction in human immunodeficiency virus type 1 replication. J. Virol. 2007;81:12382–12393. doi: 10.1128/JVI.01543-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Migueles SA, et al. HIV-specific CD8+ T cell proliferation is coupled to perforin expression and is maintained in nonprogressors. Nat. Immunol. 2002;3:1061–1068. doi: 10.1038/ni845. [DOI] [PubMed] [Google Scholar]
- 58.Horton H, et al. Preservation of T cell proliferation restricted by protective HLA alleles is critical for immune control of HIV-1 infection. J. Immunol. 2006;177:7406–7415. doi: 10.4049/jimmunol.177.10.7406. [DOI] [PubMed] [Google Scholar]
- 59.Almeida JR, et al. Superior control of HIV-1 replication by CD8+ T cells is reflected by their avidity, polyfunctionality, and clonal turnover. J. Exp. Med. 2007;204:2473–2485. doi: 10.1084/jem.20070784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Arora J, et al. HLA heterozygote advantage against HIV-1 is driven by quantitative and qualitative differences in HLA allele-specific peptide presentation. Mol. Biol. Evol. 2020;37:639–650. doi: 10.1093/molbev/msz249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Arora J, et al. HIV peptidome-wide association study reveals patient-specific epitope repertoires associated with HIV control. Proc. Natl Acad. Sci. USA. 2019;116:944–949. doi: 10.1073/pnas.1812548116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gaiha GD, et al. Structural topology defines protective CD8+ T cell epitopes in the HIV proteome. Science. 2019;364:480–484. doi: 10.1126/science.aav5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Apps R, et al. Influence of HLA-C expression level on HIV control. Science. 2013;340:87–91. doi: 10.1126/science.1232685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kulkarni S, et al. Differential microRNA regulation of HLA-C expression and its association with HIV control. Nature. 2011;472:495–498. doi: 10.1038/nature09914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ramsuran V, et al. Epigenetic regulation of differential HLA-A allelic expression levels. Hum. Mol. Genet. 2015;24:4268–4275. doi: 10.1093/hmg/ddv158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ramsuran V, et al. Elevated HLA-A expression impairs HIV control through inhibition of NKG2A-expressing cells. Science. 2018;359:86–90. doi: 10.1126/science.aam8825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Martin MP, Carrington M. Immunogenetics of HIV disease. Immunol. Rev. 2013;254:245–264. doi: 10.1111/imr.12071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pende D, et al. Killer Ig-like receptors (KIRs): their role in NK cell modulation and developments leading to their clinical exploitation. Front. Immunol. 2019;10:1179. doi: 10.3389/fimmu.2019.01179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Martin MP, et al. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat. Genet. 2002;31:429–434. doi: 10.1038/ng934. [DOI] [PubMed] [Google Scholar]
- 70.Novembre J, Galvani AP, Slatkin M. The geographic spread of the CCR5 Δ32 HIV-resistance allele. PLoS Biol. 2005;3:1954–1962. doi: 10.1371/journal.pbio.0030339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hladik F, et al. Combined effect of CCR5-Delta32 heterozygosity and the CCR5 promoter polymorphism -2459 A/G on CCR5 expression and resistance to human immunodeficiency virus type 1 transmission. J. Virol. 2005;79:11677–11684. doi: 10.1128/JVI.79.18.11677-11684.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fätkenheuer G, et al. Efficacy of short-term monotherapy with maraviroc, a new CCR5 antagonist, in patients infected with HIV-1. Nat. Med. 2005;11:1170–1172. doi: 10.1038/nm1319. [DOI] [PubMed] [Google Scholar]
- 73.Wang H, Yang H. Gene-edited babies: what went wrong and what could go wrong. PLoS Biol. 2019;17:e3000224. doi: 10.1371/journal.pbio.3000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hutter G, et al. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N. Engl. J. Med. 2009;360:692–698. doi: 10.1056/NEJMoa0802905. [DOI] [PubMed] [Google Scholar]
- 75.Gupta RK, et al. Evidence for HIV-1 cure after CCR5Δ32/Δ32 allogeneic haemopoietic stem-cell transplantation 30 months post analytical treatment interruption: a case report. Lancet HIV. 2020;7:e340–e347. doi: 10.1016/S2352-3018(20)30069-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tebas P, et al. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N. Engl. J. Med. 2014;370:901–910. doi: 10.1056/NEJMoa1300662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Smoleń-Dzirba J, et al. HIV-1 infection in persons homozygous for CCR5-Δ32 allele: the next case and the review. AIDS Rev. 2017;19:219–230. [PubMed] [Google Scholar]
- 78.Michael NL, et al. The role of viral phenotype and CCR-5 gene defects in HIV-1 transmission and disease progression. Nat. Med. 1997;3:338–340. doi: 10.1038/nm0397-338. [DOI] [PubMed] [Google Scholar]
- 79.Kulkarni S, et al. CCR5AS lncRNA variation differentially regulates CCR5, influencing HIV disease outcome. Nat. Immunol. 2019;20:824–834. doi: 10.1038/s41590-019-0406-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Carlson JM, Le AQ, Shahid A, Brumme ZL. HIV-1 adaptation to HLA: a window into virus-host immune interactions. Trends Microbiol. 2015;23:212–224. doi: 10.1016/j.tim.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 81.Alter G, et al. HIV-1 adaptation to NK-cell-mediated immune pressure. Nature. 2011;476:96–100. doi: 10.1038/nature10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hölzemer A, et al. Selection of an HLA-C*03:04-Restricted HIV-1 p24 Gag sequence variant is associated with viral escape from KIR2DL3+ natural killer cells: data from an observational cohort in South Africa. PLoS Med. 2015;12:e1001900. doi: 10.1371/journal.pmed.1001900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Snoeck J, Fellay J, Bartha I, Douek DC, Telenti A. Mapping of positive selection sites in the HIV-1 genome in the context of RNA and protein structural constraints. Retrovirology. 2011;8:87. doi: 10.1186/1742-4690-8-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bartha I, et al. A genome-to-genome analysis of associations between human genetic variation, HIV-1 sequence diversity, and viral control. eLife. 2013;2:e01123. doi: 10.7554/eLife.01123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Palmer DS, et al. Mapping the drivers of within-host pathogen evolution using massive data sets. Nat. Commun. 2019;10:3017. doi: 10.1038/s41467-019-10724-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fellay J, Pedergnana V. Exploring the interactions between the human and viral genomes. Hum. Genet. 2020;139:777–781. doi: 10.1007/s00439-019-02089-3. [DOI] [PubMed] [Google Scholar]
- 87.Ansari MA, et al. Genome-to-genome analysis highlights the effect of the human innate and adaptive immune systems on the hepatitis C virus. Nat. Genet. 2017;49:666–673. doi: 10.1038/ng.3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chaturvedi N, et al. Adaptation of hepatitis C virus to interferon lambda polymorphism across multiple viral genotypes. eLife. 2019;8:e42542. doi: 10.7554/eLife.42542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rüeger S, et al. The influence of human genetic variation on Epstein-Barr virus sequence diversity. Sci. Rep. 2021;11:4586. doi: 10.1038/s41598-021-84070-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Eisinger RW, Dieffenbach CW, Fauci AS. HIV viral load and transmissibility of HIV infection: undetectable equals untransmittable. JAMA. 2019;321:451–452. doi: 10.1001/jama.2018.21167. [DOI] [PubMed] [Google Scholar]
- 91.Joint United Nations Programme on HIV/AIDS. 90–90–90: an ambitious treatment target to help end the AIDS epidemic (UNAIDS, 2014).
- 92.Lubomirov R, et al. Association of pharmacogenetic markers with premature discontinuation of first-line anti-HIV therapy: an observational cohort study. J. Infect. Dis. 2011;203:246–257. doi: 10.1093/infdis/jiq043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mallal S, et al. Association between presence of HLA-B*5701, HLA-DR7, and HLA-DQ3 and hypersensitivity to HIV-1 reverse-transcriptase inhibitor abacavir. Lancet. 2002;359:727–732. doi: 10.1016/S0140-6736(02)07873-X. [DOI] [PubMed] [Google Scholar]
- 94.Hetherington S, et al. Genetic variations in HLA-B region and hypersensitivity reactions to abacavir. Lancet. 2002;359:1121–1122. doi: 10.1016/S0140-6736(02)08158-8. [DOI] [PubMed] [Google Scholar]
- 95.Norcross MA, et al. Abacavir induces loading of novel self-peptides into HLA-B*57: 01: an autoimmune model for HLA-associated drug hypersensitivity. AIDS. 2012;26:F21–F29. doi: 10.1097/QAD.0b013e328355fe8f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rotger M, et al. Predictive value of known and novel alleles of CYP2B6 for efavirenz plasma concentrations in HIV-infected individuals. Clin. Pharmacol. Ther. 2007;81:557–566. doi: 10.1038/sj.clpt.6100072. [DOI] [PubMed] [Google Scholar]
- 97.Arab-Alameddine M, et al. Pharmacogenetics-based population pharmacokinetic analysis of efavirenz in HIV-1-infected individuals. Clin. Pharmacol. Ther. 2009;85:485–494. doi: 10.1038/clpt.2008.271. [DOI] [PubMed] [Google Scholar]
- 98.Haas DW, et al. Pharmacogenetics of efavirenz and central nervous system side effects: an Adult AIDS Clinical Trials Group study. AIDS. 2004;18:2391–2400. [PubMed] [Google Scholar]
- 99.di Iulio J, et al. In vivo analysis of efavirenz metabolism in individuals with impaired CYP2A6 function. Pharmacogenet. Genomics. 2009;19:300–309. doi: 10.1097/FPC.0b013e328328d577. [DOI] [PubMed] [Google Scholar]
- 100.Lubomirov R, et al. Pharmacogenetics-based population pharmacokinetic analysis of etravirine in HIV-1 infected individuals. Pharmacogenet. Genomics. 2013;23:9–18. doi: 10.1097/FPC.0b013e32835ade82. [DOI] [PubMed] [Google Scholar]
- 101.Lubomirov R, et al. ADME pharmacogenetics: investigation of the pharmacokinetics of the antiretroviral agent lopinavir coformulated with ritonavir. Pharmacogenet. Genomics. 2010;20:217–230. doi: 10.1097/FPC.0b013e328336eee4. [DOI] [PubMed] [Google Scholar]
- 102.Zanger UM, Klein K. Pharmacogenetics of cytochrome P450 2B6 (CYP2B6): advances on polymorphisms, mechanisms, and clinical relevance. Front. Genet. 2013;4:24. doi: 10.3389/fgene.2013.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nordling L. How the genomics revolution could finally help Africa. Nature. 2017;544:20–22. doi: 10.1038/544020a. [DOI] [PubMed] [Google Scholar]
- 104.Kanters S, et al. Comparative efficacy and safety of first-line antiretroviral therapy for the treatment of HIV infection: a systematic review and network meta-analysis. Lancet HIV. 2016;3:e510–e520. doi: 10.1016/S2352-3018(16)30091-1. [DOI] [PubMed] [Google Scholar]
- 105.Currier JS, et al. Coronary heart disease in HIV-infected individuals. J. Acquir. Immune Defic. Syndr. 2003;33:506–512. doi: 10.1097/00126334-200308010-00012. [DOI] [PubMed] [Google Scholar]
- 106.Obel N, et al. Ischemic heart disease in HIV-infected and HIV-uninfected individuals: a population-based cohort study. Clin. Infect. Dis. 2007;44:1625–1631. doi: 10.1086/518285. [DOI] [PubMed] [Google Scholar]
- 107.Lang S, et al. Increased risk of myocardial infarction in HIV-infected patients in France, relative to the general population. AIDS. 2010;24:1228–1230. doi: 10.1097/QAD.0b013e328339192f. [DOI] [PubMed] [Google Scholar]
- 108.Périard D, et al. Atherogenic dyslipidemia in HIV-infected individuals treated with protease inhibitors. The Swiss HIV cohort study. Circulation. 1999;100:700–705. doi: 10.1161/01.CIR.100.7.700. [DOI] [PubMed] [Google Scholar]
- 109.Nadkarni GN, et al. The burden of dialysis-requiring acute kidney injury among hospitalized adults with HIV infection: a nationwide inpatient sample analysis. AIDS. 2015;29:1061–1066. doi: 10.1097/QAD.0000000000000653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Scherzer R, et al. Association of tenofovir exposure with kidney disease risk in HIV infection. AIDS. 2012;26:867–875. doi: 10.1097/QAD.0b013e328351f68f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Li Y, et al. Combination antiretroviral therapy is associated with reduction in liver fibrosis scores in HIV-1-infected subjects. Medicine. 2016;95:e2660. doi: 10.1097/MD.0000000000002660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rotger M, et al. Impact of single nucleotide polymorphisms and of clinical risk factors on new-onset diabetes mellitus in HIV-infected individuals. Clin. Infect. Dis. 2010;51:1090–1098. doi: 10.1086/656630. [DOI] [PubMed] [Google Scholar]
- 113.Rotger M, et al. Contribution of genetic background, traditional risk factors, and HIV-related factors to coronary artery disease events in HIV-positive persons. Clin. Infect. Dis. 2013;57:112–121. doi: 10.1093/cid/cit196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dietrich LG, et al. Contribution of genetic background and data collection on adverse events of anti-human immunodeficiency virus (HIV) drugs (D:A:D) clinical risk score to chronic kidney disease in Swiss HIV-infected persons with normal baseline estimated glomerular filtration rate. Clin. Infect. Dis. 2020;70:890–897. doi: 10.1093/cid/ciz280. [DOI] [PubMed] [Google Scholar]
- 115.Dietrich LG, et al. Rapid progression of kidney dysfunction in swiss people living with HIV: contribution of polygenic risk score and D:A:D clinical risk score. J. Infect. Dis. 2021;223:2145–2153. doi: 10.1093/infdis/jiaa695. [DOI] [PubMed] [Google Scholar]
- 116.Chang H, et al. Genetic architecture of cardiometabolic risks in people living with HIV. BMC Med. 2020;18:288. doi: 10.1186/s12916-020-01762-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Popejoy AB, Fullerton SM. Genomics is failing on diversity. Nature. 2016;538:161–164. doi: 10.1038/538161a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Rotman, D. DNA databases are too white: this man aims to fix that. MIT Technol. Rev.http://www.technologyreview.com/2018/10/15/139472/dna-databases-are-too-white-this-man-aims-to-fix-that/ (2018).
- 119.Vukcevic D, et al. Imputation of KIR types from SNP variation data. Am. J. Hum. Genet. 2015;97:593–607. doi: 10.1016/j.ajhg.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kawaguchi S, Higasa K, Shimizu M, Yamada R, Matsuda F. HLA-HD: an accurate HLA typing algorithm for next-generation sequencing data. Hum. Mutat. 2017;38:788–797. doi: 10.1002/humu.23230. [DOI] [PubMed] [Google Scholar]
- 121.Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Roe D, Kuang R. Accurate and efficient KIR gene and haplotype inference from genome sequencing reads with novel K-mer signatures. Front. Immunol. 2020;11:583013. doi: 10.3389/fimmu.2020.583013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sarkar I, Hauber I, Hauber J, Buchholz F. HIV-1 proviral DNA excision using an evolved recombinase. Science. 2007;316:1912–1915. doi: 10.1126/science.1141453. [DOI] [PubMed] [Google Scholar]
- 124.Hu W, et al. RNA-directed gene editing specifically eradicates latent and prevents new HIV-1 infection. Proc. Natl Acad. Sci. USA. 2014;111:11461–11466. doi: 10.1073/pnas.1405186111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ge D, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 126.Thomas DL, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Thye T, et al. Genome-wide association analyses identifies a susceptibility locus for tuberculosis on chromosome 18q11.2. Nat. Genet. 2010;42:739–741. doi: 10.1038/ng.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Malaria Genomic Epidemiology Network. A novel locus of resistance to severe malaria in a region of ancient balancing selection. Nature. 2015;526:253–257. doi: 10.1038/nature15390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhang Q, et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370:eabd4570. doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sul JH, Martin LS, Eskin E. Population structure in genetic studies: confounding factors and mixed models. PLoS Genet. 2018;14:e1007309. doi: 10.1371/journal.pgen.1007309. [DOI] [PMC free article] [PubMed] [Google Scholar]